Abstract
Metagenomic analysis of food is becoming more routine and can provide important information pertaining to the shelf life potential and the safety of these products. However, less information is available on the microbiomes associated with low water activity foods. Pine nuts and sesame seeds, and food products which contain these ingredients, have been associated with recalls due to contamination with bacterial foodborne pathogens. The objective of this study was to identify the microbial community of pine nuts and sesame seeds using targeted 16S rRNA sequencing technology. Ten different brands of each seed type were assessed, and core microbiomes were determined. A total of 21 and 16 unique taxa with proportional abundances >1% in at least one brand were identified in the pine nuts and sesame seeds, respectively. Members of the core pine nut microbiome included the genera Alishewanella, Aminivibrio, Mycoplasma, Streptococcus, and unassigned OTUs in the families of Desulfobacteraceae and Xanthomonadaceae. For sesame seeds, the core microbiome included Aminivibrio, Chryseolina, Okibacterium, and unassigned OTUs in the family Flavobacteriaceae. The microbiomes of these seeds revealed that these products are dominated by environmental bacterial genera commonly isolated from soil, water, and plants; bacterial genera containing species known as commensal organisms were also identified. Understanding these microbiomes can aid in the risk assessment of these products by identifying food spoilage potential and community members which may co-enrich with foodborne bacterial pathogens.
Introduction
Next generation sequencing, including high-throughput metagenomics approaches, has revolutionized the study of the microbial ecology of foods. A better understanding of the resident microorganisms in food products can provide insights into the quality, shelf-life, and the potential risks and safety concerns. The survival of bacterial foodborne pathogens in a food is influenced not only by the chemical composition, physiological properties, and intrinsic and extrinsic factors, but also by the native microbial population. Many published studies have utilized metagenomics sequencing to analyze the microbial populations of fresh produce, cheeses, meats, fish, and fermented foods [1]. However, compared to other food categories, the microbiomes of low water activity foods (aw <0.80) are less defined but of interest due to the ability of some foodborne pathogens to persist in these foods for long periods of time. Understanding the types and relative populations of native microbiota in low water activity foods will provide quality and safety insights.
Only one published study has utilized high throughput metagenomics sequencing to examine the microbial population of a low water activity food: masala spice [2]. This study utilized targeted 16S rRNA sequencing to examine the microbiomes of different types of masala spices, all containing 8–17 individual spices including coriander, cumin, cinnamon, cloves, ginger, and nutmeg. It was determined that the masala spices had a high level of species diversity which varied by type (i.e., meat, chicken, garam) and the total number of combined spices, yet a core microbiota of Proteobacteria, Firmicutes, Actinobacteria, and Bacteroidetes could be resolved. This study has provided insights into microbial quality, spoilage and shelf-life potential, and the core resident bacterial taxa in masala spice. The microbiomes of other low water activity foods, including nuts, seeds, legumes, dried herbs, and other spices, have yet to be defined by high throughput sequencing technology.
Of current interest are the microbiomes of seeds and multi-commodity low water activity products made with seeds. Recent foodborne outbreaks and recalls in North America have been associated with pine nuts (the seeds of pines) [3–5], sesame seeds [6–8], and products made with these seeds [9–11]. In 2011, a multistate outbreak associated with Salmonella enterica-contamination of imported pine nuts in the U.S. resulted in 53 illnesses and two hospitalizations [5,12]. The implicated manufacturer recalled more than 21,000 pounds of pine nuts. In addition, three multistate salmonellosis outbreaks have occurred in the U.S. due to contaminated tahini (roasted sesame seed paste, often with oil). In 2011, consumption of contaminated tahini at three restaurants resulted in 23 illnesses with a 100% hospitalization rate [12,13]. In 2013, imported tahini was implicated in an outbreak which resulted in 17 illnesses, one hospitalization, and one death [12,14]. Recently, in 2019, imported tahini was also implicated in an outbreak which resulted in 6 illnesses and 1 hospitalization [11].
The aforementioned recalls and foodborne outbreaks associated with seeds highlight the importance of acquiring microbial population data on these food products. Since the microbiome of a food product can influence both its quality and safety, knowing the core microbiome, and observations of deviations from this baseline, can aid in food safety assessments. Therefore, the aim of this study was to elucidate the microbiomes of pine nuts and sesame seeds, including core microbiota.
Materials and methods
Seed samples used in this study
Ten different brands of hulled sesame seeds (Sesamum indicum L.) and hulled pine nuts (Pinus pinea L.) were acquired from the following vendors: Amazon (online distributor), Jewel (local retail grocer, Chicago, IL area), and Whole Foods (local retail grocer, Chicago, IL area). The growing and packaging/distribution locations of the different seeds are listed in S1 Table. Sesame seeds and pine nuts were stored in sealed original packaging at ambient for up to 1 mo prior to use.
Total DNA extraction and 16S rRNA gene amplification
Triplicate 10-g samples of each sesame seed and pine nut brand were portioned into stomacher bags with 10 mL Butterfield’s Phosphate Buffer (BPB, pH 7.2; Thermofisher Scientific, Waltham, MA). Samples (n = 60) were homogenized for 1 min using a Seward 400 stomacher (Seward Laboratory Systems Inc., Davie, FL). Total DNA was extracted from 1 mL of each homogenate using the Qiagen DNeasy Blood and Tissue Kit (Qiagen Inc., Germantown, MD) according to the manufacturer’s instructions. DNA was quantified using the Qubit dsDNA BR Assay Kit (Invitrogen Carlsbad, CA). PCR was performed to amplify the V4 region of the16S rRNA genes from each sample as previously described [15].
Library construction and sequencing
The 16s rRNA fragments were indexed using the Nextera XT Kit (Illumina, San Diego, CA) according to the manufacturer’s instructions as previously described [15]. The library, containing 60 samples, was diluted to 10 pM, spiked with 10% of 12.5 pM PhiX, and sequenced on a MiSeq with 600 cycles and V3 chemistry.
16S rRNA amplicon sequence analysis
Raw paired-end sequences were merged into consensus fragments and quality filtered as previously described [16]. The 16S rRNA gene sequences were taxonomically profiled to the genus-level using the k-mer-based classifier Kraken2 [17] and the Ribosomal Database Project 16S database [18]. Relative abundances were determined using Bracken 2.5 [19]. The relative abundances were averaged for the triplicate samples for each brand of pine nut and sesame seed.
Metagenomic diversity analysis
The alpha diversity of the microbial population in the samples was estimated using Shannon, Simpson, inverse Simpson, and Chao1 indices using the vegan package 2.5–6 [20] in R 3.6.2. Significant differences between the indices for the pine nuts and sesame seeds were determined using Kruskal-Wallis non-parametric test. Principal coordinates analysis (PCoA) using the weighted UniFrac community distance metric [21] was used to compare the total diversity among different samples. Beta diversity was also evaluated using Bray-Curtis dissimilarity matrix; a multilevel pairwise comparison using Adonis (~Permanova) was calculated using package pairwiseAdonis [22]. A p-value less than 0.05 was considered significant.
Results and discussion
Microbial diversity in the pine nut and sesame seed microbiomes
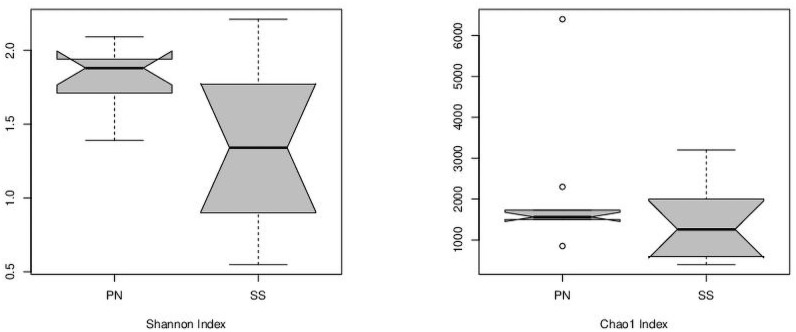
The alpha diversity of the pine nut and sesame seed samples were not significantly different as measured by Shannon and Chao1 indices, although there were differences observed between brands of the same seed type (Fig 1, S2 Table). The total unique taxa (OTU) identifications in the pine nut brands ranged from 57 to 141 with an average of 89, whereas identifications in the sesame seed brands ranged from 65 to 249 with an average of 128 (S2 Table). An overall correlation between seed growing locations and the number of taxa was determined. For pine nuts, the lowest number of taxa was observed in brands B (57), C (69), and G (61) for which the seed growing locations were Korea/Russia/Vietnam, Turkey, and Russia, respectively. The other brands had higher taxa identifications (81 to 141) and the seed growing locations were all in China. Brand A had the lowest diversity as measured by Shannon index, while brand F had the highest diversity. For sesame seeds, brands A (65), E (87), and H (69) had the lowest number of taxa identifications for which the seed growing locations were China, Mexico, and unlisted. The other brands with higher taxa identifications (97 to 249) had seed growing locations of India (with two brands having unlisted locations). Brand G had the lowest diversity, while brand E had the highest.
Fig 1. The alpha diversity of the microbial communities in sesame seed (SS; n = 10) and pine nut (PN; n = 10) microbiomes as measured by Shannon and Chao1 indices.
Approximately 90% of all pine nuts in the U.S. are imported from other countries, mainly China. Seven out of the 10 brands of pine nuts used in this study were imported from China, most likely accounting for the low microbial alpha diversity between the 10 different brands. Pine nuts grow in cones on pine trees, often high off the ground. The composition and nutrition contents of pine nuts harvested from different geographical locations can vary widely [23,24], suggesting that climate, soil type, and region-specific growing and harvesting practices play a role in pine nut composition. Similarly, the microbiome of pine nuts would also depend on these variables. Sesame seeds grow in pods on a plant closer to the ground. The more variable alpha diversity of the microbiome of sesame seeds may be due to the soil type, vicinity to wild animals and other crops, and to the diverse growing locations of the 10 different brands (i.e., China, India, Mexico, and unlisted).
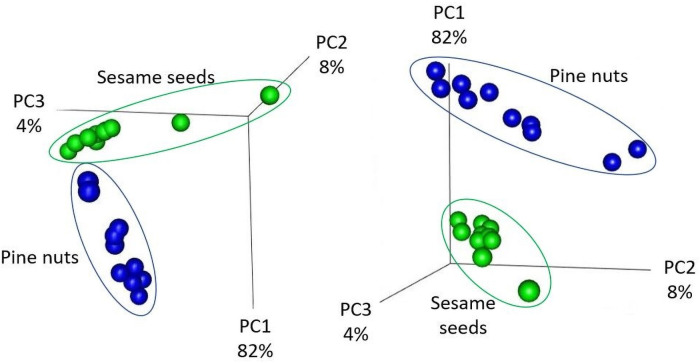
Principal coordinates analysis (PCoA) was utilized to visualize the microbial beta diversity in the 10 pine nut brands and the 10 sesame seed brands. Results revealed that the same seed type clustered together (Fig 2). The first component represented 82% of the variation in beta diversity and was sufficient to separate the microbiome of the pine nuts from the sesame seeds. Interestingly, even though both pine nuts and sesame seeds are from the same food category (i.e., seeds), the microbiota of each are clearly separated. The second and third components, representing 8 and 4% of the beta diversity, separated the different brands within the same seed type. A pairwise comparison of the microbiomes between the different pine nut brands and between the different sesame seed brands revealed no significant differences (p = 1; data not shown).
Fig 2. Principal coordinates analysis (PCoA) of the seed samples used in this study using the weighted UniFrac community distance metric.
The 10 sesame seed brands (green) and pine nut brands (blue) cluster together.
The pine nut microbiome
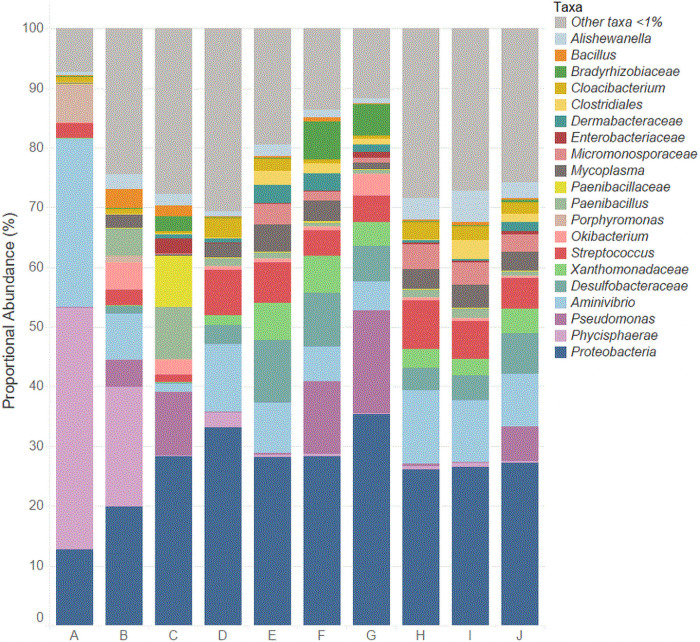
In the 10 pine nut brands, 21 unique taxa were present with a proportional abundance >1% in at least one brand (Fig 3). The 21 taxa included 10 genera as well as unassigned OTUs in eight families, two classes (Clostridiales and Phycisphaerae), and one phylum (Proteobacteria). Nineteen of those taxa had proportional abundances >3% in at least one brand, and only 11 were >5% in at least one brand. The taxa with the largest proportional abundances included Aminivibrio (28.3% in brand A), Phycisphaerae (20.1 and 40.1% in brand A and B, respectively), Pseudomonas (17.3% in brand G) and Proteobacteria (35.4%, brand G). When comparing the microbiota of the 10 pine nut brands, of notable difference was the large abundance of Phycisphaerae in brands A and B, yet the abundance of this taxon in six of the other brands was <1% and it was not identified in brand C. Another notable difference included the large abundance of Paenibacillus (8.7%) and unassigned OTUs in the family Paenibacillaceae (8.5%) in brand C. This was also the only brand where pine nuts were cultivated in Turkey.
Fig 3. Relative abundance of unique taxa in the 10 brands of pine nuts.
Core microbiome
The core microbiome of the pine nuts, defined as taxa with proportional abundances >1% and present in at least seven of the 10 brands, included Alishewanella (1.3–5.3%, 7 brands), Aminivibrio (1.5–28.3%, all 10 brands), Mycoplasma (1.0–4.6%, 8 brands), Streptococcus (1.2–6.8%, all 10 brands), unassigned genera in the families of Desulfobacteraceae (1.5–8.2%, 8 brands) and Xanthomonadaceae (1.7–6.2%, 7 brands), and unassigned OTUs in the phylum Proteobacteria (12.7–35.4%, all 10 brands). Aminivibrio and Streptococcus were the only genera present in all 10 brands with proportional abundances >1%.
Environmental microbiota
Of the 24 unique taxa identified in the pine nut brands with proportional abundance >1% in at least one brand, 14 of these taxa are considered ubiquitous environmental microbiota. Of the environmental taxa, the identified genera include Alishewanella, Aminivibrio, Bacillus, Cloacibacterium, Okibacterium, Paenibacillus, Pseudomonas, and Streptococcus. Alishewanella, a member of the core microbiome, was identified in all 10 pine nut brands at proportional abundances of 0.6–5.3%, with the greatest abundances observed in brands H and I. Alishewanella is a relatively new bacterial taxon and since its discovery in 1992 has been isolated from water and landfill soil [25,26]. Alishewanella is thought to play a role in the bioremediation process of chromate and sulfate [25] which provides insight into its existence in landfill and waste soil. Alishewanella has also been isolated from fermented foods [27] indicating its possible role in the fermentation process. This is the first occurrence where Alishewanella has been isolated in a seed product.
Aminivibrio was identified in all 10 pine nut brands at proportional abundances as low as 1.5% (brand C) to as high as 28.3% (brand A). Aminivibrio is a strictly anaerobic amino acid and organic acid-degrading microorganism which has been isolated from potato starch processing wastewater as well as the soil of rice fields [28,29]. Species of Aminivibrio are known to co-culture with hydrogen-utilizing methanogens [30] present in soil and waste of ruminants. As with Alishewanella, Aminivibrio was most likely in the soil where the pine nuts were cultivated.
Another ubiquitous environmental genus, Bacillus, was identified in all 10 pine nut brands, yet at very low abundances (<1%) in all brands except B and C where abundances were 3.1 and 1.8%, respectively. Bacillus species have been isolated recently from different soil types, including forest soil in China [31]. Since seven out of the 10 pine nut brands were cultivated in China, the finding of Bacillus is not unexpected. Bacillus species has also been identified in a wide variety of food products including dairy and powdered products [32,33], and powdered food products. Bacillus was also discovered to be a dominant microorganism in masala spice mixes, with proportional abundances as high as 13% [2]. Species in this genera are also known to be spoilage microorganisms as spores can survive heat treatment and thus lead to proliferation post-processing in many foods, including dairy products [34]. As such, identification of Bacillus in a food can provide information on its quality.
Cloacibacterium is a relatively new genus of bacteria found in diverse aquatic environments including sludge and wastewater [35,36]. In municipal wastewater and sludge, Cloacibacterium plays a role in breaking down the complex organic matter in these environments [37]. This genus was identified in all 10 pine nut brands with proportional abundances of 0.5–3.5%. the lowest abundance was determined in brand C (0.5%), where the pine nuts were cultivated in Turkey, and the highest abundances were in brands D, H, and I (3.5, 3.0, and 2.4%, respectively) where all pine nuts were cultivated in China.
Okibacterium is a relatively new plant-associated genus of bacteria containing two species. The species of Okibacterium have been isolated from plant roots and the seeds of plants [38,39]. Recently, Okibacterium was also isolated from ice wine during its fermentation process and was determined to be active based on endoenzymatic assays [40]. It is possible that Okibacterium was part of the microbiota of the berries used to make the ice wine. This genus was identified in all 10 pine nut brands with highest abundances in brands B (4.6%) and G (3.6%), both of which had the common growing location of Russia.
Paenibacillus was also identified in all 10 of the brands of pine nuts at proportional abundances ranging from 0.3–8.7%. Paenibacillus species are often isolated from soil and are associated with plant roots [41]. Paenibacillus has also been isolated from the rhizosphere of pine trees [42,43] and from pine litter [44]. Therefore, the finding of Paenibacillus in all ten of the pine nut brands is not surprising. This bacterial genus is commonly utilized for agricultural purposes. For example, certain Paenibacillus species are commonly used in biocontrol measures in agriculture [45] as well as a component in biofertilizers to promote plant growth [46]. Interestingly, Paenibacillus can inhibit the growth of the foodborne pathogen Salmonella during typical enrichment procedures [47], highlighting the need to optimize the enrichment protocols for pine nuts when assessing contamination with Salmonella.
Another ubiquitous environmental genera, Pseudomonas, was identified in all 10 pine nut brands at proportional abundances ranging from 0.8–17.3%. Large abundances of Pseudomonas were identified in brands C, F, and G (10.7, 12.2, and 17.3%, respectively). Since these three brands had different pine nut growing locations (i.e., Turkey, China, and Russia), Pseudomonas appears not to be location dependent and is part of the ubiquitous microbiota in the pine nut rhizosphere. Many of the species of Pseudomonas have been isolated from water and plants [48] and are beneficial to agriculture [49]. However, like Bacillus, Pseudomonas can also be an indicator of spoilage in foods, particularly in meats, dairy products, and vegetables [50–52]. Interestingly, also like Bacillus, Pseudomonas was identified at a high proportional abundance (19%) in Garam masala spice mixes [2].
Streptococcus is a prevalent microorganism found in a variety of environments including plants, soil, and different foods products [53] and species of this genus are also used in food fermentations [54]. Streptococcus was identified in all 10 pine nut brands ranging from 1.2% (brand C) to 8.2% (brand H). This bacterial genus has been previously identified in almonds [55]. Streptococcus is considered an indicator of soil contamination and also poor hygiene as it is present in fecal material [56]. As with Pseudomonas, Streptococcus appears not to be location dependent and is part of the ubiquitous microbiota in the pine nut rhizosphere.
Commensal microbiota
Commensals are microbes that are generally associated with host organisms and not often found in the environment. Commensal microbiota typically exist for the mutual benefit to the host; however, they can also cause disease and infection. In the pine nut brands, three commensal taxa were identified: Mycoplasma, Porphyromonas, and Dermabacteraceae. Mycoplasma was identified in all 10 of the pine nut brands in abundances ranging from 0.2% (brand A) to 4.6% (brand E). Mycoplasma are bacteria which lack a cell wall and survive by tightly adhering to hosts including humans, animals, plants, and insects [57]. This taxon is widespread in the environment and can be saprophytic or parasitic in nature. In immune compromised humans, certain species of Mycoplasma can invade the respiratory tract and cause diseases including pneumonia [57]. On plants, Mycoplasma-associated disease can be economically costly. Due to its association with plants and animals, the finding of Mycoplasma in all of the pine nut brands is not unanticipated.
Porphyromonas is a bacterial taxon identified in 9 out of 10 pine nut brands ranging from <0.1% (brands C, E, G, and J) to 6.3% (brand A); the taxon was not identified in brand F. Species of Porphyromonas are commensal and found in the oral microbiome of animals, most notably humans [58]. Although this taxon is present in healthy individuals, certain species of Porphyromonas can cause human periodontal disease [59]. In addition, Porphyromonas is present in human and animal fecal material [60], possibly explaining its identification in the pine nut brands in this study.
The bacterial family Dermabacteraceae includes the commensal genera Brachybacterium, Dermabacter, Devriesea, and Helcobacillus. Unclassified OTUs of Dermabacteraceae were identified in 8 of the 10 pine nut brands; it was not identified in brands A and C. the highest abundances were identified in brands E and F (3.1 and 3.0%, respectively), where both brands of pine nuts were cultivated in China. Species in this taxon have been isolated from both the environment [61] and human wounds [62]. Interestingly, Brachybacterium, a genus within Dermabacteraceae, has also been isolated from the surfaces of cheeses [63] from fermented seafood [64].
Taxa containing potential human foodborne pathogens
Taxa identified in the pine nut brands containing potential human foodborne pathogens include the genus Bacillus and the family Enterobacteriaceae. As mentioned previously, Bacillus was identified in all 10 pine nut brands at abundances of <1–3.1%. In addition to its ubiquitous environmental presence, Bacillus also contains species of human foodborne pathogens, most notably Bacillus cereus [65]. Unclassified OTUs of Enterobacteriaceae were identified in 6 out of 10 pine nut brands at abundances of <1%, with the exception of brand C with the unique growing location of Turkey, where the abundance was 2.5%. The family Enterobacteriaceae contains many genera of Gram-negative bacteria, some of which can be associated with foodborne illness including Escherichia, Klebsiella, Salmonella, and Shigella [66]. Enterobacteriaceae have been identified as part of the microbiomes of a variety of other food products including spice mixes, fresh produce, and dairy products [2,67–69]. In addition, the presence of Enterobacteriaceae in a food matrix is known to hinder the enrichment of the foodborne pathogen Listeria monocytogenes [70], highlighting the need for efficient enrichment procedures when Enterobacteriaceae are present in foods.
Notable between-brand differences
While the relative abundances of each taxa fluctuated between the pine nut brands, the most striking difference in the microbiota observed between brands was in high relative abundance of unclassified OTUs of the class Phycisphaerae in brands A and B (40.6 and 20.1%, respectively). The taxon Phycisphaerae includes water-dwelling bacteria which have been identified from aqueous environments [71,72]. The finding of Phycisphaerae in brands A and B most likely indicates that the water used for cultivation of the pine nuts contains that class of bacteria. Interestingly, Phycisphaerae was not identified in brand C, which was the only brand in which the pine nuts were cultivated outside of Asia.
The sesame seed microbiome
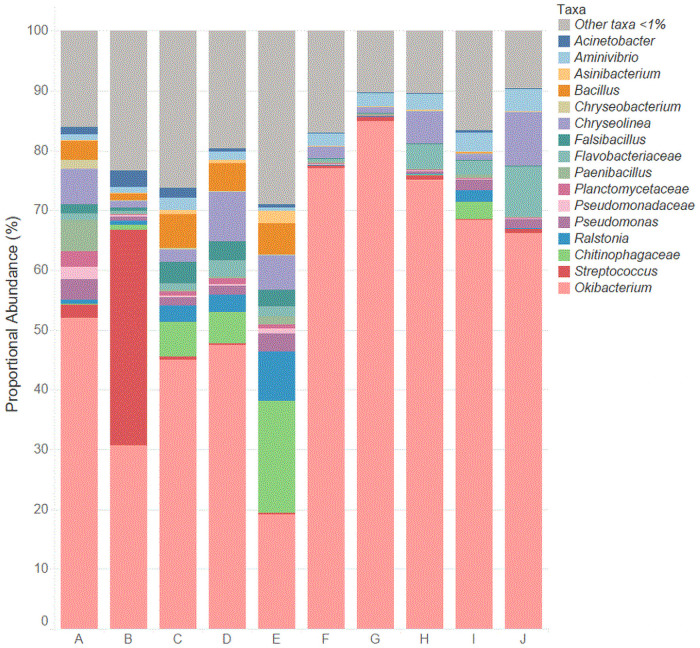
In the 10 sesame seed brands, 16 unique taxa were present with a proportional abundance >1% in at least one brand (Fig 4). These 16 taxa included 12 genera as well as unassigned OTUs in three families (Chitinophagaceae, Flavobacteriaceae, and Pseudomonadaceae) of one phylum (Plantomycetes). Out of the 16 taxa, eight had proportional abundances >5% in at least one brand and only three (Okibacterium, Streptococcus, and Chitinophagaceae) had abundances >10% in at least one brand. Okibacterium was the only taxon present at >1% in all 10 brands of sesame seeds and ranged from 19.2% (brand E) to 84.9% (brand G). Other taxa with high proportional abundances included Streptococcus (36.0% in brand B) and Chitinophagaceae (18.8% in brand E). When comparing the microbiota of the 10 different brands of sesame seeds, it was observed 13 taxa had proportional abundances >1% in brand A, however only 2 taxa (Okibacterium and Aminivibrio) were >1% in brand G.
Fig 4. Relative abundance of unique taxa in the 10 brands of sesame seeds.
Core microbiome
The core microbiome of the sesame seeds, defined as taxa with proportional abundances >1% and present in at least seven of the 10 brands, included Aminivibrio (1.0–3.7%, 8 brands), Chryseolina (1.0–9.1%, 7 brands), Flavobacteriaceae (1.1–8.6%, 7 brands), and Okibacterium (19.2–84.9%, all 10 brands). Okibacterium was the only member of the core microbiome identified in all 10 sesame seed brands.
Environmental microbiota
All of the 16 unique taxa with proportional abundances >1% in at least one brand which were identified in the sesame seeds are considered ubiquitous environmental microbiota. The genus level identifications include Acinetobacter, Aminivibrio, Asinibacterium, Bacillus, Chryseobacterium, Chryseolina, Falsibacillus, Okibacterium, Paenibacillus, Pseudomonas, Ralstonia, and Streptococcus. The genera Aminivibrio, Bacillus, Okibacterium, Paenibacillus, Pseudomonas, and Streptococcus were also identified as members of the pine nut microbiome with proportional abundances >1% in at least one of the 10 brands. Sesame seeds had much higher abundances of Okibacterium (19.2–84.9%) compared to pine nuts (0.2–4.6%). In addition, Streptococcus abundances varied widely in the sesame seeds (<1–36.0%) compared to the more uniform distribution in pine nuts (1.2–8.2%).
Acinetobacter, a ubiquitous environmental genus, was identified in the sesame seed brands at proportional abundances ranging from 0.1% (brands F, G, and J) to 2.8% (brand B). Species of Acinetobacter have been isolated from soil and water [73,74] and are often the focus of biotechnology research due to their ability to degrade certain pollutants. Acinetobacter can proliferate in many diverse environments and can become the predominate taxon under well-aerated conditions [75]. Therefore, Acinetobacter was most likely present in the soil where the sesame seeds were cultivated.
Another environmental genus, Asinibacterium, was identified at a proportional abundance of 2.2% in brand E, yet not identified in brands A, G, or H. Brand E was the only brand where sesame seeds were cultivated in Mexico. However, brands A, G, and H all had a different cultivation location and therefore no correlation between cultivation location and Asinibacterium identification could not be determined. Asinibacterium has been isolated from heavy metal-contaminated subsurface sediments [76] and can thus potentially be used for bioremediation purposes. In addition, Asinibacterium has also been isolated from one other low aw food product: donkey milk powder [77].
Chryseolina is an environmental taxon that has been repeatedly isolated from the soil and is considered a part of the soil microbiome [78]. This genus was identified in all of the sesame seed brands in proportional abundances ranging from <1% (brand G) to 9.1% (brand J). Although not much is known about this genus, Chryseolina was most likely present in the soil where the sesame seeds were cultivated.
The environmental bacterial genus, Falsibacillus, was identified in four sesame seed brands at abundances >1% (brands A, C, D, and E). Falsibacillus are considered rhizobacteria and have only ever been isolated from soil and plants [79]. The presence of this taxon in some of the sesame seed brands is therefore not surprising.
Commensal microbiota
The genera Chryseobacterium and Ralstonia, in addition to their ubiquitous environmental presence, are also considered commensal microorganisms. Chryseobacterium has been identified in plants and soil [80]. Species of this genus have also been identified as commensal organisms of insects including moths and mosquitoes [81]. Chryseobacterium was identified in sesame seed brand A at an abundance of 1.4%; however, this taxon was identified at <1% in all other brands. Similar to Asinibacterium, it appears that this taxon was most likely present in the soil where the sesame seeds of brand A were cultivated.
Ralstonia is a genus of bacteria which has been identified in soil, water, and plants [82] and are also commensals of humans [83]. Certain species of Ralstonia are members of the human oral cavity and the upper respiratory tract of healthy individuals [83]. Ralstonia can revert from a commensal organism to a pathogen in people who are immune compromised. Species of Ralstonia are also devastating plant pathogens and cause lethal wilting disease in over 200 plant species [84]. Although Ralstonia was identified in all 10 sesame seeds brands, this taxon was at abundances >1% in only four brands (C, D, E, and I). Brand E sesame seeds had the highest proportional abundance of Ralstonia (8.3%) and was also the only brand where the sesame seeds were cultivated in Mexico.
Taxa containing potential human foodborne pathogens
Similar to the pine nut brands, Bacillus was also identified in the sesame seed brands with proportional abundances ranging from <1% (brands F, G, H, I, and J) to 5.8% (brand C). In the sesame seed brands, Bacillus was the only taxon identified which contains human foodborne pathogens. Although this genus contains species which can cause foodborne illness, Bacillus is also a ubiquitous environmental pathogen and has been isolated from soil [85] and a variety of food products [32,33].
Notable between-brand differences
The total number of unique taxa in each of the brands of sesame seeds ranged from two (brand G) to 13 (brand A). In brand G, only Okibacterium and Aminivibrio were identified with proportional abundances >1%; this brand was one of four brands in which the sesame seeds were cultivated in India. Interestingly, brand A was the only brand in which the sesame seeds were cultivated in China (although some of the brands had unlisted growing and harvesting locations), indicating the difference in the soil microbiome in this location. Another notable difference is the high abundance of Streptococcus in brand B (36.0%) compared to the abundance in all other brands (0.2–2.2%). As mentioned previously, Streptococcus is considered an indicator of soil contamination as this taxon is present in fecal material [56].
Conclusions
The microbiomes associated with different food products are important in that they define the overall safety and quality. Metagenomics research is becoming more prevalent in food safety research, however there is a dearth of information related to low water activity foods. This study examined the microbiomes of ten different brands of pine nuts and sesame seeds and defined the associated core microbiomes and types of microbiota present. It is noted that this study did not examine species-level identifications and therefore no foodborne pathogens were identified in this study. Future studies could further explore the bacterial species associated with seeds. Overall, the results from this study provide insights into food safety issues including presence of spoilage organisms which may affect shelf life potential and identification of bacterial genera which may co-enrich with foodborne pathogens, possibly hindering detection.
Supporting information
(XLSX)
(XLSX)
Acknowledgments
The authors thank Christine Eckert and Kristin Pfeiffer for laboratory assistance.
Data Availability
All 16S rRNA sequences have been submitted to NCBI, Accession number PRJNA638124. Due to the random sampling nature of this study, the specific brand names are not disclosed for the pine nuts and sesame seeds. Alternatively, information on the growing and packaging/distribution locations of the different seeds are given in S1 Table.
Funding Statement
Megan Fay was supported by the Oak Ridge Institute for Science and Education (ORISE) Research Participation Program through an interagency agreement between the U.S. Department of Energy and the U.S. Food and Drug Administration. The sponsors had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.De Filippis F, Parente E, Ercolini D. Recent Past, Present, and Future of the Food Microbiome. Annual Review of Food Science and Technology. 2018;9(1):589–608. [DOI] [PubMed] [Google Scholar]
- 2.Jarvis KG, Daquigan N, White JR, Morin PM, Howard LM, Manetas JE, et al. Microbiomes Associated With Foods From Plant and Animal Sources. Front Microbiol. 2018;9:2540. doi: 10.3389/fmicb.2018.02540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.FDA. Superior Nut & Candy Co., Inc. Recalls Pine Nuts Because of Possible Health Risk 2015 [https://wayback.archive-it.org/7993/20170723005727/https://www.fda.gov/Safety/Recalls/ArchiveRecalls/2015/ucm443666.htm.
- 4.FSN. Waymouth Farms Recalls Raw Pine Nuts for Possible Salmonella Contamination 2015 [https://www.foodsafetynews.com/2015/04/waymouth-farms-recalls-good-sense-brand-pine-nuts-for-possible-salmonella-contamination/.
- 5.CDC. Multistate outbreak of human Salmonella Enteritidis infections linked to Turkish pine nuts 2011 [https://www.cdc.gov/salmonella/2011/pine-nuts-11-17-2011.html.
- 6.FSN. Salmonella Risk Spurs Sesame Seed Recall; Few Details Provided 2018 [https://www.foodsafetynews.com/2018/01/salmonella-risk-spurs-sesame-seed-recall-few-details-provided/.
- 7.Falkenstein D. Lian How Brand Sesame Seed Recall Due to Salmonella 2010 [https://www.foodpoisonjournal.com/foodborne-illness-outbreaks/lian-how-brand-sesame-seed-recall-due-to-salmonella/.
- 8.CTV. Sesame Seeds from HelloFresh Recalled Due to Salmonella 2018 [https://winnipeg.ctvnews.ca/sesame-seeds-from-hellofresh-recalled-due-to-salmonella-1.3778446.
- 9.FDA. House of Thaller recalls selected pine nut hummus products because of possible health risk 2017 [https://www.fda.gov/Safety/Recalls/ucm563822.htm.
- 10.FDA. Brodt Zenatti Holding LLC. Recalls Karawan Brand Tahini & SoCo Brand Tahini Because of Possible Health Risk 2019 [https://wayback.archive-it.org/7993/20191212073654/https://www.fda.gov/safety/recalls-market-withdrawals-safety-alerts/brodt-zenatti-holding-llc-recalls-karawan-brand-tahini-soco-brand-tahini-because-possible-health-0.
- 11.CDC. Outbreak of Salmonella infections linked to Karawan brand tahini (final update) 2019 [https://www.cdc.gov/salmonella/concord-05-19/index.html.
- 12.CDC. National Outbreak Reporting System (NORS) 207 [https://wwwn.cdc.gov/norsdashboard/.
- 13.CDC. Multistate outbreak of Salmonlla serotype Bovismorbificans infections associated with hummus and tahini- United States, 2011. MMWR Morb Mortal Wkly Rep. 2012;61(46):944–7. [PubMed] [Google Scholar]
- 14.CDC. Multistate outbreak of Salmonella Montivideo and Salmonella Mbandaka infections linked to tahini sesame paste (final update) 2013 [http://www.cdc.gov/salmonella/montevideo-tahini-05-13/.
- 15.Salazar JK, Carstens CK, Ramachandran P, Shazer AG, Narula SS, Reed E, et al. Metagenomics of pasteurized and unpasteurized gouda cheese using targeted 16S rDNA sequencing. BMC Microbiol. 2018;18(1):189. doi: 10.1186/s12866-018-1323-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ottesen A, Ramachandran P, Reed E, White JR, Hasan N, Subramanian P, et al. Enrichment dynamics of Listeria monocytogenes and the associated microbiome from naturally contaminated ice cream linked to a listeriosis outbreak. BMC Microbiol. 2016;16(1):275. doi: 10.1186/s12866-016-0894-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wood DE, Lu J, Langmead B. Improved metagenomic anaysis with Kraken 2. Genome Biol. 2019;20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, et al. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42:633–42. doi: 10.1093/nar/gkt1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu J, Breitwieser FP, Thielen P, Salzberg SL. Bracken: estimating species abundance in metagenomics data. PeerJ Computer Sci. 2017;3:e104. [Google Scholar]
- 20.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, et al. Vegan: Community Ecoloy Package. R package version 2.5–6. 2019 [https://CRAN.R-project.org/package=vegan.
- 21.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71(12):8228–35. doi: 10.1128/AEM.71.12.8228-8235.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinez Arbizu P. pairwiseAdonis: Pairwise multilevel comparison using adonis. R package version 0.4 2020 [https://github.com/pmartinezarbizu/pairwiseAdonis.
- 23.Lutz M, Alvarez K, Loewe V. Chemical composition of pine nut (Pinus pinea L.) grown in three geographical macrozones in Chile. CyTA, Journal of Food. 2017;15(2). [Google Scholar]
- 24.Zuleta A, Weisstaub A, Giacomino MS, Dyner L, Loewe MV, Del Rio R, et al. An ancient crop revisited: Chemical composition of mediterranean pine nuts grown in six countries. Ital J Food Sci. 2018;30(1):170–83. [Google Scholar]
- 25.Xia X, Li J, Liao S, Zhou G, Wang H, Li L, et al. Draft genomic sequence of a chromate- and sulfate-reducing Alishewanella strain with the ability to bioremediate Cr and Cd contamination. Stand Genomic Sci. 2016;11:48. doi: 10.1186/s40793-016-0169-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salah ZB, Rout SP, Humphreys PN. Draft Whole-Genome Sequence of the Alkaliphilic Alishewanella aestuarii Strain HH-ZS, Isolated from Historical Lime Kiln Waste-Contaminated Soil. Genome Announc. 2016;4(6). doi: 10.1128/genomeA.01447-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim MS, Roh SW, Nam YD, Chang HW, Kim KH, Jung MJ, et al. Alishewanella jeotgali sp. nov., isolated from traditional fermented food, and emended description of the genus Alishewanella. Int J Syst Evol Microbiol. 2009;59(Pt 9):2313–6. doi: 10.1099/ijs.0.007260-0 [DOI] [PubMed] [Google Scholar]
- 28.Honda T, Fujita T, Tonouchi A. Aminivibrio pyruvatiphilus gen. nov., sp. nov., an anaerobic, amino-acid-degrading bacterium from soil of a Japanese rice field. Int J Syst Evol Microbiol. 2013;63(Pt 10):3679–86. doi: 10.1099/ijs.0.052225-0 [DOI] [PubMed] [Google Scholar]
- 29.Antwi P, Li J, Opoku Boadi P, Meng J, Shi E, Xue C, et al. Functional bacterial and archaeal diversity revealed by 16S rRNA gene pyrosequencing during potato starch processing wastewater treatment in an UASB. Bioresour Technol. 2017;235:348–57. doi: 10.1016/j.biortech.2017.03.141 [DOI] [PubMed] [Google Scholar]
- 30.Harb M, Xiong Y, Guest J, Amy G, Hong P. Differences in microbial communities and performance between suspended and attached growth anaerobic membrane bioreactors treating synthetic municipal wastewater. Environ Sci: Water Res Technol. 2015;1:800–13. [Google Scholar]
- 31.Zheng X, Liu G, Wang Z, Wang J, Zhang H, Liu B. Bacillus dafuensis sp. Nov., Isolated from a Forest Soil in China. Curr Microbiol. 2020. doi: 10.1007/s00284-020-02014-2 [DOI] [PubMed] [Google Scholar]
- 32.Vidic J, Chaix C, Manzano M, Heyndrickx M. Food Sensing: Detection of Bacillus cereus Spores in Dairy Products. Biosensors (Basel). 2020;10(3). doi: 10.3390/bios10030015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heini N, Stephan R, Ehling-Schulz M, Johler S. Characterization of Bacillus cereus group isolates from powdered food products. Int J Food Microbiol. 2018;283:59–64. doi: 10.1016/j.ijfoodmicro.2018.06.019 [DOI] [PubMed] [Google Scholar]
- 34.Gram L, Ravn L, Rasch M, Bruhn JB, Christensen AB, Givskov M. Food spoilage--interactions between food spoilage bacteria. Int J Food Microbiol. 2002;78(1–2):79–97. doi: 10.1016/s0168-1605(02)00233-7 [DOI] [PubMed] [Google Scholar]
- 35.Allen TD, Lawson PA, Collins MD, Falsen E, Tanner RS. Cloacibacterium normanense gen. nov., sp. nov., a novel bacterium in the family Flavobacteriaceae isolated from municipal wastewater. Int J Syst Evol Microbiol. 2006;56(Pt 6):1311–6. doi: 10.1099/ijs.0.64218-0 [DOI] [PubMed] [Google Scholar]
- 36.Chun BH, Lee Y, Jin HM, Jeon CO. Cloacibacterium caeni sp. nov., isolated from activated sludge. Int J Syst Evol Microbiol. 2017;67(6):1688–92. doi: 10.1099/ijsem.0.001841 [DOI] [PubMed] [Google Scholar]
- 37.Nouha K, Kumar RS, Tyagi RD. Heavy metals removal from wastewater using extracellular polymeric substances produced by Cloacibacterium normanense in wastewater sludge supplemented with crude glycerol and study of extracellular polymeric substances extraction by different methods. Bioresour Technol. 2016;212:120–9. doi: 10.1016/j.biortech.2016.04.021 [DOI] [PubMed] [Google Scholar]
- 38.Evtushenko LI, Dorofeeva LV, Krausova VI, Gavrish EY, Yashina SG, Takeuchi M. Okibacterium fritillariae gen. nov., sp. nov., a novel genus of the family Microbacteriaceae. Int J Syst Evol Microbiol. 2002;52(Pt 3):987–93. doi: 10.1099/00207713-52-3-987 [DOI] [PubMed] [Google Scholar]
- 39.Wang HF, Zhang YG, Li L, Liu WH, Hozzein WN, Chen JY, et al. Okibacterium endophyticum sp. nov., a novel endophytic actinobacterium isolated from roots of Salsola affinis C. A. Mey. Antonie Van Leeuwenhoek. 2015;107(3):835–43. doi: 10.1007/s10482-014-0376-0 [DOI] [PubMed] [Google Scholar]
- 40.Buckova M, Puskarova A, Zenisova K, Krakova L, Piknova L, Kuchta T, et al. Novel insights into microbial community dynamics during the fermentation of Central European ice wine. Int J Food Microbiol. 2018;266:42–51. doi: 10.1016/j.ijfoodmicro.2017.11.010 [DOI] [PubMed] [Google Scholar]
- 41.Kampfer P, Busse HJ, McInroy JA, Hu CH, Kloepper JW, Glaeser SP. Paenibacillus nebraskensis sp. nov., isolated from the root surface of field-grown maize. Int J Syst Evol Microbiol. 2017;67(12):4956–61. doi: 10.1099/ijsem.0.002357 [DOI] [PubMed] [Google Scholar]
- 42.Kim BC, Lee KH, Kim MN, Kim EM, Min SR, Kim HS, et al. Paenibacillus pini sp. nov., a cellulolytic bacterium isolated from the rhizosphere of pine tree. J Microbiol. 2009;47(6):699–704. doi: 10.1007/s12275-009-0343-z [DOI] [PubMed] [Google Scholar]
- 43.Yuki M, Oshima K, Suda W, Oshida Y, Kitamura K, Iida T, et al. Draft Genome Sequence of Paenibacillus pini JCM 16418T, Isolated from the Rhizosphere of Pine Tree. Genome Announc. 2014;2(2). doi: 10.1128/genomeA.00210-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee HJ, Shin SY, Whang KS. Paenibacillus pinistramenti sp. nov., isolated from pine litter. Antonie Van Leeuwenhoek. 2020;113(2):155–63. doi: 10.1007/s10482-019-01325-0 [DOI] [PubMed] [Google Scholar]
- 45.Haggag WM. Colonization of exopolysaccharide-producing Paenibacillus polymyxa on peanut roots for enhancing resistance against crown rot disease. Afr J Biotechnol. 2007;6:1568–77. [Google Scholar]
- 46.Ker K, Seguin P, Driscoll BT, Fyles JW, Smith DL. Swithgrass establishment and seedling year production can be improved by inoclation with rhizosphere endophytes. Biomass Bioenergy. 2014;47:295–301. [Google Scholar]
- 47.Pettengill JB, McAvoy E, White JR, Allard M, Brown E, Ottesen A. Using metagenomic analyses to estimate the consequences of enrichment bias for pathogen detection. BMC Res Notes. 2012;5:378. doi: 10.1186/1756-0500-5-378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lopes LD, Davis EW 2nd, Pereira ESMC, Weisberg AJ, Bresciani L, Chang JH, et al. Tropical soils are a reservoir for fluorescent Pseudomonas spp. biodiversity. Environ Microbiol. 2018;20(1):62–74. doi: 10.1111/1462-2920.13957 [DOI] [PubMed] [Google Scholar]
- 49.Mercado-Blanco J, Bakker PA. Interactions between plants and beneficial Pseudomonas spp.: exploiting bacterial traits for crop protection. Antonie Van Leeuwenhoek. 2007;92(4):367–89. doi: 10.1007/s10482-007-9167-1 [DOI] [PubMed] [Google Scholar]
- 50.Mellor GE, Bentley JA, Dykes GA. Evidence for a role of biosurfactants produced by Pseudomonas fluorescens in the spoilage of fresh aerobically stored chicken meat. Food Microbiol. 2011;28(5):1101–4. doi: 10.1016/j.fm.2011.02.003 [DOI] [PubMed] [Google Scholar]
- 51.Federico B, Pinto L, Quintieri L, Carito A, Calabrese N, Caputo L. Efficacy of lactoferricin B in controlling ready-to-eat vegetable spoilage caused by Pseudomonas spp. Int J Food Microbiol. 2015;215:179–86. doi: 10.1016/j.ijfoodmicro.2015.09.017 [DOI] [PubMed] [Google Scholar]
- 52.Ternstrom A, Lindberg AM, Molin G. Classification of the spoilage flora of raw and pasteurized bovine milk, with special reference to Pseudomonas and Bacillus. J Appl Bacteriol. 1993;75(1):25–34. doi: 10.1111/j.1365-2672.1993.tb03403.x [DOI] [PubMed] [Google Scholar]
- 53.Michaylova M, Minkova S, Kimura K, Sasaki T, Isawa K. Isolation and characterization of Lactobacillus delbrueckii ssp. bulgaricus and Streptococcus thermophilus from plants in Bulgaria. FEMS Microbiol Lett. 2007;269(1):160–9. doi: 10.1111/j.1574-6968.2007.00631.x [DOI] [PubMed] [Google Scholar]
- 54.El Demerdash HA, Oxmann J, Heller KJ, Geis A. Yoghurt fermentation at elevated temperatures by strains of Streptococcus thermophilus expressing a small heat-shock protein: application of a two-plasmid system for constructing food-grade strains of Streptococcus thermophilus. Biotechnol J. 2006;1(4):398–404. doi: 10.1002/biot.200600018 [DOI] [PubMed] [Google Scholar]
- 55.King AD Jr., Miller MJ, Eldridge LC. Almond harvesting, processing, and microbial flora. Appl Microbiol. 1970;20(2):208–14. doi: 10.1128/am.20.2.208-214.1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cullen GA, Little TW. Isolation of Streptococcus uberis from the rumen of cows and from soil. Vet Rec. 1969;85(5):115–8. [DOI] [PubMed] [Google Scholar]
- 57.Razin S, Jacobs E. Mycoplasma adhesion. J Gen Microbiol. 1992;138(3):407–22. doi: 10.1099/00221287-138-3-407 [DOI] [PubMed] [Google Scholar]
- 58.Segata N, Haake SK, Mannon P, Lemon KP, Waldron L, Gevers D, et al. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 2012;13(6):R42. doi: 10.1186/gb-2012-13-6-r42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fiorillo L, Cervino G, Laino L, D’Amico C, Mauceri R, Tozum TF, et al. Porphyromonas gingivalis, Periodontal and Systemic Implications: A Systematic Review. Dent J (Basel). 2019;7(4). doi: 10.3390/dj7040114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cabral JP. Water microbiology. Bacterial pathogens and water. Int J Environ Res Public Health. 2010;7(10):3657–703. doi: 10.3390/ijerph7103657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Singh H, Du J, Yang JE, Shik Yin C, Kook M, Yi TH. Brachybacterium horti sp. nov., isolated from garden soil. Int J Syst Evol Microbiol. 2016;66(1):189–95. doi: 10.1099/ijsem.0.000696 [DOI] [PubMed] [Google Scholar]
- 62.Leal SM Jr., Jones M, Gilligan PH. Clinical Significance of Commensal Gram-Positive Rods Routinely Isolated from Patient Samples. J Clin Microbiol. 2016;54(12):2928–36. doi: 10.1128/JCM.01393-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schubert K, Ludwig W, Springer N, Kroppenstedt RM, Accolas JP, Fiedler F. Two coryneform bacteria isolated from the surface of French Gruyere and Beaufort cheeses are new species of the genus Brachybacterium: Brachybacterium alimentarium sp. nov. and Brachybacterium tyrofermentans sp. nov. Int J Syst Bacteriol. 1996;46(1):81–7. doi: 10.1099/00207713-46-1-81 [DOI] [PubMed] [Google Scholar]
- 64.Park SK, Kim MS, Jung MJ, Nam YD, Park EJ, Roh SW, et al. Brachybacterium squillarum sp. nov., isolated from salt-fermented seafood. Int J Syst Evol Microbiol. 2011;61(Pt 5):1118–22. doi: 10.1099/ijs.0.022517-0 [DOI] [PubMed] [Google Scholar]
- 65.Kramer JM, Gilbert RJ. Bacillus cereus and other Bacillus species. In: Doyle MP, editor. Foodborne Bacterial Pathogens. New York: Marcel Dekker; 1989. p. 21–70. [Google Scholar]
- 66.Brenner DJ, Farmer J. Enterobacteriaceae. In: Whitman WB, Rainey F, Kampfer P, Trujillo M, Chun P, DeVos B, et al., editors. Bergey’s Manual of Systematics of Archaea and Bacteria 2015. [Google Scholar]
- 67.Lopez-Velasco G, Welbaum GE, Boyer RR, Mane SP, Ponder MA. Changes in spinach phylloepiphytic bacteria communities following minimal processing and refrigerated storage described using pyrosequencing of 16S rRNA amplicons. J Appl Microbiol. 2011;110(5):1203–14. doi: 10.1111/j.1365-2672.2011.04969.x [DOI] [PubMed] [Google Scholar]
- 68.Jackson CR, Randolph KC, Osborn SL, Tyler HL. Culture dependent and independent analysis of bacterial communities associated with commercial salad leaf vegetables. BMC Microbiol. 2013;13:274. doi: 10.1186/1471-2180-13-274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Escobar-Zepeda A, Sanchez-Flores A, Quirasco Baruch M. Metagenomic analysis of a Mexican ripened cheese reveals a unique complex microbiota. Food Microbiol. 2016;57:116–27. doi: 10.1016/j.fm.2016.02.004 [DOI] [PubMed] [Google Scholar]
- 70.Al-Zeyara SA, Jarvis B, Mackey BM. The inhibitory effect of natural microflora of food on growth of Listeria monocytogenes in enrichment broths. Int J Food Microbiol. 2011;145(1):98–105. doi: 10.1016/j.ijfoodmicro.2010.11.036 [DOI] [PubMed] [Google Scholar]
- 71.Fukunaga Y, Kurahashi M, Sakiyama Y, Ohuchi M, Yokota A, Harayama S. Phycisphaera mikurensis gen. nov., sp. nov., isolated from a marine alga, and proposal of Phycisphaeraceae fam. nov., Phycisphaerales ord. nov. and Phycisphaerae classis nov. in the phylum Planctomycetes. J Gen Appl Microbiol. 2009;55(4):267–75. doi: 10.2323/jgam.55.267 [DOI] [PubMed] [Google Scholar]
- 72.Qu J, Zhang Q, Zhang N, Shen L, Liu P. Microbial community diversity in water and sediment of an eutrophic lake during harmful algal bloom using MiSeq Illumina technology. IPCBEE. 2015;87:67–72. [Google Scholar]
- 73.Choi JY, Ko G, Jheong W, Huys G, Seifert H, Dijkshoorn L, et al. Acinetobacter kookii sp. nov., isolated from soil. Int J Syst Evol Microbiol. 2013;63(Pt 12):4402–6. doi: 10.1099/ijs.0.047969-0 [DOI] [PubMed] [Google Scholar]
- 74.Krizova L, Maixnerova M, Sedo O, Nemec A. Acinetobacter albensis sp. nov., isolated from natural soil and water ecosystems. Int J Syst Evol Microbiol. 2015;65(11):3905–12. doi: 10.1099/ijsem.0.000511 [DOI] [PubMed] [Google Scholar]
- 75.Abdelhaleem D. Acinetobacter: environmental and biotechnological applications. Afr J Biotechnol. 2003;2(4):71–4. [Google Scholar]
- 76.Brzoska RM, Huntemann M, Clum A, Chen A, Kyrpides N, Palaniappan K, et al. Complete Genome Sequence for Asinibacterium sp. Strain OR53 and Draft Genome Sequence for Asinibacterium sp. Strain OR43, Two Bacteria Tolerant to Uranium. Microbiol Resour Announc. 2019;8(14). doi: 10.1128/MRA.01701-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee DG, Park JM, Kang H, Hong SY, Lee KR, Chang HB, et al. Asinibacterium lactis gen. nov., sp. nov., a member of the family Chitinophagaceae, isolated from donkey (Equus asinus) milk. Int J Syst Evol Microbiol. 2013;63(Pt 9):3180–5. doi: 10.1099/ijs.0.047639-0 [DOI] [PubMed] [Google Scholar]
- 78.Zheng J, Chen J, Pan G, Wang G, Liu X, Zhang X, et al. A long-term hybrid poplar plantation on cropland reduces soil organic carbon mineralization and shifts microbial community abundancne and composition. Appl Soil Ecology. 2017;111:94–104. [Google Scholar]
- 79.Shi SB, Liu C, Jiang MG, Li GD, Yang LF, Wu JF, et al. Falsibacillus albus sp. nov., isolated from mangrove soil. Int J Syst Evol Microbiol. 2019;69(5):1411–6. doi: 10.1099/ijsem.0.003328 [DOI] [PubMed] [Google Scholar]
- 80.Cho SH, Lee KS, Shin DS, Han JH, Park KS, Lee CH, et al. Four new species of Chryseobacterium from the rhizosphere of coastal sand dune plants, Chryseobacterium elymi sp. nov., Chryseobacterium hagamense sp. nov., Chryseobacterium lathyri sp. nov. and Chryseobacterium rhizosphaerae sp. nov. Syst Appl Microbiol. 2010;33(3):122–7. doi: 10.1016/j.syapm.2009.12.004 [DOI] [PubMed] [Google Scholar]
- 81.Reetha BMA, Mohan M. Diversity of commensal bacteria from mid-gut of pink stem borer (Sesamia inferens [Walker])-Lepidoptera insect populations of India. J Asia-Pacific Entomol. 2018;21(3):937–43. [Google Scholar]
- 82.Singh D, Sinha S, Yadav DK, Chaudhary G. Detection of Ralstonia solanacearum from asymptomatic tomato plants, irrigation water, and soil through non-selective enrichment medium with hrp gene-based bio-PCR. Curr Microbiol. 2014;69(2):127–34. doi: 10.1007/s00284-014-0566-z [DOI] [PubMed] [Google Scholar]
- 83.Stelzmueller I, Biebl M, Wiesmayr S, Eller M, Hoeller E, Fille M, et al. Ralstonia pickettii- innocent bystander or a potential threat? Clin Microbiol Infect. 2006;12:99–101. doi: 10.1111/j.1469-0691.2005.01309.x [DOI] [PubMed] [Google Scholar]
- 84.Denny TP. Ralstonia solanacearum--a plant pathogen in touch with its host. Trends Microbiol. 2000;8(11):486–9. doi: 10.1016/s0966-842x(00)01860-6 [DOI] [PubMed] [Google Scholar]
- 85.Hempel PP, Yao M, Yannarell S, Shevchenko O, Vogt F, Donofrio N, et al. Complete Genome Sequence of Bacillus velezensis Strain S4, Isolated from Biochar-Treated Soil. Microbiol Resour Announc. 2020;9(20). doi: 10.1128/MRA.00352-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
Data Availability Statement
All 16S rRNA sequences have been submitted to NCBI, Accession number PRJNA638124. Due to the random sampling nature of this study, the specific brand names are not disclosed for the pine nuts and sesame seeds. Alternatively, information on the growing and packaging/distribution locations of the different seeds are given in S1 Table.