Abstract
Motile cilia (also interchangeably called “flagella”) are conserved organelles extending from the surface of many animal cells and play essential functions in eukaryotes including cell motility and environmental sensing. Large motor complexes, the ciliary dyneins, are present on ciliary outer-doublet microtubules and drive movement of cilia. Ciliary dyneins are classified into two general types; the outer dynein arms (ODAs) and the inner dynein arms (IDAs). While ODAs are important for generation of force and regulation of ciliary beat frequency, IDAs are essential for control of the size and shape of the bend, features collectively referred to as waveform. Also, recent studies have revealed unexpected links between IDA components and human diseases. In spite of their importance, however, studies on IDAs have been difficult since they are very complex and composed for several types of IDA motors, each unique in composition and location in the axoneme. Thanks in part to genetic, biochemical and structural analysis of Chlamydomonas reinhardtii, we are beginning to understand the organization and function of the ciliary IDAs. In this review, we summarize the composition of Chlamydomonas IDAs particularly focusing on each subunit, and discuss the assembly, conservation, and functional role(s) of these IDA subunits. Further, we raise several additional questions/challenges regarding IDAs, and discuss future perspectives of IDA studies.
Keywords: flagella, cilia, subunit, inner-arm dynein, IDA, motility, Chlamydomonas
1. Introduction
Motile cilia (also called flagella) are antenna-like organelles protruding from many eukaryotic cells. For the single-celled eukaryotes such as Paramecium and Euglena, these organelles are essential for both swimming and sensing the external environments (Vincensini et al., 2011). For the multicellular eukaryotes including vertebrates, these organelles play indispensable functions in organ homeostasis, development and fertility (Fliegauf et al., 2007; Satir & Christensen, 2007). The basic structure of these motile organelles is highly conserved among eukaryotes, most all containing the “9+2” axoneme. The axoneme cross section is built from a total of 233 tubulin protofilaments, in which 9 outer doublet microtubules surround 2 central pair microtubules [233 = 23×9 (outer doublets) + 13×2 (a central pair); 13 and 233 are Fibonacci numbers (Mughal & Weaire, 2017; Swinton & Ochu, 2016)]. This elegant ciliary axonemal structure has attracted the attention of cell biologists for several decades.
The mechanism of repetitive bending motion of motile cilia had been a mystery until Gibbons and Rowe found a motor protein potentially generating force for ciliary motility (Gibbons & Rowe, 1965). They named this motor protein “dynein” after the unit name of force, dyne. Ciliary dyneins are classified into two groups based on their location in the axoneme; ODAs protruding outward from doublet microtubules and IDAs protruding inward from doublet microtubules (Figure 1a, 1b; Table 1, 2). Subsequently, Satir provided experimental and structural evidence for a sliding microtubule model for ciliary bending (Satir, 1968; Satir et al., 2014). Then, Summers and Gibbons succeeded in directly observing axonemal microtubule sliding in vitro (Summers & Gibbons, 1971). This result directly supported a dynein-driven, sliding microtubule model for ciliary bending. Consistently, Shingyoji et al (Shingyoji et al., 1977) and Brokaw (Brokaw, 1991) demonstrated that ciliary microtubules slide during bending. Ciliary microtubule sliding was further confirmed using the Tetrahymena ciliary axonemes as a model (Sale & Satir, 1976, 1977).
Figure 1. Potential arrangements of IDAs in Chlamydomonas cilia.
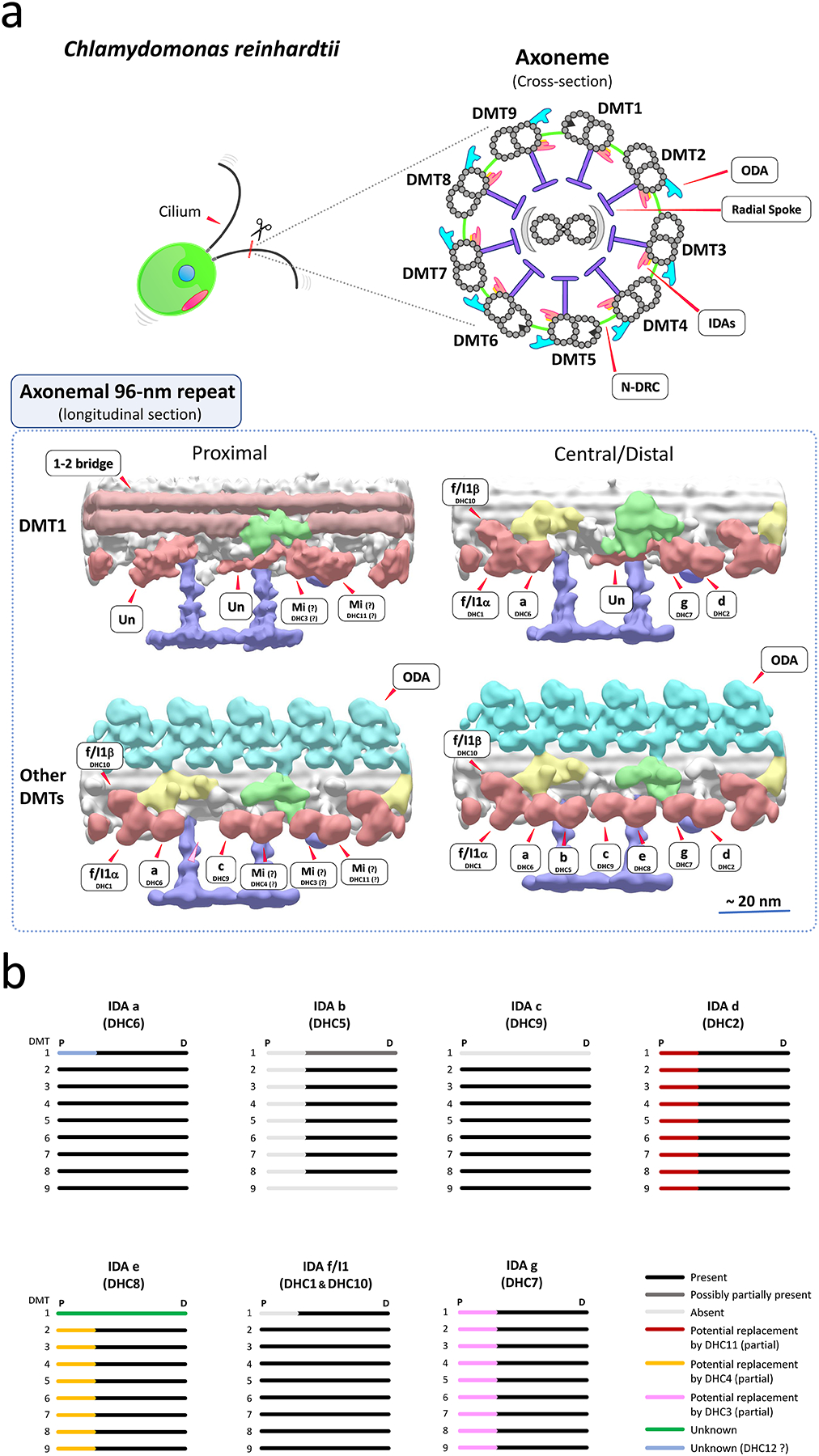
(a) (top) Drawings of a Chlamydomonas cell and ciliary axonemal cross-section. (bottom) Hypothesized models of IDA arrangements on surface-rendering images of the 96-nm repeats of Chlamydomonas ciliary doublet microtubules (DMTs). The images are the results of sub-tomogram averages from reconstructed cryo-electron tomograms. Surface-rendering images of the proximal and central/distal regions of DMT1 and other averaged DMTs (DMTs 2–8 or 2–9) are shown. In Chlamydomonas, DMT1 totally lacks ODAs, and also has particularly interesting features including the arrangement of IDAs (Bui et al., 2012; Hoops & Witman, 1983). In the proximal region of ciliary axonemes, the arrangement of IDAs differs from that in the central/distal region, lacking IDA b (DHC5) and possibly minor IDAs replacing some major IDAs (Bui et al., 2012). Approximate locations of ODAs, IDAs, the IC/LC complex of IDA f/I1, N-DRC, radial spokes and the 1–2 bridges are shown in water blue, old-rose red, yellow, green, purple, and light brown, respectively. A light pink arrowhead indicates the missing IDA b location in the proximal portion of the DMT2-9 average. Mi, minor IDA; Un, unknown density likely an IDA species (Bui et al., 2012). The tomograms were reconstructed and published in the previous publication (Bui et al., 2012), and refined for this review. The accession IDs of the density maps (Bui et al., 2012) in EMDataBank (http://www.emdatabank.org/) used to make this figure are as follows: DMT1 proximal region, EMD-2119; DMT1 central region, EMD-2113; DMT2-9 proximal region, EMD-2131; DMT2-8 central/distal region, EMD-2132. (b) A summary of proposed arrangements of IDAs in Chlamydomonas cilia. The figure is adapted/refined from (Bui et al., 2012). Chlamydomonas cilia have heterogeneity in the arrangement of IDAs among DMTs, and this heterogeneity could contribute to generation of proper ciliary waveform. IDA d and e may be partially replaced by minor IDAs DHC11 and DHC4 in the proximal portion of the axonemes (Bui et al., 2012). Also, replacement of IDA g by DHC3 was proposed in (Bustamante-Marin et al., 2020). DMT1 has unique organization of IDAs throughout the axonemes (Bui et al., 2012; Hoops & Witman, 1983). P, Proximal; D, Distal.
Table 1.
Composition of IDAs in Chlamydomonas reinhardtii
| IDA Composition of Chlamydomonas reinhardtiia | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Species | a | b | c | d | e | f/I1 | g | DHC3 | DHC4 | DHC11 | DHC12 |
| IDA type | Major | Major | Major | Major | Major | Major | Major | Minor | Minor | Minor | Minor |
| Head type | Single-headed | Single-headed | Single-headed | Single-headed | Single-headed | Double-headed | Single-headed | Single-headed | Single-headed | Single-headed | Single-headed |
| MT rotation | + | − | + | + | + | +/−b | + | N/Ac | N/A | N/A | N/A |
| MT bending | − | − | − | + | − | − | + | N/A | N/A | N/A | N/A |
| HC gene | DHC6 | DHC5 | DHC9 (IDA9) | DHC2 | DHC8 |
DHC1 (PF9/IDA1) DHC10 (IDA2) |
DHC7 | DHC3 | DHC4 | DHC11 | DHC12 (DHC1a) |
| Other name | I2’ | I3’ | I2A | I2’ | I2B | None | I3 | None | None | None | None |
| ICs/LCs | IC140 | ||||||||||
| IC138 | |||||||||||
| IC97 | |||||||||||
| p44 | |||||||||||
| Actin | Actin (NAP)d | Actin | Actin | Actin | Actin (NAP) | (Actin)e | (Actin) | (Actin) | (Actin) | ||
| p38 | |||||||||||
| FAP120 | |||||||||||
| p28 | p28 | p28 | (p28) | (p28) | |||||||
| MOT7 | |||||||||||
| Centrin | Centrin | Centrin | (Centrin) | (Centrin) | |||||||
| TCTEX1 | |||||||||||
| TCTEX2b | |||||||||||
| LC7a | |||||||||||
| LC7b | |||||||||||
| LC8 | |||||||||||
This table is based on (Hom et al., 2011; Kagami & Kamiya, 1992; Kamiya & Yagi, 2014; Kikushima & Kamiya, 2008; Piperno, 1995; Yagi et al., 2009).
Negligible rotation at high IDA f/I1 densities, while erratic rotation was observable at low IDA f/I1 densities (Kotani et al., 2007).
Not analyzed previously.
NAP serves as a subunit of IDAs b and g in the absence of actin [in the ida5 mutant (Kato-Minoura et al., 1998)].
Parentheses for LCs of minor IDAs mean that they are not fully experimentally determined (Kamiya & Yagi, 2014).
Table 2.
Subunits of IDAs in Chlamydomonas reinhardtii
| IDA subunits of Chlamydomonas reinhardtiia | |||||
|---|---|---|---|---|---|
| Name | Description | Predicted pI/Mwb | Domain/Motifc | Chromosomed | Accession No/IDe |
| DHC | |||||
| DHC1/PF9/IDA1 | HC of IDA f/I1 | 5.33/522885.80 | AAA/Coiled-coil | 12 | Phytozome: Cre12.g484250.t1.1 |
| DHC2 | HC of IDA d | 5.46/467004.81 | AAA/Coiled-coil | 9 | Phytozome: Cre09.g392282.t1.1 |
| DHC3 | HC of minor IDA | 6.20/585030.81 | AAA/Coiled-coil | 6 | Phytozome: Cre06.g265950.t1.1 |
| DHC4 | HC of minor IDA | 6.13/524768.01 | AAA/Coiled-coil | 2 | Phytozome: Cre02.g107350.t1.1 |
| DHC5 | HC of IDA b | 5.81/463901.76 | AAA/Coiled-coil | 2 | Phytozome: Cre02.g107050.t1.1 |
| DHC6 | HC of IDA a | 5.51/477687.67 | AAA/Coiled-coil | 5 | Phytozome: Cre05.g244250.t1.2 |
| DHC7 | HC of IDA g | 5.42/468333.64 | AAA/Coiled-coil | 14 | Phytozome: Cre14.g627576.t1.1 |
| DHC8 | HC of IDA e | 5.56/470226.38 | AAA/Coiled-coil | 16 | Phytozome: Cre16.g685450.t1.1 |
| DHC9/IDA9 | HC of IDA c | 5.62/464742.71 | Coiled-coil (AAA)f | 2 | Phytozome: Cre02.g141606.t1.1 |
| DHC10/IDA2 | HC of IDA f/I1 | 5.95/510666.80 | AAA/Coiled-coil | 14 | Phytozome: Cre14.g624950.t1.1 |
| DHC11 | HC of minor IDA | 5.58/517580.00 | Coiled-coil (AAA) | 12 | Phytozome: Cre12.g555950.t1.2 |
| DHC12/DHC1a | Probable minor HC of IDA | 6.17/653333.60 | AAA/Coiled-coil | 6 | Phytozome: Cre06.g297850.t1.1 |
| IC | |||||
| IC140/DIC3/IDA7 | IC of IDA f/I1 | 4.84/109816.66 | WD40/Coiled-coil | 16 | NCBI: XP_001695786.1 |
| IC138/DIC4/BOP5 | IC of IDA f/I1 | 5.62/111159.34 | WD40 | 12 | NCBI: XP_001696921.1 |
| IC97/DII6/FAP94 | IC of IDA f/I1 | 5.46/81498.37 | Coiled-coil | 14 | NCBI: ACN22075 |
| LC | |||||
| Actin/DII4/IDA5 | LC of most likely all the single-headed IDAs | 5.30/41836.06 | Actin | 13 | Phytozome: Cre13.g603700.t1.2 |
| Centrin/DLE2/VFL2 | LC of IDAs b, e, g and probably of DHC3 and DHC4 | 4.70/19458.94 | EF hand | 11 | Phytozome: Cre11.g468450.t1.2 |
| LC7a/DLR1/ODA15 | LC of IDA f/I1 and ODA | 6.91/11927.71 | Roadblock/LC7 | 8 | Phytozome: Cre08.g376550.t1.2 |
| LC7b/DLR2 | LC of IDA f/I1 and ODA | 5.40/11118.70 | Roadblock/LC7 | 12 | Phytozome: Cre12.g546400.t1.2 |
| LC8/DLL1/FLA14 | LC of IDA f/I1, ODA, and the IFT dynein | 6.89/10321.77 | Dynein light chain type 1 | 3 | Phytozome: Cre03.g181150.t1.1 |
| NAP/DII5 | Replace actin in ida5 for IDAs b and g | 5.52/41553.34 | Actin | 3 | Phytozome: Cre03.g176833.t1.1 |
| p28/DII1/IDA4 | LC of IDAs a, c, d and probably of DHC11 and DHC12 | 6.68/28652.72 | Coiled-coil | 12 | NCBI: CAA88139 |
| TCTEX1/DLT3 | LC of IDA f/I1 | 5.25/12668.45 | (Tctex-1)g | 1 | Phytozome: Cre01.g004250.t1.2 |
| TCTEX2b/DLT4 | LC of IDA f/I1 | 5.67/13693.37 | (Tctex-1) | 9 | NCBI: DAA05278 |
| Accessary subunits | |||||
| FAP120/DII7 | Accessary subunit of IDA f/I1 | 4.94/31861.12 | Ankyrin repeat /Coiled-coil | 2 | NCBI: BAN15819 |
| MOT7 | Accessary subunit of IDA f/I1 | 4.91/26212.92 | (BLUF)h | 1 | Phytozome: Cre01.g038750.t1.2 |
| p44/DII3i | Accessary subunit of IDA d | 5.46/46091.86 | TPR/Coiled-coil | 8 | NCBI: AB353122.2 |
| p38/DII2/FAP146 | Accessary subunit of IDA d | 5.29/40923.19 | None | 3 | NCBI: BAG07147 |
This table is based on (Hom et al., 2011; Kamiya & Yagi, 2014; Yagi et al., 2009).
Calculated by ExPASy algorithm (http://web.expasy.org/compute_pi/)(Wilkins et al., 1999).
Predicted by SMART analysis (http://smart.embl-heidelberg.de/)(Letunic et al., 2021; Schultz et al., 1998).
The localizing chromosome of each subunit is based on Phytozome Chlamydomonas reinhardtii v5.5.
Protein ID/accession number in NCBI or Phytozome Chlamydomonas reinhardtii v5.5.
In some dynein sequences, AAA motifs were not detected in the default SMART settings but predicted in the pfam analyses (https://pfam.xfam.org/)(Punta et al., 2012).
Tctex-1 family was predicted in the pfam analyses (https://pfam.xfam.org/)(Punta et al., 2012).
A potential orthologue of MOT7 in Ciona intestinalis, DYBLUP, has a BLUF domain in its sequence (Kutomi et al., 2021).
Based on the updated p44 cDNA/protein.
Ciliary motility is required for normal human development and health in adults. In 1976, Afzelius analyzed cilia from human patients displaying bronchitis, sinusitis, male infertility and situs inversus, and found that the cilia were immotile in many cases and the ciliary dynein arms were missing (Afzelius, 1976). This disease was called “immotile cilia syndrome” and is now referred to as “primary cilia dyskinesia” or “PCD” (Brown & Witman, 2014; Horani et al., 2016; Knowles et al., 2016). Since this original report, there has been much focus on PCD, and additional diseases that result from failure in assembly of the immotile sensory cilia called the primary cilia (Brown & Witman, 2014; Reiter & Leroux, 2017). Further advances in understanding PCD will require more detailed analyses and understanding of conserved axonemal components including IDAs.
Since the importance of ciliary motility in human health was revealed, research has intensified on discovery of conserved components and mechanisms of assembly of the cilium. To this end, Chlamydomonas reinhardtii is a particularly powerful model genetic organism for discovery of conserved genes and mechanisms (Dutcher, 2014; Kamiya & Yagi, 2014; Ostrowski et al., 2011). Notable advances using Chlamydomonas include discovery of intra-flagellar transport (IFT)(Kozminski et al., 1993), definition of the cytoplasmic “preassembly” of ciliary dyneins (Fowkes & Mitchell, 1998), identification of the molecular basis of the 96-nm ciliary axonemal repeat (Oda et al., 2014), and a high-resolution structure of the meshwork of inner proteins of doublet microtubules (Ma et al., 2019). Chlamydomonas mutants have also revealed the conserved components of the axonemal radial spokes (Gui et al., 2021; Pigino et al., 2011; Poghosyan et al., 2020), nexin-dynein regulatory complex (N-DRC)(Bower et al., 2013; Heuser et al., 2009), calmodulin spoke complex (CSC)(Dymek et al., 2011; Dymek & Smith, 2007) and central pair complex (Loreng & Smith, 2017). In addition, Chlamydomonas has contributed to understanding of the basal bodies/centrioles (Dutcher & O’Toole, 2016) and the function of the Bardet-Biedl Syndrome (BBS) proteins in cilia (Lechtreck et al., 2013; Li et al., 2004; Liu & Lechtreck, 2018; Nachury et al., 2007).
Ciliary dynein composition and regulatory mechanism(s) have also been extensively studied in this organism (Alford et al., 2012; Kamiya & Yagi, 2014; King, 2016; Wakabayashi, 2012; Yagi & Kamiya, 2012), and in Chlamydomonas all of the heavy chain (HC) genes of the dynein family were identified in the genome (Hom et al., 2011; Merchant et al., 2007; Mitchell, 1994; Porter et al., 1996; Yagi et al., 2009). Chlamydomonas has also revealed that each dynein heavy chain plays a specialized role in control of ciliary movement [reviewed in (Kamiya, 2002; Kamiya & Yagi, 2014)]. For example, by comparing the ciliary motility between mutants deficient in ODAs or IDAs, Brokaw and Kamiya showed that ODAs are particularly important for control of ciliary beat frequency, while IDAs control normal ciliary waveform (Brokaw & Kamiya, 1987). In addition, mutants lacking all ODAs (oda strains) can still move albeit at reduce beat frequency (Kamiya, 1988; Mitchell & Rosenbaum, 1985). In contrast, failure in assembly of multiple IDAs (e.g. the pf23 mutant) results in paralyzed cilia or in defective motility (Huang et al., 1979; Yamamoto et al., 2017; Yamamoto et al., 2020). Notably, most of the proteins associated with IDAs, including IDA subunits, are conserved in eukaryotic organisms that bear motile cilia (Wickstead & Gull, 2007), and recent studies revealed unexpected links between IDAs and human diseases [e.g. (Ben Khelifa et al., 2014; Bustamante-Marin et al., 2020)].
In this review, we discuss the organization of IDAs in Chlamydomonas focusing on each subunit, and also discuss potential functions of these IDA subunits in other eukaryotes. Chlamydomonas IDAs are very complex including multiple IDA species distinct in composition and location in the axoneme (Figure 1, Table 1), thus we first list and review the subunits identified to date in Chlamydomonas in an itemized style in the next section (Section 2)(Figure 2, 3; Table 2), and in the later sections (Sections 3 and 4) discuss remaining questions/challenges, future direction and perspective of IDA studies.
Figure 2. Domain structure of IDA HCs in Chlamydomonas.
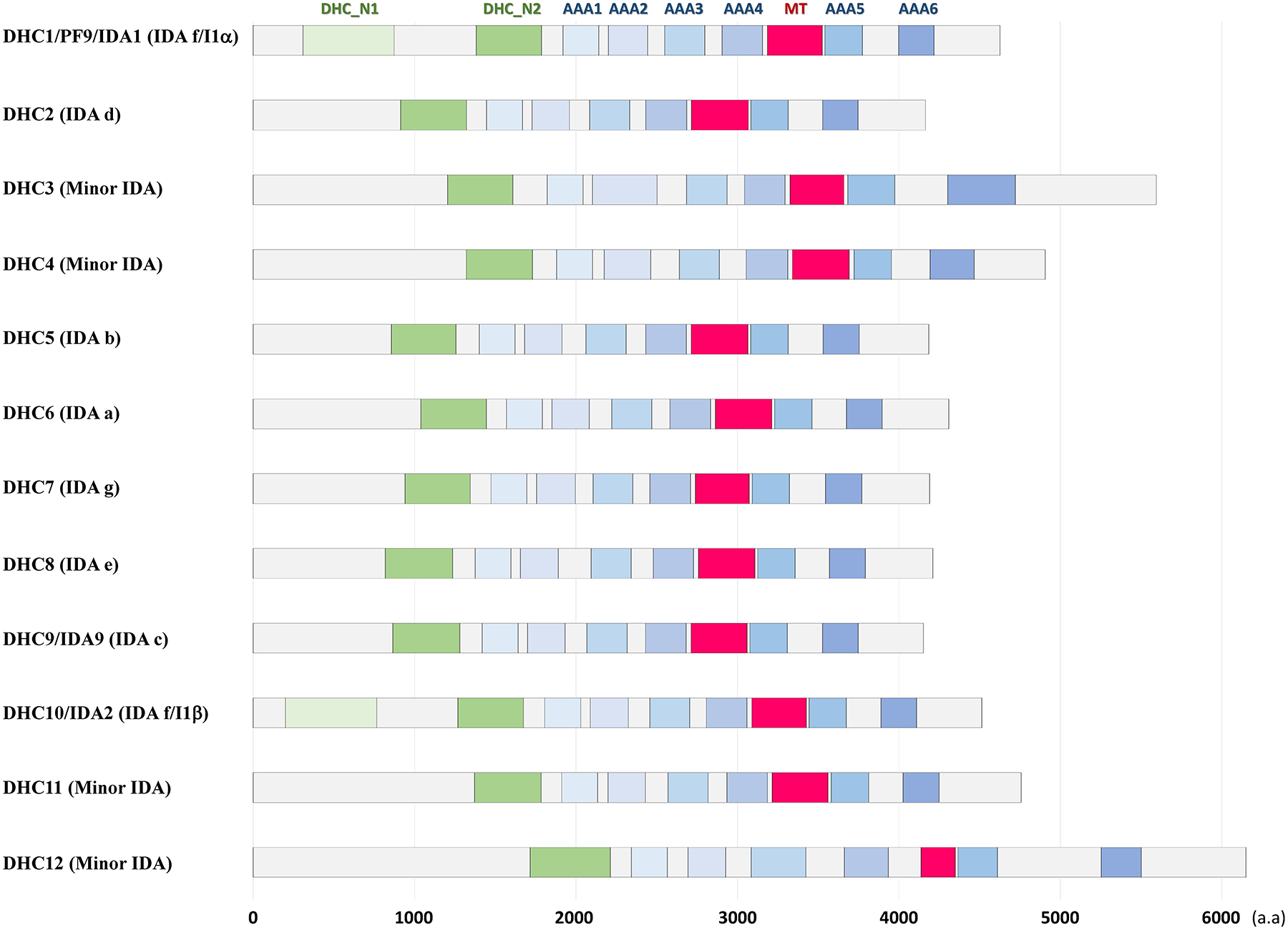
Domains/motifs in DHCs of Chlamydomonas ciliary IDAs. “Dynein heavy chain, N-terminal region 1 (DHC_N1)”, “dynein heavy chain, N-terminal region 2 (DHC_N2)”, and “microtubule-binding stalk of dynein motor (MT)” domains were predicted using the pfam analyses (https://pfam.xfam.org/)(Punta et al., 2012). Appropriate locations of 6 “ATPases-associated-with-diverse-cellular-activities (AAA)” domains/rings in the IDA DHCs were predicted by aligning the IDA sequences with the Tripneustes gratilla ODAβ DHC [X59603.1 (NCBI)](Mocz & Gibbons, 2001). Note that two minor IDAs, DHC3 and DHC12, have longer molecular length than other IDAs. The accession Nos/IDs in the Phytozome Chlamydomonas v5.5 (https://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Creinhardtii) used for these domain predictions were as follows: DHC1/PF9/IDA1, Cre12.g484250.t1.1; DHC2, Cre09.g392282.t1.1; DHC3, Cre06.g265950.t1.1; DHC4, Cre02.g107350.t1.1; DHC5, Cre02.g107050.t1.1; DHC6, Cre05.g244250.t1.2; DHC7, Cre14.g627576.t1.1; DHC8, Cre16.g685450.t1.1; DHC9/IDA9, Cre02.g141606.t1.1; DHC10/IDA2, Cre14.g624950.t1.1; DHC11, Cre12.g555950.t1.2; DHC12, Cre06.g297850.t1.1.
Figure 3. Domain structure of IDA ICs/LCs/accessary subunits in Chlamydomonas.
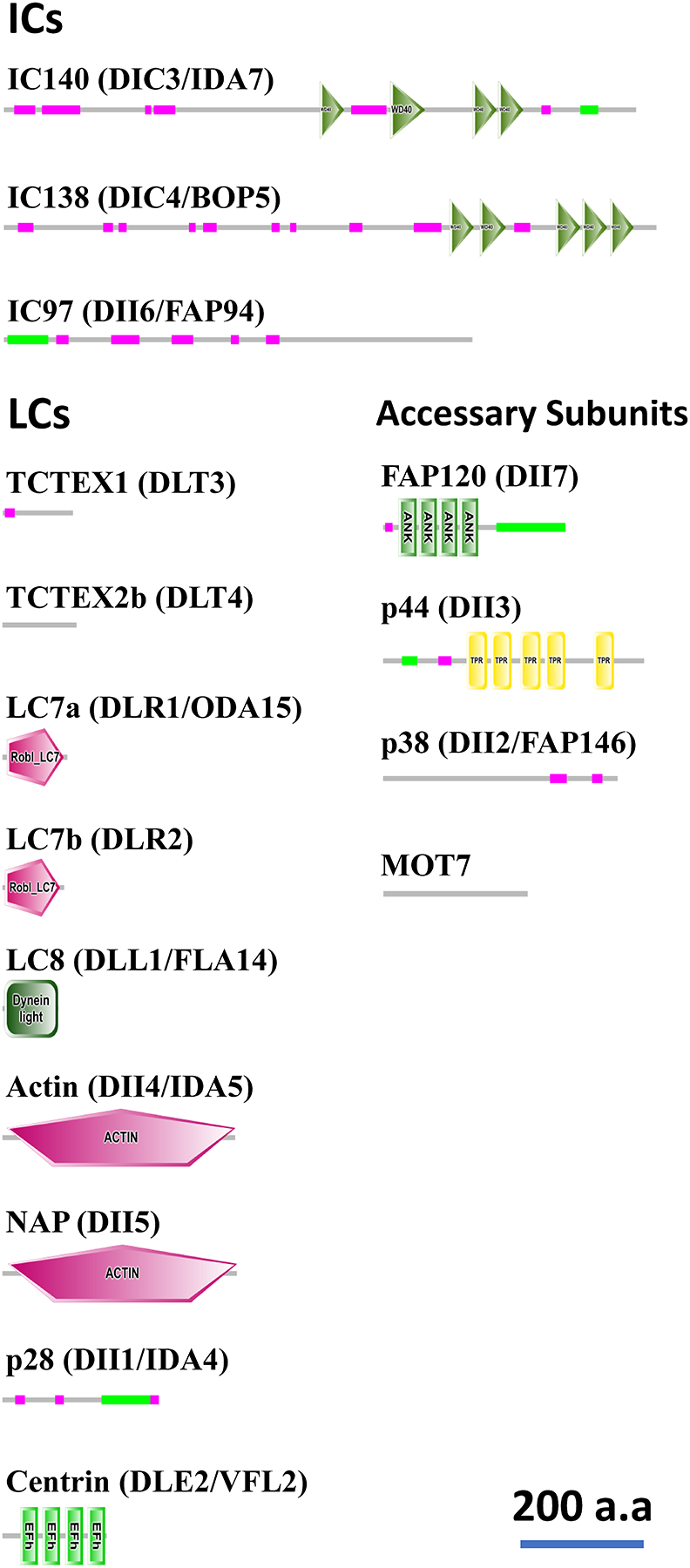
Domains/motifs in ICs/LCs/accessary subunits of Chlamydomonas ciliary IDAs were predicted using the SMART analyses (http://smart.embl-heidelberg.de/)(Letunic et al., 2021; Schultz et al., 1998) with the default setting and outputs with scale modification were shown. The green thin lines indicate the coiled-coil regions, and the pink thin lines indicate the low-complexity regions. While seven WD repeats were identified in the original descriptions both for the IC140 and IC138 molecules (Hendrickson et al., 2004; Yang & Sale, 1998), fewer repeats were predicted by SMART analyses in this figure due to the threshold. The domain structures of FAP120 and p44 were re-analyzed from (Ikeda et al., 2009; Yamamoto et al., 2008). The accession Nos/IDs in NCBI (https://www.ncbi.nlm.nih.gov/) or Phytozome Chlamydomonas v5.5 (https://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Creinhardtii) used for these domain predictions were as follows: IC140/DIC3/IDA7, XP_001695786.1 (NCBI); IC138/DIC4/BOP5, XP_001696921.1 (NCBI); IC97/DII6/FAP94, ACN22075 (NCBI); TCTEX1/DLT3, Cre01.g004250.t1.2 (Phytozome); TCTEX2b/DLT4, DAA05278 (NCBI); LC7a/DLR1/ODA15, Cre08.g376550.t1.2 (Phytozome); LC7b/DLR2, Cre12.g546400.t1.2 (Phytozome); LC8/DLL1/FLA14, Cre03.g181150.t1.1 (Phytozome); Actin/DII4/IDA5, Cre13.g603700.t1.2 (Phytozome); NAP/DII5, Cre03.g176833.t1.1 (Phytozome); p28/DII1/IDA4, CAA88139 (NCBI); Centrin/DLE2/VFL2, Cre11.g468450.t1.2 (Phytozome); FAP120/DII3, BAN15819 (NCBI); p44/DII3, AB353122.2 (NCBI); p38/DII2/FAP146, BAG07147 (NCBI); MOT7, Cre01.g038750.t1.2 (Phytozome).
2. Subunits of IDAs identified in Chlamydomonas, their potential functions, and homologues/orthologues in other eukaryotes
In this section, we categorize Chlamydomonas IDA subunits into four groups (dynein heavy chains (DHCs), intermediate chains (ICs), light chains (LCs), and accessary subunits), and discuss their potential function(s) in ciliary assembly and regulation.
2-1. DHCs
DHCs are essential subunits of ciliary dyneins since they convert chemical energy of ATP to mechanical force to drive ciliary motility. Based on DHC composition and location in the axoneme, Chlamydomonas IDAs were first classified into five groups, I1, I2, I2’, I3, I3’ (Huang et al., 1979; Piperno et al., 1990)(Figure 1a, 1b). I1 dynein was shown to contain two DHCs (HCα and HCβ, two-headed/dimeric dynein), while other species were each shown to contain one DHC (single-headed/monomeric dynein)(Goodenough et al., 1987; Piperno et al., 1990). Using ion-exchange chromatography, Kagami and Kamiya later found that IDAs consist of seven major types (IDA species “a” to “g”), each containing one or two distinct DHCs and total of eight distinct DHCs (Kagami & Kamiya, 1992). In addition, four minor types have also been identified: three of the DHCs are specifically located in the proximal end of the axoneme (Bui et al., 2012; Yagi et al., 2009)(Figure 1a, 1b, 2; Table 1). Notably, the minor IDAs replace specific major types of IDAs in the 96-nm axonemal repeat (Bui et al., 2012; Kamiya & Yagi, 2014). Currently the major IDAs are referred to as IDAs a, b, c, d, e, f/I1 and g (Kamiya & Yagi, 2014; Yagi & Kamiya, 2012).
Among the sixteen DHC genes present in Chlamydomonas genome (Merchant et al., 2007; Porter et al., 1996; Wickstead & Gull, 2007; Yagi et al., 2009), twelve genes (DHC1 - DHC12) encode IDA HCs (Figure 2; Table 1, 2). Of the twelve DHC genes for IDAs, eight DHC genes encode the major IDA types (DHC1 for I1/fα, DHC2 for d, DHC5 for b, DHC6 for a, DHC7 for g, DHC8 for e, DHC9 for c, and DHC10 for I1/fβ)(Hom et al., 2011; Yagi et al., 2005; Yagi et al., 2009)(Figure 1a, 1b, 2). The minor DHCs are encoded by DHC3, DHC4, DHC11, and DHC12 (Figure 1a, 1b, 2)(Hom et al., 2011; Kamiya & Yagi, 2014; Yagi et al., 2005; Yagi et al., 2009). Of the remaining four DHC genes, three encode ODA DHCs (DHC13 - DHC15)(Hom et al., 2011; Liu et al., 2008; Sakakibara et al., 1991; Sakakibara et al., 1993) and one encodes the DHC responsible for retrograde IFT (DHC16)(Engel et al., 2012; Pazour et al., 1999).
Distinct localization of these IDAs was determined by structural comparison of wild-type and mutant axonemes defective in assembly of specific IDA species and revealed the precise location of each IDA in the axoneme 96-nm repeat (Bui et al., 2008, 2009; Bui et al., 2012; Heuser, Barber, et al., 2012; Mastronarde et al., 1992; Nicastro et al., 2006; Piperno et al., 1990; Porter et al., 1992; Yagi et al., 2005; Yamamoto et al., 2010)(Figure 1a, 1b). In addition, asymmetric localization of IDAs across the axonemes was observed, and such asymmetry is predicted to contribute to the asymmetric ciliary bending pattern (Bui et al., 2009; Bui et al., 2012; King et al., 1994; Lin et al., 2019; Piperno & Ramanis, 1991). The minor IDA species (DHC3, DHC4, and DHC11) are located at the proximal axoneme where they replace major IDAs (Bui et al., 2012; Yagi et al., 2009)(Figure 1a, 1b). It has been postulated that the minor IDAs located at the proximal axoneme play a role in ciliary bend initiation. Localization of the remaining minor IDA species, DHC12, has not been determined.
Each IDA species exhibits distinct mechanical and enzymatic properties as assessed by cell movement, in vitro motor assays and ATPase activity. For example, motility analysis of a mutant lacking IDA f/I1 HC (ida1/pf9 lacking DHC1 or ida2 lacking DHC10)(Figure 2; Supporting Table S1) reveals slower cell motility and altered ciliary waveform compared to wild-type (Kamiya et al., 1991; Kotani et al., 2007; Myster et al., 1997; Perrone et al., 2000; Toba et al., 2011). The slow swimming and altered waveform indicate that the IDAs function to regulate ciliary waveform (also discussed later in the IC138 section). Intriguingly, mutants lacking IDA f/I1 also show reduced phototactic ability compared to wild-type, suggesting some role(s) of this dynein species in the phototactic response/signaling pathway (King & Dutcher, 1997; Okita et al., 2005; VanderWaal et al., 2011). A mutant lacking IDA c (ida9 lacking DHC9)(Figure 2; Supporting Table S1) shows only slight swimming defect in normal conditions but greatly reduced swimming ability in viscous media, suggesting the role of IDA c is to generate force under viscous conditions (Yagi et al., 2005).
Genetic defects in humans leading to a loss of specific ciliary IDA DHCs can result in defective sperm flagellar assembly or defective sperm motility. For example, defects in human DNAH1 gene, a potential orthologue of Chlamydomonas DHC2 (IDA d)(Figure 2)(Kollmar, 2016; Morris et al., 2006) are linked to infertility with anomalies of the sperm flagella including dysplasia of the sperm fibrous sheath (Ben Khelifa et al., 2014; Imtiaz et al., 2015; Wang et al., 2017). Also, human DNAH2 gene, a potential orthologue of Chlamydomonas IDA f/I1β HC (DHC10)(Kollmar, 2016) is a candidate gene of one type of the teratozoospermia, multiple morphological abnormalities of the sperm flagella (MMAF)(Li et al., 2019). In addition, defects in human DNAH6 gene, a potential orthologue of Chlamydomonas DHC7 (IDA g) or minor IDA DHC3 (Bustamante-Marin et al., 2020; Kollmar, 2016) have been reported to cause azoospermia, suggesting the important role of IDA g in spermatogenesis and sperm motility (Gershoni et al., 2017; Y. Li et al., 2016).
2-2. ICs
Three IDA ICs discussed below, all subunits of IDA f/I1, have been identified in Chlamydomonas. All these ICs are important for either assembly or regulation of IDA f/I1.
a. IC140 (DIC3/IDA7)
IC140, also called DIC3 or IDA7, is one of the three ICs of IDA f/I1, having seven WD40 repeats in the C-terminal half of its structure (Perrone et al., 1998; Yang & Sale, 1998)(Figure 3; Table 1, 2). The mutant missing this IC, ida7, shows slower swimming velocity and altered waveform compared to wild-type (Perrone et al., 1998)(Supporting Table S1). Importantly, in ida7 IDA f/I1 fails to assemble in the ciliary axoneme, strongly suggesting the roles of IC140 in assembly or docking of IDA f/I1 (Perrone et al., 1998). Consistently, IDA f/I1 fails to preassemble in the cytoplasm of ida7 cells (Viswanadha et al., 2014). Based on chemical crosslinking, IC140 and another IC of IDA f/I1, IC138 (see below), are thought to directly interact with tubulin (Hendrickson et al., 2013). A potential mammalian orthologue of IC140, WDR63 (Table 3), is not required for fertility in mice (Young et al., 2015). However, a recent study showed a potential and intriguing relationship between an intragenic deletion of WDR63 and occipital encephalocele in humans (Hofmeister et al., 2018)(Table 3).
Table 3.
BLAST best-hit search of Chlamydomonas IDA IC/LC/accessary subunits in human genome
| BLAST comparisona | ||
|---|---|---|
| Chlamydomonas reinhardtii | Homo sapiensb | Potential related/associated disease in humans |
| Actin/DII4/IDA5 | ACTG1 (0.0)c, d | Various |
| Centrin/DLE2/VFL2 | CETN2 (2e-55) | - |
| FAP120/DII7 | N/Ae | - |
| IC140/DIC3/IDA7 | WDR63/DNAI3 (6e-55) | Occipital encephalocele (Hofmeister et al., 2018) |
| IC138/DIC4/BOP5 | WDR78/DNAI4 (2e-68) | - |
| IC97/DII6/FAP94 | CASC1/CFAP94 (2e-11) | Pulmonary carcinoma (Galbiati et al., 2006) |
| LC7a/DLR1/ODA15 | DYNLRB2 (1e-40) | - |
| LC7b/DLR2 | DYNLRB2 (3e-20) | - |
| LC8/DLL1/FLA14 | DYNLL2 (2e-56) | - |
| MOT7 | TEX47 (6e-14) | Hypothyroidism (Eriksson et al., 2012) |
| NAP/DII5 | N/Af | - |
| p28/DII1/IDA4 | DNALI1 (1e-93) | - |
| p38/DII2/FAP146 | ZMYND12 (1e-19) | - |
| p44/DII3 | TTC29 (2e-13) | Asthenoteratospermia/Asthenozoospermia (Liu et al., 2019; Lorès et al., 2019) |
| TCTEX1/DLT3 | DYNLT1 (2e-42) | - |
| TCTEX2b/DLT4 | TCTEX1D2/DYNLT2B (5e-35) | Jeune asphyxiating thoracic dystrophy (Schmidts et al., 2015) |
For the presence/absence of the potential homologous DHCs of Chlamydomonas IDAs in humans, see the previous papers (Hom et al., 2011; Kollmar, 2016; Morris et al., 2006). For potential links between human IDA DHCs and diseases, see Section 2-1 in the main text.
Blast search using the Chlamydomonas sequences as queries against the Ensembl genome browser 102 human database (http://www.ensembl.org/Homo_sapiens/Info/Index).
Gene names and BLAST e-Values are shown in the columns.
Letters in bold in human proteins mean that the reciprocal BLAST search against Chlamydomonas Phytozome genome database (v5.5) identified the corresponding dynein subunit as the best hit in Chlamydomonas reinhardtii.
Because of the presence of ANK repeats, the exact human orthologue(s) of FAP120 in humans is hard to determine (Ikeda et al., 2009).
From the detailed phylogenetic analyses, homologues of NAP/DII5 are only present in algae phylogenetically close to Chlamydomonas (Kato-Minoura et al., 2003; Kato-Minoura et al., 1998).
b. IC138 (DIC4/BOP5)
IC138, also called DIC4 or BOP5, is the second IC of IDA f/I1, and like IC140, contains seven WD40 repeats in the C-terminal half of its structure (Hendrickson et al., 2004)(Figure 3; Table 1, 2). Unlike IC140, IC138 is not essential for axonemal assembly of the IDA f/I1 complex (Hendrickson et al., 2004; VanderWaal et al., 2011). However, IC138 is required for assembly of the IC138 subcomplex containing at least four subunits of IDA f/I1, including IC138, IC97, LC7b, and FAP120 (Bower et al., 2009; Hendrickson et al., 2004; Heuser, Barber, et al., 2012; Ikeda et al., 2009). In addition, IC138 was shown to be a phospho-protein important for regulation of ciliary motility (Elam et al., 2011; Gokhale et al., 2009; King & Dutcher, 1997; VanderWaal et al., 2011; Wirschell et al., 2007). Analyses of bop5 alleles lacking IC138 show phenotypes similar to mutants lacking the entire IDA f/I1 complex (ida1/pf9 and ida2) with slower motility and altered waveform. These observations suggest a regulatory role of IC138 in IDA f/I1 activity (Hendrickson et al., 2004; VanderWaal et al., 2011)(Supporting Table S1).
A model of regulation for IDA f/I1 has been proposed, where IDA f/I1 activity correlates with IC138 phosphorylation (Wirschell et al., 2007). The key axonemal kinase/phosphatase that phosphorylates/de-phosphorylates IC138 are currently not determined, but casein kinase 1 (CK1) and protein phosphatase 2A (PP2A)(Supporting Table S1) have been proposed as strong candidates since they are located in the axoneme (Elam et al., 2011; Gokhale et al., 2009). Recently, CK1 has been shown to be stabilized by the IDA f/I1 “tether” structure, which intriguingly binds ciliary microtubule and f/I1 heads (Fu et al., 2018)(Supporting Table S1). Also, an axonemal complex that appears to support and determine the relative position of IC138 and these kinase/phosphatase in cilia has been reported as the “modifiers of inner arms (MIA) complex” (King & Dutcher, 1997; Yamamoto et al., 2013)(Supporting Table S1). The core of the MIA complex consists of two coiled-coil axonemal proteins, flagellar-associated protein 100 (FAP100) and FAP73 (Yamamoto et al., 2013). Intriguingly, potential mammalian homologues of FAP100 and FAP73, coiled-coil domain containing 37 (CCDC37), CCDC38, and CCDC42a/b are all implicated in some aspects of the chronic obstructive pulmonary disease (COPD) and/or the lung cancer, suggesting the importance of IC138 and IDA f/I1 in normal lung function (Aravind Kumar et al., 2018; Kwon et al., 2012; Menche et al., 2014; Soler Artigas et al., 2011; Tessema et al., 2015; Tian et al., 2017; Wain et al., 2014). A potential mammalian orthologue of IC138, WDR78 (Table 3), was identified, suggesting ciliary IC138 function is conserved from Chlamydomonas to multi-cellular eukaryotes. WDR78 is strongly expressed in testis and thymus, while the expression is weak in brain (Young et al., 2015). A recent study has revealed that WDR78 is indispensable for ciliary motility and ciliary IDA f/I1 assembly in mammals, confirming the conserved importance of this IDA subunit/species (Zhang et al., 2019). An alternative splicing form of WDR78 is reported to be influenced by zinc-finger Ran-binding domain containing protein 2 (ZRANB2), a human splicing factor (Yang et al., 2013).
c. IC97 (DII6/FAP94)
IC97, also called DII6 or FAP94, is the third IC of IDA f/I1 and contains the Cancer Susceptibility Candidate 1, N-terminal (Casc1_N) domain (Wirschell et al., 2009)(Figure 3; Table 1, 2). IC97 is a component of IC138 subcomplex of IDA f/I1 and possibly functions in concert with IC138 to regulate IDA f/I1 (Bower et al., 2009; Heuser, Barber, et al., 2012). In the bop5 mutant deficient in IC138 (Supporting Table S1), IC97 fails to assemble in the axoneme, indicating IC138 is required for IC97 stability in cilia (Hendrickson et al., 2004; Heuser, Barber, et al., 2012; VanderWaal et al., 2011). IC97 was also shown to bind α- and β- tubulins directly in axonemes (Wirschell et al., 2009). However, to date, no detailed analysis of Chlamydomonas ic97 mutant has been reported. Since meta-insertional mutant library projects (Chlamydomonas Library Project :CLiP) have been completed and the mutants having deficient in the IC97 gene seem to be available on request (https://www.chlamylibrary.org/)(X. Li et al., 2016), it would be interesting to determine the exact role(s) of IC97 in regulation of IDA f/I1 and ciliary motility. A potential mammalian orthologue of IC97, CASC1 (Table 3), was reported to be involved in or related to pulmonary adenoma (Liu et al., 2006).
2-3. LCs
Chlamydomonas has large variety in the LC composition between the two-headed IDA f/I1 and other remaining single-headed IDAs. Among the LCs we present in this subsection below, first five LCs (TCTEX1, TCTEX2b, LC7a, LC7b, and LC8) are all LCs of IDA f/I1, while the others (actin, NAP, p28 and centrin) are LCs of single-headed IDAs.
a. TCTEX1 (DLT3)
TCTEX1, also called DLT3, is a LC of IDA f/I1 that contains the Tctex-1 domain in its structure (Harrison et al., 1998)(Figure 3; Table 1, 2). This protein was first identified as a cytoplasmic dynein LC, but later was revealed to be also an IDA f/I1 component (DiBella et al., 2001; Harrison et al., 1998). The structure of the TCTEX1 dimer was solved by the nuclear magnetic resonance (NMR) and revealed its similarity to the LC8 (described below) dimers (Wu et al., 2001; Wu et al., 2005). To date, no tctex1 mutant in Chlamydomonas has been reported. Thus, the function of this LC is unclear. In mammals, TCTEX1 (also called DYNLT1)(Table 3) is a potential candidate of the distorter/sterility factor in the non-Mendelian transmission of mouse t-haplotypes (King et al., 1996; Lader et al., 1989). TCTEX1/DYNLT1 has also been implicated in ciliary resorption (Li et al., 2011).
b. TCTEX2b (DLT4)
TCTEX2b, also called DLT4, is an IDA f/I1 LC and contains a Tctex-1 domain in its structure (DiBella, Smith, et al., 2004)(Figure 3; Table 1, 2). TCTEX2b co-purifies with IDA f/I1 (DiBella, Smith, et al., 2004). TCTEX2b is not required for IDA f/I1 assembly in the axonemes, but the stability of the IDA f/I1 complex is reduced when axonemes are suspended in high-salt buffers (DiBella, Smith, et al., 2004). Analysis of the double mutant pf16-D2, deficient in both PF16 and TCTEX2b, revealed a role for the TCTEX2b in regulation of IDA f/I1 activity and ciliary motility (DiBella, Smith, et al., 2004; Hom et al., 2011)(Supporting Table S1). A potential mammalian orthologue of TCTEX2b, TCTEX1D2 (Table 3), is implicated in its association with short-rib polydactyly syndrome (SRPS) proteins (Gholkar et al., 2015; Mukhopadhyay, 2015). Mutations in TCTEX1D2 gene was also shown to cause Jeune asphyxiating thoracic dystrophy in humans by impairing the retrograde IFT machinery (Table 3), suggesting TCTEX1D2 is an important component of IFT dynein (Schmidts et al., 2015).
c. LC7a (DLR1/ODA15)
LC7a, also called DLR1 or ODA15, is a LC of IDA f/I1 and ODAs, and contains the Roadblock/LC7 (Robl_LC7) domain in its structure (Pazour & Witman, 2000)(Figure 3; Table 1, 2). A mutant lacking LC7a, oda15, assembles only ~ 30% of ODAs compared to wild-type in cilia (DiBella, Sakato, et al., 2004)(Supporting Table S1). This result suggests LC7a plays a role in docking or stability of ODAs. In addition, oda15 shows assembly defect in IDA f/I1, suggesting LC7a also plays a role in docking and stability of IDA f/I1 (DiBella, Sakato, et al., 2004)(Supporting Table S1). Potential mammalian homologues of LC7a, DYNLRB1 and DYNLRB2 (Table 3), are both subunits of human cytoplasmic dynein-1 and dynein-2 (Jiang et al., 2001). DYNLRB1 has been shown to interact with Rab6 family small GTPases and to possibly modulate their GTPase activities (Wanschers et al., 2008). DYNLRB2 has been implicated in its involvement in leukemia virus infection in mice (Opazo et al., 2017).
d. LC7b (DLR2)
LC7b, also called DLR2, is a LC of IDA f/I1 and ODAs, and contains the Robl_LC7 domain in its structure (DiBella, Sakato, et al., 2004)(Figure 3; Table 1, 2). LC7b is a component of the IC138 subcomplex of IDA f/I1 (Bower et al., 2009). The bop5 allele expressing a C-terminally truncated form of IC138 (bop5-1) fails to assemble LC7b, suggesting the C-terminal region of IC138 is essential for stability of LC7b in cilia (Supporting Table S1). In contrast, LC7a is assembled in the bop5-1 axoneme (Hendrickson et al., 2004). Thus, despite the high sequence homology between these two LCs, they seem to serve different functions. To date, no mutant lacking LC7b has been reported, and the exact function of LC7b is unclear. However, a null allele of IC138 (bop5-2) lacks IC138 subcomplex (IC138, IC97, FAP120 and LC7b) but still assembles IDA f/I1 in ciliary axoneme, suggesting LC7b is dispensable for IDA f/I1 assembly (Bower et al., 2009; Heuser, Barber, et al., 2012; Ikeda et al., 2009)(Supporting Table S1). Thus, LC7b is predicted to have some regulatory role for IDA f/I1 activity.
e. LC8 (DLL1/FLA14)
LC8, also called DLL1 or FLA14, is a LC of IDA f/I1, ODAs and the IFT dynein (cytoplasmic dynein 2)(DiBella et al., 2001; Pazour et al., 1998)(Figure 3; Table 1, 2). LC8 is also a component of ciliary radial spokes involved in the assembly of radial spoke structures (Gui et al., 2021; Gupta et al., 2012; Yang et al., 2001; Yang et al., 2009). LC8 contains a “Dynein light chain type 1 (Dynein light)” domain in its structure, and LC8 sequence/structure has been studied in detail (Barbar, 2008; King & Patel-King, 1995). Because of the molecular versatility in ciliary function, a mutant of LC8, fla14, shows very short immotile cilia due to the impaired IFT, also having defects in radial spokes, ODAs, IDAs, and ciliary beak-like structures (Gupta et al., 2012; Pazour et al., 1998; Yang et al., 2009)(Supporting Table S1). A mutant pf14-3 (also called pf5) expresses LC8 with an extension at the C-terminus (Yang et al., 2009). pf14-3 cells have short cilia that show paralyzed or sporadic-twitching motility (Yang et al., 2009). Two minor radial-spoke proteins (RSP13 and RSP15) are missing in pf14-3 axonemes. In addition, IC97 (described before) is lacking in pf14-3 axonemes, suggesting a structural interaction between IC97 and LC8 (Yang et al., 2009).
LC8 is highly conserved with mammalian homologues DYNLL1 and DYNLL2 (Table 3). The LC8 proteins are regarded “hub” proteins (Barbar, 2008), and have been implicated in various cellular functions including nuclear transport, apoptosis and mitosis (Rapali et al., 2011).
f. Actin (DII4/IDA5)
Chlamydomonas has one conventional actin gene, IDA5 also called DII4 (Kato-Minoura et al., 1997; Kato-Minoura et al., 2003)(Figure 3; Table 1, 2). Surprisingly at the time, researchers identified an actin-like protein in cilia (Behnke et al., 1971; Piperno & Luck, 1979, 1981). Subsequent studies revealed that actin is a subunit of nearly all of the single-headed IDA species including IDAs a, b, c, d, e, and g, and most likely DHC3, DHC4, DHC11, and DHC12 (Kagami & Kamiya, 1992; Kamiya & Yagi, 2014; LeDizet & Piperno, 1995b; Piperno, 1988; Yagi et al., 2009). A mutant having defect in the ACTIN gene, ida5 shows slower cell motility and altered ciliary waveform. The motility phenotype is due to failure in assembly of IDAs a, c, d and e (Kato et al., 1993; Kato-Minoura et al., 1997)(Supporting Table S1). Thus, actin is indispensable for axonemal docking of these IDA species. Actin interacts with an additional IDA LC, p28 or centrin (LeDizet & Piperno, 1995b; Yanagisawa & Kamiya, 2001). Recently, it was determined that the elongated axonemal proteins FAP59/CCDC39 and FAP172/CCDC40 define the ciliary axonemal 96-nm repeat structure in Chlamydomonas (Oda et al., 2014). Since actin is required for docking of a subset of single-headed IDAs, it is possible that actin interacts with the FAP59/FAP172 for localization of these IDAs (Figure 1a, 1b). This model has not yet been tested, and this would be an interesting future challenge (discussed later in Section 3). Further, surprisingly, polymerized actin filaments have recently been shown to form in primary cilia and possibly function in ciliary disassembly (Kiesel et al., 2020; Phua et al., 2017). Predictably, actin or actin-like (e.g. observed in dynactin) filaments will be found in motile cilia.
g. NAP (DII5)
Analysis of the actin mutant ida5 has revealed a novel actin-like protein (NAP) replaces actin as a LC of IDAs b and g (Kato-Minoura et al., 1997; Kato-Minoura et al., 1998)(Figure 3; Table 1, 2). NAP contains an actin domain (Figure 3), but has only 65% identity and 80% similarity to Chlamydomonas conventional actin. Moreover, NAP is only weakly expressed under normal conditions in wild-type cells. However, when the conventional actin gene is disrupted in ida5, expression of NAP is strongly up-regulated, and NAP substitutes for actin in docking IDAs b and g (Hirono et al., 2003; Kato-Minoura et al., 1998)(Supporting Table S1). From the biochemical analyses, NAP has a low ability to polymerize (Kato-Minoura, 2011). However, NAP can provide “f-actin” functions in vivo (Onishi et al., 2016), and can partially replace conventional actin for successful ciliogenesis (Jack et al., 2019). The homologues of NAP have been only found in some volvocine algae such as Volvox and Gonium, which are phylogenetically close to Chlamydomonas (Kato-Minoura et al., 2003).
h. p28 (DII1/IDA4)
p28, also called DII1 or IDA4, is a LC in IDAs a, c and d, and most likely of DHC11 and DHC12 (Kamiya & Yagi, 2014; LeDizet & Piperno, 1995a, 1995b; Piperno et al., 1992)(Figure 3; Table 1, 2). p28 and centrin (described below) each bind to separate single-headed IDAs in a complementary manner (Kagami & Kamiya, 1992; LeDizet & Piperno, 1995b; Yanagisawa & Kamiya, 2001). Cilia from the mutant lacking p28, ida4 (Supporting Table S1), fail to assemble IDAs a, c and d, indicating a role of p28 in the axonemal docking (LeDizet & Piperno, 1995a). p28 directly interacts with the N-terminal fragments of the DHCs of these IDAs and also with actin (described above). From the stoichiometric analysis, two p28 molecules were thought to interact with one actin monomer and bind to each one of the DHCs (IDAs DHC6/a, DHC9/c, DHC2/d, and most likely DHC11 and DHC12)(Kamiya & Yagi, 2014; Yanagisawa & Kamiya, 2001). Recently, a p28 homodimer interacting with DHC9 and actin was identified in cilia (Gui et al., 2021). A mammalian orthologue of p28, DNALI1 (Table 3), is also a LC of axonemal dyneins (Rashid et al., 2006).
i. Centrin (DLE2/VFL2)
Centrin, also called DLE2 or VFL2, is a LC of IDAs b, e and g, and most likely of DHC3 and DHC4 as well (Kamiya & Yagi, 2014; Piperno et al., 1992; Taillon et al., 1992)(Figure 3; Table 1, 2). Centrin contains four EF-hand domains (Figure 3). In Chlamydomonas centrin is found in the nucleus-basal body connector, the distal striated fiber between basal bodies, and in the ciliary transition zone (Dutcher & O’Toole, 2016; Ruiz-Binder et al., 2002; Salisbury et al., 1988; Sanders & Salisbury, 1989; Wright et al., 1989). A null mutant of centrin in Chlamydomonas is not available. However, a mutant expressing mutated centrin (vfl2) displays various numbers of cilia ranging from 0 to 6 with aberrant swimming patterns (Hayashi et al., 1998; Taillon et al., 1992)(Supporting Table S1). Thus, along with its role as a subunit in a subset of IDAs, centrin plays important roles in ciliogenesis and normal basal-body positioning (Dutcher & O’Toole, 2016). From stoichiometric analysis, a centrin monomer is predicted to directly interact with actin and the N-terminal fragment of a subset of the IDA DHCs (IDAs DHC5/b, DHC8/e, DHC7/g, and most likely DHC3 and DHC4)(Kagami & Kamiya, 1992; Yagi et al., 2009; Yanagisawa & Kamiya, 2001). A mammalian orthologue of centrin, CENT2 (Table 3), has been shown to play a role in cilia, and CENT2 deficient mice show ciliopathic phenotypes including dysosmia and hydrocephalus (Ying et al., 2014). CENT2 has also been reported to regulate ciliogenesis of primary cilia (Prosser & Morrison, 2015).
2-4. Accessary subunits
In addition to DHCs, ICs and LCs described above in this section, some IDA species have unique accessary subunits (FAP120 and MOT7 for IDA f/I1, and p44 and p38 for IDA d) that are weakly associated to these IDAs and/or not present in other similar Chlamydomonas IDA species.
a. FAP120 (DII7)
FAP120, also called DII7, is an accessary subunit of IDA f/I1, with four ankyrin (ANK) repeats in the N-terminal half and a coiled-coil domain in the C-terminal half (Ikeda et al., 2009)(Figure 3; Table 1, 2). Although FAP120 does not co-elute with IDA f/I1 in ion-exchange chromatography, FAP120 is absent or greatly reduced in the axonemes from mutants lacking IDA f/I1 (ida1/pf9, ida2, and ida7). This result strongly suggests that FAP120 is an IDA f/I1 component (Heuser, Barber, et al., 2012; Ikeda et al., 2009). In addition, analyses of bop5 mutants (ic138 alleles) revealed that the level of FAP120 reduction in axonemes correlates with loss of IC138, further supporting that FAP120 is a component of the IC138 subcomplex (Ikeda et al., 2009). The bop5 allele expressing a C-terminal truncated IC138 (bop5-1) lacks FAP120 in their axonemes (Ikeda et al., 2009). Thus, the C-terminal fragment of IC138 missing in bop5-1 mutant seems to be required for axonemal localization of FAP120.
Due to the presence of ANK repeats in its structure, an exact orthologue of FAP120 in mammals is difficult to determine (Table 3). By the basic local alignment search tool (BLAST) analyses using FAP120 protein sequence as a query on human non-redundant protein sequences in the national center for biotechnology information (NCBI) database, the most homologous protein to FAP120 in humans is ANK2, an ankyrin-2 protein. However, from the expression pattern and molecular size, this protein does not seem to be the exact orthologue of FAP120 in humans, and further study will be needed to determine the human orthologue of FAP120. The cDNA sequence of FAP120 has been deposited in DNA Data Bank of Japan (DDBJ) under the accession No. AB818238.
b. MOT7
An orthologue of Chlamydomonas MOT7, a dynein-associated BLUF-domain-containing protein (DYBLUP), has recently been identified as a subunit of IDA f/I1 in Ciona intestinalis (sea squirt)(Kutomi et al., 2021). An intriguing feature of DYBLUP is the presence of the “sensors of blue-light using FAD (BLUF)” domain, which senses blue light using the flavin adenine dinucleotide (FAD) or flavin mononucleotide (FMN) as a co-factor, although the prediction value of the BLUF domain varies depending on the organisms (Gomelsky & Klug, 2002; Park & Tame, 2017). In Ciona, DYBLUP is predicted to bind the head of IDA f/I1’s HCβ. One idea under investigation is that IDA f/I1 can be regulated directly by light through DYBLUP, an entirely novel mechanism of regulation of dynein motor activity. In Chlamydomonas, although the BLUF domain is not predicted in the MOT7 molecule with high probability like in Ciona DYBLUP (Figure 3), MOT7 was shown to interact with FAP44, an IDA f/I1-head interacting protein, strongly suggesting MOT7 interacts with an IDA f/I1’s head in Chlamydomonas cilia, connecting the f/I1’s head and the f/I1-tether complex (Kutomi et al., 2021)(Figure 3; Table 1, 2). Since Chlamydomonas mutants lacking IDA f/I1 components or the MIA complex (ida1/pf9, ida2, bop5, mot7, mia1, and mia2)(Supporting Table S1) display abnormal phototaxis and/or photoshock response, IDA f/I1 is thought to function in phototaxis/photoshock responses (King & Dutcher, 1997; Okita et al., 2005; VanderWaal et al., 2011). One idea, yet to be tested, is that MOT7 is a light-sensing protein involved in the dynein regulation and/or in signaling pathway of phototaxis/photoshock responses in Chlamydomonas as a subunit of IDA f/I1. A potential mammalian orthologue of DYBLUP/MOT7, C7orf62/TEX47 (Table 3), has been reported to have potential link to hypothyroidism (Eriksson et al., 2012).
c. p44 (DII3)
p44, also called DII3, is an accessary subunit of IDA d (Yamamoto et al., 2008)(Figure 3; Supporting Figure S1; Table 1, 2). p44 has a small coiled-coil region near the N-terminal end and several tetratricopeptide repeat (TPR) motifs in middle to C-terminal half of its structure. p44 was first identified as a 44-kDa protein co-purified with IDA d (Kagami & Kamiya, 1992; Kato et al., 1993), and the identity of the protein was revealed based on the domain structure (Yamamoto et al., 2008). Since no mutant having defect in the DII3 gene is available in Chlamydomonas, the exact function of p44 is unclear. However, it has been proposed that p44 plays a role in docking IDA d in the axoneme (Yamamoto et al., 2008). Recently, re-analysis of the Chlamydomonas p44 cDNA (Yamamoto et al., 2008), based on the newest Chlamydomonas genome database (Phytozome v5.5), filled a small gap in the sequence near the C-terminus region (Supporting Figure S1). The p44 cDNA/protein sequence was updated accordingly in DDBJ (Accession No. AB353122.2). A potential mammalian orthologue of p44, TTC29/NYD-SP14 (Table 3), is expressed primarily in the spermatocytes in testis in rats, and the expression of TTC29/NYD-SP14 is reported to be regulated by follicle-stimulating hormone (Ohta et al., 2012). TTC29/NYD-SP14 is also reported to possibly function in the IFT machinery (Brooks, 2014). In addition, recent studies of human/mouse TTC29/NYD-SP14 show that defects in this protein cause the male infertility with the asthenoteratospermia/asthenozoospermia, suggesting this protein is important for ciliary motility and/or ciliogenesis (Liu et al., 2019; Lorès et al., 2019)(Table 3).
d. p38 (DII2/FAP146)
p38, also called DII2 or FAP146, is an accessary subunit of IDA d, and p38 does not display obvious motifs or domains (Yamamoto et al., 2006)(Figure 3; Table 1, 2). Together with p44, p38 was first identified as a 38-kDa protein that co-purified with IDA d (Kagami & Kamiya, 1992; Kato et al., 1993). Like p44, the axonemal amount of p38 in mutants lacking IDA d (ida4 and ida5) was greatly reduced compared to wild-type (Yamamoto et al., 2006, 2008). The exact function of p38 is unclear, but a complex of p44 and p38 was proposed to play a role in docking IDA d on the axoneme (Yamamoto et al., 2008). p38 was shown to be localized in cilia and also near the basal-body region in Chlamydomonas (Lechtreck et al., 2009; Yamamoto et al., 2006). Knockdown of a potential Trypanosoma orthologue of p38, TAX-1 (Tb09.211.1790), has been shown to cause both motility and growth defects in bloodstream (Broadhead et al., 2006). A potential mammalian orthologue of p38, ZMYND12 (Table 3), has also been identified (Yamamoto et al., 2006).
3. Remaining questions and challenges
In this review we focused on the subunit composition and the potential function(s) of each subunit of ciliary IDAs in Chlamydomonas. Although many aspects of IDAs, including potential function(s) of each subunit in ciliary assembly/regulation, have been revealed by studies using Chlamydomonas, various key questions remain.
3.1. What is the regulatory mechanism(s) of single-headed IDA species?
While the regulatory mechanism of IDA f/I1 has been extensively studied in Chlamydomonas and functional models have been proposed (Wirschell et al., 2007), few reports have described regulatory mechanism(s) of the single-headed IDAs. Recently, it was reported that DRC3 connected N-DRC to IDA g for control of ciliary motility (Awata et al., 2015). Also, the CSC complex is located near the base of IDAs d and g, and thought to stabilize and regulate these IDAs (Dymek et al., 2011; Heuser, Dymek, et al., 2012). Similarly, FAP57/IDA8/BOP2 (Supporting Table S1) is important for axonemal stability of IDAs d and g on specific doublet microtubules (Lin et al., 2019). In addition, FAP206, a novel ciliary protein, has been identified as a possible docking adapter for radial spoke 2, also interacting with IDA c. FAP206/CSC may regulate and stabilize IDA c, potentially transmitting a regulatory signal from radial spoke to IDA c to regulate ciliary motility (Vasudevan et al., 2015). However, how the various single-headed IDAs coordinate their activities with each other and with ODAs remains a challenging and open question.
3.2. What is the high-resolution/atomic structure of the individual IDA motors?
Presumably the axonemal dyneins will display similar motor structures to cytoplasmic dyneins (Carter et al., 2011; Kon et al., 2012), but currently we do not know if this is really the case. Although axonemal dynein structures have been resolved by cryo-electron tomography at the resolution of ~ 20Å [e.g. (Bui et al., 2008, 2009; Bui et al., 2012; Nicastro et al., 2006; Oda et al., 2014)], however, to date only detailed high-resolution structures of ODAs have been reported (Mali et al., 2021; Walton et al., 2021). Such structural data of IDAs is essential for further definition of the regulation and mechanism of IDAs.
3.3. What determines the localization of IDAs on axonemal doublet microtubules?
Although recent studies have revealed part of the molecular basis for the axonemal 96-nm repeat and IDA docking in Chlamydomonas (Awata et al., 2015; Dymek et al., 2011; Heuser et al., 2009; Lin et al., 2019; Ma et al., 2019; Oda et al., 2014), how each IDA is localized and docked in the 96-nm repeat is not fully understood. One interesting observation is that among the major single-headed IDA species, those with p28 (IDAs a, c and d) and with centrin (IDAs b, e and g) as a subunit are always located side by side in the axonemes (IDAs a and b, IDAs c and e, and IDAs d and g)(Figure 1a; Table 1). Thus, the p28 and centrin subunits may play a role in docking these single-headed IDAs in the axoneme. Additionally, it has been proposed that the p28 and centrin bearing single-headed IDAs potentially form pseudo-dimers that function as dimers in cilia (Bui et al., 2012). This “pseudo-dimer” hypothesis is partially supported by the in vitro observation that the motility of the isolated IDA c is facilitated by mixing with isolated IDA e, which are located adjacent to each other in the axoneme (Shimizu et al., 2014)(Figure 1a). Also, IDA species causing the clockwise microtubule translocation in vitro at a right angle to the direction of translocation include only IDAs d and g, which are located next to each other in the axoneme (Kikushima & Kamiya, 2008)(Figure 1a). However, tests of these ideas require additional work, and the binding site(s) on microtubules of these LCs of the single-headed IDAs, as well as of ICs/LCs of the two-headed IDA f/I1 must both be biochemically and structurally analyzed. Predictably, high-resolution/atomic structure(s) of IDA docking sites on doublet microtubules observed by the single-particle cryo-electron microscopy [like (Gui et al., 2021; Ma et al., 2019)] will reveal IDA docking mechanisms.
3.4. Are there any functional/compositional differences of IDAs among eukaryotes?
Although function and composition of IDAs have been extensively studied in Chlamydomonas, as described above, conservation of IDAs between Chlamydomonas and other eukaryotes with motile cilia are not yet fully understood. Recent studies revealed some unexpected and unique features of IDAs in other eukaryotes compared to Chlamydomonas. For example, in Trypanosoma axonemes, the IC/LC complexes of IDA f/I1 seem to be much larger than those in Chlamydomonas, and these large IDA f/I1 IC/LC complexes apparently extend to the adjacent doublet microtubules like N-DRC complexes (Imhof et al., 2019). Also, in cilia from Drosophila chordotonal neurons, IDAs f/I1 and g may not assemble, while other IDAs can assemble properly (Zur Lage et al., 2019). In addition, a recent report indicated that sperm cilia from two species of Ephemeroptera (mayfly), Ecdyonurus venosus and Cloeon dipterum lack ODAs and bear a unique arrangement of IDAs (Mencarelli et al., 2014). These sperm cilia have an unusual “9+9+0” axonemal organization that contains accessory microtubules, but lacks a central pair complex (Mencarelli et al., 2014). Predictably, these cilia generate lower beat frequency compared to Chlamydomonas. In addition, the axonemes from these sperm display a 32-nm periodicity rather than a 96-nm repeat along the outer doublet microtubules. Moreover, these axonemes appear to contain one species of DHC suggesting Ephemeroptera has only one species of IDA. If this is true, then one type of IDA can drive progressive motility and rhythmic beating in these sperm axonemes (Mencarelli et al., 2014). It will be important to identify the IDA DHCs in Chlamydomonas and vertebrates, that is homologous to the insect dynein.
3.5. What are the evolutionary origins of IDAs?
One approach is to determine the IDA structure of the last eukaryotic common ancestor (LECA), which is thought to possess cilia (Mitchell, 2017; Satir et al., 2008). The Chlamydomonas mutant lacking most of IDAs is paralyzed (Huang et al., 1979; Yamamoto et al., 2017; Yamamoto et al., 2020), while mutants lacking all ODAs can still produce ciliary movement at reduced beat frequency (Kamiya, 1988; Mitchell & Rosenbaum, 1985). One interesting idea is that IDA and ODA have evolved together to amplify the efficiency and power for ciliary beating. However, there are exceptions: the motile cilia of some organisms/organs, including the eel sperm and Physcomitrella, only assemble IDAs, whereas those of some other organisms (e.g. in the diatom Thalassiosira) only assemble ODAs (Desai et al., 2015; King, 2012; Kollmar, 2016; Wickstead & Gull, 2007; Woolley, 1997). In recent analyses of DHCs across various eukaryotes, a step-wise evolution of ciliary dyneins in LECA was proposed in which: Ia) ODA was duplicated to generate IDA f/I1; or Ib) IDA f/I1 was duplicated to generate ODA; II) duplication of IDA f/I1, or one DHC of IDA f/I1, generated the adjacent single-headed IDA(s); and III) duplication of the single-headed IDA generated other single-headed IDA(s)(Kollmar, 2016). Investigators have also suggested that LECA expressed only three subgroups of single-headed IDAs in addition to ODA and IDA f/I1 (Kollmar, 2016; Rajagopalan & Wilkes, 2016; Wickstead & Gull, 2007). In addition, it has been proposed that the N-terminal regions of several Chlamydomonas IDA DHCs [DHC5 (IDA b), DHC8 (IDA e), and DHC4 (minor IDA)] were swapped or recombined to generate tail regions capable of interacting with centrin, whereas the paralogous DHCs in other ciliated organisms have the tail regions interacting with p28 (Kollmar, 2016).
4. Conclusions and perspective
In this review, we summarized our current knowledge of Chlamydomonas IDAs focusing on each subunit, discussed their molecular function(s), and raised some future questions. For more than 40 years, Chlamydomonas served as an outstanding organism for study of the molecular function, composition, regulation and structure of IDAs. Chlamydomonas will continue to play an important role in our understanding of IDAs as essential components of ciliary motility. The remaining questions of IDAs described in this review await new work by cell biologists, biophysicists and clinicians.
Supplementary Material
Supporting Figure S1. Updated cDNA/protein sequence of Chlamydomonas p44 with a gap near the C-terminus filled
(a) Updated cDNA/protein sequence of Chlamydomonas p44 from (Yamamoto et al., 2008). As described before (Yamamoto et al., 2008), 5 TPR motifs (yellow dotted lines) and a coiled-coil motif (a green dotted line) were identified by SMART (http://smart.embl-heidelberg.de/)(Letunic et al., 2021; Schultz et al., 1998) analysis. The nucleotide translation was performed using an on-line translation tool (http://reverse-complement.com/translate-protein/ROOT/). The start and stop codons are boxed in red. Chlamydomonas polyadenylation signal is boxed in purple. (b) Refined sequence alignment of the potential orthologues of p44 based on the previous study (Yamamoto et al., 2008). 5 TPR motifs and a coiled-coil motif in Chlamydomonas p44 were highlighted in the yellow and green lines, respectively. Protein sequences were aligned using ClustalW (https://www.genome.jp/tools-bin/clustalw) and subsequently formatted with BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). The accession Nos/IDs in NCBI (https://www.ncbi.nlm.nih.gov/)/ UniProtKB (https://www.uniprot.org/help/uniprotkb)/ KEGG (https://www.genome.jp/kegg/genome.html) used for this comparison were as follows: Chlamydomonas p44, AB353122.2 (NCBI); mouse TTC29, E9QLU4 (UniProtKB); human TTC29, Q8NA56 (UniProtKB); zebrafish TTC29, E9QCZ1 (UniProtKB); Tetrahymena p44, XP_001014763.3 (NCBI); Trypanosoma p44, Tb927.3.1990 (KEGG).
Supporting Table S1. IDA-related mutants of Chlamydomonas reinhardtii
Acknowledgements
We apologize to our colleagues whose studies were not included in this review due to the space limits. We thank Dr. Toshiki Yagi (Prefectural University of Hiroshima) for valuable discussion and information on IDAs. RY was/is a recipient of a grant from Uehara Memorial Foundation, a grant from Itoh-Chubei Foundation, the JSPS Grant-in-Aid for Young Scientists (B)(JP17K15117) and for Scientific Research (C)(JP20K06622). JH and WSS are supported by NIH grant GM051173. TK is supported by the JSPS Grant-in-Aid for Scientific Research (B)(JP26291034 and JP17H03665).
Footnotes
Conflict of Interest
The authors declare there is no conflict of interest in this work.
References
- Afzelius BA (1976). A human syndrome caused by immotile cilia. Science, 193(4250), 317–319. [DOI] [PubMed] [Google Scholar]
- Alford LM, Wirschell M, Yamamoto R, & Sale WS (2012). 11 - Control of Axonemal Inner Dynein Arms A2 - King, Stephen M. In Dyneins (pp. 312–335). Academic Press. [Google Scholar]
- Aravind Kumar M, Singh V, Naushad SM, Shanker U, & Lakshmi Narasu M (2018). Microarray-based SNP genotyping to identify genetic risk factors of triple-negative breast cancer (TNBC) in South Indian population. Mol Cell Biochem, 442(1–2), 1–10. [DOI] [PubMed] [Google Scholar]
- Awata J, Song K, Lin J, King SM, Sanderson MJ, Nicastro D, & Witman GB (2015). DRC3 connects the N-DRC to dynein g to regulate flagellar waveform. Mol Biol Cell, 26(15), 2788–2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbar E (2008). Dynein light chain LC8 is a dimerization hub essential in diverse protein networks. Biochemistry, 47(2), 503–508. [DOI] [PubMed] [Google Scholar]
- Behnke O, Forer A, & Emmersen J (1971). Actin in sperm tails and meiotic spindles. Nature, 234(5329), 408–410. [DOI] [PubMed] [Google Scholar]
- Ben Khelifa M, Coutton C, Zouari R, Karaouzene T, Rendu J, Bidart M, Yassine S, Pierre V, Delaroche J, Hennebicq S, Grunwald D, Escalier D, Pernet-Gallay K, Jouk PS, Thierry-Mieg N, Toure A, Arnoult C, & Ray PF (2014). Mutations in DNAH1, which encodes an inner arm heavy chain dynein, lead to male infertility from multiple morphological abnormalities of the sperm flagella. Am J Hum Genet, 94(1), 95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower R, Tritschler D, Vanderwaal K, Perrone CA, Mueller J, Fox L, Sale WS, & Porter ME (2013). The N-DRC forms a conserved biochemical complex that maintains outer doublet alignment and limits microtubule sliding in motile axonemes. Mol Biol Cell, 24(8), 1134–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower R, VanderWaal K, O’Toole E, Fox L, Perrone C, Mueller J, Wirschell M, Kamiya R, Sale WS, & Porter ME (2009). IC138 defines a subdomain at the base of the I1 dynein that regulates microtubule sliding and flagellar motility. Mol Biol Cell, 20(13), 3055–3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadhead R, Dawe HR, Farr H, Griffiths S, Hart SR, Portman N, Shaw MK, Ginger ML, Gaskell SJ, McKean PG, & Gull K (2006). Flagellar motility is required for the viability of the bloodstream trypanosome. Nature, 440(7081), 224–227. [DOI] [PubMed] [Google Scholar]
- Brokaw CJ (1991). Microtubule sliding in swimming sperm flagella: direct and indirect measurements on sea urchin and tunicate spermatozoa. J Cell Biol, 114(6), 1201–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brokaw CJ, & Kamiya R (1987). Bending patterns of Chlamydomonas flagella: IV. Mutants with defects in inner and outer dynein arms indicate differences in dynein arm function. Cell Motil Cytoskeleton, 8(1), 68–75. [DOI] [PubMed] [Google Scholar]
- Brooks ER (2014). Control of intraflagellar transport: studies of the planar cell polarity effector Fuz, the small GTPase Rsg1, and the novel protein TTC29. The University of Texas at Austin, Doctoral Thesis, 1–158. [Google Scholar]
- Brown JM, & Witman GB (2014). Cilia and Diseases. Bioscience, 64(12), 1126–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui KH, Sakakibara H, Movassagh T, Oiwa K, & Ishikawa T (2008). Molecular architecture of inner dynein arms in situ in Chlamydomonas reinhardtii flagella. J Cell Biol, 183(5), 923–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui KH, Sakakibara H, Movassagh T, Oiwa K, & Ishikawa T (2009). Asymmetry of inner dynein arms and inter-doublet links in Chlamydomonas flagella. J Cell Biol, 186(3), 437–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui KH, Yagi T, Yamamoto R, Kamiya R, & Ishikawa T (2012). Polarity and asymmetry in the arrangement of dynein and related structures in the Chlamydomonas axoneme. J Cell Biol, 198(5), 913–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante-Marin XM, Horani A, Stoyanova M, Charng WL, Bottier M, Sears PR, Yin WN, Daniels LA, Bowen H, Conrad DF, Knowles MR, Ostrowski LE, Zariwala MA, & Dutcher SK (2020). Mutation of CFAP57, a protein required for the asymmetric targeting of a subset of inner dynein arms in Chlamydomonas, causes primary ciliary dyskinesia. PLoS Genet, 16(8), e1008691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AP, Cho C, Jin L, & Vale RD (2011). Crystal structure of the dynein motor domain. Science, 331(6021), 1159–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai PB, Freshour JR, & Mitchell DR (2015). Chlamydomonas axonemal dynein assembly locus ODA8 encodes a conserved flagellar protein needed for cytoplasmic maturation of outer dynein arm complexes. Cytoskeleton (Hoboken), 72(1), 16–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiBella LM, Benashski SE, Tedford HW, Harrison A, Patel-King RS, & King SM (2001). The Tctex1/Tctex2 class of dynein light chains. Dimerization, differential expression, and interaction with the LC8 protein family. J Biol Chem, 276(17), 14366–14373. [DOI] [PubMed] [Google Scholar]
- DiBella LM, Sakato M, Patel-King RS, Pazour GJ, & King SM (2004). The LC7 light chains of Chlamydomonas flagellar dyneins interact with components required for both motor assembly and regulation. Mol Biol Cell, 15(10), 4633–4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiBella LM, Smith EF, Patel-King RS, Wakabayashi K, & King SM (2004). A novel Tctex2-related light chain is required for stability of inner dynein arm I1 and motor function in the Chlamydomonas flagellum. J Biol Chem, 279(20), 21666–21676. [DOI] [PubMed] [Google Scholar]
- Dutcher SK (2014). The awesome power of dikaryons for studying flagella and basal bodies in Chlamydomonas reinhardtii. Cytoskeleton (Hoboken), 71(2), 79–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher SK, & O’Toole ET (2016). The basal bodies of Chlamydomonas reinhardtii. Cilia, 5, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymek EE, Heuser T, Nicastro D, & Smith EF (2011). The CSC is required for complete radial spoke assembly and wild-type ciliary motility. Mol Biol Cell, 22(14), 2520–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymek EE, & Smith EF (2007). A conserved CaM- and radial spoke associated complex mediates regulation of flagellar dynein activity. J Cell Biol, 179(3), 515–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elam CA, Wirschell M, Yamamoto R, Fox LA, York K, Kamiya R, Dutcher SK, & Sale WS (2011). An axonemal PP2A B-subunit is required for PP2A localization and flagellar motility. Cytoskeleton (Hoboken), 68(7), 363–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel BD, Ishikawa H, Wemmer KA, Geimer S, Wakabayashi K, Hirono M, Craige B, Pazour GJ, Witman GB, Kamiya R, & Marshall WF (2012). The role of retrograde intraflagellar transport in flagellar assembly, maintenance, and function. J Cell Biol, 199(1), 151–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson N, Tung JY, Kiefer AK, Hinds DA, Francke U, Mountain JL, & Do CB (2012). Novel associations for hypothyroidism include known autoimmune risk loci. PLoS One, 7(4), e34442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliegauf M, Benzing T, & Omran H (2007). When cilia go bad: cilia defects and ciliopathies. Nat Rev Mol Cell Biol, 8(11), 880–893. [DOI] [PubMed] [Google Scholar]
- Fowkes ME, & Mitchell DR (1998). The role of preassembled cytoplasmic complexes in assembly of flagellar dynein subunits. Mol Biol Cell, 9(9), 2337–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu G, Wang Q, Phan N, Urbanska P, Joachimiak E, Lin J, Wloga D, & Nicastro D (2018). The I1 dynein-associated tether and tether head complex is a conserved regulator of ciliary motility. Mol Biol Cell, 29(9), 1048–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbiati F, Pettinicchio A, Dragani TA, & Manenti G (2006). Allelic effects of mouse Pas1 candidate genes in human lung cancer cell lines. Cancer Lett, 244(2), 176–181. [DOI] [PubMed] [Google Scholar]
- Gershoni M, Hauser R, Yogev L, Lehavi O, Azem F, Yavetz H, Pietrokovski S, & Kleiman SE (2017). A familial study of azoospermic men identifies three novel causative mutations in three new human azoospermia genes. Genetics In Medicine, 19, 998. [DOI] [PubMed] [Google Scholar]
- Gholkar AA, Senese S, Lo YC, Capri J, Deardorff WJ, Dharmarajan H, Contreras E, Hodara E, Whitelegge JP, Jackson PK, & Torres JZ (2015). Tctex1d2 associates with short-rib polydactyly syndrome proteins and is required for ciliogenesis. Cell Cycle, 14(7), 1116–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons IR, & Rowe AJ (1965). Dynein: A Protein with Adenosine Triphosphatase Activity from Cilia. Science, 149(3682), 424–426. [DOI] [PubMed] [Google Scholar]
- Gokhale A, Wirschell M, & Sale WS (2009). Regulation of dynein-driven microtubule sliding by the axonemal protein kinase CK1 in Chlamydomonas flagella. J Cell Biol, 186(6), 817–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomelsky M, & Klug G (2002). BLUF: a novel FAD-binding domain involved in sensory transduction in microorganisms. Trends Biochem Sci, 27(10), 497–500. [DOI] [PubMed] [Google Scholar]
- Goodenough UW, Gebhart B, Mermall V, Mitchell DR, & Heuser JE (1987). High-pressure liquid chromatography fractionation of Chlamydomonas dynein extracts and characterization of inner-arm dynein subunits. J Mol Biol, 194(3), 481–494. [DOI] [PubMed] [Google Scholar]
- Gui M, Ma M, Sze-Tu E, Wang X, Koh F, Zhong ED, Berger B, Davis JH, Dutcher SK, Zhang R, & Brown A (2021). Structures of radial spokes and associated complexes important for ciliary motility. Nat Struct Mol Biol, 28(1), 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Diener DR, Sivadas P, Rosenbaum JL, & Yang P (2012). The versatile molecular complex component LC8 promotes several distinct steps of flagellar assembly. J Cell Biol, 198(1), 115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison A, Olds-Clarke P, & King SM (1998). Identification of the t complex-encoded cytoplasmic dynein light chain tctex1 in inner arm I1 supports the involvement of flagellar dyneins in meiotic drive. J Cell Biol, 140(5), 1137–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Yagi T, Yoshimura K, & Kamiya R (1998). Real-time observation of Ca2+-induced basal body reorientation in Chlamydomonas. Cell Motil Cytoskeleton, 41(1), 49–56. [DOI] [PubMed] [Google Scholar]
- Hendrickson TW, Goss JL, Seaton CA, & Rohrs HW (2013). The IC138 and IC140 intermediate chains of the I1 axonemal dynein complex bind directly to tubulin. Biochim Biophys Acta, 1833(12), 3265–3271. [DOI] [PubMed] [Google Scholar]
- Hendrickson TW, Perrone CA, Griffin P, Wuichet K, Mueller J, Yang P, Porter ME, & Sale WS (2004). IC138 is a WD-repeat dynein intermediate chain required for light chain assembly and regulation of flagellar bending. Mol Biol Cell, 15(12), 5431–5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser T, Barber CF, Lin J, Krell J, Rebesco M, Porter ME, & Nicastro D (2012). Cryoelectron tomography reveals doublet-specific structures and unique interactions in the I1 dynein. Proc Natl Acad Sci U S A, 109(30), E2067–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser T, Dymek EE, Lin J, Smith EF, & Nicastro D (2012). The CSC connects three major axonemal complexes involved in dynein regulation. Mol Biol Cell, 23(16), 3143–3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser T, Raytchev M, Krell J, Porter ME, & Nicastro D (2009). The dynein regulatory complex is the nexin link and a major regulatory node in cilia and flagella. J Cell Biol, 187(6), 921–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirono M, Uryu S, Ohara A, Kato-Minoura T, & Kamiya R (2003). Expression of conventional and unconventional actins in Chlamydomonas reinhardtii upon deflagellation and sexual adhesion. Eukaryot Cell, 2(3), 486–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmeister W, Pettersson M, Kurtoglu D, Armenio M, Eisfeldt J, Papadogiannakis N, Gustavsson P, & Lindstrand A (2018). Targeted copy number screening highlights an intragenic deletion of WDR63 as the likely cause of human occipital encephalocele and abnormal CNS development in zebrafish. Hum Mutat, 39(4), 495–505. [DOI] [PubMed] [Google Scholar]
- Hom EF, Witman GB, Harris EH, Dutcher SK, Kamiya R, Mitchell DR, Pazour GJ, Porter ME, Sale WS, Wirschell M, Yagi T, & King SM (2011). A unified taxonomy for ciliary dyneins. Cytoskeleton (Hoboken), 68(10), 555–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoops HJ, & Witman GB (1983). Outer doublet heterogeneity reveals structural polarity related to beat direction in Chlamydomonas flagella. J Cell Biol, 97(3), 902–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horani A, Ferkol TW, Dutcher SK, & Brody SL (2016). Genetics and biology of primary ciliary dyskinesia. Paediatr Respir Rev, 18, 18–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Piperno G, & Luck DJ (1979). Paralyzed flagella mutants of Chlamydomonas reinhardtii. Defective for axonemal doublet microtubule arms. J Biol Chem, 254(8), 3091–3099. [PubMed] [Google Scholar]
- Ikeda K, Yamamoto R, Wirschell M, Yagi T, Bower R, Porter ME, Sale WS, & Kamiya R (2009). A novel ankyrin-repeat protein interacts with the regulatory proteins of inner arm dynein f (I1) of Chlamydomonas reinhardtii. Cell Motil Cytoskeleton, 66(8), 448–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imhof S, Zhang J, Wang H, Bui KH, Nguyen H, Atanasov I, Hui WH, Yang SK, Zhou ZH, & Hill KL (2019). Cryo electron tomography with volta phase plate reveals novel structural foundations of the 96-nm axonemal repeat in the pathogen Trypanosoma brucei. Elife, 8, e52058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imtiaz F, Allam R, Ramzan K, & Al-Sayed M (2015). Variation in DNAH1 may contribute to primary ciliary dyskinesia. BMC Med Genet, 16, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack B, Mueller DM, Fee AC, Tetlow AL, & Avasthi P (2019). Partially Redundant Actin Genes in Chlamydomonas Control Transition Zone Organization and Flagellum-Directed Traffic. Cell Rep, 27(8), 2459–2467. e2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Yu L, Huang X, Chen X, Li D, Zhang Y, Tang L, & Zhao S (2001). Identification of two novel human dynein light chain genes, DNLC2A and DNLC2B, and their expression changes in hepatocellular carcinoma tissues from 68 Chinese patients. Gene, 281(1–2), 103–113. [DOI] [PubMed] [Google Scholar]
- Kagami O, & Kamiya R (1992). Translocation and rotation of microtubules caused by multiple species of Chlamydomonas inner-arm dynein. J Cell Sci, 103(3), 653–664. [Google Scholar]
- Kamiya R (1988). Mutations at twelve independent loci result in absence of outer dynein arms in Chylamydomonas reinhardtii. J Cell Biol, 107(6), 2253–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya R (2002). Functional diversity of axonemal dyneins as studied in Chlamydomonas mutants. Int Rev Cytol, 219, 115–155. [DOI] [PubMed] [Google Scholar]
- Kamiya R, Kurimoto E, & Muto E (1991). Two types of Chlamydomonas flagellar mutants missing different components of inner-arm dynein. J Cell Biol, 112(3), 441–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya R, & Yagi T (2014). Functional diversity of axonemal dyneins as assessed by in vitro and in vivo motility assays of Chlamydomonas mutants. Zoolog Sci, 31(10), 633–644. [DOI] [PubMed] [Google Scholar]
- Kato T, Kagami O, Yagi T, & Kamiya R (1993). Isolation of two species of Chlamydomonas reinhardtii flagellar mutants, ida5 and ida6, that lack a newly identified heavy chain of the inner dynein arm. Cell Struct Funct, 18(6), 371–377. [DOI] [PubMed] [Google Scholar]
- Kato-Minoura T (2011). Extremely low polymerizability of a highly-divergent Chlamydomonas actin (NAP). Biochem Biophys Res Commun, 412(4), 723–727. [DOI] [PubMed] [Google Scholar]
- Kato-Minoura T, Hirono M, & Kamiya R (1997). Chlamydomonas inner-arm dynein mutant, ida5, has a mutation in an actin-encoding gene. J Cell Biol, 137(3), 649–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato-Minoura T, Okumura M, Hirono M, & Kamiya R (2003). A novel family of unconventional actins in volvocalean algae. J Mol Evol, 57(5), 555–561. [DOI] [PubMed] [Google Scholar]
- Kato-Minoura T, Uryu S, Hirono M, & Kamiya R (1998). Highly divergent actin expressed in a Chlamydomonas mutant lacking the conventional actin gene. Biochem Biophys Res Commun, 251(1), 71–76. [DOI] [PubMed] [Google Scholar]
- Kiesel P, Alvarez Viar G, Tsoy N, Maraspini R, Gorilak P, Varga V, Honigmann A, & Pigino G (2020). The molecular structure of mammalian primary cilia revealed by cryo-electron tomography. Nat Struct Mol Biol, 12(12), 1115–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikushima K, & Kamiya R (2008). Clockwise translocation of microtubules by flagellar inner-arm dyneins in vitro. Biophys J, 94(10), 4014–4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SJ, & Dutcher SK (1997). Phosphoregulation of an inner dynein arm complex in Chlamydomonas reinhardtii is altered in phototactic mutant strains. J Cell Biol, 136(1), 177–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SJ, Inwood WB, O’Toole ET, Power J, & Dutcher SK (1994). The bop2–1 mutation reveals radial asymmetry in the inner dynein arm region of Chlamydomonas reinhardtii. J Cell Biol, 126(5), 1255–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SM (2012). Integrated control of axonemal dynein AAA(+) motors. J Struct Biol, 179(2), 222–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SM (2016). Axonemal Dynein Arms. Cold Spring Harb Perspect Biol, 8(11), a028100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SM, Dillman JF 3rd, Benashski SE, Lye RJ, Patel-King RS, & Pfister KK (1996). The mouse t-complex-encoded protein Tctex-1 is a light chain of brain cytoplasmic dynein. J Biol Chem, 271(50), 32281–32287. [DOI] [PubMed] [Google Scholar]
- King SM, & Patel-King RS (1995). The M(r) = 8,000 and 11,000 outer arm dynein light chains from Chlamydomonas flagella have cytoplasmic homologues. J Biol Chem, 270(19), 11445–11452. [DOI] [PubMed] [Google Scholar]
- Knowles MR, Zariwala M, & Leigh M (2016). Primary Ciliary Dyskinesia. Clin Chest Med, 37(3), 449–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollmar M (2016). Fine-Tuning Motile Cilia and Flagella: Evolution of the Dynein Motor Proteins from Plants to Humans at High Resolution. Mol Biol Evol, 33(12), 3249–3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon T, Oyama T, Shimo-Kon R, Imamula K, Shima T, Sutoh K, & Kurisu G (2012). The 2.8 A crystal structure of the dynein motor domain. Nature, 484(7394), 345–350. [DOI] [PubMed] [Google Scholar]
- Kotani N, Sakakibara H, Burgess SA, Kojima H, & Oiwa K (2007). Mechanical properties of inner-arm dynein-f (dynein I1) studied with in vitro motility assays. Biophys J, 93(3), 886–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski KG, Johnson KA, Forscher P, & Rosenbaum JL (1993). A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc Natl Acad Sci U S A, 90(12), 5519–5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutomi O, Yamamoto R, Hirose K, Mizuno K, Nakagiri Y, Imai H, Noga A, Obbineni JM, Zimmermann N, Nakajima M, Shibata D, Shibata M, Shiba K, Kita M, Kigoshi H, Tanaka Y, Yamasaki Y, Asahina Y, Song C, Nomura M, Nomura M, Nakajima A, Nakachi M, Yamada L, Nakazawa S, Sawada H, Murata K, Mitsuoka K, Ishikawa T, Wakabayashi K. i., Kon T, & Inaba K (2021). A dynein-associated photoreceptor protein prevents ciliary acclimation to blue light. Science Advances, 7(9), eabf3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YJ, Lee SJ, Koh JS, Kim SH, Lee HW, Kang MC, Bae JB, Kim YJ, & Park JH (2012). Genome-wide analysis of DNA methylation and the gene expression change in lung cancer. J Thorac Oncol, 7(1), 20–33. [DOI] [PubMed] [Google Scholar]
- Lader E, Ha HS, O’Neill M, Artzt K, & Bennett D (1989). tctex-1: a candidate gene family for a mouse t complex sterility locus. Cell, 58(5), 969–979. [DOI] [PubMed] [Google Scholar]
- Lechtreck KF, Brown JM, Sampaio JL, Craft JM, Shevchenko A, Evans JE, & Witman GB (2013). Cycling of the signaling protein phospholipase D through cilia requires the BBSome only for the export phase. J Cell Biol, 201(2), 249–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck KF, Luro S, Awata J, & Witman GB (2009). HA-tagging of putative flagellar proteins in Chlamydomonas reinhardtii identifies a novel protein of intraflagellar transport complex B. Cell Motil Cytoskeleton, 66(8), 469–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeDizet M, & Piperno G (1995a). ida4–1, ida4–2, and ida4–3 are intron splicing mutations affecting the locus encoding p28, a light chain of Chlamydomonas axonemal inner dynein arms. Mol Biol Cell, 6(6), 713–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeDizet M, & Piperno G (1995b). The light chain p28 associates with a subset of inner dynein arm heavy chains in Chlamydomonas axonemes. Mol Biol Cell, 6(6), 697–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Khedkar S, & Bork P (2021). SMART: recent updates, new developments and status in 2020. Nucleic Acids Res, 49(D1), D458–d460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Saito M, Chuang JZ, Tseng YY, Dedesma C, Tomizawa K, Kaitsuka T, & Sung CH (2011). Ciliary transition zone activation of phosphorylated Tctex-1 controls ciliary resorption, S-phase entry and fate of neural progenitors. Nat Cell Biol, 13(4), 402–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JB, Gerdes JM, Haycraft CJ, Fan Y, Teslovich TM, May-Simera H, Li H, Blacque OE, Li L, Leitch CC, Lewis RA, Green JS, Parfrey PS, Leroux MR, Davidson WS, Beales PL, Guay-Woodford LM, Yoder BK, Stormo GD, Katsanis N, & Dutcher SK (2004). Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell, 117(4), 541–552. [DOI] [PubMed] [Google Scholar]
- Li X, Zhang R, Patena W, Gang SS, Blum SR, Ivanova N, Yue R, Robertson JM, Lefebvre PA, Fitz-Gibbon ST, Grossman AR, & Jonikas MC (2016). An indexed, mapped mutant library enables reverse genetics studies of biological processes in Chlamydomonas reinhardtii. Plant Cell, 28(2), 367–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Sha Y, Wang X, Ding L, Liu W, Ji Z, Mei L, Huang X, Lin S, Kong S, Lu J, Qin W, Zhang X, Zhuang J, Tang Y, & Lu Z (2019). DNAH2 is a novel candidate gene associated with multiple morphological abnormalities of the sperm flagella. Clin Genet, 95(5), 590–600. [DOI] [PubMed] [Google Scholar]
- Li Y, Yagi H, Onuoha EO, Damerla RR, Francis R, Furutani Y, Tariq M, King SM, Hendricks G, Cui C, Saydmohammed M, Lee DM, Zahid M, Sami I, Leatherbury L, Pazour GJ, Ware SM, Nakanishi T, Goldmuntz E, Tsang M, & Lo CW (2016). DNAH6 and Its Interactions with PCD Genes in Heterotaxy and Primary Ciliary Dyskinesia. PLoS Genet, 12(2), e1005821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Le TV, Augspurger K, Tritschler D, Bower R, Fu G, Perrone C, O’Toole ET, Mills KV, Dymek E, Smith E, Nicastro D, & Porter ME (2019). FAP57/WDR65 targets assembly of a subset of inner arm dyneins and connects to regulatory hubs in cilia. Mol Biol Cell, 30(21), 2659–2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, He X, Liu W, Yang S, Wang L, Li W, Wu H, Tang S, Ni X, Wang J, Gao Y, Tian S, Zhang L, Cong J, Zhang Z, Tan Q, Zhang J, Li H, Zhong Y, Lv M, Li J, Jin L, Cao Y, & Zhang F (2019). Bi-allelic Mutations in TTC29 Cause Male Subfertility with Asthenoteratospermia in Humans and Mice. Am J Hum Genet, 105(6), 1168–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, & Lechtreck KF (2018). The Bardet-Biedl syndrome protein complex is an adapter expanding the cargo range of intraflagellar transport trains for ciliary export. Proc Natl Acad Sci U S A, 115(5), E934–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Wang Y, Vikis H, Maciag A, Wang D, Lu Y, Liu Y, & You M (2006). Candidate lung tumor susceptibility genes identified through whole-genome association analyses in inbred mice. Nat Genet, 38(8), 888–895. [DOI] [PubMed] [Google Scholar]
- Liu Z, Takazaki H, Nakazawa Y, Sakato M, Yagi T, Yasunaga T, King SM, & Kamiya R (2008). Partially functional outer-arm dynein in a novel Chlamydomonas mutant expressing a truncated gamma heavy chain. Eukaryot Cell, 7(7), 1136–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreng TD, & Smith EF (2017). The Central Apparatus of Cilia and Eukaryotic Flagella. Cold Spring Harb Perspect Biol, 9(2), a028118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorès P, Dacheux D, Kherraf ZE, Nsota Mbango JF, Coutton C, Stouvenel L, Ialy-Radio C, Amiri-Yekta A, Whitfield M, Schmitt A, Cazin C, Givelet M, Ferreux L, Fourati Ben Mustapha S, Halouani L, Marrakchi O, Daneshipour A, El Khouri E, Do Cruzeiro M, Favier M, Guillonneau F, Chaudhry M, Sakheli Z, Wolf JP, Patrat C, Gacon G, Savinov SN, Hosseini SH, Robinson DR, Zouari R, Ziyyat A, Arnoult C, Dulioust E, Bonhivers M, Ray PF, & Touré A (2019). Mutations in TTC29, Encoding an Evolutionarily Conserved Axonemal Protein, Result in Asthenozoospermia and Male Infertility. Am J Hum Genet, 105(6), 1148–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M, Stoyanova M, Rademacher G, Dutcher SK, Brown A, & Zhang R (2019). Structure of the Decorated Ciliary Doublet Microtubule. Cell, 179(4), 909–922. e912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali GR, Ali FA, Lau CK, Begum F, Boulanger J, Howe JD, Chen ZA, Rappsilber J, Skehel M, & Carter AP (2021). Shulin packages axonemal outer dynein arms for ciliary targeting. Science, 371(6532), 910–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN, O’Toole ET, McDonald KL, McIntosh JR, & Porter ME (1992). Arrangement of inner dynein arms in wild-type and mutant flagella of Chlamydomonas. J Cell Biol, 118(5), 1145–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mencarelli C, Mercati D, Dallai R, & Lupetti P (2014). Ultrastructure of the sperm axoneme and molecular analysis of axonemal dynein in Ephemeroptera (Insecta). Cytoskeleton (Hoboken), 71(5), 328–339. [DOI] [PubMed] [Google Scholar]
- Menche J, Sharma A, Cho MH, Mayer RJ, Rennard SI, Celli B, Miller BE, Locantore N, Tal-Singer R, Ghosh S, Larminie C, Bradley G, Riley JH, Agusti A, Silverman EK, & Barabasi AL (2014). A diVIsive Shuffling Approach (VIStA) for gene expression analysis to identify subtypes in Chronic Obstructive Pulmonary Disease. BMC Syst Biol, 8 Suppl 2, S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Marechal-Drouard L, Marshall WF, Qu LH, Nelson DR, Sanderfoot AA, Spalding MH, Kapitonov VV, Ren Q, Ferris P, Lindquist E, Shapiro H, Lucas SM, Grimwood J, Schmutz J, Cardol P, Cerutti H, Chanfreau G, Chen CL, Cognat V, Croft MT, Dent R, Dutcher S, Fernandez E, Fukuzawa H, Gonzalez-Ballester D, Gonzalez-Halphen D, Hallmann A, Hanikenne M, Hippler M, Inwood W, Jabbari K, Kalanon M, Kuras R, Lefebvre PA, Lemaire SD, Lobanov AV, Lohr M, Manuell A, Meier I, Mets L, Mittag M, Mittelmeier T, Moroney JV, Moseley J, Napoli C, Nedelcu AM, Niyogi K, Novoselov SV, Paulsen IT, Pazour G, Purton S, Ral JP, Riano-Pachon DM, Riekhof W, Rymarquis L, Schroda M, Stern D, Umen J, Willows R, Wilson N, Zimmer SL, Allmer J, Balk J, Bisova K, Chen CJ, Elias M, Gendler K, Hauser C, Lamb MR, Ledford H, Long JC, Minagawa J, Page MD, Pan J, Pootakham W, Roje S, Rose A, Stahlberg E, Terauchi AM, Yang P, Ball S, Bowler C, Dieckmann CL, Gladyshev VN, Green P, Jorgensen R, Mayfield S, Mueller-Roeber B, Rajamani S, Sayre RT, Brokstein P, Dubchak I, Goodstein D, Hornick L, Huang YW, Jhaveri J, Luo Y, Martinez D, Ngau WC, Otillar B, Poliakov A, Porter A, Szajkowski L, Werner G, Zhou K, Grigoriev IV, Rokhsar DS, & Grossman AR (2007). The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science, 318(5848), 245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DR (1994). Cell and molecular biology of flagellar dyneins. Int Rev Cytol, 155, 141–180. [DOI] [PubMed] [Google Scholar]
- Mitchell DR (2017). Evolution of Cilia. Cold Spring Harb Perspect Biol, 9(1), a028290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DR, & Rosenbaum JL (1985). A motile Chlamydomonas flagellar mutant that lacks outer dynein arms. J Cell Biol, 100(4), 1228–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocz G, & Gibbons IR (2001). Model for the motor component of dynein heavy chain based on homology to the AAA family of oligomeric ATPases. Structure, 9(2), 93–103. [DOI] [PubMed] [Google Scholar]
- Morris RL, Hoffman MP, Obar RA, McCafferty SS, Gibbons IR, Leone AD, Cool J, Allgood EL, Musante AM, Judkins KM, Rossetti BJ, Rawson AP, & Burgess DR (2006). Analysis of cytoskeletal and motility proteins in the sea urchin genome assembly. Dev Biol, 300(1), 219–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mughal A, & Weaire D (2017). Phyllotaxis, disk packing, and Fibonacci numbers. Phys Rev E, 95(2–1), 022401. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S (2015). TCTEX1D2, a potential link to skeletal ciliopathies. Cell Cycle, 14(3), 293–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myster SH, Knott JA, O’Toole E, & Porter ME (1997). The Chlamydomonas Dhc1 gene encodes a dynein heavy chain subunit required for assembly of the I1 inner arm complex. Mol Biol Cell, 8(4), 607–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury MV, Loktev AV, Zhang Q, Westlake CJ, Peranen J, Merdes A, Slusarski DC, Scheller RH, Bazan JF, Sheffield VC, & Jackson PK (2007). A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell, 129(6), 1201–1213. [DOI] [PubMed] [Google Scholar]
- Nicastro D, Schwartz C, Pierson J, Gaudette R, Porter ME, & McIntosh JR (2006). The molecular architecture of axonemes revealed by cryoelectron tomography. Science, 313(5789), 944–948. [DOI] [PubMed] [Google Scholar]
- Oda T, Yanagisawa H, Kamiya R, & Kikkawa M (2014). A molecular ruler determines the repeat length in eukaryotic cilia and flagella. Science, 346(6211), 857–860. [DOI] [PubMed] [Google Scholar]
- Ohta M, Ohyama K, Asano A, Yokota S, Khalid AM, & Yamano Y (2012). Regulation of rat tetratricopeptide repeat domain 29 gene expression by follicle-stimulating hormone. Biosci Biotechnol Biochem, 76(8), 1540–1543. [DOI] [PubMed] [Google Scholar]
- Okita N, Isogai N, Hirono M, Kamiya R, & Yoshimura K (2005). Phototactic activity in Chlamydomonas ‘non-phototactic’ mutants deficient in Ca2+-dependent control of flagellar dominance or in inner-arm dynein. J Cell Sci, 118(3), 529–537. [DOI] [PubMed] [Google Scholar]
- Onishi M, Pringle JR, & Cross FR (2016). Evidence That an Unconventional Actin Can Provide Essential F-Actin Function and That a Surveillance System Monitors F-Actin Integrity in Chlamydomonas. Genetics, 202(3), 977–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opazo T, Garcés A, Tapia D, Barraza F, Bravo A, Schwenke T, Cancino J, & Arriagada G (2017). Functional Evidence of the Involvement of the Dynein Light Chain DYNLRB2 in Murine Leukemia Virus Infection. J Virol, 91(10), e00129–00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski LE, Dutcher SK, & Lo CW (2011). Cilia and models for studying structure and function. Proc Am Thorac Soc, 8(5), 423–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, & Tame JRH (2017). Seeing the light with BLUF proteins. Biophys Rev, 9(2), 169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, Dickert BL, & Witman GB (1999). The DHC1b (DHC2) isoform of cytoplasmic dynein is required for flagellar assembly. J Cell Biol, 144(3), 473–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, Wilkerson CG, & Witman GB (1998). A dynein light chain is essential for the retrograde particle movement of intraflagellar transport (IFT). J Cell Biol, 141(4), 979–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, & Witman GB (2000). Forward and reverse genetic analysis of microtubule motors in Chlamydomonas. Methods, 22(4), 285–298. [DOI] [PubMed] [Google Scholar]
- Perrone CA, Myster SH, Bower R, O’Toole ET, & Porter ME (2000). Insights into the structural organization of the I1 inner arm dynein from a domain analysis of the 1beta dynein heavy chain. Mol Biol Cell, 11(7), 2297–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone CA, Yang P, O’Toole E, Sale WS, & Porter ME (1998). The Chlamydomonas IDA7 locus encodes a 140-kDa dynein intermediate chain required to assemble the I1 inner arm complex. Mol Biol Cell, 9(12), 3351–3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phua SC, Chiba S, Suzuki M, Su E, Roberson EC, Pusapati GV, Schurmans S, Setou M, Rohatgi R, Reiter JF, Ikegami K, & Inoue T (2017). Dynamic Remodeling of Membrane Composition Drives Cell Cycle through Primary Cilia Excision. Cell, 168(1–2), 264–279. e215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigino G, Bui KH, Maheshwari A, Lupetti P, Diener D, & Ishikawa T (2011). Cryoelectron tomography of radial spokes in cilia and flagella. J Cell Biol, 195(4), 673–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G (1988). Isolation of a sixth dynein subunit adenosine triphosphatase of Chlamydomonas axonemes. J Cell Biol, 106(1), 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G (1995). Regulation of dynein activity within Chlamydomonas flagella. Cell Motil Cytoskeleton, 32(2), 103–105. [DOI] [PubMed] [Google Scholar]
- Piperno G, & Luck DJ (1979). An actin-like protein is a component of axonemes from Chlamydomonas flagella. J Biol Chem, 254(7), 2187–2190. [PubMed] [Google Scholar]
- Piperno G, & Luck DJ (1981). Inner arm dyneins from flagella of Chlamydomonas reinhardtii. Cell, 27(2), 331–340. [DOI] [PubMed] [Google Scholar]
- Piperno G, Mead K, & Shestak W (1992). The inner dynein arms I2 interact with a “dynein regulatory complex” in Chlamydomonas flagella. J Cell Biol, 118(6), 1455–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, & Ramanis Z (1991). The proximal portion of Chlamydomonas flagella contains a distinct set of inner dynein arms. J Cell Biol, 112(4), 701–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, Ramanis Z, Smith EF, & Sale WS (1990). Three distinct inner dynein arms in Chlamydomonas flagella: molecular composition and location in the axoneme. J Cell Biol, 110(2), 379–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poghosyan E, Iacovache I, Faltova L, Leitner A, Yang P, Diener DR, Aebersold R, Zuber B, & Ishikawa T (2020). The structure and symmetry of the radial spoke protein complex in Chlamydomonas flagella. J Cell Sci, 133(16), jcs245233. [DOI] [PubMed] [Google Scholar]
- Porter ME, Knott JA, Myster SH, & Farlow SJ (1996). The dynein gene family in Chlamydomonas reinhardtii. Genetics, 144(2), 569–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter ME, Power J, & Dutcher SK (1992). Extragenic suppressors of paralyzed flagellar mutations in Chlamydomonas reinhardtii identify loci that alter the inner dynein arms. J Cell Biol, 118(5), 1163–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser SL, & Morrison CG (2015). Centrin2 regulates CP110 removal in primary cilium formation. J Cell Biol, 208(6), 693–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer EL, Eddy SR, Bateman A, & Finn RD (2012). The Pfam protein families database. Nucleic Acids Res, 40(Database issue), D290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan V, & Wilkes DE (2016). Evolution of the Dynein Heavy Chain Family in Ciliates. J Eukaryot Microbiol, 63(1), 138–141. [DOI] [PubMed] [Google Scholar]
- Rapali P, Szenes A, Radnai L, Bakos A, Pal G, & Nyitray L (2011). DYNLL/LC8: a light chain subunit of the dynein motor complex and beyond. FEBS J, 278(17), 2980–2996. [DOI] [PubMed] [Google Scholar]
- Rashid S, Breckle R, Hupe M, Geisler S, Doerwald N, & Neesen J (2006). The murine Dnali1 gene encodes a flagellar protein that interacts with the cytoplasmic dynein heavy chain 1. Mol Reprod Dev, 73(6), 784–794. [DOI] [PubMed] [Google Scholar]
- Reiter JF, & Leroux MR (2017). Genes and molecular pathways underpinning ciliopathies. Nat Rev Mol Cell Biol, 18(9), 533–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Binder NE, Geimer S, & Melkonian M (2002). In vivo localization of centrin in the green alga Chlamydomonas reinhardtii. Cell Motil Cytoskeleton, 52(1), 43–55. [DOI] [PubMed] [Google Scholar]
- Sakakibara H, Mitchell DR, & Kamiya R (1991). A Chlamydomonas outer arm dynein mutant missing the alpha heavy chain. J Cell Biol, 113(3), 615–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara H, Takada S, King SM, Witman GB, & Kamiya R (1993). A Chlamydomonas outer arm dynein mutant with a truncated beta heavy chain. J Cell Biol, 122(3), 653–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sale WS, & Satir P (1976). Splayed Tetrahymena cilia. A system for analyzing sliding and axonemal spoke arrangements. J Cell Biol, 71(2), 589–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sale WS, & Satir P (1977). Direction of active sliding of microtubules in Tetrahymena cilia. Proc Natl Acad Sci U S A, 74(5), 2045–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury JL, Baron AT, & Sanders MA (1988). The centrin-based cytoskeleton of Chlamydomonas reinhardtii: distribution in interphase and mitotic cells. J Cell Biol, 107(2), 635–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders MA, & Salisbury JL (1989). Centrin-mediated microtubule severing during flagellar excision in Chlamydomonas reinhardtii. J Cell Biol, 108(5), 1751–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satir P (1968). Studies on cilia. 3. Further studies on the cilium tip and a “sliding filament” model of ciliary motility. J Cell Biol, 39(1), 77–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satir P, & Christensen ST (2007). Overview of structure and function of mammalian cilia. Annu Rev Physiol, 69, 377–400. [DOI] [PubMed] [Google Scholar]
- Satir P, Heuser T, & Sale WS (2014). A Structural Basis for How Motile Cilia Beat. Bioscience, 64(12), 1073–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satir P, Mitchell DR, & Jekely G (2008). How did the cilium evolve? Curr Top Dev Biol, 85, 63–82. [DOI] [PubMed] [Google Scholar]
- Schmidts M, Hou Y, Cortes CR, Mans DA, Huber C, Boldt K, Patel M, van Reeuwijk J, Plaza JM, van Beersum SE, Yap ZM, Letteboer SJ, Taylor SP, Herridge W, Johnson CA, Scambler PJ, Ueffing M, Kayserili H, Krakow D, King SM, Beales PL, Al-Gazali L, Wicking C, Cormier-Daire V, Roepman R, Mitchison HM, & Witman GB (2015). TCTEX1D2 mutations underlie Jeune asphyxiating thoracic dystrophy with impaired retrograde intraflagellar transport. Nat Commun, 6, 7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, & Ponting CP (1998). SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci U S A, 95(11), 5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu Y, Sakakibara H, Kojima H, & Oiwa K (2014). Slow axonemal dynein e facilitates the motility of faster dynein c. Biophys J, 106(10), 2157–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingyoji C, Murakami A, & Takahashi K (1977). Local reactivation of Triton-extracted flagella by iontophoretic application of ATP. Nature, 265(5591), 269–270. [DOI] [PubMed] [Google Scholar]
- Soler Artigas M, Loth DW, Wain LV, Gharib SA, Obeidat M, Tang W, Zhai G, Zhao JH, Smith AV, Huffman JE, Albrecht E, Jackson CM, Evans DM, Cadby G, Fornage M, Manichaikul A, Lopez LM, Johnson T, Aldrich MC, Aspelund T, Barroso I, Campbell H, Cassano PA, Couper DJ, Eiriksdottir G, Franceschini N, Garcia M, Gieger C, Gislason GK, Grkovic I, Hammond CJ, Hancock DB, Harris TB, Ramasamy A, Heckbert SR, Heliovaara M, Homuth G, Hysi PG, James AL, Jankovic S, Joubert BR, Karrasch S, Klopp N, Koch B, Kritchevsky SB, Launer LJ, Liu Y, Loehr LR, Lohman K, Loos RJ, Lumley T, Al Balushi KA, Ang WQ, Barr RG, Beilby J, Blakey JD, Boban M, Boraska V, Brisman J, Britton JR, Brusselle GG, Cooper C, Curjuric I, Dahgam S, Deary IJ, Ebrahim S, Eijgelsheim M, Francks C, Gaysina D, Granell R, Gu X, Hankinson JL, Hardy R, Harris SE, Henderson J, Henry A, Hingorani AD, Hofman A, Holt PG, Hui J, Hunter ML, Imboden M, Jameson KA, Kerr SM, Kolcic I, Kronenberg F, Liu JZ, Marchini J, McKeever T, Morris AD, Olin AC, Porteous DJ, Postma DS, Rich SS, Ring SM, Rivadeneira F, Rochat T, Sayer AA, Sayers I, Sly PD, Smith GD, Sood A, Starr JM, Uitterlinden AG, Vonk JM, Wannamethee SG, Whincup PH, Wijmenga C, Williams OD, Wong A, Mangino M, Marciante KD, McArdle WL, Meibohm B, Morrison AC, North KE, Omenaas E, Palmer LJ, Pietilainen KH, Pin I, Pola Sbreve Ek O, Pouta A, Psaty BM, Hartikainen AL, Rantanen T, Ripatti S, Rotter JI, Rudan I, Rudnicka AR, Schulz H, Shin SY, Spector TD, Surakka I, Vitart V, Volzke H, Wareham NJ, Warrington NM, Wichmann HE, Wild SH, Wilk JB, Wjst M, Wright AF, Zgaga L, Zemunik T, Pennell CE, Nyberg F, Kuh D, Holloway JW, Boezen HM, Lawlor DA, Morris RW, Probst-Hensch N, Kaprio J, Wilson JF, Hayward C, Kahonen M, Heinrich J, Musk AW, Jarvis DL, Glaser S, Jarvelin MR, Ch Stricker BH, Elliott P, O’Connor GT, Strachan DP, London SJ, Hall IP, Gudnason V, & Tobin MD (2011). Genome-wide association and large-scale follow up identifies 16 new loci influencing lung function. Nat Genet, 43(11), 1082–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers KE, & Gibbons IR (1971). Adenosine triphosphate-induced sliding of tubules in trypsin-treated flagella of sea-urchin sperm. Proc Natl Acad Sci U S A, 68(12), 3092–3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinton J, & Ochu E (2016). Novel Fibonacci and non-Fibonacci structure in the sunflower: results of a citizen science experiment. R Soc Open Sci, 3(5), 160091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taillon BE, Adler SA, Suhan JP, & Jarvik JW (1992). Mutational analysis of centrin: an EF-hand protein associated with three distinct contractile fibers in the basal body apparatus of Chlamydomonas. J Cell Biol, 119(6), 1613–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessema M, Yingling CM, Picchi MA, Wu G, Liu Y, Weissfeld JL, Siegfried JM, Tesfaigzi Y, & Belinsky SA (2015). Epigenetic Repression of CCDC37 and MAP1B Links Chronic Obstructive Pulmonary Disease to Lung Cancer. J Thorac Oncol, 10(8), 1181–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian F, Zhao J, Fan X, & Kang Z (2017). Weighted gene co-expression network analysis in identification of metastasis-related genes of lung squamous cell carcinoma based on the Cancer Genome Atlas database. J Thorac Dis, 9(1), 42–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toba S, Fox LA, Sakakibara H, Porter ME, Oiwa K, & Sale WS (2011). Distinct roles of 1alpha and 1beta heavy chains of the inner arm dynein I1 of Chlamydomonas flagella. Mol Biol Cell, 22(3), 342–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanderWaal KE, Yamamoto R, Wakabayashi K, Fox L, Kamiya R, Dutcher SK, Bayly PV, Sale WS, & Porter ME (2011). bop5 Mutations reveal new roles for the IC138 phosphoprotein in the regulation of flagellar motility and asymmetric waveforms. Mol Biol Cell, 22(16), 2862–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan KK, Song K, Alford LM, Sale WS, Dymek EE, Smith EF, Hennessey T, Joachimiak E, Urbanska P, Wloga D, Dentler W, Nicastro D, & Gaertig J (2015). FAP206 is a microtubule-docking adapter for ciliary radial spoke 2 and dynein c. Mol Biol Cell, 26(4), 696–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincensini L, Blisnick T, & Bastin P (2011). 1001 model organisms to study cilia and flagella. Biol Cell, 103(3), 109–130. [DOI] [PubMed] [Google Scholar]
- Viswanadha R, Hunter EL, Yamamoto R, Wirschell M, Alford LM, Dutcher SK, & Sale WS (2014). The ciliary inner dynein arm, I1 dynein, is assembled in the cytoplasm and transported by IFT before axonemal docking. Cytoskeleton (Hoboken), 71(10), 573–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wain LV, Sayers I, Soler Artigas M, Portelli MA, Zeggini E, Obeidat M, Sin DD, Bosse Y, Nickle D, Brandsma CA, Malarstig A, Vangjeli C, Jelinsky SA, John S, Kilty I, McKeever T, Shrine NR, Cook JP, Patel S, Spector TD, Hollox EJ, Hall IP, & Tobin MD (2014). Whole exome re-sequencing implicates CCDC38 and cilia structure and function in resistance to smoking related airflow obstruction. PLoS Genet, 10(5), e1004314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi K. i. (2012). 10 - Regulation of Axonemal Outer-Arm Dyneins in Cilia A2 - King, Stephen M. In Dyneins (pp. 296–311). Academic Press. [Google Scholar]
- Walton T, Wu H, & Brown A (2021). Structure of a microtubule-bound axonemal dynein. Nat Commun, 12(1), 477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Jin H, Han F, Cui Y, Chen J, Yang C, Zhu P, Wang W, Jiao G, Hao C, & Gao Z (2017). Homozygous DNAH1 frameshift mutation causes multiple morphological anomalies of the sperm flagella in Chinese. Clin Genet, 91(2), 313–321. [DOI] [PubMed] [Google Scholar]
- Wanschers B, van de Vorstenbosch R, Wijers M, Wieringa B, King SM, & Fransen J (2008). Rab6 family proteins interact with the dynein light chain protein DYNLRB1. Cell Motil Cytoskeleton, 65(3), 183–196. [DOI] [PubMed] [Google Scholar]
- Wickstead B, & Gull K (2007). Dyneins across eukaryotes: a comparative genomic analysis. Traffic, 8(12), 1708–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, & Hochstrasser DF (1999). Protein identification and analysis tools in the ExPASy server. Methods Mol Biol, 112, 531–552. [DOI] [PubMed] [Google Scholar]
- Wirschell M, Hendrickson T, & Sale WS (2007). Keeping an eye on I1: I1 dynein as a model for flagellar dynein assembly and regulation. Cell Motil Cytoskeleton, 64(8), 569–579. [DOI] [PubMed] [Google Scholar]
- Wirschell M, Yang C, Yang P, Fox L, Yanagisawa HA, Kamiya R, Witman GB, Porter ME, & Sale WS (2009). IC97 is a novel intermediate chain of I1 dynein that interacts with tubulin and regulates interdoublet sliding. Mol Biol Cell, 20(13), 3044–3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolley DM (1997). Studies on the eel sperm flagellum. I. The structure of the inner dynein arm complex. J Cell Sci, 110(1), 85–94. [DOI] [PubMed] [Google Scholar]
- Wright RL, Adler SA, Spanier JG, & Jarvik JW (1989). Nucleus-basal body connector in Chlamydomonas: evidence for a role in basal body segregation and against essential roles in mitosis or in determining cell polarity. Cell Motil Cytoskeleton, 14(4), 516–526. [DOI] [PubMed] [Google Scholar]
- Wu H, Maciejewski MW, Benashski SE, Mullen GP, & King SM (2001). 1H, 15N and 13C resonance assignments for the Tctex1 dynein light chain from Chlamydomonas flagella. J Biomol NMR, 20(1), 89–90. [DOI] [PubMed] [Google Scholar]
- Wu H, Maciejewski MW, Takebe S, & King SM (2005). Solution structure of the Tctex1 dimer reveals a mechanism for dynein-cargo interactions. Structure, 13(2), 213–223. [DOI] [PubMed] [Google Scholar]
- Yagi T, & Kamiya R (2012). 9 - Genetic Approaches to Axonemal Dynein Function in Chlamydomonas and Other Organisms A2 - King, Stephen M. In Dyneins (pp. 272–295). Academic Press. [Google Scholar]
- Yagi T, Minoura I, Fujiwara A, Saito R, Yasunaga T, Hirono M, & Kamiya R (2005). An axonemal dynein particularly important for flagellar movement at high viscosity. Implications from a new Chlamydomonas mutant deficient in the dynein heavy chain gene DHC9. J Biol Chem, 280(50), 41412–41420. [DOI] [PubMed] [Google Scholar]
- Yagi T, Uematsu K, Liu Z, & Kamiya R (2009). Identification of dyneins that localize exclusively to the proximal portion of Chlamydomonas flagella. J Cell Sci, 122(9), 1306–1314. [DOI] [PubMed] [Google Scholar]
- Yamamoto R, Hirono M, & Kamiya R (2010). Discrete PIH proteins function in the cytoplasmic preassembly of different subsets of axonemal dyneins. J Cell Biol, 190(1), 65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto R, Obbineni JM, Alford LM, Ide T, Owa M, Hwang J, Kon T, Inaba K, James N, King SM, Ishikawa T, Sale WS, & Dutcher SK (2017). Chlamydomonas DYX1C1/PF23 is essential for axonemal assembly and proper morphology of inner dynein arms. PLoS Genet, 13(9), e1006996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto R, Song K, Yanagisawa HA, Fox L, Yagi T, Wirschell M, Hirono M, Kamiya R, Nicastro D, & Sale WS (2013). The MIA complex is a conserved and novel dynein regulator essential for normal ciliary motility. J Cell Biol, 201(2), 263–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto R, Yanagi S, Nagao M, Yamasaki Y, Tanaka Y, Sale WS, Yagi T, & Kon T (2020). Mutations in PIH proteins MOT48, TWI1 and PF13 define common and unique steps for preassembly of each, different ciliary dynein. PLoS Genet, 16(11), e1009126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto R, Yanagisawa HA, Yagi T, & Kamiya R (2006). A novel subunit of axonemal dynein conserved among lower and higher eukaryotes. FEBS Lett, 580(27), 6357–6360. [DOI] [PubMed] [Google Scholar]
- Yamamoto R, Yanagisawa HA, Yagi T, & Kamiya R (2008). Novel 44-kilodalton subunit of axonemal Dynein conserved from chlamydomonas to mammals. Eukaryot Cell, 7(1), 154–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa HA, & Kamiya R (2001). Association between actin and light chains in Chlamydomonas flagellar inner-arm dyneins. Biochem Biophys Res Commun, 288(2), 443–447. [DOI] [PubMed] [Google Scholar]
- Yang P, Diener DR, Rosenbaum JL, & Sale WS (2001). Localization of calmodulin and dynein light chain LC8 in flagellar radial spokes. J Cell Biol, 153(6), 1315–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, & Sale WS (1998). The Mr 140,000 intermediate chain of Chlamydomonas flagellar inner arm dynein is a WD-repeat protein implicated in dynein arm anchoring. Mol Biol Cell, 9(12), 3335–3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Yang C, Wirschell M, & Davis S (2009). Novel LC8 mutations have disparate effects on the assembly and stability of flagellar complexes. J Biol Chem, 284(45), 31412–31421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YH, Markus MA, Mangs AH, Raitskin O, Sperling R, & Morris BJ (2013). ZRANB2 localizes to supraspliceosomes and influences the alternative splicing of multiple genes in the transcriptome. Mol Biol Rep, 40(9), 5381–5395. [DOI] [PubMed] [Google Scholar]
- Ying G, Avasthi P, Irwin M, Gerstner CD, Frederick JM, Lucero MT, & Baehr W (2014). Centrin 2 is required for mouse olfactory ciliary trafficking and development of ependymal cilia planar polarity. J Neurosci, 34(18), 6377–6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SA, Miyata H, Satouh Y, Kato H, Nozawa K, Isotani A, Aitken RJ, Baker MA, & Ikawa M (2015). CRISPR/Cas9-Mediated Rapid Generation of Multiple Mouse Lines Identified Ccdc63 as Essential for Spermiogenesis. Int J Mol Sci, 16(10), 24732–24750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen Y, Zheng J, Wang J, Duan S, Zhang W, Yan X, & Zhu X (2019). Vertebrate Dynein-f depends on Wdr78 for axonemal localization and is essential for ciliary beat. J Mol Cell Biol, 11(5), 383–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zur Lage P, Newton FG, & Jarman AP (2019). Survey of the Ciliary Motility Machinery of Drosophila Sperm and Ciliated Mechanosensory Neurons Reveals Unexpected Cell-Type Specific Variations: A Model for Motile Ciliopathies. Front Genet, 10, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Figure S1. Updated cDNA/protein sequence of Chlamydomonas p44 with a gap near the C-terminus filled
(a) Updated cDNA/protein sequence of Chlamydomonas p44 from (Yamamoto et al., 2008). As described before (Yamamoto et al., 2008), 5 TPR motifs (yellow dotted lines) and a coiled-coil motif (a green dotted line) were identified by SMART (http://smart.embl-heidelberg.de/)(Letunic et al., 2021; Schultz et al., 1998) analysis. The nucleotide translation was performed using an on-line translation tool (http://reverse-complement.com/translate-protein/ROOT/). The start and stop codons are boxed in red. Chlamydomonas polyadenylation signal is boxed in purple. (b) Refined sequence alignment of the potential orthologues of p44 based on the previous study (Yamamoto et al., 2008). 5 TPR motifs and a coiled-coil motif in Chlamydomonas p44 were highlighted in the yellow and green lines, respectively. Protein sequences were aligned using ClustalW (https://www.genome.jp/tools-bin/clustalw) and subsequently formatted with BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). The accession Nos/IDs in NCBI (https://www.ncbi.nlm.nih.gov/)/ UniProtKB (https://www.uniprot.org/help/uniprotkb)/ KEGG (https://www.genome.jp/kegg/genome.html) used for this comparison were as follows: Chlamydomonas p44, AB353122.2 (NCBI); mouse TTC29, E9QLU4 (UniProtKB); human TTC29, Q8NA56 (UniProtKB); zebrafish TTC29, E9QCZ1 (UniProtKB); Tetrahymena p44, XP_001014763.3 (NCBI); Trypanosoma p44, Tb927.3.1990 (KEGG).
Supporting Table S1. IDA-related mutants of Chlamydomonas reinhardtii


