Abstract
Since emerging from Wuhan, China, in December of 2019, the coronavirus (SARS-CoV-2) has been causing devastating severe respiratory infections in humans worldwide. With the disease spreading faster than the medical community could contain it, death tolls increased at an alarming rate worldwide, causing the World Health Organization to officially sanction the SARS-CoV-2 outbreak as a pandemic, leading to a state of worldwide lockdown for the majority of the year 2020. There have been reports of new strains of the virus emerging in various parts of the world, with some strains displaying even greater infectivity and transmissibility. Areas of the emerging variant of concern arise from countries like the United Kingdom, South Africa, Brazil, and India. These mutations carry a lineage from N501Y, D614G, N439K, Y453F, and others, which are globally dominated by clades 20A, 20B, and 20C. This literature review intends to identify and report SARS-CoV-2 variants that are currently evolving and their disease implications.
Keywords: 501Y.V1, 501Y.V2, 501.V3, ACE2, B.1.1.7, COVID-19, D614G, E484K, genetic variations, N439K, N501Y, SARS-CoV-2, spike mutations, variants of concern, Y453F
Introduction
The novel coronavirus of 2019 (COVID-19) pandemic is primarily due to the respiratory illness caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and has affected millions of individuals worldwide.1,2 As of 11 May 2021, according to the World Health Organization (WHO), cumulative confirmed cases and deaths due to COVID-19 were 158 million and 3.2 million, respectively. 3 The single-stranded SARS-CoV-2 is a ribonucleic acid (RNA) virus with a genetic configuration similar to that of the SARS outbreak from 2002 to 2004, SARS-CoV-1.1,2,4 The structural proteins involved in SARS-CoV-2 are the RNA-containing nucleocapsid (N) protein and the three viral envelope proteins, which are S (spike), E (envelope), and M (membrane).1,2,4 The mechanism of action of SARS-CoV-2 is cell entry via attachment of the spike protein to angiotensin-converting enzyme 2 (ACE2) receptors located on human cells. 1 The virus can be transmitted via respiratory droplets and aerosols. 1 A unique property of SARS-CoV-2 that has made it highly pathogenic and increased its susceptibility for transmission is the polybasic cleavage site. 5 This cleavage site on SARS-CoV-2 allows the activation of multiple spike proteins simultaneously, which effectively increases the pathogenicity, virulence, and species-to-species transmission. 5
Since the pandemic began in China in December 2019, thousands of variants of SARS-CoV-2 have emerged.4,6,7 The WHO defined the SARS-CoV-2 variant of concern (VOC) as a variant with increased transmissibility, virulence, and decreased response to available diagnostics, vaccines, and therapeutics. 8 The first major variant was observed in September in the United Kingdom (UK).6,9 The variant, termed Variant of Concern 202012/01 (VOC 202012/01), causes point mutations of asparagine to tyrosine in the receptor-binding domain (RBD) of the spike protein.6,9–11 This N501Y mutation became a growing concern due to the virus being able to adhere to the ACE2 receptor more strongly.9,10 Currently, the N501Y variant has been detected in over 40 countries outside of the UK.9,10 The second major variant, that emerged in the last quarter of 2020, was the 501Y.V2 variant in South Africa.9,10 This strain had a similar mechanism of action to the N501Y in the UK.9,10 The 501Y.V2 variant has been detected in many countries outside of South Africa and is characterized by mutations in the S protein, including residues in the RBD—K417N, E484K, and N501Y.9,12 Also, another variant with multiple mutations emerged in Denmark termed the “Cluster 5.” 13 These point mutations occurred among four different amino acids in the spike protein. 13 Additionally, this Cluster 5 variant is immune to neutralizing antibodies. 13 The emergence of variants has highlighted the importance of early identification due to the potential for higher infectivity, transmissibility, and risks of mortality.14,15 This paper aims to report on SARS-CoV-2 genetic variants of concern and their implication on the continuously evolving COVID-19 disease process.
Methodology
An electronic literature search was performed predominantly using databases such as PubMed, Google Scholar, EBSCOhost, Mendeley, and MedLine Plus. The search was limited to applicable journals and articles published from the initial emergence of the virus, 1 January 2020, until 11 May 2021. A manuscript was selected if it was relevant to the topic of genetic mutations or variants of SARS-CoV-2. Listed keywords were sought to narrow and navigate the search process. They include but were not limited to COVID-19, SARS-CoV-2, Variants of Concern, genetic variations, spike mutations, N501Y, D614G, N439K, Y453F, E484K, B.1.1.7, 501Y.V1, 501Y.V2, 501.V3, and ACE2.
Variants of SARS-CoV-2
SARS-CoV-2 infection utilizes the ACE2 receptor and the transmembrane serine protease (TMPRSS2) to spread disease by infecting human respiratory cells. 16 The entry of SARS-CoV-2 into the human body depends on the attachment of the viral spike (S) proteins to cellular receptors, like ACE2.17–19 Also, S protein priming by human cell proteases, such as TMPRSS2, plays a role in the pathogenic entry of coronaviruses. 17 Differences in the interactions between proteins encoded by ACE2 human alleles and SARS-CoV-2 S proteins are being evaluated to better understand the prognosis of COVID-19 disease. 20 In other words, ACE2 and TMPRSS2 are important determinants for the pathophysiology of SARS-CoV-2 mutation(s), especially given the current manifestations in various variants, expressions, and epigenetic aspects observed in COVID-19 patients.21,22
Variants of SARS-CoV-2 are further being assessed, to determine whether a specific variant’s transmissibility, clinical presentation, severity, or impact on countermeasures (i.e., diagnostics, therapeutics, and vaccines) hinders the disease process.9,14,15,23 Therefore, priority has been placed on tracking the following mutations circulating worldwide: D614G (B.1 lineage), N501Y (several lineages), E484K (several lineages), K417 (several lineages), L452R (several lineages), Q677 (several lineages), and others. 9 D614G, a mutation of SARS-CoV-2 which emerged in early 2020, has a substitution in the gene encoding the S protein. 9 The D614G was the dominant form for several months worldwide, with increased infectivity and transmission.7,9,24 This mutation replaced the initial SARS-CoV-2 strain originally identified in China (NC_045512) 24 and has not been shown to produce severe illness or alter the efficacy of the present laboratory diagnostics, therapeutics, vaccines, or public health preventative measures.9,14,15 N439K was found to be less infectious than variant D614G; however, N439K still showed infectivity of COVID-19, making it a variant of interest and concern. 25 Receptor-binding motif (RBM) mutation is widespread among the second most commonly identified mutation in the RBD as of late 2020, N439K, which arose from lineage B.1 from the D614G mutated background.9,26 N439K RBM mutation has independently emerged in multiple lineages, showing increased affinity in the spike protein for ACE2, resisting several monoclonal antibodies (a therapeutic approach), and escaping some polyclonal responses. 26 “Cluster 5” or Y453F variant, which emerged in August of 2020, may potentially evade the immunity of convalescent individuals, further suggesting that this variant may challenge the vaccine strategy should it spread. 27 Therefore, emphasis is placed on characterizing the transmission capacity and the effect of this new variant. 27
Originating from N501Y, the last VOC, the year 2020, month 12, variant 01, referred to as SARS-CoV-2 VOC 202012/01 or B.1.1.7, emerged in the UK, containing 23 nucleotide substitutions from the initial SARS-CoV-2.9,15,28–30 This variant has shown increased transmissibility within the population.15,31 Furthermore, VOC 202012/01 has a deletion at the position 69/70del that affects the performance of diagnostic polymerase chain reaction (PCR) assays with an S gene target which is not expected to pose a significant concern, as most facilities globally use PCR assays with multiple targets.9,28 Since December 2020, this variant (B.1.1.7) has been detected in 110 countries/territories/areas. 9 Variant 501Y.V2, yet another subtype of N501Y mutation, is different from the UK 202012/01 variant. 9 The South African originating 501Y.V2 has caused the rapid displacement of other lineages circulating in Eastern Cape, Western Cape, and KwaZulu-Natal provinces, which may suggest increased transmissibility.13,32
As time evolves, so do the various mutations of SARS-CoV-2. Mutation E484K, connected with several lineages, has been shown to avoid select antibodies; whereas K417 mutation, also seen among several lineages which include B.1.351 and the P.1 of Brazil (an offshoot of B.1.1.28, but a close relative of the B.1.351), may bind more tightly to cells. 8 E484K has given rise to the B.1.525/B.1.526 lineage seen spreading among New York residents, perhaps because this version is more capable of evading antibodies, as well as binding more tightly to human cells due to the co-S477N mutation.8,33,34
Surging in late 2020, mutation L452R also referred to as the CAL.20C, a variant with lineages B.1.427 and B.1.429, has not yet been shown to be more infectious, though the numbers of confirmed cases are increasing throughout the state of California.9,35,36 Furthermore, one unique lineage of interest with increased infectivity and immune escape is B.1.617, the “double mutant” that carries two prominent mutations: E484Q and L452R. This E484Q has a similar translocated location as E484K, which allows the virus to evade select types of antibodies. However, B.1.617, the predominant variant in India identified in October 2020, is responsible for the spread of deadly cases in the western state of Maharashtra and is vastly spreading in the UK, USA, and other locations. Another variant found to be spreading in Bengal, B.1.618, also known as “triple mutant,” is suspected to have evolved from B.1.617 and has the V382L mutation in addition to E484Q and L452R.9,37–39
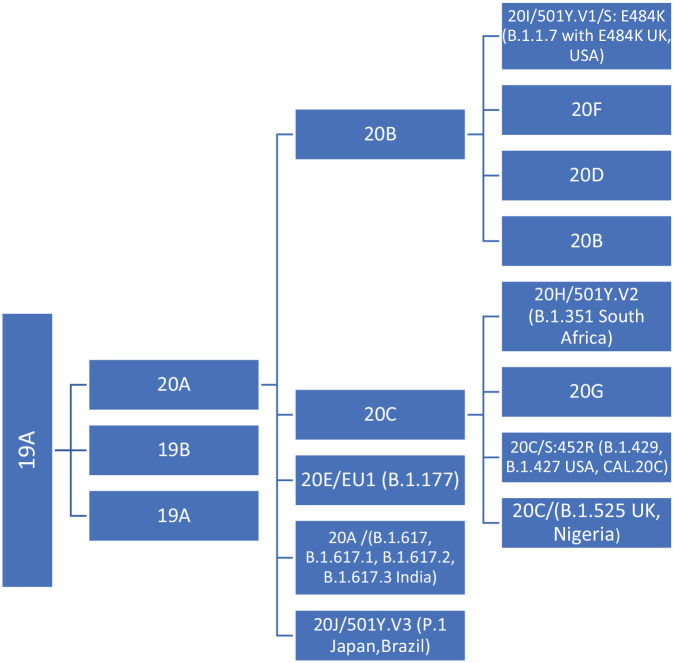
The emergence of variants, particularly from the lineage of B.1.1.7, has raised global scientific concern.9,40 These clade and lineage nomenclatures in Table 1 aid in the genomic epidemiology of SARS-CoV-2 and tracking of the variants as shown in Figure 1. Therefore, multiple sources and the Global Initiative on Sharing Avian Influenza Data (GISAID), an open-access genomics consortium, were used to extract information to further identify genome sequences of SARS-CoV-2. 41 The marker mutations of six phylogenetic groupings comprise the nomenclature system for major clades: S, L, V, G, GH, and GR. 41 Initially, the human coronaviruses (hCoVs) split early from L (originating in Wuhan, China) to S; L into V and G; and later G into GH and GR. 41 The clades identified in Table 1 are further detailed by the tool referred to as the Phylogenetic Assignment of Name Global Outbreak LINeages (PANGOLIN), providing a comprehensive understanding of the COVID-19 pandemic. 41 Furthermore, clades are defined by the year they emerged and are assigned a new alphabetical letter based on discovery: 19A (first appearing in 2019), 19B (appearing after 19A), 20A (new emergence at the beginning of 2020), 20B, and 20C.41–43
Table 1.
Clade and lineage nomenclature to understand genomic epidemiology of active coronaviruses.
| Clades/Nomenclature/Phylogenetic Groupings*/** | Initial clade identified** | PANGOLIN lineage** | Variant mutation adaptation*** | Notes* |
|---|---|---|---|---|
| L | 19A | B | Wuhan, China December 2019 Disappearing worldwide |
|
| S | 19B | A | 1st mutation of L strain (beginning of 2020) Seen in Spain and restricted areas in the USA |
|
| V | 19A | B | Appearance mid-January 2020 Disappearing worldwide |
|
| G | 20A | B.1.525 B.1.160 B.1.617.1 B.1.617.2 B.1.258 B.1.221 |
N439K | Appearance mid-January 2020 Most widespread to date Increasing in prevalence |
| GR | 20B 20B 20B 20D 20F 20I/501Y.V1 |
B.1.1 B.1.1.277 B.1.1.302 B.1.1.1 D.2 B.1.1.7 |
N501Y D614G |
G mutated to GR at the end of February 2020. Most prevalent in Europe, Italy, and South America Increasing in prevalence |
| GH | 20C 20C 20C 20C 20G 20H/501Y.V2 20J/501Y.V3 |
B.1.427 B.1.429 B.1.526 B.1.367 B.1.2 B.1.351 P.1 |
N501Y D614G |
G mutated to GH at the end of February 2020. Most prevalent in North America, France, and Germany Increasing in prevalence D6146 mutation spread first around the world***** N501Y evolved into several lineages - P.1 (Brazil), B.1.351 (South Africa), and B.1.1.7 (UK)***** |
| GV | 20E (EU1) | B.1.177 | ||
| Others****/***** | Y453F | Mink**** Cluster 5**** |
||
| E484K | Evolved into many lineages that can avoid several antibodies***** | |||
| K417 | The virus binds more tightly to cells and includes several lineages like P.1 and B.1.351***** | |||
| L452R | The variant of concern particularly in California***** | |||
| Q677 | Found in USA lineages, but has not yet proven to be more infectious***** |
Figure 1.
Chart showing the track of SARS-CoV-2 variants. Data recreated from Nextstrain. 43
Note. 19A and 19B emerged in Wuhan and led the outbreak early in the course of disease transmissibility. 20A originated from 19A and influenced the European outbreak, which has since spread globally. 20B, 20C, 20D, and 20E are all derived from 20A. 20B and 20C are globally distributed, whereas 20D has been seen mainly in South Africa, South America, and southern Europe. Furthermore, 20E is localized in Europe. 20F comes from 20B and is located primarily in Australia. 20G arises from 20C and is detected primarily in the USA. 20H and 20I include two variants of concern (VOCs) emerging from N501Y; 501Y.V2 in South Africa and 501Y.V1 in the UK while 20J has the VOC 501Y.V3 or P1 which emerged from Brazil and Japan. The Indian VOC B.1.617 originated from 20A.
Discussion
SARS-CoV-2, like many coronaviruses, is surrounded by a membrane.1,2 To gain access to the host, the virus uses glycoproteins to fuse its membrane to that of the cell where replication takes place. The spike protein is a glycoprotein composed of a chain of 1273 amino acids. 19 Three spike molecules combine to form a functional unit known as a trimer. 19 In the case of SARS-CoV-2, there are approximately 26 trimers on a virus, one of which binds to the ACE2 protein initiating the release of the viral genome through fusion.17–19
Viruses are constantly mutating, and this results in the upsurge of new variants. With the ongoing spread comes the expansion of SARS-CoV-2 as it adapts and acquires selective advantages. Currently, multiple variants are circulating globally. From the lineage nomenclature described in Table 1, a newfound lineage B.1.1.7 has resulted in numerous genetic mutations. 9 This strain was first reported in the UK, accounting for the majority of cases in England, and in comparison to the other lineages spreads quickly.7,9,44,45 In addition to this lineage, several other variants have been noted and their implications could have potential consequences for efficacious spread, disease severity, evasion of detection by specific diagnostic tests, changes in therapeutic agents, and vaccine-induced immunity.9,14,46 It is essential that the scientific community closely track the following mutations:
N501Y rapidly spreads as part of the B.1.1.7 and 501.V2 clades with an affinity for ACE2 receptors in humans.9,47 This has been studied and shown to be indicative of natural selection, resulting in high transmissibility within a population.48,49 Furthermore, a newer variant referred to as B.1.1.248 lineage has several mutations in the S protein, including N501Y and E484K. 50 This isolate has similarities with the isolate from the UK and South Africa. The variant isolate of E484K belonging to B.1.1.248, reported in the first week of 2021 in Brazil, also known as 501.V3 or P.1, is not identical to the new isolate identified in Japan, even though it is believed to have been carried via passengers aboard a flight from Brazil to Tokyo, Japan. 51 As a result, safety protocols should be enforced.
D614G, like N501Y, is also suggestive of natural selection and has been seen to infect many geographic regions.48,51,52 It is proposed that this mutation resulted from viruses harboring 614G leading to an outbreak of the D614G mutation, 53 where the (D) stands for aspartic acid at residue 614 of the spike viral protein that converts to glycine (G). 54 When a phylodynamic analysis was conducted of 25,000 sequences in the UK, it was found that those with 614G spread faster and contributed to more phylogenetic clusters than those with 614D. 54 The increased transmission rate of 614G viruses was confirmed in mouse models. Moreover, spike D614G (G614 virus) from hamsters infected with SARS-CoV-2 revealed higher infectious titers in the upper versus lower respiratory tract, supporting the evidence of increased transmission within humans. 55 In design, the current vaccines available within the USA are based on the original D614 sequence. 55 Sera from hamsters with D614G showed higher neutralization titers, which indicates that the efficacy of vaccines in clinical trials to protect against COVID-19 will not be compromised. 55
N439K has a single amino acid change and has decreased sensitivity to neutralizing antibodies. 56 According to various studies, B.1.617 is the prevalent variant devastating India, which carries L452R spike mutation, and another, referred to as a “double mutant” 8 Y453F has an RBD which has seen an increased ACE2 binding. 57 This permits an immune escape from monoclonal antibodies, leading to an escape from the REGN10933. 58 Another strain has emerged in South Africa with B.1.3519,59 lineage (Figure 1), also known as 20H / 501Y.V2, but has not been shown to have as many mutations nor contain the deletion 69/70 as seen with B.1.1.7.9,49 Emerging data suggest that the UK VOC B.1.1.7 may be associated with an increased risk of death.9,31 A diverse set of lineages with two main clusters from the 20G and the 20C lineage was identified in California.9,35 The larger cluster consisted of a novel variant defined by five mutations descended from cluster 20C and is designated as CAL.20C (20C/S:452R; /B.1.429) (Figure 1). 35 As of 22 January 2021, CAL.20C has been reported in 26 states in the USA and other countries.9,35 Furthermore, a new variant strain known as B.1.526 is reported to be spreading at an alarming rate in New York City since November 2020. 9 Spike mutation E484K is present in about half of this lineage, with a smaller fraction having S477N instead of E484K.9,32,60,61
Pathogenicity study showed that the cell surface Toll-like receptors (TLRs), especially TLR4, are probably involved in the recognition of molecular patterns from SARS-CoV-2 to induce inflammatory responses. Also, the S1 subunit of SARS-CoV-2 was shown to possess greater mutability potential when compared with the equivalent peptides found in MERS-CoV and SARS-CoV, which may explain its transmissibility in humans and across species with ease.46,62 Thus, the S protein is a major structural protein of SARS-CoV-221,22,62 and the variants are a result of its modifications. This protein sequence is the target of the current vaccines produced by Pfizer/BioNTech and Moderna/National Institute of Allergy and Infectious Diseases (NIAID).63–65 The scientific and medical communities are confident that the vaccine will be effective against the new variants.64,65 However, it is uncertain whether the strength of the vaccine could potentially be reduced given that many mutations would be needed to completely escape the antibodies.64,65 Currently, many trials and studies are being conducted to test whether the efficacy of the vaccines is uncompromised due to the variants, and should they be compromised, the vaccines would have to be redesigned.45,66,67 The efficacy of ChAdOx1 nCoV-19 (AstraZeneca vaccine) against the B.1.1.7 variant of SARS-CoV-2 has been reported to be equivalent to the efficacy of the vaccine against other lineages. 68 Similarly, the mRNA-based SARS-CoV-2 vaccines from Moderna (mRNA-1273) and Pfizer-BioNTech (BNT162b2) elicit antibody responses against the RBD, which is the major target of neutralizing antibodies, in a manner that resembled natural infection. 65 However, the study further showed that variants that carry K417N/T, E484K, and N501Y mutations, such as the UK (B1.1.7/501Y.V1), South African (501Y.V2), and Brazil (B1.1.28/501.V3) variants, can reduce the neutralization potency of vaccine plasma. 65
Therefore, close surveillance of the S protein must take place as the virus mutates to make certain the effectiveness of the authorized vaccines.64,65 Also, vaccines against COVID-19 need to be updated regularly and the immunity monitored to compensate for viral evolution that may arise from the mutations. 65 In any instance, preventative measures such as social distancing, face mask use, and regular handwashing with soaps and water, or the use of hand sanitizers must be upheld to slow the spread and help countries reduce hospitalizations and deaths. Therefore, additional research and investigations are required to fully comprehend the impact of each specific SARS-CoV-2 variant. Because of its complexity, this requires time, concerted effort, and collaboration by all concerned to effectively tackle and eliminate the disease caused by the virus.9,14
Conclusion
The infectious SARS-CoV-2 (COVID-19) virus is responsible for millions of casualties worldwide. It is the cause of a global pandemic, and its mutations form genetic variants that are yet to be fully understood. Furthermore, it is crucial to understand the new variants, to assess their impact on the rate of transmission, resilience, and mortality. The recent upward trend of COVID-19 cases can be attributed to these variants and their ability to transmit adeptly. Their pathological makeup makes them more resilient than the original strain and able to bind to receptor proteins more efficiently. However, further research is required to fully understand these variants and increase the accuracy of treatments to save lives. The public should consider vaccination along with the prescribed preventative measures, such as wearing a face mask, washing hands frequently, and practicing social distancing for the best chance of avoiding contracting the virus.
Footnotes
Author contributions: All authors substantially contributed to the conception, drafting, and final approval of the manuscript.
Conflict of interest statement: The authors declare that there is no conflict of interest.
Funding: The authors received no financial support for the research, authorship, and/or publication of this article.
Ethical statement: This study did not require an ethical board approval because it did not contain human or animal trials.
ORCID iD: Adekunle Sanyaolu  https://orcid.org/0000-0002-6265-665X
https://orcid.org/0000-0002-6265-665X
Contributor Information
Adekunle Sanyaolu, Federal Ministry of Health, Department of Public Health, New Federal Secretariat Complex, Phase III, Ahmadu Bello Way, Central Business District, FCT, Abuja, Nigeria.
Chuku Okorie, Union County College, Plainfield Campus, NJ, USA.
Aleksandra Marinkovic, Saint James School of Medicine, Anguilla, BWI.
Nafees Haider, All Saints University School of Medicine, Dominica.
Abu Fahad Abbasi, Loyola University Medical Center, Maywood, IL, USA.
Urooj Jaferi, All Saints University School of Medicine, Dominica.
Stephanie Prakash, Saint James School of Medicine, Anguilla, BWI.
Vyshnavy Balendra, Saint James School of Medicine, Anguilla, BWI.
References
- 1. Gabutti G, d’Anchera E, Sandri F, et al. Coronavirus: update related to the current outbreak of COVID-19. Infect Dis Ther 2020; 9: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yuki K, Fujiogi M, Koutsogiannaki S. COVID-19 pathophysiology: a review. Clin Immunol 2020; 215: 108427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. WHO. Coronavirus disease 2019 (COVID-19): dashboard. World Health Organization. https://covid19.who.int/ (2021, accessed 11 May 2021). [PubMed]
- 4. Srivastava S, Banu S, Singh P, et al. SARS-CoV-2 genomics: an Indian perspective on sequencing viral variants. J Biosci 2021; 46: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Winstone H, Lista MJ, Reid AC, et al. The polybasic cleavage site in SARS-CoV-2 spike modulates viral sensitivity to type I interferon and IFITM2. J Virol 2021; 95: e02422-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Biswas SK, Mudi SR. Genetic variation in SARS-CoV-2 may explain variable severity of COVID-19. Med Hypotheses 2020; 143: 109877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Koyama T, Platt D, Parida L. Variant analysis of SARS-CoV-2 genomes. Bull World Health Organ 2020; 98: 495–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. WHO. COVID-19 weekly epidemiological update: proposed working definitions of SARS-CoV-2 variants of interest and variants of concern. WHO Health Organization. https://www.who.int/publications/m/item/covid-19-weekly-epidemiological-update (2021, accessed 11 May 2021). [Google Scholar]
- 9. Corum J, Zimmer C. Coronavirus variants and mutations. The New York Times, 26 April. https://www.nytimes.com/interactive/2021/health/coronavirus-variant-tracker.html (2021, accessed 10 May 2021).
- 10. Davies NG, Barnard RC, Jarvis CI, et al. Estimated transmissibility and severity of novel SARS-CoV-2 variant of concern 202012/01 in England. medRxiv 2021. [Google Scholar]
- 11. Volz E, Mishra S, Chand M, et al. Transmission of SARS-CoV-2 lineage B.1.1.7 in England: insights from linking epidemiological and genetic data. medRxiv 2021. [Google Scholar]
- 12. Tegally H, Wilkinson E, Giovanetti M, et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv 2020. [Google Scholar]
- 13. WHO. SARS-CoV-2 mink-associated variant strain – Denmark. World Health Organization. https://www.who.int/csr/don/03-december-2020-mink-associated-sars-cov2-denmark/en/ (2020, accessed 10 January 2021).
- 14. Abdool Karim SS, de Oliveira T. New SARS-CoV-2 variants - clinical, public, health, and vaccine implications. NEJM. https://www.nejm.org/doi/pdf/10.1056/NEJMc2100362?articleTools=true (2021, accessed 10 May 2021). [DOI] [PMC free article] [PubMed]
- 15. Challen R, Read JM, Dyson L, et al. Risk of mortality in patients infected with SARS-CoV-2 variant of concern 202012/1: matched cohort study. BMJ 2021; 372: n579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Secolin R, de Araujo TK, Gonsales MC, et al. Genetic variability in COVID-19-related genes in the Brazilian population. Hum Genome Var 2021; 8: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020; 181: 271–280.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sun K, Gu L, Ma L, et al. Atlas of ACE2 gene expression reveals novel insights into transmission of SARS-CoV-2. Heliyon 2021; 7: e05850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Duan L, Zheng Q, Zhang H, et al. The SARS-CoV-2 spike glycoprotein biosynthesis, structure, function, and antigenicity: Implications for the design of spike-based vaccine immunogens. Front Immunol 2020; 11: 576622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hussain M, Jabeen N, Raza F, et al. Structural variations in human ACE2 may influence its binding with SARS-CoV-2 spike protein. J Med Virol 2020; 92: 1580–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li Q, Wu J, Nie J, et al. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell 2020; 182: 1284–1294.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Choudhary S, Sreenivasulu K, Mitra P, et al. Role of genetic variants and gene expression in the susceptibility and severity of COVID-19. Ann Lab Med 2021; 41: 129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nassir AA, Musanabaganwa C, Mwikarago I. Mutation landscape of SARS-CoV-2 in Africa. bioRxiv 2020. [Google Scholar]
- 24. Tomaszewski T, DeVries RS, Dong M, et al. New pathways of mutational change in SARS-CoV-2 proteomes involve regions of intrinsic disorder important for virus replication and release. Evol Bioinform Online. 2020; 16: 1176934320965149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhou W, Xu C, Wang P, et al. N439K variant in spike protein may alter the infection efficiency and antigenicity of SARS-CoV-2 based on molecular dynamics simulation. bioRxiv. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Thomson EC, Rosen LE, Shepherd JG, et al. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediate mediated. Cell 2021; 184: 1171–1187.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bayarri-Olmos R, Rosbjerg A, Johnsen LB, et al. The SARS-CoV-2 Y453F mink variant displays a pronounced increase in ACE-2 affinity but does not challenge antibody neutralization. J Biol Chem 2021; 296: 100536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rambaut A, Loman N, Pybus O, et al. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. Virological 2021. https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563 (2021).
- 29. Leung K, Shum MHH, Leung GM, et al. Early empirical assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. medRxiv 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Horby P, Huntley C, Davies N, et al. NERVTAG note on B.1.1.7 severity. SAGE. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/955239/NERVTAG_paper_on_variant_of_concern__VOC__B.1.1.7.pdf (2021, accessed 28 January 2021). [Google Scholar]
- 31. Iacobucci G. COVID-19: new UK variant may be linked to increased death rate, early data indicate. BMJ 2021; 372: n230. [DOI] [PubMed] [Google Scholar]
- 32. Giandhari J, Pillay S, Wilkinson E, et al. Early transmission of SARS-CoV-2 in South Africa: an epidemiological and phylogenetic report. medRxiv 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. West AP, Jr, Barnes CO, Yang Z, et al. SARS-CoV-2 lineage B.1.526 emerging in the New York region detected by software utility created to query the spike mutational landscape. bioRXiv 2021. [Google Scholar]
- 34. West AP, Jr, Wertheim JO, Wang JC, et al. Detection and characterization of the SARS-CoV-2 lineage B. 1.526 in New York. bioRxiv 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang W, Davis BD, Chen SS, et al. Emergence of a novel SARS-CoV-2 variant in southern California. JAMA 2021; 325: 1324–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Deng X, Garcia-Knight MA, Khalid MM, et al. Transmission, infectivity, and antibody neutralization of an emerging SARS-CoV-2 variant in California carrying a L452R spike protein mutation. medRxiv 2021. [Google Scholar]
- 37. Cherian S, Potdar V, Jadhav S, et al. Convergent evolution of SARS-CoV-2 spike mutations, L452R, E484Q, and P681R, in the second wave of COVID-19 in Maharashtra, India. bioRxiv 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ferreira I, Datir R, Papa G, et al. SARS-CoV-2 B.1.617 emergence and sensitivity to vaccine-elicited antibodies. bioRxiv 2021. [Google Scholar]
- 39. Vaidyanathan G. Coronavirus variants are spreading in India — what scientists know so far: variants including B.1.617 have been linked to India’s surge in infections. Researchers are hurrying to determine how much of a threat they pose. Nature 2021; 593: 321–322. [DOI] [PubMed] [Google Scholar]
- 40. Lauring AS, Hodcroft EB. Genetic variants of SARS-CoV-2—what do they mean? JAM. 2021; 325: 529–531. [DOI] [PubMed] [Google Scholar]
- 41. SARS-CoV-2: Clade and lineage nomenclature, July 4, 2020. GISAID. https://www.gisaid.org/references/statements-clarifications/clade-and-lineage-nomenclature-aids-in-genomic-epidemiology-of-active-hcov-19-viruses/ (2021, accessed 12 January 2021).
- 42. Università di Bologna. The six strains of SARS-CoV-2. Science Daily. https://www.sciencedaily.com/releases/2020/08/200803105246.htm (2020, accessed 12 January 2021).
- 43. Genomic epidemiology of novel coronavirus - global subsampling. Nextstrain. https://nextstrain.org/sars-cov-2/ (2021, accessed 12 January 2021).
- 44. Lee R. B.1.1.7: what we know about the novel SARS-CoV-2 variant. The American Society for Microbiology. https://asm.org/Articles/2021/January/B-1-1-7-What-We-Know-About-the-Novel-SARS-CoV-2-Va (2021, accessed 28 January 2021).
- 45. Muik A, Wallisch AK, Sänger B, et al. Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine-elicited human sera. bioRxiv 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Patra R, Das NC, Mukherjee S. Targeting human TLRs to combat COVID-19: a solution? J Med Virol 2021; 93: 615–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tu H, Avenarius MR, Kubatko L, et al. Distinct patterns of emergence of SARS-CoV-2 spike variants including N501Y in clinical samples in Columbus Ohio. bioRxiv 2021. [Google Scholar]
- 48. van Dorp L, Tan CCS, Lam SD, et al. Recurrent mutations in SARS-CoV-2 genomes isolated from mink point to rapid host-adaptation. bioRxiv 2020. [Google Scholar]
- 49. Galloway SE, Paul P, MacCannell DR, et al. Emergence of SARS-CoV-2 B.1.1.7 lineage - United States, December 29, 2020 - January 12, 2021. MMWR. https://www.cdc.gov/mmwr/volumes/70/wr/pdfs/mm7003e2-H.pdf (2021, accessed January 22, 2021). [DOI] [PMC free article] [PubMed]
- 50. NIID. Brief report: new variant strain of SARS-CoV-2 identified in travelers from Brazil. National Institute of Infectious Diseases, Japan. https://www.niid.go.jp/niid/en/2019-ncov-e/10108-covid19-33-en.html (2021, accessed 28 January 2021).
- 51. Chowdhury J, Scarr S, Wardell J. How the novel coronavirus has evolved. Reuters. https://graphics.reuters.com/HEALTH-CORONAVIRUS/EVOLUTION/yxmpjqkdzvr/ (2020, accessed 19 January 2021).
- 52. Tegally H, Wilkinson E, Lessells RR, et al. Major new lineages of SARS-CoV-2 emerge and spread in South Africa during lockdown. medRxiv 2020. [Google Scholar]
- 53. Lorenzo-Redondo R, Nam HH, Roberts SC, et al. A clade of SARS-CoV-2 viruses associated with lower viral loads in patient upper airways. EBioMedicine 2020; 62: 103112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang L, Jackson CB, Mou H, et al. The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. bioRxiv 2020. [Google Scholar]
- 55. Plante JA, Liu Y, Liu J, et al. Spike mutation D614G alters SARS-CoV-2 fitness and neutralization susceptibility. bioRxiv 2020. [Google Scholar]
- 56. Weisblum Y, Schmidt F, Zhang F, et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. bioRxiv 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hayashi T, Yaegashi N, Konishi I. Effect of RBD mutation (Y453F) in spike glycoprotein of SARS-CoV-2 on neutralizing antibody affinity. bioRxiv 2020. [PubMed] [Google Scholar]
- 58. Starr TN, Greaney AJ, Addetia A, et al. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. bioRxiv 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cocks T, Winning A. Previous coronavirus infection may offer less protection from new variant. Reuters. https://www.reuters.com/article/uk-health-coronavirus-safrica-variant/previous-coronavirus-infection-may-offer-less-protection-from-new-variant-idUSKBN29O0LX (2021, accessed 22 January 2021).
- 60. Pango lineages. Lineage B.1.526. Pango. https://cov-lineages.org/lineages/lineage_B.1.526.html (2021, accessed 25 February 2021).
- 61. Annavajhala MK, Mohri H, Zucker JE, et al. A novel SARS-CoV-2 variant of concern, B.1.526, identified in New York. medRxiv 2021. [Google Scholar]
- 62. Choudhury A, Mukherjee S. In silico studies on the comparative characterization of the interactions of SARS-CoV-2 spike glycoprotein with ACE-2 receptor homologs and human TLRs. J. Med. Virol 2020; 92: 2105–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. FDA. COVID-19 vaccines. U.S. Food & Drug Administration. https://www.fda.gov/emergency-preparedness-and-response/coronavirus-disease-2019-covid-19/covid-19-vaccines (2021, accessed 11 May 2021). [PubMed]
- 64. FDA. Coronavirus (COVID-19) update: FDA issues policies to guide medical product developers addressing virus variants. U.S. Food and Drug Administration. https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-issues-policies-guide-medical-product-developers-addressing-virus (2021, accessed 11 May 2021). [Google Scholar]
- 65. Wang Z, Schmidt F, Weisblum Y, et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. bioRxiv 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Dai L, Gao GF. Viral targets for vaccines against COVID-19. Nat Rev Immunol 2021; 21: 73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Johnson CY, McGinley L, Achenbach J. New coronavirus variants accelerate race to make sure vaccines keep up. The Washington Post. https://www.washingtonpost.com/health/2021/01/25/covid-vaccine-virus-variant/ (2021, accessed 28 January 2021).
- 68. Emary KR, Golubchik T, Aley PK, et al. Efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine against SARS-CoV-2 VOC 202012/01 (B.1.1. 7). Lancet 2021; 397: 1351–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]