Abstract
Computer Tomography (CT) detection can effectively overcome the problems of traditional detection of Corona Virus Disease 2019 (COVID-19), such as lagging detection results and wrong diagnosis results, which lead to the increase of disease infection rate and prevalence rate. The novel coronavirus pneumonia is a significant difference between the positive and negative patients with asymptomatic infections. To effectively improve the accuracy of doctors' manual judgment of positive and negative COVID-19, this paper proposes a deep classification network model of the novel coronavirus pneumonia based on convolution and deconvolution local enhancement. Through convolution and deconvolution operation, the contrast between the local lesion region and the abdominal cavity of COVID-19 is enhanced. Besides, the middle-level features that can effectively distinguish the image types are obtained. By transforming the novel coronavirus detection problem into the region of interest (ROI) feature classification problem, it can effectively determine whether the feature vector in each feature channel contains the image features of COVID-19. This paper uses an open-source COVID-CT dataset provided by Petuum researchers from the University of California, San Diego, which is collected from 143 novel coronavirus pneumonia patients and the corresponding features are preserved. The complete dataset (including original image and enhanced image) contains 1460 images. Among them, 1022 (70%) and 438 (30%) are used to train and test the performance of the proposed model, respectively. The proposed model verifies the classification precision in different convolution layers and learning rates. Besides, it is compared with most state-of-the-art models. It is found that the proposed algorithm has good classification performance. The corresponding sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and precision are 0.98, 0.96, 0.98, and 0.97, respectively.
Keywords: COVID-19 classification network, Convolution, Deconvolution, ROI extraction, Enhanced CT features
Graphical abstract
1. Introduction
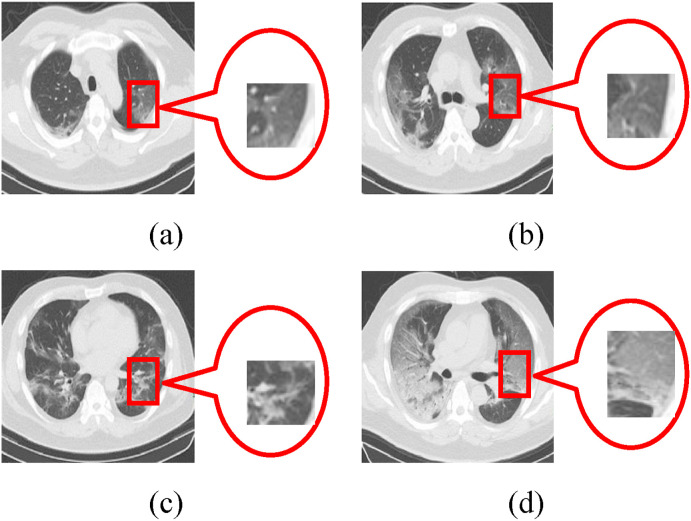
Corona Virus Disease 2019 (COVID-19) [1,2], which broke out in December 2019, is the seventh known coronavirus that can infect human beings. The International Committee on Taxonomy of viruses has been named severe acute respiratory syndrome coronavirus 2 (SARS-COV-2). As of May 2021, Beijing time, there are more than 157.5 million confirmed cases and 3.2 million deaths worldwide, as shown in Fig. 1 (a) [3,4].
Fig. 1.
The COVID-19 CT images from multi-angles. (a) The process of COVID-19 infection; (b) The CT image from multi-angles.
Due to the short-term shortage of nucleic acid detection kits, the positive result of novel coronavirus detection lag behind, which makes some patients with COVID-19 do not receive timely treatment. It not only causes relatively typical lung lesions, but also increases the infection rate of novel coronavirus pneumonia [5,6]. As a faster and more convenient means of examination, imaging examination [[7], [8], [9]] can effectively reduce the impact of patients that infected with COVID-19 and plays an important role in the treatment of novel coronavirus. At present, X-ray [[10], [11], [12]] and Computer Tomography (CT) [[13], [14], [15]] images are the main screening ways. The X-ray image is formed based on different X-rays absorbed by different tissues of the human body. Narin A et al. [10], Hemdan E E D et al. [11], and Parnian Afshar et al. [12] detect the infection status of COVID-19 on the X-ray image, which can help the diagnosis of novel pneumonia to some extent. However, due to the overlapping of tissue structures in an X-ray image, it is difficult to distinguish the negative and positive of COVID-19.
The horizontal lung CT image can effectively solve this problem. It can effectively judge the tumor region of the chest wall and bronchial cartilage calcification according to the image structure of bilateral lung middle bronchus and chest, as shown in Fig. 1(b) [16]. Here, lung consolidation shadow is a process in which the alveoli are filled with exudate. Central local consolidation is the feature overlap of the old lung lesions and the COVID-19. Ground glass shadow refers to the thin cloud-like shadow with slightly increased density in chest CT scan, which can grow diffusely or only locally. Actually, the diagnosis of COVID-19 is mainly judged by ground glass shadow. It can not only overcome the limitations of nucleic acid detection technology that may cause a false-negative result, but also implement better treatment for asymptomatic or negative CT image of patients infected with COVID-19. Chua F et al. [17] systematically introduce the related symptoms of infection with COVID-19. Through the comparison of data and information, it concludes that the use of CT image to judge the status of COVID-19 meets the needs of current disease diagnosis. Fang Y et al. [18] and Bernheim A et al. [19] find that the severity of the disease gradually increased by analyzing extensive CT image. Therefore, how to realize the monitoring of COVID-19 infection has become an important way to control the spread of the epidemic effectively and efficiently.
Effective measures can be taken to solve the problem of the high infection rate of novel coronavirus for patients that diagnosed with COVID-19 infection. Doctors usually make an artificial judgment according to the CT image [20,21] of the lung and judge whether there is a substantial lesion by observing the exudation of coronavirus from granulocytes. However, there is a high demand for doctors' theoretical knowledge and practical ability. In this case, machine learning plays a more and more important role [[22], [23], [24]]. Here, the deep neural network can obtain a diagnosis model with high precision, which can get the characteristics of the novel coronavirus infection through self-learning. The Deep network model [[25], [26], [27], [28]] simulates the process of human brain neurons transmitting signals to obtain information, which can effectively extract the image features. The convolutional neural network (CNN) has been widely used and has become a classic classification model.
For example, Mohamed Loey et al. [29] propose an algorithm for classifying COVID-19 image using a deep transfer learning model. This method enhances CT image through deep revolutionary neural networks to generate more mixed data for training. On the basis, Gifani P et al. [30] propose an automatic learning method based on an integrated deep transfer learning system. This algorithm is used to diagnose the negative and positive results of COVID-19 by using the optimal combination of deep transfer learning outputs. Hall Lo et al. [31] propose a COVID-19 classification algorithm based on Visual Geometry Group16 (VGG16) model. The proposed method changes the last layer of the VGG16 model into a trainable part. The final classification result is obtained through a fully connected layer composed of the global average pooling. The above-proposed algorithms [[29], [30], [31]] can get high performance at a low computational cost. However, the computational efficiency in the open dynamic environment is low, which can not meet the precision and strength required by the diagnosis of COVID-19.
To solve the problem, Feng Shi et al. [32] propose a VB-Net algorithm, which combines the V-Net model with the bottleneck layer. This method extracts the shrinking and expanding paths, which integrates the fine-grained COVID-19 image features. Thus, it can reduce the number of feature mapping channels and effectively increase the convolution speed. Eduardo Luz et al. [33] place the target class in the leaf node of the tree by setting up hierarchical classification. The information is transmitted by the classifier on the middle node and the classification task of COVID-19 is carried out at the root node of the tree. Similarly, Sanhita Basu et al. [34] propose a new extended transfer learning algorithm. The feature of the COVID-19 image is detected by grad cam and the final classification result is obtained. Singh D et al. [35] create a new CNN model to tune the initial parameters through multi-objective differential evolution. The classification precision of this algorithm is improved, but its efficiency is still slightly insufficient. To solve this problem, Polsinelli M et al. [36] propose a processing strategy to enable GPU acceleration in the nvidacuda core environment, which can further improve the performance. Besides, it reduces the requirements of deep learning on equipment space and speed. He X et al. [37] combine contrast self-supervised learning with transfer learning cooperatively, which can learn powerful feature representation. On the basis, Zhao J et al. [38] develop a diagnosis method of COVID-19 based on multi task learning and self-supervised learning. Bai H X et al. [39] establish an artificial intelligence system to classify the negative and positive of COVID-19. To evaluate the performance of radiologists, a two-layer fully connected neural network is used to gather the slices together.
For the above methods, the classification performance needs to be improved for a medical image dataset with a large sample distribution offset. When the learning depth deepens, the gradient dispersion phenomenon will appear, which results in local convergence and the occurrence of the overfitting phenomenon. To effectively reduce the problem of gradient disappearance and information confusion caused by the similarity of multiple lesions, this paper proposes a novel classification algorithm for COVID-19 lung CT image based on a deep convolutional network, which deepens the contrast by convolution and deconvolution operation. Besides, it can effectively detect novel coronavirus pneumonia and provide a new computer-aided method for epidemic diagnosis. Compared with the traditional CNN model, the proposed model has the following innovations:
-
●
The proposed model enhances the contrast between the local lesion region and the abdominal cavity of COVID-19 by convolution and deconvolution [40]. Thus, it can effectively overcome the problem of similar pixel values between the lesions and the normal background.
-
●
The proposed model transforms the novel coronavirus detection into a feature classification problem of the region of interest (ROI) [41], which can effectively determine whether the feature vector in each feature channel contains the image features of COVID-19.
-
●
The proposed algorithm obtains the stable expression of potential local COVID-19 features at all levels of the lung CT image. Thus, it can overcome gradient vanishing and realizes the reuse of feature information.
The paper is organized as follows. In Section II, the ROI localization of lung CT image and the enhancement of COVID-19 feature information by convolution and deconvolution are introduced. Section III introduces the framework of the proposed deep convolution network model and the corresponding training process. Section IV introduces the experimental result and analysis. The proposed deep network is compared with the other state-of-art classification algorithms. Finally, the conclusion is given in Section V.
2. Image preprocessing
2.1. The CT image characteristics of COVID-19 patients
For COVID-19, there are some problems such as alveolar swelling, thickening of the alveolar septum, and exudation of alveolar septal fluid [42,43]. Therefore, observing the suspected COVID-19 patients with lung CT image can effectively identify the status of virus infection.
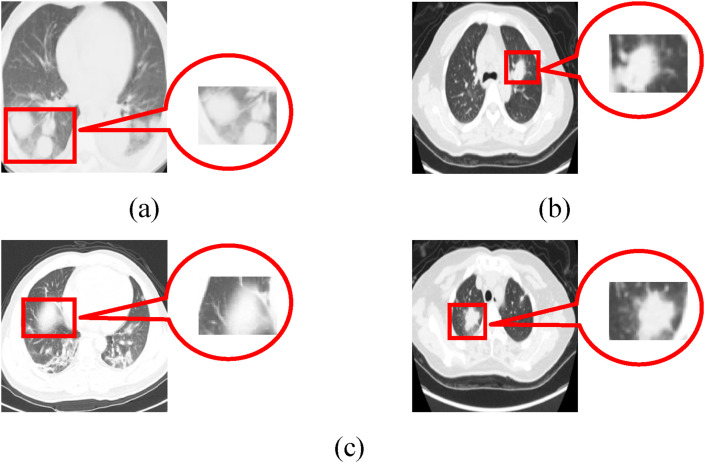
The CT image of COVID-19 patients shows single or multiple subsegmental or segmental speckle ground glass shadow at the early stage, as shown in Fig. 2 (a). In the advanced stage, the novel coronavirus has spread along the alveoli, and the large strain spread from the center to the periphery by the bronchioles. Compared with the early stage of infection, the number of lesions increases, the scope of bacterial infection expands, and the virus accumulates in multiple lobes. There are coexisting phenomena of fireworks like ground glass shadow and consolidation shadow or stripe shadow, as shown in Fig. 2(b)(c). Besides, a small number of patients are likely to be accompanied by fibrosis. Severe patients with diffuse lung lesions are often accompanied by large consolidation shadow, ground glass shadow, cord shadow, and air bronchogram. A few patients even have white lung, pleural effusion, and lymph node enlargement, as shown in Fig. 2(d).
Fig. 2.
The CT image of COVID-19 patients at different stages. (a) Ground glass shadow; (b) Fireworks like ground glass shadow; (c) Consolidation shadow; (d) White lung.
Due to different field offsets caused by medical equipment and the patient's own activities and breathing, there are some problems such as uneven gray and signal-to-noise ratio in the lung CT image. Besides, the image features between the novel coronavirus and other diseases are similar [44]. For the CT image of the acquired immune deficiency syndrome (AIDS), the image features also show that the boundary is fuzzy and the surrounding is accompanied by hazy ground glass shadow, as shown in Fig. 3 . Considering the multiplicity and complexity that exist in COVID-19 diseases, it is necessary to preprocess the CT image and enhance the local feature contrast of the COVID-19 lesion region.
Fig. 3.
Comparison of image features between the AIDS and COVID-19 diseases. (a) Image feature of AIDS; (b) Image feature of COVID-19; (c) Image feature of the coexistence of AIDS and COVID-19.
2.2. The contrast enhancement of the lesion region
To enhance the contrast between the normal background and the ROI of the lesion, it is necessary to recognize the region infected with COVID-19 of the lung CT image. According to a lot of statistics, the proportion of lung parenchyma in CT image is small, but the position of both lungs is relatively fixed. Besides, the distribution of the left and right lung parenchyma in the human thoracic cavity is relatively stable [[45], [46], [47]].
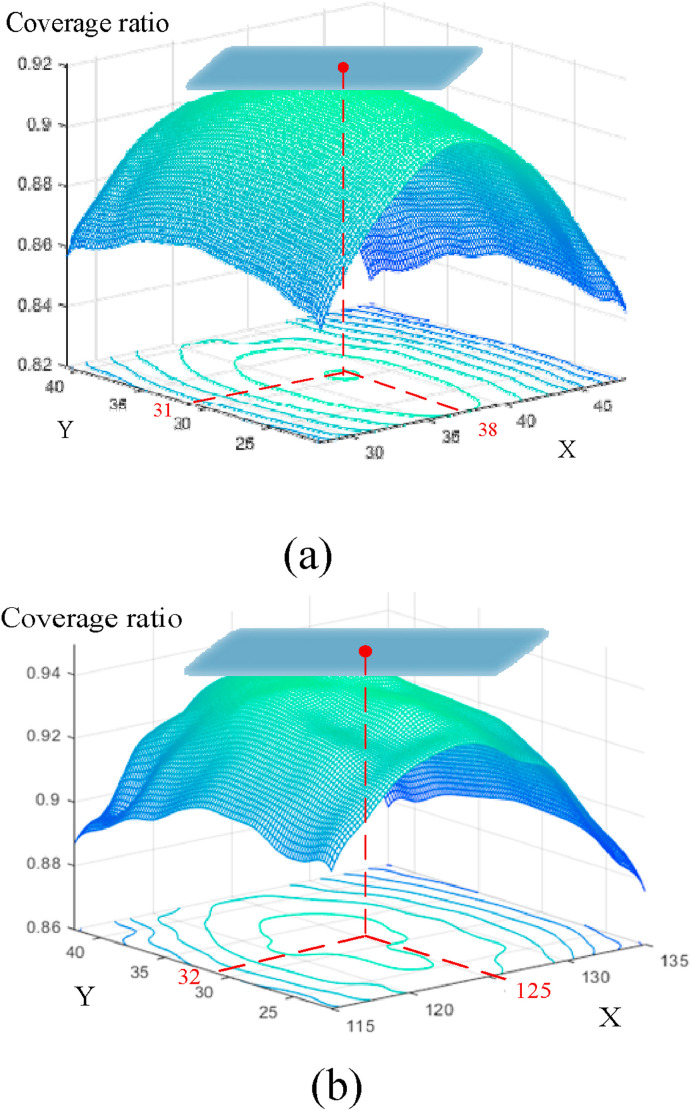
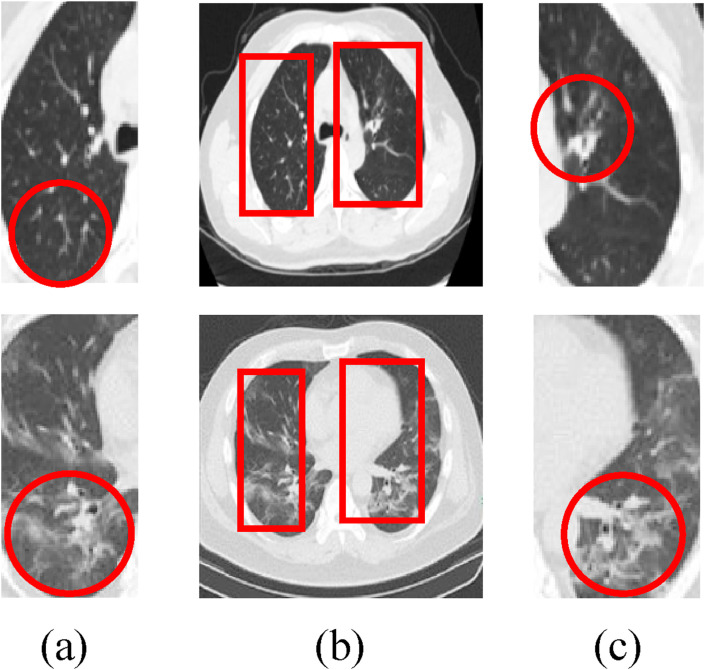
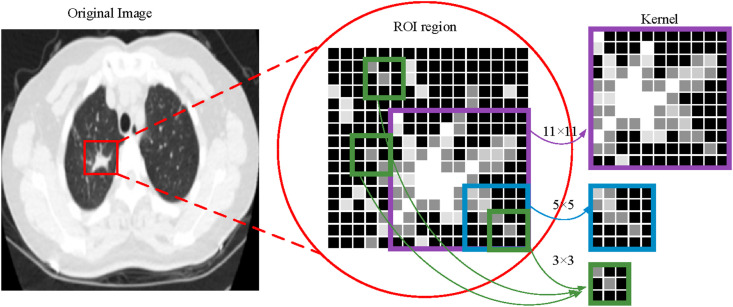
In this paper, the bilateral lung regions of 333 patients with COVID-19 and 397 normal people are analyzed, as shown in Fig. 4 . For image size 227*227, the extracted regions [38, 31, 65, 156] and [125, 32, 70, 158] in the left and right lungs can represent the effective distribution infected with COVID-19. The corresponding coverage ratio of the lesion region can reach 91% and 94% in the left and right lungs, respectively. Here, the first and last two numbers of the matrix represent the coordinates of the upper left corner and the width (height) of the extracted region, respectively. Fig. 5 shows the result of ROI extraction from lung CT image [48], which contains more complete image features.
Fig. 4.
The coverage ratio of the effective distribution infected with COVID-19. (a) Left lung; (b) Right lung.
Fig. 5.
The ROI extraction. (a) The ROI of left lung; (b) The extracted region of the lung CT image; (c) The ROI of the right lung.
For the COVID-19 CT image, the ground glass shadow may overlap with bronchial branches, bilateral pulmonary veins, and arteries. Besides, the characteristics of the fireworks like glass shadow and capillaries in the CT image are also similar. Therefore, this paper proposes a deconvolution network model based on the ROI to enhance the infection characteristics of the novel coronaviruses [49,50].
The lung CT image contains two ROI to be enhanced [51,52], where and are the left and right lung regions, respectively. is the convolution kernel size of (in this paper, is set). Suppose that the ROI is composed of feature channels . By finding the feature vector in each feature channel and the characteristic distribution kernel , the convoluted image is obtained
| (1) |
where and .
Through the maximum convolution kernel, the stable expression of potential local features at all levels of the image is obtained, which can effectively divide the normal background and the lesion region. To obtain more complex feature information and enhance the virus image, the deconvolution network is also considered in this paper [53].
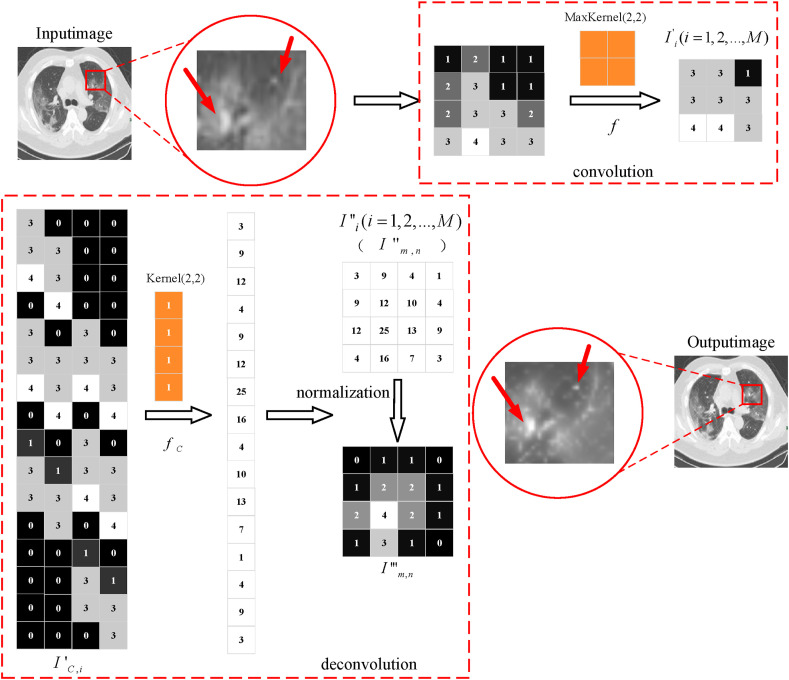
As shown in Fig. 6 , the obtained 3 × 3 local feature region is mapped to a 16 × 4 local sparse feature matrix . The local feature enhancement matrix with the same size as the original channel is obtained by multiplying it with the 4 × 1 deconvolution distribution kernel . Finally, the local deconvoluted image is obtained .
Fig. 6.
The process chart of the contrast enhancement.
Due to the existence of singular sample in the dataset, the training time will be increased, which may cause the problem that the network cannot converge. In this paper, the ROI extraction operation is used to obtain the lung region containing the lesion. Besides, the abdominal background is compressed to 0.8 times of the original image by enhancement algorithm. Then, the statistical distribution of samples is unified by matrix normalization. The concrete computational process is as follows:
| (2) |
where is the eigenvalue of row and column of , is the maximum eigenvalue of local deconvoluted eigenmatrix , is the maximum eigenvalue of pre-trained convolution matrix, is the local characteristic matrix after deconvolution normalization. (In this paper, is the original image, is the convoluted image, is the deconvoluted image, and is the matrix normalized image. is the local characteristic channel, and are the matrix index.).
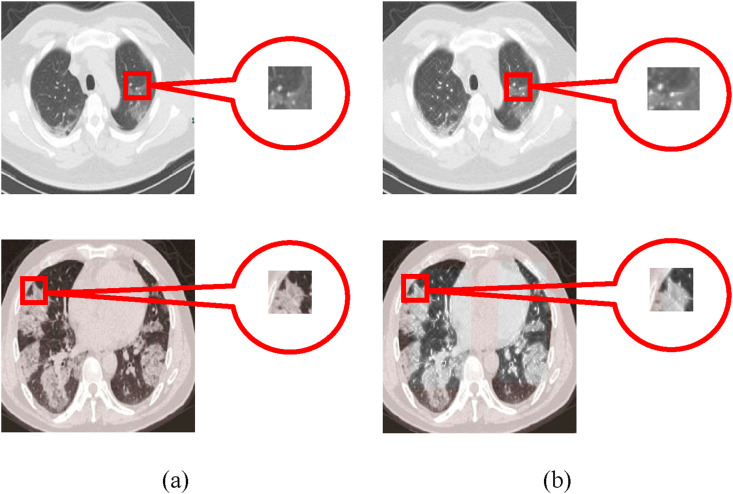
While maintaining the original distribution features of ROI, the matrix normalization can effectively reduce the impact on smaller features and achieve more accurate regional enhancement results, as shown in Fig. 7 . Thus, the contrast between the background region of the abdominal cavity and lesion is enhanced, which effectively overcomes the problem of similar pixel values, i.e., between the lung ground glass shadow and the thoracic cavity.
Fig. 7.
The contrast enhancement of the lesion region. (a) The original image; (b) The ROI enhanced image.
3. Deep learning network based on COVID-19 CT image
3.1. Convolutional neural network (CNN)
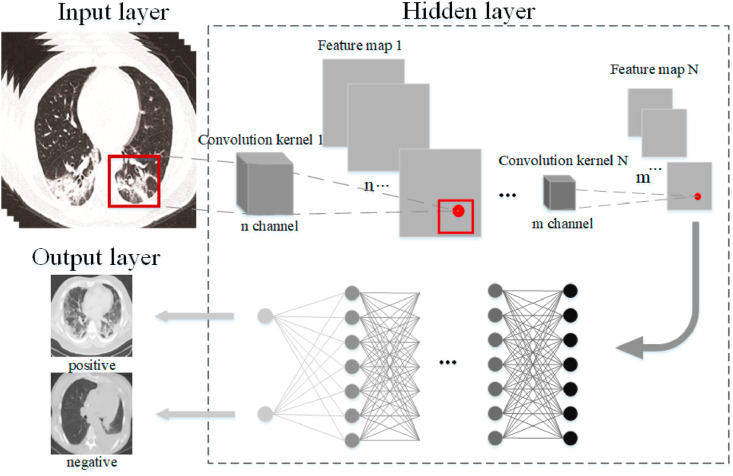
Inspired by the human visual nervous system, the CNN model [[54], [55], [56]] uses a convolution kernel to obtain feature information in the local receptive domain. It can reduce the amount of calculation and effectively maintain the hierarchical network structure, as shown in Fig. 8 . The traditional neural model is composed of the input layer, hidden layer, and output layer. Based on the network, the CNN model adds convolution layer and pooling layer in front of the fully connected layer.
Fig. 8.
The structure of the CNN model.
The weight sharing in the CNN model can effectively reduce the complexity and the number of weights. By combining local sensing region and sampling operation, this model makes full use of the local features, which can ensure the translation invariance and rotation invariance to a certain extent. Because the CNN model has achieved a good result in the field of feature extraction and classification, this paper improves the structure of the traditional CNN. The proposed model is used to extract the features of the CT image to accurately classify whether the patients belong to the infection category. Thus, the classification of negative and positive patients with COVID-19 infection can be realized.
3.2. The proposed model
To effectively improve the detection efficiency of patients with COVID-19 and reduce the virus infection rate, it needs a scheme that can obtain diagnostic precision and reduce the nucleic acid detection cost. Lung CT image is one of the most common ways to diagnose whether a patient has COVID-19. This technique can analyze the lung tissue structure and lesion morphology of suspected COVID-19 patients. In this paper, the deep convolution neural network based on a lung CT image is used to judge the suspected COVID-19 patients [57,58].
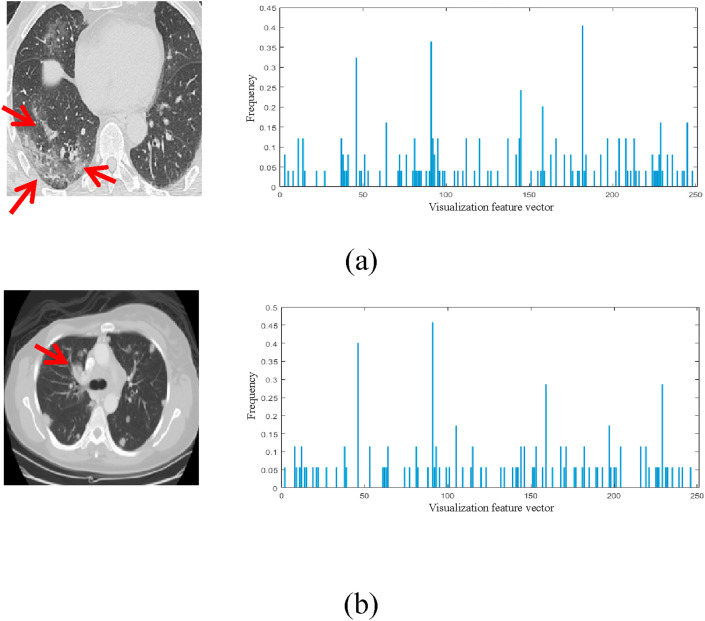
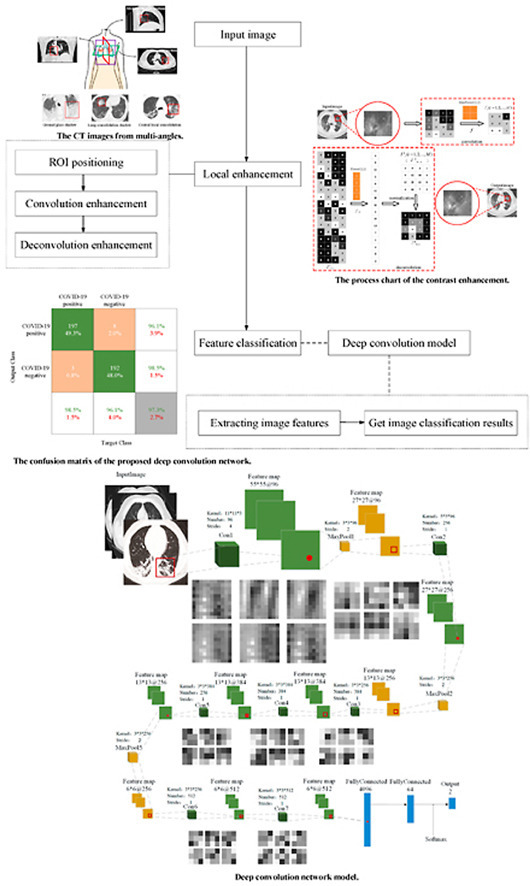
Firstly, the determinant of the matrix is obtained by convolution, and the corresponding visual vector describing the features of the COVID-19 image is obtained. Fig. 9 shows the positive and negative visual feature vectors of COVID-19, respectively. Here, feature differences of novel coronavirus pneumonia are extracted from the two categories (positive and negative) of visual vectors. Besides, their respective proportions are manually selected to balance the number of two categories. In this paper, 250 visual feature vectors are used to describe the image information of COVID-19, which represents the characteristic frequencies that can correctly distinguish the negative and positive of COVID-19. The final classification model is obtained through the subsequent deep convolution network. The deep convolution network model can be used to effectively handle the image features of the lung CT image infected with COVID-19, and assist doctors to determine whether the patients are infected with the novel coronavirus. The deep convolution network model based on COVID-19 proposed in this paper is shown in Fig. 10 .
Fig. 9.
The visualization of the COVID-19 feature vector. (a) The COVID-19 positive image; (b) The COVID-19 negative image. (The first and the second columns represent the original image and the corresponding visual feature vector distribution, respectively.)
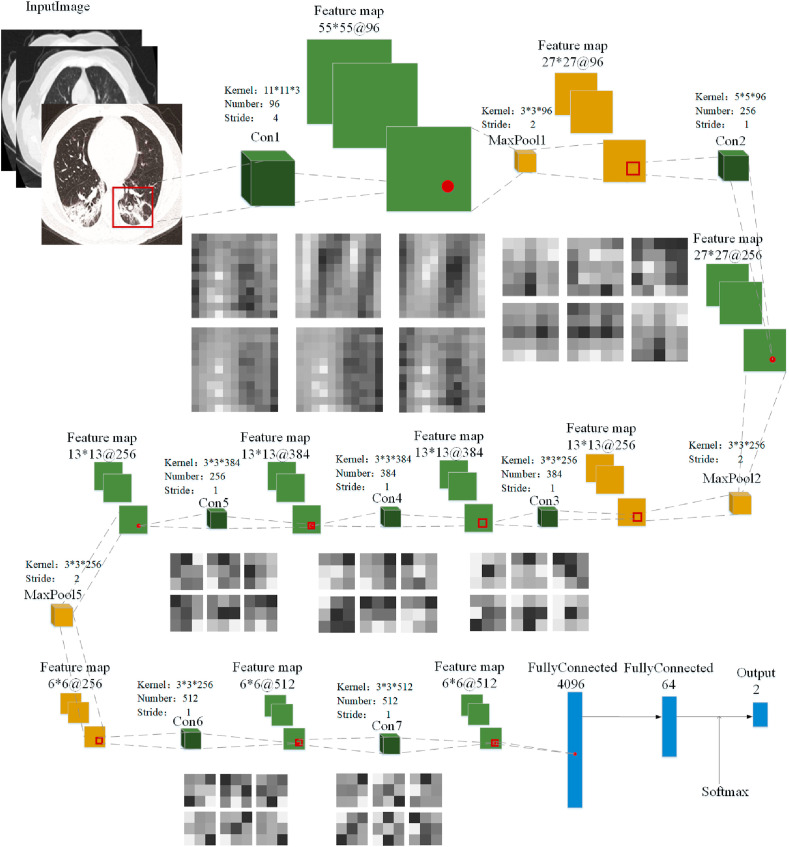
Fig. 10.
Deep convolution network model.
To effectively classify patients infected with and without COVID-19, the deep convolution network [59,60] includes seven convolution layers, three pooling layers, and three full connection layers. Among them, represents the input image layer, represents the convolution layer, represents the maximum pooling layer, represents the full connection layer, represents the feature map, and represents the output result layer.
3.2.1. The convolution and pooling layer
In this paper, three kernels 11 × 11, 5 × 5, and 3 × 3 are set in the convolution layer [61,62]. Fig. 11 shows the extracted feature map with different convolution kernels. Here, the convolution kernels 11 × 11 and 5 × 5 are mainly used to obtain the same receptive domain. Then, a convolution kernel 3 × 3 is used to replace the single nonlinear activation layer, which increases the discrimination ability of the network model. For instance, three consecutive convolution operations 3 × 3 can achieve the feature extraction of the convolution kernel 7 × 7. Thus, it can decrease times of calculation (M is the number of feature channels) and effectively reduce the amount of calculation by 45%. Similarly, two consecutive convolution operations 3 × 3 are equivalent to the feature extraction of the convolution kernel 5 × 5, which can effectively reduce the amount of computation by 28%.
Fig. 11.
The feature map with convolution kernels of different sizes.
By using a smaller convolution kernel, a more discriminative mapping function in a deep convolution network can be obtained. Therefore, the proposed convolution layer can reduce the number of calculation parameters while maintaining the range of the receptive domain, so that a point on the feature map can correspond to the feature region on the input map efficiently.
After convolution operation, the maximum pooling layer is used to reduce the size of the feature map, which can effectively reduce the training parameters and enhance the generalization ability of the model. In this paper, the size 2 × 2 of the pooling layer is set, which can obtain the stable expression of novel coronavirus pneumonia at all levels of the lung CT image.
3.2.2. The activation and normalization layer
Besides, both the convolution layer and the full connection layer contain an activation layer and a normalization layer . To overcome the slow convergence speed and “gradient explosion” of the neural network, the batch processing of the COVID-19 image is normalized. The proposed model can effectively enhance the training efficiency and reduce the sensitivity of the model to the initial weights.
In this paper, is used to specify the modified linear elements and perform threshold calculation on the eigenmatrix, that is . Due to the highly nonlinear characteristics of the deep neural network, the insufficient average operation, and the regularization, the over-fitting phenomenon is easy to occur. Besides, the model in different training sets will lead to sample misclassification. To solve this problem, this paper uses local response normalization . The normalized value is obtained by replacing it with the element of the adjacent feature channel in the normalization window. The calculation formula is as follows:
| (3) |
where , , and are the super parameters of normalization operation [63], which set to 2, to 0.0001, to 0.75. is the sum of elements of the feature channel , is the size of the feature channel. By local response normalization , the linear characteristics of the model are obtained. Therefore, the proposed model can improve the anti-interference ability for the counter sample.
3.2.3. The full connection layer
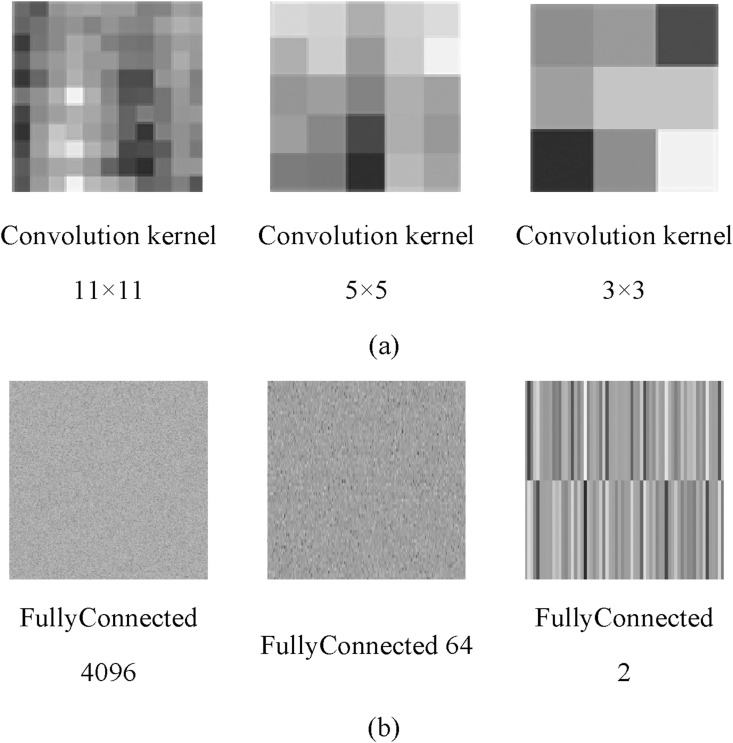
Fig. 12 shows the kernel image of each layer using different convolution layers. Fig. 12(a) shows the convolution kernel of different sizes in the proposed deep convolution network. Fig. 12(b) shows the characteristic map corresponding to three fully connected layers.
Fig. 12.
The kernel image in different convolution layers. (a) Convolution kernels of different sizes; (b) Characteristic map of fully connected layers.
By multiplying the weight matrix and adding the bias vector, the full connection layer combines all the local information in the negative and positive classification results. The last full connection layer (classification layer) combines the novel coronavirus features to recognize the larger pattern of the COVID-19 dataset and realize the negative and positive classification function.
The unit activation function of the classification layer is as follows:
| (4) |
| (5) |
where is the matrix element, is the conditional probability and is the corresponding probability of the current class, and are the prior probabilities.
To avoid the overfitting problem caused by too many training times, this paper compares the obtained eigenvalues with the input eigenvalues by the backpropagation algorithm. If the interpolation is larger than the set allowable error rate , this paper sets as 0.001. The proposed deep model combines the idea of ROI detection and image classification network and transforms the problem of COVID-19 detection into ROI feature classification. Thus, image features of the novel coronavirus pneumonia can be judged only if the feature vectors in each feature channel are contained. The dense network is used to connect the deep and shallow feature information effectively, which is to overcome the problem of gradient vanishing, realize the circulation, and enhance the reuse of feature information.
4. Experimental results and analysis
4.1. Data sources and evaluation indicators
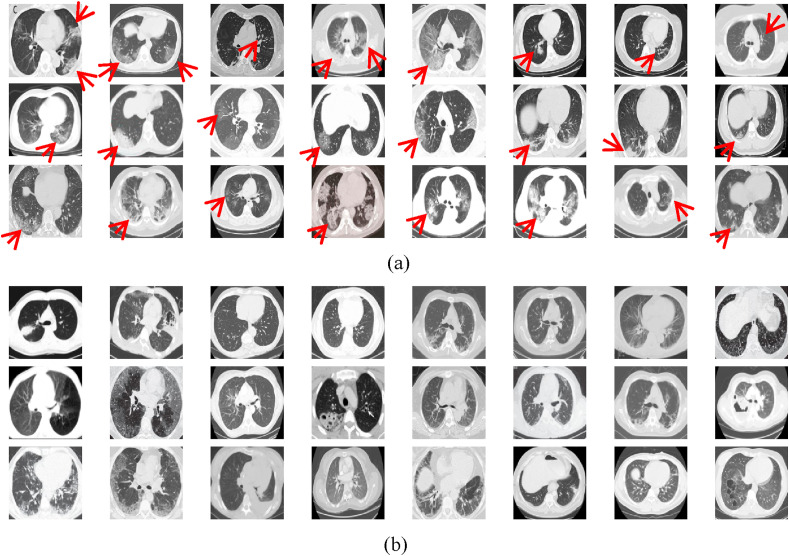
This paper uses an open-source COVID-CT dataset [64] provided by Petuum researchers from the University of California, San Diego, whose purpose is to help analyze CT image of COVID-19 patients. The dataset is collected from 143 novel coronavirus pneumonia patients and the corresponding features are preserved. The CT image of 333 cases with the positive detection of COVID-19 and 397 cases with the positive detection is provided. The partial negative and positive images of COVID-19 are shown in Fig. 13 . The image sizes of these patients are different, the minimum height is 153, the maximum height is 1853, and the average is 491. For fairness, the image size is set to 227 × 227 in this paper.
Fig. 13.
The COVID-CT dataset. (a) COVID-19 positive image; (b) COVID-19 negative image.
As shown in Table 1 , 70% of the local enhanced lung image and the original image mixed dataset are used to train the deep convolution network, and the remaining 30% is used to test. To effectively evaluate the proposed method, sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and precision are used as follows:
| (6) |
| (7) |
| (8) |
| (9) |
| (10) |
where represents the output of the proposed model, represents the correct classification label, represents totals of the images in the dataset and is the number of the input image, and represent the number of the CT image whose results are the same as or different from the labels, respectively. and are positive and negative results, respectively.
Table 1.
The statistics of the setting dataset in this paper.
| Positive COVID-19 | Negative COVID-19 | Total number | |
|---|---|---|---|
| Training set | 466 | 556 | 1022 (70%) |
| Testing set | 200 | 238 | 438 (30%) |
| Total number of CT image | 666 | 794 | 1460 |
4.2. The process of the proposed method
Considering the COVID-19 features in the lung CT image, seven convolution layers, three pooling layers, and three fully connected layers are designed for the proposed model. The corresponding architecture of the deep convolution network is shown in Appendix A. The concrete process is as follows:
Step 1
Obtain the region of interest (ROI) of both lungs and achieve the range containing the characteristic information of novel coronavirus;
Step 2
Realize the contrast between the COVID-19 local lesion region and the abdominal cavity through convolution and deconvolution operation
Step 3
Connect the deep and shallow information to determine whether the feature vector contains the COVID-19 information
Step 4
Transform the novel coronavirus detection into ROI feature region classification problem, refer to Appendix A.
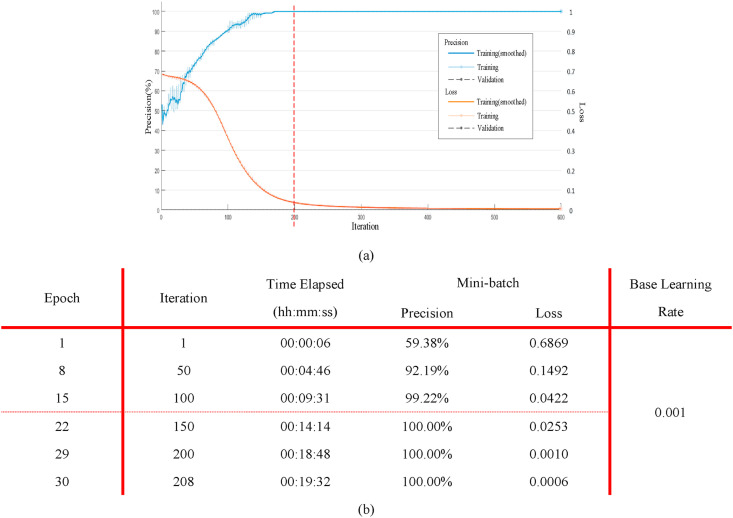
Fig. 14(a) shows the precision (blue) and loss (Orange) of the proposed model when the learning rate is set to 0.0001 (all training samples are trained for 200 times). It can be seen from the figure that when the number of iterations is 200 (the epoch is 30), the precision is close to 100% and the loss is low. Therefore, this paper sets epoch 30 and the corresponding training process is shown in Fig. 14(b).
Fig. 14.
The training process of the deep convolution model. (a) The learning rate and loss curve of the proposed deep convolution model in the training process; (b) The corresponding parameters of the proposed model using different iterations.
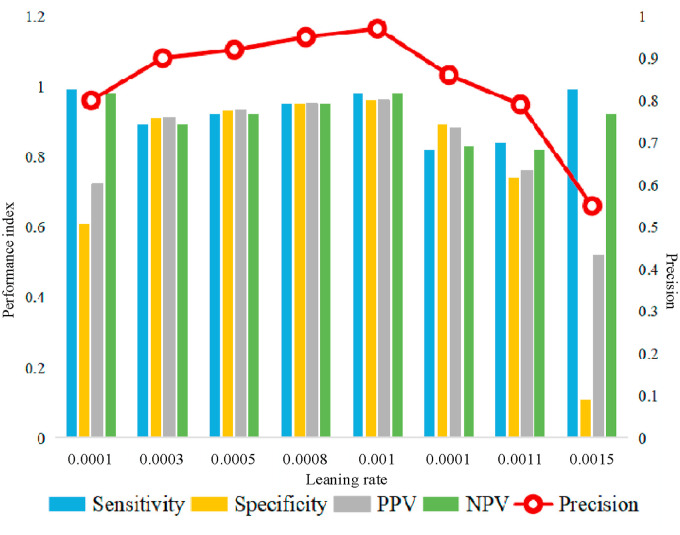
Fig. 15 shows the indexes of sensitivity, specificity, PPV, NPV, and precision using different learning rates between 0.0001 and 0.0015. When the learning rate is less than 0.001, the network fitting speed is too slow and it is easy to arrive local extreme. When the learning rate is set to be greater than 0.001, the network may not converge and it appears instability. Therefore, the proposed model uses 0.001 as the learning rate in the training process.
Fig. 15.
Index values under different learning rates.
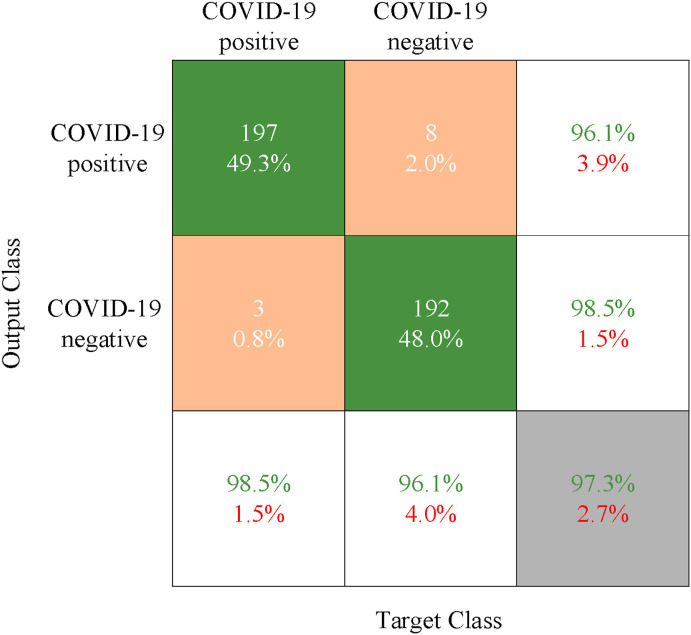
Fig. 16 shows the classification result using the proposed deep convolution network. Here, each row represents the classification result of the real patient (Output Class), that is, the true attribution classification of the novel coronavirus pneumonia image. Each column is to judge whether the result of the COVID-19 is positive or negative (Target Class), that is, the predicted classification result. In the confusion matrix, the green, orange, and gray blocks indicate the correct, wrong, and final classification result, respectively. The green and red numbers in the white and gray blocks indicate the proportion of samples with real tags correctly and wrongly classified, respectively. The experimental result shows that the precision of the proposed deep convolution model is up to about 97%.
Fig. 16.
The confusion matrix of the proposed deep convolution network.
4.3. Analysis of the proposed method
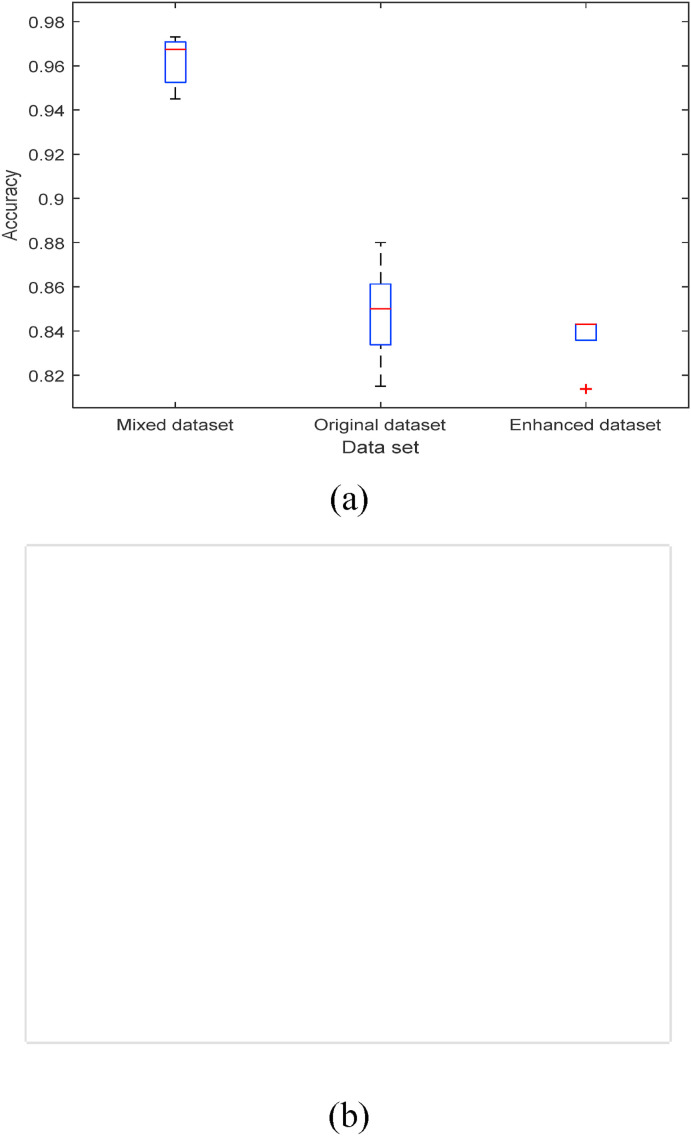
To enhance the image features of patients with positive COVID-19 in the lung CT, the contrast between the normal abdominal region and ROI is increased. As shown in Fig. 16, experiments show that the result of training the deep convolution model by mixing the enhanced CT image and the original image are better than those by using only one dataset. The precision can be up to 97%, as shown in Fig. 17 (a). Since the size of the mixed dataset is twice that of the origin or enhanced one, the running time of the model is about twice that of the normal dataset, as shown in Fig. 17(b).
Fig. 17.
The classification precision under the different datasets. (a) The precision of the model test under the different datasets; (b) The time required to test different datasets.
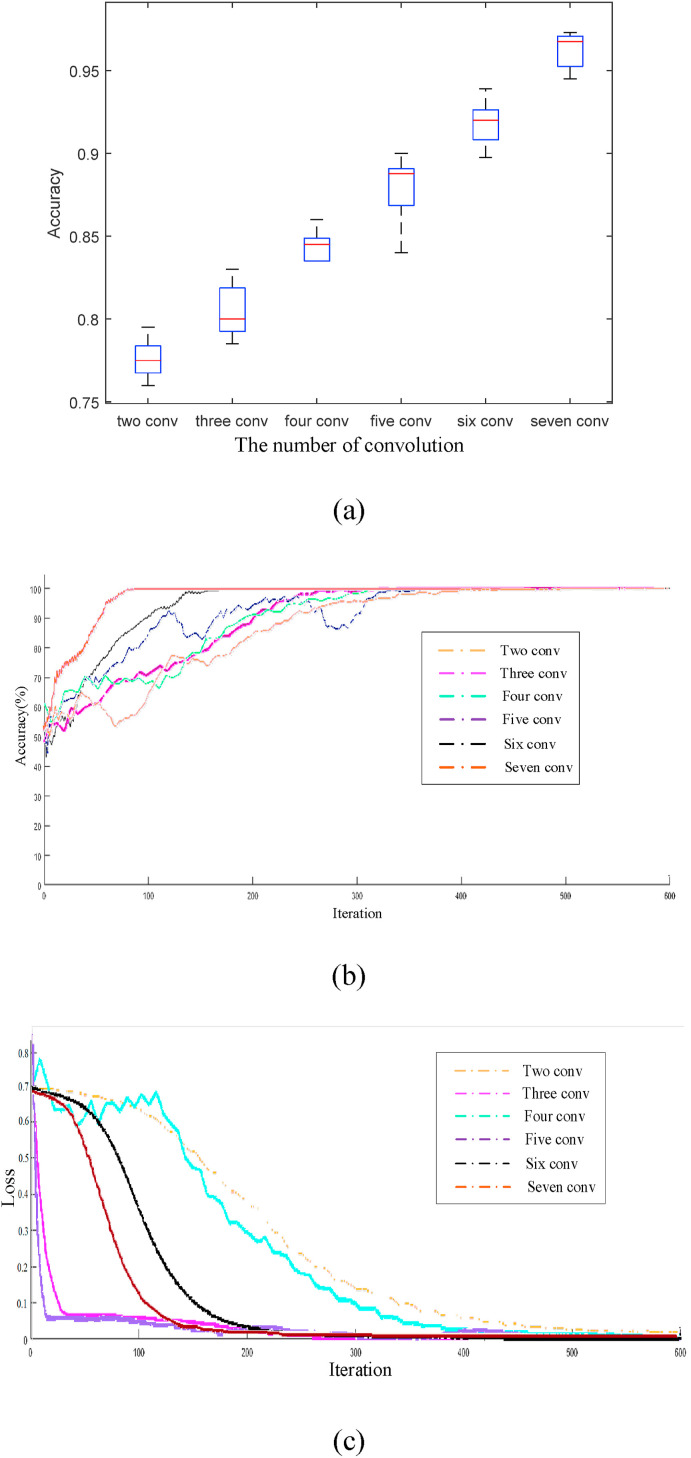
To further verify the effect of different convolution layers in the proposed model, Fig. 18 (a) shows the corresponding classification precision. Because the COVID-19 suspected patients may also be accompanied by other lung diseases, the lesions are similar in the image features. Therefore, to correctly distinguish the negative and positive images of COVID-19, more local receptive regions are needed to represent the features of the novel coronavirus. Because the spatial relationship between the image exists locally, it is necessary to set more neurons to obtain local feature information. Thus, the information of different local features will be combined in the high-level full connection layer to obtain the global perception. With the increase of convolution network layers, more local feature information can be obtained and the corresponding classification precision increases. The classification precision of the proposed model is up to 97%. With the increase of the iterations using different convolution layers, the changes in precision and loss rate are shown in Fig. 18(b)(c). Besides, with the increase of the number of layers, a more consistent feature map with the real information is obtained.
Fig. 18.
The result of the different convolution layers. (a) The Classification precision of the different convolution layers; (b) The classification precision; (c) The loss rate.
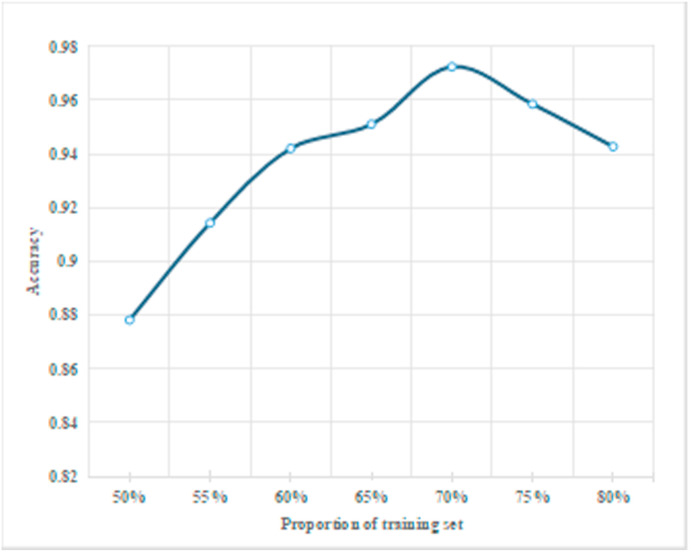
During the experiment, the precision of the proposed deep convolution model is different when setting different proportions of the training and test dataset, as shown in Fig. 19 . It can be seen that the precision of the proposed classification model increases with the increase of the proportion of the training dataset. This is because when there are fewer datasets for training, the existing image features learned by the deep convolution network can not achieve the classification effect. Besides, with the increase of the proportion of the training dataset, the model can obtain more and more COVID-19 features and the final classification precision is higher. When the dataset proportion is set to 70%, the classification precision reaches the highest 97%. This is due to the fact that with the increase of the training dataset, the characteristics of novel coronavirus patients obtained are redundant, leading to the overfitting of feature information. Therefore, the training dataset of this paper accounts for 70% of the whole dataset.
Fig. 19.
The classification precision under different proportions of the training dataset.
To further verify the effectiveness of the proposed model, Table 2 describes other state-of-the-art methods used to determine the classification result of the negative and positive COVID19 disease in the COVID-19 CT dataset. For fairness, all datasets and experimental environments are consistent. The convolution and deconvolution in the proposed method are used to enhance the contrast between the normal background and the COVID-19 lesion region. Therefore, the proposed deep convolution network has a higher classification precision. Thus, the proposed algorithm is dedicated to providing a highly robust automatic segmentation tool for COVID-19 detection through the depth model, which will be considered to be applied to the actual negative and positive detection.
Table 2.
Comparison of the proposed deep convolution network with other models in the COVID-19 CT dataset.
| Method | Sensitivity | Specificity | PPV | NPV | Precision |
|---|---|---|---|---|---|
| CNN | 0.87 | 0.875 | 0.882 | 0.862 | 0.872 |
| Loey M et al. [29] | 0.78 | 0.88 | – | – | 0.83 |
| Gifani P et al. [30] | – | – | 0.85 | 0.86 | 0.86 |
| Polsinelli M et al. [36] | 0.88 | 0.82 | 0.85 | 0.85 | 0.85 |
| Zhao J et al. [38] | – | – | – | – | 0.89 |
| Singh D et al. [359] | 0.91 | 0.89 | 0.90 | 0.91 | 0.90 |
| Parnian Afshar et al. [12] | 0.90 | 0.96 | – | – | 0.96 |
| He X et al. [37] | – | – | – | – | 0.96 |
| Bai H X et al. [39] | 0.95 | 0.96 | – | – | 0.96 |
| The proposed method | 0.98 | 0.96 | 0.96 | 0.98 | 0.97 |
5. Conclusion
In this paper, the deep network model is proposed to effectively locate the specific region of bilateral lung infection. Firstly, convolution and deconvolution methods are used to enhance the image features of the localized ROI lesions, which is to preprocess the COVID-19 image. Then, the deep convolution network method is used to classify the negative and positive COVID19 disease. To effectively overcome the problem of the network degradation caused by simply stacking convolution modules, this paper designs the self normalized convolution operation, which is used to suppress the network degradation through the residual module. Besides, to improve the ability of nonlinear transformation and learning depth, multiple continuous 3 × 3 convolution kernels are used to replace the large kernel. The proposed model obtains a better classification precision compared with most state-of-the-art models. The corresponding sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and precision are 0.98, 0.96, 0.98, and 0.97, respectively. It proves that the proposed method can effectively diagnose the novel coronavirus pneumonia infected by COVID-19 disease.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the Natural Science Foundations of China under Grant 61801202.
Biographies

Lingling Fang, born in 1985, Ph.D., associate professor of Liaoning Normal University. She is now in Computer Engineering, School of computer and information technology, Liaoning Normal University. In June 2013, She is graduated from Suzhou University with a doctor's degree in computer application. Her current research interests include: video and image information processing based on partial differential equations and multi-scale analysis.

Xin Wang, born in 1998, graduate students in Liaoning Normal University. Her research interests include medical image processing and image segmentation.
Appendix A. Deep Convolution Model
| 1 | ‘input' | Image Input | 227x227x3 images with ‘zerocenter’ normalization | Input layer |
|---|---|---|---|---|
| 2 | ‘conv1′ | Convolution | 96 11x11x3 convolutions with stride [44] and padding [0 0 0 0] | The 1st layer |
| 3 | ‘relu1′ | ReLU | ReLU | |
| 4 | ‘norm1′ | Cross Channel Normalization | cross channel normalization with 5 channels per element | |
| 5 | ‘pool1′ | Max Pooling | 3x3 max pooling with stride [22] and padding [0 0 0 0] | |
| 6 | ‘conv' | Convolution | 256 5x5x48 convolutions with stride [11] and padding [2 2 2 2] | The 2nd layer |
| 7 | ‘relu2′ | ReLU | ReLU | |
| 8 | ‘norm2′ | Cross Channel Normalization | cross channel normalization with 5 channels per element | |
| 9 | ‘pool2′ | Max Pooling | 3x3 max pooling with stride [22] and padding [0 0 0 0] | |
| 10 | ‘conv3′ | Convolution | 384 3x3x256 convolutions with stride [11] and padding [1 1 1 1] | The 3rd layer |
| 11 | ‘relu3′ | ReLU | ReLU | |
| 12 | ‘conv4′ | Convolution | 384 3x3x192 convolutions with stride [11] and padding [1 1 1 1] | The 4th layer |
| 13 | ‘relu4′ | ReLU | ReLU | |
| 14 | ‘conv5′ | Convolution | 256 3x3x192 convolutions with stride [11] and padding [1 1 1 1] | The 5th layer |
| 15 | ‘relu5′ | ReLU | ReLU | |
| 16 | ‘pool5′ | Max Pooling | 3x3 max pooling with stride [22] and padding [0 0 0 0] | |
| 17 | ‘conv6′ | Convolution | 512 3x3 convolutions with stride [11] and padding [1 1 1 1] | The 6th layer |
| 18 | ‘relu6′ | ReLU | ReLU | |
| 19 | ‘conv7′ | Convolution | 512 3x3 convolutions with stride [11] and padding [1 1 1 1] | The 7th layer |
| 20 | ‘relu7′ | ReLU | ReLU |
References
- 1.Kraemer Moritz U.G., Scarpino Samuel V., Vukosi Marivate, et al. Data curation during a pandemic and lessons learned from COVID-19. Nat. Comput. Sci. Jan. 2021;1(1):9–10. doi: 10.1038/s43588-020-00015-6. [DOI] [PubMed] [Google Scholar]
- 2.Harsh Panwar, Gupta P.K. Siddiqui Mohammad Khubeb, et all, “A Deep Learning and Grad-CAM based Color Visualization Approach for Fast Detection of COVID-19 Cases using Chest X-ray and CT-Scan Images,” Chaos. Solitons Fractals. Aug. 2020;140 doi: 10.1016/j.chaos.2020.110190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ismael Aras M., Abdulkadir Şengür. Deep learning approaches for COVID-19 detection based on chest X-ray images. Expert Syst. Appl. Jan. 2021;164 doi: 10.1016/J.ESWA.2020.114054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abdulhadee Yakoh, Umaporn Pimpitak, Sirirat Rengpipat, et al. ” Biosensors and Bioelectronics; Dec. 2020. Paper-based Electrochemical Biosensor for Diagnosing COVID-19: Detection of SARS-CoV-2 Antibodies and Antigen. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rai Praveen, Krishna Kumar Ballamoole, Kumar Deekshit Vijaya, et al. Detection technologies and recent developments in the diagnosis of COVID-19 infection. Appl. Microbiol. Biotechnol. Jan. 2021:1–15. doi: 10.1007/S00253-020-11061-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cady Nathaniel C., Natalya Tokranova, Armond Minor, et al. Multiplexed detection and quantification of human antibody response to COVID-19 infection using a plasmon enhanced biosensor platform. Biosens. Bioelectron. Jan. 2021;171 doi: 10.1016/J.BIOS.2020.112679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manivel Vijay, Lesnewski Andrew, Shamim Simin, et al. CLUE: COVID‐19 lung ultrasound in emergency department. Emerg. Med. Australasia (EMA) May. 2020;32(4):694–696. doi: 10.1111/1742-6723.13546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Shuyi, Zhang Yunfei, Shen Jie. Clinical potential of UTE‐MRI for assessing COVID ‐19: patient‐ and lesion‐based comparative analysis. J. Magn. Reson. Imag. Jun. 2020;52(2):397–406. doi: 10.1002/jmri.27208. PhD, MD. PhD. B.S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Apostolopoulos Ioannis D., Aznaouridis Sokratis I., Tzani Mpesiana A. Extracting possibly representative COVID-19 biomarkers from X-ray images with deep learning approach and image data related to pulmonary diseases. J. Med. Biol. Eng. Jun. 2020;40(3):462–469. doi: 10.1007/s40846-020-00529-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Narin A., Kaya C., Pamuk Z. Jan. 2020. Automatic Detection of Coronavirus Disease (Covid-19) Using X-Ray Images and Deep Convolutional Neural Networks; p. 10849. arXiv preprint arXiv. 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hemdan E.E.D., Shouman M.A., Karar M.E. Jan. 2020. Covidx-net: A Framework of Deep Learning Classifiers to Diagnose Covid-19 in X-Ray Images; p. 11055.https://arxiv.org/abs/2003.11055 arXiv preprint arXiv. 2003. [Google Scholar]
- 12.Afshar Parnian, Heidarian Shahin, Naderkhani Farnoosh, Oikonomou Anastasia, Konstantinos N., Plataniotis Arash Mohammadi. COVID-CAPS: a capsule network-based framework for identification of COVID-19 cases from X-ray images. Pattern Recogn. Lett. Jan. 2020;138:638–643. doi: 10.1016/j.patrec.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Tao, Zhao Yongguo, Zhu Lin, Liu Guangliang, Ma Zhengguang, Zheng Jianghua. vol. 5. Nov. 2020. pp. 199–203. (Lung CT Image Aided Detection COVID-19 Based on Alexnet Network). Sichuan University.Proceedings of the 5th International Conference on Communication, Image and Signal Processing (CCISP 2020) [DOI] [Google Scholar]
- 14.Luo Lin, Luo Zhendong, Jia Yizhen, et al. CT differential diagnosis of COVID-19 and non-COVID-19 in symptomatic suspects: a practical scoring method. BMC Pulm. Med. May. 2020;20(11):719–739. doi: 10.1186/s12890-020-1170-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Kunwei, Fang Yijie, Li Wenjuan, et al. CT image visual quantitative evaluation and clinical classification of coronavirus disease (COVID-19) Eur. Radiol. Aug. 2020;30(8):1–10. doi: 10.1007/s00330-020-06817-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao Jing, Liu Jun-Qiang, Wen Heng-Jun, et al. The unsynchronized changes of CT image and nucleic acid detection in COVID-19: reports the two cases from Gansu, China. Respir. Res. Apr. 2020;21(10):558–570. doi: 10.1186/s12931-020-01363-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chua F., Armstrong-James D., Desai S.R., et al. The role of CT in case ascertainment and management of COVID-19 pneumonia in the UK: insights from high-incidence regions. The Lancet Respir. Med. May. 2020;8(5):438–440. doi: 10.1016/S2213-2600(20)30132-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang Y., Zhang H., Xu Y., et al. CT manifestations of two cases of 2019 novel coronavirus (2019-nCoV) pneumonia. Radiology. Apr. 2020;295(1):208–209. doi: 10.1148/radiol.2020200280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bernheim A., Mei X., Huang M., et al. Chest CT findings in coronavirus disease-19 (COVID-19): relationship to duration of infection. Radiology. Apr. 2020;295(3) doi: 10.1148/radiol.2020200463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krzysztof Misztal, Agnieszka Pocha, Kozica Martyna Durak, et al. “The importance of standardisation – COVID-19 CT & Radiograph Image Data Stock for deep learning purpose. Comput. Biol. Med. Nov. 2020;127 doi: 10.1016/j.compbiomed.2020.104092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fattane Shirani, Azin Shayganfar, Somayeh Hajiahmadi. COVID-19 pneumonia: a pictorial review of CT findings and differential diagnosis. Egypt. J. Radiol. Nucl. Med. Jan. 2021;52(1):263–268. [Google Scholar]
- 22.Khurana Shikhar, Chopra Rohan, Khurana Bharti. Automated processing of social media content for radiologists: applied deep learning to radiological content on twitter during COVID-19 pandemic. Emerg. Radiol. Apr. 2021:1–7. doi: 10.1007/s10140-020-01885-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beck Bo Ram, Shin Bonggun, Choi Yoonjung, et al. Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug-target interaction deep learning model. Comput. Struct. Biotechnol. J. Apr. 2020;18:784–790. doi: 10.1016/j.csbj.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan B.B., Yang H. Analysis of identifying COVID-19 with deep learning model. J. Phys. Conf. Aug. 2020;1601 doi: 10.1088/1742-6596/1601/5/052021. [DOI] [Google Scholar]
- 25.Khaled Bayoudh, Fayçal Hamdaoui, Abdellatif Mtibaa. Hybrid-COVID: a novel hybrid 2D/3D CNN based on cross-domain adaptation approach for COVID-19 screening from chest X-ray images. Phys. Eng. Sci. Med. Dec. 2020;43(4):1–17. doi: 10.1007/s13246-020-00957-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morteza Heidari, Seyedehnafiseh Mirniaharikandehei, Abolfazl Zargari Khuzani, et al. Improving the performance of CNN to predict the likelihood of COVID-19 using chest X-ray images with preprocessing algorithms. Int. J. Med. Inf. Dec. 2020;144 doi: 10.1016/j.ijmedinf.2020.104284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matteo Polsinelli, Cinque Luigi, Giuseppe Placidi. A light CNN for detecting COVID-19 from CT scans of the chest. Pattern Recogn. Lett. Dec. 2020;140:95–100. doi: 10.1016/j.patrec.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohata E.F., Bezerra G.M., das Chagas J.V.S., et al. Automatic detection of COVID-19 infection using chest X-ray images through transfer learning. IEEE/CAA J. Automatica Sinica. Nov. 2020;8(1):239–248. [Google Scholar]
- 29.Loey M., Manogaran G., Khalifa N.E.M. A deep transfer learning model with classical data augmentation and cgan to detect covid-19 from chest ct radiography digital images. Neural Comput. Appl. Oct. 2020:1–13. doi: 10.20944/preprints202004.0252.v3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gifani P., Shalbaf A., Vafaeezadeh M. Automated detection of COVID-19 using ensemble of transfer learning with deep convolutional neural network based on CT scans. Int. J. Computer Assist. Radiol. Surg. Nov. 2020:1–9. doi: 10.1007/s11548-020-02286-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hall L.O., Paul R., Goldgof D.B., et al. Dec. 2020. Finding Covid-19 from Chest X-Rays Using Deep Learning on a Small Dataset. arXiv preprint arXiv:2004.02060. [DOI] [Google Scholar]
- 32.Shi F., Xia L., Shan F., et al. Large-scale screening to distinguish between COVID-19 and community-acquired pneumonia using infection size-aware classification. Phys. Med. Biol. Jan. 2021;66(6) doi: 10.1088/1361-6560/abe838. [DOI] [PubMed] [Google Scholar]
- 33.Hu C., Fang C., Lu Y., et al. Selective oxidation of diethylamine on CuO/ZSM-5 catalysts: the role of cooperative catalysis of CuO and surface acid sites. Ind. Eng. Chem. Res. Dec. 2020;59(20):9432–9439. [Google Scholar]
- 34.Basu S., Mitra S., Saha N. IEEE; May. 2020. Deep Learning for Screening Covid-19 Using Chest X-Ray Images; pp. 2521–2527. 2020 IEEE Symposium Series on Computational Intelligence (SSCI) [DOI] [Google Scholar]
- 35.Singh D., Kumar V., Kaur M. Classification of COVID-19 patients from chest CT images using multi-objective differential evolution–based convolutional neural networks. Eur. J. Clin. Microbiol. Infect. Dis. Jul. 2020;39(7):1379–1389. doi: 10.1007/s10096-020-03901-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Polsinelli M., Cinque L., Placidi G. A light cnn for detecting covid-19 from ct scans of the chest. Pattern Recogn. Lett. Dec. 2020;140:95–100. doi: 10.1016/j.patrec.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He X., Yang X., Zhang S., et al. Jul. 2020. Sample-Efficient Deep Learning for Covid-19 Diagnosis based on ct Scans; p. 430. MedRxiv. [DOI] [Google Scholar]
- 38.Zhao J., Zhang Y., He X., et al. Jan. 2020. Covid-ct-dataset: a Ct Scan Dataset about Covid-19; p. 490. arXiv preprint arXiv, vol. 2003, no. 13865. [Google Scholar]
- 39.Bai H.X., Wang R., Xiong Z., et al. Artificial intelligence augmentation of radiologist performance in distinguishing COVID-19 from pneumonia of other origin at chest CT. Radiology. Sept. 2020;296(3):E156–E165. doi: 10.1148/radiol.2020201491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eggo Rosalind M., Jeanette Dawa, Kucharski Adam J., et al. The importance of local context in COVID-19 models. Nat. Comput. Sci. Jan. 2021;1(1):6–8. doi: 10.1038/s43588-020-00014-7. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Xiaorui, Zhang Wenfang, et al. A robust watermarking scheme based on ROI and IWT forRemote consultation of COVID-19. Comput. Mater. Continua (CMC) Jul. 2020;64(3):1435–1452. [Google Scholar]
- 42.Kakani Preeti, Sorensen Andrea, Quinton Jacob K., et al. Patient characteristics associated with telemedicine use at a large academic health system before and after COVID-19. J. Gen. Intern. Med. Jan. 2021:1–3. doi: 10.1007/s11606-020-06544-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krishnan Arunkumar, Hamilton James P., Alqahtani Saleh A., et al. A narrative review of coronavirus disease 2019 (COVID-19): clinical, epidemiological characteristics, and systemic manifestations. Intern. Emerg. Med. Jan. 2021:1–16. doi: 10.1007/S11739-020-02616-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paidi Ramesh K., Jana Malabendu, Mishra Rama K., et al. ACE-2-interacting domain of SARS-CoV-2 (AIDS) peptide suppresses inflammation to reduce fever and protect lungs and heart in mice: implications for COVID-19 therapy. J. Neuroimmune Pharmacol. Jan. 2021:1–12. doi: 10.1007/S11481-020-09979-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liao X., Zhao J., Jiao C., et al. A segmentation method for lung parenchyma image sequences based on superpixels and a self-generating neural forest. PloS One. Dec. 2016;11(8) doi: 10.1371/journal.pone.0160556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yi S., Jiang J., Song J., et al. “Can PET/CT show heterogeneous distribution of tumor's proliferative and infiltrative area in malignant solitary pulmonary nodule? J. Nucl. Med. Jan. 2018;59 https://jnm.snmjournals.org/content/59/supplement_1/1357.short [Google Scholar]
- 47.Buzan Maria T.A., Andreas Wetscherek, Claus Peter Heussel, et al. Texture analysis using proton density and T2 relaxation in patients with histological usual interstitial pneumonia (UIP) or nonspecific interstitial pneumonia (NSIP) PloS One. Sep. 2017;12(5) doi: 10.1371/journal.pone.0177689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Basu Indira, Radhika Nagappan, Fox Lewis Shivani, et al. Evaluation of extraction and amplification assays for the detection of SARS-CoV-2 at Auckland Hospital laboratory during the COVID-19 outbreak in New Zealand. J. Virol Methods. Jan. 2021;289:114042. doi: 10.1016/J.JVIROMET.2020.114042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tao Yifeng, Lei Haoyun, Lee Adrian V., Ma Jian, Russell Schwartz. Neural network deconvolution method for resolving pathway-level progression of tumor clonal expression programs with application to breast cancer brain metastases. Front. Physiol. Sep. 2020;11 doi: 10.3389/fphys.2020.01055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chithra P.L., Dheepa G. “Di‐phase midway convolution and deconvolution network for brain tumor segmentation in MRI images. Int. J. Imag. Syst. Technol. Aug. 2020;30(3):674–686. [Google Scholar]
- 51.McIlwain Sean J., Wu Zhijie, Wetzel Molly, et al. Enhancing top-down proteomics data analysis by combining deconvolution results through a machine learning strategy. J. Am. Soc. Mass Spectrom. May. 2020;31(5):1104–1113. doi: 10.1021/jasms.0c00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morgan Rhianna K., Psaras Alexandra Maria, Quinea Lassiter, Kelsey Raymer, Brooks Tracy A. “G-quadruplex deconvolution with physiological mimicry enhances primary screening: optimizing the FRET Melt2 assay,” Biochimica et biophysica acta. Gene Regul. Mech. Jan. 2020;1863(1) doi: 10.1016/j.bbagrm.2019.194478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiang Xiaolei, Liao Erchong, Liu Xiaofeng. Blind image deconvolution via enhancing significant segments. Neural Process. Lett. Sep. 2019:1–16. doi: 10.1007/s11063-019-10123-8. [DOI] [Google Scholar]
- 54.Kim J., Sangjun O., Kim Y., et al. Convolutional neural network with biologically inspired retinal structure. Procedia Comput. Sci. Jan. 2016;88:145–154. doi: 10.1016/j.procs.2016.07.418. [DOI] [Google Scholar]
- 55.Simonyan K., Zisserman A. vol. 1409. Jan. 2014. Very Deep Convolutional Networks for Large-Scale Image Recognition; p. 1556.https://arxiv.org/abs/1409.1556 arXiv preprint arXiv. [Google Scholar]
- 56.Szegedy C., Vanhoucke V., Ioffe S., et al. Jan. 2016. Rethinking the Inception Architecture for Computer Vision; pp. 2818–2826. Proceedings of the IEEE conference on computer vision and pattern recognition. [DOI] [Google Scholar]
- 57.Yasar Huseyin, Ceylan Murat. A new deep learning pipeline to detect Covid-19 on chest X-ray images using local binary pattern, dual tree complex wavelet transform and convolutional neural networks. Appl. Intell. Nov. 2020:1–24. doi: 10.1007/s10489-020-02019-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ali Abbasian Ardakani, Alireza Rajabzadeh Kanafi, Acharya U Rajendra, et al. Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: results of 10 convolutional neural networks. Comput. Biol. Med. Jun. 2020;121 doi: 10.1016/j.compbiomed.2020.103795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krizhevsky A., Sutskever I., Hinton G. NIPS. Curran Associates Inc.; Jan. 2012. ImageNet Classification with Deep Convolutional Neural Networks. 2012. [DOI] [Google Scholar]
- 60.Zhang Kaishuo, Robinson Neethu, Lee Seong-Whan, et al. Adaptive transfer learning for EEG motor imagery classification with deep Convolutional Neural Network. Neural Network. Jan. 2021;136:1–10. doi: 10.1016/j.neunet.2020.12.013. [DOI] [PubMed] [Google Scholar]
- 61.Wang Shui-Hua, Varthanan Govindaraj Vishnu, Manuel Gó Juan, et al. Covid-19 classification by FGCNet with deep feature fusion from graph convolutional network and convolutional neural network. Inf. Fusion. Jan. 2020;67:208–229. doi: 10.1016/j.inffus.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dureja A., Pahwa P. Medical image retrieval for detecting pneumonia using binary classification with deep convolutional neural networks. J. Inf. Optim. Sci. Aug. 2020;41(6):1419–1431. [Google Scholar]
- 63.Krizhevsky A., Sutskever I., Hinton G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. Jan. 2012;25:1097–1105. [Google Scholar]
- 64.Zhao J., Zhang Y., He X., et al. Jan. 2020. Covid-ct-dataset: a Ct Scan Dataset about Covid-19; p. 13865.https://europepmc.org/article/ppr/ppr269030 arXiv preprint arXiv:2003. [Google Scholar]