Abstract
Ophiocordyceps sinensis has been a source of valuable materials in traditional Asian medicine for over two thousand years. With recent global warming and overharvest, however, the availability of these wild fungi has decreased dramatically. While fruiting body of O. sinensis has been artificially cultivated, the molecular mechanisms that govern the induction of fruiting body at the transcriptional and post-transcriptional levels are unclear. In this study, we carried out both mRNA and small RNA sequencing to identify crucial genes and miRNA-like RNAs (milRNAs) involved in the development of fruiting body. A total of 2875 differentially expressed genes (DEGs), and 71 differentially expressed milRNAs (DEMs) were identified among the mycoparasite complex, the sclerotium (ST) and the fruiting body stage. Functional enrichment and Gene Set Enrichment Analysis indicated that the ST had increased oxidative stress and energy metabolism and that mitogen-activated protein kinase signaling might induce the formation of fruiting body. Integrated analysis of DEGs and DEMs revealed that n_os_milR16, n_os_milR21, n_os_milR34, and n_os_milR90 could be candidate milRNAs that regulate the induction of fruiting body. This study provides transcriptome-wide insight into the molecular basis of fruiting body formation in O. Sinensis and identifies potential candidate genes for improving induction rate.
Subject terms: Gene expression, Gene regulation, Genetics, Molecular biology
Introduction
Ophiocordyceps sinensis (Berk.) G. H. Sung, J. M. Sung, Hywel‐Jones & Spatafora consists of a sclerotium, holding the carcass of Hepialidae insect larvae, and stroma, the fruiting body of which is also the primary fungal structure used for taxonomic identification1,2. As one of the most valuable fungal traditional medicinal materials, O. sinensis has been widely used to treat lung inflammation, night sweats, asthma, nocturnal emissions, and other diseases for over two thousand years3,4. Due to environmental conditions, the fruiting body of O. sinensis forms only after 3–5 years in the wild5. In recent years, overharvest due to huge market demand has led to the rapid reduction of wild O. sinensis populations, promoting the need for and development of artificial cultivation of O. sinensis. Even under optimized artificial cultivation conditions, it takes the fruiting body more than one year to mature due to its complex life cycle6–8. However, the induction of fruiting body is still inefficient, and the high cost of large-scale artificial cultivation of this fungus is unsustainable in China.
The development of the fruiting body inside ascomycetes is a complex cellular differentiation process that requires special environmental conditions and is controlled by many developmentally-regulated genes. With the advancement of whole-genome data of O. sinensis, the molecular mechanisms of its growth and development have gradually been revealed9,10. Four mating-type genes and 121 other genes that may be involved in fruiting body development were discovered via the O. sinensis 454-EST database, which indicated that the mitogen-activated protein kinase (MAPK) signaling pathway was likely involved in the development of fruiting body11. Recently, transcriptome analyses have compared the transcripts of six O. sinensis developmental stages12, suggested that fungi in primordium differentiation and sexual maturation display similar gene expression patterns. Moreover, half of the genes related to mating showed the highest expression in the ST stage, indicating that fruiting in this fungus is initiated in the ST stage12. Previously, we compared transcript expression in three other stages of the fungal life cycle (asexual mycelium, developing fruiting body, and mature fruiting body). Four fifths MAPKKK genes and MAPK binding proteins were upregulated in the fruiting body compared with the mycelium, indicating the development of fruiting body in O. sinensis may be dependent on the MAPK signaling pathway13. These results provided initial indications for further study of the mechanism of induction.
MicroRNAs (miRNAs) are a class of endogenous small single-stranded RNAs that are composed of approximately 19–24 nucleotides, and which play important roles in post-transcriptional regulation of gene expression in eukaryotes14. Although there are still no miRNAs identified in miRbase that are encoded by fungi, novel miRNA-like RNAs (milRNAs) have been predicted in the O. sinensis genome15,16. Recently, some studies have shown that small RNAs play vital roles in fungal sexual development. The small RNA-mediated RNA interference mechanism plays an important role in the fine-tuning of the transcriptome during ascospore formation in Fusarium graminearum17. In Cordyceps militaris, disruption and overexpression of candidates milR4 and milR16 confirmed that milRNAs in C. militaris regulate fruiting body formation18. These studies suggested that milRNAs may play important roles in the regulation of development in O. sinensis.
In this study, three important stages of O. sinensis fruiting body formation were examined by RNA sequencing data. An integrated mRNA and miRNA transcriptome analysis was conducted before and after the sclerotium developmental stage (Fig. 1). Important factors and target genes associated with fruiting body induction during the development of O. sinensis were identified, providing a basic molecular mechanism to aid in facilitating large-scale artificial cultivation of O. sinensis.
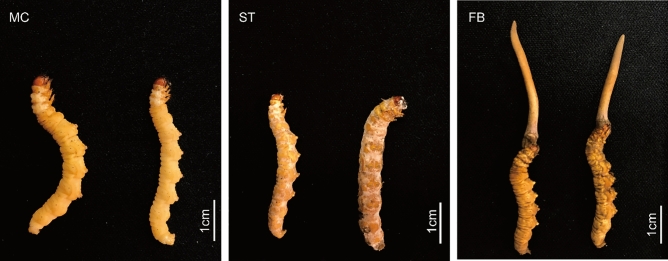
Figure 1.
The morphologies of O. sinensis at three different developmental stages (MC mycoparasite complex, ST sclerotium, FB fruiting body).
Results
Overview of transcriptome and small RNA sequencing
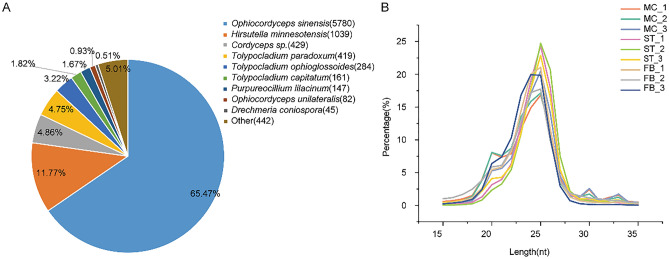
After transcriptome sequencing and filtering, approximately 63.92 Gb of clean reads were obtained, with an average of 6.54 Gb for each sample from nine cDNA libraries, for which the Q30 base percentage was greater than 93.54% (Table S1). The clean reads from each sample were compared with the specified reference genome, for which the efficiency of alignment varied from 91.52 to 97.59%. The above results indicate that the sequences obtained in this study was of good quality and could be used for subsequent analysis. A total of 9921 transcripts were obtained, including 1008 that mapped to predicted new genes. Gene function annotations showed that 8851 genes had significant matches in COG, GO, KEGG, KOG, Pfam, Swissport, eggNOG, or NR databases, with 3241 (36.62%), 6129 (69.25%), 3359 (37.95%), 4490 (50.73%), 6361 (71.87%), 5298 (59.86%), 7962 (89.96%), and 8837 (99.84%) genes, respectively. In addition to the functional annotation of O. sinensis, 11.77% of the genes also had high homology with Hirsutella minnesotensis (Fig. 2A).
Figure 2.
(A) Number of genes annotated in NR databases. (B) Length distribution and frequency of small RNAs (sRNAs) in the nine O. sinensis libraries.
After removing adaptors and low-quality reads, 142.96 M clean reads of small RNAs were generated, with each sample yielding greater than 11.05 M. The statistical results are shown in Table S2 with an overview of small RNA classification and annotation. The normalized clean reads were used for the analysis of small RNA distribution; the length distribution map of small RNA sequences demonstrated that the length of these small RNAs was 15–35 nucleotides (nt) (Fig. 2B). In general, most of the clean reads were 23–26 nt in length, with reads of 25 nt being the highest.
DEGs and DEMs expression analysis of O. sinensis at differential development stages
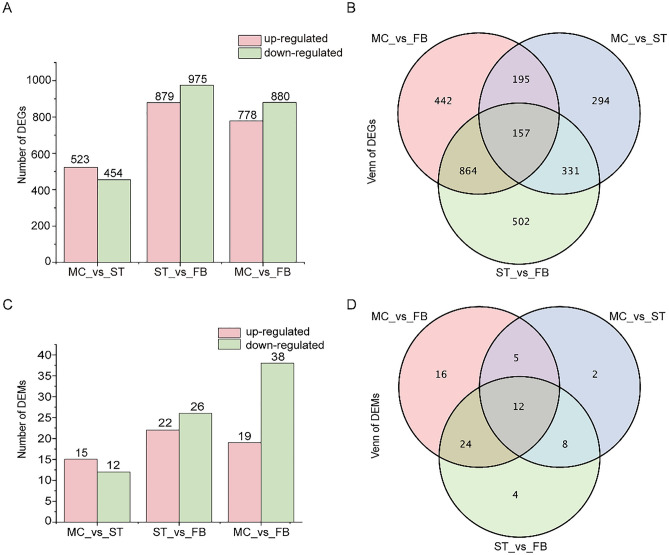
To investigate the changes in gene expression levels in O. sinensis during the development of fruiting body, DESeq2 software was used to compare the gene expression of samples at different stages based on clean reads. In the three comparison groups, we identified a total of 2875 DEGs. The initial stage (mycoparasite complex, MC) represented a control condition, the numbers of DEGs in the sclerotium (ST) stage and fruiting body (FB) periods were 977 and 1658, respectively. 1854 DEGs were also screened between the ST and FB stages (Fig. 3A). There were only 157 co-expressed genes in all three stages, and the most significant gene changes occurred during the FB stage (Fig. 3B). These differential genes included Cytochrome P450 monooxygenase (gene-G6O67_005633), Catalase (gene-G6O67_006909), Glucokinase (gene-G6O67_001528), and Phosphoenolpyruvate carboxykinase (gene-G6O67_008067) (Table S3), which are key enzyme genes in many metabolic pathways.
Figure 3.
Comparative analysis of differentially expressed genes (DEGs) and milRNA (DEMs) at the MC, ST, and FB stage. (A) Numbers and (B) Venn diagram of DEGs between two samples (MC_vs_ST, ST_vs_FB, ST_vs_FB). (C) Numbers and (D) Venn diagram of DEMs between two samples (MC_vs_ST, ST_vs_FB, ST_vs_FB).
To investigate the known and putatively novel miRNAs expressed at the three stages of O. sinensis, we first compared the known mature miRNAs and miRNA precursors in miRBase; no conserved miRNAs were identified. However, a total of 106 novel milRNAs were identified in the nine small RNA libraries using the miRDeep2 program (Table S4). Differential expression analysis of the miRNAs between these three samples was performed based on normalized read counts (TPM) for each identified miRNA. We obtained 27, 48, and 57 differentially expressed milRNAs (DEMs) in MC vs ST, ST vs FB, and MC vs FB comparisons, respectively (Fig. 3C). More DEMs were downregulated during fruiting body development. Only 12 DEMs were co-expressed in all three stages. Characterizing the differential expression of miRNAs is important in predicting the occurrence and development of fruiting body in O. sinensis (Fig. 3D).
Functional annotation and classification of DEGs
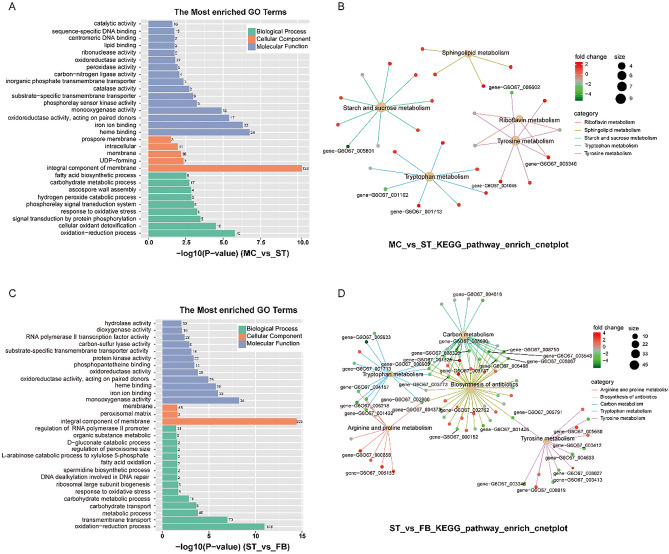
To infer the biological functions affected by DEGs at the three stages (MC, ST, and FB), we performed GO functional analysis. In the two developmental processes, 477 and 1027 DEGs were classified into 47 terms of three major biological processes (biological processes, cellular components, and molecular functions), respectively. The enrichment results of the three major biological functions of GO are shown in Fig. 4A,B (P value ≤ 0.03). The most dominant terms included the oxidation–reduction process, integral components of membranes, oxidoreductase activity, monooxygenase activity, and iron ion binding. KEGG pathway enrichment analysis was also conducted; 97 and 220 DEGs were enriched, corresponding to 77 and 103 pathways, respectively. In the process of ossification of O. sinensis, “Starch and sucrose metabolism”, “Tryptophan metabolism”, “Tyrosine metabolism”, and “Sphingolipid metabolism” pathways were significantly enriched (Fig. 4C). In the FB formation stage, the degree of enrichment of “Biosynthesis of antibiotics” and “Carbon metabolism” varied greatly (Fig. 4D). These metabolic pathways may closely relate to the formation of sclerotia and fruiting body. All DEGs, as well as GO and KEGG analysis results, are shown in Table S3. However, some DEGs encoded functionally unknown proteins, which might relate to O. sinensis growth and development; further studies will be required to verify their functionalities.
Figure 4.
Gene Ontology and KEGG pathway enrichment of DEGs. (A) The most enriched GO terms and (B) KEGG pathway cnetplot of MC_vs_ST. (C) The most enriched GO terms and (D) KEGG pathway cnet plot of ST_vs_FB (GO P value ≤ 0.03, top five KEGG pathway category, the shown genes log2|FC|≥ 2).
Analysis of GSEA significant enrichment
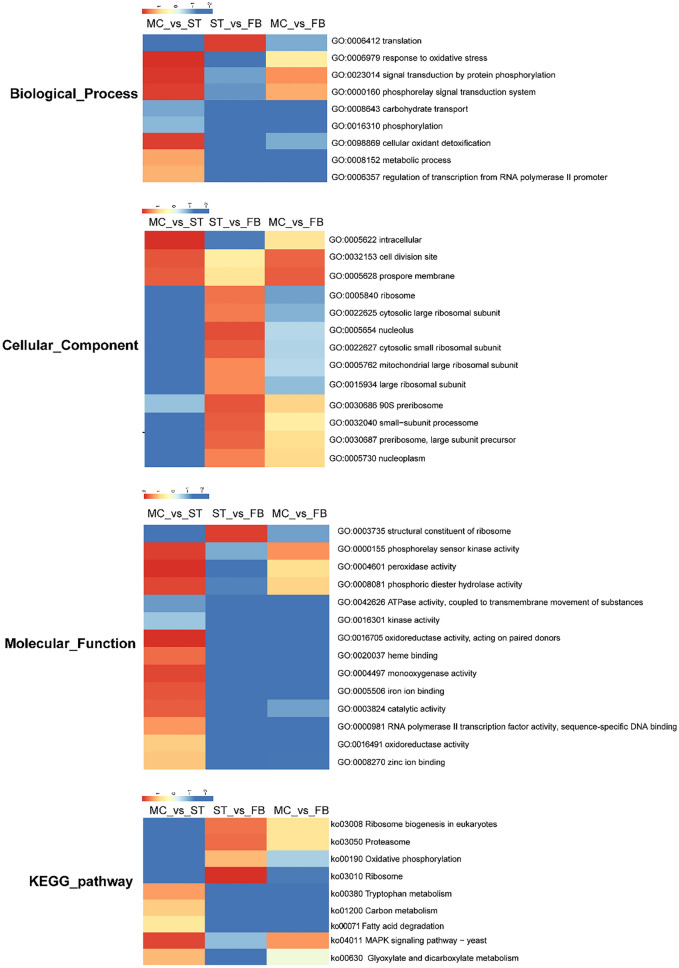
While GO- and KEGG-based analyses have compared the DEGs from functional categories, effective statistical methods were not used to analyze the overall trend of gene expression. Therefore, GSEA was adapted to analyze the enrichment of genes differentially expressed in each GO term and KEGG pathway. In this study, normalized enrichment scores were used to draw a cluster heat map (P value < 0.05) (Fig. 5). In the process of sclerotium formation, phosphorylation-related signaling (phosphorelay signal transduction system (GO:0000160), signal transduction by protein phosphorylation (GO:0023014), phosphorelay sensor kinase activity (GO:0023014), and oxidative phosphorylation (ko00190)) and oxidation-related (response to oxidative stress (GO:0000155), cellular oxidant detoxification (GO:0098869) peroxidase activity (GO:0004601), and monooxygenase activity (GO:0004497)) were significantly upregulated. During the fruiting body growth stage, the expression trend of ribosome-related terms and pathways were increased significantly, including the nucleolus (GO:0005730), ribosome (GO:0005840, ko030100), 90S preribosome (GO:0030686), and ribosome biogenesis in eukaryotes (ko03008). However, carbohydrate transport (GO:0008643), fatty acid degradation (ko00071), glyoxylate and dicarboxylate metabolism (ko00630), and carbon metabolism related to energy metabolism were downregulated.
Figure 5.
GSEA analysis heat map of O. sinensis at different development stages, include the biological process, cellular component, molecular function, and KEGG pathway (P < 0.05).
Some prominent pathways are listed in Table 1, such as the MAPK signaling pathway. DEGs in this pathway include mitogen-activated protein kinase kinase (pbs2), glycerol-3-phosphate dehydrogenase (gld1), catalase (cat1), and other important genes encoding enzymes. The expression of Cat was upregulated with log2(fold change) of 2.312 and downregulated with log2(fold change) of 2.160, respectively. In the citrate cycle, the genes encoding the enzymes malate dehydrogenase (mdh1), phosphoenolpyruvate carboxykinase (pck1), and succinate-CoA ligase (lsc2), which catalyze the oxidation of citric acid for energy, were highest in the ST stage, upregulated with log2(FC) of 2.237, 3.607, and 3.025, respectively, compared with the FB stage, and were slightly higher than in the MC stage.
Table 1.
DEGs involved in metabolic pathways. Significant differences in expression were evaluated using one-way ANOVA (ST was used as the control group, *P < 0.01).
| Gene ID | Annotation | FPKM (Average value) | ||
|---|---|---|---|---|
| MC | ST | FB | ||
| MAPK-related genes | ||||
| gene-G6O67_001149 | ATF/CREB family transcription factor | 159.450 | 296.989 | 310.307 |
| gene-G6O67_001860 | Mitogen-activated protein kinase kinase | 38.493 | 72.839 | 117.395 |
| gene-G6O67_006243 | Histidine kinase | 35.787 | 77.025 | 31.850 |
| gene-G6O67_006845 | Pheromone receptor | 10.897 | 33.218 | 15.188 |
| gene-G6O67_005494 | Glycerol-3-phosphate dehydrogenase (NAD +) | 105.461 | 88.457 | 16.160* |
| gene-G6O67_007807 | Ras homolog gene family | 7.340 | 12.113 | 5.378* |
| gene-G6O67_001713 | Catalase | 290.274* | 1624.276 | 362.997* |
| Amino acid-related genes | ||||
| gene-G6O67_003346 | Tyrosinase | 29.075* | 427.065 | 1.401* |
| gene-G6O67_004157 | Acetamidase | 36.780* | 94.229 | 23.372* |
| gene-G6O67_001432 | Amidase signature domain protein | 74.558 | 36.214 | 4.771* |
| gene-G6O67_006218 | Amidase | 84.452 | 101.630 | 4.214* |
| gene-G6O67_000733 | Acetyl-CoA acetyltransferase | 153.290 | 166.490 | 50.872* |
| gene-G6O67_001178 | Glutaryl-CoA dehydrogenase | 57.967 | 66.856 | 33.046 |
| gene-G6O67_002860 | Aldehyde dehydrogenase family | 645.414* | 1378.017 | 351.529* |
| gene-G6O67_005633 | Cytochrome P450 monooxygenase | 88.415 | 163.465 | 2.668* |
| gene-G6O67_001713 | Catalase | 290.274* | 1624.276 | 362.997* |
| gene-G6O67_006145 | Catalase-peroxidase | 251.388 | 239.481 | 92.286* |
| Energy-related metabolism genes | ||||
| gene-G6O67_000383 | Isocitrate dehydrogenase | 87.656 | 48.958 | 102.003* |
| gene-G6O67_003773 | Isocitrate lyase | 42.254* | 67.603 | 13.513* |
| gene-G6O67_001528 | Glucokinase | 17.177 | 11.438 | 176.097* |
| gene-G6O67_001884 | Transaldolase | 259.266 | 281.600 | 113.116* |
| gene-G6O67_003014 | Acyl-CoA dehydrogenase | 73.135 | 72.973 | 37.604 |
| gene-G6O67_003548 | Citrate synthase | 326.182 | 61.291 | 0.000 |
| gene-G6O67_003742 | NAD(P)-binding domain protein | 140.674 | 148.164 | 67.100 |
| gene-G6O67_006486 | Malate dehydrogenase | 788.497 | 1088.906 | 236.372* |
| gene-G6O67_006510 | Isocitrate dehydrogenase NADP | 63.750 | 30.549 | 72.962 |
| gene-G6O67_007408 | Succinate dehydrogenase cytochrome b small subunit | 269.799 | 242.459 | 111.577 |
| gene-G6O67_007964 | Acyl-CoA dehydrogenase domain protein | 77.659 | 45.968 | 17.089 |
| gene-G6O67_008750 | Succinyl-CoA synthetase beta chain | 87.758* | 209.074 | 26.275* |
| gene-G6O67_008752 | Alpha subunit of GDP-forming succinate-CoA ligase | 118.068 | 145.249 | 42.014* |
| gene-G6O67_002224 | Succinate dehydrogenase iron-sulfur protein | 281.527 | 115.061 | 248.025 |
| gene-G6O67_004996 | Gluconolactonase precursor | 16.812 | 5.929 | 8.262* |
| gene-G6O67_008325 | Fructose-1,6-bisphosphatase I | 140.215 | 107.728 | 8.231 |
| gene-G6O67_008067 | Phosphoenolpyruvate carboxykinase | 929.438 | 1282.465 | 96.630* |
Integrated analysis of DEGs and DEMs
To explore the regulatory relationship between milRNAs and mRNAs, 1096 potential target genes of the milRNAs were predicted, with 112 target genes obtained from the 33 DEMs in MC vs ST, and 456 target genes from the 27 DEMs in ST vs FB. To understand the functions of these genes targeted by DEMs, GO annotation, and KEGG enrichment was performed. Target genes were classified into cell cycle-related, cyanoamino acid metabolism, and energy metabolism-related pathways (Fig. 6A,B). These results indicated that milRNAs played important roles in the growth process of O. sinensis.
Figure 6.
(A, B) KEGG enrichment analysis of differentially expressed genes (DEGs) targeted by differentially expressed milRNAs (DEMs). The regulatory relationship between DEMs and their targets DEGs in O. sinensis was determined. (C, D) The interaction network between DEMs and their differentially expressed targets (MC_vs_ST and ST_vs_FB). Circles indicate target genes, yellow triangles indicate upregulated milRNAs, and green triangles indicate downregulated milRNAs.
There were 38 and 75 DEM-DEG relationship pairs found in MC and FB stage with ST as a control, respectively (Table S5). The network regulation diagram drawn by Cytoscape of some functionally annotated target genes indicated that one DEM could regulate more than one DEG, with both positive and negative correlation. Most milRNAs had more than one possible target gene, while different milRNAs could also regulate the same targets. As miRNAs regulate gene expression mainly by promoting cleavage of the target mRNAs or regulating transcription factors (TFs), we focused on negatively correlated pairs. According to the target regulation map in Fig. 6C,D, key enzyme genes in β oxidation gene-G6O67_007081 (3-hydroxyacyl-CoA dehydrogenase, targeted by n_os_milR90) and ecological adapting-related gene gene-G6O67_007081 (tat pathway signal sequence, targeted by n_os_milR16) were upregulated. From the ST to FB stage, gene-G6O67_006617 (ABC transporter) and gene-G6O67_008466 (SET domain protein) were significantly downregulated by n_os_milR34, with a log2(fold change) of 5.106 and 3.096, respectively. According to the target gene annotation and regulatory network, n_os_milR16, n_os_milR21, n_os_milR34, and n_os_milR90 represent candidate milRNAs to affect fruiting body development.
Validation of the DEGs and DEMs by RT-qPCR
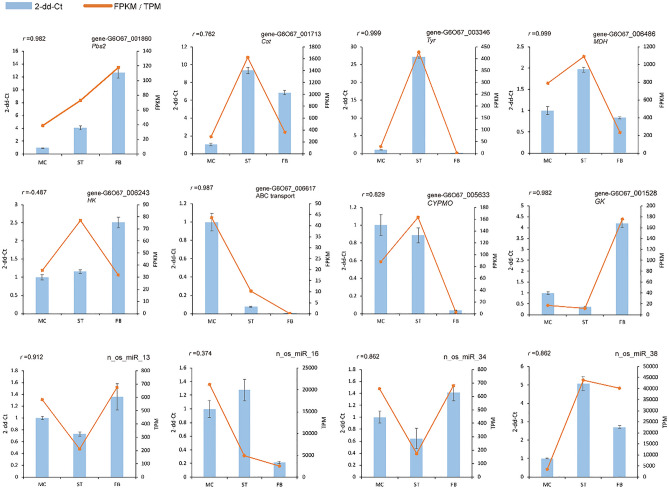
To confirm the reliability of the sequencing data, a total of eight DEGs and four DEMs were randomly selected to validate the RNA-Seq and small RNA expression profiles. As expected, qRT-PCR results showed that most of these mRNAs and miRNAs shared a similar expression with those from the sequencing data. Pearson correlation also showed that most of the relative expression levels were strongly correlated with FPKM/TPM, 83.33% r2 > 0.8 (Fig. 7), which confirm the reliability of the transcriptome sequencing data described above.
Figure 7.
Quantitative RT-PCR (qRT-PCR) validation of DEGs and DEMs at different developmental stages (MC, ST, and FB). The x-axis represents sample names, the left y-axis represents relative expression level (2-dd-Ct), and the right y-axis represents RNA-Seq results (FPKM/TPM). ‘r’ indicates the Pearson correlation coefficient.
Discussion
In order to determine the mechanism of induction of fruiting body in O. sinensis and analyze the expression of key genes, we performed an integrated mRNA and milRNA profiling of three developmental stages of O. sinensis using high-throughput sequencing. Our results provide new insights into the developmental regulation and mechanism of induction of fruiting body.
In the research of the developmental molecular mechanism of the fruiting body among Neurospora crassa, Sordaria macrospora, Aspergillus nidulans, and other model ascomycetes, PKA and MAPK19–24 were identified as pivotal signaling pathways. The development of C. militaris (FB) and O. sinensis (FB) belong to the genus Cordyceps is also mainly regulated by these signaling pathways11,25,26. Transcriptional profiling indicates that the development of fruiting body was shown to be more dependent on the MAPK signaling pathway than on PKA signaling27. Previous research has shown that the MAPK pathway participates in many physiological and developmental processes, including osmotic and oxidative stress, cell and sexual cycle regulation, and virulence26,28. MAPKs have also been generally conserved in all species studied thus far, and have very similar organization and functions20. Therefore, understanding the regulatory changes of MAPK signaling pathway is of great significance for revealing the differentiation and growth mechanism of the O. sinensis fruiting body.
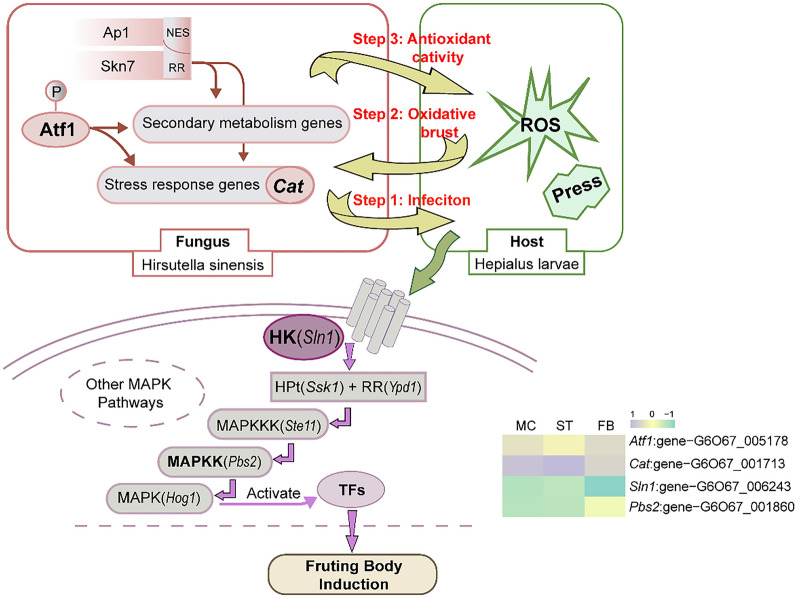
The signal transduction processes in which MAP kinases are involved starts with the sensing of environmental stimuli by two-component signal transduction systems (TCS) and receptor tyrosine kinases (RTKs). TCS consists of three components or signal transducers: a histidine kinase (HK, such as sln1), a response regulator (such as ypd1), and a histidine-containing phospho-transmitter (HPt, such as ssk1 or skn7)20,23,24,26,29,30. From the MC to ST stages, the host Hepialus larvae acts as a growth container for O. sinensis mycelium, splitting and proliferating continuously in it31. The increase in body filling leads to increased pressure in the surrounding cells, which induces autophosphorylation of sln1 , then subsequently transmitting the stress signal from sln1 to ssk1 through ypd1, thereby activating MAPK22,32,33. On the other hand, oxidative stress can activate different signal transduction pathways, either via the localization of specific regulators to the nucleus upon stress, followed by subsequent activation of detoxification genes expressions, or phosphorylation-driven intervention of the MAPK pathway20,34,35. In the MAPK signaling pathway, phosphorylation occurs at every step of signal transduction36. The phosphorylation of HK or RTKs directly activates a MAPKKK, which in turn activates a MAPKK via the phosphorylation of serine/threonine residues. This latter protein phosphorylates one or several MAPKs on serine/threonine/tyrosine residues, which finally gives rise to the activation of TFs that induce or repress genes involved in cellular adaptation or response to the sensed stimuli37,38. Moreover, DEG analysis also showed that the degree of phosphorylation was highest at the ST stage. Hence, high oxidative stress may activate the MAPK signaling pathway to regulate the formation of O. sinensis fruiting body, the inferred induction mechanism is shown in Fig. 8.
Figure 8.
The inferred mechanism of O. sinensis fruiting body induction. atf1, a transcript factor belongs to the ATF/CREB protein family; cat1, catalase; sln1, a histidine kinase; pbs2, mitogen-activated protein kinase kinase.
Although Li et al. have proposed in the transcriptome study on the sexual development of O. sinensis, the high-expression mating-genes indicating fruiting body was initiated in the ST stage. Here, we conducted by collecting the latest two stages of ST, added the important stage of the mycoparasite complex, verified this conclusion from the perspective of MAPK signal pathway. MAPK signaling in O. sinensis was most active at the ST stage, but it decreased during the FB period. We speculate that the MAPK signaling pathway is closely involved in the ability of O. sinensis to successfully bud and differentiate into fruiting body. Oxidative stress and inflation pressure are the major factors that stimulate MAPK signaling and induce downstream gene expression in fruiting body differentiation, morphogenetic processes, filamentous growth, mating, and osmolyte synthesis.
After fungal infection, the host produces huge amounts of reactive oxygen species (ROS)39. Through the mRNA-Seq of O. sinensis, we found that a large number of genes related to oxidative activity were highly expressed in the sclerotium stage (Figs. 3, 4, Table S2). The most enriched DEG was the cat1 which plays a central role in defense against oxidative stress. Catalase can catalyze the breakdown of H2O2 into O2 and H2O to protect proteins from oxidation by oxygen40. O. sinensis has the highest degree of ROS expression and the strongest oxidative stress capacity in the sclerotia stage. Therefore, O. sinsensis can eliminate active oxygen through catalase to ensure the survival of fungal cells.
The large amount of reactive oxygen produced by fungi infecting the host can not only induce transcriptional activation of stress response genes but can also activate the biosynthesis of certain secondary metabolites41,42. In Aspergillus, the antioxidant enzymes were inferred as the first line of defense against excessive ROS formation, while the synthesis of secondary metabolites as the second line of defense against ROS damage43. For example, aflatoxins have oxidative stress-inducing properties, which can be inhibited by phenolic antioxidants41,44. Through analysis of the chemical components of O. sinensis, a large number of antioxidant metabolites were found, such as cordycepic acid, phenols, and vitamin B (riboflavin)45, and pathways involved in the biosynthesis of antibiotics were relatively active in the sclerotium stage with a high oxygen environment. Therefore, we speculate that O. sinensis can overcomes oxidative breakdown through two ways of antioxidant activity and the production of secondary metabolites as a scavenging for reactive oxygen. Based on the changes in molecular regulation during the development of O. sinensis, the overlap between oxidative stress and secondary metabolism may be interpreted as an adaptive mechanism resulting from the molecular dialog between the host and the fungus39,41 (Fig. 8).
Most fungal vegetative hyphae do not continuously form fruiting body, but require special nutrient conditions to enable them to acquire a certain stage of “competence” before differentiating the fruiting body46. It has been proposed that carbohydrates are stored during vegetative growth to be utilized as a carbon source for sexual development, as well as in preparation for the subsequent fruiting process47,48. In our study, energy metabolism, especially carbon metabolism, was most active in the ST stage, prior to the fruiting body formation. In many species, the fruiting body is formed preferentially at much lower nutrient concentrations than those promoting vegetative growth, which may explain why energy metabolism decreases during the FB stage.
With the development of high-throughput sequencing technology, the existence of miRNA or milRNAs in O. sinensis have already been shown. Here, 106 novel milRNAs were identified from our sRNA-Seq data, which greatly enrich the miRNA database of this Cordyceps fungi16,18. miRNAs regulate gene expression mainly by promoting cleavage of the target mRNAs or by regulating TFs; therefore, DEM-DEG target gene relationship pairs can reflect the regulatory function of milRNAs more accurately. For example, the n_os_milR34gene-G6O67_006617 pair, encoding ATP-binding cassette (ABC) transporters, might provide protection against plant defense compounds and fungicides by ATP-driven efflux mechanisms. The defense function of O. sinensis was reduced during the FB phase (Fig. 7, Table S6), consistent with the transcriptome analysis (Table S3), implying that one of the factors that induce fruiting body formation might be the reduction of self-defense via the ABC transporter. However, the authenticity of these novel milRNAs requires further verification.
In conclusion, transcriptome and small RNA sequencing analyses of O. sinensis were carried out at various development stages. Our results indicated that the critical period for the successful formation of fruiting body was in the sclerotium stage. High oxidative stress and expansion pressure in the sclerotium stage stimulates the MAPK signaling pathway and induces downstream gene expression to promote transcription of genes involved in fruiting body differentiation, morphogenesis, filamentous growth, mating, and the osmotic pressure response. Based on changes in molecular regulation during the development of O. sinensis, an overlap between the antioxidant defense system, the catalase enzyme system, and secondary metabolites has been described. Further investigation will be required to verify the function of milRNAs whose expression changed the most during FB stage. Additional functional analysis of DEGs and DEMs would provide critical clues to reveal the molecular mechanism in the development of fruiting body. These target genes or milRNAs may be useful for improving the cultivation of O. sinensis.
Materials and methods
Fungi materials
Fungi at three developmental stages were collected before and after the induction of fruiting body (Fig. 1). Larvae of the bat moth completely infested by Hirsutella sinensis were designated as the mycoparasite complex (MC). Mummified larvae coated with mycelia before stroma development were designated as the sclerotium (ST). Samples of the fruiting bodies were designated as the whole O. sinensis with fruiting body (FB). Fresh samples were harvested from the artificial cultivation workshop at Chengdu Eastern Sunshine Co. Ltd, and stored at -80℃ until further processing. The strains of H. sinensis were preserved at Sichuan Center of Industrial Culture Collection under the accession number: SICC 5.02.
RNA extraction, library construction, and sequencing
Total RNA was extracted using the RNeasy Plant Mini Kit (Qiagen, Germany) according to the manufacturer’s protocol. RNA concentration and integrity were evaluated using a Nanodrop2000 (Thermo Fisher Scientific, Wilmington, DE) and Bioanalyzer 2100 system (Agilent Technologies, CA, USA). OD values between 1.8–2.2 and RIN ≥ 7.0 were required, and the concentration of the RNA was not less than 250 ng/μl.
For transcriptome sequencing, 1 μg of total RNA per group was used as input material for RNA sample preparation using a NEBNext Ultra Directional RNA Library Prep Kit for Illumina (NEB, USA). For small RNA sequencing, 5 μg of total RNA was ligated to 5′-RNA and 3′-RNA adaptors according to the NEBNext Multiplex Small RNA Library Prep Set for Illumina protocol (NEB, USA). RNAs were reverse transcribed to cDNAs to obtain a cDNA library, followed by PCR amplification. Two kinds of libraries for sequencing were generated; index codes were added to attribute sequences to each sample, and then samples were sequenced by Biomarker Technology Co., Ltd. (Beijing, China) on an Illumina NovaSeq6000 platform with 125 bp paired-end and 50 bp single-end reads, respectively. Three biological replicates were performed for each sample.
Analysis of differentially expressed genes (DEGs)
To control the quality of RNA-Seq raw data, the Fast QC Toolkit v0.11.9 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) was used to remove adaptor sequences and low-quality reads. The expression level of each transcript was measured as the number of clean reads mapped to its reference sequence. Clean reads from each sample were mapped to the reference genome of O. sinensis (NCBI accession number: PRJNA608258) using HISAT2 v2.0.4 (http://daehwankimlab.github.io/hisat2/). StringTie v2.1.2 (https://ccb.jhu.edu/software/stringtie/) was employed to calculate expression levels of genes49. Fragments per kilobases of exon per million fragments mapped (FPKM) values were used to normalize the expression level, and differential expression analysis was performed using the DESeq2 v1.30.1 R package (https://bioconductor.org/packages/release/bioc/html/DESeq2.html)50. A False Discovery Rate (FDR) ≤ 0.05 & |log2(fold change, FC)|≥ 1 were set as thresholds for DEG selection.
Identification of miRNAs and target gene prediction
As there are no miRNAs in miRbase 21.0 (http://www.mirbase.org/) confirmed to be encoded by fungi, approaches to identify animal or plant miRNAs were employed to identify fungal miRNAs or milRNAs50. Small RNA raw data in fastq format were first processed through cutadapt and fastp to obtain clean data, excluding reads with an “N” content ≥ 10%, reads without a 3′-adaptor sequences, low-quality reads, and sequences shorter than 18 nt or longer than 30 nt. Bowtie software was used to map the unannotated reads to the reference genome51. Mapped reads were aligned with mature miRNA sequences in the miRbase database to identify known miRNAs. miDeep2 (https://www.mdc-berlin.de/content/mirdeep2-documentation) was used to predict new miRNAs from unidentified miRNA reads52. Moreover, miRNA target genes were predicted using miRanda and targetscan scripts with default parameters53. The expression levels of miRNAs in each sample were normalized using the TPM algorithm. Differentially expressed miRNAs (DEMs) between samples were identified using the DESeq2 R package, with a significance threshold set to FDR ≤ 0.05 and |log2 (fold change)|≥ 1. Cytoscape v3.8.2 (https://cytoscape.org/) software was used to construct a DEMs-DEGs regulatory network54. The intersection of “candidate target genes” and s “DEGs” was referred to as “differential target genes”.
Functional annotation and KEGG pathway enrichment
Gene ontology (GO, http://geneontology.org/) functional annotations and Kyoto Encyclopedia of Genes and Genomes (KEGG, https://www.genome.jp/kegg/) signaling pathway annotations for DEGs and DEMs in O. sinensis were performed55,56. GO and KEGG annotations were subjected to Fisher’s exact tests (FDR < 0.05) and KOBAS3.0 (http://kobas.cbi.pku.edu.cn/kobas3) with an enrichment P value < 0.05, respectively. Moreover, Gene Set Enrichment Analysis (GSEA)57 v4.1.0 (http://www.gsea-msigdb.org/gsea/index.jsp) was performed to fully understand the gene expression trends of the gene sets identified in the KEGG or GO term analyses.
Validation of quantitative real-time PCR (qRT-PCR)
To validate the RNA-Seq and miRNA profiling results, quantitative real-time PCR (qRT-PCR) was performed using a CFX96 Real-time system (Bio-RAD, USA) and carried out with 2X Ultra SYBR Mixture (TransGen, Beijing, China) according to the manufacturer’s instructions. Total RNA samples were the same as the Illumina HiSeq sequencing input samples. Approximately 1 µg RNA from each sample was used to synthesize single-stranded miRNA and cDNA via reverse transcription using the miRcute miRNA first-strand cDNA synthesis kit (TIANGEN, Beijing, China). Ten miRNA-specific and target-gene primers are listed in Table S6. miRNA and target gene expression levels were calculated using the 2−ΔΔCt method and normalized to the levels of 18S ribosomal RNA (rRNA)58, each reaction was performed in triplicate59,60.
Supplementary Information
Acknowledgements
This study was supported by the Natural Sciences Foundation of China Science (Nos. 81872959, 81373920, 30801522). Sichuan Province Youth Innovation Team Fund (No. 19CXTD0055), China Scholarship Fund (No. 201708570027), the Open Research Fund of Chengdu University of Traditional Chinese Medicine Key Laboratory of Systematic Research of Distinctive Chinese Medicine Resources in Southwest China, Xinglin Scholar Research Premotion Project of Chengdu University of TCM.
Author contributions
H.Z. and J.G. conceived and designed the experiments; H.Z. performed the experiments, analyzed the data and wrote the paper; X.T., P.Y., J.B. and J.Y. collected materials and data analysis.
Data availability
All data were deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive under the accessions GSE160504 (RNA-Seq) and GSE160506 (small RNA sequencing).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-91718-x.
References
- 1.Shrestha B, Zhang W, Zhang Y, Liu X. What is the Chinese caterpillar fungus Ophiocordyceps sinensis (Ophiocordycipitaceae)? Mycology. 2010;1:228–236. doi: 10.1080/21501203.2010.536791. [DOI] [Google Scholar]
- 2.Sung G-H, et al. Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Stud. Mycol. 2007;57:5–59. doi: 10.3114/sim.2007.57.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu J, et al. The mechanisms of pharmacological activities of Ophiocordyceps sinensis Fungi. Phyther. Res. 2016;1583:1572–1583. doi: 10.1002/ptr.5673. [DOI] [PubMed] [Google Scholar]
- 4.Panda AK, Swain KC. Traditional uses and medicinal potential of Cordyceps sinensis of Sikkim. J. Ayurveda Integr. Med. 2011;2:9–13. doi: 10.4103/0975-9476.78183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong C, Guo S, Wang W, Liu X. Cordyceps industry in China. Mycology. 2015;6:121–129. doi: 10.1080/21501203.2015.1043967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qin QL, et al. Obstacles and approaches in artificial cultivation of Chinese cordyceps. Mycology. 2018;9:7–9. doi: 10.1080/21501203.2018.1442132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai-Hong D, et al. Cordyceps industry in China: current status, challenges and perspectives—Jinhu declaration for cordyceps industry development. Mycosystema. 2016;35:1–15. [Google Scholar]
- 8.Li X, et al. A breakthrough in the artificial cultivation of Chinese cordyceps on a large-scale and its impact on science, the economy, and industry. Crit. Rev. Biotechnol. 2019;39:181–191. doi: 10.1080/07388551.2018.1531820. [DOI] [PubMed] [Google Scholar]
- 9.Hu X, et al. Genome survey uncovers the secrets of sex and lifestyle in caterpillar fungus. Chin. Sci. Bull. 2013;58:2846–2854. doi: 10.1007/s11434-013-5929-5. [DOI] [Google Scholar]
- 10.Xia EH, et al. The caterpillar fungus, Ophiocordyceps sinensis, genome provides insights into highland adaptation of fungal pathogenicity. Sci. Rep. 2017;7:1–11. doi: 10.1038/s41598-016-0028-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiang L, et al. Transcriptome analysis of the Ophiocordyceps sinensis fruiting body reveals putative genes involved in fruiting body development and cordycepin biosynthesis. Genomics. 2014;103:154–159. doi: 10.1016/j.ygeno.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 12.Li X, et al. Developmental transcriptomics of Chinese cordyceps reveals gene regulatory network and expression profiles of sexual development-related genes. BMC Genom. 2019;20:1–18. doi: 10.1186/s12864-018-5379-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tong X, et al. Comparative transcriptome analysis revealed genes involved in the fruiting body development of Ophiocordyceps sinensis. PeerJ. 2020;2020:1–24. doi: 10.7717/peerj.8379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reis RS. The entangled history of animal and plant microRNAs. Funct. Integr. Genom. 2017;17:127–134. doi: 10.1007/s10142-016-0513-0. [DOI] [PubMed] [Google Scholar]
- 15.Griffiths-Jones, S. miRBase: microRNA Sequences and Annotation. Curr. Protoc. Bioinform.12, 1–10 (2010). [DOI] [PubMed]
- 16.Zhang W, et al. Identification of microRNA-like RNAs in Ophiocordyceps sinensis. Sci. China Life Sci. 2019;62:349–356. doi: 10.1007/s11427-017-9277-9. [DOI] [PubMed] [Google Scholar]
- 17.Son H, Park AR, Lim JY, Shin C, Lee YW. Genome-wide exonic small interference RNA-mediated gene silencing regulates sexual reproduction in the homothallic fungus Fusarium graminearum. PLoS Genet. 2017;13:1–27. doi: 10.1371/journal.pgen.1006595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shao Y, et al. MilR4 and milR16 mediated fruiting body development in the medicinal fungus Cordyceps militaris. Front. Microbiol. 2019;10:1–11. doi: 10.3389/fmicb.2019.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang G, et al. The mitogen-activated protein kinase GlSlt2 regulates fungal growth, fruiting body development, cell wall integrity, oxidative stress and ganoderic acid biosynthesis in Ganoderma lucidum. Fungal Genet. Biol. 2017;104:6–15. doi: 10.1016/j.fgb.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 20.Martínez-Soto D, Ruiz-Herrera J. Functional analysis of the MAPK pathways in fungi. Rev. Iberoam. Micol. 2017;34:192–202. doi: 10.1016/j.riam.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 21.Pangilinan J, Dilworth D, Sandor L, Pöggeler S, Gabaldón T. Comparative genomics and transcriptomics to filamentous ascomycetes. Genetics. 2019;213:1545–1563. doi: 10.1534/genetics.119.302749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pöggeler, S., Nowrousian, M. & Kück, U. Fruiting-Body Development in Ascomycetes. Differentiation and Sexuality.1, 325–355 (2006). 10.1007/3-540-28135-5.
- 23.Lichius A, et al. Importance of MAP kinases during protoperithecial morphogenesis in Neurospora crassa. PLoS ONE. 2012;7:e42565. doi: 10.1371/journal.pone.0042565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bayram Ö, et al. The Aspergillus nidulans MAPK module AnSte11-Ste50-Ste7-Fus3 controls development and secondary metabolism. PLoS Genet. 2012;8:e1002816. doi: 10.1371/journal.pgen.1002816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wen T, Li M, Kang J, He J. A molecular genetic study on fruiting-body formation of Cordyceps militaris. Afr. J. Microbiol. Res. 2012;6:5215–5221. doi: 10.5897/AJMR12.729. [DOI] [Google Scholar]
- 26.Mazzola D, et al. Inhibition of Yap2 activity by MAPKAP kinase Rck1 affects yeast tolerance to cadmium. FEBS Lett. 2015;589:2841–2849. doi: 10.1016/j.febslet.2015.07.049. [DOI] [PubMed] [Google Scholar]
- 27.Zheng P, et al. Genome sequence of the insect pathogenic fungus Cordyceps militaris, a valued traditional Chinese medicine. Genome Biol. 2011;12:R116. doi: 10.1186/gb-2011-12-11-r116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzuki, et al. The role of AtfA and HOG MAPK pathway in stress tolerance in conidia of Aspergillus fumigatus. Fungal Genet. Biol. 2014;73:138–149. doi: 10.1016/j.fgb.2014.10.011. [DOI] [PubMed] [Google Scholar]
- 29.Catlett NL, Yoder OC, Turgeon BG. Whole-genome analysis of two-component signal transduction genes in fungal pathogens. Eukaryot. Cell. 2003;2:1151–1161. doi: 10.1128/EC.2.6.1151-1161.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szeto CYY, Leung GS, Kwan HS. Le.MAPK and its interacting partner, Le.DRMIP, in fruiting body development in Lentinula edodes. Gene. 2007;393:87–93. doi: 10.1016/j.gene.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y, Li E, Wang C, Li Y, Liu X. Ophiocordyceps sinensis, the flagship fungus of china: terminology, life strategy and ecology. Mycology. 2012;3:2–10. [Google Scholar]
- 32.Szeto CYY, Wong QWL, Leung GS, Kwan HS. Isolation and transcript analysis of two-component histidine kinase gene Lenik1 in Shiitake mushroom, Lentinula edodes. Mycol. Res. 2008;112:108–116. doi: 10.1016/j.mycres.2007.08.024. [DOI] [PubMed] [Google Scholar]
- 33.Wei W, et al. Nox Complex signal and MAPK cascade pathway are cross-linked and essential for pathogenicity and conidiation of mycoparasite Coniothyrium minitans. Sci. Rep. 2016;6:24325. doi: 10.1038/srep24325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alvaro CG, Thorner J. Heterotrimeric G protein-coupled receptor signaling in yeast mating pheromone response. J. Biol. Chem. 2016;291:7785–7798. doi: 10.1074/jbc.R116.714980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown AJP, et al. Stress adaptation in a pathogenic fungus. J. Exp. Biol. 2014;217:144–155. doi: 10.1242/jeb.088930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kholodenko BN. Negative feedback and ultrasensitivity can bring about oscillations in the mitogen-activated protein kinase cascades. Eur. J. Biochem. 2000;267:1583–1588. doi: 10.1046/j.1432-1327.2000.01197.x. [DOI] [PubMed] [Google Scholar]
- 37.Hagiwara D, Sakamoto K, Abe K, Gomi K. Signaling pathways for stress responses and adaptation in Aspergillus species: stress biology in the post-genomic era. Biosci. Biotechnol. Biochem. 2016;80:1667–1680. doi: 10.1080/09168451.2016.1162085. [DOI] [PubMed] [Google Scholar]
- 38.Printen JA, Sprague GF. Protein-protein interactions in the yeast pheromone response pathway: Ste5p interacts with all members of the MAP kinase cascade. Genetics. 1994;138:609–619. doi: 10.1093/genetics/138.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ponts N, Pinson-Gadais L, Verdal-Bonnin MN, Barreau C, Richard-Forget F. Accumulation of deoxynivalenol and its 15-acetylated form is significantly modulated by oxidative stress in liquid cultures of Fusarium graminearum. FEMS Microbiol. Lett. 2006;258:102–107. doi: 10.1111/j.1574-6968.2006.00200.x. [DOI] [PubMed] [Google Scholar]
- 40.Lushchak VI, Gospodaryov DV. Catalases protect cellular proteins from oxidative modification in Saccharomyces cerevisiae. Cell Biol. Int. 2005;29:187–192. doi: 10.1016/j.cellbi.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 41.Montibus M, Pinson-Gadais L, Richard-Forget F, Barreau C, Ponts N. Coupling of transcriptional response to oxidative stress and secondary metabolism regulation in filamentous fungi. Crit. Rev. Microbiol. 2015;41:295–308. doi: 10.3109/1040841X.2013.829416. [DOI] [PubMed] [Google Scholar]
- 42.Reverberi M, Ricelli A, Zjalic S, Fabbri AA, Fanelli C. Natural functions of mycotoxins and control of their biosynthesis in fungi. Appl. Microbiol. Biotechnol. 2010;87:899–911. doi: 10.1007/s00253-010-2657-5. [DOI] [PubMed] [Google Scholar]
- 43.Fountain JC, et al. Responses of Aspergillus flavus to oxidative stress are related to fungal development regulator, antioxidant enzyme, and secondary metabolite biosynthetic gene expression. Front. Microbiol. 2016;7:1–16. doi: 10.3389/fmicb.2016.02048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reverberi M, et al. Aoyap1 regulates OTA synthesis by controlling cell redox balance in Aspergillus ochraceus. Appl. Microbiol. Biotechnol. 2012;95:1293–1304. doi: 10.1007/s00253-012-3985-4. [DOI] [PubMed] [Google Scholar]
- 45.Zhao J, Xie J, Wang LY, Li SP. Advanced development in chemical analysis of Cordyceps. J. Pharm. Biomed. Anal. 2014;87:271–289. doi: 10.1016/j.jpba.2013.04.025. [DOI] [PubMed] [Google Scholar]
- 46.Moore-Landecker & Elizabeth Physiology and biochemistry of ascocarp induction and development. Mycol. Res. 1992;96:705–716. doi: 10.1016/S0953-7562(09)80438-3. [DOI] [Google Scholar]
- 47.Wei H, et al. A putative high affinity hexose transporter, hxtA, of Aspergillus nidulans is induced in vegetative hyphae upon starvation and in ascogenous hyphae during cleistothecium formation. Fungal Genet. Biol. 2004;41:148–156. doi: 10.1016/j.fgb.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 48.Han KH, et al. Environmental factors affecting development of Aspergillus nidulans. J. Microbiol. 2003;41:34–40. [Google Scholar]
- 49.Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016;11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Friedländer MR, MacKowiak SD, Li N, Chen W, Rajewsky N. MiRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Witkos M, Koscianska T, Krzyzosiak J. Practical aspects of microRNA target prediction. Curr. Mol. Med. 2011;11:93–109. doi: 10.2174/156652411794859250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Du Z, Zhou X, Ling Y, Zhang Z, Su Z. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010;38:W64–W70. doi: 10.1093/nar/gkq310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhong X, et al. Transcriptome analysis of Ophiocordyceps sinensis before and after infection of Thitarodes larvae. Fungal Biol. 2016;120:819–826. doi: 10.1016/j.funbio.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 59.Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP. Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods. 2007;3:1–12. doi: 10.1186/1746-4811-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Livak K, Schmittgen T. Analysis of relative gene expression data using real-time quantitative PCR. Methods. 2002;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data were deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive under the accessions GSE160504 (RNA-Seq) and GSE160506 (small RNA sequencing).