Abstract
Background:
Exposure to violence (ETV) or chronic stress may influence asthma through unclear mechanisms.
Methods:
Epigenome-wide association study (EWAS) of ETV or chronic stress measures and DNA methylation in nasal epithelium from 487 Puerto Ricans aged 9 to 20 years who participated in the Epigenetic Variation and Childhood Asthma in Puerto Ricans study [EVA-PR]). We assessed four measures of ETV and chronic stress in children (ETV scale, gun violence, and perceived stress) and their mothers (perceived stress). Each EWAS was conducted using linear regression, with CpGs as dependent variables and the stress/violence measure as a predictor, adjusting for age, sex, the top five principal components, and SVA latent factors. We then selected the top 100 CpGs (by P-value) associated with each stress/violence measure in EVA-PR and conducted a meta-analysis of the selected CpGs and atopic asthma using data from EVA-PR and two additional cohorts (Project Viva and PIAMA).
Results:
Three CpGs (in SNN, PTPRN2, and LINC01164) were associated with maternal perceived stress or gun violence (P=1.28 to 3.36 ×10−7), but not with atopic asthma, in EVA-PR. In a meta-analysis of three cohorts, which included the top CpGs associated with stress/violence measures in EVA-PR, 12 CpGs (in STARD3NL, SLC35F4, TSR3, CDC42SE2, KLHL25, PLCB1, BUD13, OR2B3, GALR1, TMEM196, TEAD4 and ANAPC13) were associated with atopic asthma at FDR-P < 0.05.
Conclusions:
Pending confirmation in longitudinal studies, our findings suggest that nasal epithelial methylation markers associated with measures of ETV and chronic stress may be linked to atopic asthma in children and adolescents.
Keywords: violence, stress, epigenetics, childhood asthma
INTRODUCTION
The U.S. has the highest rate of firearm-related deaths of all industrialized countries1. As recently reviewed, exposure to violence (ETV, including gun violence) affects stress responses, and both ETV and chronic stress may influence asthma pathogenesis2,3. Prolonged ETV can lead to chronic and frequent activation of the body’s stress responses, disrupting neuroendocrine, autonomic nervous system, and immune responses. Chronic stress, both related and unrelated to ETV, may cause or worsen asthma by altering circulating levels of and response to catecholamines and glucocorticoids, as well as by affecting immune responses, leading to airway inflammation and airflow obstruction2–4. ETV and chronic stress may also cause or worsen asthma through indirect effects mediated by other risk factors2. For example, community violence may decrease outdoor physical activity, leading to obesity5.
Puerto Ricans are often exposed to violence in their communities6–8. ETV is associated with asthma in Puerto Rican children8,9, in whom ETV is also associated with DNA methylation of the promoter of the gene for the pituitary adenylate cyclase activating polypeptide 1 type 1 receptor (ADCYAP1R1) in white blood cells8. Moreover, such methylation has been associated with childhood asthma in Puerto Ricans.
Based on prior findings2,3,8,9, we hypothesized that both ETV and chronic stress would increase asthma risk through DNA methylation of genes regulating autonomic, neuroendocrine, and immune responses in airway epithelium, a tissue relevant to asthma. To examine this hypothesis, we first tested for association between ETV or chronic stress measures and genome-wide DNA methylation in nasal epithelium, as DNA methylation in nasal (airway) epithelium is well correlated with that in bronchial (airway) epithelium10–13, and epigenetic regulation of airway epithelial gene expression has been implicated in asthma pathogenesis14. We then examined whether the top CpGs associated with ETV or chronic stress were also associated with atopic asthma in Puerto Rican children and in two independent cohorts. We focused on atopic asthma, both because atopic asthma is the most common type of childhood asthma and because we previously showed that DNA methylation profiles can accurately classify children according to the presence of atopic asthma14.
METHODS
Study population
Subject recruitment and study procedures for the Epigenetic Variation and Childhood Asthma in Puerto Ricans study (EVA-PR) have been previously described14. In brief, EVA-PR is a case-control study of asthma in subjects aged 9 to 20 years. Participants with and without asthma were recruited from households in San Juan (PR) from February 2014 through May 2017, using multistage probability sampling; 638 households had ≥1 eligible child, and 543 (85.1%) children (one per household) agreed to participate. The study was approved by the institutional review boards of the University of Puerto Rico (San Juan, PR) and the University of Pittsburgh (Pittsburgh, PA). Written parental consent and assent were obtained from participants <18 years old, and consent was obtained from participants ≥18 years old.
The study protocol included administration of questionnaires on ETV and chronic stress in the child and her/his mother, measurement of serum allergen-specific IgEs, and collection of nasal epithelial samples for DNA and RNA extraction. Participating children completed a modified version of the ETV Scale15–17, a questionnaire used to assess ETV in children ≥8 years. The ETV Scale measures both witnessing and direct victimization for five events: shoving, punching, or kicking; knife attacks; shootings; hearing gunshots; and witnessing verbal abuse of the child’s primary caregiver. An ETV score (range=0–15) is obtained by summing the scores for each section. Internal consistency and test-retest reliability and validity have been established for the English and Spanish versions. The child’s exposure to gun violence was derived from the ETV scale9. Participating children also completed the Checklist of Children’s Distress Symptoms (CCDS), a 28-item scale to assess stress symptoms in the previous six months as a result of ETV16,18. Answers for each question in the CCDS range from 1 to 5. An overall score is obtained by summing the scores for all 28 questions, and then dividing by the number of questions, so that the score ranges between 1 and 5. Mothers of participating children completed the perceived stress scale (PSS) questionnaire, which includes 4 items and measures the degree to which mothers believed that their lives were unpredictable, uncontrollable, or overwhelming in the preceding month19. Each question in the maternal PSS ranges between 0 and 4, and a total score ranges between 0 and 1619.
Serum IgEs to each of five common allergens in Puerto Rico (dust mite [Der p 1], cockroach [Bla g 2], cat dander [Fel d 1], dog dander [Can f 1], and mouse urinary protein [Mus m 1]) were measured using the UniCAP 100 system (Pharmacia & Upjohn, Kalamazoo, Mich). Atopy was defined as an IgE ≥0.35 IU/mL to ≥1 of the allergens tested. Asthma was defined as a physician’s diagnosis plus ≥1 episode of wheeze in the previous year. Atopic asthma was defined as the presence of both atopy and asthma.
Whole-genome methylation in nasal epithelium was measured using Infinium HumanMethylation450 BeadChip arrays (Illumina, San Diego, CA). Methylation β-values were calculated as a percentage: β=M/(M+U+α), where M and U represent methylated and unmethylated signal intensities, respectively, and α is an arbitrary offset to stabilize β-values where fluorescent intensities are low. The β-values were used in all downstream analyses. We performed the same preprocessing and quality control procedures as in our previous study14. In brief, Raw IDAT files were loaded using the R package minfi20. Samples with low detection values (>10% CpG probes with a detection P >0.01) were removed. The R package Enmix21 was used to perform background correction and normalization. Cross-reactive (n=26,098) and SNP-containing probes22 (n=15,834), sex chromosomal probes (n=9,490), and low-quality probes (>10% of samples with detection P >0.01; n=21,174) were also removed. After further filtering, CpG sites with an overall mean β-value of greater than 0.9 or less than 0.1 were removed, leaving 227,901 CpG probes in the analysis.
RNA-Seq was performed using the Illumina NextSeq 500 platform, with paired-end reads at 75 cycles and 80 million reads per sample; reads were aligned to reference human genome (hg19)23 and TPM (Transcripts Per Kilobase Million) were used as proxy for gene expression level. After QC, 16,737 genes were retained for the analyses of gene expression, which focused on genes in cis (within 1 Mb on each side of the transcription start site) with the top CpGs from the methylation analyses. Genome-wide genotyping was conducted using the HumanOmni2.5 BeadChip platform, (Illumina, San Diego, CA), as previously described.
Study cohorts included in the meta-analysis of atopic asthma
PIAMA
PIAMA is a birth cohort study of children born in the Netherlands in 1996 and 1997. The study protocol has been previously described 24. The Medical Ethical Committees of the participating institutions approved the study, and the parents and legal guardians of all participants, as well as the participants themselves, gave written informed consent. At age 16 years, nasal epithelial cells were collected at two study centers (Groningen and Utrecht)25 by brushing the lateral area underneath the right inferior turbinate. Serum IgEs to each of four common allergens (house dust mite, cat, dactylis (grass) and birch) were measured, and atopy was defined as an IgE ≥0.35 IU/mL to any of the allergens tested. Asthma was defined as physician-diagnosed asthma and either wheeze in the previous year or use of medications for respiratory or lung problems. Atopic asthma was defined as the presence of both asthma and atopy.
In total, 479 nasal epithelial samples were hybridized to the Infinium HumanMethylation450 BeadChip arrays. DNA methylation data were pre-processed with Bioconductor package minfi20. Samples with call rate <99% were removed. 65 SNP probes were used to check for concordance between paired DNA samples (nasal and blood DNA samples from the same subjects were hybridized in the same experiments); paired samples with Pearson correlation coefficient <0.9 were excluded, as were probes on sex chromosomes, probes that mapped to multiple loci, 65 SNP-probes, and probes containing SNPs at the target CpG sites with a MAF>5%22. “DASEN”26 was used to perform signal correction and normalization. After QC, 455 samples and 436,824 probes remained in the analysis, as previously described27. Of these 455 samples, 246 samples corresponded to subjects with atopic asthma and non-atopic control subjects, and were thus included in the meta-analysis.
Project Viva
Project Viva is a prospective pre-birth cohort study of mother-child pairs recruited between 1999 and 2002 during the mothers’ first prenatal visits at Atrius Harvard Vanguard Medical Associates, a medical practice in eastern Massachusetts (U.S.)28. The Institutional Review Board of Harvard Pilgrim Health Care reviewed and approved all study protocols. Asthma and atopy were assessed at the same time-point as collection of nasal samples. Atopy was defined as ≥1 IgE ≥0.35 IU/mL to the allergens tested (Dermatophagoides farinae, cat dander, dog dander, Alternaria or Aspergillus species, rye grass, ragweed, and oak and silver birch).
Nasal swabs were collected from the anterior nares, previously demonstrated to yield respiratory epithelial cells29. Epigenome-wide methylation was measured on DNA extracted from nasal samples, using Infinium MethylationEPIC BeadChip arrays (Illumina). DNA methylation data was imported into the R statistical software for preprocessing using minfi20. QC was first performed at the sample level, excluding samples that: had overall low intensity (<10.5; n=3), were mismatched on recorded sex (n=4), or had mixed genotype distributions on the measured SNP probes (n=59) indicating possible sample contamination (n=8). In addition, 35 technical duplicates were excluded. Of 547 high-quality samples, 156 corresponded to subjects with atopic asthma and non-atopic controls and were thus retained for the meta-analysis. QC was then performed at the probe level by computing a detection P-value relative to control probes. In total, 40,377 cross-reactive probes previously identified in the MethylationEPIC BeadChip30 were excluded, leaving 719,075 high-quality probes included in the analysis. DNA methylation heterogeneity was estimated by including potential cell-type variation, using 10 principal components derived via ReFACTor 31.
Statistical analysis
We analyzed four measures of ETV and chronic stress. As in prior work, the child’s ETV scale was analyzed as continuous, the child’s exposure to gun violence was analyzed as binary (having heard gunshots at least twice vs. no more than once)9, the child’s perceived stress scale was analyzed as binary (upper quartile [≥2.6 points] vs. lowest three quartiles [<2.6 points])32, and maternal perceived stress (assessed by the PSS) was analyzed as binary (upper quartile [>7 points] vs. lowest three quartiles [≤7 points])32. Of the 487 participants in EVA-PR who had nasal genome-wide methylation (GWM) data and complete data on relevant covariates, 470 had ETV data, 475 had data on gun violence, 471 had CCDS data, and 476 had maternal PSS data.
We conducted epigenome-wide association studies (EWAS) of ETV or chronic stress measures among subjects in EVA-PR using linear regression models, as follows:
| (1) |
where CpG is the β-value for a CpG methylation, with known batches adjusted by R function Combat33, BMI z-score is calculated using CDC growth charts, residential distance to a road (a proxy for traffic-related air pollution) is a binary covariate (as in prior work, < 444.7 vs. ≥ 444.7 meters or quartiles 1–3 vs. quartile 4), annual household income is a binary covariate (≥ $15,000 vs. < $15,000 per year, near the median household income in Puerto Rico in 2008–2009), 5 PCs are the top five principal components from genome-wide genotypic data, and latent factors (LFs) are estimated by sva34 (which capture unknown data heterogeneity).
Because of limited statistical power in EVA-PR and lack of a replication cohort with data on stress/violence and nasal DNA methylation, we selected the top 100 CpG sites from each of the four EWAS (one for each measure of ETV or chronic stress) in EVA-PR for the analysis of atopic asthma, in order to reduce multiple testing. For this analysis in EVA-PR, we included 272 participants (167 subjects with atopic asthma and 105 non-atopic controls). Next, we used linear regression modeling to test whether the top methylation signals for ETV or chronic stress were also associated with atopic asthma in each of the three study cohorts (EVA-PR, Viva, and PIAMA), as follows:
| (2) |
We then conducted a meta-analysis of the three EWAS of atopic asthma. In this meta-analysis, P-values from each of the three independent studies were taken as input, with effect direction considered. First, a Z-score was calculated based on the P-value and direction of effect in each study, as follows:
where is the Z-score for study i, is the P-value for study i, is the direction of effect for the study i, and gives the percentile of a standard normal distribution. Combined coefficients were calculated by averaging the study-specific coefficients, with weights reflecting the standard errors (SE) from each study, as follows:
where is the combined coefficient, is the study-specific coefficient, and is the weight for study i. An overall P-value was then calculated as:
where is the overall P-value. The false discovery rate (FDR) approach was used to adjust for multiple testing in the meta-analysis of the selected CpG sites and atopic asthma.
To assess the biological relevance of the top CpG sites for atopic asthma, we conducted an expression quantitative trait methylation (eQTM) analysis, to test whether those CpGs affected the expression of cis-genes (i.e. those with transcription start sites located within 1 Mb of the CpG, on each side) in EVA-PR. This analysis was adjusted for age, sex, asthma and atopy status, the top five PCs, RNA sample sorting protocol (i.e., whole-cells or CD326-positive nasal epithelial cells)37, methylation and RNA-Seq batch, and LFs estimated from sva34.
RESULTS
The main characteristics of the EVA-PR participants included in the four EWAS of ETV and stress measures (ETV, gun violence, CCDS and PSS) are shown in Supplementary Table S1. In reviewing the results for each of these EWAS, the Q-Q plots showed no evidence of inflation (Figure 1). In these EWAS, methylation of cg18961589 in LINC01164 (on chromosome 10) was associated with gun violence exposure (β=0.03, P=1.28×10−7 at FDR-P < 0.05), and methylation of cg03402351 in SNN (on chromosome 16, β=0.04, P=1.69×10−7) and cg19064846 in PTPRN2 (on chromosome 7, β=0.03, P=3.36×10−7) were associated with maternal perceived stress at FDR-P < 0.05 (Figure 2).
Figure 1. Q-Q plots of four epigenome-wide analyses of stress/violence measures in nasal epithelium from participants in the Epigenetic Variation of Childhood Asthma in Puerto Ricans study (EVA-PR).
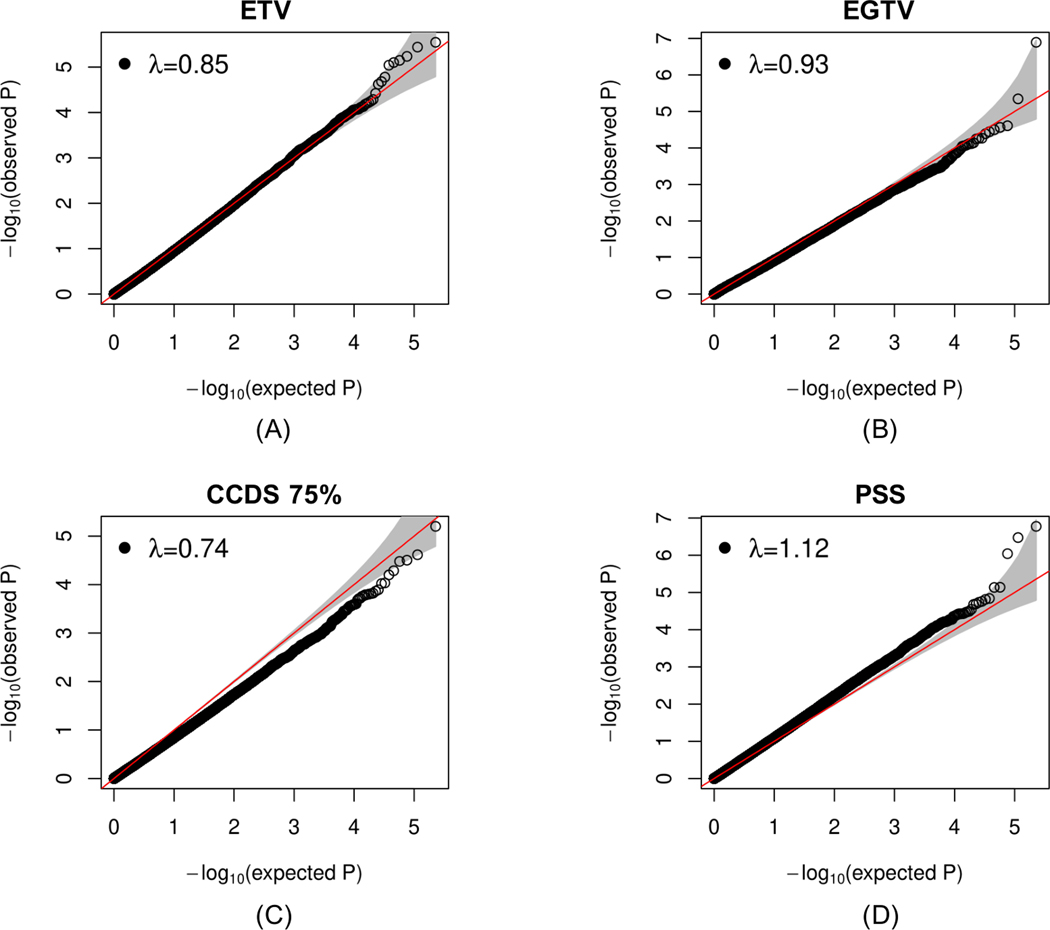
ETV: Exposure to violence. ETGV: Exposure to gun violence. CCDS: Checklist of Children’s Distress Symptoms. PSS: Perceived Stress Scale.
Figure 2. Manhattan plots for the epigenome-wide association studies (EWAS) of four ETV/stress measures in nasal epithelial samples from participants in the Epigenetic Variation of Childhood Asthma in Puerto Ricans study (EVA-PR).
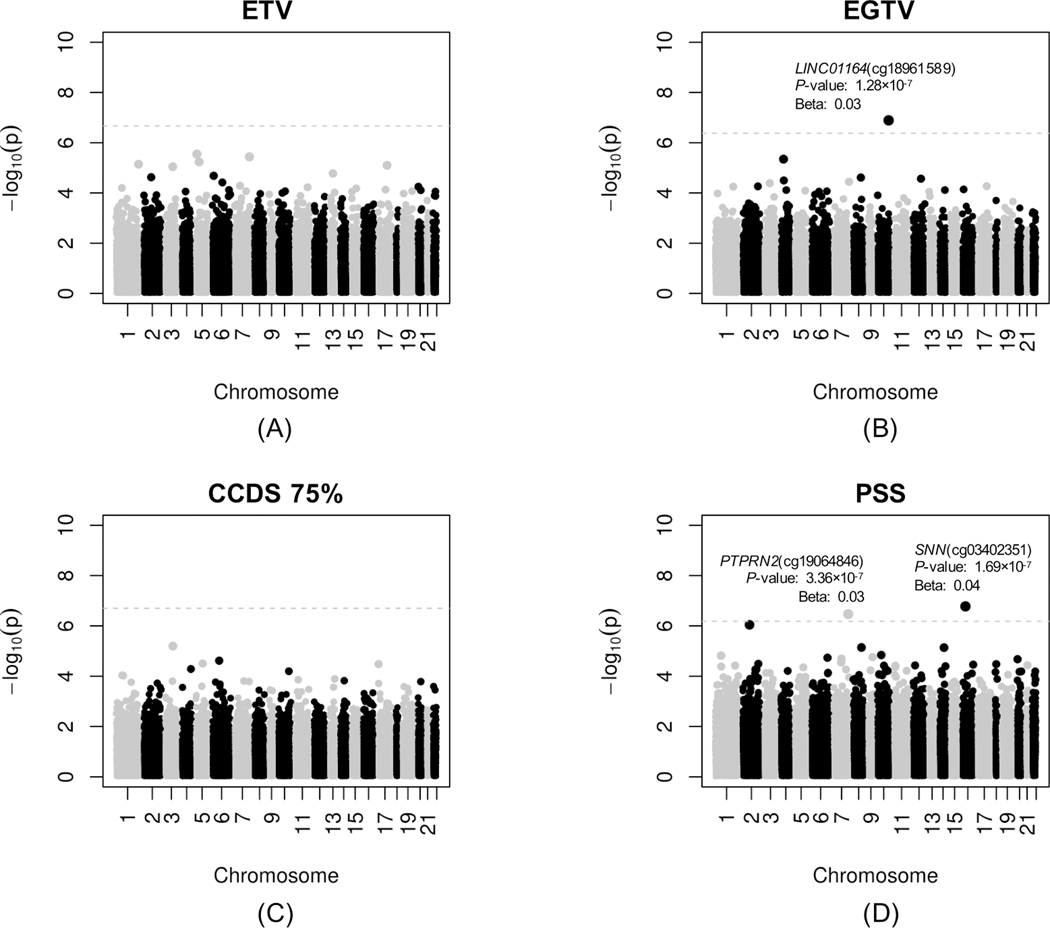
(A) Exposure to Violence scale (ETV), (B) Exposure to gun violence (ETGV), (C) Checklist of Child Distress Symptoms (CCDS), and (D) maternal perceived stress symptoms (PSS). The chromosomal position of each CpG site is displayed along the X-axis and the negative logarithm of the association P-value is displayed on the Y-axis. The dotted line represents the threshold for genome-wide significance line (false-discovery rate-adjusted P < 0.05).
We then selected the top 100 CpG sites (by P-value) from each of the four EWAS in EVA-PR for testing for association with atopic asthma in a meta-analysis including data from EVA-PR, PIAMA, and Project Viva. A total of 336 CpG sites were included in this meta-analysis, as 4 CpG sites overlapped across EWAS and 60 CpG sites were missing in Project Viva; see supplementary tables S2–S3). The main characteristics of participants in the three cohorts included in the meta-analysis of atopic asthma and nasal epithelial DNA methylation are shown in Table 1. As expected, EVA-PR (a case-control study of asthma) had a larger proportion of participants with atopic asthma than PIAMA or Viva (both unselected for asthma). The studies were conducted in different geographic locations, ranging from Puerto Rico to Boston to the Netherlands. While all participants in EVA-PR were Puerto Rican and most participants in PIAMA were European, Viva was ethnically diverse.
Table 1.
Summary of characteristics of participants in the studies included in the meta-analysis of CpG sites (associated with stress or violence measures in EVA-PR) and atopic asthma.
| EVA-PR (n=272) | PIAMA (n=246) | Project Viva (n=156) | |
|---|---|---|---|
| Age in years (mean ± SD) | 15.5±3.1 | 16.3±0.2 | 12.9±0.6 |
| Male gender (n, %) | 144(52.9) | 111 (47.0) | 79(50.6) |
| Race/ethnicity | |||
| White | 239(97.2) | 110(70.5) | |
| Black | 21(13.5) | ||
| Hispanic or Latino | 272 (100) | 8(5.1) | |
| Asian | 5(3.2) | ||
| More than one race/unknown | 7 (2.8) | 12(7.7) | |
| Atopic asthma (n, %) | 167(61.4) | 27(11.0) | 36(23.1) |
| Study sites | San Juan (Puerto Rico) | The Netherlands | Boston (MA) |
| Methylation platform | Infinium HumanMethylation450 BeadChips | Infinium HumanMethylation450 BeadChips | Infinium MethylationEPIC BeadChip |
We first performed separate analyses of the 336 CpG sites and atopic asthma in each of the three study cohorts (EVA-PR, PIAMA and Viva), and then combined the results from the three cohorts in a meta-analysis. In this meta-analysis, we identified 7 CpGs (cg02695349 in STARD3NL, cg18146152 in TSR3, cg03541903 in CDC42SE2, cg21376795 in KLHL25, cg26772788 in GALR1, cg03729152 in TMEM196 and cg16848072 in ANAPC13) associated with higher odds of atopic asthma, and 5 CpGs (cg04990977 in SLC35F4, cg27178677 in PLCB1, cg17076485 in BUD13, cg17335499 in OR2B3, and cg01039401 in TEAD4) associated with lower odds of atopic asthma, all at FDR-P < 0.05.
We then conducted a cis-expression quantitative trait methylation (cis-eQTM) analysis of the 12 CpGs associated with atopic asthma in nasal epithelial cells from participants in EVA-PR.. In this analysis, we identified 16 nominally significant cis-acting eQTM probes in nasal epithelium (Supplementary Table 4).
Our EWAS of ETV or stress measures in EVA-PR included children with and without asthma. We did not adjust for asthma status in these analyses, as this could have attenuated our effect estimates for the analysis of atopic asthma. In a sensitivity analysis, we repeated the four EWAS of ETV or stress measures after additional adjustment for asthma status, obtaining similar results (supplementary figures S1 and S2). Moreover, when the top 100 CpGs from each of these four EWAS (adjusted for asthma status) were tested for association with atopic asthma in a meta-analysis of the three study cohorts, 7 of the 12 CpGs from the primary analysis were also significant at FDR-P <0.05 (in STARD3NL, TSR3, CDC42SE2, PLCB1, OR2B3, GALR1 and TMEM196).
DISCUSSION
ETV and chronic stress have been linked to childhood asthma2. We previously showed that ETV and gun violence are associated with childhood asthma in Puerto Ricans, a high-risk group 8,9,35. To our knowledge, this is the first report of an association between chronic stress or ETV and DNA methylation in nasal epithelium, as well as the first association study of stress- or violence-related methylation markers in nasal epithelium and atopic asthma.
Among Puerto Rican children, exposure to gun violence was significantly associated with increased methylation of a CpG site (cg18961589) in the gene for long intergenic non-protein coding 1164 (LINC01164). LINC01164 is most highly expressed in brain tissue from GTEx36 (Supplementary Figure S3), and SNPs in this gene have been associated with cognitive function and educational attainment37. Although cg18961589 was not significantly associated with atopic asthma in the current analysis, methylation of another CpG site in LINC01164 (cg15491439) was significantly associated with atopic asthma in our recent EWAS in EVA-PR (FDR-P = 4.20×10−4)14. Moreover, this long non-coding RNA is adjacent (~128Kbp) to PPP2R2D, a gene that codes for a regulatory subunit of the PPP2A phosphatase family that has been linked to T-cell proliferation and apoptosis in mice38.
Increased methylation of cg03402351 in SNN (stannin) and increased methylation of cg19064846 in PTPRN2 (receptor-type tyrosine-protein phosphatase N2) in nasal epithelium were associated with maternal perceived stress but not with atopic asthma. Both SNN and PTPRN2 are highly expressed in brain tissue from GTEx36. PTPRN2 methylation and expression have been associated with Parkinson’s Disease40–42, and a meta-analysis of epigenome-wide data showed that a differentially methylated region in PTPRN2 was associated with childhood asthma 43.
In the meta-analysis of selected stress- or violence-associated CpG sites in the study cohorts, we identified 12 CpG sites associated with atopic asthma (in genes STARD3NL, TSR3, CDC42SE2, KLHL25, GALR1, TMEM196, ANAPC13, SLC35F4, PLCB1, BUD13, OR2B3, and TEAD4). Of note, expression levels of STARD3NL (log2FC [fold-change] = −0.16; FDR-P = 6.24×10−6), TSR3 (log2FC=0.07; FDR-P=0.04), CDC42SE2 (log2FC= −0.18; FDR-P=2.62×10−4), KLHL25 (log2FC =0.21; FDR-P=3.11×10−6), PLCB1 (log2FC= −0.31; FDR-P=0.01), and ANAPC13 (log2FC = −0.13; FDR-P=1.27×10−3) were associated with atopic asthma in our recent transcriptome-wide association study using nasal epithelial samples from Puerto Ricans44.
PLCB1 has been associated with bronchodilator response in children with asthma45, and is differentially expressed in children with treatment-resistant asthma compared with children with controlled persistent asthma or healthy controls46. Silencing PLCB1 can attenuate lipopolysaccharide-induced endothelial cell inflammation by inhibiting expression of proinflammatory cytokines47. In the brain, PLCB1 is expressed mainly in the cortex, hippocampus and amygdala; the protein it codes for, PLCB1, is a mediator of synaptic plasticity that plays an important role in cognitive behavior and emotions48 and may influence the pathogenesis of stress-related disorders. Indeed, expression of PLCB1 is decreased in the frontal cortex of rats subjected to chronic stress, and this downregulation is reversed by quetiapine, a drug used to treat major depression49. PLCB1 has also been implicated in depression, bipolar disorder, epilepsy, and schizophrenia50.
Four SNPs in TMEM196 have been shown to be associated with decreased odds of nonsteroidal anti-inflammatory drug (NSAID)-exacerbated respiratory disease (NERD) in asthma 51. Moreover, TMEM196 has been shown to regulate autophagy and apoptosis of cancer and inflammatory cells 52. Of note, disruption of the balance between autophagy and apoptosis induces hyperplasia of the airways, a typical feature of nasal polyps and airway eosinophilia in NERD 53–55.
We recognize several study limitations. First, we had limited statistical power to examine ETV or chronic stress and DNA methylation in EVA-PR and lacked a cohort with data on stress/ETV for replication analyses. Although top methylation markers in the EWAS of stress/violence measures were associated with atopic asthma in the meta-analysis of three study cohorts, no such marker achieved genome-wide significance for ETV or stress measures in EVA-PR. Thus, false positive results cannot be excluded pending replication studies. Second, we cannot assess temporal relationships or causality in this cross-sectional study. Third, covariates correlated with stress or violence may partly explain our results. However, our EWAS of stress or violence measures in EVA-PR was adjusted for indicators of socioeconomic status (annual income), adiposity (BMI), and traffic-related air pollution. Moreover, some of the top genes associated with atopic asthma have been implicated in neuropsychiatric function or disorders (i.e. PLCB1). Fourth, our statistical power may have been reduced by variability in race/ethnicity, geographic location, and exposure to psychosocial stressors across the cohorts included in the meta-analysis.
In summary, we show that measures of chronic stress or violence are associated with nasal DNA methylation markers in Puerto Rican youth. Moreover, we show that some stress- or violence-related methylation markers are also associated with atopic asthma in a meta-analysis of data from three cohorts of youth in diverse racial/ethnic groups. Although our findings must await confirmation in other studies, they offer “proof of concept” and support longitudinal studies of chronic stress or violence, DNA methylation in nasal epithelium, and stress- or violence-linked methylation markers and asthma in children.
Supplementary Material
Table 2.
CpG sites associated with atopic asthma (at FDR-P < 0.05) in a meta-analysis of data from three cohorts (EVA-PR, Project Viva, and PIAMA)
| CpG | Gene | Chr | Position | EVA-PR | Project Viva | PIAMA | β meta-analysis | P value meta-analysis | FDR_P meta-analysis | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β | P value | β | P value | β | P value | |||||||
| cg02695349 | STARD3NL | chr7 | 38269241 | 0.0205 | 1.04E-02 | 0.0488 | 1.10E-05 | 0.0138 | 7.44E-02 | 0.0237 | 1.40E-06 | 2.80E-04 |
| cg04990977 | SLC35F4 | chr14 | 58221774 | −0.0478 | 4.67E-06 | −0.0134 | 1.87E-02 | −0.0170 | 2.62E-02 | −0.0199 | 1.66E-06 | 2.80E-04 |
| cg18146152 | TSR3 | chr16 | 1400772 | −0.0042 | 5.24E-01 | 0.0073 | 8.90E-03 | 0.0131 | 5.79E-05 | 0.0084 | 2.62E-05 | 2.93E-03 |
| cg03541903 | CDC42SE2 | chr5 | 130651434 | 0.0071 | 5.39E-01 | 0.0095 | 8.64E-03 | 0.0209 | 1.90E-03 | 0.0117 | 1.24E-04 | 1.05E-02 |
| cg21376795 | KLHL25 | chr15 | 86302121 | 0.0050 | 3.25E-01 | 0.0027 | 4.01E-01 | 0.0154 | 7.41E-06 | 0.0080 | 1.74E-04 | 1.17E-02 |
| cg27178677 | PLCB1 | chr20 | 8834803 | −0.0328 | 3.60E-05 | 0.0028 | 7.65E-01 | −0.0119 | 2.03E-02 | −0.0144 | 2.30E-04 | 1.29E-02 |
| cg17076485 | BUD13 | chr11 | 116580712 | −0.0112 | 1.18E-01 | −0.0014 | 8.88E-01 | −0.0225 | 2.28E-04 | −0.0150 | 4.09E-04 | 1.96E-02 |
| cg17335499 | OR2B3 | chr6 | 29054325 | −0.0179 | 9.49E-03 | −0.0085 | 1.07E-02 | −0.0040 | 2.90E-01 | −0.0078 | 7.54E-04 | 3.13E-02 |
| cg26772788 | GALR1 | chr18 | 75609054 | 0.0042 | 4.81E-01 | 0.0045 | 3.84E-01 | 0.0224 | 3.35E-05 | 0.0106 | 8.39E-04 | 3.13E-02 |
| cg03729152 | TMEM196 | chr7 | 19961027 | 0.0247 | 3.27E-03 | 0.0054 | 2.28E-01 | 0.0148 | 2.00E-02 | 0.0110 | 9.56E-04 | 3.15E-02 |
| cg01039401 | TEAD4 | chr12 | 3070406 | −0.0612 | 1.80E-05 | 0.0036 | 8.14E-01 | −0.0192 | 1.15E-01 | −0.0260 | 1.03E-03 | 3.15E-02 |
| cg16848072 | ANAPC13 | chr3 | 134182665 | 0.0074 | 1.91E-01 | 0.0044 | 1.58E-01 | 0.0100 | 6.30E-03 | 0.0068 | 1.72E-03 | 4.80E-02 |
Acknowledgments:
We thank all the participating children and families.
Sources of Funding: This study was supported by grants HL079966, HL117191, and MD011764 (PI: Celedón JC) from the National Heart, Lung, and Blood Institute (NHLBI) of the U.S. National Institutes of Health (NIH). Dr. Yan’s contribution was supported by NIH grant HL138098. Dr. Forno’s contribution was supported by NIH grant HL149693. Research operations at the University of Puerto Rico were additionally supported by grant U54MD007587 from the National Institute on Minority Health and Health Disparities (NIMHD) and the National Institute of Allergy and Infectious Diseases (NIAID) of the U.S. NIH. PIAMA was supported by The Netherlands Organization for Health Research and Development; The Netherlands Organization for Scientific Research; The Netherlands Lung Foundation (with methylation studies supported by AF 4.1.14.001); The Netherlands Ministry of Spatial Planning, Housing, and the Environment; and The Netherlands Ministry of Health, Welfare, and Sport. Project Viva is supported by grants from the U.S. NIH (R01 HL 111108, R01 NR013945, R01 HD 034568, UH3 OD023286, and R01 AI102960).
Disclosures of interest: Dr. Celedón has received research materials from GSK and Merck (inhaled steroids) and Pharmavite (vitamin D and placebo capsules), in order to provide medications free of cost to participants in NIH-funded studies, unrelated to the current work.
Footnotes
The other authors declare no conflicts of interest.
REFERENCES
- 1.Hahn RA, Bilukha O, Crosby A, et al. Firearms laws and the reduction of violence: a systematic review. Am J Prev Med 2005;28:40–71. [DOI] [PubMed] [Google Scholar]
- 2.Landeo-Gutierrez J, Forno E, Miller GE, Celedon JC. Exposure to Violence, Psychosocial Stress, and Asthma. Am J Respir Crit Care Med 2020;201:917–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Landeo-Gutierrez J, Celedon JC. Chronic stress and asthma in adolescents. Ann Allergy Asthma Immunol 2020; 125(4):393–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenberg SL, Miller GE, Brehm JM, Celedon JC. Stress and asthma: novel insights on genetic, epigenetic, and immunologic mechanisms. J Allergy Clin Immunol 2014;134:1009–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen RT, Celedon JC. Community Violence and Health Disparities in Asthma. J Pediatr 2016;173:13–5. [DOI] [PubMed] [Google Scholar]
- 6.Rosser FJ, Forno E, Cooper PJ, Celedon JC. Asthma in Hispanics. An 8-year update. Am J Respir Crit Care Med 2014;189:1316–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martinez-Taboas A, Canino G, Wang MQ, Garcia P, Bravo M. Prevalence and victimization correlates of pathological dissociation in a community sample of youths. Journal of traumatic stress 2006;19:439–48. [DOI] [PubMed] [Google Scholar]
- 8.Chen W, Boutaoui N, Brehm JM, et al. ADCYAP1R1 and asthma in Puerto Rican children. Am J Respir Crit Care Med 2013;187:584–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramratnam S, Han Y, Rosas-Salazar C, et al. Exposure to gun violence and asthma among children in Puerto Rico. Respir Med 2015; 109(8):975–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lopez-Guisa JM, Powers C, File D, Cochrane E, Jimenez N, Debley JS. Airway epithelial cells from asthmatic children differentially express proremodeling factors. J Allergy Clin Immunol 2012;129:990–7 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baccarelli A, Rusconi F, Bollati V, et al. Nasal cell DNA methylation, inflammation, lung function and wheezing in children with asthma. Epigenomics 2012;4:91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poole A, Urbanek C, Eng C, et al. Dissecting childhood asthma with nasal transcriptomics distinguishes subphenotypes of disease. J Allergy Clin Immunol 2014;133:670–8 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao C, Biagini Myers JM, Ji H, et al. Vanin-1 expression and methylation discriminate pediatric asthma corticosteroid treatment response. J Allergy Clin Immunol 2015; 136(4):923–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forno E, Wang T, Qi C, et al. DNA methylation in nasal epithelium, atopy, and atopic asthma in children: a genome-wide study. Lancet Respir Med 2019;7:336–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sternthal MJ, Jun HJ, Earls F, Wright RJ. Community violence and urban childhood asthma: a multilevel analysis. Eur Respir J 2010;36:1400–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suglia SF, Ryan L, Wright RJ. Creation of a community violence exposure scale: accounting for what, who, where, and how often. Journal of traumatic stress 2008;21:479–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomson CC, Roberts K, Curran A, Ryan L, Wright RJ. Caretaker-child concordance for child’s exposure to violence in a preadolescent inner-city population. Arch Pediatr Adolesc Med 2002;156:818–23. [DOI] [PubMed] [Google Scholar]
- 18.Suglia SF, Ryan L, Bellinger DC, Enlow MB, Wright RJ. Children’s exposure to violence and distress symptoms: influence of caretakers’ psychological functioning. Int J Behav Med 2011;18:35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen S, Kamarck T, Mermelstein R. A global measure of perceived stress. J Health Soc Behav 1983;24:385–96. [PubMed] [Google Scholar]
- 20.Aryee MJ, Jaffe AE, Corrada-Bravo H, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 2014;30:1363–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu Z, Niu L, Li L, Taylor JA. ENmix: a novel background correction method for Illumina HumanMethylation450 BeadChip. Nucleic acids research 2016;44:e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen YA, Lemire M, Choufani S, et al. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics 2013;8:203–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brunekreef B, Smit J, de Jongste J, et al. The prevention and incidence of asthma and mite allergy (PIAMA) birth cohort study: design and first results. Pediatr Allergy Immunol 2002;13:55–60. [DOI] [PubMed] [Google Scholar]
- 25.Xu CJ, Soderhall C, Bustamante M, et al. DNA methylation in childhood asthma: an epigenome-wide meta-analysis. Lancet Respir Med 2018;6:379–88. [DOI] [PubMed] [Google Scholar]
- 26.Pidsley R, CC YW, Volta M, Lunnon K, Mill J, Schalkwyk LC. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics 2013;14:293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qi C, Jiang Y, Yang IV, et al. Nasal DNA methylation profiling of asthma and rhinitis. J Allergy Clin Immunol 2020; 145(6):1655–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oken E, Baccarelli AA, Gold DR, et al. Cohort profile: project viva. Int J Epidemiol 2015;44:37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lai PS, Liang L, Cibas ES, et al. Alternate methods of nasal epithelial cell sampling for airway genomic studies. J Allergy Clin Immunol 2015;136:1120–3 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pidsley R, Zotenko E, Peters TJ, et al. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol 2016;17:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rahmani E, Zaitlen N, Baran Y, et al. Correcting for cell-type heterogeneity in DNA methylation: a comprehensive evaluation. Nat Methods 2017;14:218–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brehm JM, Ramratnam SK, Tse SM, et al. Stress and Bronchodilator Response in Children with Asthma. Am J Respir Crit Care Med 2015;192:47–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007;8:118–27. [DOI] [PubMed] [Google Scholar]
- 34.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012;28:882–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cohen RT, Canino GJ, Bird HR, Celedon JC. Violence, abuse, and asthma in Puerto Rican children. Am J Respir Crit Care Med 2008;178:453–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Consortium GT. The Genotype-Tissue Expression (GTEx) project. Nat Genet 2013;45:580–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee JJ, Wedow R, Okbay A, et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat Genet 2018;50:1112–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou P, Shaffer DR, Alvarez Arias DA, et al. In vivo discovery of immunotherapy targets in the tumour microenvironment. Nature 2014;506:52–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buck-Koehntop BA, Mascioni A, Buffy JJ, Veglia G. Structure, dynamics, and membrane topology of stannin: a mediator of neuronal cell apoptosis induced by trimethyltin chloride. J Mol Biol 2005;354:652–65. [DOI] [PubMed] [Google Scholar]
- 40.Chuang YH, Lu AT, Paul KC, et al. Longitudinal Epigenome-Wide Methylation Study of Cognitive Decline and Motor Progression in Parkinson’s Disease. Journal of Parkinson’s disease 2019;9:389–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grunblatt E, Mandel S, Jacob-Hirsch J, et al. Gene expression profiling of parkinsonian substantia nigra pars compacta; alterations in ubiquitin-proteasome, heat shock protein, iron and oxidative stress regulated proteins, cell adhesion/cellular matrix and vesicle trafficking genes. Journal of neural transmission 2004;111:1543–73. [DOI] [PubMed] [Google Scholar]
- 42.Sandor C, Robertson P, Lang C, et al. Transcriptomic profiling of purified patient-derived dopamine neurons identifies convergent perturbations and therapeutics for Parkinson’s disease. Hum Mol Genet 2017;26:552–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.den Dekker HT, Burrows K, Felix JF, et al. Newborn DNA-methylation, childhood lung function, and the risks of asthma and COPD across the life course. Eur Respir J 2019;53. [DOI] [PubMed] [Google Scholar]
- 44.Forno E, Zhang R, Jiang Y, et al. Transcriptome-wide and differential expression network analyses of childhood asthma in nasal epithelium. J Allergy Clin Immunol 2020; 146(3):671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mak ACY, White MJ, Eckalbar WL, et al. Whole-Genome Sequencing of Pharmacogenetic Drug Response in Racially Diverse Children with Asthma. Am J Respir Crit Care Med 2018;197:1552–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Persson H, Kwon AT, Ramilowski JA, et al. Transcriptome analysis of controlled and therapy-resistant childhood asthma reveals distinct gene expression profiles. J Allergy Clin Immunol 2015;136:638–48. [DOI] [PubMed] [Google Scholar]
- 47.Lin YJ, Chang JS, Liu X, et al. Genetic variants in PLCB4/PLCB1 as susceptibility loci for coronary artery aneurysm formation in Kawasaki disease in Han Chinese in Taiwan. Sci Rep 2015;5:14762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cabana-Dominguez J, Roncero C, Pineda-Cirera L, et al. Association of the PLCB1 gene with drug dependence. Sci Rep 2017;7:10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Orsetti M, Di Brisco F, Rinaldi M, Dallorto D, Ghi P. Some molecular effectors of antidepressant action of quetiapine revealed by DNA microarray in the frontal cortex of anhedonic rats. Pharmacogenet Genomics 2009;19:600–12. [DOI] [PubMed] [Google Scholar]
- 50.Yang YR, Kang DS, Lee C, et al. Primary phospholipase C and brain disorders. Adv Biol Regul 2016;61:80–5. [DOI] [PubMed] [Google Scholar]
- 51.Lee JU, Chang HS, Baek DG, Shin HD, Park CS, Park JS. Associations between TMEM196 polymorphisms and NSAID-exacerbated respiratory disease in asthma. Pharmacogenet Genomics 2019;29:69–75. [DOI] [PubMed] [Google Scholar]
- 52.Liu WB, Han F, Jiang X, et al. TMEM196 acts as a novel functional tumour suppressor inactivated by DNA methylation and is a potential prognostic biomarker in lung cancer. Oncotarget 2015;6:21225–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fruth K, Schramek E, Docter D, et al. Dysregulated survivin expression in nasal polyps of individuals with aspirin exacerbated respiratory disease. American journal of rhinology & allergy 2012;26:380–4. [DOI] [PubMed] [Google Scholar]
- 54.Kowalski ML, Grzegorczyk J, Pawliczak R, Kornatowski T, Wagrowska-Danilewicz M, Danilewicz M. Decreased apoptosis and distinct profile of infiltrating cells in the nasal polyps of patients with aspirin hypersensitivity. Allergy 2002;57:493–500. [DOI] [PubMed] [Google Scholar]
- 55.Farooque SP, Lee TH. Aspirin-sensitive respiratory disease. Annu Rev Physiol 2009;71:465–87. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


