Abstract
Epigenetics, especially histone marks, functions beyond the DNA sequences to regulate gene expression. Depletion of NSD1, which catalyzes H3K36me2, leads to both up- and down-regulation of gene expression, indicating NSD1 is associated with both active and repressed gene expression. It's known that NSD1 regulates the deposition and expansion of H3K27me3, a repressive mark for gene expression, to keep active gene transcription. However, how NSD1 functions to repress gene expression is largely unknown. Here, we find that, when NSD1 is knocked out in mouse embryonic stem cells (mESCs), H3K27ac increases correlatively with the decrease of H3K36me2 at active enhancers, which is associated with mesoderm differentiation genes, leading to elevated gene expression. Mechanistically, NSD1 recruits HDAC1, the deacetylase of H3K27ac, to chromatin. Moreover, HDAC1 knockout (KO) recapitulates the increase of H3K27ac at active enhancers as the NSD1 depletion. Together, we propose that NSD1 deposits H3K36me2 and recruits HDAC1 at active enhancers to serve as a ‘safeguard’, preventing further activation of active enhancer-associated genes.
INTRODUCTION
In eukaryotic cells, DNA is assembled into chromatin in a basic repeating unit, the nucleosome, which contains one histone H3–H4 tetramer and two histone H2A–H2B dimers wrapped around 147 base pairs (bp) of DNA (1,2). Histone post-translational modifications, mainly including methylation, ubiquitylation, phosphorylation and acetylation, have been proposed to alter the chromatin structure and to participate in the regulation of gene expression and cellular processes (3–5).
Histone H3K36 can be post-translationally modified via methylation to form mono-, di- and tri-methylation (H3K36me1/H3K36me2/H3K36me3) (6–8). H3K36 methylation is usually associated with transcriptional activation. In gene body regions, there is a progressive shift from H3K36me1 to H3K36me3 through the promoter to the 3′-end in active genes (9). Besides, H3K36me2 is also enriched in intergenic regions. The initial understanding of the roles of H3K36 methylation is largely based on the studies of Set2, which is the only methyltransferase responsible for all forms of H3K36 methylation in yeast (10,11). In human cells, ASH1L, NSD1-3, SMYD2, SETMAR and SETD3, are reported to catalyze H3K36me1/me2, whereas SETD2 is the only characterized enzyme specific for H3K36me3 so far (9).
In yeast, RNA polymerase II (RNA Pol II) decorates histones with H3K36me3, which serves as a mark for histone deacetylation to prevent aberrant transcriptional initiation (12,13). In human cells, this prevention is mediated not only by deacetylation but also by an interplay between H3K9 and H3K4 methylation (14,15). Moreover, Dnmt3a and Dnmt3b bind to H3K36me3 and methylate DNA, demonstrating a role for H3K36me3 in the repression of spurious transcriptional initiation via DNA methylation (16–18). Several models are proposed for the function of H3K36me3 in gene regulation besides transcription initiation, such as alternative pre-mRNA splicing (19), cryptic transcription (10), RNA m6A modification (20), 3D chromosome organization (21), X-chromosome dosage compensation (22), and DNA damage response (23).
On the other hand, H3K36me2 is enriched at both intergenic regions and gene bodies (6). H3K36me2, which is less studied, is proposed to contribute to gene regulation through the interplay with other histone marks and DNA methylation (24). The H3K36 methylation may cross-talk with other histone marks, such as H3K27 methylation, to contribute to the gene regulation (25,26). Ezh2, the methyltransferase responsible for H3K27 methylation, contains a specific binding pocket, that distinguishes methylated and unmodified H3K36 residues, shifting the enzymatic activities to methylate the unmodified nucleosomes (27,28). Additionally, NSD1-mediated H3K36me2 represses the expansion of H3K27me3, regulating the demarcation of H3K27me3 across the genome of mouse embryonic stem cells (mESCs) (26). This interplay connects H3K36me2 with active gene expression. NSD1 mutations or depletions have been identified in neuroblastoma, carcinomas of the gastrointestinal tract, head and neck tumors, gliomas, pediatric acute myelogenous leukemia and Sotos syndrome (29–32). Moreover, a global increase of H3K36me2 abundance is sufficient to activate abnormal transcription, which could then contribute to cancer development (33). However, the molecular mechanism by which H3K36me2 regulates gene expression is poorly understood.
In this study, we investigated how gene expression is modulated by NSD1-dependent H3K36me2 in mESCs. We found that loss of NSD1 led to both up- and down-regulation of gene expression. While the increased H3K27me3 was proposed to be responsible for the down-regulation of gene expression, our data revealed that, correlated with the decrease of H3K36me2, H3K27ac was increased at active enhancers upon NSD1 depletion, contributing to elevated expressions of genes associated with mesoderm differentiation. Moreover, NSD1 recruited HDAC1 to reduce H3K27ac at active enhancers. The HDAC1 knockout thus recapitulated the increase of H3K27ac at active enhancers in NSD1 KO mESCs. Together, our observations described that NSD1-dependent H3K36me2 marked active enhancers, serving as a ‘safeguard’ to prevent further activation of enhancers by recruiting HDAC1. These data provide new insights into the functions of H3K36me2 in the regulation of gene expression.
MATERIALS AND METHODS
Cell culture and cell lines
All J1 mESCs were cultured in DMEM supplemented with 15% fetal bovine serum (FBS), 1% antibiotic solution (penicillin/streptomycin), 1% glutamax, 1% MEM nonessential amino acids, 1% sodium pyruvate, 0.1 mM β-mercaptoethanol and 1000 U/ml recombinant leukemia inhibitory factor (LIF). Cells were cultured on 0.1% gelatin coated plates with fresh medium. T/C28a2 and HEK293T cells were cultured in DMEM supplemented with 1% Glutamax, 10% FBS and 1% antibiotic solution (penicillin/streptomycin). All cells were grown at 37°C and 5% CO2.
NSD1 knockout and HDAC1 knockout cell lines were constructed from two independent sgRNAs, which were cloned into pSpCas9(BB)-2A-Puro (34) from Feng Zhang (Addgene plasmid #48139). Individual clones were picked under the microscope for the following genotyping.
Cell proliferation assay and alkaline phosphatase staining
500 cells were seeded into each well of a 96-well plate, and the cell viability was measured after 0, 24, 48, 72 and 96 h, respectively by CellTiter-Blue cell viability assay kit (Promega, Cat. #G8081).
106 mESCs were plated in each well of a 24-well plate. Cells were fixed with 1% paraformaldehyde for 5 min at room temperature and then were stained by INT/BCIP (Brown) kit (Sangon Biotech, Cat.# C500033-0005) for 15 min at 37°C. T/C28a2 cells were used as the negative controls.
MNase digestion
4 × 106 cells were harvested and incubated in nuclear extraction buffer A (85 mM KCl, 10 mM Tris pH7.5, 0.2 mM spermidine, 0.2 mM EDTA, 160 mM sucrose, and 250 μM PMSF) for 5 min on ice. Samples were then lysed in nuclear extraction buffer B (buffer A supplemented with 0.1% NP-40) for 5 min on ice. Nuclei were washed with digestion buffer (50 mM Tris pH7.5, 20 mM KCl, 0.32 M sucrose, 4 mM MgCl2 and 3 mM CaCl2) and digested with MNase (2000 units/ml) at 37°C for 0, 1, 2.5, 5, 10, 20 min, respectively. The reactions were stopped with an equal volume of 2× stop buffer (0.2% SDS and 10 mM EDTA) at 37°C for 10 min. Samples were treated with 20 μg/ml RNase A at 37°C for 30 min, then 100 μg/ml proteinase K at 55°C for 1 h. DNA was purified with phenol/chloroform/isoamyl alcohol and precipitated with 1/2 volume of 7.5 M ammonium acetate and 2 volume of 100% ethanol. DNA was resuspended with 20 μl TE buffer and then analyzed by 1.5% agarose gel.
Embryoid body formation and RT-PCR
mESCs in good condition were diluted to 10 000 cells/ml in medium without LIF. Then cells were seeded into 30 μl/drop (300 cells/drop) onto the lid of the 15 cm-Petri dish. Around 120 drops were seeded per lid. Cells were washed off the lid, resuspended into 10 ml medium without LIF, then poured into the low-attachment 10 cm dish. Medium were changed every 2 days. Samples were collected at 0, 3, 5 and 7 days. RNA was isolated by UNIQ-10 column total RNA purification kit (Sangon Biotech, Cat.# B511361-0100) and 500 ng RNA was used for reverse transcription by EasyScript® One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen, Cat.# AE311-02). Two-step qPCR (95°C 30 s; 95°C 5 s and 60°C 15 s for 40 cycles) was performed in 10 μl reaction with 0.1 μl 10 mM primers and 5 μl Hieff qPCR SYBR Green Master Mix (YEASEN, Cat. #11201ES08) using Quantagene q225 qPCRsystem (Kubo Technology, Beijing). β-actin was used as a control to normalize the expression of other genes.
Antibodies
Rabbit polyclonal anti-Histone H3 (Abcam, Cat. #ab1791); Rabbit polyclonal anti-Histone H3K4me3 (Abcam, Cat. #ab8580); Rabbit monoclonal anti-Histone H3K27me3 (Cell Signaling Technology, Cat. #9733); Rabbit polyclonal anti-Histone H3K27ac (Abcam, Cat. #ab4729); Rabbit monoclonal anti-H3K36me2 (Cell Signaling Technology, Cat. #2901); Rabbit polyclonal anti-H3K36me3 (Active motif, Cat. #61101); Rabbit polyclonal anti-Histone H3K4me1 (Abcam, Cat. #ab8895); Rabbit monoclonal anti-HDAC1 (Cell Signaling Technology, Cat. #34589); Mouse monoclonal anti-β-Tubulin (Cell Signaling Technology, Cat. #86298); Rabbit polyclonal anti-Brd4 (Abcam, Cat. #ab75898); Mouse monoclonal anti-FLAG (GenScript, Cat. #A00187); Rabbit polyclonal anti-NSD1(Abbexa, Cat. #135901).
ChIP-qPCR
mESCs were fixed with 1% paraformaldehyde in 10 ml medium for 10 mins at room temperature and quenched with 0.5 ml of 2.5 M glycine for 5 min. Cells were then washed twice by cold TBS. The pellets were resuspended in cell lysis buffer (10 mM Tris pH7.5, 10 mM NaCl and 0.5% NP-40) and incubated on ice for 10 mins. Lysates were washed by 1/2 volume (cell lysis buffer) of MNase digestion buffer (20 mM Tris pH 7.5, 15 mM NaCl, 60 mM KCl and 1 mM CaCl2). After resuspended in 500 μl MNase digestion buffer supplemented with protease inhibitor cocktail, the mixtures were incubated at 37°C, 1000 rpm for 20 min in the presence of 1000 units of MNase (NEB, Cat. #M0247S). Digestion was stopped by 500 μl 2× Stop/ChIP buffer I (100 mM Tris pH8.1, 20 mM EDTA, 200 mM NaCl, 2% Triton X-100 and 0.2% sodium deoxycholate) and lysates were sonicated for 5 min (5 cycles of 30 s-on, 30 s-off) in the Diagenode Bioruptor. Samples were then centrifuged at 15 000 rpm and 4°C for 10 min before the supernatants were collected. The concentration of chromatin was detected by Qubit assay (Vazyme, Cat. #EQ121-02-AA). 1% input was saved and 950 μl of supernatant was incubated with 2 μg of Rabbit monoclonal anti-H3K36me2 (Cell Signaling Technology, Cat. #2901), Rabbit monoclonal anti-Histone H3K27me3 (Cell Signaling Technology, Cat. #9733) or Rabbit polyclonal anti-Histone H3K27ac (Abcam, Cat. #ab4729) on a rocker at 4°C overnight. 30 μl of 1× ChIP buffer pre-washed protein G beads (Sangon Biotech, Cat. #D110561) were added to samples and incubated at 4°C for 3 h on a rotator. Beads were washed alternately by 1× ChIP buffer, high salt buffer (ChIP buffer + 0.5 M NaCl), Tris/LiCl buffer (10 mM Tris pH8.0, 0.25 M LiCl, 0.5% NP-40, 0.5% sodium deoxycholate and 1 mM EDTA) and TE buffer (50 mM Tris pH8.0 and 10 mM EDTA) twice. Every washing step was performed on the rotator for 5 min. The washed beads were eluted twice by incubating at 65°C, 1200 rpm shaking for 15 min with 50 μl 1× elution buffer (10 mM Tris pH8.0, 10 mM EDTA, 150 mM NaCl, 5 mM DTT and 1% SDS). Two eluents were pooled together and reverse-crosslinked at 65°C overnight. The mixtures were incubated with 2 μl of DNase-free RNase A (10 mg/ml) at 37°C for 1 h and 10 μl of proteinase K (20 mg/ml) at 37°C for 2 h. DNA was purified by the Min-Elute PCR purification kit (Qiagen, Cat. #28006). Diluted ChIP products were used for qPCR to analyze the relative enrichments.
CUT&RUN
CUT&RUN was performed as previously described with small modifications (35). In brief, mESCs were harvested and washed with Wash buffer (20 mM HEPES pH7.5, 150 mM NaCl, 0.5 mM Spermidine and 1× Roche Complete Protease Inhibitor EDTA-Free tablet), 10 μl pre-washed Concanavalin A coated beads in binding buffer (20 mM HEPES pH 7.9, 10 mM KCl, 1 mM CaCl2 and 1 mM MnCl2) were added per sample. Samples were incubated for 10 min at room temperature. Beads-bound nuclei were incubated with 1:100 diluted primary antibody at 4°C overnight and washed with Dig-wash buffer (500 μl 5% Digitonin in 40 ml Wash buffer) before incubated with 1:100 diluted secondary antibody at room temperature for 30 min. After washing with Dig-wash buffer, 700 ng/ml pA-MNase was added and rotated at 4°C for 1 h. Beads were washed twice with Dig-wash buffer and put into wet ice at 0°C, mixed with 2 μl 100 mM CaCl2, and incubated at 0°C for 30 min. Digestion was stopped with 2× stop buffer (340 mM NaCl, 20 mM EDTA, 4 mM EGTA, 0.02% digitonin, 0.05 mg/ml glycogen and 2 pg/ml heterologous spike-in yeast DNA) and incubated at 37°C for 10 min to release fragmented DNA. DNA was purified by Min-Elute PCR purification kit (Qiagen, Cat. #28006) and then used for library construction using VAHTS Universal Pro DNA Library Prep Kit for Illumina (Vazyme, Cat. #ND608).
CUT&Tag
CUT&Tag was performed as previously described (36) using Rabbit monoclonal anti-HDAC1 (Cell Signaling Technology, Cat. #34589), Rabbit monoclonal anti-H3K36me2 (Cell Signaling Technology, Cat. #2901), and Rabbit polyclonal anti-Histone H3K27ac (Abcam, Cat.# ab4729) respectively.
RNA-seq
Total RNA from 106 cells was purified by RNeasy plus mini kit (Qiagen, Cat. #74134). DNA was digested by DNase I and rRNA was depleted by Ribo-Zero™ Magnetic Kit. After RNA fragmentation, cDNA was generated with random primers and followed by second-strand cDNA synthesis. Adapters were integrated into 3′ adenylated fragments for library construction. Amplified DNA was purified by AMPure XP Beads (Beckman, Cat.# A63880) and sequenced.
ATAC-seq
106 mESCs were collected, lysed by lysis buffer (10 mM Tris pH 7.4, 10 mM NaCl, 3 mM MgCl2 and 0.1% NP-40), and separated into 50 μl with 50 000 cells. The nuclei were resuspended in 50 μl transposition reaction mix (10 μl of 5× DMF reaction buffer: 50% DMF, 50 mM Tris pH 7.5, 20 ng/μl Tn5 transposome and 250 mM MgCl2) and incubated at 55°C for 10 min. DNA was purified by the Min-Elute PCR purification kit (Qiagen, Cat. #28006). Library amplification was carried out using transposed DNA, i5 primer, i7 primer and 2× HiFi polymerase mix. PCR reaction was incubated at 72°C for 5 min, and then 98°C for 30 s, 14 cycles of 98°C for 15 s and 60°C for 30 s, followed by 72°C for 5 min. Libraries were purified by AMPure XP beads (Beckman, Cat.# A63880) and sequenced.
NSD1 protein complex purification
FLAG-tagged NSD1 was overexpressed in mESCs using virus generated from MSCV-NSD1 expression vector, a kind gift from Dr. Juerg Schwaller. Cells were lysed by lysis buffer (50 mM HEPES pH7.4, 200 mM NaCl, 0.5% NP-40, 10% glycerol and 1 mM EDTA), homogenized 14 strokes by a B type pestle (glass Dounce homogenizer), and then incubated on ice for 10 min. 10% input was saved after centrifugation twice. The supernatant was incubated with pre-washed anti-FLAG beads (Sangon Biotech, Cat. #D111139) and rotated at 4°C overnight. Beads were washed five times with washing buffer (50 mM HEPES pH7.4, 100 mM NaCl, 0.01% NP-40, 10% glycerol and 1 mM EDTA) at 4°C for 5 min. SDS sample loading buffer was added to washed beads and boiled for 5 min to elute the bound proteins. Proteins were resolved on SDS-PAGE and subjected to analysis by mass spectrometry (37) or western blot using the indicated antibodies.
Separation of cell fractionation
3 × 106 cells were collected and resuspended with 200 μl Hypo buffer (20 mM HEPES pH8.0, 5 mM KCl, 1.5 mM MgCl2 and 0.1 mM DTT). 100 μl mix was saved as whole-cell extraction. The other 100 μl mix was incubated on ice for 20 min and centrifuged at 12 000 rpm and 4°C for 10 min. The nuclear pellets were washed twice by Hypo buffer and then boiled in 100 μl 2× SDS sample loading buffer.
Epigenomic data processing and visualization
Adaptors and low-quality reads were removed by Trim Galore (version 0.6.4) (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore) with the parameter ‘–paired’. Trimmed reads were then mapped to the mouse reference genome mm9 using Bowtie2 (38) (version 2.3.5.1). PCR duplicates were removed by GATK4 (version 4.1.4.0) (https://github.com/broadinstitute/gatk) with the parameter ‘–REMOVE_DUPLICATES = true’. MACS2 (39) (version 2.2.6) was used to call broad peaks for H3K27me3, H3K36me3 and H3K4me1, and narrow peaks for H3K27ac, H3K36me2, H3K4me3 and HDAC1.
For epigenomic signals visualization, BEDtools (40) (version 2.92.2) and bedGraphToBigWig (version 4) (https://www.encodeproject.org/software/bedgraphtobigwig/) were used to normalize mapped reads and calculate the coverage of features across the genome with the following parameters ‘genomecov –scaleFactor 10,000,00/(the number of reads mapped to Escherichia coli or Saccharomyces cerevisiae genome)’. Function computeMatrix and plotHeatmap in deepTools (41) (version 3.4.3) were used to draw heatmaps with reference-point mode. Normalized signals were visualized in Integrative Genomics Viewer (IGV) (42) (version 2.6.3). Peak distribution across the genome was generated by R package ChIPseeker (43) (version 1.22.1). For downstream differential peaks analysis, average reads density across the peak was used. Differential peaks analysis was performed by limma (44) (version 3.42.2) with normalized signals. HOMER (45) (version 4.11) was used with ‘findMotifsGenome.pl -mset vertebrates’ to find transcription factor motifs in differential peak regions. Gene Ontology (GO) analysis was performed by clusterProfiler (46) (version 3.14.3). The correlations between two repeats were summarized in Supplementary Table S1. The correlations between two KO cell lines after merging the two replicates were summarized in Supplementary Table S2. A 1 Kb sliding window across the whole genome was used to calculate the Pearson product moment correlation.
RNA-seq data processing and analysis
Trim Galore was used to remove sequencing reads with adaptors or low-quality bases with the parameter ‘–paired’. STAR (47) (version 2.5.4b) was used to map the filtered reads to mouse reference genome mm9. Expression matrixes were counted by featureCounts (48) (version 2.0.0) and normalized as reads per kilobase per million mapped reads (RPKM) by R package edgeR (49) (version 3.28.1). The Venn plots were plotted by R package VennDiagram (version 1.6.20) (https://github.com/cran/VennDiagram). edgeR was used to find genes with different expression levels, which were selected by the threshold log2(foldchange) > 2 and P value < 0.01. The volcano plot showing differential genes distribution was performed by ggplot2 (50) (version 3.3.2).
Primers
Primers used in this study were summarized in Supplementary Table S3.
RESULTS
Similar numbers of genes are up- and down-regulated in NSD1 KO mESCs
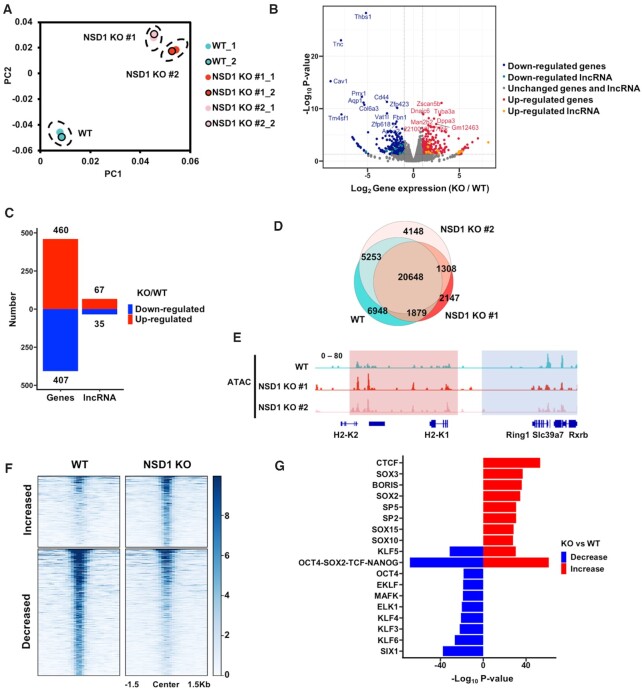
To uncover the functions of NSD1-catalyzed H3K36me2 in the regulation of gene expression, we utilized CRISPR/Cas9 system and knocked out NSD1 in mESCs by two independent sgRNAs. Two single clones were generated with homozygous mutations (Supplementary Figure S1A). The total levels of H3K36me2 decreased and H3K27me3 increased in NSD1 KO cells as previously reported (26). The total levels of other tested histone modifications, including H3K36me3, H3K27ac and H3K4me3, were not changed to notable levels as detected by Western blotting (Supplementary Figure S1B). Please note, the Western blotting results were only semi-quantitative for the total levels and didn’t reflect the changes of exact chromatic locations of these histone marks. Upon NSD1 KO, cell proliferation was slightly decreased without affecting the activity of alkaline phosphatases in mESCs (Supplementary Figure S1C and D). We further compared the gene expression profiles in NSD1 KO cells by RNA seq. Two independent replicates of each cell line were sequenced and exhibited good correlations (Figure 1A). In addition, the two NSD1 KO clones were similar to each other, showing a consistent gene expression profile. We then merged the RNA seq results of two NSD1 KO clones to get the consistently changed coding genes and long non-coding RNAs (lncRNAs), which were selected with a log2(foldchange) > 2 and P value <0.01 (Figure 1B). Because H3K36me2 could inhibit repressive histone mark H3K27me3, H3K36me2 was proposed to be associated with active gene expression. Surprisingly, we found that a similar number or even slightly a greater number of genes were up-regulated in NSD1 KO mESCs, when compared with down-regulated genes (Figure 1C and Supplementary Figure S1E). 460 and 407 coding genes were up- and down-regulated in NSD1 KO cells, respectively. Moreover, 67 and 35 long non-coding RNAs were up- and down-regulated in NSD1 KO cells, respectively. These data indicate that NSD1 KO is correlated with both active and repressed gene expression.
Figure 1.
NSD1 KO in mESCs leads to both increased and decreased gene expression and chromatin accessibilities. (A) The principal component analysis (PCA) plot of the gene expression data from wild-type (WT), and NSD1 KO mESCs. NSD1 KO clones were generated from two independent sgRNAs. Two replicates of each cell line were presented. (B) Gene expression levels in wild-type and NSD1 KO mESCs. To get the reproducible results, RNA sequencing data from two replicates of each cell line were merged and two NSD1 KO cell lines were further merged. Red dots, the up-regulated genes with over 2-fold increased expression levels in NSD1 KO mESCs and P value <0.01. Blue dots, the down-regulated genes with over 2-fold decreased expression levels in NSD1 KO mESCs and P value <0.01. Yellow and cyan dots, the up-regulated and down-regulated long non-coding RNAs (lncRNAs) which were defined with the same criteria as coding genes, respectively. (C) The numbers of up- and down-regulated coding genes and lncRNAs in NSD1 KO mESCs were summarized. The criteria for the definitions of increased and decreased genes were the same as in (B). (D) Venn diagram illustrating the overlap of ATAC-seq peaks in WT and NSD1 KO mESCs. To generate reproducible results, sequencing data of two replicates from each cell line were merged. Fisher's exact statistical tests were used to analyze the overlaps of each samples. P values were <0.001 between every two samples respectively. (E) Integrative Genomics Viewer (IGV) tracks representing the signals of ATAC-seq in wild-type and two NSD1 KO mESCs. Blue box, the ATAC-seq signals were decreased in two NSD1 KO cell lines. Red box, the ATAC-seq signals were increased in two NSD1 KO cell lines. Two replicates of each cell line were merged. (F) Heatmaps illustrating ATAC-seq signals at peaks with changed reads densities. Average reads densities were calculated at each peak and peaks with different signals were selected as the P value <0.05. 503 and 897 peaks with increased and decreased ATAC-seq signals were in NSD1 KO mESCs, respectively. ATAC-seq signals from 1.5 kb upstream to 1.5 kb downstream of the peak regions (in rows) were shown on a per-peak basis (in columns). (G) Bar graph showing the enrichments of transcription factors binding motifs at altered ATAC-seq peaks. Peaks with increased and decreased ATAC-seq signals were selected as in (F) and the enrichments of vertebrates’ transcription factors motifs were analyzed at these peaks by HOMER. The top 10 motifs ranked by P value were shown.
To further demonstrate the alternations of gene expression, we analyzed the chromatin accessibility in NSD1 KO mESCs. We first compared the structure of chromatin in wild-type and NSD1 KO cells by digestion with micrococcal nuclease (MNase), which depletes the linker DNA between nucleosomes (Supplementary Figure S1F). We found that the chromatin of wild-type and NSD1 KO cells showed similar sensitivity to MNase digestion, suggesting the compaction of chromatin was not dramatically altered in NSD1 KO cells. We then conducted ATAC-seq (assay for transposase-accessible chromatin using sequencing) to profile the open status of chromatin in all cell lines with two independent repeats. Since the two repeats of each cell line correlated well across the genome, we merged two repeats for further analysis (Supplementary Table S1). The ATAC-seq peaks in wild-type and NSD1 KO cells were largely overlapped, consisting with the MNase digestion results (Figure 1D). The individual browser tack views revealed that the accessibilities of chromatin were both elevated (indicated by the red box) and reduced (indicated by the blue box) in NSD1 KO cells (Figure 1E). To extensively and reliably compare the chromatin status between wild-type and NSD1 KO cells, we merged two NSD1 KO clones and plotted ATAC-seq signals at all peak regions with changed ATAC-seq signals (P < 0.05) between wild-type and NSD1 KO cells (Figure 1F). Compared with wild-type cells, NSD1 KO cells showed a similar number of peaks with significantly increased (503) and decreased (897) accessibilities. Moreover, we annotated the transcription factor (TF) motifs at the increased and decreased peaks by HOMER, and found that the enriched TF motifs were largely different between increased and decreased peaks (Figure 1G). Together, these data suggest that, through distinct regulatory mechanisms, knocking out of NSD1 affected active and repressed genes.
H3K27ac is increased at active enhancers in NSD1 KO mESCs
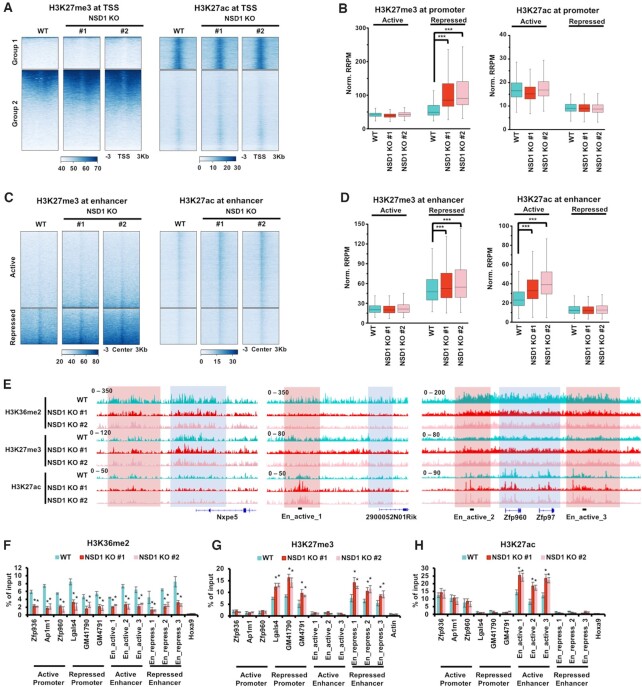
We speculated that the epigenome was extensively altered in NSD1 KO mESCs. So, we profiled H3K36me2, H3K27me3 and H3K27ac in wild-type and NSD1 KO cells by CUT&RUN. To compare the enrichment of sequencing reads in different cells, we used internal S. cerevisiae spike-in reads to normalize the detected reads from mESCs. Since H3K27me3 and H3K27ac antagonized each other in promoter and enhancer regions, we separated the analysis into promoter regions and enhancer regions. We analyzed the enrichments of H3K27me3 and H3K27ac spanning 3 kb from the transcription start sites (TSS), that were determined by NCBI RefSeq (Figure 2A). Genes were clustered into two clusters by their enrichments of H3K27me3 and H3K27ac. Genes in Group 1 were associated with low H3K27me3 and high H3K27ac, and genes in Group 2 were enriched with high H3K27me3 and low H3K27ac. Not surprisingly, the enrichment of H3K27me3 at Group 2 genes was increased in NSD1 KO cells. Conversely, H3K27ac was not changed to a detectable level around the TSS in neither group of genes. In addition, H3K36me2 was decreased at both groups of genes when NSD1 was knocked out (Supplementary Figure S2A). Chromatin accessibilities, as detected by ATAC-seq, were decreased at both groups of genes in NSD1 KO cells. To more definitively compare the changes of histone marks in wild-type and NSD1 KO cells, we sought to calculate the total enrichments of histone marks at active and repressed promoters. We defined promoters as 2 Kb upstream and 500 bp downstream of TSS, which were determined by NCBI RefSeq. Active promoters were further classified by the high enrichment of H3K27ac and low enrichment of H3K27me3, and repressed promoters were defined by an opposite enrichment. Consistent with the observation from heatmaps, in NSD1 KO cells, H3K27me3 was increased at repressed promoters, and H3K27ac was not significantly changed in neither active nor repressed promoters (Figure 2B). Additionally, H3K36me2 and chromatin accessibilities were significantly decreased in both active and repressed promoters (Supplementary Figure S2B).
Figure 2.
H3K27ac is increased at active enhancers in NSD1 KO mESCs. (A) Heatmaps illustrating H3K27me3 and H3K27ac levels within 3 kb from transcription start sits of all genes determined by NCBI RefSeq. Genes were clustered into two groups by H3K27me3 and H3K27ac signals, showing high H3K27ac in group 1 and high H3K27me3 in group 2. Sequencing results of two independent replicates were merged for analysis. (B) H3K27ac was not changed at promoters and H3K27me3 was increased at repressed promoters in NSD1 KO mESCs. Promoters were defined as 2 kb upstream and 500 bp downstream of NCBI RefSeq TSS. Active promoters were classified by a high H3K27ac and low H3K27me3, whereas repressed promoters were with low H3K27ac and high H3K27me3. Average reads densities of H3K27ac and H3K27me3 at classified promoters were calculated. *** P < 0.001 as determined by Student's t-test. (C) Heatmaps showing the enrichments of H3K27me3 and H3K27ac at 3 kb regions spanning the active and repressed enhancers. Enhancers were determined as H3K4me1 peaks without overlapped H3K4me3 peaks or annotated promoters. Active enhancers were further classified by high H3K27ac and low H3K27me3, whereas repressed enhancers were identified by low H3K27ac and high H3K27me3. (D) H3K27ac and H3K27me3 were increased at active enhancers and repressed enhancers in NSD1 KO mESCs, respectively. Average reads densities of H3K27ac and H3K27me3 at classified enhancers were calculated. *** P < 0.001 as determined by Student's t-test. (E) IGV tracks illustrating the enrichments of H3K36me2, H3K27me3, and H3K27ac in wild-type and two NSD1 KO mESCs. H3K27me3 levels were high and H3K27ac levels were low at Nxpe5 locus, exhibiting a repressed status. H3K27ac was high and H3K27me3 was low at 2900052N01Rik, Zfp960 and Zfp97 loci, showing an active status. Red box, the intergenic regions. Blue box, promoter and gene regions. H3K36me2 was enriched at both intergenic and gene body regions. The primers used in ChIP-qPCR for active enhancers were marked. (F–H) The enrichments of H3K36me2 (F), H3K27me3 (G), and H3K27ac (H) at the classified promoters and enhancers were analyzed via ChIP-qPCR. The data were represented by the mean ± SD (N = 3 independent replicates). Three loci of active promoters, repressed promoters, active enhancers, and repressed enhancers were analyzed respectively. The repressed Hoxa9 locus was used as the negative controls for H3K36me2 and H3K27ac. The active Actin locus was used as the negative control for H3K27me3. The primers used for active enhancers were marked as in (E). * P < 0.05 as determined by Student's t-test.
The increased H3K27me3 may lead to repressed gene expression but not the elevation of gene expression. To further explore the underlying mechanism of elevated gene expression in mESCs, we analyzed the enrichments of H3K27me3 and H3K27ac at enhancer regions. As previously reported (51), we identified enhancers as genomic loci with H3K4me1 peaks but not overlapped with annotated promoters or H3K4me3 peaks. Active enhancers were further classified by high levels of H3K27ac and low levels of H3K27me3, whereas repressed enhancers were identified by low H3K27ac and high H3K27me3 enrichments. As expected, H3K36me2 was decreased at both active and repressed enhancers when NSD1 was knocked out (Supplementary Figure S2C and D). H3K27me3 was increased at repressed enhancers in NSD1 KO mESCs. Importantly, H3K27ac, which was the active gene expression mark, was elevated at active enhancers in NSD1 KO cells (Figure 2C and D). Consistent with the increased H3K27ac at active enhancers and increased H3K27me3 at repressed enhancers, chromatin accessibilities were increased at active enhancers and decreased at repressed enhancers in NSD1 KO cells (Supplementary Figure S2C and D). In addition, the increase of H3K27ac at active enhancers and increase of H3K27me3 at repressed enhancers and promoters were revealed by IGV views (Figure 2E). At the Nxpe5 locus, H3K27me3 was high and H3K27ac was low at the promoters and intergenic regions, exhibiting a repressed status. H3K36me2 was decreased at the Nxpe5 locus when NSD1 was knocked out. While H3K27ac was not altered around the Nxpe5 locus, the enrichments of H3K27me3 in both promoter regions and intergenic regions were increased in NSD1 KO cells. Besides, H3K27me3 was low and H3K27ac was high around the 2900052N01Rik, Zfp960 and Zfp97 loci, showing an active status. When NSD1 was knocked out, H3K36me2 was decreased at the 2900052N01Rik, Zfp960 and Zfp97 loci. In NSD1 KO cells, H3K27ac was increased in intergenic regions but not the promoter regions, whereas H3K27me3 was not altered around the 2900052N01Rik, Zfp960 and Zfp97 loci. Moreover, through ChIP-qPCR, we confirmed the increase of H3K27me3 at repressed promoters and enhancers, the increase of H3K27ac at active enhancers, and the decrease of H3K36me2 in NSD1 KO cells (Figure 2F–H). Three genomic loci of active and repressed promoters and enhancers were tested respectively. Actin locus was used as the negative control for the enrichment of H3K27me3. Hoxa9 locus was tested as the negative control for the enrichments of H3K27ac and H3K36me2.
Active enhancers with elevated H3K27ac are associated with increased expression of genes enriched in mesoderm differentiation
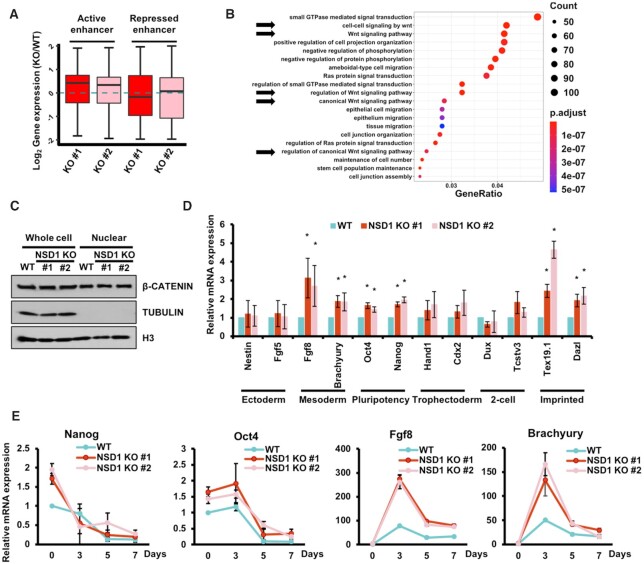
Since we detected increased H3K27ac at enhancers, we further elucidated the effects of elevated H3K27ac on the expression of associated genes. To investigate the changes between enhancer activation and gene expression, we assigned enhancers to the closest promoters within 500 kb as described (52). Notably, the expression levels of genes associated with repressed enhancers were lower than the genes associated with active enhancers (Supplementary Figure S3A). Additionally, the distances of associated active enhancers to the closest TSS were gradually increased from up-regulated, unchanged, to down-regulated genes, further confirming the classification of active and repressed enhancers (Supplementary Figure S3B). Compared with the parental cell line, the expression levels of genes associated with active enhancers were significantly increased in both of the two NSD1 KO cell lines (Figure 3A). In addition, the expression levels of repressed enhancers-associated genes were not altered between wild-type and NSD1 KO cells.
Figure 3.
Active enhancers with elevated H3K27ac are associated with mesoderm differentiation. (A) The expression levels of active enhancers-associated genes were increased in NSD1 KO mESCs. Active and repressed enhancers were assigned to the closest promoter within 500 Kb. Compared to wild-type cells, the relative expression levels of associated genes in NSD1 KO cells were calculated. (B) GO analysis result of genes associated with active enhancers which exhibited elevated H3K27ac in NSD1 KO mESCs. The top 20 GO terms ranked by P values were shown. Arrows indicated the GO terms linked to Wnt signals. (C) The canonical Wnt signal was not altered in NSD1 KO mESCs. Whole-cell lysates and nuclear fractions from parental and NSD1 KO mESCs were analyzed by the indicated antibodies. Tubulin which was in the cytoplasmic fraction was used as the negative control for the nuclear fraction. (D) RT-PCR analysis of the marker genes in wild-type and NSD1 KO mESCs. Gene expressions were normalized to Actin and the expression levels in wild-type cells were further normalized as 1. The data were represented by the mean ± SD (N = 3 independent replicates). * P < 0.05 as determined by Student's t-test. (E) Two mesoderm marker genes were expressed higher in NSD1 KO mESCs during random differentiation. mESCs were cultured as embryoid bodies to induce random differentiation.
We then performed Gene Ontology (GO) analysis using genes that were assigned by active enhancers with increased H3K27ac. We found that these 2,228 genes were largely enriched in Wnt signaling pathways, which played important roles in the regulation of mesoderm layer differentiation (Figure 3B). This data suggest that these altered genes were involved in the mESC differentiation. To further clarify whether the observed increase of Wnt signaling-associated genes was regulated by the active enhancers or was affected by the activation of Wnt signal, we analyzed the total amount and nuclear import of the key activator in Wnt signal activation, β-catenin (Figure 3C). The results showed that the total amount of β-catenin was not affected in either whole cell extract or nuclear fraction when NSD1 was knocked out, indicating that the activity of Wnt signal itself is not disturbed. We then examined the expression levels of pluripotency, ectoderm, mesoderm, trophectoderm, 2-cell, and imprinted markers in wild-type and NSD1 KO cells. Two marker genes of each category were analyzed by RT-PCR (Figure 3D). Gene expressions of both two tested pluripotency (Nanog and Oct4), mesoderm (Fgf8 and Brachyury), and imprinted (Tex19.1 and Dazl) marker genes were increased. The increase of mesoderm and imprinted mark genes was consistent with the elevated activation of Wnt signal-associated genes (53,54). Moreover, we assigned enhancers to the closest promoter within 500 Kb (52) and linked the enhancers into mesoderm, ectoderm, and trophectoderm markers genes as defined before (55). We then compared the changes of enrichments of H3K27ac at these enhancers. The H3K27ac at enhancers associated with mesoderm markers were increased in NSD1 KO cells, whereas H3K27ac at enhancers associated with ectoderm and trophectoderm markers was not changed respectively (Supplementary Figure S3C). Previous analysis in NSD1 knockdown mESCs didn’t show the increased expressions of pluripotency marker genes (26). This difference may be because NSD1 KO completely depleted NSD1 in cells, leading to a severer interruption of gene expression. Moreover, we tested the differentiation of wild-type and NSD1 KO mESCs by embryoid body (EB) formation (Figure 3E). Nanog and Oct4, the marker genes of pluripotency, were decreased in both wild-type and NSD1 KO cells during the EB formation, indicating a successful differentiation of mESCs. Fgf8 and Brachyury, the marker genes of mesoderm differentiation, were higher in NSD1 KO cells throughout the differentiation process. Together, these data suggest that increased H3K27ac at active enhancers affects, at least in part, the Wnt-associated genes to regulate the differentiation of mESCs into mesoderm cells.
NSD1 binds and recruits HDAC1 to chromatin in mESCs
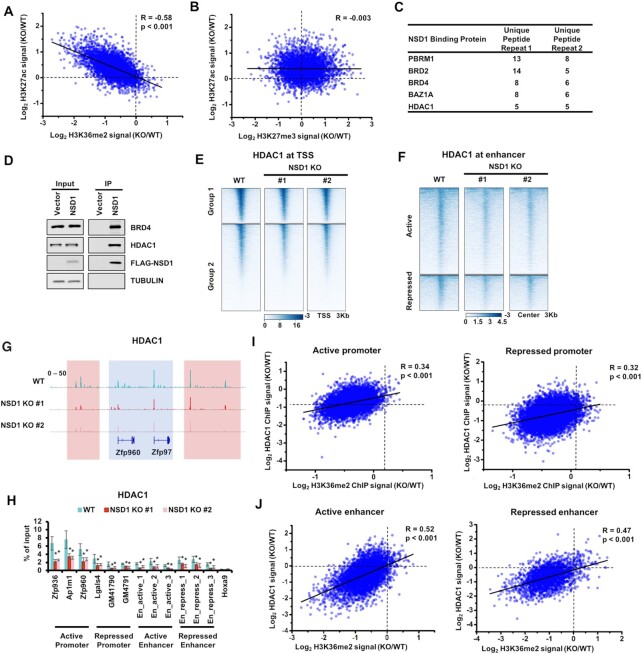
The increased H3K27ac at active enhancers promoted us to more closely evaluate the correlations between H3K27ac and H3K36me2 at enhancers. We plotted the alternations of H3K27ac and H3K36me2 at active enhancers that were within the H3K36me2 peaks in wild-type cells (Figure 4A). We observed a strong correlation between the changes of H3K27ac and H3K36me2, suggesting that the changes of H3K27ac were directedly associated with H3K36me2. Since H3K27ac and H3K27me3 were the post-translational modification on the same H3K27 residue and may play as a Yin-Yang process to influence each other, it's possible that the alternations of H3K27ac were a consequence of H3K27me3 changes caused by NSD1 KO. To elucidate this possibility, we plotted the alternations of H3K27ac and H3K27me3 at enhancers within the H3K36me2 peaks in wild-type cells (Figure 4B). The correlation was quite low between changes of H3K27ac and H3K27me3. It's unlikely that the increase of H3K27ac is caused by the alterations of H3K27me3.
Figure 4.
Depletion of NSD1 impairs the recruitment of HDAC1. (A) Correlations between the changes of H3K27ac and H3K36me2 at active enhancers that were within the H3K36me2 peaks in wild-type cells. Each dot indicated a single enhancer. R, correlation coefficients that were assessed by Pearson product moment correlation. (B) Same as in (A), except changes of H3K27ac and H3K27me3 were plotted. (C) Bromodomain-containing proteins and histone acetylation-associated proteins were enriched in NSD1 purification. Proteins identified in both of two replicates were shown. (D) Over-expressed NSD1 bound with BRD4 and HDAC1. NSD1 was purified from mESCs (negative control), mESCs expressing FLAG-tagged NSD1. Proteins from input and immunoprecipitated (IP) samples were analyzed by Western blotting using the indicated antibodies. (E) Heatmaps illustrating HDAC1 levels within 3 kb from transcription start sits of all genes which were determined by NCBI RefSeq. Genes were clustered by their enrichments of H3K27ac and H3K27me3. Group1 genes were with high H3K27ac and low H3K27me3. Group 2 genes were with low H3K27ac and high H3K27me3. Sequencing results of two independent replicates were merged for analysis. (F) Heatmaps showing the enrichments of HDAC1 at 3 Kb regions spanning the active and repressed enhancers. (G) IGV tracks presenting the enrichments of HDAC1 in wild-type and two NSD1 KO mESCs. The active Zfp960 and Zfp97 loci were shown. HDAC1 was enriched high in wild-type cells and decreased in NSD1 KO cells. Red box, the intergenic regions. Blue box, promoter and gene regions. (H) The enrichments of HDAC1 at the classified promoters and enhancers were analyzed via ChIP-qPCR. Three loci of active promoters, repressed promoters, active enhancers, and repressed enhancers were analyzed respectively. The repressed Hoxa9 locus was used as the negative control. The data are represented by the mean ± SD (N = 3 independent replicates). * P < 0.05 as determined by Student's t-test. (I) Correlations between the changes of HDAC1 and H3K36me2 at promoters that were within the H3K36me2 peaks in wild-type cells. Each dot indicated a single promoter. R, correlation coefficients that were assessed by Pearson product moment correlation. (J) Same as in (I), except the enhancers within the H3K36me2 peaks in wild-type cells were used for plotting.
We then sought to explore the underlying mechanism of the observed increase of H3K27ac in NSD1 KO cells. We overexpressed FLAG-tagged NSD1 in mESCs and performed immunoprecipitation (IP) coupled mass spectrometry analysis. Two independent repeats were carried out (Figure 4C). As previously reported (56,57), we detected several bromodomain-containing proteins in the purification of NSD1, including PBRM1, BRD2, BRD4 and BAZ1A, confirming the success of IP experiments. We further evaluated proteins associated with NSD1 for their enrichments of histone acetylation-associated proteins, including acetyltransferases and deacetylases. Interestingly, we only detected HDAC1 using this criterion. We further confirmed the interaction of HDAC1 and BRD4 with NSD1 by Western blotting (Figure 4D).
It's possible that the over-expression of NSD1 led to a false-positive binding between HDAC1 and NSD1. We then analyzed whether NSD1 KO affected the protein level and chromatin localization of HDAC1. As determined by Western blotting, the total levels of HDAC1 were not altered when NSD1 was knocked out (Supplementary Figure S4A). To profile the genome-wide distribution of HDAC1, we conducted HDAC1 CUT&Tag in wild-type and NSD1 KO cells. Two independent sequencing replicates of each cell line were correlated well (Supplementary Table S1) and the sequencing results of two NSD1 knockout clones were highly correlated (Supplementary Table S2). In addition, HDAC1 peaks were enriched at both promoters and intergenic regions as previously reported (58) (Supplementary Figure S4B). For comparison between the enrichment of sequencing reads in different samples, internal E. coli spike-in reads were used to normalize the detected reads from mESCs. We then calculated the enrichments of HDAC1 at promoters and enhancers respectively. The enrichments of HDAC1 were significantly decreased at promoters and enhancers, despite the promoters and enhancers were active or repressed (Figure 4E and F, and Supplementary Figure S4C and D). Moreover, IGV views showed HDAC1 was decreased at promoters (indicated by the blue box) and intergenic regions (indicated by the red box) in NSD1 KO cells (Figure 4G). These reductions of HDAC1 were further confirmed by ChIP-qPCR at three loci of active promoter, repressed promoter, active enhancer, and repressed enhancer respectively (Figure 4H). The enrichment of HDAC1 at Hoxa9 was used as the negative control. More importantly, the alternations of HDAC1 were correlated well with changes of H3K36me2 at both enhancers and promoters which were within H3K36me2 peaks in wild-type cells, despite the enhancers and promoters were active or repressed (Figure 4I and J). Together, these data suggest that depletion of NSD1 leads to the impaired recruitment of HDAC1 at both promoters and enhancers.
Active enhancers with increased H3K27ac in HDAC1 and NSD1 KO mESCs are largely overlapped
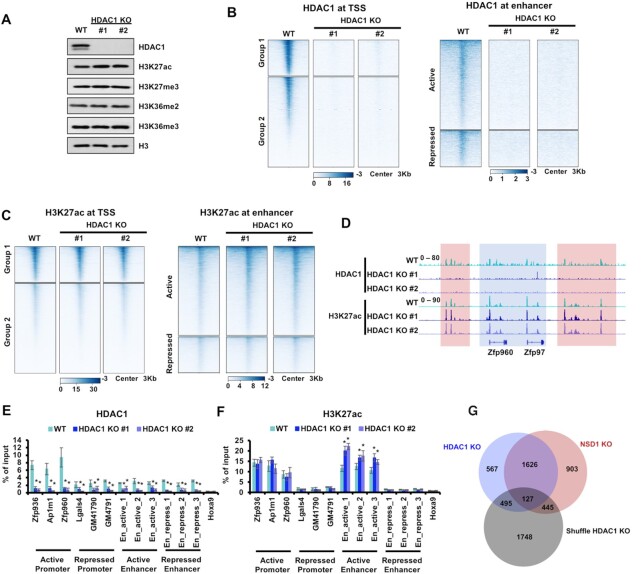
NSD1 KO reduced HDAC1 at both promoters and enhancers. However, we only detected an increase of H3K27ac at the active enhancers in NSD1 KO cells. It's possible that other HDACs were enriched at these loci and played as complementary regulators for the H3K27ac so that depletion of HDAC1 only increased H3K27ac at active enhancers. To test this hypothesis, we knocked out HDAC1 in mESCs with two independent sgRNAs (Supplementary Figure S5A). The total levels of H3K27ac were increased in both of the two HDAC1 KO clones. Other tested histone marks, including H3K27me3, H3K36me2 and H3K36me3, were not changed obviously in HDAC1 KO cells (Figure 5A). In HDAC1 KO cells, cell proliferations were slightly increased and the alkaline phosphatase activities were not changed (Supplementary Figure S5B and C).
Figure 5.
HDAC1 KO increases H3K27ac at active enhancers. (A) Western blot showing the indicated protein levels in parental and HDAC1 KO mESCs. HDAC1 KO led to slightly increased H3K27ac. Cell extracts were analyzed via Western blotting using the indicated antibodies. (B) Heatmaps illustrating HDAC1 levels around TSS and enhancers. Left, 3 Kb windows from TSS of all genes as determined by NCBI RefSeq. Genes were clustered by their enrichments of H3K27ac and H3K27me3. Group1 genes were with high H3K27ac and low H3K27me3. Group 2 genes were with low H3K27ac and high H3K27me3. Right, 3 Kb regions spanning the active and repressed enhancers. (C) Same as in (B), except H3K27ac levels were illustrated. (D) IGV tracks presenting the enrichments of HDAC1 and H3K27ac in wild-type and two HDAC1 KO mESCs. The active Zfp960 and Zfp97 loci were shown. HDAC1 was enriched high in wild-type cells and abolished in HDAC1 KO cells. H3K27ac was increased in intergenic regions but not the promoter regions when HDAC1 was knocked out. Red box, the intergenic regions. Blue box, promoter and gene regions. (E and F) The enrichments of HDAC1 and H3K27ac at the classified promoters and enhancers were analyzed via ChIP-qPCR. Three loci of active promoters, repressed promoters, active enhancers, and repressed enhancers were analyzed respectively. The repressed Hoxa9 locus was used as the negative control for H3K27ac and HDAC1. The data were represented by the mean ± SD (N = 3 independent replicates). * P < 0.05 as determined by Student's t-test. (G) Venn diagram illustrating the overlap of active enhancers with increased H3K27ac in HDAC1 and NSD1 KO mESCs. Active enhancers used as the negative control were generated by shuffling the active enhancers, that were with increased H3K27ac in HDAC1 KO cells, within all active enhancers. To generate reproducible results, sequencing data of two independent clones were merged. Fisher's exact statistical tests were performed between every two samples. Only the overlap between HDAC1 and NSD1 KO showed a P value less than 0.001. Other overlaps were not significant with P values more than 0.05.
To elucidate how HDAC1 KO affected H3K27ac genome-widely, we performed H3K27ac and HDAC1 CUT&Tag in wild-type and HDAC1 KO cells. For the comparison of sequencing results among different cell lines, internal E. coli spike-in reads were used to normalize the detected reads from mESCs. As expected, the signals of HDAC1 were significantly depleted at the promoters and enhancers in HDAC1 KO cells, further confirming the specificity of HDAC1 CUT&Tag results (Figure 5B and Supplementary Figure S6A and B). More importantly, the enrichment of H3K27ac was only increased at the active enhancers but not the promoters or repressed enhancers, which were similar to the alternations of H3K27ac in NSD1 KO cells (Figure 5C and Supplementary Figure S6C and D). IGV views showed that H3K27ac was increased at intergenic regions (red box) and unchanged at promoters (blue box) in HDAC1 KO cells, whereas HDAC1 signals were abolished in HDAC1 KO cells (Figure 5D). Meanwhile, we conducted ChIP-qPCR to further analyze the changes of H3K27ac and HDAC1 at promoters and enhancers, of which three active and repressed loci were tested (Figure 5E and F). Hoxa9 locus which was not enriched with HDAC1 or H3K27ac was used as the negative control. HDAC1 signals were depleted at all tested loci in HDAC1 KO cells. The enrichments of H3K27ac were elevated at three tested active enhancers but not at other tested loci. Furthermore, we compared the active enhancers with increased H3K27ac in HDAC1 and NSD1 KO cells by merging two independent KO clones (Figure 5G). To generate a negative control for this comparison, we shuffled the active enhancers, that were with increased H3K27ac in HDAC1 KO cells, within all active enhancers. H3K27ac was increased at 3101 and 2815 active enhancers in NSD1 and HDAC1 KO cells respectively, whereas over 50% of active enhancers with increased H3K27ac (1753) were overlapped between NSD1 KO and HDAC1 KO cells. All these data indicate that HDAC1 knockout increases H3K27ac at active enhancers but not the promoters, which recapitulates the changes of H3K27ac in NSD1 KO cells.
DISCUSSION
An outstanding question in epigenetics studies is how histone marks regulate gene expression. The interplay between different histone modifications adds a second layer of regulation on gene expression, leading to the huge increase in complexity of gene regulation. Our data are in agreement with previous enzymatic activity and boundary expansion analyses between the NSD1-deposited H3K36me2 and PRC2-mediated H3K27me3. More importantly, we peruse the up-regulation of gene expression in NSD1 KO mESCs, and find NSD1 recruited HDAC1 as the regulator for increased H3K27ac at active enhancers, that are associated with elevated gene expression. We propose that NSD1-mediated H3K36me2 modulates HDAC1 by preventing the over-acetylation of active enhancers.
Histone acetylation is generally associated with active gene expression. Besides, histone deacetylation is mostly coupled with transcriptional repression (59). HDACs, which deacetylate histones co-transcriptionally, are highly conserved from yeast to human (60). There are four classes of HDACs defined by their homology with yeast orthologs: Rpd3-like proteins (HDAC1-3, and 8; Class I), Hda1-like proteins (HDAC4-7, 9 and 10; Class II), Sir2-like proteins (SIRT1-7; Class III) and HDAC11 (Class IV) (61). HDACs of Class III use NAD+ to generate nicotinamide and metabolite 2′-O-acetyl-ADP-ribose during the deacetylation, and HDACs of other classes catalyze the hydrolysis of acetylated histones in a zinc-dependent manner (61–63). HDACs play critical roles in many cellular processes so that disturbed expression of HDACs has been found in multiple cancer tumorigenesis and drug resistance (64,65). HDACs deacetylate histones leading to transcriptional repression, however, they are enriched at active genes but not silent genes and positively correlated with transcription (58). HDACs display different preferences regarding the lysine residues in histones. More importantly, HDACs are distributed across the genome with similarities and unique characteristics, leading to both complemental and independent regulations of histone acetylation and gene expression. HDAC1 is enriched with higher levels at active promoters when compared with active enhancers. We observed that, when HDAC1 was indirectly abolished from chromatin by NSD1 KO or directly depleted by KO, the H3K27ac was not significantly changed at active promoters. Several HDACs are enriched at active promoters, including HDAC1–3 and HDAC6, whereas other HDACs may play complementary roles at these sites to maintain the restriction of H3K27ac.
NSD1 is misregulated in various diseases, like Sotos syndrome, Myelodysplastic syndrome, acute myeloid leukemia (AML), prostate cancer, neuroblastoma, breast cancer, and head and neck squamous cell carcinomas (HNSCCs) (29,30,32,66). In HNSCCs, as expected, the increase of H3K27me3 is detected if NSD1 is deficient. However, a subsequent loss of H3K27ac at enhancers, correlating with reduced expression of their target genes, is identified in NSD1 KO HNSCCs (66). This does not contradict to our observations. It's possible that these NSD1 KO cells are based on the HNSCC tumor cells, which contain other mutations to regulate H3K27ac and gene expression in cells. Our results are based on the wild-type mESCs with NSD1 KO as the only mutation. Moreover, except for the interactions detected between HDAC1 and NSD1 by immunoprecipitation, we identified a high overlap of active enhancers with elevated H3K27ac between NSD1 KO and HDAC1 KO cells.
Like NSD1, other H3K36me2 methyltransferases are mutated in tumors. NSD2 translocates from its original locus on chromosome 4 with the immunoglobulin heavy chain locus on chromosome 14, leading to the elevated expression of NSD2 in around 15–20% of multiple myeloma (MM) patients (67). Decreased H3K36me2 and increased H3K27me3 are observed in MM cells with low NSD2 expression. Compared with MM cells with high NSD2 expression, a greater number of genes (∼3-fold than the increased genes) are decreased in MM cells with low NSD2 expression. In addition, H3K27ac peak numbers decrease in MM cells with low expression of NSD2. This gene expression and H3K27 alternation are different from the NSD1 KO cells, in which a similar number of genes are up- and down-regulated and H3K27ac is increased at the active enhancers. NSD1 and NSD2 both catalyze the methylation of H3K36me2. Nevertheless, they may function in the regulation of gene expression through different molecular mechanisms. It would be very interesting to dissect the differences among all the histone methyltransferases for H3K36me2, including ASH1L, NSD1-3, SMYD2, SETMAR and SETD3. The genome-wide distributions of H3K36me2 enzymes are revealed by their enzymatic product H3K36me2, which is an indirect profiling result. To reveal the specific targeting sites on chromatin, these H3K36me2 enzymes are mainly analyzed by manipulating the enzyme to determine a consequent H3K36me2 alternation. It's highly possible that other histone methyltransferases responsible for H3K36me2 were disturbed when the target enzyme is altered, leading to a further/consequent change of H3K36me2. The emerging needs for directly profiling H3K36me2 enzymes would lead to a clear resolution of how these enzymes are distributed on the chromatin to regulate histone modifications and, subsequent, gene expression.
The diseases of NSD1 misregulation are difficult to treat. Inhibitors specific to H3K36me2 methyltransferases are understudied because the enzymatic pockets of different H3K36 methyltransferases are similar (68). Our findings connect NSD1 with HDAC1, which is druggable. To present, there are lots of inhibitors developed for HDACs, some of which are under clinical trials (69). Our finding would open a new direction for the treatments of diseases with NSD1 mutations or misregulations. Further studies were needed for the oncogenes associated with active enhancers. It would be very interesting to determine to what extent inhibitions of HDAC1 would change the expressions of these oncogenes and tumor development.
DATA AVAILABILITY
Raw data have been deposited in the GEO database with the series accession GSE169049.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Juerg Schwaller for sharing the plasmid to overexpress NSD1. We thank Hangjun Wu in the Center of Cryo-Electron Microscopy (CCEM), Zhejiang University for his technical assistance on the computing node setup. We thank Tianjiao Du for her assistance in the analysis of ATAC-seq data.
Author contributions: Conceptualization, Y.F., Y.T., Y.Z., Y.Y. and D.F.; Methodology, Y.F., Y.T., Y.Z. and Y.P.; Investigation, Y.F., Y.T., Y.Z., J.J., Z.S., W.Z., J.C. and D.F.; Writing – Original Draft, Y.F., Y.T. and D.F.; Writing – Review & Editing, Y.F., Y.T., Y.Z., Y.P. and D.F.; Funding Acquisition, D.F.; Supervision, D.F.
Contributor Information
Yuan Fang, Zhejiang Provincial Key Laboratory for Cancer Molecular Cell Biology, Life Sciences Institute, Zhejiang University, Hangzhou, Zhejiang 310058, China.
Yin Tang, Zhejiang Provincial Key Laboratory for Cancer Molecular Cell Biology, Life Sciences Institute, Zhejiang University, Hangzhou, Zhejiang 310058, China.
Yanjun Zhang, Zhejiang Provincial Key Laboratory for Cancer Molecular Cell Biology, Life Sciences Institute, Zhejiang University, Hangzhou, Zhejiang 310058, China.
Yixin Pan, Department of Medical Oncology, Key Laboratory of Cancer Prevention and Intervention, Ministry of Education, The Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, Zhejiang, China.
Junqi Jia, Zhejiang Provincial Key Laboratory for Cancer Molecular Cell Biology, Life Sciences Institute, Zhejiang University, Hangzhou, Zhejiang 310058, China.
Zhongxing Sun, Zhejiang Provincial Key Laboratory for Cancer Molecular Cell Biology, Life Sciences Institute, Zhejiang University, Hangzhou, Zhejiang 310058, China.
Weiwu Zeng, Zhejiang Provincial Key Laboratory for Cancer Molecular Cell Biology, Life Sciences Institute, Zhejiang University, Hangzhou, Zhejiang 310058, China.
Jiaqi Chen, Department of Medical Oncology, Key Laboratory of Cancer Prevention and Intervention, Ministry of Education, The Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, Zhejiang, China.
Ying Yuan, Department of Medical Oncology, Key Laboratory of Cancer Prevention and Intervention, Ministry of Education, The Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, Zhejiang, China.
Dong Fang, Zhejiang Provincial Key Laboratory for Cancer Molecular Cell Biology, Life Sciences Institute, Zhejiang University, Hangzhou, Zhejiang 310058, China; Department of Medical Oncology, Key Laboratory of Cancer Prevention and Intervention, Ministry of Education, The Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, Zhejiang, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [81874153]; Yixin Pan was supported by the Innovation research grant program for 8-year-system medical students at Zhejiang University [119000-5405A1]. Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Strahl B.D., Allis C.D.. The language of covalent histone modifications. Nature. 2000; 403:41–45. [DOI] [PubMed] [Google Scholar]
- 2. Jenuwein T., Allis C.D.. Translating the histone code. Science. 2001; 293:1074–1080. [DOI] [PubMed] [Google Scholar]
- 3. Zhang Y., Reinberg D.. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 2001; 15:2343–2360. [DOI] [PubMed] [Google Scholar]
- 4. Agalioti T., Chen G., Thanos D.. Deciphering the transcriptional histone acetylation code for a human gene. Cell. 2002; 111:381–392. [DOI] [PubMed] [Google Scholar]
- 5. Lee J.S., Smith E., Shilatifard A.. The language of histone crosstalk. Cell. 2010; 142:682–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rao B., Shibata Y., Strahl B.D., Lieb J.D.. Dimethylation of histone H3 at lysine 36 demarcates regulatory and nonregulatory chromatin genome-wide. Mol. Cell. Biol. 2005; 25:9447–9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vakoc C.R., Sachdeva M.M., Wang H., Blobel G.A.. Profile of histone lysine methylation across transcribed mammalian chromatin. Mol. Cell. Biol. 2006; 26:9185–9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xiao T., Shibata Y., Rao B., Laribee R.N., O’Rourke R., Buck M.J., Greenblatt J.F., Krogan N.J., Lieb J.D., Strahl B.D.. The RNA polymerase II kinase Ctk1 regulates positioning of a 5′ histone methylation boundary along genes. Mol. Cell. Biol. 2007; 27:721–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bannister A.J., Schneider R., Myers F.A., Thorne A.W., Crane-Robinson C., Kouzarides T.. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J. Biol. Chem. 2005; 280:17732–17736. [DOI] [PubMed] [Google Scholar]
- 10. Keogh M.C., Kurdistani S.K., Morris S.A., Ahn S.H., Podolny V., Collins S.R., Schuldiner M., Chin K., Punna T., Thompson N.J.et al.. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005; 123:593–605. [DOI] [PubMed] [Google Scholar]
- 11. Strahl B.D., Grant P.A., Briggs S.D., Sun Z.W., Bone J.R., Caldwell J.A., Mollah S., Cook R.G., Shabanowitz J., Hunt D.F.et al.. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol. Cell. Biol. 2002; 22:1298–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kizer K.O., Phatnani H.P., Shibata Y., Hall H., Greenleaf A.L., Strahl B.D.. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol. Cell. Biol. 2005; 25:3305–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xiao T., Hall H., Kizer K.O., Shibata Y., Hall M.C., Borchers C.H., Strahl B.D.. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev. 2003; 17:654–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fang R., Barbera A.J., Xu Y., Rutenberg M., Leonor T., Bi Q., Lan F., Mei P., Yuan G.C., Lian C.et al.. Human LSD2/KDM1b/AOF1 regulates gene transcription by modulating intragenic H3K4me2 methylation. Mol. Cell. 2010; 39:222–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huang C., Zhu B.. Roles of H3K36-specific histone methyltransferases in transcription: antagonizing silencing and safeguarding transcription fidelity. Biophys Rep. 2018; 4:170–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rondelet G., Dal Maso T., Willems L., Wouters J.. Structural basis for recognition of histone H3K36me3 nucleosome by human de novo DNA methyltransferases 3A and 3B. J. Struct. Biol. 2016; 194:357–367. [DOI] [PubMed] [Google Scholar]
- 17. Dhayalan A., Rajavelu A., Rathert P., Tamas R., Jurkowska R.Z., Ragozin S., Jeltsch A.. The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J. Biol. Chem. 2010; 285:26114–26120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rinaldi L., Datta D., Serrat J., Morey L., Solanas G., Avgustinova A., Blanco E., Pons J.I., Matallanas D., Von Kriegsheim A.et al.. Dnmt3a and Dnmt3b Associate with Enhancers to Regulate Human Epidermal Stem Cell Homeostasis. Cell Stem Cell. 2016; 19:491–501. [DOI] [PubMed] [Google Scholar]
- 19. Leung C.S., Douglass S.M., Morselli M., Obusan M.B., Pavlyukov M.S., Pellegrini M., Johnson T.L.. H3K36 methylation and the chromodomain protein Eaf3 are required for proper cotranscriptional spliceosome assembly. Cell Rep. 2019; 27:3760–3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang H., Weng H., Zhou K., Wu T., Zhao B.S., Sun M., Chen Z., Deng X., Xiao G., Auer F.et al.. Histone H3 trimethylation at lysine 36 guides m(6)A RNA modification co-transcriptionally. Nature. 2019; 567:414–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chantalat S., Depaux A., Hery P., Barral S., Thuret J.Y., Dimitrov S., Gerard M.. Histone H3 trimethylation at lysine 36 is associated with constitutive and facultative heterochromatin. Genome Res. 2011; 21:1426–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bell O., Conrad T., Kind J., Wirbelauer C., Akhtar A., Schubeler D.. Transcription-coupled methylation of histone H3 at lysine 36 regulates dosage compensation by enhancing recruitment of the MSL complex in Drosophila melanogaster. Mol. Cell. Biol. 2008; 28:3401–3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li F., Mao G., Tong D., Huang J., Gu L., Yang W., Li G.M.. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSalpha. Cell. 2013; 153:590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Weinberg D.N., Papillon-Cavanagh S., Chen H., Yue Y., Chen X., Rajagopalan K.N., Horth C., McGuire J.T., Xu X., Nikbakht H.et al.. The histone mark H3K36me2 recruits DNMT3A and shapes the intergenic DNA methylation landscape. Nature. 2019; 573:281–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jani K.S., Jain S.U., Ge E.J., Diehl K.L., Lundgren S.M., Muller M.M., Lewis P.W., Muir T.W.. Histone H3 tail binds a unique sensing pocket in EZH2 to activate the PRC2 methyltransferase. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:8295–8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Streubel G., Watson A., Jammula S.G., Scelfo A., Fitzpatrick D.J., Oliviero G., McCole R., Conway E., Glancy E., Negri G.L.et al.. The H3K36me2 methyltransferase Nsd1 demarcates PRC2-mediated H3K27me2 and H3K27me3 domains in embryonic stem cells. Mol. Cell. 2018; 70:371–379. [DOI] [PubMed] [Google Scholar]
- 27. Schmitges F.W., Prusty A.B., Faty M., Stutzer A., Lingaraju G.M., Aiwazian J., Sack R., Hess D., Li L., Zhou S.et al.. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol. Cell. 2011; 42:330–341. [DOI] [PubMed] [Google Scholar]
- 28. Yuan W., Xu M., Huang C., Liu N., Chen S., Zhu B.. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J. Biol. Chem. 2011; 286:7983–7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kurotaki N., Imaizumi K., Harada N., Masuno M., Kondoh T., Nagai T., Ohashi H., Naritomi K., Tsukahara M., Makita Y.et al.. Haploinsufficiency of NSD1 causes Sotos syndrome. Nat. Genet. 2002; 30:365–366. [DOI] [PubMed] [Google Scholar]
- 30. Papillon-Cavanagh S., Lu C., Gayden T., Mikael L.G., Bechet D., Karamboulas C., Ailles L., Karamchandani J., Marchione D.M., Garcia B.A.et al.. Impaired H3K36 methylation defines a subset of head and neck squamous cell carcinomas. Nat. Genet. 2017; 49:180–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cancer Genome Atlas Research, N. Ley T.J., Miller C., Ding L., Raphael B.J., Mungall A.J., Robertson A., Hoadley K., Triche T.J. Jr, Laird P.W.et al.. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013; 368:2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wagner E.J., Carpenter P.B.. Understanding the language of Lys36 methylation at histone H3. Nat. Rev. Mol. Cell Biol. 2012; 13:115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kuo A.J., Cheung P., Chen K., Zee B.M., Kioi M., Lauring J., Xi Y., Park B.H., Shi X., Garcia B.A.et al.. NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming. Mol. Cell. 2011; 44:609–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F.. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013; 8:2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Skene P.J., Henikoff J.G., Henikoff S.. Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat. Protoc. 2018; 13:1006–1019. [DOI] [PubMed] [Google Scholar]
- 36. Kaya-Okur H.S., Wu S.J., Codomo C.A., Pledger E.S., Bryson T.D., Henikoff J.G., Ahmad K., Henikoff S.. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 2019; 10:1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhou H., Madden B.J., Muddiman D.C., Zhang Z.. Chromatin assembly factor 1 interacts with histone H3 methylated at lysine 79 in the processes of epigenetic silencing and DNA repair. Biochemistry. 2006; 45:2852–2861. [DOI] [PubMed] [Google Scholar]
- 38. Langdon W.B. Performance of genetic programming optimised Bowtie2 on genome comparison and analytic testing (GCAT) benchmarks. BioData Min. 2015; 8:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W.et al.. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Quinlan A.R. BEDTools: the Swiss-Army tool for genome feature analysis. Curr. Protoc. Bioinformatics. 2014; 47:11.12.1–11.12.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ramirez F., Dundar F., Diehl S., Gruning B.A., Manke T.. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014; 42:W187–W191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Thorvaldsdottir H., Robinson J.T., Mesirov J.P.. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 2013; 14:178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yu G., Wang L.G., He Q.Y.. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015; 31:2382–2383. [DOI] [PubMed] [Google Scholar]
- 44. Ritchie M.E., Phipson B., Wu D., Hu Y.F., Law C.W., Shi W., Smyth G.K.. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015; 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yu G., Wang L.G., Han Y., He Q.Y.. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012; 16:284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liao Y., Smyth G.K., Shi W.. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014; 30:923–930. [DOI] [PubMed] [Google Scholar]
- 49. Robinson M.D., McCarthy D.J., Smyth G.K.. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010; 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Villanueva R.A.M., Chen Z.J.. ggplot2: elegant graphics for data analysis, 2nd edition. Meas-Interdiscip Res. 2019; 17:160–167. [Google Scholar]
- 51. Calo E., Wysocka J.. Modification of enhancer chromatin: what, how, and why?. Mol. Cell. 2013; 49:825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Consortium E.P., Birney E., Stamatoyannopoulos J.A., Dutta A., Guigo R., Gingeras T.R., Margulies E.H., Weng Z., Snyder M., Dermitzakis E.T.et al.. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007; 447:799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kishimoto K., Furukawa K.T., Luz-Madrigal A., Yamaoka A., Matsuoka C., Habu M., Alev C., Zorn A.M., Morimoto M.. Bidirectional Wnt signaling between endoderm and mesoderm confers tracheal identity in mouse and human cells. Nat. Commun. 2020; 11:4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Theka I., Sottile F., Cammisa M., Bonnin S., Sanchez-Delgado M., Di Vicino U., Neguembor M.V., Arumugam K., Aulicino F., Monk D.et al.. Wnt/beta-catenin signaling pathway safeguards epigenetic stability and homeostasis of mouse embryonic stem cells. Sci. Rep. 2019; 9:948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Maguire C.T., Demarest B.L., Hill J.T., Palmer J.D., Brothman A.R., Yost H.J., Condic M.L.. Genome-wide analysis reveals the unique stem cell identity of human amniocytes. PLoS One. 2013; 8:e53372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bennett R.L., Swaroop A., Troche C., Licht J.D.. The role of nuclear receptor-binding SET domain family histone lysine methyltransferases in cancer. Cold Spring Harb. Perspect. Med. 2017; 7:a026708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rahman S., Sowa M.E., Ottinger M., Smith J.A., Shi Y., Harper J.W., Howley P.M.. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol. Cell. Biol. 2011; 31:2641–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang Z., Zang C., Cui K., Schones D.E., Barski A., Peng W., Zhao K.. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009; 138:1019–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang Z., Zang C., Rosenfeld J.A., Schones D.E., Barski A., Cuddapah S., Cui K., Roh T.Y., Peng W., Zhang M.Q.et al.. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 2008; 40:897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kadosh D., Struhl K.. Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Genes Dev. 1998; 12:797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Seto E., Yoshida M.. Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb. Perspect. Biol. 2014; 6:a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lombardi P.M., Cole K.E., Dowling D.P., Christianson D.W.. Structure, mechanism, and inhibition of histone deacetylases and related metalloenzymes. Curr. Opin. Struct. Biol. 2011; 21:735–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Vaquero A., Sternglanz R., Reinberg D.. NAD+-dependent deacetylation of H4 lysine 16 by class III HDACs. Oncogene. 2007; 26:5505–5520. [DOI] [PubMed] [Google Scholar]
- 64. Bose P., Dai Y., Grant S.. Histone deacetylase inhibitor (HDACI) mechanisms of action: emerging insights. Pharmacol. Ther. 2014; 143:323–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Suraweera A., O’Byrne K.J., Richard D.J.. Combination therapy with histone deacetylase inhibitors (HDACi) for the treatment of cancer: achieving the full therapeutic potential of HDACi. Front. Oncol. 2018; 8:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Farhangdoost N., Horth C., Hu B., Bareke E., Chen X., Li Y., Coradin M., Garcia B.A., Lu C., Majewski J.. Chromatin dysregulation associated with NSD1 mutation in head and neck squamous cell carcinoma. Cell Rep. 2021; 34:108769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lhoumaud P., Badri S., Rodriguez-Hernaez J., Sakellaropoulos T., Sethia G., Kloetgen A., Cornwell M., Bhattacharyya S., Ay F., Bonneau R.et al.. NSD2 overexpression drives clustered chromatin and transcriptional changes in a subset of insulated domains. Nat. Commun. 2019; 10:4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Huang H., Howard C.A., Zari S., Cho H.J., Shukla S., Li H., Ndoj J., Gonzalez-Alonso P., Nikolaidis C., Abbott J.et al.. Covalent inhibition of NSD1 histone methyltransferase. Nat. Chem. Biol. 2020; 16:1403–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. West A.C., Johnstone R.W.. New and emerging HDAC inhibitors for cancer treatment. J. Clin. Invest. 2014; 124:30–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data have been deposited in the GEO database with the series accession GSE169049.