Abstract
The zinc uptake regulator (Zur) is a member of the Fur (ferric uptake regulator) family transcriptional regulators that plays important roles in zinc homeostasis and virulence of bacteria. Upon zinc perception, Zur binds to the promoters of zinc responsive genes and controls their transcription. However, the mechanism underlying zinc-mediated Zur activation remains unclear. Here we report a 2.2-Å crystal structure of apo Zur from the phytopathogen Xanthomonas campestris pv. campestris (XcZur), which reveals the molecular mechanism that XcZur exists in a closed inactive state before regulatory zinc binding. Subsequently, we present a 1.9-Å crystal structure of holo XcZur, which, by contrast, adopts an open state that has enough capacity to bind DNA. Structural comparison and hydrogen deuterium exchange mass spectrometry (HDX-MS) analyses uncover that binding of a zinc atom in the regulatory site, formed by the hinge region, the dimerization domain and the DNA binding domain, drives a closed-to-open conformational change that is essential for XcZur activation. Moreover, key residues responsible for DNA recognition are identified by site-directed mutagenesis. This work provides important insights into zinc-induced XcZur activation and valuable discussions on the mechanism of DNA recognition.
Graphical Abstract
Graphical Abstract.
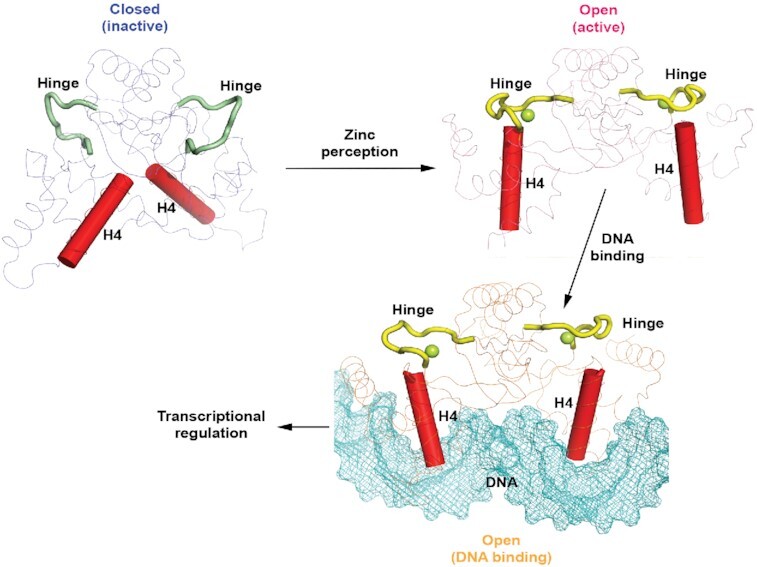
A proposed model of zinc-perception-induced DNA binding for XcZur. In the model, apo XcZur is in a closed inactive state. Zinc perception in the regulatory zinc binding site induces a 180° rotation of the hinge region to drive a closed-to-open conformational change of the XcZur dimer. Holo XcZur thus bears an active state, in which the putative DNA binding elements, especially the recognition helix (H4), are precisely positioned and poised for DNA binding. Subsequently, holo XcZur binds with DNA in a mechanism of mutual induced fit and regulates DNA transcription.
INTRODUCTION
As a chemically stable trace element, zinc is essential for almost all living organisms and required for the structural stability and/or catalytic activity of many zinc-dependent metalloproteins (1,2). However, zinc in excess inhibits NADH oxidase activity of the respiratory chain and is toxic to cells (3,4). Consequently, to survive, cells must develop mechanisms to sense this metal ion and tightly regulate its intracellular level (5). In bacteria, maintenance of zinc homeostasis depends primarily on the zinc uptake regulator (Zur) and the MerR family and ArsR/SmtB family regulators (ZntR, SmtB, CzrA) (6,7). Zur is a zinc sensor that not only represses the zinc-uptake systems but also activates the zinc-export systems, in a zinc-dependent manner (8,9). Zinc depletion leads to inactivation of Zur and de-repression of zinc responsive genes encoding the zinc importer system (6). Corresponding to its critical role in regulation of zinc homeostasis, Zur homologs have been identified in a number of bacteria, including Escherichia coli (10), Bacillus subtilis (11), Listeria monocytogenes (12), Staphylococcus aureus (13), Xanthomonas campestris (8), Mycobacterium tuberculosis (14), Streptomyces coelicolor (15), Streptococcus suis (16), Yersinia pestis (17), Corynebacterium glutamicum (18), Pseudomonas aeruginosa (19) and Salmonella enterica (20).
Zur belongs to the Fur (ferric uptake regulator) family of transcriptional regulators that modulate the expression of metal responsive genes, in response to the fluxes of a specific metal. Since the first discovery of Fur in E. coli, >16 000 members have been documented in Pfam (PF01475) (21). The Fur family regulators are typically divided into three functional groups, i.e. the Fur group members that sense ferric and participate in iron homeostasis and virulence (22–24), the Zur group members that sense zinc and are involved in zinc uptake, virulence and rearrangement of ribosomal proteins (6,25), and the PerR group members that sense peroxides and play important roles in regulating the peroxide stress response (26,27). In addition, Fur family regulators that respond to other metals include sensors of manganese (Mur) (28,29) and nickel (Nur) (30,31). To couple metal perception and DNA binding, Fur family proteins conservatively fold into a two-domain architecture and frequently utilize two or more metal binding sites for metal-dependent conformational regulation (32). Available Fur family structures reveal a dimeric assembly and a modular domain organization for each monomer, which contains an N-terminal DNA binding domain (DBD), a C-terminal dimerization domain (DD), and a hinge region linking the two domains (32). The capability for a Fur protein to bind its target DNA depends on the exact conformation of the protein, which in turn depends on the type and number of binding metals as well as where the metals bind to.
The molecular mechanisms by which metal binding induces conformational changes of Fur proteins have been studied in some details. Evolutionary pressure seems to have forced Fur proteins to adopt similar active sates so that structural elements required for DNA binding, especially the recognition helix and the β-hairpin wing in the DBD, are precisely positioned (24,31,33–40). Conversely, inactive Fur proteins can adopt very distinct conformations and often vary between individual proteins and their different metal-binding states (38,41–48). Structural comparisons between inactive and active states reveal an allosteric mechanism for activation of Fur and PerR group members, in which binding with appropriate metal ion in the correct metal binding site is the requisite (31,34,38,47). Typically, Fur family proteins possess two or three metal binding sites. Site 1 is a common structural site with four conserved cysteines for zinc coordination; site 2 is thought to be the major metal sensor located between DBD and DD; while the function of DD-located site 3 is less clear (49–51). It appears that metal binding in site 2 may affect the mutual orientation of the two domains and regulate DNA binding. Despite these advances, the mechanism and structural basis of zinc-induced Zur activation remains unclear, due partly to lack of structural information on the inactive state at which only one zinc is bound (apo state). Only three structures to date have been determined for the Zur group proteins (MtZur from M. tuberculosis, ScZur from S. coelicolor and EcZur from E. coli) and they all contain two or three zinc ions (36,37,43). EcZur binds to its cognate DNA as a dimer of dimers, which is stabilized by a pair of asymmetric salt bridges between D49 and R52 (37). Consequently, it is entirely unknown which conformation apo Zur is most likely to adopt and what structural changes are associated with zinc binding.
Our previous studies characterized a Zur protein (XcZur) in Xanthomonas campestris pv. campestris (Xcc), the causal agent of black rot disease of cruciferous crops worldwide (8). The studies demonstrated that XcZur is essential for zinc homeostasis and full virulence of Xcc (8). To regulate the intracellular zinc level, XcZur acts as an activator for the expression of the zinc-export gene, XC2976, and a repressor for the expression of the zinc-uptake genes, XC0267, XC2472 and XC3788 (52). Particularly, the proteins XC0267 and XC2472 are components of an YciABC-like low-affinity zinc-uptake system; XC3788 exhibits 38% similarity to the ZnuC, a component of a high-affinity zinc-uptake system; while XC2976 displays 67% similarity to the zinc efflux pump CzcD of Ralstonia metallidurans. In addition, XcZur is involved in hypersensitive response (HR) and positively regulates the transcription of genes associated with HR and pathogenicity (53). Among Zur group members, XcZur shares 42% sequence identity with EcZur (8), indicating a similar structure and biochemical function. However, sequence alignment between XcZur and MtZur or ScZur reveals limited conservation (∼30% identity only).
To understand the fundamental mechanisms of Zur activation and zinc perception, we determined the crystal structures of an inactive apo XcZur bound to a single zinc ion and an active holo XcZur in complex with two zinc ions. These structures reveal drastic conformational changes associated with XcZur bound to a zinc ion in the regulatory site. In conjunction with results from hydrogen deuterium exchange mass spectrometry (HDX-MS) analyses of XcZur in different conformational states, our work provides a mechanism and structural basis for zinc-induced XcZur activation as well as DNA recognition.
MATERIALS AND METHODS
Sequence alignment and phylogenetic analysis
The primary sequences of XcZur homologs were acquired by a three-iteration PSI-BLAST search (54) from the PDB (protein data bank) database. A total of 15 non-redundant sequences from 21 structures were identified and their NCBI accession numbers are as follows: EcZur, 4MTD_A; MtZur, 2O03_A; ScZur, 3MWM_A; ScNur, 3EYY_A; CjPerR, 6DK4_A; LiPerR, 5NL9_A; SpPerR, 4I7H_A; BsPerR, 2FE3_A; CjFur, 4ETS_A; HpFur, 2XIG_A; RlMur, 5FD6_A; MgFur, 4RAY_A; FtFur, 5NBC_A; VcFur, 2W57_A; PaFur, 1MZB_A.
The T-Coffee server (55) was used to perform the multiple-sequence alignment (MSA). Then, MEGA version 7 was applied to conduct phylogenetic analysis for the MSA using the bootstrap neighbor joining method (56). For test of phylogeny, 10 000 replications were used in the bootstrap test (57). The Poisson correction method (58) was used to calculate evolutionary distances. Results derived from MSA were also subjected to figure production of sequence alignment by ESPript version 3.0 (59).
Cloning and expression
The coding sequence of XcZur was amplified by standard polymerase chain reaction (PCR) from the genomic DNA of Xcc strain 8004. The amplified DNA fragment was inserted into the pRSFDuet-1 vector (Novagen) between BamHI and XhoI restriction sites. After confirmation by DNA sequencing, the recombinant plasmid was introduced into E. coli strain BL21(DE3) for protein expression. Transformed cells were grown at 37°C in Luria-Bertani medium containing kanamycin. When the OD600 reached 0.6–0.8, protein expression was induced by adding 0.5 mM Isopropyl β-d-1-thiogalactopyranoside (IPTG) followed by 18–20 h incubation at 16°C. The construct of XcZur-Δ16 was generated and expressed in a similar way. The final protein product from the recombinant pRSFDuet-1 vector is expected to contain a 6× His tag at the N-terminus.
Protein purification
The cells were harvested by centrifugation at 4200 rpm for 15 min, resuspended with 30 ml lysis buffer containing 20 mM Tris–HCl (pH 8.0), 150 mM NaCl, 30 mM imidazole, and then homogenized using a low-temperature ultra-high-pressure cell disrupter (JNBIO, Guangzhou, China). After centrifugation at 12 000 rpm for 1 h, the resulting supernatant was loaded onto a Ni-NTA agarose column (GE Healthcare). The eluted protein was subjected onto a HiTrap Heparin column (GE Healthcare) equilibrated with 20 mM Tris–HCl (pH 8.0), 75 mM NaCl. The protein was eluted by a linear NaCl gradient and further purified by size exclusion chromatography using Superdex 200 columns (GE Healthcare) in a buffer containing 20 mM Tris (pH 8.0) and 150 mM NaCl. The protein was concentrated to approximate 10 mg/ml and stored at −80°C.
Crystallization and structure determination
Crystals were obtained by the hanging drop vapor diffusion method, by mixing 1 μl of protein sample with 1 μl of reservoir solution at 16°C. Best crystals of apo XcZur grew in a reservoir solution containing 0.2 M calcium acetate hydrate, 0.1 M sodium cacodylate trihydrate pH 6.5, 18% (w/v) polyethylene glycol 8000, 0.3 M glycyl-glycyl-glycine. To avoid zinc precipitation in basic conditions, holo XcZur was buffer exchanged into a solution containing 100 mM MES (pH 6.0) and 150mM NaCl and then incubated with excess zinc acetate (molar ratio of metal to protein equals to 1.2:1). Best crystals of holo XcZur grew in a reservoir solution containing 0.2 M sodium malonate pH 5.0, 20% (w/v) polyethylene glycol 3,350.
The native and single-wavelength anomalous dispersion (SAD) datasets of apo XcZur were collected on beamline BL19U1 (60) at the Shanghai Synchrotron Radiation Facility (SSRF). Datasets were processed and scaled using XDS (61). Two sets of data were merged to improve the anomalous signal. The initial phase was determined by Phenix.autosol (62) using the SAD method, during which NCS of 4 molecules in an asymmetric unit was used for density improvement. Native data in higher resolution was then used for model building in Phenix.autobuild (63), refinement and validation. The whole model of the structure was manually built in COOT (64) and iteratively refined by Phenix.refine (65). In the final model, more than 96.4% residues fall in the favored region in the Ramachandran plot and the final Rwork/Rfree are 0.219/0.276.
The SAD dataset of holo XcZur was collected on beamline BL17U1 at SSRF (66), processed and scaled using XDS (61). The initial phase was determined by Phenix.autosol (62) through the SAD method and the initial model was built by Phenix.autobuild (63). The whole model was manually built in COOT (64) and iteratively refined by Phenix.refine (65). In the final model, more than 96.4% residues fall in the favored region in the Ramachandran plot and the final Rwork/Rfree are 0.213/0.252.
Data collection and refinement statistics are summarized in Table 1. Structural figures were produced by PyMOL (The PyMOL Molecular Graphics System, Schrödinger, LLC).
Table 1.
Data collection and structure refinement statistics
| Apo XcZur | Holo XcZur | ||
|---|---|---|---|
| Parameters | Zn-SAD | Native | Zn-SAD |
| Data collection statistics | |||
| Cell parameters | |||
| a (Å) | 153.43 | 153.64 | 34.56 |
| b (Å) | 77.91 | 78.05 | 122.9 |
| c (Å) | 73.09 | 73.12 | 86.89 |
| α, β, γ (°) | 90,0 103.08, 90.0 | 90,0 103,06 90.0 | 90, 90, 90 |
| Space group | C2 | C2 | C2221 |
| Wavelength used (Å) | 0.9785 | 0.9785 | 0.9792 |
| Resolution (Å) | 50.0–2.40 (2.49–2.40)c | 50.00–2.20 (2.34–2.20)c | 22.57–1.85 (1.92–1.85)c |
| No. of all reflections | 413 164 | 141 407 | 186 417 |
| No. of unique reflections | 32 172 | 42 394 | 15 694 |
| Completeness (%) | 99.7 (99.6) | 98.7 (96.9) | 95.5 (86.7) |
| Average I/σ (I) | 17.4 (4.93) | 13.01 (2.26) | 15.35 (1.69) |
| R merge a (%) | 6.2 (20.5) | 4.8 (38.7) | 4.0 (52.9) |
| Refinement statistics | |||
| No. of reflections used (σ(F) > 0) | 42 394 | 15 557 | |
| R work b (%) | 21.94 | 21.26 | |
| R free b (%) | 27.55 | 25.17 | |
| r.m.s.d. bond distance (Å) | 0.008 | 0.015 | |
| r.m.s.d. bond angle (°) | 1.430 | 1.315 | |
| Average B-value (Å2) | |||
| Average B-value for protein atoms | 40.06 | 50.14 | |
| Average B-value for solvent atoms | 41.18 | 46.17 | |
| No. of atoms | |||
| No. of protein atoms | 4550 | 1086 | |
| No. of solvent atoms | 364 | 70 | |
| Ramachandran plot | |||
| Res. in favored regions (%) | 96.43 | 96.35 | |
| Res. in outlier regions (%) | 0 | 0 | |
a R merge = ΣhΣi|Ih,i−Ih|/ΣhΣiIh,i, where Ih is the mean intensity of the i observations of symmetry related reflections of h.
b R work = ∑(||Fp(obs)|−|Fp(calc)||)/∑|Fp(obs)|; Rfree is an R factor for a pre-selected subset (5%) of reflections that was not included in refinement. Fp, structure factor of protein.
cNumbers in parentheses are corresponding values for the highest resolution shell.
Site-specific mutagenesis
The 19 mutant clones were constructed by a PCR-based strategy using the Fast Mutagenesis System (TransGen Biotech, Beijing, China). The corresponding primers were listed in Supplementary Table S1. Each mutant clone, verified by DNA sequencing, was transferred into BL21(DE3) and expressed using the same procedure for the wild type clone. The mutant protein was purified tandemly by using the Ni-NTA agarose column (GE Healthcare), the HiTrap Q column (GE Healthcare) and the Superdex 200 Increase prepacked column (GE Healthcare).
Analytical ultracentrifugation (AUC)
Sedimentation Velocity Analytical Ultracentrifugation (SV-AUC) experiments were carried out at 20°C in an XL-I analytical ultracentrifuge (Beckman Coulter). Absorbance (XcZur and XcZur-DNA complexes: 280 nm; DNA: 260 nm) and Raleigh interference optics were used to simultaneously record the radial concentration as a function of time until the lightest sedimenting component cleared the optical window (7 h). All samples (400 μl) were centrifuged at 50 000 rpm for 8 h in an An50Ti rotor using 12 mm double-sector aluminum centerpieces. Apo XcZur was prepared in the interaction buffer A (20 mM HEPES pH 7.4, 150 mM NaCl), other samples were in the interaction buffer B (20 mM HEPES pH 7.4, 150 mM NaCl, 0.18 mM EDTA, 0.2 mM Zn). Interference profiles were recorded every 6 min. Data analysis was conducted by the software SEDFIT 11.7 (67), GUSSI and SEDPHAT. Theoretical sedimentation coefficients were calculated by using HydroPro 7c (68) with a hydrated radius of 3.1 Å for the atomic elements.
Electrophoretic mobility shift assay (EMSA)
DNA oligonucleotides (Supplementary Table S2) were synthesized by Shanghai mapbioo (China) with the forward strand labelled by FAM (fluorophore 6-carboxy-fluorescein) and annealed in a resolving buffer containing 50 mM Tris–HCl (pH 7.5) and 250 mM KCl. For each assay, FAM-labelled DNA (10 nM) was incubated with protein in a reaction buffer containing 10 mM Tris–HCl (pH 7.5), 50 mM KCl, 5 mM MgCl2, 1 mM DTT, 0.1% NP-40, 10% glycerol at room temperature for 30 min. Then the samples were subjected to a native 6% polyacrylamide gel at 150 V and 4°C in a running buffer of 40 mM Tris–HCl pH 8.0. The gel was scanned using the PharosFX Personal Molecular Image (Bio-Rad). All assays were performed in triplicate.
Hydrogen/deuterium exchange mass spectrometry (HDX-MS)
For HDX-MS, 8 μg/μl of XcZur in the presence and absence of ligand was prepared. For each sample, deuterium labeling was carried out for 1 min and 5 min to capture different exchange times, in a procedure described previously (69). To increase peptide coverage of XcZur (Supplementary Figure S1), protein digestion was accomplished by separately adding 6 μl of 1 μM pepsin or protease from Aspergillus saitoi Type XIII on ice for 10 min. After centrifugation, the resulting soluble peptides were mixed and analyzed by the ACQUITY UPLC 1.7 μm BEH C18 1.0 μm × 50 mm column (Waters) in a 0–100% gradient of acetonitrile, supplemented with 1% (v/v) formic acid, over a course of 20 min. Mass spectrometry analysis was performed on Q Exactive Orbitrap mass spectrometer (Thermo, CA). Data analysis was performed by the HDExaminer (Sierra Analytics) and Thermo Xcalibur Qual Browser (Thermo, CA). According to HDX-MS analyses, the peptides are listed by the amplitude of rate changes caused by zinc or DNA binding, and eight peptides with most significant changes are selected and mapped onto the crystal structure of holo XcZur. On the other hand, eight peptides with most significant changes of solvent accessibility are also mapped onto the structure of holo XcZur.
Metal content analysis by ICP-MS
The metal content of protein samples was determined by inductively coupled plasma mass spectrometry (ICP-MS) using Plasma Quant MS from Analytik Jena AG. All protein samples were soaked in 65% HNO3 and incubated overnight at room temperature. Then the protein samples dissolved in 65% HNO3 were digested in Graphite furnace at a temperature of up to 140°C. The final reaction was diluted to 4% HNO3, and the metal concentration of the protein was determined by ICP-MS. The protein samples for metal content determination include XcZur, EDTA treated XcZur, XcZur-Δ16, EDTA treated XcZur-Δ16, XcZurH99A and XcZurC125S, and all samples were measured in three replicates.
Biolayer interferometry (BLI)
The DNA binding affinity of XcZur was determined by biolayer interferometry (BLI) on ForteBio Octet K2 two-channel system by using the streptavidin (SA) sensors. Binding study was performed in 96-well microplates in a volume of 200 μl per well at 25°C with constant shaking at 1000 rpm. The assay buffer contained 20 mM HEPES pH 7.4, 150 mM NaCl, 0.18 mM EDTA, 0.2 mM Zn and 0.02% Tween-20. The analysis was carried out using a set of eight appropriate XcZur concentrations (P0267 and P2472: 0, 9.4–600 nM; P3788: 0, 15.6 to 1000 nM). The sequence for each run was as follows: SA sensors prewet (10 min), equilibration (180 s), binding of 10 nM biotinylated–DNA (90 s), baseline stabilization (180 s), XcZur association (300 s) and XcZur dissociation (500 s). For each run a new biosensor was used. Global fitting was used for data analysis by the ForteBio software, specifying the 1:1 kinetic model and correcting both association and dissociation curves.
RESULTS
XcZur is a Zur group regulator with specific sequence features
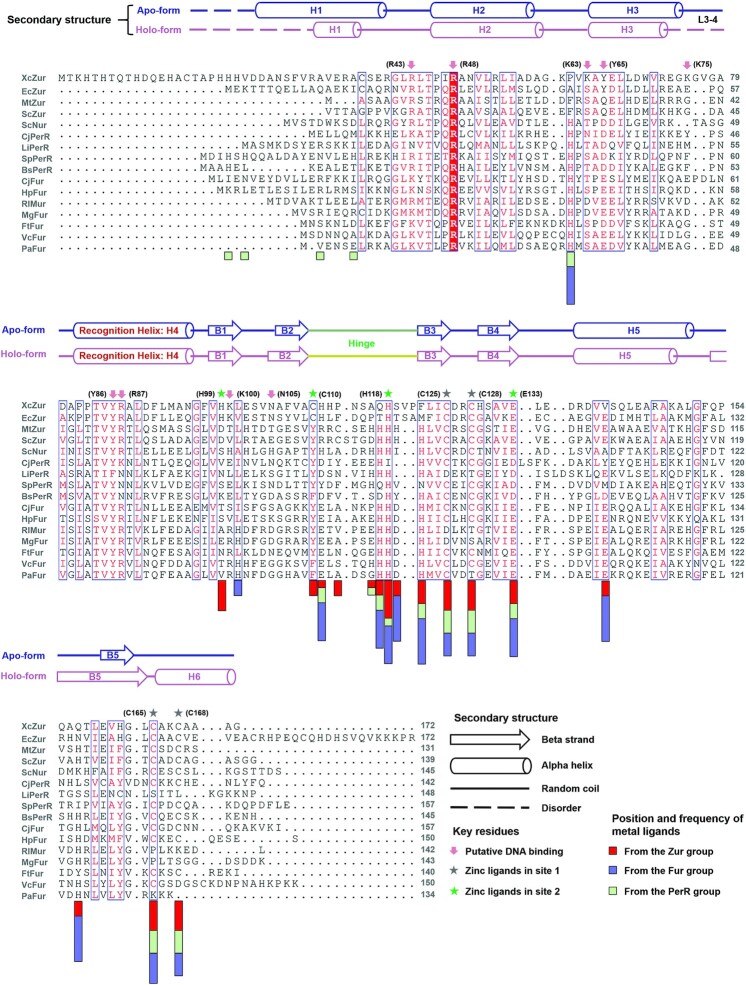
The amino acid sequence of XcZur was subjected to a three-iteration PSI-BLAST search (54) and led to the identification of 15 sequences with significant regions of similarity from PDB. As can be expected, all identified proteins belong to the Fur family of transcriptional regulators. The neighbor-joining method (56) was used to perform phylogenetic analysis shown in Supplementary Figure S2A. The phylogenetic tree includes three main clades, corresponding exactly to the three functional groups of Fur family proteins. Notably, the four selected Zur proteins (XcZur, EcZur, MtZur and ScZur) and a nickel uptake regulator (Nur) are located in the same clade (referred to as Zur group) but separated from other Fur family regulators (Fur group and PerR group) on the evolutionary tree. Sequence alignment of the 16 Fur family proteins were accomplished by combining multiple methods and shown in Figure 1.The Fur family regulators possess a distinctive core domain with ∼130 residues in length, corresponding to a fragment of XcZur encompassing residues C37 to C165. Nevertheless, the overall amino acid sequence identity among the Fur family members is not extensive and there is only one invariant residue existing in the family (R48, numbered in XcZur sequence) (Figure 1). Limited overall sequence identity can be also found in the Zur group members (Figure 1), although within which XcZur displays a sequence identity of 42% with EcZur (8). XcZur is unique in that it has an additional extension N-terminal to the relatively conserved core domain. While the overall sequence identities of Fur family proteins are relatively low, three of the four ligands in the structural metal binding site (C125, C128, C165) and three ligands in the regulatory site (C110, H118, E133) are well conserved (Figure 1).
Figure 1.
Structural elements and conservation of Fur family proteins. Sequence alignment of XcZur with fifteen representative Fur family proteins. Conserved amino acids are highlighted in blue box. Secondary structure elements of apo and holo XcZur are shown above the alignment. The position and frequency of metal ligands are indicated by red (Zur group), blue (Fur group) and green (PerR group) boxes, under the alignment. The key residues for DNA binding, zinc coordination in site 1 and in site 2 are displayed as red arrow, gray stars and green stars, respectively.
Zinc perception is essentially required for the DNA binding capacity of XcZur
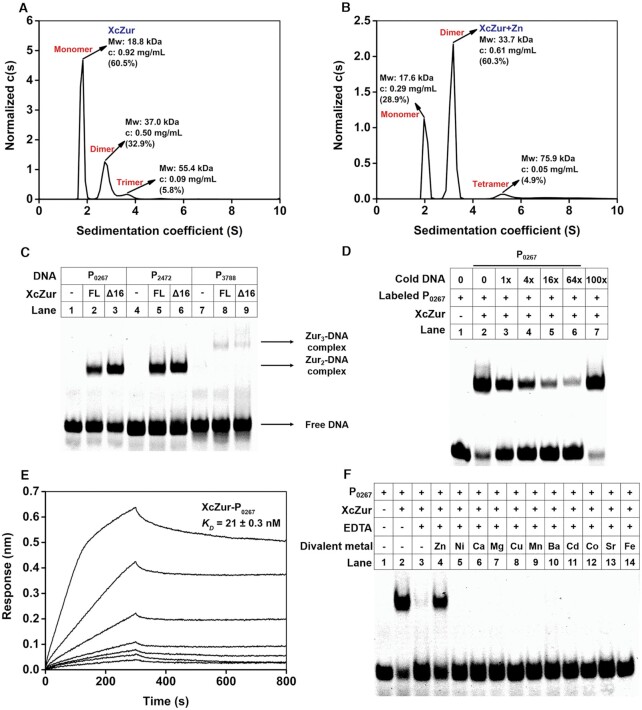
It is reported that Fur family proteins behave typically as homodimers in solution, however, previous NMR study revealed that E. coli Fur exists in solution under several oligomeric states, including both dimeric and monomeric states (41). To investigate the oligomeric state of XcZur in solution, purified XcZur was subjected to SV-AUC analyses. Purified XcZur behaved mainly as monomers (60.5%) and dimers (32.9%), with a trace amount of trimers (5.8%) (Figure 2A). Zinc addition altered the oligomeric states to 60.3% of dimers, 28.9% of monomers and 4.9% of tetramers (Figure 2B). These results demonstrate that XcZur can form both monomers and dimers in solution and zinc perception appears to be able to significantly stabilize the dimeric form.
Figure 2.
Biochemical characterization of XcZur. (A) Sedimentation coefficient distribution c(s) of XcZur determined by SV-AUC. The calculated value of the protein oligomeric state of XcZur is indicated. (B) Sedimentation coefficient distribution c(s) of XcZur+Zn determined by SV-AUC. The calculated value of the protein oligomeric state of XcZur is indicated. (C) EMSA for XcZur and XcZur-Δ16 with P0267, P2472 and P3788. (D) Competitive binding assay. The FAM-labelled P0267 probe and XcZur were incubated with varying amounts of unlabeled DNA. Lane1, DNA probe; lane 2, standard binding reaction; lanes 3–6, standard binding reaction plus 1- to 64-fold (4-fold increase) molar excess of unlabeled P0267, lane 7, standard binding reaction plus 100-fold molar excess irrelevant unlabeled DNA. (E) Affinity determination of XcZur binding with P0267 by BLI. The BLI responses for association and dissociation, and the calculated KD are shown. (F) Effect of EDTA, zinc and other divalent metals on the binding of XcZur to P0267. Each EMSA experiment was repeated three times and similar results were obtained.
Previous DNaseI footprinting studies revealed that XcZur binds with specific DNA fragments (referred to as P0267, P2472, P3788 and P2976) from the promoters of four zinc homeostasis genes (52). According to the results derived from EMSA, XcZur bound to three of the four DNA fragments, P0267, P2472 and P3788, with the former two as the most preferred targets (Figure 2C). These three DNA fragments are well conserved with a pseudo-palindromic core sequence (Supplementary Figure S2B). To demonstrate the specificity of XcZur–DNA interaction, FAM-labeled P0267 and XcZur were incubated with varying amounts of unlabeled DNA (cold DNA). Competitive binding of excess cold P0267, but not that of a control DNA with a random sequence, to XcZur confirmed the binding specificity (Figure 2D). Surprisingly, no stable association was detected between XcZur and P2976, suggesting requirement of other nucleotides in the promoter for protein-DNA interaction (Supplementary Figure S3). The DNA binding affinity of XcZur was determined by BLI (Figure 2E and Supplementary Figure S4). The resulting dissociation constants (KD) of XcZur for P0267, P2472 and P3788 were approximate 22, 38 and 63 nM, respectively. These dissociation constants are in a similar range with the DNA-binding affinities of many Fur family regulators and consistent with our EMSA results (Figure 2C). To determine the stoichiometries of the three XcZur-DNA complexes, we subjected them to SV-AUC analyses. The stoichiometries for the XcZur-P0267, XcZur-P2472 and XcZur-P3788 complexes were 2:1, 2:1 and 3:1, respectively (Supplementary Figure S5).
To test whether zinc perception is required for the DNA binding activity of XcZur, we titrated the regulator with P0267 in the presence of the zinc chelator EDTA by EMSA. Addition of 3 mM of EDTA abrogated DNA binding (Figure 2F), suggesting that a metal ion is required for the DNA binding activity of XcZur. Importantly, further addition of zinc to a concentration of 0.3 mM in the presence of 3 mM EDTA almost completely restored the DNA binding capacity of XcZur (Figure 2F). Since there are 5 mM of magnesium ions in the binding buffer, most of the EDTA can be used by magnesium ions and 0.3 mM of zinc ions can be sufficient to activate the DNA binding activity of XcZur. In sharp contrast, however, addition of the other 10 divalent metal ions, including the most common Fur ligands nickel, ferric and manganese, could not restore the DNA binding ability (Figure 2F). Consistently, ICP-MS revealed that XcZur can bind zinc ions but not nickel ions (Supplementary Table S3). These observations strongly indicate that zinc binding is essentially required for the DNA binding capability of XcZur.
Apo- and holo-XcZur structures reveal distinct competencies for DNA binding
To facilitate structural studies, we employed an approach combining disorder prediction, sequence alignment with other Fur family proteins and DNA binding assay to define the domain boundaries in XcZur. The flexible N-terminal 16 residues unique to XcZur are dispensable for DNA binding (Figure 2C). We thus purified the N-terminal extension deleted XcZur (T17-G172, hereafter referred to as XcZur) and crystallized this core domain in its apo form. To assess possible conformational changes associated with zinc binding, we also crystallized XcZur in complex with additionally supplemented zinc (holo form). Both structures were determined by the SAD method, and the final atomic models were refined at 2.2 and 1.9 Å resolutions, respectively, for the apo and holo form of XcZur (Table 1).
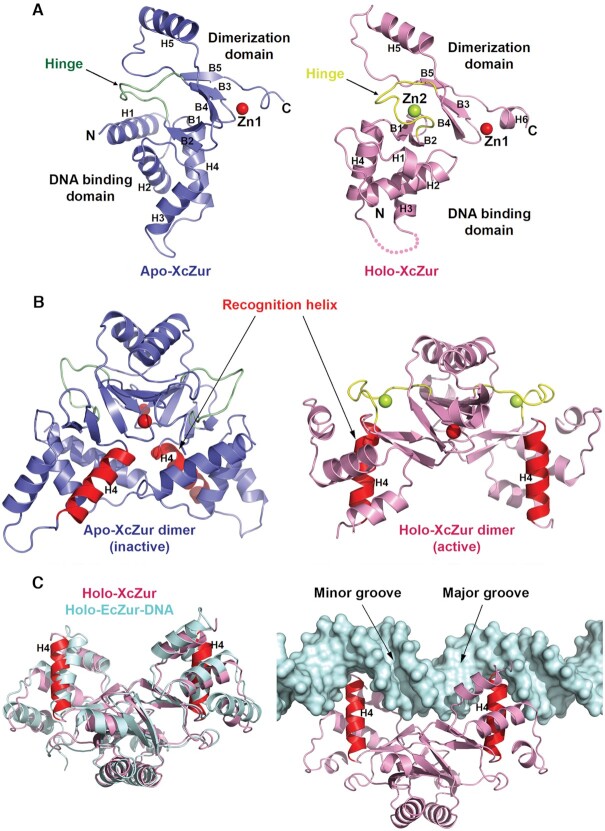
Similar to the reported structures of Fur family proteins, XcZur adopts a two-domain architecture connected by a partially flexible hinge, with its N-terminal DBD containing four α helices and a β hairpin and its C-terminal DD comprising a three-stranded β sheet and a long α helix (Figures 1 and 3A). Although both apo and holo XcZur fold into a conserved two-domain structure and consist of almost identical secondary structure elements (Figures 1 and 3A), structural comparison reveals significant conformational variation (Figure 3A). Evidently, apo XcZur binds only one zinc ion (Zn1) while holo XcZur binds two (Zn1 and Zn2). In addition, zinc binding to the regulatory site, formed by the hinge region and the two domains, results in drastic structural changes of the connecting hinge and completely different orientations of the two domains.
Figure 3.
Apo- and holo-XcZur structures reveal distinct competencies for DNA binding. (A) Cartoon representation of the apo- and holo-XcZur monomers. The apo-XcZur structure is shown in blue, with the hinge region highlighted in green (left panel). The holo-XcZur structure is shown in pink, with the hinge highlighted in yellow (right panel). Zn1 and Zn2 are shown in red and limon spheres, respectively. Secondary structural elements are labeled. (B) Cartoon representation of the apo- (left panel) and holo-XcZur (right panel) dimers. Recognition helix (H4) is highlighted in red. (C) Structural comparison of the holo-XcZur dimer with the holo-EcZur structure from the EcZur-DNA complex. Left panel shows a superimposition of holo XcZur (pink) with holo EcZur (cyan). Right panel displays a docking of holo XcZur into the EcZur-box (cyan surface) in the same orientation of EcZur. Minor and major grooves of the DNA are indicated.
Examination of crystal packing identified a dimeric assembly of both apo and holo XcZur structures, consistent with the observations that XcZur can exist as homodimers in solution (Figure 2A and 2B) and that most available structures of the Fur family proteins are in a dimeric form. Notably, the apo and holo XcZur dimers represent two different conformations, termed respectively the ‘closed’ and ‘open’ conformations, according to different size of the DNA binding groove and accessibility (Figure 3B). In particular, apo-XcZur dimer is in a closed and inactive state in which the recognition helices cannot accommodate DNA binding. On the contrary, holo-XcZur dimer is in an open and active state, and the recognition helices of holo XcZur are precisely positioned and poised to contact DNA. Supporting this notion, structural comparisons reveal that the overall structure of EcZur in complex with DNA (37) is most similar to that of holo XcZur (Figure 3C), but totally different from that of apo XcZur. Superimposition of the EcZur dimer with holo- and apo-XcZur dimer gives a root mean square deviation (r.m.s.d.) of 1.713 Å (229 aligned Cα atoms) and 9.873 Å (246 aligned Cα atoms), respectively. Moreover, holo-XcZur dimer can be positioned into the E. coli Zur-box in an orientation similar but not identical to EcZur, with limited steric clashes (Figure 3C). These findings indicate that holo XcZur adopts a conformation intermediate between that of apo XcZur and that of the DNA bound EcZur. In other words, zinc perception is able to activate XcZur for the subsequent DNA binding.
Zinc perception induces an inactive-to-active conformational transition of XcZur
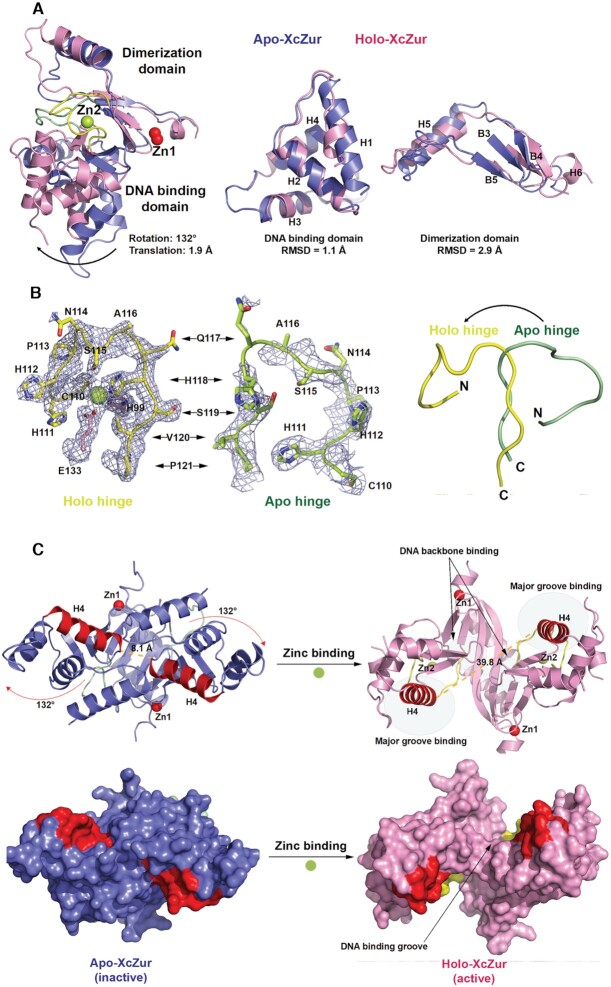
To accurately located the site of the conformational rearrangement, we compared the results of two structural alignments between the apo- and holo-XcZur monomers. The first alignment was based on the individual DNA binding domain of XcZur whereas the other alignment focused on the individual dimerization domain. Intriguingly, the comparison revealed that the DBD structures of apo and holo XcZur are very similar, with a r.m.s.d. of 1.1 Å for 69 aligned Cα atoms, and their DDs share moderate structural similarity (2.9 Å r.m.s.d. for 44 aligned Cα atoms) (Figure 4A). These observations indicate a rigid body rotation of one domain related to the other upon zinc binding. When the DDs of apo and holo XcZur were aligned, the DBDs exhibited a rotation of 132° accompanied by a translation of 1.9 Å along the rotation axis (Figure 4A). In this process, the hinge region, where zinc perception takes place, is proposed to be the critical site responsible for the dramatic conformational transition. Obviously, zinc perception in the hinge region caused an approximate 180° rotation of this structural element, especially at its N-terminal portion (residues C110 to Q117) (Figure 4B), which is expected to subsequently alter the orientation of the DBD with respect to the DD (Figure 4A).
Figure 4.
Zinc binding induces an inactive-to-active conformational transition of XcZur. (A) Structural comparison of apo (blue) and holo XcZur (pink) on the whole monomer (left panel), the DNA binding domain (middle panel), and the dimerization domain (right panel). The representation and color scheme are the same as in Figure 3A. (B) Structural comparison of the apo-XcZur hinge (green) and holo-XcZur hinge (yellow). Left and middle panels show the 2Fo-Fc electron density maps, contoured at 1σ, of the apo- and holo-XcZur hinges, respectively. Right panel displays the overall structure comparison of apo- and holo-XcZur hinges. Residues in the hinge are shown as colored sticks. The bound Zn2 atom in the hinge is shown as a limon sphere. (C) The inactive-to-active conformational transition of XcZur induced by zinc binding. Upper panel shows the cartoon representation of the apo- (blue) and holo-XcZur (pink) dimers in which the recognition helix (H4) is highlighted in red. Structural elements responsible for DNA binding are indicated. Lower panel displays the surface representation of apo- and holo-XcZur dimers in which the recognition helix is highlighted in red. The putative DNA binding groove is indicated.
The results from previous structural studies on Fur family regulators support an ‘open-to-closed’ conformational change mechanism upon metal-dependent activation (24,31,33–37,41–46,70), however, our structural observations reveal a completely different ‘closed-to-open’ transition model. As a result of this dramatic change, the C-termini (residue N94) of the recognition helices are now 39.8 Å apart from each other in the open holo XcZur dimer, compared to 8.1 Å in the closed apo XcZur dimer (Figure 4C). Ultimately, this closed-to-open transition of the DNA binding groove exposed the H4 recognition helices, the C-terminal portions of H1 and the β-hairpin wings (B1 and B2) in the DBD (Figures 1 and 4C). Importantly, these structural elements were reported to contribute to major groove, minor groove and nonspecific DNA contacts, respectively (37,38). These observations indicate that zinc perception of XcZur in the hinge region induces an inactive-to-active conformational transition that precisely rearranges the structural elements responsible for target DNA binding.
Structural basis for the zinc perception of XcZur
Similar to most Fur family metalloregulators, XcZur bears two types of zinc binding sites, a structural site (site 1) and a regulatory site (site 2). The structural zinc binding site locates in the C-terminal DD whereas the regulatory site resides between the two domains (Figures 3A and 5). At site 1, the structural zinc ion (Zn1) is coordinated by four cysteines (C125, C128, C165 and C168) from two CXXC sequence motifs. The C125-XX-C128 motif is derived from the loop between β strands B3 and B4 and the C165-XX-C168 motif comes from helix H6 and its preceding N-terminal loop (Figures 5A and B). Obviously, Zn1 functions as an organizing center for packing of almost all structural elements in DD (B3-B5 and H6) and zinc perception in site 2 has no detectable effect on Zn1 coordination (Figures 5A and B). Coordination of the structural zinc seems to be extremely stable, as evidenced by the result from ICP-MS that treatment of EDTA (10 mM) along cannot remove this zinc (Supplementary Table S3). Previous study revealed that the structural zinc can only be removed by protein denature in addition to EDTA (71). In agreement with its critical structural role in maintaining the correct DD conformation, substitution of any of the four zinc ligands in site 1 led to complete loss of DNA binding of XcZur (Figure 5E).
Figure 5.
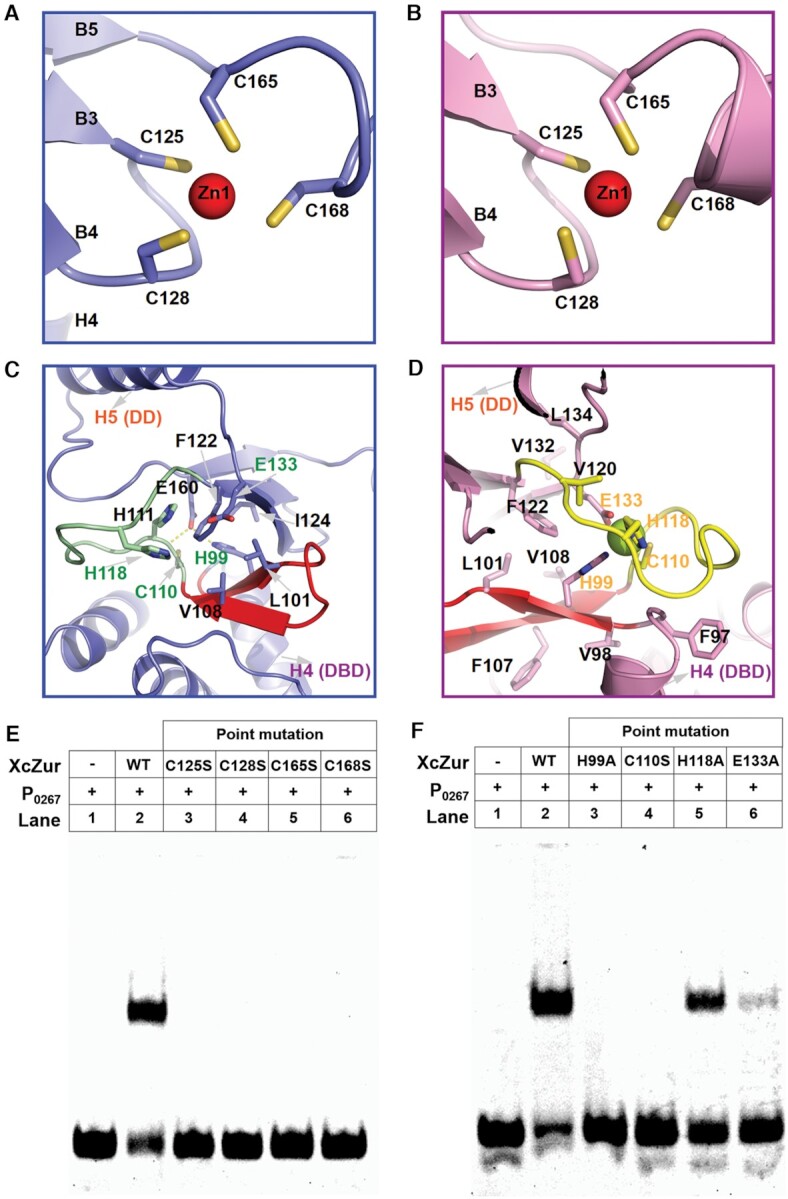
The structural and regulatory zinc binding sites of XcZur. (A) Close-up view of the structural zinc binding site (site 1) of apo XcZur. The zinc atom is represented by a red sphere and the corresponding coordination ligands are shown in colored sticks. (B) Close-up view of the structural zinc binding site of holo XcZur. (C) Close-up view of the regulatory zinc binding site (site 2) of apo XcZur. Key residues in this site are shown in colored sticks. The characteristic β hairpin in DBD are shown in red. (D) Close-up view of the regulatory zinc binding site of holo XcZur. (E) Binding activity of wild type and site 1-related mutants of XcZur with P0267. (F) Binding activity of wild type and site 2-related mutants of XcZur with P0267.
In apo XcZur, the hinge packs against a highly curved ten-stranded β sheet, with five β strands from each monomer (Figure 5C). Despite the fact that this large β sheet brings the XcZur dimer into an intact entity, interactions between the hinge and the β sheet are extensive but suboptimal, because the mixed polar and nonpolar residues at the interface have not made effective contacts (Figure 5C). In fact, zinc ion (Zn2) perception in site 2 optimizes these interactions. In holo XcZur, binding of the regulatory zinc is coupled to disruption of the ten-stranded β sheet and formation of two β sandwiches instead between the β hairpin from the DBD and the three β strands from the DD (Figure 5D). Most of the hydrophobic residues are buried in the core of the new β sandwich to link together the hydrophobic cores of the two domains, whereas polar residues are either involved in Zn2 perception or exposed to the solvent (Figure 5D). At one end of the β sandwich, the regulatory zinc ion is coordinated by four ligands, H99 from the hairpin of DBD, E133 from strand B4 of DD, C110 and H118 from the hinge region (Figures 1 and 5D). These contacts sequester the hinge and contribute to formation and stabilization of the holo conformation. To confirm the importance of Zn2 perception in site 2, we generated four missense mutations in XcZur and individually examined their interactions with DNA by EMSA. Consistent with our structural observation, the H99A and C110S mutations resulted in complete abrogation of DNA binding whereas the E133A mutation substantially reduced interactions with DNA (Figure 5F). Notably, the H118A mutation gave a moderate effect on the DNA binding ability of XcZur (Figure 5F), which might be explained by the existence of possible zinc ligands nearby in the hinge (C110-HHPNSAQ-H118-SV-P121, underlined) (72).
To map the position and frequency of the metal ligands in Fur family metalloregulators, we examined and counted on 23 structures from the aforementioned 15 XcZur homologs and XcZur (Figure 1, Supplementary Tables S4 and S5). Our study revealed that the metal ligands locate primarily at the hinge region and three β strands in its close vicinity, i.e. B3 and B4 in the DD and B1 in the DBD. Other ligands reside in H5, H6 and B5 from DD, and in the L2-3 loop and H1 from DBD. Notably, most members from three Fur groups utilize the structural site to bind a zinc ion. By contrast, H99 and C110 of site 2, which are essentially required for the DNA binding activity, are distinctive to XcZur and EcZur, confirming the importance of this regulatory site in specific zinc perception and functional activation of XcZur and EcZur.
Putative DNA-binding mode of XcZur
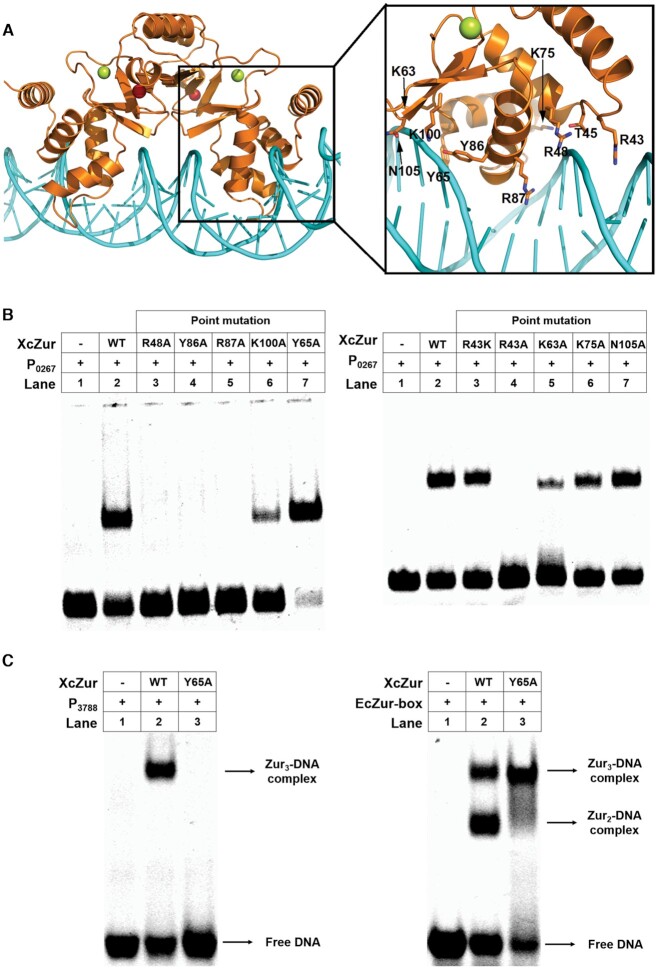
To characterize the DNA-binding mode of XcZur, we constructed a model for the complex between XcZur and the P0267 DNA by the NPdock server (73). In the model, the caliper-like XcZur dimer employs two recognition helices, each from one monomer, to contact DNA in the major groove, by principally using residues Y86 and R87 (Figure 6A). Residues in the L1-2 loop (R43 and T45), the N-terminal portion of H2 (R48), N- and C-termini of H3 (K63, Y65 and K75) and the DBD β hairpin (K100 and N105) also appear to contribute to DNA recognition (Figure 6A). We next used mutagenesis to test whether these residues play key roles in DNA binding. All mutants, except for T45A, were successfully expressed and purified to homogeneity. As shown in Figure 6B, the Y86A and R87A mutations at the recognition helix essentially abolished DNA binding, whereas the K75A and N105A mutations had little or no effect on formation of the protein-DNA complex. Intriguingly, replacement of Y65 by alanine in H3, corresponding to a base-contacting residue Y45 in EcZur (37), greatly enhanced DNA binding. Consistent with the observation that MgFur and EcZur apply a positive-charged residue for minor groove contact (37,38), the corresponding R43A mutation failed to bind DNA while the R43K mutation had no detectable effect. Notably, alanine substitution of the only invariant residue R48 among Fur family proteins resulted in a complete loss of DNA recognition (Figures 1 and 6B), revealing an indispensable role of the protein-backbone interactions. In support, the K63A and K100A mutations, which might also impact the interactions between XcZur and the phosphate backbone, significantly impaired DNA binding. Taken together, contacts from major groove, minor groove and DNA backbone all play important roles in XcZur-DNA recognition.
Figure 6.
Putative DNA-binding residues of XcZur. (A) A structure model for the XcZur-DNA complex and a close-up view of putative DNA binding residues. XcZur and DNA are colored in orange and cyan, respectively. The Zn1 and Zn2 atoms are shown as red and limon spheres, respectively. Putative DNA contacting residues are highlighted as colored sticks. (B) Mutational effects of putative DNA binding residues on the DNA binding activity of XcZur. (C) EMSA for XcZur and XcZurY65A with P3788 and EcZur-box.
As described above, XcZur prefers to bind its target DNA in a XcZur2–DNA mode, resembling that of MgFur (38). This result is in contrast to a recent report in which EcZur mainly forms a high affinity (EcZur2)2–DNA complex in a polymeric mode (37). The discrepancy may be in part due to different DNA fragments used, since we observed that XcZur could also forms a XcZur3–DNA complex with the Zur-box of EcZur and with P3788 (Figures 2C and 6C). Interestingly, XcZur could also forms a XcZur2–DNA complex with the EcZur-box (Figure 6C).
In solution evidence for zinc-mediated conformational changes of XcZur
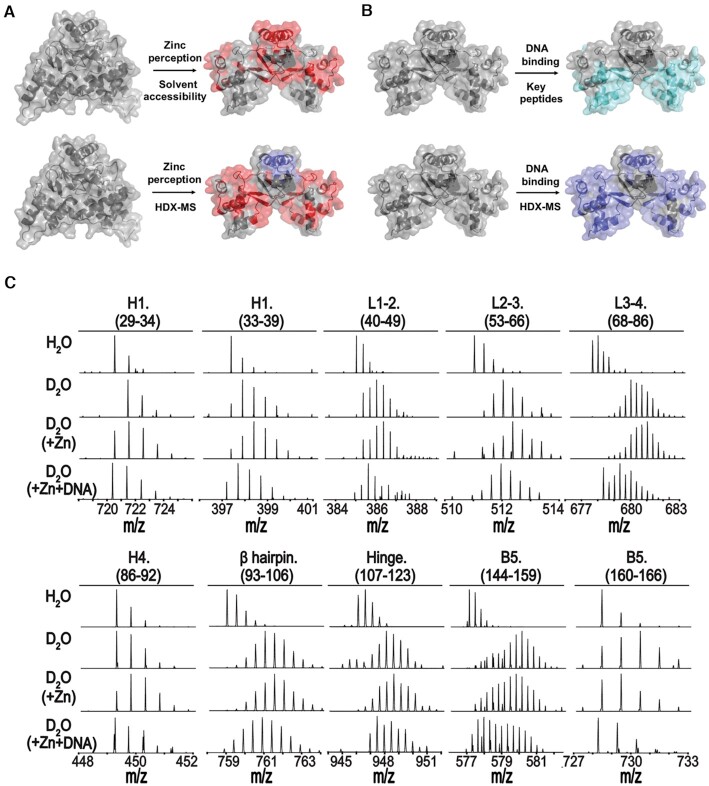
To further investigate conformational changes of XcZur upon zinc and DNA binding in solution, we performed HDX-MS for apo XcZur, holo XcZur and holo-XcZur–DNA complex (Supplementary Table S6). HDX-MS revealed that zinc perception increased deuterium exchange of DBD, especially H1, the L1-2 and L2-3 loops, H4, the β hairpin and hinge region (Figures 7A and C), suggesting obvious conformational alterations which might contribute to exposure of key structural elements for subsequent DNA binding. Mapping peptides with significant changes of deuterium exchange or solvent accessibility to XcZur structures revealed a very similar ‘exposed’ pattern in DBD, but distinct patterns in DD, especially the last β strand (Figure 7A, Supplementary Table S6). This may be explained by the fact that, besides the closed-to-open conformational change, zinc perception of XcZur in solution involves an additional monomer-to-dimer transition to severely decrease the deuterium exchange of DD and create the ‘hidden’ pattern in HDX-MS (Figures 2A, B and 7A).
Figure 7.
In solution evidence for zinc-mediated conformational changes of XcZur. (A) Peptides with significant changes in solvent accessibility (upper panel) or deuterium exchange (lower panel) upon zinc perception are mapped onto the crystal structures. (B) Peptides that contain residues important to DNA binding are mapped onto the holo-XcZur structure (upper panel). Peptides with remarkable changes in deuterium exchange during DNA binding are mapped onto the holo-XcZur structure (lower panel). For (A) and (B), the more exposed and hidden peptides are colored in red and blue, respectively. Key peptides for DNA recognition are colored in cyan. (C) Representative mass spectra for XcZur peptides from HDX-MS.
Notably, target DNA binding decreased deuterium exchange in XcZur encompassing nearly the whole protein sequence (Figures 7B and C, Supplementary Figure S6), indicating the formation of a stable protein-DNA complex. Particularly, peptides with significantly decreased deuterium exchange in DBD matched nicely with those involved in DNA recognition (Figure 6).
DISCUSSION
XcZur is a global transcriptional regulator essential for zinc homeostasis and full virulence of Xanthomonas (8,53). Our biochemical studies reveal that zinc perception is necessary and sufficient for the DNA binding capacity of XcZur. Detailed structural analyses demonstrate that XcZur utilizes two sites for zinc perception and conformational activation. The Zn1 atom bound in the structural site 1 plays an indispensable role in stabilizing the proper fold of the dimerization domain. The Zn2 atom bound in the regulatory site 2 functions in productive positioning of the DNA binding domain into an active conformation, which is capable for DNA binding. It is generally accepted that, similar to BsPerR (Supplementary Figure S7A), Fur family proteins employ an open-to-closed mechanism for conformational activation (24,31,33–37,41–46,70). However, by comparing the XcZur structures in the apo form and the holo form, we discovered that Zn2 perception in site 2 induces a closed-to-open conformational change to activate the transcriptional regulator (Supplementary Figure S7A).
Unlike BsPerR and XcZur, most Fur family proteins have only the inactive- or active-state structure determined. To explore the possible mechanisms that underlie Fur family protein activation, we compared structures in the inactive state and the active state, respectively. Structure comparisons reveal that Fur family proteins appear to employ a conserved caliper-like active conformation (Supplementary Figure S7B). In contrast, inactive conformations in Fur family members are diverse (Supplementary Figure S7C) and can be categorized into at least three forms, the closed form, the linear form, and the intermediate form between the two. Notably, the recognition helices of Fur family regulators in the all three forms cannot mediate DNA contact without a pronounced conformational change of the dimeric structure. The closed form can be also termed as the V-shaped form because the dimeric structure resembles a V-shaped letter (45,48). Obviously, apo XcZur belongs to the closed form, although it cannot be reasonably aligned with the other two structures (Supplementary Figure S7C). Half of the inactive structures, such as apo BsPerR (42), MtZur (43), oxidized BsPerR (44) and SpPerR (46), exhibits a linear form with an extended conformation. The intermediate form, which brings a closed-like monomer and a linear-like monomer together in a homodimer, possesses a chimeric character from both forms. In particular, apo MgFur (38) and LiPerR (47) are included in the intermediate form. With regard to the three inactive forms, it can be safely assumed that only Fur proteins with a linear or intermediate inactive form could experience an open-to-closed conformational activation process upon metal binding. On the other hand, the closed-to-open activation mechanism of XcZur is likely applicable to other Fur family proteins with a similar closed inactive conformation, although their structures of active conformation are yet to be determined.
Our structural observations suggest that appropriate binding of a specific metal in the regulatory site, which is formed by residues from the hinge region and the two domains, is a key determinant underlying the conformational activation of Fur family proteins. Supporting this notion, most of the inactive dimer structures have at least one monomer in the apo state in which only the structural site is occupied by a metal (38,42,44,45,47,48), and the absence of a metal in the regulatory site probably renders the two recognition helices of a Fur dimer unable to contact DNA. SpPerR does bear a nickel atom bound in the hinge region, however, coordination of the nickel atom by additional residues from the most N-terminal helix effectively sequesters the protein in a linear inactive conformation (46). This indicates that the metal coordinating residues used in the regulatory site play important roles in regulation of Fur protein conformations. Given that the unique N-terminal extension of XcZur is rich in metal-ligand candidates (7 histidines, 1 cysteine, 1 glutamate and 1 aspartate in a 23-residue fragment), this region is highly possible to participate in metal coordination and conformational regulation, as observed in SpRerR (46). We further discovered that an acidic residue corresponding to E133 in β strand B4 of XcZur is required for maintenance of the active state in Fur family proteins. Particularly, this highly conserved residue (with only aspartate and glutamate being tolerated, Figure 1) is used for metal coordination in the regulatory site of all active Fur structures, and the E133A mutation almost completely abrogates DNA binding (Figure 5F). In keeping with this, the MtZur structure with zinc occupation in the regulatory site but not involved the glutamate ligand folds into a linear inactive conformation (43). In this regard, involvement of this acidic residue in metal perception of the regulatory site can be used as a hallmark of the active state.
Our previous study found that XcZur binding might induce local distortions corresponding to DNase I hypersensitive nucleotides in the XcZur-protected regions of target DNAs (52). Consistently, MgFur and EcZur use a combination of base readout (major groove contacts) and DNA-shape readout (minor groove contacts) mechanisms to recognize the distorted DNA (37,38). On the other hand, formation of the XcZur-DNA complex may in turn result in certain conformational alterations of the protein (Figures 7B and C, Supplementary Figure S6). As mentioned earlier, XcZur utilizes respectively the dimeric mode, the polymeric mode and mixed mode (with both dimers and polymers) to recognize P0267/P2472, P3788 and the EcZur-box (Figures 2C and 6C). Despite that XcZur can form XcZur3-DNA complex with both P3788 and EcZur-box, the structures of XcZur in these two complexes are likely to be distinct. According to our mutagenesis studies, the Y65A mutation, which affects a tyrosine residue important for base contact in EcZur (37), eliminates P3788 binding but retains EcZur-box recognition in the XcZur3-DNA complex mode (Figure 6C). According to the results from the present and previous studies (37,38,52), DNA binding of XcZur is likely to be a process of induced fit. The mechanism of induced fit can be used to explain why XcZur is able to bind different target DNAs with such diverse modes. In fact, XcZur prefers binding P0267 and P2472, the promoters of low-affinity zinc-uptake systems, to binding P3788, the promoter of high-affinity zinc-uptake system (Figure 2C and Supplementary Figure S8). This indicates that the P0267/P2472 and P3788 might serve as a tuner and an on-off switch, respectively, for the control of zinc uptake by XcZur (Supplementary Figure S8). Our previous in vivo and in vitro assays showed that XcZur represses transcription from the promoters of XC0267, XC2472 and XC3788, but activates transcription from the XC2976 promoter (52). The current EMSA results may provide a possible explanation for the distinct functions of XcZur. Unlike binding with promoter sequences between the -10 and -35 regions of XC0267/XC2472/XC3788 to possibly compete RNA polymerase contact, the binding of XcZur to the P2976-containing fragment requires DNA sequences outside of the promoter region and may facilitate the recruitment of RNA polymerase to transcriptionally activate the zinc-export gene.
In summary, our study provides important insights into zinc-mediated conformational activation of a zinc uptake transcriptional regulator in Xanthomonas (Figure 8). Our structural and biophysical analyses reveal that zinc perception in the regulator site of XcZur induces a closed-to-open conformational change to activate the transcriptional regulator, and to which subsequent DNA binding likely employs a mechanism of induced fit. These mechanisms may also apply to other Fur family regulators.
Figure 8.
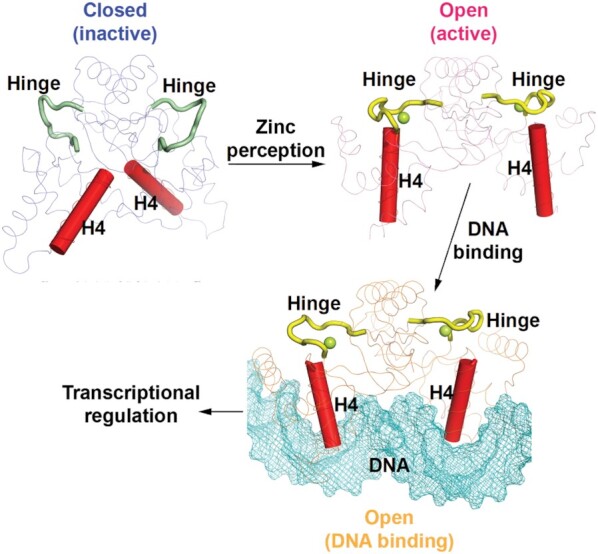
A proposed model of zinc-perception-induced DNA binding for XcZur. In the model, apo XcZur is in a closed inactive state. Zinc perception in the regulatory zinc binding site induces a 180° rotation of the hinge region to drive a closed-to-open conformational change of the XcZur dimer. Holo XcZur thus bears an active state, in which the putative DNA binding elements, especially the recognition helix (H4), are precisely positioned and poised for DNA binding. Subsequently, holo XcZur binds with DNA in a mechanism of mutual induced fit and regulates DNA transcription.
DATA AVAILABILITY
Atomic coordinates and structure factors for the present crystal structures have been deposited to the Protein Data Bank under accession codes 7DH7 and 7DH8.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the staffs from beamlines BL17U1 and BL19U1 at SSRF, and Dr Liming Yan from Tsinghua University, for their assistance with data collection. We also thank Dr Hai-Bo Liu (Guangxi University) and Dr Feng Yang (Guangxi Normal University) for assistance with determination of binding affinity; Cuiyan Zhou (Tsinghua University) for assistance with AUC data analysis; Dr Jixing Xia (College of Life Science and Technology) and Jun Zhang (School of Marine Sciences) at Guangxi University for help with ICP-MS analysis; Wei Hu at Guangxi University for assistance in HDX-MS analysis.
Contributor Information
Fenmei Liu, State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources, College of Life Science and Technology, Guangxi Key Laboratory for Sugarcane Biology, Guangxi University, Nanning 530004, China.
Zihui Su, State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources, College of Life Science and Technology, Guangxi Key Laboratory for Sugarcane Biology, Guangxi University, Nanning 530004, China.
Peng Chen, State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources, College of Life Science and Technology, Guangxi Key Laboratory for Sugarcane Biology, Guangxi University, Nanning 530004, China.
Xiaolin Tian, Protein Chemistry and Proteomics Facility, Protein Research Technology Center, Tsinghua University, Beijing 100084, China.
Lijie Wu, iHuman Institute, ShanghaiTech University, Shanghai 201210, China.
Dong-Jie Tang, State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources, College of Life Science and Technology, Guangxi Key Laboratory for Sugarcane Biology, Guangxi University, Nanning 530004, China.
Peifang Li, State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources, College of Life Science and Technology, Guangxi Key Laboratory for Sugarcane Biology, Guangxi University, Nanning 530004, China.
Haiteng Deng, Protein Chemistry and Proteomics Facility, Protein Research Technology Center, Tsinghua University, Beijing 100084, China.
Pengfei Ding, Department of Chemistry and Biochemistry, University of Maryland, Baltimore County, Baltimore, MD 21250, USA.
Qiang Fu, State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources, College of Life Science and Technology, Guangxi Key Laboratory for Sugarcane Biology, Guangxi University, Nanning 530004, China.
Ji-Liang Tang, State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources, College of Life Science and Technology, Guangxi Key Laboratory for Sugarcane Biology, Guangxi University, Nanning 530004, China.
Zhenhua Ming, State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources, College of Life Science and Technology, Guangxi Key Laboratory for Sugarcane Biology, Guangxi University, Nanning 530004, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31700052]; Ba Gui Scholar Program of Guangxi Zhuang Autonomous Region of China [2014A002]; Guangxi Natural Science Foundation [2020GXNSFFA297007]; State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources [SKLCUSA-a201806]; Guangxi Key Laboratory for Sugarcane Biology [GXKLSCB-20190304]. Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Berg J.M., Shi Y.. The galvanization of biology: a growing appreciation for the roles of zinc. Science. 1996; 271:1081–1085. [DOI] [PubMed] [Google Scholar]
- 2. Lu D., Boyd B., Lingwood C.A.. Identification of the key protein for zinc uptake in Hemophilusinfluenzae. J. Biol. Chem. 1997; 272:29033–29038. [DOI] [PubMed] [Google Scholar]
- 3. Kasahara M., Anraku Y.. Succinate- and NADH oxidase systems of Escherichia coli membrane vesicles. Mechanism of selective inhibition of the systems by zinc ions. J. Biochem. 1974; 76:967–976. [PubMed] [Google Scholar]
- 4. Beard S.J., Hughes M.N., Poole R.K.. Inhibition of the cytochrome bd-terminated NADH oxidase system in Escherichia coli K-12 by divalent metal cations. FEMS Microbiol. Lett. 1995; 131:205–210. [DOI] [PubMed] [Google Scholar]
- 5. Outten C.E., O’Halloran T.V.. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science. 2001; 292:2488–2492. [DOI] [PubMed] [Google Scholar]
- 6. Hantke K. Bacterial zinc uptake and regulators. Curr. Opin. Microbiol. 2005; 8:196–202. [DOI] [PubMed] [Google Scholar]
- 7. Giedroc D.P., Arunkumar A.I.. Metal sensor proteins: nature's metalloregulated allosteric switches. Dalton Trans. 2007; 29:3107–3120. [DOI] [PubMed] [Google Scholar]
- 8. Tang D.J., Li X.J., He Y.Q., Feng J.X., Chen B., Tang J.L.. The zinc uptake regulator Zur is essential for the full virulence of Xanthomonas campestris pv. campestris. Mol. Plant Microbe Interact. 2005; 18:652–658. [DOI] [PubMed] [Google Scholar]
- 9. Choi S.H., Lee K.L., Shin J.H., Cho Y.B., Cha S.S., Roe J.H.. Zinc-dependent regulation of zinc import and export genes by Zur. Nat. Commun. 2017; 8:15812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Patzer S.I., Hantke K.. The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol. Microbiol. 1998; 28:1199–1210. [DOI] [PubMed] [Google Scholar]
- 11. Gaballa A., Helmann J.D.. Identification of a zinc-specific metalloregulatory protein, Zur, controlling zinc transport operons in Bacillus subtilis. J. Bacteriol. 1998; 180:5815–5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dalet K., Gouin E., Cenatiempo Y., Cossart P., Hechard Y.. Characterisation of a new operon encoding a Zur-like protein and an associated ABC zinc permease in Listeria monocytogenes. FEMS Microbiol. Lett. 1999; 174:111–116. [DOI] [PubMed] [Google Scholar]
- 13. Lindsay J.A., Foster S.J.. zur: a Zn(2+)-responsive regulatory element of Staphylococcus aureus. Microbiology (Reading). 2001; 147:1259–1266. [DOI] [PubMed] [Google Scholar]
- 14. Maciag A., Dainese E., Rodriguez G.M., Milano A., Provvedi R., Pasca M.R., Smith I., Palu G., Riccardi G., Manganelli R.. Global analysis of the Mycobacterium tuberculosis Zur (FurB) regulon. J. Bacteriol. 2007; 189:730–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shin J.H., Oh S.Y., Kim S.J., Roe J.H.. The zinc-responsive regulator Zur controls a zinc uptake system and some ribosomal proteins in Streptomyces coelicolor A3(2). J. Bacteriol. 2007; 189:4070–4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Feng Y., Li M., Zhang H., Zheng B., Han H., Wang C., Yan J., Tang J., Gao G.F.. Functional definition and global regulation of Zur, a zinc uptake regulator in a Streptococcus suisserotype 2 strain causing streptococcal toxic shock syndrome. J. Bacteriol. 2008; 190:7567–7578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li Y., Qiu Y., Gao H., Guo Z., Han Y., Song Y., Du Z., Wang X., Zhou D., Yang R.. Characterization of Zur-dependent genes and direct Zur targets in Yersinia pestis. BMC Microbiol. 2009; 9:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schroder J., Jochmann N., Rodionov D.A., Tauch A.. The Zur regulon of Corynebacterium glutamicum ATCC 13032. BMC Genomics. 2010; 11:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ellison M.L., Farrow J.M. 3rd, Parrish W., Danell A.S., Pesci E.C.. The transcriptional regulator Np20 is the zinc uptake regulator in Pseudomonas aeruginosa. PLoS One. 2013; 8:e75389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Osman D., Foster A.W., Chen J., Svedaite K., Steed J.W., Lurie-Luke E., Huggins T.G., Robinson N.J.. Fine control of metal concentrations is necessary for cells to discern zinc from cobalt. Nat. Commun. 2017; 8:1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. El-Gebali S., Mistry J., Bateman A., Eddy S.R., Luciani A., Potter S.C., Qureshi M., Richardson L.J., Salazar G.A., Smart A.et al.. The Pfam protein families database in 2019. Nucleic Acids Res. 2019; 47:D427–D432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hantke K. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 2001; 4:172–177. [DOI] [PubMed] [Google Scholar]
- 23. Vasil M.L., Ochsner U.A.. The response of Pseudomonas aeruginosa to iron: genetics, biochemistry and virulence. Mol. Microbiol. 1999; 34:399–413. [DOI] [PubMed] [Google Scholar]
- 24. Pohl E., Haller J.C., Mijovilovich A., Meyer-Klaucke W., Garman E., Vasil M.L.. Architecture of a protein central to iron homeostasis: crystal structure and spectroscopic analysis of the ferric uptake regulator. Mol. Microbiol. 2003; 47:903–915. [DOI] [PubMed] [Google Scholar]
- 25. Panina E.M., Mironov A.A., Gelfand M.S.. Comparative genomics of bacterial zinc regulons: enhanced ion transport, pathogenesis, and rearrangement of ribosomal proteins. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:9912–9917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Herbig A.F., Helmann J.D.. Roles of metal ions and hydrogen peroxide in modulating the interaction of the Bacillus subtilis PerR peroxide regulon repressor with operator DNA. Mol. Microbiol. 2001; 41:849–859. [DOI] [PubMed] [Google Scholar]
- 27. Lee J.W., Helmann J.D.. The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature. 2006; 440:363–367. [DOI] [PubMed] [Google Scholar]
- 28. Diaz-Mireles E., Wexler M., Sawers G., Bellini D., Todd J.D., Johnston A.W.B.. The Fur-like protein Mur of Rhizobium leguminosarum is a Mn(2+)-responsive transcriptional regulator. Microbiology (Reading). 2004; 150:1447–1456. [DOI] [PubMed] [Google Scholar]
- 29. Diaz-Mireles E., Wexler M., Todd J.D., Bellini D., Johnston A.W.B., Sawers R.G.. The manganese-responsive repressor Mur of Rhizobium leguminosarum is a member of the Fur-superfamily that recognizes an unusual operator sequence. Microbiology (Reading). 2005; 151:4071–4078. [DOI] [PubMed] [Google Scholar]
- 30. Ahn B.E., Cha J., Lee E.J., Han A.R., Thompson C.J., Roe J.H.. Nur, a nickel-responsive regulator of the Fur family, regulates superoxide dismutases and nickel transport in Streptomyces coelicolor. Mol. Microbiol. 2006; 59:1848–1858. [DOI] [PubMed] [Google Scholar]
- 31. An Y.J., Ahn B.E., Han A.R., Kim H.M., Chung K.M., Shin J.H., Cho Y.B., Roe J.H., Cha S.S.. Structural basis for the specialization of Nur, a nickel-specific Fur homolog, in metal sensing and DNA recognition. Nucleic Acids Res. 2009; 37:3442–3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee J.W., Helmann J.D.. Functional specialization within the Fur family of metalloregulators. Biometals. 2007; 20:485–499. [DOI] [PubMed] [Google Scholar]
- 33. Sheikh M.A., Taylor G.L.. Crystal structure of the Vibrio cholerae ferric uptake regulator (Fur) reveals insights into metal co-ordination. Mol. Microbiol. 2009; 72:1208–1220. [DOI] [PubMed] [Google Scholar]
- 34. Jacquamet L., Traore D.A., Ferrer J.L., Proux O., Testemale D., Hazemann J.L., Nazarenko E., El Ghazouani A., Caux-Thang C., Duarte V.et al.. Structural characterization of the active form of PerR: insights into the metal-induced activation of PerR and Fur proteins for DNA binding. Mol. Microbiol. 2009; 73:20–31. [DOI] [PubMed] [Google Scholar]
- 35. Dian C., Vitale S., Leonard G.A., Bahlawane C., Fauquant C., Leduc D., Muller C., de Reuse H., Michaud-Soret I., Terradot L.. The structure of the Helicobacter pylori ferric uptake regulator Fur reveals three functional metal binding sites. Mol. Microbiol. 2011; 79:1260–1275. [DOI] [PubMed] [Google Scholar]
- 36. Shin J.H., Jung H.J., An Y.J., Cho Y.B., Cha S.S., Roe J.H.. Graded expression of zinc-responsive genes through two regulatory zinc-binding sites in Zur. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:5045–5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gilston B.A., Wang S., Marcus M.D., Canalizo-Hernandez M.A., Swindell E.P., Xue Y., Mondragon A., O’Halloran T.V.. Structural and mechanistic basis of zinc regulation across the E. coli Zur regulon. PLoS Biol. 2014; 12:e1001987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Deng Z., Wang Q., Liu Z., Zhang M., Machado A.C., Chiu T.P., Feng C., Zhang Q., Yu L., Qi L.et al.. Mechanistic insights into metal ion activation and operator recognition by the ferric uptake regulator. Nat. Commun. 2015; 6:7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sarvan S., Charih F., Butcher J., Brunzelle J.S., Stintzi A., Couture J.F.. Crystal structure of Campylobacter jejuni peroxide regulator. FEBS Lett. 2018; 592:2351–2360. [DOI] [PubMed] [Google Scholar]
- 40. Perard J., Nader S., Levert M., Arnaud L., Carpentier P., Siebert C., Blanquet F., Cavazza C., Renesto P., Schneider D.et al.. Structural and functional studies of the metalloregulator Fur identify a promoter-binding mechanism and its role in Francisella tularensis virulence. Commun Biol. 2018; 1:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pecqueur L., D’Autreaux B., Dupuy J., Nicolet Y., Jacquamet L., Brutscher B., Michaud-Soret I., Bersch B.. Structural changes of Escherichia coli ferric uptake regulator during metal-dependent dimerization and activation explored by NMR and X-ray crystallography. J. Biol. Chem. 2006; 281:21286–21295. [DOI] [PubMed] [Google Scholar]
- 42. Traore D.A., El Ghazouani A., Ilango S., Dupuy J., Jacquamet L., Ferrer J.L., Caux-Thang C., Duarte V., Latour J.M.. Crystal structure of the apo-PerR-Zn protein from Bacillus subtilis. Mol. Microbiol. 2006; 61:1211–1219. [DOI] [PubMed] [Google Scholar]
- 43. Lucarelli D., Russo S., Garman E., Milano A., Meyer-Klaucke W., Pohl E.. Crystal structure and function of the zinc uptake regulator FurB from Mycobacterium tuberculosis. J. Biol. Chem. 2007; 282:9914–9922. [DOI] [PubMed] [Google Scholar]
- 44. Traore D.A., El Ghazouani A., Jacquamet L., Borel F., Ferrer J.L., Lascoux D., Ravanat J.L., Jaquinod M., Blondin G., Caux-Thang C.et al.. Structural and functional characterization of 2-oxo-histidine in oxidized PerR protein. Nat. Chem. Biol. 2009; 5:53–59. [DOI] [PubMed] [Google Scholar]
- 45. Butcher J., Sarvan S., Brunzelle J.S., Couture J.F., Stintzi A.. Structure and regulon of Campylobacter jejuni ferric uptake regulator Fur define apo-Fur regulation. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:10047–10052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Makthal N., Rastegari S., Sanson M., Ma Z., Olsen R.J., Helmann J.D., Musser J.M., Kumaraswami M.. Crystal structure of peroxide stress regulator from Streptococcus pyogenes provides functional insights into the mechanism of oxidative stress sensing. J. Biol. Chem. 2013; 288:18311–18324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kebouchi M., Saul F., Taher R., Landier A., Beaudeau B., Dubrac S., Weber P., Haouz A., Picardeau M., Benaroudj N.. Structure and function of the Leptospira interrogans peroxide stress regulator (PerR), an atypical PerR devoid of a structural metal-binding site. J. Biol. Chem. 2018; 293:497–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sarvan S., Charih F., Askoura M., Butcher J., Brunzelle J.S., Stintzi A., Couture J.F.. Functional insights into the interplay between DNA interaction and metal coordination in ferric uptake regulators. Sci. Rep. 2018; 8:7140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lucarelli D., Vasil M.L., Meyer-Klaucke W., Pohl E.. The metal-dependent regulators FurA and FurB from Mycobacterium Tuberculosis. Int. J. Mol. Sci. 2008; 9:1548–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vitale S., Fauquant C., Lascoux D., Schauer K., Saint-Pierre C., Michaud-Soret I.. A ZnS(4) structural zinc site in the Helicobacter pylori ferric uptake regulator. Biochemistry. 2009; 48:5582–5591. [DOI] [PubMed] [Google Scholar]
- 51. Mikhaylina A., Ksibe A.Z., Scanlan D.J., Blindauer C.A.. Bacterial zinc uptake regulator proteins and their regulons. Biochem. Soc. Trans. 2018; 46:983–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Huang D.L., Tang D.J., Liao Q., Li H.C., Chen Q., He Y.Q., Feng J.X., Jiang B.L., Lu G.T., Chen B.et al.. The Zur of Xanthomonas campestris functions as a repressor and an activator of putative zinc homeostasis genes via recognizing two distinct sequences within its target promoters. Nucleic Acids Res. 2008; 36:4295–4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Huang D.L., Tang D.J., Liao Q., Li X.Q., He Y.Q., Feng J.X., Jiang B.L., Lu G.T., Tang J.L.. The Zur of Xanthomonas campestris is involved in hypersensitive response and positively regulates the expression of the hrp cluster via hrpX but not hrpG. Mol. Plant Microbe Interact. 2009; 22:321–329. [DOI] [PubMed] [Google Scholar]
- 54. Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J.. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997; 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Di Tommaso P., Moretti S., Xenarios I., Orobitg M., Montanyola A., Chang J.M., Taly J.F., Notredame C.. T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 2011; 39:W13–W17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Saitou N., Nei M.. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987; 4:406–425. [DOI] [PubMed] [Google Scholar]
- 57. Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985; 39:783–791. [DOI] [PubMed] [Google Scholar]
- 58. Zuckerkandl E., Pauling L.. Bryson V., Vogel H.J.. Evolving Genes and Proteins. 1965; Academic Press; 97–166. [Google Scholar]
- 59. Robert X., Gouet P.. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014; 42:W320–W324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang W.Z., Tang J.C., Wang S.S., Wang Z.J., Qin W.M., He J.H.. The protein complex crystallography beamline (BL19U1) at the Shanghai Synchrotron Radiation Facility. Nucl. Sci. Tech. 2019; 30:170. [Google Scholar]
- 61. Kabsch W. Xds. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W.et al.. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Terwilliger T.C., Grosse-Kunstleve R.W., Afonine P.V., Moriarty N.W., Zwart P.H., Hung L.W., Read R.J., Adams P.D.. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D. Biol. Crystallogr. 2008; 64:61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Adams P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W., Read R.J., Sacchettini J.C., Sauter N.K., Terwilliger T.C.. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D. Biol. Crystallogr. 2002; 58:1948–1954. [DOI] [PubMed] [Google Scholar]
- 66. Wang Z., Pan Q., Yang L., Zhou H., Xu C., Yu F., Wang Q., Huang S., He J.. Automatic crystal centring procedure at the SSRF macromolecular crystallography beamline. J. Synchrotron Radiat. 2016; 23:1323–1332. [DOI] [PubMed] [Google Scholar]
- 67. Brown P.H., Schuck P.. Macromolecular size-and-shape distributions by sedimentation velocity analytical ultracentrifugation. Biophys. J. 2006; 90:4651–4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Garcia de la Torre J., Huertas M.L., Carrasco B.. HYDRONMR: prediction of NMR relaxation of globular proteins from atomic-level structures and hydrodynamic calculations. J. Magn. Reson. 2000; 147:138–146. [DOI] [PubMed] [Google Scholar]
- 69. Jia Z., Yan L., Ren Z., Wu L., Wang J., Guo J., Zheng L., Ming Z., Zhang L., Lou Z.et al.. Delicate structural coordination of the Severe Acute Respiratory Syndrome coronavirus Nsp13 upon ATP hydrolysis. Nucleic Acids Res. 2019; 47:6538–6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lin C.S., Chao S.Y., Hammel M., Nix J.C., Tseng H.L., Tsou C.C., Fei C.H., Chiou H.S., Jeng U.S., Lin Y.S.et al.. Distinct structural features of the peroxide response regulator from group A Streptococcus drive DNA binding. PLoS One. 2014; 9:e89027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Althaus E.W., Outten C.E., Olson K.E., Cao H., O’Halloran T.V.. The ferric uptake regulation (Fur) repressor is a zinc metalloprotein. Biochemistry. 1999; 38:6559–6569. [DOI] [PubMed] [Google Scholar]
- 72. Vallee B.L., Auld D.S.. Cocatalytic zinc motifs in enzyme catalysis. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:2715–2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tuszynska I., Magnus M., Jonak K., Dawson W., Bujnicki J.M.. NPDock: a web server for protein-nucleic acid docking. Nucleic Acids Res. 2015; 43:W425–W430. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors for the present crystal structures have been deposited to the Protein Data Bank under accession codes 7DH7 and 7DH8.