Summary
Our daily life depends on muscle contraction, a process that is controlled by the neuromuscular junction (NMJ). However, mechanisms of NMJ assembly remain unclear. Here, we show that Rapsn, a protein critical to NMJ formation, undergoes liquid-liquid phase separation (LLPS) and condensates into liquid-like assemblies. Such assemblies can recruit acetylcholine receptors (AChRs), cytoskeletal proteins and signaling proteins for postsynaptic differentiation. Rapsn LLPS requires multivalent binding of tetratricopeptide repeats (TPRs) and is increased by Musk signaling. The capacity of Rapsn to condensate and to co-condensate with interaction proteins is compromised by mutations of congenital myasthenic syndrome (CMS). NMJ formation is impaired in mutant mice carrying a CMS-associated, LLPS-deficient mutation. These results reveal a critical role of Rapsn LLPS in forming a synaptic semi-membraneless compartment for NMJ formation.
In brief
Rapsn is critical to acetylcholine receptor (AChR) clustering and NMJ formation. In this study, Xing et al. show that rapsn undergoes phase separation, resulting condensates recruit the AChR and signaling proteins to form membraneless compartments. These processes are compromised by congenital myasthenic syndrome mutations of rapsn.
Introduction
Muscle contraction enables us to breathe, drink and eat, walk and any movement of the body parts. The control of it requires proper function of the neuromuscular junction (NMJ), a cholinergic synapse between motor nerve terminals and muscle fibers (Li et al., 2018; Sanes and Lichtman, 1999; Tintignac et al., 2015; Wu et al., 2010). Action potentials stimulate motor nerve terminals to release acetylcholine (ACh) that activates ACh receptors (AChRs) to depolarize the muscle fibers and triggers calcium release from the sarcoplasmic reticulum to initiate muscle contraction. AChRs are packed at the postjunctional membrane at a concentration of 10,000–20,000/μm2 (Fambrough et al., 1973; Fertuck and Salpeter, 1976). Also enriched beneath the postjunctional membrane are a plethora of cytoplasmic, signaling proteins and cytoskeletal proteins that are critical to induction and/or maintenance of AChR expression at the NMJ (Li et al., 2018; Wu et al., 2010). Reduced density or impaired function of the AChR is implicated in neurological disorders including myasthenia gravis, amyotrophic lateral sclerosis and congenital myasthenic syndromes (CMSs) (Cappello and Francolini, 2017; Engel et al., 2015; Gilhus et al., 2019; Li et al., 2018). Unlike synapses in the brain that are innumerably formed onto dendrites and somas of a neuron, the NMJ is positioned in the middle of a muscle fiber, occupying 0.01% ~ 0.1% of surface. How the AChR becomes concentrated has riveted neuroscientists of many generations. Prior to innervation by motor nerves, muscle fibers develop nascent, aneural AChR clusters (Lin et al., 2001; Yang et al., 2001; Yang et al., 2000) which mark a central region of muscle fibers to attract incoming motor nerve axons (Flanagan-Steet et al., 2005; Liu et al., 2008). After innervation, nerve terminals release agrin that binds to LRP4 to activate Musk in muscle cells (DeChiara et al., 1996; Glass et al., 1996; Jennings et al., 1993; Kim et al., 2008; McMahan, 1990; Zhang et al., 2008); ensuing signaling leads to the formation of large AChR clusters. At the same time, the activation of muscle fibers by ACh disperses extrasynaptic AChR clusters (Lin et al., 2005; Misgeld et al., 2005; Shi et al., 2012). Evidently, these complex interplays require that necessary signaling proteins overcome the enormous cytoplasmic volume to be concentrated beneath the postjunctional membrane, i.e., into a membraneless compartment that is not enclosed by lipid membranes, a process that is not well understood.
Rapsn was identified as a co-purifying protein of the AChR from electric organs of torpedo (Neubig and Cohen, 1979; Sobel et al., 1978). It plays a critical role of Rapsn in NMJ formation and maintenance. Rapsn null mutant mice fail to form aneural AChR clusters and the NMJ (Gautam et al., 1995; Li et al., 2016; Xing et al., 2019). In accord, numerous mutations have been identified in patients with CMS (Milone et al., 2009). In heterologous cells, Rapsn alone is able to form puncta and to recruit co-transfected AChRs into the puncta (Froehner et al., 1990; Li et al., 2016; Phillips et al., 1991); Rapsn mutant myotubes fail to form AChR clusters without altering AChR protein level (Fuhrer et al., 1999; Xing et al., 2019). As a classic adapter protein, Rapsn is believed to anchor the AChR by binding to cytoskeleton-associated proteins such as β-dystroglycan, α-actinin, and MACF1 (Bartoli et al., 2001; Dobbins et al., 2008; Oury et al., 2019). In addition, Rapsn could interact with and thus engage signaling proteins to regulate NMJ formation (Li et al., 2018; Wu et al., 2010; Xing et al., 2020). For example, its interaction with calpain could counteract the inhibitory effect of muscle activation on AChR clustering (Chen et al., 2007). Recent studies suggest that Rapsn may serve as an enzyme whose activity is necessary for the NMJ formation (Li et al., 2016; Xing et al., 2019). Nevertheless, mechanisms of how Rapsn initiates and maintains AChR clustering remains unclear.
Phase separation is key to the formation of membraneless signaling complexes and supramolecular assemblies (Boeynaems et al., 2018; Chen et al., 2020; Shin and Brangwynne, 2017; Wu et al., 2020). Via a process called liquid-liquid phase separation (LLPS), signaling as well as structural proteins undergo phase transition into liquid-like condensates that stably exist within a liquid milieu. Such membraneless, coherent structures contribute to spatiotemporal regulation of gene expression (Boija et al., 2018), assembly of cytoskeleton and tight junction (Beutel et al., 2019; Hernandez-Vega et al., 2017; Schwayer et al., 2019; Woodruff et al., 2017), regulation of enzyme activities (Guo et al., 2020; Zhu et al., 2020), and autophagic degradation (Wilfling et al., 2020; Zhang et al., 2018). Aberrant LLPS or liquid-solid phase transition has been implicated in neurodegenerative disorders (Alberti and Dormann, 2019; Hofweber et al., 2018; Molliex et al., 2015; Murakami et al., 2015). Recently LLPS has been implicated in active zone formation, synaptic vesicle clustering and postsynaptic density (PSD) formation (Bai et al., 2020; Feng et al., 2019; McDonald et al., 2020; Milovanovic et al., 2018; Wu et al., 2019; Zeng et al., 2018; Zeng et al., 2019; Zeng et al., 2016). Postsynaptic proteins PSD-95, SynGAP, Shank, GKAP and Homer do not condensate individually, but when mixed, PSD-95 and SynGAP undergo LLPS. Shank, GKAP and Homer, when mixed, were also able to condensate and co-condensate with PSD-95 and SynGAP (Feng et al., 2019; Zeng et al., 2018; Zeng et al., 2019; Zeng et al., 2016), suggesting phase separation as a possible mechanism of PSD formation.
Here, we show that Rapsn is able to condensate into liquid-like compartments in vitro, in HEK293T cells and in muscles via phase separation. Rapsn co-condensates AChR subunits as well as cytoskeletal proteins, demonstrating that via LLPS, Rapsn serves as a vehicle to recruit different cargo proteins. Remarkably, the capacity of raspyn to phase separate and to co-condensate with cargo proteins is compromised by CMS mutations, providing genetic evidence for an in-vivo role of LLPS. Rapsn does not contain an intrinsically disordered region that is required for LLPS (Boeynaems et al., 2018; Shin and Brangwynne, 2017). We show that Rapsn LLPS is driven by the TPR (tetratricopeptide repeat)-containing domain which possesses multivalent binding property and is promoted by Musk-mediated tyrosine phosphorylation. Together, these results suggest that Rapsn LLPS into a semi-membraneless compartment underlies NMJ formation and maintenance.
Results
Phase separation of Rapsn into condensates in vitro
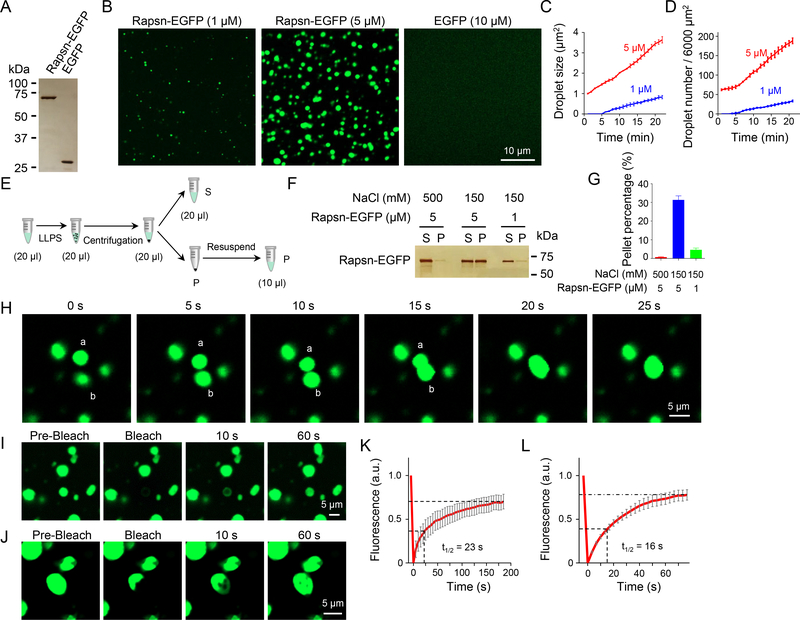
To determine whether Rapsn can phase separate into liquid-like condensates, enhanced green fluorescent protein (EGFP)-tagged Rapsn (Rapsn-EGFP) was purified from transfected HEK293T cells (Figure 1A). Time-lapse microscopic analysis indicates that Rapsn-EGFP was able to spontaneously form spherical, condensed droplets in the physiological buffer (Figures 1A and 1B, Video S1). Quantitatively, droplet numbers and sizes gradually increased over time (Figures 1C and 1D), and in a concentration-dependent manner because more and larger droplets were formed by 5 μM Rapsn-EGFP than by 1 μM (Figures 1B–1D). In contrast, as a control, no droplets were detectable with EGFP even at 10 μM (Figure 1B). To further eliminate a possible role of EGFP, we generated a recombinant Rapsn without the EGFP tag (Figure S1A), and labeled it with a red dye (Mix-n-Stain™ CF555, CF555-Rapsn) (Zeng et al., 2018). As shown in Figures S1B–S1D, CF555-Rapsn could also form droplets. These results indicated that Rapsn was able to phase separate into condensates.
Figure 1. LLPS of Rapsn-EGFP into condensates in vitro.
(A-D) LLPS of Rapsn-EGFP, but not EGFP, into condensed droplets in a concentration-dependent manner. (A) Silver staining showing Rapsn-EGFP and EGFP. (B) Rapsn-EGFP, but not EGFP, was able to phase separate into condensed droplets. Rapsn-EGFP (1 μM and 5 μM) and EGFP (10 μM) were diluted into physiological buffer, and after 20 min, representative images were acquired. (C, D) Quantification of droplet size and number at indicated times.
(E) Schematic diagram showing separation of condensed phase from aqueous phase by centrifugation.
(F) High salt and lower protein concentration reduced the amount of Rapsn in pellets. Shown were representative silver staining.
(G) Quantification of Rapsn-EGFP in pellets in (F). Data was shown as mean ± SEM; n = 3.
(H) Fusion of two Rapsn-EGFP droplets.
(I-L) Rapsn-EGFP in condensed droplets extensively exchanged with surrounding aqueous phase or within droplets. FRAP analysis of a Rapsn-EGFP droplet (I) or a part of droplet (J). (K, L) Quantification of fluorescence recovery in (I, J).
Data was shown as mean ± SEM; n = or > 3.
See also Figure S1.
To demonstrate that Rapsn indeed condensates into droplets from aqueous phase, Rapsn solution was subjected to a centrifugation (14,000 x g, 15 min) (Figure 1E), the amounts of Rapsn in condensed phase and aqueous phase were visualized by silver staining. Both Rapsn-EGFP and Rapsn were detectable in the pellets after centrifugation (Figures 1F and S1E), also in a concentration-dependent manner (Figures 1G and S1F). In addition, Rapsn LLPS was dependent on salt concentration and inhibited in the presence of 500 mM NaCl. At 150 mM NaCl, 30% of Rapsn was detected in condensed phase; and, when LLPS was inhibited in the presence of 500 mM NaCl, Rapsn was barely detectable in condensed phase (Figures 1G and S1F). These results provide evidence that Rapsn phase separated into condensed phase directly from an aqueous phase.
Condensed droplets of Rapsn had liquid properties. First, two droplets could fuse upon contact (Figure 1H, Video S1). Second, when a droplet was photobleached, the fluorescence was able to recover rapidly, with a time constant of t1/2 = 23 s (Figures 1I and 1K, Video S2), suggesting a dynamic exchange of Rapsn between aqueous and condensed phases. This notion is supported by the observation that when a region of a droplet was photobleached, fluorescence recovery in the bleached area occurred at a faster rate (t1/2 = 16 s) (Figures 1J and 1L, Video S3). These results indicate an extensive exchange of Rapsn proteins between condensates and surrounding buffer, and within the condensates.
Rapsn phase separation into subcellular compartments in HEK293T cells, myotubes and muscles
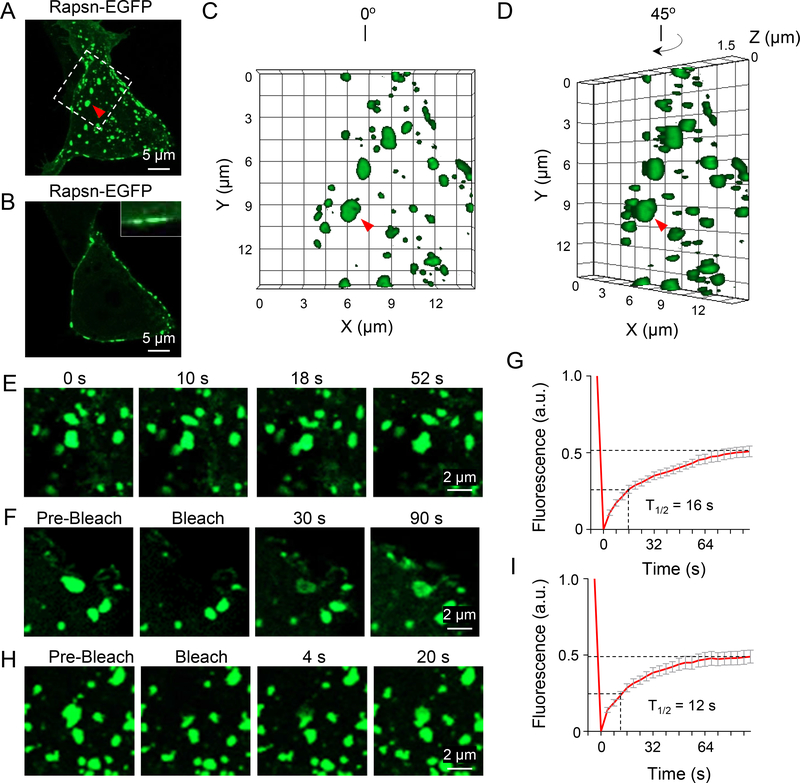
Rapsn is known to form puncta in heterologous cells (Froehner et al., 1990; Phillips et al., 1991). To examine whether these puncta are phase separated membraneless compartments, HEK293T cells were transfected with Rapsn-EGFP, and examined 12 hr after transfection. Confocal analysis by collapsed Z-stack images showed that Rapsn-EGFP formed puncta in HEK293T cells (Figure 2A), consistent with previous work (Li et al., 2016; Xing et al., 2019). Single plane analysis at the middle of cells indicated that these puncta were attached to plasma membrane (Figure 2B), presumably due to myristoylation (Frail et al., 1988). Rapsn puncta were spherical or elliptical in HEK293T cells as revealed by 3D reconstruction analysis (Figures 2C and 2D). As shown in Video S4, Rapsn puncta were changing in size and shape constantly; adjacent puncta could coalesce into one upon contact (Figure 2E, Video S4, red arrow). In addition, the fluorescence of photobleached puncta could rapidly recover within seconds (t1/2 = 16 s) (Figures 2F and 2G, Video S4, white arrow). When a part of puncta was photobleached, the fluorescence was recovered at a faster rate (t1/2 = 12 s) (Figures 2H and 2I). Similar results were observed in HEK293T cells expressing mCherry-tagged Rapsn (Figures S2A–2C). These results indicate that Rapsn puncta in HEK293T cells are membraneless subcellular compartments, formed by LLPS, and that the exchange of Rapsn between the compartments and the environment and within the compartment is dynamic.
Figure 2. Rapsn LLPS into liquid-like compartments in HEK293T cells.
(A-D) Formation of membrane-attached Rapsn-EGFP puncta in transfected HEK293T cells. (A) Representative 3D projection image showing circular or oval Rapsn-EGFP puncta; (B) Single panel image showing membrane-attached Rapsn-EGFP puncta; (C) High-magnification image showing highlighted region in (A); (D) View of the same image of (C) at different angle. White arrow, membrane-attached puncta. Red triangle, same puncta in (A), (C) and (D).
(E) Fusion of two Rapsn-EGFP puncta in HEK293T cells.
(F-I) Dynamic exchange of Rapsn-EGFP between puncta and surrounding milieu, and within puncta in HEK293T cells. (F) FRAP analysis of a Rapsn-EGFP puncta, and (H) quantification of fluorescence recovery; (G) FRAP analysis of a part of puncta and (I) quantification of fluorescence recovery.
Data was shown as mean ± SEM; n = or > 3.
See also Figure S2.
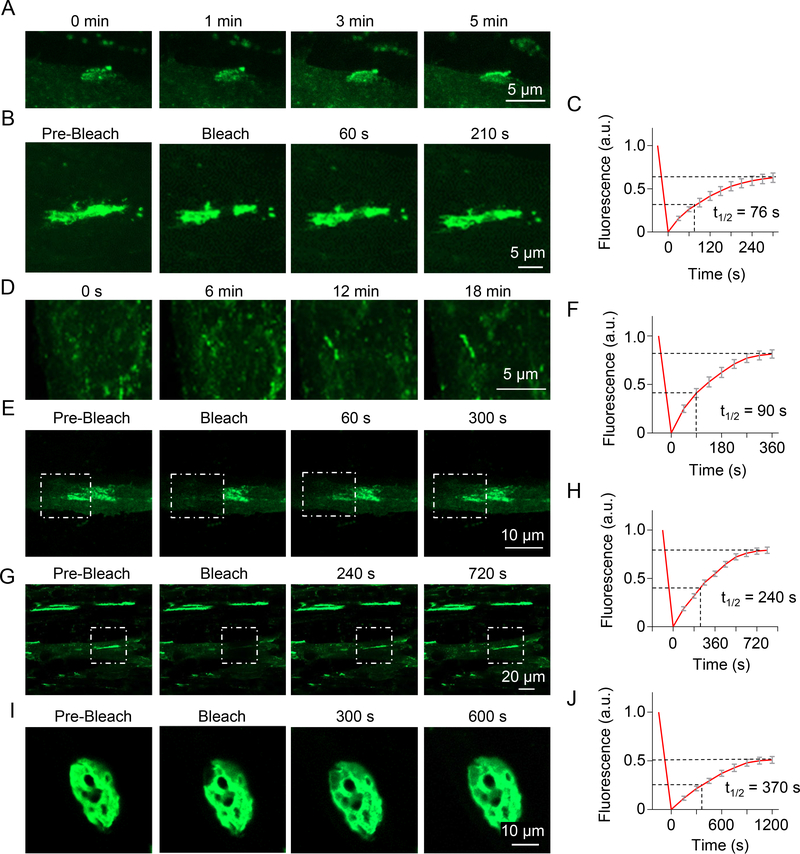
To determine whether Rapsn LLPS occurs in myotubes, we performed time-lapse imaging. To avoid potential non-specific effects of Rapsn overexpression which prevents the formation of large AChR clusters (Han et al., 1999; Yoshihara and Hall, 1993), Rapsn-EGFP was transfected in Rapsn −/− myoblasts (Fuhrer et al., 1999). Resulting myotubes were examined for Rapsn-EGFP clusters. As shown in Figure 3A, in the absence of agrin, small Rapsn aggregates fused to form a large cluster within minutes. When a part of spontaneous cluster was photobleached, the fluorescence recovered quickly (Figures 3B and 3C, Video S5), suggesting that the clusters display liquid-like property. Next, we treated myotubes with agrin and examined agrin-induced clusters of Rapsn-EGFP. They grew by fusing small aggregates (Figure 3D). When partially photobleached, the fluorescence recovered within minutes (Figures 3E and 3F, Video S6). When a cluster is photobleached in entirety, the fluorescence wcould also recover albeit at a slower rate (Figures 3G and 3H, Video S7). These observations support the notion that Rapsn form membraneless subcellular compartments via LLPS in myotubes.
Figure 3. Rapsn LLPS into liquid-like compartments in myotubes and in muscles.
(A) Fusion of small spontaneous Rapsn-EGFP aggregates into large, continuous clusters in myotubes. Arrow, cluster during fusion.
(B) Dynamic property of Rapsn-EGFP protein within spontaneous clusters.
(C) Quantification of fluorescence recovery in (B).
(D) Fusion of agrin-induced Rapsn-EGFP clusters in myotubes.
(E-H) Dynamic exchange of Rapsn-EGFP between cluster and surrounding milieu, and within cluster in myotubes. (E) FRAP analysis of a agrin-induced Rapsn-EGFP cluster, and (F) quantification of fluorescence recovery; (G) FRAP analysis of a part of cluster and (H) quantification of fluorescence recovery.
(I, J) Dynamic property of Rapsn-EGFP in living muscles.
Data was shown as mean ± SEM; n = or > 3.
Finally, we determined whether Rapsn LLPS occurs in live muscle fibers. Rapsn-EGFP was electroporated in vivo into tibialis anterior (TA) muscles at postnatal day 10 (P10). Muscles were isolated at P22 in oxygenated Ringer’s solution and examined for in vivo EGFP clusters. As shown in Figures 3I and 3J, there was a fast recovery of fluorescence of the photobleached area of Rapsn-EGFP clusters in live muscles. Together, these results from HEK293T cells, myotubes and muscles demonstrate that Rapsn-EGFP clusters are dynamic and support a role of Rapsn LLPS in NMJ development. It is interesting to note that the FRAP recovery rate of Rapsn-EGFP was slower in myotubes and muscles than that in HEK293T cells and Rapsn localization pattern in myotubes and muscles is quite different with that of HEK293T cells. This may be because that Rapsn condensates in muscles as well as myotubes recruit muscle- or myotube-specific cytoskeletal components or other components that may constrain Rapsn. Moreover, during cluster or synapse development, Rapsn condensates may transit to a solid phase, a phenomenon that has been observed for active zone proteins, ELKS-1 and SYD-2 (McDonald et al., 2020).
Multivalent binding of TPRs critical to Rapsn LLPS
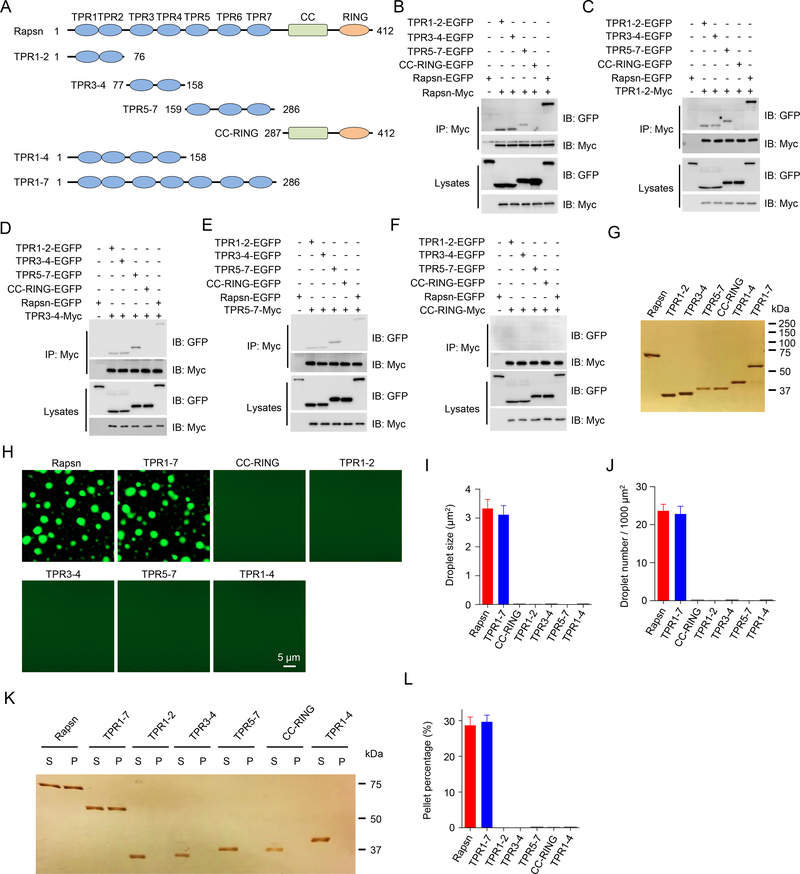
LLPS could be mediated by proteins with intrinsic disordered regions (Milovanovic et al., 2018). However, Rapsn does not have an intrinsic disordered domain (IUPred2A, https://iupred2a.elte.hu). Alternatively, LLPS could occur via a multi-protein complex such as those formed by PSD95 and SynGAP (Zeng et al., 2016) and by RIM/RIM-BP (Wu et al., 2019). Yet, Rapsn itself is sufficient for LLPS. Rapsn has multiple TPR domains, a common motif for protein-protein interaction (Perez-Riba and Itzhaki, 2019). Because two to three TPRs could form a concave groove for interaction (Perez-Riba and Itzhaki, 2019) and Rapsn has seven TPRs, we posited that the TPR region of Rapsn may possess multiple interaction motifs. To test this hypothesis, we first generated recombinant proteins containing two or three TPR domains: TPR1–2, TPR3–4, TPR5–7 and CC-RING (Figure 4A) and examined their ability to bind Rapsn. In cells co-transfected with Rapsn-Myc, as shown in Figure 4B, Rapsn-EGFP was detected in the immunocomplex precipitated by anti-Myc beads. Note that Rapsn-EGFP was not detected in cells that were not co-transfected with Rapsn-Myc (Figure 4B), indicating the specificity of the interaction. These results suggest that the Rapsn proteins were able to self-associate, in agreement with previous work (Li et al., 2016; Ramarao et al., 2001; Xing et al., 2019). Interestingly, Rapsn-Myc could also co-precipitate TPR1–2, TPR3–4, and TPR5–7, but not CC-RING (Figure 4B), indicating that TPR1–2, TPR3–4, and TPR5–7, but not CC-RING, bind with full-length Rapsn. To determine whether these TPR combinations bind to each other, HEK293T cells were co-transfected with a Myc-tagged TPR protein and EGFP-tagged Rapsn or a truncation mutant. Subsequent co-immunoprecipitation assays indicated that TPR1–2-Myc (Figure 4C), TRP3–4-Myc (Figure 4D), and TPR5–7-Myc (Figure 4E) could bind to full-length Rapsn and each of the three TPR proteins. In contrast, CC-RING-Myc failed to interact with full-length or any of the TPR motifs (Figure 4F). These results suggest that two or more TPR motifs were able to form a binding motif for Rapsn and self-association. To test this hypothesis further, we generated additional TPR combinations: TPR2–3, TPR4–5, TPR5–6, and TPR6–7 (Figure S3A). As shown in Figure S3B, TPR2–3, TPR4–5, and TPR5–6, but not TPR6–7 could bind with full-length Rapsn. Moreover, these combinations could interact with one another (Figures S3C, S3D and S3F). However, TPR6–7 did not interact with full length Rapsn or any of the TPR combination (Figure S3E). These results suggest that Rapsn contains multiple self-interaction binding sites via various combination of TPR motifs, revealing a potential molecular mechanism of Rapsn LLPS.
Figure 4. Multivalent binding of TPR domains for Rapsn LLPS.
(A) Schematic domain structures of Rapsn and truncation mutants.
(B) Binding of Rapsn with TPR1–2, TPR3–4, TPR5–7, and full-length Rapsn, but not CC-RING.
(C-F) TPR1–2 (C), TPR3–4 (D), TPR5–7 (E), but not CC-RING (F), were able to self-interact and bind each other.
(G) Silver staining showing purified EGFP-tagged full-length and truncated Rapsn proteins.
(H) LLPS of TPR1–7 and full-length Rapsn, but not CC-RING, TPR1–2, TPR3–4, TPR5–7, and TPR1–4.
(I, J) Quantification of droplet size (I) and number (J) in (H).
(K) Representative silver staining image showing WT and TPR1–7, but not TPR1–2, TPR3–4, TPR5–7, CC-RING, and TPR1–4, were able to condensate into pellets after centrifugation.
(L) Quantification of the percentage of proteins in pellets in (K).
Data was shown as mean ± SEM; n = or > 3.
See also Figure S3.
Next, we determined whether TPR-containing proteins alone or in combination undergo LLPS in vitro. As shown in Figures 4G and 4H, TPR1–7 was able to condensate to form droplets. The droplet size and number of TPR1–7-EGFP were comparable to those by full-length Rapsn-EGFP (Figures 4I and 4J), suggesting TPR1–7 was sufficient for phase separation. This notion was supported by centrifugation assays where similar amounts of TPR1–7 and full-length Rapsn were detected in condensed phase (Figures 4K and 4L). In contrast, CC-RING failed to form detectable droplets or condensate into pellets (Figures 4H, 4K and 4L), indicating a necessary role of TPR1–7 for Rapsn LLPS. Note that none of TPR1–2, TPR3–4, TPR5–7, and TPR1–4 was able to form droplets or condensate into pellets (Figures 4H–4L), although each of them was able to interact with full length Rapsn. These results indicate that Rapsn LLPS requires the multivalent binding of the TPR region.
Recruitment of cargo proteins into Rapsn condensates
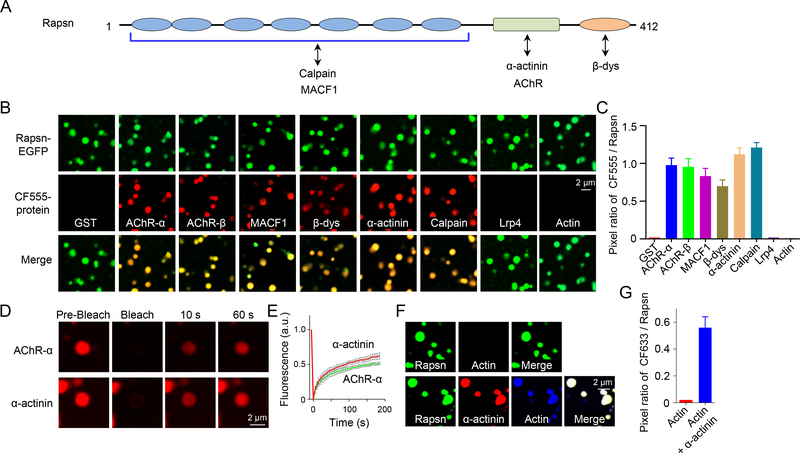
After finding that Rapsn phase separated into membraneless compartments in cells (Figures 2 and 3), we hypothesized that Rapsn LLPS may serve as a vehicle to carry interaction or cargo proteins into its condensates. First, we generated GST fusion proteins containing intracellular regions (loop3/4) between 3rd and 4th transmembrane domains of AChR-α and -β subunits, which are required for Rapsn interaction (Lee et al., 2009). Resulting GST fusion proteins, referred to AChR-α and AChR-β hereafter, were purified (Figure S4A), and labeled with CF555. AChR-α or AChR-β alone was unable to phase separate into droplets (Figure S4B); however, when mixed with equimolar Rapsn-EGFP, they were enriched in Rapsn droplets (Figures 5A–5C), suggesting recruitment of AChR-α or AChR-β by Rapsn LLPS. Consistently, AChR-α or AChR-β alone could not be detected in condensed phase but could when mixed with Rapsn (Figures S5C–S5F). As a negative control, GST was unable to be enriched into Rapsn droplets and could not form condensed pellets when mixed with Rapsn-EGFP (Figures 5B, 5C, S5A and S5B), suggesting the specificity of Rapsn-mediated recruitment.
Figure 5. Recruitment of cargo proteins into Rapsn condensates.
(A) Schematic domain structure of Rapsn and interaction proteins.
(B, C) AChR-α, AChR-β, MACF1, β-dystroglycan, α-actinin or calpain, but not GST, LRP4, or actin, were recruited into Rapsn LLPS-mediated condensates.
(D) FRAP analysis of AChR-α and α-actinin enriched in Rapsn droplets showing dynamic protein exchange.
(E) Quantification of fluorescence recovery of AChR-α and α-actinin in (D).
(F) Recruitment of actin into Rapsn condensed droplets in the presence of α-actinin.
(G) Quantification of the pixel ratio of CF633 / Rapsn-EGFP in (F).
Data was shown as mean ± SEM; n = or > 3.
See also Figures S4 and S5.
Rapsn has been shown to associate with intracellular proteins that regulate the cytoskeleton or are involved in signaling transduction (Xing et al., 2020). Next, we determined whether these proteins could be recruited into Rapsn condensates. We focused on proteins that directly interact with Rapsn such as β-dystroglycan (Bartoli et al., 2001), α-actinin (Dobbins et al., 2008), MACF1 (Antolik et al., 2007; Oury et al., 2019), and calpain (Chen et al., 2007) (Figure 5A). Respective recombinant proteins were purified and labeled with CF555 (Figure S4A). They alone did not form condensates (Figure S4B) but were detectable in Rapsn droplets (Figures 5B and 5C). In agreement, they were detected in condensed phase when mixed with Rapsn, but not alone (Figures S5G–S5N). These results suggest that Rapsn LLPS could enrich these interaction proteins into condensates. When CF555-AChR-α or α-actinin in Rapsn condensates was photobleached, the fluorescence rapidly recovered (Figures 5D and 5E), indicating that cargo proteins in Rapsn condensates are dynamic and exhibit rapid protein exchange between condensates and surrounding buffer.
Co-immunoprecipitation evidence indicates that Rapsn associates with actin in electric organs and in muscle cells (Li et al., 2016; Walker et al., 1984). However, CF555- or CF633-labeled actin was not detectable in Rapsn droplets and in condensed phase (Figures 5B, 5C, 5F, 5G, S5Q and S5R), perhaps because that actin may not interact with Rapsn directly. Interestingly, in the presence α-actinin, a protein that directly interacts with actin (Sjoblom et al., 2008), CF633-labeled actin was detectable in Rapsn droplets (Figures 5F and 5G), indicating that Rapsn may be able to recruit partners of its interaction proteins. Consistently, LRP4 was not detectable in Rapsn droplets and condensed phase (Figures 5B, 5C, S5O and S5P) although it was presented in Rapsn aggregates in myotubes (Figure S5S), suggesting that LRP4 may be recruited indirectly into Rapsn aggregates.
Promotion of Rapsn LLPS by Musk signaling
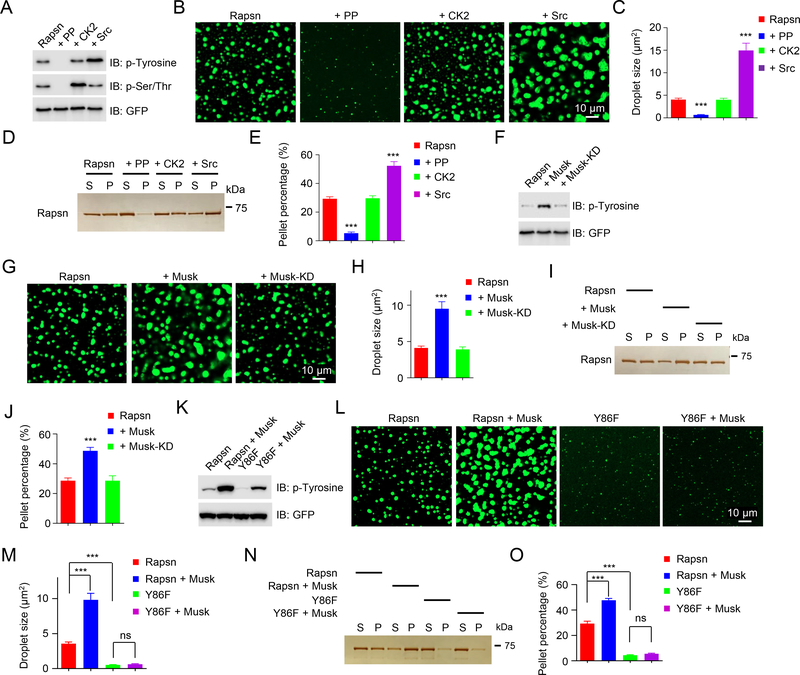
Rapsn becomes tyrosine phosphorylated, which is necessary for aggregation and AChR clustering by agrin (Xing et al., 2019; Xing et al., 2020). Therefore, we determined whether Rapsn LLPS is regulated by tyrosine phosphorylation. First, Rapsn-EGFP was purified from transfected HEK293T cells where it was tyrosine phosphorylated (Lee et al., 2008; Xing et al., 2019) (Figure 6A). Treatment with lambda phosphatase (PP) reduced the phospho-tyrosine level of Rapsn (Figure 6A) and its ability to condensate (Figures 6B–E), suggesting a potential regulation by tyrosine phosphorylation. In contrast, Src, a proto-oncogene tyrosine-protein kinase, increased phospho-tyrosine of Rapsn (Figure 6A) and LLPS (both droplet formation and condensation into pellets) (Figures 6B–6E). Because PP also reduced phospho-serine/threonine (Figure 6A), we determined whether serine/threonine phosphorylation was involved in Rapsn LLPS. Rapsn was incubated with casein kinase 2 (CK2), a serine/threonine kinase that binds to Rapsn and has been implicated in NMJ formation (Cheusova et al., 2006; Eiber et al., 2019; Herrmann et al., 2015), which increased phospho-serine/threonine of Rapsn (Figure 6A). However, it had little effect on Rapsn droplet formation and condensation into pellets (Figures 6B–6E). Together, these results suggest that Rapsn LLPS is promoted by tyrosine phosphorylation.
Figure 6. Promotion of Rapsn LLPS by Musk signaling.
(A-E) Promotion of Rapsn LLPS by tyrosine phosphorylation. Rapsn-EGFP was treated with PP, CK2, or Src. (A) Phosphorylation levels of Rapsn-EGFP. (B, C) PP reduces and Src increases droplet formation. (B) Representative blots. (C) Quantitative data. ***, P < 0.001; One-way ANOVA. (D, E) PP reduces and Src increases Rapsn in pellets. (D) Representative silver staining. (E) Quantitative data. n = 3; ***, P < 0.001; One-way ANOVA.
(F-J) Enhanced Rapsn LLPS by Musk-induced tyrosine phosphorylation. Rapsn-EGFP was purified from HEK293T cells cotransfected with Musk or Musk-KD.
(F) Tyrosine phosphorylation of Rapsn. (G, H) Increased droplet formation by Musk cotransfection. ***, P < 0.001; One-way ANOVA.
(I, J) Increased Rapsn in pellets by Musk cotransfection. n = 3; ***, P < 0.001; One-way ANOVA.
(K-O) Reduced Rapsn LLPS by Y86F mutation. Rapsn or Y86F-EGFP was transfected into HEK293T cells alone or with Musk and purified for LLPS. (K) Tyrosine phosphorylation of Rapsn. (L) Representative images of droplets. (M) Quantification of droplet size in (L). n = 3; ***, P < 0.001; One-way ANOVA. (N) Reduced Y86F Rapsn in pellets. (O) Quantitative data in (M). n = 3; ***, P < 0.001; One-way ANOVA.
Data was shown as mean ± SEM; n = or > 3.
To examine whether Rapsn LLPS is regulated by Musk-induced tyrosine phosphorylation, we co-transfected Rapsn-EGFP with Musk into HEK293T cells. The co-transfection increased tyrosine phosphorylation of Rapsn, compared with Rapsn-EGFP alone (Figure 6F), consistent with previous work (Lee et al., 2008; Xing et al., 2019). In parallel, Rapsn LLPS was increased (Figures 6G–6J). This effect was not observed with Rapsn from HEK293T cells co-transfected with kinase dead Musk mutant (Musk-KD) (Figures 6F–6J). Previously, we showed that Y86 of Rapsn becomes tyrosine-phosphorylated and its mutation, Y86F, reduces phosphorylation and self-association (Xing et al., 2019) (Figure 6K). Interestingly, droplet size and level of Y86F in condensed phase were reduced, compared with WT Rapsn (Figures 6L–6O). In addition, the Y86F mutation reduced Musk-promoted Rapsn LLPS in both droplet size and condensed phase (Figures 6L–6O). All together, these results suggested that Musk signaling stimulates AChR clustering by promoting Rapsn LLPS.
Decreased LLPS and cargo-carrying abilities of CMS mutants
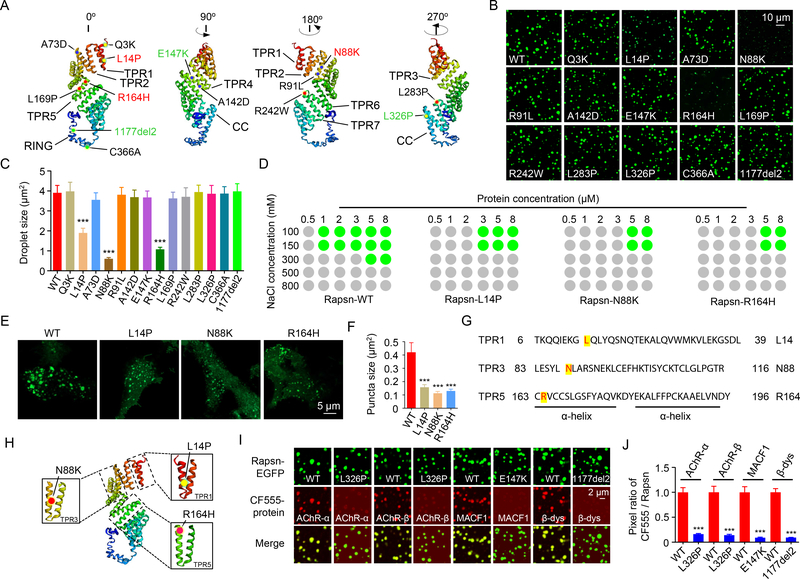
Numerous Rapsn mutations have been identified in patients with CMS (Milone et al., 2009). Because a crystal structure of Rapsn is not available, we modeled Rapsn’s structure based on a virtual computer program I-TASSER (Roy et al., 2010). As shown in Figure 7A, the seven TPR motifs are grouped in the N-terminal region, followed by the CC domain and a seemingly loose RING domain. CMS mutations are distributed within the TPR region, the CC and RING domains as well as in junction regions between these domains (Figure 7A). We wondered whether CMS-associated mutations may alter Rapsn LLPS. Remarkably, the size of droplets formed by L14P, N88K or R164H was smaller than that of Rapsn WT at 5 μM (Figures 7B, 7C), suggesting a compromised ability for LLPS. Rapsn LLPS was not altered by eleven other CMS mutations including those in the junction regions and within the CC and RING domains (Figures 7B and 7C) (Table S1).
Figure 7. Decreased LLPS and cargo-carrying abilities by CMS mutations.
(A) Modeled structure of Rapsn and localizations of CMS mutations. Red, mutations that reduce Rapsn LLPS; Green, mutations that decreased cargo recruitment.
(B) Representative images showing condensed droplets formed by LLPS of WT or indicated mutant Rapsn (5 μM).
(C) Quantification of droplet size in (B). n = 3; ***, P < 0.001; One-way ANOVA.
(D) Phase diagrams showing droplet formation of WT or three mutant Rapsn (L14P, N88K and R164H) with different protein concentrations in 25 mM Tris, pH7.4, 5 mM DTT, NaCl (ranging from 100mM – 800mM). Green dots, phase separation. Gray dots, no phase separation.
(E) Mutant Rapsn (L14P, N88K and R164H) forming smaller puncta, compared with WT Rapsn in transfected HEK293T cells.
(F) Quantification of puncta size in (E). n = 10; ***, P < 0.001; One-way ANOVA.
(G, H) Location of LLPS inhibiting mutations in the first α-helix.
(I) The recruitment of cargo proteins into Rapsn condensates were inhibited by CMS-related mutations.
(J) Quantification of the pixel ratio of CF555 / Rapsn-EGFP in (I). n = 3; ***, P < 0.001; One-way ANOVA.
Data was shown as mean ± SEM; n = or > 3.
See also Table S1.
We determined the minimal concentrations of Rapsn WT and L14P, N88K or R164H mutants in forming droplets and their sensitivity to increasing NaCl concentrations. Rapsn WT was able to phase separate into droplets at 1 μM in the presence of 100 or 150 mM NaCl (Figure 7D), two concentrations close to physiological salt concentration. However, under these conditions, none of the three Rapsn mutants at 1 μM were able to form droplets (Figure 7D). The minimal concentrations for L14P, N88K and R164H to form droplets increased to 3 μM and 5 μM, respectively in the presence of 100 or 150 mM NaCl (Figure 7D). On the other hand, increasing NaCl concentrations inhibited LLPS of both WT and mutant Rapsn (Figure 7D). However, Rapsn WT at 5 μM protein concentration was able to phase separate into droplets at 300 mM NaCl where LLPS was not observed for Rapsn mutants under the same conditions, even when protein concentrations were increased to 8 μM (Figure 7D). Moreover, the puncta size formed by L14P, N88K or R164H was smaller than those formed by WT Rapsn in HEK293T cells (Figures 7E and 7F), indicating reduced LLPS in cells. It is worthy noticing that L14P, N88K and R164H were all localized in TPR motifs (TPR1, TPR3, and TPR5, respectively) (Figures 7G and 7H), consistent with the notion that the multivalent binding of TPR motifs is critical to Rapsn LLPS (Figures 4 and S3). Intriguingly, the three mutations are located within the first α-helix (Figures 7G and 7H).
Of 14 CMS-associated mutations, eleven had no effect on Rapsn LLPS. We determined whether they alter the ability of Rapsn to recruit cargo proteins into condensates. As shown in Figures 7I and 7J, AChR-α and AChR-β were hardly detectable in the droplets formed by L326P Rapsn, in contrast to robust CF555 signal in droplets formed by WT Rapsn. These results indicate a critical role of L326 in carrying AChR-α and AChR-β into droplets. On the other hand, compared with WT Rapsn, E147K reduced the amount of MACF1 in the droplets whereas 1177del2 inhibited the recruitment of β-dystroglycan (Figures 7I and 7J). The effects of L326P, L147K, and 1177del2 on cargo carrying ability was cargo-specific and they had no effect on co-condensation with other cargo proteins (Table S1). Together, these results suggest two ways by which CMS-associated mutations impair NMJ formation - 1) preventing Rapsn from forming condensates and 2) impairing the ability of Rapsn to carry cargo proteins into condensates, shedding light on mechanisms of CMS pathology.
Diminished aneural and nerve-induced AChR clusters in R164H knockin mutant mice
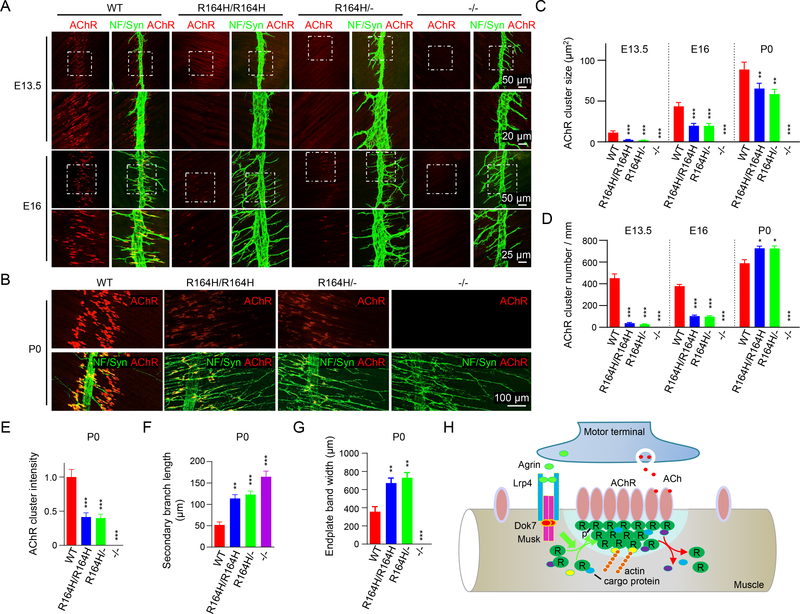
We demonstrated that the N88K mutation prevented mice from forming proper NMJs (Xing et al., 2019), providing genetic evidence for a role of Rapsn LLPS in NMJ formation. To further test this hypothesis, we generated a knockin mouse strain carrying R164H (Figures S6A and S6B), a mutation in the fifth TRP motif that inhibits Rapsn LLPS (Figure 7). Self-association of R164H Rapsn was reduced, compared with WT (Figures S7A and S7B). The mutation had little effect on Rapsn mRNA and protein levels (Figures S6C and S6D) but prevented Rapsn from rescuing AChR clustering deficits in Rapsn −/− myotubes (Figures S7C and S7D). In WT mice, muscle fibers were dotted with primitive aneural AChR clusters in the central region at embryonic day 13.5 (E13.5) (a phenomenon called pre-patterning, prior to NMJ formation) (Li et al., 2018; Lin et al., 2001; Yang et al., 2001; Yang et al., 2000) (Figure 8A). However, in R164H/R164H mice, aneural AChR clusters were barely detectable, a phenotype like that of Rapsn null mice (−/−) (Figure 8A). Because R164H mice were generated by CRISPR-Cas9, we characterized R164H/- mice in which one chromosome carried R164H and the other carries null allele. Similar phenotypes were observed in R164H/- mice and R164H/R164H mice (Figure 8A), suggesting that NMJ deficits were not an off-target effect. These results suggest that the R164H mutation impaired the formation of aneural AChR clusters.
Figure 8. Diminished aneural and nerve-induced AChR clusters in R164H knockin mutant mice.
(A, B) Diminished aneural and nerve-induced AChR clusters in R164H knockin mutant mice. Diaphragms of indicated genotypes at different ages were stained whole-mount with Flour 594-α-BTX (red) to label AChR clusters and with anti-NF/Syn (green) antibodies to label motor nerve terminals. Areas in rectangles in top panels are enlarged and shown in lower panels in A.
(C-G) Quantitative data of AChR cluster size (C) and number (D), AChR intensity (E), secondary nerve branch length (F), and endplate band width (G). Data were shown as mean ± SEM; n = 3; *, P < 0.05, **, P < 0.01; ***, P < 0.001; One-way ANOVA.
(H) Working model. Rapsn condensates to form a liquid-like semi-membraneless compartment. In so doing, Rapsn recruits cargo proteins that regulate AChR clustering. These condensates may serve as a hub that promotes cytoskeletal interaction and are promoted by agrin-LRP4-Musk signaling.
See also Figures S6 and S7.
In WT mice, innervation occurs ~E14.5, induces larger clusters and disperses aneural clusters. As shown in Figures 8A, 8C and 8D, both the number and size of AChR clusters were reduced in R164H/R164H and R164H/- mice at age of E16, compared with WT controls. At P0, AChR clusters appear as oval plaques that are distributed within a restricted central region in WT mice (Figure 8B). However, in R164H/R164H and R164H/- mice, AChR clusters were smaller in size with reduced intensity and distributed in wider center area (Figures 8B, 8C and 8E), indicating that R164 is necessary for nerve-induced AChR clustering. Note that R164H had little effect on Rapsn stability or interaction with AChR, actin, or other binding partners including β-dystroglycan, calpain, MACF1, and α-actinin (Figures S7E–S7M). These results provide further genetic evidence for a role of Rapsn LLPS in NMJ formation. In addition to postsynaptic deficits, motor nerve terminals extensively arborized in R164H mutant mice (Figures 8B, 8D, 8F and 8G), a phenotype commonly observed in mutant mice of Rapsn, LRP4, Musk or Dok7 (DeChiara et al., 1996; Gautam et al., 1995; Okada et al., 2006; Weatherbee et al., 2006). The cause of presynaptic deficits is unclear but may be due to a compensatory mechanism for postsynaptic deficits and/or loss of a retrograde signaling directly or indirectly downstream of the agrin pathway (DeChiara et al., 1996).
Discussion
We provide evidence that Rapsn undergoes LLPS into dynamic condensates in vitro, in HEK293T cells, and in muscles (Figures 1–3). Rapsn condensates can co-condensate AChRs as well as cytoskeletal and signaling proteins (Figures 5B and 5C). Remarkably, the capacity of Rapsn to phase separate and to co-condensate with cargo proteins is compromised by CMS-associated mutations (Figure 7). In particular, N88K, a prevalent CMS-related mutation in Rapsn, reduced LLPS of Rapsn; NMJ formation is impaired in N88K mutant mice (Xing et al., 2019). Here we show that R164H, another CMS mutation, inhibits Rapsn LLPS and prevents proper NMJ assembly (Figures 7 and 8), providing genetic evidence for Rapsn LLPS in NMJ formation and revealing potential pathological mechanisms of neuromuscular disorders.
Our results support a working model that Rapsn condensates to form a membraneless subcellular compartment to orchestrate NMJ assembly (Figure 8H), through dual manners. First, Rapsn compartments co-condensates AChR subunits and cytoskeletal proteins, directly or indirectly (Figure 5B), suggesting Rapsn LLPS serves as a structural platform for AChR clustering and anchoring AChRs to cytoskeleton. Because its ability to condensate without an extrinsic regulator, Rapsn is sufficient to initiate AChR clustering in muscles prior to innervation by motor neurons. Interestingly, Rapsn is more conserved than many NMJ proteins across the species; for example, C. elegans express Rapsn, but not agrin, LRP4 or Musk. Therefore, Rapsn may play a key role in synaptogenesis in species before complex regulatory mechanisms evolve. Second, Rapsn condensates may function as a signaling hub to favor AChR clustering. LLPS is more robust by Rapsn purified from cells that expressed WT, but not kinase dead mutant Musk (Figures 6G–6J), suggesting that Rapsn LLPS could be promoted by the agrin-LRP4-Musk signaling. In addition, signaling molecules including protein kinase A (PKA) (Choi et al., 2012), casein kinase 2 (Herrmann et al., 2015), and HSP90β (Luo et al., 2008) could be recruited to the condensates through direct or indirect binding. Bringing together Rapsn (an E3 ligase) and AChR subunits, Rapsn LLPS may facilitate AChR neddylation that was required for AChR stability (Li et al., 2016; Xing et al., 2020). Consistent with this notion, N88K mutation, which reduces LLPS of Rapsn, also reduces AChR neddylation (Xing et al., 2019).
Rapsn is a major target of CMS, accounting for 15% of total CMS cases (Milone et al., 2009). Rapsn mutations can reduce Rapsn ability to induce AChR clustering (Cossins et al., 2006; Maselli et al., 2007; Maselli et al., 2003; Ohno et al., 2002; Ohno et al., 2003). However, little is known regarding underlying molecular mechanisms. Here, L14P, N88K and R164H diminish the ability of Rapsn to condensate in solution and in cells (Figure 7). On the other hand, three other mutations, L326P, E147K, and 1177del2 have no effect on Rapsn LLPS, but prevent Rapsn from carrying interacting proteins into condensates (Figure 7). These results reveal a novel pathological mechanism of CMS mutations, i.e., by disrupting the formation of semi-membraneless compartment or recruitment of signaling proteins into the compartment. It is worthy pointing out that some CMS mutations had no effect on Rapsn LLPS or co-condensation. This may reflect a failure to interact with other cargos that were not tested, or yet another mechanism that is distinct from altering LLPS or cargo interaction.
Unlike the PSD of brain synapses, the NMJ does not have a postsynaptic electron density structure. LLPS by PSD proteins requires interactions of two or three organizers. For example, PSD-95, SynGAP, Shank, GKAP and Homer do not condensate individually; however, PSD-95 and SynGAP, or Shank, homer and GAKP, when mixed, can phase separate (Zeng et al., 2018; Zeng et al., 2016). Resulting condensates can recruit other PSD-associated proteins and cluster glutamatergic receptors (Zeng et al., 2018). Evidently, Rapsn alone is able to phase separate to form droplets, through multivalent binding of TPR domain. A TPR motif is composed of 34 amino acids that could mediate oligomerization of TPR-containing proteins (Blatch and Lassle, 1999; Zeytuni and Zarivach, 2012). Indeed, Rapsn self-associates through TPRs (Ramarao et al., 2001; Xing et al., 2019). We show that two or more of the TPRs except TPR6–7 can interact with one another or self-associate; such multivalent binding is critical to Rapsn LLPS. TPR domains exist in many proteins critical to neural development including FKBP52 for neuronal differentiation (Quinta and Galigniana, 2012), postsynaptic scaffold proteins of the TANC family that bind PSD95 (Han et al., 2010), and TRIP8b, an adaptor protein for channel trafficking (Lewis et al., 2011). Our results suggest that LLPS may be involved in many aspects of neuronal functions.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Lin Mei (lin.mei@case.edu).
Materials availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and code availability
The data supporting the current study are available from the Lead Contact on request. This study did not generate source code.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Rapsn null mutant mice were kindly provided by Dr. Peter Noakes (Gautam et al., 1995). The R164H mutant mice were generated by the transgenic and targeting facility of Case Western Reserve University using CRISPR-Cas9. Briefly, donor DNA, AGGCC CTGCG CTATG CCCAC AACAA CGATG ACACC ATGCT GGAGT GTCAC GTCTG CTGCA GCCTG GGCAG TTTCT ACGCC CAGGT CAAGG TGGGC CTGGT, was synthesized and PAGE purified by Integrated DNA Technologies. Cas9 protein (CP01–20) and sgRNA containing targeting sequence, TGCCC AGGCT GCAGC AGACA, were from PNA bio. Mixture containing Cas9 protein (100 ng / μl), sgRNA (50 ng / μl), and donor DNA (100 ng / μl) were injected into C57BL/6J fertilized eggs and the survival two-cell stage embryos were transferred to pseudo-pregnant C57BL/6J females. Mice carrying R164H mutation were screened by PCR analysis. Primers: 5’- ACA CCA TGC TGG AGT GCC GT −3’ and 5’- TTC TCA GGG AGC CTC AAA TC −3’ were used to verify WT genomic DNA, and primers: 5’- AGG CTG CAG CAG ACG TGA −3’ and 5’- ATG GGC AAT GCT TTC CTG GG −3’ were used to verify R164H mutant genomic DNA. Mice were housed in cages in a room with 12 hr light-dark cycle with ad libitum access to water and rodent chow diet (Diet P3000). Animal protocols have been approved by the Institutional Animal Care and Use Committee of Case Western Reserve University.
METHOD DETAILS
Protein expression, purification, and fluorescence labeling
Constructs for protein expression were generated through standard molecular cloning methods. Rapsn and truncation mutants, α-actinin, HSP90β, β-catenin, MACF1, actin, EGFP and calpain were cloned into modified pCDNA3.1 backbone containing Myc tag, His tag and a home-made thrombin digestion site, and transfected into HEK293T cells by polyethylenimine transfection (PEI, linear MW 25,000, Polysciences, Cat#: 23966–1) (Zhang et al., 2012; Zhang et al., 2019). Three days after transfection, cells were harvested and sonicated in buffer containing 500 mM NaCl, 25 mM Tris, pH 7.4, 5 mM DTT, protease inhibitors (Complete EDTA-free, Sigma, Cat#: 11873580001), and phosphatase inhibitors (PhosSTOP, Sigma, Cat#: 4906845001). Lysates were centrifuged at 4°C, 14,000 x g for 30 min, and supernatants were collected and centrifuged again at 4°C, 14,000 x g for 30 min. They were incubated with Ni-NTA resins for 4 hr at 4°C; resins were washed with same buffer for three times and then incubated with the protease thrombin (working condition: 1 U protease thrombin in 1 X cleavage buffer containing 20 mM sodium citrate, pH6.5, 20 mM NaCl, 0.01% PEG-8000, and 5% glycerol) to digest the tags and to elute conjugated proteins. Eluted proteins were further purified by size exclusion chromatography. For expression and purification of AChR-α, AChR-β, plectin1f, β-dys, respective cDNAs were cloned into pGEX-6P-1 vector with a GST tag. Escherichia coli BL21 cells were transformed with respective constructs and cultured in Lysogeny broth (LB) medium (supplied with ampicillin) at 18°C overnight. After the addition of 0.1 mM IPTG (final concentration) to induce protein expression, bacteria were cultured for another 9 hr, harvested and incubated with lysozyme in PBS for 20 min. Bacteria were lysed by sonication with 50 % output and lysates were incubated with glutathione beads at 4°C for 1 hr. Beads were washed three times in PBS buffer and bound proteins were eluted by reduced glutathione and further purified by size exclusion chromatography in buffer containing 500 mM NaCl, 25 mM Tris (pH 7.4), and 5 mM DTT. Some proteins were labeled with CF555 tag using a kit (Mix-n-Stain™ CF™ 555, Sigma, MX555S100; Mix-n-Stain™ CF™ 633, Sigma, MX633S100) as described by the manufacturer.
Phase separation of proteins in vitro and imaging
Purified proteins were stored in high salt buffer (500 mM NaCl, 25 mM tris, pH 7.4, 5 mM DTT), and diluted into the physiological salt buffer (150 mM NaCl, 25 mM tris, pH 7.4, 5 mM DTT) to examine LLPS at room temperature. For co-condensation assay, 5 μM CF555 labeled cargo proteins were incubated with equimolar Rapsn-EGFP, and recruitment of cargo proteins was indicated by enrichment of CF555 signal into Rapsn droplets. For imaging droplets or condensates, diluted proteins were loaded on the glass bottom of culture dishes (35mm Dish, 14mm Glass diameter, uncoated; MatTek, P35G-1.5–14-C), and covered with a cover slide to reduce solution evaporation. Images were collected by confocal Zeiss LSM 810 using a 60 X oil objective.
Co-immunoprecipitation assay
The procedures for co-immunoprecipitation were performed as described previously (Zhang et al., 2008). Briefly, proteins were expressed in HEK293T cells by PEI transfection. 48 hr after transfection, cells were washed once with ice-cold PBS and lysed in buffer containing 5% glycerol, 150 mM NaCl, 25 mM Tris, pH7.4, 1% Triton X-100, protease inhibitors and phosphatase inhibitors. Lysates were centrifuged at 14,000g for 20 min at 4 °C, and the supernatants were incubated with beads overnight. Beads were pelleted by centrifugation at 4,000 x g for 1 min, 4 °C, and washed with a buffer containing 5% glycerol, 150 mM NaCl, 25 mM Tris, pH7.4, 0.2% Triton X-100, protease inhibitors and phosphatase inhibitors for three times. Precipitated proteins were eluted by 1 X SDS boiling and detected by western blotting.
FRAP assay
Fluorescence signals were bleached using 100% intensity laser beam. Because fluorescence decays during image collection, two droplets or puncta were selected for single experiment. One was bleached and the other one was not bleached and acted as experimental controls. The ratio of bleached area / non-bleached control was calculated at different time points. The pre-bleach signal was normalized to 100 %. For FRAP analysis of in vitro droplets, all data were collected with 20–40 min after proteins diluted in the physiological buffer to eliminate the possibility that aged droplets affect fluorescence recovery. For FRAP of AChR clusters in live muscles, muscles with tendons were isolated in oxygenated Ringer’s solution (137 mM NaCl, 5 mM KCl, 12 mM NaHCO3,1mM NaH2PO4,1mM MgCl2, 2mM CaCl2, 11 mM D-glucose, pH 7.3, perfused with 95% O2 and 5% CO2) and mounted on culture dishes with glass bottom for live imaging. FRAP assay was performed on Zeiss LSM 810 confocal.
Immunofluorescence staining
The procedures for immunostaining diaphragms were described previously (Xing et al., 2019; Zhao et al., 2018). Briefly, embryonic or P0 pups were sacrificed and fixed in 4% paraformaldehyde in PBS for 24 hr, and then diaphragms were dissected and rinsed in 0.1 M glycine in PBS for 1hr, room temperature, followed by three-time washing in 0.5% PBST (0.5% Triton X-100 in PBS). Samples were then blocked in the blocking buffer (5% BSA, 5% goat serum, 0.5% Triton X-100 in PBS) for 1 hr, room temperature, and incubated with primary antibodies diluted in blocking buffer, 4 °C, overnight. Next day, samples were washed three times in washing buffer (0.5% Triton X-100 in PBS), and then incubated with fluorescence-labeled secondary antibodies and Flour-594 conjugated α-Bungarotoxin diluted in blocking buffer, room temperature, for 1–2 hr. After that, samples were washed and mounted with Vectashield mounting medium (H1000, Vector Laboratories, Burlingame, CA). Images were collected with LSM 810.
Live cell imaging and quantification of Rapsn-EGFP concentration in cells
For live cell imaging, HEK293T or C2C12 cells were cultured on 35mm dish with glass bottom plate (35 mm Dish, 14 mm Glass diameter, collagen coated; MatTek, P35GCOL-0–14-C), and transfected with Rapsn-EGFP or Rapsn-mCherry. For HEK293T cells, 12 hr after transfection, images were taken with confocal Zeiss LSM 810, supplemented with incubator with controllable temperature and CO2 concentration (37 °C, 5% CO2). For C2C12 cells, after differentiation into myotubes, agrin was added to culture cells and images were taken simultaneously. To quantify Rapsn-EGFP concentration in HEK293T cells, the pixels of purified Rapsn-EGFP were calibrated using fluorescence microscopy and established the linear fitting of protein concentration and mean pixel intensity values. Occasionally, Rapsn became aggregated in ER or Golgi in proximal regions of nuclei. These aggregates were usually large and not associated to the plasma membrane. Cells with such aggregates were excluded from analysis. To avoid the potential impact of mitosis-associated cytoskeletal alteration on Rapsn LLPS, aggregates in mitotic cells were also excluded from analysis. Z-stack images were taken for all cells under same settings, and cell volumes were measured by imageJ plugins Volumest. The overall pixels were calculated by a sum-projection. The mean intensity was calculated as the ratio of overall pixel / cell volume.
Isolation of phase-separated condensates from aqueous phase
Proteins were stored in the buffer containing 500 mM NaCl, 25 mM tris, pH 7.4, 5 mM DTT, and pre-cleared via centrifugation at 14,000 x g for 1 min before using. To isolate phase-separated pellets with aqueous phase, proteins were diluted into low salt buffer (150 mM NaCl, 25 mM tris, pH 7.4, 5 mM DTT) with designed concentrations and combinations, and after incubation for 20 min at room temperature, samples were subjected to centrifugation at 14,000 x g for 15 min at room temperature. The aqueous phase/supernatant (designed as S) was transferred to a new tube and condensed phase/pellet were washed once with low salt buffer and re-suspended with half volume of supernatant fraction of high salt buffer (designed as P). Proteins in both S and P fractions were resolved by SDS-PAGE and analyzed by silver staining. Band intensities were quantified by ImageJ software.
Silver staining
The procedures for silver staining were performed as described previously (Shevchenko et al., 1996). Briefly, proteins were separated by SDS-PAGE (10% or 12 %), and gels were fixed in solution containing 40% ethanol, 10% acetic acid in water for 30 min, and fixed gel was rinsed with 50% methanol for 10 min, followed by water washing for 10 min. The gel was then incubated with the sensitizing solution containing 0.02% Na2S2O3 for 1 min, and rinsed in water for 2 X 1 min. Sensitized gel was then incubated in 0.1% silver nitrate solution for 20 min at 4 °C. After that, gels were rinsed in water for 2 X 1 min and incubated in developing solution containing 2% Na2CO3 and 0.04% formalin to visualize protein bands.
CK2 and Src kinase assay
The procedures for CK2 and Src kinase assay were described by the manufacturers (CK2, New England Biolabs, Cat#: P6010S; Src, Cell signaling technology, Cat#: 7496).
Real time PCR
Total RNA was purified from mouse muscles using TRIzol (Thermo Fisher Scientific, 15596026) (Rio et al., 2010), and was measured and reverse transcribed to cDNAs according to manufacturer’s protocol (Promega, GoScript reverse transcription kit, A2801) with Oligo dT primers. A 20 μl reaction system containing SYBR GreenER qPCR mix, gene-specific primers and cDNA was used to perform Real time PCR in StepOnePlus™ Real-Time PCR System (Thermo Fisher Scientific, 4376600) using 2hr cycling procedure, 95°C (10min), followed by 40 cycles of 95°C (15 s)- 60°C (60 s). Following primers were used. 5’-GGC AGG ACC AGA CAA AGC AA-3’ and 5’-CGAGTGAGCTGTTACCAAGCA-3’ for Rapsn; 5’-AAG GTC ATC CCA GAG CTG AA-3’ and 5’-CTG CTT CAC CAC CTT CTT GA-3’ for GAPDH.
Statistical analysis
Data were presented as mean ± SEM, unless otherwise indicated. Statistical analysis was performed by Unpaired Student T-test, One-way ANOVA or Two-way ANOVA. Statistical difference was considered when P < 0.05.
Supplementary Material
Video showing in vitro Rapsn droplet formation (5 μM), Related to Figure 1. Arrow, droplet fusion. Scale bar, 10 μm.
Video showing FRAP analysis of a Rapsn droplet (white arrow), Related to Figure 1. Scale bar, 5 μm.
Video showing FRAP analysis of a part of Rapsn droplet (white arrow), Related to Figure 1. Scale bar, 5 μm.
Video showing FRAP analysis of a Rapsn puncta (white arrow) and fusion of puncta (red arrow) in HEK293T cell, Related to Figure 2. Scale bar, 5 μm.
Video showing the fluorescence recovery of a part of photobleached spontaneous Rapsn-EGFP cluster (white arrow) in myotubes, Related to Figure 3. The cluster was photobleached before the video started.
Video showing the fluorescence recovery of a part of photobleached agrin-induced Rapsn-EGFP cluster (white arrow) in myotubes, Related to Figure 3.
Video showing the fluorescence recovery of a photobleached Rapsn-EGFP cluster (white arrow) in myotubes, Related to Figure 3.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-GFP | Cell signaling technology | Cat#: 2555; RRID: AB_10692764 |
| Anti-p-Tyr-100 | Cell signaling technology | Cat #: 9411; RRID: AB_331228 |
| Anti-p-Thr/Ser | Cell signaling technology | Cat #: 9631; RRID: AB_330308 |
| Anti-Actin | Cell signaling technology | Cat #: 4967; RRID: AB_330288 |
| Anti-synapsin | Cell signaling technology | Cat #: 5297; RRID: AB_2616578 |
| Anti-neurofilament | Cell signaling technology | Cat #: 2837S; RRID: AB_823575 |
| Anti-Flag | Sigma | Cat #: F7425; |
| Anti-HA | Sigma | Cat#: H6908; RRID: AB_260070 |
| Anti-Myc | Sigma | Cat#: C3956; RRID: AB_439680 |
| Mouse anti-Flag M2 affinity gel | Sigma | Cat #: A2220; RRID: AB_10063035 |
| Anti-GAPDH | Santa Cruz | Cat #: sc-32233; RRID: AB_627679 |
| Mouse monoclonal anti-HA Agarose | Thermo Fisher Scientific | Cat#: 26181; RRID: AB_2537081 |
| Mouse monoclonal anti-Myc beads | Thermo Fisher Scientific | Cat#: 20169; RRID: AB_2537081 |
| Flour-594 conjugated α-Bungarotoxin | Thermo Fisher Scientific | Cat #: B13423 |
| Chemicals, peptides, and recombinant proteins | ||
| Agrin | R and D Systems | Cat#: 550-AG-100 |
| Rapsyn-EGFP (Rapsyn: aa 1M-412V, UniProt: P12672) | This paper | N/A (Custom-made) |
| Rapsyn (aa 1M-412V, UniProt: P12672) | This paper | N/A (Custom-made) |
| Rapsyn TPR1–2-EGFP (TPR1–2: aa 1M-76L. Uniprot: P12672) | This paper | N/A (Custom-made) |
| Rapsyn TPR 3–4-EGFP (TPR3–4: aa 77E-158D. UniProt: P12672) | This paper | N/A (Custom-made) |
| Rapsyn TPR 5–7-EGFP (TPR5–7: aa 159T-286V. UniProt: P12672) | This paper | N/A (Custom-made) |
| Rapsyn CC-RING-EGFP (TPR5–7: aa 296W-412V. UniProt: P12672) | This paper | N/A (Custom-made) |
| Rapsyn TPR1–4-EGFP (TPR1–4: aa 1M-158D. UniProt: P12672) | This paper | N/A (Custom-made) |
| Rapsyn TPR1–7-EGFP (TPR1–7: aa 1M-286V. UniProt: P12672) | This paper | N/A (Custom-made) |
| α-actinin2 (aa, 1M-894L, UniProt: Q9JI91) | This paper | N/A (Custom-made) |
| Calpain 2 (aa 1M-700L. UniProt: O085029) | This paper | N/A (Custom-made) |
| Actin, α1 (aa 1M-377F. UniProt: P68134) | This paper | N/A (Custom-made) |
| GST-AChR-α (aa 317N-428H UniProt: P04756) | This paper | N/A (Custom-made) |
| GST-AChR-β (aa 333H-469R. UniProt: P09690) | This paper | N/A (Custom-made) |
| GST-MACF1 (aa 1M-293I. UniProt: Q9QXZ0) | This paper | N/A (Custom-made) |
| GST-β-dystroglycan (aa 773Y-893P. UniProt: Q62165) | This paper | N/A (Custom-made) |
| Q3K-EGFP | This paper | N/A (Custom-made) |
| L14P-EGFP | This paper | N/A (Custom-made) |
| A73D-EGFP | This paper | N/A (Custom-made) |
| N88K-EGFP | This paper | N/A (Custom-made) |
| R91L-EGFP | This paper | N/A (Custom-made) |
| E147K-EGFP | This paper | N/A (Custom-made) |
| R164H-EGFP | This paper | N/A (Custom-made) |
| L169P-EGFP | This paper | N/A (Custom-made) |
| R242W-EGFP | This paper | N/A (Custom-made) |
| L283P-EGFP | This paper | N/A (Custom-made) |
| L326P-EGFP | This paper | N/A (Custom-made) |
| C366A-EGFP | This paper | N/A (Custom-made) |
| 1172del2-EGFP | This paper | N/A (Custom-made) |
| Y86F-EGFP | This paper | N/A (Custom-made) |
| EGFP | This paper | N/A (Custom-made) |
| Critical commercial assays | ||
| Mix-n-Stain™ CF™ 633 Antibody Labeling Kit (50–100μg) | Sigma | Cat#: MX633S100 |
| Mix-n-Stain™ CF™ 555 Antibody Labeling Kit (50–100μg) | Sigma | Cat#: MX555S100 |
| Thrombin protease | Sigma | Cat#: GE27–0846-01 |
| CK2 Kinase assay | New England Biolabs | Cat#: P6010S |
| Lambda protein phosphatase | New England Biolabs | Cat#: P0753S |
| Src Kinase assay | Cell signaling technology | Cat#: 7496 |
| Experimental models: cell lines | ||
| HEK293T | ATCC | Cat#: CRL-3216; RRID: CVCL_0063 |
| C2C12 | ATCC | Cat#: CRL-1772; RRID: CVCL_0188 |
| Rapsyn-/- myoblast | PMID: 10414969 | |
| Experimental models: organisms/strains | ||
| Rapsyn mutant mice (rapsyn -/-) | PMID: 7675108 | |
| R164H | This paper | N/A (Custom-made) |
| Recombinant DNA | ||
| pGEX-6P-1 | Sigma | Cat#: GE28–9546-48 |
| pGEX-6P-1-AChR-α (aa 317–428) | This paper | N/A (Custom-made) |
| pGEX-6P-1-AChR-β (aa 333–469) | This paper | N/A (Custom-made) |
| pGEX-6P-1-MACF1 (aa 1–293) | This paper | N/A (Custom-made) |
| pGEX-6P-1-β-dystroglycan (aa 773–893) | This paper | N/A (Custom-made) |
| pEGX-6P-1-LRP4 (aa 1748–1905) | This paper | N/A (Custom-made) |
| pCDNA3.1-rapsyn | This paper | N/A (Custom-made) |
| pCDNA3.1-TPR1–2 | This paper | N/A (Custom-made) |
| pCDNA3.1-TPR3–4 | This paper | N/A (Custom-made) |
| pCDNA3.1-TPR5–7 | This paper | N/A (Custom-made) |
| pCDNA3.1-TPR1–4 | This paper | N/A (Custom-made) |
| pCDNA3.1-TPR1–7 | This paper | N/A (Custom-made) |
| pCDNA3.1-CC-RING | This paper | N/A (Custom-made) |
| pCDNA3.1-TPR2–3 | This paper | N/A (Custom-made) |
| pCDNA3.1-TPR4–5 | This paper | N/A (Custom-made) |
| pCDNA3.1-TPR6–7 | This paper | N/A (Custom-made) |
| pCDNA3.1-TPR5–6 | This paper | N/A (Custom-made) |
| pEGFP-N1-rapsyn | This paper | N/A (Custom-made) |
| pEGFP-N1-TPR1–2 | This paper | N/A (Custom-made) |
| pEGFP-N1-TPR3–4 | This paper | N/A (Custom-made) |
| pEGFP-N1-TPR5–7 | This paper | N/A (Custom-made) |
| pEGFP-N1-TPR1–4 | This paper | N/A (Custom-made) |
| pEGFP-N1-TPR1–7 | This paper | N/A (Custom-made) |
| pEGFP-N1-CC-RING | This paper | N/A (Custom-made) |
| pEGFP-N1-TPR2–3 | This paper | N/A (Custom-made) |
| pEGFP-N1-TPR4–5 | This paper | N/A (Custom-made) |
| pEGFP-N1-TPR6–7 | This paper | N/A (Custom-made) |
| pEGFP-N1-TPR5–6 | This paper | N/A (Custom-made) |
| Software and algorithms | ||
| ImageJ | National Institutes of Health | https://imagej.nih.gov/ij/download.html |
| Zen Software | Zeiss | https://www.zeiss.com/microscopy/us/products/microscope-software/zen-lite.html |
| Adobe illustrator | Adobe | https://www.adobe.com/products/illustrator/free-trial-download.html |
| Prism 7 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
Highlights.
Rapsn phase separates into dynamic condensates in vitro, in cells and in muscles
Rapsn co-condensation to recruit cargo proteins to membraneless compartments
Rapsn condensates as a signaling platform
Disease mutations impair rapsn condensates & co-condensates, NMJ formation
Acknowledgments
We thank Dr. Rajendra Boggavarapu for helping with purification of proteins; members of Mei/Xiong Lab for discussion. This study was supported by grants from Veterans Affairs and NIH to L.M and W.C.X.
Footnotes
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alberti S, and Dormann D (2019). Liquid-Liquid Phase Separation in Disease. Annual review of genetics 53, 171–194. [DOI] [PubMed] [Google Scholar]
- Antolik C, Catino DH, O’Neill AM, Resneck WG, Ursitti JA, and Bloch RJ (2007). The actin binding domain of ACF7 binds directly to the tetratricopeptide repeat domains of Rapsn. Neuroscience 145, 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai G, Wang Y, and Zhang M (2020). Gephyrin-mediated formation of inhibitory postsynaptic density sheet via phase separation. Cell research. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartoli M, Ramarao MK, and Cohen JB (2001). Interactions of the Rapsn RING-H2 domain with dystroglycan. The Journal of biological chemistry 276, 24911–24917. [DOI] [PubMed] [Google Scholar]
- Beutel O, Maraspini R, Pombo-Garcia K, Martin-Lemaitre C, and Honigmann A (2019). Phase Separation of Zonula Occludens Proteins Drives Formation of Tight Junctions. Cell 179, 923–936 e911. [DOI] [PubMed] [Google Scholar]
- Blatch GL, and Lassle M (1999). The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. BioEssays : news and reviews in molecular, cellular and developmental biology 21, 932–939. [DOI] [PubMed] [Google Scholar]
- Boeynaems S, Alberti S, Fawzi NL, Mittag T, Polymenidou M, Rousseau F, Schymkowitz J, Shorter J, Wolozin B, Van Den Bosch L, et al. (2018). Protein Phase Separation: A New Phase in Cell Biology. Trends in cell biology 28, 420–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boija A, Klein IA, Sabari BR, Dall’Agnese A, Coffey EL, Zamudio AV, Li CH, Shrinivas K, Manteiga JC, Hannett NM, et al. (2018). Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell 175, 1842–1855 e1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappello V, and Francolini M (2017). Neuromuscular Junction Dismantling in Amyotrophic Lateral Sclerosis. International journal of molecular sciences 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Qian L, Yang ZH, Huang Y, Ngo ST, Ruan NJ, Wang J, Schneider C, Noakes PG, Ding YQ, et al. (2007). Rapsn interaction with calpain stabilizes AChR clusters at the neuromuscular junction. Neuron 55, 247–260. [DOI] [PubMed] [Google Scholar]
- Chen X, Wu X, Wu H, and Zhang M (2020). Phase separation at the synapse. Nature neuroscience 23, 301–310. [DOI] [PubMed] [Google Scholar]
- Cheusova T, Khan MA, Schubert SW, Gavin AC, Buchou T, Jacob G, Sticht H, Allende J, Boldyreff B, Brenner HR, et al. (2006). Casein kinase 2-dependent serine phosphorylation of Musk regulates acetylcholine receptor aggregation at the neuromuscular junction. Genes & development 20, 1800–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KR, Berrera M, Reischl M, Strack S, Albrizio M, Roder IV, Wagner A, Petersen Y, Hafner M, Zaccolo M, et al. (2012). Rapsn mediates subsynaptic anchoring of PKA type I and stabilisation of acetylcholine receptor in vivo. Journal of cell science 125, 714–723. [DOI] [PubMed] [Google Scholar]
- Cossins J, Burke G, Maxwell S, Spearman H, Man S, Kuks J, Vincent A, Palace J, Fuhrer C, and Beeson D (2006). Diverse molecular mechanisms involved in AChR deficiency due to Rapsn mutations. Brain : a journal of neurology 129, 2773–2783. [DOI] [PubMed] [Google Scholar]
- DeChiara TM, Bowen DC, Valenzuela DM, Simmons MV, Poueymirou WT, Thomas S, Kinetz E, Compton DL, Rojas E, Park JS, et al. (1996). The receptor tyrosine kinase Musk is required for neuromuscular junction formation in vivo. Cell 85, 501–512. [DOI] [PubMed] [Google Scholar]
- Dobbins GC, Luo S, Yang Z, Xiong WC, and Mei L (2008). alpha-Actinin interacts with Rapsn in agrin-stimulated AChR clustering. Molecular brain 1, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiber N, Rehman M, Kravic B, Rudolf R, Sandri M, and Hashemolhosseini S (2019). Loss of Protein Kinase Csnk2b/CK2beta at Neuromuscular Junctions Affects Morphology and Dynamics of Aggregated Nicotinic Acetylcholine Receptors, Neuromuscular Transmission, and Synaptic Gene Expression. Cells 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel AG, Shen XM, Selcen D, and Sine SM (2015). Congenital myasthenic syndromes: pathogenesis, diagnosis, and treatment. The Lancet Neurology 14, 420–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fambrough DM, Drachman DB, and Satyamurti S (1973). Neuromuscular junction in myasthenia gravis: decreased acetylcholine receptors. Science 182, 293–295. [DOI] [PubMed] [Google Scholar]
- Feng Z, Chen X, Zeng M, and Zhang M (2019). Phase separation as a mechanism for assembling dynamic postsynaptic density signalling complexes. Current opinion in neurobiology 57, 1–8. [DOI] [PubMed] [Google Scholar]
- Fertuck HC, and Salpeter MM (1976). Quantitation of junctional and extrajunctional acetylcholine receptors by electron microscope autoradiography after 125I-alpha-bungarotoxin binding at mouse neuromuscular junctions. The Journal of cell biology 69, 144–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan-Steet H, Fox MA, Meyer D, and Sanes JR (2005). Neuromuscular synapses can form in vivo by incorporation of initially aneural postsynaptic specializations. Development 132, 4471–4481. [DOI] [PubMed] [Google Scholar]
- Frail DE, McLaughlin LL, Mudd J, and Merlie JP (1988). Identification of the mouse muscle 43,000-dalton acetylcholine receptor-associated protein (Rapsn) by cDNA cloning. The Journal of biological chemistry 263, 15602–15607. [PubMed] [Google Scholar]
- Froehner SC, Luetje CW, Scotland PB, and Patrick J (1990). The postsynaptic 43K protein clusters muscle nicotinic acetylcholine receptors in Xenopus oocytes. Neuron 5, 403–410. [DOI] [PubMed] [Google Scholar]
- Fuhrer C, Gautam M, Sugiyama JE, and Hall ZW (1999). Roles of Rapsn and agrin in interaction of postsynaptic proteins with acetylcholine receptors. The Journal of neuroscience : the official journal of the Society for Neuroscience 19, 6405–6416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautam M, Noakes PG, Mudd J, Nichol M, Chu GC, Sanes JR, and Merlie JP (1995). Failure of postsynaptic specialization to develop at neuromuscular junctions of Rapsn-deficient mice. Nature 377, 232–236. [DOI] [PubMed] [Google Scholar]
- Gilhus NE, Tzartos S, Evoli A, Palace J, Burns TM, and Verschuuren J (2019). Myasthenia gravis. Nature reviews Disease primers 5, 30. [DOI] [PubMed] [Google Scholar]
- Glass DJ, Bowen DC, Stitt TN, Radziejewski C, Bruno J, Ryan TE, Gies DR, Shah S, Mattsson K, Burden SJ, et al. (1996). Agrin acts via a Musk receptor complex. Cell 85, 513–523. [DOI] [PubMed] [Google Scholar]
- Guo C, Che Z, Yue J, Xie P, Hao S, Xie W, Luo Z, and Lin C (2020). ENL initiates multivalent phase separation of the super elongation complex (SEC) in controlling rapid transcriptional activation. Science advances 6, eaay4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Noakes PG, and Phillips WD (1999). Overexpression of Rapsn inhibits agrin-induced acetylcholine receptor clustering in muscle cells. Journal of neurocytology 28, 763–775. [DOI] [PubMed] [Google Scholar]
- Han S, Nam J, Li Y, Kim S, Cho SH, Cho YS, Choi SY, Choi J, Han K, Kim Y, et al. (2010). Regulation of dendritic spines, spatial memory, and embryonic development by the TANC family of PSD-95-interacting proteins. The Journal of neuroscience : the official journal of the Society for Neuroscience 30, 15102–15112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Vega A, Braun M, Scharrel L, Jahnel M, Wegmann S, Hyman BT, Alberti S, Diez S, and Hyman AA (2017). Local Nucleation of Microtubule Bundles through Tubulin Concentration into a Condensed Tau Phase. Cell reports 20, 2304–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann D, Straubinger M, and Hashemolhosseini S (2015). Protein kinase CK2 interacts at the neuromuscular synapse with Rapsn, Rac1, 14–3-3gamma, and Dok-7 proteins and phosphorylates the latter two. The Journal of biological chemistry 290, 22370–22384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofweber M, Hutten S, Bourgeois B, Spreitzer E, Niedner-Boblenz A, Schifferer M, Ruepp MD, Simons M, Niessing D, Madl T, et al. (2018). Phase Separation of FUS Is Suppressed by Its Nuclear Import Receptor and Arginine Methylation. Cell 173, 706–719 e713. [DOI] [PubMed] [Google Scholar]
- Jennings CG, Dyer SM, and Burden SJ (1993). Muscle-specific trk-related receptor with a kringle domain defines a distinct class of receptor tyrosine kinases. Proceedings of the National Academy of Sciences of the United States of America 90, 2895–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Stiegler AL, Cameron TO, Hallock PT, Gomez AM, Huang JH, Hubbard SR, Dustin ML, and Burden SJ (2008). Lrp4 is a receptor for Agrin and forms a complex with Musk. Cell 135, 334–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Rudell J, and Ferns M (2009). Rapsn interacts with the muscle acetylcholine receptor via alpha-helical domains in the alpha, beta, and epsilon subunit intracellular loops. Neuroscience 163, 222–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Rudell J, Yechikhov S, Taylor R, Swope S, and Ferns M (2008). Rapsn carboxyl terminal domains mediate muscle specific kinase-induced phosphorylation of the muscle acetylcholine receptor. Neuroscience 153, 997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis AS, Vaidya SP, Blaiss CA, Liu Z, Stoub TR, Brager DH, Chen X, Bender RA, Estep CM, Popov AB, et al. (2011). Deletion of the hyperpolarization-activated cyclic nucleotide-gated channel auxiliary subunit TRIP8b impairs hippocampal Ih localization and function and promotes antidepressant behavior in mice. The Journal of neuroscience : the official journal of the Society for Neuroscience 31, 7424–7440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Cao Y, Wu H, Ye X, Zhu Z, Xing G, Shen C, Barik A, Zhang B, Xie X, et al. (2016). Enzymatic Activity of the Scaffold Protein Rapsn for Synapse Formation. Neuron 92, 1007–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Xiong WC, and Mei L (2018). Neuromuscular Junction Formation, Aging, and Disorders. Annual review of physiology 80, 159–188. [DOI] [PubMed] [Google Scholar]
- Lin W, Burgess RW, Dominguez B, Pfaff SL, Sanes JR, and Lee KF (2001). Distinct roles of nerve and muscle in postsynaptic differentiation of the neuromuscular synapse. Nature 410, 1057–1064. [DOI] [PubMed] [Google Scholar]
- Lin W, Dominguez B, Yang J, Aryal P, Brandon EP, Gage FH, and Lee KF (2005). Neurotransmitter acetylcholine negatively regulates neuromuscular synapse formation by a Cdk5-dependent mechanism. Neuron 46, 569–579. [DOI] [PubMed] [Google Scholar]
- Liu Y, Padgett D, Takahashi M, Li H, Sayeed A, Teichert RW, Olivera BM, McArdle JJ, Green WN, and Lin W (2008). Essential roles of the acetylcholine receptor gamma-subunit in neuromuscular synaptic patterning. Development 135, 1957–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, Zhang B, Dong XP, Tao Y, Ting A, Zhou Z, Meixiong J, Luo J, Chiu FC, Xiong WC, et al. (2008). HSP90 beta regulates Rapsn turnover and subsequent AChR cluster formation and maintenance. Neuron 60, 97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maselli R, Dris H, Schnier J, Cockrell J, and Wollmann R (2007). Congenital myasthenic syndrome caused by two non-N88K Rapsn mutations. Clinical genetics 72, 63–65. [DOI] [PubMed] [Google Scholar]
- Maselli RA, Dunne V, Pascual-Pascual SI, Bowe C, Agius M, Frank R, and Wollmann RL (2003). Rapsn mutations in myasthenic syndrome due to impaired receptor clustering. Muscle & nerve 28, 293–301. [DOI] [PubMed] [Google Scholar]
- McDonald NA, Fetter RD, and Shen K (2020). Assembly of synaptic active zones requires phase separation of scaffold molecules. Nature 588, 454–458. [DOI] [PubMed] [Google Scholar]
- McMahan UJ (1990). The agrin hypothesis. Cold Spring Harbor symposia on quantitative biology 55, 407–418. [DOI] [PubMed] [Google Scholar]
- Milone M, Shen XM, Selcen D, Ohno K, Brengman J, Iannaccone ST, Harper CM, and Engel AG (2009). Myasthenic syndrome due to defects in Rapsn: Clinical and molecular findings in 39 patients. Neurology 73, 228–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milovanovic D, Wu Y, Bian X, and De Camilli P (2018). A liquid phase of synapsin and lipid vesicles. Science 361, 604–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misgeld T, Kummer TT, Lichtman JW, and Sanes JR (2005). Agrin promotes synaptic differentiation by counteracting an inhibitory effect of neurotransmitter. Proceedings of the National Academy of Sciences of the United States of America 102, 11088–11093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, and Taylor JP (2015). Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami T, Qamar S, Lin JQ, Schierle GS, Rees E, Miyashita A, Costa AR, Dodd RB, Chan FT, Michel CH, et al. (2015). ALS/FTD Mutation-Induced Phase Transition of FUS Liquid Droplets and Reversible Hydrogels into Irreversible Hydrogels Impairs RNP Granule Function. Neuron 88, 678–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubig RR, and Cohen JB (1979). Equilibrium binding of [3H]tubocurarine and [3H]acetylcholine by Torpedo postsynaptic membranes: stoichiometry and ligand interactions. Biochemistry 18, 5464–5475. [DOI] [PubMed] [Google Scholar]
- Ohno K, Engel AG, Shen XM, Selcen D, Brengman J, Harper CM, Tsujino A, and Milone M (2002). Rapsn mutations in humans cause endplate acetylcholine-receptor deficiency and myasthenic syndrome. American journal of human genetics 70, 875–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno K, Sadeh M, Blatt I, Brengman JM, and Engel AG (2003). E-box mutations in the RAPSN promoter region in eight cases with congenital myasthenic syndrome. Human molecular genetics 12, 739–748. [DOI] [PubMed] [Google Scholar]
- Okada K, Inoue A, Okada M, Murata Y, Kakuta S, Jigami T, Kubo S, Shiraishi H, Eguchi K, Motomura M, et al. (2006). The muscle protein Dok-7 is essential for neuromuscular synaptogenesis. Science 312, 1802–1805. [DOI] [PubMed] [Google Scholar]
- Oury J, Liu Y, Topf A, Todorovic S, Hoedt E, Preethish-Kumar V, Neubert TA, Lin W, Lochmuller H, and Burden SJ (2019). MACF1 links Rapsn to microtubule- and actin-binding proteins to maintain neuromuscular synapses. The Journal of cell biology 218, 1686–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riba A, and Itzhaki LS (2019). The tetratricopeptide-repeat motif is a versatile platform that enables diverse modes of molecular recognition. Current opinion in structural biology 54, 43–49. [DOI] [PubMed] [Google Scholar]
- Phillips WD, Kopta C, Blount P, Gardner PD, Steinbach JH, and Merlie JP (1991). ACh receptor-rich membrane domains organized in fibroblasts by recombinant 43-kildalton protein. Science 251, 568–570. [DOI] [PubMed] [Google Scholar]
- Quinta HR, and Galigniana MD (2012). The neuroregenerative mechanism mediated by the Hsp90-binding immunophilin FKBP52 resembles the early steps of neuronal differentiation. British journal of pharmacology 166, 637–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramarao MK, Bianchetta MJ, Lanken J, and Cohen JB (2001). Role of Rapsn tetratricopeptide repeat and coiled-coil domains in self-association and nicotinic acetylcholine receptor clustering. The Journal of biological chemistry 276, 7475–7483. [DOI] [PubMed] [Google Scholar]
- Rio DC, Ares M Jr., Hannon GJ, and Nilsen TW (2010). Purification of RNA using TRIzol (TRI reagent). Cold Spring Harbor protocols 2010, pdb prot5439. [DOI] [PubMed] [Google Scholar]
- Roy A, Kucukural A, and Zhang Y (2010). I-TASSER: a unified platform for automated protein structure and function prediction. Nature protocols 5, 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes JR, and Lichtman JW (1999). Development of the vertebrate neuromuscular junction. Annual review of neuroscience 22, 389–442. [DOI] [PubMed] [Google Scholar]
- Schwayer C, Shamipour S, Pranjic-Ferscha K, Schauer A, Balda M, Tada M, Matter K, and Heisenberg CP (2019). Mechanosensation of Tight Junctions Depends on ZO-1 Phase Separation and Flow. Cell 179, 937–952 e918. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, and Mann M (1996). Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Analytical chemistry 68, 850–858. [DOI] [PubMed] [Google Scholar]
- Shi L, Fu AK, and Ip NY (2012). Molecular mechanisms underlying maturation and maintenance of the vertebrate neuromuscular junction. Trends in neurosciences 35, 441–453. [DOI] [PubMed] [Google Scholar]
- Shin Y, and Brangwynne CP (2017). Liquid phase condensation in cell physiology and disease. Science 357. [DOI] [PubMed] [Google Scholar]
- Sjoblom B, Salmazo A, and Djinovic-Carugo K (2008). Alpha-actinin structure and regulation. Cellular and molecular life sciences : CMLS 65, 2688–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel A, Heidmann T, Hofler J, and Changeux JP (1978). Distinct protein components from Torpedo marmorata membranes carry the acetylcholine receptor site and the binding site for local anesthetics and histrionicotoxin. Proceedings of the National Academy of Sciences of the United States of America 75, 510–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tintignac LA, Brenner HR, and Ruegg MA (2015). Mechanisms Regulating Neuromuscular Junction Development and Function and Causes of Muscle Wasting. Physiological reviews 95, 809–852. [DOI] [PubMed] [Google Scholar]
- Walker JH, Boustead CM, and Witzemann V (1984). The 43-K protein, v1, associated with acetylcholine receptor containing membrane fragments is an actin-binding protein. The EMBO journal 3, 2287–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatherbee SD, Anderson KV, and Niswander LA (2006). LDL-receptor-related protein 4 is crucial for formation of the neuromuscular junction. Development 133, 4993–5000. [DOI] [PubMed] [Google Scholar]
- Wilfling F, Lee CW, Erdmann PS, Zheng Y, Sherpa D, Jentsch S, Pfander B, Schulman BA, and Baumeister W (2020). A Selective Autophagy Pathway for Phase-Separated Endocytic Protein Deposits. Molecular cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff JB, Ferreira Gomes B, Widlund PO, Mahamid J, Honigmann A, and Hyman AA (2017). The Centrosome Is a Selective Condensate that Nucleates Microtubules by Concentrating Tubulin. Cell 169, 1066–1077 e1010. [DOI] [PubMed] [Google Scholar]
- Wu H, Xiong WC, and Mei L (2010). To build a synapse: signaling pathways in neuromuscular junction assembly. Development 137, 1017–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Cai Q, Feng Z, and Zhang M (2020). Liquid-Liquid Phase Separation in Neuronal Development and Synaptic Signaling. Developmental cell 55, 18–29. [DOI] [PubMed] [Google Scholar]
- Wu X, Cai Q, Shen Z, Chen X, Zeng M, Du S, and Zhang M (2019). RIM and RIM-BP Form Presynaptic Active-Zone-like Condensates via Phase Separation. Molecular cell 73, 971–984 e975. [DOI] [PubMed] [Google Scholar]
- Xing G, Jing H, Zhang L, Cao Y, Li L, Zhao K, Dong Z, Chen W, Wang H, Cao R, et al. (2019). A mechanism in agrin signaling revealed by a prevalent Rapsn mutation in congenital myasthenic syndrome. eLife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing G, Xiong WC, and Mei L (2020). Rapsn as a signaling and scaffolding molecule in neuromuscular junction formation and maintenance. Neurosci Lett 731, 135013. [DOI] [PubMed] [Google Scholar]
- Yang X, Arber S, William C, Li L, Tanabe Y, Jessell TM, Birchmeier C, and Burden SJ (2001). Patterning of muscle acetylcholine receptor gene expression in the absence of motor innervation. Neuron 30, 399–410. [DOI] [PubMed] [Google Scholar]
- Yang X, Li W, Prescott ED, Burden SJ, and Wang JC (2000). DNA topoisomerase IIbeta and neural development. Science 287, 131–134. [DOI] [PubMed] [Google Scholar]
- Yoshihara CM, and Hall ZW (1993). Increased expression of the 43-kD protein disrupts acetylcholine receptor clustering in myotubes. The Journal of cell biology 122, 169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng M, Chen X, Guan D, Xu J, Wu H, Tong P, and Zhang M (2018). Reconstituted Postsynaptic Density as a Molecular Platform for Understanding Synapse Formation and Plasticity. Cell 174, 1172–1187 e1116. [DOI] [PubMed] [Google Scholar]
- Zeng M, Diaz-Alonso J, Ye F, Chen X, Xu J, Ji Z, Nicoll RA, and Zhang M (2019). Phase Separation-Mediated TARP/MAGUK Complex Condensation and AMPA Receptor Synaptic Transmission. Neuron 104, 529–543 e526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng M, Shang Y, Araki Y, Guo T, Huganir RL, and Zhang M (2016). Phase Transition in Postsynaptic Densities Underlies Formation of Synaptic Complexes and Synaptic Plasticity. Cell 166, 1163–1175 e1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeytuni N, and Zarivach R (2012). Structural and functional discussion of the tetra-trico-peptide repeat, a protein interaction module. Structure 20, 397–405. [DOI] [PubMed] [Google Scholar]
- Zhang B, Liang C, Bates R, Yin Y, Xiong WC, and Mei L (2012). Wnt proteins regulate acetylcholine receptor clustering in muscle cells. Molecular brain 5, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Luo S, Wang Q, Suzuki T, Xiong WC, and Mei L (2008). LRP4 serves as a coreceptor of agrin. Neuron 60, 285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Wang Z, Du Z, and Zhang H (2018). mTOR Regulates Phase Separation of PGL Granules to Modulate Their Autophagic Degradation. Cell 174, 1492–1506 e1422. [DOI] [PubMed] [Google Scholar]
- Zhang H, Sathyamurthy A, Liu F, Li L, Zhang L, Dong Z, Cui W, Sun X, Zhao K, Wang H, et al. (2019). Agrin-Lrp4-Ror2 signaling regulates adult hippocampal neurogenesis in mice. eLife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Shen C, Li L, Wu H, Xing G, Dong Z, Jing H, Chen W, Zhang H, Tan Z, et al. (2018). Sarcoglycan Alpha Mitigates Neuromuscular Junction Decline in Aged Mice by Stabilizing LRP4. The Journal of neuroscience : the official journal of the Society for Neuroscience 38, 8860–8873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Xie J, Kong W, Xie J, Li Y, Du L, Zheng Q, Sun L, Guan M, Li H, et al. (2020). Phase Separation of Disease-Associated SHP2 Mutants Underlies MAPK Hyperactivation. Cell 183, 490–502 e418. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video showing in vitro Rapsn droplet formation (5 μM), Related to Figure 1. Arrow, droplet fusion. Scale bar, 10 μm.
Video showing FRAP analysis of a Rapsn droplet (white arrow), Related to Figure 1. Scale bar, 5 μm.
Video showing FRAP analysis of a part of Rapsn droplet (white arrow), Related to Figure 1. Scale bar, 5 μm.
Video showing FRAP analysis of a Rapsn puncta (white arrow) and fusion of puncta (red arrow) in HEK293T cell, Related to Figure 2. Scale bar, 5 μm.
Video showing the fluorescence recovery of a part of photobleached spontaneous Rapsn-EGFP cluster (white arrow) in myotubes, Related to Figure 3. The cluster was photobleached before the video started.
Video showing the fluorescence recovery of a part of photobleached agrin-induced Rapsn-EGFP cluster (white arrow) in myotubes, Related to Figure 3.
Video showing the fluorescence recovery of a photobleached Rapsn-EGFP cluster (white arrow) in myotubes, Related to Figure 3.
Data Availability Statement
The data supporting the current study are available from the Lead Contact on request. This study did not generate source code.