Abstract
Purpose
To develop microchannel-based preparation of curcumin (Cur)-loaded hybrid nanoparticles using enzyme-targeted peptides and star-shaped polycyclic lipids as carriers, and to accomplish a desirable targeted drug delivery via these nanoparticles, which could improve the bioavailability and antitumor effects of Cur.
Methods
The amphiphilic tri-chaintricarballylic acid-poly (ε-caprolactone)-methoxypolyethylene glycol (Tri-CL-mPEG) and the enzyme-targeted tetra-chain pentaerythritol-poly (ε-caprolactone)-polypeptide (PET-CL-P) were synthesized. The Cur-loaded enzyme-targeted hybrid nano-delivery systems (Cur-P-NPs) were prepared by using the microfluidic continuous granulation technology. The physicochemical properties, release behavior in vitro, and stability of these Cur-P-NPs were investigated. Their cytotoxicity, cellular uptake, anti-proliferative efficacy in vitro, biodistribution, and antitumor effects in vivo were also studied.
Results
The particle size of the prepared Cur-P-NPs was 146.1 ± 1.940 nm, polydispersity index was 0.175 ± 0.014, zeta potential was 10.1 ± 0.300 mV, encapsulation rate was 74.66 ± 0.671%, and drug loading capacity was 5.38 ± 0.316%. The stability of Cur-P-NPs was adequate, and the in vitro release rate increased with the decrease of the environmental pH. Seven days post incubation, the cumulative release values of Cur were 52.78%, 67.39%, and 98.12% at pH 7.4, pH 6.8 and pH 5.0, respectively. Cur-P-NPs exhibited better cell entry and antiproliferation efficacy against U251 cells than the Cur-solution and Cur-NPs and were safe for use. Cur-P-NPs specifically targeted tumor tissues and inhibited their growth (78.63% tumor growth inhibition rate) with low toxic effects on normal tissues.
Conclusion
The enzyme-targeted hybrid nanoparticles prepared in the study clearly have the tumor-targeting ability. Cur-P-NPs can effectively improve the bioavailability of Cur and have potential applications in drug delivery and tumor management.
Keywords: enzyme targeting, nanoparticle fabrication, mouse model, growth inhibition
Introduction
Cancer has become a major threat to human life and health,1 and chemotherapy remains the main treatment modality for cancers.2 Curcumin (Cur) is an o-methoxyphenol derivative from turmeric.3 Extensive clinical trials have shown that Cur has therapeutic potential against a variety of diseases, including cancer,4,5 cardiovascular disease,6,7 inflammatory bowel disease,8 and Alzheimer’s disease.9 However, the low water solubility, low bioavailability, and rapid in vivo metabolism of Cur have severely limited its clinical applications.10–13 Nano-delivery systems that have been developed in recent years can effectively increase the solubility of insoluble drugs,14 thereby improving the targeting ability15,16 and bioavailability of these drugs.17,18
Studies have shown that the solubility,19 long circulation in vivo,20 active targeting ability,15 and cell penetration ability21 of these nanocarriers have to be significantly improved to achieve efficient drug delivery. Star-shaped polymers have good drug-carrying ability,22,23 and polyethylene glycol (PEG) modification helps in reducing both the self-polymerization of particles in plasma and recognition by the reticuloendothelial system to prolong the circulation time in vivo.24–26 PEGylated star-shaped polymers are ideal drug carriers. Nanoparticles enter tumor tissue mainly through the enhanced permeability and retention (EPR) effect with low targeting efficiency.27 To this end, a series of active tumor targeting nanoparticles (enzyme response, pH response, REDOX response) have been constructed based on the tumor microenvironment (low pH, specific enzymes, etc.).18,20 The diversity of specific enzymes in tumor microenvironment has also received increasing attention.28 Matrix metalloproteinases (MMPs) are active and overexpressed in tumor cells but play the role of a silencer in normal cells.29 Therefore, they can be used as a specific target to achieve active drug delivery in tumor therapy.30 The cell internalization efficiency of drug-loaded nanocarriers is another important factor affecting the antitumor efficacy. Cell-penetrating peptides (CPPs) are capable to carry bioactive substances into cells.31,32 New functional peptides constructed by MMPS targeted peptides and CPPs enable both capacities of active targeting and cell penetration. Star-shaped polymers modified by the new functional peptides are ideal carriers for drug delivery. However, star-shaped polymers modified to contain both peptide and PEG have excessive molecular weights (MWs) and undergo changes in nanoparticle size, which affect the in-vivo distribution and drug release rate achieved. In addition, the hydrophilic/lipophilic modifiability of the drug-carrying polymers is reduced, and the drug-carrying species are thereby limited. Therefore, new drug delivery platforms with wide applicability and tunability are required.
Hybrid nanoparticles act as multifunctional drug delivery systems, owing to the heterogeneous assembly of different polymers that retain their specific properties.33 This delivery system combines the advantages of multiple nanoparticles (with different particle sizes, potentials, and morphologies) for drug delivery, exhibiting collective properties different from those of single nanoparticles.34 Nanoparticles of uniform size can be continuously prepared using microchannels.35 However, this has not been utilized for the preparation of hybrid nanoparticles.
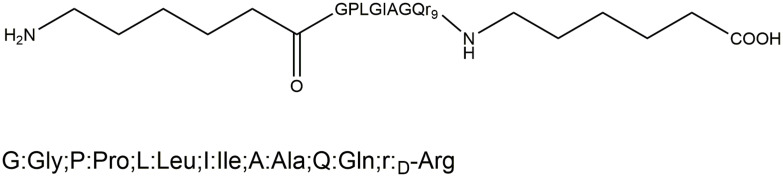
Therefore, in this study, peptide-modified star-shaped polymers (PET-CL-P) and PEGylated star-shaped polymers (Tri-CL-mPEG) were synthesized as the carrier materials. ACP-GPLGIAGr9-ACP was selected as the functional peptide. The targeting element used was protein endonuclease specific shear peptide (GPLGIAG), which can be specifically recognized and sheared by the MMP-2 and MMP-9.36 After shearing, the cationic CPP polyarginine r9 could enhance the nanoparticle penetration into cells.37 Thereafter, with Cur as the model drug, we prepared Cur-loaded enzyme-targeted hybrid nano-delivery systems (Cur-P-NPs) using a microchannel continuous granulation technology. Furthermore, the in vitro and in vivo antitumor properties of the Cur-P-NPs were also investigated.
Materials and Methods
Materials
Curcumin and (ACP)-GPLGIAGQr9-(ACP) were purchased from Hangzhou Guang Lin Biological Pharmaceutical Co. Ltd. (Hangzhou, China) and China Peptides Co. Ltd. (Shanghai, China), respectively. 1,2,3-Propanetricarboxylic acid was obtained from Tokyo Chemical Industry Co., Ltd (Tokyo, Japan). N,N’-Dicyclohexylcarbodiimide (DCC), ε-caprolactone (ε-PCL), 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (EDC), stannous 2-ethylhexanoate [Sn(Oct)2], mPEG (MW=1900), pentaerythritol (PET), and 4-dimethylaminopyridine (DMAP) were purchased from Sigma-Aldrich (Shanghai, China).
Dialysis bags (MWCO=14 kDa) and Dulbecco’s modified Eagle’s medium (DMEM) were purchased from Gene Star Co. (Shanghai, China) and Thermo Fisher Scientific Suzhou Co., Ltd (Suzhou, China), respectively. Fetal bovine serum (FBS) was purchased from Zhejiang Tianhang Biotechnology Co.,Ltd. (Huzhou, China). Mouse embryonic fibroblast L929 cells and human glioma U251 cells were provided by Zhejiang Provincial Center for Disease Control and Prevention (Hangzhou, China).
Synthesis and Characterization of Tri-CL-mPEG and PET-CL-P
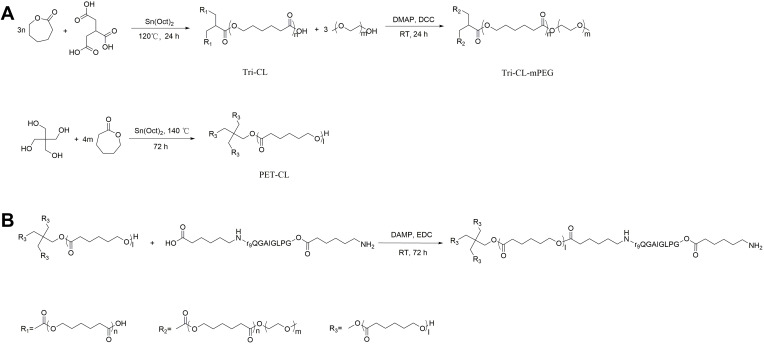
Tri-CL-mPEG was synthesized as previously described,38 and the reaction procedure is shown in Figure 1A. Specifically, 1,2,3-propanetricarboxylic acid, ε-caprolactone and Sn(Oct)2 were allowed to react in a nitrogen atmosphere for 24 h at 120°C. Tri-CL was obtained by dichloromethane dissolution and ice–ether precipitation. Tri-CL and mPEG were allowed to react at 25°C for 24 h in the presence of DCC and DMAP in a nitrogen atmosphere to obtain the amphiphilic triple chain polymers Tri-CL-mPEG.
Figure 1.
(A) Synthesis of Tri-CL-mPEG, (B) Synthesis of PET-CL-P.
Abbreviations: Tri-CL-mPEG, tricarballylic acid-poly (ε-caprolactone)-methoxypolyethylene glycol; PET-CL-P, pentaerythritol-poly (ε-caprolactone)-polypeptide.
PET-CL-P was synthesized in two steps, and the reaction procedure is shown in Figure 1B. The reaction was carried out as follows: pentaerythritol (2 mmol, 272.3 mg) and ε-lactone (120 mmol, 13.70 g) were stirred and dissolved in a three-necked flask. Next, Sn(Oct)2 (0.5% wt) was added to the flask, and the reaction was carried out at 140°C for 72 h in a nitrogen atmosphere. At the end of the reaction, dichloromethane and ice–ether (v:v 1:10) were added sequentially to precipitate and filter the crude product. The crude product was purified using dichloromethane and ice ether (v:v 1:10) and dried under vacuum at 25°C to obtain PET-CL.
The peptides ACP-GPLGIAGQr9-ACP (0.04266 mmol, 100 mg), DMAP (0.4266 mmol, 52.12 mg), and EDC (0.4266 mmol, 81.78 mg) were dissolved in 20 mL dimethylformamide (DMF), and then activated in an ice bath under a nitrogen atmosphere for 2 h. Subsequently, PET-CL (0.04266 mmol, 507.7 mg) was added. After stirring at 25°C for 96 h, the reaction solution was loaded into a dialysis bag (MW = 14 kDa) and dialyzed for 120 h. PET-CL-P was finally obtained by freeze-drying.
Next, the carrier materials were characterized. The weight-average molecular weight (MW), number-average molecular weight (Mn), and polydispersity index (PDI) of the polymers were determined by gel permeation chromatography (GPC) (LC-20AT, Shimadzu, Tokyo, Japan).23 The structures of the synthetic polymers were confirmed by 1H-nuclear magnetic resonance spectroscopy (1H NMR) (AVANCE-III, 500 MHz, Bruker Daltonics, Bremen, Germany) and Fourier transform infrared spectroscopy (FT-IR) (FT-IR, Nicolet 6700, Thermo Fisher Scientific, Waltham, MA, USA).
Preparation and Characterization of Nanoparticles
Preparation of Nanoparticles
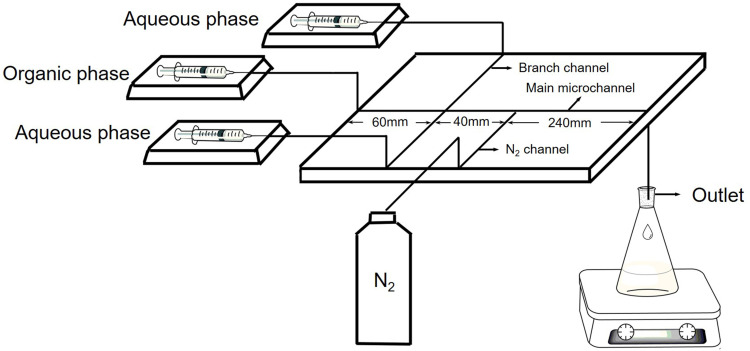
Cur-loaded enzyme-targeted hybrid nano-delivery systems (Cur-P-NPs) were prepared by co-encapsulating Cur with the amphiphilic tri-chain polymer Tri-CL-mPEG and the targeting peptide-modified tetra-chain polymer PET-CL-P. Cur, Tri-CL-mPEG, and PET-CL-P were dissolved in acetone (oil phase) and purified water (aqueous phase) and injected into the microchannel system (Figure 2) with a precision syringe pump at a fixed rate. The nanoparticle suspensions were collected at the outlet and centrifuged (4000 g, 4°C) for 20 min to obtain the nanoparticle solution. The solution was centrifuged (25,000 g, 4°C) for 60 minutes and the precipitate was lyophilized to obtain Cur-P-NPs. Blank enzyme-targeted hybridized nanoparticles (blank-P-NPs) were prepared as described above, except without Cur in the oil phase.
Figure 2.
Schematic of the microfluidic device. The width of the main microchannel is 360 μm; the width of the branch channel is 590 μm; and the width of the N2 channel is 470 μm.
Cur-loaded hybridized nanoparticles (Cur-NPs) were prepared from Tri-CL-mPEG and PET-CL co-coated with Cur in a microchannel as mentioned above. Blank hybridized nanoparticles (blank-NPs) were prepared in the same manner as Cur-NPs (without Cur in the oil phase).
The optimal parameters for the preparation of nanoparticles using a microchannel are as follows: aqueous phase flow rate of 1.2 mL/min, oil phase flow rate of 0.6 mL/min, Cur concentration of 1.0 mg/mL, material ratio of 1:1, and Cur to polymers ratio of 1:15.
Physicochemical Properties of Hybridized Nanoparticles
The drug content in nanoparticles was determined using an ultraviolet (UV) spectrophotometer (TU-1900, Beijing Purkinje General Instrument Co., Ltd, Beijing, China) at a wavelength of 420 nm. Drug loading (DL%) and drug entrapment efficiency (EE%) were calculated as follows:
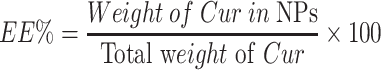 |
(1) |
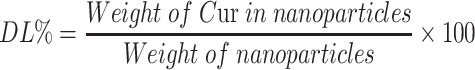 |
(2) |
The particle size, PDI, and zeta potential were determined using a Zetasizer Nano-ZS90 instrument (Malvern, UK). The morphologies of the Cur-P-NPs were observed by transmission electron microscopy (TEM, JEM-1010, MA, USA). The surface properties were characterized by X-ray powder diffraction (XRD) (X’ Pert PRO, PNAlytical, Holland).
In vitro Release of Nanoparticles
To simulate the release of nanoparticles in the in vivo environment, the release behaviors of Cur-P-NPs were determined in phosphate buffered saline (PBS) (0.01 M) at three pH levels: pH 7.4 (simulating physiological conditions), pH 6.8 (simulating tumor tissue), and pH 5.0 (simulating lysosomes and late endosomes in tumor cells).39 For each level, 5 mL of Cur-P-NPs solution was placed in a dialysis bag (MW = 14kDa), which was in turn placed in 100 mL of PBS in a shaker at 100 rpm and 37°C. At each set time point, 5 mL of the release medium was removed to determine the Cur content, and an equal amount of fresh medium was added. The cumulative release rate was calculated, and Cur content was determined using a TU-1900 UV spectrophotometer at 420 nm. All the experiments were carried out in triplicate.
Stability of Nanoparticles
To assess nanoparticle storage stability, Cur-P-NP solutions were sealed under shade and stored in a refrigerator at 4°C; samples were checked after 1, 8, 15, 22, and 30 days.
To investigate the stability of Cur-P-NPs at body temperature and their delivery, in vivo conditions were simulated with 0.01 M PBS (pH 7.4) and high-sugar cell culture medium (DMEM) containing 10% FBS (DMEM+), respectively. The Cur-P-NP solution was diluted with the medium at a ratio of 1:4, incubated at 37°C, and sampled after 1, 2, 3, 8, 24, 48, 72, and 168 h.
The particle size, zeta potential, and PDI of all three replicates of each sample were determined.
In vitro Cytotoxicity Study
The cytotoxicity of blank-NPs and blank-P-NPs was evaluated by MTT assay using L929 cells. The cells were seeded at a density of 1×105 cells/well in 96-well plates. After incubating for 24 h, 100 μL of sterilized solution containing either blank-NPs, blank-P-NPs, or medium only was added and incubated for 24 h. Complete medium (100 μL) containing 10 μL of 5 mg/mL MTT was then added, and co-cultured at 37°C for 4 hours. Next, the medium was discarded, and 100-µL dimethyl sulfoxide (DMSO) was added to each well. All the samples were prepared in triplicate. The absorbance intensity of each sample was measured at 570 nm, and cell viability (%) was calculated.
In vitro Anti‑Proliferation Efficacy
The anti-proliferative effects of the Cur-solution (Cur dissolved in DMSO:H2O at a 1:1000 ratio [v:v]), Cur-NPs, and Cur-P-NPs against cancer cells was assessed by MTT assay using the U251 cell line. The cells were seeded at a density of 1×105 cells/well in 96-well plates and incubated with 200 μL of sterilized Cur or the medium for 24 h and 48 h. Analyses were performed as mentioned above for the in vitro cytotoxicity study.
Cellular Uptake Study
Cellular uptake of Cur-NPs was analyzed using the L929 and U251 cell line. The cells were seeded into a 6-well plate (2 × 105 cells/well) and incubated for 24 h at 37°C. Next, Cur-Solution, Cur-NPs, Cur-P-NPs (containing 40 μg/mL Cur), and medium only were added and further incubated for 3 hours at 37°C. The cells were then washed with PBS and observed under fluorescence microscopy (Eclipse Ti-S, Nikon, Tokyo, Japan).
For flow cytometric measurements, the U251 cells were pre-incubated in 6-well plates (106 cells/well) in DMEM containing 5% FBS for 24 h at 37°C. Then, the medium was discarded, and 2 mL DMEM, Cur-Solution, Cur-NPs, or Cur-P-NPs were added to the cells for 3 h. The concentration of Cur was 40 μg/mL. After being washed with PBS, the cells were carefully detached using trypsin, collected by centrifugation at 100 g for 5 min, and resuspended in PBS (0.5 mL); flow cytometry measurements were obtained using a flow cytometer (CytoFLEX S; Beckman Coulter, Brea, CA).
In vivo Pharmacodynamic and Safety Studies
Animal Studies
BALB/c nude mice (age, 5–6 weeks; weight, 20 ± 2 g) were obtained from the Shanghai SLAC Laboratory Animal Co., Ltd (Shanghai, China). U251 cell suspensions (5 × 107 cells/mL, 0.2 mL) were injected subcutaneously into the left forelimb of each mouse to obtain a glioma xenograft model. Biodistribution and antitumor activity of the nanoparticles were evaluated after tumor formation (~100 mm3). All the animal experiments were conducted in accordance with the institutional guidelines for the care and use of laboratory animals at Zhejiang University of Technology, Hangzhou, China and the National Institutes of Health Guide for Care and Use of Laboratory Animals (Publication No. 85–23, revised 1996). All the animal experiments have been approved by Zhejiang University of Technology.
Biodistribution Studies
Tumor-bearing nude mice (n = 36; 12×3 groups) were injected with 0.2 mL Cur-Solution, Cur-NPs, or Cur-P-NPs via the tail vein. The concentration of Cur was equivalent to 50 μg/mL. Half of the mice in each group was sacrificed at 1 h, and the other half was sacrificed at 24 h. The tumor and main organs were harvested, weighed, and homogenized in PBS. Cur was extracted using acetonitrile by vortexing for 30 s and sonicating for 5 min. After centrifugation at 7000 g for 10 min, the supernatant was dried by nitrogen flow at 40°C. The resulting solid samples were dissolved again using acetonitrile. After vortexing and centrifugation at 11,000 g for 5 min, Cur content was measured by high-performance liquid chromatography (Ultimate 3000, Thermo Scientific, Germering, Germany) at 420 nm.
In vivo Antitumor Activity Study
Twelve tumor xenograft mice were selected and divided into four groups randomly to inject saline, Cur-Solution, Cur-NPs, or Cur-P-NPs via the tail vein. The dosage was 2 mg/kg, once every two days for 19 days. Weights and tumor volumes were recorded.
Histological Analysis
After 19 days, the mice were sacrificed. The tumor and main organs were harvested and fixed in 4% paraformaldehyde. Pathological sections of these organs were obtained and stained with hematoxylin-eosin (H&E).
Statistical Analysis
All statistical analyses were performed using SPSS 19.0 (SPSS, IBM, Armonk, NY, USA). t-tests were used for comparisons between groups. In the in-vitro anti‑proliferation tests, the half maximal inhibitory concentration (IC50) of each sample was calculated. Results showing P < 0.05 were considered statistically different; those with P < 0.01 were considered statistically significant; and those with P < 0.001 were considered extremely significant. The results are expressed as mean ± standard error.
Results and Discussion
Characterization of the Carrier Materials
Tri-CL-mPEG and PET-CL-P were successfully synthesized. The FT-IR and 1H NMR spectra of Tri-CL, Tri-CL-mPEG, PET-CL, and PET-CL-P are given in the Supplementary Material text sections and Figures S1–6, and the Mn, MW, and PDI are shown in Table 1.
Table 1.
Molecular Weights of the Copolymers
| Copolymer | Mn (g/mol) | MW (g/mol) | PDI |
|---|---|---|---|
| Tri-CL | 12,731.7 | 16,223.6 | 1.25 |
| Tri-CL-mPEG | 18,755.6 | 26,921.0 | 1.44 |
| PET-CL | 11,754.3 | 15,110.2 | 1.29 |
| PET-CL-P | 12,627.1 | 15,723.6 | 1.25 |
Abbreviations: MW, weight-average molecular weight; Mn, number-average molecular weight; PDI, polydispersity index; Tri-CL, tricarballylic acid-poly (ε-caprolactone); Tri-CL-mPEG, tricarballylic acid-poly (ε-caprolactone)-methoxypolyethylene glycol; PET-CL, pentaerythritol-poly (ε-caprolactone); PET-CL-P, pentaerythritol-poly (ε-caprolactone)-polypeptide.
Physicochemical Properties of Nanoparticles
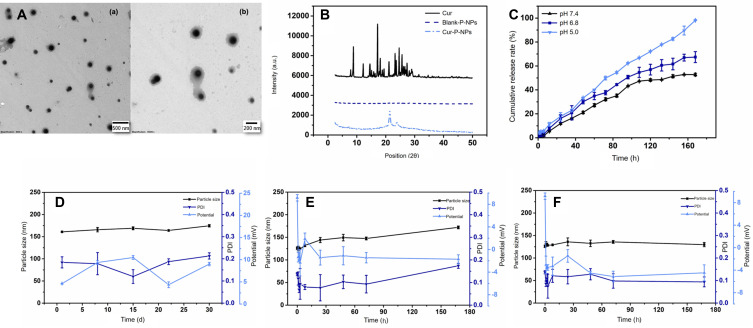
The particle size, zeta potential, encapsulation rate, and DL of the prepared nanoparticles are shown in Table 2. The nanoparticles showed adequate drug delivery, and their distributions were concentrated.40 The DL and encapsulation rate of Cur-P-NPs were higher than those of Cur-NPs. Cur-P-NPs showed a flip in zeta potential unlike Cur-NPs, which may be due to the cationic CPP polyarginine nine-peptide r9 in the peptide block.41 The TEM micrograph (Figure 3A) shows that the Cur-P-NPs were uniformly dispersed with spherical structures, about 150 nm in size, similar to the results of the particle size determination (146 nm) test. The XRD results are shown in Figure 3B. Cur had a large number of high intensity spikes. The curve for blank-P-NPs had no spikes and that for Cur-P-NPs had two small spikes; no peaks characteristic of Cur were observed. These results indicate that Cur was completely wrapped in the nanoparticles.
Table 2.
Characterization of Blank-NPs, Cur-NPs, Blank-P-NPs, and Cur-P-NPs
| Particle Size (nm) | PDI | Zeta Potential (mV) | EE (%) | DL (%) | |
|---|---|---|---|---|---|
| Blank-NPs | 142.4 ± 0.1414 | 0.164 ± 0.045 | −8.76 ± 0.311 | - | - |
| Cur-NPs | 148.1 ± 5.869 | 0.211 ± 0.016 | −10.0 ± 0.304 | 71.19 ± 0.529 | 4.25 ± 0.438 |
| Blank-P-NPs | 150.9 ± 2.121 | 0.227 ± 0.021 | 10.1 ± 0.247 | - | - |
| Cur-P-NPs | 148.3 ± 2.192 | 0.199 ± 0.053 | 12.4 ± 0.0707 | 74.66 ± 0.671 | 5.38 ± 0.316 |
Abbreviations: Blank-NPs, blank hybridized nanoparticles; Cur-NP, Cur-loaded hybridized nanoparticles; Blank-P-NPs, blank enzyme-targeted hybridized nanoparticles; Cur-P-NPs, Cur-loaded enzyme-targeted hybrid nano-delivery systems; EE, entrapment efficiency; PDI, polydispersity index; DL, drug loading.
Figure 3.
Characterization of NPs: (A) TEM graphs of Cur-P-NPs: magnification: (a) 8000×; (b) 15,000×, (B) XRD graphs of Cur, blank-P-NPs, and Cur-P-NPs, (C) Release curves of Cur-P-NPs in pH 7.4/6.8/5.0 PBS, (D) Stability of Cur-P-NPs at 4°C, (E) Stability of Cur-P-NPs at 37°C in pH 7.4 PBS, (F) Stability of Cur-P-NPs at 37°C in DMEM+.
Abbreviations: TEM, transmission electron microscopy; Cur-P-NPs, Cur-loaded enzyme-targeted hybrid nano-delivery systems; XRD, X-ray powder diffraction; Cur, curcumin; Blank-P-NPs, blank enzyme-targeted hybridized nanoparticles; DMEM, Dulbecco’s modified Eagle medium.
In vitro Release
The release rate accelerated with decreasing pH, as per the release curves (Figure 3C). The cumulative release rates at pH 7.4, pH 6.8, and pH 5.0 PBS were 21.12%, 30.06%, and 33.31%, respectively, at 48 h, and 52.78%, 67.39%, and 98.12%, respectively, at 168 h. The release behavior of the Cur-P-NPs was similar to that observed in other studies.42 These results indicate that Cur was released faster and more completely in the tumor microenvironment (weakly acidic conditions) and in the lysosomes and endosomes (pH approximately 5.0) than in the simulated normal internal environment (pH 7.4), suggesting that tumor cells would be affected more than the normal cells by the nanoparticles. This could effectively reduce damage to normal tissues during treatment.
Stability of Nanoparticles
The storage stability of the Cur-P-NPs is shown in Figure 3D. After 30 days of storage at 4°C, the particle size increased slightly, and the PDI and zeta potential fluctuated within a small range. The stabilities of Cur-P-NPs in PBS and DMEM+ at 37°C are shown in Figure 3E and F, respectively. The particle size of the Cur-P-NPs slightly increased in the medium; however, their stability was not affected. The zeta potential of the Cur-P-NPs flipped in both media, from +9.15 mV to a negative potential. This might have been caused by the negative electrical properties of the medium (PBS: −0.880 mV; DMEM+: −3.80 mV). No significant nanoparticle or drug precipitation was observed in the medium during the 7-day investigation period. Thus, Cur-P-NPs can maintain effective morphology and solubility both in vitro and in vivo.
In vitro Cytotoxicity
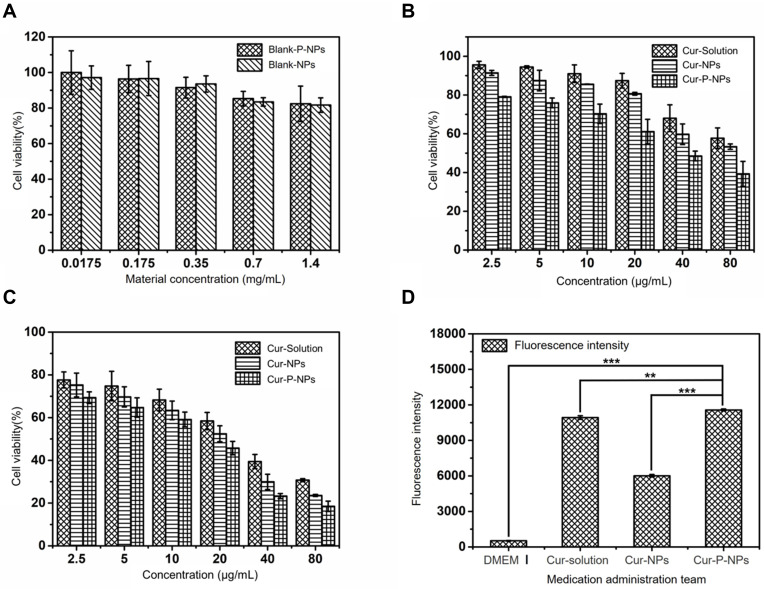
Results of the cytotoxicity tests for blank-NPs and blank-P-NPs against L929 cells are shown in Figure 4A. The cell viabilities were 97.11 ± 6.62%, 96.59 ± 9.60%, 93.55 ± 4.60%, 83.43 ± 2.35%, and 81.68 ± 4.08% when the concentrations of the blank-NPs were 0.0175, 0.175, 0.35, 0.7, and 1.4 mg/mL, respectively. When treated with the same concentrations of blank-P-NPs, the cell viabilities were 99.98 ± 12.22%, 96.34 ± 7.68%, 91.51 ± 5.83%, 85.30 ± 4.08%, and 82.41 ± 9.95%, respectively. In both cases, the cell viabilities were higher than 80% across this entire concentration range.
Figure 4.
(A) Viabilities of L929 cells treated with blank-NPs and blank-P-NPs, (B) Anti-proliferative effects of Cur-solution, Cur-NPs, and Cur-P-NPs against U251 cells (24 h), (C) Anti-proliferation effects of Cur-solution, Cur-NPs, and Cur-P-NPs against U251 cells (48 h), (D) Fluorescence intensity of U251 cells. (**P < 0.01, ***P < 0.001).
Abbreviations: Blank-NPs, blank hybridized nanoparticles; Blank-P-NPs, blank enzyme-targeted hybridized nanoparticles; Cur-Solution, curcumin dissolved in DMSO:H2O at 1:1000 (v:v); Cur-NPs, Cur-loaded hybridized nanoparticles; Cur-P-NPs, Cur-loaded enzyme-targeted hybrid nano-delivery systems; DMEM, Dulbecco’s modified Eagle’s medium.
In vitro Anti‑Proliferation Efficacy
The anti-proliferation test results are shown in Figure 4B and C. The inhibitory effects of all three agents on U251 cells were concentration dependent. After 24 h of incubation, the IC50s of Cur-solution, Cur-NPs, and Cur-P-NPs were 111.510, 95.955, and 39.547 μg/mL, respectively. After 48 h of incubation, the IC50s of the Cur-solution, Cur-NPs, and Cur-P-NPs were 25.457, 16.568, and 11.447 μg/mL, respectively. These results indicate that the anti-proliferative effect of Cur-P-NPs on U251 cells was superior to that of the Cur-solution and Cur-NPs. Tumor cell growth inhibition was positively correlated with drug concentration and duration of action.
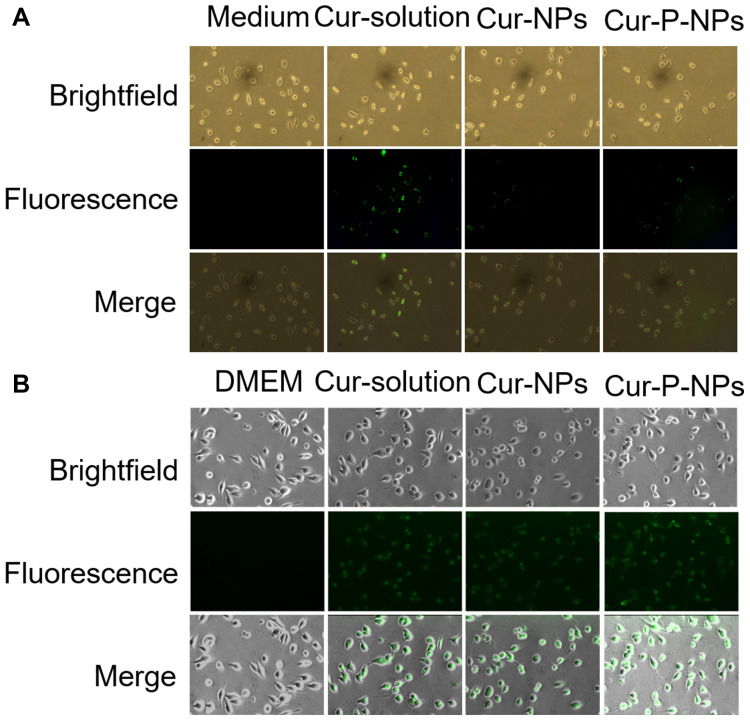
Cellular Uptake
Results of the cellular uptake studies are shown in Figures 4D and 5. The L929 cells in the medium group did not show green fluorescence. The intracellular fluorescence intensity was greater in the Cur-solution group than in the others. This may be because the hydrophobic Cur can easily cross the cell membranes and enter cells. However in the Cur-NP and Cur-P-NP group, Cur entry into cells required either drug release or endocytosis, with the entry being much slower than that of the Cur-solution. The absence of significant difference between Cur-NPs and Cur-P-NPs is attributed to the low expression of MMPs in L929 cells.
Figure 5.
Cellular uptake of (A) L929 cells, (B) U251 cells.
Abbreviations: Cur-Solution, curcumin dissolved in DMSO:H2O at 1:1000 (v:v); Cur-NPs, Cur-loaded hybridized nanoparticles; Cur-P-NPs, Cur-loaded enzyme-targeted hybrid nano-delivery systems; DMEM, Dulbecco’s modified Eagle’s medium.
The mean fluorescence intensities of DMEM (with 5% FBS), Cur-solution, Cur-NPs, and Cur-P-NPs after co-culture with U251 cells for 3 h were 518.33 ± 23.78, 10,927.43 ± 149.53, 6010.90 ± 105.45, and 11,571.30 ± 82.50, respectively. The cells in the DMEM group did not show green fluorescence; most of the U251 cells in this group were shuttle-shaped with better cell morphology than those in the other groups. The intracellular fluorescence intensity was the greatest in the Cur-P-NP group and the weakest in the Cur-NP group.
Nanoparticles deliver drugs into cells by endocytosis.40 The fluorescence intensity of cells treated with Cur-P-NPs was 1.93 times higher than that of Cur-NP-treated cells. The probable cause is that MMP-2 and MMP-9 overexpressed on the surface of U251 cells could specifically recognize and degrade the targeting peptides in the Cur-P-NPs. After enzymatic cleavage, the cell entry efficiency of these particles was significantly increased by the penetrating peptide. Meanwhile, cell membranes are negatively charged, and Cur-P-NPs are positively charged, thereby increasing drug uptake through electrostatic adsorption. In contrast, Cur-NPs are negatively charged and are electrostatically repulsed by cell membranes, resulting in a decrease in cellular uptake efficiency.43
The fluorescence intensity of cells treated with the Cur-solution was higher than that of Cur-P-NP-treated cells; this may have been caused by the full exposure of the cells to the lipid-soluble Cur in the solution, which may then have entered the cells faster by diffusion. The flow cytometry results were consistent with those obtained with fluorescence microscopy. The results indicate that Cur-P-NPs can effectively increase the uptake of Cur by U251 cells and enhance its ability to kill tumor cells. This is consistent with the in vitro anti-proliferation results discussed above.
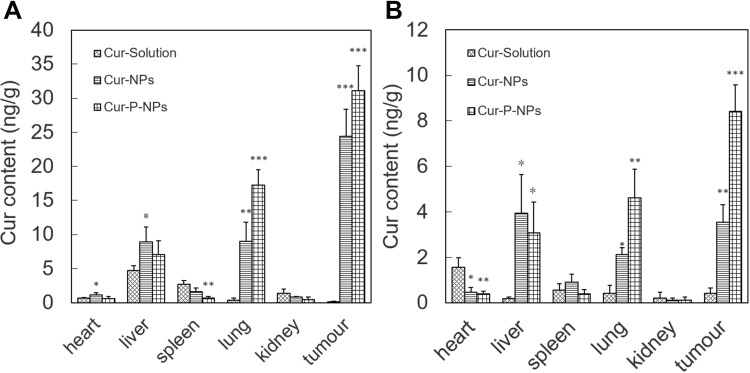
Biodistribution in vivo
The distributions of Cur at 1 h and 24 h are shown in Figure 6A and B, respectively. Cur was detected mainly in the liver and spleen 1 h after injection of the Cur-solution. No significant uptake was observed in tumors, indicating poor bioavailability of the active constituent in the Cur-solution. In contrast, the tumor Cur content was 24.4 and 31.1 ng/g 1 h after injection of Cur-NPs and Cur-P-NPs, respectively. Cur-P-NPs exhibited better tumor targeting ability. This could be attributed to targeting modification using the GPLGIAGQr9 polypeptide, which was specifically recognized and degraded by MMPs. After degradation, r9 was presumably exposed and cellular uptake improved. In samples collected 24 h after injection, the concentrations of Cur in the major organs and tumors decreased in all groups. However, in the Cur-NP and Cur-P-NP groups, considerable amounts of Cur were still present in the tumor tissue, at levels of 3.55 and 8.42 ng/g, respectively.
Figure 6.
Distribution of Cur in mice after IV administration (A) 1 h, (B) 24 h (*** P < 0.001, **P < 0.01, *P < 0.05 compared to Cur-Solution).
Abbreviations: Cur-Solution, curcumin dissolved in DMSO:H2O at a 1:1000 ratio (v:v); Cur-NPs, Cur-loaded hybridized nanoparticles; Cur-P-NPs, curcumin-loaded hybrid nano-delivery systems.
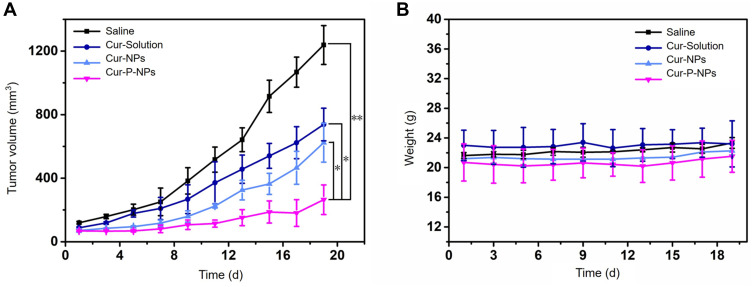
In vivo Antitumor Activity Assay
The tumor volumes of the U251 tumor-bearing mice are shown in Figure 7A. When treated with saline, Cur-solution, Cur-NPs, and Cur-P-NPs for 19 days, the tumor volumes were 1238.23 ± 351.61, 738.22 ± 286.54, 623.56 ± 219.36, and 264.59 ± 121.28 mm3, respectively. The tumor inhibition rates were 78.63% in the Cur-P-NP group, 40.38% in the Cur-solution group, and 49.64% in the Cur-NP group. Cur-P-NPs had the best tumor growth inhibitory effect.
Figure 7.
(A) In vivo tumor growth curves of U251-bearing nude mice treated with saline, Cur-solution, Cur-NPs, and Cur-P-NPs. (*P < 0.05; **P < 0.01), (B) Body weight changes of U251-bearing nude mice treated with saline, Cur-solution, Cur-NPs, and Cur-P-NPs.
Abbreviations: Cur-solution, curcumin dissolved in DMSO:H2O at 1:1000 (v:v); Cur-NPs, Cur-loaded hybridized nanoparticles; Cur-P-NPs, Cur-loaded enzyme-targeted hybrid nano-delivery systems.
No significant changes in body weight of the tumor xenograft mice were observed in any group during drug administration for 19 days (Figure 7B), indicating that the drug and carrier materials were safe for use in mice.
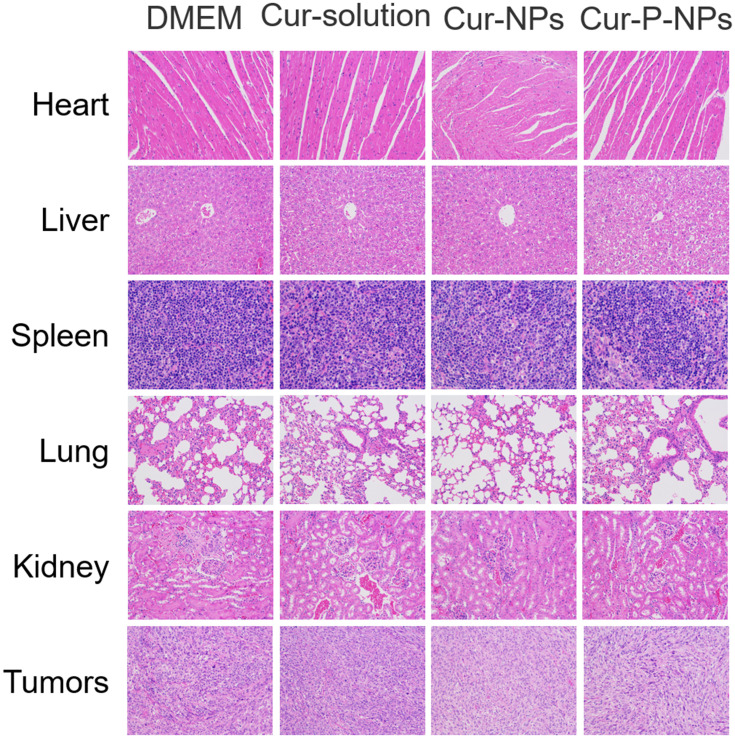
Histological Analysis
Sections of E-stained heart, liver, spleen, lung, kidney, and tumor were used to evaluate the toxicity of the formulation in vivo (Figure 8). The heart cells in all four groups were closely arranged with clear transverse lines, and some of the heart cells in the Cur-NP group had deep eosin-stained cytoplasms. The liver cells of the four groups showed different degrees of nuclear fixation, in the following rank order: Cur-solution and Cur-NP group > saline group > Cur-P-NP group. The splenocytes in the four groups were similar in structure, and the morphology of the splenic vesicles was normal. The alveolar structure was reduced in the saline, Cur-solution, and Cur-NP groups; in contrast, it was normal in the Cur-P-NP group. Inflammatory cell infiltration was observed in the kidney sections of all four groups, and it was relatively mild in the Cur-P-NP group. The tumor cells were closely arranged with round nuclei after treatment with saline. In the Cur-P-NP group, the tumor cells showed different morphology, with pyknotic nuclei and an indistinct cytoplasmic interface.
Figure 8.
H&E-stained organ slices from U251-bearing nude mice treated with different formulations after 19 days (400×); histological results showing the in vivo biocompatibility.
Abbreviation: H&E, hematoxylin-eosin.
These results indicate that Cur-P-NPs could effectively destroy tumor cells with less damage to normal tissues because of specificity. The Cur-NPs and Cur-solution treatments resulted in some degree of damage to normal organs, assumed to be due to the lack of active targeting.
Conclusion
In this study, Cur-P-NPs with good biocompatibility and stability were successfully prepared in microchannels for the first time. In vitro release studies of Cur-P-NPs in different pH environments revealed that the drug could be released faster in the tumor microenvironment (pH 6.5). The release rate was maximized in the lysosomes and endosomes (pH 5.0). Cur-P-NPs could kill tumor cells efficiently, with reduced toxic side effects on normal tissues. Cur-P-NPs had stronger antitumor activity and cell entry efficiency than Cur-NPs. Cur-P-NPs could effectively improve cellular uptake of Cur, and inhibit tumor growth effectively (up to 78.63%). These findings highlight the potential application of Cur-P-NPs in antitumor therapy in future.
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (No. 22078297), Zhejiang Provincial Natural Science Foundation of China (LY19B060012), Taizhou Science and Technology Project (1901ky49), Special Research Fund of Hospital Pharmacy of Zhejiang Pharmaceutical Association (2019ZYY44, 2016ZYY35).
Abbreviations
ACP-GPLGIAGr9-ACP:
Disclosure
Anqin Li is an employee of Zhejiang Share Bio-Pharm Co., Ltd, The authors report no other potential conflicts of interest.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi: 10.3322/caac.21590 [DOI] [PubMed] [Google Scholar]
- 2.Martin-Romano P, Ammari S, El-Dakdoukti Y, et al. Chemotherapy beyond immune checkpoint inhibitors in patients with metastatic colorectal cancer. Eur J Cancer. 2020;137:117–126. doi: 10.1016/j.ejca.2020.06.030 [DOI] [PubMed] [Google Scholar]
- 3.Akbari S, Kariznavi E, Jannati M, Elyasi S, Tayarani-Najaran Z. Curcumin as a preventive or therapeutic measure for chemotherapy and radiotherapy induced adverse reaction: a comprehensive review. Food Chem Toxicol. 2020;145:111699. doi: 10.1016/j.fct.2020.111699 [DOI] [PubMed] [Google Scholar]
- 4.Lee YH, Song NY, Suh J, et al. Curcumin suppresses oncogenicity of human colon cancer cells by covalently modifying the cysteine 67 residue of SIRT1. Cancer Lett. 2018;431:219–229. doi: 10.1016/j.canlet.2018.05.036 [DOI] [PubMed] [Google Scholar]
- 5.Tamaddoni A, Mohammadi E, Sedaghat F, Qujeq D, As’Habi A. The anticancer effects of curcumin via targeting the mammalian target of rapamycin complex 1 (mTORC1) signaling pathway. Pharmacol Res. 2020;156:104798. doi: 10.1016/j.phrs.2020.104798 [DOI] [PubMed] [Google Scholar]
- 6.Pourbagher-Shahri AM, Farkhondeh T, Ashrafizadeh M, Talebi M, Samargahndian S. Curcumin and cardiovascular diseases: focus on cellular targets and cascades. Biomed Pharmacother. 2021;136:111214. doi: 10.1016/j.biopha.2020.111214 [DOI] [PubMed] [Google Scholar]
- 7.Hadi A, Pourmasoumi M, Ghaedi E, Sahebkar A. The effect of Curcumin/Turmeric on blood pressure modulation: a systematic review and meta-analysis. Pharmacol Res. 2019;150:104505. doi: 10.1016/j.phrs.2019.104505 [DOI] [PubMed] [Google Scholar]
- 8.Hasanzadeh S, Read MI, Bland AR, Majeed M, Jamialahmadi T, Sahebkar A. Curcumin: an inflammasome silencer. Pharmacol Res. 2020;159:104921. doi: 10.1016/j.phrs.2020.104921 [DOI] [PubMed] [Google Scholar]
- 9.Teter B, Morihara T, Lim GP, et al. Curcumin restores innate immune Alzheimer’s disease risk gene expression to ameliorate Alzheimer pathogenesis. Neurobiol Dis. 2019;127:432–448. doi: 10.1016/j.nbd.2019.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sabet S, Rashidinejad A, Qazi HJ, Mcgillivray DJ. An efficient small intestine-targeted curcumin delivery system based on the positive-negative-negative colloidal interactions. Food Hydrocoll. 2020;111:106375. doi: 10.1016/j.foodhyd.2020.106375 [DOI] [Google Scholar]
- 11.Hussein Y, Loutfy SA, Kamoun EA, El-Moslamy SH, Radwan EM, Elbehairi SEI. Enhanced anti-cancer activity by localized delivery of curcumin form PVA/CNCs hydrogel membranes: preparation and in vitro bioevaluation. Int J Biol Macromol. 2020;170:107–122. doi: 10.1016/j.ijbiomac.2020.12.133 [DOI] [PubMed] [Google Scholar]
- 12.Yadav P, Bandyopadhyay A, Chakraborty A, Sarkar K. Enhancement of anticancer activity and drug delivery of chitosan-curcumin nanoparticle via molecular docking and simulation analysis. CarbohydrPolym. 2018;182:188–198. doi: 10.1016/j.carbpol.2017.10.102 [DOI] [PubMed] [Google Scholar]
- 13.Lubtow MM, Nelke LC, Seifert J, et al. Drug induced micellization into ultra-high capacity and stable curcumin nanoformulations: physico-chemical characterization and evaluation in 2D and 3D in vitro models. J Control Release. 2019;303:162–180. doi: 10.1016/j.jconrel.2019.04.014 [DOI] [PubMed] [Google Scholar]
- 14.Bahari LA, Javadzadeh Y, Jalali MB, Johari P, Nokhodchi A, Shokri J. Nano-suspension coating as a technique to modulate the drug release from controlled porosity osmotic pumps for a soluble agent. Colloids Surf B Biointerfaces. 2017;153:27–33. doi: 10.1016/j.colsurfb.2017.02.007 [DOI] [PubMed] [Google Scholar]
- 15.Moradi Kashkooli F, Soltani M, Souri M. Controlled anti-cancer drug release through advanced nano-drug delivery systems: static and dynamic targeting strategies. J Control Release. 2020;327:316–349. doi: 10.1016/j.jconrel.2020.08.012 [DOI] [PubMed] [Google Scholar]
- 16.Kim M, Sahu A, Kim GB, et al. Comparison of in vivo targeting ability between cRGD and collagen-targeting peptide conjugated nano-carriers for atherosclerosis. J Control Release. 2018;269:337–346. doi: 10.1016/j.jconrel.2017.11.033 [DOI] [PubMed] [Google Scholar]
- 17.Ren T, Hu M, Cheng Y, et al. Piperine-loaded nanoparticles with enhanced dissolution and oral bioavailability for epilepsy control. Eur J Pharm Sci. 2019;137:104988. doi: 10.1016/j.ejps.2019.104988 [DOI] [PubMed] [Google Scholar]
- 18.Chen M, Zhou X, Chen R, et al. Nano-carriers for delivery and targeting of active ingredients of Chinese medicine for hepatocellular carcinoma therapy. Mater Today. 2018;25:66–87. doi: 10.1016/j.mattod.2018.10.040 [DOI] [Google Scholar]
- 19.Ren X, Cheng S, Liang Y, Yu X, Ning R. Mesoporous silica nanospheres as nanocarriers for poorly soluble drug itraconazole with high loading capacity and enhanced bioavailability. Microporous Mesoporous Mater. 2020;305:110389. doi: 10.1016/j.micromeso.2020.110389 [DOI] [Google Scholar]
- 20.Tanziela T, Shaikh S, Jiang H, Lu Z, Wang X. Efficient encapsulation of biocompatible nanoparticles in exosomes for cancer theranostics. Nano Today. 2020;35:100964. doi: 10.1016/j.nantod.2020.100964 [DOI] [Google Scholar]
- 21.Choi H, Liu T, Qiao H, et al. Biomimetic nano-surfactant stabilizes sub-50 nanometer phospholipid particles enabling high paclitaxel payload and deep tumor penetration. Biomaterials. 2018;181:240–251. doi: 10.1016/j.biomaterials.2018.07.034 [DOI] [PubMed] [Google Scholar]
- 22.Kostka L, Kotrchova L, Subr V, et al. HPMA-based star polymer biomaterials with tuneable structure and biodegradability tailored for advanced drug delivery to solid tumours. Biomaterials. 2020;235:119728. doi: 10.1016/j.biomaterials.2019.119728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yao X, Huang P, Nie Z. Cyclodextrin-based polymer materials: from controlled synthesis to applications. Prog Polym Sci. 2019;93:1–35. doi: 10.1016/j.progpolymsci.2019.03.004 [DOI] [Google Scholar]
- 24.Behl A, Parmar VS, Malhotra S, Chhillar AK. Biodegradable diblock copolymeric PEG-PCL nanoparticles: synthesis, characterization and applications as anticancer drug delivery agents. Polymer. 2020;207:122901. doi: 10.1016/j.polymer.2020.122901 [DOI] [Google Scholar]
- 25.Hira SK, Mitra K, Srivastava P, et al. Doxorubicin loaded pH responsive biodegradable ABA-type Amphiphilic PEG-b-aliphatic Polyketal-b-PEG block copolymer for therapy against aggressive murine lymphoma. Nanomedicine. 2020;24:102128. doi: 10.1016/j.nano.2019.102128 [DOI] [PubMed] [Google Scholar]
- 26.Repp L, Rasoulianboroujeni M, Lee HJ, Kwon GS. Acyl and oligo(lactic acid) prodrugs for PEG-b-PLA and PEG-b-PCL nano-assemblies for injection. J Control Release. 2020;330:1004–1015. doi: 10.1016/j.jconrel.2020.11.008 [DOI] [PubMed] [Google Scholar]
- 27.Golombek SK, May JN, Theek B, et al. Tumor targeting via EPR: strategies to enhance patient responses. Adv Drug Deliv Rev. 2018;130:17–38. doi: 10.1016/j.addr.2018.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahalanobish S, Saha S, Dutta S, Sil PC. Matrix metalloproteinase: an upcoming therapeutic approach for idiopathic pulmonary fibrosis. Pharmacol Res. 2020;152:104591. doi: 10.1016/j.phrs.2019.104591 [DOI] [PubMed] [Google Scholar]
- 29.Sette P, Amankulor N, Li A, et al. GBM-Targeted oHSV armed with matrix metalloproteinase 9 enhances anti-tumor activity and animal survival. Mol Ther-Oncolyt. 2019:15. doi: 10.1016/j.omto.2019.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iaccarino G, Profeta M, Vecchione R, Netti P. Matrix metalloproteinase-cleavable nanocapsules for tumor-activated drug release. Acta Biomater. 2019;89:89. doi: 10.1016/j.actbio.2019.02.043 [DOI] [PubMed] [Google Scholar]
- 31.Jauset T, Beaulieu ME. Bioactive cell penetrating peptides and proteins in cancer: a bright future ahead. CurrOpinPharmacol. 2019;47:133–140. doi: 10.1016/j.coph.2019.03.014 [DOI] [PubMed] [Google Scholar]
- 32.Khan MM, Filipczak N, Torchilin VP. Cell penetrating peptides: a versatile vector for co-delivery of drug and genes in cancer. J Control Release. 2020;330:1220–1228. doi: 10.1016/j.jconrel.2020.11.028 [DOI] [PubMed] [Google Scholar]
- 33.Platel R, Vaure L, Palleau E, et al. Synthesis of hybrid colloidal nanoparticles for a generic approach to 3D electrostatic directed assembly: application to anti-counterfeiting. J Colloid Interface Sci. 2021;582(Pt B):1243–1250. doi: 10.1016/j.jcis.2020.08.098 [DOI] [PubMed] [Google Scholar]
- 34.Ferreira Soares DC, Domingues SC, Viana DB, Tebaldi ML. Polymer-hybrid nanoparticles: current advances in biomedical applications. Biomed Pharmacother. 2020;131:110695. doi: 10.1016/j.biopha.2020.110695 [DOI] [PubMed] [Google Scholar]
- 35.Abdelkarim M, Abd Ellah NH, Elsabahy M, Abdelgawad M, Abouelmagd SA. Microchannel geometry vs flow parameters for controlling nanoprecipitation of polymeric nanoparticles. Colloids Surf APhysicochemEng Asp. 2021;611:125774. doi: 10.1016/j.colsurfa.2020.125774 [DOI] [Google Scholar]
- 36.Kratz F, Drevs J, Bing G, et al. Development and in vitro efficacy of novel MMP2 and MMP9 specific doxorubicin albumin conjugates. Bioorg Med Chem Lett. 2001;11(15):2001–2006. doi: 10.1016/s0960-894x(01)00354-7 [DOI] [PubMed] [Google Scholar]
- 37.Wang HX, Yang XZ, Sun CY, Mao CQ, Zhu YH, Wang J. Matrix metalloproteinase 2-responsive micelle for siRNA delivery. Biomaterials. 2014;35(26):7622–7634. doi: 10.1016/j.biomaterials.2014.05.050 [DOI] [PubMed] [Google Scholar]
- 38.Wu W, Wu J, Fu Q, et al. Elaboration and characterization of curcumin-loaded Tri-CL-mPEG three-arm copolymeric nanoparticles by a microchannel technology. Int J Nanomedicine. 2019;14:4683–4695. doi: 10.2147/IJN.S198217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao D, Lo PC. Polymeric micelles encapsulating pH-responsive doxorubicin prodrug and glutathione-activated zinc(II) phthalocyanine for combined chemotherapy and photodynamic therapy. J Control Release. 2018;282:46–61. doi: 10.1016/j.jconrel.2018.04.030 [DOI] [PubMed] [Google Scholar]
- 40.Bareford LM, Swaan PW. Endocytic mechanisms for targeted drug delivery. Adv Drug Deliv Rev. 2007;59(8):748–758. doi: 10.1016/j.addr.2007.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meloni BP, Brookes LM, Clark VW, et al. Poly-arginine and arginine-rich peptides are neuroprotective in stroke models. J Cereb Blood Flow Metab. 2015;35(6):993–1004. doi: 10.1038/jcbfm.2015.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi C, Guo X, Qu Q, Tang Z, Wang Y, Zhou S. Actively targeted delivery of anticancer drug to tumor cells by redox-responsive star-shaped micelles. Biomaterials. 2014;35(30):8711–8722. doi: 10.1016/j.biomaterials.2014.06.036 [DOI] [PubMed] [Google Scholar]
- 43.Medel S, Syrova Z, Kovacik L, et al. Curcumin-bortezomib loaded polymeric nanoparticles for synergistic cancer therapy. Eur Polym J. 2017;93:116–131. doi: 10.1016/j.eurpolymj.2017.05.036 [DOI] [Google Scholar]