Abstract
Aim: Numerous drugs are being widely prescribed for COVID-19 treatment without any direct evidence for the drug safety/efficacy in patients across diverse ethnic populations. Materials & methods: We analyzed whole genomes of 1029 Indian individuals (IndiGen) to understand the extent of drug–gene (pharmacogenetic), drug–drug and drug–drug–gene interactions associated with COVID-19 therapy in the Indian population. Results: We identified 30 clinically significant pharmacogenetic variants and 73 predicted deleterious pharmacogenetic variants. COVID-19-associated pharmacogenes were substantially overlapped with those of metabolic disorder therapeutics. CYP3A4, ABCB1 and ALB are the most shared pharmacogenes. Fifteen COVID-19 therapeutics were predicted as likely drug–drug interaction candidates when used with four CYP inhibitor drugs. Conclusion: Our findings provide actionable insights for future validation studies and improved clinical decisions for COVID-19 therapy in Indians.
Keywords: : COVID-19 therapies, drug–drug–gene interactions, drug–drug interactions, Indian population, pharmacogenomics
It has been well established that genetic variants among several other factors significantly explain inter-individual differences in therapeutic response [1,2]. Several examples of large-scale ethnic differences in treatment response have been attributed to differential prevalence of these pharmacogenetic (PGx) variants among distinct populations [3,4]. As the novel COVID-19 pandemic continues to present unprecedented challenges to healthcare systems across the world, an array of new vaccines and repurposed or novel therapeutics are being developed, undergoing clinical trials or are currently in use in different parts of the world [5]. Given the rapid worldwide spread of this novel disease, many of these therapeutics, although unapproved or lacking adequate direct evidence for efficacy and safety, are being widely used in COVID-19 patients across age groups, ethnic backgrounds and diverse underlying health conditions [1]. It is therefore critical to harness parallel efforts toward understanding the pharmacogenomics of COVID-19 therapies and develop population-specific PGx maps for the widely used drugs. Defining population-specific actionable PGx biomarkers for COVID-19 therapy can potentially help clinicians choose appropriate treatment regimens and improve overall treatment outcomes. A few recent studies have analyzed and reviewed the clinical implications of human genome interactions with repurposed drugs [3,6] most of which have genotype-guided dosing guidelines [7]. Moreover, it is also crucial to assess drug interactions and the associated adverse events [7].
Some of the common examples of adverse drug reactions in COVID-19 patients involve the use of experimental drugs like chloroquine, hydroxychloroquine, azithromycin and lopinavir/ritonavir which can cause patients to develop long QT syndrome and torsades de pointes [2,3]. Some of them already have pharmacogenetic markers listed on their US FDA-recommended drug labels like G6PD gene for chloroquine and hydroxychloroquine and IFNL3 for ritonavir [4,8,9]. Similarly, variants in ITPA gene are reported to have protective effects against hemolytic anemia, which is a well-known adverse effect of ribavirin, a widely prescribed RNA polymerase inhibitor drug in COVID-19 patients that is originally indicated for Hepatitis C infection [10]. In addition to these drug–gene interactions, drug–drug interactions (DDIs) are also a key factor in impaired COVID-19 treatment response owing to the use of several drugs in tandem to aid the recovery of patients from multiple short-term and long-term COVID-19 complications in addition to their pre-existing conditions [11].
Since its emergence in Wuhan, China in December 2019, millions of individuals have been infected with COVID-19 across the world [12]. India, the world’s second most populous country, is also witnessing a significant number of cases and deaths [13]. In the wake of such massive outbreaks in India, the limited medical resources call for rapid and robust tools that can guide clinicians toward effective clinical management of COVID-19 patients. Defining the PGx landscape of COVID-19 drugs in the Indian population can potentially guide clinicians toward adopting effective therapeutic regimens for COVID-19 patients.
Whole genome sequencing of large and diverse human populations across the world over the last two decades have generated great insights about human genetic diversity, evolution and migration [14]. In the last 5 years, several large-scale national genomic sequencing initiatives have been established with the aim of advancing genomic medicine and generating population-specific evidence for its wider adoption in clinical practice [15]. Recently, India has initiated its own national genome sequencing initiative, the IndiGen Project, which in its first phase has successfully sequenced over 1029 Indian whole genomes belonging to diverse ethnic groups in India [16]. The IndiGen dataset along with the South Asian subpopulations included in the 1000 Genomes project (n = 489) collectively represent one of the most comprehensive genomic representations of the geographic and ethnic diversity of the Indian population available as of today. Systematic mining of PGx variants from such population-scale genomic data can generate critical actionable insights for the population as a whole.
The current study aims to understand the extent of drug–gene (PGx), DDI and drug–drug–gene interactions (DDGI) associated with COVID-19 in the Indian population. We provide a comprehensive population-specific PGx landscape for drugs that are widely used for COVID-19 treatment using large-scale genomic variation datasets representing the Indian population. We also highlight actionable PGx markers that are relevant for Indian COVID-19 patients which can enable clinicians to make informed prescription decisions and potentially improve overall therapeutic outcomes. To the best of our knowledge, this is the first comprehensive study of PGx associations, DDIs and DDGIs associated with therapeutics used in COVID-19 in the Indian population.
Materials & methods
Study population & datasets
The genetic variants and their allele frequencies in the cosmopolitan Indian population were obtained from Variant Calling Format of 1029 whole genome sequences of unrelated Indian individuals, sequenced across India as a part of IndiGen [16] study. The participants of the IndiGen study spread across most states in India [16]. The variants are annotated according to the GRCh38 human reference genome and genotype information of 55,898,112 genetic variations consisting of SNVs and indels.
Quality control
We performed genotype and individual level missingness tests (95%) and Hardy–Weinberg disequilibrium test (p < 5 × 10-7) using PLINK v1.09 [17] and obtained 53,672,515 SNVs which were used for further analysis.
Pharmacogenomic variant analysis workflow
Variant annotation
ANNOVAR [18] was used to annotate variants using dbsnp v150 as the reference variation database and RefGene for gene annotations, and also used to compile the allele frequencies of the variants from global variation projects such as 1000 Genomes Phase 3 (1KGP3) database [19], gnomAD database [20], Qatar and Greater Middle East (GME) variome database [21].
Prediction of potential deleterious variants
The functional impact of exonic variants were predicted using SIFT [22], PolyPhen2 [23] and MutationTaster [24]. The exonic nonsynonymous variants that were predicted deleterious (SIFT: Damaging; PolyPhen2: Probably Damaging and MutationTaster2: Disease_causing) by at least two of these tools were taken for downstream analysis.
Data collection
Information about therapeutics used in the management of COVID-19 and their clinical trial information was curated from PharmGKB [25] and DrugBank [26] databases (accessed on 17 August 2020). PharmGKB has a list of 54 therapeutics involved in COVID-19 clinical trials that inhibit viral replication or viral entry and anticytokine/anti-inflammatory drugs. DrugBank database has published a list of 38 drugs which are experimentally unapproved treatments for COVID-19. All these drugs were overlapped and obtained a unique list of 89 drugs that are widely used for COVID-19 treatment (Table 1 & Supplementary Table 1).
Table 1. List of proposed COVID-19 drugs.
| Drug | DrugBank ID | Category | PharmGKB variants (Level 1 & 2) (n) | DrugBank targets (n) | DrugBank enzymes (n) | DrugBank transporters/carriers (n) |
|---|---|---|---|---|---|---|
| Anticytokine/anti-inflammatory | ||||||
| Tocilizumab | DB06273 | Interleukin inhibitor, monoclonal antibodies (anti IL-6) | 1 | 1 | ||
| Sarilumab | DB11767 | Interleukin inhibitor, monoclonal antibodies (anti IL-6) | 6 | 1 | ||
| Anakinra | DB00026 | Interleukin inhibitor (anti IL-1) | 1 | |||
| Siltuximab | DB09036 | Interleukin inhibitor, monoclonal antibodies (anti IL-6) | 1 | 1 | ||
| Leflunomide | DB01097 | Immunomodulators | 3 | 2 | 1 | |
| Clazakizumab | DB12849 | Interleukin inhibitor, monoclonal antibodies (anti IL-6) | ||||
| Prazosin | DB00457 | Cardiovascular agents | 6 | 4 | 1 | |
| Canakinumab | DB06168 | Interleukin inhibitor, monoclonal antibodies (anti IL-1β) | 1 | |||
| Naltrexone | DB00704 | Analgesics | 4 | 1 | ||
| Ketamine | DB01221 | Anesthetics | 11 | 5 | ||
| Sirukumab | DB11803 | Interleukin inhibitor, monoclonal antibodies (anti IL-6) | ||||
| Fluoxetine | DB00472 | Antidepressants | 1 | 7 | 8 | 3 |
| Astegolimab | Interleukin inhibitor, monoclonal antibodies (anti IL-33) | |||||
| Ulinastatin | DB12038 | Protease inhibitor | ||||
| Mavrilimumab | DB12534 | Monoclonal antibody (anti GM-CSF) | ||||
| Axatilimab | Monoclonal antibody (anti GM-CSF) | |||||
| Lenzilumab | DB15148 | Monoclonal antibody (anti GM-CSF) | ||||
| Sargramostim | DB00020 | Immunomodulators | 5 | |||
| Tofacitinib | DB08895 | Immunomodulators | 4 | 2 | 1 | |
| Leronlimab | DB05941 | Monoclonal antibodies (anti CCR5) | 1 | |||
| Eculizumab | DB01257 | Monoclonal antibodies (anti C5) | 1 | |||
| Dexamethasone | DB01234 | Corticosteroids | 5 | 15 | 7 | |
| Apremilast | DB05676 | Immunomodulators | 1 | 3 | 1 | |
| Cenicriviroc | DB11758 | Antiviral agents | ||||
| Icatibant | DB06196 | Analgesics | 2 | |||
| Razuprotafib | Angiopoietin modulator | |||||
| Naproxen | DB00788 | Analgesics | 2 | 10 | 6 | |
| Baricitinib | DB11817 | Immunomodulator | 4 | 1 | 7 | |
| Nicotine | DB00184 | Cholinergics | 8 | 13 | 13 | 5 |
| Disulfiram | DB00822 | Acetyl aldehyde dehydrogenase inhibitors | 2 | 3 | 1 | |
| Inhibit viral entry | ||||||
| Hydroxychloroquine | DB01611 | Antiprotozoals | 3 | 3 | 3 | |
| Chloroquine | DB00608 | Antiprotozoals | 6 | 5 | 2 | |
| Camostate mesylate (Camostat) | DB13729 | Protease inhibitor | 4 | |||
| Umifenovir | DB13609 | Antiviral agents | 10 | |||
| DAS181 | DB15313 | Recombinant proteins | ||||
| Losartan | DB00678 | Cardiovascular agents | 1 | 9 | 6 | |
| Isotretinoin | DB00982 | Vitamin A derivative | 2 | 1 | 1 | |
| Telmisartan | DB00966 | Cardiovascular agents | 2 | 2 | 4 | |
| Ramipril | DB00178 | Cardiovascular agents | 2 | 1 | 2 | |
| Nicotine | DB00184 | Cholinergics | 8 | 13 | 13 | 5 |
| Inhibit viral replication | ||||||
| Remdesivir | DB14761 | Antiviral agents | 3 | 6 | ||
| Favipiravir | DB12466 | Antiviral agents | 4 | 5 | ||
| Ribavirin | DB00811 | Antiviral agents | 13 | 2 | 2 | 2 |
| Darunavir | DB01264 | Antiviral agents | 2 | 4 | ||
| Clevudine | DB06683 | Antiviral agents | ||||
| Lopinavir | DB01601 | Antiviral agents | 1 | 6 | 6 | |
| Ritonavir | DB00503 | Antiviral agents | 1 | 1 | 9 | 11 |
| Interferon alfa-2b, recombinant | DB00105 | Immunomodulator | 1 | 2 | 1 | |
| Famotidine | DB00927 | Gastrointestinal agents | 1 | 1 | 4 | |
| Rintatolimod | Immunomodulator | |||||
| EIDD-2801 | DB15661 | Experimental unapproved treatment for COVID-19 | ||||
| Peginterferon lambda-1a | DB14767 | |||||
| AT-527 | ||||||
| Merimepodib | DB04862 | Antiviral agents, immunomodulator | ||||
| Disulfiram | DB00822 | Acetyl aldehyde dehydrogenase inhibitors | 2 | 3 | 1 | |
| Others | ||||||
| Deferoxamine | DB00746 | Chelating agents | 1 | 1 | ||
| Tranexamic acid | DB00302 | Hemostatics | 1 | 1 | ||
| Ruxolitinib | DB08877 | Antineoplastic and immunomodulating agents | 2 | 1 | ||
| Sirolimus | DB00877 | Immunomodulators | 1 | 3 | 3 | 3 |
| Enoxaparin | DB01225 | Cardiovascular agents | 2 | 1 | ||
| Fluvoxamine | DB00176 | Antidepressants | 1 | 2 | 7 | 2 |
| Chlorhexidine | DB00878 | Antiseptics | 1 | |||
| Acalabrutinib | DB11703 | Antineoplastic and immunomodulating agents | 1 | 2 | 1 | |
| AT-001 | DB15121 | |||||
| Dapagliflozin | DB06292 | Oral hypoglycemic agents | 1 | 9 | 1 | |
| Progesterone | DB00396 | Steroidal hormones | 10 | 10 | 10 | |
| Acetylcysteine | DB06151 | Mucolytics | 10 | 2 | ||
| Heparin | DB01109 | Hemostatics | 12 | 1 | 1 | |
| Dornase alfa | DB00003 | Mucolytics | ||||
| Nitric oxide | DB00435 | Cardiovascular agents | 3 | 4 | ||
| Galidesivir | DB11676 | Experimental unapproved treatment for COVID-19 | ||||
| Human interferon beta | DB14999 | Immunomodulator | 1 | 1 | 1 | |
| Triazavirin | DB15622 | Antiviral agents | ||||
| TMC-310911 | DB15623 | Antiviral agents | ||||
| AZD1222 | DB15656 | Experimental unapproved treatment for COVID-19 | ||||
| Fingolimod | DB08868 | Immunomodulators | 5 | 3 | 4 | |
| Methylprednisolone | DB00959 | Corticosteroids | 2 | 7 | 2 | |
| Bevacizumab | DB00112 | Antineoplastic and immunomodulating agents | 9 | |||
| Azithromycin | DB00207 | Antibacterial agents | 1 | 1 | 2 | |
| N4-Hydroxycytidine | DB15660 | Experimental unapproved treatment for COVID-19 | ||||
| Elbasvir | DB11574 | Antiviral agents | 4 | 1 | ||
| GS-441524 | DB15686 | Antiviral agents | ||||
| Tridecactide | DB15687 | Experimental unapproved treatment for COVID-19 | ||||
| Metenkefalin | DB12668 | Experimental unapproved treatment for COVID-19 | 2 | 1 | ||
| Vazegepant | DB15688 | Experimental unapproved treatment for COVID-19 | 1 | |||
| Ibuprofen | DB01050 | Analgesics | 2 | 10 | 9 | 9 |
| Anti-SARS-CoV-2 REGN-COV2 | DB15691 | Experimental unapproved treatment for COVID-19 | ||||
| COVID-19 convalescent plasma | DB15692 | Experimental unapproved treatment for COVID-19 | ||||
| INO-4800 | DB15693 | Experimental unapproved treatment for COVID-19 | ||||
| Colchicine | DB01394 | Musculoskeletal system | 1 | 4 | 2 | |
| LY-CoV555 | DB15718 | Experimental unapproved treatment for COVID-19 | ||||
Annotation of pharmacogenetic variants from PharmGKB & DrugBank
The latest release of clinical annotations of PGx variants were obtained from PharmGKB database (release dated 5 August 2020) which included 4077 annotations linked to SNVs and 491 linked to haplotype variants. COVID-19 therapy associated PGx annotations were further shortlisted using the list of 89 drugs and population-specific allele frequencies were further estimated for the relevant PGx variants.
Stargazer [27], a tool for genotyping PGx genes from next generation sequencing data, was used to call star alleles in the PGx genes from the whole genome data of IndiGen and 1KGP3 database. Then, allele frequency (AF) was calculated to evaluate the prevalence of these variants.
A comprehensive list of pharmacogenes were downloaded from DrugBank and overlapped with our list of drugs to fetch associated genes. The predicted deleterious variants in the IndiGen dataset were overlapped with this list to generate a list of potential deleterious PGx variants in the Indian population.
Construction of drug pathways & visualization
Pharmacogenes related to the PGx variants, those were functionally disrupted in the Indian population with an allele frequency of more than 1% were fetched. A Sankey diagram, depicting the drug function disruption pathway was generated using flourish studio [28] by mapping these genes to the associated drugs in our list and DrugBank database.
Statistical analysis
Fisher’s exact test was used to compare the Indian allele frequencies with the global populations (1KGP3-ALL and gnomAD-ALL) and also other regional populations (QATAR-ALL and GME-ALL) to assess the significant differences in allele frequencies with respect to Indian population.
Drug–drug interaction analysis
The COVID-19 associated drug-gene interactions were visualized in the form of a network using Cytoscape [29]. The drug–gene associations were obtained from the DrugBank database. The gene labels are proportional to the degree of the node whereas the drug labels are sized according to a score that estimates the proportion of shared PGx genes associated with each drug. The score was calculated as the cumulative average sum of gene degrees associated with each drug.
where Denzyme - i, DTarget - i and DTransporter - i are the degrees of enzyme i, target i and transporter i associated with the drug and Nenzymes, NTargets and NTrasnporters are the total number of enzymes, targets and transporters associated with each drug. The final scores were normalized to the maximum score obtained for each node type (drug/gene).
The drug–gene associations of drugs used for treating metabolic disorders were obtained from the DrugBank database and the overlaps were plotted using Venn Diagrams [30]. The details of the drug categories and the associations are listed in Supplementary Table 5. The inhibitor status for COVID-19 drugs were obtained by overlapping the list of 89 drugs with the Flockhart table of drug–gene interactions [31]. The list of potential DDIs for the CYP inhibitor drugs used in COVID-19 therapy were obtained using the drug–gene interactions in the DrugBank database and clinical guideline annotations from PharmGKB.
Results
Drugs involved in COVID-19 therapy
In this study, we identified 89 drugs that are currently being used clinically or are undergoing clinical trials for COVID-19 as listed in the PharmGKB [25] and DrugBank [26] databases (Table 1 & Supplementary Table 1). These drugs are classified into four groups: anticytokine/anti-inflammatory (n = 30), inhibiting viral entry (n = 10), inhibiting viral replication (n = 15) and others (n = 36). While ten drugs out of 89 have at least one high-confidence PharmGKB clinical annotation (Level 1 & Level 2), 60 drugs have at least one associated pharmacogene listed by the DrugBank database in the role of a metabolizing enzyme, target, transporter or carrier. A total of 27 out of 89 drugs do not have any pharmacogenetic information available as some of these are new/experimental drugs.
Clinically significant PharmGKB variants associated with COVID-19 treatment among Indians
Toward cataloging clinically relevant COVID-19-associated PGx variants predominant in the Indian population, we obtained a total of 330 variant-drug clinical annotations that overlapped with our drug list in the PharmGKB database. A total of 29 of these associations involve nine drugs and encompass 16 single nucleotide variants (SNVs) and 14 haplotypes in 17 genes which have Level 1 & Level 2 evidence, and are considered clinically significant. Comparative analysis of the allele frequencies of these variants among Indians and other global populations included in gnomAD [20], 1000 Genomes Project [19], GME [21] and Qatar [32] indicate remarkable inter-population differences (Figure 1 & Supplementary Table 2). For example, the variant rs12979860 in gene IFNL3/IFNL4 showed up to threefold difference in prevalence ranging from 20% among Indians to 60% among African populations. Similarly, the prevalence of the variant rs578776 in CHRNA3 gene is 26% among Ashkenazi Jewish population compared with 77% among East Asians. Indian allele frequencies for every variant were found to be significantly different (p < 0.05, Fisher’s test) from the global average represented by at least one among gnomAD (gnomAD-ALL) and 1000 Genomes projects (1KGP3-ALL).
Figure 1. Allele frequencies of PharmGKB variants associated with COVID-19 drugs.
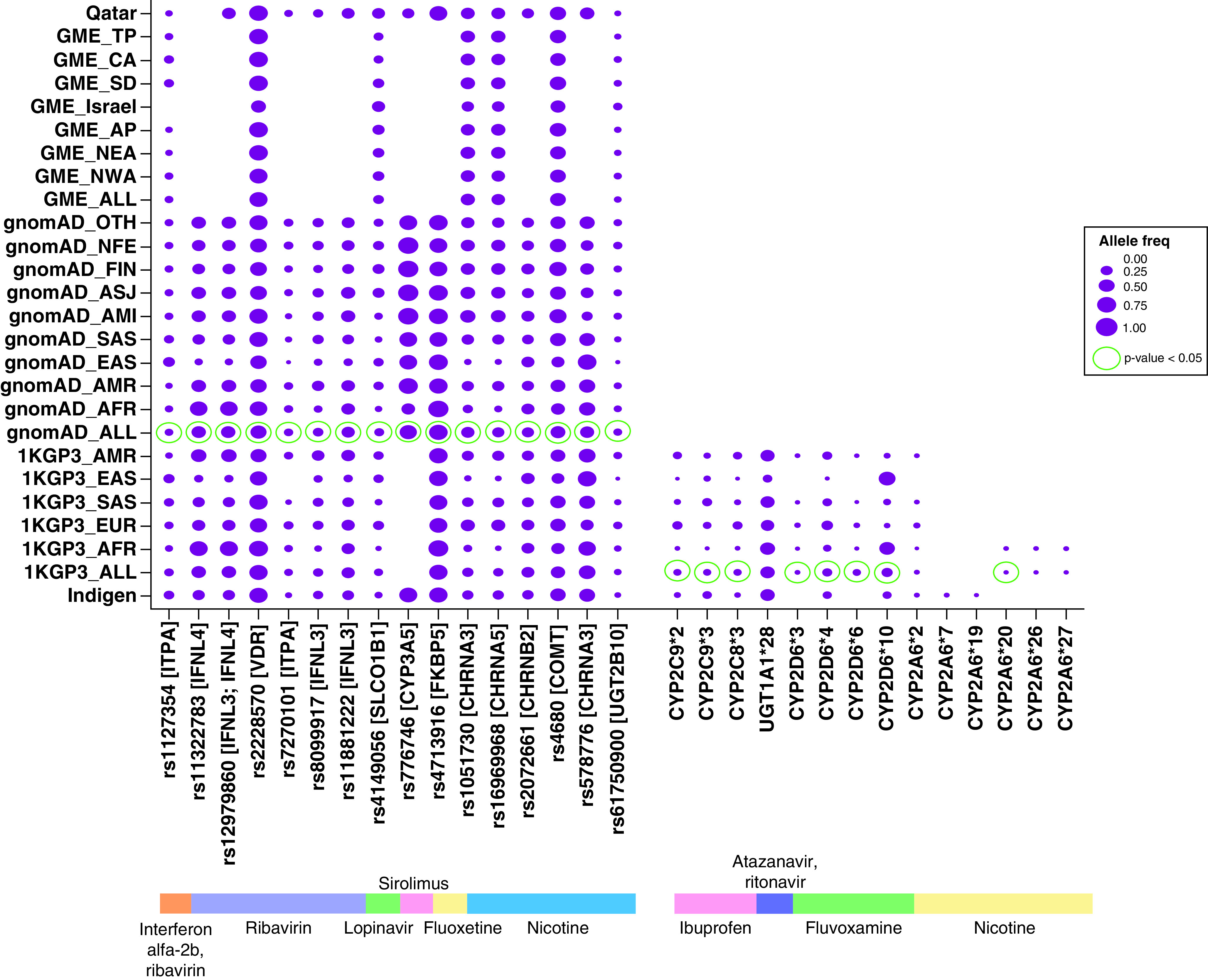
Comparison of Indian allele frequencies of clinically relevant PGx variants with populations in 1000 Genomes dataset, gnomAD database, GME database and Qatar database. PGx variants in Indians which yielded significant p-value (p < 0.05) in the Fisher’s exact test comparing Indian allele frequency with other databases are highlighted in green outer circle.
GME: Greater Middle East; PGx: Pharmacogenetic.
A total of 12 out of the 29 annotations involved antiviral drug ribavirin in association with four genes (IFNL3, IFNL4, ITPA, VDR). Two of these variants, rs12979860 and rs8099917 in IFNL3 and IFNL4 genes with the highest level of evidence (Level 1A/1B) have clinical guideline annotations issued by the Clinical Pharmacogenetics Implementation Consortium [33]. As per the guideline, our analysis shows that 37% of the Indian population carry at least one of the variants and are therefore unlikely to have a favorable response to ribavirin. Another variant belonging to ITPA gene, rs1127354, shows twofold significantly higher prevalence among Indians compared with others (IndiGen: 0.12, gnomAD-ALL: 0.06) and is associated with conferring protective effects against ribavirin-induced hemolytic anemia in patients with chronic hepatitis C. This variant is also associated with the dosage of ribavirin when accompanied by interferon alfa-2b. Similarly, the variant rs2228570 in VDR gene associated with decreased response to treatment with peginterferon alfa-1b and ribavirin shows higher prevalence in Indians when compared with the global populations (IndiGen: 0.74; gnomAD-ALL: 0.66).
The variant rs4149056 in SLCO1B1 gene associated with increased plasma lopinavir concentrations shows lesser prevalence among Indians when compared with frequencies that are twofold higher in the global population and fourfold higher in Qataris (IndiGen: 0.05; gnomAD-ALL:0.12; QATAR: 0.24). Similarly, eight out of 29 annotations were associated with nicotine involving five genes (UGT2B10, CHRNA3, CHRNA5, CHRNB2 and COMT). One of these variants rs578776 in CHRNA3 gene associated with toxicity and metabolism of nicotine shows higher prevalence among Indians when compared with global average frequencies (IndiGen: 0.5122; gnomAD-ALL: 0.3939) with varied prevalence among other subpopulations. We also observed that two haplotypes CYP2A6*7 and CYP2A6*19 that are associated with decreased metabolism of nicotine are sparsely present in India (IndiGen: 0.0087, 0.0082) while it could not be detected in the 1000 Genomes populations.
In concordance with the global prevalence, UGT1A1*28 haplotype that is associated with increased risk for hyperbilirubinemia in case of joint administration of atazanavir and ritonavir drugs in HIV patients was found to be present at a high allele frequency of 40% among Indians (IndiGen: 0.40; gnomAD-ALL: 0.39). In case of haplotypes CYP2C9 *2 and *3 which are associated with decreased metabolism of ibuprofen, the Indian allele frequencies for the former is lower (IndiGen: 0.0306; 1KGP3-ALL: 0.0584) while the latter is higher (IndiGen: 0.11; 1KGP3-ALL: 0.06) in comparison with global averages.
CYP2D6*10 haplotype has a lower allele frequency of 7% in Indians compared with the global average (1KGP3-ALL: 0.1759) and is associated with increased plasma concentration of antidepressant drug, fluvoxamine with increased risk of developing gastrointestinal side effects. Similarly, CYP2D6*4 allele that is associated with the decreased clearance of fluvoxamine drug has a higher allele frequency of 9% in Indians compared with the global population (1KGP3-ALL: 0.006).
Potentially deleterious PGx variants associated with COVID-19 therapy among Indians
In an attempt to identify a comprehensive set of hitherto unknown population-specific COVID-19 associated PGx variants in the Indian population, we performed a systematic survey of potential functional disruption in genes associated with COVID-19 drugs as per DrugBank database. A total of 222 genes were found to be associated with 60 drugs involved in the role of enzymes, targets, transporters or carriers. Consensus functional predictions based on SIFT [22], Polyphen-2 [23] and MutationTaster [24] tools identified 1386 potentially deleterious variants disrupting the function of 211 genes associated with 43 COVID-19 drugs. A total of 73 of these variants are prevalent in the Indian population at over 1% effect allele frequency (Figure 2 & Supplementary Table 3). The variant rs60140950 in SLCO1B3 transporter gene that has been previously associated with altered transporter expression of associated statins [34], is prevalent in 5% of the Indian population (IndiGen: 0.053; gnomAD-ALL: 0.10). Variants in the gene NR3C1 are associated with methylprednisolone potency [35]. We report a potentially deleterious variant rs6190 in the same gene with 2% allele frequency in the Indian population (IndiGen: 0.02; gnomAD-ALL: 0.02). Our analysis also highlighted a drug resistance-associated variant in ABCC4 gene [35,36], rs11568658, that potentially affects response to two COVID-19 drugs ibuprofen and remdesivir whose prevalence is twice higher in Indians in comparison to the global population (IndiGen: 0.078; gnomAD-ALL: 0.03).
Figure 2. Allele frequencies of most common potentially deleterious nonsynonymous variants in Indians involved in COVID-19 drug transport, metabolism and targeting.
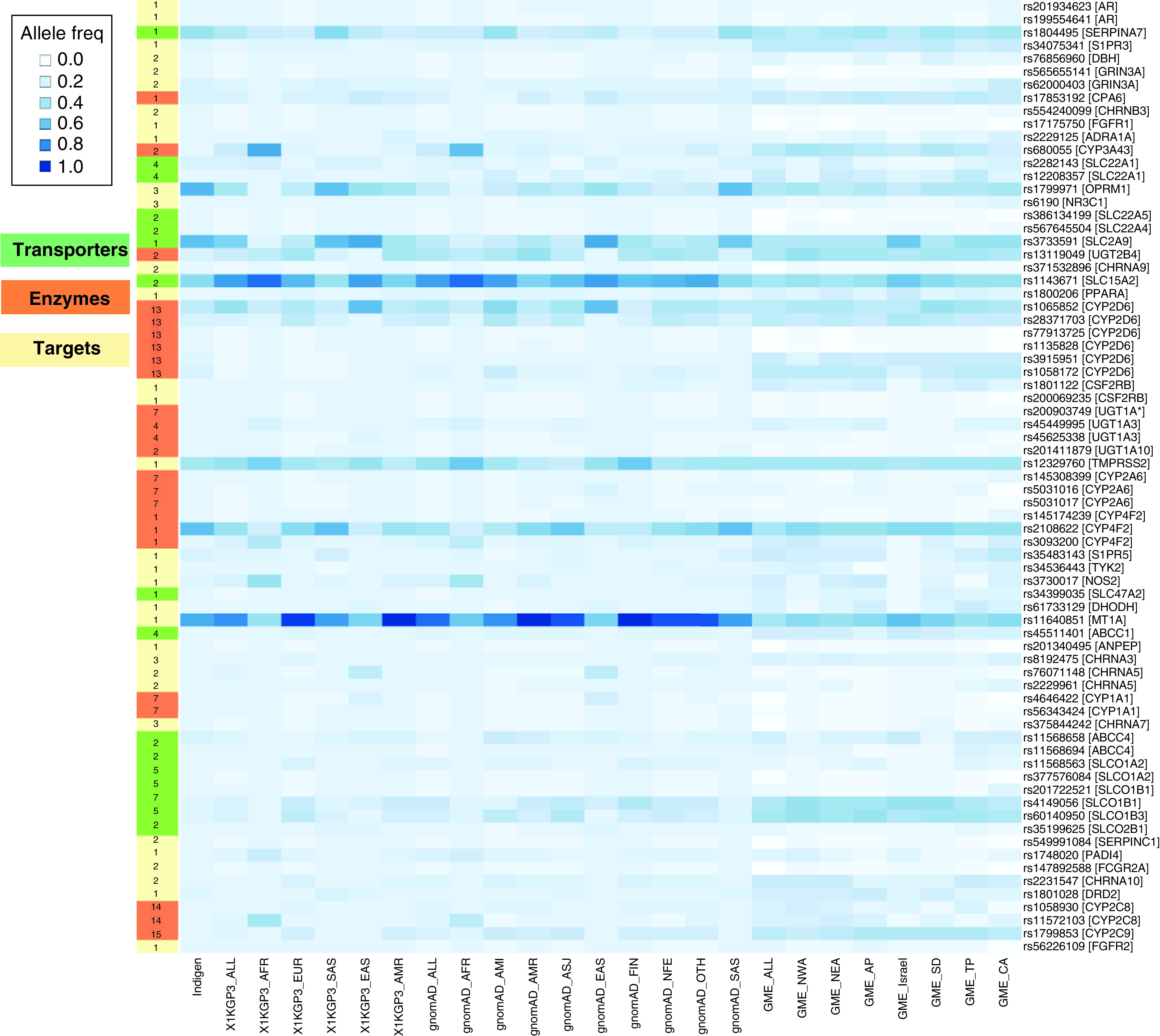
Allele frequencies are compared with 1000 genomes database, gnomAD database, GME database and Qatar database. Y-axis represents the variant [Gene Name] and x-axis represents the population and subpopulation. Gene function category is color coded on the left with the number of drugs associated with each gene.
GME: Greater Middle East.
Drug pathways disrupted in the Indian population
Toward deriving pathway level insights into the key pharmacokinetic/pharmacodynamic functions associated with response to COVID-19 therapy that are frequently disrupted in the Indian population, we performed a drug pathway analysis focused on 60 drugs associated with 52 genes that are disrupted in at least 1% of the population (Figure 3). The results are presented as a Sankey flow chart allowing one to visualize the individual drug–gene relationships organized into multiple levels/columns based on the gene functions (transporter, enzyme, target) along with information regarding the extent of overall drug functional loss. The analysis reveals that six drugs, icatibant, naproxen, remdesivir, tranexamic acid, dapagliflozin and metenkefalin showed at least 50% disruption of its overall function via impaired transport, target or metabolism. We also observed that three drugs, tranexamic acid, heparin and metenkefalin have complete disruption of at least one of their individual functions, namely enzymes, transport or targets.
Figure 3. Drug pathway map representing the pharmacogenes functionally disrupted in the Indian population.
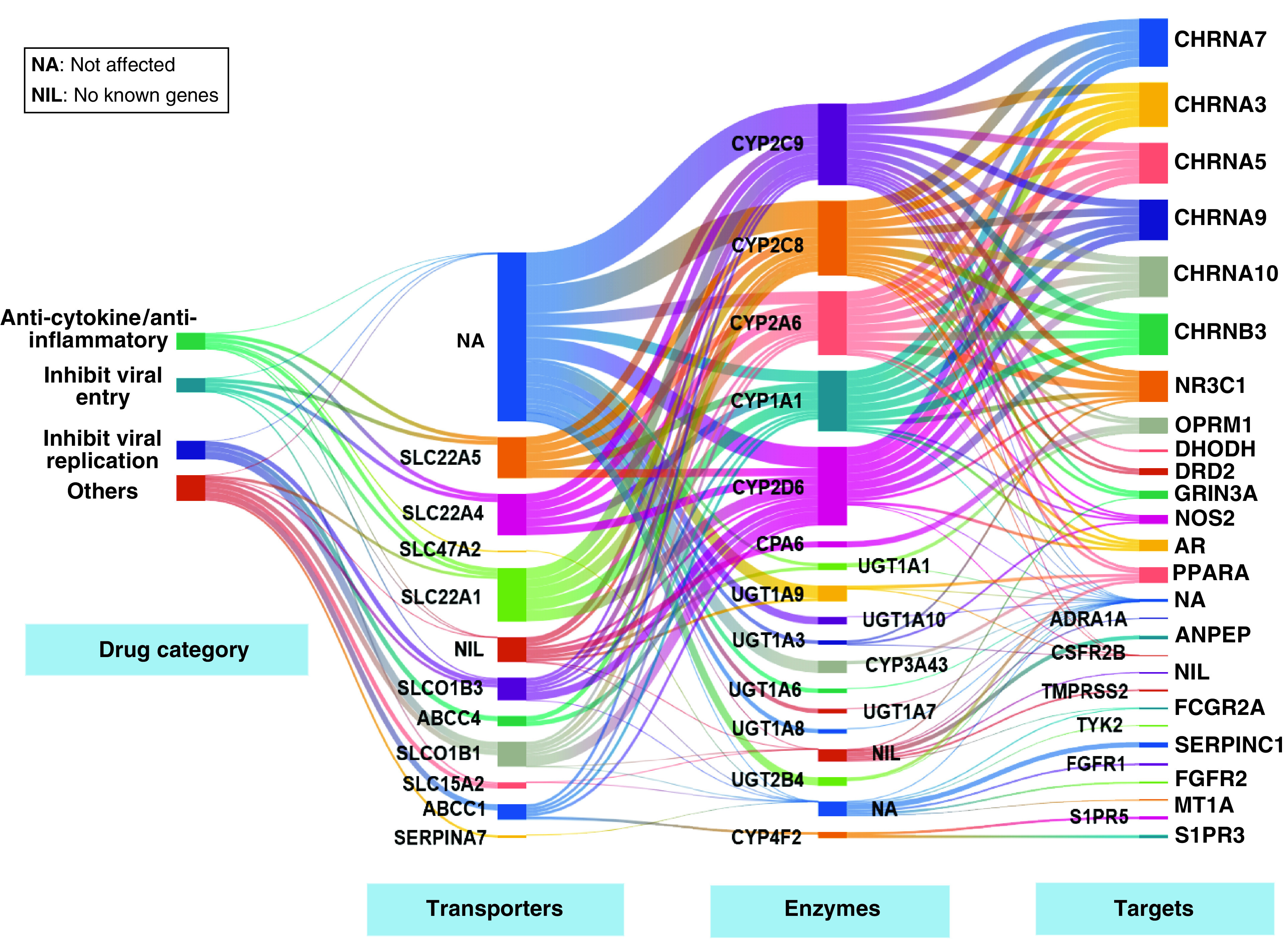
The first column represents the broad drug category associated with the potentially deleterious variants in the Indian population. The second, third and fourth columns represent the pharmacogenes belonging to the classes: transporter/carriers, enzymes and targets respectively. The line width represents the degree of functional loss of the given drug in terms of the pharmacogenes classes.
Remdesivir, which is studied in 12 clinical trials for COVID-19 treatment has half of the genes involved in its metabolism and transport disrupted in at least 1% of the Indian population. Hydroxychloroquine, used widely in the treatment of COVID-19 may have altered metabolism and transport function among Indians, as three of its associated genes namely CYP2D6, CYP2C8 and SLCO1A2 are commonly (over 10, 1 and 1%, respectively) impaired in the population. This drug is also reported to have adverse effects such as hemolytic anemia, cardiomyopathy, neutropenia, gastrointestinal disturbances, retinopathy, rash and QT prolongation [3]. Metenkefalin, an investigational endogenous opioid being studied for treatment of COVID-19 is found to have 66% of function disruption as its sole metabolizing enzyme CPA6 along with one out of its two target genes (OPRM1) has a potentially deleterious variant rs1799971 with an allele frequency of 40% in Indians, which is known to affect the metabolism of other opioids [3,37].
DDIs in COVID-19 therapy
Toward demonstrating the pharmacogenetic basis for DDIs triggered by polypharmacy during COVID-19 therapy, we performed a network analysis involving COVID-19 drugs and their shared drug targets, metabolizing enzymes and transporters/carriers. Our analysis showed that CYP3A4, followed by CYP2C9, CYP1A2 and CYP2C8 were the most shared enzymes associated with a total of 77 COVID-19 drugs suggesting potential risk for interactions if the related drugs are co-administered (Figure 4A & Supplementary Table 4). An overlap with the Flockhart table of CYP–drug interactions [31] indicates that three COVID-19 drugs, fluoxetine, ritonavir and fluvoxamine act as strong inhibitors of CYP2D6, CYP3A4/5/7 and CYP1A2 respectively (Figure 4B). These inhibitor drugs can therefore compete with other substrate drugs for the associated enzyme (CYP2D6 [n = 10], CYP3A4 [n = 30], CYP3A5 [n = 9], CYP3A7 [n = 4] and CYP1A2 [n = 13]) and cause >fivefold increase in their plasma area under the curve (AUC) values or more than 80% decrease in their clearance. In the case of transporters, ABCB1 was identified as one of the most shared transporters that has been associated with 16 drugs in our list. Association of azithromycin with common ABCB1 variants, rs2032582 (IndiGen: Not Available, 1KGP3-SAS: 0.36) and rs1045642 (IndiGen: 0.40) have been reported earlier causing up to twofold differences in peak azithromycin levels [38]. This is particularly concerning for a QT-prolonging agent as it may further increase the risks for cardiac toxicity when combined with similar drugs such as hydroxychloroquine/chloroquine during COVID-19 therapy [38,39]. Our analysis also highlighted that drugs such as isotretinoin, azithromycin, baricitinib, tofacitinib, apremilast and fluvoxamine have increased likelihood for DDIs by virtue of the proportion of shared PGx genes associated with them.
Figure 4. Pharmacogenetic of predicted drug–drug interactions and drug–drug–gene interactions in COVID-19 therapy.
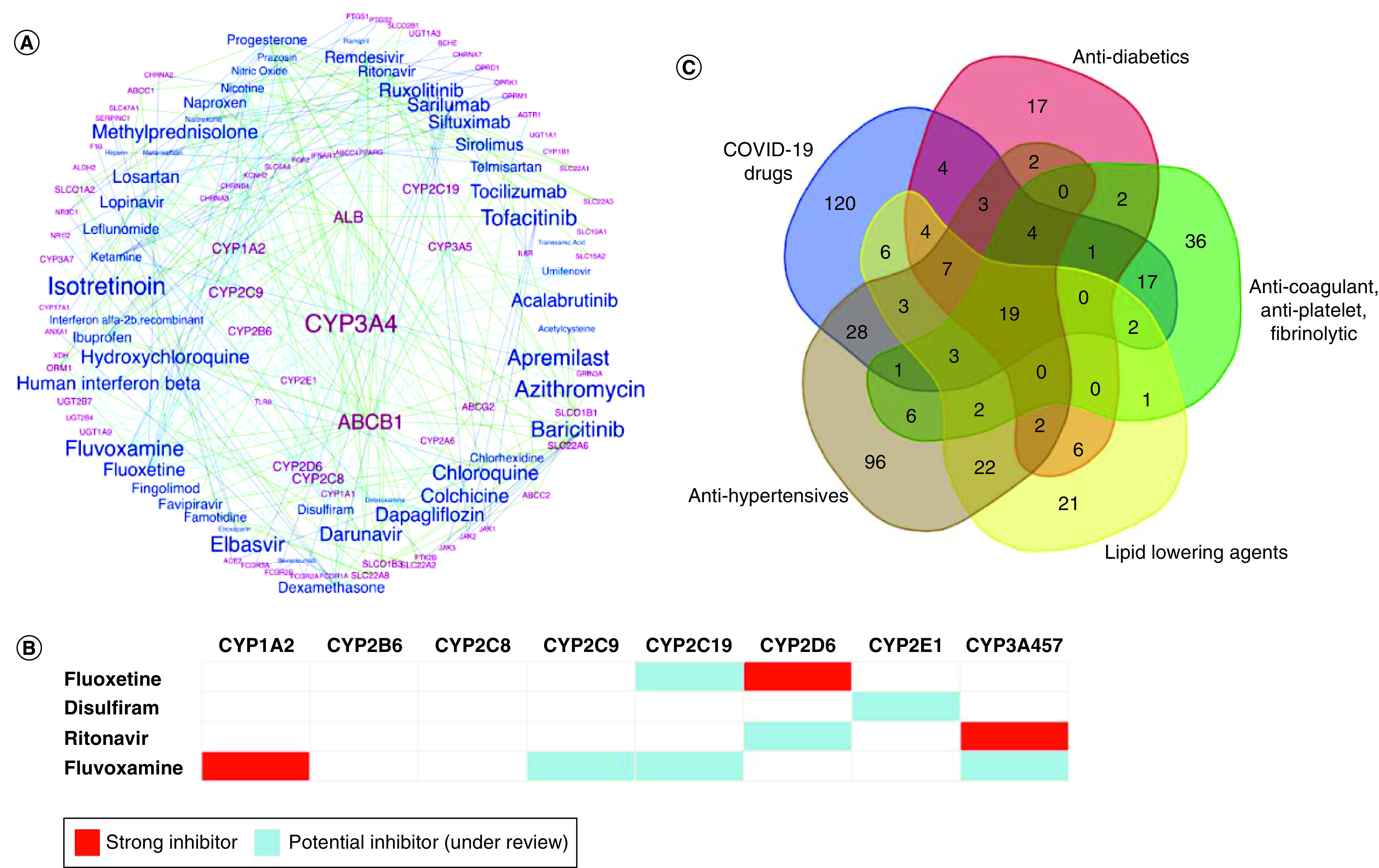
(A) A network representation of the drug–gene interactions involved in COVID-19 therapy. The drugs and gene labels are highlighted in blue and pink colours, respectively. The label sizes of the genes are proportional to the number of drug connections while that of the drugs are proportional to the proportion of shared PGx genes associated with each drug (See Methods for details). (B) A Venn diagram representing the overlap of genes associated with COVID-19 therapy and treatment of metabolic disorders. (C) A list of COVID-19 drugs showing their inhibitor status for the major CYP enzymes as per the Flockhart table of drug–gene interactions.
PGx: Pharmacogenetic.
The mounting evidence for increased risk of severe COVID-19 infection among patients with comorbidities mostly involving metabolic disorders also prompted us to compare the shared pharmacogenes between our list of COVID-19 drugs and the widely prescribed antidiabetics, lipid lowering agents, anticoagulants, antiplatelets, fibrinolytics and antihypertensives (Supplementary Table 5). Overall, 19 genes, primarily enzymes and transporters, were found to be shared by all the drug categories. Our results also highlight that 45% of genes associated with lipid lowering agents, 34% of antihypertensives associated genes, 49% of genes associated with anticoagulant, antiplatelet and fibrinolytic and 59% of antidiabetics were found to be shared with COVID-19 drugs (Figure 4C).
The knowledge of key CYP inhibitors among COVID-19 drugs also allowed us to compile a candidate list of potential DDIs by identifying drugs that are solely metabolized by the relevant enzymes or have clinical guideline annotations in PharmGKB (Table 2). The former approach identified a total of 36 drugs including 15 COVID-19 drugs, five antidiabetics, 13 antihypertensives, two lipid lowering agents and two cardiac drugs that can cause potential DDIs involving the four inhibitor drugs (fluvoxamine, ritonavir, fluoxetine and disulfiram) described earlier (Figure 4B). The latter approach helped us highlight an additional 80 drugs across diverse drug categories that may require careful monitoring when used with the associated COVID-19 drug as per the PharmGKB guidelines (Supplementary Table 5).
Table 2. List of potential drug–drug interactions in COVID-19 therapy.
| Drug | Reported DDGI | COVID-19 drugs | Antidiabetics | Antihypertensives | Lipid lowering agents | Anticoagulant/antiplatelet/fibrinolytic | Dosing guidelines (count) | Ref. | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Drug | Gene | Drug | Gene | Drug | Gene | Drug | Gene | Drug | Gene | ||||
|
Ritonavir inhibits CYP3A4 CYP3A5 CYP3A7 CYP2D6 |
Voriconazole | Tocilizumab Sarilumab Siltuximab Baricitinib Isotretinoin Ruxolitinib Azithromycin |
3A4 | Linagliptin | 3A4 | Indapamide Aliskiren Hydralazine Ivabradine Lacidipine |
3A4 | Fish oils | 3A4 | Argatroban | 3A4/5 |
3A4/5 (1) 2D6 (37) |
[38] |
| Darunavir | 3A4 2D6 |
Saxagliptin | 3A4 3A5 |
Lercanidipine | 3A4/5/7 2D6 |
||||||||
| Sirolimus | 3A457 | Ranolazine | 3A4 2D6 |
||||||||||
| Acalabrutinib | 3A45 | Tadalafil | 3A4 3A5 |
||||||||||
|
Fluvoxamine inhibits CYP1A2 CYP3A4 CYP2C9 CYP2C19 |
Lansoprazole | Tocilizumab Sarilumab Siltuximab Baricitinib Isotretinoin Ruxolitinib Azithromycin |
3A4 | Linagliptin | 3A4 | Indapamide Aliskiren Hydralazine Ivabradine Lacidipine |
3A4 | Fish oils | 3A4 | Acenocoumarol | 1A2 3A4 2C9 2C19 |
3A4 (1) 2C9 (21) 2C19 (13) |
[38] |
| Leflunomide | 1A2 2C9 |
Gliclazide | 2C9 2C19 |
Macitentan | 3A4 2C19 |
Rosuvastatin | 2C9 | ||||||
| Interferon alfa-2b Famotidine Human interferon beta |
1A2 | Glimepiride | 2C9 | Treprostinil Valsartan |
2C9 | ||||||||
| Tofacitinib | 2C19 3A4 |
Bosentan | 2C9 3A4 |
||||||||||
|
Fluoxetine inhibits CYP2D6 CYP2C19 |
2D6 (37) 2C19 (13) 2D6 + 2C19 (10) |
||||||||||||
|
Disulfiram inhibits CYP2E1 |
Isosorbide dinitrate | 2E1 | |||||||||||
DDGI: Drug–drug–gene interaction.
It is known that genetic polymorphisms can influence the magnitude of DDIs. For example, the inhibitory effect of fluvoxamine (CYP1A2/2C9/2C19/3A457 inhibitor) on the biotransformation of chloroguanide (CYP2C19 substrate) is greater in normal CYP2C19 metabolizers compared with poor metabolizers [40]. DDIs involving drugs that use multiple biotransformation pathways can also be additionally influenced by genetic polymorphisms in the associated enzymes [40,41]. For example, Bahar et al. showed that co-administration of CYP3A4 inhibitor, ritonavir with antifungal agent voriconazole (a substrate of CYP3A4 and CYP2C19) caused 54% increase in plasma AUC levels in normal CYP2C19 metabolizers while it caused 807% increase in CYP2C19 poor metabolizers [42]. Similar gene-dependent effects were reported for fluvoxamine when used with gastric proton pump inhibitor, lansoprazole where normal and intermediate CYP2C19 metabolizers showed major interactions (normal metabolizer [NM]: 283% increase in AUC and intermediate metabolizer [IM]: 150% increase in AUC) compared with minimal effect in case of poor metabolizers (4% increase in AUC) [42]. These results are particularly significant for specific populations like India with substantial prevalence of CYP2C19 variants (*2: 0.03 and *3: 0.1).
Discussion
The current study has systematically identified and cataloged the prevalence of 30 clinically significant PGx variations along with 73 predicted deleterious PGx variants associated with response to COVID-19 therapy in the Indian population. Remarkable population-scale allele frequency differences were observed for most variants justifying the need for formulating country-level policies for the use of COVID-19 therapies in distinct populations. The availability of population-scale Indian whole genomes provided us a unique opportunity to provide a comprehensive summary of the clinically significant CYP star alleles in addition to other rare and common SNVs in the population.
Analysis of Indian allele frequencies for clinically actionable PGx variants suggest that a third of Indian COVID-19 patients are likely to respond less favorably to peginterferon alpha and ribavirin combination therapy, that is being adopted especially to treat COVID-19-associated severe pneumonia [43]. Carriers of rs12979860 and rs8099917 variants have increased likelihood for lower sustained virological response rate during treatment with peginterferon alpha and ribavirin in chronic hepatitis C and B patients [43,44]. We also show that a large majority of Indian patients are at an increased risk of ribavirin-induced anemia as only 22 and 4% of the population carry the protective variants, rs1127354 and rs7270101, respectively, in the ITPA gene. The inability to tolerate higher doses of ribavirin in such patients may further reduce their chances of attaining sustained virological response. Another key actionable PGx finding for treating Indian COVID-19 patients relates to the antidepressant drug, fluvoxamine, which is being repurposed for treating the cytokine storm associated with COVID-19 [45]. We observed that 2.2% of Indians are CYP2D6 poor metabolizers who are recommended 25–50% reduced dosage of fluvoxamine or an alternate CYP2D6-independent drug to reduce possible adverse reactions [46].
The pharmacokinetic/dynamic investigations also helped us prioritize a list of drug candidates for potential DDIs in COVID-19 therapy. The concomitant use of strong CYP inhibitors identified in this study with the associated substrate drugs should be carefully monitored. For example, drugs such as interferon alfa-2b (recombinant), famotidine and human interferon beta which utilize CYP1A2 as their sole metabolizing enzyme are more likely to show altered exposure when administered with CYP1A2 inhibitor, fluvoxamine. Similarly, regimens containing CYP3A4/5/7 inhibitor ritonavir could elicit an impaired response to a large set of exclusively CYP3A4/5/7-metabolized drugs such as ruxolitinib, acalabrutinib, baricitinib, azithromycin, tocilizumab, sarilumab, siltuximab and isotretinoin.
Given the increased risk posed by polypharmacy in severe COVID-19 patients with metabolic disorders, our predicted list of potential DDIs associated with metabolic disorder therapy also warrants systematic clinical monitoring and validation studies. For example, the effect of fluvoxamine on elevated risk of over-anticoagulation during acenocoumarol maintenance treatment has already been reported [46,47]. Similarly, the use of strong CYP3A4/5/7 inhibitors ritonavir and/or fluvoxamine with saxagliptin or linagliptin in diabetes patients is also expected to cause altered exposure thereby requiring dose adjustments [46–48]. In addition, PGx studies of DDIs and DDGIs suggest that the strength of these predicted interactions could be altered to a great extent based on the patient’s metabolizer status (rapid/normal/intermediate/poor) for the affected as well as alternate enzymes. Accordingly, a large proportion of Indian patients are likely to show varied response to CYP2C19-mediated DDGIs given the high cumulative prevalence of CYP2C19 *2 (AF: 36%) and *3 (AF: 0.6%) variants.
Conclusion
The current study has performed a comprehensive assessment of known and predicted pharmacogenetic variants, DDIs and DDGIs associated with COVID-19 therapy in the Indian population. We highlight the most clinically significant associations along with predicted associations and interactions that can be prioritized for clinical monitoring and validation. Given the inexorable spread of the pandemic in the Indian subcontinent, the insights from the current study can be utilized toward planning large-scale nation-level COVID-19 clinical trials toward ensuring improved therapeutic outcomes by maximizing the efficacy and safety of treatment regimens.
Future perspective
As routine and individual PGx testing is not feasible in current clinical settings, a population scale PGx analysis for specific therapies would be of utmost clinical significance. This becomes even more important in pandemics like COVID-19 where the illness is acute and hence individual pharmacogenetic testing and its implications remain a large limitation. With widely available population genome datasets, an extensive PGx analysis can help in expediting decision making for choice of empirical therapies on a population scale for various diseases, for better clinical outcomes with minimal adverse effects. Similar analysis on population datasets can provide further understanding of current therapy structures for COVID-19, combination therapies and post COVID-19 therapy evaluation of efficacy and adverse effects.
Summary points.
Background
Numerous experimental and repurposed drugs are undergoing clinical trials or are being widely prescribed for COVID-19 treatment without adequate direct evidence for its safety/efficacy in patients across diverse ethnic populations.
The recently launched population-scale whole genome sequencing of Indian genomes (IndiGen project) provides a unique opportunity to explore the landscape of pharmacogenetic (PGx) variants associated with differential COVID-19 response among Indians which is currently one of the worst affected countries in the world.
Identification of clinically significant & predicted deleterious PGx variants among Indians
We identified 30 clinically significant PGx variations along with 73 predicted deleterious PGx variants associated with pharmacological response to COVID-19 therapies in the Indian population.
Our analysis shows that a large majority of Indian patients are at an increased risk of ribavirin-induced anemia as only 22 and 4% of the population carry the protective variants.
A total of 2.2% of Indians are CYP2D6 poor metabolizers who are recommended 25–50% reduced dosage of fluvoxamine or an alternate CYP2D6-independent drug to reduce possible adverse reactions.
Drug–drug & drug–drug–gene interactions analysis
CYP3A4, CYP2C9, CYP1A2, CYP2C8, ABCB1 and ALB were identified as the most shared PGx genes in COVID-19 therapy.
A total of 15 therapeutics used in the treatment of COVID-19 were predicted as likely candidates for potential drug–drug interactions (DDIs) with four CYP inhibitor drugs, fluvoxamine, ritonavir, fluoxetine and disulfiram.
Our results also highlight the high prevalence of CYP2C19 alleles that are potentially associated with differential DDIs and drug–drug–gene interactions in COVID-19 therapy.
Conclusion
This is the first comprehensive study of PGx associations, DDIs and drug–drug–gene interactions associated with therapeutics used in COVID-19 in the Indian population providing timely and useful insights for clinical decisions and monitoring as well as for planning validation studies.
Supplementary Material
Supplementary data
To view the supplementary data that accompany this paper please visit the journal website at: www.futuremedicine.com/doi/suppl/10.2217/pgs-2021-0028
Author contributions
S Sahana, A Sivadas, V Scaria conceived the study. S Sahana, M Mangla and A Sivadas performed data curation, bioinformatics and statistical analysis. A Sivadas, M Mangla and V Scaria provided critical feedback on analyzed data. S Sahana and A Sivadas wrote the manuscript. S Sahana, A Sivadas, M Mangla, V Scaria and S Sivasubbu read, edited and confirmed the final version of the manuscript. All the remaining authors contributed in collection, sequencing and analysis of the genome sequence data.
Ethical conduct of research
This study was approved by the Institutional Human Ethics Committee (IHEC) of CSIR-Institute of Genomics and Integrative Biology.
Footnotes
Financial & competing interests disclosure
This work was supported by The Council of Scientific and Industrial Research, India [MLP1809 and MLP2001]. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
- 1.Sanders JM, Monogue ML, Jodlowski TZ, Cutrell JB. Pharmacologic treatments for coronavirus disease 2019 (COVID-19): A Review. JAMA 323(18), 1824–1836 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Kalil AC. Treating COVID-19 –off-label drug use, compassionate use, and randomized clinical trials during pandemics. JAMA 323(19), 1897–1898 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Takahashi T, Luzum JA, Nicol MR, Jacobson PA. Pharmacogenomics of COVID-19 therapies. NPJ Genom. Med. 5, 35 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.da Rocha J, da Rocha J, Othman H et al. G6PD variant distribution in sub-Saharan Africa and potential risks of using chloroquine/hydroxychloroquine based treatments for COVID-19. medRxiv (2020) (Epub ahead of print). [Google Scholar]
- 5.van der Graaf PH, Giacomini KM. COVID-19: a defining moment for clinical pharmacology? Clin. Pharmacol. Ther. 108(1), 11–15 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Badary OA. Pharmacogenomics and COVID-19: clinical implications of human genome interactions with repurposed drugs. Pharmacogenomics J. 21, 275–284 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zubiaur P, Koller D, Saiz-Rodríguez M, Navares-Gómez M, Abad-Santos F. Important pharmacogenetic information for drugs prescribed during the SARS-CoV-2 infection (COVID-19). Clin. Transl. Sci. 13(6), 1023–1033 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kohlrausch FB, de Cássia Estrela R, Barroso PF, Suarez-Kurtz G. The impact of SLCO1B1 polymorphisms on the plasma concentration of lopinavir and ritonavir in HIV-infected men. Br. J. Clin. Pharmacol. 69(1), 95–98 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lo C, Nguyen S, Yang C et al. Pharmacogenomics in Asian subpopulations and impacts on commonly prescribed medications. Clin. Transl. Sci. 13(5), 861–870 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chi X, Wang M, Pan Y et al. Inosine triphosphate pyrophosphatase polymorphisms are predictors of anemia in Chinese patients with chronic hepatitis C during therapy with ribavirin and interferon. J. Gastroenterol. Hepatol. 35(1), 97–103 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Wang L-Y, Cui J-J, OuYang Q-Y et al. Genetic profiles in pharmacogenes indicate personalized drug therapy for COVID-19. medRxiv (2020) (Epub ahead of print). [Google Scholar]
- 12.Kamel Boulos MN, Geraghty EM. Geographical tracking and mapping of coronavirus disease COVID-19/severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) epidemic and associated events around the world: how 21st century GIS technologies are supporting the global fight against outbreaks and epidemics. Int. J. Health Geogr. 19(1), 8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chaubey G. Coronavirus (SARS-CoV-2) and mortality rate in india: the winning edge. Front Public Health 8, 397 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sivadas A, Scaria V. Population-scale genomics – enabling precision public health. : Advances in genetics Kumar DBT (Ed.). Academic Press, Volume 103, 119–161 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Stark Z, Dolman L, Manolio TA et al. Integrating genomics into healthcare: a global responsibility. Am. J. Hum. Genet. 104(1), 13–20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jain A, Bhoyar RC, Pandhare K et al. IndiGenomes: a comprehensive resource of genetic variants from over 1000 Indian genomes. Nucleic Acids Res. 49(D1), D1225–D1232 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Purcell S, Neale B, Todd-Brown K et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81(3), 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38(16), e164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Auton A, Abecasis GR, Altshuler DM et al. A global reference for human genetic variation. Nature 526(7571), 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bahar, Muh A. Pharmacogenetics of drug–drug interaction and drug–drug–gene interaction: a systematic review on CYP2C9, CYP2C19 and CYP2D6. Pharmacogenomics 18.7 701–739 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Scott EM, Halees A, Itan Y et al. Characterization of Greater Middle Eastern genetic variation for enhanced disease gene discovery. Nat. Genet. 48(9), 1071–1076 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 31(13), 3812–3814 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Current Protocols in Human Genetics 76(1), 7–20 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat. Methods 11(4), 361–362 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Hewett M. PharmGKB: the pharmacogenetics knowledge base. Nucleic Acids Res. 30(1), 163–165 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wishart DS, Knox C, Guo AC et al. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 36(Database issue), D901–906 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee S-B, Wheeler MM, Patterson K et al. Stargazer: a software tool for calling star alleles from next-generation sequencing data using CYP2D6 as a model. Genet. Med. 21(2), 361–372 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flourish Studio, Kiln Enterprises Ltd. (2020). https://flourish.studio/
- 29.Shannon P, Markiel A, Ozier O et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13(11), 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sterck L. ‘Draw venn diagram’. http://bioinformatics.psb.ugent.be/webtools/Venn/
- 31.Flockhart DA. Drug interactions: cytochrome P450 drug interaction table. Indiana University School of Medicine; (2007). https://drug-interactions.medicine.iu.edu [Google Scholar]
- 32.Koshy R, Ranawat A, Scaria V. al mena: a comprehensive resource of human genetic variants integrating genomes and exomes from Arab, Middle Eastern and North African populations. J. Hum. Genet. 62(10), 889–894(2017). [DOI] [PubMed] [Google Scholar]
- 33.Relling MV, Klein TE, Gammal RS, Whirl-Carrillo M, Hoffman JM, Caudle KE. The Clinical Pharmacogenetics Implementation Consortium: 10 years later. Clin. Pharmacol. Ther. 107(1), 171–175 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wagner JB, Abdel-Rahman S, Gaedigk A et al. Impact of SLCO1B1 genetic variation on rosuvastatin systemic exposure in pediatric hypercholesterolemia. Clin. Transl. Sci. 13(3), 628–637 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cazzola M, Rogliani P, Calzetta L, Matera MG. Pharmacogenomic response of inhaled corticosteroids for the treatment of asthma: considerations for therapy. Pharmgenomics. Pers. Med. 13, 261–271 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsukamoto M, Yamashita M, Nishi T, Nakagawa H. A human ABC transporter ABCC4 gene SNP (rs11568658, 559 G >T, G187W) reduces ABCC4-dependent drug resistance. Cells 8(1), 39 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seguí HA, Melin K, Quiñones DS, Duconge J. A review of the pharmacogenomics of buprenorphine for the treatment of opioid use disorder. J. Transl. Genet. Genom. 4, 263–277 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He X-J, Zhao L-M, Qiu F, Sun Y-X, Li-Ling J. Influence of ABCB1 gene polymorphisms on the pharmacokinetics of azithromycin among healthy Chinese Han ethnic subjects. Pharmacol. Rep. 61(5), 843–850 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Rosenberg ES, Dufort EM, Udo T et al. Association of treatment with hydroxychloroquine or azithromycin with in-hospital mortality in patients with COVID-19 in New York State. JAMA 323(24), 2493–2502 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeppesen U, Rasmussen BB, Brøsen K. Fluvoxamine inhibits the CYP2C19-catalyzed bioactivation of chloroguanide. Clin. Pharmacol. Ther. 62(3), 279–286 (1997). [DOI] [PubMed] [Google Scholar]
- 41.Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 138(1), 103–141 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Bahar MA, Setiawan D, Hak E, Wilffert B. Pharmacogenetics of drug–drug interaction and drug–drug–gene interaction: a systematic review on CYP2C9, CYP2C19 and CYP2D6. (Pharmacogenomics 18(7), 701–739 (2017). [DOI] [PubMed] [Google Scholar]
- 43.El-Lababidi RM, Mooty M, Bonilla M-F, Salem NM. Treatment of severe pneumonia due to COVID-19 with peginterferon alfa 2a. IDCases 21, e00837 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ragheb MM, Nemr NA, Kishk RM et al. Strong prediction of virological response to combination therapy by IL28B gene variants rs12979860 and rs8099917 in chronic hepatitis C genotype 4. Liver Int. 34(6), 890–895 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Fitzgerald PJ. Noradrenergic and serotonergic drugs may have opposing effects on COVID-19 cytokine storm and associated psychological effects. Med. Hypotheses 144, 109985 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hicks JK, Bishop JR, Sangkuhl K et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clin. Pharmacol. Ther. 98(2), 127–134 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teichert M, Visser LE, Uitterlinden AG et al. Selective serotonin re-uptake inhibiting antidepressants and the risk of overanticoagulation during acenocoumarol maintenance treatment. Br. J. Clin. Pharmacol. 72(5), 798–805 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.May M, Schindler C. Clinically and pharmacologically relevant interactions of antidiabetic drugs. Ther. Adv. Endocrinol. Metab. 7(2), 69–83 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


