Abstract
Specific capture of chromatin fractions with distinct and well-defined features has emerged as both challenging and a key strategy towards a comprehensive understanding of genome biology. In this context, we developed aniFOUND (accelerated native isolation of factors on unscheduled nascent DNA), an antibody-free method, which can label, capture, map and characterise nascent chromatin fragments that are synthesized in response to specific cues outside S-phase. We used the ‘unscheduled’ DNA synthesis (UDS) that takes place during the repair of UV-induced DNA lesions and coupled the captured chromatin to high-throughput analytical technologies. By mass-spectrometry we identified several factors with no previously known role in UVC-DNA damage response (DDR) as well as known DDR proteins. We experimentally validated the repair-dependent recruitment of the chromatin remodeller RSF1 and the cohesin-loader NIPBL at sites of UVC-induced photolesions. Developing aniFOUND-seq, a protocol for mapping UDS activity with high resolution, allowed us to monitor the landscape of UVC repair-synthesis events genome wide. We further resolved repair efficacy of the rather unexplored repeated genome, in particular rDNA and telomeres. In summary, aniFOUND delineates the proteome composition and genomic landscape of chromatin loci with specific features by integrating state-of-the-art ‘omics’ technologies to promote a comprehensive view of their function.
INTRODUCTION
During development but also throughout the whole life of living organisms, chromatin is in a state of continuous changes required to sustain vital biological cellular processes. The complex nature of these events is reflected on the sophisticated organization of chromatin structure and its spatial and functional features (1). Although in the recent years a number of sophisticated proteomic based approaches (2) have been developed, novel technologically advanced strategies to isolate specific chromatin loci are still necessary. A small series of methods dedicated in identifying proteins associated with certain chromatin states, such as the newly replicated DNA (3–5) have greatly contributed to our understanding of the mechanisms of chromatin replication (6,7). Nevertheless, DNA synthesis is not exclusive to replication as it occurs also outside S-phase, for instance during DNA repair. Given that DNA damage is a constant threat to the integrity of the genome and it requires a prompt and well-coordinated repair to take place in chromatin for its elimination, examining DNA synthesis coupled to repair and its associated proteome/genomic landscape is crucial towards an in depth elucidation of the mechanisms of chromatin repair.
Considering that cancer is among the main causes of human death, especially in countries with high life expectancies, scrutinizing every aspect of the factors and processes that lead to its development and uncovering new diagnostic, prognostic and treatment options is of pivotal importance. To this end, nucleotide excision repair (NER)-associated DNA damage responses (NER-DDR) have attracted prominent interest in cancer research. Exposure to environmental agents such as ultraviolet (UV) irradiation, cigarette smoke and several chemotherapeutics currently in clinical use (8), induce helix distorting DNA lesions that trigger a multi-layered cellular NER-DDR for their repair (9). Increased numbers of mutations due to an overwhelmed or defective NER are causatively linked to cutaneous melanoma carcinogenesis as well as basal and squamous cell carcinoma and certain lung cancers (10–12). Moreover, research on NER-DDR may aid in deepening our understanding and provide new strategies for treatment of Xeroderma Pigmentosum, Cockayne Syndrome, and Trichothiodystrophy, which are rare human disorders caused by NER defects (12).
The era of high throughput technologies has allowed new insights into the NER field. Several strategies have been developed for analysing the effects of NER-inducing genotoxic factors on the whole chromatin-associated proteome (13,14) as well as the cell's PTMome (post-translation modifications) (15–17). In addition, a multiomic strategy entailing proteomic screens and a functional genomics screen has been used to address transcription-related DNA damage responses (13). In parallel, novel methods have been developed for deep sequencing of damaged DNA (18–20) and the DNA that is excised from damaged sites (21). Despite the interesting findings emerging from these approaches, important questions remain unanswered, in particular in the context of chromatin and its coupling to the repair.
There are two sub-pathways of NER, the transcription coupled NER (TC-NER), which is triggered by an actively transcribing RNA polymerase II (Pol II) and removes DNA lesions from the transcribed strand of genes and gene regulatory regions (21,22) and the global genomic NER (GG-NER) that removes lesions from the entire genome. The two sub-pathways mainly differ at the step of damage recognition. Following the removal by the NER core machinery of DNA oligomers containing helix distorting DNA lesions, unscheduled DNA synthesis (UDS) takes place and DNA polymerases fill in the resulting single stranded DNA gap (of length of ∼30 nucleotides) using the non-damaged complementary strand as a template (12). Here, we took advantage of this property of NER and of previously reported Click chemistry-based protocols for isolation of replication-derived nascent chromatin (3,4) to develop aniFOUND (accelerated native isolation of factors on unscheduled nascent DNA), using UVC as a specific NER-inducing genotoxic factor. aniFOUND is a novel unbiased (antibody-free) method for the specific labelling, enrichment and purification of repaired, newly-synthesized chromatin. By coupling nascent UDS labelling to proteomic and Next Generation Sequencing (NGS) technologies, this method provides an in-depth view of the repaired chromatin-associated proteome composition and its genome-wide distribution.
MATERIALS AND METHODS
Cells and cell propagation
1BR.3 (23) and VH10 (24) normal and NER-deficient XPA (24) hTert immortalized human skin fibroblast cell lines were used in this study. The cell lines were maintained under standard conditions in DMEM with high glucose and sodium pyruvate (Gibco) supplemented with 10% fetal bovine serum (FBS) (Gibco), 100 units/ml of penicillin and 100 μg/ml of streptomycin (Gibco) at 37°C in a 5% CO2 humidified incubator.
Isolation of the UVC-UDS-associated chromatin
For one pull-down experiment 120 million human fibroblasts were used. Cells were plated in 150 mm dishes and grown until 100% confluency. Then they were maintained for three more days in culture medium supplemented with 0.5% FBS. In all cell culture preparations that were subjected to MS, 30 min-long pre-treatment with 10 mM HU was applied before irradiation. After washing with PBS the cells were irradiated with 20 or 30 J/m2 UVC (254 nm, TUV Lamp, Philips) as indicated (doses that induce sufficient amounts of DNA lesions for the subsequent analyses), and incubated in FBS-free culture medium containing 10 μM EdU (3,4) and 10 mM hydroxyurea (HU) for 4 h. Next, the cells were washed with PBS, scraped and pelleted in the presence of 1 mM PMSF. The following steps, from nuclei isolation till capturing, were adapted from Leung et al. (4) with minor changes. Briefly, the nuclei were isolated by resuspending the cell pellet in nuclear extraction buffer (NEB) and rotating for 15 min at 4°C. The pelleted nuclei were washed with PBS and subjected to Click-reaction by resuspending in Click-reaction mix supplemented with 1 mM THPTA and rotating for 1 h at room temperature (RT). Next the nuclei were pelleted, washed with PBS, resuspended in 1ml B1 buffer supplemented with protease inhibitors (PIs), incubated for 15 min and sonicated twice for 10 s with 1 W and once for 120 s (with 10 s breaks after every 10 s of sonication) with 4 W in an VC 70 sonicator (Sonics & Materials). Between each sonication cycle the chromatin was centrifuged, resuspended in a fresh B1 buffer and incubated for 15 min on ice. This resulted in fragment sizes between 300 and 600 bp (Supplementary Figure S1). After the last sonication cycle, the chromatin was pelleted, the supernatant was kept and an equal amount of buffer B2 (supplemented with PIs) and 40 μl MyOne T1 Dynabeads were added to it. The mixture was rotated overnight at 4°C and the next day the beads were washed three times with buffer B2. The captured chromatin was eluted by boiling the beads for 5 min in 0.1% SDS. Additional information is available as a Supplementary Protocol.
Immunocytochemistry
The following antibodies were used for immunocytochemistry: anti-CPD (CosmoBio, cat. no. NMDND001), anti-γH2AX (Abcam, cat. no. ab2893), anti-XPG (Novus Biologicals, cat. no. NB100-74611), anti-RSF1 (Abcam, cat. no. ab109002), anti-NIPBL (SantaCruz, cat. no. SC-374625), anti-PCNA (Abcam, cat. no. ab15497). Cells that were grown on coverslips were washed with PBS and were locally irradiated with 30 or 100 J/m2 UVC through a 5 or 8 μm pore polycarbonate membrane filter (Merck, cat. no. TMTP04700, TETP04700) or mock irradiated and left for recovery in the presence or absence of EdU. After recovery the coverslips were washed with ice-cold PBS and incubated in CSK buffer for 5 min on ice. Next, cells were fixed with 4% paraformaldehyde for 10 min and were washed three times with PBS. If co-staining with anti-CPD was to be done, an incubation step in 37°C in the presence of 0.1 N HCl for 10 min was included after the fixation followed by two PBS washes. Then, cells were blocked in 10% FBS or 3% BSA (Sigma-Aldrich, cat. no. A9647), diluted in PBS for 20 min at RT. For EdU staining, the cells were subjected to Click-reaction by incubation in PBS containing 25 μM Alexa Fluor Azide (Thermo Fisher, cat. no. A10270), 10 mM sodium l(+)-ascorbate (Applichem, cat. no. A5048) and 4 mM CuSO4 (Applichem, cat. no. A3327) for 1 h at RT in dark. Cells were washed three times with PBS–Tween (0.05%) and for co-staining with antibodies cells were incubated with the appropriate antibody overnight at 4°C protected from light. Next day cells were washed three times with wash buffer, incubated for 1 h with Alexa Fluor secondary antibody at RT, washed twice, stained with Dapi for 5 min, washed twice again and mounted with Mowiol (Fluka, 81381. polyvinyl alcohol 4–88).
Images were acquired with a LEICA DM2000 microscope equipped with the DFC345 FX camera and pseudocolour was applied using the LAS V4.12 software. Imaging conditions, i.e. exposure time, brightness and contrast remained identical between different conditions. ImageJ was used for the quantification of the signal. The ImageJ images were colour threshold adjusted for grey-scaled dapi staining and then converted to binary. After filling holes, nuclear particles were analysed and added to ROI manager. Red channel was overlaid with regions of interest and Mean Intensity of each nucleus was measured on three photos for each condition.
FACS sorting
Cells were trypsinized, collected and washed with ice-cold PBS. 1.5 million cells were resuspended in 500 μl PBS containing 0.1% glucose and were fixed with 5 ml 70% ethanol for 1 day at –20°C. After rehydration they were shaken for 40 min in the presence of 50 μg/ml propidium iodide and 20 μg/ml RNase A in the dark. Samples were measured in a BD FACS CANTO II flow cytometer (Becton Dickinson) and analysed with BD FACSDiva Software v6.0 (Becton Dickinson).
Dot blot/slot blot
Labelled DNA for input was extracted either as described in Note 8 of the supplementary protocol 1 or by treatment with Proteinase K and extraction with the QIAquick PCR Purification Kit (Qiagen, cat. no. 28104). The captured and eluted DNA was extracted by the MinElute Reaction Cleanup Kit (Qiagen, cat. no. 28204). DNA was quantified with Nanodrop or Qubit and samples were boiled at 95°C for 10 min to denature DNA. Samples were returned onto ice immediately and SSC buffer was added. Single stranded DNA was loaded on a nitrocellulose membrane (Li-Cor, cat. no. 926-31092) and the blots/slots were washed three times with a mixture of 30% SSC and 70% TE. The membrane was baked at 80°C for 1 h, blocked overnight with Odyssey Blocking Buffer (Li-Cor, cat. no. 927–40 000) and incubated with Streptavidin Alexa Fluor (Thermo-Fisher, cat. no. S32357) for 1 h in the dark. The washed membrane was scanned in an Odyssey scanner (Li-Cor).
Western Blot
Samples were boiled for 10 min in a Laemmli sample buffer and loaded in 4–12% gradient polyacrylamide gels (Thermo-Fisher, cat. no. NP0321PK2). The proteins were transferred onto a PVDF membrane (Merck, cat. no. IPFL00010) by electrophoresis at 130 V for 1 h at 4°C. The membrane was blocked with Odyssey Blocking Buffer (Li-Cor, cat. no. 927-40000) for 1 h at RT. Then the part of the membrane corresponding to histones molecular weight was incubated overnight at 4°C with the appropriate antibodies: H2A-ub (Millipore, cat. no. 05-678), H2B (Millipore, cat. no. 07-371), H4 (Abcam, cat no ab10158). Next day. the membrane was washed with PBS–Tween, incubated with the relevant secondary antibodies (Li-Cor) for 1 h at RT, washed with PBS–Tween and scanned in an Odyssey scanner (Li-Cor).
Sample preparation for Mass Spectrometry
Samples were digested following filter-aided sample preparation (FASP) method published by Wisniewski et al. (25). Samples were mixed with 200 μl of 8 M urea in 0.1 M Tris/HCl (pH 8.5) and transferred onto a Vivacon 500 10 kDa MW cut-off filter and centrifuged at a constant 14 000 × g. This step was repeated once more and then the flow-through solvent was discarded. Alkylation step was performed when 100 μl of 1.5 mg/ml iodoacetamide was added and incubated for 20 min in the dark at room temperature. The filter was centrifuged for 10 min. Wash steps were performed with 100 μl of 8 M urea (three times) and 100 μl of 0.05 M ammonium bicarbonate in H2O (three times), each step the filter was centrifuged until dryness. Vials containing the flow-through were exchanged with clean ones before adding 80 μl ammonium bicarbonate buffer containing 1 μg trypsin/LysC (Promega) onto the filter. Trypsin digestion was performed for 16 h at 37°C with shaking. Following digestion, 40 μl of water was added to the filter, and the peptides were eluted by centrifugation for 10 min at 14 000 × g. The part containing the peptides was collected and dried down by speed-vac-assisted solvent removal and reconstituted in a solution of 2% (v/v) ACN and 0.1% (v/v) formic acid. The peptide solution was incubated for 3 min in a sonication water bath. Peptide concentration was determined by Nanodrop absorbance measurement at 280 nm.
2.5 μg peptides were pre-concentrated with a flow of 3 μl/min for 10 min using a C18 trap column (Acclaim PepMap100, 100 μm × 2 cm, Thermo Scientific) and then loaded onto a 50 cm long C18 column (75 μm ID, particle size 2 μm, 100 Å, Acclaim PepMap100 RSLC, Thermo Scientific). The binary pumps of the HPLC (RSLCnano, Thermo Scientific) consisted of Solution A (2% (v/v) ACN in 0.1% (v/v) formic acid) and Solution B (80% (v/v) ACN in 0.1% (v/v) formic acid). The peptides were separated using a linear gradient of 4% B up to 40% B in 210 min with a flow rate of 300 nl/min. The column was placed in an oven operating at 35°C. The eluted peptides were ionized by a nanospray source and detected by an LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) operating in a data dependent mode (DDA). Full scan MS spectra were acquired in the orbitrap (m/z 300–1600) in profile mode with the resolution set to 60,000 at m/z 400 and automatic gain control target at 106 ions. The six most intense ions were sequentially isolated for collision-induced (CID) MS/MS fragmentation and detection in the linear ion trap. Dynamic exclusion was set to 1 min and activated for 90 s. Ions with single charge states were excluded. Lockmass of m/z 445.120025 was used for continuous internal calibration. XCalibur (Thermo Scientific) was used to control the system and acquire the raw files.
Protein identification, quantification and data analysis
The mass spectral files (.RAW files) were processed using MaxQuant software (1.5.8.3). Default parameters were used for protein identification and quantification. Trypsin specificity with two missed cleavages was allowed and minimum peptide length was set to seven amino acids. Cysteine carbamidomethylation was set as fixed, and methionine oxidation, deamidation of asparagine and glutamine and N-terminal acetylation were set as variable modifications. A maximum of five modifications per peptide was set. The false discovery rate both for peptide and protein were set to 5%. For the calculation of the protein abundances, label-free quantification (LFQ) was performed with both ‘second peptide’ and ‘match between run’ options enabled. The complete human database was downloaded from Uniprot 05/17. Statistical analysis was performed using Perseus (version 1.5.3.2) (26). Proteins identified as ‘contaminants’, ‘reverse’ and ‘only identified by site’ were filtered out. The LFQ intensities were transformed to logarithmic and missing values were imputed—replaced by random numbers drawn from a normal distribution. The three replicas were grouped for each set of conditions (treated and control conditions for both experimental set A and B) and a two-sided Student's t-test of the grouped proteins within each experimental set was performed using FDR values for truncation.
RNA sequencing
Total RNA was isolated in duplicate from two 1BR.3 fibroblasts cultures using Trizol (ThermoFisher Scientific). The Agilent RNA 6000 Nano kit with the bioanalyser from Agilent were used for analyzing RNA samples in terms of quantity and quality in the BSRC ‘Alexander Fleming’ Genomics Facility. RNA samples with RNA integrity number (RIN) >7 were used for library construction using the 3′ mRNA-Seq Library Prep Kit Protocol for Ion Torrent (QuantSeq-LEXOGEN™) according to manufacturer's instructions. The DNA High Sensitivity Kit was used along with the bioanalyser for assessing the quantity and quality of the libraries. Next, libraries were pooled and templated using the Ion PI™ HiQ OT2 200 Kit (ThermoFisher Scientific) on Ion One Touch System. Sequencing was performed in BSRC ‘Alexander Fleming’ Genomics Facility using the Ion PI™ HiQ Sequencing 200 Kit and Ion Proton PI™ V2 chips (ThermoFisher Scientific) on an Ion Proton™ System, according to the manufacturer's instructions. Mapping of sequencing reads to hg19 human reference genome was performed using hisat2 (27) and gene expression quantification was performed by the Bioconductor package metaseqR2 (28), using the 3' UTR pipeline with default settings. Genes with at least one read in both replicates were considered actively expressed.
NGS library construction
Biotinylated chromatin was treated with Proteinase K and DNA was extracted by phenol–chloroform. RNA molecules were degraded by treatment with RNAse A. DNA was sheared in a Covaris S2 sonicator and selection of the fragments between 100 and 200 bp was done by running in agarose gel and DNA extraction with the QIAquick Gel Extraction Kit. The biotinylated DNA was isolated by incubation with Dynabeads MyOne Streptavidin T1 magnetic beads for 20 min at RT. The next steps, up to PCR amplification, were done on beads. The ends of the DNA fragments were repaired by incubation with T4 DNA polymerase, Klenow Fragment and T4 DNA Polynucleotide Kinase, then A-tailing was done by incubation with dATPs in the presence of Klenow 3–5 exo and TruSeq adapters were ligated with Quick Ligase. The DNA was amplified in an end-point PCR instrument using the minimum number of cycles for which PCR products could be seen. Finally, the libraries were cleaned with AMPure XP beads. For the libraries of input DNA, the sheared and size-selected biotinylated DNA was subjected to standard Illumina protocols as previously described (29).
NGS sequencing and bioinformatics analysis
Both aniFOUND-seq and input libraries were sequenced as single-end reads at Genecore-EMBL with an Illumina HiSeq 2000 instrument. To analyse the NGS datasets in an automated and reproducible manner, custom bioinformatics pipelines were developed.
Quality control and read alignment
Quality control of raw FASTQ files was performed using fastQC version 0.11.5 (www.bioinformatics.babraham.ac.uk/projects/fastqc/). To avoid read length biases in the downstream analysis (see ‘Differential repair enrichment analysis in the repeated genome’, and ‘Telomeric content enrichment analysis’ methodologies), the reads that were longer than 50 bases were initially trimmed at the 3′ end, until a constant read length of 50 bases was reached for all samples. Remains of adapters were clipped and low-quality bases were trimmed from both ends of the sequenced reads (Phred score 20) using cutadapt version 2.4 version 0.7.12 (30), allowing a minimum of 10 bases per read. High-quality reads were mapped against the UCSC hg19 reference genome using bwa-mem (31) with default settings, and allowing 2 mismatches between the subject and the reference sequences, to account for sequencing errors and SNPs between the 1BR.3 cell line and the sequenced genome.
Read density analysis
To produce average read density profiles, read density heatmaps, read density boxplots and UCSC Genome Browser tracks (32), uniquely aligned and deduplicated reads were used. Specifically, to define a ‘uniquely’ aligned set, hits with mapping quality score less than 10 were filtered out using samtools version 1.9 (33), and chimeric and secondary alignments were filtered out using the ‘XA’ and ‘SA’ tags. Duplicated alignments were eliminated using samtools markdup -r. Replicates were downsampled to the minimum sample read depth per biological condition and merged in order to create a consensus dataset. Genomic annotations of hg19 RefSeq transcription start sites (TSSs) and enhancers were retrieved by UCSC table browser (34) and FANTOM5 project (35), while active and inactive characterization of TSSs, enhancers, and asPROMPTs was applied using previously published RNA Pol II-ser2P, H3K27ac and H3K27me3 ChIP-seq data from VH10 hTERT-immortalized human skin fibroblasts (22,29) and CAGE-seq data (35) from dermal and skin fibroblasts, as described in Liakos et al. (22). All TSSs were extended to 2 kb on each direction, subdivided to 20 bp genomic bins, and were used as a reference for read counting. Heatmaps were generated using seqMINER, while average profiles were generated using custom R scripts and ggplot2.
Boxplots of reads per million values (RPM) of each extended TSS region were generated using custom R scripts, representing RPM distributions of actively transcribed, or inactive regions. To apply per sample comparisons between the active and inactive sets distributions, a permutation strategy was applied. Briefly, for each distribution comparison and for each active/inactive set, 10,000 samplings of 100 data points were randomly generated, and 95% confidence intervals of mean differences between active and inactive regions were calculated. Effect sizes of log2 counts between active and inactive sets were calculated using Cohen's method (CES).
Genome browser-compatible files (bigWig) were generated using deeptools bamCoverage with counts per million normalization (cpm).
Genomic annotation of sequenced reads
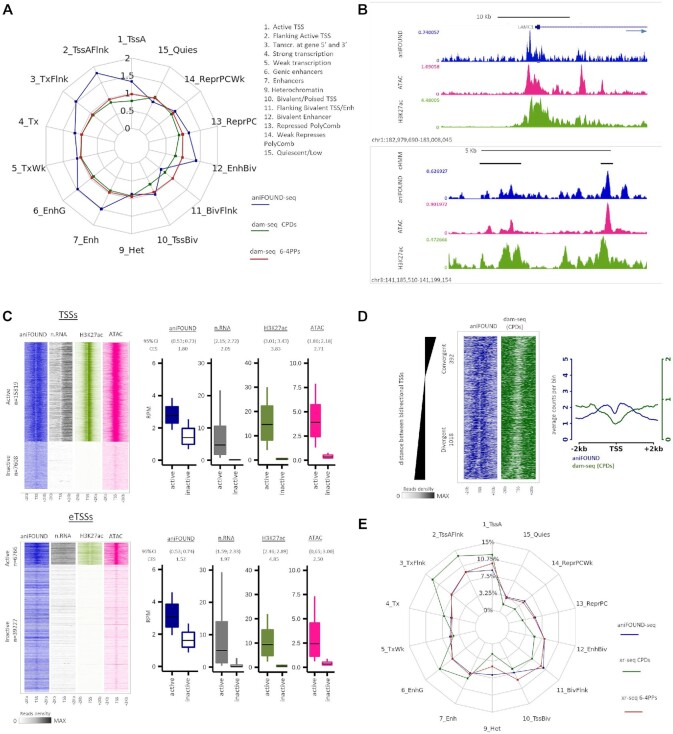
To annotate the aniFOUND-seq, XR-seq and damage-seq samples in respect to their genomic origin, genomic annotations from roadmap epigenomics project were used, and in particular the NHDF-Ad_Adult_Dermal_Fibroblasts core 15-state model, by excluding the 8th chromatin state, ‘ZNF genes & repeats’, since they were analysed in a separate analysis module (see below). For each biological condition, only uniquely aligned and deduplicated reads were used and replicates were merged as described above, and each high-quality alignment was centered and assigned to a unique chromatin state. All resulting region counts were aggregated per chromatin state and normalized using the total counts of each dataset, as also the total genome coverage of each chromatin state. These values were either visualized as ratios, normalized by their corresponding input dataset (Figure 5A), or as percentages of the total annotations (Figure 5E).
Figure 5.
Genome-wide distribution of aniFOUND-seq signal. (A) Repair and damage ratios in different chromatin states. The chromatin states are defined according to the 15-state ChromHMM annotation (see Materials and Methods). Repair ratios are calculated by aniFOUND-seq reads normalized by their input reads for each state. Similarly, damage ratios have been resulted from normalized dam-seq (20) by their inputs (see Materials and Methods). (B) UCSC Genome Browser snapshots of the signals of aniFOUND, H3K27 and ATAC-seq. Upper panel: depiction of a gene TSS and its flanking regions. The direction of transcription is shown by arrow. Lower panel: enhancers located in an area free of genes. The top track (cHMM) shows the ChromHMM states; black bars correspond to enhancers. (C) aniFOUND signal in active and inactive transcription start sites. Left panels: heat maps with the signal of aniFOUND, nRNA, H3K27 and ATAC-seq 2 kb around the transcription start sites of active and inactive genes (TSSs) and enhancers (eTSSs). The designation of human genes in active and inactive state was done as previously described (22). Right panels: Box plots with the signal distributions of the gene sets shown in the corresponding heat maps of the left panel. Boxes show the 25th–75th percentiles and error bars show data range to the larger and smaller values. For each active/inactive set, 10,000 samplings of 100 data points were randomly generated, and 95% confidence intervals of mean differences between active and inactive regions were calculated. Effect sizes of log2 counts between active and inactive sets were calculated using Cohen's method (CES). (D) DNA damage and repair on bidirectional promoters. Left panel: heat maps of aniFOUND and dam-seq around the TSSs of bidirectional genes. The sorting was done based on the distance between the TSSs of the two bidirectional genes (see Materials and Methods). Right panel: Aggregate plots of aniFOUND and dam-seq around the TSSs for the gene sets shown in the left panel. (E) Distribution of aniFOUND and XR-seq signals among chromatin states. For XR-seq all the available data sets up to 4 h after irradiation were merged (5 min, 20 min, 1 h, 2 h and 4 h for the 6-4PPs and 1 and 4 h for CPDs). The states are defined according to the 15-state ChromHMM annotation. For each library the number of reads that correspond to a chromatin state has been corrected for the length of the state. Y-axis shows the percentage of the corrected reads that fall in each state. For the hypothetical library in which all states were equally represented, a polygon with all its sides positioned at around 7% (≈ 100%/14 states) would be resulted. aniFOUND: aniFOUND-seq; dam-seq (6-4 PPs): HS-Damage-seq (6-4 PPs) (0 h after irradiation with 20 J/m2 UVC (20)); dam-seq (CPDs): HS-Damage-seq (CPDs) (0 h after irradiation with 10 J/m2 UVC (20)); nRNA: nascent RNA-seq (2 h after irradiation with 20 J/m2 UVC (29)); ATAC: ATAC-seq (2 h after irradiation with 15 J/m2 UVC (22)); H3K27ac: ChIP-seq of H3K27ac (2 h after irradiation with 15 J/m2 UVC (22)); XR-seq (CPDs): NHF1 CPD XR-seq (merged data sets of 1 and 4 h after irradiation with 10 J/m2 UVC (58)); XR-seq (6-4PPs): NHF1 (6-4)PP XR-seq (merged data sets of 5 min, 20 min, 1 h, 2 h and 4 h after irradiation with 20 J/m2 UVC (58)).
Differential repair enrichment analysis in the repeated genome
To determine potential differential patterns of repair prevalence along the repeated genome, a differential enrichment analysis between the aniFOUND-seq and the input libraries was performed. In summary, quality filtered, adapter-free, and trimmed to the same length FASTQ reads were used as an input to RepeatMasker software. To efficiently run the algorithm, FASTQ files were first converted to FASTA files and were split to 300,000 sequence chunks. RepeatMasker was run with parameters: -e crossmatch -pa 30 -q -low -species human -a -inv -lcambig -html -source -gff -excln -u -nopost to produce pairwise alignment files of repeat elements against the examined fasta sequences, using RepBase (36) and Dfam (37) as repeat species reference. The output of the software was further processed using ProcessRepeats to produce repeat specific annotation files, containing information about the alignment of every repeat species against each sequenced read. All annotation files of each library were summarized to produce a count-like matrix, containing the number of total repeat species occurrences in each of the examined samples. A total of 1279 unique repeat species were identified in all datasets and Counts were summarized to a total of 68 repeat families of origin.
The final {repeat families × samples} count matrix was further processed by DESeq2 (38) to perform differential enrichment analysis. The input dataset was used as a reference sample, and size factors and dispersion were estimated using default settings. A test for significance of coefficients in a negative binomial Generalized Linear Model (GLM), was applied using the abovementioned estimated size factors. Only results with a P-adjusted value threshold lower than 0.05 were reported.
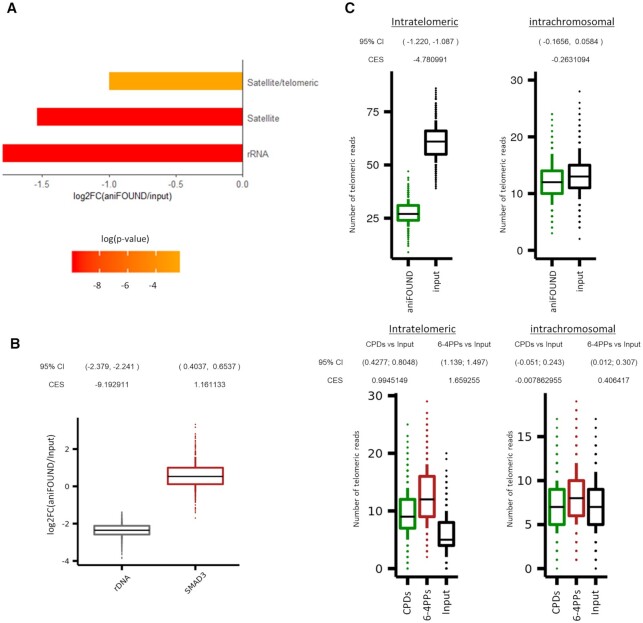
To further examine the degree of NER repair activity at rDNA regions, a differential enrichment analysis between aniFOUND-seq and input datasets at rDNA repeats was conducted as follows. Initially, the hg19 human reference genome FASTA file was extended by adding the 45S pre-ribosomal N5 (RNA45SN5) NCBI sequence (https://www.ncbi.nlm.nih.gov/nuccore/NR_046235) as a new chromosome, using the >NR_046235.3 identifier. High-quality aniFOUND-seq FASTQ reads were mapped against the extended genome index using bwa mem (31) with quality filter -T 0. Low quality and duplicated alignment were not filtered out, and replicates were downscaled to a similar read depth (19,000,000 reads), merged to a single file and sampled 1000 times to produce 100,000 alignment chunks that were in turn summarized at the NR_046235.3 chromosome and SMAD3 gene to produce boxplots of log2 count ratios between the aniFOUND-seq and input datasets. To apply a statistical comparison between the two count distributions for each examined element, 1000 samplings of 100 data points were randomly generated, and 95% confidence intervals of mean log2 differences between PD and INPUT regions were calculated as described in the ‘Read density analysis‘ paragraph. Effect sizes of log2 counts between PD and INPUT sets were calculated using Cohen's method (CES).
Telomeric content enrichment analysis
In order to examine the extent of NER repair and the UVC-induced DNA damage burden on telomere sequences, an analysis pipeline was developed to compare the occurrence of TGAGGG repeats in both aniFOUND and damage-seq libraries and their input libraries. Initially, high-quality reads were mapped against the UCSC hg19 reference genome using bwa-mem with quality threshold -T 0, in order to minimize the fraction of the unmapped reads (31). Both mapped and unmapped reads might contain TGAGGG repeats, but only the unmapped sequences are considered as telomeric (39). Replicates were downsampled to the minimum sample read depth, merged per biological condition, and downsampled to a similar read depth level of 16,000,000 reads. The resulting BAM files were randomly sampled for 1000 times, to generate BAM files of 100,000 reads that were further processed. Specifically, each subsample file was examined for TGAGGG enrichment, using TelomereHunter (39) with default parameters, and without using a control sample. Candidate telomeric reads were classified into three categories: (a) Intrachromosomal reads, which comprise of telomeric repeats that are mapped to the chromosomal regions of the genome, except for the first and last band. The particular regions were considered ‘pseudo’ telomeric and were used as a control set. (b) Subtelomeric reads, which consist of telomeric reads aligned to the first or last band of a chromosome. (c) Unmapped telomeric reads, which were categorized as intratelomeric and were considered as the ‘true’ telomeric content. The outputs of all the telomeric quantifications were summarized, to produce a telomeric content occurrence distribution for each of the ‘true’ and ‘pseudo’ telomeric categories, for each dataset. To compare the intratelomeric and intrachromosomal distributions between aniFOUND or HS-damage-seq datasets, as also their corresponding input libraries, a similar approach to calculate confidence intervals and effect sizes as described in the ‘Read density analysis‘ paragraph was applied. HS-Damage-seq samples used in this analysis include: (i) damage-seq (CPDs): HS-Damage-seq (CPDs) (0 h after irradiation with 10 J/m2 UVC (20), (ii) damage-seq (6-4PPs): HS-Damage-seq (6-4 PPs) (0 h after irradiation with 20 J/m2 UVC (20), (iii) damage-seq (input): NHF1_input (20).
RESULTS AND DISCUSSION
aniFOUND isolates chromatin associated specifically with UVC-induced, unscheduled DNA synthesis
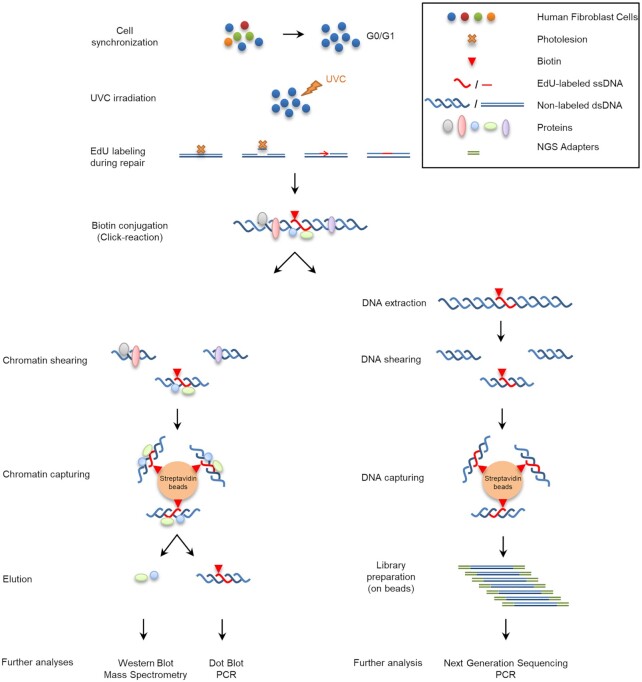
To develop aniFOUND, we exploited UDS, which occurs during the repair of UVC-induced DNA lesions by NER and provides a direct measure of repair efficacy (40). We used 5-ethynyl-2′-deoxyuridine (EdU) and Click chemistry to label and biotinylate the newly synthesized repaired DNA in cells. EdU-labelled native chromatin was subsequently isolated by pull-down with streptavidin and subjected to high-throughput analyses, such as mass spectrometry and NGS as illustrated and described in detail in Figure 1. The complete elimination of active DNA synthesis apart from NER-derived UDS is a key step in aniFOUND. As replication-associated DNA synthesis is the major contributor to nucleotide incorporation into DNA when cells are not synchronized outside of S phase, we arrested human skin fibroblasts in G0/G1 by both contact inhibition and serum starvation (Supplementary Figure S2). Additionally, to sufficiently diminish replication-derived DNA synthesis from the fraction of the cells that still escape to the S-phase, cultures were treated with 10 mM hydroxyurea (HU) during the DNA labelling step (Supplementary Figure S3 and S4). We found no effect of HU at this concentration on DNA synthesis during NER (EdU incorporation, Supplementary S5) in line with earlier findings (41). Under these conditions, UVC-induced UDS was the major source of detectable DNA synthesis (Supplementary Figure S4). By using established protocols (42), we confirmed that, under our conditions, only newly synthesized DNA undergoing NER was labelled, as labelling occurred specifically at the sites of locally-induced DNA damage. This is depicted in Figure 2A, illustrating the colocalization of cyclobutane pyrimidine dimers (CPDs)—a type of UV-induced lesion—and EdU. We further verified that labelled nucleotide incorporation did not occur in the NER deficient XPA cells, as shown in Supplementary Figure S6.
Figure 1.
Schematic depiction of aniFOUND. An asynchronous population of fibroblasts is synchronized to G0/G1 phase by serum starvation and contact inhibition. DNA photolesions are formed by UVC-irradiation and left to be repaired in the presence of EdU. HU is added in this step to eliminate the replication of any escaper cells. Since no replication occurs, UDS is the only source of DNA synthesis. Biotin molecules are conjugated to EdU, the chromatin or the extracted DNA is sheared and the biotin-labelled fragments are isolated with streptavidin beads. The captured material can be used for several further analyses.
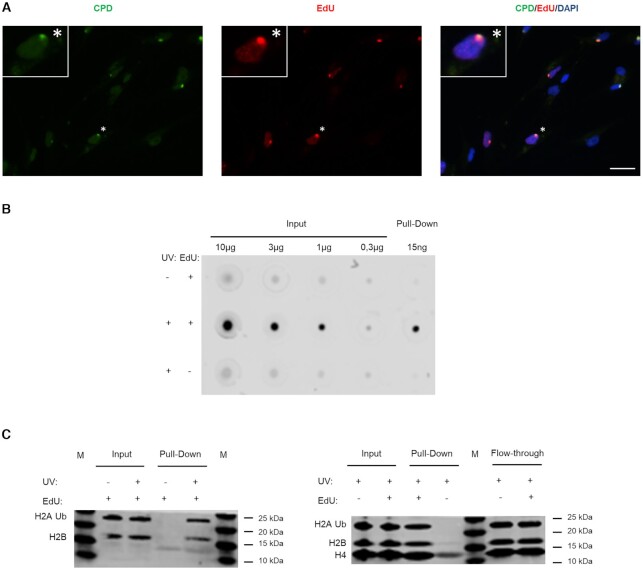
Figure 2.
Specific labelling and isolation of the UDS-associated chromatin. (A) 1BR.3 fibroblasts were locally irradiated with 100 J/m2 UVC using 8 μm pore filters and incubated for 4 h in no serum medium containing EdU and HU. The coverslips were incubated with Alexa-azide for labelling the incorporated EdU, and with anti-CPD. Scale bar: 25 μm. (B) VH10 fibroblasts were synchronized for aniFOUND and were UVC- or mock-irradiated with 30 J/m2. They were kept in the presence of HU and in the presence/absence of EdU for 4 h. Next, cells were subjected to aniFOUND and the DNA from the isolated material (as well as from input) was extracted, quantified and immobilized on a nitrocellulose membrane. The membrane was incubated with Streptavidin-Alexa for detecting the incorporated EdU. C1 Dynabeads were used for this experiment. The blot was cropped to advance clarity. (C) Fibroblasts were synchronized for aniFOUND and then were either UVC- or mock-irradiated with 30 J/m2 and kept for 4 h in the presence of EdU and HU (1BR.3 fibroblasts; left panel) or were irradiated and kept for 4 h in the presence of HU and in the presence/absence of EdU (VH10 fibroblasts; right panel). Next, cells were subjected to aniFOUND and the isolated material was used for western blot. Material from 1 million cells was loaded for inputs while for the pull-down the whole amount was loaded. The membrane was labelled with the indicated antibodies using two discriminable secondary antibodies with different wavelengths.
We then used the above cell synchronization protocol to specifically isolate UDS-associated chromatin under native conditions. The experimental pull down of interest, termed aniFOUND-UVC, was performed with cells that had been UV-irradiated and left to recover in the presence of EdU (+UV/+EdU). We employed a short range of UVC irradiation doses (20–30 J/m2) to induce sufficient amounts of DNA damage for the subsequent analyses while allowing the majority of cells to recover from the stress. The EdU concentration (10 μm) that we used enabled efficient labelling of UDS-specific nucleotide incorporation and nuclear fluorescent intensity (Figure 2, Supplementary Figures S4 and S6), in agreement with a previous report (40) and it is considered the optimal EdU concentration for Click chemistry-based enrichment protocols (3,4). To distinguish any non-specific material, we used two different negative controls. A non-irradiated but labelled control (–UV/+EdU) to distinguish any newly synthesized but non-UDS-derived chromatin and an irradiated but non-labelled control (+UV/–EdU) to identify any non-specific binding on the beads. In both experimental sets, the isolated aniFOUND-UVC chromatin (+UV/+EdU) was found to be highly enriched in EdU-labelled nucleotides in comparison to the two negative controls –UV/+EdU and +UV/–EdU (Figure 2B), validating the specificity of the purification procedure for unscheduled, non-replicative, newly synthesized DNA. The aniFOUND-UVC chromatin was enriched for core histones (Figure 2C), including the damage-associated ubiquitinated form of H2A (43). Taken together, our findings show that the isolated material is appropriate for both proteomic and genomic analyses.
aniFOUND coupled to proteomic analysis
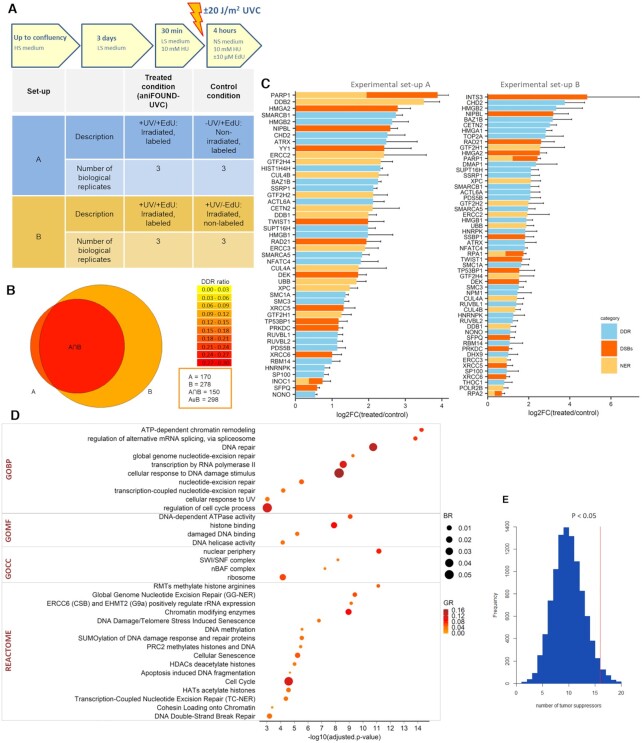
We first employed mass spectrometry (aniFOUND-MS) for the isolation and identification of aniFOUND-UVC-associated proteins. aniFOUND-MS is described in detail in Supplementary Protocol 1. We carried out two experimental set-ups, set A and set B, with identical treatment condition (+UV/+EdU: 20 J/m2 of UVC irradiation and 4 hour-long recovery in the presence of EdU, which we call aniFOUND-UVC) (Figure 3A; Supplementary Figure S7) but different non-treated control conditions, as described in the previous paragraph. For each of the two experimental set-ups, we conducted three biological replicates for the aniFOUND-UVC samples and the corresponding control samples. The isolated chromatin samples were analysed separately by protein-mass spectrometry following a label free quantification protocol. By carrying out two separate statistical analyses (one for each set-up), we found two protein lists significantly enriched in the aniFOUND-UVC samples (+UV/+EdU) compared to the two negative controls. To take full advantage of the outcome of both sets, the protein lists were merged and all proteins enriched in any of the two negative control conditions (statistically significant or not in A [–UV/+EdU] or B [+UV/-EdU]) were filtered out. This resulted in 182 and 308 proteins for set A and set B, respectively. The union of the two lists gave 323 enriched proteins, constituting the aniFOUND-UVC protein list, while the intersection gave a strict aniFOUND list consisted of 167 proteins. Both lists contain a considerable fraction of known DDR factors (Figure 3B; Supplementary Table S1; Supplementary Figure S8).
Figure 3.
Characterization of the proteins resulted by aniFOUND-MS. (A) Description of the two experimental sets that were used for the aniFOUND-MS. 1BR.3 fibroblasts were treated as shown in the upper panel. The conducted experiments are described in the lower panel. HS: High serum-containing medium (10%); LS: low serum-containing medium (0.5%); NS: no serum medium. (B) Venn diagram for the identified proteins of the two experimental sets. The areas of the circles are proportional to the number of the identified proteins. The colour of each compartment represents the ratio of the identified DDR-related genes (DDR ratio). (C) Proteins of both experimental sets that are significantly enriched in the treated conditions (two-sided Student's t-test of the grouped proteins was performed using FDR values for truncation; FDR < 0.1; s0 > 0.1) and fall under DDR-related GO terms. On the x-axis the log2-fold change and standard errors of the treated compared to control condition are shown (in each experimental set the treated condition has been compared to its coupled control condition). With yellow are shown the proteins that are annotated to ‘nucleotide excision repair’ (GO:0006289), with red the proteins that are annotated to ‘Double strand break repair’ (GO:0006302) and with blue the ones that are annotated to DDR-related GO terms. (D) Gene Ontology and Reactome terms that are overrepresented in the aniFOUND-UVC protein list. As background universe all the expressed genes in the 1BR.3 cell line, as resulted by RNA-seq analysis (see Materials and Methods), were used. Gene Ontology: BP (Biological Process); CC (Cellular Component); MF (Molecular Function). (E) Frequency histogram of randomization tests for the number of tumour suppressors (according to OncoKB (46)) that are contained in random lists of equal size to the strict aniFOUND list and are annotated to GOCC ‘chromatin’ (GO:0005717). The randomization test was conducted 10,000 times. The red line shows the number of tumour suppressors contained in the strict aniFOUND protein list (observed value) resulted from the intersection of experimental sets A and B. P-values were calculated as the ratio of tests that resulted in values equal or greater than the observed value divided by the total number of tests (*P< 0.05).
The aniFOUND-UVC protein list was significantly enriched in nucleus- (GO:0005634; P = 8.44E−66) and chromatin-associated proteins (GO:0000785; P = 7.36E−20), as expected (Supplementary Table S2). In contrast, the proteins enriched in the two negative control conditions, were enriched for proteins such as cytoplasm- (GO:0005737; P = 4.05E−11) and mitochondria-associated proteins (GO:0005739; P = 2.08E−06).
The specificity of the aniFOUND-MS method was further validated by comparison to the results of two relevant mass spectrometry studies. More specifically, 42% of the proteins in the aniFOUND-UVC list were present in the list of proteins identified by a multiomic analysis of transcription-related DDR of Boeing et al. (13). From these 11.8% are annotated to the GO term ‘Nucleotide excision repair’ and 25.7% to DDR-related GO terms. Moreover, 28.2% of our proteins were enriched on DNA lesions induced by cisplatin (14), a chemotherapeutic that induces damage repaired mainly by NER (20.9% of the common proteins between the two lists is annotated to DDR-related GO terms).
aniFOUND-MS results in enrichment of DDR-related and other proteins and pathways
Gene ontology analysis of the aniFOUND-UVC protein list revealed proteins that are annotated to GO terms related to DNA damage responses (18.3%; 59 proteins) and to GO ‘nucleotide-excision repair’ (5.3%; 17 proteins). It further identified proteins annotated to GO term ‘double-strand break (DSB) repair’ (5.3%; 17 proteins) (Figure 3C) in line with previously reported findings derived from different omics strategies (13). Given that HU induces DSBs in replicating cells, the fact that we have minimized the S-phase fraction and we have also treated the control conditions with HU, makes rather difficult the rare HU-derived DSB events to give rise in statistically significant DDR factors. It should be noted that these numbers of proteins correspond strictly to the ones annotated under the respective GO terms and do not necessarily reflect all proteins that are mentioned in the literature to be implicated in DDR, NER or DSBs.
All the significant ones as well as the most relevant to DDR terms that resulted from gene ontology and pathway overrepresentation analysis of the aniFOUND-UVC protein list are depicted in Figure 3D and Supplementary Table S2. A parallel ontology analysis performed on the clusters of highly interconnected proteins confirmed that the identified factors can be divided into four categories, with the largest associated with chromatin organization and DNA repair (Supplementary Figure S9; blue cluster). An outline of the epigenetic role of proteins associated with the UDS-chromatin is further presented in Supplementary Figure S10.
The aniFOUND-UVC protein list was also enriched in several protein complexes according to the CORUM database. For example, complexes related to ‘cellular response to DNA damage stimulus’ (GO:0006974) and ‘DNA repair’ (GO:0006281) were identified, including complexes containing DDB1/2/Cullin4A, ERCC2/ERCC3, RUVBL1/2 or XRCC5/6 (Supplementary Table S3). Of note, the last two groups of complexes are principally reported to be associated with DSBs, providing support to the notion that a number of protein complexes participate in both DSB- and UVC-related DDR. In addition to the core DNA damage and repair complexes, aniFOUND uncovered whole protein complexes that are involved in other relevant biological processes, such as the reorganization of chromatin and the post-translational modification of proteins. All these complexes are presented, along with citations documenting their implication in DDR, in Supplementary Table S3.
By searching for clusters of highly interconnected proteins in the protein-protein interactions (PPI) networks built on the aniFOUND-UVC protein list we identified putative clusters consisting of nuclear pore complex (NPC) proteins (Supplementary Figure S11A) and others consisting of ribonucleoproteins (Supplementary Figure S11B). Both classes are implicated in DDR and linked to tumorigenesis, but very little is known about their putative role specifically in UV damage repair. In support of our findings, a recent study showed that Nup84 mutant strains, a component of the yeast nuclear pore complex and homologue of human Nup107 (present in aniFOUND-UVC list), display NER defects and increased UV sensitivity during S phase (44).
Taking into account that UV irradiation and NER are relevant to mutagenesis that may lead to cancer (10,12,45), we further explored whether aniFOUND-MS can be a practically useful tool in providing information related to cancer research. Indeed, the strict aniFOUND-UVC protein list comprise significantly more tumour suppressor proteins [as defined by oncoKB (46)] than random chromatin-associated proteins lists (Figure 3E). By searching for highly interconnected clusters between aniFOUND proteins and melanoma driver genes in PPI networks, we find that proteins such as TP53 as well as ARID2, which is part of the BAF-complex, may also be of high importance in UV-induced melanoma carcinogenesis (Supplementary Figure S12).
Identification and validation of novel UVC-induced DDR players
To experimentally validate the recruitment of aniFOUND-MS proteins on repaired nascent chromatin or in its vicinity, we induced local UV damage (LUDs) in cells and tested the localization of selected candidates by immunofluorescence (47). Here, we present evidence for the recruitment of two of the proteins that have not been reported to be implicated in NER, remodelling and spacing factor 1 (RSF1) and nipped-B-like protein (NIPBL) (Figure 4; Supplementary Figure S13).
Figure 4.
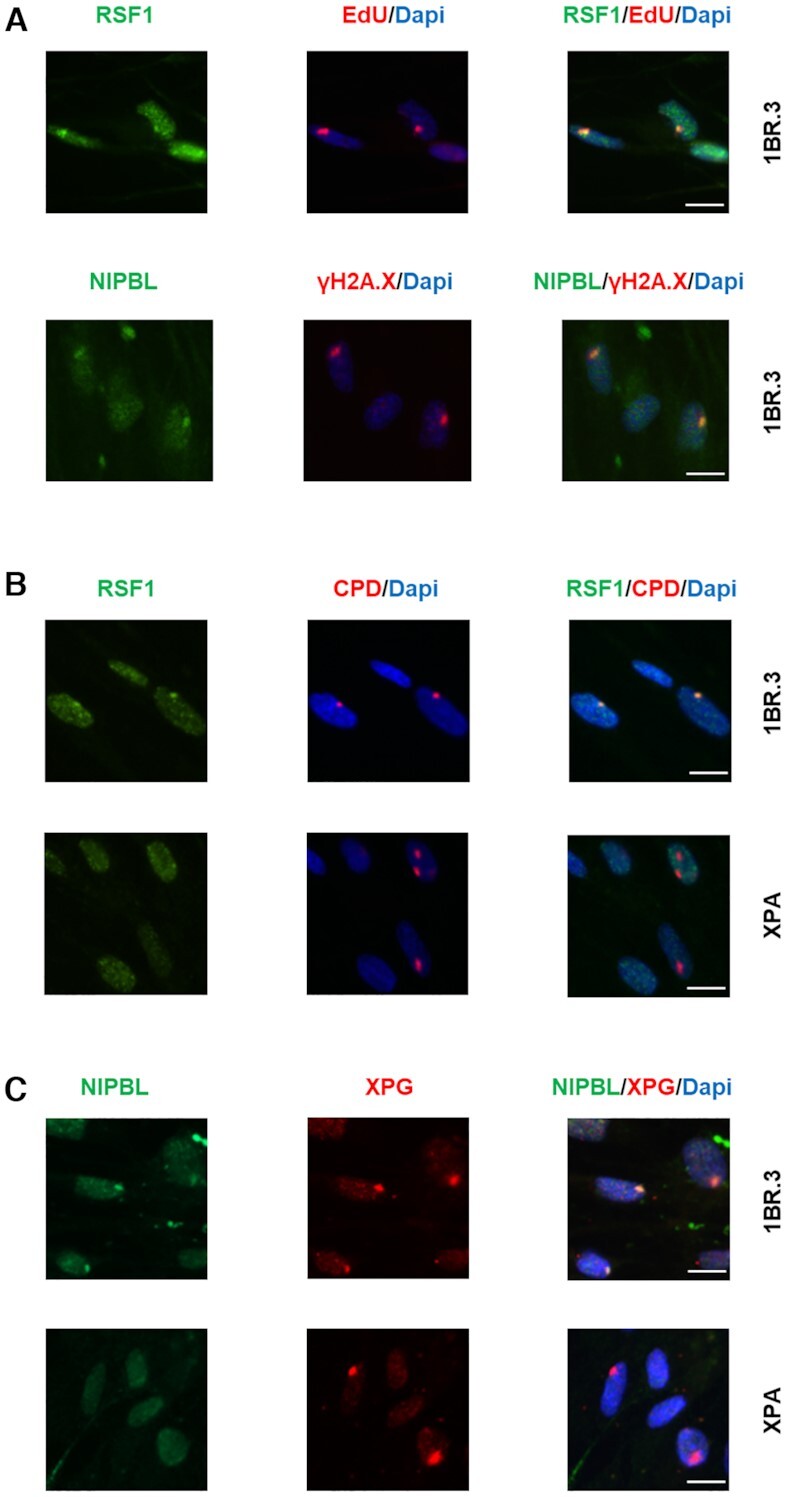
RSF1 and NIPBL are recruited to sites of UV damage. (A) RSF1 and NIPBL are recruited at sites of UV damage in wild type cells upon 30 J/m2 irradiation. Fibroblasts were locally irradiated using 5 μm pore filters and left for 4 h in no serum containing medium with HU (10 mM) and with (upper panel) or without (lower panel) EdU. After fixation cells were labelled with Alexa-azide and anti-RSF1 (upper panel) or with anti-γH2A.X and anti-NIPBL (lower panel). (B, C) RSF1 (B) and NIPBL (C) is recruited at the sites of DNA damage in wild type cells (upper panel) but not in XPA deficient cells (lower panel) upon 100 J/m2 UV irradiation. Fibroblasts were locally irradiated using 5 μm pore filters and left for 1 hour in no serum containing medium with HU (10 mM). After fixation cells were labelled with anti-CPDs and anti-RSF1 (B) or with anti-XPG and anti-NIPBL (C). Scale bar: 12.5 μm.
RSF1 is a chromatin-remodelling factor frequently overexpressed in a number of cancers; it is shown to participate in DSB-DDR and to accumulate at DSB foci (48,49). RSF1 is reported to form complex with SMARCA5 (also present in the aniFOUND-UVC protein list), which is a member of the SWI/SNF remodelling factors and is implicated in NER-related DDR (50). NIPBL is one of the causal genes (mutated in 60% of cases) of Cornelia de Lange Syndrome (51), a rare developmental disorder with characteristic facial features, stunted growth, and mental retardation, as well as multiple other systemic abnormalities. NIPBL is essential for cohesin loading to chromatin and has been detected at DSBs (52,53). Notably, the core members of cohesin (SMC1A, SMC3 and RAD21), which are also known to participate in DDR, and specifically in NER-DDR in Caenorhabditis elegans (54), as well as certain regulatory proteins (STAG2 and PDS5B) were also present in the aniFOUND-UVC protein list (55). Recently, based on transcription-associated analyses, NIPBL was suggested as a Cockayne Syndrome marker (56). Nevertheless, to the best of our knowledge, no evidence exists for the recruitment of either RSF1 or NIPBL at sites of UV photolesions. Figure 4A illustrates the recruitment of RSF1 and NIPBL at LUDs (defined by EdU or γH2A.X staining) in NER-proficient (wild type- WT) cells at 4 h post-UVC irradiation (30 J/m2). This recruitment is not dependent on HU (Supplementary Figure S13). Next, we examined RSF1 and NIPBL accumulation at LUDs in both WT (1BR.3) and Xeroderma pigmentosum A (XPA) cells, which carry mutations in the XPA gene, an early core NER factor required for NER pre-incision complex assembly. Cells were locally UVC irradiated (with 100 J/m2, a dose high enough to ensure the detection of DDR factors at the damage spots) and left to recover for 1 h (Figure 4B and C). In these experiments, LUDs were defined by anti-CPD staining or by staining for XPG, a core NER structure-specific endonuclease, which is assembled at NER complexes independently of XPA. Figure 4B illustrates RSF1 and CPD colocalization in 1BR.3 cells, but no recruitment of RSF1 at LUDs in XPA-deficient cells. In line with these results, it was reported that lack of RSF1 confers weak sensitivity to UVC (48). Similar to the picture seen with RSF1, NIPBL colocalized with XPG in WT, but not in NER-deficient cells (Figure 4C). Taken together, our data show that RSF1 and NIPBL associate with UDS-enriched chromatin in response to UV irradiation and are likely involved in NER-DDR in human cells. We further provide important insights into their spatio-temporal involvement in the cascade of NER events, as their dependence on XPA suggests that they are recruited at a later step of the process, probably after the incision of the damage-containing oligonucleotide has taken place.
aniFOUND is coupled to genomic analysis
In parallel to MS, we coupled aniFOUND to NGS to map the landscape and dynamics of DNA repair/synthesis via NER. We termed this protocol ‘aniFOUND-seq’ (Figure 1, Supplementary Protocol 2). Serum starved human fibroblasts were irradiated, left to recover for 4 h in the presence of EdU and HU and the nascent DNA was biotinylated, as described for aniFOUND-MS. Next, the DNA was extracted, fragmented, and the labelled fraction was isolated by streptavidin pull-down, enabling high-stringency purification. On-bead NGS library construction was performed and NER-associated DNA was sequenced and mapped to the genome. We also isolated DNA from control samples (–UV/+EDU, as described above), however the amount was negligible.
To obtain information on the genome wide distribution of loci associated with UVC-induced UDS, we calculated the overlap of aniFOUND-seq sequences, obtained from two experiments (Supplementary Figure S14), with chromatin states defined by the 15-state ChromHMM annotation (57) (Figure 5A). We excluded from this analysis repeat sequences, as they were analysed separately (see below). In contrast to the rather uniform formation of the UV-induced photolesions (CPDs and 6-4PPs) immediately after irradiation, as mapped by the high-sensitivity damage sequencing methodology (20) (HS-Damage-seq) (Figure 5A, HS-damage-seq), the aggregated UVC-UDS activity was unequally distributed to the different chromatin states (Figure 5A, aniFOUND-seq; Supplementary Figure S15A). Active transcription start sites (TSSs) and their flanking regions (states 1, 2 and 3), as well as enhancer-associated regions (states 6, 7 and 12) showed elevated repair-synthesis. These results suggest faster NER-gap filling activity during the 4-h recovery period in actively transcribed genes and regulatory regions compared to repressed and quiescent regions. Our data are in agreement with and complete previous reports showing how NER activity is implemented with altered speeds in different genomic regions (20,21).
We next analysed the repair patterns revealed by aniFOUND-seq specifically around TSSs (see Materials and Methods). Inspection of the aniFOUND-seq signal and comparison with ATAC-seq, and H3K27ac ChIP-seq data (22,29) highlighted enhanced levels of UDS at regions with increased chromatin accessibility (22), in particular, at TSSs and the flanking regions of actively expressed genes (protein-coding and lnc-RNA genes, Materials and Methods) (Figure 5B; Supplementary Figure S15B, upper panel) and at highly accessible enhancer TSSs (eTSSs) (22) (Figure 5B; Supplementary Figure S15B, lower panel). Similarly, as illustrated in Figure 5C and Supplementary Figure S15C, the aniFOUND signal was clearly prominent around active TSSs and eTSSs compared to inactive ones (subsets of active/inactive genes/enhancers defined in Materials and Methods and as described previously by Liakos et al. (22)). We note a characteristic pattern more pronounced at TSSs (Figure 5C), matching previously observed global increases of nascent-RNA reads in both the sense (mRNA) and antisense (PROMPT - PROMoter uPstream transcripts) directions and confirming that NER occurs quickly and efficiently at open and actively transcribing regions (22). Nonetheless, a lower but detectable UDS signal was also detected at non-transcribed regions during the first 4 h of recovery (Figure 5C, signal not detected in nascent RNA- and H3K27ac-seq; Supplementary Figure S15C).
Given that the underlying distribution of DNA damage would affect the localization of DNA repair-synthesis events, we further plotted the UDS signal along with the damage signal (HS-Damage-seq data (20)) around active TSSs of bidirectional transcripts (Figure 5D; Supplementary Figure S15D). Active bidirectional promoters (TSSs of active mRNA-mRNA pairs transcribed in the opposite direction) were sorted according to their InterCage distance between the sense and antisense TSSs (see Materials and Methods; (22)), and further categorized as convergent (when overlapping transcription occurred) or divergent (when overlapping transcription did not occur) (Figure 5D; Supplementary Figure S15D; left panels). Using this set-up, we found that the observed UVC-UDS pattern around TSSs (Figure 5C) follows the shape of UV-damage formation. The same is also seen in the aggregate plot where the valley in the UDS curve coincides with the absence of UV-lesions (Figure 5D; Supplementary Figure S15D; right panels). Taken together these results confirm that aniFOUND-seq can exclusively isolate and map in high resolution newly synthesized chromatin patches at sites of UV lesions that have undergone NER.
Therefore, we next thought to relate NER-synthesis events (aniFOUND-UVC) with NER excision events as established by XR-seq (21,58) (Figure 5E; Supplementary Figure S15A). To do this, we took into consideration the particular features of the two methodologies, XR-seq and aniFOUND-seq: (a) aniFOUND maps the newly synthesized DNA in chromatin after repair has taken place, while XR-seq maps the damaged DNA fragments excised during the early steps of NER; (b) aniFOUND cumulatively captures repair-synthesis events, while XR-seq captures 10-min-long excision activity; (c) aniFOUND captures total UDS activity, which is associated with the repair of both CPDs and 6-4 PPs, whereas XR-seq focuses on one type of lesion per experiment. In view of these differences and to make aniFOUND-seq and XR-seq data as comparable as possible, we merged the available XR-seq datasets corresponding to different time-points (Figure 5E, from 5-min to 4-h recovery) of 6-4PP repair (58). Similarly, for CPDs, we merged the two available time-points (1 and 4 h). The spider plot (Figure 5E) demonstrates that the distribution of UDS signal (aniFOUND-seq) in different ChromHMM states (57) is very similar to that of 6-4PP signal (21), in line with the fact that the majority of 6-4PPs are repaired during the first 4 hours after damage induction (Figure 5E). On the other hand, the elevated levels of XR-seq CPD signal compared to aniFOUND-seq signal in states related to active transcription (states 1, 2, 3, 4, 6 and 7) and the reduced levels of CPD repair in repressed chromatin and heterochromatin (states 9–14) are in agreement with CPDs being preferentially repaired by TC-NER early in the post-UV recovery process (21,58). Focusing on the repair activity around TSSs and eTSSs, we find that during the first 4 h of repair both methods detect increased signal in active regions, contrasting with lower repair in inactive loci (Supplementary Figure S16). We thus conclude that aniFOUND-seq, which has major differences and unique features in comparison to the established method, is a valid and fast method to accurately map the cumulative pattern of DNA repair-synthesis events genome-wide.
aniFOUND-seq estimation of NER efficacy on repetitive sequences
We next used the aniFOUND-seq data to study UDS activity at repetitive sequences, a considerable and under-explored part of the genome. We annotated the reads of both aniFOUND-seq and input libraries to repeat elements according to RepeatMasker (www.repeatmasker.org). Next, we calculated differential enrichment between aniFOUND-seq and input for all annotated repetitive families (see Materials and Methods) to find the repeat families preferentially or not subjected to UDS during the 4-h post-UV recovery. Although a small number of repeat families showed some gain in aniFOUND-seq signal, this was found to be insignificant (P-adjusted value threshold higher than 0.05, Materials and Methods). Figure 6A (and Supplementary Figure S17A) depicts only the repeat families that are significantly differentially represented in aniFOUND-seq libraries (P-adjusted value threshold lower than 0.05). This analysis revealed rDNA to be less frequently repaired compared to the genome overall and thus we decided to further validate the underrepresentation of these repeat elements by implementing an rDNA-centric analysis. We rebuilt the hg19 human reference genome by adding a single copy of the 45 S pre-RNA sequence and compared the content of ribosomic reads between the repaired fraction (pull-down) and the genome (input). We also included the SMAD3 gene in this analysis pipeline, as a genomic area that is transcribed and thus expected to be over-represented in the repaired fraction. In Figure 6B (and Supplementary Figure S17B), it is shown that ribosomes are under-represented in the repair fraction, further supporting the results of Figure 6A. There are contradictory findings in the literature concerning rDNA repair: although recent work reported that rDNA is not subject to TC-NER (59), another report showed considerable repair of rDNA via TC-NER during the first 3 hours of recovery and attributes this to the relocation of damaged rDNA to the nucleolar periphery, where access to the repair machinery is enabled (60). In contrast, some studies assert that rDNA is slowly repaired, possibly due to insufficient access to damaged DNA by repair factors (61,62). Our data reveal a slow cumulative repair-synthesis of rDNA during the first 4 h after damage induction.
Figure 6.
aniFOUND-seq on repeats. (A) Differentially represented repeat families between aniFOUND-seq and input libraries. The bars show the log2 ratio of the aniFOUND-seq library reads over the input library reads that are annotated to the same repeat family. The repeat families are defined according to the classification system of Repbase. The color of each bar denotes its adjusted p-value. Only families with an adjusted P-value lower than 0.05 are shown. (B) Distributions of mapped read ratios on rDNA and SMAD3 gene between aniFOUND and input libraries. On Y-axis, the logarithmized fold change of 1000 random samples is shown. For each random sample the reads were aligned on an extended reference genome consisting of the UCSC hg19 and a single copy of the human rDNA (NR_046235) sequence (see Materials and Methods). Effect sizes refer to the difference from zero of the distributions depicted by the box plots and were calculated by using Cohen's method (CES). (C) Random samples of UDS (upper panel) and DNA damage (lower panel) signal on telomeres as estimated with aniFOUND-seq and damage-seq, respectively (see Materials and Methods). Y-axis shows the number of telomeric reads that resulted from 1000 TelomerHunter runs on samples with 100,000 alignments each (see online methods). For both aniFOUND-seq and damage-seq, pull-down and input libraries have been plotted. 95% Confidence Intervals (95% CI) of log2 differences between pull-down and input libraries were calculated using 10,000 samples of 100 data points from each examined library. Effect sizes were calculated using Cohen's method (CES). damage-seq (CPDs): HS-Damage-seq (CPDs) [0 h after irradiation with 10 J/m2 UVC (20)]; damage-seq (6-4PPs): HS-Damage-seq (6-4 PPs) [0 h after irradiation with 20 J/m2 UVC (20)]; damage-seq (input): NHF1_input (20).
Telomeres are typical repetitive regions consisting of tandem 6 nucleotide-long sequences. Even though both their vulnerability to DNA damage and the cell's ability to repair them are tightly linked to aging and cancer, it is not yet clear whether they are prone to UVC and if they are repaired by the cell to the same extent as the rest of the genome. We used TelomereHunter, an established bioinformatics tool (39), to count the telomeric reads in aniFOUND-seq and input libraries as an indirect estimation of telomeric repair through determination of the telomeric content. In Figure 6C (upper panel) (and Supplementary Figure S17C), it is clear that telomeric reads are under-represented in aniFOUND-seq showing that telomeres are subjected to UVC-derived UDS at significantly lower frequency compared to the rest of the genome. In contrast, the intrachromosomal telomeric reads (telomeric-like reads that are mapped at chromosomal regions far from chromosome ends) do not differ between aniFOUND-seq and input libraries.
In order to examine whether this decreased activity could arise from lower damage burden, we performed the same analysis on HS-damage-seq datasets at 0 hours after irradiation (20). We found that the damage burden (both CPDs and 6-4PPs) is higher in telomeric regions compared to the overall genome (Figure 6C; lower panel). Again, there was either no significant difference or a considerable smaller size effect in the intrachromosomal telomeric reads, for CPDs and 6-4PPs, respectively. There are two models for damage formation and repair of bulky adducts on telomeres. As stated by the first, telomeres are susceptible to UVC-induced DNA damage, but damage removal is almost absent (63); according to the second, telomeres are partly protected from UVC, and CPD and 6-4PP removal is respectively faster and equal, compared to the rest of the genome (64). Our data reveal that under our experimental conditions, the UVC-induced UDS at telomeres during the first 4 h after irradiation is lower than in the rest of the genome and that this reduced repair-synthesis activity is not due to reduced occurrence of DNA damage.
DISCUSSION/CONCLUSION
Here, we report the development of aniFOUND, a new method for the specific capture of chromatin areas with well-defined origin. aniFOUND is a comprehensive and high-resolution method that can integrate proteome characterization and genome-wide mapping of chromatin loci associated with NER-derived non-replicative DNA synthesis, providing a thorough understanding of UVC-DDR mechanisms in chromatin. To the best of our knowledge, this is the first attempt for the isolation and study of the chromatin fraction associated with UDS.
We coupled aniFOUND to MS analysis for the identification of histone and non-histone proteins that reside in UDS-associated chromatin and relied on native conditions to remove proteins indirectly or non-specifically binding to crosslinked chromatin. Importantly, aniFOUND is an antibody-free approach, specifically addressing the non-replicative chromatin synthesis, as it eliminates the technical challenges deriving from potentially limited abundances of peptide epitopes and it prevents purifying unrelated macro-complexes via target proteins often known to play multiple biological roles (2). We identified a set of 323 proteins that describe the UVC-UDS’ome’. The aniFOUND-UVC list contains proteins and protein complexes that can be divided into three main categories: those with a known role in UVC-DDR (both TC-NER and GG-NER), those implicated in DDR, and a novel subset of potentially important missing players with no reported role in DDR. We experimentally validated the recruitment of two proteins, RSF1 and NIPBL, from the second category at sites of NER-derived UDS. Our findings underscore the efficacy of aniFOUND-MS to describe in great detail the fraction of the proteome associated with repaired chromatin, and highlights previously unidentified components involved in UVC-DDR and the restoration of UVC-damaged chromatin.
Furthermore, the labelled DNA was subjected to aniFOUND-seq, a novel protocol for deep sequencing of the repaired DNA segments. We applied aniFOUND-seq to map repair-synthesis activity genome-wide, with particular emphasis given to promoters, enhancers and repeats. We specifically designed analysis pipelines (Materials and Methods) for the assessment of NER-UDS activity in specific chromosomal regions. Repair efficacy during the first 4 h after damage induction was assessed for rDNA and telomeres, for which contradictory explanatory models have been suggested. As far as we know, this is the first time that NGS-based approaches have been adapted to tackle these issues in telomeric DNA. Thus, aniFOUND’s unbiased (antibody-free) manner of detecting DNA repair-synthesis activity may offer advantages for refining the spatio-temporal understanding of genome maintenance mechanisms requiring UDS after damage.
We thus consider aniFOUND a very powerful and versatile research tool for the in-depth description of UDS-associated chromatin. Its flexible nature (in terms of both damage types detected and the potential repair assessment period) renders it suitable for capturing of the whole repair activity during shorter or longer time windows, thus allowing alternative perspectives of repaired/synthesized chromatin to be captured and analysed. aniFOUND has a potentially broad spectrum of applications, as genotoxic factors other than UVC (e.g. chemotherapeutics) can be used for triggering and analysing NER-DDR. In this regard, differences in the DNA repair efficiency per se and its spatiotemporal distribution between tumour cells with diverse chemotherapeutic resistance could be easily scrutinized. Furthermore, the development of aniFOUND was based on the labelling of short nascent DNA fragments generated following excision of the damage-containing DNA; given the ongoing need for methods suitable for studying pathways that could benefit from UDS labelling (65), our method can be an appropriate starting point in this direction. In the broader perspective the extendable nature of aniFOUND makes it a precious basis for tools dedicated in studying biological processes associated to every type of nascent chromatin that occurs outside the S-phase.
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD012997.
The primary sequencing data have been deposited to the BioSample database with accession ID PRJNA632708.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Matthieu D. Lavigne, Pantelis Hatzis, Anastasios Liakos, for critical discussions and reading of the manuscript. Giorgos Panayotou, Panagiotis Moulos, Giorgos Pavlopoulos, Antonis Giakountis, Athanassios Margaritis, Pinelopi Nikolopoulou and the members of the Fousteri lab for productive discussions. Vladimir Benes and the Genecore facility (EMBL, Germany) for the sequencing support, Vaggelis Harokopos from the Genomics Facility at BSRC Fleming for performing RNA-seq library preparation and sequencing, Sofia Grammenoudi from the Flow Cytometry Facility at BSRC Fleming and Mihalis Verykokakis for their help with the FACS experiments.
Contributor Information
Georgios C Stefos, Institute for Fundamental Biomedical Research, BSRC ‘Alexander Fleming’, Vari 16672, Greece.
Eszter Szantai, Institute for Fundamental Biomedical Research, BSRC ‘Alexander Fleming’, Vari 16672, Greece.
Dimitris Konstantopoulos, Institute for Fundamental Biomedical Research, BSRC ‘Alexander Fleming’, Vari 16672, Greece.
Martina Samiotaki, Proteomics Facility, BSRC ‘Alexander Fleming’, Vari 16672, Greece.
Maria Fousteri, Institute for Fundamental Biomedical Research, BSRC ‘Alexander Fleming’, Vari 16672, Greece.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Research Council [ERC-StG-2012/Agreement-309612 to M.F.], ARISTEIA [2429 to M.F.] co funded by National Strategic Reference Framework 2007–2013; National sources and KRIPIS II [MIS 5002562]. Funding for open access charge: KRIPIS II [MIS 5002562].
Conflict of interest statement. None declared.
REFERENCES
- 1. Yadav T., Quivy J.P., Almouzni G.. Chromatin plasticity: a versatile landscape that underlies cell fate and identity. Science. 2018; 361:1332–1336. [DOI] [PubMed] [Google Scholar]
- 2. Gauchier M., Mierlo G.V, Vermeulen M.. Purification and enrichment of specific chromatin loci. Nat. Methods. 2020; 17:380–389. [DOI] [PubMed] [Google Scholar]
- 3. Sirbu B.M., McDonald W.H., Dungrawala H., Badu-Nkansah A., Kavanaugh G.M., Chen Y., Tabb D.L., Cortez D.. Identification of proteins at active, stalled, and collapsed replication forks using isolation of proteins on nascent DNA (iPOND) coupled with mass spectrometry. J. Biol. Chem. 2013; 288:31458–31467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Thomas Leung K.H., El Hassan M.A., Bremner R.. A rapid and efficient method to purify proteins at replication forks under native conditions. BioTechniques. 2013; 55:204–206. [DOI] [PubMed] [Google Scholar]
- 5. Alabert C., Bukowski-Wills J.C., Lee S.B., Kustatscher G., Nakamura K., De Lima Alves F., Menard P., Mejlvang J., Rappsilber J., Groth A.. Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat. Cell Biol. 2014; 16:281–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ribeyre C., Zellweger R., Chauvin M., Bec N., Larroque C., Lopes M., Constantinou A.. Nascent DNA proteomics reveals a chromatin remodeler required for topoisomerase I loading at replication forks. Cell Rep. 2016; 15:300–309. [DOI] [PubMed] [Google Scholar]
- 7. Bj Rås K., Sousa M.M.L., Sharma A., Fonseca D.M., S Gaard C.K., Bj Rås M., Otterlei M.. Monitoring of the spatial and temporal dynamics of BER/SSBR pathway proteins, including MYH, UNG2, MPG, NTH1 and NEIL1-3, during DNA replication. Nucleic Acids Res. 2017; 45:8291–8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gavande N.S., Vandervere-Carozza P.S., Hinshaw H.D., Jalal S.I., Sears C.R., Pawelczak K.S., Turchi J.J.. DNA repair targeted therapy: the past or future of cancer treatment. Pharmacol. Ther. 2016; 160:65–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Marteijn J.A., Lans H., Vermeulen W., Hoeijmakers J.H.J.. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat. Rev. Mol. Cell Biol. 2014; 15:465–481. [DOI] [PubMed] [Google Scholar]
- 10. Pleasance E.D., Cheetham R.K., Stephens P.J., McBride D.J., Humphray S.J., Greenman C.D., Varela I., Lin M.-L., Ordóñez G.R., Bignell G.R.et al.. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010; 463:191–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Haradhvala N.J., Polak P., Stojanov P., Covington K.R., Shinbrot E., Hess J.M., Rheinbay E., Kim J., Maruvka Y.E., Braunstein L.Z.et al.. Mutational strand asymmetries in cancer genomes reveal mechanisms of DNA damage and repair. Cell. 2016; 164:538–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liakos A., Lavigne M.D., Fousteri M.. Nucleotide excision repair: from neurodegeneration to cancer. Adv. Exp. Med. Biol. 2017; 1007:17–39. [DOI] [PubMed] [Google Scholar]
- 13. Boeing S., Williamson L., Encheva V., Gori I., Saunders R.E., Instrell R., Aygün O., Rodriguez-Martinez M., Weems J.C., Kelly G.P.et al.. Multiomic analysis of the UV-induced DNA damage response. Cell Rep. 2016; 15:1597–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ming X., Groehler A., Michaelson-Richie E.D., Villalta P.W., Campbell C., Tretyakova N.Y.. Mass spectrometry based proteomics study of cisplatin-induced DNA-protein cross-linking in human fibrosarcoma (HT1080) cells. Chem. Res. Toxicol. 2017; 30:980–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xu H., Chen X., Ying N., Wang M., Xu X., Shi R., Hua Y.. Mass spectrometry-based quantification of the cellular response to ultraviolet radiation in HeLa cells. PLoS One. 2017; 12:e0186806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Konstantakou E.G., Velentzas A.D., Anagnostopoulos A.K., Giannopoulou A.F., Anastasiadou E., Papassideri I.S., Voutsinas G.E., Tsangaris G.T., Stravopodis D.J.. Unraveling the human protein atlas of metastatic melanoma in the course of ultraviolet radiation-derived photo-therapy. J. Proteomics. 2018; 188:119–138. [DOI] [PubMed] [Google Scholar]
- 17. Heidelberger J.B., Wagner S.A., Beli P.. Mass spectrometry-based proteomics forinvestigating DNA damage-associated protein ubiquitylation. Front. Genet. 2016; 7:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bryan D.S., Ransom M., Adane B., York K., Hesselberth J.R.. High resolution mapping of modified DNA nucleobases using excision repair enzymes. Genome Res. 2014; 24:1534–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. García-Nieto P.E., Schwartz E.K., King D.A., Paulsen J., Collas P., Herrera R.E., Morrison A.J.. Carcinogen susceptibility is regulated by genome architecture and predicts cancer mutagenesis. EMBO J. 2017; 36:2829–2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hu J., Adebali O., Adar S., Sancar A.. Dynamic maps of UV damage formation and repair for the human genome. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:6758–6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hu J., Adar S., Selby C.P., Lieb J.D., Sancar A.. Genome-wide analysis of human global and transcription-coupled excision repair of UV damage at single-nucleotide resolution. Genes Dev. 2015; 29:948–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liakos A., Konstantopoulos D., Lavigne M.D., Fousteri M.. Continuous transcription initiation guarantees robust repair of all transcribed genes and regulatory regions. Nat. Commun. 2020; 11:916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Theron T., Fousteri M.I., Volker M., Harries L.W., Botta E., Stefanini M., Fujimoto M., Andressoo J.-O., Mitchell J., Jaspers N.G.J.et al.. Transcription-associated breaks in xeroderma pigmentosum group D cells from patients with combined features of xeroderma pigmentosum and cockayne syndrome. Mol. Cell. Biol. 2005; 25:8368–8378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Overmeer R.M., Gourdin A.M., Giglia-Mari A., Kool H., Houtsmuller A.B., Siegal G., Fousteri M.I., Mullenders L.H.F., Vermeulen W.. Replication factor C recruits DNA polymerase delta to sites of nucleotide excision repair but is not required for PCNA recruitment. Mol. Cell. Biol. 2010; 30:4828–4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wiśniewski J.R., Zougman, A., Nagaraj N., Mann M.. Universal sample preparation method for proteome analysis. Nat. Methods. 2009; 6:359–362. [DOI] [PubMed] [Google Scholar]
- 26. Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J.. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016; 13:731–740. [DOI] [PubMed] [Google Scholar]
- 27. Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L.. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019; 37:907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fanidis D., Moulos P.. Integrative, normalization-insusceptible statistical analysis of RNA-Seq data, with improved differential expression and unbiased downstream functional analysis. Brief. Bioinform. 2020; bbaa156. [DOI] [PubMed] [Google Scholar]
- 29. Lavigne M.D., Fousteri M., Konstantopoulos D., Ntakou-Zamplara K.Z., Liakos A., Fousteri M.. Global unleashing of transcription elongation waves in response to genotoxic stress restricts somatic mutation rate. Nat. Commun. 2017; 8:2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011; 17:10–12. [Google Scholar]
- 31. Li H., Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler a.D.. The human genome browser at UCSC. Genome Res. 2002; 12:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.. The sequence alignment/map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Karolchik D., Hinrichs A.S., Furey T.S., Roskin K.M., Sugnet C.W., Haussler D., Kent W.J.. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004; 32:493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Abugessaisa I., Noguchi S., Hasegawa A., Harshbarger J., Kondo A., Lizio M., Severin J., Carninci P., Kawaji H., Kasukawa T.. Data Descriptor: FANTOM 5 CAGE profiles of human and mouse reprocessed for GRCh 38 and GRCm 38 genome assemblies. Sci Data. 2017; 4:170107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bao W., Kojima K.K., Kohany O.. Repbase update, a database of repetitive elements in eukaryotic genomes. Mob. DNA. 2015; 6:4–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hubley R., Finn R.D., Clements J., Eddy S.R., Jones T.A., Bao W., Smit A.F.A., Wheeler T.J.. The Dfam database of repetitive DNA families. Nucleic Acids Res. 2016; 44:81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Feuerbach L., Sieverling L., Deeg K.I., Ginsbach P., Hutter B., Buchhalter I., Northcott P.A., Mughal S.S., Chudasama P., Glimm H.et al.. TelomereHunter - In silico estimation of telomere content and composition from cancer genomes. BMC Bioinformatics. 2019; 20:272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jia N., Nakazawa Y., Guo C., Shimada M., Sethi M., Takahashi Y., Ueda H., Nagayama Y., Ogi T.. A rapid, comprehensive system for assaying DNA repair activity and cytotoxic effects of DNA-damaging reagents. Nat. Protoc. 2014; 10:12–24. [DOI] [PubMed] [Google Scholar]
- 41. Katz E.J., Sirover M.A.. DNA nucleotide excision-repair synthesis is dependent of pertubations of deoxynucleoside triphosphate pool size. Mutat. Res. 1987; 183:249–256. [DOI] [PubMed] [Google Scholar]
- 42. Limsirichaikul S., Niimi A., Fawcett H., Lehmann A., Yamashita S., Ogi T.. A rapid non-radioactive technique for measurement of repair synthesis in primary human fibroblasts by incorporation of ethynyl deoxyuridine (EdU). Nucleic Acids Res. 2009; 37:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Guerrero-Santoro J., Kapetanaki M.G., Hsieh C.L., Gorbachinsky I., Levine A.S., Rapic V.. The Cullin 4B – based UV-damaged DNA-binding protein ligase binds to UV-damaged chromatin and ubiquitinates histone H2A. Cancer Res. 2008; 68:5014–5022. [DOI] [PubMed] [Google Scholar]
- 44. Gaillard H., Santos-Pereira J.M., Aguilera A.. The Nup84 complex coordinates the DNA damage. Nucleic Acids Res. 2019; 47:4054–4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ma J., Setton J., Lee N.Y., Riaz N., Powell S.N.. The therapeutic significance of mutational signatures from DNA repair deficiency in cancer. Nat. Commun. 2018; 9:3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chakravarty D., Gao J., Phillips S.M., Kundra R., Zhang H., Wang J., Rudolph J.E, Yaeger R., Soumerai T., Nissan M.H.et al.. OncoKB: a precision oncology knowledge base. JCO Precis. Oncol. 2017; 2017:PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Volker M., Mone M.J., Karmakar P., Hoffen A.V, Schul W., Vermeulen W., Hoeijmakers J.H.J., Driel R.V, Zeeland A.A.V, Mullenders L.H.F.. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell. 2001; 8:213–224. [DOI] [PubMed] [Google Scholar]
- 48. Pessina F., Lowndes N.F.. The RSF1 histone-remodelling factor facilitates DNA double-strand break repair by recruiting centromeric and Fanconi anaemia proteins. PLoS Biol. 2014; 12:e1001856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Helfricht A., Van Attikum H.. Remodeling and spacing factor 1 (RSF1): A rising star in DNA repair. Epigenomics. 2014; 6:261–265. [DOI] [PubMed] [Google Scholar]
- 50. Aydin Ö.Z., Marteijn J.A., Ribeiro-Silva C., Rodríguez López A., Wijgers N., Smeenk G., Van Attikum H., Poot R.A., Vermeulen W., Lans H.. Human ISWI complexes are targeted by SMARCA5 ATPase and SLIDE domains to help resolve lesion-stalled transcription. Nucleic Acids Res. 2014; 42:8473–8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mannini L., Cucco F., Quarantotti V., Krantz I.D., Musio A.. Mutation spectrum and genotype – phenotype correlation in Cornelia de Lange syndrome. Hum. Mutat. 2013; 34:1589–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Oka Y., Suzuki K., Yamauchi M., Mitsutake N., Yamashita S.. Recruitment of the cohesin loading factor NIPBL to DNA double-strand breaks depends on MDC1, RNF168 and HP1γ in human cells. Biochem. Biophys. Res. Commun. 2011; 411:762–767. [DOI] [PubMed] [Google Scholar]
- 53. Bot C., Pfeiffer A., Giordano F., Edara D.M., Dantuma N.P., Ström L.. Independent mechanisms recruit the cohesin loader protein NIPBL to sites of DNA damage. J. Cell Sci. 2017; 130:1134–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lans H., Marteijn J.A., Schumacher B., Hoeijmakers J.H.J., Jansen G., Vermeulen W.. Involvement of global genome repair, transcription coupled repair, and chromatin remodeling in UV DNA damage response changes during developm. PLos Genet. 2010; 6:e1000941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Litwin I., Wysocki R.. New insights into cohesin loading. Curr. Genet. 2017; 64:53–61. [DOI] [PubMed] [Google Scholar]
- 56. Epanchintsev A., Rauschendorf M.A., Costanzo F., Calmels N., Obringer C., Sarasin A., Coin F., Laugel V., Egly J.M.. Defective transcription of ATF3 responsive genes, a marker for Cockayne syndrome. Sci. Rep. 2020; 10:1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-moussavi A., Kheradpour P., Zhang Z., Wang J.et al.. Integrative analysis of 111 reference human epigenomes. Nature. 2015; 518:317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Adar S., Hu J., Lieb J.D., Sancar A.. Genome-wide kinetics of DNA excision repair in relation to chromatin state and mutagenesis. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E2124–E2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yang Y., Hu J., Selby C.P., Li W., Yimit A., Jiang Y., Sancar A.. cro Single-nucleotide resolution analysis of nucleotide excision repair of ribosomal DNA in humans and mice. J. Biol. Chem. 2019; 294:210–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Daniel L., Cerutti E., Donnio L., Nonnekens J., Carrat C., Zahova S., Rnap C.. Mechanistic insights in transcription-coupled nucleotide excision repair of ribosomal DNA. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:E6770–E6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Christians F.C., Hanawalt P.C.. Repair in ribosomal RNA genes is deficient in xeroderma pigmentosum group C and in Cockayne's syndrome cells. Mutat. Res. 1994; 323:179–187. [DOI] [PubMed] [Google Scholar]
- 62. Fritz L.K., Suquet C., Smerdon M.J.. Strand breaks are repaired efficiently in human ribosomal genes. J. Biol. Chem. 1996; 271:12972–12976. [DOI] [PubMed] [Google Scholar]
- 63. Rochette P.J., Brash D.E.. Human telomeres are hypersensitive to UV-induced DNA damage and refractory to repair. PLoS Genet. 2010; 6:e1000926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Parikh D., Fouquerel E., Murphy C.T., Wang H., Opresko P.L.. excision repair of photoproducts. Nat. Commun. 2015; 6:8214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wienholz F., Vermeulen W., Marteijn J.A., Tc-ner N.E.R.. Amplification of unscheduled DNA synthesis signal enables fluorescence-based single cell quantification of transcription-coupled nucleotide excision repair. Nucleic Acids Res. 2017; 45:e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD012997.
The primary sequencing data have been deposited to the BioSample database with accession ID PRJNA632708.