Summary
Severe COVID-19 is accompanied by rampant immune dysregulation in the lung and periphery, with immune cells of both compartments contributing to systemic distress. The extent to which immune cells of the lung and blood enter similar or distinct pathological states during severe disease remains unknown. Here, we leveraged 96 publicly available single-cell RNA sequencing datasets to elucidate common and compartment-specific features of severe to critical COVID-19 at the levels of transcript expression, biological pathways, and ligand-receptor signaling networks. Comparing severe patients to milder and healthy donors, we identified distinct differential gene expression signatures between compartments and a core set of co-directionally regulated surface markers. A majority of severity-enriched pathways were shared, whereas TNF and interferon responses were polarized. Severity-specific ligand-receptor networks appeared to be differentially active in both compartments. Overall, our results describe a nuanced response during severe COVID-19 where compartment plays a role in dictating the pathological state of immune cells.
Subject areas: Pathophysiology, Immunology, Complex system biology, Transcriptomics
Graphical abstract

Highlights
-
•
Transcriptomic comparisons of lung and blood immune cell compartments in COVID-19
-
•
96 single-cell public datasets from severe, mild-to-moderate, and healthy donors
-
•
Severity-specific cell surface markers and pathways shared between compartments
-
•
Putative ligand-receptor signaling dialogs within and between compartments
Pathophysiology; Immunology; Complex system biology; Transcriptomics
Introduction
Within months of the identification of the novel coronavirus SARS-CoV-2 in Wuhan, China, in December 2019, the virus had spread to every major country on Earth (Baj et al., 2020; Khafaie and Rahim, 2020; Wu et al., 2020). The pandemic disease caused by SARS-CoV-2, termed coronavirus disease 2019 (COVID-19), has diverse clinical presentations, ranging from asymptomatic infection to mild symptomatic infection with possible pneumonia, severe respiratory distress, critical forms of respiratory failure, disseminated inflammation, or multiple organ failure (Baj et al., 2020; Chan et al., 2020; Wu et al., 2020; Wu and McGoogan, 2020). A hallmark of severe and critical COVID-19 cases is a rampant dysregulation of the immune system concomitant with the development of a hypoxemic respiratory condition widely characterized as acute respiratory distress syndrome (ARDS) (Gattinoni et al., 2020; Ramanathan et al., 2020; Wilson et al., 2020; Wu et al., 2020; Xu et al., 2020b). These observations were validated by early serological profiles of patients with severe COVID-19, which largely resembled the cytokine profile of ARDS driven by diverse etiologies (Wilson et al., 2020) and have been characterized as “cytokine release syndrome” (Blanco-Melo et al., 2020; Del Valle et al., 2020; Huang et al., 2020; Mehta et al., 2020; Wilson et al., 2020). With this information a basic understanding of the immune response to COVID-19 was established; however, a more granular analysis of the immunological features distinguishing severe from mild patients is needed to better inform treatment options for this disease.
In recent months, observations in hospitals across the world have led to consistent descriptions of COVID-19 severity at a clinical level (Arentz et al., 2020; Grasselli et al., 2020; Zhou et al., 2020), yet the biological underpinnings of immune system hyperactivation in severe COVID-19 are still being defined. Cells of the immune system are already known to be transcriptionally distinct between the lung and periphery during baseline health (Travaglini et al., 2020), suggesting a model for two immune subsystems that may respond differently during infection. Bulk RNA sequencing and single-cell RNA sequencing (scRNA-seq) studies have identified stark transcriptional differences between bronchoalveolar lavage fluid (BALF) and peripheral blood mononuclear cell (PBMC) samples in hospitalized patients with COVID-19, indicating that immunological responses may be highly compartment specific (Daamen et al., 2020; Gardinassi et al., 2020; Xiong et al., 2020; Xu et al., 2020a). However, the critical question of whether severity-specific dysregulatory transcriptional states are conserved between compartments has not yet been addressed. The role of interferons and their subsequent responses is particularly a conundrum, as several groups have shown a downregulation of interferon signaling in the blood but an upregulation of interferons in the lungs of patients with severe COVID-19 (Broggi et al., 2020; Grajales-Reyes and Colonna, 2020; Hadjadj et al., 2020). Although understanding of the transcriptional responses of immune cells during COVID-19 across the spectrum of disease severity is growing, it remains unknown whether immunological dynamics between the lung and blood compartments affect the course of the disease and whether mechanistic differences between compartments could play a role in the efficacy of therapeutic strategies.
In this study, we examined and compared the immunological features of COVID-19 in the lung and periphery, focusing on severity-specific transcriptional states. We re-analyzed 96 publicly available scRNA-seq datasets from eight published studies containing either BALF or PBMC samples across the spectrum of COVID-19 severity, allowing us to make comparisons between severe-to-critical patients, mild-to-moderate patients, and healthy donors. Leveraging differential expression data, we compared features of COVID-19 severity between the lung and periphery at the level of individual transcripts, biological pathways, and active ligand-receptor signaling networks. Our integrative analysis adds to a growing body of knowledge around lung and peripheral responses during acute respiratory infections and contributes to a finer understanding of the mechanisms that drive ARDS-related immune dysregulation in severe-to-critical COVID-19. Our findings may guide future work informing potential interventional strategies to improve patient outcomes as the COVID-19 pandemic continues to unfold.
Results
scRNA-seq datasets contain comparable immune cell types in the lung microenvironment and peripheral circulation
Severe COVID-19 has been shown to result in profound immune dysregulation at both a local (pulmonary) and systemic (circulatory) level. Here, we performed an analysis of publicly available scRNA-seq datasets to identify features of COVID-19 severity that are common or specific to these compartments. We used identical methods to separately analyze scRNA-seq datasets from BALF (three cohorts) and peripheral PBMCs (four cohorts) obtained from patients with severe-to-critical (referred to as “severe”) and mild-to-moderate (referred to as “mild”) COVID-19 as well as healthy control subjects. All BALF and PBMC cohorts were obtained from separate prior studies (Arunachalam et al., 2020; Lee et al., 2020; Liao et al., 2020; Schulte-Schrepping et al., 2020; Wilk et al., 2020). A BALF cross-control cohort was created by merging data from two additional studies (Morse et al., 2019; Mould et al., 2021). Detailed information on datasets and clinical characteristics of patient donors are available in Table 1. We primarily focused on comparing the Liao et al. BALF and Lee et al. PBMC datasets and used the other datasets for validation (see STAR Methods).
Table 1.
Overview of the publicly available datasets used in this study
| Author | Liao et al. | Wauters et al. | Morse et al. | Mold et al. | Arunachalam et al. | Wilk et al. | Lee et al. | Schulte-Schrepping et al. |
|---|---|---|---|---|---|---|---|---|
| PMID | 32398875 | 33473155 | 31221805 | 33079572 | 32788292 | 32514174 | 32651212 | 32810438 |
| Accession no. | GEO: GSE145926 | EGA: EGAS00001004717 | GEO: GSM3660650 | GEO: GSE151928 | GEO: GSE155673 | GEO: GSE150728 | GEO: GSE149689 | EGA: EGAS00001004571 |
| Compartment | BALF | BALF | BALF | BALF | PBMC | PBMC | PBMC | PBMC |
| Healthy control subjects | 3 | 0 | 1 | 10 | 5 | 6 | 4 | 0 |
| Mild COVID-19 patients | 3 | 2 | 0 | 0 | 3 | 0 | 3 | 5 |
| Severe COVID-19 patients | 6 | 20 | 0 | 0 | 4 | 7 | 4 | 10 |
| Total donors (n) | 12 | 22 | 1 | 10 | 12 | 13 | 11 | 15 |
After preprocessing raw gene-barcode matrices from each donor in the study, all BALF and PBMC donor datasets were merged and integrated using Harmony (Korsunsky et al., 2019). The cells were then clustered using Seurat (Stuart et al., 2019). The resulting 21 clusters were first labeled by a coarse cell type annotation. The clusters corresponding to myeloid cells, plasma/B/proliferating cells, NK/T cells, and dendritic cells were re-integrated in four separate groups and sub-clustered in separate feature spaces. Following annotation of the sub-clusters, cell type labels were transferred back to the original shared space resulting in 20 distinct cell types that were visualized by uniform manifold approximation and projection (UMAP) as shown in Figure 1A. UMAPs of the sub-clusters and key marker genes used for annotation are shown in Figure S1. In total, 96 datasets from lung and peripheral sample donors were successfully integrated via Harmony, as shown in Figures 1B and 1C. Subdividing the total pool of cells into three categories based on the disease status of the donors (severe COVID-19, mild COVID-19, and healthy controls) showed that most clusters appeared in all three conditions (Figure 1D) and no differences between disease states were readily apparent.
Figure 1.
Single-cell RNA sequencing reveals comparable cell populations in bronchoalveolar lavage fluid (BALF) and peripheral blood mononuclear cell (PBMC) isolates in healthy controls and across the spectrum of COVID-19 severity
(A) UMAP and annotation of 20 cell types identified from BALF and PBMC samples from healthy, mild COVID-19, and severe COVID-19 donors. Abbreviations: AM, alveolar macrophage; Mono, monocyte; cDC, conventional dendritic cell; MP, mononuclear phagocyte; NK, natural killer cell; pDC, plasmacytoid dendritic cell.
(B) UMAP breakdown according to cohort of origin demonstrating the successful integration of the seven cohorts used in this study. Abbreviations: BALFxControl, BALF cross-control cohort.
(C) UMAP breakdown of the cells according to their compartment of origin. (Left) Lung-derived BALF cells. (Right) Blood-derived PBMCs. Cells are colored according to their donor cohorts, which strongly overlap as shown in (B).
(D) UMAP breakdown across the spectrum of healthy donors, mild COVID-19 patient donors, and severe COVID-19 patient donors demonstrating that most cell types annotated in (A) are recovered across the spectrum of disease severity.
See also Figure S1.
The lung and blood immune compartments demonstrate both common and compartment-specific perturbations of gene expression in severe COVID-19
Severe COVID-19 is characterized by rampant immune dysregulation in the lung and blood, leading us to ask to what extent severity-specific transcriptional changes are conserved between consanguineous populations of immune cells at the site of local infection and in the peripheral circulation. We began by comparing patients with severe COVID-19 with healthy controls and identified differentially expressed genes (DEGs) in most of the cell types common to BALF and PBMCs (Figure 2A). To discern the extent of conserved differential gene expression, we made paired comparisons between BALF and PBMC cohorts to identify DEGs unique to BALF, those unique to PBMCs, those regulated in opposite directions in both compartments (contra-directional), and those regulated in the same direction in both compartments (co-directional/overlapping). The relative proportions of these genes served as a metric to evaluate transcriptional similarity between the lung and blood compartments (see STAR Methods). Only a small proportion of all differentially expressed transcripts demonstrated co-directional overlap between the lung and blood in the same cell types, indicating strong dissimilarity between these gene sets that was robust across the datasets we compared (Figure 2B). In order to obtain a meaningful benchmark for these results, we also made paired PBMC-to-PBMC cohort comparisons (Figure S3A). The resulting PBMC-PBMC comparisons tended to reveal higher degrees of overlap than BALF-PBMC comparisons, pointing to disparities between compartments at the level of differentially expressed transcripts between severe patients and controls.
Figure 2.
Transcriptional signatures of COVID-19 severity demonstrate minimal overlap between lung and blood immune compartments in comparable cell types
(A) Differentially expressed genes (DEGs) between severe patients and healthy controls were identified in cell types originating from BALF and PBMC donors. Volcano plots of DEGs are shown for CD14 monocytes of the lung (left) from the Liao et al. cohort and blood (right) from the Lee et al. cohort. Genes denoted by red dots met a threshold of p < 0.05 and |log2FC > 1|. FC = fold change (ratio of severe expression to control expression).
(B) Degree of transcriptional similarity between the lung and blood compartments assessed using overlap metrics for severe versus healthy control DEGs. Rose plots indicate the fraction of total DEGs significant in the BALF compartment only (purple), the PBMC compartment only (blue), significant in both compartments but regulated in opposite directions (green), or significant and co-directional in both compartments (yellow, representing compartmental overlap). Concentric circles represent fractions of the total number of DEGs from 0 to 1. Each rose plot compares the Liao BALF data to a different PBMC cohort.
(C) Example volcano plots of DEGs between severe and mild patients for CD14 monocytes of the lung (left) and blood (right) using the same cohorts as (A).
(D) Degree of transcriptional similarity between the lung and blood compartments in shared cell types assessed using severe versus mild DEGs. Each rose plot compares the Liao BALF data to a different PBMC cohort.
See also Figures S2–S4, as well as Figure S5 for cohort sequencing depths.
To compare dysregulatory transcriptional programs associated with severe as opposed to mild disease courses, we next analyzed DEGs between severe and mild patients (Figure 2C). Again, severity-specific DEGs overlapped in cells of the lung and periphery by a narrow margin (Figures 2D and S2A). Comparing PBMC cohorts to each other (Figure S3B) again showed higher degrees of overlap than inter-compartmental comparisons. By contrast, the two BALF cohorts showed very little overlap when directly compared (Figure S3C), which we attributed to the very few mild patients (n = 2) in the Wauters et al. BALF cohort. Across all of the cell types analyzed, the sets of transcripts uniquely perturbed in patients with severe compared with mild COVID-19 showed only a few commonalities between the lung and periphery. This generally dissimilar trend followed when using an alternative approach to measure dissimilarity by directly conducting differential gene expression between cells of different compartments (Figure S4A, see STAR Methods).
To identify the most strongly evidenced differentially expressed transcripts common to both compartments, we used a DEG classification scheme with more stringent p value and fold change thresholds and explicitly accounted for donor-to-donor variation in the statistical test (see STAR Methods). The stringently selected DEGs that were observed in both the Liao BALF dataset and at least two PBMC datasets are given in Table S1. Declines in HLA class II-related gene expression (HLA-DPs, HLA-DRs), upregulation of calprotectin and calgranulin genes (S100A8 and S10012), and upregulation of the clusterin gene CLU were common features of severe patients in both the lung and periphery.
A minimal set of surface markers is shared between the lung and blood in severe COVID-19
Cellular surface markers corresponding to clinical categories of COVID-19 have not yet been rigorously characterized. Furthermore, the extent to which surface proteins on blood leukocytes reflect the immunological state of immune cells in the lung during a viral infection remains largely unknown. Given our observation of strong differences between the lung and periphery at the level of differentially regulated individual transcripts, we sought to determine if any transcripts coding for cell surface markers show similar regulatory patterns between the compartments. The sets of DEGs studied in Figure 2 (adjusted p < 0.05 and |log2FC > 1|) were filtered to find transcripts coding for cell surface proteins. Comparing severe patients to healthy control subjects, we found that myeloid cells tended to share differentially regulated surface transcripts, whereas lymphoid cells did not. Compartmentally conserved downregulated transcripts in severe donors included HLA-DPB1 on cDC2s and CD74 on CD14 monocytes (Figure 3A). We found that the CLU transcript was upregulated in severe donors from both compartments in several cell types, including CD14 monocytes (Figure 3B). These transcript patterns were robust across pairwise comparisons between cohorts. In general, most myeloid cell types shared at least 5% of differentially regulated surface transcripts between compartments (Figure 3C).
Figure 3.
Cell surface marker transcripts associated with COVID-19 severity demonstrate overlap between the lung and periphery predominantly in myeloid cells
(A) (Left) Transcript expression levels of the cell surface marker HLA-DPB1 in healthy controls (blue violins), mild COVID-19 patients (purple violins), and severe COVID-19 patients (red violins) in cDC2 cells derived from the Liao BALF cohort (top) and Lee PBMC cohort (bottom). (Right) Expression levels of the surface marker CD74 in CD14 monocytes from the same BALF (top) and PBMC (bottom) cohorts. ∗p < 0.05 and |log2FC > 1|.
(B) Transcript expression levels of the surface marker CLU in CD14 monocytes derived from BALF (top) and PBMCs (bottom).
(C) Compartmental similarity of cell surface markers differentially expressed between severe COVID-19 patients and healthy control subjects following the format of Figure 2, where yellow segments indicate compartmental overlap.
(D) Compartmental similarity of surface markers differentially expressed between severe and mild COVID-19 patients.
We next investigated differences in expression of surface marker transcripts between severe and mild patients. A number of surface markers were markers of severe disease only; these transcripts were differentially regulated between severe and mild patients but not between mild patients and healthy controls. The HLA-DPB1, CD74, and CLU transcripts discussed above exhibited this property in the same respective cell types, as shown in Figures 3A and 3B. In addition to HLA-DPB1, many other HLA class II transcripts were downregulated in a severity-specific manner. Across the cohorts studied, myeloid cells, NK cells, and B cells showed high degrees of overlap in severe versus mild surface markers (Figure 3D). Of interest, T cell populations did not appear to share surface markers between compartments in any of the analyses in Figure 3. Overall, the most robust compartmentally conserved surface markers identified were downregulated transcripts for antigen presentation in cDC2s, monocytes, and B cells.
A fraction of severity-specific pathways is polarized between the lung and blood including type I/III and II interferon responses and TNF-α signaling
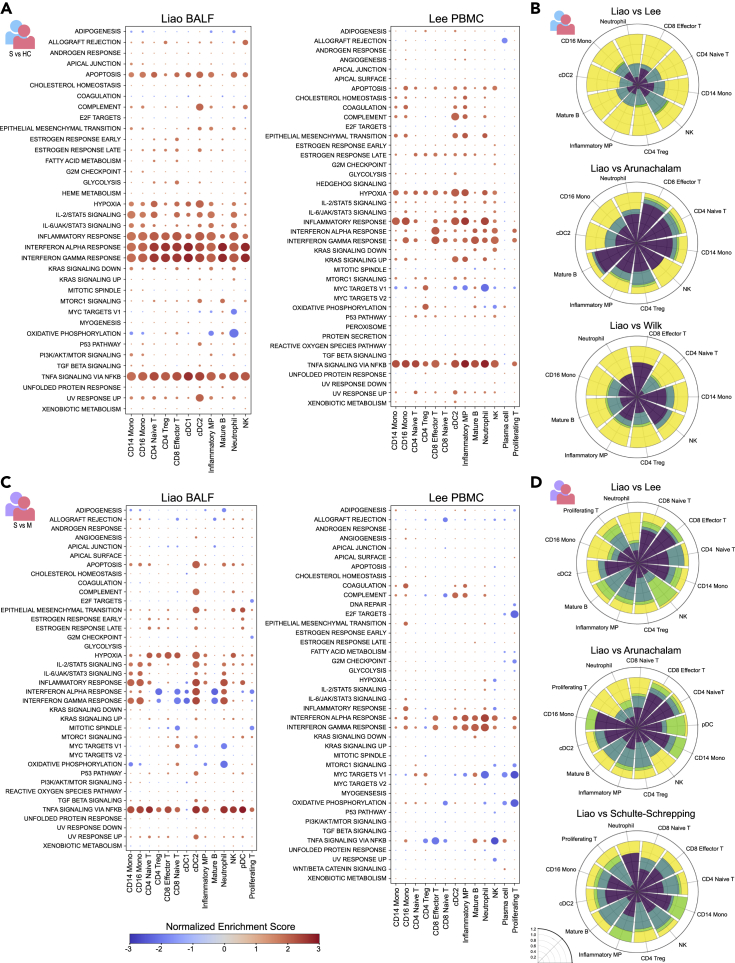
To probe the local and systemic immune responses to severe COVID-19 at a broader scale, we sought to utilize the large number of differentially expressed transcripts to identify biological pathways that are altered across the spectrum of disease severity in the lung and periphery. Applying gene set enrichment analysis (GSEA) to severe versus healthy control DEGs in the Liao BALF cohort revealed broadly enriched pathways including the IL-2/STAT5 signaling, IL-6/JAK/STAT3 signaling, IFN-α response, IFN-γ response, and TNF-α signaling via NF-κB pathways, indicating that these pathways are generally active in severe disease across nearly every immune cell type (Figure 4A). In the Lee PBMC cohort, many of the same enriched pathways were observed, including IFN-α response, IFN-γ response, and TNF-α signaling via NF-κB (Figure 4A). Of interest, the majority of differentially regulated pathways appeared to be conserved between the lung and blood for most cell types (Figure 4B), and this result was consistent across cohorts (Figures 4B and S7).
Figure 4.
Pathway-level signatures of severe COVID-19 demonstrate varying degrees of overlap between lung and blood immune compartments
(A) (Left) Hallmark gene sets detected by gene set enrichment analysis (GSEA) indicate enrichment of molecular pathways in severe patients compared with control subjects for BALF cell types from the Liao et al. cohort. Normalized enrichment score is shown by a blue-red color scale and dot size is proportional to -log10(p), with the smallest dot size indicating non-significant adjusted p values (p < 0.05). (Right) Enrichment of molecular pathways in severe patients compared with control subjects for PBMC cell types from the Lee et al. cohort.
(B) Degree of compartmental similarity of significantly enriched or depleted pathways between severe patients and healthy control subjects in cell types shared between lung and blood. The Liao BALF cohort was compared with three PBMC cohorts. Rose plots follow the format of the plots in Figure 2, where yellow segments indicate compartmental overlap and green segments represent polarization.
(C) Enrichment of molecular pathways in severe compared with mild patients for BALF cell types from the Liao cohort (left) and PBMC cell types from the Lee cohort (right).
(D) Degree of compartmental similarity of significantly enriched or depleted pathways between severe and mild patients in cell types shared between lung and blood. The Liao BALF cohort was compared with three PBMC cohorts.
See also Figures S4, S7, and S8.
To examine pathway-level regulation specific to severe disease courses, we applied the same analysis to DEGs between patients with severe and mild COVID-19 (Figure 4C). In the lung, a number of broadly enriched pathways including IL-2/STAT5 signaling and TNF-α signaling via NF-κB were observed (Figure 4C). Strikingly, type I/III and type II interferon responses showed mixed enrichment and depletion across lung cell types, with enrichment tending to occur in myeloid cells (CD14 and CD16 monocytes, cDC2s, neutrophils) and depletion tending to occur in lymphoid cells (T cells, B cells, and pDCs). In the blood, more consistent enrichment of IFN-α and IFN-γ responses was observed, without a clear distinction between myeloid cells and lymphoid cells (Figure 4C). In contrast to the lung, the TNF-α signaling pathway was significantly depleted in multiple blood cell types including CD4 Treg, CD8 effector and naive T, and NK cells. Pooling data from all the available cohorts, we observed a similar effect in which the TNF pathway was more strongly active in the blood compared with lung for healthy and mild donors but more active in the lung than in the blood for severe donors (Figure S4B, see STAR Methods). As many of the cell types studied via GSEA showed significant enrichment of both the “interferon alpha response” and “interferon gamma response” hallmark pathways, we sought to identify whether these pathways had been induced by the same gene sets. The degree of overlap (Jaccard index of the GSEA leading edge, see STAR Methods) was less than 50% in the large majority of cell types and conditions (Figure S8). On the whole, alterations in pathway-level activity differentiating severe from mild patients demonstrated greater than 20% conservation in most cell types across the cohorts compared in this analysis, but a substantial fraction of contra-directionally regulated or “polarized” pathways including TNF-α signaling and interferon responses were detected across all of the cohorts studied (Figures 4D, S7A, and S7B).
Ligand-receptor analysis suggests differentially active signaling between the lung and blood immune environments
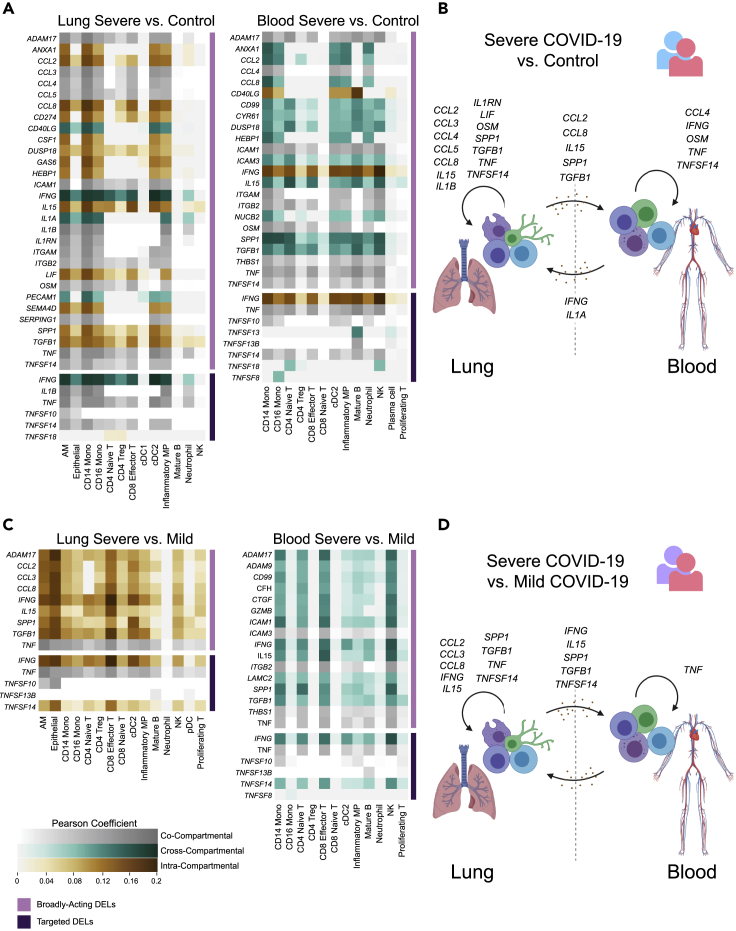
Following the identification of common and compartment-specific pathway regulation in the lung and periphery of patients with severe COVID-19, we sought to examine transcriptional signatures of cytokine signaling by constructing a putative network of cell-cell interactions across and within compartments. Specifically, we aimed to identify DEG signatures or “footprints” in each compartment that could be induced by ligand-receptor interactions, then to identify which compartment(s) upregulate the inducer ligands during severe disease. We leveraged this information to predict whether transcriptional signatures were likely induced through intra-compartmental signaling or whether the data were consistent with cross-compartmental signaling, termed “cross talk” (see DISCUSSION for alternative mechanisms). Briefly, differentially expressed ligands (DELs) were identified using NicheNet through (1) a data-driven unbiased approach searching for ligands differentially expressed between the conditions of interest that act broadly to induce differential gene expression "footprints" in over one-third of cell types in a receiver compartment and (2) a targeted approach to find specific transcripts of interest (see STAR Methods). Following the identification of DELs, we categorized each according to whether their “footprint” in a given compartment could be explained by the DEL's upregulation in the same compartment (suggesting intra-compartmental signaling), the opposite compartment (suggesting cross talk), or both compartments.
The DEL profile for differential gene expression between severe patients and healthy controls is shown in Figure 5A and is schematically represented in Figure 5B. TNF-induced differential expression signatures along with upregulation of TNF occurred in both compartments, suggesting intra-compartmental signaling. Upregulation of genes including TGFB1, CCL2, and SPP1 was observed only in the lung. However, over one-third of cell types in the blood bore differential gene expression signatures induced by these ligands, indicating potential lung-to-blood cross talk via these secreted factors. On the other hand, gene expression signatures downstream of IFN-γ and IL-1α were widespread in the lung, but IFNG and IL1A were only upregulated in the blood, suggesting blood-to-lung cross talk.
Figure 5.
Ligand-receptor analysis reveals putative signaling networks involved in severe COVID-19 including cross-compartmental signaling between the lung and blood
(A) Cross-compartmental, intra-compartmental, and co-compartmental activity of differentially expressed ligands (DELs) upregulated in severe patients compared to control subjects. (Left) Heatmap intensity indicates the extent to which DELs (rows) explain differential expression programs observed in each lung “receiver” cell type (columns) using a NicheNet-defined Pearson correlation coefficient. Heatmap colors show DELs originating in the lung only (intra-compartmental, brown), lung and blood (co-compartmental, gray), and blood only (cross-compartmental, teal). (Right) Signaling interactions acting on blood “receiver” cells based on DELs originating in the blood only (intra-compartmental, brown), lung and blood (co-compartmental, gray), and lung only (cross-compartmental, teal). Broadly acting DELs identified through an unbiased approach are adjacent to the lavender bar, whereas interferon, inflammasome-related, and TNF superfamily DELs are adjacent to the violet bar.
(B) Schematic representation of putative signaling interactions detected within the lung compartment, within the blood compartment, and between compartments using differential expression between severe patients and healthy control subjects.
(C) Compartmental activity of DELs upregulated between severe and mild patients following the format of (A).
(D) Schematic of putative signaling interactions using differential expression between severe and mild patients following the format of (B).
Using the same analysis pipeline to examine differential gene expression programs between severe and mild disease (Figure 5C, schematically represented in Figure 5D) showed broadly acting signaling activity in both compartments. Most of the gene expression signatures in the blood were only traceable to DELs in the lung, with the exception of TNF. In the lung, a number of broadly acting ligands including CCL2, CCL3, CCL8, and IL15 appeared to enact their severity-specific functions locally. Other factors such as SPP1 and IFNG induced severity-specific signatures in both compartments but were only upregulated in the blood. Altogether, this analysis revealed that the lung and blood compartments are marked by distinct broadly active receptor-ligand interactions. In addition, certain ligands including IFNG were differentially expressed in a single compartment, yet induced gene expression in both compartments, suggesting a possible mechanism of lung-blood cross talk. Furthermore, ligand-receptor analysis provided orthogonal evidence for a modulated type II interferon response between severe and mild patients in over one-third of the cell types in both compartments.
Discussion
Severe COVID-19 is characterized by extreme states of immune dysregulation affecting immune cell populations at the local site of infection and in the peripheral circulation, from which a coordinated response is necessary to resolve a viral infection. Given the distinct stimuli and environmental contexts affecting immune cells in the lung and blood, we sought to understand how states of dysregulation in severe-to-critical COVID-19 may differ or demonstrate conservation between these compartments. We leveraged publicly available scRNA-seq data from 96 patients and controls who contributed either peripheral blood or BALF samples to compare transcriptomic responses across the spectrum of COVID-19 severity in consanguineous cell types, with the main analyses focusing on differentially regulated transcripts between severe COVID-19 donors and either mild COVID-19 donors or healthy controls.
We identified a small degree of overlap between the lung and blood at the level of individual differentially expressed genes in the majority of the common cell types tested. The degree of similarity between BALF and PBMC cohorts was lesser than if two PBMC cohorts were compared directly, indicating that distinct transcriptional mechanisms are likely at play. Filtering the overlapping gene sets to preserve the most strongly evidenced differentially expressed transcripts revealed that severe COVID-19 is characterized by compartmentally conserved downregulation of MHC class II genes, upregulation of calprotectin and calgranulin-related genes, and upregulation of clusterin. Of importance, our analysis predicted that MHC class II molecules, the invariant chain, and clusterin may be useful for immunophenotyping patients with COVID-19, as the status of these markers in the blood may also reflect the status of these markers in the lung. The severity-specific decreases we observed in HLA-DRs, HLA-DPBs, and CD74 across antigen-presenting cell types agree with reports of decreased antigen presentation in patients with severe COVID-19 (Bost et al., 2020; Giamarellos-Bourboulis et al., 2020; Kuri-Cervantes et al., 2020; Schulte-Schrepping et al., 2020; Wilk et al., 2020; Xu et al., 2020a). However, the upregulation of CLU has not been as widely reported and this work adds to the growing understanding of the role of this gene in severe COVID-19.
Despite compartmental differences at the level of individual transcripts, differentially expressed gene sets converged on a core set of pathways that appeared to be highly conserved, with the exception of a small number that were polarized between the lung and periphery. The conserved pathways tended to include apoptosis, hypoxia, IL-2 signaling, IL-6 signaling, and hypoxia. The polarized pathways, ostensibly of more interest, included the response to type I/III and type II interferons as well as TNF-α signaling. The pattern of type I/III and type II interferon signatures observed in this study raise the question of why myeloid cells in the lung show signs of responding more sensitively to interferons during a severe COVID infection than a mild one, while lung lymphoid cells become desensitized. It also remains unknown why interferon responses are homogenously enriched in the blood but show mixed enrichment and depletion in the lung. The broad enrichment of the TNF-α signaling via NF-κB pathway across all cells in the lung but depletion of the pathway in T and NK cells in the blood is an interesting feature that also indicated a lymphoid-myeloid dependence. These results suggest that, as immune cells experience a barrage of dysregulatory cues during severe COVID-19, their compartment-specific behavior depends partly on their lineage. This effect could be the result of a dysregulated myeloid-lymphoid axis in which compartment-specific cues cause lymphoid cells to become unresponsive to certain stimuli while myeloid cells respond uniformly across compartments.
To probe cytokine responses further using an orthogonal tool, we conducted ligand-receptor analysis on cells of the lung and blood compartments together and in isolation. This analysis revealed severity-specific ligand-receptor interactions with increased activity in severe patients compared with milder patients. A subset of these ligands was predicted to act broadly on immune cells of both the lung and periphery, including IFNG, IL15, SPP1, TGFB1, and TNF. These ligands may serve as pan-regulators of the severity-specific immune response during COVID-19. Other ligands, including CCL2, CCL3, and CCL8 appeared to be broadly active in the lung alone. Of interest, some of the gene expression signatures observed in the blood were explained only through upregulation of their matching ligand in the lung, suggesting potential cross talk between compartments. An alternative explanation to compartmental cross talk could be that certain ligand proteins follow transient expression kinetics in the blood compartment yet leave long-lasting transcriptional signatures on blood immune cells. Of note, severity-specific type I and III interferon signaling networks were not detected in either compartment, potentially pointing to a deficiency of these factors that has been the subject of ongoing investigation (Arunachalam et al., 2020; Broggi et al., 2020; Grajales-Reyes and Colonna, 2020; Hadjadj et al., 2020; Lee et al., 2020; Major et al., 2020; Schulte-Schrepping et al., 2020; Silvin et al., 2020). Alternatively, the IFNA, IFNB, and IFNL transcripts may not be well captured via scRNA-seq.
Altogether, the results of this work suggest both common and unique mechanisms for severity-associated immune dysregulation in the lung and blood compartments. This study also suggests that severity-specific interferon and TNF responses to SARS-CoV-2 infection depend on the lineage of immune cells as well as their compartment. Consideration of the distinct transcriptional states of immune cell populations in the lung and blood will likely be crucial in the development of immunomodulatory COVID-19 therapies such as immune blockades and interferon supplementation to aid in patient recovery.
Limitations of the study
The study is subject to several limitations. First, our study does not include a cohort in which cells of the lung and the blood have been analyzed in the same patients, although this type of cohort would be ideal for validating our predictions. Next, the ability to use healthy BALF data as a reference point remains limited without conducting differential gene expression tests between cohorts collected by separate investigators in separate studies. In addition, the set of publicly available mild COVID-19 patient BALF scRNA-seq samples remains small and limits our ability to make entirely orthogonal comparisons between the lung and blood in this study. Finally, this study does not contain experimental validation of the putative ligand-receptor networks predicted to be active in the lung and blood or the potential cross talk between these networks.
STAR★Methods
Key resources table
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Kalon Overholt (overholt@mit.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
Multi-donor datasets from eight separate studies were used in this work and these can be found under the following accession numbers: GEO: GSE145926 (Liao et al., 2020), GEO: GSE149689 (Lee et al., 2020), EGA: EGAS00001004717 (Wauters et al., 2021), GEO: GSM3660650 (Morse et al., 2019), EGA: EGAS00001004571 (Schulte-Schrepping et al., 2020), GEO: GSE150728 (Wilk et al., 2020), GEO: GSE155673 (Arunachalam et al., 2020), GEO: GSE151928 (Mould et al., 2021). See Table 1 and the key resources table for additional information on publicly available data. All of the code used for analysis in this study is available in a public GitHub repository at https://github.com/uberholzer/2021_iScience_Overholt_Krog_COVID.
Experimental model and subject details
The sample sizes and clinical status of human subjects analyzed in this study are given in Table 1. The age, sex or gender, and other demographic data for these subjects are available in the source publications (Arunachalam et al., 2020; Lee et al., 2020; Liao et al., 2020; Morse et al., 2019; Mould et al., 2021; Schulte-Schrepping et al., 2020; Wilk et al., 2020).
Methods details
Patient severity definitions
We provide the following operational definitions of COVID-19 clinical severity levels in order to harmonize the use of patient data from 6 separate studies as well as control data from an additional 2 studies. We define “mild” COVID-19 patients as SARS-CoV-2+ individuals experiencing mild or moderate symptomatic infection as noted by the original investigators at the time of sample collection. “Severe” patients are defined as SARS-CoV-2+ individuals experiencing severe or critical disease as noted by the original investigators at the time of sample collection. We also refer to this group as severe-to-critical patients in this manuscript. Severe-to-critical patients may or may not have required invasive mechanical ventilation or been diagnosed with ARDS. For patients who were sampled at multiple timepoints in the disease course by the original investigators, we used only the sample collected most proximally to infection, i.e. the earliest time point, to better capture infection-related immune dynamics.
Data acquisition
All single cell RNA-sequencing data used in this analysis were obtained from publicly available datasets. A summary of the 96 single-cell datasets used in this paper obtained from 8 separate studies (cohorts) is given in Table 1. In this analysis, we frequently focused on comparing two particular cohorts, the Liao et al. BALF cohort (Liao et al., 2020) and the Lee et al. PBMC cohort (Lee et al., 2020), since these cohorts both contained mild and severe COVID-19 patients as well as healthy control subjects. We noted that the number of severe patients in the Liao and Lee cohorts who were in critical condition at the time of cell sampling was roughly comparable (3 of 4 severe patients in the Lee cohort and 5 out of 6 severe patients in the Liao cohort were mechanically ventilated).
To set up a more comprehensive analysis and mitigate confounders in our investigation that could arise from batch effects, we incorporated 4 cohorts consisting of COVID-19 patients and healthy controls when provided (Arunachalam et al., 2020; Schulte-Schrepping et al., 2020; Wauters et al., 2021; Wilk et al., 2020) as well as an additional cohort for cross-control assembled from 2 studies containing healthy BALF donors (Morse et al., 2019; Mould et al., 2021). These additional BALF and PBMC cohorts, shown in Table 1, were used to provide additional granularity and increase the number of possible BALF-PBMC comparisons to be made. Among these additional cohorts, only the Arunachalam et al. PBMC cohort contained healthy, mild, and severe donors all from the same study (Arunachalam et al., 2020). Notably, the severe patients in the Arunachalam study were not as directly comparable to the Liao BALF cohort as the donors from the Lee study (only 1 of 4 severe patients in Arunachalam et al. was admitted to the intensive care unit). The makeup of the cohorts that we utilized in the present study as well as the data accession numbers and publication PubMed IDs are available in Table 1.
Data preprocessing, dimensionality reduction, multi-donor integration, and clustering
Gene-barcode matrices obtained from the GEO were preprocessed using Seurat (v. 3.1.5) (Stuart et al., 2019) in R (v. 4.0.2). Matrices were filtered to preserve cells with a unique molecular identifier (UMI) count over 1,000, gene count between 200 and 6,000, and expression of less than 10% mitochondrial RNA. Cells were additionally filtered to preserve cells expressing less than 20% 18S RNA and less than 20% 28S RNA. Next, expected doublets were removed in each matrix using DoubletFinder (v. 2.0.3) (McGinnis et al., 2019) setting an expected doublet formation rate of 7.5% and automatically generating parameters using the paramSweep option. All of the doublet-filtered gene-barcode matrices for the donors used in this study were then merged into a single object, which was log-normalized using the ‘NormalizeData’ function and the 2,000 most highly variable genes (features) were identified via the ‘FindVariableFeatures’ function with the variance stabilizing transformation method. The highly variable genes were scaled using the ‘ScaleData’ function regressing the percentage of mitochondrial transcripts and UMI count depth, and the list of highly variable genes was filtered to remove mitochondrial, ribosomal, and rRNA transcripts. The merged dataset was dimensionality-reduced using principal component analysis (PCA) acting on the number of principal components (PCs) calculated above, then integrated using the standard Harmony (v. 3.8) integration workflow (Korsunsky et al., 2019) using the highly variable gene list with theta = 2. The number of PCs to use for Harmony was determined using the point at which the “first difference” in standard deviation between two PCs was less than 0.05% of the total standard deviation from the first 100 PCs, then adding 5 PCs to this number. Clustering was conducted by first performing UMAP on the calculated number of PCs using the ‘harmony’ reduction, finding nearest neighbors using the ‘FindNeighbors’ function in Seurat acting on this number of dimensions, then running the Louvain algorithm via the ‘FindClusters’ function with a resolution parameter of 0.5. The resulting jointly integrated and clustered datasets from BALF and PBMC samples were visualized together by UMAP. We verified that large-scale dataset integration using Harmony is in agreement with best current practices for data integration (Luecken et al., 2020).
Cell type annotation and iterative data integration
Cell type annotation was conducted using the resultant clusters. Clusters were annotated according to scaled average expression levels of canonical marker genes identified in the original papers for the BALF and PBMC datasets, the broader literature, and the Human Protein Atlas (Regev et al., 2017). Clusters deemed identical by similar presence of marker genes were merged during annotation. Following one round of coarse cell type annotation, major clusters of interest were isolated, separately re-integrated, and sub-clustered. We performed re-integration and sub-clustering separately on four disjoint groupings of cells: myeloid cells; plasma cells, B cells, and proliferating cells; NK and T cells; and dendritic cells. Re-integration was peformed using Harmony as explained above, but for myeloid cells the PCs were calculated using the point where the first difference in standard deviation was less than 0.1% to produce the cleanest clustering result. Cell types were then annotated using scaled expression levels of canonical marker genes as explained above. Figure S1 shows the levels of key marker genes for the sub-cluster analysis.
Cluster quality control
We took multiple steps to improve the purity of sub-clusters. Following cell type annotation, clusters labeled as putative doublets (positive for multiple lineage markers) were removed from the analysis. Additionally, cells containing more than 5% total hemoglobin (HBA, HBB, HBD) transcripts or more than 1% PPBP transcripts were considered to be erythrocytes, platelets, or contaminated cells and these cells were removed prior to downstream analysis. Additionally, cells were removed from the analysis if they had coarsely clustered with myeloid cells, NK/T cells, or dendritic cells but had total immunoglobulin percentages (IGH-, IGK-, IGL-) greater than 5%, since these might represent plasma cells of uncertain status or contaminated cells. Filtering on immunoglobulin genes was only conducted after sub-clustering since these genes provided important information to delineate sub-clusters and improve purity.
Differential expression analysis between severity conditions (main pipeline, Figures 2, 3, 4, and 5)
Differential expression analysis was performed in Seurat using the ‘FindMarkers’ function utilizing model-based analysis of single-cell transcriptomics (MAST) statistical framework through the “MAST” R package (v. 3.11) (Finak et al., 2015) using the cellular detection rate (UMIs per cell) as a covariate. A number of differential expression schemes were used for the different analyses in this paper. For the main differential expression tests (Figures 2 and 3 and underlying the results of Figures 4 and 5), we isolated cells belonging to one cohort only, then performed differential expression between cell populations pooled across donors from the disease severity conditions of interest. To examine differential gene expression across the spectrum of disease severity, we conducted three types of differential expression tests: severe vs. mild, mild vs. control, and severe vs. control. Significantly differentially expressed genes (DEGs) are were classified by MAST adjusted p < 0.05, although more stringent filtering was applied in some downstream analyses (described in the next section).
Identification of stringent DEGs between severity conditions (Table S1)
To identify the highest-confidence DEGs using stringent methods, the same differential expression analysis was employed as in the main pipeline with modifications to the MAST algorithm and DEG classification thresholds. In addition to using the UMI count as a numerical covariate in MAST, the donor from whom the cells originated was incorporated as a categorical covariate in the ‘latent.vars’ variable of the ‘FindMarkers’ function. This allowed us to explicitly model donor-to-donor variation using a fixed effects model. Additionally, genes were only classified as “stringent DEGs” if they showed adjusted p < 0.01, and |log2FC| > 1.5 (FC = fold change) and were present in at least 25% of the cells in one of the categories tested.
Differential expression analysis between compartments (Figure S4)
In the supplementary analyses of Figure S4, we desired to study transcriptional differences between compartments for a given cell type in a given severity condition (e.g. severe neutrophils in BALF vs. severe neutrophils in PBMCs). To do this, it was necessary to conduct differential gene expression tests between donors who had been sequenced as part of different studies. We used the following method to attempt to mitigate the issue of batch effect. First, all BALF donors from the severity condition of interest were combined into a single pool and all PBMC donors from that condition were combined into a separate pool. Next, we conducted differential gene expression between these pools in a given cell type using the ‘FindMarkers’ function in Seurat implementing the MAST algorithm. In addition to using the UMI count depth as a numerical covariate in MAST, we also used the study of origin as a categorical covariate in the ‘latent.vars’ variable of the ‘FindMarkers’ function. This allowed us to explicitly model the batch effects between studies using a fixed effects model. As a result, the DEG list was shorter than if the same approach had been used without considering the study of origin as a covariate. In Figure S4, we considered DEGs that were robust to study-to-study batch variation with adjusted p < 0.01, |log2FC| > 1, and expression in at least 10% of cells in one of the conditions tested.
Surface marker identification
The identification of cell surface markers indicative of severe disease was performed by cross-referencing the previously established DEGs for all cell types in a given dataset with entries in the Cell Surface Protein Atlas (CSPA) (Bausch-Fluck et al., 2015). DEGs were considered to be differentially expressed surface markers if they were included in the ‘high confidence’ CSPA category and showed significant differential expression (adjusted p < 0.05 and |log2FC| > 1). Differentially expressed surface markers were further examined for constitutive expression in cell types of interest by cross referencing with the Immune Cell Atlas human bulk RNA sequencing data (Regev et al., 2017).
Gene set enrichment analysis (GSEA)
Gene set enrichment analysis (GSEA) (Subramanian et al., 2005) was performed using the “fgsea” package (v. 1.12.0) (Korotkevich et al., 2016) in R using the ‘multilevel’ option. Differentially expressed gene lists generated using MAST were ranked by log2(FC), since -log10(p) values became arbitrarily large. GSEA results were interpreted according to normalized enrichment score (NES) and an adjusted p value, with a p < 0.05 threshold for defining significance. Pathways with positive NES were defined throughout the text as “enriched” and pathways with negative NES were defined as “depleted”.
Leading edge analysis (Subramanian et al., 2005) was conducted for cell types in which the “interferon alpha response” and “interferon gamma response” were both enriched or both depleted to determine the extent to which the same genes contributed to these pathways. The leading edge subsets for both pathways were compared with identify unique and common genes. Overlap was quantified using the Jaccard similarity index (intersection of leading edge subsets/union of leading edge subsets). The Jaccard index and the genes uniquely contributing to either pathway were visualized for each cell type (Figure S8).
Compartmental comparisons for main pipeline (Figures 2, 3, 4, and 5)
In Figures 2, 3, 4, and 5, we established DEGs separately for each cohort between three conditions of COVID-19 status (healthy, mild, and severe). Compartmental comparisons were performed for all DEGs, surface marker DEGs, and significantly enriched/depleted pathways using the following scheme. Following separate statistical tests for each compartment, the intersecting set of genes or pathways that were significantly upregulated in both compartments or significantly downregulated in both compartments was identified and termed “co-directional”. The intersecting set of genes or pathways that were significantly regulated in different directions between compartments was identified and termed “contradirectional”. The remaining genes not part of the intersection were termed “BALF only” or “PBMC only”. A modified Jaccard similarity index was developed to describe the degree of overlap between co-directional and contradirectional genes or pathways between compartments. Briefly, the co-directional Jaccard index was defined as the number of co-directional intersection elements divided by the number of elements in the union. The contra-directional Jaccard index was defined similarly. The relative fractions of co-directional, contra-directional, BALF only, and PBMC only genes or pathways summed to 1, such that the degree of overlap could be shown in a polar rose plot. Rose plots were created in Python (v. 3.7.3) using the “plotly” (v. 4.10.0) package (Plotly Technologies Inc., 2015).
To generate the rose plots in Figures 2 and 3, we considered only DEGs with adjusted p < 0.05 and |log2FC| > 1 that were expressed in at least 10% of cells in one of the conditions tested. Additionally, we filtered mitochondrial, ribosomal, hemoglobin, immunoglobulin, lncRNA, and T cell receptor genes out of the list as these genes are often assumed to represent technical artifacts of little biological relevance (Wauters et al., 2021). However, we did not filter these genes prior to conducting GSEA as the GSEA algorithm requires the total gene set as input (Subramanian et al., 2005).
Analysis of between-compartment DEGs
In Figure S4, we established DEGs between compartments for a given cell type in a given severity condition. For example, a differential expression test was conducted between severe neutrophils in the lung and severe neutrophils in blood. These DEG lists were first filtered as described above to remove genes that could represent technical artifacts. For each condition tested (healthy, mild, severe), rose plots were constructed representing the fraction of differentially expressed genes out of the total number of genes tested (after filtering) in that cell type. Next, we conducted GSEA on the unfiltered DEG lists. The differential expression test represented fold change as the ratio of expression in BALF to expression in PBMCs; hence, GSEA results with a positive NES represent pathways enriched in the lung while those with a negative NES represent pathways enriched in the blood. Finally, we sought to characterize the biological roles of DEGs between the lung and blood during severe disease but not during baseline health, or during severe but not mild disease. To do this, the “set difference” between severe DEGs and either mild or healthy control DEGs was extracted, and this gene set was subjected to gene ontology analysis using EnrichR (described below).
Gene ontology (GO) analysis
Further analysis of significantly differentially expressed genes was performed using the “EnrichR” (v. 2.1) (Kuleshov et al., 2016) package in R. Gene ontology (GO) analysis was conducted on the set difference between severe DEGs and either mild or healthy control DEGs. The set difference was calculated for upregulated (log2FC > 0) and downregulated (log2FC < 0) separately. The GO analysis was conducted using the “biological function” annotations representing large scale biological programs. GO results were ranked based on the EnrichR ‘Combined Score’ metric and significance was determined using an adjusted p value threshold of p < 0.05.
Ligand-receptor network analysis
To investigate cell-cell interactions potentially contributing to the observed differential gene expression in severe patients, we employed the ligand-receptor interaction tool NicheNet via the “nichenetr” package (v. 0.1.0) in R (Browaeys et al., 2020). Studying differential expression in a compartment of interest entailed first designating a “receiver cell” population in the compartment. Next, ligand-expressing “sender cells” were defined as all cells within the compartment of interest (to investigate intra-compartmental signaling) or all cells in the other compartment (to investigate cross-compartmental signaling). Briefly, we generated a list of ligands expressed in the assigned sender population with the potential to induce the observed differential gene expression in each receiver cell population in the compartment of interest. This procedure (described below) was iterated over all receiver populations in the compartment of interest. The list of ligands inducing differential expression in the compartment of interest was filtered using two approaches: an unbiased data-driven approach filtering for broadly-acting differentially expressed ligands (DELs) explaining differential gene expression in over one-third of the cell types in the compartment, and a targeted approach filtering specifically for interferon DELs, inflammasome-activated DELs, and TNF superfamily DELs.
For the unbiased approach, all ligands identified by NicheNet to act on cells in given compartment were assigned a NicheNet-calculated Pearson correlation coefficient quantifying how strongly the ligand activity explained differential gene expression in each cell type for a given comparison (severe vs. mild, severe vs. healthy control, mild vs. healthy control). Only ligands with a Pearson coefficient greater than 0.08 in over one-third of the cell types in the receiver compartment were preserved. This list of ligands was filtered further to preserve only ligands that were differentially upregulated (adjusted p < 0.05, log2FC > 0) in at least one cell type in the sender compartment of interest. These ligands were classified as broadly-acting DELs. For the targeted approach, ligands of interest were found by searching the list of DELs for transcripts beginning with ‘IFN’, ‘IL1’, ‘IL18’, and ‘TNF’.
DELs explaining differential gene expression in a given compartment might: 1) originate in sender cells of the same compartment only (termed “intra-compartmental signaling”), 2) originate in sender cells of the opposite compartment only (termed “cross-compartmental signaling”), or 3) originate in sender cells of both compartments (termed “co-compartmental signaling”). We used these classifications of DELs to construct putative ligand-receptor interaction networks.
Iterative NicheNet procedure
The following standard NicheNet procedure (Browaeys et al., 2020) was looped iteratively through the receiver cell populations in the receiver compartment of interest. Differentially expressed target genes between conditions of interest (severe vs. mild, severe vs. healthy control, mild vs. healthy control) in each receiver cell population were identified using the ‘FindMarkers’ function in Seurat with criteria of p < 0.05, average natural log FC > 0.25, and expression in over 10% of the receiver cell population in severe patients. Concurrently, a list of potential receptors expressed in over 10% of cells in the severe patient receiver population was generated using NicheNet. A list of “sender” cells was created comprising all cell types in: 1) the receiver compartment, or 2) the opposite compartment. For each sender cell population, potential ligands were inferred using the NicheNet “high-confidence” ligand-receptor network applied to genes expressed in over 10% of the “severe” cells in the sender population. All ligands identified through this procedure were subjected to downstream analysis to identify DELs.
Quantification and statistical analysis
Differential gene expression was analyzed using the “MAST” package statistical framework through the ‘FindMarkers’ function in Seurat, always implementing the UMI count per cell as a numerical covariate as a proxy for the cellular detection rate. Additional categorical covariates were implemented where indicated. Differential expression statistical significance was qualified using the MAST false discovery rate (FDR) adjusted p value, always using a significance threshold of at least p < 0.05. More stringent p value thresholds are indicated where they are used. Pathway enrichment was analyzed using the ‘multilevel’ GSEA method in the “fgsea” package. Statistical significance of normalized enrichment scores was qualified using the fgsea FDR adjusted p value, with a significance threshold of p < 0.05. Differential gene expression relevant to ligand-receptor interactions was evaluated using the NichNet pipeline implementing the Wilcoxon rank-sum test through the ‘FindMarkers’ function in Seurat. Statistical significance was qualified using the ‘FindMarkers’ Bonferroni-adjusted p value, with a significance threshold of p < 0.05. GO analysis was conducted using the “EnrichR” package, and statistical significance was qualified using the EnrichR adjusted p value based on Fisher's exact test with a significance threshold of p < 0.05.
Acknowledgments
The authors gratefully acknowledge Paul Blainey, Cal Gunnarsson, Bianca Lepe, Megan Tse, Andy Kim, and Harvey Yang for providing feedback during the conceptual phase. The authors also thank Josh Peters, Krista Pullen, Inma Barrasa, Kristen Overholt, Charalampos Lazaris, and Lily Xu for helpful discussions. Figures in this article were partially created using BioRender.com. The authors would like to thank the MIT Department of Biological Engineering for support throughout the research effort. This material is based upon work supported by the National Science Foundation Graduate Research Fellowship under Grant No. 1745302, funding K.J.O.
Author contributions
K.J.O. and J.R.K. conceptualized the research; K.J.O. and J.R.K contributed equally to the analysis and sought guidance and supervision from B.D.B. throughout the investigation; K.J.O., J.R.K., I.Z., and B.D.B. contributed ideas and wrote and edited the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: July 23, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102738.
Supplemental information
References
- Arentz M., Yim E., Klaff L., Lokhandwala S., Riedo F.X., Chong M., Lee M. Characteristics and outcomes of 21 critically Ill patients with COVID-19 in Washington state. JAMA. 2020;323:1612–1614. doi: 10.1001/jama.2020.4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arunachalam P.S., Wimmers F., Mok C.K.P., Perera R.A.P.M., Scott M., Hagan T., Sigal N., Feng Y., Bristow L., Tak-Yin Tsang O. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369:eabc6261. doi: 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baj J., Karakuła-Juchnowicz H., Teresiński G., Buszewicz G., Ciesielka M., Sitarz E., Forma A., Karakuła K., Flieger W., Portincasa P., Maciejewski R. COVID-19: specific and non-specific clinical manifestations and symptoms: the current state of knowledge. J. Clin. Med. 2020;9:1753. doi: 10.3390/jcm9061753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bausch-Fluck D., Hofmann A., Bock T., Frei A.P., Cerciello F., Jacobs A., Moest H., Omasits U., Gundry R.L., Yoon C. A mass spectrometric-derived cell surface protein atlas. PLoS One. 2015;10 doi: 10.1371/journal.pone.0121314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bost P., Giladi A., Liu Y., Bendjelal Y., Xu G., David E., Blecher-Gonen R., Cohen M., Medaglia C., Li H. Host-viral infection maps reveal signatures of severe COVID-19 patients. Cell. 2020;181:1475–1488.e12. doi: 10.1016/j.cell.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broggi A., Ghosh S., Sposito B., Spreafico R., Balzarini F., Lo Cascio A., Clementi N., De Santis M., Mancini N., Granucci F., Zanoni I. Type III interferons disrupt the lung epithelial barrier upon viral recognition. Science. 2020;369:706–712. doi: 10.1126/science.abc3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browaeys R., Saelens W., Saeys Y. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat. Methods. 2020;17:159–162. doi: 10.1038/s41592-019-0667-5. [DOI] [PubMed] [Google Scholar]
- Chan J.F.W., Yuan S., Kok K.H., To K.K.W., Chu H., Yang J., Xing F., Liu J., Yip C.C.Y., Poon R.W.S. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daamen A.R., Bachali P., Owen K.A., Kingsmore K.M., Hubbard E.L., Labonte A.C., Robl R., Shrotri S., Grammer A.C., Lipsky P.E. Comprehensive transcriptomic analysis of COVID-19 blood, lung, and airway. bioRxiv. 2020 doi: 10.1101/2020.05.28.121889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Valle D.M., Kim-Schulze S., Huang H.-H., Beckmann N.D., Nirenberg S., Wang B., Lavin Y., Swartz T.H., Madduri D., Stock A. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020;3:1–8. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finak G., McDavid A., Yajima M., Deng J., Gersuk V., Shalek A.K., Slichter C.K., Miller H.W., McElrath M.J., Prlic M. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 2015;16 doi: 10.1186/s13059-015-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardinassi L.G., Souza C.O.S., Sales-Campos H., Fonseca S.G. Immune and metabolic signatures of COVID-19 revealed by transcriptomics data reuse. Front. Immunol. 2020;11:1636. doi: 10.3389/fimmu.2020.01636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattinoni L., Chiumello D., Rossi S. COVID-19 pneumonia: ARDS or not? Crit. Care. 2020;24:154. doi: 10.1186/s13054-020-02880-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giamarellos-Bourboulis E.J., Netea M.G., Rovina N., Akinosoglou K., Antoniadou A., Antonakos N., Damoraki G., Gkavogianni T., Adami M.E., Katsaounou P. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe. 2020;27:992–1000.e3. doi: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grajales-Reyes G.E., Colonna M. Interferon responses in viral pneumonias. Science. 2020:626–627. doi: 10.1126/science.abd2208. https://science.sciencemag.org/content/369/6504/626.summary [DOI] [PubMed] [Google Scholar]
- Grasselli G., Zangrillo A., Zanella A., Antonelli M., Cabrini L., Castelli A., Cereda D., Coluccello A., Foti G., Fumagalli R. Baseline characteristics and outcomes of 1591 patients infected with SARS-CoV-2 admitted to ICUs of the lombardy region, Italy. JAMA. 2020;323:1574–1581. doi: 10.1001/jama.2020.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., Péré H., Charbit B., Bondet V., Chenevier-Gobeaux C. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khafaie M.A., Rahim F. Cross-country comparison of case fatality rates of Covid-19/SARS-CoV-2. Osong Public Health Res. Perspect. 2020;11:74–80. doi: 10.24171/j.phrp.2020.11.2.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotkevich G., Sukhov V., Sergushichev A. Fast gene set enrichment analysis. bioRxiv. 2016:060012. doi: 10.1101/060012. [DOI] [Google Scholar]
- Korsunsky I., Millard N., Fan J., Slowikowski K., Zhang F., Wei K., Baglaenko Y., Brenner M., Loh P.R., Raychaudhuri S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods. 2019;16:1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44 doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuri-Cervantes L., Pampena M.B., Meng W., Rosenfeld A.M., Ittner C.A.G., Weisman A.R., Agyekum R.S., Mathew D., Baxter A.E., Vella L.A. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.abd7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.S., Park S., Jeong H.W., Ahn J.Y., Choi S.J., Lee H., Choi B., Nam S.K., Sa M., Kwon J.S. Immunophenotyping of covid-19 and influenza highlights the role of type i interferons in development of severe covid-19. Sci. Immunol. 2020;5:1554. doi: 10.1126/sciimmunol.abd1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Cheng L., Li J., Wang X., Wang F. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- Luecken M.D., Büttner M., Chaichoompu K., Danese A., Interlandi M., Mueller M.F., Strobl D.C., Zappia L., Dugas M., Colomé-Tatché M., Theis F.J. Benchmarking atlas-level data integration in single-cell genomics. bioRxiv. 2020;1:5. doi: 10.1101/2020.05.22.111161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major J., Crotta S., Llorian M., McCabe T.M., Gad H.H., Priestnall S.L., Hartmann R., Wack A. Type I and III interferons disrupt lung epithelial repair during recovery from viral infection. Science. 2020;369:712–717. doi: 10.1126/science.abc2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis C.S., Murrow L.M., Gartner Z.J. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 2019;8:329–337.e4. doi: 10.1016/j.cels.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse C., Tabib T., Sembrat J., Buschur K.L., Bittar H.T., Valenzi E., Jiang Y., Kass D.J., Gibson K., Chen W. Proliferating SPP1/MERTK-expressing macrophages in idiopathic pulmonary fibrosis. Eur. Respir. J. 2019;54 doi: 10.1183/13993003.02441-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mould K.J., Moore C.M., McManus S.A., McCubbrey A.L., McClendon J.D., Griesmer C.L., Henson P.M., Janssen W.J. Airspace macrophages and monocytes exist in transcriptionally distinct subsets in healthy adults. Am. J. Respir. Crit. Care Med. 2021;203:946–956. doi: 10.1164/RCCM.202005-1989OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotly Technologies Inc . Plotly Technologies Inc; 2015. Collaborative Data Science.https://plot.ly [Google Scholar]
- Ramanathan K., Antognini D., Combes A., Paden M., Zakhary B., Ogino M., MacLaren G., Brodie D., Shekar K. Planning and provision of ECMO services for severe ARDS during the COVID-19 pandemic and other outbreaks of emerging infectious diseases. Lancet Respir. Med. 2020;8:518–526. doi: 10.1016/S2213-2600(20)30121-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regev A., Teichmann S.A., Lander E.S., Amit I., Benoist C., Birney E., Bodenmiller B., Campbell P., Carninci P., Clatworthy M. The human cell atlas. Elife. 2017;6 doi: 10.7554/eLife.27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte-Schrepping J., Reusch N., Paclik D., Baßler K., Schlickeiser S., Zhang B., Krämer B., Krammer T., Brumhard S., Bonaguro L. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell. 2020;182:1419. doi: 10.1016/j.cell.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvin A., Chapuis N., Dunsmore G., Goubet A.G., Dubuisson A., Derosa L., Almire C., Hénon C., Kosmider O., Droin N. Elevated calprotectin and abnormal myeloid cell subsets discriminate severe from mild COVID-19. Cell. 2020;182:1401. doi: 10.1016/j.cell.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travaglini K.J., Nabhan A.N., Penland L., Sinha R., Gillich A., Sit R.V., Chang S., Conley S.D., Mori Y., Seita J. A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature. 2020;587:619. doi: 10.1038/s41586-020-2922-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wauters E., Van Mol P., Garg A.D., Jansen S., Van Herck Y., Vanderbeke L., Bassez A., Boeckx B., Malengier-Devlies B., Timmerman A. Discriminating mild from critical COVID-19 by innate and adaptive immune single-cell profiling of bronchoalveolar lavages. Cell Res. 2021;31:272–290. doi: 10.1038/s41422-020-00455-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk A.J., Rustagi A., Zhao N.Q., Roque J., Martínez-Colón G.J., McKechnie J.L., Ivison G.T., Ranganath T., Vergara R., Hollis T. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020;26:1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J.G., Simpson L.J., Ferreira A.-M., Rustagi A., Roque J.A., Asuni A., Ranganath T., Grant P.M., Subramanian A.K., Rosenberg-Hasson Y. Cytokine profile in plasma of severe COVID-19 does not differ from ARDS and sepsis. JCI Insight. 2020;5 doi: 10.1172/jci.insight.140289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72314 cases from the Chinese center for disease control and prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- Xiong Y., Liu Y., Cao L., Wang D., Guo M., Jiang A., Guo D., Hu W., Yang J., Tang Z. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg. Microbes Infect. 2020;9:761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G., Qi F., Li H., Yang Q., Wang H., Wang X., Liu X., Zhao J., Liao X., Liu Y. The differential immune responses to COVID-19 in peripheral and lung revealed by single-cell RNA sequencing. Cell Discov. 2020;6:1–14. doi: 10.1038/s41421-020-00225-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., Liu S., Zhao P., Liu H., Zhu L. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Multi-donor datasets from eight separate studies were used in this work and these can be found under the following accession numbers: GEO: GSE145926 (Liao et al., 2020), GEO: GSE149689 (Lee et al., 2020), EGA: EGAS00001004717 (Wauters et al., 2021), GEO: GSM3660650 (Morse et al., 2019), EGA: EGAS00001004571 (Schulte-Schrepping et al., 2020), GEO: GSE150728 (Wilk et al., 2020), GEO: GSE155673 (Arunachalam et al., 2020), GEO: GSE151928 (Mould et al., 2021). See Table 1 and the key resources table for additional information on publicly available data. All of the code used for analysis in this study is available in a public GitHub repository at https://github.com/uberholzer/2021_iScience_Overholt_Krog_COVID.