Abstract
Background:
Classical methods for detecting left ventricular hypertrophy (LVH) using 12-lead electrocardiograms (ECGs) are insensitive. Deep learning models using ECG to infer cardiac magnetic resonance (CMR)-derived LV mass may improve LVH detection.
Methods and Results:
Within 32,239 individuals of the UK Biobank prospective cohort who underwent CMR and 12-lead ECG, we trained a convolutional neural network to predict CMR-derived LV mass using 12-lead ECGs (“LVM-AI”). In independent test sets (UK Biobank [n=4,903] and Mass General Brigham [MGB, n=1,371]), we assessed correlation between LVM-AI predicted and CMR-derived LV mass and compared LVH discrimination using LVM-AI versus traditional ECG-based rules (i.e., Sokolow-Lyon, Cornell, aVL, or any ECG rule). In the UK Biobank and an ambulatory MGB cohort (MGB Outcomes, n=28,612), we assessed associations between LVM-AI predicted LVH and incident cardiovascular outcomes using age- and sex-adjusted Cox regression. LVM-AI predicted LV mass correlated with CMR-derived LV mass in both test sets, although correlation was greater in the UK Biobank (r=0.79) versus MGB (r=0.60, p<0.001 for both). When compared to any ECG rule, LVM-AI demonstrated similar LVH discrimination in the UK Biobank (LVM-AI c-statistic 0.653; 95% CI, 0.608–0.698 vs any ECG rule c-statistic 0.618; 95% CI, 0.574–0.663, p=0.11), and superior discrimination in MGB (0.621; 95% CI, 0.592–0.649 vs 0.588; 95% CI, 0.564–0.611, p=0.02). LVM-AI predicted LVH was associated with incident atrial fibrillation, myocardial infarction, heart failure, and ventricular arrhythmias.
Conclusions:
Deep learning-inferred LV mass estimates from 12-lead ECGs correlate with CMR-derived LV mass, associate with incident cardiovascular disease, and may improve LVH discrimination compared to traditional ECG rules.
Keywords: machine learning, left ventricular hypertrophy, electrocardiogram
Introduction
Left ventricular hypertrophy (LVH) is defined as pathologically increased LV mass1 and predicts adverse cardiovascular events including atrial fibrillation (AF)2 and heart failure (HF).3 Electrocardiograms (ECGs) are common, inexpensive, and have been used to infer the presence of LVH for decades using amplitude-based rules.4,5 Yet studies consistently demonstrate that ECG-based LVH rules have limited sensitivity.6 Cardiac magnetic resonance (CMR) provides accurate and reproducible quantification of cardiac structure and now represents the gold-standard for LVH diagnosis.7
Deep learning architectures are a subset of machine learning algorithms capable of modeling multiple nonlinear interactions present within complex data.8 A potential role for deep learning on clinical data is to leverage information present within data types available at scale to infer rich structural features typically available only through complex, expensive, or invasive diagnostics.9 CMR is costly, time-consuming, and not universally available. Conversely, ECG is inexpensive, ubiquitous, and may contain sufficiently rich information to infer cardiac structure.
Recent work has demonstrated that LV mass estimation using deep learning on 12-lead ECG is feasible,9,10 but previous studies have utilized echocardiogram-based LV mass,9,10 were not designed to assess for associations between predicted LV mass and incident cardiovascular outcomes, and developed models within modestly sized and retrospectively ascertained healthcare-related datasets which are subject to selection bias and may have limited generalizability.
In this study, we analyzed a unique dataset of over 35,000 individuals in the UK Biobank prospective cohort study who underwent acquisition of both 12-lead ECG and CMR, and trained a deep learning model to infer CMR-derived LV mass using 12-lead ECG (“LVM-AI”). We then compared the performance of LVM-AI to established ECG-based criteria4,5 for LVH diagnosis in independent test sets from the UK Biobank and an external healthcare system (Mass General Brigham, MGB), and assessed whether LVM-AI predicted LVH was associated with incident cardiovascular events.
Methods
Data availability
UK Biobank data are publicly available by application (www.ukbiobank.ac.uk). MGB data contain protected health information and cannot be shared publicly. The code underlying LVM-AI is accessible at https://github.com/broadinstitute/ml4h/tree/master/model_zoo/left_ventricular_mass_from_ecg_student_and_mri_teacher.
Derivation sample
The UK Biobank is a prospective cohort of 502,629 participants recruited between 2006–2010.11 Briefly, approximately 9.2 million individuals aged 40–69 living within 25 miles of the 22 assessment centers in England, Wales, and Scotland were invited, and 5.4% participated in the baseline assessment. Extensive questionnaires, physical measures, and biological samples were collected at recruitment, with multimodal imaging obtained in a large subset. All participants are followed for health outcomes through linkage to national datasets. Participants provided written informed consent. The UK Biobank was approved by the UK Biobank Research Ethics Committee (reference number 11/NW/0382). Use of UK Biobank (application 7089) and MGB data were approved by the local MGB Institutional Review Board.
Baseline assessment
For this analysis, we included all individuals who underwent both resting 12-lead ECG and CMR contemporaneously during the UK Biobank imaging assessment. Demographics including age, sex, and race, and physical measurements including height, weight, and body mass index (BMI) were obtained at the imaging assessment or study visit most closely preceding.
The UK Biobank CMR protocol has been described previously.12 Briefly, all CMRs were acquired on a clinical wide-bore 1.5 Tesla scanner (MAGNETOM Aera, Syngo Platform VD13A, Siemens Healthcare, Erlangen, Germany) and used balanced steady-state free precession with typical parameters.
Data processing
Resting 12-lead ECG data were downloaded as XML files and converted into arrays of lead amplitude sequence data. CMR images were downloaded as DICOM slabs and converted into arrays of three-dimensional voxel data. We utilized a validated deep learning model (ML4Hseg)13 to extract LV mass from the CMR images, which served as the ground truth for LVM-AI. A conceptual overview of the study is shown in Figure 1.
Figure 1.
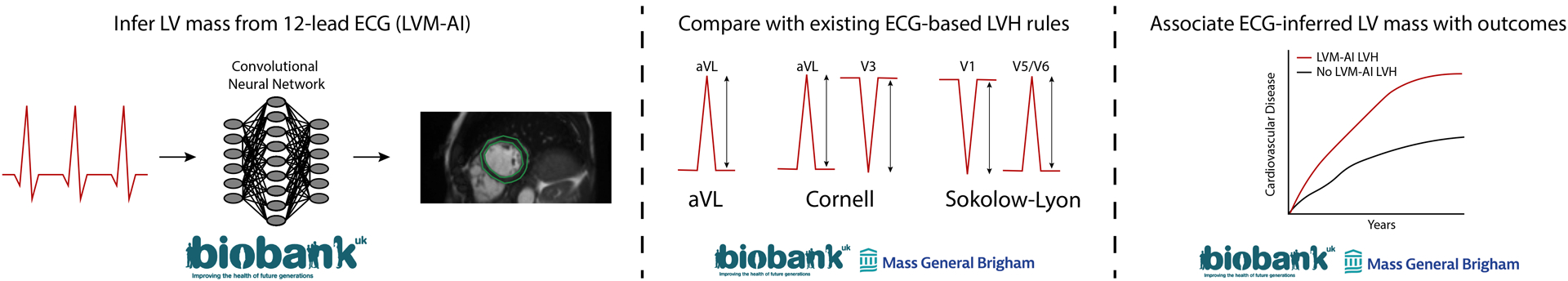
Study conceptual overview
We developed LVM-AI – a deep learning model to infer CMR-derived LV mass using 12-lead ECG – in the UK Biobank. Then, we compared LVM-AI to traditional ECG-based rules for diagnosing CMR-derived LVH in independent test sets within the UK Biobank and Mass General Brigham (MGB). Lastly, we assessed for associations between LVM-AI predicted LVH and incident cardiovascular disease in the UK Biobank and an MGB-based ambulatory cohort.
LVM-AI
After setting aside a random sample (n=4,903) as an internal test set (“UK Biobank Test”, Figure 2), we trained LVM-AI within 32,239 individuals with paired CMR and 12-lead ECG. LVM-AI is a one-dimensional convolutional neural network designed to infer LV mass using 12-lead ECG (Supplemental Figure I). LVM-AI was provided with the entire 10 seconds of the 12-lead ECG waveform as well as participant age, sex, and BMI. Given the clinical importance of diagnosing LVH (i.e., elevated LV mass), LVM-AI utilized a loss function giving additional weight to errors at the high extreme of the LV mass distribution.14
Figure 2.
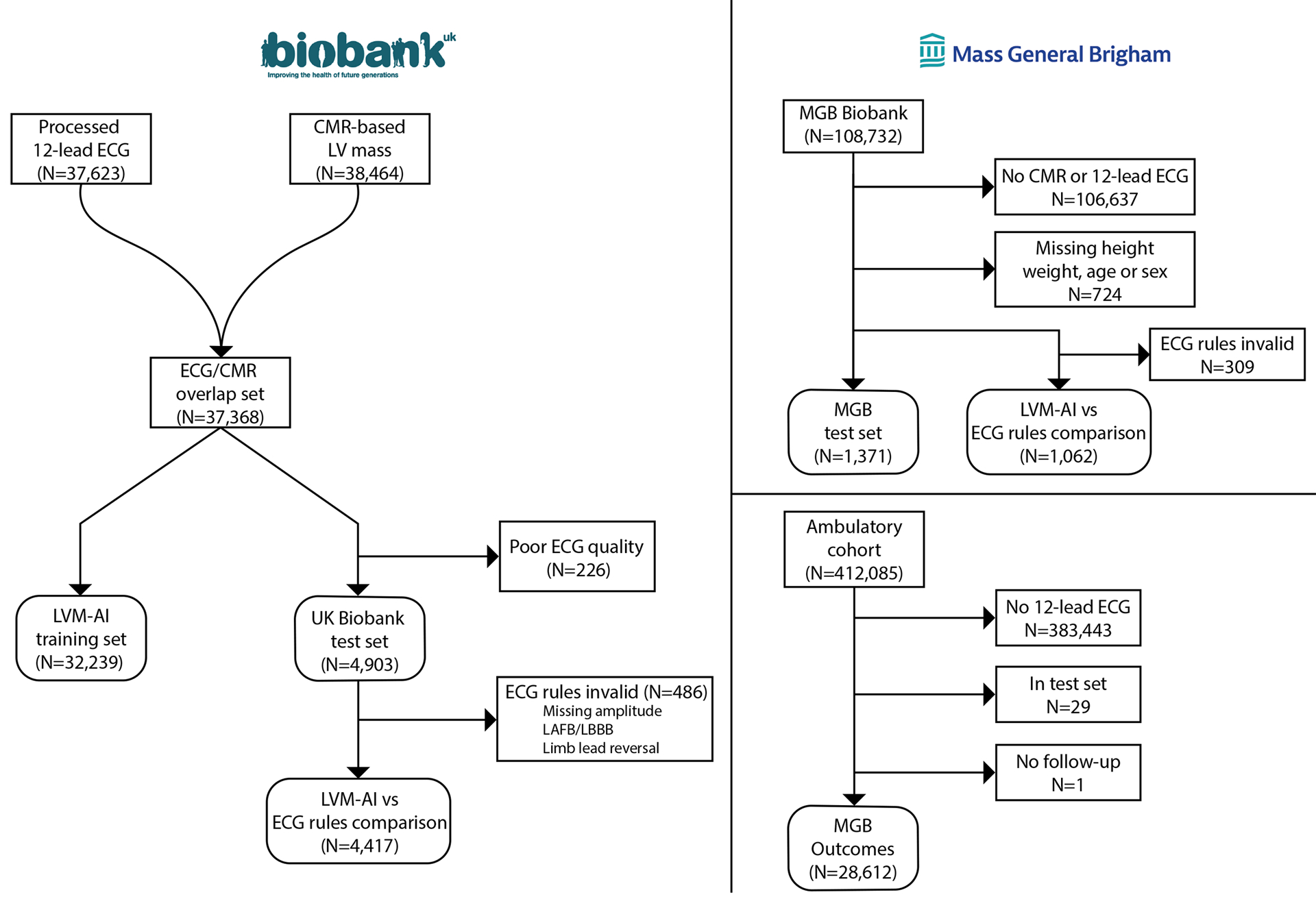
Overview of training and test samples
The training sample consisted of individuals in the UK Biobank with 12-lead ECG and CMR images available. LVM-AI was evaluated in a UK Biobank test set as well as an external healthcare-based MGB dataset. In both test sets, we compared LVM-AI to traditional ECG-based rules for diagnosing CMR-derived LVH. Associations between LVM-AI predicted LV mass index and incident cardiovascular events was performed in the UK Biobank and a separate MGB-based ambulatory cohort (MGB Outcomes).
ECG rules
We sought to compare LVM-AI to the aVL (R wave in lead aVL > 1.1mV),4 Sokolow-Lyon,5 and Cornell voltage4 rules for diagnosing LVH using 12-lead ECG (Supplemental Table I). Although ECG rules adjusted for additional clinical factors have been proposed,15 we focused on the original rules since they are commonly used.6 However, in secondary analyses we assessed the performance of a Cornell voltage product adjusted for age, sex, BMI, and hypertension.15
To apply ECG rules, we extracted lead-specific R and S wave amplitudes from the 12-lead ECG XML files. To validate ECG rule calculation, we developed a plotting function16 to reconstruct 12-lead ECG waveforms for visual interpretation (Supplemental Figure II). Two cardiologists (SK and JPP) manually assessed the accuracy of extracted amplitudes and presence of LVH by each rule. Both per-lead (98.7–100%) and per-rule (98.7–99.3%) accuracy met pre-specified criteria (>90%). Interrater agreement on a per-lead (Gwet’s AC117 0.98) and per-rule (range 0.91–0.96) basis was excellent (Supplemental Tables II-III). Given that ECG-based LVH rules have not been validated in the setting of left bundle branch block, left anterior fascicular block, and limb lead reversal, we excluded these tracings from ECG rule analyses.6
Disease associations
We assessed for associations between LVM-AI predicted LV mass and incident AF, myocardial infarction (MI), HF, and ventricular arrhythmias (VA) within 35,350 participants with follow-up clinical data available after the imaging assessment. Diseases were defined using self-report and inpatient ICD-9/10 codes (updated through 2020-03-31, Supplemental Table IV). Follow-up started at ECG acquisition and spanned until the earliest of an event, death, or last follow-up. Last follow-up was dependent upon the availability of linked hospital data, and was therefore defined as March 31, 2020 for participants enrolled in England (99.6%), October 31, 2016 for participants enrolled in Scotland (0.2%), and February 29, 2016 for participants enrolled in Wales (0.2%).
External validation
We tested LVM-AI in the external MGB healthcare-related dataset (Figure 2). First, we assessed correlation between LVM-AI predicted LV mass and CMR-derived LV mass in a sample of individuals from the MGB Biobank with both ECG and CMR performed within one year of each other (“MGB Test”). Second, we compared the accuracy of LVM-AI predicted LVH to traditional ECG-based rules in MGB Test. Third, we assessed for associations between LVM-AI predicted LVH and incident AF, MI, HF, and VA in a previously described MGB-based dataset independent of MGB Test (“MGB Outcomes”).18 Disease definitions have been previously validated (Supplemental Table IV).18,19 External validation methods are described in detail in the Supplemental Methods.
Statistical analyses
We calculated the Pearson correlation and mean absolute error between LVM-AI predicted LV mass and CMR-derived LV mass. We assessed agreement using Bland-Altman plots.20 We quantified calibration using the mean within-individual difference between LVM-AI predicted and CMR-derived LV mass. To compare LVM-AI to traditional ECG rules, we calculated sensitivity, specificity, positive predictive value, negative predictive value, and c-statistic of each using CMR-derived LVH as the reference. We also generated contingency tables and performed net reclassification analyses. In all analyses, LVH was defined as indexed LV mass >72g/m2 (men) and >55 g/m2 (women).21 The sex-specific 90th percentile of LV mass index was a secondary LVH definition. Indexing for body surface area was performed using the DuBois formula.22 Confidence intervals were generated using the exact method and test characteristics were compared using 1,000-sample bootstrapping.
To assess LVM-AI behavior, we produced saliency maps depicting areas of the ECG having the largest gradients (i.e., greatest influence on LV mass predictions). For each individual, we additionally identified the ECG lead having the highest absolute gradient, as a surrogate for the most influential ECG lead on that individual’s LV mass estimate.
We assessed associations between LVM-AI predicted LV mass index and incident AF, MI, HF, and VA using Cox proportional hazards models adjusted for age and sex. The proportional hazards assumption was assessed by inspecting Schoenfeld residuals. Substantial deviations from proportional hazards (observed for age and sex only) were modeled using interaction terms including strata of person-time. We plotted cumulative risk of events within strata of LVM-AI predicted LVH using the Kaplan-Meier method. Given evidence that anatomic LVH may provide complementary prognostic information to ECG rule-based LVH,23 we fit analogous Cox proportional hazards models using a) the ECG rules, and b) both the ECG rules and LVM-AI predicted LVH as exposures of interest.
We performed sensitivity analyses to assess robustness of our results. First, we trained a version of LVM-AI taking ECG alone as input. Second, we trained a version of LVM-AI utilizing an unweighted logcosh function regression loss (a loss function giving equal weight to errors at either extreme of the LV mass distribution). Third, we compared LVM-AI to a modified Cornell voltage product adjusted for age, sex, BMI, and hypertension.15
Analyses were performed using Python v3.8 (including ‘TensorFlow’),24,25 the ML4H codebase,16 and R v4.0 (packages ‘data.table’, ‘ggplot2’, ‘epiR’, ‘pROC’, ‘nricens’).26 P-values <0.05 were considered statistically significant.
Results
LVM-AI derivation
A total of 37,142 individuals had both CMR-derived LV mass and 12-lead ECG available. Setting aside 4,903 individuals for UK Biobank Test, we trained LVM-AI on a total of 32,239 participants (Figure 2). Individuals in the training set had a mean age of 64.2±7.5 years and 52% were female. The mean CMR-derived LV mass index was 47.0±9.6 g/m2. Other characteristics are shown in Table 1.
Table 1.
Baseline characteristics of training and test sets
| UKBB training set (N=32,239) | UKBB test set (N=4,903) | MGB test set (N=1,371) | MGB Ambulatory (N=28,612) | |
|---|---|---|---|---|
| Age | 64.2±7.5 | 63.6±7.7 | 55.5±14.6 | 62.3±10.4 |
| Female | 16,591 (51.5%) | 2,573 (52.5%) | 630 (46.0%) | 15,012 (52.5%) |
| Race/Ethnicity | - | - | - | - |
| White | 31,166 (96.7%) | 4,763 (97.1%) | 1,178 (86.0%) | 24,812 (86.7%) |
| Black | 221 (0.7%) | 24 (0.5%) | 85 (6.2%) | 1,055 (3.7%) |
| Hispanic or Latino | - | - | 31 (2.3%) | 794 (2.8%) |
| Asian or Pacific Islander | 449 (1.4%) | 54 (1.1%) | 31 (2.3%) | 625 (2.2%) |
| Mixed | 153 (0.5%) | 21 (0.4%) | - | - |
| Other | 163 (0.5%) | 24 (0.5%) | 20 (1.5%) | 406 (1.4%) |
| Unknown | 87 (0.3%) | 17 (0.3%) | 26 (1.9%) | 920 (3.2%) |
| Systolic blood pressure (mmHg) | 138±18 | 137±18 | 126±19 | 131±18 |
| Diastolic blood pressure (mmHg) | 79±10 | 79±10 | 76±12 | 76±10 |
| HTN | 9,893 (30.7%) | 1,413 (28.9%) | 333 (24.3%) | 11,807 (41.3%) |
| Diabetes | 1214 (3.8%) | 150 (3.1%) | 261 (19.0%) | 5,170 (18.1%) |
| Heart failure | 177 (0.55%) | 24 (0.49%) | 173 (12.6%) | 2,499 (8.7%) |
| Myocardial infarction | 652 (2.0%) | 91 (1.9%) | 181 (13.2%) | 3,278 (11.5%) |
| CMR-derived LV mass (g) | 89.1±24.8 | 89.3±24.3 | 120.8±48.7 | - |
| CMR-derived LV mass index (g/m2) | 47.0±9.6 | 47.3±9.4 | 61.1±21.5 | - |
LVM-AI validation
Individuals in UK Biobank Test (mean age 63.6±7.7, 53% female) were similar in composition to the training set, whereas individuals in MGB Test (mean age 55.5±14.6, 46% female) had substantially greater cardiac comorbidity (Table 1). The mean CMR-derived LV mass index was 47.3±9.4 g/m2 in UK Biobank Test and 61.1±21.5 g/m2 in MGB Test. The prevalence of CMR-derived LVH was 126/4,903 (2.6%) in UK Biobank Test and 454/1,327 (34%) in MGB Test.
In UK Biobank Test, when compared to CMR-derived LV mass, LVM-AI demonstrated good correlation (r=0.79; 95% CI, 0.78–0.80, p<0.001), accuracy (mean absolute error 12.6g; 95% CI, 12.3–12.9), and calibration (within-individual mean difference −3.1g; 95% CI, −3.5 to −2.6, Figure 3). Within MGB Test, LVM-AI demonstrated moderate correlation (r=0.48; 95% CI, 0.44–0.52, p<0.001) but poor accuracy (mean absolute error 117.0g, 95% CI, 111.7–122.3), due largely to systematic overestimation (within-individual mean difference −111.8g; 95% CI, −117.3 to −106.2). After linear recalibration (Supplemental Methods III), correlation (r=0.60, 95% CI 0.57–0.64, p<0.001) and accuracy (mean absolute error 28.4g; 95% CI, 27.1–29.8) were improved (Figure 3). In both test sets, correlation was slightly lower for indexed LV mass (UK Biobank r=0.63, 95% CI 0.61–0.64; MGB r=0.51, 95% CI, 0.47–0.55, Figure 3). Bland-Altman plots demonstrated greater agreement in UK Biobank Test (95% limits of agreement −15.1–18.6 g/m2) than MGB Test (95% limits of agreement −36.3–36.3 g/m2), as well as a tendency to make conservative estimation errors in MGB (Supplemental Figure III). Sex-stratified distributions of actual and predicted LV mass are shown in Supplemental Figure IV. Plots depicting learned embeddings from LVM-AI are shown in Supplemental Figure V. Saliency maps demonstrated that components of the ECG waveform plausibly relevant for LV mass estimation (e.g., p wave, early portion of QRS complex) had the greatest impact on LV mass estimates (Figure 4 and Supplemental Figure VI). On an individual basis, the ECG lead exerting the greatest influence on LV mass estimates was most frequently V5 (97.4%), followed by V4 (2.4%) then V1 (0.3%).
Figure 3.
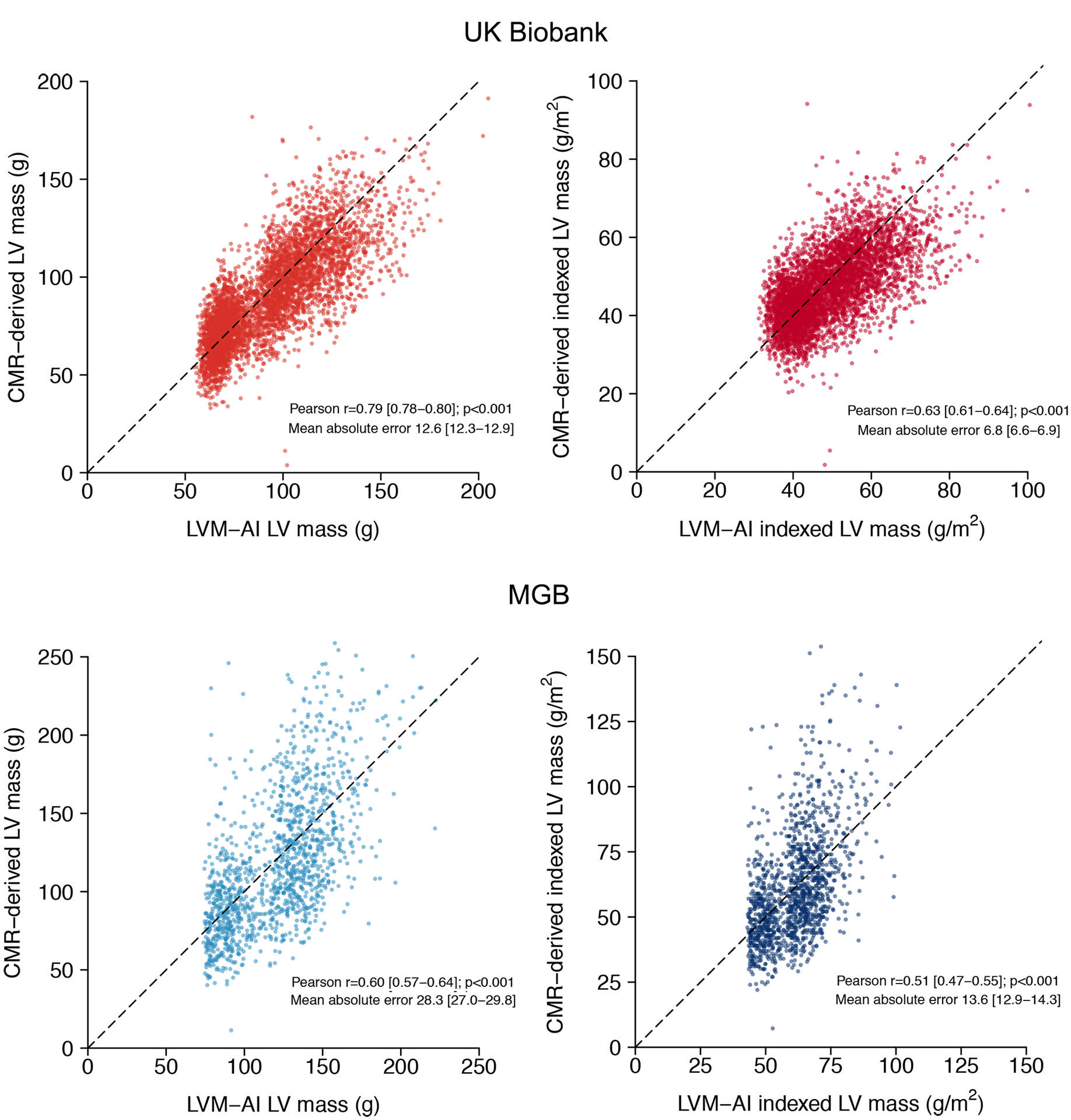
Correlation between LVM-AI predicted and CMR-derived LV mass in test sets
Depicted is the correlation between LVM-AI predicted (x-axis) and CMR-derived (y-axis) LV mass in UK Biobank (upper panels) and MGB (lower panels) test sets. Left panels compare unindexed LV mass and right panels compare indexed LV mass. Lower panels reflect LVM-AI predicted LV mass values after linear recalibration (see text). The diagonal line represents perfect correlation. The Pearson correlation coefficient and mean absolute error are shown on each plot.
Figure 4.
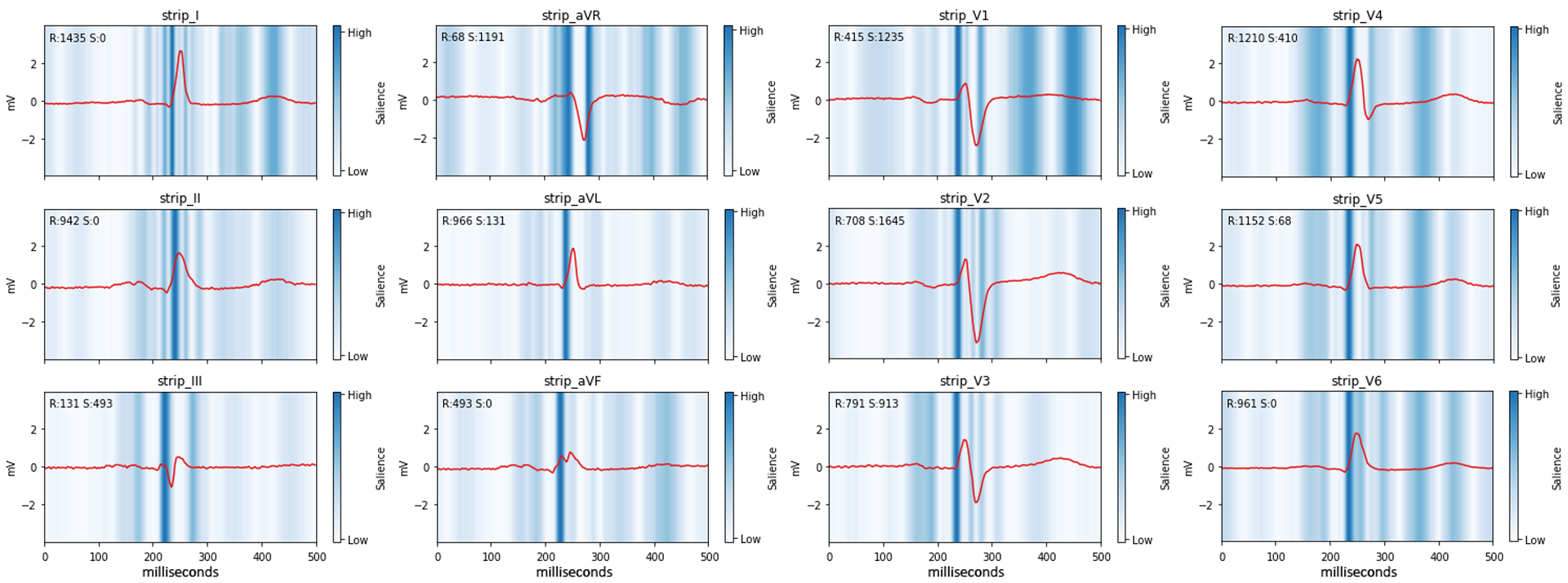
LVM-AI saliency maps
Depicted is a 12-lead ECG tracing and saliency map from a single individual correctly classified by LVM-AI as having LVH despite not meeting any ECG-based LVH rules. The red waveform is the individual’s ECG tracing. Blue shades depict the magnitude of the gradient of predicted LV mass with respect to the ECG waveform amplitude, where darker shades illustrate regions of the waveform exerting greater influence on predicted LV mass. Measured R and S wave amplitudes (μV) are depicted on the top left of each plot.
In total, 4,417 (90.0%) in UK Biobank Test and 1,062 (77.5%) in MGB Test had a 12-lead ECG suitable for ECG rule calculation (Figure 2). LVM-AI demonstrated moderate LVH discrimination (UK Biobank c-statistic 0.653; 95% CI, 0.608–0.698; MGB 0.621; 95% CI 0.592–0.649), which was favorable when compared individually to Sokolow-Lyon, Cornell, and aVL criteria (p<0.001 for all, Figure 5 and Supplemental Table V). LVH discrimination using LVM-AI was similar to the presence of any ECG rule in UK Biobank Test (c-statistic 0.618; 95% CI, 0.574–0.663; p=0.11), but significantly greater than any ECG rule in MGB Test (0.588; 95% CI, 0.564–0.611; p=0.02).
Figure 5.
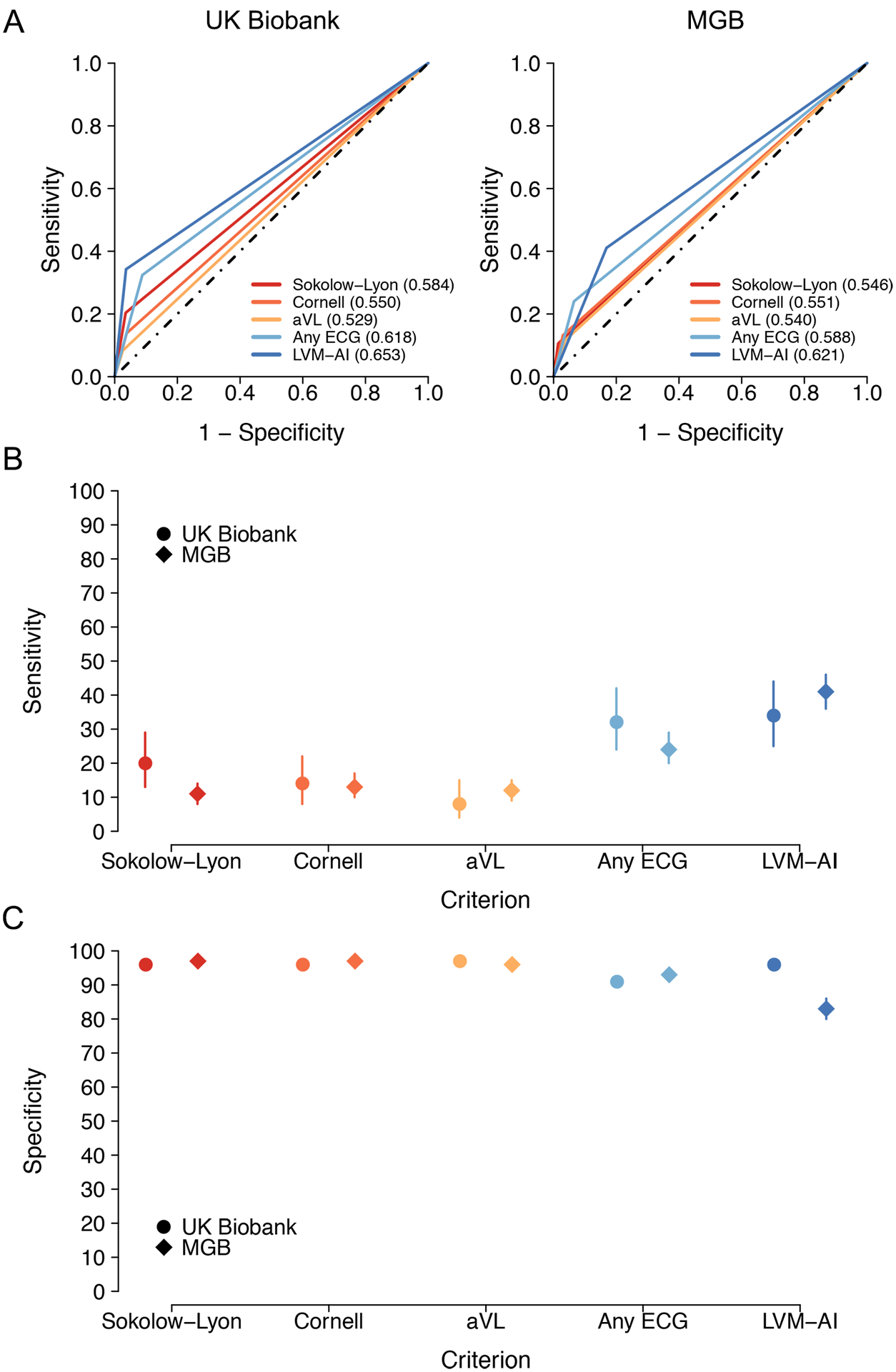
Diagnostic test performance of LVM-AI versus ECG rules for LVH
Depicted are diagnostic test characteristics for LVH using LVM-AI predicted LV mass (dark blue) versus traditional ECG rules including Sokolow-Lyon (red), Cornell (orange), aVL (yellow), and any ECG rule (light blue) in both UK Biobank Test (circles) and MGB Test (diamonds). Panels A depict receiver operating characteristic curves for LVH, Panel B compares sensitivity for LVH, and Panel C compares specificity for LVH. LVH is defined as CMR-derived indexed left ventricular mass >72g/m2 in men and >55 g/m2 in women.21
When compared to any ECG rule, LVM-AI had greater sensitivity and specificity in UK Biobank Test (sensitivity 34%; 95% CI 25–44 vs 32%; 95% CI 24–42; specificity 96%; 95% CI, 96–97 vs 91%; 95% CI, 90–92), and greater sensitivity but lower specificity in MGB Test (sensitivity 41%; 95% CI, 36–46 vs 24%; 95% CI 20–29; specificity 83%; 95% CI, 80–86 vs 93%; 95% CI, 91–95, Figure 5). When compared to individual ECG rules, LVM-AI had greater sensitivity, with comparable or moderately lower specificity (Figure 5 and Supplemental Table V). Net reclassification improvement using LVM-AI versus any ECG rule was 0.071 (95% CI, −0.016 to 0.17) in UK Biobank Test, and 0.067 (95% CI, 0.0072–0.13) in MGB Test (Supplemental Table VI), with increased case detection in both sets (UK Biobank 1.9%; 95% CI −7.6–13; MGB 17%, 95% CI 12–22).
LVM-AI and incident events
In the UK Biobank and MGB Outcomes samples, LVM-AI predicted LVH was associated with incident AF (hazard ratio [HR] 1.84; 95% CI, 1.29–2.63 in UK Biobank; 1.34; 95% CI, 1.21–1.50 in MGB), MI (HR 1.80; 95% CI, 1.09–2.96; 1.29; 95% CI, 1.15–1.46), HF (3.97; 95% CI, 2.70–5.84; 1.49; 95% CI, 1.37–1.62), and VA (3.16; 95% CI, 1.62–6.18; 1.71; 95% CI, 1.47–1.99). Associations were similar using LVH defined as the 90th percentile of LV mass index and using LV mass index as a continuous variable (Table 2 and Supplemental Figure VII). Cumulative risk curves stratified by presence of LVM-AI predicted LVH are shown in Figure 6 and Supplemental Figure VIII.
Table 2.
Associations between LVM-AI predicted LV mass index and incident disease
| Hazard ratio for covariate (95% CI)* | |||||
|---|---|---|---|---|---|
| N events/N total† | Follow-up, yrs (Q1,Q3) | LVMI (per 1 SD) | LVH (UKBB cutoff) | LVH (90th percentile) | |
| UK Biobank | |||||
| Atrial fibrillation | 376/34242 | 2.3 (1.3,3.7) | 1.30 (1.18–1.43) | 1.84 (1.29–2.63) | 1.45 (1.08–1.95) |
| Myocardial infarction | 193/34454 | 2.3 (1.3,3.8) | 1.38 (1.19–1.59) | 1.80 (1.09–2.96) | 1.51 (1.01–2.27) |
| Heart failure | 182/35077 | 2.3 (1.3,3.8) | 1.50 (1.40–1.60) | 3.97 (2.69–5.84) | 3.44 (2.47–4.79) |
| Ventricular arrhythmias | 69/35213 | 2.3 (1.3,3.8) | 1.43 (1.25–1.64) | 3.16 (1.62–6.18) | 3.05 (1.76–5.27) |
| Mass General Brigham | |||||
| Atrial fibrillation | 4661/28612 | 11.3 (6.4,14.2) | 1.08 (1.02–1.14) | 1.34 (1.21–1.50) | 1.20 (1.10–1.30) |
| Myocardial infarction | 2134/25334 | 11.9 (7.4,14.3) | 1.20 (1.11–1.30) | 1.29 (1.15–1.46) | 1.44 (1.22–1.71) |
| Heart failure | 4042/26113 | 11.4 (6.7,14.3) | 1.22 (1.15–1.29) | 1.49 (1.37–1.62) | 1.70 (1.52–1.89) |
| Ventricular arrhythmias | 1165/27547 | 10.6 (7.8,14.3) | 1.35 (1.22–1.50) | 1.71 (1.47–1.99) | 2.22 (1.82–2.70) |
Hazard ratios obtained using Cox proportional hazards models adjusted for age and sex
Includes individuals without the prevalent condition at imaging assessment (UK Biobank) or start of cohort follow-up (MGB)
Figure 6.
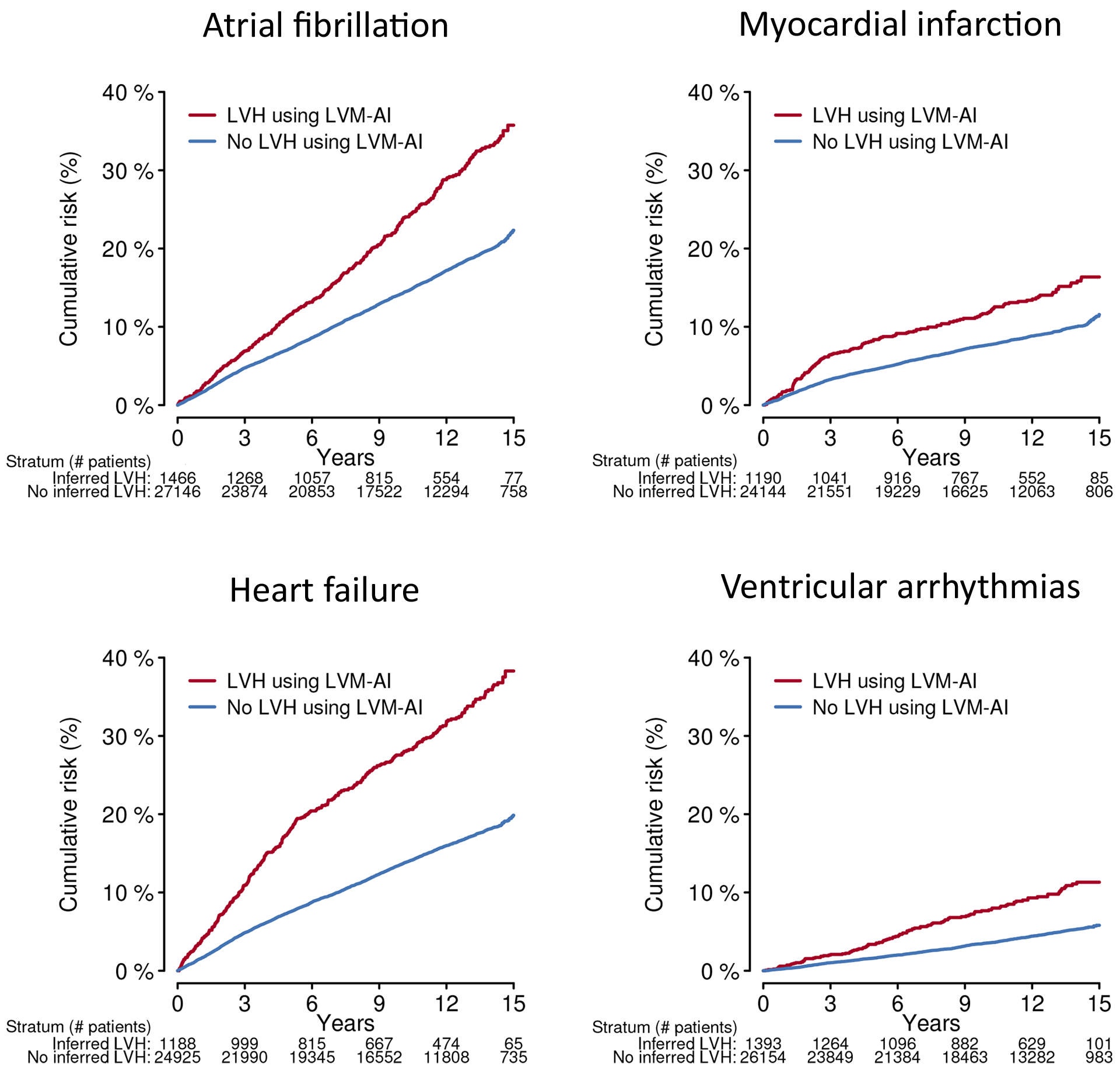
Cumulative risk of cardiovascular events stratified by LVM-AI predicted LVH in MGB Test
Depicted are the cumulative risks of atrial fibrillation, myocardial infarction, heart failure, and ventricular arrhythmias in MGB Test, stratified by presence (red) versus absence (blue) of LVM-AI predicted LVH. The number remaining at risk within each stratum is listed below each plot. LVH was defined as LVM-AI predicted LV mass index >72g/m2 in men and >55 g/m2 in women.21
In secondary analyses, associations between ECG rule-based LVH and incident events varied by specific rule, although the presence of LVH by any ECG rule was consistently associated with incident atrial fibrillation and heart failure (Supplemental Table VII). In models including both LVM-AI predicted and ECG rule-based LVH, LVM-AI predicted LVH was independently associated with incident events (Supplemental Table VIII). Cumulative risk curves stratified by the presence of LVH using LVM-AI and any ECG rule are shown in Supplemental Figure IX.
Sensitivity analyses
An age, sex, BMI, and hypertension-adjusted Cornell voltage product had lower sensitivity and specificity than LVM-AI (Supplemental Table IX and Supplemental Figure X). In the UK Biobank, a version of LVM-AI trained using ECG alone (i.e., without age, sex, or BMI) had similar correlation with CMR-derived LV mass (r=0.72; 95% CI, 0.70–0.73) and diagnostic performance for LVH (c-statistic 0.654; 95% CI, 0.608–0.700]). In contrast, LVM-AI trained using an unweighted loss function had slightly higher correlation (r=0.82; 95% CI, 0.81–0.83), but substantially worse LVH diagnostic performance (c-statistic 0.528; 95% CI, 0.505–0.552). Performance of the secondary LVM-AI models is summarized in Supplemental Figures XI–XII.
Discussion
In a prospective community-based sample of over 30,000 individuals with CMR and 12-lead ECG, we developed LVM-AI, a deep learning model that estimates CMR-derived LV mass using 12-lead ECG waveforms. When assessed in two independent test sets, LVM-AI appeared more sensitive for the presence of LVH on CMR as compared to traditional ECG rules applied individually or in aggregate. Importantly, LVM-AI predicted LVH was consistently associated with incident cardiovascular events. Our findings demonstrate the potential of deep learning on medical data available at scale to recapitulate structural information otherwise obtainable only through advanced imaging, as well as the potential to transfer such models across disparate clinical settings.
Our findings support and extend previous work using deep learning to infer cardiac structure from ECG. Tison et al.9 and Kwon et al.10 used neural networks to predict the presence of increased LV mass on echocardiography. Differences in LVH discrimination observed in Tison et al. (c-statistic 0.870) and Kwon et al. (0.868) versus our study (0.654) may reflect the impact of sample composition on model performance. Specifically, retrospective ascertainment of ECGs (for inference) and echocardiograms (for LVH definition) performed for clinical reasons in prior studies may enrich for pathology, and potentially introduce selection bias. It is also possible that differences in in performance may be related to varying model architectures. Nevertheless, we submit that training on prospectively collected ECG and CMR (the gold-standard for LV mass measurement),7 has the potential to reduce model bias.
Our results suggest that deep learning-based LV mass estimation may improve the yield of LVH screening using 12-lead ECG. Since antihypertensive treatment is a low-cost, well-tolerated intervention that can lead to LVH regression and improved outcomes,27 it is critical for ECG-based LVH screening tools to be sufficiently sensitive. To this end, LVM-AI demonstrated increased case detection when compared to ECG rules applied individually or in aggregate. At the same time, overall discrimination for CMR-derived LVH using LVM-AI remained modest, and improved methods for discriminating LVH are warranted. Nevertheless, even modest improvements in performance may be substantial when applied at scale and leveraging the potential for automation. Whether deep learning models explicitly trained to exhibit certain test characteristics (e.g., very high sensitivity) are better suited for specific clinical applications merits further study.
Our results demonstrate that deep learning models can be transferred across populations with varying characteristics, although performance appears to decline. We transferred LVM-AI, which was trained in a prospective community-based cohort, to an independent healthcare-related dataset in which the prevalence of LVH and related comorbidities were frequently over twice as high. Although initial model predictions in MGB required linear recalibration, LVM-AI predicted LV mass correlated with CMR-derived LV mass and LVM-AI discriminated CMR-based LVH better than traditional ECG rules. Nevertheless, model accuracy, calibration, and agreement with CMR-derived LV mass were noticeably lower within MGB, suggesting that model generalizability may have been constrained by limited diversity in patient characteristics within the UK Biobank training set, or alternatively overfitting. Future work is warranted to further evaluate expected declines in model performance when transferring across settings, and whether training on data from multiple settings leads to more generalizable models.
The current study underscores the potential for deep learning models using raw ECG data to produce clinically relevant output. Recent studies have shown that ECG-based deep learning models can discriminate individuals at higher risk for short-term outcomes including death28 or AF.29 In our study, the presence of LVM-AI predicted LVH was associated with substantially increased risks of incident cardiovascular events over many years. Effect sizes for AF and VA were similar to those reported previously using imaging-based LVH,1,3 whereas those for HF and MI were slightly lower.2,30 Notably, effect sizes were larger in the UK Biobank as opposed to MGB, which may reflect less accurate CMR-derived LV mass estimation in MGB, or a higher risk population in MGB, in which the relative effect of LV mass on outcomes may be smaller. Interestingly, when added to the presence of ECG rule-based LVH, LVM-AI – a surrogate for anatomic LVH – remained independently associated with outcomes. Such findings demonstrate the added prognostic value of LVM-AI and are consistent with previous reports suggesting that ECG-based LVH may be an electrophysiologic risk marker comprising elements independent of ventricular anatomy.23 We anticipate that future deep learning models may add even further value if they can characterize additional aspects of cardiac structure and function that can be difficult to quantify using current imaging techniques.
Our findings must be interpreted in the context of study design. First, LVM-AI was trained within the UK Biobank, a sample enriched for health and socioeconomic status and having a relatively low prevalence of LVH, which may have impacted model generalizability. Nevertheless, we observed reasonable accuracy in an external healthcare-related dataset, although we note initial LVM-AI estimates were poorly calibrated and required linear adjustment. On balance, our findings demonstrate that portability is feasible, but model training within populations most similar to those in which implementation is intended may optimize performance. Second, in the absence of manually annotated CMR images, we utilized a segmentation algorithm to derive CMR-based LV mass. Although the algorithm is accurate,13 imperfect LV mass estimates may have impacted the performance of LVM-AI. Third, LVM-AI is a black box model. However, saliency maps demonstrated that components of the ECG waveform plausibly relevant for LV mass estimation had the greatest impact on predicted LV mass.
In summary, using prospectively collected ECG and CMR data from a sizeable community-based cohort, we developed LVM-AI, a deep learning algorithm that estimates CMR-derived LV mass with fair accuracy using 12-lead ECG. We validated LVM-AI in two independent samples including a healthcare dataset, and demonstrated improved diagnostic performance compared to traditional ECG-based rules applied individually or in aggregate. LVM-AI predicted LVH was associated with increased risk of cardiovascular events independently of ECG rule-based LVH. Our findings highlight the utility of deep learning to leverage clinical data available at scale to infer cardiac structural information otherwise requiring dedicated imaging to characterize.
Supplementary Material
Disclosures:
Dr. Pirruccello has consulted for Maze Therapeutics. Dr. Philippakis receives research support from Bayer AG, IBM, Intel, and Verily, and has consulted for Novartis and Rakuten. Dr. Ho receives research support from Bayer AG and Gilead Sciences, and has received research supplies from EcoNugenics. Dr. Friedman receives research support from Bayer AG and IBM. Dr. Anderson receives research support from Bayer AG and has consulted for ApoPharma, Inc. Dr. Batra receives research support from Bayer AG and IBM, and consults for Novartis. Dr. Lubitz receives research support from Bristol Myers Squibb/Pfizer, Bayer AG, Boehringer Ingelheim, and Fitbit, and has consulted for Bristol Myers Squibb/Pfizer and Bayer AG, and participates in a research collaboration with IBM. Dr. Ellinor receives research support from Bayer AG, and has consulted for Bayer AG, Novartis, MyoKardia and Quest Diagnostics.
Sources of Funding:
Dr. Khurshid is supported by NIH (T32HL007208). Dr. Pirruccello is supported by a John S. LaDue Memorial Fellowship. Dr. Ho is supported by NIH (R01HL134893/R01HL140224/K24HL153669). Dr. Lubitz is supported by NIH (1R01HL139731) and American Heart Association (AHA) (18SFRN34250007). Dr. Ellinor is supported by NIH (1R01HL092577/R01HL128914/K24HL105780), AHA (18SFRN34110082), and the Foundation Leducq (14CVD01). Dr. Anderson is supported by NIH (R01NS103924) and AHA (18SFRN34250007).
Footnotes
Supplemental Material:
Methods
Tables I-IX
Figures I-XII
References
- 1.Bluemke DA, Kronmal RA, Lima JAC, Liu K, Olson J, Burke GL, Folsom AR. The relationship of left ventricular mass and geometry to incident cardiovascular events: the MESA (Multi-Ethnic Study of Atherosclerosis) study. J Am Coll Cardiol. 2008;52:2148–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chrispin J, Jain A, Soliman EZ, Guallar E, Alonso A, Heckbert SR, Bluemke DA, Lima JAC, Nazarian S. Association of electrocardiographic and imaging surrogates of left ventricular hypertrophy with incident atrial fibrillation: MESA (Multi-Ethnic Study of Atherosclerosis). J Am Coll Cardiol. 2014;63:2007–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kawel-Boehm N, Kronmal R, Eng J, Folsom A, Burke G, Carr JJ, Shea S, Lima JAC, Bluemke DA. Left Ventricular Mass at MRI and Long-term Risk of Cardiovascular Events: The Multi-Ethnic Study of Atherosclerosis (MESA). Radiology. 2019;293:107–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casale PN, Devereux RB, Kligfield P, Eisenberg RR, Miller DH, Chaudhary BS, Phillips MC. Electrocardiographic detection of left ventricular hypertrophy: development and prospective validation of improved criteria. J Am Coll Cardiol. 1985;6:572–580. [DOI] [PubMed] [Google Scholar]
- 5.Sokolow M, Lyon TP. The ventricular complex in left ventricular hypertrophy as obtained by unipolar precordial and limb leads. Am Heart J. 1949;37:161–186. [DOI] [PubMed] [Google Scholar]
- 6.Pewsner D, Jüni P, Egger M, Battaglia M, Sundström J, Bachmann LM. Accuracy of electrocardiography in diagnosis of left ventricular hypertrophy in arterial hypertension: systematic review. BMJ. 2007;335:711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lenstrup M, Kjaergaard J, Petersen CL, Kjaer A, Hassager C. Evaluation of left ventricular mass measured by 3D echocardiography using magnetic resonance imaging as gold standard. Scand J Clin Lab Invest. 2006;66:647–657. [DOI] [PubMed] [Google Scholar]
- 8.Deo RC. Machine Learning in Medicine. Circulation. 2015;132:1920–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tison GH, Zhang J, Delling FN, Deo RC. Automated and Interpretable Patient ECG Profiles for Disease Detection, Tracking, and Discovery. Circ Cardiovasc Qual Outcomes. 2019;12:e005289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwon J-M, Jeon K-H, Kim HM, Kim MJ, Lim SM, Kim K-H, Song PS, Park J, Choi RK, Oh B-H. Comparing the performance of artificial intelligence and conventional diagnosis criteria for detecting left ventricular hypertrophy using electrocardiography. Europace. 2020;22:412–419. [DOI] [PubMed] [Google Scholar]
- 11.Littlejohns TJ, Sudlow C, Allen NE, Collins R. UK Biobank: opportunities for cardiovascular research. Eur Heart J. 2019;40:1158–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petersen SE, Matthews PM, Francis JM, Robson MD, Zemrak F, Boubertakh R, Young AA, Hudson S, Weale P, Garratt S, Collins R, Piechnik S, Neubauer S. UK Biobank’s cardiovascular magnetic resonance protocol. J Cardiovasc Magn Reson. 2016;18:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khurshid S, Friedman SF, Pirruccello JP, Di Achille P, Diamant N, Anderson CD, Ellinor PT, Batra P, Ho JE, Philippakis AA, Lubitz SA. Deep learning to estimate cardiac magnetic resonance–derived left ventricular mass. Cardiovasc Digit Health J. 2021;S2666693621000232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodrigues J, Zellner A. Weighted balanced loss function and estimation of the mean time to failure. Communications in Statistics - Theory and Methods. 1994;23:3609–3616. [Google Scholar]
- 15.Norman JE, Levy D. Adjustment of ECG left ventricular hypertrophy criteria for body mass index and age improves classification accuracy. The effects of hypertension and obesity. J Electrocardiol. 1996;29 Suppl:241–247. [DOI] [PubMed] [Google Scholar]
- 16.ML4CVD Group. Machine Learning for Health (ML4H). https://github.com/broadinstitute/ml. GitHub. 2020; [Google Scholar]
- 17.Gwet KL. Computing inter-rater reliability and its variance in the presence of high agreement. Br J Math Stat Psychol. 2008;61:29–48. [DOI] [PubMed] [Google Scholar]
- 18.Hulme OL, Khurshid S, Weng L-C, Anderson CD, Wang EY, Ashburner JM, Ko D, McManus DD, Benjamin EJ, Ellinor PT, Trinquart L, Lubitz SA. Development and Validation of a Prediction Model for Atrial Fibrillation Using Electronic Health Records. JACC Clin Electrophysiol. 2019;5:1331–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khurshid S, Choi SH, Weng L-C, Wang EY, Trinquart L, Benjamin EJ, Ellinor PT, Lubitz SA. Frequency of Cardiac Rhythm Abnormalities in a Half Million Adults. Circ Arrhythm Electrophysiol. 2018;11:e006273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bland JM, Altman DG. Measuring agreement in method comparison studies. stat methods med res. 1999;8:135–160. [DOI] [PubMed] [Google Scholar]
- 21.Petersen SE, Aung N, Sanghvi MM, Zemrak F, Fung K, Paiva JM, Francis JM, Khanji MY, Lukaschuk E, Lee AM, Carapella V, Kim YJ, Leeson P, Piechnik SK, Neubauer S. Reference ranges for cardiac structure and function using cardiovascular magnetic resonance (CMR) in Caucasians from the UK Biobank population cohort. J Cardiovasc Magn Reson. 2017;19:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Du Bois D, Du Bois EF. A formula to estimate the approximate surface area if height and weight be known. 1916. Nutrition. 1989;5:303–311; discussion 312–313. [PubMed] [Google Scholar]
- 23.Leigh JA, O’Neal WT, Soliman EZ. Electrocardiographic Left Ventricular Hypertrophy as a Predictor of Cardiovascular Disease Independent of Left Ventricular Anatomy in Subjects Aged ≥65 Years. Am J Cardiol. 2016;117:1831–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Python Core Team. (2015). Python: A dynamic, open source programming language. Python Software Foundation. https://www.python.org/. [Google Scholar]
- 25.Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C, Corrado GS. TensorFlow: Large-scale machine learning on heterogeneous systems. 2015; [Google Scholar]
- 26.R Core Team (2015). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/. [Google Scholar]
- 27.Okin PM, Devereux RB, Jern S, Kjeldsen SE, Julius S, Nieminen MS, Snapinn S, Harris KE, Aurup P, Edelman JM, Wedel H, Lindholm LH, Dahlöf B, LIFE Study Investigators. Regression of electrocardiographic left ventricular hypertrophy during antihypertensive treatment and the prediction of major cardiovascular events. JAMA. 2004;292:2343–2349. [DOI] [PubMed] [Google Scholar]
- 28.Raghunath S, Ulloa Cerna AE, Jing L, vanMaanen DP, Stough J, Hartzel DN, Leader JB, Kirchner HL, Stumpe MC, Hafez A, Nemani A, Carbonati T, Johnson KW, Young K, Good CW, Pfeifer JM, Patel AA, Delisle BP, Alsaid A, Beer D, Haggerty CM, Fornwalt BK. Prediction of mortality from 12-lead electrocardiogram voltage data using a deep neural network. Nat Med [Internet]. 2020. [cited 2020 May 20];Available from: http://www.nature.com/articles/s41591-020-0870-z [DOI] [PubMed] [Google Scholar]
- 29.Raghunath S, Pfeifer JM, Ulloa-Cerna AE, Nemani A, Carbonati T, Jing L, vanMaanen DP, Hartzel DN, Ruhl JA, Lagerman BF, Rocha DB, Stoudt NJ, Schneider G, Johnson KW, Zimmerman N, Leader JB, Kirchner HL, Griessenauer CJ, Hafez A, Good CW, Fornwalt BK, Haggerty CM. Deep Neural Networks Can Predict New-Onset Atrial Fibrillation From the 12-Lead Electrocardiogram and Help Identify Those at Risk of AF-Related Stroke. Circulation. 2021;CIRCULATIONAHA.120.047829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haider AW, Larson MG, Benjamin EJ, Levy D. Increased left ventricular mass and hypertrophy are associated with increased risk for sudden death. J Am Coll Cardiol. 1998;32:1454–1459. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
UK Biobank data are publicly available by application (www.ukbiobank.ac.uk). MGB data contain protected health information and cannot be shared publicly. The code underlying LVM-AI is accessible at https://github.com/broadinstitute/ml4h/tree/master/model_zoo/left_ventricular_mass_from_ecg_student_and_mri_teacher.


