Abstract
Purpose
Recombinant rotavirus A vaccines are being developed as an alternative to existing live oral attenuated vaccines. One of the main problems in the production of such vaccines is the genetic diversity of the strains that are in circulation. The goal of this study was to create an antigen panel for modern broad-spectrum recombinant rotavirus A vaccine.
Materials and Methods
The antigens of rotavirus were cloned and expressed in Escherichia coli. Antigenic specificity was investigated by Western blot analysis, which was performed using commercial polyclonal antisera to several RVA strains. Phylogenetic analysis was based on the amino acid sequences of the VP8* protein fragment of human RVA isolates representing genotypes P[4], P[6], and P[8].
Results
A universal panel of antigens was established, including consensus and conserved sequences of structural proteins VP8*, VP5*, and VP7, which are the main targets of neutralizing antibodies. For the first time, a consensus approach was used in the design of extended antigens based on VP8* (genotypes P[4], P[6], and P[8]) and VP5* (genotype P[8]) proteins' fragments. In addition, a gene coding the protein (ep-875) containing several copies of conserved short neutralizing epitopes of VP8*, VP7, and VP5* was created. Western blot analysis demonstrated that three synthetic VP8*-based antigens were not recognized by commercial antiserum against rotavirus strains isolated more than 35 years ago, but the specific activity of the VP5* and ep-875 antigens was confirmed. The problems of serological mismatch of vaccine strains and antigens with currently circulating strains are discussed.
Conclusion
Five antigens representing sequences of structural proteins belonging to different genotypes can be used in various combinations (from mono- to pentavalent mixtures) for the development of an effective broad-spectrum rotavirus vaccine.
Keywords: Rotavirus vaccine, Antigens, Consensus sequence, Genetic variation
Introduction
Acute intestinal infections are among the most common infectious diseases in young children. Group A rotaviruses (rotavirus A, RVA) remain the most common cause of severe gastroenteritis and mortality in children under 5 years of age in the world, despite the widespread introduction of vaccination [1,2,3]. Currently, two live attenuated vaccines licensed in 2006 and 2008—monovalent Rotarix (GlaxoSmithKline Biologicals, Rixensart, Belgium) and pentavalent RotaTeq (Merck & Co. Inc., Kenilworth, NJ, USA)— are extensively used to immunize children in more than 100 countries. Live attenuated rotavirus vaccines are quite effective in countries with high-income levels. However, their high cost, the risk of complications, and low effectiveness in developing countries, together with high infant mortality, indicate the need for fresh approaches to the development of new rotavirus vaccines. It is assumed that some serious side effects of the use of attenuated vaccines, such as intestinal intussusception, are associated with replication of the oral vaccine and can be overcome by using non-replicating parenteral vaccines [4]. It is also suggested that the low efficacy of live attenuated vaccines due to high titers of maternal antibodies can be overcome with a recombinant vaccine [5]. Modern recombinant vaccines that contain epitopes or extended fragments of structural proteins are safe and allow a parenteral route for administration [6].
VP7 (G-protein) and VP4 (P-protein), which are cleaved by trypsin-like proteases into VP8* and VP5* during infection, are the main rotavirus antigens that induce the production of virus-neutralizing antibodies [7,8,9,10,11,12,13,14,15,16]. Their names are used for binary classification, reflecting combinations of G and P genotypes of rotavirus. There are 14 G types, 17 P types of human RVA, and about 90 G-P combinations [17]. It is worth noting that a significant number of strains circulating in the world today do not belong to either an existing or a developing vaccine candidate, and contain a significant number of amino acid substitutions in the antigenic sites of proteins VP4 and VP7.
Although the problems of low efficacy of live attenuated vaccines in countries with high infant mortality are likely to be related to a number of factors [18], it should be noted in particular that there is a significant difference in serotypes between vaccine strains and actual rotavirus strains. For example, according to meta-analysis, strains with type G1P [8] represented more than 70% of rotavirus infections in North America, Europe, and Australia, but only about 30% of infections in South America and Asia, and 23% in Africa, between 1989 and 2004. In addition, African strains with the P[6] genotype represented one-third of all strains identified [19]. After the mass introduction of vaccines in various countries, there was the replacement of previously widespread RVA strains with new ones (including rare genotypes), an increase in the diversity of genotypes, and an increase in the number of mutations in the antigenic determinants of the VP4 and VP7 proteins of strains in circulation compared to vaccine strains [20,21,22,23]. The lower efficacy of vaccines against emerging RVA strains has been reported [20]. It is assumed that the long-term use of the Rotarix (genotype G1P1A[8]) and RotaTeq (genotypes G1, G2, G3, G4, and P1A[8]) vaccines may lead to the selection of strains that can avoid vaccine-induced immunity [24]. The diversity of genotypes and their significant geographical variability with seasonal and interannual fluctuations, as well as the variability of antigenic determinants, represent major challenges to the development of a broad-spectrum vaccine.
Recent developments in recombinant RVA vaccines include virus-like particle vaccines, recombinant subunit vaccines, and vaccines based on viral and live bacterial vectors [18]. There is a candidate vaccine based on the recombinant VP8* protein fragment (65–223 amino acid residues) of the human isolate Wa with the G1P[8] genotype. The fragment was obtained by expression in Escherichia coli. This vaccine passed the first phase of clinical trials and demonstrated a good safety profile and immunogenicity in the presence of an adjuvant (aluminum hydroxide). However, a poor immune response to heterologous RVA strains suggested that subunit vaccines should include different rotavirus genotypes in order to provide broader protection [6]. In response to this, a trivalent recombinant vaccine (P2-VP8) has been developed. It includes VP8* antigens of three strains—Wa (P[8]), DS-1 (P[4]), and 1076 (P[6]) —the most common P-genotypes in the world. At the moment, it is undergoing clinical trials [25]. RVA strains isolated more than 35 years ago were used to generate the P[8], P[4], and P[6] genotype sequences of the P2-VP8 candidate vaccine. The efficacy of this vaccine compared with strains currently in circulation requires further research [26].
In this work, we developed and created a universal panel of antigens that includes consensus and conserved sequences of structural proteins VP8*, VP5*, and VP7, which play a key role in the induction of an effective protective immune response. For the first time, consensus sequences (instead of the sequence of a specific viral isolate) of protein fragments VP8*and VP5* were used for expression in E. coli as part of genetically engineered constructs.
In addition, a gene coding the protein-containing repeated conserved epitopes of VP8*, VP7, and VP5* proteins was constructed. Five synthetic antigens representing consensus and conserved sequences of three structural proteins (VP8*, VP5*, and VP7) belonging to different genotypes can be used in various combinations (from mono- to pentavalent mixtures), which provides encouragement for the possibility of developing an effective broad-spectrum rotavirus vaccine.
Materials and Methods
Recombinant antigens design
Synthetic genes VP8*-P4, P6, P8, and VP5* were designed as described in the Results section, assembled from overlapping oligonucleotides by the Evrogen company (Moscow, Russia) and cloned into a pQE-30 expression vector (Qiagen, Hilden, Germany) between BamHI and HindIII restriction sites. Synthetic gene ep875 was constructed by our group.
Recombinant antigens expression and purification
E. coli strains M15 and SG13009 were employed to express cloned rotavirus antigens. Recombinant proteins were purified by metal affinity chromatography according to the manufacturer's protocol (Qiagen). Proteins were studied by electrophoresis in odium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE; 8%–20%). Gels were imaged and analyzed using the ChemiDoc XRS+ System, with Image Lab Software (Bio-Rad Laboratories, Hercules, CA, USA).
Western-blot analysis
Proteins were separated by SDS-PAGE in an 8%–20% gradient gel and transferred onto a Hybond-P polyvinylidene fluoride membrane (Amersham) using a Pierce Power Blotter electric transfer system (Thermo Scientific). The membranes were next treated with commercial polyclonal antisera to five RVA strains: Wa-G1P[8], Ds-1 G2P[4], Ito-G3P[8], Hochi-G4P[8], 69M-G8P[10] (Cat#MBS316568; MyBioSource Inc., San Diego, CA, USA), at a 1:500 dilution and then with secondary anti-species antibodies conjugated with horseradish peroxidase (Cat#713-035-003; Jackson ImmunoResearch Laboratories Inc., West Grove, PA, USA) in a 1:10,000 dilution. The membranes were next treated with ECL substrate (Amersham), and the chemiluminescence signals were registered using the ChemiDoc XRS+ system (Bio-Rad Laboratories).
Phylogenetic analysis
Multiple amino acid alignments were carried out using ClustalW ver. 2.1 software (http://bioedit.software.informer.com/download/?df3445). In addition to synthetic consensus sequences from human RVA P[4], P[6], and P[8] genotypes, sequences representing some “old” isolates (1974–1987) were chosen, as well as the “modern” (2012–2020) sequences discovered in geographically distant regions and sequences typical for known genetic lineages of rotavirus genogroup P[II] [27]. Japanese isolate AU1 (1982), belonging to genotype P[9], 2021.10.2.123was used as a phylogenetic outgroup. The PhyML 3.0 version of the maximum likelihood algorithm was used for analysis [28]. Nodes with less than 50% support were collapsed. Graphical tree presentation was achieved using the MEGA7 program [29]. Amino acid sequence identities were determined employing the Lipman-Pearson algorithm implemented in the MegAlign ver. 7.1.0 program (DNASTAR Lasergene package; DNASTAR Inc., Madison, WI, USA) [30].
Results
Consensus sequences P[4], P[6], and P[8] represent concanavalin-like domains (“spike head”) of human RVA VP8* protein (159 amino acid residues) that correspond to residues 65–223 of UniProt sequence P11193. The main design principle was the wide geographical distribution of the corresponding isolates. Most of the isolates used for analysis were discovered between 2008 and 2014. The VP5* fragment representing the consensus sequence to the most widespread genotype, P[8], was chosen since it is known that, in contrast to variable VP8*, the VP5* protein is more conserved and induces a broad heterotypic neutralization antibody response [9,15]. A sequence of conserved VP5* “stalk and body” (232 amino acid residues) corresponded to residues 247–478 of UniProt sequence P11193. A similar criterion was used to create a consensus of the VP5* protein fragment and this time the interval for most of the selected isolates covered the years 1974 to 2011. Amino acid sequences obtained (Supplement 1) were subjected to reverse translation in silico and nucleotide sequences were optimized in several steps, as described previously [31].
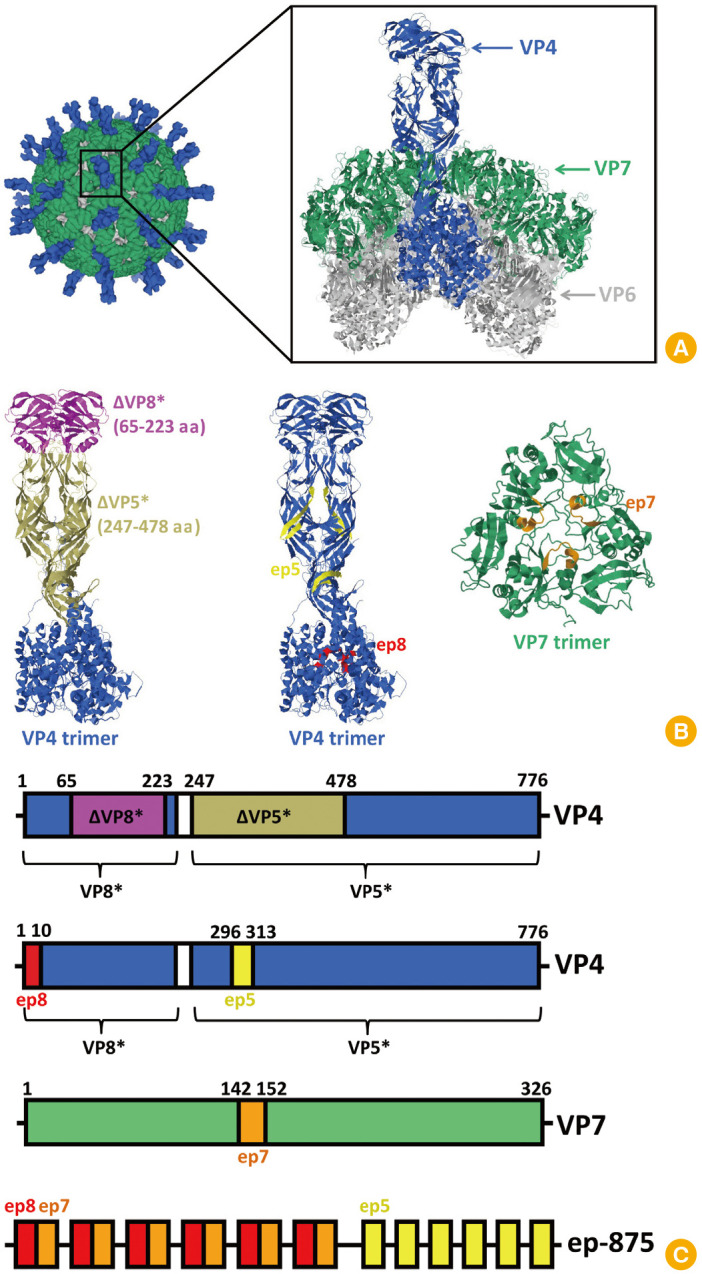
A gene coding the fusion protein (ep-875) containing repeated conserved short neutralizing epitopes of VP8*, VP7, and VP5* proteins was constructed [32,33,34,35] (Fig. 1B, C). Conserved neutralizing epitopes were selected based on analysis of data published earlier [8,10,11,14,15,32], as well as information obtained from analysis of the protein sequences deposited in Genbank. The VP8* protein epitope (ep8, MASLIYRQLL 1–10 amino acid) is highly conserved. For the genotypes P[4], P[8], and P[6], similarity index values were 100%, 95%, and more than 90%, respectively. For the genotypes rarely found in the human population, this indicator exceeds 96%. Amino acid sequence identities were determined employing the Lipman-Pearson algorithm implemented in the MegAlign ver. 7.1.0 program (DNASTAR Inc.) [30]. Previously, it was shown that a recombinant fusion protein (TrxP2(VP81–10)3) containing three copies of the VP8* epitope, thioredoxin, and the tetanus toxin T-cell epitope (P2) induced significantly higher antibodies titers in mice compared with the TrxP2 fusion protein (VP81–10, single copy of the same epitope) or a chemically synthesized peptide (VP81–10) administered together with a carrier protein and T-cell epitope. In addition, the recombinant protein containing three copies of the epitope induced significantly higher antibody titers with virus-neutralizing activity than the full-length recombinant VP8* protein [36]. The selected epitope of the VP5* protein (ep5, KAANYQYNYLRDGEQVTA 296–313 amino acid) has a site for binding to the cellular integrin receptor. The epitope is conserved in strains of the world's most common genotype, P[8] (similarity index values >90%). The short epitope of the VP7 protein (ep7, MKYDQNLELDM 142–152 amino acid) is conserved in strains of genotype G[1], which occurs in combination with 10 out of 17 P-genotypes found in humans: P[1], P[3], P[4], P[6], P[8]-P[11], P[14], and P[19] (similarity index values >80%).
Fig. 1. (A) Cryo-electron microscopy reconstruction of rotavirus particle and high-resolution structure of outer and middle layers (Protein Data Bank [PDB]: 4V7Q [32,33,34,35]). (B) High-resolution structures of VP4 and VP7 trimers showing the location of fragments ΔVP8* (65–223 amino acid [aa]), ΔVP5* (247–478 aa) and location of epitopes ep8, ep5, ep7 (PDB: 4V7Q [32,34,35]). (C) Linear diagrams of VP4 and VP7 showing color-coded fragments and epitopes (not to scale), and linear diagram of recombinant antigen ep-875 (not to scale). VP4 fragment (232–247 aa) removed during VP4 in vivo proteolysis is highlighted in white.
Nucleotide sequence coding for the epitope from the VP5 protein (ep5, KAANYQYNYLRDGEQVTA) was amplified by polymerase chain reaction (PCR) using primers ep5-p (5′-tttagatctgtcgacaaagctgctaactaccagtacaactacctgcg-3′) and ep5-m (5′-tttctcgagagcggtaacctgttcaccgtcacgcaggtagttgtact-3') and sequence coding for the fusion epitope from VP8 and VP7 proteins (ep87, MASLIYRQLLMKYDQNLELDM)—with primers ep87-p (5′-tttagatctgtcgacatggcttctctgatctaccgtcagctgctgatgaaatacg-3′) and ep87-m (5′- tttctcgagcatgtccagttccaggttctggtcgtatttcatcagcagctg-3′). PCR products were digested with BglII-XhoI restriction enzymes and ligated into the pQE30 vector at the same sites. The resulting plasmids pQE-ep5x1 and pQE-ep87x1 contained a single copy of the corresponding epitope and were used for subsequent multiplication of epitopes. To duplicate epitopes, the constructs pQE-ep5x1 and pQE-ep87x1 were digested at the XhoI-XbaI sites. The inserts were cut from the same vectors using SalI-XbaI sites. The “sticky” ends formed after hydrolysis of XhoI and SalI are complementary to each other (“compatible ends”). After ligation, both sites were no longer recognized by their “own” restrictases, since only five of the six nucleotides of each palindrome were retained in the recombinant sequence. When such cloning was repeated (any of the newly obtained plasmids pQE-ep5x2 or pQE-ep87x2 was used as the source for both the vector and the insert), the number of epitopes was doubled again (up to four copies). Final constructs pQE-ep5x6 and pQE-ep87x6 carried six copies of each epitope. To obtain expression vector coding for the polyepitope protein ep875, plasmid pQE-ep87x6 was cut by XhoI-XbaI and ligated with SalI-XbaI fragment excised from plasmid pQE-ep5x6. Thus, protein ep875 contained six copies of epitope ep87 (N-terminus) fused to six copies of epitope ep5 (C-terminus).
The recombinant antigens were expressed in E. coli and analyzed by electrophoretic analysis. Fig. 2A demonstrates the results of the expression of five proteins, VP8*-[P8], VP8*-[P6], VP8*-[P4], VP5*, and ep-875, after chromatographic purification. The molecular weight of each recombinant protein during expression and purification corresponded to the calculated value: 18 kDa for VP8*-[P8] and VP8*-[P4], 18.5 kDa for VP8*-[P6], 28 kDa for VP5*, and 29 kDa for ep-875. The protein yield varied from 11 to 35 mg per 1 L of the culture medium.
Fig. 2. Characterization of rotavirus A (RVA) recombinant antigens. Lanes: VP8*-[P8] (1), VP8*-[P6] (2), VP8*-[P4] (3), ep-875 (4), VP5* (5). (A) Electrophoresis analysis, 8%–20% SDS-PAGE (sodium dodecyl sulphate–polyacrylamide gel electrophoresis), staining with Coomassie G-250. (B) Western blot membrane stained by Ponceau S. (C) Western blot analysis with commercial polyclonal antisera to 5 RVA strains: Wa-G1P[8], Ds-1 G2P[4], Ito-G3P[8], HOCHI-G4P[8], 69M-G8P[10] (Cat# MBS316568; MyBioSource Inc., San Diego, CA, USA) as primary antibodies and secondary anti-species antibodies conjugated with horseradish peroxidase. Positions of molecular weights (kDa) markers are indicated in the right.
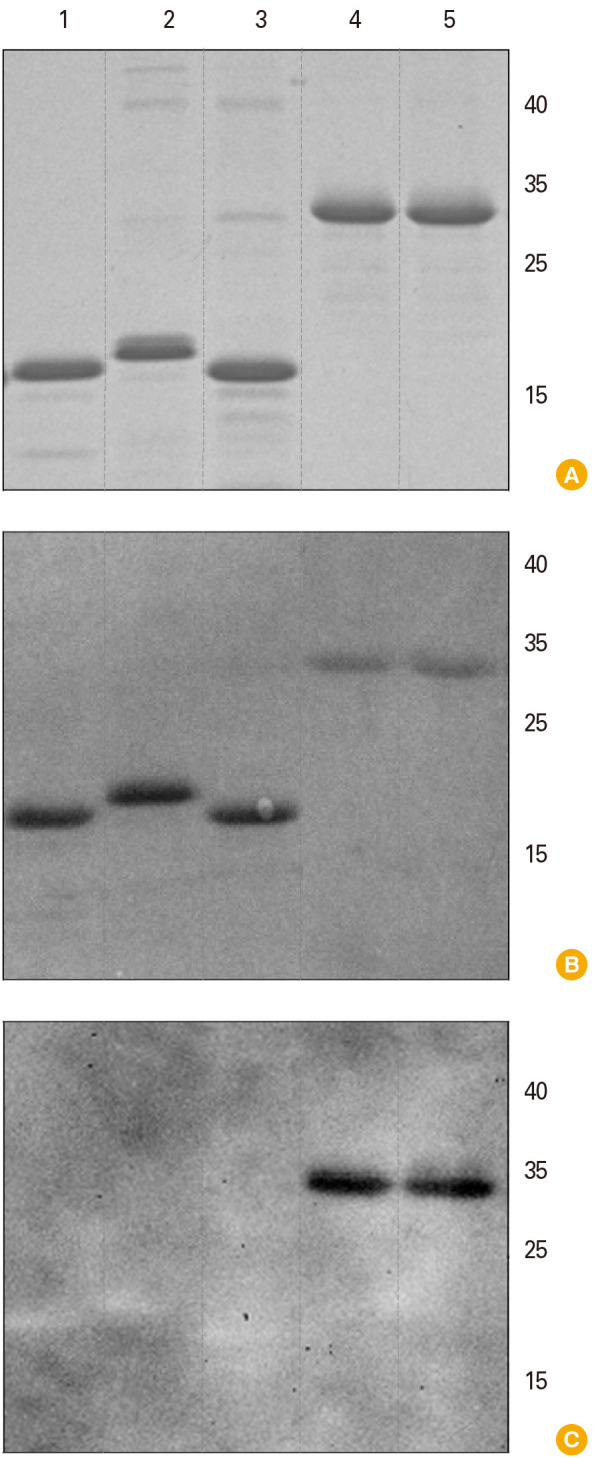
Antigenic specificity was investigated by Western blot analysis, which was performed using commercial polyclonal antisera to five RVA strains: Wa-G1P[8], Ds-1 G2P[4], Ito-G3P[8], Hochi-G4P[8], and 69M-G8P[10]. The results of Western blot analysis are presented in Fig. 2B and C. VP8* antigen proteins of the three genotypes P[4], P[6], and P[8] did not interact with commercial antiserum to rotavirus strains isolated more than 35 years ago, including P[8] isolate Wa (1974), P[8] isolate Ito (1979), P[8] isolate Hochi (1980), P[4] isolate DS-1 (1976), and P[10] isolate 69M (1984) (Fig. 2C, lanes 1–3). At the same time, VP5* and ep-875 antigens were recognized by this antiserum (Fig. 2C, lanes 4, 5).
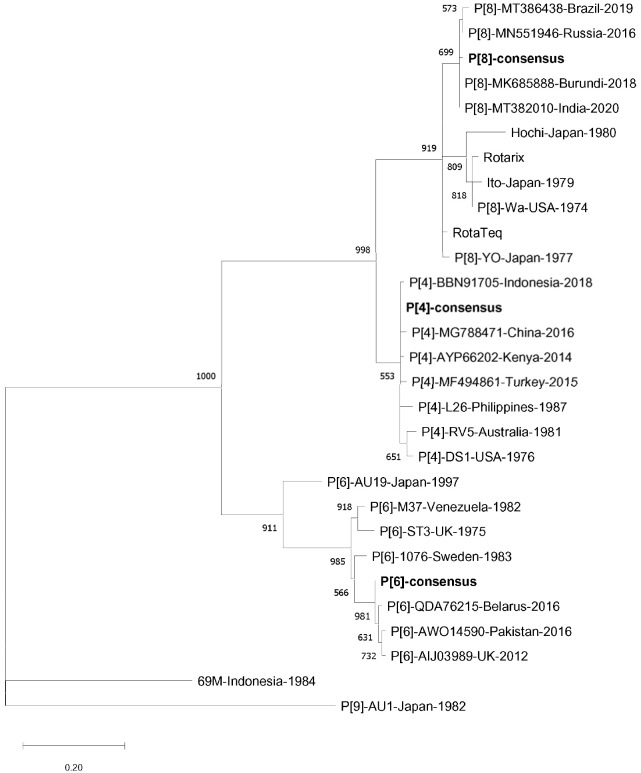
As a follow-up to the results obtained, we performed a phylogenetic analysis based on the amino acid sequences of the VP8* protein fragment (residues 65–223) of human RVA isolates representing genotypes P[4], P[6], and P[8]. It demonstrated that “old” isolates, together with the common rotavirus vaccine strains, formed clusters that were clearly distinct from actual isolates; this conclusion was supported by high bootstrap values for P[8] and P[6] genotypes [28] (Fig. 3). In the case of P[4], genotype isolates collected in 1976 and 1981 were separated with a convincing bootstrap value (651 out of 1,000 replicates); genetic distances among the other P[4] isolates were too small (Fig. 3).
Fig. 3. Phylogenetic tree based on the amino acid sequences of the VP8* protein fragment (residues 65–223) of human rotavirus A isolates representing genotypes P[4], P[6], and P[8]. Synthetic consensus sequences are shown in bold. The tree was reconstructed using the PhyML 3.0 version of maximum likelihood algorithm [28]. Bootstrap values (>50%, 1,000 replicates) are displayed next to the corresponding nodes. GenBank accession numbers of the previously characterized isolates are indicated on the right of the tree with the exception of Hochi (BAF80174), Ito (BAA77545), Wa (AGJ72856), Rotarix (JN849113), RotaTeq (GU565044), YO (AB008279), L26 (M58292), RV5 (M32559), DS1 (AEG25325), AU19 (AB770153), M37 (L20877), ST3 (L33895), 1076 (M88480), 69M (AAA47336), and AU1 (D10970). The scale bar indicates the number of substitutions per residue.
Analysis of amino acid sequence identities between the human RVA P[4], P[6], and P[8] consensus sequences designed in this work, common vaccine strains (Rotarix and RotaTeq) and some representative “old” isolates revealed significant differences. In particular, for the genotype P[8] isolates Wa (1974), Ito (1979), and Hochi (1980), similarity index values were 91.2%, 90.6%, and 84.3%, respectively; for P[6] isolate 1076 (1983)—93.7% and P[4] isolate DS-1 (1976)—96.2%. Identities between actual isolates (2012–2020) and synthetic consensus sequences varied between 98.1% and 100%. Comparison of the consensus sequence VP5* with natural ones gave the following results: for the P[8] isolates including Wa (1974), Ito (1979), and Hochi (1980), the index of similarity varied between 96.1% and 100%.
Discussion
Structural protein VP4 forms asymmetric trimeric spikes. These spikes are located on the surface of viral particles, and there are 60 copies (180 molecules) of these. During an infection, the VP4 trimer undergoes proteolysis into VP5* and VP8*. It is suggested that one of the VP8* subunits in the trimer may be removed by proteolysis [32]. The surface of the virion consists of 260 trimers of glycoprotein VP7 (780 molecules). Proteins VP4 and VP7 comprise the outer layer and are important rotavirus antigens, containing different neutralizing epitopes [32,33,34,35] (Fig. 1).
Here, for creating a panel of antigens for a broad-spectrum vaccine against RVA, two approaches were developed. A consensus approach was used to design RVA antigens based on fragments of VP8* and VP5* proteins (Fig. 1B). The concept of a consensus sequence is being applied in the development of vaccines for the induction of broad-spectrum immune responses, especially for viruses with a high degree of genetic variability [36]. This strategy allows the creation of a synthetic antigen with minimal genetic differences compared to strains currently in circulation, which can help to overcome the high genetic diversity of RVA. The second approach was based on the selection of conserved short neutralizing epitopes of the VP8*, VP5*, and VP7 proteins, followed by the production of a fusion protein containing several repeats of these epitopes to induce a stronger antibody response. A number of studies have shown that epitope repeats in a recombinant protein can enhance the response of antibodies specific to the epitope and contribute to the creation of an effective recombinant vaccine [37,38,39].
A consensus approach was used in the design of extended antigens based on VP8* (genotypes P[4], P[6], and P[8]) and VP5* (genotype P[8]) proteins' fragments. In addition, a gene coding the protein (ep-875) containing several copies of conserved short neutralizing epitopes of VP8*, VP7, and VP5* was created. Western blot analysis demonstrated that three synthetic VP8*-based antigens were not recognized by commercial antiserum against rotavirus strains isolated more than 35 years ago, but the specific activity of the VP5* and ep-875 antigens was confirmed.
Evolutionary changes, the growing number of point mutations of VP8* sequences, the effect of variability in the VP8* antigenic epitopes on vaccine efficacy and the discrepancy between the RVA lines used in licensed or development vaccines and circulating strains are all problems that are widely discussed. The results of the current study suggest that a mismatch between the vaccine VP8* antigen sequences and actual isolates may be the reason for the ineffectiveness of a vaccine based on only one antigen. A recent study demonstrated that monoclonal antibodies to VP5* provide a strong heterotypic response. The authors considered that recombinant VP5* may be effective for developing a broad-spectrum vaccine [15].
Analysis of amino acid sequence identities between the human RVA P[4], P[6], and P[8] consensus sequences designed in this work, common vaccine strains (Rotarix and RotaTeq) and some representative “old” isolates revealed significant differences. VP5* fragment (247–478 amino acid residues) representing the consensus sequence to the most wide-spread genotype P[8] was chosen since it is known that in contrast to variable VP8*, the VP5* protein is more conserved. Therefore, it can serve as a good complement to the antigen panel.
A limitation of this study is the lack of available commercial antisera against new RVA isolates and blood serum samples from patients with a confirmed diagnosis. When designing the antigen panel for creating a broad-spectrum vaccine, we used a consensus sequence approach, suggesting that maximum compliance of the synthetic antigen with actual strains could lead to the development of a more effective vaccine. However, methods for creating consensus sequences are highly dependent on different parameters and the effectiveness of the antigens obtained should be established through further research.
In conclusion, the results of this study together with recently published findings [26], raise the question of the degree of applicability of current rotavirus vaccines, and they also can be useful for the development of new rotavirus vaccines. Five antigens representing sequences of structural proteins belonging to different genotypes can be used in various combinations (from mono- to pentavalent mixtures) for the development of an effective broad-spectrum rotavirus vaccine.
Footnotes
This work was supported by the “National Immunobiological Company” (contract 1-24/18), the Russian Science Foundation (grant no., 18-14-00044) (approaches to antigens design) and Interdisciplinary Scientific and Educational School of Moscow University (Molecular Technologies of the Living Systems and Synthetic Biology).
Supplementary Materials
Supplementary materials are available at Clinical and Experimental Vaccine Research website (http://www.ecevr.org).
Consensus amino acid sequences of antigens VP8* (three genotypes) and VP5* that were employed for “reverse translation” in silico with subsequent cloning of synthetic genes into a vector for expression in Escherichia coli. (A) VP8*-P[4], (B) VP8*-P[6], (C) VP8*-P[8], (D) VP5*.
References
- 1.Tate JE, Burton AH, Boschi-Pinto C, Parashar UD, World Health Organization?Coordinated Global Rotavirus Surveillance Network Global, regional, and national estimates of rotavirus mortality in children <5 years of age, 2000-2013. Clin Infect Dis. 2016;62 Suppl 2:S96–S105. doi: 10.1093/cid/civ1013. [DOI] [PubMed] [Google Scholar]
- 2.GBD Diarrhoeal Diseases Collaborators. Estimates of global, regional, and national morbidity, mortality, and aetiologies of diarrhoeal diseases: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Infect Dis. 2017;17:909–948. doi: 10.1016/S1473-3099(17)30276-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Troeger C, Khalil IA, Rao PC, et al. Rotavirus vaccination and the global burden of rotavirus diarrhea among children younger than 5 years. JAMA Pediatr. 2018;172:958–965. doi: 10.1001/jamapediatrics.2018.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burnett E, Parashar U, Tate J. Rotavirus vaccines: effectiveness, safety, and future directions. Paediatr Drugs. 2018;20:223–233. doi: 10.1007/s40272-018-0283-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang H, Luo G, Zeng Y, et al. The distinct impact of maternal antibodies on the immunogenicity of live and recombinant rotavirus vaccines. Vaccine. 2019;37:4061–4067. doi: 10.1016/j.vaccine.2019.05.086. [DOI] [PubMed] [Google Scholar]
- 6.Groome MJ, Koen A, Fix A, et al. Safety and immunogenicity of a parenteral P2-VP8-P[8] subunit rotavirus vaccine in toddlers and infants in South Africa: a randomised, double-blind, placebo-controlled trial. Lancet Infect Dis. 2017;17:843–853. doi: 10.1016/S1473-3099(17)30242-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coulson BS, Tursi JM, McAdam WJ, Bishop RF. Derivation of neutralizing monoclonal antibodies to human rotaviruses and evidence that an immunodominant neutralization site is shared between serotypes 1 and 3. Virology. 1986;154:302–312. doi: 10.1016/0042-6822(86)90456-3. [DOI] [PubMed] [Google Scholar]
- 8.Dyall-Smith ML, Lazdins I, Tregear GW, Holmes IH. Location of the major antigenic sites involved in rotavirus serotype-specific neutralization. Proc Natl Acad Sci U S A. 1986;83:3465–3468. doi: 10.1073/pnas.83.10.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mackow ER, Shaw RD, Matsui SM, Vo PT, Dang MN, Greenberg HB. The rhesus rotavirus gene encoding protein VP3: location of amino acids involved in homologous and heterologous rotavirus neutralization and identification of a putative fusion region. Proc Natl Acad Sci U S A. 1988;85:645–649. doi: 10.1073/pnas.85.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taniguchi K, Maloy WL, Nishikawa K, et al. Identification of cross-reactive and serotype 2-specific neutralization epitopes on VP3 of human rotavirus. J Virol. 1988;62:2421–2426. doi: 10.1128/jvi.62.7.2421-2426.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coulson BS, Kirkwood C. Relation of VP7 amino acid sequence to monoclonal antibody neutralization of rotavirus and rotavirus monotype. J Virol. 1991;65:5968–5974. doi: 10.1128/jvi.65.11.5968-5974.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larralde G, Li BG, Kapikian AZ, Gorziglia M. Serotype-specific epitope(s) present on the VP8 subunit of rotavirus VP4 protein. J Virol. 1991;65:3213–3218. doi: 10.1128/jvi.65.6.3213-3218.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobayashi N, Taniguchi K, Urasawa S. Identification of operationally overlapping and independent cross-reactive neutralization regions on human rotavirus VP4. J Gen Virol. 1990;71(Pt 11):2615–2623. doi: 10.1099/0022-1317-71-11-2615. [DOI] [PubMed] [Google Scholar]
- 14.Kovacs-Nolan J, Yoo D, Mine Y. Fine mapping of sequential neutralization epitopes on the subunit protein VP8 of human rotavirus. Biochem J. 2003;376(Pt 1):269–275. doi: 10.1042/BJ20021969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nair N, Feng N, Blum LK, et al. VP4- and VP7-specific antibodies mediate heterotypic immunity to rotavirus in humans. Sci Transl Med. 2017;9:eaam5434. doi: 10.1126/scitranslmed.aam5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng N, Hu L, Ding S, et al. Human VP8* mAbs neutralize rotavirus selectively in human intestinal epithelial cells. J Clin Invest. 2019;129:3839–3851. doi: 10.1172/JCI128382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doro R, Laszlo B, Martella V, et al. Review of global rotavirus strain prevalence data from six years post vaccine licensure surveillance: is there evidence of strain selection from vaccine pressure? Infect Genet Evol. 2014;28:446–461. doi: 10.1016/j.meegid.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kondakova OA, Nikitin NA, Trifonova EA, Atabekov JG, Karpova OV. Rotavirus vaccines: new strategies and approaches. Mosc Univ Biol Sci Bull. 2017;72:169–178. [Google Scholar]
- 19.Santos N, Hoshino Y. Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Rev Med Virol. 2005;15:29–56. doi: 10.1002/rmv.448. [DOI] [PubMed] [Google Scholar]
- 20.Folorunso OS, Sebolai OM. Overview of the development, impacts, and challenges of live-attenuated oral rotavirus vaccines. Vaccines (Basel) 2020;8:341. doi: 10.3390/vaccines8030341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bowen MD, Mijatovic-Rustempasic S, Esona MD, et al. Rotavirus strain trends during the postlicensure vaccine era: United States, 2008-2013. J Infect Dis. 2016;214:732–738. doi: 10.1093/infdis/jiw233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogden KM, Tan Y, Akopov A, et al. multiple introductions and antigenic mismatch with vaccines may contribute to increased predominance of G12P[8] rotaviruses in the United States. J Virol. 2018;93:e01476-18. doi: 10.1128/JVI.01476-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harastani HH, Reslan L, Sabra A, et al. Genetic diversity of human rotavirus a among hospitalized children under-5 years in Lebanon. Front Immunol. 2020;11:317. doi: 10.3389/fimmu.2020.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matthijnssens J, Bilcke J, Ciarlet M, et al. Rotavirus disease and vaccination: impact on genotype diversity. Future Microbiol. 2009;4:1303–1316. doi: 10.2217/fmb.09.96. [DOI] [PubMed] [Google Scholar]
- 25.Groome MJ, Fairlie L, Morrison J, et al. Safety and immunogenicity of a parenteral trivalent P2-VP8 subunit rotavirus vaccine: a multisite, randomised, double-blind, placebo-controlled trial. Lancet Infect Dis. 2020;20:851–863. doi: 10.1016/S1473-3099(20)30001-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Velasquez DE, Jiang B. Evolution of P[8], P[4], and P[6] VP8* genes of human rotaviruses globally reported during 1974 and 2017: possible implications for rotavirus vaccines in development. Hum Vaccin Immunother. 2019;15:3003–3008. doi: 10.1080/21645515.2019.1619400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y, Huang P, Tan M, et al. Rotavirus VP8*: phylogeny, host range, and interaction with histo-blood group antigens. J Virol. 2012;86:9899–9910. doi: 10.1128/JVI.00979-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 29.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lipman DJ, Pearson WR. Rapid and sensitive protein similarity searches. Science. 1985;227:1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- 31.Ryabchevskaya EM, Evtushenko EA, Granovskiy DL, et al. Two approaches for the stabilization of Bacillus anthracis recombinant protective antigen. Hum Vaccin Immunother. 2021;17:560–565. doi: 10.1080/21645515.2020.1772632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Settembre EC, Chen JZ, Dormitzer PR, Grigorieff N, Harrison SC. Atomic model of an infectious rotavirus particle. EMBO J. 2011;30:408–416. doi: 10.1038/emboj.2010.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sehnal DR, Rose AS, Koca J, Burley S, Velankar S. Mol*: towards a common library and tools for web molecular graphics; Proceedings of the Workshop on Molecular Graphics and Visual Analysis of Molecular Data; 2018 Jun 4; Brno, Czech Republic. Goslar: The Eurographics Association; 2018. pp. 29–33. [Google Scholar]
- 34.Berman HM, Westbrook J, Feng Z, et al. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jmol: an open-source Java viewer for chemical structures in 3D [Internet] [place unknown]: Jmol; c2021. [cited 2021 Jan 2]. Available from: http://www.jmol.org. [Google Scholar]
- 36.Carter DM, Darby CA, Lefoley BC, et al. Design and characterization of a computationally optimized broadly reactive hemagglutinin vaccine for H1N1 influenza viruses. J Virol. 2016;90:4720–4734. doi: 10.1128/JVI.03152-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kovacs-Nolan J, Mine Y. Tandem copies of a human rotavirus VP8 epitope can induce specific neutralizing antibodies in BALB/c mice. Biochim Biophys Acta. 2006;1760:1884–1893. doi: 10.1016/j.bbagen.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 38.Park SY, Kim HJ, Seo JY, Choi EY, Oh SW. Production of an epitope-specific antibody using recombinant repetitive oligonucleotides. Anim Cells Syst. 2014;18:259–266. [Google Scholar]
- 39.Zuqiang L, Zuquang W, Yinghua C. Repeated epitope in the recombinant epitope-peptide could enhance ELDKWA-epitope-specific antibody response. Prog Nat Sci. 2005;15:89–92. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Consensus amino acid sequences of antigens VP8* (three genotypes) and VP5* that were employed for “reverse translation” in silico with subsequent cloning of synthetic genes into a vector for expression in Escherichia coli. (A) VP8*-P[4], (B) VP8*-P[6], (C) VP8*-P[8], (D) VP5*.