Abstract
The value of utilizing a multi-gene pharmacogenetic panel to tailor pharmacotherapy is contingent on the prevalence of prescribed medications with an actionable pharmacogenetic association. The Clinical Pharmacogenetics Implementation Consortium (CPIC) has categorized over 35 gene-drug pairs as ‘Level A’, for which there is sufficiently strong evidence to recommend that genetic information be used to guide drug prescribing. The opportunity to use genetic information to tailor pharmacotherapy among adult patients was determined by elucidating the exposure to CPIC Level A drugs among 11 Implementing Genomics In Practice Network (IGNITE)-affiliated health systems across the U.S. Inpatient and/or outpatient electronic-prescribing data were collected between 1/1/2011 and 12/31/2016 for patients ≥ 18 years of age who had at least one medical encounter that was eligible for drug prescribing in a calendar year. A median of approximately 7.2 million adult patients was available for assessment of drug prescribing per year. From 2011 to 2016, the annual estimated prevalence of exposure to at least one CPIC Level A drug prescribed to unique patients ranged between 15,719 (95% confidence interval [CI]: 15,658–15,781) in 2011 to 17,335 (CI: 17,283–17,386) in 2016 per 100,000 patients. The estimated annual exposure to at least two drugs was above 7,200 per 100,000 patients in most years of the study, reaching an apex of 7,660 (CI: 7,632–7,687) per 100,000 patients in 2014. An estimated 4,748 per 100,000 prescribing events were potentially eligible for a genotype-guided intervention. Results from this study show that a significant portion of adults treated at medical institutions across the U.S. is exposed to medications for which genetic information, if available, should be used to guide prescribing.
Keywords: Pharmacogenetics, Pharmacogenomics, Clinical Pharmacogenetics Implementation Consortium, Drug Exposure
INTRODUCTION
Clinical pharmacogenetics is an important component of precision medicine, with the goal of using genetic information to inform prescribing decisions to maximize drug efficacy and reduce adverse effects.(1) More than 100 commercially-available drugs in the U.S. contain pharmacogenetic information in their FDA-approved labeling for which data support therapeutic management recommendations, indicate a potential impact on drug safety or response, or demonstrate potential impact on pharmacokinetic properties.(2, 3) Additionally, the Clinical Pharmacogenetics Implementation Consortium (CPIC) publishes evidence-based, peer-reviewed guidelines for how to translate genetic test results into actionable prescribing decisions for affected drugs.(4–6) CPIC has categorized over 35 gene-drug pairs as ‘Level A,’ for which the preponderance of evidence is sufficiently strong to recommend that genetic information, if available, be used to guide drug prescribing.(5) CPIC Level A drugs span virtually all major therapeutic areas and are routinely prescribed in acute and chronic care settings (e.g., certain antidepressants(7), codeine(8), ondansetron(9), simvastatin(10), tramadol(8), and warfarin(11)). Some CPIC Level A drugs are indicated for less prevalent diseases but are associated with severe and potentially life-threatening reactions (e.g., fluoropyrimidines(12) and thiopurines(13)) that can be predicted in part by pharmacogenetic testing.
Although there are examples of early adopters using single gene or multi-gene panel testing, integration of pharmacogenetics into routine clinical practice is not yet common.(14–17) Adoption has lagged expectations despite several studies demonstrating that pharmacogenetic variants are common, with more than 90% of patients having at least one variant that could impact a drug prescribing event.(18–20) Barriers to implementing multi-gene panel tests include provider knowledge gaps, costs and insurance coverage, inadequate information technology infrastructure, and limited understanding of the clinical and economic impact across health care populations.(21) Importantly, population impact is contingent on the prescribing frequencies of drugs influenced by pharmacogenes across health systems.
The main objective of this study was to systematically evaluate the opportunity to use genetic information to tailor CPIC Level A pharmacotherapy among adult patients across large and diverse health care systems in the United States, thus addressing a pragmatic barrier to the implementation of pharmacogenetics in the clinical setting. We also examined the prescribing patterns of alternatives to selected CPIC Level A drugs (simvastatin, warfarin, and clopidogrel), whose utilization is reported to be in decline thus potentially affecting the utility of pharmacogenetic testing.
METHODS
Setting and Data Collection
The National Institutes of Health-funded IGNITE (Implementing GeNomics In pracTicE) Network was established in 2013 to support the development, investigation, and dissemination of genomic medicine practice models.(22) This longitudinal study of CPIC Level A drug prescribing prevalence was conducted across five IGNITE-funded health systems and six affiliate members participating in the IGNITE Pharmacogenetics Working Group (Supplementary Materials and Table S1).(23) Institutions obtained approval from their respective Institutional Review Boards for data extraction and pooling of de-identified, aggregate data.
Collection of demographic and drug prescribing information was guided by a structured data-dictionary. Each participating site extracted drug prescribing data (Table S2) from e-prescription records per calendar year for patients ≥ 18 years of age in the inpatient and/or outpatient setting between 1/1/2011 and 12/31/2016, or a subset of years depending on data availability. Drug prescribing data were obtained for 47 target drugs; 36 CPIC Level A drugs and 11 alternative medications (Table S2). The number of unique patients who had at least one medical encounter with prescribing potential per calendar year was determined for the purpose of calculating the prevalence of drug exposure across an eligible population. Non-eligible encounters (e.g., physical therapy encounters or anatomical pathology consultations where a medical record number is generated) were excluded to prevent under estimation of drug exposure rates. Drug exposures were determined using prescription data among unique patients and did not include duration of treatment. Medications prescribed ‘as needed’ were included in data analysis. A data collation script was executed at all sites to standardize the aggregation of de-identified patient level prescribing data. The estimated prevalence of actionable exposures was determined as described in the Supplementary Materials.
Statistical Analysis
For demographic characteristics at each site, summary statistics were calculated on an annual basis and prevalence estimates were summarized as the median across calendar year. Site-specific summaries were combined to obtain overall summaries (Table 1).
Table 1:
Characteristics of patient populations across 11 sites observed from 2011 to 2016
| Number of sites | 11 |
| Academic Medical Centersa | 9 |
| Community Hospitals | 2 |
| Age in years | |
| 25th percentile (Median [range]) | 39 [36.5, 40.8] |
| 50th percentile (Median [range]) | 54.3 [51.5, 55.8] |
| 75th percentile (Median [range]) | 65 [64.1, 66.8] |
| Female sex % (median [range]) | 56.8 [51.7, 60.9] |
| Race | |
| White % (Median [range]) | 64.1 [27.8, 90.0] |
| African American % (Median [range]) | 26.8 [6.2, 60.6] |
| Asian % (Median [range]) | 1.2 [0.1,1.9] |
| American Indian or Alaska Native % (Median [range]) |
0.2 [0.1, 0.4] |
| Pacific Islander % (Median [range]) | 0.1 [0.0, 0.2] |
| Other/Unknown % (Median [range]) | 5.9 [1.8, 14.4] |
| Unique Patients with Encounters per Year | |
| Median [range] | 248,533 [44,476, 3,200,408] |
| Sum of Medians Across Sites | 7,204,434 |
| Unique Patients with Target Prescriptionsb per Year | |
| Median [range] | 55,781 [5,036, 282,397] |
| Sum of Medians Across Sites | 844,307 |
Summary statistics were derived from site-level, across-year medians. For example, the median [range] of unique patients with encounters was derived by calculating site-specific median number of encounters per year across observed years and then by calculating the median [range] of the site-specific median values. For the 25th percentile of age summary, at each site, we calculated the 25th percentile of age each year and then used the median of those values. The median [range] reported in the table is the across-site median [range] of the site-specific median values for the 25th percentiles.
One participating site (Site 11) is considered an Academic Medical Center and a Community Hospital.
Target prescriptions defined as CPIC Level A drugs or alternative medications within the class.
Prescribing patterns over time
Not every site was able to provide standard prescribing data for every year due to shifts in e-prescribing systems over time. Thus, logistic regression was used to fit models describing annual prevalence from 2011 to 2016 for each of the following: a) at least one CPIC Level A medication, b) > 2, > 3, or > 4 CPIC Level A medications, c) distinct CPIC Level A drug classes (e.g., antiplatelet P2Y12 inhibitors, statins, anticoagulants), d) individual CPIC Level A medications, and e) medications combined with the relevant associated pharmacogenes. Because of site-to-site variability in sample size (Table S1), two distinct weighting procedures were considered that combined site-year prevalence across sites to estimate an overall prevalence for each year: 1) by-site weighting, which weights each site equally, and 2) by-patient weighting, which weights sites in proportion to the number of patients with encounters. The exposures reported in the main text are based primarily on by-site weighting.
Prescribing patterns by demographic characteristics
To examine the prescribing patterns by gender, race, and age, we fit and summarized logistic regression models similar to those described above and in the Supplementary Materials. To estimate prescribing patterns across the age distribution, age was substituted for the year variable using restricted cubic spline functions to permit non-linear age trends. To capture prescribing patterns for each gender and race, site-specific estimates were combined from models that included indicator variables for the demographic subgroups.
RESULTS
The median of unique patients eligible for drug prescribing at each IGNITE-affiliated health system ranged from nearly 45,000 to 3.2 million patients per year (Table 1, Table S1). Summing the medians, an estimated 7.2 million adult patients were eligible for drug prescribing annually. Among the entire cohort, 56.8% (range 51.7–60.9%) were female and 26.8% (range 6.2–60.6%) self-declared African American race. The median of unique patients prescribed a CPIC Level A drug or alternative (i.e., target drugs) at each site ranged from 5,036 to 282,397 per year, and when summed an estimated 844,307 patients were prescribed at least one target drug annually.
CPIC Level A Drug Exposure
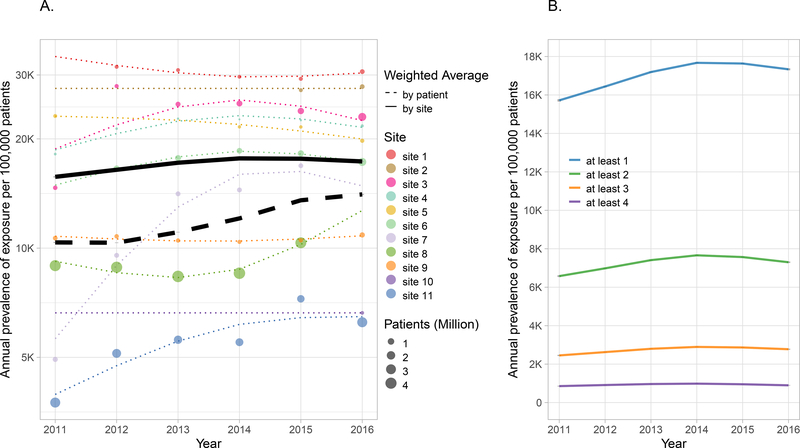
The prevalence of at least one CPIC Level A drug prescription among unique patients by site weighting (i.e., each site weighted equally) tended to increase from approximately 15,719 (95% confidence interval [CI]: 15,658–15,781) per 100,000 patients in 2011 to 17,335 (CI: 17,283–17,386) per 100,000 in 2016 (Figure 1A), with an apex of 17,671 (CI: 17,632–17,711) per 100,000 patients in 2014. This increasing trend was more pronounced when analyzed by patient weighting (i.e., each site weighted in proportion to its size), increasing from 10,349 (CI: 10,333–10,364) to 14,062 (CI: 14,021–14,103) per 100,000 patients between 2011 and 2016. There was substantial variability in drug exposure across sites, ranging from less than 7,500 to over 25,000 per 100,000 patients prescribed a CPIC Level A medication. The estimated annual exposure to at least two CPIC Level A drugs was above 7,200 per 100,000 patients in later years and as high as 7,660 (CI: 7,632–7,687) per 100,000 patients in 2014 (Figure 1B).
Figure 1. Annual prevalence of exposure to at least one CPIC Level A drug by site (A) and to more or more CPIC Level A medications (B).
A) Exposure (log scale) to at least one CPIC Level A drug for each site from 2011–2016. Each colored circle represents the exposure for the corresponding site. Circles are absent for years where data are not available. The size of the circle is proportional to the number of patients eligible for drug prescribing during the calendar year. The dotted colored lines are the prevalence of exposure estimated from the model fit. The mean prevalence of exposure for the entire cohort weighted by site is represented by the solid black line and weighted by encounters is represented by the dotted black line. The 95% confidence bands for the two means are represented by gray shading but may be too narrow to be observed. B) Mean site-weighted prevalence stratified by at least one, two, three or four CPIC Level A drugs from 2011–2016 plotted on a linear scale. Note that confidence intervals are represented by gray shading but may be too narrow to be observed.
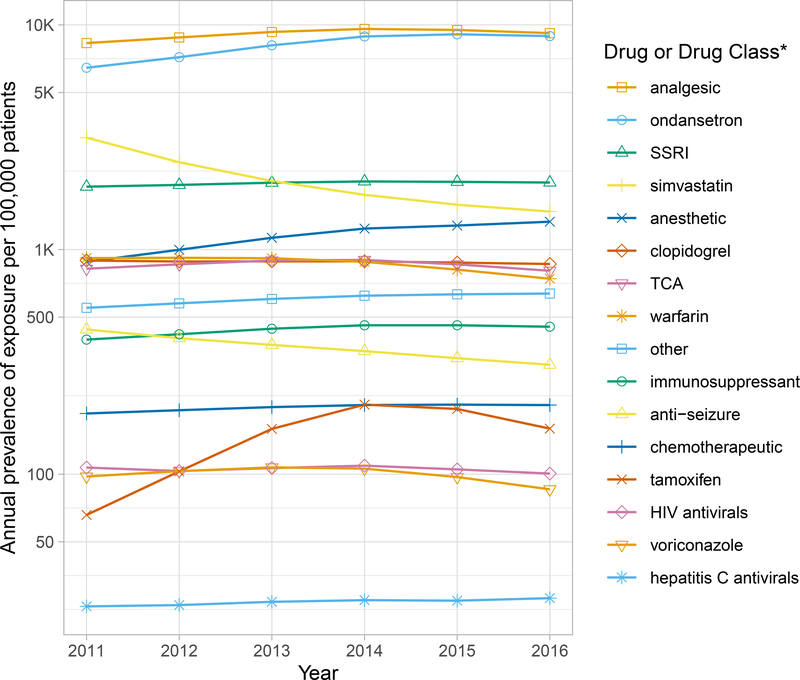
For the majority of CPIC Level A drugs or drug classes, the prescribing prevalence remained the same or slightly increased between 2011 and 2016 (Figure 2). Medications influenced by CYP2D6, CYP2C19, SLCO1B1, RYR1, CACNA1S, CYP2C9, VKORC1, or HLA-B*58:01 were prescribed to a greater extent compared to other gene-drug pairs (Figure S1), with the prevalence of medications influenced by CYP2D6 being far more common than all others. In 2016, the estimated prescribing prevalence of medications influenced by CYP2D6 was 14,117 per 100,000 patients while prescribing prevalence for CYP2C19, the next highest gene exposure, was 3,026 per 100,000 patients. The estimated prescribing prevalence of medications influenced by SLCO1B1, CYP2C9, or RYR1/CACNA1S was greater than 1,000 per 100,000 patients annually, and for VKORC1 or HLA-B*58:01 the prescribing prevalence was greater than 500 per 100,000 patients annually (Table 2, Figure S1). Opioid analgesics and ondansetron, which are metabolized by CYP2D6, were the most highly prescribed drugs, with an estimated annual exposure of 9,112 and 8,495 per 100,000 patients, respectively (Figure 2). Selective serotonin reuptake inhibitors (SSRIs) were also highly prescribed, with an estimated 1,971 per 100,000 patients exposed to SSRIs annually.
Figure 2. Annual Prevalence of Exposure Stratified by CPIC Level A Drug or Drug Class.
Mean site-weighted prevalence of exposure (log scale) to a CPIC Level A drug or drug class from 2011–2016. Analgesic includes codeine, oxycodone, tramadol; Selective serotonin reuptake inhibitors (SSRIs) includes citalopram, escitalopram, fluvoxamine, paroxetine; Anesthetic includes desflurane, isoflurane, sevoflurane, succinylcholine; Tricyclic antidepressant includes amitriptyline, nortriptyline; Other includes allopurinol, ivacaftor, rasburicase; Immunosuppressant includes azathioprine, mercaptopurine, tacrolimus, thioguanine; Anti-seizure includes carbamazepine, phenytoin; Chemotherapeutic includes capecitabine, fluorouracil, irinotecan; HIV antivirals includes abacavir, atazanavir; Hepatitis C antivirals includes peginterferon alfa-2a/2b, ribavirin.
Table 2:
Annual Estimated Prevalence of Gene-Drug Interactions
| Medication | Annual prescription prevalence per 100,000 patientsa | 95% CI lower | 95% CI upper | Gene | Actionable phenotypeb | Annual estimated gene-drug interaction per 100,000 patients | 95% CI lower | 95% CI upper |
|---|---|---|---|---|---|---|---|---|
| Ondansetron | 8,495 | 8,474 | 8,516 | CYP2D6 | UM | 309 | 308 | 309 |
| Oxycodone | 6,647 | 6,627 | 6,667 | CYP2D6 | PM, IM, UM | 1,137 | 1,134 | 1,141 |
| Tramadol | 2,506 | 2,490 | 2,522 | CYP2D6 | PM, IM, UM | 438 | 435 | 441 |
| Simvastatin | 2,066 | 2,056 | 2,076 | SLCO1B1 | PF, DF | 510 | 507 | 512 |
| Codeine | 1,091 | 1,083 | 1,099 | CYP2D6 | PM, IM, UM | 189 | 188 | 191 |
| Succinylcholine | 1,020 | 1,011 | 1,029 | CACNA1S | Positivec | < 1 | < 1 | < 1 |
| Succinylcholine | 1,020 | 1,011 | 1,029 | RYR1 | Positivec | 1 | 1 | 1 |
| Citalopram | 1,016 | 1,008 | 1,023 | CYP2C19 | PM, RM, UM | 339 | 336 | 341 |
| Clopidogrel | 879 | 872 | 886 | CYP2C19 | PM, IM | 285 | 283 | 288 |
| Warfarin | 851 | 844 | 857 | CYP2C9 | PM, IM | 277 | 275 | 279 |
| Warfarin | 851 | 844 | 857 | VKORC1 | Carrierd | 473 | 469 | 476 |
| Escitalopram | 677 | 671 | 682 | CYP2C19 | PM, RM, UM | 227 | 225 | 229 |
| Amitriptyline | 674 | 668 | 681 | CYP2C19 | PM, RM, UM | 222 | 220 | 225 |
| Amitriptyline | 674 | 668 | 681 | CYP2D6 | PM, IM, UM | 117 | 116 | 118 |
| Allopurinol | 629 | 622 | 636 | HLA-B*58:01 | Positivec | 26 | 26 | 26 |
Only drugs with a prevalence ≥ 500 per 100,000 patients included. See Supplemental Table 3 for the other CPIC Level A drugs.
Actionable phenotype defined as a phenotype that would prompt a prescribing action according to CPIC guidance.
Positive is defined as harboring an actionable genetic variant.
Carrier is defined as a VKORC1 c-1639G>A heterozygote or homozygote. Abbreviations are as follows: UM=ultrarapid metabolizer, RM=rapid metabolizer, IM=intermediate metabolizer, PM=poor metabolizer, DF=decreased function, PF=poor function
Exposure to CPIC Level A Cardiovascular Drugs and Alternative Agents
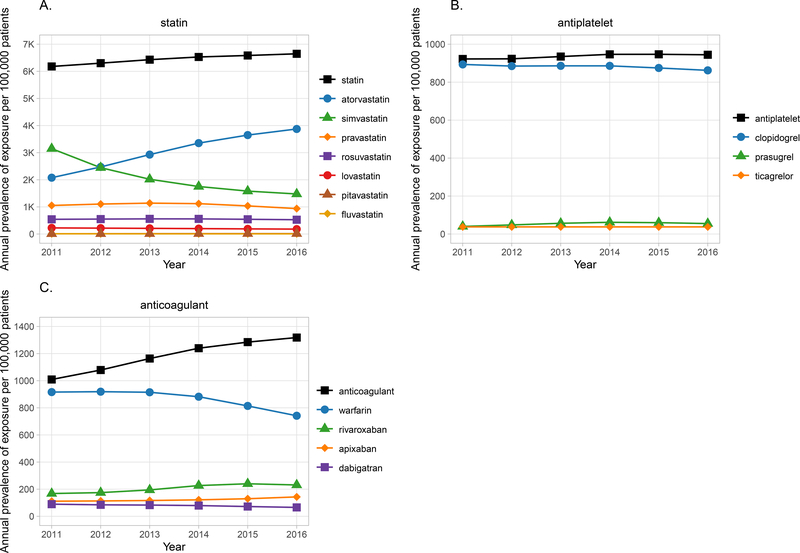
Notably, exposure to CPIC Level A cardiovascular drugs decreased over the study period even though the overall prescribing prevalence for statins, oral anticoagulants, and antiplatelet P2Y12 inhibitors increased between 2011 and 2016 (Figure 3). Simvastatin, the most commonly prescribed statin in 2011, declined in use from 3,148 per 100,000 patients in 2011 to 1,477 per 100,000 patients in 2016, and atorvastatin became the most commonly prescribed statin 2012 (Figure 3). There was a decline in use of warfarin (from 916 per 100,000 patients in 2011 to 742 per 100,000 patients in 2016) and clopidogrel (from 886 per 100,000 patients in 2011 to 862 per 100,000 patients in 2016), accompanied by an increase in use of alternative agents (Figure 3). However, both remained the most commonly prescribed agents in their respective classes even at the end of five-year observational period.
Figure 3. Annual prevalence of exposure to A) statins, B) anticoagulants, and C) antiplatelets.
A) Top line represents all statins with exposure to individual statins shown in the lines below. Exposure to any statin increased over time, whereas exposure to simvastatin decreased and exposure to atorvastatin increased. B) Top line represents all oral anticoagulants with exposure to individual anticoagulants shown in the lines below. Exposure to warfarin decreased over time; however, warfarin remained the most commonly prescribed anticoagulant. C) Top line represents all oral antiplatelet P2Y12 inhibitors with exposure to individual agents illustrated with the lines below. Exposure to clopidogrel remained higher than for other agents. Note that confidence intervals are represented by gray shading but may be too narrow to be observed.
CPIC Level A Drug Exposure by Race and Age
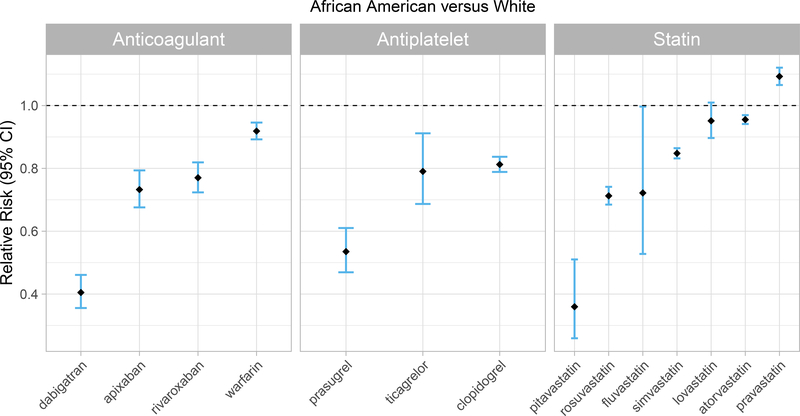
P2Y12 inhibitors and anticoagulants were generally less commonly prescribed in African Americans compared to whites; however, this trend was more pronounced for the newer, alternative agents than for the older CPIC Level A medications (Figure 4). Specifically, the prescribing prevalence for clopidogrel was 19% (CI: 16%−21%) lower, but for prasugrel was 46% (CI: 39%−54%) lower, in African Americans compared to whites. Warfarin prescribing prevalence was 8% (CI: 5%−11%) lower in African Americans than whites, but for rivaroxaban and dabigatran was 23% (CI: 20%−28%) and 60% (CI: 54%−64%) lower, respectively. Similarly, the prevalence of simvastatin was 15% (CI: 14%−17%) lower, but for rosuvastatin was 29% (CI: 26%−32%) lower, in African Americans versus whites.
Figure 4. Cardiovascular drug exposure compared between races.
Cardiovascular CPIC Level A drug exposure and alternatives compared between African American and white. Odds ratios were converted to risk ratios.
The prevalence of exposure to at least one CPIC Level A drug (in aggregate) was generally consistent across age groups (Figure S2). However, there were substantial differences in exposure between age groups for certain CPIC Level A drugs. The prevalence of ondansetron and oxycodone prescriptions were highest among those less than 30 years of age, while exposure to warfarin, simvastatin and clopidogrel started to increase at 35 to 40 years of age reaching an apex at approximately 80 years of age. Exposure to amitriptyline was more prevalent among middle-aged patients, reaching an apex at approximately 50 years of age.
Estimated Prevalence of Actionable Exposures
Using population-based pharmacogenetic variant frequencies in combination with our cohort race/ethnicity data, the prevalence of prescribing a drug to those with an actionable phenotype was estimated for each gene-drug pair (Table 2, Table S3). For the entire cohort, an estimated 4,748 per 100,000 prescribing events were potentially eligible for a genotype-guided intervention, if genotype information was available. The more commonly prescribed drugs, defined as having a prevalence of ≥ 500 per 100,000 patients (Table 2), predominantly had higher estimated actionable exposures. For example, 438 per 100,000 tramadol prescriptions were estimated to occur in those harboring a genetic variant that could influence prescribing decisions (e.g., CYP2D6 poor, intermediate, and ultrarapid metabolizers).(8) Although there was a lower exposure to fluoropyrimidines and thiopurines (Table S3), 12 per 100,000 fluoropyrimidine prescriptions and 21 per 100,000 thiopurine prescriptions were estimated to occur in those carrying a genetic variant that increases the risk of a severe, life-threatening, gene-drug interaction.
DISCUSSION
In a longitudinal, multi-center study of 11 health systems, drugs with a high level of evidence for pharmacogenetic guidance were prescribed in nearly 20% of all adult patients. Medications influenced by CYP2D6 or CYP2C19 were prescribed to a greater extent than other gene-drug pairs, though the prevalence of medications influenced by SLCO1B1, RYR1, CACNA1S, CYP2C9, VKORC1, or HLA-B*58:01 was not insubstantial at greater than 500 per 100,000 patients. This observation was persistent across a five-year period, suggesting that there are ample opportunities for implementing a multi-gene pharmacogenetic panel to guide drug prescribing among adult patients.
Exposure to many of the drug classes (e.g., opioids, antidepressants, and anesthetics) was observed across a broad age range. Ondansetron and oxycodone prescribing were more common in those younger than 30 years of age. A potential explanation is that those patients may have had a higher proportion of encounters for acute care where these medications are often prescribed. There was a significant increase in exposure to cardiovascular drugs among older adults, particularly starting around 35 to 40 years of age. Our observations are similar to other studies showing opioids were more commonly prescribed to younger adults, with cardiovascular drug exposure increasing with age.(24–26) While several other studies have also demonstrated a high prevalence of CPIC Level A drug prescribing, to our knowledge this was the first study to do so across a broad adult age range and diverse health systems.(27)
Multiple exposure analysis, weighted by site, showed that in later years of the study almost 8% of patients were exposed to at least two CPIC Level A drugs annually. Thus, not only did our study demonstrate that a substantial portion of patients across U.S. health systems were exposed to at least one CPIC Level A drug, but also that results from a multi-gene pharmacogenetic panel would likely be reusable for 8% of tested patients annually. Because unique patients were not followed across years, we likely underestimated the percentage of patients exposed to at least two CPIC Level A drugs over the study period.(28) A similar population-wide study evaluating opportunities for pharmacogenetic testing among approximately eight million patients within the U.S. Veterans Health Administration found that 25% of patients were exposed to at least two newly prescribed CPIC Level A drugs over a 6-year period.(18)
The prevalence of exposure to multiple drugs that are influenced by various pharmacogenetic variants supports the use of a multi-gene pharmacogenetic panel to tailor pharmacotherapy, rather than single gene testing.(18, 27, 28) Implementation models have been developed for pharmacogenetic panel testing, including preemptive models where testing is performed before drug prescribing.(14) Other models are reactive, where the prescribing of a medication with an actionable pharmacogenetic association prompts pharmacogenetic panel testing.(29) Genotyping of key genetic alterations (e.g., single nucleotide polymorphisms) remains the most common method for pharmacogenetic panel testing, though some newer sequencing platforms report pharmacogenetic alterations.(30) DNA sequencing is becoming broadly applicable in the clinical setting (e.g., oncology or diagnosis of genomic syndromes), and pharmacogenetic data obtained from sequencing results is likely to become more common.
With the availability of newer agents that are less impacted by pharmacogenes, it has been argued that prescribing of drugs addressed by CPIC guidelines may decline, thus limiting the utility of pharmacogenetic testing.(31) To specifically examine changes in the prescribing prevalence of CPIC Level A drugs in relation to alternative medications, we focused on cardiovascular agents. Consistent with previous data, use of simvastatin, warfarin, and clopidogrel declined over the five-year observational period as use of newer alternative agents increased.(32–34) However, these drugs remained the most commonly (i.e., warfarin and clopidogrel) or among the most commonly (i.e., simvastatin) prescribed agents in their respective classes. Of interest, in initial years, the site weighted prevalence of warfarin, apixaban, dabigatran, and rivaroxaban, when summed, exceeded total exposure to oral anticoagulants as a drug class. These data suggest there was frequent drug switching, most likely from warfarin to a direct-acting oral anticoagulant, and thus, many patients were exposed to both drugs in a given year.
Also consistent with previous studies,(35, 36) we observed that African Americans were less likely than whites to be prescribed newer cardiovascular agents, though differences in the frequency of comorbidities that impact drug prescribing might partially account for our observations. African American race and lower household income have been associated with lower use of newer agents, suggesting that both race and socioeconomic status affect access to novel therapies.(35, 36) Multi-gene pharmacogenetic testing may be especially beneficial for minority and lower income populations who are more likely to be exposed to CPIC Level A drugs than to newer alternative agents.
Taking into consideration the prevalence of CPIC Level A prescribing and race-based pharmacogenetic variant frequencies, we estimated that 4,748 per 100,000 prescriptions were to patients with an actionable phenotype. However, drug indication, dosage, and duration of therapy were not collected as part of this study, and CPIC recommendations were inclusive of all strength categories, which together could have resulted in an overestimation of opportunities for genotype-guided interventions. For example, CPIC guidelines for clopidogrel are specific for patients who have acute coronary syndrome and undergo coronary intervention.(37) Amitriptyline can be used at lower doses for neuropathic pain, and in such instances CYP2D6 poor metabolism is not recommended to guide prescribing.(38) Several CPIC Level A medications may be prescribed for a short duration, which could limit the clinical impact of gene-drug interactions predictive of toxicities due to supratherapeutic drug concentrations. However, gene-drug interactions predictive of inefficacy are of clinical importance even for short durations of therapy such as in the perioperative setting.(39) Furthermore, for several CPIC Level A drugs associated with severe, life-threatening adverse events, applicability of genetic results is independent of diagnosis or duration of therapy, and in certain instances (e.g., carbamazepine and fluoropyrimidines) no safe dose has been established for particular phenotypes.(12, 13, 40)
Prior studies have demonstrated that over 90% of individuals carry at least one actionable pharmacogenetic variant, with higher estimates among minority populations.(19, 20) High prevalence of actionable genetic variants in combination with exposure to CPIC Level A drugs and opportunities to avoid gene-drug interactions supports the clinical implementation of a multi-gene pharmacogenetic panel. “Normal” phenotype results may also be highly informative for pharmacotherapy decisions (e.g., supporting use of clopidogrel vs. alternative therapy).(37) Furthermore, emerging studies have shown potential cost effectiveness of multi-gene panel testing,(41–44) in part due to ample opportunities to reuse genetic test results.(45) Pharmacogenetic testing has also been proposed as a strategy to increase adherence to some medications, including statins, which might improve pharmacotherapy outcomes.(46–48) Despite the growing body of evidence supporting the utility of multi-gene panel testing, additional large clinical outcome and cost effectiveness studies will likely be needed to promote greater payer coverage of multi-gene pharmacogenetic testing costs.(31)
There are several limitations to this study. With the exception of cardiovascular drugs, only CPIC Level A medications were extracted from electronic health records. Therefore, there is an incomplete understanding of the prescribing patterns in relation to other medications, particularly those in the same drug class not influenced by pharmacogenetic variants. Diverse practice settings, including drug formularies, among sites may have confounded prescribing practices. Determining drug exposures over longer time horizons was not possible, including the reusability of pharmacogenetic test results beyond one year. Quantifying the number of patients who entered or exited health systems over the study period was not possible; this information would shed light on the potential portability of genetic information. One large academic health system was able to provide drug prescribing data only in the ambulatory setting which could have influenced the exposure to CPIC Level A drugs, particularly those commonly used for acute care. Associating drug prescribing with encounter type was not possible at all sites, which limited our ability to elucidate prescribing prevalence among inpatient and ambulatory settings. The prevalence of actionable pharmacogenetic phenotypes was estimated from CPIC resources, rather than direct genotyping, and clinical outcomes of potential gene-drug interactions were not confirmed by performing chart reviews. Finally, the sample of health systems used for this study may not fully represent all the settings where clinical care is delivered in the US which could influence both the point estimates and confidence intervals reported.
At the time of study inception there were 36 drugs classified as CPIC Level A. As evidence continues to emerge, additional drugs have since been classified as CPIC Level A including atomoxetine(49) and nonsteroidal anti-inflammatory drugs(50) (e.g., celecoxib and ibuprofen). The CPIC guideline for proton pump inhibitors (PPIs) was recently published(51) and a CPIC guideline for aminoglycosides is being drafted. Because these drugs were not included in this study, we may have underestimated pharmacogenetic testing opportunities.
A significant portion of adults in our study cohort was exposed to medications for which genetic information, if available, should be used to guide prescribing according to CPIC. These findings demonstrate the opportunity to implement a multi-gene pharmacogenetic panel to tailor pharmacotherapy across a wide spectrum of health systems. Additional research investigating the impact of pharmacogenetic variants on drug response, clinical outcomes, and cost-effectiveness is needed. Taken together, our data supports a multi-gene panel-based pharmacogenetic testing approach, particularly for populations with high exposure to drugs with strong pharmacogenetic evidence such as CPIC Level A drugs.
Supplementary Material
STUDY HIGHLIGHTS.
What is the current knowledge on the topic?
A high prevalence of exposure to medications with an actionable pharmacogenetic association has been demonstrated in select patient populations, but little is known about exposure to CPIC Level A drugs among diverse health and population settings.
What question did this study address?
The prevalence of CPIC Level A drug prescribing among diverse health care systems affiliated with the Implementing Genomics In Practice Network.
What does this study add to our knowledge?
A significant portion of adults across a broad age range and diverse health systems is exposed to medications with an actionable pharmacogenetic association.
How might this change clinical pharmacology or translational science?
Our findings demonstrate the opportunity to implement a multi-gene pharmacogenetic panel to tailor pharmacotherapy across a wide spectrum of health systems.
Acknowledgements:
University of Colorado contributions were supported by the Health Data Compass Data Warehouse project (healthdatacompass.org)
Funding: NIH IGNITE Network (http://ignite-genomics.org/) through grants U01HG007269, U01HG007253, U01HG007762, and U01HG007775; Moffitt Cancer Center NIH P30-CA076292, ASHP Foundation, and DeBartolo Family Personalized Medicine Institute (JKH); Leon Lowenstein Foundation (PE); NIH R01HL092173 and K24HL133373 (NAL) and Clinical and Translational Science Award UL1TR000165 (NAL, JC); T32HG008961 (BD); U01TR001427 (LHC); and UL1TR000445 (JFP).
Footnotes
Conflict of Interest: The authors declared no competing interests for this work.
Disclaimer: As an Associate Editor of Clinical Pharmacology & Therapeutics, Sara L. Van Driest was not involved in the review or decision process for this paper.
REFERENCES
- (1).Relling MV & Evans WE Pharmacogenomics in the clinic. Nature 526, 343–50 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Administration U.F.a.D. Table of pharmacogenomic biomarkers in drug labeling. (US Food and Drug Administration; ). [Google Scholar]
- (3).Administration U.F.a.D. Table of Pharmacogenetic Associations. (US Food and Drug Administration; ). [Google Scholar]
- (4).Relling MV & Klein TE CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin Pharmacol Ther 89, 464–7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Caudle KE et al. Incorporation of pharmacogenomics into routine clinical practice: the Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline development process. Curr Drug Metab 15, 209–17 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Relling MV, Klein TE, Gammal RS, Whirl-Carrillo M, Hoffman JM & Caudle KE The Clinical Pharmacogenetics Implementation Consortium: 10 Years Later. Clin Pharmacol Ther 107, 171–5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Hicks JK et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and CYP2C19 Genotypes and Dosing of Selective Serotonin Reuptake Inhibitors. Clin Pharmacol Ther 98, 127–34 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Crews KR et al. Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450 2D6 genotype and codeine therapy: 2014 update. Clin Pharmacol Ther 95, 376–82 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Bell GC et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 genotype and use of ondansetron and tropisetron. Clin Pharmacol Ther 102, 213–8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Ramsey LB et al. The clinical pharmacogenetics implementation consortium guideline for SLCO1B1 and simvastatin-induced myopathy: 2014 update. Clin Pharmacol Ther 96, 423–8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Johnson JA et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for Pharmacogenetics-Guided Warfarin Dosing: 2017 Update. Clin Pharmacol Ther 102, 397–404 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Amstutz U et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for Dihydropyrimidine Dehydrogenase Genotype and Fluoropyrimidine Dosing: 2017 Update. Clin Pharmacol Ther 103, 210–6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Relling MV et al. Clinical Pharmacogenetics Implementation Consortium Guideline for Thiopurine Dosing Based on TPMT and NUDT15 Genotypes: 2018 Update. Clin Pharmacol Ther 105, 1095–105 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Dunnenberger HM et al. Preemptive clinical pharmacogenetics implementation: current programs in five US medical centers. Annu Rev Pharmacol Toxicol 55, 89–106 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Empey PE et al. Multisite Investigation of Strategies for the Implementation of CYP2C19 Genotype-Guided Antiplatelet Therapy. Clin Pharmacol Ther 104, 664–74 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Cavallari LH et al. Multi-site investigation of strategies for the clinical implementation of CYP2D6 genotyping to guide drug prescribing. Genet Med 21, 2255–63 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Hicks JK et al. Implementation of Clinical Pharmacogenomics within a Large Health System: From Electronic Health Record Decision Support to Consultation Services. Pharmacotherapy 36, 940–8 (2016). [DOI] [PubMed] [Google Scholar]
- (18).Chanfreau-Coffinier C et al. Projected Prevalence of Actionable Pharmacogenetic Variants and Level A Drugs Prescribed Among US Veterans Health Administration Pharmacy Users. JAMA Netw Open 2, e195345 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Bush WS et al. Genetic variation among 82 pharmacogenes: The PGRNseq data from the eMERGE network. Clin Pharmacol Ther 100, 160–9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Van Driest SL et al. Clinically actionable genotypes among 10,000 patients with preemptive pharmacogenomic testing. Clin Pharmacol Ther 95, 423–31 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Levy KD et al. Opportunities to implement a sustainable genomic medicine program: lessons learned from the IGNITE Network. Genet Med 21, 743–7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Weitzel KW et al. The IGNITE network: a model for genomic medicine implementation and research. BMC Med Genomics 9, 1 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Cavallari LH et al. The IGNITE Pharmacogenetics Working Group: An Opportunity for Building Evidence with Pharmacogenetic Implementation in a Real-World Setting. Clin Transl Sci 10, 143–6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Zhong W et al. Age and sex patterns of drug prescribing in a defined American population. Mayo Clin Proc 88, 697–707 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Kantor ED, Rehm CD, Haas JS, Chan AT & Giovannucci EL Trends in Prescription Drug Use Among Adults in the United States From 1999–2012. JAMA 314, 1818–31 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Martin CB, Hales CM, Gu Q & Ogden CL Prescription Drug Use in the United States, 2015–2016. 2019. [PubMed] [Google Scholar]
- (27).Samwald M et al. Incidence of Exposure of Patients in the United States to Multiple Drugs for Which Pharmacogenomic Guidelines Are Available. PLoS One 11, e0164972 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Schildcrout JS et al. Optimizing drug outcomes through pharmacogenetics: a case for preemptive genotyping. Clin Pharmacol Ther 92, 235–42 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Eadon MT et al. Implementation of a pharmacogenomics consult service to support the INGENIOUS trial. Clin Pharmacol Ther 100, 63–6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Sangkuhl K et al. Pharmacogenomics Clinical Annotation Tool (PharmCAT). Clin Pharmacol Ther 107, 203–10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Keeling NJ, Rosenthal MM, West-Strum D, Patel AS, Haidar CE & Hoffman JM Preemptive pharmacogenetic testing: exploring the knowledge and perspectives of US payers. Genet Med 21, 1224–32 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Zhu J, Alexander GC, Nazarian S, Segal JB & Wu AW Trends and Variation in Oral Anticoagulant Choice in Patients with Atrial Fibrillation, 2010–2017. Pharmacotherapy 38, 907–20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Mortensen MB, Falk E & Schmidt M Twenty-Year Nationwide Trends in Statin Utilization and Expenditure in Denmark. Circ Cardiovasc Qual Outcomes 10, (2017). [DOI] [PubMed] [Google Scholar]
- (34).Dayoub EJ et al. Trends in Platelet Adenosine Diphosphate P2Y12 Receptor Inhibitor Use and Adherence Among Antiplatelet-Naive Patients After Percutaneous Coronary Intervention, 2008–2016. JAMA Intern Med 178, 943–50 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Essien UR et al. Association of Race/Ethnicity With Oral Anticoagulant Use in Patients With Atrial Fibrillation: Findings From the Outcomes Registry for Better Informed Treatment of Atrial Fibrillation II. JAMA Cardiol 3, 1174–82 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Nathan AS et al. Racial, Ethnic, and Socioeconomic Inequities in the Prescription of Direct Oral Anticoagulants in Patients With Venous Thromboembolism in the United States. Circ Cardiovasc Qual Outcomes 12, e005600 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Scott SA et al. Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin Pharmacol Ther 94, 317–23 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Hicks JK et al. Clinical pharmacogenetics implementation consortium guideline (CPIC) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 update. Clin Pharmacol Ther 102, 37–44 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Truong TM et al. The ImPreSS Trial: Implementation of Point-of-Care Pharmacogenomic Decision Support in Perioperative Care. Clin Pharmacol Ther 106, 1179–83 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Phillips EJ et al. Clinical Pharmacogenetics Implementation Consortium Guideline for HLA Genotype and Use of Carbamazepine and Oxcarbazepine: 2017 Update. Clin Pharmacol Ther 103, 574–81 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Hart MR, Garrison LP Jr., Doyle DL, Jarvik GP, Watkins J & Devine B Projected Cost-Effectiveness for 2 Gene-Drug Pairs Using a Multigene Panel for Patients Undergoing Percutaneous Coronary Intervention. Value Health 22, 1231–9 (2019). [DOI] [PubMed] [Google Scholar]
- (42).Dong OM et al. Cost-Effectiveness of Multigene Pharmacogenetic Testing in Patients With Acute Coronary Syndrome After Percutaneous Coronary Intervention. Value Health 23, 61–73 (2020). [DOI] [PubMed] [Google Scholar]
- (43).Weitzel KW, Cavallari LH & Lesko LJ Preemptive Panel-Based Pharmacogenetic Testing: The Time is Now. Pharm Res 34, 1551–5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Graves JA, Garbett S, Zhou Z & Peterson JF The Value of Pharmacogenomic Information. In: Economic Dimensions of Personalized and Precision Medicine (eds. Berndt ER, Goldman DP and Rowe JW) 53–86 (National Bureau of Economic Research, 2018). [Google Scholar]
- (45).Roden DM et al. Benefit of Preemptive Pharmacogenetic Information on Clinical Outcome. Clin Pharmacol Ther 103, 787–94 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Li JH et al. Genetically guided statin therapy on statin perceptions, adherence, and cholesterol lowering: a pilot implementation study in primary care patients. J Pers Med 4, 147–62 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Peyser B et al. Effects of Delivering SLCO1B1 Pharmacogenetic Information in Randomized Trial and Observational Settings. Circ Genom Precis Med 11, e002228 (2018). [DOI] [PubMed] [Google Scholar]
- (48).Fagerness J et al. Pharmacogenetic-guided psychiatric intervention associated with increased adherence and cost savings. Am J Manag Care 20, e146–56 (2014). [PubMed] [Google Scholar]
- (49).Brown JT et al. Clinical Pharmacogenetics Implementation Consortium Guideline for Cytochrome P450 (CYP)2D6 Genotype and Atomoxetine Therapy. Clin Pharmacol Ther 106, 94–102 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Theken KN et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2C9 and Nonsteroidal Anti-inflammatory Drugs. Clin Pharmacol Ther, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Lima JJ et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2C19 and Proton Pump Inhibitor Dosing. Clin Pharmacol Ther, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.