Abstract
Animals susceptible to chronic social defeat stress (CSDS) exhibit depression-related behaviors, and show aberrant transcription across several limbic brain regions. The nucleus accumbens (NAc) in particular shows unique susceptible versus resilient phenotypes at the transcriptional, neuroanatomical, and physiological levels. Early life stress (ELS) promotes susceptibility to CSDS in adulthood, but associated enduring changes in transcriptional control mechanisms in NAc have not yet been investigated. Here, we examined long-lasting changes in histone modifications induced in NAc by ELS and studied their underlying mechanisms in mediating heightened lifelong stress susceptibility in male and female mice. We identify dimethylation of lysine 79 of histone H3 (H3K79me2) and the enzymes that control this modification, selectively in D2-type medium spiny neurons, as crucial for the expression of ELS-induced stress susceptibility. We also map the site-specific regulation of this histone mark genome-wide, and reveal the transcriptional networks it modulates. Finally, we demonstrate the potential clinical relevance of this epigenetic mechanism by showing that systemic delivery of a small molecule inhibitor of H3K79me2 deposition reverses ELS-induced behavioral deficits.
Introduction
The nucleus accumbens (NAc) integrates information across multiple limbic brain regions to shape reward- and stress-related behavioral output. It helps tune motivation towards positive stimuli and away from aversive ones1,2, and responds to stressful environments to help guide this tuning3. As such, it plays an important role in gating the effects of stress on depression-related behavioral abnormalities.
Stress activates numerous inputs to the NAc4–6, the differential activation of which influences susceptible versus resilient behavioral outcomes7,8. Downstream of this differential activation, physiological and transcriptional changes within the NAc itself are associated with susceptibility versus resilience9–15. All of these changes, though, are recorded after chronic stress – that is, upon the behavioral expression of either susceptibility or resilience.
A life history of stress is the strongest known risk factor for depression in humans. In particular, early life stress (ELS) increases the risk for adult depression by up to three-fold, depending on its timing, intensity, and specific features16,17. In animal models, the enduring impact of early life stress has been documented widely18–25. Importantly, ELS has been shown to increase the likelihood of behavioral susceptibility to stress later in life26,27, and to have particularly strong effects in the NAc, at both the transcriptional and physiological levels28–31.
To understand how ELS alters transcriptional regulation in NAc to increase susceptibility to depression-related outcomes in adulthood, we utilized a mouse model of ELS timed during a specific period in pre-weaning development27. This procedure is useful to disentangle chromatin mechanisms regulating future susceptibility from ongoing behavioral changes as it does not cause life-long behavioral abnormalities on its own, but increases the susceptibility of the animals to a second hit of stress later in life. Recent work by our laboratory characterized transcriptional responses to ELS and adult stress in several reward-related regions31, providing an important characterization of ELS priming of depression throughout the limbic brain.
Despite this insight, little is known about the underlying mechanisms that help drive lasting changes in gene transcription induced by ELS. To understand this question, we examined here chromatin modifications, which have the power to prime gene expression changes long after an initial insult. We performed mass spectrometry (MS) on adult NAc tissue of post-ELS male and female mice, and cross-referenced the results with transcriptional data about expression levels of epigenetic writers and erasers from Peña et al.31. From this convergent analysis, we identified dimethylation of histone H3 lysine 79 (H3K79me2) as a critical regulator of ELS-induced adult transcription in the NAc. We found further that deposition of this mark in this brain region is regulated throughout normal development, and is dysregulated after ELS via transcriptional alterations to its writer (Dot1l) and eraser (Kdm2b) enzymes, which we go on to show mediate the ability of ELS to enhance stress susceptibility to adult chronic social defeat stress (CSDS).
We next sought to understand the cell type-specificity of these effects. The NAc is composed predominantly of GABAergic projection neurons called medium spiny neurons (MSNs). Two subpopulations of MSNs are known based on their predominant expression of D1 dopamine receptors (Drd1) or D2 dopamine receptors (Drd2), and these cell types mediate distinct responses to stress: activation of D1 MSNs promotes resilience to CSDS, whereas activation of D2 MSNs promotes susceptibility10,26,32–34. The Drd2 gene is also an important correlate of ELS-induced susceptibility to depression in humans28,35. We show, through cell type-specific interrogation of the effects of ELS on Dot1l and Kdm2b, that this mechanism is selective for D2 MSNs in NAc. Finally, we provide preclinical translation of these findings by showing that systemic administration of a small molecule inhibitor of DOT1L reverses the increase in stress susceptibility induced by ELS.
Results
Mass spectrometry of histones in NAc reveals persistent chromatin modifications following ELS in male and female mice
To determine the enduring nature of chromatin modifications following ELS, we performed serial histone MS on NAc tissue of standard-reared (Std) and ELS mice. ELS consisted of maternal separation and low homecage nesting material from postnatal day 10 (P10) to P17. NAc samples were collected on the day of weaning (P21), in adolescence (P35), and in adulthood (P70-P80). Principal component analysis efficiently separated samples by rearing on PC1/PC2 and by sex on PC3/PC4 (Supplementary Figure 1A).
Our MS dataset demonstrated regulation of several histone modifications at each timepoint. Many modifications changed significantly across postnatal development into adulthood in all groups regardless of ELS exposure or sex (Figure 1A). ELS altered a number of histone modifications relative to their standard-reared counterparts (Figure 1B, Supplementary Table 1). We focused on marks whose regulation was more significant in ELS than Std animals for both sexes, as noted in the red boxes in Figure 1A, and which reached significance in ELS animals of at least one sex. No modifications were significantly altered by ELS among female mice at any age, and the difference between male and female histone changes after ELS is in line with the distinct and often opposite patterns of male and female transcription in human depression36 and mouse models31,36. In male mice, ELS significantly enriched for H3K9me1 at P21, and suppressed H3K79me1 and H3K79me2 in adulthood (Figure 1B). While the magnitude of these changes was moderate, they fall well within the range of normal values expected in MS experiments.
Figure 1. Chromatin modifications after ELS point to persistent epigenetic changes.
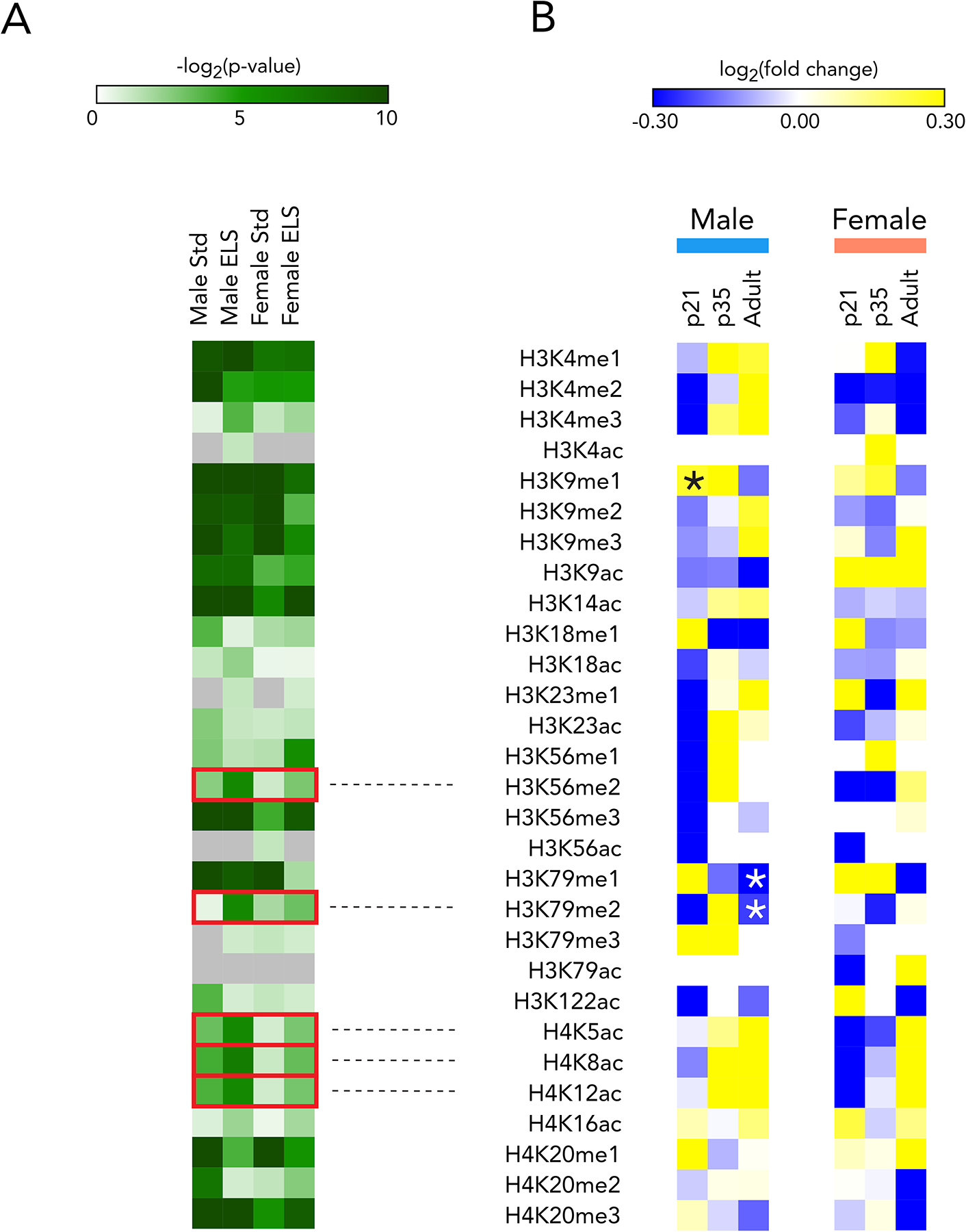
(A) p-values for one-way ANOVA of each mark (main effect of time). Boxes represent marks for which significance increases in ELS over Std animals in both sexes, and reaches p<0.05 in ELS animals of at least one sex
(B) Mass spectrometry of male and female NAc at p21, p35, and adulthood. Fold change is ELS/Std at each timepoint with stars representing significant regulation. n=3 for each group, with each sample containing pooled bilateral punches from two animals
We next interrogated ELS-induced changes in expression levels of histone modifying enzymes in NAc from available RNAseq data31 to determine whether these transcriptional changes mirrored the altered histone modifications (Supplementary Table 2, Supplementary Figure 1C). Among the 120 epigenetic enzymes, 29 were significantly regulated in either male or female NAc after ELS. Of these 29, only three were significantly – and strongly – regulated in both sexes. The most regulated was Dot1l, which encodes the histone methyltransferase that catalyzes H3K79 mono-, di-, and tri-methylation.
Combined, these observations revealed the prominent and lasting regulation of H3K79 methylation, and transcription of its writer enzyme DOT1L, in the NAc after ELS. Further evidence is provided in Supplementary Figure 1B for the importance of this mark in networks of other histone modifications in this brain region: in both males and females, H3K79me2 shows more positive correlation with other histone modifications in ELS animals than it does with other histone modifications in Std animals. While these co-regulated marks bear further investigation, we focused the present study on understanding the behavioral implications and transcriptional control patterns of H3K79 methylation, with the goal of defining a discrete post-ELS target.
The developmental trajectory of Dot1l and Kdm2b transcription in D2 MSNs of NAc is altered by ELS
We investigated the transcriptional regulation of the writer (Dot1l) and eraser (Kdm2b) of H3K79 methylation in NAc. In qPCR of whole NAc tissue from P21, P35, and adulthood of ELS and Std animals, we found a significant interaction effect by two-way ANOVA of age and rearing for Dot1l expression in males (F2,49 = 3.48, p = 0.04) (Figure 2A) and females (F2,31 = 4.41, p = 0.02) (Figure 2D). In males, ELS increases the variability of Dot1l expression in whole NAc during adolescence (P35) and causes a persistent, though modest elevation in adulthood (t-test; p < 0.0001, t=5.648). Dot1l expression trended higher in adult female NAc, the opposite of what was seen in our RNAseq dataset (Supplementary Figure 1C), although the data were highly variable which might explain the discrepant findings. We also found a significant increase in Kdm2b expression in adult ELS males (p = 0.006) (Figure 2A) and ELS females (p = 0.04) (Figure 2D).
Figure 2. ELS alters the expression of Dot1l and Kdm2b over development, most intensely in D2-MSNs of males.
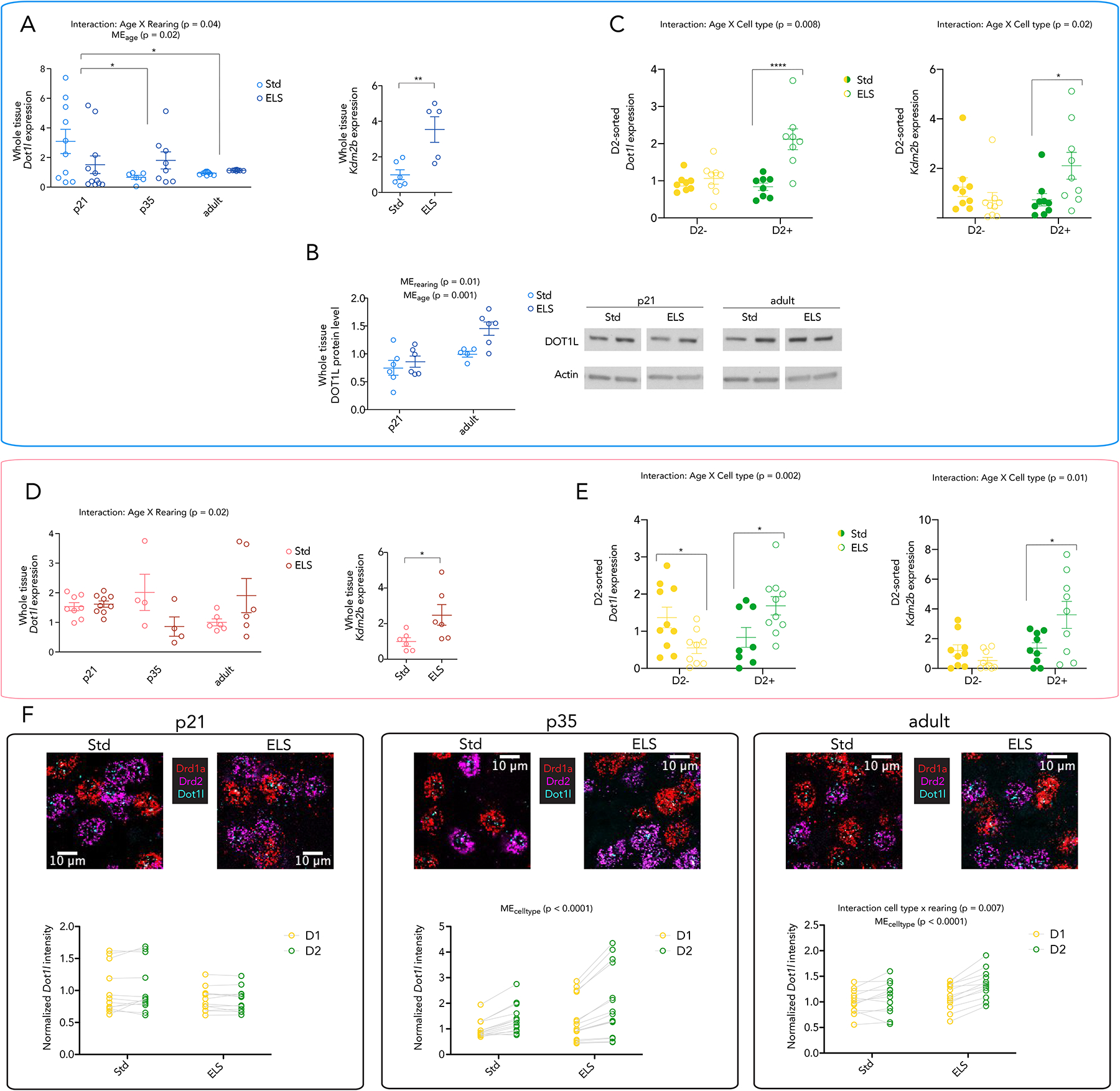
(A) Whole NAc expression of Dot1l and Kdm2b in Std and ELS males at p21, p35, and adulthood. Expression is normalized to Std adult. Significance in left panel by 2-way ANOVA, bars represent Bonferroni’s posstest, * < 0.05. Right panel shows significance by two-sided ttest, ** < 0.01
(B) Whole NAc protein levels of DOT1L at p21 and adulthood (left panel) with original blot shown (right panel). Expression is normalized to Std adult. Significance by 2-way ANOVA, bars represent Bonferroni’s posttest, * < 0.05, ** < 0.01, *** < 0.001
(C) Expression of Dot1l and Kdm2b in D2+ and D2− nuclei of Std and ELS adult males. Significance by 2-way ANOVA, bars represent Bonferroni’s posttest, * < 0.05, ****<0.0001
(D) Whole NAc expression of Dot1l and Kdm2b in Std and ELS females at p21, p35, and adulthood. Expression is normalized to Std adult. Significance in left panel by 2-way ANOVA, in right panel by two-sided ttest
(E) Expression of Dot1l and Kdm2b in D2+ and D2− nuclei of Std and ELS adult females. Significance by 2-way ANOVA, bars represent Bonferroni’s posttest, * < 0.05
(F) Quantification of Dot1l intensity in D1- and D2-MSNs in NAc of males at age p21, p35, and in adulthood. Significance by repeated measures 2-way ANOVA
We examined DOT1L protein levels in male NAc, and found a main effect of rearing (2-way ANOVA, p = 0.01). We also observed an overall increase in DOT1L protein from P21 to adulthood (2-way ANOVA, main effect of age, p = 0.001) (Figure 2B) that is not reflected in the qPCR results in Figure 2A. This indicates, perhaps, that Dot1l is under tighter transcriptional and translational regulation in adulthood, supported by the reduced variance in RNA and protein levels of Dot1l expression, as compared to the P21 timepoint.
Given the small fold change of Dot1l in whole tissue, opposite regulation in females, as well as the enrichment of Dot1l in male D2 MSNs at baseline as evidenced in a recently published dataset38, we hypothesized that the transcriptional regulation of H3K79-related enzymes by ELS may be enriched in the D2 MSN population of the NAc, with selective analysis of such cells revealing more consistent effects of ELS in both sexes.
To probe this hypothesis, we FACS-isolated D2+ and D2− nuclei from NAc of adult ELS and Std males and females. At baseline, Dot1l displayed similar enrichment in D2 MSNs in both sexes. After ELS, we found a significant induction of Dot1l (two-way ANOVA; main effect of rearing F1,14 = 20.97, p = 0.0004; main effect of cell type, F1,14 = 6.254, p = 0.03; interaction rearing × cell type, F1,14 = 9.229, p = 0.009) and Kdm2b (two-way ANOVA; interaction rearing × cell type, F1,32 = 6.041, p = 0.02) in D2+ nuclei of ELS males (Figure 2C). Similar results were seen in females, with an interaction effect of rearing and cell type for the expression of Dot1l (F1,33 = 11.58, p = 0.002), and significant regulation of Kdm2b expression (two-way ANOVA; main effect of cell type, F1,32 = 9.088, p=0.005; F1,32 = 7.367, p = 0.01) (Figure 2E). These effects were not seen in D2− nuclei.
To investigate the cell type-specificity of Dot1l regulation over time, we performed RNA in situ hybridization on NAc sections of P21, P35, and adult Std and ELS male mice (Figure 2F). Probes were designed for Drd1a, Drd2, and Dot1l mRNA, and Dot1l intensity was measured in the two MSN subtypes (see Methods). At P21, no significant regulation was observed either by rearing or by cell type, and the variability of Dot1l expression seen by qPCR (Figure 2A) was reflected in both cell types of Std animals, as was the lower expression of Dot1l in ELS animals at this early time point. At both P35 and adult timepoints, effects of cell type are observed, indicating that this effect of ELS builds (incubates) across development. Dot1l enrichment in D2 MSNs emerges over development for Std animals, and this enrichment is further magnified by ELS and sustained into adulthood, with an interaction effect of cell type and rearing in adulthood (2-way repeated measures ANOVA, p = 0.007).
D2 MSN specific manipulation of Dot1l and Kdm2b in NAc controls ELS-induced susceptibility to adult CSDS
To understand the implications of this D2-enriched epigenetic mechanism for behavior, we tested whether Dot1l expression in NAc mediates the increased behavioral susceptibility to adult stress seen after ELS. We sought to modulate this effect via cell type-specific viral-mediated manipulation of Dot1l with a microRNA (miRNA) or an overexpression vector selectively introduced into D2 MSNs of post-ELS adult mice. The miRNA was designed as a 64 bp insert, cloned into a Cre-dependent vector, and packaged into Herpes simplex virus (HSV). Knockdown of Dot1l was confirmed by qPCR of whole NAc (Supplementary Figure 2A, p = 0.03) 5 days after viral infection. Even though the manipulation was cell type-specific, we observed a decrease in Dot1l mRNA at the whole tissue level. For overexpression, Dot1l was cloned into a Cre-dependent vector and packaged into an HSV. Increased Dot1l expression was confirmed by qPCR on whole NAc (Supplementary Figure 2B, p = 0.01).
These cell type-specific manipulations proved capable of altering several behavioral outcomes in response to CSDS, including social interaction, open field exploration (a measure of anxiety-like behavior), and immobility in a forced swim test (a measure of passive coping with stress). Knockdown (KD) of Dot1l in NAc D2 MSNs of adult mice that had experienced ELS reversed the behavioral susceptibility that ELS animals typically exhibit, which manifests as impaired social interaction (t-test; t14=2.675, p =0.02), reduced open field exploration (t-test; t14=4.051, p=0.001), and increased forced swim immobility (t-test; t14=3.745, p=0.002) (Figure 3A). Compared to ELS-GFP animals, ELS-Dot1l KD animals showed improved social interaction (t-test; t11=3.651, p=0.004), increased exploration of the center of an open field (t-test; t11=3.854, p=0.003), and reduced immobility during forced swimming (t-test; t11=3.223, p=0.008). Furthermore, we show that the effect on social interaction becomes more pronounced over the week after CSDS and that Dot1l KD in standard-reared animals does not affect this behavior (Supplementary Figure 2H). Conversely, Dot1l overexpression (OE) in NAc D2 MSNs of standard-reared animals replicated the ELS-induced behavioral phenotype, which showed impaired social interaction (), as well as less (Figure 3A). Std-Dot1l OE animals showed impaired social interaction (t-test; t14=3.383, p=0.005), reduced open field exploration (t-test; t14=5.908, p<0.0001), and increased forced swim immobility as compared to Std-GFP animals (t-test; t14=2.795, p=0.01). Behavioral scores for individual animals across these three paradigms were significantly correlated (Figure 3B). For the first time, we also demonstrated that ELS induces deficits in reversal learning (Figure 3C) without affecting initial acquisition of the task (Supplementary Figure 2I), indicating a deficit in cognitive flexibility in ELS animals (two-way ANOVA; main effect of day, F3,48 = 13.63, p < 0.0001; main effect of rearing, F1,48 = 14.44, p = 0.0004). Furthermore, the effect on reversal learning is recapitulated by overexpression of Dot1l in Std animals (two-way ANOVA; main effect of day, F3,48 = 7.257, p = 0.0004; main effect of rearing, F1,48 = 16.62, p = 0.0002) (Figure 3C, Supplementary Figure 2I).
Figure 3. D2-MSN-specific manipulation of Dot1l and Kdm2b alters ELS-induced behavior.
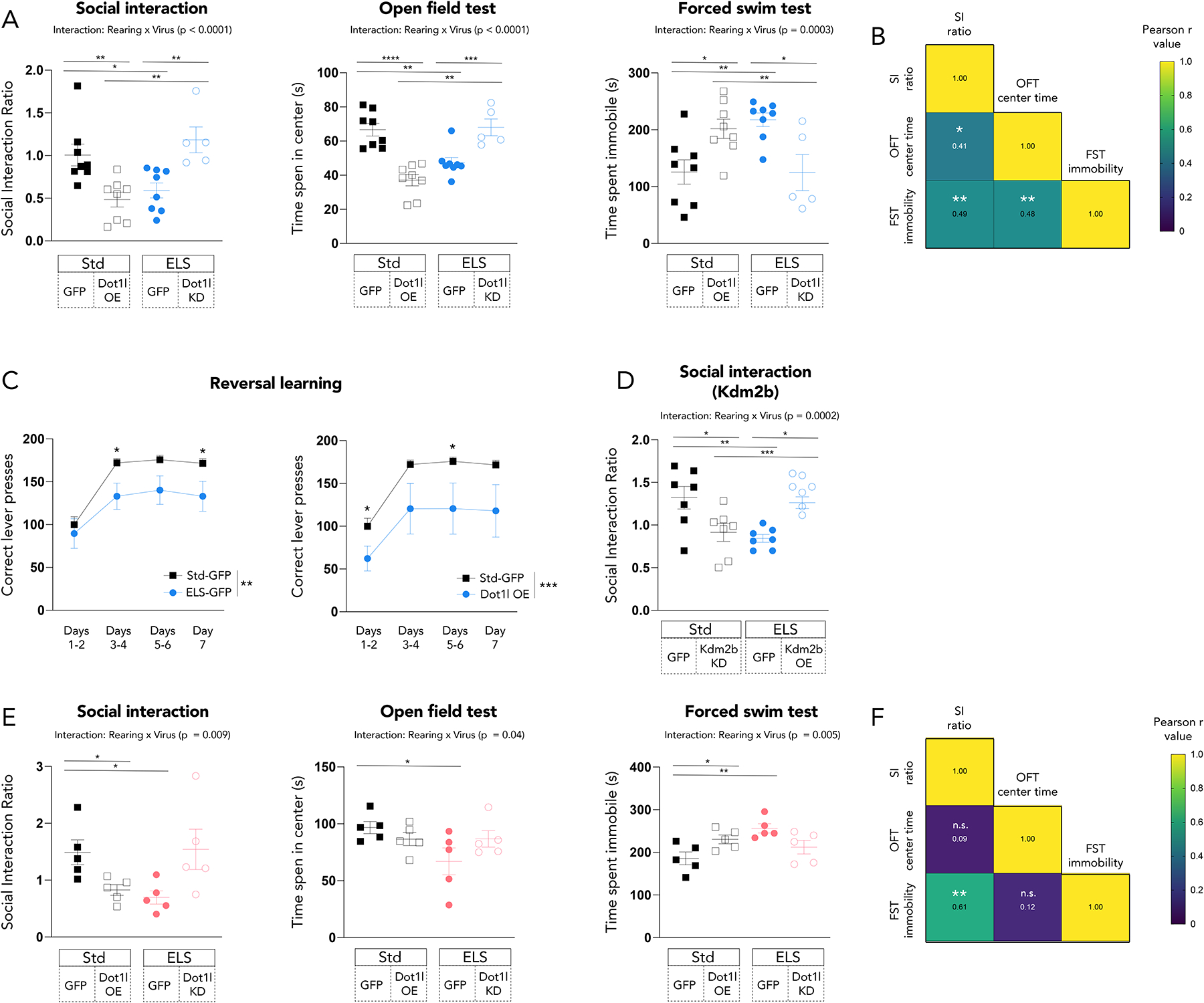
(A) Manipulation of Dot1l in D2 MSNs in males alters stress- and depression-relevant behaviors after CSDS at 24 hours after the last defeat. Significance by by t-test,*<0.05, ** < 0.01, **** < 0.0001
(B) Correlation of absolute values of individual male animals’ behavioral scores. Color and text of squares represent Pearson r values and stars represent significance, *<0.05, **<0.01
(C) ELS and overexpression of Dot1l both produce deficits in reversal learning. Significance by 2-way ANOVA, bars represent main effect of group and individual stars represent Bonferroni‘s posttest, *<0.05, **<0.01, ***<0.001
(D) Manipulation of Kdm2b in D2 MSNs in males alters ELS-induced reduction of social interaction after CSDS at 24 hours after the last defeat. Significance by by t-test,*<0.05, ** < 0.01, **** < 0.0001
(E) Manipulation of Dot1l in D2 MSNs in females alters stress- and depression-relevant behaviors after CSDS at 24 hours after the last defeat. Significance by by t-test,*<0.05, ** < 0.01, **** < 0.0001
(F) Correlation of absolute values of individual female animals’ behavioral scores. Color and text of squares represent Pearson r values and stars represent significance, **<0.01
We further probed this NAc D2 MSN-specific mechanism by virally altering expression of Kdm2b exclusively in this cell type. Two Cre-dependent vectors were prepared, and both were packaged into HSVs. One vector contained a Kdm2b transgene, while the other contained a miRNA designed to target Kdm2b; validation of these effects is shown in Supplementary Figures 2C & 2D (p < 0.01, 0.001, respectively). In line with our manipulations of Dot1l, we found that overexpression of Kdm2b in D2 MSNs of NAc reversed (t-test; t12=5.104, p=0.0003) the ELS-induced heightened susceptibility (t-test; t12=3.406, p=0.005) to CSDS in male mice 24 hours after the last defeat, while knockdown of Kdm2b in D2 MSNs increased susceptibility of stress-naïve mice (t-test; t12=2.397, p=0.03) (Figure 3D).
Manipulation of Dot1l in D2 MSNs of adult ELS females also modulates behavioral susceptibility to CSDS (Figure 3E). The results in females moved in the same direction but were less pronounced than the results in males. ELS itself altered social interaction (t-test; t8=2.605, p=0.03), open field exploration (t-test; t8=2.298, p=0.05), and forced swim immobility (t-test; t8=3.815, p=0.005). Dot1l overexpression also reduced social interaction (t-test; t8=2.299, p=0.05) and open field exploration (t-test; t8=2.442, p=0.04). Behavior in ELS-Dot1l KD animals did not significantly differ from the behavior of Std animals. Behavioral results in the social interaction and forced swim tests were significantly correlated (Figure 3F).
By contrast to the above results in D2 MSNs, Dot1l manipulation in D1 MSNs did not produce the same behavioral alterations, with ELS male and female mice showing increased susceptibility a week after CSDS regardless of whether Dot1l was knocked down or not (Supplementary Figure 2E). Dot1l overexpression in D2 MSNs of the PFC did not induce susceptibility to stress (Supplementary Figure 2F), providing regional specificity for Dot1l’s influence in the NAc. Furthemore, manipulation of another stress-relevant gene, Snca (which encodes α-synuclein), in D2-MSNs of the NAc did not induce social interaction deficits (Supplementary Figure 2G). These findings demonstrate cell-, region-, and gene-specificity for a particular stress mechanism modulated by Dot1l and Kdm2b in NAc D2 MSNs.
Together, these results indicate that elevated Dot1l or decreased Kdm2b in D2 MSNs of the NAc increases stress susceptibility, while diminished Dot1l or increased Kdm2b in these D2 MSNs reverses ELS-induced enhancement of stress susceptibility in adulthood.
RNA sequencing of D2 MSNs uncovers transcriptional signatures of ELS and of Dot1l regulation in NAc
To address the transcriptional effects downstream of this epigenetic mechanism, we performed RNA sequencing (RNAseq) on FACS-sorted D2 MSNs from virally-manipulated NAc tissue. Our experiment contained four groups: Std-GFP, ELS-GFP, Std-Dot1l overexpression (Std-OE), and ELS-Dot1l knockdown (Std-miR). We hypothesized that the Std-GFP and ELS-miR groups would show a more Std-like profile, while ELS-GFP and Std-OE groups would show a more ELS-like profile.
At a genome-wide level, as analyzed by rank-rank hypergeometric overlap (RRHO), ELS was strikingly similar to Dot1l overexpression in D2 MSNs (rho = 0.62) (Figure 4A). Using differential expression analysis, we found genes regulated in each of these groups, and observed a large and robustly significant (p < 10−50) overlap in genes regulated by ELS and by Dot1l overexpression in D2 MSNs (Figure 4B). More dramatically, these overlapping genes are regulated in the same direction and to a similar degree in both conditions (Figure 4D). Enrichment analysis of these overlapping genes using g:Profiler39 showed regulation of MSN signaling pathways (“Protein kinase A binding”, “Protein phosphatase 2A binding”) as well as cytoskeletal elements and regulation of axonal growth (“Axon ensheathment in central nervous system”, “Cell projection organization”) (Figure 4C). These terms point to specific functional outcomes of Dot1l-induced transcription in D2 MSNs.
Figure 4. D2-MSN-specific overexpression of Dot1l mimics ELS-induced transcription, while knockdown of Dot1l reverses ELS-induced transcription.
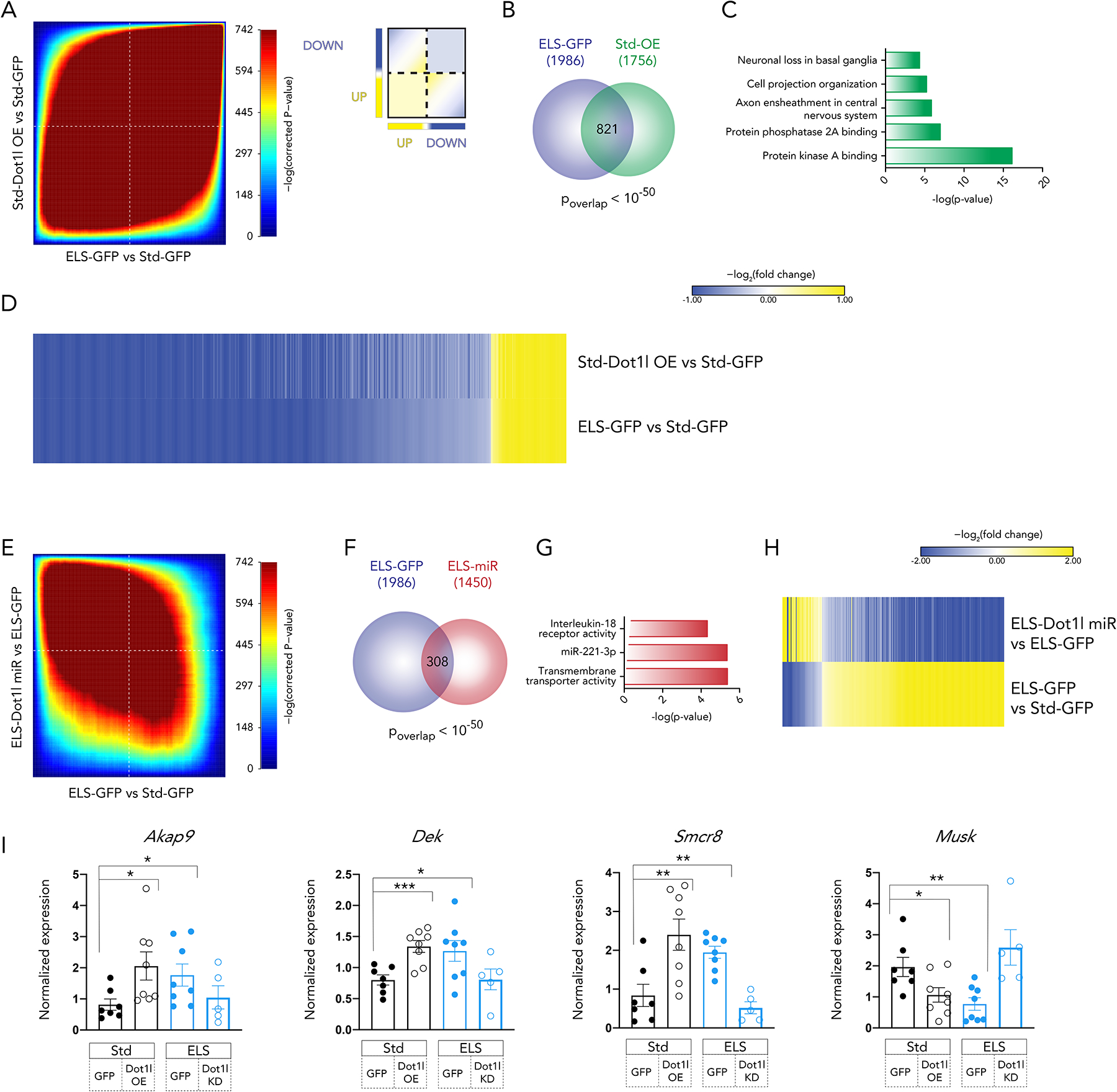
(A) Rank-rank hypergeometric overlap (RRHO) comparing transcriptional changes after ELS and Dot1l overexpression. Inset key for interpretation.
(B) Overlap of differentially expressed genes after ELS and Dot1l overexpression (|log2fold change| > 0.38, p < 0.05)
(C) Functional enrichment of 821 overlapping genes using gProfiler
(D) Fold change of 821 overlapping genes in each comparison
(E) RRHO comparing transcriptional changes after ELS and ELS + Dot1l knockdown.
(F) Overlap of differentially expressed genes after ELS and ELS + Dot1l knockdown in NAc D2 MSNs (|log2fold change| > 0.38, p < 0.05)
(G) Functional enrichment of 308 overlapping genes using gProfiler
(H) Fold change of 308 overlapping genes in each comparison
(I) qPCR of highly regulated RNAseq genes in animals from a separate cohort. Significance by ttest (*<0.05, **<0.01, ***<0.001)
Conversely, at the genome-wide level, a striking inverse relationship between the ELS-induced transcriptome and the Dot1l KD-induced transcriptome was observed, with the RRHO analysis indicating a rho value of −0.44 (Figure 4E). Significant overlap was also found in differentially expressed genes from the ELS-GFP vs Std-GFP and ELS-miR vs ELS-GFP conditions (p < 10−50) (Figure 4F), and the opposite regulation of genes noted at the genome-wide level was also detected in the subset of significantly regulated genes (Figure 4H). Together, these results indicate that knockdown of Dot1l in NAc D2 MSNs after ELS reverses the D2 MSN-specific transcriptional response induced by ELS itself. g:Profiler analysis of the overlapping genes revealed enrichment for interleukin-18 signaling (Figure 4G), which has been implicated in neuroinflammation via MAPK and STAT3 regulation40,41 and shown to be elevated in the serum of patients with a number of psychiatric disorders42,43.
A direct link between behavioral outcomes and these transcriptional responses was drawn by looking at expression of several top regulated genes from the RNAseq dataset (Figure 4I, Supplementary Figure 2J) in a separate cohort of animals whose behavior is presented above. Expression of each of these four genes correlated with at least one of the three behavioral scores presented in Figure 3A (Supplementary Figure 2K). Dek expression correlated with social interaction (r = −0.41, p = 0.03) and with forced swim immobility (r = 0.51, p = 0.005). Smcr8 expression correlated with social interaction (r = −0.44, p = 0.02) as did Akap9 expression (r = −0.39, p = 0.04), and Musk expression correlated with open field center time (r = 0.37, p = 0.05). The expression of these genes was assayed in unsorted NAc tissue, given the demonstration in Supplementary Figure 3 that a cell type-specific manipulation (D2-specific alteration of Dot1l expression) is detected at the whole tissue level (bulk H3K79me2 levels).
ChIP sequencing of H3K79me2 provides insight into the transcriptional effects of ELS and CSDS in NAc
In order to clarify the link between Dot1l regulation and transcriptional signatures, we performed ChIP sequencing (ChIPseq) for H3K79me2 in adult NAc tissue of Std and male ELS mice. Whole tissue was used as opposed to sorted D2 MSNs because it is not feasible to obtain enough input chromatin from sorted cells without pooling an excessive number of animals. We confirmed that the H3K79me2 antibody pulled down 24% and 40% over input in the Disc1 and Gapdh promoters, respectively (compared to 0.1% and 0.6% input for IgG). In an intergenic region, where the mark is predicted to be less abundant, H3K79me2 pulled down only 7% of input (Supplementary Figure 4A). These loci were determined based on publicly available H3K79me2 ChIPseq data from mouse liver (GSM1000152). See Supplementary Figure 4B–D for an exploration of H3K79me2 peaks in the NAc of Std adult males. We see a modest inverse relationship of peak size to gene length, and no relationship to level of RNA expression or to enrichment for enhancer sites. This is consistent with H3K79me2’s documented baseline function in other tissues of primarily marking the 3’ end of gene bodies.
46% of ChIPseq regions differentially regulated by ELS were found in gene bodies and, in these sites, H3K79me2 accumulated in the 3’ direction from the transcription start site (TSS) (Figure 5A). Our results indicated a genomic distribution of H3K79me2 predominantly over gene bodies (Figure 5B), as has been documented previously in other tissues44,45.
Figure 5. H3K79me2 genome-wide pattern after ELS resembles transcription after two stresses more than one stress, and ‘ELS-like’ transcription in D2-MSNs more than ‘Std-like’ transcription.
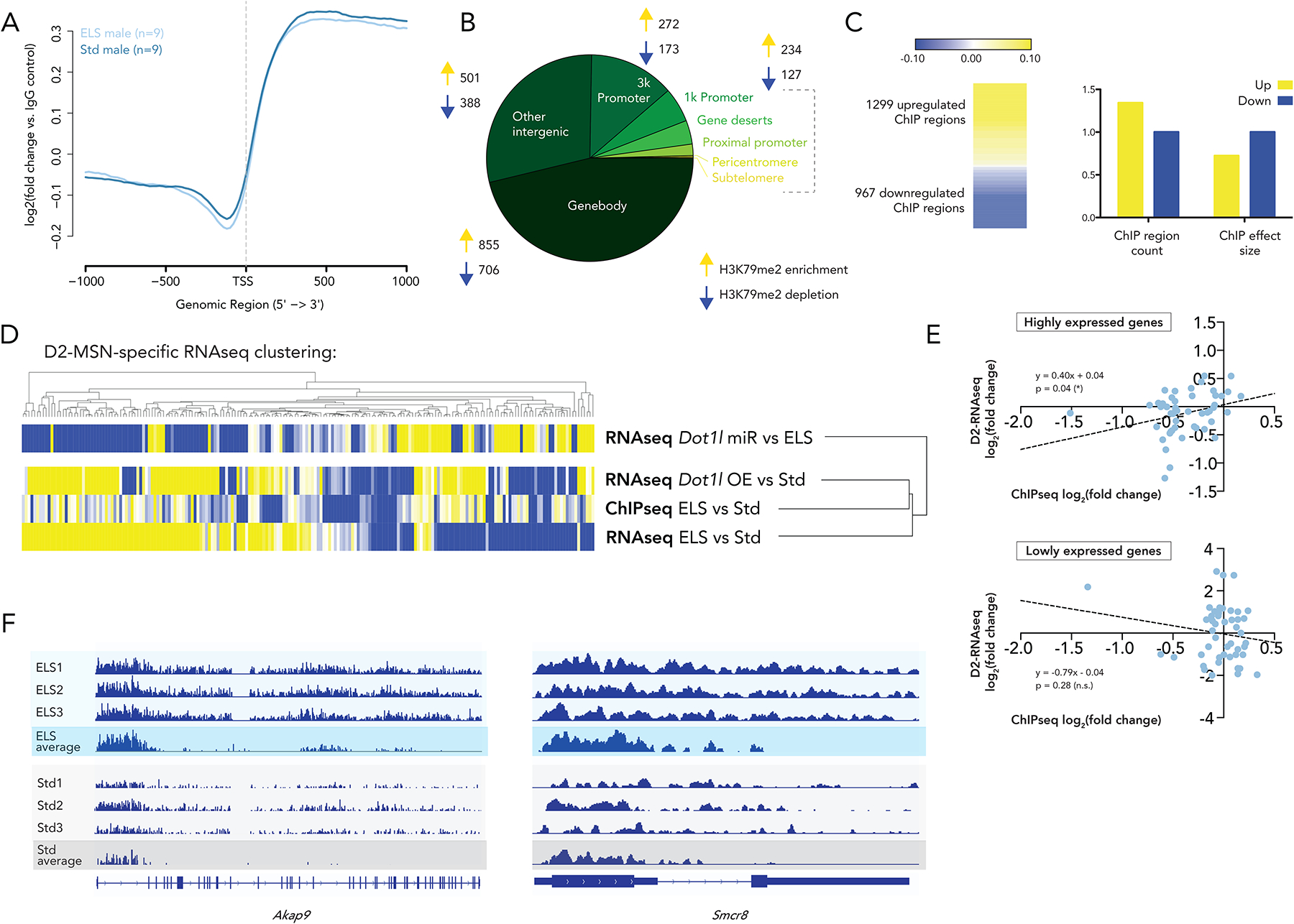
(A) Genome-wide distribution of read counts shows an enrichment of H3K79me2 in gene bodies. Values represented are log2(fold change) of H3K79me2 compared to an IgG control
(B) Genomic distribution of differentially enriched H3K79me2 regions after ELS shows predomination in gene bodies
(C) Up and downregulated regions show a greater number of H3K79me2-enriched peaks after ELS, but a larger effect size for depleted regions
(D) Heatmaps representing fold change in ChIPseq data and various comparisons from D2-MSN specific RNAseq, showing genes significantly regulated (pAdj < 0.05) in the ChIPseq dataset. Dendrograms represent Euclidean distance clustering
(E) Linear regression of RNAseq and ChIPseq log2(fold change) enrichment (ELS vs Std) of the 25% most highly and lowly expressed genes
(F) ChIPseq tracks for Akap9 and Smcr8 showing representative individual samples as well as group averages
The first point we addressed with this dataset was the discrepancy between the significantly elevated Dot1l mRNA and protein levels in D2 MSNs after ELS (Figures 2A, 2B) and the significantly decreased levels of H3K79me2 in whole NAc under these same conditions (Figure 1B). We see from the MS data that this is not due to increased conversion to H3K79me3. With our ChIPseq analysis, we found that 1299 regions were enriched for H3K79me2 after ELS, while 967 were depleted for it (Figure 5C). Moreover, when examining the effect size (number of regions × average log2foldchange per region in ELS/Std), depletion of H3K79me2 has a 38% larger magnitude than enrichment of H3K79me2 after ELS. These findings indicate that, while Dot1l is increased after ELS and this increase is associated with deposition of H3K29me2 at a larger number of genomic sites, a larger amount of H3K79me2 is lost at a subset of sites perhaps through the coordinated induction of Kdm2b. These results helped to clarify the paradox between qPCR and MS data.
ChIPseq genes with more H3K79me2 deposition after ELS enrich predominantly for steroid hormone biosynthesis and metabolism. The upregulated genes also enrich for many more predicted transcription factors than the downregulated genes (109 vs 14), suggesting that H3K79me2 marks the landscape of a coordinated transcriptional program (Supplementary Table 3). The top two most significant transcription factor predictions – NEUROD2 (pAdj = 1.18 × 10−9) and NEUROG1 (pAdj = 1.49 × 10−9) – are highly relevant to neuronal function and are likely involved in the downstream cellular effects of H3K79 methylation.
Downregulated genes, on the other hand, enrich overwhelmingly for flavonoid processing pathways and glucuronidation. This is an interesting finding, as flavonoids have been extensively implicated in stress resilience, and glucuronidation represents a mechanism for increasing flavonoid solubility and thus facilitating their diffusion. Downregulation of this pathway may broadly decrease flavonoid availability and work in conjunction with other functional changes to D2 MSNs indicated by the RNAseq data to increase stress susceptibility.
To understand the relationship of post-ELS H3K79me2 distribution to post-ELS transcription, we selected significantly regulated ChIPseq genomic regions (pAdj ≤ 0.05) and interrogated their fold change in a NAc D2 MSN-specific RNAseq dataset (Figure 5D). 189 of the 440 genomic regions significantly regulated in the ChIPseq dataset were represented in the RNAseq data, and were examined in three comparisons. Two of the RNAseq comparisons were ELS-like (ELS vs Std and Dot1l OE vs Std), while one comparison represented a Std-like condition (ELS-Dot1l miR vs ELS). Employing a Euclidean distance clustering method (Broad Institute, Morpheus tool), we clustered fold change from the ChIPseq data with fold change from the three comparisons within the RNAseq data. The H3K79me2 ChIPseq signal clustered with the ELS-like conditions and not with the Std-like condition. In other words, the pattern of deposition of H3K79me2 is more similar to the transcriptional effects, in D2 MSNs, of ELS than it is to Std transcription.
To further probe the relationship of the H3K79me2 ChIPseq signal to ELS-induced transcription, we correlated ChIPseq log2(fold change) of ELS vs Std with RNAseq log2(fold change) of ELS vs Std. We broke this analysis up by gene expression level across ELS and Std conditions, and found a significant correlation between H3K79me2 deposition and RNA expression at the 25% most highly expressed genes that was not present in the 25% most lowly expressed genes (Figure 5E). This result is consistent with our understanding of H3K79me2 as altering gene expression by interacting with machinery of active transcription44,45.
We show representative tracks at two of these highly expressed genes, Akap9 and Smcr8, which we showed in Figure 4 to be correlated with behavioral score. Across individual samples and as a population average, we see enrichment of H3K79me2 in the 3’ gene body of transcripts upregulated by ELS.
IP administration of a small molecule inhibitor of DOT1L rescues behavior after ELS
Having elucidated the transcriptional networks regulated by H3K79me2, we aimed to recapitulate the behavioral results of our cell type-specific viral-mediated gene manipulations using a more therapeutically accessible approach. This involved IP injection of pinometostat, a small molecule inhibitor of DOT1L, currently under phase II clinical investigation for treatment of pediatric mixed lineage leukemia (MLL). Though this approach is systemic and not brain region- or cell type-specific, we leverage the fact that, especially after ELS, DOT1L is enriched in NAc D2 MSNs.
Dosage of pinometostat was determined based on previous work, in which 35 mg/kg in rats and 40 mg/kg in mice was effective for cancer treatment46, well below the dosage at which treatment-limiting side effects (apnea, elevated transaminases) were seen in a clinical population47. We chose a dose much lower than the anti-cancer one, but one that has been demonstrated to reduce H3K79 methylation with a half-life longer than 1 hour48.
Twice daily administration of pinometostat (10 mg/kg IP) throughout the 10-day course of CSDS reduced H3K79me2 levels in the NAc (Figure 6A, t-test; p = 0.001, t=4.132). This pharmacological manipulation was sufficient to reverse the intensified stress susceptibility exhibited by ELS mice. Compared to ELS-saline controls, ELS-pinometostat animals showed a social interaction ratio after CSDS similar to that of Std-saline animals (Figure 6B). Notably, this 10 day course of administration did not alter animal weight or baseline locomotor activity (Supplementary Figure 5).
Figure 6. IP pinometostat administration during CSDS reverses ELS-induced social interaction deficit.
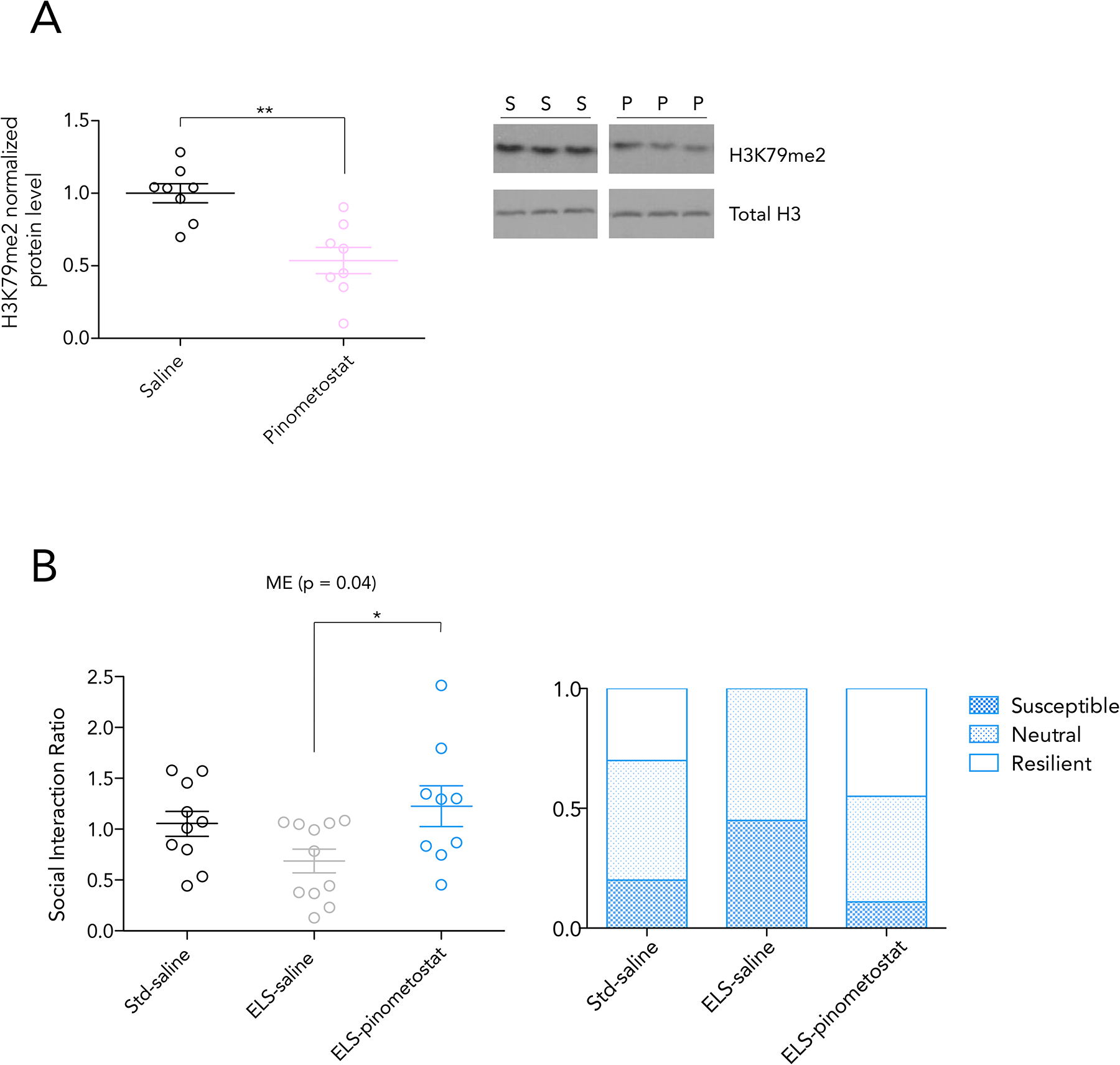
A) H3K79me2 protein level by ELISA in the NAc after 10 days of twice daily IP pinometostat administration at 10mg/kg reduces
B) Social interaction ratio after 10 days of CSDS, with administration of saline or pinometostat via IP injection. Significance by 1-way ANOVA, bars represent Bonferroni’s posttest, * < 0.05. Ratio of behavioral phenotypes shown in the right panel
Discussion
This study characterizes a newly discovered mechanism by which stress early in life reprograms D2 MSNs in the NAc to increase susceptibility to a second hit of stress in adulthood. We focus on H3K79me2 and its writer and eraser enzymes, DOT1L and KDM2B, respectively, based on two converging avenues of open-ended exploration: 1) proteomic analysis of histone modifications in NAc that are altered during development and in adulthood in response to ELS, and 2) transcriptomic data of all histone-modifying enzymes in NAc in both male and female mice after ELS. These analyses pointed to H3K79me2 and Dot1l as the most highly and consistently regulated in this brain region. We show that the regulation of this mark on a genome-wide scale is complex and involves its up- or down-regulation at hundreds of loci, which are likely carried out through interactions of DOT1L and KDM2B with several partners that have been shown to modulate their activity44,45.
We demonstrate that transcriptional changes in Dot1l and Kdm2b expression predominate in D2 MSNs of the NAc, and are not detectable in D1 MSNs. Next, using viral-mediated gene transfer to overexpress or knockdown Dot1l or Kdm2b selectively in D2 MSNs, we demonstrate that suppression of Dot1l or induction of Kdm2b reverses the ability of ELS to increase stress susceptibility in adulthood, whereas induction of Dot1l or suppression of Kdm2b mimics ELS by rendering standard-reared mice more susceptible to adult stress. Manipulation of these enzymes in D1 MSNs or in PFC neurons has no discernable effect. The NAc D2 MSN manipulations reveal that broad increases in H3K79me2 promotes susceptibility, while broad decreases promote resilience. Of course, the site-specificity of these increases in H3K79me2 is what determines the nuanced effects of ELS, where we notably see induction of both Dot1l and Kdm2b. To probe this site-specificity, we use RNAseq of D2 MSNs and ChIPseq for H3K79me2 to define the transcriptional consequences of ELS on the NAc via regulation of this chromatin mark.
It is notable that ELS induces both Dot1l and Kdm2b in D2 MSNs of the adult NAc, and that the induction of the two enzymes is associated with decreased total levels of H3K79me2 in this brain region. As increased transcription of Dot1l and Kdm2b appears to be coordinated, we hypothesize that their co-induction plays an essential role in the complex effects that ELS has on genome-wide H3K79me2 patterns, with more genomic loci showing enrichment for the mark, but many other loci showing larger reductions consistent with the overall decrease in total nuclear levels of H3K79me2 seen by MS. Further work is needed to understand the factors that control these differential effects of elevated DOT1L and KDM2B across the genome. The MS data indicate that the decrease in H3K79me2 is not due simply to conversion to the monomethyl state (which is also significantly decreased in post-ELS adult mice) or to the trimethyl state (which remains very sparse in both Std and ELS adult mice). The MS findings also reveal several other histone modifications that are highly coordinated with ELS-induced changes in H3K29me2 and point to networks of histone marks for future study, which may provide further insight into the complex regulation observed for H3K79me2, Dot1l, and Kdm2b and their downstream functional effects. The inability to obtain MS data from D2 MSNs specifically, which is currently not feasible, is a potential confound of these interpretations and something that now also warrants further study when permitted by technical advances.
Transcriptomic analysis of the effects of ELS, ELS plus Dot1l knockdown, or Dot1l overexpression alone on D2 MSNs reveals striking patterns. In this cell population, we demonstrate a robust similarity of transcriptional effects of ELS and Dot1l overexpression. Conversely, we find a robust inverse relationship between the effects of ELS alone and those of ELS followed by Dot1l knockdown. The dramatic nature of these patterns supports the behavioral data, namely, a central role for Dot1l in D2 MSNs in mediating the lasting effects of ELS seen in adulthood. Overall, the transcriptional programs controlled by ELS and by Dot1l enrich for intracellular signaling pathways, cytoskeletal rearrangements, interleukin signaling, and processes of axonal development. All of these represent important stress-relevant pathways which may be downstream of the H3K79me2 chromatin modification and will now guide efforts to study their functional consequences.
To further probe the downstream effects of Dot1l and Kdm2b co-induction after ELS, we performed ChIPseq on adult NAc tissue of ELS and Std males. We see loss of H3K79me2 at genes relating to flavonoid processing, a process which has been widely implicated in depression susceptibility and rescue of depressive symptoms. Indeed, several flavonoids have been shown to exhibit antidepressant activity49–51. This metabolic pathway may be permanently disturbed by ELS, contributing to behavioral vulnerabilities later in life, and may converge on the same JAK/STAT signaling pathway as does interleukin signaling52–55. By contrast, we see an enrichment of H3K79me2 at the binding sites for several transcription factors, such as NEUROD2 and NEUROG1, which likely help orchestrate transcriptional effects seen in the various ELS RNAseq datasets.
A series of clustering analyses comparing the ChIPseq signal to RNAseq datasets derived from whole NAc as well as from sorted D2 MSNs revealed important characteristics of the observed genome-wide H3K79me2 pattern. We see that the ChIPseq signal more closely resembles the D2 MSN-specific signal of ELS/Dot1l overexpression than it does that of Dot1l knockdown. The results also indicate that the ChIPseq signal more closely resembles transcription after two stresses (ELS + CSDS) than transcription after ELS alone, suggesting that this mark delineates CSDS-responsive genes that are primed by ELS. As such, it defines a maladaptive program of transcriptional priming and offers a target for clinical attempts to unhitch the detrimental effects of ELS later in life.
Although downregulation of H3K79me2 and induction of Dot1l and Kdm2b in NAc occurs after ELS in both male and female mice, the pattern of this regulation and its behavioral consequences are in general more robust in males as compared to females. This observation is consistent with our findings that, while ELS increases stress susceptibility in male and female mice, the associated transcriptional changes observed in NAc and other limbic regions are highly distinct31. This will be an important consideration in further research of this phenomenon and in the pursuit of any clinical applications.
As an initial effort to test the preclinical potential of this work, we investigated the effect of pinometostat, a highly-specific, small molecule inhibitor of DOT1L, which is now in advanced clinical trials for MLL. No deleterious behavioral effects of pinometostat have been reported in clinical studies to date, and our work represents the first exploration of drug’s actions in neuropsychiatric disease models. We administered pinometostat IP to ELS and Std male mice undergoing CSDS in adulthood. A 10-day, twice-daily course of injections – which decreased levels of H3K79me2 in NAc – was sufficient to reverse the susceptibility-priming effects of ELS on adult animals, without producing detectable untoward side effects. While this opportunistic experiment does not address the brain region- or cell type-specificity of Dot1l-H3K79me2 actions, or its utility in females, it does support the possible viability of pursuing this novel ELS-induced epigenetic mechanism for therapeutic purposes.
Methods
Transgenic animal lines
All mice were bred on a C57BL/6J background. Mice used for whole cell manipulations and FACS isolation for RNAseq heterozygously expressed Cre recombinase under the promoter of either Drd1 or Drd2.
Behavioral paradigms
All animal protocols were approved by IACUC and carried out in accordance with their guidelines. Mice were maintained on a 12 hour light-dark cycle, and given access to ad libitum food and water.
ELS consisted of maternal separation for 4 hours a day from P10-P17 as described27. Pups were separated to a cage with sawdust rather than corncob bedding, and 50% of homecage nesting material was removed on P10. All mice, Std and ELS, were weaned at P21, with male and female littermates separated.
CSDS was performed on male mice after viral surgeries as documented previously14,56 at ages 10–12 weeks. Mice were placed in a large hamster cage along with a CD1 male, separated by a transparent, perforated plastic barrier. CD1 males were ordered as retired breeders from Charles River, and were screened for aggression for three days prior to CSDS in daily 3-minute bouts with C57BL/6J mice. Target mice were moved across the barrier to the CD1 side for 5 minutes per session, with two sessions for each of the 4 days of defeat. In the case of subthreshold defeat, only one session was carried out per day. If mice became too physically injured, they were moved back to the other side before the full 5 minutes had elapsed. Mice were cycled through different aggressors throughout the 4 days of the protocol, and after the last bout of CSDS, target C57BL/6J mice were singly housed. A very similar protocol was used for CSDS in female mice57.
CSDS for mice in the pinometostat experiment – when time was not limited by viral expression – was carried out according to the original 10-day protocol, with once daily 5-minute exposures to the aggressor.
Social interaction testing was performed as described previously56 24 hours after single housing in a room separate from that where the CSDS took place. Briefly, target mice were given 150 seconds to explore an arena including an interaction zone containing a plastic enclosure with wire mesh allowing interaction at floor level. Next, the target mouse was allowed to explore the same arena with a novel CD1 (not encountered during CSDS) occupying the enclosure.
Open field test was measured in an open arena of 44×44 cm with a center area of 34×34 cm over a period of 480 seconds. Forced swim immobility was assessed in 600 mL of room temperature water in 1 L glass beakers over a period of 360 seconds. All behaviors were video recorded and analyzed using Ethovision 10.0 software (Noldus).
Reversal learning was conducted in mouse operant chambers (Interior dimensions: Interior: 55.69 × 38.1 × 40.64 cm; exterior dimensions: 63.5 × 43.18 × 44.45 cm, and walls: 1.9 cm) from Med Associates (St. Albans, VT, United States). Chambers were enclosed in light and sound attenuating cubicles equipped with white house lights as well as fans to provide ventilation and to mask external noise. Each operant chamber contained two retractable levers, located on the right and left sides of a central reward magazine calibrated to deliver ~50 μL of water. Adult male mice were water deprived and given 4 hours of water access during the 4 days prior to the beginning of behavioral training. They received a single operant session every day and were given 2 hours of water access following each daily session throughout the course of the experiment. During the single 40 minute pre-training session, mice explored the operant chamber and learned to introduce their noses into the central reward magazine to get water rewards, which were delivered every 60 seconds. Levers were retracted throughout.
The task itself consisted of an acquisition and a reversal phase. During acquisition, mice learned to press the left lever to get access to a water reward on an FR1 schedule. The number of correct (left) and incorrect (right) lever presses along with the number of earned rewards were recorded for each session, with a 75% correct criterion for learning. During reversal, mice are expected to learn that the rewarded lever is now on the right. Again, correct (left) and incorrect (right) lever presses along with the number of earned rewards were recorded.
Viral reagents
Cre-dependent HSV vectors were prepared using the p1006. This plasmid contains a floxed STOP site, allowing for recombination and expression in the presence of Cre. Transgenes are cloned downstream of a CMV promoter. Four p1006 plasmids were cloned – two miRNAs and two mRNA clones. Both miRNAs were prepared using the BLOCK-iT™ Pol II miR RNAi Expression Vector Kit (Thermofisher, cat #K4935–00) with forward and reverse sequences for a Dot1l miR (FWD: 5’-TGC TGA TGA GCA GCA TCA TGG TGC TTG TTT TGG CCA CTG ACT GAC AAG CAC CAA TGC TGC TCA T-3’, REV: 5’-CCT GAT GAG CAG CAT TGG TGC TTG TCA GTC AGT GGC CAA AAC AAG CAC CAT GAT GCT GCT CAT C-3’) and a Kdm2b miR (FWD: 5’- TGC TGA CAA ACT GGG ACA TGC TCA TCG TTT TGG CCA CTG ACT GAC GAT GAG CAT CCC AGT TTG T-3’, REV: 5’-CCT GAC AAA CTG GGA TGC TCA TCG TCA GTC AGT GGC CAA AAC GAT GAG CAT GTC CCA GTT TGT C-3’). A lacZ miR control was used as well (FWD: 5’-TGC TGA AAT CGC TGA TTT GTG TAG TCG TTT TGG CCA CTG ACT GAC GAC TAC ACA TCA GCG ATT T-3’, REV: 5’-CCT GAA ATC GCT GAT GTG TAG TCG TCA GTC AGT GGC CAA AAC GAC TAC ACA AAT CAG CGA TTT C-3’). Overexpression vectors were prepared with gene clones for Dot1l (OriGene, cat #MC224665) and Kdm2b (GenScript, cat #OMu14453).
Viral-mediated gene transfer
Mice were anesthetized with ketamine (100 mg/kg) and xylazine (10 mg/kg) in an IP bolus, and head-fixed in a stereotaxic apparatus for small animals (Kopf Instruments). Coordinates from Bregma were determined using a mouse brain atlas. Using a 0.33 mm Hamilton syringe at 10° from normal, virus was injected (0.5 μl at 0.1 μl/min) at the following coordinates relative to Bregma: anterior/posterior +1.6 mm, medial/lateral +1.5 mm, dorsal/ventral −4.4 mm. Syringe was kept in place for 5 minutes after injection before being retracted. Surgical site was treated with bupivacaine for local analgesia.
Tissue collection
For all experiments, NAc tissue was harvested as follows. After cervical dislocation, mouse brains were dissected and placed in a metal brain block on ice. Razor blades were placed at 1 mm separation, and two bilateral 14-gauge punches per animal were taken from the 1 mm slice containing the NAc. Punches were immediately flash-frozen and stored at −80C until further use.
Nuclear isolation for mass spectrometry and nuclear FACS
Nuclei were isolated from frozen tissue. Punches were processed with a handheld tissue homogenizer in a dissection buffer (60 mM KCl, 300 mM sucrose, 15 mM NaCl, 15 mM Tris-HCl pH 8, 5 mM MgCl2, 0.1 mM EGTA, 1 mM DTT). Samples were centrifuged and cells were collected in the pellet, resuspended on ice in lysis buffer (dissection buffer + 0.4% NP40), and centrifuged to collect nuclei. Frozen nuclei were shipped to UPenn for MS. Nuclei for FACS were resuspended in a 0.4% BSA PBS solution and sorted on a BD FACSAria II 3-laser machine using a 100 μm nozzle, with nuclei sorted into Trizol LS and flash-frozen. Gates from a representative sort are visualized in Supplementary Figure 6.
Mass spectrometry
Histone proteins were extracted as described previously58. Each group had n=3, with each sample containing pooled bilateral NAc punches from two animals. Briefly, histones were acid extracted from nuclei with 0.2 M H2SO4 for 2 hours and precipitated with 33% trichloroacetic acid (TCA) overnight. The pellets, containing histone proteins, were dissolved in 30 μl of 50 mM NH4HCO3, pH 8.0 plus 5 μl of acetonitrile. 5 μl of propionic anhydride were mixed with the histone sample and incubated for 20 minutes at room temperature. The reaction was performed twice to ensure derivatization completion. Histones were then digested with trypsin (enzyme:sample ratio 1:20, overnight, room temperature) in 50 mM NH4HCO3. The derivatization procedure was repeated after digestion to derivatize peptide N-termini. Samples were then desalted with C18 Stage-tips.
After drying, samples were resuspended in 10 μl of water + 0.1% formic acid. 2 μl of histone peptide solution were injected onto a 75 μm ID × 25 cm analytical column mounted onto an EasyLC 1000 nanoHPLC (Thermo Scientific). The HPLC gradient was as follows: 2% to 28% solvent B (A = 0.1% formic acid; B = 95% MeCN, 0.1% formic acid) over 45 minutes, from 28% to 80% solvent B in 5 minutes, 80% B for 10 minutes at a flow-rate of 300 nL/min. nLC was coupled to a Q-Exactive HF-X mass spectrometer (Thermo Scientific). Data were acquired using a data-independent acquisition (DIA) method59. Specifically, a full scan MS spectrum (m/z 300−1100) was acquired in the Orbitrap with a resolution of 120,000 (at 200 m/z) and an AGC target of 5×10e5. MS/MS was performed with a resolution of 7,500 with AGC target of 5×10e4. MS/MS was acquired using higher collision dissociation (HCD) with normalized collision energy of 27.
Data were analyzed by using EpiProfile 2.060. EpiProfile extracts the ion chromatogram of the (un)modified histone peptides. The peptide relative ratio was calculated using the total area under the extracted ion chromatograms of all peptides with the same aa sequence (including all of its modified forms) as 100%. For isobaric peptides, the relative ratio of two isobaric forms was estimated by averaging the ratio for each fragment ion with different mass between the two species. Statistical regulation of histone marks was assessed by using either a homoscedastic or heteroscedastic t-test (2-tails) depending on the significance of the F-test. Statistical significance was assessed when p-value was smaller than 0.05.
Whole cell FACS of virally infected D2 MSNs and cell type-specific RNA sequencing
Whole cells were isolated from fresh tissue 5 days after viral surgery. Brains were bathed in an ice cold choline chloride solution before bilateral 2 mm 14-gauge punches of NAc were taken and placed in a digestion buffer (0.2 M kynurenic acid, 1 mM MgCl2 6H2O, 8 mM D-glucose, in HBSS and HEPES), then enzymatically digested at 37°C with papain (Worthington Biochemical, cat #LS003118) for 45 min. Tissue was then triturated with successively smaller pipette tips until a single cell suspension was obtained and filtered through a 40um filter. Cells were centrifuged and resuspended in an ovomucoid solution to inhibit further papain digestion. Whole cells were then separated from debris by centrifugation on an ovomucoid-albumin gradient.
FACS was performed on a BD FACSAria 3-laser machine using a 100 μm nozzle, with whole cells sorted into Trizol LS and flash-frozen. RNA was isolated from frozen whole cells in Trizol using the Direct-zol™ RNA Miniprep Kit (Zymo Research, cat #R2050) and prepared for sequencing with the SMARTer® Stranded Total RNA Kit (Takara Biotech, cat #634875). Samples were sequenced with Genewiz on an Illumina HISeq 4000 machine using a 2×150bp paired-end read configuration to a minimum depth of 12M reads (only two of the samples, though, had fewer than 30M reads).
QC was performed using FASTQC software (see URLs cited at end of Methods). Reads were aligned using the HISAT2 program (see URLs), and count matrices were generated using the featureCounts function of the Subreads program (see URLs).
Differential expression was analyzed using the DESeq2 package (see URLs) and the following cutoffs were used for determining differentially regulated genes: |log2foldchange| > 0.38, and a significance cutoff. For overlap and ontology analyses a cutoff of p-value < 0.05 was used, while all individually analyzed genes met a cutoff of pAdj < 0.05. Significance of gene list overlaps was determined using the GeneOverlap package (see URLs) for R, and RRHOs were obtained using the RRHO package (see URLs).
Preparation of chromatin and ChIP sequencing
Chromatin was isolated as described previously61 from frozen tissue, after fixation in 1% formaldehyde and quenching in 2M glycine. Fixed tissue punches were mechanically processed by being passed through a 22-guage needle 10 times. Cells were lysed for 15 minutes (5 mM PIPES pH 8, 85 mM KCl, 0.5% NP40, protease inhibitors), followed by a 10 minute nuclear lysis (50 mM Tris-HCl pH 8, 10 mM EDTA, 1% SDS, protease inhibitors). Chromatin was sheared using a sonicator (Bioruptor, Diagenode) with 25 cyles of 30 seconds on/off. Sheared chromatin was centrigured at high speed and resuspended in RIPA buffer. Magnetic M-280 Dynabeads Sheep anti-Rabbit IgG (Thermofisher, cat #11202D) were conjugated to H3K79me2 antibody (Abcam, cat #ab3594) by rotation at 4°C for 6 hours. Conjugated beads and chromatin were incubated together with rotation at 4°C overnight. H3K79me2-associated DNA was washed in order with a low salt buffer (0.1% SDS, 1% Triton ×100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl pH 8), a high salt buffer (0.1% SDS, 1% Triton ×100, 2 mM EDTA, 500 mM NaCl, 20 mM Tris-HCl pH 8), a LiCl buffer (150 mM LiCl, 1% NP40, 1% NaDOC, 1 mM EDTA, 10 mM Tris-HCl pH 8), and TE + 50 mM NaCl. DNA was eluted in ChIP elution buffer (1% SDS, 100 mM NaHCO3) and de-crosslinked overnight at 65°C.
Sequencing libraries were prepared using the NEBNext® Ultra™ II DNA Library Prep Kit (NEB, cat #E7645) and sheared to an average size of 300–400 bp. Libraries were submitted to sequencing with Genewiz on an Illumina HISeq 4000 machine with a 2×150 bp paired-end read configuration to a minimum depth of 30M reads. The bedtools genomecov package was used for genome partitioning, and the diffReps algorithm was used to calculate differential expression (see URLs).
Euclidean and k-means clustering was performed on fold change values from the D2-RNAseq data and our ChIPseq data, using genes significantly enriched or depleted for H3K79me2 (pAdj < 0.05) after ELS using the Broad Institute Morpheus tool (https://software.broadinstitute.org/morpheus).
Correlation of ChIPseq fold change and D2-RNAseq fold change was performed on a subset of the above genes used for clustering. The top 25% most highly expressed and lowly expressed genes were chosen based on their expression across ELS and Std conditions, using the ‘baseMean’ value output by the DESeq2 algorithm.
Std H3K79me2 peaks for the analysis in Supplementary Figure 3 were generated using the MACS algorithm. Fold enrichment of these peaks over input was compared to gene length as well as gene expression level, the latter using the ‘baseMean’ output of the DESeq2 RNASeq comparison of Std and ELS. Peaks were also overlapped with a list of putative enhancers generated from several in-house ChIPseq experiments on NAc.
Analysis of existing RNA-sequencing data
Analysis of experimentally matching NAc RNAseq data (available in Gene Expression Omnibus GSE89692) was as previously described26 with differential gene expression determined by DESeq2 with at least 25% fold-change threshold applied.
Preparation of RNA and qPCR
RNA from whole tissue was prepared using the RNeasy Mini Kit (Qiagen, cat #74104) and from sorted tissue using the Direct-zol™ RNA Miniprep Kit (Zymo Research, cat #R2050). In both cases, it was converted to cDNA using the iScript™ cDNA Synthesis Kit (Bio Rad, cat #1708890). qPCR for Dot1l , Kdm2b, Akap9, Dek, Musk, Smcr8 and the Hprt1 control gene were performed using the below primers.
Dot1l:
FWD: 5’-GCG GAA CCG TTG GAG GTA AT-3’
REV : 5’-TTC ACA GTG GCT CCA TGT CC-3’
Kdm2b:
FWD: 5’-GGA CTT TGC AAA CGG ATC TGC-3’
REV: 5’-_TTT CCT CCA CGT CCG ACA AG-3’
Hprt1:
FWD: 5’-GCA GTA CAG CCC CAA AAT GG-3’
REV: 5’-GGT CCT TTT CAC CAG CAA GCT-3’
Akap9:
FWD: 5’- GAA ACG ATG CCT TGC TTC GG-3’
REV: 5’- TGA GTA GTG GAC CCT GAC TGT-3’
Dek:
FWD: 5’- GCG ACT TCT ACA CAG CGG C-3’
REV: 5’- GAC CCT TCC CTT GTG TCA CTG-3’
Musk:
FWD: 5’- ACT TCC AAA AGC CCC TGT CA-3’
REV: 5’- GAA AAC CAC GGA GGA GGG AC-3’
Smcr8:
FWD: 5’- CGT GGA CTT CGG AGT GGA AA-3’
REV: 5’- GGA GAG GCC ACT CTT TGT AGG-3’
RNAscope® in situ hybridization analysis
Brains of ELS and Std mice were flash-frozen in 2-methylbutane on dry ice, and sliced at −20°C in a cryostat at 20 μm. Slices were fixed in 4% PFA for 15 minutes and prepared using the RNAscope® Assay to hybridize the following probes to RNA transcripts and fluorophores: Mm-Drd1a-C3 (cat #406491-C3), Mm-Drd2 (cat #406501), Mm-Dot1l-C2 (cat #533431-C2). Images were taken within two weeks of staining on a Zeiss LSM780 confocal microscope, and analyzed using a macro to find intensity of Drd1, Drd2, and Dot1l staining for each DAPI-positive nucleus. Nuclei above the 75th percentile of Drd1 expression were deemed Drd1+, and likewise for Drd2+ nuclei. Average Dot1l intensity was calculated for a total of 4 animals per group with at least 3 slices analyzed per animal, and all values were normalized to Std D1 Dot1l intensity within timepoint and rearing. Images were captured at 512×512 pixels, and 140×140 pixel cutouts are shown in the figures.
Western blotting
Protein was isolated from flash-frozen brain samples. Two NAc punches were pooled bilaterally from one animal, homogenized in 80 μl of RIPA buffer using a handheld homogenizer for 20 seconds. Samples were then incubated at 4°C for 30 minutes with agitation, then sonicated using a Bioruptor for 5 cycles of 20 seconds on/off. After 15 minutes of centrifugation at 14,000g at 4°C, supernatant was collected and protein concentration was quantified using the BCA assay (Thermo Pierce). 11 μg of each sample were mixed with Laemmli buffer and ß-mercaptoethanol, and the solution was heated at 95°C for five minutes. Samples were removed from heat and centrifuged for 30 seconds before loading on a Criterion 4–15% Trish-HCL 1.0 mm precast gel (Bio-Rad). Proteins were separated based on molecular weight with SDS-PAGE followed by membrane transfer to PVDF membranes (Bio-Rad, cat #162–0175) at 100 V for one hour. Protein transfer was completed in SDS-free transfer buffer containing 20% methanol at 4°C. To reduce non-specific binding, membranes were blocked in TrisBuffered Saline (TBS) containing 5% bovine serum albumin and 0.1% Tween-20 for 1 hour at room temperature. Subsequently, membranes were incubated in primary antibody in blocking solution overnight at 4°C (Dot1l, Abcam, cat #ab64077; Actin, MP Biomedical, cat #691001). Following multiple washes, membranes were incubated in peroxidase-based secondary antibodies (Vector Labs) in blocking solution at room temperature for 2 hours. Membranes were washed again, and then developed with chemiluminescent substrate (Thermo Scientific). Primary antibodies were used at a 1:1,000 dilution and secondary antibodies were used at a 1:50,000 dilution. Protein expression was quantified using Image J software (National Institute of Health). In order to compensate for any irregularities in the gel or imaging, the space immediately below the protein of interest was measured as well and its intensity was subtracted as a control. Dot1l expression was normalized to actin, while H3K79me2 expression was normalized to total H3.
Statistics
All statistics were performed using Prism version 5.0 (GraphPad Software) with the exception of 1-way ANOVAs from Figure 1, which were performed using R’s aov() function. Any 2-way ANOVA result indicating a significant interaction was followed up with a 1-way ANOVA and Bonferroni post-test post-test. Outlier detection was performed for all results using a Grubbs test with an alpha value of 0.05. Statistical outliers were excluded from analysis.
Supplementary Material
Acknowledgments
This work was supported by funding from the National Institutes of Health (P50 MH096890 and R01 MH051399 to EJN) and the Hope for Depression Research Foundation.
Footnotes
Declaration of Interests
There are no conflicts of interest to report.
Data Availability
RNA sequencing and ChIP sequencing datasets that supported this study (Figures 4 and 5) have been deposited in the Gene Expression Omnibus under the accession code GSE133889 (reviewer access token: gxergksqdjybdqx).
URLs
FASTQC: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
HISAT2: https://ccb.jhu.edu/software/hisat2/index.shtml
DESeq2: https://bioconductor.org/packages/release/bioc/html/DESeq2.html
Subread: http://subread.sourceforge.net/
GeneOverlap: https://www.bioconductor.org/packages/release/bioc/html/GeneOverlap.html
RRHO: https://systems.crump.ucla.edu/rankrank/rankranksimple.php
Genomecov: https://bedtools.readthedocs.io/en/latest/content/tools/genomecov.html
References
- 1.Reynolds SM & Berridge KC Emotional environments retune the valence of appetitive versus fearful functions in nucleus accumbens. Nat. Neurosci 11, 423–425 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richard JM & Berridge KC Nucleus Accumbens Dopamine/Glutamate Interaction Switches Modes to Generate Desire versus Dread: D1 Alone for Appetitive Eating But D1 and D2 Together for Fear. J. Neurosci 31, 12866–12879 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walsh JJ et al. Stress and CRF gate neural activation of BDNF in the mesolimbic reward pathway. Nat. Neurosci 17, 27–29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brancato A et al. Sub-chronic variable stress induces sex-specific effects on glutamatergic synapses in the nucleus accumbens. Neuroscience 350, 180–189 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lipski WJ, Dibble SM, Rinaman L & Grace AA Psychogenic Stress Activates C-Fos in Nucleus Accumbens-Projecting Neurons of the Hippocampal Ventral Subiculum. Int. J. Neuropsychopharmacol 20, 855–860 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gill KM & Grace AA Differential effects of acute and repeated stress on hippocampus and amygdala inputs to the nucleus accumbens shell. Int. J. Neuropsychopharmacol 16, 2013–2025 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao J-L et al. Mesolimbic Dopamine Neurons in the Brain Reward Circuit Mediate Susceptibility to Social Defeat and Antidepressant Action. J. Neurosci 30, 16453–16458 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bagot RC et al. Ventral hippocampal afferents to the nucleus accumbens regulate susceptibility to depression. Nat. Commun 6, 7062 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krishnan V et al. Molecular Adaptations Underlying Susceptibility and Resistance to Social Defeat in Brain Reward Regions. Cell 131, 391–404 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Francis T et al. Nucleus Accumbens Medium Spiny Neuron Subtypes Mediate Depression-Related Outcomes to Social Defeat Stress. Biol. Psychiatry 77, 212–222 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hodes GE et al. Sex Differences in Nucleus Accumbens Transcriptome Profiles Associated with Susceptibility versus Resilience to Subchronic Variable Stress. J. Neurosci 35, 16362–16376 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dias C et al. β-catenin mediates stress resilience through Dicer1/microRNA regulation. Nature 516, 51–5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calipari ES et al. In vivo imaging identifies temporal signature of D1 and D2 medium spiny neurons in cocaine reward. Proc. Natl. Acad. Sci. U. S. A 113, 2726–31 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bagot RC et al. Circuit-wide Transcriptional Profiling Reveals Brain Region-Specific Gene Networks Regulating Depression Susceptibility. Neuron 1–15 (2016). doi: 10.1016/j.neuron.2016.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wook Koo J et al. Essential Role of Mesolimbic Brain-Derived Neurotrophic Factor in Chronic Social Stress–Induced Depressive Behaviors. Biol. Psychiatry 80, 469–478 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mandelli L, Petrelli C & Serretti A The role of specific early trauma in adult depression: A meta-analysis of published literature. Childhood trauma and adult depression. Eur. Psychiatry 30, 665–680 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Herbison CE, Allen K, Robinson M, Newnham J & Pennell C The impact of life stress on adult depression and anxiety is dependent on gender and timing of exposure. Dev. Psychopathol 29, 1443–1454 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Delpech J-C et al. Early life stress perturbs the maturation of microglia in the developing hippocampus. Brain. Behav. Immun 57, 79–93 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abbink MR, Naninck EFG, Lucassen PJ & Korosi A Early-life stress diminishes the increase in neurogenesis after exercise in adult female mice. Hippocampus 27, 839–844 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Hoeijmakers L et al. Early-life stress lastingly alters the neuroinflammatory response to amyloid pathology in an Alzheimer’s disease mouse model. Brain. Behav. Immun 63, 160–175 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Feifel AJ, Shair HN & Schmauss C Lasting effects of early life stress in mice: interaction of maternal environment and infant genes. Genes, Brain Behav. 16, 768–780 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Wang X-D et al. Forebrain CRF1 Modulates Early-Life Stress-Programmed Cognitive Deficits. J. Neurosci 31, 13625–13634 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yajima H et al. Early-life stress induces cognitive disorder in middle-aged mice. Neurobiol. Aging 64, 139–146 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Singh-Taylor A, Korosi A, Molet J, Gunn BG & Baram TZ Synaptic rewiring of stress-sensitive neurons by early-life experience: A mechanism for resilience? Neurobiol. Stress 1, 109–115 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yam KY et al. Exposure to chronic early-life stress lastingly alters the adipose tissue, the leptin system and changes the vulnerability to western-style diet later in life in mice. Psychoneuroendocrinology 77, 186–195 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y et al. Dopamine Receptor D2 and Associated microRNAs Are Involved in Stress Susceptibility and Resistance to Escitalopram Treatment. Int. J. Neuropsychopharmacol 18, pyv025–pyv025 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peña CJ et al. Early life stress confers lifelong stress susceptibility in mice via ventral tegmental area OTX2. Science (80-. ). 356, 1185–1188 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lovic V et al. Early postnatal experience and DRD2 genotype affect dopamine receptor expression in the rat ventral striatum. Behav. Brain Res 237, 278–282 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y et al. Dopamine Receptor D2 and Associated microRNAs Are Involved in Stress Susceptibility and Resistance to Escitalopram Treatment. Int. J. Neuropsychopharmacol 18, pyv025–pyv025 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romano-López A et al. Maternal separation and early stress cause long-lasting effects on dopaminergic and endocannabinergic systems and alters dendritic morphology in the nucleus accumbens and frontal cortex in rats. Dev. Neurobiol 76, 819–831 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Pena CJ et al. Early life stress alters transcriptomic patterning across reward circuitry in male and female mice. Nat. Commun 10, 5098 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gerfen CR et al. D1 and D2 dopamine receptor-regulated gene expression of striatonigral and striatopallidal neurons. Science 250, 1429–32 (1990). [DOI] [PubMed] [Google Scholar]
- 33.Kreitzer AC & Malenka RC Endocannabinoid-mediated rescue of striatal LTD and motor deficits in Parkinson’s disease models. Nature 445, 643–647 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Chandra R et al. Reduced Slc6a15 in Nucleus Accumbens D2-Neurons Underlies Stress Susceptibility. J. Neurosci 37, 6527–6538 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cerdá M, Sagdeo A, Johnson J & Galea S Genetic and environmental influences on psychiatric comorbidity: a systematic review. J. Affect. Disord 126, 14–38 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Labonté B et al. Sex-specific transcriptional signatures in human depression. Nat. Med 23, 1102–1111 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seney ML et al. Opposite Molecular Signatures of Depression in Men and Women. Biol. Psychiatry 84, 18–27 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kronman H et al. Biology and Bias in Cell Type-Specific RNAseq of Nucleus Accumbens Medium Spiny Neurons. Sci. Rep 9, 8350 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reimand J et al. g:Profiler—a web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res. 44, W83–W89 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song L, Pei L, Yao S, Wu Y & Shang Y NLRP3 Inflammasome in Neurological Diseases, from Functions to Therapies. Front. Cell. Neurosci 11, 63 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alboni S et al. Interleukin 18 activates MAPKs and STAT3 but not NF-κB in hippocampal HT-22 cells. Brain. Behav. Immun 40, 85–94 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Businaro R et al. Interleukin-18 modulation in autism spectrum disorders. J. Neuroinflammation 13, 2 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fan N et al. Relationship of serum levels of TNF-α, IL-6 and IL-18 and schizophrenia-like symptoms in chronic ketamine abusers. Schizophr. Res 169, 10–15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vlaming H & van Leeuwen F The upstreams and downstreams of H3K79 methylation by DOT1L. Chromosoma 1–13 (2016). doi: 10.1007/s00412-015-0570-5 [DOI] [PubMed] [Google Scholar]
- 45.Kerry J et al. MLL-AF4 Spreading Identifies Binding Sites that Are Distinct from Super-Enhancers and that Govern Sensitivity to DOT1L Inhibition in Leukemia. Cell Rep. 18, 482–495 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waters NJ Preclinical Pharmacokinetics and Pharmacodynamics of Pinometostat (EPZ-5676), a First-in-Class, Small Molecule S-Adenosyl Methionine Competitive Inhibitor of DOT1L. Eur. J. Drug Metab. Pharmacokinet 42, 891–901 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Shukla N et al. Final Report of Phase 1 Study of the DOT1L Inhibitor, Pinometostat (EPZ-5676), in Children with Relapsed or Refractory MLL-r Acute Leukemia. Blood 128, (2016). [Google Scholar]
- 48.Waters NJ et al. Exploring drug delivery for the DOT1L inhibitor pinometostat (EPZ-5676): Subcutaneous administration as an alternative to continuous IV infusion, in the pursuit of an epigenetic target. J. Control. Release 220, 758–765 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Guan L-P & Liu B-Y Antidepressant-like effects and mechanisms of flavonoids and related analogues. Eur. J. Med. Chem 121, 47–57 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Yan T et al. Brain-derived Neurotrophic Factor Signaling Mediates the Antidepressant-like Effect of the Total Flavonoids of Alpiniae Oxyphyllae Fructus in Chronic Unpredictable Mild Stress Mice. Phyther. Res 30, 1493–1502 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Hritcu L et al. Antidepressant Flavonoids and Their Relationship with Oxidative Stress. Oxid. Med. Cell. Longev 2017, 5762172 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yan X, Qi M, Li P, Zhan Y & Shao H Apigenin in cancer therapy: anti-cancer effects and mechanisms of action. Cell Biosci. 7, 50 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baba AB, Nivetha R, Chattopadhyay I & Nagini S Blueberry and malvidin inhibit cell cycle progression and induce mitochondrial-mediated apoptosis by abrogating the JAK/STAT-3 signalling pathway. Food Chem. Toxicol 109, 534–543 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Arumuggam N, Bhowmick NA & Rupasinghe HPV A Review: Phytochemicals Targeting JAK/STAT Signaling and IDO Expression in Cancer. Phyther. Res 29, 805–817 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Senggunprai L, Kukongviriyapan V, Prawan A & Kukongviriyapan U Quercetin and EGCG Exhibit Chemopreventive Effects in Cholangiocarcinoma Cells via Suppression of JAK/STAT Signaling Pathway. Phyther. Res 28, 841–848 (2014). [DOI] [PubMed] [Google Scholar]
- 56.Golden SA, Iii HEC, Berton O & Russo SJ A standardized protocol for repeated social defeat stress in mice. 6, 1183–1192 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takahashi A et al. Establishment of a repeated social defeat stress model in female mice. Sci. Rep 7, 12838 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sidoli S, Bhanu NV, Karch KR, Wang X & Garcia BA Complete Workflow for Analysis of Histone Post-translational Modifications Using Bottom-up Mass Spectrometry: From Histone Extraction to Data Analysis. J. Vis. Exp (2016). doi: 10.3791/54112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sidoli S, Simithy J, Karch KR, Kulej K & Garcia BA Low Resolution Data-Independent Acquisition in an LTQ-Orbitrap Allows for Simplified and Fully Untargeted Analysis of Histone Modifications. Anal. Chem 87, 11448–54 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yuan Z-F et al. EpiProfile 2.0: A Computational Platform for Processing Epi-Proteomics Mass Spectrometry Data. J. Proteome Res 17, 2533–2541 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cates HM et al. Transcription Factor E2F3a in Nucleus Accumbens Affects Cocaine Action via Transcription and Alternative Splicing. Biol. Psychiatry 84, 167–179 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


