Abstract
The limitations of conventional diagnostic procedures, such as real-time PCR-based methods and serological tests, have led the scientific community to innovate alternative nucleic acid detection approaches for SARS-CoV-2 RNA, thereby addressing the dire need for increased testing. Such approaches aim to provide rapid, accurate, cost-effective, sensitive, and high-throughput detection of SARS-CoV-2 RNA, on multiple specimen types, and without specialized equipment and expertise. The CRISPR-Cas13 system functions as a sequence-specific RNA-sensing tool that has recently been harnessed to develop simplified and flexible testing formats. This review recapitulates technical advances in the most recent CRISPR-Cas13-based methods for SARS-CoV-2/COVID-19 diagnosis. The challenges and opportunities for implementing mass testing using these novel CRISPR-Cas13 platforms are critically analyzed.
Keywords: SARS-CoV-2/COVID-19, CRISPR-Cas13, Collateral cleavage, Sherlock, Rapid detection, Point-of-care testing
Introduction
As authorities fight to contain SARS-CoV-2 [Severe Acute Respiratory Syndrome (SARS)-associated Coronavirus 2], the cause of COVID-19, experts warn that the death toll will almost certainly rise. Globally, as of 10 March 2021, more than 117,332,262 confirmed cases and 2,605,356 deaths caused by SARS-CoV-2 infection were reported in more than 210 countries (https://covid19.who.int/).
One of the most significant limitations of the response to the current outbreak has been in our ability to chart the path that the SARS-CoV-2 virus follows and to determine who has it and who does not. Current estimates indicate that SARS-COV-2 viral infection has an incubation period of 2–14 days, with potential for asymptomatic transmission [1]. Thus, rapid and early detection of the SARS-CoV-2 virus is a necessity for contact tracing, quarantine of confirmed cases, and enhanced surveillance, which undoubtedly depends on data obtained from widespread testing.
The conventional method used for diagnosing SARS-COV-2 is by real-time quantitative reverse transcription PCR (RT-PCR). Although this method is effective, it requires qualified personnel to run the tests and interpret the results. It also employs expensive equipment and lacks standardized protocols, hindering field-deployable viral diagnostics. Besides, it is time-consuming and cannot be used to screen large cohorts of people rapidly [2], [3].
On the other hand, the sensitivity and reliability of RT-PCR have been questioned due to the return of negative results for some patients who were highly suspected of having COVID-19 [4]. Hence, a significant drawback of current RT-PCR-based diagnostics is that they cannot detect the virus immediately after infection and need time for the viral load to rise, leading to false-negative results for recently infected individuals [5]. As a result, although samples collected from patients with symptoms seem to present relatively high viral titers, the testing of asymptomatic patients and testing before quarantine release may need extremely sensitive tests to confirm negative results during re-examinations and to avoid the potential risk of viral transmission [6], [7]. Therefore, there is an urgent need to implement new alternatives for rapid molecular diagnostics in patients and species of live animals suspected of SARS-COV-2 infection, especially in places where SARS-CoV-2/COVID-19 testing capabilities are limited.
This review describes current advances regarding diverse CRISPR-Cas13 systems, including SHERLOCK, SHINE, CREST, SENSR, CARMEN and the quantitative direct detection methods, in the development of fast, accurate, and portable diagnostic tests to address the dire need for increased SARS-COV-2 testing.
CRISPR-Cas13-based systems
Since their initial discovery, six types and 22 subtypes of the clustered regularly interspaced short palindromic repeats (CRISPR) systems have been discovered and explored [8]. CRISPR-Cas systems are divided into two classes: class I systems, which rely on multiple CRISPR-associated (Cas) proteins that mediate the target interference, and class II systems, which involve single, multidomain effector proteins [9]. The class II CRISPR-Cas systems include types II, V, and VI. Types II and V comprise endonucleases that operate at the DNA level [10], [11]. By contrast, type VI CRISPR-Cas systems are RNA-guided and RNA-targeting machineries that provide prokaryotes with immunity against RNA [12]. All type VI CRISPR-Cas systems have a single effector protein known as the Cas13 effector, previously known as the C2c2 CRISPR-Cas system [12], [13]. CRISPR-Cas13 systems comprise just two components: the programmable single-effector RNA-guided RNase Cas13, which possesses a ribonuclease (endoRNase) activity provided by its two Higher Eukaryotic and Prokaryotic Nucleotide-binding domains (HEPN); and a 64–66-nt pre-CRISPR-RNA (crRNA), which recognizes a 24–30-nt sequence (mature crRNA) on the target RNA, with a preference for targets with a protospacer flanking site (PFS) motif [13], [14], [15]. Once activated by the specific binding of the target RNA, these systems confer collateral ‘promiscuous’ activity and non-specifically degrade the RNA transcripts [12], [16].
Cas13a from the gram-negative bacteria Leptotrichia shahii (LshCas13a) was the first Cas13 ortholog to be harnessed for programmable RNA-targeting activities [12]. Since the first description of LshCas13a, several studies have identified more variants of Cas13 proteins belonging to different Cas13 families, classified into four type VI subtypes (A–D) [17], [18], [19]. These Cas13 variants exhibit variable efficiencies for transcriptome targeting and engineering. For example, Cas13a from Leptotrichia wadei (LwaCas13a) has been reported to mediate more robust RNA-targeting activity than the LshCas13a system, but requires a fusion to a stabilizer, such as the msfGFP protein, for efficient interference activity [19]. Subsequent studies have identified that the Cas13b ortholog from Prevotella sp. P5–125 (PspCas13b) is more suitable for RNA targeting in mammalian cells than the LwaCas13a protein, and PspCas13b does not require a stabilizer protein for its activity [18]. An additional Cas13d subtype was recently identified, type VI-D, which shows minimal sequence identity with previous Cas13 effectors [20]. Interestingly, Cas13d is among the smallest CRISPR-Cas single effectors, with 20–30% less mass than all previously reported type VI Cas13 endoRNases [20]. Hence, these Cas13 effector proteins provide a rich resource for new RNA-targeting technologies and have been developed recently for RNA knockdown, detection, editing, splicing, and viral delivery [16], [18], [19], [21].
CRISPR-Cas13-based diagnosis
SHERLOCK
The natural RNA-guided ribonuclease activity of the Cas13 protein and its nonspecific cleavage of collateral single-stranded RNAs (ssRNAs) in trans allowed the Zhang group and colleagues from the Broad Institute of MIT and Harvard to develop and optimize a method termed Specific High-sensitivity Enzymatic Reporter unLOCKing (SHERLOCK) [16]. This method was developed mainly to achieve the sensitive and specific detection of viral nucleic acid sequences in both synthetic and patient-derived samples [16].
SHERLOCK starts with an isothermal target nucleic acid pre-amplification step, in which either a DNA or an RNA target input is replicated by loop-mediated isothermal amplification (LAMP), most often using recombinase polymerase amplification (RPA) [22] (Fig. 1 I). Subsequently, the amplified targets are converted to RNA via T7 transcription. The specificity of Cas13 is conferred by crRNA–target pairing, and further sensitivity is achieved through signal amplification by the collateral cleavage activity of Cas13 on the RNA reporters that are added to the reaction, which can be captured on a colorimetric lateral-flow strip (by biotin–fluorescein RNA reporters) or visualized by fluorescence signal (by molecular beacon fluorescent reporters) (Fig. 1 I) [16], [22], [23]. Thus, SHERLOCK allows rapid (in as little as one hour with a setup time of less than 15 minutes) and reliable detection of nucleic acids at concentrations down to approximately 2 attomolar (aM; 10–18 molar) and with single base-pair mismatch specificity [22].
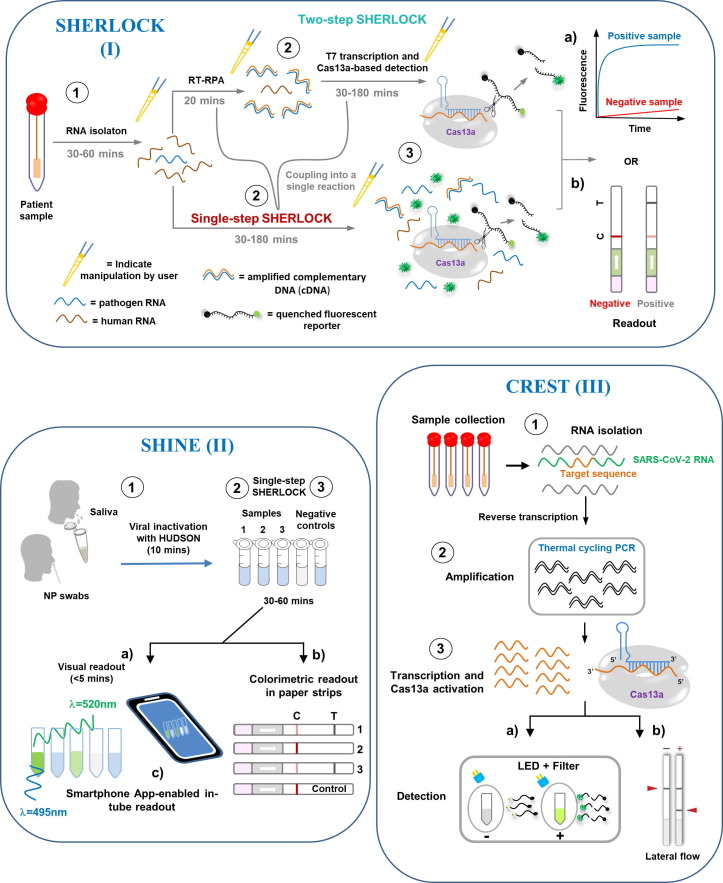
Figure 1.
Detection of SARS-COV-2 by CRISPR-Cas13 technology. ① After collecting clinical samples in a point-of-care setting (at the patient’s bedside or in the field), the RNA is extracted (except when using HUDSON methods such as SHINE). ②Then, reverse transcription into cDNA is performed, followed by amplification of the specific target sequences (while also adding T7 promoter sequences to the 5′ terminus) by RT-PCR, LAMP, or recombinase polymerase amplification (RPA) through a combination of T7 transcription (CREST, SHERLOCK, SHINE, and SENSR). In the case of CREST, the amplification step uses cost-effective Taq polymerase and portable thermocyclers (mini16 thermocycler) instead of isothermal reactions. ③ The CRISPR-Cas13 − crRNA RNP complex is activated by binding to a complementary target RNA. This activation triggers collateral cleavage of a nonspecific ssRNA reporter in trans, by the higher eukaryotic and prokaryotic nucleotide-binding domains (denoted by scissors). Thus, sensing differs according to the desired output detection method. (a) In SHERLOCK, SHINE, CREST, SENSR, and the Quantitative Direct Detection method, cleavage of the quenched reporters (which are not fluorescent in their native state due to the proximity of the conjugated fluorophore and the quencher) by the activated Cas effectors produces a signal, which indicates the presence of a specified nucleic acid target sequence. The SARS-CoV-2 genome can then be detected through an in-tube fluorescent readout. In this case, fluorescence can be seen to accumulate over time in an in vitro transcription (IVT)-coupled cleavage reaction for a specific guide RNA (gRNA). In CREST, Cas13 activation is followed by fluorescence detection of a de-quenched poly-U cleavage reporter visualized by a blue light emitting diode (LED) (~495 nm) and orange filter, or another fluorescence detection system. (b) Otherwise, in SHERLOCK, CREST, SHINE, and SENSR, the SARS-CoV-2 RNA can also be detected by visualization on a lateral flow test strip using a one-dimensional lateral flow immunochromatography system, in which a reporter can be coupled to a 6-carboxyfluorescein (6-FAM) fluorophore and biotin. Here, a streptavidin-impregnated line captures biotin in the absence of the target nucleic acid. In the presence of the target, the reporter is cleaved, leading to the separation of 6-FAM and biotin, which allows the reporter to move beyond the first streptavidin line to a second detection line where an anti-6-FAM capture antibody is found. (c) By taking advantage of fluorescence visualizers such as mobile phone cameras and smartphone applications (instead of portable plate readers), CREST and SHINE allow for a binary interpretation of results. Notably, the Quantitative Direct Detection method employs a mobile phone fluorescent microscope using a 488 nm diode laser, a green fluorescence interference filter, and a Pixel 4 XL phone camera (black box). A pipette indicates steps involving user manipulation. Times shown are the suggested incubation intervals. C, control line; T, test line. crRNA, CRISPR RNA; NP, nasopharyngeal swab; ssRNA, single-stranded RNA.
To distinguish between pathogenic RNA viruses that cause similar symptoms, however, it is necessary to detect multiple sequences at once. As a result, the SHERLOCK method has been further optimized to incorporate multiplex detection in clinical settings through a simplified and more specific protocol (SHERLOCKv2) [24]. Thus, employing the SHERLOCKv2 platform, scientists have been able to detect viral particles at concentrations down to 1.25 × 103 copies/mL (2.1 aM) and to distinguish reliably between Zika virus and Dengue virus in body fluids such as urine, saliva, serum, plasma, and whole blood [16], [24]. (Zika virus and Dengue virus are closely related flaviviruses that also possess a positive-sense RNA genome and may cause encephalitis.) This example highlights the wide applicability of SHERLOCK-based platforms to detect, for example, both single-stranded negative-sense RNA viruses, including Filoviridae family members such as the Ebola virus [25], and single-stranded positive-sense RNA viruses, including Coronaviridae family members such as SARS-CoV-2.
Shortly after the pandemic started, Zhang and colleagues described the SARS-CoV-2/SHERLOCK protocol (v.20200321) used to detect the novel coronavirus, which was developed by employing synthetic SARS-CoV-2 virus RNA fragments that were based on the full genomic sequences of SARS-COV-2 strains that are available in databases [26]. SHERLOCK/SARS-CoV-2 showed high sensitivity in detecting both the S gene, which encodes the spike (S) proteins of the virus, and the Orf1ab gene, which encodes orf1ab polyproteins. The limit of detection (LOD) for this protocol was between 20 and 200 aM (10–100 SARS-CoV-2 RNA copies per microliter of input), within the first 40 minutes of amplification. The use of RPA amplification primers and LwaCas13a guide RNAs (gRNAs) minimized off-target detection of related human respiratory virus genomes [26].
Patchsung and colleagues [27] recently performed SHERLOCK detection on a total of 534 clinical samples of SARS-CoV-2 RNA extracted from nasopharyngeal and throat swabs from infected patients in Thailand. The SHERLOCK system was 100% specific and 100% sensitive with fluorescence readout (LOD of 42 SARS-CoV-2 RNA copies per reaction, corresponding to a Ct of 33.5 on the RT-qPCR analysis), and 100% specific and 97% sensitive with lateral-flow readout [27]. The novelty of the SHERLOCK system that was designed by Patchsung and colleagues is the incorporation of an internal control for contamination by ribonucleases (RNases) in a single lateral flow strip. Each flow strip incorporates a RNase-responsive RNA reporter that is resistant to Cas13a cleavage, but that remains susceptible to RNase I-, RNase A- and RNase T1-mediated cleavage. This control feature is a valuable addition to a test for the detection of SARS-CoV-2, which may be used in settings with limited resources where there is a higher risk of contamination by RNases [27].
Among the main advantages of detection methods that are based on CRISPR-Cas13 (SHERLOCK) are: 1) rapid turnaround time, 2) limited infrastructure requirements, 3) isothermal amplification that avoids the need for thermocycling, 4) specificity of the target sequence at the single-nucleotide level, and 5) the possibility of adaptation to allow detection through visual readout based on lateral flow immunochromatography [16], [22].
Nevertheless, SHERLOCK typically uses a target amplification step, followed by CRISPR-mediated nucleic acid detection to recognize SARS-CoV-2 [22]. These two processes make SHERLOCK a complex diagnostic platform because they depend on an RNA extraction step and multiple liquid-handling steps that increase the risk of cross-contamination of samples [28]. Although these factors could limit the scalability of CRISPR-Cas13 (SHERLOCK)-based diagnostics, the method could nevertheless become a great option for testing in non-clinical settings, as well as in clinical settings where there is a need for rapid, inexpensive, accurate, and highly sensitive identification of SARS-COV-2 in multiple specimen types, without the need for specialized infrastructure.
SHINE
During the current outbreak, there is an unprecedented demand for the reagents and sophisticated equipment necessary for SARS-CoV-2/COVID-19 diagnostic tests. Amidst global supply shortages, it is necessary to innovate strategies to overcome such problems. On this point, Myhrvold et al. [23] previously established an approach to release viral nucleic acids from clinical specimens directly and to protect them from degradation, thus bypassing the need for nucleic acid extraction. This method, named HUDSON (Heating Unextracted Diagnostic Samples to Obliterate Nucleases), employs heat and chemical reduction to inactivate the pervasive ribonucleases found in body fluids. Then, viral particles are lysed by disruption of the viral envelope, resulting in the release of nucleic acids into solution [23]. HUDSON-treated biological samples can be directly added to isothermal amplification reaction mixtures, with no dilution or purification step, without inhibiting subsequent amplification or detection [23]. Therefore, a lab environment and professionally trained personnel are not essential for this protocol [16], [23].
Building on this method, Arizti-Sanz and colleagues [29] coupled HUDSON and CRISPR-based programmability into one step to create SHINE (SHERLOCK and HUDSON Integration to Navigate Epidemics), a scalable diagnostics tool for detecting viral RNA from unextracted patient samples that has minimal equipment requirements and quick turnaround time [29] (Fig. 1 II).
SHINE has been validated on 50 nasopharyngeal clinical samples, and shown to have 90% sensitivity [with SARS-CoV-2 RNA detected at ten copies per microliter (cp/μl)] and 100% specificity when compared to RT-PCR, and with a mean response time of 50 minutes [29]. The main advantages of SHINE are that it decreases user manipulations and assay time, and reduces the risk of sample contamination. Employing SHINE, Arizti-Sanz and colleagues [29] detected SARS-CoV-2 RNA in HUDSON-treated patient samples (in nasopharyngeal swabs and saliva) with either a paper-based colorimetric readout or an in-tube fluorescent readout, which can be performed with portable equipment and with reduced risk of sample contamination. The in-tube fluorescence readout and smartphone application used with SHINE (Fig. 1 II) allowed scalable, high-throughput testing and automated interpretation of results. Thus, by reducing specialized personnel, infrastructure, and time to obtain outcomes, without sacrificing sensitivity or specificity [29], this platform has the potential to provide point-of-care diagnostic tests for SARS-COV-2.
CREST
Rauch and colleagues [30] recently developed another CRISPR-based SARS-CoV-2 RNA detection method named CREST (Cas13-based, Rugged, Equitable, Scalable Testing). This method was developed as an option for recurrent testing that could overcome issues with the supply of reagents and specialized equipment, and that is easy to deploy at sites with minimal infrastructure [30]. CREST addresses three of the main hurdles that limit the scalability of Cas13-based testing at sites with a minimal supply of resources: 1) reagent accessibility, 2) equipment availability, and 3) cost. By taking advantage of widely available reagents (that are stable at room temperature), low-cost thermocyclers (instead of isothermal reactions), and easy-to-use fluorescent visualizers, this method bypasses the need for highly trained personnel and experienced molecular diagnostic laboratories [30].
The CREST method employs standard sample collection, RNA extraction, and reverse transcription (Fig. 1 III). However, the amplification step uses cost-effective Taq polymerases and portable thermocyclers. This has been made possible by creating inexpensive, Bluetooth-enabled thermocyclers (which can even be powered by a 9 V battery and run using mobile device applications) and simple plastic-filter-based, LED visualizers [30]. In the CREST platform, transcription and Cas13 activity are followed by fluorescence detection of de-quenched poly-U cleavage reporter, visualized with a blue LED (~495 nm) and orange filter or other fluorescence detection system. Thus, the use of expensive antibodies or antibody-conjugates for lateral flow immunochromatography, is avoided (Fig. 1 III). Results can be registered with a smartphone camera and uploaded to the cloud, enabling a massive deployment of point-of-care testing that is capable of addressing the current global demand for SARS-CoV-2/COVID-19 testing.
The RNA extraction step that is required for CREST, which uses commercial kits, is a potential bottleneck that might limit the accessibility and scalability of this method for detecting SARSCoV-2. However, the developers overcame this limitation by coupling to a method called PEARL (Precipitation Enhanced Analyte RetrievaL), a fast and accessible approach for the isolation of RNA, DNA, and proteins [31].
Using CREST, Rauch and colleagues [30] detected the presence of the N1, N2, and N3 sites in the SARS-CoV-2 nucleocapsid (N) gene, with an associated cost of reagents per test of US$ 5.90 (a single sample run in triplicate) compared to the PCR test cost of US$ 14 apiece. Thus, CREST is a relatively cheap format that allows portable diagnostic tests to assess suspected cases without sophisticated equipment. The sensitivity of CREST was equivalent to the gold standard RT-qPCR method commonly employed for SARS-CoV-2 detection [30], and it can detect just 10 copies of a target RNA molecule per microliter. Although still being evaluated, the CREST method has great potential to identify SARS-CoV-2 and other pathogens using accessible reagents and affordable equipment.
SENSR
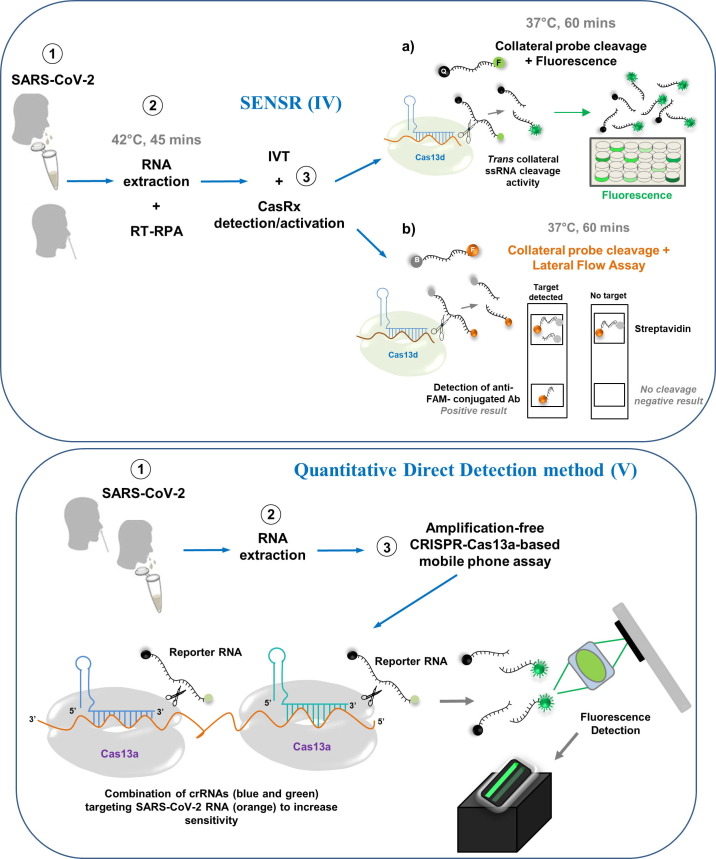
Recently, Brogan and colleagues [32] provided proof-of-principle that the Cas13d effector can be adapted as a point-of-care diagnostic through an approach termed Sensitive Enzymatic Nucleic-acid Sequence Reporter (SENSR) [32], which was derived from protocols developed initially for SHERLOCK.
SENSR is a two-step protocol that requires an initial isothermal pre-amplification reaction (45 minutes) combined with RT-RPA to produce a short double-stranded DNA amplicon containing a T7 promoter sequence [32] (Fig. 1 IV). This CRISPR-based nucleic acid molecular diagnostic tool harnesses the off-target collateral cleavage property of Cas13d ribonuclease derived from Ruminococcus flavefaciens (CasRx) to detect highly conserved SARS-CoV-2 viral sequences, in both synthetic templates and infected patient isolates [32] (Fig. 1 IV). Results are obtained via fluorescence-based readout or paper-based lateral flow assay, and the total reaction time is just two hours [32].
The LOD for SENSR was determined by fluorescence readout to be ~ 100 copies/µl (with variability between the gRNAs employed), indicating that SENSR also exhibits attomolar sensitivity and that CasRx can robustly detect and report the presence of synthetic SARS-CoV-2 RNA (E- and N-gene target templates) [32], with potential similar to those of other CRISPR-based systems [16], [29], [30], [33].
To determine the capability of SENSR to detect SARS-CoV-2 in infected patient samples, Brogan and colleagues [32] performed fluorescence detection analysis on 42 RT-qPCR validated positive (n = 21) and negative (n = 21) patient isolates [32]. SENSR yielded no false positives among the negative patient samples, demonstrating 100% specificity (0/21). In RT-qPCR analysis, a lower Ct value suggests a higher viral load within a patient sample. In this regard, SENSR detected SARS-CoV-2 RNA up to a maximum Ct value of ≤ 28, with a moderate false-negative rate of 25% (4/16) [32].
To examine whether SENSR can function as a point-of-care diagnostic test, Brogan and colleagues [32] also performed lateral flow analysis on 12 positive samples, observing 92% (11/12) concordance with SENSR fluorescence analysis. These data suggest that SENSR can be successfully adapted for the rapid detection of SARS-CoV-2 in patient samples [32]. Nevertheless, the scalability of SENSR for the identification of SARS-CoV-2 and other viral pathogens on a large scale, by multiplexed configuration, needs to be determined. Before thinking about this approach, it would be desirable for SENSR to be compatible with a one-step molecular diagnostic [32].
Cas13-based direct-detection assay: A quantitative approach
Recently, Doudna’s group developed a ground-breaking method for the direct detection of viral SARS-CoV-2 RNA that employs CRISPR-Cas13a without a pre-amplification step for the viral genome (Fig. 1 V). Unlike previous CRISPR-Cas13 diagnostics, this novel platform, which is based on LbuCas13a (the Cas13a homolog from Leptotrichia buccalis), yields quantitative RNA measurements rather than only a positive or negative result (change in color). The detection of viral RNA takes place directly in the sample without additional manipulations [34]. The strategy to increase the activity of Cas13a was through the use of multiple crRNAs along the N gene of SARS-CoV-2, which work in tandem to increase the sensitivity of the test. The rationale was that a single target RNA could activate multiple Cas13a ribonucleoproteins (RNPs). If each RNP is directed to different regions of the same viral target RNA, this could effectively double the active enzyme concentration [34]. Thus, to demonstrate the advantage of crRNA combinations, two crRNAs were pooled in the same reaction, keeping the total concentration of Cas13a RNPs constant but divided equally between RNPs made with each crRNA. Interestingly, when both crRNAs were combined, both target detection and reaction sensitivity were increased markedly when measured with a fixed in vitro transcription (IVT) target RNA concentration (480 fM) [34]. This approach allowed detection of ~ 100 copies/µl of SARS-CoV-2 viral RNA within 30 minutes, measuring changes in fluorescence over time rather than detecting only endpoint fluorescence [34].
In recent years, notably, smartphones have been adapted as diagnostic technologies that can replace some of the functionality of conventional laboratory equipment, simplifying diagnostic workflow by automating readout [35]. In this regard, the simplicity and portability of this assay were demonstrated by using a mobile-phone-based fluorescence microscope [34]. This microscope measures the fluorescent signal generated by the Cas13a direct-detection assay through a compact device, which includes low-cost laser illumination and collection optics [34] (Fig. 1 V). This compact device was approximately an order of magnitude more sensitive than a plate reader thanks to reduced measurement noise and the ability to collect more time points [34].
The functionality of this platform was validated with SARS-CoV-2 positive patient samples extracted from nasal swabs (with Ct values from 14.37 to 22.13). The accuracy was perfect, with positive results being detected within the first 5 minutes of the assay, suggesting that the fluorescence signal detected directly with Cas13a was proportional to the concentration of target RNA in the sample. Hence, this platform can provide a speedy results quickly for patients with high viral loads [34].
CARMEN-Cas13
Emerging diseases represent significant threats to human health, so it is necessary to have molecular tests available for the diagnosis and surveillance of the vast majority of disease-causing pathogens [36]. In this respect, the recent viral outbreak has highlighted the challenges of detecting known and putative unknown coronaviruses, and of differentiating between related viruses (for example, influenza (flu) virus and other respiratory viruses) that cause infections with similar symptoms [36].
The development of SHERLOCK creatively laid a foundation that could be highly multiplexed and easily scaled for simultaneous pathogen detection [24]. Multiplexing is an inherent and powerful advantage of Cas13-based approaches, allowing such methods to: 1) target a single viral genome at multiple sites; 2) differentiate between related viruses or serotypes; and 3) detect different mutations within SARS-COV-2 genomes, facilitating the simultaneous detection of co-circulating variants of SARS-CoV-2 or other coronavirus family members with high fidelity [23], [24], [37].
Sabeti’s group developed a CRISPR-based molecular diagnostics platform named CARMEN (Combinatorial Arrayed Reactions for Multiplexed Evaluation of Nucleic acids), which uses miniaturized and self-organizing microfluidic technology to enable massive multiplexing of any circulating pathogen in parallel, quickly and inexpensively [36]. In the CARMEN platform, nanoliter droplets containing CRISPR-based nucleic acid detection reagents self-organize in a microwell-array system to pair with droplets of amplified samples, testing each sample against each crRNA in replicate (Fig. 2 ).
Figure 2.
Schematic of nucleic acid detection by CARMEN–Cas13. (a) Identification of multiple circulating pathogens in human and animal species. (b) Depiction of the CARMEN–Cas13 workflow. ① After sample collection, RNA extraction, and amplification of the nucleic acid input, amplified targets are converted to RNA via T7 transcription and detected by Cas13 − crRNA RNP complexes. The resulting collateral cleavage activity of Cas13 produces a signal using a cleavage reporter RNA. In parallel, detection mixes are assembled, color-coded, and emulsified. ② Droplets from each emulsion are pooled into a single tube and mixed by pipetting. ③ The droplets are loaded onto a chip in a single pipetting step, during which the droplets self-organize into pairs. ④ Each microwell is imaged by fluorescence microscopy to identify the color code and to map the position of each droplet on the chip. ⑤ Droplets are merged, initiating the detection reaction. ⑥ After incubation, the detection reaction in each microwell can be monitored over time by fluorescence microscopy.
Ackerman et al. [36] employed CARMEN-Cas13 to test dozens of samples for more than 4500 crRNA–target pairs on a single array. This allowed the simultaneous differentiation of the samples for all 169 human-associated viruses, among them influenza A strains (haemagglutinin H1–H16 and neuraminidase N1–N9 subtypes), dozens of drug-resistance-associated mutations in HIV (six drug-resistance mutations that are prevalent in antiviral-naive patient populations), and the novel coronavirus SARS-CoV-2 [36]. Interestingly, the authors were able to incorporate a new test for SARS-CoV-2 into the coronavirus panel already on the array comprising human coronaviruses 229E, NL63, and HKU1 Middle East respiratory syndrome (MERS) coronavirus [36].
CARMEN-Cas13 offers reduced sample consumption per test, fewer pipetting steps, and shorter turnaround time, and decreases reagent cost per test by more than 300-fold. Furthermore, as we have seen for SARS-CoV-2, this platform can be optimized and scaled rapidly to detect newly discovered pathogen sequences by incorporating the specific amplification primers or crRNAs for the novel target sequences [36]. Hence, the multiplexing and high-throughput capabilities of the CARMEN–Cas13 platform make it possible to detect and differentiate viral sequences in large cohorts of people, allowing better epidemiological surveillance.
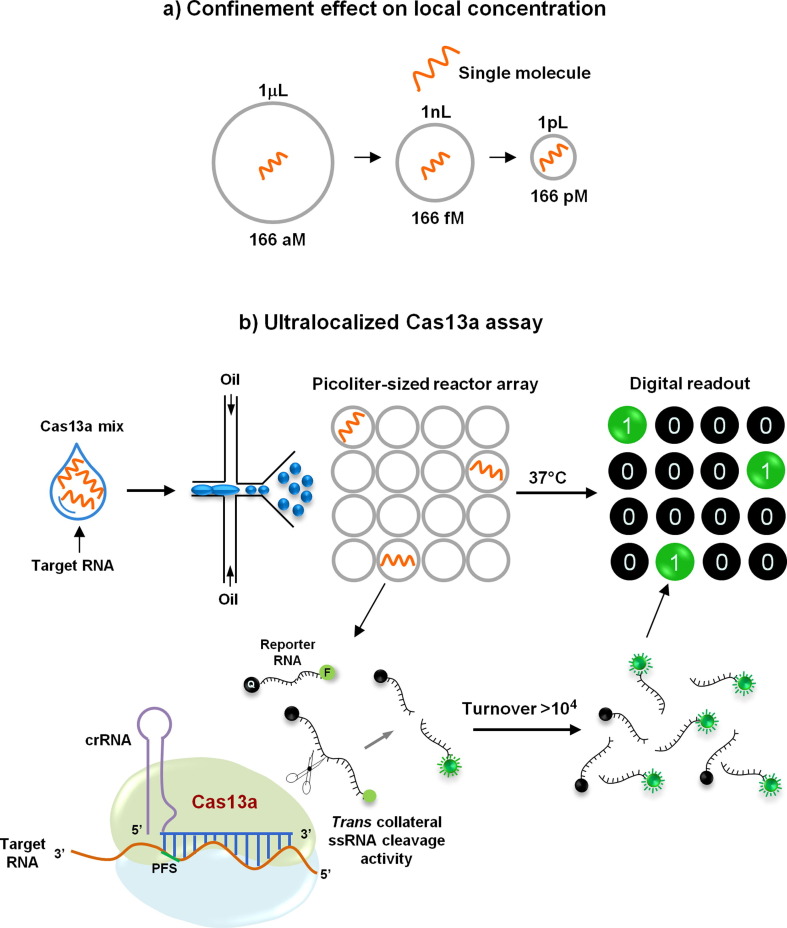
Ultralocalized Cas13a assay
The local concentration of a single molecule increases inversely with decreasing analytical volume. Exploiting this, Tian and colleagues very recently developed a droplet-digital Cas13a assay named ultralocalized Cas13a assay by harnessing the natural confinement effect for highly efficient reaction or detection [38] (Fig. 3 a). They employed a simple droplet microfluidic platform to enhance the local concentration of target sequence and RNA reporter for the Cas13a system in cell-sized volumes [38] (Fig. 3 b). In their method, target RNAs together with a Cas13a mixture are emulsified with oil into thousands of picoliter-sized droplet reactors, in which the fluorescent signal accumulated from a single RNA target activated by collateral cleavage by Cas13a is sufficient to illuminate a picoliter-sized droplet [38] (Fig. 3 b). After recognition by a crRNA, a single-target RNA can induce the cleavage of over 104 quenched fluorescent RNA reporters, yielding a fluorescently positive droplet. As a result, the ultralocalized Cas13a assay allows absolute digital quantitation (according to the Poisson distribution) of single unlabeled RNA molecules without the need for reverse transcription and sequence amplification [38].
Figure 3.
Schematic of the ultralocalized Cas13a assay. (a) Confinement effect on local concentration. The presence of a single molecule in a bulk volume, such as one microliter, yields an aM level molecular concentration. If the single-molecule is confined in a volume of 1nL or 1pL, its local concentration will increase to 1.66 fM or 1.66 pM, respectively [38]. (b) Workflow for the ultralocalized Cas13a assay. Target RNAs (orange) and a Cas13 mix are emulsified with oil into thousands of picoliter-sized droplets. Once recognized by a ~ 28-base CRISPR RNA (crRNA) (blue), one target RNA will induce the cleavage of 104 quenched fluorescent RNA reporters (F, fluorescence; Q, quencher), yielding a fluorescently positive droplet. The high-yielding fluorescent signal confined in an ultrasmall volume could be at once accumulated to illuminate a droplet. In the absence of the RNA target, no fluorescence is detected. If a single RNA target molecule is sufficient to ‘illuminate’ a droplet, digital counting of single RNA molecules can be achieved by assigning each positive droplet as ‘one’ and each negative droplet as ‘zero’ [38]. PFS, protospacer flanking site; ssRNA, single-stranded RNA.
The applicability of this novel approach was demonstrated not only for precise counting of cell-free microRNAs from clinical serum samples but also for the ultrasensitive detection of 16S rRNA from uropathogens and for the accurate diagnosis of SARS-CoV-2 (via detection of the N gene) from 40 nasopharyngeal swabs specimens collected from suspected COVID-19 patients with 100% concordance with RTqPCR results [38]. The effectiveness of this assay was evaluated using serially diluted SARS-CoV-2 RNA standards with N gene crRNA, and the authors observed a reproducible detection limit down to ~6 copies/μl and linearity (R2 = 0.99987) over 104-fold enhancement [38].
The sensitivity, the single-molecule quantitation capability, and the broad applicability of this method suggest that it is a promising RNA diagnostic tool with potential value in responses to outbreaks of infectious diseases such as COVID-19 [38].
Point-of-care testing by CRISPR-Cas13 sensors: Challenges and opportunities
For now, there is no targeted therapy for SARS-COV-2, and there are limited vaccines available, so that the best way to deal with the contagion is to control the source of infection and to achieve widespread early diagnosis [39]. In the face of an outbreak, healthcare system laboratories must have the ability to perform the tests required to detect the disease-causing pathogen in different biological samples.
Point-of-care tests avoid the need to send samples to centralized facilities, thereby enabling communities without laboratory infrastructure access to rapid diagnostics [40]. CRISPR-Cas13-based technologies can be applied as fast, accurate, and portable diagnostic assays for emerging infectious diseases. Thus, the CRISPR-Cas13 system can be used, in combination with point-of-care devices, as highly effective nucleic acid detectors that allow tests to be conducted and interpreted in various settings [40], including in low resource communities where non-specialist equipment and staff must be used.
As the COVID-19 pandemic accelerates worldwide, many low- and middle-income countries (LMICs) are still struggling to access the diagnostic tests they urgently need to bring the disease under control. Supplies (reagents or analytical equipment) can take months to deliver, with stocks frequently sold to the highest bidder [41]. Another hurdle is that many LMICs have no domestic capacity to manufacture diagnostic tests, and thus they have a high dependence on imports [41].
In the face of this health emergency and to overcome these limitations, developers of new diagnostic platforms need to seek approvals by the United States Food and Drug Administration (US FDA), or other international regulatory agencies, for their use in point-of-care tests under ‘Emergency Use Authorizations’. The CRISPR-Cas13 (SHERLOCK)-based test is the only new platform to have received such approval to date [42]. Further accurate, and portable diagnostic tests for the novel coronavirus could be implemented in the clinical setting once they obtain ‘Emergency Use Authorization’, providing a powerful complement to current standard RT-qPCR detection.
On the other hand, the Foundation for Innovative New Diagnostics (FIND) [43], a Geneva-based not-for-profit that facilitates the development and delivery of diagnostics to low-income countries, is inviting assay developers to submit their inventions for independent evaluation in collaboration with the World Health Organization (WHO) [44]. FIND has prioritized the review of rapid diagnostic tests according to: 1) regulatory status and time to market, 2) the manufacturing and distribution capacity of the supplier, and 3) clinical and analytical performance [43]. FIND also provides online training on carrying out diagnostic tests and advice about the most appropriate tests for particular uses in different countries [43]. This prioritization could be an opportunity that LMICs could use to access point-of-care testing that is based on the new CRISPR-Cas13 diagnostic platforms for massive deployment. CRISPR-Cas13-based diagnosis could be implemented to detect SARS-COV-2/COVID-19 and to help with rapid decision-making, principally in the weakest health systems where access to clinical laboratories can be challenging.
In addition, CRISPR-based diagnostic tests could be used at the main ports of entry and border crossings for rapid and accurate identification of cases of this new coronavirus (even in asymptomatic people who are subjected only to body temperature measurements or rapid diagnosis in serology), potentially prompting a strategy for action by health authorities that could limit disease spread [39], [40]. When performing these tests in a point-of-care setting, however, it will be necessary to provide the advice necessary to allow these tests to be handled by different personnel or used with varying biological sample qualities. Coordination between public and academic laboratories will also be needed to accelerate the implementation of next-generation CRISPR-based diagnostics for coronaviruses detection using a multi-disciplinary and highly collaborative approach involving scientists, clinicians, engineers, and public health practitioners. Such an approach could facilitate not only early detection of SARS-COV-2 cases in humans, but also detection of the virus in many species of live animals (e.g., pangolins and mink). The screening of large cohorts of people will provide real-world scenarios to test the fidelity of CRISPR-based platforms in diagnosing SARS-CoV-2 and other pathogens that could cause public health crises.
Concluding remarks
Regular testing can ascertain foci of infection and reduce false-negative rates [45], but comes with significant challenges, especially in many low- and middle-income countries where access to clinical laboratories can be an issue and when the pressure to get results quickly increases. To address the dire need for increased testing, researchers of several disciplines have quickly developed novel laboratory solutions to optimize the SARS-CoV-2/COVID-19 testing pipeline, adapting available commercial reagents, shortening protocol durations and skipping RNA extraction steps [29], [30], [32].
During the current COVID-19 outbreak, CRISPR-Cas13-based platforms have been recognized as novel molecular tools that have compelling programmability, specificity, and efficiency. Creative ways to take advantage of the CRISPR-Cas13 effector in different testing formats have been adapted or newly developed. Remarkable progress in CRISPR-Cas13-based approaches has provided rapid detection (<1h), with high sensitivity (attomolar limits of detection), and high-throughput [36] of SARS-CoV-2 RNA, on multiple specimen types, without the need for specialized infrastructure [26], [27], [29], [30], [32], [34].
Through these recently developed CRISPR-Cas13 based testing platforms, it is possible to integrate target recognition and sense capabilities into a single device through paper-based sensors or even single-tube reactions with easy-to-interpret accessible readout methods (such as a smartphone-based reader) [26], [29], [30], [32], [34]. Most of these CRISPR sensing methods could even be adapted to monitor the viral load of COVID-19 patients or to evaluate therapeutic efficacy in patients receiving antiviral therapy. They could also be reconfigured to detect new viruses that threaten human health.
These novel diagnostic modalities could enhance the current efforts to identify virus reservoirs, to elucidate transmission routes, and to design strategies to limit the spread of the disease until an effective countermeasure to this new coronavirus becomes available to everyone.
Acknowledgments
The author would like to thank Dr Javier T. Granados-Riveron for helpful discussion of this manuscript. G.A.-J. is supported by grant SEP-CONACyT-CB-286421 from the Mexican Council of Science and Technology (CONACyT).
References
- 1.Phelan A.L., Katz R., Gostin L.O. The novel coronavirus originating in Wuhan, China: challenges for global health governance. JAMA. 2020;323:709–710. doi: 10.1001/jama.2020.1097. [DOI] [PubMed] [Google Scholar]
- 2.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Esbin M.N., Whitney O.N., Chong S., Maurer A., Darzacq X., Tjian R. Overcoming the bottleneck to widespread testing: a rapid review of nucleic acid testing approaches for COVID-19 detection. RNA. 2020;26:771–783. doi: 10.1261/rna.076232.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 5.Long Q.X., Tang X.J., Shi Q.L., Li Q., Deng H.J., Yuan J., et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat Med. 2020;26:1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- 6.Lan L., Xu D., Ye G., Xia C., Wang S., Li Y., Xu H. Positive RT-PCR test results in patients recovered from COVID-19. JAMA. 2020;323:1502–1503. doi: 10.1001/jama.2020.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 8.Jolany Vangah S., Katalani C., Booneh H.A., Hajizade A., Sijercic A., Ahmadian G. CRISPR-based diagnosis of infectious and noninfectious diseases. Biol Proced Online. 2020;22:22. doi: 10.1186/s12575-020-00135-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koonin E.V., Makarova K.S., Zhang F. Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol. 2017;37:67–78. doi: 10.1016/j.mib.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Makarova K.S., Haft D.H., Barrangou R., Brouns S.J.J., Charpentier E., Horvath P., et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shmakov S., Abudayyah O.O., Makarova K.S., Wolf Y.I., Gootenberg J.S., Semenova E., et al. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol Cell. 2015;60:385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abudayyeh O.O., Gootenberg J.S., Konermann S., Joung J., Slaymaker I.M., Cox D.B.T., et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:aaf5573. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.East-Seletsky A., O'Connell M.R., Knight S.C., Burstein D., Cate J.H.D., Tjian R., Doudna J.A. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature. 2016;538:270–273. doi: 10.1038/nature19802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu L., Li X., Wang J., Wang M., Chen P., Yin M., et al. Two distant catalytic sites are responsible for C2c2 RNase activities. Cell. 2017;168:121–134. doi: 10.1016/j.cell.2016.12.031. [DOI] [PubMed] [Google Scholar]
- 15.Liu L., Li X., Ma J., Li Z., You L., Wang J., et al. The molecular architecture for RNA-guided RNA cleavage by Cas13a. Cell. 2017;170:714–726. doi: 10.1016/j.cell.2017.06.050. [DOI] [PubMed] [Google Scholar]
- 16.Gootenberg J.S., Abudayyeh O.O., Lee J.W., Essletzbichler P., Dy A.J., Joung J., et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan W.X., Chong S., Zhang H., Makarova K.S., Koonin E.V., Cheng D.R., Scott D.A. Cas13d is a compact RNA-targeting type VI CRISPR effector positively modulated by a WYL-domain-containing accessory protein. Mol Cell. 2018;70:327–339. doi: 10.1016/j.molcel.2018.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cox D.B.T., Gootenberg J.S., Abudayyeh O.O., Franklin B., Kellner M.J., Joung J., Zhang F. RNA editing with CRISPR-Cas13. Science. 2017;358:1019–1027. doi: 10.1126/science.aaq0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abudayyeh O.O., Gootenberg J.S., Essletzbichler P., Han S., Joung J., Belanto J.J., et al. RNA targeting with CRISPR-Cas13. Nature. 2017;550:280–284. doi: 10.1038/nature24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang C., Konermann S., Brideau N.J., Lofty P., Wu X., Novivk S.J., et al. Structural basis for the RNA-guided ribonuclease activity of CRISPR-Cas13d. Cell. 2018;175:212–223. doi: 10.1016/j.cell.2018.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Konermann S., Lofty P., Brideau N.J., Oki J., Shokhirev M.N., Hsu P.D. Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell. 2018;173:665–676. doi: 10.1016/j.cell.2018.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kellner M.J., Koob J.G., Gootenberg J.S., Abudayyeh O.O., Zhang F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat Protoc. 2019;14:2986–3012. doi: 10.1038/s41596-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Myhrvold C., Freije C.A., Gootenberg J.S., Abudayyeh O.O., Metsky H.C., Durbin A.F., et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science. 2018;360:444–448. doi: 10.1126/science.aas8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gootenberg J.S., Abudayyeh O.O., Kellner M.J., Joung J., Collins J.J., Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360:439–444. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qin P., Park M., Alfson K.J., Tamhankar M., Carrion R., Patterson J.L., et al. Rapid and fully microfluidic Ebola virus detection with CRISPR Cas13a. ACS Sens. 2019;4:1048–1054. doi: 10.1021/acssensors.9b00239. [DOI] [PubMed] [Google Scholar]
- 26.Zhang F, Abudayyeh OO, Gootenberg JS. A protocol for detection of COVID-19 using CRISPR diagnostics. https://www.broadinstitute.org/files/publications/special/COVID-19%20detection%20(updated).pdf. Published March 21, 2020. Accessed August 23, 2020.
- 27.Patchsung M., Jantarug K., Pattama A., Aphicho K., Suraritdechachai S., Meesawat P., et al. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat Biomed Eng. 2020;4:1140–1149. doi: 10.1038/s41551-020-00603-x. [DOI] [PubMed] [Google Scholar]
- 28.Joung J., Ladha A., Saito M., Kim N.G., Woolley A.E., Segel M., et al. Detection of SARS-CoV-2 with SHERLOCK one-pot testing. N Engl J Med. 2020;383:1492–1494. doi: 10.1056/NEJMc2026172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arizti-Sanz J., Freije C.A., Stanton A.C., Petros B.A., Boehm C.K., Siddiqui S., et al. Streamlined inactivation, amplification, and Cas13-based detection of SARS-CoV-2. Nat Commun. 2020;11:5921. doi: 10.1038/s41467-020-19097-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rauch N.J., Valois E., Solley S.C., Braig F., Lach R.S., Audouard M., et al. A scalable, easy-to-deploy, protocol for Cas13-based detection of SARS-CoV-2 genetic material. J Clin Microbiol. 2021;59:e02402–e2420. doi: 10.1128/JCM.02402-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ponce-Rojas J.C., Costello M.S., Proctor D.A., Kosik K.S., Wilson M.Z., Arias C., Acosta-Alvear D. A fast and accessible method for the isolation of RNA, DNA, and protein to facilitate the detection of SARS-CoV-2. Clin Microbiol. 2021;59:e02403–e2420. doi: 10.1128/JCM.02403-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brogan DJ, Chaverra-Rodriguez D, Lin CP, Smidler AL, Yang T, Alcantara LM, et al. A sensitive, rapid, and portable CasRx-based diagnostic assay for SARS-CoV-2. medRxiv 2020; 2020.10.14.20212795. [DOI] [PMC free article] [PubMed]
- 33.Chen J.S., Ma E., Harrington L.B., Da Costa M., Tian X., Palefsky J.M., Doudna J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360:436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fozouni P., Son S., de León Derby M.D., Knott G.J., Gray C.N., D'Ambrosio M.V., et al. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell. 2021;184:323–333. doi: 10.1016/j.cell.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malekjahani A., Sindhwani S., Syed A.M., Chan W.C.W. Engineering steps for mobile point-of-care diagnostic devices. Acc Chem Res. 2019;52:2406–2414. doi: 10.1021/acs.accounts.9b00200. [DOI] [PubMed] [Google Scholar]
- 36.Ackerman C.M., Myhrvold C., Thakku S.G., Freije C.A., Metsky H.C., Yang D.K., et al. Massively multiplexed nucleic acid detection with Cas13. Nature. 2020;582:277–282. doi: 10.1038/s41586-020-2279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chertow D.S. Next-generation diagnostics with CRISPR. Science. 2018;360:381–382. doi: 10.1126/science.aat4982. [DOI] [PubMed] [Google Scholar]
- 38.Tian T., Shu B., Jiang Y., Ye M., Liu L., Guo Z., et al. An ultralocalized Cas13a assay enables universal and nucleic acid amplification-free single-molecule RNA diagnostics. ACS Nano. 2021;15:1167–1178. doi: 10.1021/acsnano.0c08165. [DOI] [PubMed] [Google Scholar]
- 39.Udugama B., Kadhiresan P., Kozlowski H.N., Malekjahani A., Osborne M., Li V.Y.C., et al. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020;14:3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- 40.van Dongen J.E., Berendsen J.T.W., Steenbergen R.D.M., Wolthuis R.M.F., Eijkel J.C.T., Segerink L.I. Point-of-care CRISPR/Cas nucleic acid detection: recent advances, challenges and opportunities. Biosens Bioelectron. 2020;166 doi: 10.1016/j.bios.2020.112445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peplow M. Developing countries face diagnostic challenges as the COVID-19 pandemic surges. https://cen.acs.org/analytical-chemistry/diagnostics/Developing-countries-face-diagnostic-challenges/98/i27. Published June 26, 2020. Accessed September 8, 2020.
- 42.U.S. Food & Drug Administration. Emergency use authorizations for medical devices. https://www.fda.gov/medical-devices/emergency-situations-medical-devices/emergency-use-authorizations-medical-devices. Accessed October 26, 2020.
- 43.Foundation for the Innovation of Research Diagnostics (FIND). COVID-19 diagnostics & testing. https://www.finddx.org/covid-19/. Accessed November 30, 2020.
- 44.Sheridan C. Fast, portable tests come online to curb coronavirus pandemic. Nat Biotechnol. 2020;38:515–518. doi: 10.1038/d41587-020-00010-2. [DOI] [PubMed] [Google Scholar]
- 45.Jarvis KF, Kelley JB. Temporal dynamics of viral load and false negative rate influence the levels of testing necessary to combat COVID19 spread. medRxiv 2020; 2020.08.12.20173831. [DOI] [PMC free article] [PubMed]