Abstract
Purpose:
Fractures in vertebral bodies are among the most common complications of osteoporosis and other bone diseases. However, studies that aim to predict future fractures and assess general spine health must manually delineate vertebral bodies and intervertebral discs in imaging studies for further radiomic analysis. This study aims to develop a deep learning system that can automatically and rapidly segment (delineate) vertebrae and discs in MR, CT, and X-ray imaging studies.
Results:
We constructed a neural network to output 2D segmentations for MR, CT, and X-ray imaging studies. We trained the network on 4490 MR, 550 CT, and 1935 X-ray imaging studies (post-data augmentation) spanning a wide variety of patient populations, bone disease statuses, and ages from 2005–2020. Evaluated using 5-fold cross validation, the network was able to produce median Dice scores > 0.95 across all modalities for vertebral bodies and intervertebral discs (on the most central slice for MR/CT and on image for X-ray). Furthermore, radiomic features (skewness, kurtosis, mean of positive value pixels, and entropy) calculated from predicted segmentation masks were highly accurate (r ≥ 0.96 across all radiomic features when compared to ground truth). Mean time to produce outputs was < 1.7 seconds across all modalities.
Conclusions:
Our network was able to rapidly produce segmentations for vertebral bodies and intervertebral discs for MR, CT, and X-ray imaging studies. Furthermore, radiomic quantities derived from these segmentations were highly accurate. Since this network produced outputs rapidly for these modalities which are commonly used, it can be put to immediate use for radiomic and clinical imaging studies assessing spine health.
Keywords: Artificial Intelligence, Radiomics, Vertebral Bodies and Intervertebral Discs, Bone Disease, Image Analysis
1. Introduction:
In the United States, millions of individuals suffer complications as a result of spine and bone disorders each year. For example, among the 10 million individuals who are diagnosed with osteoporosis, almost 50% of fractures arise from vertebral bodies [1], [2]. Complications of bone diseases, vertebral fractures in particular, present an especially high burden on individuals, resulting in reduced mobility, increased back pain, increased depression, and increased mortality among patients [3].
Recently, quantitative research efforts have been undertaken to determine how imaging data can be used to gather information about underlying bone/spine disease or diagnose diseases. Quantities derived from imaging studies, termed collectively as radiomics, have allowed researchers to accurately predict underlying bone health. For example, MR-based radiomic features differentiated metastatic from non-metastatic vertebral bodies in patients with bone marrow metastatic disease [4]. Other studies show how radiomics-based models can automatically diagnose hematologic marrow diseases from MR studies at a level comparable to trained radiologists [5].
However, an issue with radiomics analyses is that they rely on individuals to segment (i.e. outline) regions of interest for quantification. This segmentation process is time-consuming, since it is done manually for each vertebral body in an imaging study, and is a hamper to the deployment of radiomics based solutions in the clinical field. Artificial intelligence (AI) algorithms offer a way to automate the segmentation process in a rapid and accurate manner. A specific subset of AI, called deep learning (DL), offers algorithms that can extract features from images to detect and classify objects. In medical research, these DL algorithms (specifically neural networks) have been used to predict diagnoses and mark clinically relevant areas in imaging studies. Neural networks are especially well posed to solve problems in medical imaging since they are able to efficiently learn how to solve problems involving classification and image feature extraction [6].
In a DL system, it is possible to create a neural network that detects vertebral bodies within imaging studies and segments (delineates) them. These segmentations can be input to popular radiomics software and to produce quantities (e.g. skewness, kurtosis, mean pixel value, and entropy) based on the underlying pixel-values isolated within segmentation outlines, independent of modality. Prior approaches to this problem have shown DL systems can work for performing segmentation within one imaging modality [7]–[9]; however, no study has been able to show a network design that performs well across multiple modalities. Furthermore, some of these approaches only work well on one region of the spine (lumbar, thoracic, or cervical) thus limiting clinical applicability.
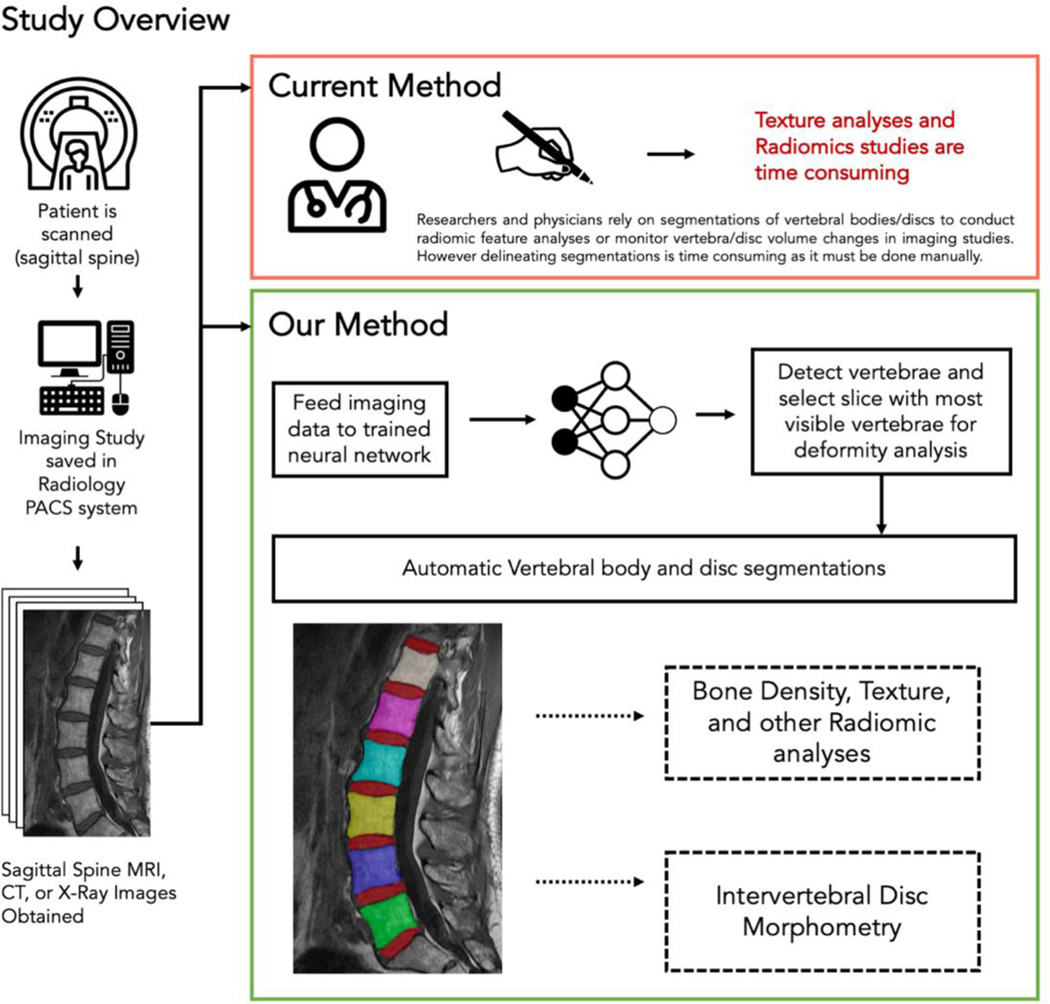
In this study, we report the development of a DL system for segmentation of vertebral bodies from MR, CT, and X-ray imaging studies. Our solution can produce accurate segmentations (evaluated by Dice scores) of vertebral bodies on a automatically selected 2D slice in a rapid manner. Additionally, we show the system can accurately segment intervertebral discs in MR imaging studies. We also demonstrate that the network is spine-region invariant and can maintain accuracy across lumbar, thoracic, and cervical vertebrae/discs. Lastly, we compare the radiomic quantities generated from predicted segmentations (skewness, kurtosis, mean of positively valued pixels, and entropy) to those from manually made segmentations to verify that this network can be used for radiomics-based studies. An overview of the capabilities and potential applications of the neural network in comparison to current methods is shown in Figure 1.
Figure 1: Basic overview of our proposed method in this study.
Currently, researchers and physicians must manually segment vertebral bodies and discs to calculate radiomic features. This process is highly time-consuming and subjective. Our method instead shows that a neural network can produce vertebral body and intervertebral disc segmentations. These segmentations can then be used for future studies that examine radiomic features/pixel values in relation to bone density, texture analyses, and diagnostic studies. Furthermore, intervertebral disc segmentations can be used to examine disc morphometry and monitor changes in disc cross-sectional area over time.
2. Materials + Methods:
2.1. Deep Learning System Overview:
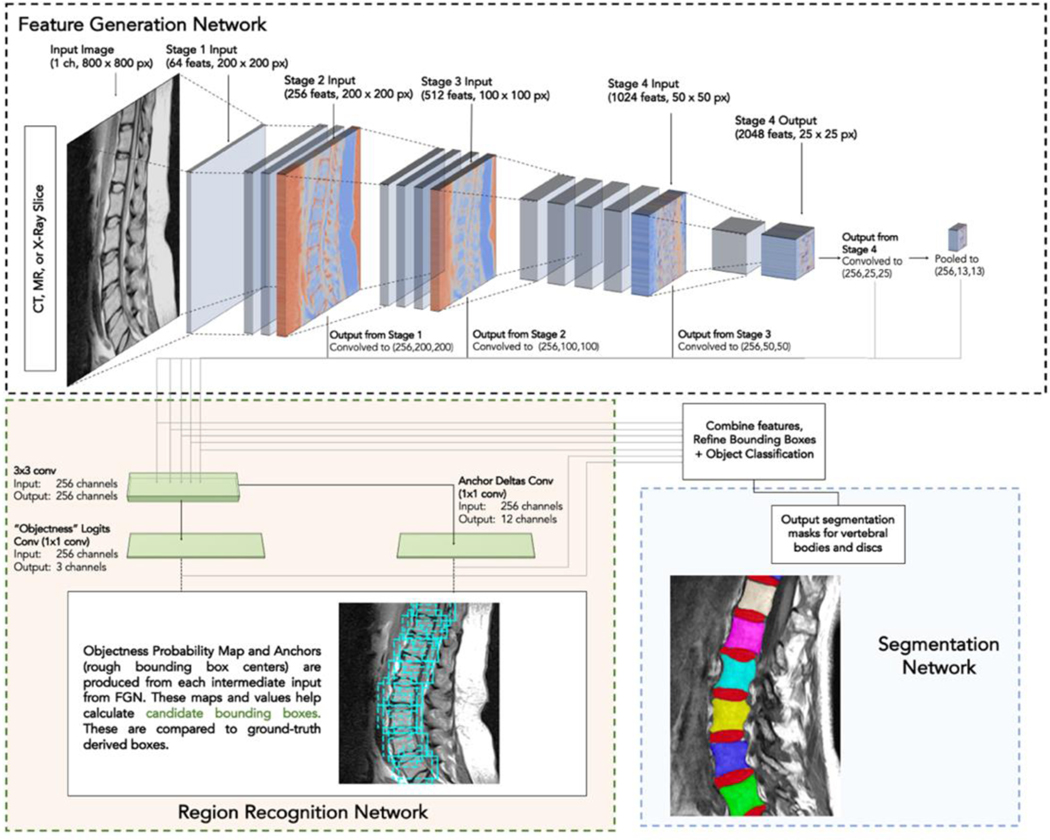
The deep learning system (a set of neural networks) works in three steps. First, intermediate features are extracted out of an input image for the network to learn on. Second, vertebral bodies and discs are localized (location denoted with a rectangular bounding box) using the intermediate features from the prior step. Lastly, each bounding box (vertebral body/disc) is segmented by the network. Outputs of the segmentation step shows which pixels correspond to a vertebral body/disc and which parts are surrounding tissue. Figure 2 shows an overview of the network design (named SpineTK) which is based on a region-based convolutional neural network architecture. Three instances of the SpineTK DL system are trained, one version for each modality the network is trained to work on (MR, CT, and X-ray). In practice, the system can receive an imaging study as input. For each slice in the imaging study, vertebral bodies bounding boxes are produced and the slice that contains the most number of visible vertebral bodies (ref Supplemental Materials/Methods: Section 3 for full description of slice selection algorithm) is selected for further analysis. Then, a segmentation mask is predicted for each vertebral body/disc in that slice. These segmentation masks can be input into most radiomics evaluation software programs. Details on components of the DL system and its individual neural networks can be found in Supplemental Materials/Methods: Section 4,6. Prior to evaluation, the DL system must be “trained”, a process by which the system learns to extract relevant image features to optimize segmentation, on imaging studies and ground-truth vertebral body/disc segmentations.
Figure 2: Basic outline of neural network.
The network is composed of three parts: a Feature Generation Network (FGN), a Region Recognition Network (RRN), and a Landmark Detection Network. The FGN performs several image transformation steps (called convolutions) in stages 1–4 to create features that can be learned on (dimensions of images after transformations are applied reported as [features(#), height(px) x width(px)] in the figure). Intermediate outputs from the FGN are each used to train the RRN which produces an “Objectness” logits map (showing probability of an approximate region containing an object) and anchor deltas (which are preliminary bounding boxes for desired objects). Then the objectness map and the preliminary bounding boxes are combined (along with intermediate features from the FGN), bounding boxes are refined, and final boxes are classified as “vertebral body”, “intervertebral disc”, or background. Next, the network delineates (segments) vertebral bodies and discs by producing a pixel-by-pixel mask (pixel value 1 = vertebral body present, 0 = background). Three networks are trained in this paper – one for each modality (MR, CT, X-ray). Network architecture adapted from Mask R-CNN [11] (RCNN = Region based Convolutional Neural Networks), layer names changed for clarity. Refer to Materials & Methods for link to implementation and ref Supplemental Figure 1 and Supplemental Materials/Methods: Section 6 for full explanation of network + full depiction of convolutional layer parameters.
2.2. Patient Population and Dataset Description:
In order to make SpineTK robust to variability in patient populations, imaging studies used to train and evaluate the network covered a large patient population across several centers with a range of underlying bone density statuses (normal, osteopenia, osteoporosis) to demonstrate the network’s efficacy in these patient populations. MR imaging studies were obtained from individuals who underwent imaging at the Hospital of the University of Pennsylvania Radiology Center and affiliated centers from January 1, 2005 to January 20, 2014. X-ray radiographs were randomly selected from the Hospital of the University of Pennsylvania radiology archives, covering a number of centers across the Pennsylvania and New Jersey areas from September 10, 2019 to June 15, 2020. CT imaging studies came from an institutional collaborator and a public segmentation challenge dataset (VerSe 2019 [10] phase 1 and 2 data primarily from Germany). All datasets included individuals with indications for back pain, fracture, or osteoporosis and were deidentified (IRB approval was obtained for this study). Images were excluded from training/evaluation due to images being corrupt (12/1153 cases MR, 10/160 cases CT, 3/492 X-ray) or images being of such low resolution that ground truth annotations could not be accurately made by human annotators (18/1153 MR, 13/160 CT, 5/492 X-ray). The total number of resulting source imaging studies were 1123 MR, 137 CT, and 484 X-ray images that were augmented prior to training the network.
MR cases were majority female (88%) aged 67 ± 11 (±1 SD) years. Approximately 17% of these individuals had osteopenia (using criteria −2.5 < T-Score < −1) and 9% had osteoporosis (T-Score ≤ −2.5). CT cases were majority female (67%) aged 65 ± 5 years. Approximately 41% of these individuals had osteopenia and 49% had osteoporosis. X-ray patients were somewhat evenly split between males and females (54% female) aged 57 ± 17 years old (no T-score data). X-ray radiographs were gathered in intraoperative settings or outpatient settings (primarily due to follow-up on prior fracture or investigating unspecified back pain). Refer to the last row of Table 1 for patient characteristics as well as imaging characteristics.
Table 1. Dataset Information.
Overview of data used to train and test the neural network. For each fold, 80% of cases are reserved to train the network to detect vertebral bodies and produce landmark annotations. 20% of cases in each fold are reserved for testing and are never used to train the network (i.e. the network has not “seen” them before). Training cases are augmented, increasing the effective training set size 5x. Additional information is provided about the number of vertebrae in the data sets that are located in the lumbar, thoracic, and cervical regions of the spine (note there were no X-rays containing cervical vertebrae in the dataset we have). Statistics on patient characteristics for age, sex, and T-Score are also reported (* CT BMD/T-Score data from institutional collaborator is incomplete and statistic is only calculated on known data). T-Score measurements were collected from DXA scans done within one year of MR/CT scan. Note, MR scanner types were not mentioned due to the large time range over which these scans were gathered (comprising of dozens of different scanner types/manufacturers). We present the imaging characteristics instead.
| Measure | Modality | Metric |
|---|---|---|
| # of imaging studies training data generated from (prior to augmentation) | MR | 898 |
| CT | 110 | |
| X-Ray | 387 | |
| # of training imaging studies (after augmentation) | MR | 4490 |
| CT | 550 | |
| X-Ray | 1935 | |
| # of testing images | MR | 225 |
| CT | 27 | |
| X-Ray | 97 | |
| # of vertebrae in each spine region (entire dataset, pre-augmentation) | MR | Lumbar: 2895; Thoracic: 1396; Cervical: 3402 |
| CT | Lumbar: 749; Thoracic: 377; Cervical: 542 | |
| X-Ray | Lumbar: 2396; Thoracic: 599; Cervical: N/A | |
| Patient characteristics | MR | Age: 67 ± 11 (1 SD); 88% Female; T-Score = 0.36 ± 2.1 (1 SD) |
| CT | Age: 65 ± 5 (1 SD); 67% Female*; T-Score = −2.48 ± 1.13 (1 SD)* | |
| X-Ray | Age: 57 ± 17 (1 SD); 54% Female; T-Score = N/A (patient scans were gathered in intraoperative settings or outpatient settings. Primary reasons for conducting scans were back pain/monitoring due to prior fracture history). | |
| Imaging Characteristics | MR | MR images T1 and T2 weighted images were acquired via a fast-spin echo sequence Magnetic Field Strength: 1.5T-3T; Slice Thickness: 3–4 mm; Repetition Time: Mean (625.77) Range (250–5000); Echo Time: Mean (11.86), Range (5.56–101.02); Pixel Size: Mean (0.55mm), Range (0.25mm-1.50mm); Matrix Size: width, height, and row measurements, Mean (487.56) Range (256–1024). |
| CT | CT scans were performed with or without contrast and imaging data were acquired with scanners in helical mode, with slice thickness of 0.5–1mm, and with a peak tube voltage of 120–140 kVp on Phillips Brilliance 64, iCT 256, & IQon scanners and Siemens Somatom Definition AS and AS+ scanners. | |
| X-Ray | Spine X-ray radiographs were acquired with a numerous equipment types and imaging conditions and were resized to 3520 × 4280 pixels (0.19mm per pixel). |
2.3. Neural Network Training and Testing:
The neural network underlying SpineTK was trained to produce segmentations using imaging studies that were annotated with manually made segmentations. Each case in the MR, CT, and X-ray datasets were segmented (using ITK-SNAP [12]) by one of 24 trained experts with several hundred hours of segmentation experience each (and segmentations are checked for accuracy by another trained expert). Annotated cases were allocated to either train the network or evaluate the network (i.e. test the network) such that the cases used to evaluate the network’s accuracy were not used to train the network.
To increase the robustness of the neural network to variations in imaging studies, data allocated to train the network was augmented (i.e. replicated with transformations) with five transformations which randomly change the rotation, contrast, brightness, crop, and vertical/horizontal flip of underlying case data (ref Supplemental Materials/Methods: Section 2 for exact parameters for augmentations). Table 1 shows the number of imaging slices prior to and after augmentation for each imaging modality, resulting in a 5x increase in the effective size of the training set.
For evaluation, 5-fold cross validation was carried out to report whether the network was easily influenced by variations in training data. In this procedure, cases were split into 5 parts prior to augmentation. 4/5 parts are used to train the network along with their augmented counterparts, and the remaining part is used to evaluate (test) the network. This allocation was repeated five times such that each part (called a “fold”) serves as testing data exactly one time, producing 5 results for each network. In the remainder of the paper, we specified results in the main text as averages of the five folds to give an accurate representation of the capabilities of the network. We also performed analyses to show that, within each fold, training and testing data did not significantly differ from each other for patient population characteristics (age, sex, T-score) using a 2-sample t-test (α = 0.05, refer to Supplemental Table 1: Row(a)).
The network was trained using NVIDIA Tesla K80 GPUs with 12 GB of RAM provided on the free-tier of the Google CoLab cloud computing service. All case images were converted to a PNG format (using pydicom v2.1.1) to load imaging data into computer memory. Pixel values were all scaled to the range of 0–255 for standardization, using OpenCV v4.4.0 and NumPy v1.19.0 [13]–[15]. Full description of training parameters and programmatic libraries used can be found in Supplemental Materials/Methods: Section 5.
2.4. Evaluation:
2.4.1. Network Evaluation Time
The ability of the network to process data quickly was measured by evaluating how long it takes to produce a segmentation output after being run on a testing dataset input. Evaluation time was averaged over all of the testing cases.
2.4.2. Slice Selection and Vertebral Body Detection Accuracy
Since it is necessary for the network to detect vertebral bodies prior to segmentation, performance of vertebral body detection was evaluated in terms of accuracy. A vertebral body counted as a true detection only when its bounding box exceeded a 70% overlap, using an Intersection over Union (IoU) metric with the ground-truth bounding box (derived from manually annotated segmentation masks). This cutoff was selected through a separate precision-recall curve analysis that showed this threshold achieved a balance between precision and sensitivity on the object detection task.
Additionally, the network must also select the correct slice (typically the most medial slice) to evaluate for segmentations. To measure the network’s ability to select the correct slice, we determined the number of slices between the slice the neural network selected for evaluation and the one our annotators selected for segmentation. We report this metric as an error distance and determine if that error distance is significantly different from zero (using a 1-sample t-test where it is favorable for the H<sub>o</sub> [ = error distance is not significantly different from 0] to not be rejected).
2.4.3. Segmentation Accuracy
Segmentation accuracy was measured in terms of a Dice similarity coefficient (DSC, aka Dice Score) which was calculated using the following formula
Where X is the set of predicted mask pixel locations, and Y is the set of ground-truth annotated pixel locations. DSCs range from 0 to 1 where 1 indicates a segmentation that matches the ground truth segmentation for all pixel locations in the segmentation masks (i.e. a 100% accurate segmentation). We evaluated the DSC for each modality and each vertebral body in the testing dataset of each fold. We also qualitatively assessed the output of segmentations for each network.
2.4.4. Radiomic Quantity Agreement
To determine whether the outputs of the network can be used for radiomics measurements, we used the predicted segmentation masks (for each vertebral body) from the network to evaluate 4 radiomic quantities: skewness, kurtosis, mean of positive valued pixels, and entropy. These values were chosen because of their demonstrated utility in prior vertebral bone health assessments ([4], [5]) and the fact that their values are highly dependent on pixels within segmentation masks. We compared these values (generated from outputs of the neural network) to the quantities generated from ground-truth segmentation masks. We then determined if the values derived from the predicted segmentations are significantly different from the values derived from the ground-truth segmentations (using a paired t-test: α = 0.05; H<sub>o</sub> = the predicted-segmentation-derived radiomics values are not significantly different from the ground-truth-derived radiomics values).
3. Results:
3.1. Network Evaluation Time:
Evaluation time (measured as the amount of seconds it takes for the network to evaluate an image from the testing dataset for each imaging modality once loaded into memory) reached an average of 1.656±0.117 s (MR: 1.566±0.098 s, CT: 1.697±0.141 s, X-ray: 1.683±0.119 s) for the segmentation task. All evaluation times are measured on NVIDIA Tesla K80s GPUs with 12 GB of RAM available via Google CoLab (free tier).
3.2. Vertebral Body Detection Accuracy:
Slice selection accuracy was evaluated across all folds for both modalities. Average accuracy across all folds for MR images was 86.5% (range: 85.3%−87.5%) with mean error distances (i.e. difference between predicted slice number and manually selected slice number) in the range of −0.04 to 0.04. For CT images, the average across all folds was 86.7% (range: 81.5%−92.6%) with mean error distances in the range - 0.04 to 0.19. At maximum, the network selected a slice that was 2 slices away from the slice the human annotator selected. All folds reported non-significant differences in slice selection compared to human annotator (all p > 0.05), indicating the automated slice selection procedure was not different compared to a human. Individual fold results can be found in Supplemental Table 1 (Row b).
For all subsequent evaluation procedures, the network was evaluated only on the same slice as the human annotators. Across all folds, modalities, and regions of the spine, the networks achieved an average of 95.2% accuracy at detecting vertebral bodies (range = 90–99%; ref Supplemental Table 1-Row c for fold-specific data). Accuracy exceeded 90% accuracy in each of the regions (lumbar, thoracic, cervical) across all folds. To analyze where the network failed to detect data, manual analysis was done on one fold. This analysis examined the vertebrae that did not reach IoU threshold and grouped these missed vertebrae into four categories. Specific numbers of vertebral body misdetections by category (vertebral body detected but below threshold, obstructive devices in scan, fused vertebrae, or imaging artifacts present) can be found in Table 2.
Table 2. Common misses.
This table outlines the common reasons for why vertebrae were missed from the object detector in the 3rd fold testing set. Main categories are outlined in the 2nd through 4th column.
| Network Modality (Type) | Vertebra detected but below IoU threshold | Obstructive devices (e.g. screws, wires) | Fused vertebrae | Imaging artifacts | Total Accuracy |
|---|---|---|---|---|---|
| MRI (Landmark) | 30 | 0 | 4 | 0 | 1505/1539 = 97.8% |
| CT (Landmark) | 0 | 2 | 5 | 8 | 312/327 = 95.4% |
| X-ray (Landmark) | 8 | 20 | 0 | 1 | 582/611 = 95.2% |
3.3. Segmentation Accuracy:
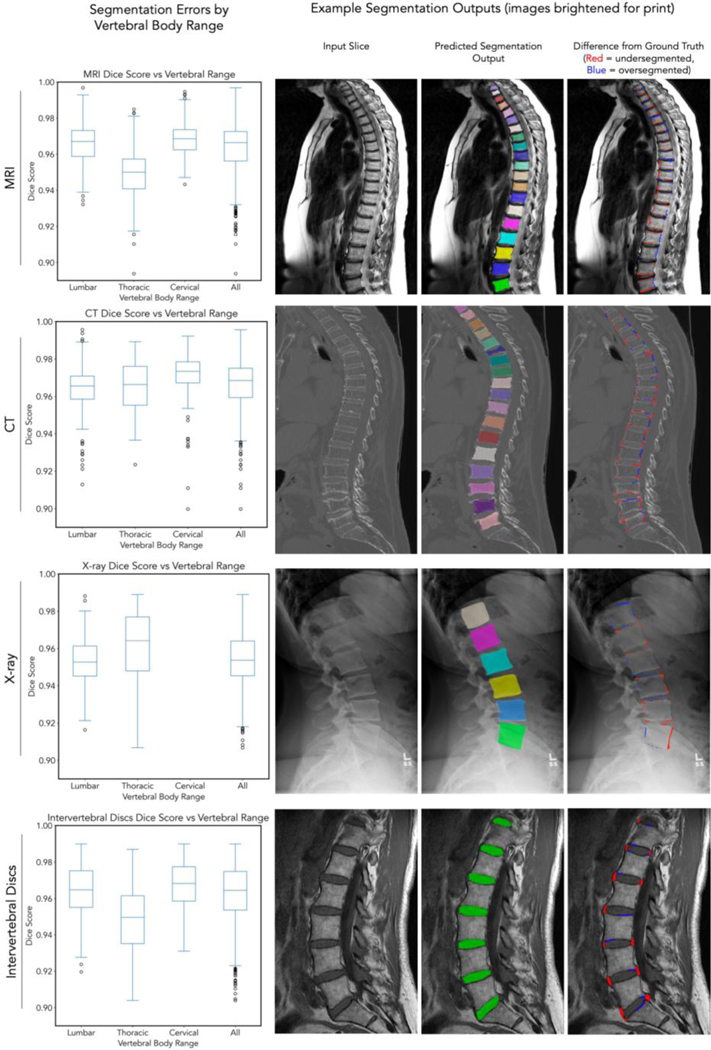
We evaluated the ability for the DL system to produce segmentations for vertebral bodies. For the MR segmentation network, the median DSC was 0.958 (IQR 25th-75th percentile: 0.944–0.963) across all regions and folds. Additionally, for the MRI segmentation network, we were able to output segmentations of intervertebral discs. The median DSC for discs was 0.959 (IQR: 0.942–0.971) across all folds and regions. For the CT segmentation network, the median DSC was 0.967 (IQR: 0.954–0.972) across all regions and folds. For the X-ray segmentation network, the median DSC was 0.957 (IQR: 0.934–0.964). Qualitatively, we report that most segmentations tend to oversegment (include more pixels than ground-truth) and undersegment (exclude pixels from ground truth) mainly in areas isolated to the edges and corners of vertebral bodies, with the vast majority of inner regions of the vertebral body being segmented correctly. Data on individual folds can be found in Supplemental Table 1 (Row d). We also visualize the results from one fold of data in Figure 3.
Figure 3: Segmentation results.
For each row, left pane corresponds to Dice score for that imaging modality across individual vertebral bodies (C1, C2 excluded because of absence from testing set). Box and whisker plots show median, 25th-75th percentile range and 1.5x IQR + outliers. Right three panels correspond to an input image, output of segmentations from the network evaluated on testing images (note the different colors that designate individual vertebral bodies as separate entities), and a difference map highlighting where segmentations differ from ground truth manually annotated for the input image. We highlight that these images contain some of the best (e.g. X-ray: cyan vertebral body DSC 0.99) and some of the worst segmentations (e.g. CT: dark purple vertebral body DSC 0.91) in our network. Supplemental Figure 2 contains vertebra-by-vertebra and intervertebral disc Dice scores. Data visualized is from fold 3.
3.4. Radiomics Quantities Accuracy:
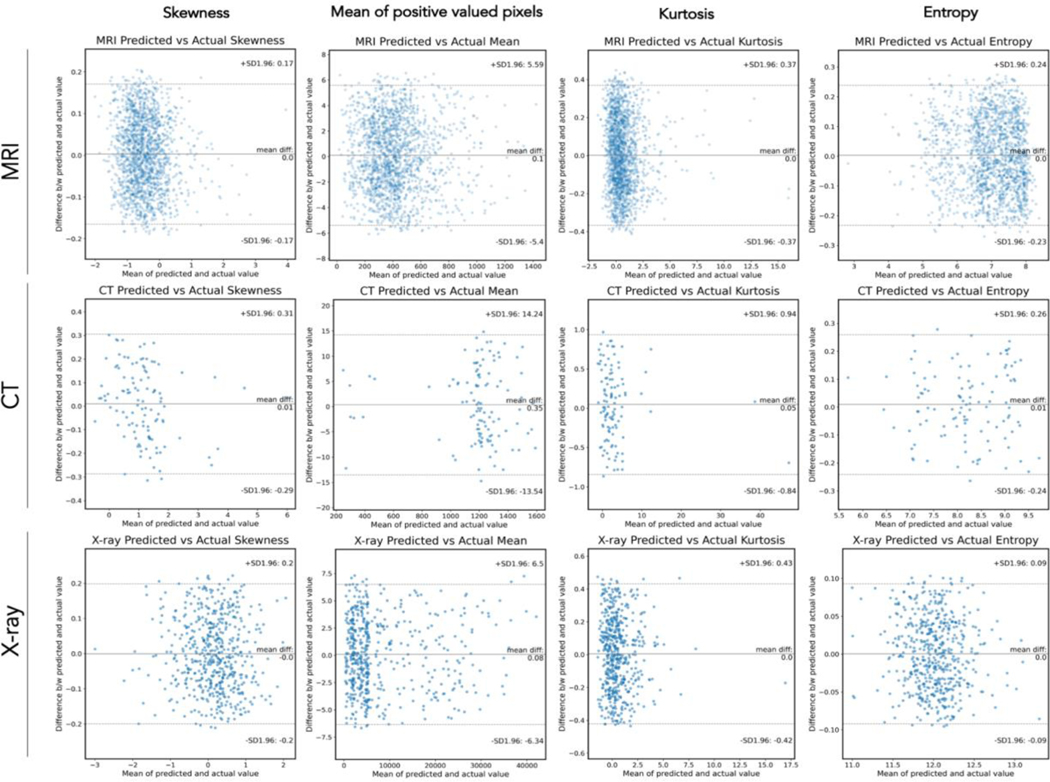
To demonstrate the applicability of segmentations produced from the network, we also calculated the following quantities from pixels included in predicted segmentation masks: kurtosis, skewness, entropy, and mean of positive valued pixels. We then compared these values (derived from predicted masks) to values generated from ground-truth masks. For the MR network (on bone), average error (i.e. average difference between radiomic values derived from predicted vs ground-truth masks) was between 0.001–0.008 for Kurtosis, 0.001–0.003 for Skewness, 0.001–0.003 for Entropy, and −0.019–0.010 for mean of positive valued pixels (MPP) across all folds (average r-values for all features ≥ 0.96, average relative errors all ≤ 5.8%). For the CT network, average error was between −0.064–0.003 for Kurtosis, 0.019–0.010 for Skewness, −0.006–0.009 for Entropy, and −0.211–0.353 for MPP across all folds (average r-values for all features ≥ 0.96, average relative errors all ≤ 6.2%). For the X-ray network, average error was between −0.003–0.013 for Kurtosis, −0.002–0.005 for Skewness, −0.003–0.002 for Entropy, and - 0.018–0.187 for MPP across all folds (average r-values for all features ≥ 0.97, average relative errors all ≤ 5.4%). Using a paired t-test, we found that values derived from predicted segmentation masks were not significantly different from the ground-truth derived values (all p > 0.05). Agreement data is summarized in Bland-Altman plots in Figure 4 for one fold of data along with standard deviations for error values. Data from each of the individual folds is summarized in Supplemental Table 1 (Row e).
Figure 4: Agreement Results for Radiomics Measurements.
For each modality (MR, CT, X-ray; rows) and each radiomic feature (skewness, mean of positive valued pixels, kurtosis, entropy), a Bland-Altmann plot is pictured to display agreement data. Central line = mean difference. Dashed upper and lower lines indicate ± 1 SD. X axis is mean of actual and predicted value, Y axis is difference between mean and predicted value.
4. Discussion & Conclusion:
We showed that the deep learning algorithm was able to successfully detect and segment vertebral bodies on a automatically selected central slice. For each version, one per modality, we evaluated the network’s ability to choose the most relevant slice for analysis, detect vertebrae, and segment vertebrae/discs. We further examined segmentation outputs to determine their utility in radiomics measures and found that radiomic quantities calculated from network segmentation masks did not significantly differ from those generated from manually made segmentation masks. To the authors’ knowledge, this is the first DL system of its type that has attempted to work on multiple modalities and has achieved the level of accuracy across the output metrics we established.
In comparison to prior works, our network outperforms them in the segmentation task. Vania et al [8] produce a U-Net architecture that achieves a 2D Dice coefficient of 0.94 for CT images only (though they do so in the axial plan; no details were provided on the training set). Our network achieves a median Dice coefficient of 0.967. Lu et al [9] demonstrate a U-Net approach on Lumbar Sagittal MR-images achieves a mean Dice coefficient of 0.93 (T2 MR, trained and tested on 4075 cases). For lumbar MR images, our network achieves a median Dice Coefficient of 0.960 (IQR: 0.951–0.966) across all folds. Cho et al [16] also use a U-Net segmentation approach on sagittal X-ray images and achieve a DSC of 0.841 (region unspecified, 629 lumbar radiographs used to train, 151 used to test). Our network achieves a median DSC of 0.957 (IQR: 0.934–0.964). Chen et al [7] take a another approach using a J-CNN and achieve a DSC of 0.910 on CT images (242 training cases, 60 test cases). Our network exceeds that metric (as mentioned above). No currently existing literature explains how segmentations can be used for radiomic quantities and (to the authors’ knowledge) this paper mentions the first results of applying segmentations to calculate radiomic features. In comparison to the aforementioned architectures, we note that our architecture may perform better due to its ability to extract image features and segment only within detected objects. These two steps reduce the feature space the segmentation network needs to learn on, leading to higher accuracy.
This network can contribute to clinical medicine and research studies through its ability to produce segmentations quickly (< 2 seconds/slice) for deformities on relatively inexpensive hardware. Furthermore, outputs from the network can be directly input into popular radiomics software packages/libraries to produce measurements automatically for further bone health analysis/diagnostic studies. Since the network produces segmentations across multiple modalities, this network can be useful to be useful to researchers and clinicians calculating radiomic quantities for a variety of diseases.
We also note that our network design can be used to repurpose this network towards other tasks. We have demonstrated that this network can be trained to segment the training data accurately in < 30 minutes (on publicly available free, compute power) once training data is annotated. Similarly, it can be used to train on other modalities and imaging planes. Additionally, we show that the network can achieve high accuracy by training on only hundreds of images annotated, due to the network’s use of image augmentation and a pre-trained feature extraction backbone. These factors enable researchers to repurpose this network’s capabilities without need for using extensive resources on manual annotation.
Generally, our network architecture currently works to produce segmentations for a single 2D slice. While this constraint is sufficient for calculating radiomic quantities in patients without scoliosis, it may not capture all vertebrae on a single central slice in patients with severe scoliosis in MR/CT imaging studies, limiting the applicability of the network architecture in those cases. Other limitations from the network primarily arise from the underlying data. Undetected vertebrae came from cases that had other obstructive devices, fused vertebrae, etc. By using more cases that have these occlusions, it may be possible to increase detection accuracy. Furthermore, for minimizing over and under segmentation errors, higher resolution scans can be used to train the network (allowing the network to gather more information about segmentation parameters to be used near edges of vertebral bodies and discs). Lastly, the data we had access to may not accurately reflect the underlying patient population for all potential use cases (especially considering that there is a significant male/female imbalance in the MR and CT datasets). These imbalances can affect results for populations underrepresented in the training dataset. This network will be evaluated and retrained on new datasets as they become available, working towards achieving comprehensive applicability in the clinical setting.
4.1. Conclusion:
We were able to create a deep learning system that can produce segmentations of vertebrae and intervertebral discs automatically, rapidly, and accurately across all regions of the spine and multiple modalities (MR, CT, X-ray). Outputs from the network can be used to calculate measurements for radiomic studies and clinical assessments of spine health. Future directions for this study include training the network to produce 3D volumes for vertebral bodies and discs and to examine its utility in a clinical care setting and performing a rigorous comparison of this model to other architectures on a standardized dataset.
Research Data for this Article:
Due to the sensitive nature of the imaging studies, participants were assured raw data would remain confidential and would not be shared. Data not available / The data that has been used is confidential. Resulting models trained in this study and code used to evaluate imaging studies will be made available upon publication.
Supplementary Material
Highlights:
A deep learning system was created that can automatically segment vertebral bodies and intervertebral discs for MR, CT, and X-ray imaging studies. It can do so across all spine regions with high accuracy.
The system achieved a median Dice Score of ≥ 0.95 (measure of 2D segmentation accuracy, best = 1) on an automatically detected central slice. It was also able to quantify radiomic features highly accurately (r ≥ 0.96 across skewness, kurtosis, mean pixel value, and entropy) across all three modalities.
This deep learning system could produce segmentations in < 1.7 seconds/slice on average using free cloud compute power and can be used immediately for radiomic studies and assessments of spine health.
Abbreviations:
- MR
Magnetic Resonance (Imaging)
- CT
Computed Tomography
- AI
Artificial Intelligence
- DL
Deep Learning
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Salaffi F. et al. , “The burden of prevalent fractures on health-related quality of life in postmenopausal women with osteoporosis: the IMOF study,” J. Rheumatol, vol. 34, no. 7, pp. 1551–1560, Jul. 2007. [PubMed] [Google Scholar]
- [2].Office of the Surgeon General (US), Bone Health and Osteoporosis: A Report of the Surgeon General. Rockville (MD): Office of the Surgeon General (US), 2004. [PubMed] [Google Scholar]
- [3].Weaver J, Sajjan S, Lewiecki EM, Harris ST, and Marvos P, “Prevalence and Cost of Subsequent Fractures Among U.S. Patients with an Incident Fracture,” J. Manag. Care Spec. Pharm, vol. 23, no. 4, pp. 461–471, Apr. 2017, doi: 10.18553/jmcp.2017.23.4.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Filograna L. et al. , “Identification of the most significant magnetic resonance imaging (MRI) radiomic features in oncological patients with vertebral bone marrow metastatic disease: a feasibility study,” Radiol. Med. (Torino), vol. 124, no. 1, pp. 50–57, Jan. 2019, doi: 10.1007/s11547-018-0935-y. [DOI] [PubMed] [Google Scholar]
- [5].Hwang E-J, Kim S, and Jung J-Y, “Bone Marrow Radiomics of T1-Weighted Lumber Spinal MRI to Identify Diffuse Hematologic Marrow Diseases: Comparison With Human Readings,” IEEE Access, vol. 8, pp. 133321–133329, 2020, doi: 10.1109/ACCESS.2020.3010006. [DOI] [Google Scholar]
- [6].Galbusera F, Casaroli G, and Bassani T, “Artificial intelligence and machine learning in spine research,” JOR Spine, vol. 2, no. 1, p. e1044, Mar. 2019, doi: 10.1002/jsp2.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chen H. et al. , “Automatic Localization and Identification of Vertebrae in Spine CT via a Joint Learning Model with Deep Neural Networks,” in Medical Image Computing and Computer-Assisted Intervention -- MICCAI 2015, vol. 9349, Navab N, Hornegger J, Wells WM, and Frangi A, Eds. Cham: Springer International Publishing, 2015, pp. 515–522. [Google Scholar]
- [8].Vania M, Mureja D, and Lee D, “Automatic spine segmentation from CT images using Convolutional Neural Network via redundant generation of class labels,” J. Comput. Des. Eng, vol. 6, no. 2, pp. 224–232, Apr. 2019, doi: 10.1016/j.jcde.2018.05.002. [DOI] [Google Scholar]
- [9].Lu J-T et al. , “DeepSPINE: Automated Lumbar Vertebral Segmentation, Disc-level Designation, and Spinal Stenosis Grading Using Deep Learning,” ArXiv180710215 Cs, July. 2018, Accessed: Sep. 01, 2020. [Online]. Available: http://arxiv.org/abs/1807.10215. [Google Scholar]
- [10].Sekuboyina A. et al. , “VerSe: A Vertebrae Labelling and Segmentation Benchmark,” ArXiv200109193 Cs Eess, June. 2020, Accessed: Aug. 26, 2020. [Online]. Available: http://arxiv.org/abs/2001.09193. [Google Scholar]
- [11].He K, Gkioxari G, Dollár P, and Girshick R, “Mask R-CNN,” ArXiv170306870 Cs, Jan. 2018, Accessed: Oct. 02, 2020. [Online]. Available: http://arxiv.org/abs/1703.06870. [Google Scholar]
- [12].Yushkevich PA et al. , “User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability,” NeuroImage, vol. 31, no. 3, pp. 1116–1128, Jul. 2006, doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
- [13].Mason D. et al. , pydicom/pydicom: pydicom 2.1.1. Zenodo, 2020. [Google Scholar]
- [14].Bradski G, “The OpenCV Library,” Dr Dobbs J. Softw. Tools, 2000. [Google Scholar]
- [15].van der Walt S, Colbert SC, and Varoquaux G, “The NumPy Array: A Structure for Efficient Numerical Computation,” Comput. Sci. Eng, vol. 13, no. 2, pp. 22–30, Mar. 2011, doi: 10.1109/MCSE.2011.37. [DOI] [Google Scholar]
- [16].Cho BH et al. , “Automated Measurement of Lumbar Lordosis on Radiographs Using Machine Learning and Computer Vision,” Glob. Spine J, vol. 10, no. 5, pp. 611–618, August. 2020, doi: 10.1177/2192568219868190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Jung A, Wada K, Tanaka S, and Graving J, imgaug. 2020. [Google Scholar]
- [18].Wu Yuxin, Kirillov A, Massa F, Lo W-Y, and Girshick R, Detectron 2. 2019. [Google Scholar]
- [19].Paszke A. et al. , “PyTorch: An Imperative Style, High-Performance Deep Learning Library,” ArXiv191201703 Cs Stat, December. 2019, Accessed: Aug. 26, 2020. [Online]. Available: http://arxiv.org/abs/1912.01703. [Google Scholar]
- [20].Virtanen P. et al. , “SciPy 1.0--Fundamental Algorithms for Scientific Computing in Python,” Nat. Methods, vol. 17, no. 3, pp. 261–272, March. 2020, doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hunter JD, “Matplotlib: A 2D Graphics Environment,” Comput. Sci. Eng, vol. 9, no. 3, pp. 90–95, 2007, doi: 10.1109/MCSE.2007.55. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.