Abstract
Type IIA topoisomerases catalyze a variety of different reactions: eukaryotic topoisomerase II relaxes DNA in an ATP-dependent reaction, whereas the bacterial representatives gyrase and topoisomerase IV (Topo IV) preferentially introduce negative supercoils into DNA (gyrase) or decatenate DNA (Topo IV). Gyrase and Topo IV perform separate, dedicated tasks during replication: gyrase removes positive supercoils in front, Topo IV removes pre-catenanes behind the replication fork. Despite their well-separated cellular functions, gyrase and Topo IV have an overlapping activity spectrum: gyrase is also able to catalyze DNA decatenation, although less efficiently than Topo IV. The balance between supercoiling and decatenation activities is different for gyrases from different organisms. Both enzymes consist of a conserved topoisomerase core and structurally divergent C-terminal domains (CTDs). Deletion of the entire CTD, mutation of a conserved motif and even by just a single point mutation within the CTD converts gyrase into a Topo IV-like enzyme, implicating the CTDs as the major determinant for function. Here, we summarize the structural and mechanistic features that make a type IIA topoisomerase a gyrase or a Topo IV, and discuss the implications for type IIA topoisomerase evolution.
INTRODUCTION
The double-helical nature of DNA poses challenges for every cell. During replication and transcription, the two strands of the DNA duplex have to be separated. Strand separation is more facile in negatively supercoiled DNA, and both processes are facilitated by the steady-state level of negative supercoiling in cellular DNA (1). The importance of this global negative supercoiling is evident from the detrimental effect of even small changes: a change of just 15% in the supercoiling density is toxic for Escherichia coli (2). The replication and transcription machineries move along the DNA, and thereby alter the topological state of the flanking DNA segments. According to the twin-domain model, negative supercoils accumulate behind the translocating machinery, whereas positive supercoils are formed in the unwound DNA ahead (3,4). The torsional stress in front of the enzymes involved inhibits further strand separation, and leads to arrest of these processes if not alleviated.
DNA topoisomerases [recently reviewed in (5)] are enzymes that maintain the steady-state level of global supercoiling and solve topological problems. Their common catalytic principle consists of the cleavage of one or two DNA strands, the manipulation of topology, and the resealing of the gap in the DNA strand(s) [reviewed in (6)]. The enzymes are classified into type I and type II topoisomerases with respect to the number of DNA strands that are cleaved. They are further divided into type IA and IB according to mechanistic differences, and into type IIA and IIB according to structural features of the enzymes. Type IIA topoisomerases include the eukaryotic topoisomerase II (Topo II) and the bacterial enzymes topoisomerase IV (Topo IV) and gyrase [reviewed in (7)]. Although these three enzymes share a highly similar core structure, they catalyze different reactions in vitro, and fulfil different tasks in the cell (Figure 1): Topo II catalyzes the ATP-dependent relaxation of positive and negative supercoils in the presence of ATP (8). Topo IV decatenates DNA, and relaxes positive and negative supercoils in the presence of ATP (9,10). In contrast to Topo II, Topo IV shows a strong preference for relaxing positive supercoils (11,12). However, in bacteria, its main task is the decatenation of entangled daughter chromosomes (10). The third representative of type IIA topoisomerases, the bacterial enzyme gyrase, is unique as it couples ATP hydrolysis to the introduction of negative supercoils. Gyrase can also decatenate DNA in the presence of ATP, and relaxes negative supercoils in the absence of ATP (13). In vivo, it is responsible for maintaining a low negative supercoiling level in the cell, and for cancelling the positive supercoils ahead of the replication fork and the transcription machinery (14,15).
Figure 1.
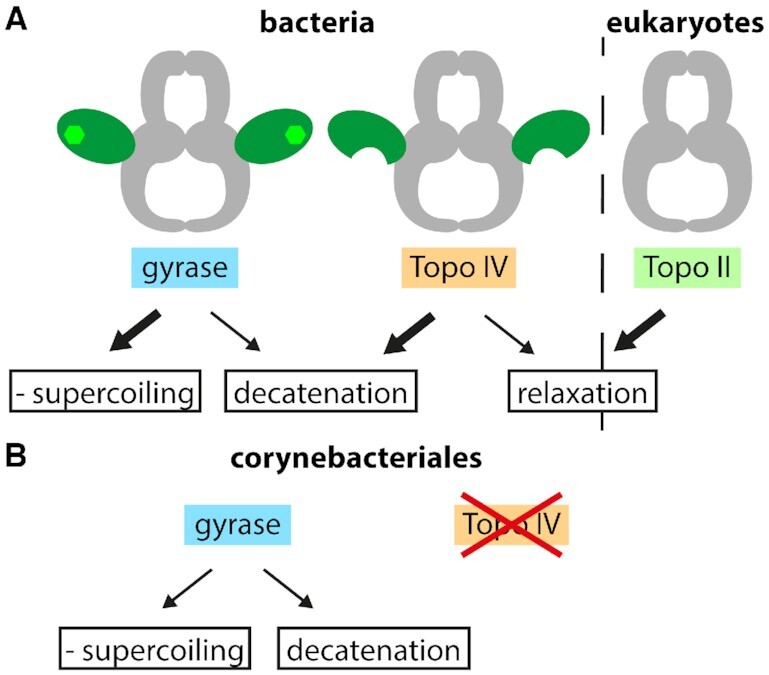
Type IIA topoisomerases and reactions catalyzed. (A) Type IIA topoisomerases comprise the bacterial enzymes gyrase and Topo IV, and the eukaryotic Topo II. The enzymes share a similar core structure (gray), but catalyze different ATP-dependent reactions in vitro: negative supercoiling and decatenation (gyrase), decatenation and relaxation (Topo IV), and relaxation (Topo II). Gyrase and Topo IV cooperate in the alleviation of torsional stress during bacterial replication. Gyrase removes positive supercoils ahead of the replication fork, whereas Topo IV removes pre-catenanes behind the fork and separates catenated DNA after replication. Gyrase also removes positive supercoils generated during transcription. (B) Some bacteria, including members of the order Corynebacteriales contain only one type IIA topoisomerase, typically a gyrase. This enzyme needs to remove positive supercoils ahead of the replication fork and decatenate replication intermediates in vivo.
Most bacteria contain two type IIA topoisomerases, gyrase and Topo IV, which cooperate in resolving topological challenges arising during DNA replication (16,17). The movement of the replisome along the DNA leads to the formation of positive supercoils ahead of the replication fork. By rotation of the fork, these positive supercoils can diffuse backwards, resulting in the formation of pre-catenanes behind the fork (18). Topo IV is an efficient decatenase, and preferentially works behind the fork to remove such pre-catenanes (19,20). Gyrase, on the other hand, is inefficient in decatenation. In vivo, it is located in front of the replication fork and removes positive supercoils (21). In this way, both enzymes divide the labor of resolving the topological problems associated with the movement of the replication fork. In some bacteria, only a single type IIA topoisomerase is present (Figure 1). These enzymes have to remove positive supercoils accumulating during transcription and replication, and to dissolve pre-catenanes behind the replication fork (see ‘Hybrid enzymes: part gyrase, part Topo IV’).
Due to their central role for bacteria, both gyrase and Topo IV are attractive drug targets. Inhibition of gyrase causes a slow stop of replication, while inhibition of Topo IV only reduces the velocity of replication elongation (22). Inhibition of both enzymes, on the other hand, causes a rapid arrest of replication. Dual inhibitors targeting both gyrase and Topo IV are thus superior to single inhibition. In addition to the more severe effect on replication, resistance development would require the concurrent appearance of mutations in both enzymes, which is far less likely than the appearance of single mutations (23). The current state of the art of gyrase and Topo IV inhibition has been reviewed elsewhere (24–26).
Gyrase and Topo IV share similar structures, but have different substrate preferences and perform different cellular activities, which raises the question: What makes a type IIA topoisomerase a gyrase or a Topo IV? In this review, we will analyze the differences and similarities of gyrase and Topo IV. In the first part, we will compare their structural features. In the second part, we will summarize how these structural features influence the interactions of these enzymes with DNA, their activities and their cellular tasks. We will conclude with a short discussion of the implications for the evolution of gyrase and Topo IV.
THE STRUCTURE OF BACTERIAL TYPE IIA TOPO-ISOMERASES
Common structural features of the type IIA topoisomerase core
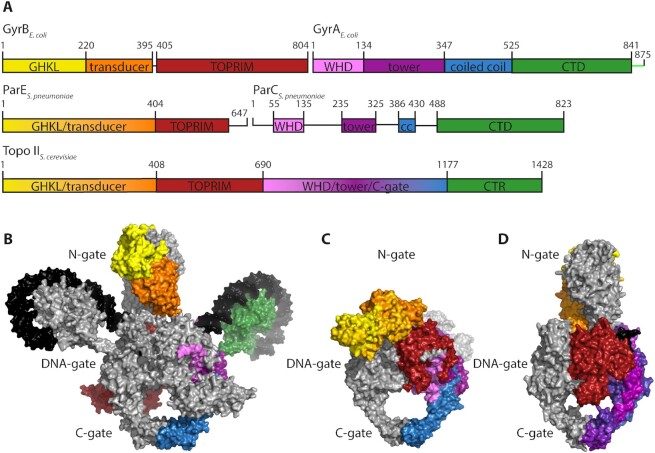
The general structure of the bacterial type IIA topoisomerase core is largely conserved. While Topo II is a dimeric enzyme, the active form of gyrase and Topo IV is a hetero-tetramer, formed by two GyrB/ParE and two GyrA/ParC subunits, respectively (1,9) (A2B2 or C2E2; Figure 2). The GyrB and ParE subunits contain the same modules, namely an N-terminal ATPase domain of the GyrB-Hsp90-histidine/serine protein kinase-MutL (GHKL) phosphotransferase superfamily, connected to a C-terminal Mg2+-binding topoisomerase-primase (TOPRIM) domain by the transducer domain (27). GyrA and ParC are also organized similarly: their N-terminal domain or breakage-reunion domain (BRD) consists of a winged-helix domain (WHD) harboring the catalytic tyrosines, a tower domain, and a coiled-coil domain (28). The C-terminal domain (CTD) is a DNA-binding domain with a β-pinwheel fold (29,30).
Figure 2.
Structural comparison of type IIA topoisomerases. (A) Schematic domain structure of Escherichia coli gyrase, Topo IV from Streptococcus pneumoniae and Topo II from Saccharomyces cerevisiae showing the GHKL-ATPase domain (yellow), transducer (orange) and TOPRIM domains (red) of GyrB/ParE/N-terminal half, and the WHD domain (light purple), tower (dark purple), the coiled coil (cc, blue) and C-terminal domain or region (CTD or CTR, green) of GyrA/ParC/C-terminal part. The C-tail of gyrase is shown in light green. (B) Cryo-EM structure of full-length E. coli gyrase with ADPNP, DNA (black) and Gepotidacin bound [PDB-ID: 6rkw (68)]. (C) Crystal structure of the topoisomerase core of Topo IV from S. pneumoniae with 34-bp DNA and Levofloxacin bound [PDB-ID: 4i3h (66)]. (D) Crystal structure of Topo II from S. cerevisiae missing the C-terminal region (CTR) with a short, linear DNA (black) and ADPNP bound [PDB-ID: 4gfh (65)]. The structures in panels (B)–(D) are colored according to the same color code as in panel A.
In the hetero-tetrameric complex, the four subunits of gyrase or Topo IV form three protein-protein interfaces, termed gates, which open and close during catalysis of topological changes (31–34). The N-gate is formed by the ATPase domains of GyrB/ParE, which dimerize upon ATP binding and make the N-gate an ATP-operated clamp (35,36). In the closed state, the ATPase domains exchange a short stretch of ∼14 amino acids at their N-terminus (37,38). This interaction stabilizes the dimer, and contributes to formation of the nucleotide binding site of the opposite GyrB/ParE (37,38). The central DNA-gate, formed by the TOPRIM domains of GyrB/ParE and the WHDs of the GyrA/ParC dimer, is the active site of the enzyme for DNA processing. Here, a double-stranded DNA segment, the G-segment, is bound, bent or distorted, and finally cleaved by the catalytic tyrosines (39,40). The third gate, termed C-gate, is formed by the globular domains at the end of the coiled-coil domains of GyrA/ParC, and is principally responsible for dimer stability (28,41,42). By this arrangement, two cavities are formed, one between the N- and DNA-gate and a second between the DNA- and C-gate (Figure 2). These cavities are thought to temporarily accommodate a second DNA segment, the T-segment, during its transport through the gap in the cleaved G-segment (see below) (41,43).
Structural differences: the CTDs
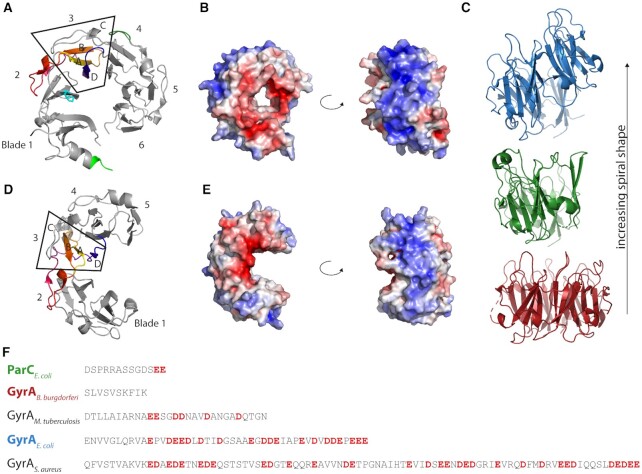
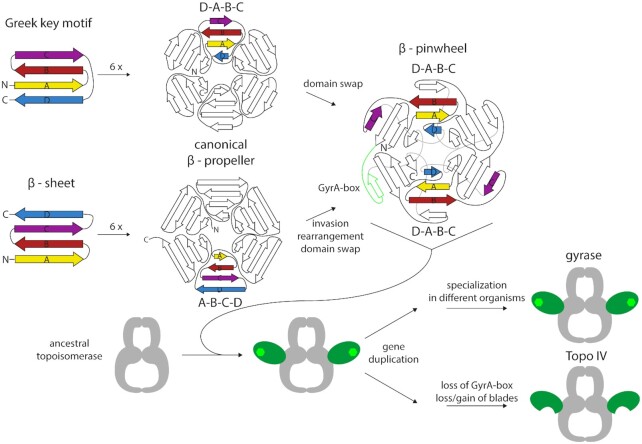
Although the topoisomerase core is highly conserved, some gyrases contain species-specific elements within the core that enable additional interactions between GyrB and GyrA subunits and modulate enzyme activity (44–46). In contrast, structures of ParC and ParE from different bacteria are virtually identical and do not show sequence-specific insertions (37,47–54). The parts that differ most between gyrase and Topo IV are the CTDs of the GyrA/ParC subunits. The GyrA CTD folds into a six-bladed β-pinwheel (Figure 3) (30). Although the global architecture of this fold is similar to a β-propeller, the order of the β-strands is different: in a β-propeller, one blade consists of a four-stranded antiparallel β-sheet with the strands arranged in the order A–B–C–D, from the N-terminal, innermost (A) to the C-terminal, outermost strand (D) (55). The strand order in the β-pinwheel is D–A–B–C, where the C-strand belongs to the adjacent blade (Figure 3A). In this arrangement, a long loop connecting strand C and D wraps around a neighboring blade. Hsiehet al. named this structure a ‘hairpin-invaded β-propeller’ (56), and compared the connection between adjacent blades achieved by the strand exchange to a ‘Velcro system’. Alternatively, the β-pinwheel may be based on the prevalent Greek key motif as a repeating unit, which follows the D–A–B–C topology (30) (see ‘Evolution of type IIA topoisomerases’). Altogether, the six blades form a closed circular structure in which blades 1 and 6 are connected by a loop containing a conserved sequence motif with the consensus sequence Q(R/K)RGG(R/K)G, termed the GyrA-box (Figure 3A) (57). Blades 2–5 contain degenerate forms of this motif (58), pointing to a possible evolutionary origin of the CTD from the duplication of a single blade (see ‘Evolution of type IIA topoisomerases’). Interestingly, in Mycobacterium tuberculosis gyrase the GyrA-box in the fifth blade of the CTD differs by only a single amino acid from the consensus, and is important for the decatenation activity of this enzyme (59). The loops of blades 1, 4, 5 and 6 form a contiguous, positively charged surface around the perimeter of the CTD, where the DNA binds (Figure 3B) (29,30,60). Most gyrases show a more or less pronounced spiral shape of their CTD, with an out-of-plane displacement of more than 10 Å in the CTD of E. coli GyrA (61). Most of this displacement, enabled by a conserved proline in a β-strand of blade 2 occurs between blades 1 and 2 (62). As a consequence, the DNA binding surface of the gyrase CTD along the perimeter follows a right-handed curve, which enables chiral wrapping of the DNA bound (Figure 3C) (60,61). The CTDs of Borrelia burgdorferi gyrase lack the conserved proline; their β-pinwheel is almost flat (30). This CTD also binds and bends DNA, but introduces less supercoils than CTDs with a spiral shape (61). The CTDs of most gyrases contain an unstructured C-terminal tail (C-tail) following the β-pinwheel, which is enriched in acidic residues and lacks positive charges (Figure 3F). Its length differs between different organisms, from zero amino acids in B. burgdorferi (no C-tail) to 35 in E. coli and 77 in Staphylococcus aureus gyrase (63). The acidic character also varies from organism to organism. The C-tail is a regulatory element (63,64) (see ‘The CTDs – binding, bending, and wrapping DNA’ in the section on ‘Mechanistic differences of gyrase and Topo IV’).
Figure 3.
Structural features of Topo IV and gyrase CTDs. (A) Crystal structure of the gyrase CTD from E. coli [PDB-ID: 1zi0 (61)]. The blades are numbered from the N- to the C-terminus; the box highlights blade 3. Strands are labeled according to the invasion hypothesis. The sequence is rainbow-colored from yellow (N-terminus) to blue (C-terminus). The β-strand-bearing proline is shown as a stick model in cyan. The crystalized part of the GyrA-box is depicted in light green, the position where the second GyrA-box is located in mycobacterial gyrase in blade 5 (here part of blade 4 due to the domain swap of strand C) is shown in dark green. (B) Electrostatic surface potential of the E. coli CTD in a front and side view (blue: positive, red: negative), calculated with PyMOL (133). (C) Crystal structure of three CTDs (colored according to panel F) showing the different spiral pitch. (D, E) Crystal structure and surface potential of the Topo IV CTD from E. coli as in panel A and B [PDB-ID: 1zvt (50)]. (F) Sequence alignment of the C-terminus for different GyrA subunits and ParC. Acidic residues in the C-terminal tail are depicted in red.
The CTDs in Topo IV show remarkable differences to the gyrase CTDs, both in sequence and structure. The ParC CTD lacks the conserved GyrA-box, although it contains degenerate forms in each blade (see ‘Evolution of type IIA topoisomerases’). Without the GyrA-box, the tight connection between the first and last blade is missing, and the ParC CTD adopts an open, C-shaped structure with a gap between these blades (Figure 3D) (50,56). The conserved proline present in the β-strand of blade 2 in gyrase is absent in most Topo IV enzymes, resulting in a planar or less spirally pitched structure (Figure 3C) (58,50). An exception is Bacillus stearothermophilus Topo IV, which contains a proline in the conserved position, and shows a spiral shape of the CTD similar to gyrase (56); see ‘Evolution of type IIA topoisomerases’). The number of blades in the Topo IV CTD is not restricted to six, but varies from zero in Chlamydia (no CTD) over five in E. coli to eight in Clostridium. The consequences of the structural differences in the CTDs for the functional properties of gyrase and Topo IV will be discussed in more detail in the section ‘The CTDs – binding, bending, and wrapping DNA’ in the second part (‘Mechanistic differences of gyrase and Topo IV’).
Different conformations of the gyrase and Topo IV hetero-tetramers
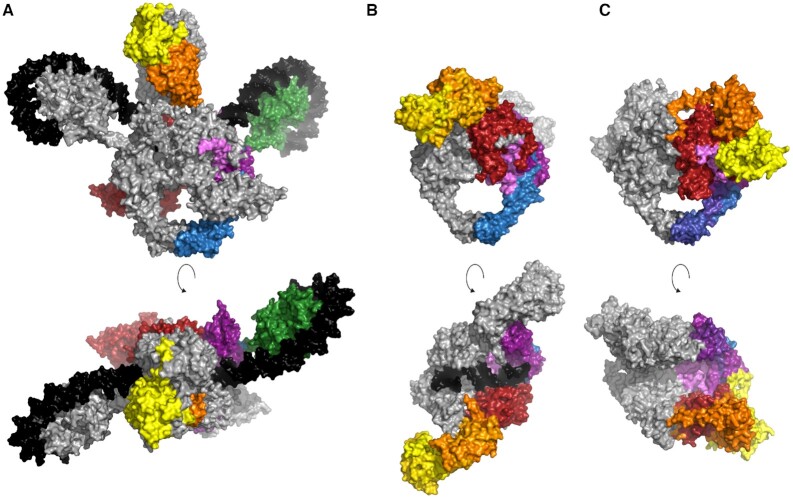
Due to their size, multi-subunit nature, conformational flexibility and heterogeneity, structure determination of complete type IIA topoisomerases has been challenging. The first structure of a type IIA topoisomerase reported was of a S. cerevisiae Topo II core, missing the C-terminal region (65). In this structure, the part that is homologous to the GyrA/ParC dimer and the parts corresponding to GyrB/ParE are positioned on top of each other, but are rotated with respect to one another. As a result of this rotation, the ATPase domain of one protomer comes into close contact with the DNA cleavage part of the second protomer (65).
From the appreciable level of sequence conservation, the shared domain architecture and similar structures of individual domains or sub-domains (28,30,37,43,50,66), an overall structure similar to Topo II has been inferred for gyrase and Topo IV heterotetramers. While this holds true for the topoisomerase core, the comparison of sequences and structures available has revealed not only differences between gyrase or Topo IV in comparison to Topo II, but also between gyrase and Topo IV; and among gyrases. The main structural difference between gyrase and Topo IV is the conformation of the GyrB/ParE subunits, and thus of the N-gate (Figure 4). A first glimpse on the structure of the complete gyrase heterotetramer, bound to DNA and nucleotide, was provided by the cryo-electron microscopy (cryo-EM) structure of the Thermus thermophilus enzyme (67). This structure captured gyrase with all three gates in the closed state. The nucleotide-bound, dimerized ATPase domains of GyrB are in an orthogonal orientation above the catalytic core, and pointing away from the DNA-gate, similar to the arrangement in Topo II. DNA is bound at the DNA-gate, and flanking regions are wrapped around both CTDs that are facing upwards, in plane with the gate. More recently, Broecket al. reported a cryo-EM structure of E. coli gyrase with a resolution of up to 3.0 Å (68) that shows a similar overall arrangement (Figure 4A). Although there is no structure available up to now with the N-gate of gyrase in the open state, it has been shown in single-molecule Förster resonance energy transfer (FRET) experiments that the two ATPase domains are only a few Ångstrøm apart in the open conformation (34).
Figure 4.
Different conformations of the heterotetrameric type IIA topoisomerases. (A) Cryo-EM structure of E. coli gyrase [PDB-ID: 6rkw, (68)] showing a closed N-gate. (B) Crystal structure of Topo IV from S. pneumoniae [PDB-ID: 4i3h, (66)] showing the wide-open conformation of the N-gate with the ATPase domains facing downward to the C-gate. (C) Crystal structure of M. tuberculosis gyrase [PDB-ID: 6gav (44)] with a wide-open conformation of the N-gate. However, the longitudinal axis of the ATPase domains is rotated with respect to the axis of the topoisomerase core in comparison to Topo IV. Top: front view, bottom: top view. The color code is the same as in Figure 2; DNA is shown in black.
In contrast to the closed state captured for gyrase bound to DNA and nucleotide, the crystal structure of S. pneumoniae Topo IV without the CTDs shows a wide-open conformation of the N-gate (66) (Figure 4B). The TOPRIM domains are in a similar orientation as in gyrase, but the ParE ATPase domains are bent downwards, facing toward the C-gate, and are far apart from each other. In this conformation, the G-segment bound at the DNA-gate and the polar side chains of the TOPRIM domain are solvent-exposed. A possible role of the TOPRIM domain in T-segment capture has been suggested (66).
In 2019, the crystal structure of gyrase from Mycobacterium tuberculosis lacking the CTDs has been determined (44) (Figure 4C). This enzyme shows a similarly wide-open conformation of the N-gate as Topo IV, despite the fact that the non-hydrolysable ATP analogue AMPPNP is bound to the active site of both GyrB subunits. The ATPase domains also point in the direction of the C-gate, but their orientation differs from the one of the ParE domains in Topo IV: the longitudinal axis of the GyrB GHKL domain is almost parallel to the two-fold axis of the cleavage core. This conformation is mediated by interactions between two motives specific for the order of Corynebacteriales, the C-loop in the ATPase domain of GyrB and the DEEE-loop in the tower domain of GyrA (69,70). The wide-open conformation has been interpreted as a possible ‘resting state’ (44).
The CTDs of gyrase and Topo IV are flexibly attached to the GyrA/ParC NTD, and constitute mobile elements that change their position relative to the topoisomerase core during the catalytic cycle. In gyrase, the CTDs face downwards towards the C-gate in the absence of DNA (71–74), but move upwards, and are aligned with the DNA-gate in the DNA-bound state (67,68,74). It is currently unclear at which stage in the catalytic cycle they return to the downward-facing state and if asymmetric intermediates exist in which one CTD faces downward, the other one upward. In Topo IV, the CTDs are located on the level of the DNA-gate in the absence of DNA (50). From the different contributions of individual blades of the CTD to different topological changes catalyzed by Topo IV, it has been proposed that the CTDs can rotate to accommodate interactions with different topological states of the DNA substrate (75).
MECHANISTIC DIFFERENCES BETWEEN GYRASE AND Topo IV
The general strand-passage mechanism of type IIA topoisomerases versus nicking-closing
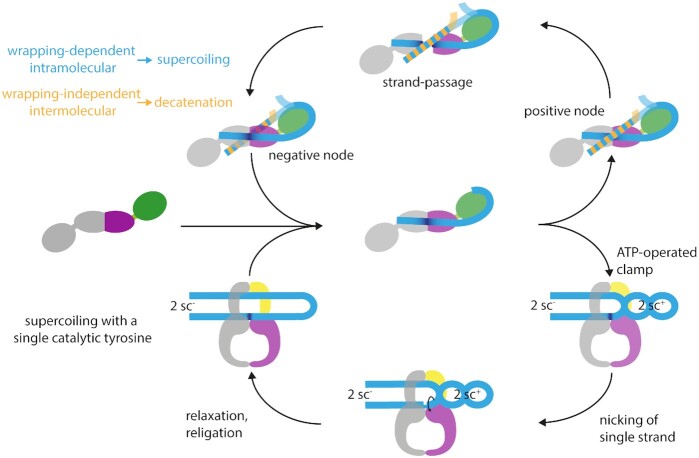
According to the current model, type IIA topoisomerases change the topological state of DNA by a strand-passage mechanism (Figure 5) (reviewed in (76)). The sequence of events leading to supercoiling, relaxation, or decatenation starts with the binding of a double-stranded G-segment at the DNA-gate. On binding, the DNA adopts A-form geometry directly at the scission sequence, while retaining B-form geometry in the remaining parts (40,77). The G-segment also becomes bent on binding, caused by the intercalation of two conserved isoleucine side chains into the DNA (40,77). In gyrase, a DNA-induced upward movement of the CTDs and a narrowing of the N-gate was observed (34,74), which may be specific for intramolecular strand passage and super-coiling. The next step in the catalytic cycle is the capture of a second double-stranded DNA segment, the T-segment, which becomes fixed above the G-segment by ATP-induced closing of the N-gate. Gyrase and Topo IV interact differently with DNA: while Topo IV can stably bind two DNA molecules, gyrase only binds a single DNA segment (78–80). Gyrase thus captures a T-segment located in the same DNA molecule as the G-segment (79,80), whereas Topo IV captures a T-segment located on a second DNA molecule (80). The different interaction with DNA thus makes the difference between intermolecular strand passage and decatenation by Topo IV, or intramolecular strand passage and supercoiling by gyrase. Subunit mixing experiments showed that binding of the second DNA molecule depends on the ParE/GyrB subunits (80). After DNA binding, both strands of the G-segment are cleaved, leading to a double-strand break with a four-base stagger. In this reaction, the catalytic tyrosines in GyrA/ParC act as nucleophiles; they remain covalently bound to the 5′-end of each DNA strand (81). The covalent enzyme-DNA intermediate formed is referred to as the cleavage complex. After opening of the DNA-gate and transport of the T-segment through the gap created, the G-segment is re-ligated. Subsequently, the T-segment can leave the enzyme, which is controlled by opening of the C-gate (32,82). Finally, after the release of ADP and phosphate, the N-gate re-opens (34,83), and the enzyme is prepared for another cycle.
Figure 5.
Possible mechanisms for type IIA topoisomerases. The upper part shows the canonical strand-passage mechanism for DNA supercoiling, depicting gyrase in top view, looking down in the DNA bound at the DNA-gate. The bottom part shows the nicking-closing mechanism, which rationalizes the ability of gyrase to negatively supercoil DNA with only one catalytic tyrosine present (84). Here, the CTDs are not shown as the structural basis for capturing two positive supercoils is unknown. In both mechanisms, the first step is the binding of the G-segment (light blue) to the DNA-gate (one subunit shown in purple, the other in gray). On binding, the G-segment becomes bent (dark blue) at the DNA-gate, and is wrapped around the CTD (green). When ATP binds, the N-gate closes, and the T-segment (blue/orange) is captured. Strand passage (top) can occur with a T-segment located on the same (blue) or a second DNA molecule (orange). Intramolecular strand passage is wrapping-dependent and leads to DNA supercoiling (blue); intermolecular strand passage is wrapping-independent and leads to DNA decatenation (orange). The two catalytic tyrosines cleave the G-segment, whereupon the DNA-gate opens, and the T-segment can pass through the gap, converting a positive into a negative node. Gyrase with a single catalytic tyrosine can supercoil DNA without strand passage, following a nicking-closing mechanism (bottom). Capture of the T-segment leads to the stabilization and segregation of two positive supercoils, resulting in the formation of two negative supercoils in the remainder of the DNA. The ATP-operated clamp inhibits the rotation of the T-segment around its helical axis. Upon induction of a nick in the G-segment by the single catalytic tyrosine, the two positive supercoils can relax, leaving two negative supercoils behind.
In contradiction to the strand-passage mechanism, gyrase in which one of the two tyrosines is replaced by a phenyl-alanine is able to supercoil DNA in the absence of double-strand cleavage and strand passage (84). This variant supercoils DNA with similar characteristics as the wildtype enzyme, introducing changes of the linking number in steps of two and undergoing the same conformational changes in the catalytic cycle (84). These observations suggest an alternative mechanism for supercoiling, in which two positive supercoils are captured by gyrase, segregated from the rest of the substrate, and relaxed by nicking of one of the strands (Figure 5) (84). The two compensatory negative supercoils, formed in the remainder of the DNA substrate on capture of the positive supercoils, remain, and relaxation of the positive supercoils thus leads to an overall decrease in the linking number by two (84) [reviewed in (85)]. DNA supercoiling by nicking-closing does not require the DNA- and C-gates to open. Nevertheless, subtle conformational changes of these interfaces may enable the associated rearrangements of the DNA (84) [reviewed in (85)]. It remains to be determined whether the nicking-closing mechanism is used for DNA supercoiling in vivo as a safeguard to avoid double-strand breaks, or whether both mechanisms are used in parallel.
In-line with the physical requirement of double-strand cleavage and strand passage for DNA decatenation, gyrase with a single catalytic tyrosine fails to decatenate DNA. The ATP-dependent relaxation activity of gyrase lacking the CTDs also depends on double-strand cleavage and strand passage (84). From these data, a similar loss of decatenation and relaxation activities for Topo IV with just a single tyrosine can be inferred. Altogether, these observations point to a different mechanistic spectrum of gyrase and Topo IV.
Sensing topology: preferences for different DNA substrates
Despite the common principles in their core mechanism, gyrase and Topo IV display large differences in their interactions with the DNA substrate, which are intimately linked to their different activity profiles. ATP-dependent relaxation by Topo IV and ATP-dependent negative supercoiling of DNA by gyrase are the result of intramolecular strand passage, whereas decatenation of DNA is brought about by intermolecular strand passage, involving two DNA molecules. Topo IV catalyzes decatenation faster than relaxation (11,86,87), whereas gyrase is more efficient in DNA supercoiling than in decatenation (86,88); decatenation by gyrase is almost two orders of magnitude slower than decatenation by Topo IV (16,89).
Gyrase and Topo IV both show strong preferences for the topology of their substrates: gyrase catalyzes the relaxation of positive supercoils much faster and more processively than the introduction of negative supercoils into relaxed DNA (12,17). Similarly, Topo IV relaxes positive supercoils much faster than negative supercoils (86,87). Nevertheless, the mechanisms for topology sensing are different for the two enzymes. The handedness of the DNA substrate is recognized at multiple stages. The first stage is binding of the G-segment. E. coli gyrase shows a 10-fold higher affinity for relaxed than for linear or negatively supercoiled DNA (90). Gyrase from M. tuberculosis binds positively supercoiled DNA with a higher affinity than negatively supercoiled DNA (91). In contrast, Topo IV has a 4-fold preference for binding to supercoiled than to linear DNA, and shows similar affinities for positive and negative supercoils (92,93). Crosslinking experiments have provided evidence for a distinct conformation of Topo IV bound to positively supercoiled DNA, with the two ParE subunits within crosslinking distance, which is not observed with other DNAs (94). A second step in topology discrimination is T-segment binding. Unlike Topo IV, gyrase does not stably interact with two DNAs simultaneously (78–80). Instead, DNA wrapping around the CTDs is key for the stable interaction with a positive supercoil, and for positioning of the T-segment above the G-segment in a defined relative orientation for supercoiling (67,68). Topo IV does not wrap DNA (58), but forms a stable complex with both G- and T-segment DNAs (80). It preferentially interacts with negative crossings occurring in positively supercoiled DNA and positive catenanes (92,95). As a third step, the topology of the substrate plays a role in DNA cleavage. Gyrases, including the M. tuberculosis enzyme, form more cleavage complexes with negatively than with positively supercoiled DNA (12,91). This preference does not depend on the CTDs, but instead it has been linked to the N-terminal part of GyrB (91). For Topo IV, the reports range from little preference under stochiometric conditions (excess enzyme) (12) to 20-fold more efficient cleavage of positively supercoiled DNA under catalytic conditions (11). Finally, the processivity of strand passage is also dependent on DNA topology. Despite the lower level of cleavage complexes on positively supercoiled DNA, gyrase catalyzes the relaxation of positive supercoils (i.e. negative supercoiling) much faster and more processively than the introduction of negative supercoils into relaxed DNA (12,17). Topo IV relaxes positive supercoils much faster than negative supercoils (86,87). Magnetic tweezers experiments revealed that, similar to gyrase, the reason for the preferred action on positively supercoiled DNA is the difference in processivity: while relaxation of positive supercoils is highly processive, the relaxation of negative supercoils is entirely distributive (11,87,92). The processivity difference has been ascribed to a more stable interaction of Topo IV CTDs with the DNA during relaxation of positively supercoiled DNA, leading to a lower rate constant for dissociation of the enzyme from positively than from negatively supercoiled DNA (50,75,87).
Despite their shared preference for positively supercoiled DNA, gyrase and Topo IV thus show substantial differences in the mechanism of topology discrimination during DNA binding, cleavage, and strand passage. While the wrapping of DNA by gyrase leads to a preferential interaction with positively supercoiled DNA and at the same time favors intramolecular strand passage, Topo IV preferentially decatenates DNA through its stable interaction with two DNA molecules, and preferentially relaxes positively supercoiled DNA, predominantly through the high processivity of this reaction. Although the GyrB/ParE subunits have been implicated in the preferred cleavage of negatively supercoiled DNA by gyrase (91) and in binding to a second DNA molecule by mycobacterial gyrase and Topo IV (80), there is mounting evidence that the CTDs are the key elements that dictate topology-dependent interactions with the DNA, and hence the different activities of gyrase and Topo IV (29,50,75,79,96,97). In-line with this hypothesis, removal of the CTDs leads to simple topoisomerases that relax DNA in the presence of ATP without a preference for negative or positive supercoils (58,96). This brings us back to a comparison of the gyrase and Topo IV CTDs to answer the question: What is so special about the CTDs?
The CTDs: binding, bending, and wrapping DNA
The important role of the CTDs in DNA binding to gyrase became evident from DNase- and hydroxyl-radical footprinting experiments: full-length gyrase protects a 140 bp-DNA segment, with a higher protection of the central 40 bp (60,98–102). Although Topo IV protects only the central region of the DNA (103,104), deletion of the CTDs in gyrase and Topo IV leads to a decrease in DNA affinity (50,74), confirming that they contribute to DNA binding in both enzymes. The CTDs of gyrase and Topo IV are in fact DNA-binding proteins on their own: the isolated CTDs bind DNA via the positively charged surface at their perimeter, which introduces a bend (30,105). The gyrase CTDs bind the DNA in a right-handed spiral, which additionally introduces writhe (29) and results in wrapping of the DNA around gyrase in a positive supercoil (61). In Topo IV, the CTDs have been implicated in stronger DNA interactions of Topo IV during the relaxation of positively supercoiled DNA and decatenation (50,75). They also contribute to bending of the G-segment by gyrase and Topo IV (34,75,105). In the following, we will dissect the role of the individual structural features of the CTD in determining the activities of gyrase and Topo IV.
Spiral shape and positive patch
Mutational studies of the CTDs revealed that the positive patch determines their DNA affinity and their capability to bend the DNA and enable wrapping (75,97). Increasing the number of positive charges on the surface of the gyrase CTD generally strengthens DNA binding. The removal of a single positive charge does not abolish binding, but leads to a loss of wrapping and of supercoiling activity (97). The effect of exchanging positive by negative charges on the surface of the Topo IV CTD on DNA binding depends on the position of the mutation: mutations close to the N-terminus do not alter the DNA affinity, whereas binding is progressively impaired when the mutations are closer to the C-terminus (75).
Through its spiral shape, the positive patch around the CTDs is also important for introducing writhe. The degree of writhe introduced varies. The linking number of DNA bound to the E. coli CTD (lacking the C-tail, see below) is changed by +0.8, whereas the CTD of B. burgdorferi only increases the linking number by 0.3 (61). This lower change in linking number has been linked to the planar shape of this CTD. In agreement with this hypothesis, mutation of the conserved proline in E. coli gyrase that enables formation of the spiral shape of the CTD leads to impaired DNA wrapping and a decrease of supercoiling activity (62). The planar Topo IV CTD also introduces only little chiral writhe (30,58). From the spiral shape of the CTD in B. stearothermophilus ParC (56) one would predict that this CTD may also be capable of introducing writhe, similar to gyrase CTDs. However, the B. stearothermophilus ParC CTD surface is less curved, and has a lower positive electrostatic potential than GyrA CTDs (56). The biochemical properties of the CTD or the reaction profile of B. stearothermophilus Topo IV have not been characterized yet.
GyrA-box
The GyrA-box in blade 1 of the CTD was discovered as a signature motif of gyrases (57,105). Deletion or substitution of the GyrA-box leads to a loss of supercoiling activity, but has no effect on the decatenation activity, similar to the removal of the complete CTD (57). According to the cryo-EM structure of E. coli gyrase, the GyrA-box faces away from the NTD and from the G-segment binding site (68). In this configuration, blade 1 is the last blade that is contacted by the wrapped DNA before it exits the CTD (68). The GyrA-box has been suggested as a key element for the local DNA geometry and for stabilizing the wrap (56). Indeed, mutations in the GyrA-box do not affect DNA binding to the isolated CTD, but the capability to bend the DNA is reduced (105). Mutations of the GyrA-box in gyrase prevent DNA wrapping (57). They also suppress DNA-induced N-gate narrowing, although the CTDs still move upwards when DNA binds (105). In the absence of the GyrA-box, DNA binding does not stimulate the ATPase activity, implicating it in coupling of DNA binding and ATP hydrolysis (105). The GyrA-box is also required for the high processivity of relaxing positively supercoiled DNA (12). It has been suggested that the strong deviation of the GyrA-box in the B. burgdorferi CTD from the consensus contributes to its inability to introduce writhe into DNA (61). However, the link between sequence and writhing has not yet been explored experimentally.
Due to the modular structure of the CTD, a sequence reminiscent of the GyrA-box is present in every blade, although these sequences have diverged from the consensus (58). In M. tuberculosis gyrase, a second GyrA-box conforming to the consensus sequence is present in blade 5 (59) (see Figure 3A). While mutations in the canonical GyrA-box lead to similar changes of activities as in E. coli gyrase, mutations in the second GyrA-box decrease the decatenation activity (59). In all gyrases, residues in the GyrA-box-like sequences in each blade contribute to the positively charged band at the CTD perimeter, with positive charges placed at regular intervals along the DNA binding surface. By removing or adding individual positive charges, Hobsonet al. have dissected the role of individual blades of the CTD for gyrase activities (97). Notably, removal of a single positive charge significantly reduces the capacity of the CTD to wrap DNA. Generally, charge removal and a concomitant weakening of DNA wrapping lead to a decrease in supercoiling activities, but enhanced decatenation, and thus generated a more Topo IV-like enzyme. Increasing the interaction with the DNA by introducing additional positive charges, on the other hand, also resulted in slower DNA supercoiling and reduced coupling, suggesting that the positively charged surface of E. coli gyrase is optimized for negative supercoiling (97). The DNA-stimulated ATPase activity of the gyrase variants correlated with the wrapping propensity. Charge addition and removal also affected the balance between supercoiling and decatenation activities. Variants with a higher ability to wrap DNA catalyzed supercoiling more efficiently than decatenation, whereas a variant with impaired wrapping was a more efficient decatenase (97). This observation is in agreement with a kinetic partitioning model proposed earlier by Nöllmannet al., who rationalized gyrase activity as a competition between wrapping and negative supercoiling on one hand, and wrapping-independent decatenation of DNA on the other hand (106).
Although the GyrA-box is a signature motif of gyrases, degenerate forms of this sequence containing positive residues are also present in Topo IV (see ‘Evolution of type IIA topoisomerases’). Voset al. showed that mutations of positive charges in different blades of Topo IV have differential effects on the interaction of Topo IV with different DNA substrate, and on different activities of Topo IV. Mutations in blades 4 and 5 led to reduced DNA binding to the isolated CTD, whereas mutations in other blades did not alter DNA affinity much (75). In the context of Topo IV, positively supercoiled DNA is bound mostly through blades 2, 3 and 4. Negatively supercoiled DNA interacts also with blades 2, and 3, and with a strong binding site on blade 5. Catenated DNAs preferentially interacts with blades 3, 4, and 5. Interestingly, reduced DNA affinity due to mutations in blade 5 was associated with an increased activity in the relaxation of negatively supercoiled DNA and in decatenation, suggesting that high-affinity binding of DNA to blade 5 is auto-inhibitory for these activities. Strikingly, residues in blade 1 located nearest to the ParC NTD contribute to bending of the G-segment DNA bound at the DNA-gate of the topoisomerase core (75). In terms of activity, mutations in the CTD either lead to global changes in Topo IV activities on any substrate (blade 1), shifted the balance between decatenation and relaxation (blade 3, blade 2), altered chiral selection and the balance between relaxation of positive and negative supercoils (blade 5), or had no effect on any of Topo IV’s activities (blade 4) (75). The strongest effect on decatenation activity was observed for mutations in blade 3. Relaxation of negative and (to a lesser extent) positive supercoils depends not only on residues in blade 3, but also on blade 2. Residues on blades 2 and 3 determine the processivity of relaxation and decatenation. The differential engagement of the CTDs with different substrates, and their contributions to G-segment bending and T-segment binding, require a reorientation of the CTDs relative to the NTD (75) that has not been probed experimentally.
C-tail
A distinct structural feature of the CTDs in gyrase, but not in Topo IV, is the presence of an unstructured, acidic C-tail. In E. coli gyrase, the C-tail acts as an auto-inhibitory element: the E. coli CTD and the GyrA subunit are not able to bind DNA or to introduce writhe, but gain these functions when the C-tail is removed (63). DNA wrapping is also restored when the gyrase heterotetramer forms, presumably through interactions of the C-tail with the GyrB subunit (63). In contrast to E. coli, the CTDs and GyrA subunits of M. tuberculosis and B. subtilis show similar or slightly increased DNA binding and bending without the C-tail (64,107). Both enzymes have a shorter C-tail than E. coli gyrase. The C-tail thus appears to be an extra regulatory element in gyrase that enables species-specific variations of supercoiling activity and the degree of supercoiling reached (63,108).
Clearly, the CTDs play a major role in determining the interactions of gyrase and Topo IV with different DNA and the different reactions these enzymes can catalyze. How important are the differences in interactions with the DNA substrate for gyrase and Topo IV activities in vivo?
Different activities in the cell
The unique hallmark reaction of gyrase, i.e. the ATP-dependent introduction of negative supercoils into DNA, is key for maintaining the negative supercoiling density of the genome (109,110). The global supercoiling state is determined by the delicate balance between opposing activities of the different topoisomerases present. DNA-dependent processes, predominantly replication and transcription, alter the local supercoiling level of the DNA (3,4). Despite the overlapping spectrum of gyrase and Topo IV activities in vitro, the in vivo tasks of these enzymes are distinct. Gyrase activity is critical during transcription, and in the beginning of and during replication, whereas Topo IV activity is essential for chromosome segregation at the end of the replication process (16,86). Gyrase has a high intrinsic affinity for positively supercoiled DNA (91), and is located ahead of the replication fork (21,111–113). By removing positive supercoils with high processivity and velocity (12), it alleviates accumulating torsional stress and ensures fork progression. In contrast, Topo IV is located on the negatively supercoiled DNA behind the replication fork, where its decatenation activity is stimulated (16,17,86,87,89). Topo IV efficiently disentangles pre-catenanes during replication elongation, and mediates chromosome decatenation at the end of the replication process to ensure chromosome segregation (19,114). The localization of Topo IV behind the fork is mediated through protein-protein interactions. Three well-examined interaction partners of Topo IV are MukB, SeqA, and FtsK. MukB interacts with Topo IV and increases the local concentration of Topo IV at the origin of replication (20,115,116). SeqA is a DNA-binding protein, which binds to hemi-methylated DNA (117). By interacting with the CTD of the ParC subunit, it recruits Topo IV to the freshly replicated DNA behind the replication fork, and increases the decatenation and relaxation activity of Topo IV (118,119). Finally, FtsK interacts with ParC, leading to an increased concentration of Topo IV at the septal ring, where it decatenates newly replicated chromosomes prior to segregation (114). FtsK also stimulates the decatenation activity of Topo IV (114).
Despite their dedicated cellular functions, Topo IV and gyrase can partially complement each other in vivo: overexpression of gyrA and gyrB partially rescues a parC and parE mutant (120), suggesting that gyrase can take over at least some of the activities of Topo IV. A gyrase variant that is supercoiling-deficient, but a more efficient decatenase than wildtype gyrase, can also rescue a parC mutant (96). Topo IV can replace gyrase during replication elongation in vitro and partially in vivo (16,121). However, the overexpression of parC and parE does not rescue a gyrB mutant (120), indicating that one or more of the cellular functions of gyrase cannot be provided by Topo IV.
The preferential relaxation of positive supercoils by Topo IV and the conversion of positive into negative supercoils by gyrase jointly remove excess positive supercoils in bacteria without relaxation of negative supercoiling required for DNA compaction and metabolism. While Topo IV cleaves negatively and positively supercoiled DNA with equal efficiencies, gyrase shows less cleavage on positively supercoiled DNA. The resulting lower inherent danger of introducing double-strand breaks in the unreplicated DNA makes gyrase a safer enzyme ahead of the replication fork than Topo IV (12). It has been a puzzle for a long time how Topo IV can resolve the ‘paradox’ that is inherent to its spectrum of activities:, i.e. how can Topo IV resolve right-handed pre-catenanes and catenanes during replication without relaxing right-handed negative supercoils and interfering with the steady-state negative supercoiling density in the cell? First single-molecule studies suggested that a preference of Topo IV for left-handed juxtapositions of G- and T-segments prevents activity on negatively supercoiled DNA. Subsequent studies determined the preferred crossing angle to approx. 85° (87). Such crossing angles are populated in right-handed catenanes, which rationalizes their dissolution by Topo IV (87). However, mathematical simulations showed that selection for left-handed DNA crossings characteristic of right-handed topologies enables Topo IV to relax positively supercoiled DNA preferentially, and to decatenate negatively supercoiled (pre-)catenanes without torsional relaxation (95). The regulation of Topo IV in space and time (through the interaction with other proteins; see before) further contributes to a preferred decatenation of replication intermediates. The preferred relaxation of positive over negatively supercoiled DNA is brought about through different processivities for these reactions, caused by different interactions of the CTDs with the DNA during catalysis (75,87,92). The distributive relaxation activity of Topo IV is reminiscent of Topo IA, a type I topo-isomerase responsible for the relaxation of negative DNA supercoils behind the translocating RNA polymerase during transcription (113,122). Inactivation of Topo IV leads to similar levels of hypernegative supercoiling as mutations of Topo IA (109), indicating that Topo IV and Topo IA may act in concert to limit the degree of negative supercoiling. Strikingly, more recent results show that both Topo IA and Topo III are not necessary when Topo IV is overexpressed, providing further evidence that Topo IV can relax negative supercoils in the wake of RNA polymerase when Topo IA is lacking (123).
Hybrid enzymes: part gyrase, part Topo IV
The division of labor between gyrase and Topo IV during replication is not possible in bacteria that possess only a single type IIA topoisomerase. These enzymes have to remove positive supercoils ahead of the fork and disentangle pre-catenanes behind the fork. One example are mycobacteria that contain only a gyrase. In-line with the physiological requirements, mycobacterial gyrases show an altered balance between intra- and intermolecular strand passage than gyrase homologs from organisms that also harbor a Topo IV, and catalyze supercoiling and decatenation with similar efficiencies (59,124,125). This balance is the result of less efficient DNA supercoiling compared to other gyrases; the decatenation activity remains lower than the one of Topo IV (124,125). Mycobacterial gyrase can thus be regarded as an evolutionary compromise to perform both tasks.
The hybrid character of mycobacterial gyrase is reflected in the fact that some of its enzymatic characteristics are more reminiscent of gyrase, whereas others are more similar to Topo IV (Table 1): the altered balance between supercoiling and decatenation may be linked to its unsual, ’Topo IV-like’ conformation, with the ATPase domains of GyrB bent downwards (44). In addition, Mycobacterium smegmatis gyrase stably binds two DNAs, similar to Topo IV (80), favoring intermolecular strand passage and decatenation. On the other hand, the CTDs of mycobacterial gyrase are gyrase-like, with six-blades in a spiral shape, and a canonical GyrA-box in blade 1, and are capable of DNA wrapping (107). As a result, the enzyme is also a gyrase with respect to its preference for positively supercoiled DNA substrates (91). In fact, mycobacterial gyrase wraps DNA more stably than E. coli gyrase (107), which, according to the kinetic competition model proposed by Nöllmannet al. (106), should favor supercoiling over wrapping-independent decatenation. On the other hand, exceedingly strong wrapping leads to a decrease in supercoiling activity of E. coli gyrase and futile cycling (97). It is conceivable that the same effect is responsible for the low supercoiling activity of mycobacterial gyrase and its higher propensity to decatenate. Notably, similar to other gyrases and unlike Topo IV, mycobacterial gyrase forms a lower level of cleavage complexes on positively than on negatively supercoiled DNA (91). Thus, despite the altered balance between supercoiling and decatenation, it remains a safe enzyme to process the unreplicated DNA in front of the replication fork with a low risk of introducing double-strand breaks (91).
Table 1.
Comparison of mycobacterial gyrase with Topo IV and other gyrases.
| Topo IV | mycobacterial gyrase | gyrase | ||
|---|---|---|---|---|
| structural features | ATPase domains | bent downa | bent downb | facing upwardsc |
| CTDs | 0–8 blades | six-bladed | six-bladed | |
| open | closed | closed | ||
| less spiral | spiral | spiral | ||
| GyrA-box | no | yes | yes | |
| interaction with DNA | DNA affinity | +sc ≈ -sc | +sc > -sc | +sc > -sc |
| # of DNAs bound | two | two | one | |
| cleavage | +sc > -sc | -sc > +sc | -sc > +sc | |
| wrapping | no | yes | yes | |
| mechanism | strand passage | decatenation: yes relaxation: yes | decatenation: yes supercoiling: ? | decatenation: yes supercoiling: not requiredd |
| reactions catalyzed | decatenation > relaxation | decatenation ≈ supercoiling | supercoiling > decatenation |
bPDB-ID 6gau, 6gav, see Figure 4C.
cAccording to single-molecule FRET data (34).
dGyrase with a single tyrosine catalyzes DNA supercoiling in the absence of strand passage by a nicking-closing mechanism. This enzyme cleaves only a single strand of the G-segment, but still catalyzes supercoiling in steps of two (84). -sc: negatively supercoiled DNA, +sc: positively supercoiled DNA.
Similar to the situation in Mycobacteria, only one type IIA topoisomerase has been identified in the thermophilic bacterium Aquifex aeolicus. Phylogenetically, this enzyme has been classified as a gyrase (126). However, the in vitro activities of this enzyme are characteristic of a Topo IV (58), making A. aeolicus the only organism known to date that lacks a functional gyrase. However, as a highly thermophilic bacterium, A. aeolicus may be a special case and may not require the introduction of negative supercoils by gyrase – in fact, the presence of such an activity could even be detrimental.
EVOLUTION OF TYPE IIA TOPOISOMERASES
The evolution of type IIA topoisomerases is most likely connected to the transition from an RNA to a DNA world, when the appearance of double-stranded DNA brought with it the topological problems resolved by these enzymes. For a long time, this hypothesis was supported by the absence of RNA topoisomerases, but with the discovery of RNA topoisomerase function for Topo III in 1996 (127), this picture has changed. It is conceivable that topoisomerase subunits were present even during the RNA world, and then jointly took over different functions later on (128). The structural similarities of type IIA topoisomerases, combined with their different functional specialization, suggest the presence of a common ancestor. The close relation of gyrase and Topo IV is also evidenced by their shared sensitivity to coumarin and quinolone inhibitors (120,129). It is unclear which of the two bacterial enzymes appeared first, gyrase or Topo IV. With its multiple cellular functions, in the maintenance of supercoiling homeostasis, and in transcription and replication, gyrase appears to be the more important enzyme of the two. In-line with this notion, gyrase is an essential enzyme present in all bacteria (with only one exception, see before), while Topo IV is not essential and not universally present (128). It was commonly believed that in organisms lacking Topo IV, the decatenation activity of gyrase could compensate for the missing Topo IV activity. In-line with this hypothesis, the gyrase from M. tuberculosis, an organisms lacking a Topo IV, shows a different balance between supercoiling and decatenation activities, and a higher tendency to decatenate DNA (124,125). Comparisons of gyrases from different organisms has shown that the balance between supercoiling and decatenation can be modulated through species-specific insertions in the topo-isomerase core (44–46). However, the CTDs are clearly a major determinant for the prevailing topoisomerase activity. This hypothesis was supported early-on by demonstrating that deletion of the gyrase CTDs converts the enzyme into a Topo IV (96). Later, it was shown that mutation of the GyrA-box is sufficient for converting a gyrase into a Topo IV (57). In fact, single point mutations in the CTD can alter the mode of DNA binding drastically, and can shift the balance between supercoiling and decatenation in gyrase (97). A single point mutation is also sufficient to generate a hybrid type IIA enzyme that acts as a gyrase on relaxed and moderately supercoiled DNA, but as a Topo IV when the DNA is highly negatively supercoiled (97). This shows that only small changes are required to change the activity profile of a gyrase into an enzyme with Topo IV-like activities.
An interesting case from an evolutionary perspective is the single type IIA topoisomerase of A. aeolicus. According to phylogenetic analyses, this enzyme is a hybrid enzyme with a gyrase-like topoisomerase core and a Topo IV-like CTD (126). This enzyme shows Topo IV-like activity in vitro, but is not capable of negative DNA supercoiling (58). Tretteret al. re-constituted a gyrase-like enzyme with supercoiling activity by mixing E. coli GyrA and A. aeolicus ParE (58). In fact, the exchange of the CTDs of A. aeolicus ParC by CTDs from Thermotoga maritima gyrase was sufficient to convert the A. aeolicus Topo IV into a gyrase. This gain-of-function experiment provides strong support for the hypothesis of a modular architecture of type IIA topoisomerases, in which the CTDs are the key determinant for the prevailing activity (58).
These considerations bring us back to the order of appearance of gyrase and Topo IV. Was the last common ancestor a gyrase or a Topo IV? The phylogenetic distribution of topoisomerases is difficult to interpret as it does not reflect the universal tree of life (128,130). Phylogenetic analyses of type IIA topoisomerase subunits put gyrase and Topo IV into one group, separate from eukaryotic Topo II (128). The structure of the phylogenetic tree suggests an early separation of Topo IV and gyrase, arguing against a development of Topo IV from gyrase (128). However, increasing experimental evidence on the central function of the CTDs supports the hypothesis that the common ancestor of gyrase and Topo IV was in fact a gyrase (Figure 6). Based on the central functional role of the CTDs for gyrase activity, this hypothesis posits that the CTDs were fused to an ancestral type IIA topoisomerase core during evolution (58,131). In fact, in B. burgdorferi, the CTD is present as a separate DNA binding protein, independent of gyrase (132). B. burgdorferi may reflect an evolutionary intermediate that provides support for the occurrence of such a fusion event in evolution. The CTD itself most likely originated from the duplication of a single blade (30,56), each of which carried a GyrA-box. The β-pinwheel topology may have originated from the assembly of blades into a canonical β-propeller, followed by invasion of a hairpin into the adjacent blade, rearrangement of the strands and a domain swap, to arrive at the observed D–A–B–C strand order (hairpin invasion hypothesis) (56). Alternatively, the β-pinwheel may have originated from the repetitive assembly of the common Greek key motif, followed by a domain swap leading to the formation of an antiparallel β-sheet between strand C of one blade and strand B of the neighboring blade (30) (Figure 6). In any case, this would make the first enzyme a gyrase, from which Topo IV appeared through another gene duplication event. During evolution, only the GyrA-box in the first blade was preserved in gyrase, whereas the other blades gained different functions in DNA binding and bending (97). Topo IV lost the canonical GyrA-box in all blades, although remnants are still present. In addition, Topo IV lost or gained individual blades of the CTD (50,58,75). In this scenario, B. stearothermophilus Topo IV with its six-bladed, spiral CTD might constitute an early evolutionary intermediate that is still close to a gyrase (56).
Figure 6.
Possible evolutionary pathway for the appearance of gyrase and Topo IV. The β-pinwheel of the CTDs was either formed by the repetitive assembly of single blades consisting of β-sheets with A–B–C–D topology into a canonical β-propeller, followed by the invasion of a hairpin formed by strands B and C into the adjacent blade, and subsequent rearrangements that convert the β-propeller into a β-pinwheel with a D–A–B–C topology, including a domain swap of strand C (bottom). Alternatively, individual blades with the D–A–B–C topology of the prevalent Greek key motif may have assembled directly into a β-pinwheel, followed by a domain swap of strand C (top). Both pathways would have generated a protein capable of binding DNA around its perimeter. Fusion of this ancestral CTD (green) to an ancestral topoisomerase core (gray) then led to the appearance of gyrase. Gene duplication then enabled branching into Topo IV and gyrase. Subsequent changes in the CTDs and the topoisomerase core enabled the specialization of gyrase in different organisms. For Topo IV, the loss of the GyrA-box and loss or gain of blades followed.
CONCLUSIONS
In this review, we have summarized how the type IIA topo-isomerase scaffold provides a common basis for two different biological tasks, decatenation and supercoiling. Supercoiling enzymes are optimized for DNA wrapping and intramolecular strand passage (gyrase), while efficient decatenases are optimized for intermolecular strand passage (Topo IV). We have also illustrated how, based on the same type IIA scaffold, gyrase can be fine-tuned to become more or less Topo IV-like. E. coli gyrase is an example for an enzyme that is highly optimized for its function as a supercoiling enzyme, with very little Topo IV-like decatenation activities. M. tuberculosis gyrase, on the other hand, is an example for a hybrid, more Topo IV-like enzyme, that is less efficient in supercoiling, but shows robust decatenation activity. Mutational studies on both gyrase and Topo IV have shown that the balance between ‘gyrase-like’ and ‘Topo IV-like’ can be tipped by as little as a single point mutation in the CTDs. Thus, gyrase and Topo IV provide an illustrative example for a continuum of overlapping functions, based on multi-layered evolutionary fine-tuning of a common core mechanism. We are only beginning to understand the molecular mechanisms that enable this spectrum of activities. While the DNA- and nucleotide-induced conformational changes that orchestrate DNA supercoiling by gyrase have been dissected in detail, less is known about the conformational changes associated with gyrase- or Topo IV-catalyzed decatenation. To arrive at a thorough mechanistic understanding of these enzymes during their different reactions, it is key to explore the potential differences in the conformational cycle of gyrase and Topo IV, differences between the catalysis of supercoiling and decatenation by gyrase, as well as catalytic nuances between gyrases from different species. The insight gained from such studies will not only define the molecular determinants that make a type IIA topoisomerase a gyrase or a Topo IV, but will also help unravel possible evolutionary relationships and pathways, and may open up novel pathways for gyrase and/or Topo IV inhibition.
Contributor Information
Jana Hirsch, University of Muenster, Institute for Physical Chemistry, Corrensstrasse 30, 48149 Muenster, Germany.
Dagmar Klostermeier, University of Muenster, Institute for Physical Chemistry, Corrensstrasse 30, 48149 Muenster, Germany.
FUNDING
Deutsche Forschungsgemeinschaft [KL1153/9-1 and KL1153/9-2 to D.K.]. The open access publication charge for this paper has been waived by Oxford University Press – NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal.
Conflict of interest statement. None declared.
REFERENCES
- 1. Mizuuchi K., O’Dea M.H., Gellert M.. DNA gyrase: subunit structure and ATPase activity of the purified enzyme. Proc. Natl. Acad. Sci U.S.A. 1978; 75:5960–5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cozzarelli N.R., Wang J.C.. DNA Topology and its Biological Effects. 1990; Cold Spring Harbor Laboratory Press. [Google Scholar]
- 3. Liu L.F., Wang J.C.. Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci USA. 1987; 84:7024–7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tsao Y.-P., Wu H.-Y., Liu L.F.. Transcription-driven supercoiling of DNA: direct biochemical evidence from in vitro studies. Cell. 1989; 56:111–118. [DOI] [PubMed] [Google Scholar]
- 5. McKie S.J., Neuman K.C., Maxwell A.. DNA topoisomerases: advances in understanding of cellular roles and multi-protein complexes via structure-function analysis. BioEssays. 2021; 4:2000286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schoeffler A.J., Berger J.M.. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q. Rev. Biophys. 2008; 41:41–101. [DOI] [PubMed] [Google Scholar]
- 7. Vos S.M., Tretter E.M., Schmidt B.H., Berger J.M.. All tangled up: how cells direct, manage and exploit topoisomerase function. Nat. Rev. Mol. Cell Biol. 2011; 12:827–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Goto T., Wang J.C.. Yeast DNA topoisomerase II. An ATP-dependent type II topoisomerase that catalyzes the catenation, decatenation, unknotting, and relaxation of double-stranded DNA rings. J. Biol. Chem. 1982; 257:5866–5872. [PubMed] [Google Scholar]
- 9. Kato J., Nishimura Y., Imamura R., Niki H., Hiraga S., Suzuki H.. New topoisomerase essential for chromosome segregation in E. coli. Cell. 1990; 63:393–404. [DOI] [PubMed] [Google Scholar]
- 10. Adams D.E., Shekhtman E.M., Zechiedrich E.L., Schmid M.B., Cozzarelli N.R.. The role of topoisomerase IV in partitioning bacterial replicons and the structure of catenated intermediates in DNA replication. Cell. 1992; 71:277–288. [DOI] [PubMed] [Google Scholar]
- 11. Crisona N.J., Strick T.R., Bensimon D., Croquette V., Cozzarelli N.R.. Preferential relaxation of positively supercoiled DNA by E. coli topoisomerase IV in single-molecule and ensemble measurements. Genes Dev. 2000; 14:2881–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ashley R.E., Dittmore A., McPherson S.A., Turnbough C.L., Neuman K.C., Osheroff N.. Activities of gyrase and topoisomerase IV on positively supercoiled DNA. Nucleic Acids Res. 2017; 45:9611–9624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kreuzer K.N., Cozzarelli N.R.. Formation and resolution of DNA catenanes by DNA gyrase. Cell. 1980; 20:245–254. [DOI] [PubMed] [Google Scholar]
- 14. Gellert M., Mizuuchi K., O’Dea M.H., Nash H.A.. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc. Natl. Acad. Sci U.S.A. 1976; 73:3872–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Higgins N.P., Peebles C.L., Sugino A., Cozzarelli N.R.. Purification of subunits of Escherichia coli DNA gyrase and reconstitution of enzymatic activity. Proc. Natl. Acad. Sci U.S.A. 1978; 75:1773–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zechiedrich E.L., Cozzarelli N.R.. Roles of topoisomerase IV and DNA gyrase in DNA unlinking during replication in Escherichia coli. Genes Dev. 1995; 9:2859–2869. [DOI] [PubMed] [Google Scholar]
- 17. Zechiedrich E.L., Khodursky A.B., Cozzarelli N.R.. Topoisomerase IV, not gyrase, decatenates products of site-specific recombination in Escherichia coli. Genes Dev. 1997; 11:2580–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Champoux J.J., Been M.D.. Topoisomerases and the Swivel Problem, Mechanistic Studies of DNA Replication and Genetic Recombination. 1980; Elsevier; 809–815. [Google Scholar]
- 19. Wang X., Reyes-Lamothe R., Sherratt D.J.. Modulation of Escherichia coli sister chromosome cohesion by topoisomerase IV. Genes Dev. 2008; 22:2426–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zawadzki P., Stracy M., Ginda K., Zawadzka K., Lesterlin C., Kapanidis A.N., Sherratt D.J.. The localization and action of topoisomerase IV in Escherichia coli chromosome segregation is coordinated by the SMC complex, MukBEF. Cell Rep. 2015; 13:2587–2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stracy M., Wollman A.J.M., Kaja E., Gapinski J., Lee J.-E., Leek V.A., McKie S.J., Mitchenall L.A., Maxwell A., Sherratt D.J.et al.. Single-molecule imaging of DNA gyrase activity in living Escherichia coli. Nucleic Acids Res. 2019; 47:210–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Khodursky A.B., Peter B.J., Schmid M.B., DeRisi J., Botstein D., Brown P.O., Cozzarelli N.R.. Analysis of topoisomerase function in bacterial replication fork movement: use of DNA microarrays. Proc. Natl. Acad. Sci U.S.A. 2000; 97:9419–9424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Strahilevitz J., Hooper D.C.. Dual targeting of topoisomerase IV and gyrase to reduce mutant selection: direct testing of the paradigm by using WCK-1734, a new fluoroquinolone, and ciprofloxacin. Antimicrob. Agents Chemother. 2005; 49:1949–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sissi C., Palumbo M.. In front of and behind the replication fork: bacterial type IIA topoisomerases. Cell. Mol. Life Sci. 2010; 67:2001–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bisacchi G.S., Manchester J.I.. A new-class antibacterial–almost. Lessons in drug discovery and development: a critical analysis of more than 50 years of effort toward ATPase inhibitors of DNA gyrase and topoisomerase IV. ACS Infect. Dis. 2015; 1:4–41. [DOI] [PubMed] [Google Scholar]
- 26. Tse-Dinh Y.-C. Targeting bacterial topoisomerases: how to counter mechanisms of resistance. Future Med. Chem. 2016; 8:1085–1100. [DOI] [PubMed] [Google Scholar]
- 27. Fu G., Wu J., Liu W., Zhu D., Hu Y., Deng J., Zhang X.-E., Bi L., Wang D.-C.. Crystal structure of DNA gyrase B′ domain sheds lights on the mechanism for T-segment navigation. Nucleic Acids Res. 2009; 37:5908–5916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cabral J.H.M., Jackson A.P., Smith C.V., Shikotra N., Maxwell A., Liddington R.C.. Crystal structure of the breakage-reunion domain of DNA gyrase. Nature. 1997; 388:903–906. [DOI] [PubMed] [Google Scholar]
- 29. Reece R.J., Maxwell A.. The C-terminal domain of the Escherichia coli DNA gyrase A subunit is a DNA-binding protein. Nucleic Acids Res. 1991; 19:1399–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Corbett K.D., Shultzaberger R.K., Berger J.M.. The C-terminal domain of DNA gyrase A adopts a DNA-bending β-pinwheel fold. Proc. Natl. Acad. Sci U.S.A. 2004; 101:7293–7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Roca J., Wang J.C.. DNA transport by a type II DNA topoisomerase: evidence in favor of a two-gate mechanism. Cell. 1994; 77:609–616. [DOI] [PubMed] [Google Scholar]
- 32. Roca J., Berger J.M., Harrison S.C., Wang J.C.. DNA transport by a type II topoisomerase: direct evidence for a two-gate mechanism. Proc. Natl. Acad. Sci U.S.A. 1996; 93:4057–4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Roca J. The path of the DNA along the dimer interface of topoisomerase II. J. Biol. Chem. 2004; 279:25783–25788. [DOI] [PubMed] [Google Scholar]
- 34. Gubaev A., Klostermeier D.. DNA-induced narrowing of the gyrase N-gate coordinates T-segment capture and strand passage. Proc. Natl. Acad. Sci U.S.A. 2011; 108:14085–14090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tingey A.P., Maxwell A.. Probing the role of the ATP-operated clamp in the strand-passage reaction of DNA gyrase. Nucleic Acids Res. 1996; 24:4868–4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Williams N.L., Howells A.J., Maxwell A.. Locking the ATP-operated clamp of DNA gyrase: probing the mechanism of strand passage. J. Mol. Biol. 2001; 306:969–984. [DOI] [PubMed] [Google Scholar]
- 37. Laponogov I., Pan X.-S., Veselkov D.A., Skamrova G.B., Umrekar T.R., Fisher L.M., Sanderson M.R.. Trapping of the transport-segment DNA by the ATPase domains of a type II topoisomerase. Nat. Commun. 2018; 9:2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Brino L., Urzhumtsev A., Mousli M., Bronner C., Mitschler A., Oudet P., Moras D.. Dimerization of Escherichia coli DNA-gyrase B provides a structural mechanism for activating the ATPase catalytic center. J. Biol. Chem. 2000; 275:9468–9475. [DOI] [PubMed] [Google Scholar]
- 39. Gubaev A., Hilbert M., Klostermeier D.. The DNA-gate of Bacillus subtilis gyrase is predominantly in the closed conformation during the DNA supercoiling reaction. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:13278–13283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dong K.C., Berger J.M.. Structural basis for gate-DNA recognition and bending by type IIA topoisomerases. Nature. 2007; 450:1201–1205. [DOI] [PubMed] [Google Scholar]
- 41. Berger J.M., Gamblin S.J., Harrison S.C., Wang J.C.. Structure and mechanism of DNA topoisomerase II. Nature. 1996; 379:225–232. [DOI] [PubMed] [Google Scholar]
- 42. Tennyson R.B., Lindsley J.E.. Type II DNA topoisomerase from Saccharomyces cerevisiae is a stable dimer. Biochemistry. 1997; 36:6107–6114. [DOI] [PubMed] [Google Scholar]
- 43. Wigley D.B., Davies G.J., Dodson E.J., Maxwell A., Dodson G.. Crystal structure of an N-terminal fragment of the DNA gyrase B protein. Nature. 1991; 351:624–629. [DOI] [PubMed] [Google Scholar]
- 44. Petrella S., Capton E., Raynal B., Giffard C., Thureau A., Bonneté F., Alzari P.M., Aubry A., Mayer C.. Overall structures of Mycobacterium tuberculosis DNA Gyrase reveal the role of a corynebacteriales GyrB-specific insert in ATPase activity. Structure. 2019; 27:579–589. [DOI] [PubMed] [Google Scholar]
- 45. Weidlich D., Klostermeier D.. Functional interactions between gyrase subunits are optimized in a species-specific manner. J. Biol. Chem. 2020; 295:2299–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schoeffler A.J., May A.P., Berger J.M.. A domain insertion in Escherichia coli GyrB adopts a novel fold that plays a critical role in gyrase function. Nucleic Acids Res. 2010; 38:7830–7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bellon S., Parsons J.D., Wei Y., Hayakawa K., Swenson L.L., Charifson P.S., Lippke J.A., Aldape R., Gross C.H.. Crystal structures of Escherichia coli topoisomerase IV ParE subunit (24 and 43 kilodaltons): a single residue dictates differences in novobiocin potency against topoisomerase IV and DNA gyrase. Antimicrob. Agents Chemother. 2004; 48:1856–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tari L.W., Li X., Trzoss M., Bensen D.C., Chen Z., Lam T., Zhang J., Lee S.J., Hough G., Phillipson D.et al.. Tricyclic GyrB/ParE (TriBE) inhibitors: a new class of broad-spectrum dual-targeting antibacterial agents. PLoS One. 2013; 8:e84409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jung H.Y., Heo Y.-S.. Crystal structures of the 43 kDa ATPase domain of Xanthomonas oryzae pv. oryzae topoisomerase IV ParE subunit and its complex with novobiocin. Crystals. 2019; 9:577. [Google Scholar]
- 50. Corbett K.D., Schoeffler A.J., Thomsen N.D., Berger J.M.. The structural basis for substrate specificity in DNA topoisomerase IV. J. Mol. Biol. 2005; 351:545–561. [DOI] [PubMed] [Google Scholar]
- 51. Carr S.B., Makris G., Phillips S.E.V., Thomas C.D.. Crystallization and preliminary X-ray diffraction analysis of two N-terminal fragments of the DNA-cleavage domain of topoisomerase IV from Staphylococcus aureus. Acta Crystallogr., Sect. F: Struct. Biol. Cryst. Commun. 2006; 62:1164–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Laponogov I., Sohi M.K., Veselkov D.A., Pan X.-S., Sawhney R., Thompson A.W., McAuley K.E., Fisher L.M., Sanderson M.R.. Structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases. Nat. Struct. Mol. Biol. 2009; 16:667. [DOI] [PubMed] [Google Scholar]
- 53. Wohlkonig A., Chan P.F., Fosberry A.P., Homes P., Huang J., Kranz M., Leydon V.R., Miles T.J., Pearson N.D., Perera R.L.et al.. Structural basis of quinolone inhibition of type IIA topoisomerases and target-mediated resistance. Nat. Struct. Mol. Biol. 2010; 17:1152. [DOI] [PubMed] [Google Scholar]
- 54. Skepper C.K., Armstrong D., Balibar C.J., Bauer D., Bellamacina C., Benton B.M., Bussiere D., Pascale G., Vicente J., Dean C.R.et al.. Topoisomerase inhibitors addressing fluoroquinolone resistance in Gram-negative bacteria. J. Med. Chem. 2020; 63:7773–7816. [DOI] [PubMed] [Google Scholar]
- 55. Paoli M. Protein folds propelled by diversity. Prog. Biophys. Mol. Biol. 2001; 76:103–130. [DOI] [PubMed] [Google Scholar]
- 56. Hsieh T.-J., Farh L., Huang W.M., Chan N.-L.. Structure of the topoisomerase IV C-terminal domain A broken beta-propeller implies a role as geometry facilitator in catalysis. J. Biol. Chem. 2004; 279:55587–55593. [DOI] [PubMed] [Google Scholar]
- 57. Kramlinger V.M., Hiasa H.. The “GyrA-box” is required for the ability of DNA gyrase to wrap DNA and catalyze the supercoiling reaction. J. Biol. Chem. 2006; 281:3738–3742. [DOI] [PubMed] [Google Scholar]
- 58. Tretter E.M., Lerman J.C., Berger J.M.. A naturally chimeric type IIA topoisomerase in Aquifex aeolicus highlights an evolutionary path for the emergence of functional paralogs. Proc. Natl. Acad. Sci U.S.A. 2010; 107:22055–22059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bouige A., Darmon A., Piton J., Roue M., Petrella S., Capton E., Forterre P., Aubry A., Mayer C.. Mycobacterium tuberculosis DNA gyrase possesses two functional GyrA-boxes. Biochem. J. 2013; 455:285–294. [DOI] [PubMed] [Google Scholar]
- 60. Liu L.F., Wang J.C.. DNA-DNA gyrase complex: the wrapping of the DNA duplex outside the enzyme. Cell. 1978; 15:979–984. [DOI] [PubMed] [Google Scholar]
- 61. Ruthenburg A.J., Graybosch D.M., Huetsch J.C., Verdine G.L.. A superhelical spiral in the Escherichia coli DNA gyrase A C-terminal domain imparts unidirectional supercoiling bias. J. Biol. Chem. 2005; 280:26177–26184. [DOI] [PubMed] [Google Scholar]
- 62. Hsieh T.-J., Yen T.-J., Lin T.-S., Chang H.-T., Huang S.-Y., Hsu C.-H., Farh L., Chan N.-L.. Twisting of the DNA-binding surface by a β-strand-bearing proline modulates DNA gyrase activity. Nucleic Acids Res. 2010; 38:4173–4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tretter E.M., Berger J.M.. Mechanisms for defining supercoiling set point of DNA gyrase orthologs I. A nonconserved acidic C-terminal tail modulates Escherichia coli gyrase activity. J. Biol. Chem. 2012; 287:18636–18644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lanz M.A., Farhat M., Klostermeier D.. The acidic C-terminal tail of the GyrA subunit moderates the DNA supercoiling activity of Bacillus subtilis gyrase. J. Biol. Chem. 2014; 289:12275–12285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Schmidt B.H., Osheroff N., Berger J.M.. Structure of a topoisomerase II-DNA-nucleotide complex reveals a new control mechanism for ATPase activity. Nat. Struct. Mol. Biol. 2012; 19:1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Laponogov I., Veselkov D.A., Crevel I.M.-T., Pan X.-S., Fisher L.M., Sanderson M.R.. Structure of an ‘open’clamp type II topoisomerase-DNA complex provides a mechanism for DNA capture and transport. Nucleic Acids Res. 2013; 41:9911–9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Papillon J., Menetret J.-F., Batisse C., Helye R., Schultz P., Potier N., Lamour V.. Structural insight into negative DNA supercoiling by DNA gyrase, a bacterial type 2A DNA topoisomerase. Nucleic Acids Res. 2013; 41:7815–7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Broeck A.V., Lotz C., Ortiz J., Lamour V.. Cryo-EM structure of the complete E. coli DNA gyrase nucleoprotein complex. Nat. Commun. 2019; 10:4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Piton J., Petrella S., Delarue M., André-Leroux G., Jarlier V., Aubry A., Mayer C.. Structural insights into the quinolone resistance mechanism of Mycobacterium tuberculosis DNA gyrase. PLoS One. 2010; 5:e12245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Agrawal A., Roué M., Spitzfaden C., Petrella S., Aubry A., Hann M., Bax B., Mayer C.. Mycobacterium tuberculosis DNA gyrase ATPase domain structures suggest a dissociative mechanism that explains how ATP hydrolysis is coupled to domain motion. Biochem. J. 2013; 456:263–273. [DOI] [PubMed] [Google Scholar]
- 71. Kirchhausen T., Wang J.C., Harrison S.C.. DNA gyrase and its complexes with DNA: direct observation by electron microscopy. Cell. 1985; 41:933–943. [DOI] [PubMed] [Google Scholar]
- 72. Costenaro L., Grossmann J.G., Ebel C., Maxwell A.. Small-angle X-ray scattering reveals the solution structure of the full-length DNA gyrase a subunit. Structure. 2005; 13:287–296. [DOI] [PubMed] [Google Scholar]
- 73. Baker N.M., Weigand S., Maar-Mathias S., Mondragon A.. Solution structures of DNA-bound gyrase. Nucleic Acids Res. 2011; 39:755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lanz M.A., Klostermeier D.. Guiding strand passage: DNA-induced movement of the gyrase C-terminal domains defines an early step in the supercoiling cycle. Nucleic Acids Res. 2011; 39:9681–9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Vos S.M., Lee I., Berger J.M.. Distinct regions of the Escherichia coli ParC C-terminal domain are required for substrate discrimination by topoisomerase IV. J. Mol. Biol. 2013; 425:3029–3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wang J.C. Moving one DNA double helix through another by a type II DNA topoisomerase: the story of a simple molecular machine. Q. Rev. Biophys. 1998; 31:107–144. [DOI] [PubMed] [Google Scholar]
- 77. Laponogov I., Pan X.-S., Veselkov D.A., McAuley K.E., Fisher L.M., Sanderson M.R.. Structural basis of gate-DNA breakage and resealing by type II topoisomerases. PLoS One. 2010; 5:e11338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Maxwell A., Gellert M.. The DNA dependence of the ATPase activity of DNA gyrase. J. Biol. Chem. 1984; 259:14472–14480. [PubMed] [Google Scholar]
- 79. Rau D.C., Gellert M., Thoma F., Maxwell A.. Structure of the DNA gyrase-DNA complex as revealed by transient electric dichroism. J. Mol. Biol. 1987; 193:555–569. [DOI] [PubMed] [Google Scholar]
- 80. Kumar R., Riley J.E., Parry D., Bates A.D., Nagaraja V.. Binding of two DNA molecules by type II topoisomerases for decatenation. Nucleic Acids Res. 2012; 40:10904–10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Mizuuchi K., Fisher L.M., O’Dea M.H., Gellert M.. DNA gyrase action involves the introduction of transient double-strand breaks into DNA. Proc. Natl. Acad. Sci U.S.A. 1980; 77:1847–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Williams N.L., Maxwell A.. Probing the two-gate mechanism of DNA gyrase using cysteine cross-linking. Biochemistry. 1999; 38:13502–13511. [DOI] [PubMed] [Google Scholar]
- 83. Hartmann S., Gubaev A., Klostermeier D.. Binding and hydrolysis of a single ATP is sufficient for N-gate closure and DNA supercoiling by gyrase. J. Mol. Biol. 2017; 429:3717–3729. [DOI] [PubMed] [Google Scholar]
- 84. Gubaev A., Weidlich D., Klostermeier D.. DNA gyrase with a single catalytic tyrosine can catalyze DNA supercoiling by a nicking-closing mechanism. Nucleic Acids Res. 2016; 44:10354–10366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Klostermeier D. Why two? On the role of (a-) symmetry in negative supercoiling of DNA by gyrase. Int. J. Mol. Sci. 2018; 19:1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ullsperger C., Cozzarelli N.R.. Contrasting enzymatic activities of topoisomerase IV and DNA gyrase from Escherichia coli. J. Biol. Chem. 1996; 271:31549–31555. [DOI] [PubMed] [Google Scholar]
- 87. Neuman K.C., Charvin G., Bensimon D., Croquette V.. Mechanisms of chiral discrimination by topoisomerase IV. Proc. Acad. Natl. Sci. Philadelphia. 2009; 106:6986–6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Marians K.J. DNA gyrase-catalyzed decatenation of multiply linked DNA dimers. J. Biol. Chem. 1987; 262:10362–10368. [PubMed] [Google Scholar]
- 89. Hiasa H., Marians K.J.. Two distinct modes of strand unlinking during θ-type DNA replication. J. Biol. Chem. 1996; 271:21529–21535. [DOI] [PubMed] [Google Scholar]
- 90. Higgins N.P., Cozzarelli N.R.. The binding of gyrase to DNA: analysis by retention by nitrocellulose filters. Nucleic Acids Res. 1982; 10:6833–6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ashley R.E., Blower T.R., Berger J.M., Osheroff N.. Recognition of DNA supercoil geometry by Mycobacterium tuberculosis gyrase. Biochemistry. 2017; 56:5440–5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Stone M.D., Bryant Z., Crisona N.J., Smith S.B., Vologodskii A., Bustamante C., Cozzarelli N.R.. Chirality sensing by Escherichia coli topoisomerase IV and the mechanism of type II topoisomerases. Proc. Natl. Acad. Sci U.S.A. 2003; 100:8654–8659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Charvin G., Strick T.R., Bensimon D., Croquette V.. Topoisomerase IV bends and overtwists DNA upon binding. Biophys. J. 2005; 89:384–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Crisona N.J., Cozzarelli N.R.. Alteration of Escherichia coli topoisomerase IV conformation upon enzyme binding to positively supercoiled DNA. J. Biol. Chem. 2006; 281:18927–18932. [DOI] [PubMed] [Google Scholar]
- 95. Rawdon E.J., Dorier J., Racko D., Millett K.C., Stasiak A.. How topoisomerase IV can efficiently unknot and decatenate negatively supercoiled DNA molecules without causing their torsional relaxation. Nucleic Acids Res. 2016; 44:4528–4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Kampranis S.C., Maxwell A.. Conversion of DNA gyrase into a conventional type II topoisomerase. Proc. Natl. Acad. Sci U.S.A. 1996; 93:14416–14421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Hobson M.J., Bryant Z., Berger J.M.. Modulated control of DNA supercoiling balance by the DNA-wrapping domain of bacterial gyrase. Nucleic Acids Res. 2020; 48:2035–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Klevan L., Wang J.C.. Deoxyribonucleic acidgyrase-deoxyribonucleic acid complex containing 140 base pairs of deoxyribonucleic acid and an α2β2 protein core. Biochemistry. 1980; 19:5229–5234. [DOI] [PubMed] [Google Scholar]
- 99. Fisher L.M., Mizuuchi K., O’Dea M.H., Ohmori H., Gellert M.. Site-specific interaction of DNA gyrase with DNA. Proc. Natl. Acad. Sci U.S.A. 1981; 78:4165–4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Kirkegaard K., Wang J.C.. Mapping the topography of DNA wrapped around gyrase by nucleolytic and chemical probing of complexes of unique DNA sequences. Cell. 1981; 23:721–729. [DOI] [PubMed] [Google Scholar]
- 101. Morrison A., Cozzarelli N.R.. Contacts between DNA gyrase and its binding site on DNA: features of symmetry and asymmetry revealed by protection from nucleases. Proc. Natl. Acad. Sci U.S.A. 1981; 78:1416–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Orphanides G., Maxwell A.. Evidence for a conformational change in the DNA gyrase-DNA complex from hydroxyl radical footprinting. Nucleic Acids Res. 1994; 22:1567–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Peng H., Marians K.J.. The interaction of Escherichia coli topoisomerase IV with DNA. J. Biol. Chem. 1995; 270:25286–25290. [DOI] [PubMed] [Google Scholar]
- 104. Lee M.P., Sander M., Hsieh T.-S.. Nuclease protection by Drosophila DNA topoisomerase II. Enzyme/DNA contacts at the strong topoisomerase II cleavage sites. J. Biol. Chem. 1989; 264:21779–21787. [PubMed] [Google Scholar]
- 105. Lanz M.A., Klostermeier D.. The GyrA-box determines the geometry of DNA bound to gyrase and couples DNA binding to the nucleotide cycle. Nucleic Acids Res. 2012; 40:10893–10903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Nöllmann M., Stone M.D., Bryant Z., Gore J., Crisona N.J., Hong S.-C., Mitelheiser S., Maxwell A., Bustamante C., Cozzarelli N.R.. Multiple modes of Escherichia coli DNA gyrase activity revealed by force and torque. Nat. Struct. Mol. Biol. 2007; 14:264–271. [DOI] [PubMed] [Google Scholar]
- 107. Tretter E.M., Berger J.M.. Mechanisms for defining supercoiling set point of DNA gyrase orthologs II. The shape of the gyra subunit C-terminal domain (CTD) is not a sole determinant for controlling supercoiling efficiency. J. Biol. Chem. 2012; 287:18645–18654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Rovinskiy N.S., Agbleke A.A., Chesnokova O.N., Higgins N.P.. Supercoil levels in E. coli and Salmonella chromosomes are regulated by the C-terminal 35-38 amino acids of GyrA. Microorganisms. 2019; 7:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Zechiedrich E.L., Khodursky A.B., Bachellier S., Schneider R., Chen D., Lilley D.M.J., Cozzarelli N.R.. Roles of topoisomerases in maintaining steady-state DNA supercoiling in Escherichia coli. J. Biol. Chem. 2000; 275:8103–8113. [DOI] [PubMed] [Google Scholar]
- 110. Rovinskiy N., Agbleke A.A., Chesnokova O., Pang Z., Higgins N.P.. Rates of gyrase supercoiling and transcription elongation control supercoil density in a bacterial chromosome. PLos Genet. 2012; 8:e1002845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Willmott C. Jr, Critchlow S.E., Eperon I.C., Maxwell A.. The complex of DNA gyrase and quinolone drugs with DNA forms a barrier to transcription by RNA polymerase. J. Mol. Biol. 1994; 242:351–363. [DOI] [PubMed] [Google Scholar]
- 112. Tadesse S., Graumann P.L.. Differential and dynamic localization of topoisomerases in Bacillus subtilis. J. Bacteriol. 2006; 188:3002–3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Ahmed W., Sala C., Hegde S.R., Jha R.K., Cole S.T., Nagaraja V.. Transcription facilitated genome-wide recruitment of topoisomerase I and DNA gyrase. PLos Genet. 2017; 13:e1006754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Espeli O., Levine C., Hassing H., Marians K.J.. Temporal regulation of topoisomerase IV activity in E. coli. Mol. Cell. 2003; 11:189–201. [DOI] [PubMed] [Google Scholar]
- 115. Li Y., Stewart N.K., Berger A.J., Vos S., Schoeffler A.J., Berger J.M., Chait B.T., Oakley M.G.. Escherichia coli condensin MukB stimulates topoisomerase IV activity by a direct physical interaction. Proc. Natl. Acad. Sci U.S.A. 2010; 107:18832–18837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Nicolas E., Upton A.L., Uphoff S., Henry O., Badrinarayanan A., Sherratt D.. The SMC complex MukBEF recruits topoisomerase IV to the origin of replication region in live Escherichia coli. mBio. 2014; 5:e01001-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Lu M., Campbell J.L., Boye E., Kleckner N.. SeqA: a negative modulator of replication initiation in E. coli. Cell. 1994; 77:413–426. [DOI] [PubMed] [Google Scholar]
- 118. Kang S., Han J.S., Park J.H., Skarstad K., Hwang D.S.. SeqA protein stimulates the relaxing and decatenating activities of topoisomerase IV. J. Biol. Chem. 2003; 278:48779–48785. [DOI] [PubMed] [Google Scholar]
- 119. Joshi M.C., Magnan D., Montminy T.P., Lies M., Stepankiw N., Bates D.. Regulation of sister chromosome cohesion by the replication fork tracking protein SeqA. PLos Genet. 2013; 9:e1003673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Kato J., Suzuki H., Ikeda H.. Purification and characterization of DNA topoisomerase IV in Escherichia coli. J. Biol. Chem. 1992; 267:25676–25684. [PubMed] [Google Scholar]
- 121. Peng H., Marians K.J.. Decatenation activity of topoisomerase IV during oriC and pBR322 DNA replication in vitro. Proc. Acad. Natl. Sci. Philadelphia. 1993; 90:8571–8575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Massé E., Drolet M.. Relaxation of transcription-induced negative supercoiling is an essential function of Escherichia coli DNA topoisomerase I. J. Biol. Chem. 1999; 274:16654–16658. [DOI] [PubMed] [Google Scholar]
- 123. Reuß D.R., Faßhauer P., Mroch P.J., Ul-Haq I., Koo B.-M., Pöhlein A., Gross C.A., Daniel R., Brantl S., Stülke J.. Topoisomerase IV can functionally replace all type 1A topoisomerases in Bacillus subtilis. Nucleic Acids Res. 2019; 47:5231–5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Manjunatha U.H., Dalal M., Chatterji M., Radha D.R., Visweswariah S.S., Nagaraja V.. Functional characterisation of mycobacterial DNA gyrase: an efficient decatenase. Nucleic Acids Res. 2002; 30:2144–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Aubry A., Fisher L.M., Jarlier V., Cambau E.. First functional characterization of a singly expressed bacterial type II topoisomerase: the enzyme from Mycobacterium tuberculosis. Biochem. Biophys. Res. Commun. 2006; 348:158–165. [DOI] [PubMed] [Google Scholar]
- 126. Deckert G., Warren P.V., Gaasterland T., Young W.G., Lenox A.L., Graham D.E., Overbeek R., Snead M.A., Keller M., Aujay M.et al.. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998; 392:353–358. [DOI] [PubMed] [Google Scholar]
- 127. Wang H., Di Gate R.J., Seeman N.C.. An RNA topoisomerase. Proc. Natl. Acad. Sci U.S.A. 1996; 93:9477–9482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Forterre P., Gribaldo S., Gadelle D., Serre M.-C.. Origin and evolution of DNA topoisomerases. Biochimie. 2007; 89:427–446. [DOI] [PubMed] [Google Scholar]
- 129. Khodursky A.B., Zechiedrich E.L., Cozzarelli N.R.. Topoisomerase IV is a target of quinolones in Escherichia coli. Proc. Natl. Acad. Sci U.S.A. 1995; 92:11801–11805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Woese C.R., Kandler O., Wheelis M.L.. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci U.S.A. 1990; 87:4576–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Dillon S.C., Dorman C.J.. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 2010; 8:185–195. [DOI] [PubMed] [Google Scholar]
- 132. Knight S.W., Samuels D.S.. Natural synthesis of a DNA-binding protein from the C-terminal domain of DNA gyrase A in Borrelia burgdorferi. EMBO J. 1999; 18:4875–4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. DeLano W.L., others Pymol: an open-source molecular graphics tool. CCP4 Newslett. Protein Crystallogr. 2002; 40:82–92. [Google Scholar]