Abstract
Chromatin structure and gene expression are dynamically controlled by post-translational modifications (PTMs) on histone proteins, including ubiquitylation, methylation, acetylation and small ubiquitin-like modifier (SUMO) conjugation. It was initially thought that histone sumoylation exclusively suppressed gene transcription, but recent advances in proteomics and genomics have uncovered its diverse functions in cotranscriptional processes, including chromatin remodeling, transcript elongation, and blocking cryptic initiation. Histone sumoylation is integral to complex signaling codes that prime additional histone PTMs as well as modifications of the RNA polymerase II carboxy-terminal domain (RNAPII-CTD) during transcription. In addition, sumoylation of histone variants is critical for the DNA double-strand break (DSB) response and for chromosome segregation during mitosis. This review describes recent findings on histone sumoylation and its coordination with other histone and RNAPII-CTD modifications in the regulation of chromatin dynamics.
INTRODUCTION
The small ubiquitin-like modifier (SUMO) is an evolutionarily conserved protein expressed in all eukaryotes (1). Humans express five SUMO paralogs, SUMO-1, -2, -3, -4 and -5, while the budding yeast Saccharomyces cerevisiae expresses a single SUMO ortholog, Smt3, that shares 48% identity and 75% similarity with human SUMO-1 (2,3). SUMO proteins modulate the functions of targeted proteins through their dynamic attachment and detachment. SUMO-1, -2, -3 and -5 (and yeast Smt3) are initially translated as C-terminally extended precursors, and the C-terminal tail is cleaved by SUMO-specific proteases to yield mature proteins ending in a pair of glycine residues; the C-terminal α-carboxylate is the site of covalent attachment to other proteins, termed sumoylation (4). In contrast to the other SUMO paralogs, the conjugation capacity of SUMO-4 is unclear because the C-terminal tail apparently cannot be processed in vivo (5).
Mature SUMO proteins are covalently attached to lysine (K) side chains of substrate proteins through the activities of an enzyme cascade similar to that in the ubiquitylation pathway (6). Briefly, the SUMO C-terminus is first activated by the heterodimeric SAE1/SAE2 (Aos1/Uba2 in S. cerevisiae) SUMO-activating enzyme (E1) and is then transferred to a cysteine in the Ubc9 SUMO-conjugating enzyme (E2). Subsequently, SUMO ligases (E3s) promote the transfer of SUMO from E2 to lysine residue(s) on target proteins. SUMO can also be assembled into polymers on substrates; in yeast, these are specifically disassembled by the Ulp2 SUMO protease. SUMO ligation alters the interactions of substrates with their binding partners; the latter proteins often have one or more SUMO‐interaction motifs (SIMs) that enhance recognition of SUMO-conjugated proteins (7). SIM-containing proteins have emerged as key ‘readers’ of protein sumoylation.
This post-translational modification (PTM) is highly dynamic as it is readily reversed by SUMO-specific proteases. Humans have nine known SUMO-specific proteases, while S. cerevisiae expresses two, Ulp1 and Ulp2 (8). Sumoylation of proteins is a critical regulator of many diverse cellular processes, including transcription, DNA replication, cell-cycle progression, mitochondrial dynamics, ribosome biogenesis, DNA repair, apoptosis and stress responses (9,10).
Chromatin structure is centered on nucleosomes, dynamically regulated multiprotein complexes that act as scaffolds for genomic DNA. Each nucleosome is composed of 145 to 147 bp of DNA wrapped around an octamer of histone proteins (two copies each of histones H2A, H2B, H3 and H4) plus a linker histone (H1) involved in higher-order chromatin compaction (11). These histones are subject to multiple PTMs, including sumoylation.
Primary sumoylation of human histone H4 as well as weak sumoylation signals from H2A, H2B and H3 were first observed in 2003 (12), and subsequent studies identified sumoylation at K12 of H4 (13) and K18 of H3 (14). Sumoylation of histone variant H2A.X (15) and H1 (16) were also reported in human cells (Table 1). In S. cerevisiae, SUMO can be conjugated to all four core histones (17), as well as the H2A variant H2A.Z (17,18) and H3 variant Cse4 (19). Known SUMO attachment sites of H2B are K6, K7, K16 and K17, while those of H4 are K5, K8, K12, K16 and K20 (17), although there are other potential sites.
Table 1.
Histone sumoylation sites and functions
| Organism | Histone | aDiscovery | Sites | Function | Refs. |
|---|---|---|---|---|---|
| H. sapiens | H2A | 2003 | Transcriptional repression or chromatin compaction | (12,15,16) | |
| H2B | 2003 | (12,16) | |||
| H3 | 2003 | K18 | (12,14–16) | ||
| H4 | 2003 | K12 | (12,13,15,16,28,29) | ||
| H2A.X | 2013 | K5, K9, K13, K15, K118, K119, K127, K133, K134 | (15) | ||
| H1 | 2009 | (16) | |||
| S. cerevisiae | H2A | 2006 | bK126 | Transcriptional repression/activation, inhibition of cryptic initiation | (17) |
| H2B | 2006 | K6, K7, K16, K17 | (17,31,40,45) | ||
| H3 | 2006 | (17) | |||
| H4 | 2006 | K5, K8, K12, K16, K20 | (17,40,45) | ||
| H2A.Z | 2006 | K126, K133 | cDSB repair | (17,18) | |
| Cse4 | 2016 | K65, K215, K216 | Cse4 incorporation or proteolysis | (19,77,78) |
aThe first detected year of histone sumoylation.
bH2A sumoylation level was not changed in an arginine substitution mutant of this site.
cDSB: double-stranded break.
Among the myriad known histone PTMs, histone sumoylation was discovered relatively recently, so less is known about its effects on chromatin organization and gene expression compared to ubiquitylation, methylation, and acetylation. Although first reported in 2003 (12), most studies on histone sumoylation have appeared within the past five years. Recent investigations utilizing biochemical and genome-wide analyses have contributed much toward our understanding of the patterns and associated functions of this modification. Further, new functions of histone sumoylation continue to be uncovered, and intriguing examples of epigenetic regulation have recently been revealed. The present review provides an overview of newly discovered functions for histone sumoylation, including the dynamic regulation of eukaryotic chromatin structure and transcription.
HISTONE SUMOYLATION AND TRANSCRIPTION
Histone sumoylation in transcriptional repression
Many SUMO target proteins are transcriptional co-activators or co-repressors (20–23), suggesting that sumoylation may have both positive and negative effects on the expression levels of diverse gene types, including constitutively expressed and inducible genes (23). The first report of histone sumoylation in human cells by Shiio and Eisenman suggested a negative effect on transcription because an engineered SUMO−H4 translational fusion associated with the transcriptional repressors histone deacetylase 1 (HDAC1) and heterochromatin protein HP1 in cells (12). The authors observed SUMO-1 attachment to acetylated H4 and enhancement of this sumoylation reaction by co-expression of the histone acetyltransferase (HAT) p300, suggesting that histone acetylation may facilitate subsequent SUMO conjugation to H4 (12); these results provided an early hint of the complex interplay of histone sumoylation with other histone PTMs. In the case of non-histone substrates, it has also been reported that SUMO-modified p300 and CREB-binding protein mediate transcriptional repression by promoting recruitment of HDAC6 (24). Despite these early results, there has been relatively limited study of transcriptional repression mechanisms by histone sumoylation until recently.
Gene expression levels are tightly regulated by co-activators and co-repressors that promote reversible switching between ‘on’ and ‘off’ states. Histone acetylation is a major driver of the transcriptionally active chromatin state, while ensuing sumoylation may provide reciprocal control to limit expression. Indeed, in follow-up studies to those noted above, histone acetylation was found to stimulate Ubc9-mediated histone sumoylation, and conversely, p300-mediated gene activation is repressed by histone-SUMO modification and subsequent HDAC6 recruitment in human cells (Figure 1A) (24). Such histone sumoylation also leads to condensed chromatin and gene silencing by facilitating the recruitment of HDAC1 and HP1. However, it has yet to be determined whether histone sumoylation affects H3K9 methylation, a marker of HP1-mediated gene repression (25).
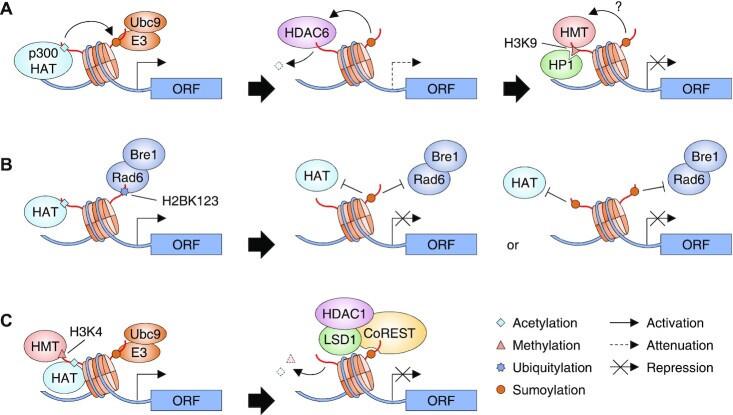
Figure 1.
Models for the functions of histone sumoylation in transcriptional repression. (A) During switching from transcriptional activation to repression in mammals, p300 HAT-mediated histone acetylation promotes histone sumoylation by activating Ubc9 and SUMO E3 ligase. Sumoylated histones then recruits both HDAC6, which attenuates transcription, and HP1, which contributes to chromatin compaction. However, it is still unclear whether histone sumoylation stimulates H3K9 methylation, a marker for HP1 binding. (B) Histone sumoylation in yeast potentially interferes with histone acetylation by HATs or H2BK123 ubiquitylation by Rad6 and Bre1, thereby inhibiting transcription. (C) LSD1−CoREST−HDAC1 complex is associated with sumoylated histone through the SUMO-interacting motif (SIM) in the CoREST subunit, allowing LSD1 and HDAC1 to reverse H3K4 methylation and histone acetylation, respectively; both of the latter histone marks normally promote transcription.
The sophisticated molecular genetic tools available for studying the yeast S. cerevisiae were employed not long after these first mammalian studies, allowing important advances in our understanding of histone sumoylation. To circumvent the lack of sumoylated histone-specific antibodies, Berger and colleagues evaluated yeast histone H2B sumoylation levels by a two-step chromatin double immunoprecipitation (ChDIP) protocol in cells expressing H2B tagged with a Flag epitope and SUMO (Smt3) tagged with an HA epitope; anti-Flag beads were used to isolate Flag-H2B in the first step and anti-HA beads against HA-SUMO were utilized in the second step (17,26). Sumoylated H2B was observed at many genomic locations, including the galactose-inducible GAL1 gene, with slightly stronger signals at subtelomeric regions.
Alanine substitutions at K6, K7, K16 and K17 of H2B (H2B-4KA) strongly reduced its conjugation to SUMO and led to modest increases in expression of several tested genes, including GAL1, under non-inducing conditions, in S. cerevisiae (17). Conversely, direct fusion of SUMO to H2B (or H3) strongly reduced expression of GAL1 under inducing conditions. Increased H2B or H4 sumoylation correlated with decreased histone acetylation (H2BK16Ac) under GAL1-inducing conditions as well as in mutants with a substitution at the ubiquitylation site H2BK123. Moreover, H3 acetylation was enhanced in a ubc9ts strain (a temperature-sensitive E2 mutant) and in cells lacking the Siz1 and Siz2 SUMO E3 ligases. Collectively, these findings suggest that histone sumoylation may be involved in transcriptional repression via inhibition of, or competition with, histone epigenetic marks for gene activation such as ubiquitylation and acetylation (Figure 1B).
Histone sumoylation and chromatin structure
Two critical issues that remained unresolved in these early studies were (i) whether histone sumoylation results exclusively in transcriptional repression or if transcriptional activation is also possible, and (ii) whether individual genes can be regulated by specific patterns of histone sumoylation. An early proteomics analysis of HeLa cells identified the K12 residue of H4 as a site of SUMO-3 conjugation (13). Chatterjee and colleagues employed a disulfide-directed protein modification strategy, which uses disulfide chemistry to crosslink two peptides (27), and generated a human histone H4K12C protein crosslinked to the C-terminus of SUMO (28). Nucleosomes assembled in vitro with this SUMO-modified version of H4 were less stable and were unable to form dinucleosomes in vitro, similar to nucleosomes bearing K16-acetylated H4, a known marker of open chromatin. These findings suggest that sumoylation at H4K12 also inhibits chromatin compaction by inhibiting inter-nucleosomal interactions.
These results would appear to be at odds with the earlier reports (12,17), which had implied a role for histone sumoylation in forming closed chromatin structures. Subsequent experiments, however, provided evidence that could reconcile these data. In particular, these analyses showed that SUMO-3-conjugated H4 stimulates lysine-specific demethylase 1 (LSD1)-mediated removal of H3K4 methylation in vitro; H3K4 methylation is a sign of active chromatin (29). To effect transcriptional repression, LSD1 associates with CoRepressor for Element 1 Silencing Transcription factor (CoREST) and HDAC1 (30). Importantly, a SIM in CoREST is required for H3K4 demethylation by LSD1, but this demethylation activity is not propagated into adjacent nucleosomes. Chatterjee and colleagues suggested that transient histone sumoylation may provide a binding platform for CoREST, LSD1 and HDAC1, allowing spatially restricted gene repression by clearance of local PTMs that would otherwise promote transcription (Figure 1C). Their model is consistent with the low level of histone sumoylation in cells (17) because subsequent elimination of SUMO modifications is required to suppress its negative effects on chromatin compaction and to establish silenced heterochromatin (29).
A very recent study identified another possible role for histone sumoylation in the regulation of chromatin structure in S. cerevisiae (31). The Remodeling the Structure of Chromatin (RSC) complex is a member of the ATP-dependent nucleosome remodeler family (32) that alters the position, occupancy, and composition of nucleosomes in chromatin; RSC activity regulates transcription (33–35), DNA replication (36), chromosome segregation (37), and DNA repair (38,39). DNA footprint analysis of nucleosome-associated Sth1, the catalytic subunit of the RSC complex, revealed that H3K14 acetylation facilitates nucleosome binding of RSC by association with the C-terminal bromodomain of Sth1 (31). Furthermore, yeast mutants with substitutions of the SUMO sites in H2B or loss of the SUMO ligases Siz1 and Siz2 displayed impaired association of RSC with nucleosomes in vivo; conversely, nucleosomes containing SUMO-fused H2B showed greater in vitro binding to RSC than did unmodified nucleosomes. This suggests that SUMO-histone conjugation promotes binding by RSC; however, the relevance of this mechanism (Figure 2) to RSC-controlled cellular processes such as transcription or replication still needs to be demonstrated.
Figure 2.
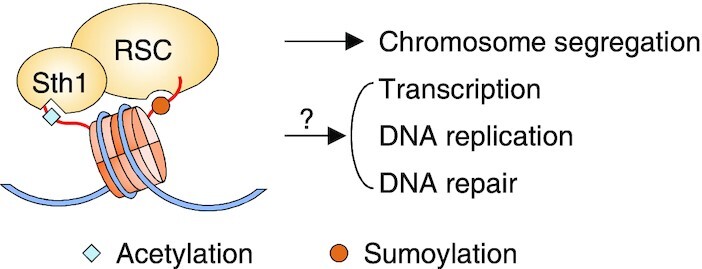
Histone sumoylation promotes chromatin binding of RSC. The Sth1 subunit of RSC recognize H3K14 acetylation, and an unknown RSC component recognizes sumoylated histones. This dual recognition has been implicated in chromosome segregation, but its function in other RSC-controlled processes has not yet been determined.
Taken together, these findings suggest that histone sumoylation can suppress the formation of certain higher-order chromatin structures and acts as a crucial signal for recruitment of factors involved in gene activation as well as repression.
Histone sumoylation is integral to multiple transcriptional regulatory cascades
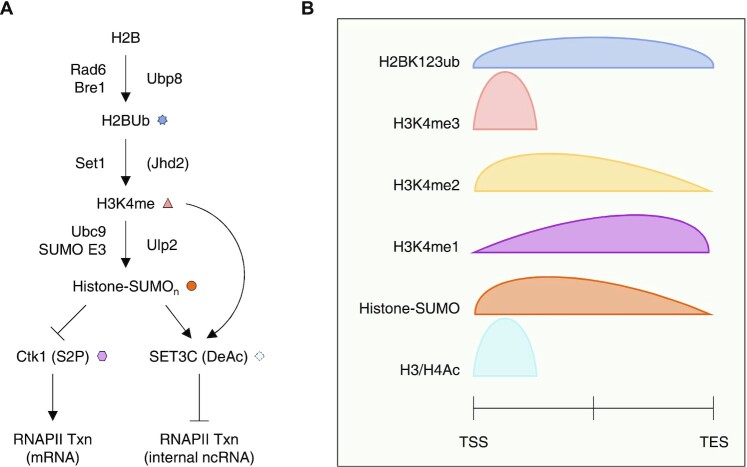
An early study reported only mono-sumoylated histones in WT and ulp2Δ yeast cells (17). However, recent work conducted under experimental conditions that preserve most sumoylated species has also identified polymeric SUMO chains on histones H2B and H4 (40). SUMO chains can act as complex signaling codes to guide subsequent protein activity, including addition or removal of other PTMs (41,42). For yeast histones, poly-SUMO chains are efficiently disassembled by the Ulp2 SUMO protease (40), so they are highly dynamic. Genome-wide localization studies have identified enrichment of SUMO-conjugated H2B and Ulp2 at the loci of constitutively transcribed genes, activated inducible genes, and genes encoding ribosomal proteins (40,43–46).
While histone modifications were once considered independent PTMs, it is now becoming clear that modifications at different sites can exhibit interdependence under specific conditions (context-dependent crosstalk), which has important implications for the control of chromatin dynamics (47). For example, yeast histone sumoylation may interfere with or counteract H2B mono-ubiquitylation (17), while other evidence suggests that H2B and H4 sumoylation may require H2B ubiquitylation mediated by Rad6 (E2) and Bre1 (E3) (40). The Ubp8 deubiquitylase-mediated removal of ubiquitin from H2B is also required for nucleosome binding to Ctk1 kinase (48). Ctk1 in turn phosphorylates serine-2 (S2) within the C-terminal domain (CTD) Y1S2P3T4S5P6S7 heptad repeat region of Rpo21/Rpb1, the largest RNA polymerase II (RNAPII) subunit; this is known to promote transcriptional elongation and couple it to mRNA 3′ end processing (49,50). The association between the nucleosome and Ctk1 is blocked by both H2B ubiquitylation and sumoylation, and Ulp2-dependent desumoylation of histone facilitates later transcriptional elongation steps by promoting Ctk1 recruitment (40). Removal of the phosphate on the CTD S5 residue by the Rtr1 phosphatase is also required for transcriptional elongation (51) (Figure 3A and B). These data describing sequential histone modification changes provided the first suggestion that histone sumoylation may also be involved in transcriptional activation.
Figure 3.
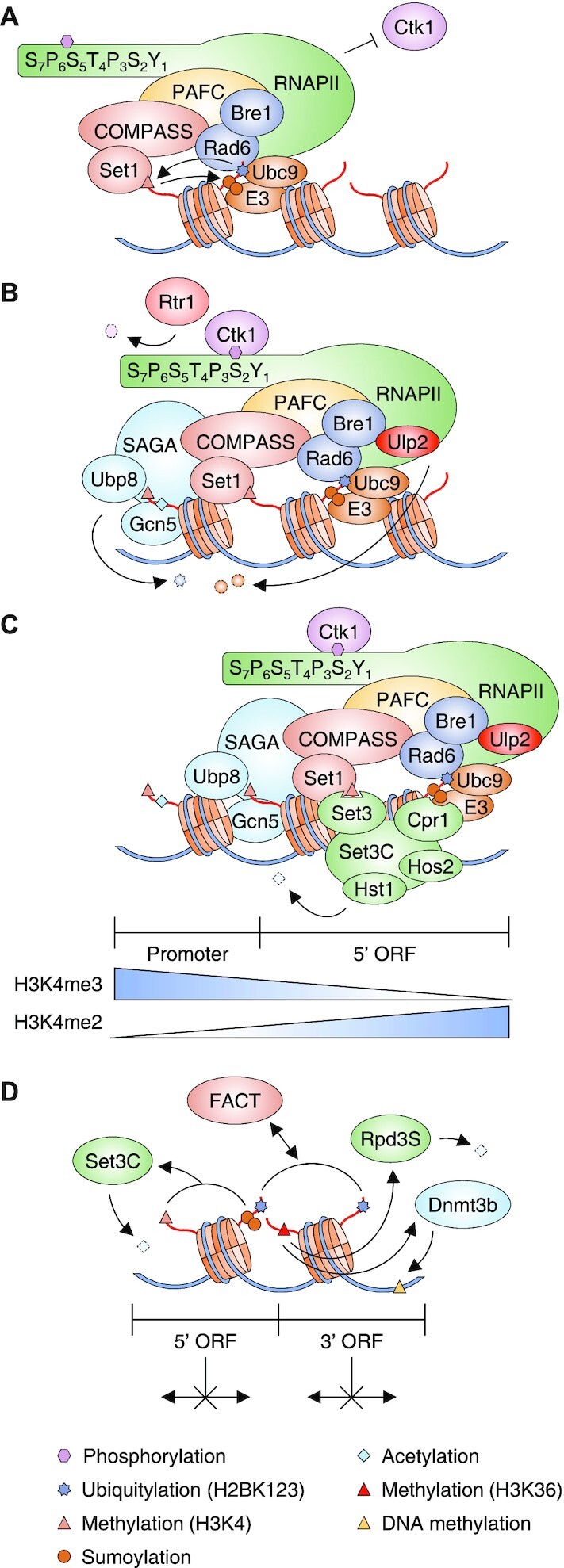
Model of histone sumoylation in the prevention of cryptic initiation. (A–C) Illustrations show the relevant components, but not the precise physical association or order of events. Triangles at the bottom indicate gradients of H3K4me3 and H3K4me2 modifications over the promoter and 5′ regions of the open reading frame (ORF). (A) At the early stage of transcription, the CTD S5 phosphorylated forms of RNAPII and PAFC are required for H2BK123 ubiquitylation by Rad6 and Bre1. H2B ubiquitylation drives two sequential modifications, COMPASS/Set1-mediated H3K4 methylation and histone poly-sumoylation by Ubc9 and a SUMO E3. Both H2B ubiquitylation and histone sumoylation inhibit Ctk1 (the major S2 kinase) association with the RNAPII transcription machinery. (B) Ubiquitin removal from histones by SAGA component Ubp8 and polySUMO disassembly by Ulp2 together facilitate Ctk1 recruitment and CTD S2 phosphorylation for subsequent transcription elongation, while Rtr1 dephosphorylates S5 in the CTD. The Gcn5 HAT, another SAGA subunit, mediates histone acetylation during transcription elongation. (C) In the transcription elongation step, repeated rounds of H2B ubiquitylation and histone sumoylation and their reversal occur while an H3K4 methylation gradient is gradually established. Recognition of H3K4me2 by Set3 and sumoylated histones by Cpr1, both subunits of the SET3C deacetylase, is required for recruitment of SET3C to the 5′ regions of ORFs. Hst1 and Hos2, the catalytic subunits of SET3C, block accumulation of hyperacetylated histones in these ORF regions. (D) Inhibition of spurious transcription initiation by cotranscriptional histone modifications. H2B ubiquitylation functions cooperatively with the FACT complex to suppress cryptic transcription of genes. H3K4 methylation and histone sumoylation facilitate histone deacetylation by SET3C in 5′ ORF regions, and H3K36 methylation promotes histone deacetylation by Rpd3S in 3′ ORF regions. In mammals, Dnmt3b-mediated DNA methylation restricts the generation of cryptic transcripts in a H3K36 methylation-dependent manner.
The prototypical example of histone crosstalk is the ‘histone trans-tail pathway’ involving H2B ubiquitylation-dependent K4 and K79 methylation of H3 (52,53). Intriguingly, H2B ubiquitylation-mediated H3K4 dimethylation (me2), but not trimethylation (me3), is also required for subsequent sumoylation at H2B and H4 during transcription (45). Methylation of H3K4 exhibits an intrinsic gradient pattern, with me3 more frequent near the promoter, me2 in the 5′ region of the open reading frame (ORF), and monomethylation (me1) in more gene-distal regions (54,55). The degree of H3K4 methylation is determined by the amount of time the Set1 methyltransferase is tethered near the nucleosome during multiple rounds of transcription (56). Notably, H3K4me2 has a function distinct from that of H3K4me3 in transcription by providing a binding site for the PHD finger within Set3, a subunit of the Set3 complex (SET3C); SET3C is a histone deacetylase that includes two active HDAC subunits, Hos2 and Hst1 (57,58). SET3C-mediated histone deacetylation in the 5′ ORF region contributes to the suppression of cryptic initiation of both sense and antisense RNA transcription from within the ORF (58,59).
In addition to SET3C binding to H3K4me2, yeast SET3C also preferentially associates both in vivo and in vitro with SUMO-modified histones via a SIM in Cpr1, another subunit of SET3C (45). Importantly, the changes in noncoding RNA (ncRNA) expression exhibited by cells lacking Set3 strongly overlapped with those in cells exclusively expressing the H2B-4KA mutant (which strongly reduces its sumoylation), implying a strong association between SET3C function and histone sumoylation (45). Notably, the H2B-4KA mutations lead to dramatic decreases in Set3 and Cpr1 occupancy at target genes and increases in spurious transcription of sense ncRNAs initiated from cryptic internal promoters.
Taken together, these results indicate that sequential histone modifications—H2B ubiquitylation, H3K4 methylation, histone sumoylation and histone deacetylation—function in a complex crosstalk pathway to prevent inappropriate internal transcription within gene coding sequences (Figures 3A–C and 4). This mechanism is distinct from other histone modification-dependent mechanisms that also contribute to the suppression of spurious transcription initiation, namely, interdependent regulation of nucleosome reassembly by H2B ubiquitylation and the FACT complex and H3K36 methylation-mediated association of the Rpd3S HDAC complex or Dnmt3b DNA methyltransferase (Figure 3D) (60–63). In summary, the role of histone sumoylation in transcription cannot be simply defined as ‘positive’ or 'negative’. Instead, crosstalk with other histone modifications both regulates transcriptional elongation and maintains transcriptional fidelity by an elaborate regulation of transcription steps (Figure 4A).
Figure 4.
Genome-wide distribution pattern of histone modifications that crosstalk with histone sumoylation in active genes. (A) Schematic diagram depicting the distinct roles of histone sumoylation in transcription and its crosstalk with other histone modifications. (B) The genomic localization of histone modifications is mapped on a generalized gene aligned from transcription start site (TSS) to transcription end site (TES). The curves represent their distribution patterns determined by genome-wide analyses in yeast.
Genome-wide maps of histone sumoylation and other histone modifications
The genome-wide localization of histone modifications associated with histone sumoylation has been examined to uncover their potential roles in yeast transcription (Figure 4A and B). H2B ubiquitylation appears at an early stage among the dynamic changes in histone PTMs and chromatin during transcription; it is preferentially enriched across transcribed regions and correlates positively with transcriptional gene activity (64–66) (Figure 4B). In turn, H3K4 methylation, which requires H2BK123 ubiquitylation, is distributed in distinct gradients relative to the transcribed DNA sequence that depend on the extent of H3K4 methylation (56,67). The genome-wide localization pattern of SUMO-modified histones correlates closely with the H3K4me2 profile on actively transcribed genes (45), supporting the idea that these two modifications contribute to the same or a similar step of chromatin-mediated transcription. Finally, a strong peak of acetylated histones H3 and H4 is detected upstream of the histone-SUMO and H3K4me2-enriched regions in active genes (55,68), consistent with histone sumoylation and H3K4me2 working in concert to recruit SET3C for histone deacetylation.
HISTONE SUMOYLATION IN DSB REPAIR
Genomic DNA suffers double-strand breaks (DSBs) throughout an organism's life due to genotoxic agents (such as γ-irradiation) or physiological processes such as meiosis (69). Therefore, DSB repair is an essential step for cell survival and the maintenance of genome integrity. Repair is mediated by either error-prone nonhomologous end-joining (NHEJ) or homologous recombination (HR), which utilizes sequences homologous to the broken DNA to guide accurate repair (70,71). In the HR repair pathway, yeast histone variant H2A.Z is required to create an open chromatin structure (72,73). Upon induction of a persistent DSB without available DNA homology for repair, H2A.Z is rapidly loaded near the break site to drive the relocation of the unrepaired chromosomal ends toward the nuclear envelope and then is slowly removed (18). While only a low level of SUMO-modified H2A.Z was originally reported in yeast cells during DSB repair (17), a subsequent investigation found that H2A.Z incorporated into nucleosomes at a persistent DSB site was sumoylated and that this sumoylation was required for DSB tethering to the nuclear periphery (18). It has not been determined how SUMO modification of H2A.Z affects DSB relocation, but a key recombination factor in the DSB response, Rad52, is a known SUMO substrate (74,75).
HISTONE SUMOYLATION AT THE CENTROMERE
Recent studies have demonstrated a specific role for sumoylation of the S. cerevisiae histone H3 variant Cse4 (human CENP-A) in mitosis. The incorporation of Cse4 into centromeric nucleosomes is required for normal kinetochore assembly and chromosome stability, and thus ultimately for faithful chromosome segregation (76). The Cse4 protein is a substrate for Siz1 and Siz2 SUMO ligases in vitro and in vivo (19), and subsequent biochemical studies revealed the major SUMO-Cse4 conjugation sites (77,78). Sumoylation at C-terminal K215/216 sites of Cse4 facilitates its association with the Cse4-specific histone chaperone Scm3 (78), promoting Cse4 deposition at centromeres (Figure 5A). The chromatin assembly factor-1 (CAF-1) complex also interacts with K215/216-sumoylated Cse4 and drives overexpressed Cse4 into incorrect, noncentromeric sites (78) (Figure 5B). However, CAF-1 can also trigger the deposition of Cse4 into the centromeric region when SCM3 gene expression is reduced (78).
Figure 5.
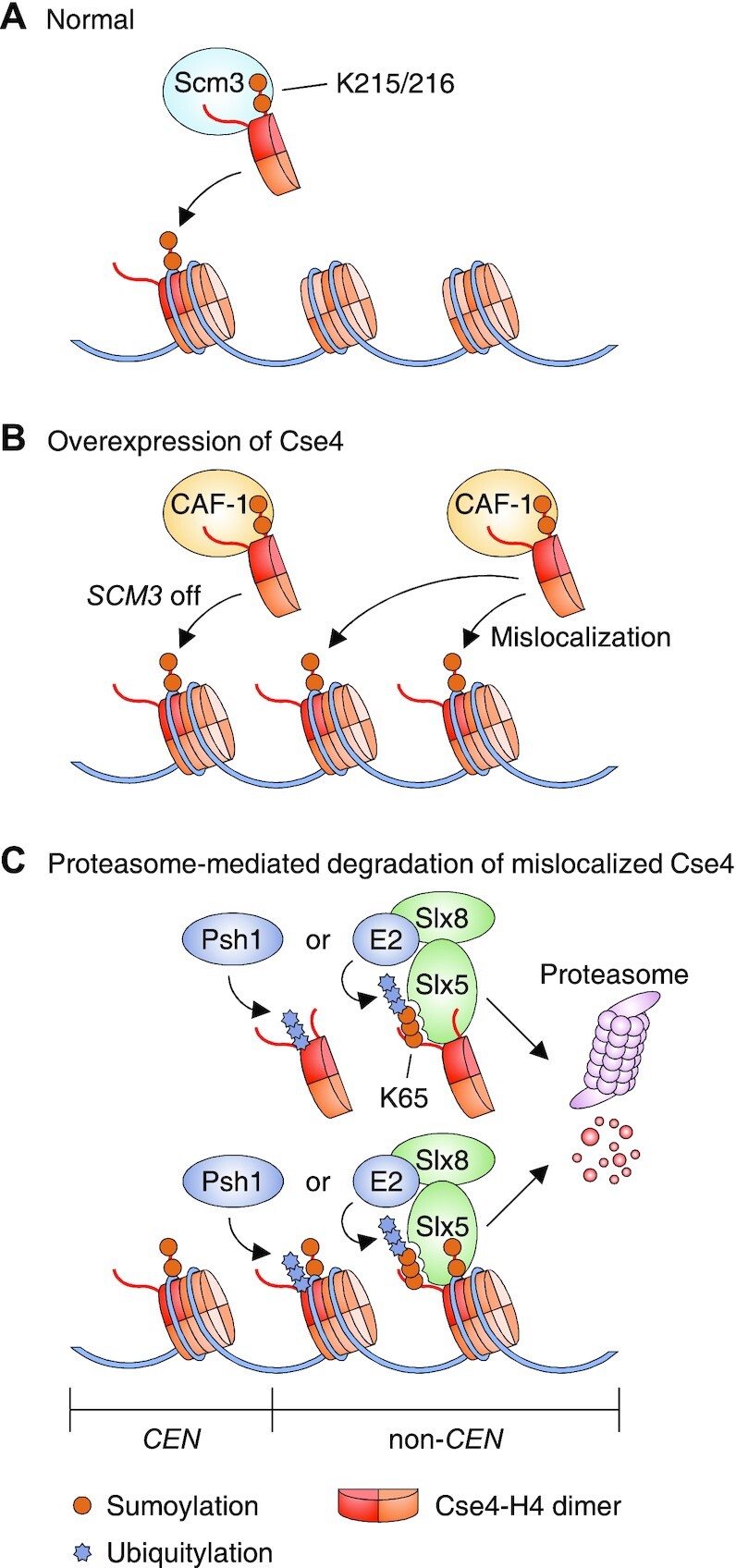
Sumoylation of Cse4 mediates its appropriate localization. (A and B) Cse4-K215/216 sumoylation triggers Scm3-dependent incorporation of Cse4−H4 dimers into the CEN regions of chromosomes in normal cells (A), while CAF-1 also interacts with K215/216-sumoylated Cse4 and promotes deposition of overexpressed Cse4−H4 dimers into non-CEN regions (and CEN domains when Scm3 levels are low) (B). The SIMs of Scm3 and CAF-1 are not yet determined. (C) Sumoylation of K65 in Cse4 limits its levels or prevents its mislocalization in a manner dependent on Slx5/Slx8-mediated ubiquitylation and proteasome-mediated proteolysis. The Psh1 ubiquitin ligase independently facilitates proteasomal degradation of mislocalized Cse4.
Interestingly, sumoylation of Cse4 at a different site, K65, prevents the aberrant spread of Cse4 into euchromatin by providing a signal for ubiquitylation by the Slx5/Slx8 SUMO-targeted ubiquitin ligase (STUbL), which leads to Cse4 degradation by the proteasome. An alternative E3 ubiquitin ligase, Psh1, can also mark mislocalized or excess Cse4 for proteasomal degradation by a SUMO-independent mechanism (19,77) (Figure 5C). A reduction in histone H4 dosage prevents the ectopic localization of overexpressed Cse4, which correlates with reduced Cse4 sumoylation (79). The data imply that noncentromeric deposition of excess Cse4 is mediated by Cse4 sumoylation in the context of H4-Cse4 dimers. Overexpression and mislocalization of CENP-A, the human ortholog of Cse4, have been observed in many cancers and lead to aneuploidy, defined as the presence of an abnormal number of chromosome copies (80–88). Therefore, further study of SUMO modification of this H3 variant may provide a better understanding of tumor development and reveal potential new strategies for cancer treatment.
PERSPECTIVE
Since the first evidence for SUMO conjugation of histone proteins ∼18 years ago (12), multiple investigators have demonstrated potential functions for histone-SUMO conjugates in the epigenetic regulation of gene expression as well as the DNA DSB response and chromosome segregation. However, mechanistic studies have been largely limited to yeast and to mammalian cells in vitro. Therefore, many of the mechanisms discussed still need to be extended by in vivo studies of mammals, including analyses of the roles of histone sumoylation in tumorigenesis.
There are several hurdles yet to overcome for a more detailed understanding of histone sumoylation in epigenetic gene regulation and other genomic control processes. Of particular note, we do not know how the Ubc9 E2 or SUMO E3 ligases are able to sumoylate histones in a chromosomal site-specific way. Although some evidence indicates such enzymes can be recruited to distinct loci, these proteins do not possess obvious domains for DNA binding or histone modification recognition, such as ubiquitin-binding or methylated lysine-binding domains (43,89). Proteins such as transcriptional activating or silencing factors may also help to localize SUMO pathway enzymes to specific chromatin sites.
Major experimental challenges include preservation and detection of the low-abundance sumoylated forms of histones and unambiguous determination of sumoylation sites (6,17). Unfortunately, overexpression of SUMO proteins by increased SUMO gene dosage, promoter swapping, or environmental changes such as heat shock can also alter the normal levels, intracellular location, or biological activity of SUMO targets as well as the complex crosstalk among protein PTMs (90). It is hoped that advances in quantitative proteomics and the development of site-specific antibodies to SUMO-conjugated histones and other sumoylated chromatin factors will provide powerful and unbiased approaches to identify sumoylated proteins and their modification sites.
Understanding the physiological impact of the complex signaling codes conferred by sumoylation and other PTMs of histones will require identifying their full range and dynamics. Moreover, SUMO proteins are themselves subject to PTMs such as phosphorylation, acetylation, and ubiquitylation (91–95), and also have internal sumoylation sites, resulting in the formation of SUMO chains (14,41). Improved mass spectrometry and peptide sequencing technology will facilitate the identification of complex PTMs, including those of SUMO (96). While the cues provided by these complex codes have not been elucidated in most instances, several studies have provided early clues about how a few of them specify downstream biological events. For instance, the BRCA1-A complex subunit RAP80 recognizes mixed SUMO-ubiquitin chains formed at sites of DNA damage and thus can recruit the BRCA1 DNA repair complex to these sites (97). Combinations of histone sumoylation and other histone modifications or the variously phosphorylated forms of RNAPII-CTD also provide potential signals to guide transcription and other chromatin-dependent processes. To understand these signals, many more single-gene and genome-level analyses will be required.
SUMO modification is essential for myriad cellular processes in all eukaryotes and is implicated in diverse diseases (98). In addition, dysfunctional RNAPII CTD phosphorylation and histone ubiquitylation, methylation, and acetylation, all of which interact with histone sumoylation, are strongly implicated in neurodegenerative diseases and cancer (99–103). Thus, in addition to revealing new insights into the epigenetic mechanisms regulating gene expression, chromatin structure, and genome stability, understanding the functions of histone sumoylation will likely provide new therapeutic strategies and drug targets for disease treatment.
DATA AVAILABILITY
All data in the referenced studies published by the present authors are available from the authors or have been deposited in public databases.
Contributor Information
Hong-Yeoul Ryu, School of Life Sciences, BK21 FOUR KNU Creative BioResearch Group, College of National Sciences, Kyungpook National University, Daegu 41566, Republic of Korea.
Mark Hochstrasser, Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT 06520, USA.
FUNDING
NIH [GM053756, GM136325 to M.H.]; National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) [2020R1C1C1009367 to H.Y.R.]. Funding for open access charge: BK21 FOUR KNU Creative BioResearch Group.
Conflict of interest statement. None declared.
REFERENCES
- 1. Johnson E.S. Protein modification by SUMO. Annu. Rev. Biochem. 2004; 73:355–382. [DOI] [PubMed] [Google Scholar]
- 2. Huang W.C., Ko T.P., Li S.S., Wang A.H.. Crystal structures of the human SUMO-2 protein at 1.6 A and 1.2 A resolution: implication on the functional differences of SUMO proteins. Eur. J. Biochem. 2004; 271:4114–4122. [DOI] [PubMed] [Google Scholar]
- 3. Liang Y.C., Lee C.C., Yao Y.L., Lai C.C., Schmitz M.L., Yang W.M.. SUMO5, a novel poly-SUMO isoform, regulates PML nuclear bodies. Sci Rep-UK. 2016; 6:26509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Johnson P.R., Hochstrasser M.. SUMO-1: ubiquitin gains weight. Trends Cell Biol. 1997; 7:408–413. [DOI] [PubMed] [Google Scholar]
- 5. Owerbach D., McKay E.M., Yeh E.T.H., Gabbay K.H., Bohren K.M.. A proline-90 residue unique to SUMO-4 prevents maturation and sumoylation. Biochem Bioph Res Co. 2005; 337:517–520. [DOI] [PubMed] [Google Scholar]
- 6. Hendriks I.A., Vertegaal A.C.. A comprehensive compilation of SUMO proteomics. Nat. Rev. Mol. Cell Biol. 2016; 17:581–595. [DOI] [PubMed] [Google Scholar]
- 7. Song J., Durrin L.K., Wilkinson T.A., Krontiris T.G., Chen Y.A.. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:14373–14378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hickey C.M., Wilson N.R., Hochstrasser M.. Function and regulation of SUMO proteases. Nat. Rev. Mol. Cell Biol. 2012; 13:755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Flotho A., Melchior F.. Sumoylation: a regulatory protein modification in health and disease. Annu. Rev. Biochem. 2013; 82:357–385. [DOI] [PubMed] [Google Scholar]
- 10. Ryu H.Y., Ahn S.H., Hochstrasser M.. SUMO and cellular adaptive mechanisms. Exp. Mol. Med. 2020; 52:931–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Strahl B.D., Allis C.D.. The language of covalent histone modifications. Nature. 2000; 403:41–45. [DOI] [PubMed] [Google Scholar]
- 12. Shiio Y., Eisenman R.N.. Histone sumoylation is associated with transcriptional repression. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:13225–13230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Galisson F., Mahrouche L., Courcelles M., Bonneil E., Meloche S., Chelbi-Alix M.K., Thibault P.. A novel proteomics approach to identify SUMOylated proteins and their modification sites in human cells. Mol. Cell. Proteomics. 2011; 10:M110 004796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hendriks I.A., D'Souza R.C.J., Yang B., Verlaan-de Vries M., Mann M., Vertegaal A.C.O.. Uncovering global SUMOylation signaling networks in a site-specific manner. Nat. Struct. Mol. Biol. 2014; 21:927–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen W.T., Alpert A., Leiter C., Gong F., Jackson S.P., Miller K.M.. Systematic identification of functional residues in mammalian histone H2AX. Mol. Cell. Biol. 2013; 33:111–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Matafora V., D’Amato A., Mori S., Blasi F., Bachi A.. Proteomics analysis of nucleolar SUMO-1 target proteins upon proteasome inhibition. Mol. Cell. Proteomics. 2009; 8:2243–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nathan D., Ingvarsdottir K., Sterner D.E., Bylebyl G.R., Dokmanovic M., Dorsey J.A., Whelan K.A., Krsmanovic M., Lane W.S., Meluh P.B.et al.. Histone sumoylation is a negative regulator in Saccharomyces cerevisiae and shows dynamic interplay with positive-acting histone modifications. Genes Dev. 2006; 20:966–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kalocsay M., Hiller N.J., Jentsch S.. Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol. Cell. 2009; 33:335–343. [DOI] [PubMed] [Google Scholar]
- 19. Ohkuni K., Takahashi Y., Fulp A., Lawrimore J., Au W.C., Pasupala N., Levy-Myers R., Warren J., Strunnikov A., Baker R.E.et al.. SUMO-targeted ubiquitin ligase (STUbL) Slx5 regulates proteolysis of centromeric histone H3 variant Cse4 and prevents its mislocalization to euchromatin. Mol. Biol. Cell. 2016; 27:1500–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gill G. Something about SUMO inhibits transcription. Curr. Opin. Genet. Dev. 2005; 15:536–541. [DOI] [PubMed] [Google Scholar]
- 21. Zhao J. Sumoylation regulates diverse biological processes. Cell. Mol. Life Sci. 2007; 64:3017–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Makhnevych T., Sydorskyy Y., Xin X., Srikumar T., Vizeacoumar F.J., Jeram S.M., Li Z., Bahr S., Andrews B.J., Boone C.et al.. Global map of SUMO function revealed by protein-protein interaction and genetic networks. Mol. Cell. 2009; 33:124–135. [DOI] [PubMed] [Google Scholar]
- 23. Chymkowitch P., Nguea P.A., Enserink J.M.. SUMO-regulated transcription: challenging the dogma. Bioessays. 2015; 37:1095–1105. [DOI] [PubMed] [Google Scholar]
- 24. Girdwood D., Bumpass D., Vaughan O.A., Thain A., Anderson L.A., Snowden A.W., Garcia-Wilson E., Perkins N.D., Hay R.T.. p300 transcriptional repression is mediated by SUMO modification. Mol. Cell. 2003; 11:1043–1054. [DOI] [PubMed] [Google Scholar]
- 25. Nathan D., Sterner D.E., Berger S.L.. Histone modifications: now summoning sumoylation. P Natl Acad Sci USA. 2003; 100:13118–13120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Trujillo K.M., Tyler R.K., Ye C.Y., Berger S.L., Osley M.A.. A genetic and molecular toolbox for analyzing histone ubiquitylation and sumoylation in yeast. Methods. 2011; 54:296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chatterjee C., McGinty R.K., Fierz B., Muir T.W.. Disulfide-directed histone ubiquitylation reveals plasticity in hDot1L activation. Nat. Chem. Biol. 2010; 6:267–269. [DOI] [PubMed] [Google Scholar]
- 28. Dhall A., Wei S., Fierz B., Woodcock C.L., Lee T.H., Chatterjee C.. Sumoylated human histone H4 prevents chromatin compaction by inhibiting long-range internucleosomal interactions. J. Biol. Chem. 2014; 289:33827–33837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dhall A., Weller C.E., Chu A., Shelton P.M.M., Chatterjee C.. Chemically sumoylated histone H4 stimulates intranucleosomal demethylation by the LSD1-CoREST complex. ACS Chem. Biol. 2017; 12:2275–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shi Y.J., Matson C., Lan F., Iwase S., Baba T., Shi Y.. Regulation of LSD1 histone demethylase activity by its associated factors. Mol. Cell. 2005; 19:857–864. [DOI] [PubMed] [Google Scholar]
- 31. Jain N., Tamborrini D., Evans B., Chaudhry S., Wilkins B.J., Neumann H.. Interaction of RSC chromatin remodeling complex with nucleosomes is modulated by H3 K14 acetylation and H2B SUMOylation in vivo. Iscience. 2020; 23:101292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cairns B.R., Lorch Y., Li Y., Zhang M.C., Lacomis L., ErdjumentBromage H., Tempst P., Du J., Laurent B., Kornberg R.D.. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996; 87:1249–1260. [DOI] [PubMed] [Google Scholar]
- 33. Spain M.M., Ansari S.A., Pathak R., Palumbo M.J., Morse R.H., Govind C.K.. The RSC complex localizes to coding sequences to regulate Pol II and histone occupancy. Mol. Cell. 2014; 56:653–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Musladin S., Krietenstein N., Korber P., Barbaric S.. The RSC chromatin remodeling complex has a crucial role in the complete remodeler set for yeast PHO5 promoter opening. Nucleic Acids Res. 2014; 42:4270–4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Floer M., Wang X., Prabhu V., Berrozpe G., Narayan S., Spagna D., Alvarez D., Kendall J., Krasnitz A., Stepansky A.et al.. A RSC/nucleosome complex determines chromatin architecture and facilitates activator binding. Cell. 2010; 141:407–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Niimi A., Chambers A.L., Downs J.A., Lehmann A.R.. A role for chromatin remodellers in replication of damaged DNA. Nucleic Acids Res. 2012; 40:7393–7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hsu J.M., Huang J., Meluh P.B., Laurent B.C.. The yeast RSC chromatin-remodeling complex is required for kinetochore function in chromosome segregation. Mol. Cell. Biol. 2003; 23:3202–3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shim E.Y., Ma H.L., Oum J.H., Yanez Y., Lee S.E.. The yeast chromatin remodeler RSC complex facilitates end joining repair of DNA double-strand breaks. Mol. Cell. Biol. 2005; 25:3934–3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shim E.Y., Hong S.J., Oum J.H., Yanez Y., Zhang Y., Lee S.E.. RSC mobilizes nucleosomes to improve accessibility of repair machinery to the damaged chromatin. Mol. Cell. Biol. 2007; 27:1602–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ryu H.Y., Su D., Wilson-Eisele N.R., Zhao D.J., Lopez-Giraldez F., Hochstrasser M.. The Ulp2 SUMO protease promotes transcription elongation through regulation of histone sumoylation. EMBO J. 2019; 38:e102003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Matic I., van Hagen M., Schimmel J., Macek B., Ogg S.C., Tatham M.H., Hay R.T., Lamond A.I., Mann M., Vertegaal A.C.O.. In vivo identification of human small ubiquitin-like modifier polymerization sites by high accuracy mass spectrometry and an in vitro to in vivo strategy. Mol. Cell. Proteomics. 2008; 7:132–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tatham M.H., Jaffray E., Vaughan O.A., Desterro J.M.P., Botting C.H., Naismith J.H., Hay R.T.. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 2001; 276:35368–35374. [DOI] [PubMed] [Google Scholar]
- 43. Rosonina E., Duncan S.M., Manley J.L.. SUMO functions in constitutive transcription and during activation of inducible genes in yeast. Genes Dev. 2010; 24:1242–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chymkowitch P., Nguea A.P., Aanes H., Koehler C.J., Thiede B., Lorenz S., Meza-Zepeda L.A., Klungland A., Enserink J.M.. Sumoylation of Rap1 mediates the recruitment of TFIID to promote transcription of ribosomal protein genes. Genome Res. 2015; 25:897–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ryu H.Y., Zhao D., Li J., Su D., Hochstrasser M.. Histone sumoylation promotes Set3 histone-deacetylase complex-mediated transcriptional regulation. Nucleic Acids Res. 2020; 48:12151–12168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ryu H.Y., Lopez-Giraldez F., Knight J., Hwang S.S., Renner C., Kreft S.G., Hochstrasser M.. Distinct adaptive mechanisms drive recovery from aneuploidy caused by loss of the Ulp2 SUMO protease. Nat. Commun. 2018; 9:5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lee J.S., Smith E., Shilatifard A.. The language of histone crosstalk. Cell. 2010; 142:682–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wyce A., Xiao T., Whelan K.A., Kosman C., Walter W., Eick D., Hughes T.R., Krogan N.J., Strahl B.D., Berger S.L.. H2B ubiquitylation acts as a barrier to Ctk1 nucleosomal recruitment prior to removal by Ubp8 within a SAGA-related complex. Mol. Cell. 2007; 27:275–288. [DOI] [PubMed] [Google Scholar]
- 49. Ahn S.H., Kim M., Buratowski S.. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell. 2004; 13:67–76. [DOI] [PubMed] [Google Scholar]
- 50. Cho E.J., Kobor M.S., Kim M., Greenblatt J., Buratowski S.. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Gene Dev. 2001; 15:3319–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mosley A.L., Pattenden S.G., Carey M., Venkatesh S., Gilmore J.M., Florens L., Workman J.L., Washburn M.P.. Rtr1 is a CTD phosphatase that regulates RNA polymerase II during the transition from serine 5 to serine 2 phosphorylation. Mol. Cell. 2009; 34:168–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shahbazian M.D., Zhang K.L., Grunstein M.. Histone H2B ubiquitylation controls processive methylation but not monomethylation by Dot1 and Set1. Mol. Cell. 2005; 19:271–277. [DOI] [PubMed] [Google Scholar]
- 53. Sun Z.W., Allis C.D.. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002; 418:104–108. [DOI] [PubMed] [Google Scholar]
- 54. Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K.. High-resolution profiling of histone methylations in the human genome. Cell. 2007; 129:823–837. [DOI] [PubMed] [Google Scholar]
- 55. Pokholok D.K., Harbison C.T., Levine S., Cole M., Hannett N.M., Lee T.I., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E.et al.. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005; 122:517–527. [DOI] [PubMed] [Google Scholar]
- 56. Soares L.M., He P.C., Chun Y., Suh H., Kim T., Buratowski S.. Determinants of Histone H3K4 methylation patterns. Mol. Cell. 2017; 68:773–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pijnappel W.W., Schaft D., Roguev A., Shevchenko A., Tekotte H., Wilm M., Rigaut G., Seraphin B., Aasland R., Stewart A.F.. The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes Dev. 2001; 15:2991–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kim T., Buratowski S.. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell. 2009; 137:259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kim T., Xu Z., Clauder-Münster S., Steinmetz L.M., Buratowski S.. Set3 HDAC mediates effects of overlapping noncoding transcription on gene induction kinetics. Cell. 2012; 150:1158–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Smolle M., Workman J.L.. Transcription-associated histone modifications and cryptic transcription. BBA-Gene Regul. Mech. 2013; 1829:84–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Teissandier A., Bourc’his D.. Gene body DNA methylation conspires with H3K36me3 to preclude aberrant transcription. EMBO J. 2017; 36:1471–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Laribee R.N., Fuchs S.M., Strahl B.D.. H2B ubiquitylation in transcriptional control: a FACT-finding mission. Gene Dev. 2007; 21:737–743. [DOI] [PubMed] [Google Scholar]
- 63. Pavri R., Zhu B., Li G.H., Trojer P., Mandal S., Shilatifard A., Reinberg D.. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006; 125:703–717. [DOI] [PubMed] [Google Scholar]
- 64. Shieh G.S., Pan C.H., Wu J.H., Sun Y.J., Wang C.C., Hsiao W.C., Lin C.Y., Tung L., Chang T.H., Fleming A.B.et al.. H2B ubiquitylation is part of chromatin architecture that marks exon-intron structure in budding yeast. BMC Genomics. 2011; 12:627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Schulze J.M., Hentrich T., Nakanishi S., Gupta A., Emberly E., Shilatifard A., Kobor M.S.. Splitting the task: Ubp8 and Ubp10 deubiquitinate different cellular pools of H2BK123. Gene Dev. 2011; 25:2242–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Schulze J.M., Jackson J., Nakanishi S., Gardner J.M., Hentrich T., Haug J., Johnston M., Jaspersen S.L., Kobor M.S., Shilatifard A.. Linking cell cycle to histone modifications: SBF and H2B monoubiquitination machinery and cell-cycle regulation of H3K79 dimethylation. Mol. Cell. 2009; 35:626–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Choudhury R., Singh S., Arumugam S., Roguev A., Stewart A.F.. The Set1 complex is dimeric and acts with Jhd2 demethylation to convey symmetrical H3K4 trimethylation. Gene Dev. 2019; 33:550–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Magraner-Pardo L., Pelechano V., Coloma M.D., Tordera V.. Dynamic remodeling of histone modifications in response to osmotic stress in Saccharomyces cerevisiae. BMC Genomics. 2014; 15:247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sun H., Treco D., Schultes N.P., Szostak J.W.. Double-strand breaks at an initiation site for meiotic gene conversion. Nature. 1989; 338:87–90. [DOI] [PubMed] [Google Scholar]
- 70. Krogh B.O., Symington L.S.. Recombination proteins in yeast. Annu. Rev. Genet. 2004; 38:233–271. [DOI] [PubMed] [Google Scholar]
- 71. San Filippo J., Sung P., Klein H.. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008; 77:229–257. [DOI] [PubMed] [Google Scholar]
- 72. Xu Y., Ayrapetov M.K., Xu C., Gursoy-Yuzugullu O., Hu Y., Price B.D.. Histone H2A.Z controls a critical chromatin remodeling step required for DNA double-strand break repair. Mol. Cell. 2012; 48:723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. van Attikum H., Fritsch O., Gasser S.M.. Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. EMBO J. 2007; 26:4113–4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sacher M., Pfander B., Hoege C., Jentsch S.. Control of Rad52 recombination activity by double-strand break-induced SUMO modification. Nat. Cell Biol. 2006; 8:1284–1290. [DOI] [PubMed] [Google Scholar]
- 75. Altmannova V., Eckert-Boulet N., Arneric M., Kolesar P., Chaloupkova R., Damborsky J., Sung P., Zhao X.L., Lisby M., Krejci L.. Rad52 SUMOylation affects the efficiency of the DNA repair. Nucleic Acids Res. 2010; 38:4708–4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Biggins S. The composition, functions, and regulation of the budding yeast kinetochore. Genetics. 2013; 194:817–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ohkuni K., Levy-Myers R., Warren J., Au W.C., Takahashi Y., Baker R.E., Basrai M.A.. N-terminal sumoylation of centromeric histone H3 variant Cse4 regulates Its proteolysis to prevent mislocalization to non-centromeric chromatin. G3 (Bethesda). 2018; 8:1215–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ohkuni K., Suva E., Au W.C., Walker R.L., Levy-Myers R., Meltzer P.S., Baker R.E., Basrai M.A.. Deposition of centromeric histone H3 variant CENP-A/Cse4 into chromatin is facilitated by its C-terminal sumoylation. Genetics. 2020; 214:839–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Eisenstatt J.R., Ohkuni K., Au W.C., Preston O., Gliford L., Suva E., Costanzo M., Boone C., Basrai M.A.. Reduced Gene Dosage of Histone H4 Prevents CENP-A Mislocalization and Chromosomal Instability in Saccharomyces cerevisiae. Genetics. 2021; doi:10.1093/genetics/iyab033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Shrestha R.L., Ahn G.S., Staples M.I., Sathyan K.M., Karpova T.S., Foltz D.R., Basrai M.A.. Mislocalization of centromeric histone H3 variant CENP-A contributes to chromosomal instability (CIN) in human cells. Oncotarget. 2017; 8:46781–46800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Amato A., Schillaci T., Lentini L., Di Leonardo A.. CENPA overexpression promotes genome instability in pRb-depleted human cells. Mol. Cancer. 2009; 8:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Athwal R.K., Walkiewicz M.P., Baek S., Fu S., Bui M., Camps J., Ried T., Sung M.H., Dalal Y.. CENP-A nucleosomes localize to transcription factor hotspots and subtelomeric sites in human cancer cells. Epigenet Chromatin. 2015; 8:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Hu Z., Huang G., Sadanandam A., Gu S.D., Lenburg M.E., Pai M., Bayani N., Blakely E.A., Gray J.W., Mao J.H.. The expression level of HJURP has an independent prognostic impact and predicts the sensitivity to radiotherapy in breast cancer. Breast Cancer Res. 2010; 12:R18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lacoste N., Woolfe A., Tachiwana H., Garea A.V., Barth T., Cantaloube S., Kurumizaka H., Imhof A., Almouzni G.. Mislocalization of the centromeric histone variant CenH3/CENP-A in human cells depends on the chaperone DAXX. Mol. Cell. 2014; 53:631–644. [DOI] [PubMed] [Google Scholar]
- 85. Li Y., Zhu Z., Zhang S.H., Yu D.H., Yu H.Y., Liu L.N., Cao X.Z., Wang L., Gao H.J., Zhu M.H.. ShRNA-targeted centromere protein a inhibits hepatocellular carcinoma growth. PLoS One. 2011; 6:e17794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Sun X., Clermont P.L., Jiao W.L., Helgason C.D., Gout P.W., Wang Y.Z., Qu S.F.. Elevated expression of the centromere protein-A(CENP-A)-encoding gene as a prognostic and predictive biomarker in human cancers. Int. J. Cancer. 2016; 139:899–907. [DOI] [PubMed] [Google Scholar]
- 87. Tomonaga T., Matsushita K., Yamaguchi S., Oohashi T., Shimada H., Ochiai T., Yoda K., Nomura F.. Overexpression and mistargeting of centromere protein-A in human primary colorectal cancer. Cancer Res. 2003; 63:3511–3516. [PubMed] [Google Scholar]
- 88. Wu Q., Qian Y.M., Zhao X.L., Wang S.M., Feng X.J., Chen X.F., Zhang S.H.. Expression and prognostic significance of centromere protein A in human lung adenocarcinoma. Lung Cancer. 2012; 77:407–414. [DOI] [PubMed] [Google Scholar]
- 89. Neyret-Kahn H., Benhamed M., Ye T., Le Gras S., Cossec J.C., Lapaquette P., Bischof O., Ouspenskaia M., Dasso M., Seeler J.et al.. Sumoylation at chromatin governs coordinated repression of a transcriptional program essential for cell growth and proliferation. Genome Res. 2013; 23:1563–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Everett R.D., Boutell C., Hale B.G.. Interplay between viruses and host sumoylation pathways. Nat. Rev. Microbiol. 2013; 11:400–411. [DOI] [PubMed] [Google Scholar]
- 91. Choudhary C., Kumar C., Gnad F., Nielsen M.L., Rehman M., Walther T.C., Olsen J.V., Mann M.. Lysine acetylation targets protein complexes and Co-Regulates major cellular functions. Science. 2009; 325:834–840. [DOI] [PubMed] [Google Scholar]
- 92. Kim W., Bennett E.J., Huttlin E.L., Guo A., Li J., Possemato A., Sowa M.E., Rad R., Rush J., Comb M.J.et al.. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell. 2011; 44:325–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Wagner S.A., Beli P., Weinert B.T., Scholz C., Kelstrup C.D., Young C., Nielsen M.L., Olsen J.V., Brakebusch C., Choudhary C.. Proteomic analyses reveal divergent ubiquitylation site patterns in murine tissues. Mol. Cell. Proteomics. 2012; 11:1578–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Danielsen J.M.R., Sylvestersen K.B., Bekker-Jensen S., Szklarczyk D., Poulsen J.W., Horn H., Jensen L.J., Mailand N., Nielsen M.L. Mass spectrometric analysis of lysine ubiquitylation reveals promiscuity at site level. Mol. Cell. Proteomics. 2011; 10:M110.003590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Olsen J.V., Vermeulen M., Santamaria A., Kumar C., Miller M.L., Jensen L.J., Gnad F., Cox J., Jensen T.S., Nigg E.A.et al.. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 2010; 3:ra3. [DOI] [PubMed] [Google Scholar]
- 96. Pedrioli P.G.A., Raught B., Zhang X.D., Rogers R., Aitchison J., Matunis M., Aebersold R.. Automated identification of SUMOylation sites using mass spectrometry and SUMmOn pattern recognition software. Nat. Methods. 2006; 3:533–539. [DOI] [PubMed] [Google Scholar]
- 97. Guzzo C.M., Berndsen C.E., Zhu J.M., Gupta V., Datta A., Greenberg R.A., Wolberger C., Matunis M.J.. DNA DAMAGE RNF4-dependent hybrid SUMO-ubiquitin chains are signals for RAP80 and thereby mediate the recruitment of BRCA1 to sites of DNA damage. Sci. Signal. 2012; 5:ra88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Yang Y., He Y., Wang X., Liang Z., He G., Zhang P., Zhu H., Xu N., Liang S.. Protein SUMOylation modification and its associations with disease. Open Biol. 2017; 7:170167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Srivastava R., Ahn S.H.. Modifications of RNA polymerase II CTD: connections to the histone code and cellular function. Biotechnol. Adv. 2015; 33:856–872. [DOI] [PubMed] [Google Scholar]
- 100. Timmermann S., Lehrmann H., Polesskaya A., Harel-Bellan A.. Histone acetylation and disease. Cell. Mol. Life Sci. 2001; 58:728–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Greer E.L., Shi Y.. Histone methylation: a dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012; 13:343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Jeusset L.M.P., McManus K.J. Developing targeted therapies that exploit aberrant histone ubiquitination in cancer. Cells. 2019; 8:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Cao J., Yan Q.. Histone ubiquitination and deubiquitination in transcription, DNA damage response, and cancer. Front. Oncol. 2012; 2:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data in the referenced studies published by the present authors are available from the authors or have been deposited in public databases.