Abstract
All perennial plants harbor diverse endophytic fungal communities, but why they tolerate these complex asymptomatic symbioses is unknown. Using a multi-pronged approach, we conclusively found that a dryland grass supports endophyte communities comprised predominantly of latent saprophytes that can enhance localized nutrient recycling after senescence. A perennial bunchgrass, Stipagrostis sabulicola, which persists along a gradient of extreme abiotic stress in the hyper-arid Namib Sand Sea, was the focal point of our study. Living tillers yielded 20 fungal endophyte taxa, 80% of which decomposed host litter during a 28-day laboratory decomposition assay. During a 6-month field experiment, tillers with endophytes decomposed twice as fast as sterilized tillers, consistent with the laboratory assay. Furthermore, profiling the community active during decomposition using next-generation sequencing revealed that 59–70% of the S. sabulicola endophyte community is comprised of latent saprophytes, and these dual-niche fungi still constitute a large proportion (58–62%) of the litter community more than a year after senescence. This study provides multiple lines of evidence that the fungal communities that initiate decomposition of standing litter develop in living plants, thus providing a plausible explanation for why plants harbor complex endophyte communities. Using frequent overnight non-rainfall moisture events (fog, dew, high humidity), these latent saprophytes can initiate decomposition of standing litter immediately after tiller senescence, thus maximizing the likelihood that plant-bound nutrients are recycled in situ and contribute to the nutrient island effect that is prevalent in drylands.
Keywords: endophyte, latent saprophyte, decomposition, fungal community, drylands, nutrient islands
Introduction
All perennial plants sampled to date harbor asymptomatic fungal endophytes in the apoplast of their stems, leaves and roots. Even in the most extreme habitats, where plant production and reproduction are severely constrained by thermal, moisture and/or nutrient stress, plants support diverse endophytic fungal communities (Rosa et al., 2009; Sun et al., 2012; Zabalgogeazcoa et al., 2013). Co-adaptive one-to-one relationships are well-known between endophytes and their plant hosts: fungi play roles in ameliorating thermal, drought, soil disturbance and herbivory-related stresses (Porras-Alfaro and Bayman, 2011), offsetting the nutritional costs associated with supporting specific endosymbionts. The question remains, however, why many plants support diverse endophyte communities instead of more specialized relationships to avail these services.
Given the energetic costs associated with supporting endosymbionts in general, and the potential challenges of navigating relationships with multiple endophytic taxa, it is still unclear what benefits might accrue to plants from hosting diverse endophytic communities. While these dynamics have been relatively unexplored among endophytes, associations between mycorrhizal fungi and their plant hosts offer a useful parallel. Mycorrhizae are similarly common associations between fungi and host plants that share nutritional and stress-related benefits, but under extreme abiotic stress (disturbance, drought, and salinity), plants may reject their mycorrhizal fungal partners (Allen, 1991). For instance, perennial grasses in the Namib Sand Sea (NSS; Figure 1) co-occur with their mycorrhizal fungi across a gradient of moisture stress, but drop these associations in the hyper-arid western regions of the NSS (Jacobson, 1997). To explore whether there are similar limits for plants supporting their endophytic symbionts, we sampled tillers of an endemic perennial grass, Stipagrostis sabulicola across this NSS gradient. We predicted that due to the moisture gradient, endophytic communities in the eastern region (∼65 mm/year) would be comparable to those in other drylands (Rosa et al., 2009; Sun et al., 2012; Massimo et al., 2015), but absent in the extreme environment of the western NSS (< 19mm/year) where plants appear to minimize their energetically costly interactions with microbial symbionts.
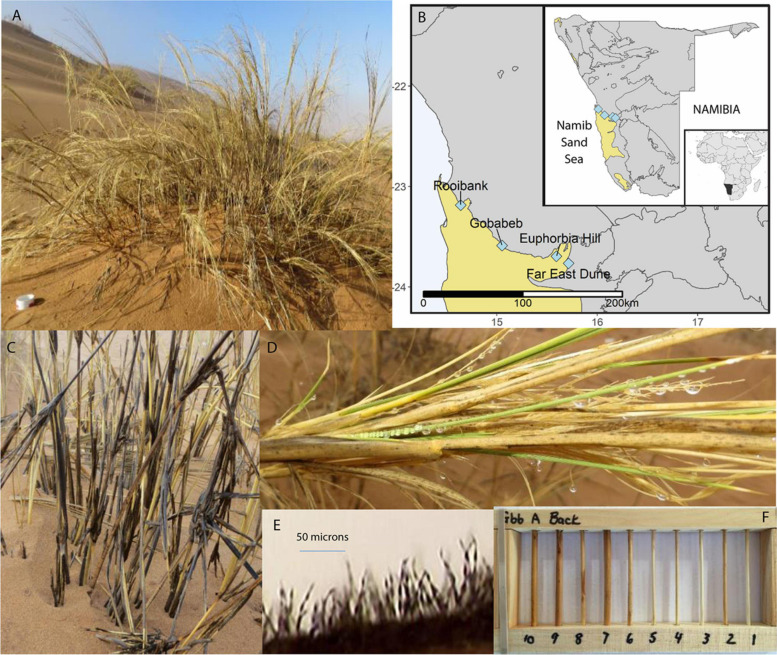
FIGURE 1.
Overview of Stipagrostis sabulicola in the Namib Sand Sea (NSS) context. (A) S. sabulicola in the NSS. Scale is 10 cm diameter PVC pipe in the foreground. (B) Endophyte sampling sites spanning the width of the NSS. Maps created using public domain shapefiles accessed through the Natural Earth database (https://www.naturalearthdata.com/) and the Digital Atlas of Namibia (http://www.uni-koeln.de/sfb389/e/e1/download/atlas_namibia/main_ namibia_atlas.html). (C) Standing senescent tillers of S. sabulicola moistened by an 8-h fog are dark gray because of fungal growth in response to these frequent brief moisture pulses. (D) Dew droplets on living tillers and inflorescence. Note gray spots on senesced leaves which are fungal colonies. (E) Sporulating fungal hyphae on tiller surface following exposure to 10 h of 100% humidity. (F) Wooden racks containing 9-cm tiller sections used during the 6-month decomposition experiment at Gobabeb.
We also explored the benefits an endophyte community might afford its perennial host. We hypothesized that plants tolerate complex endophyte communities because they serve as latent saprophytes in standing tillers, initiating the decomposition process immediately after a tiller senesces. Endophyte communities are commonly comprised of fungi that are plant litter and soil saprophytes (Osono, 2006; Purahong and Hyde, 2011), and indeed, more than half of the saprophytic fungi isolated from standing litter of S. sabulicola in a previous study were common endophytes of plants in other ecosystems (Jacobson et al., 2015). Aridity does not preclude surface litter decomposition because saprophytic fungi are active during frequent non-rainfall moisture (NRM; i.e., fog, dew, and high humidity) and small rain events in the Namib (Evans et al., 2020). Indeed, a recent study revealed that community richness and Shannon diversity indices of litter fungi did not differ among eastern and western NSS sites, although the communities themselves were distinct (Logan et al., 2021). Furthermore, NRM is an important driver of surface litter decomposition in all grasslands studied to date: arid, semi-arid, and temperate grasslands and wetlands (Kuehn et al., 1999; Dirks et al., 2010; Wang et al., 2017; Evans et al., 2020; Lodato et al., 2021). Decomposition of grass tillers and leaves comprised of lignocellulose is a complex successional process accomplished by a suite of fungal species (Deacon, 2006; Lodato et al., 2021). If much of a plant’s endophyte community is comprised of latent saprophytes, this could explain why plants support diverse fungal communities, in addition to specific taxa that might support physiological processes of the living plant. Essential macronutrients needed for plant growth are notoriously low in dryland soils (Klopatek and Stock, 1994; Abrams et al., 1997; André et al., 1997; Jacobson, 1997), and therefore a perennial plant’s ability to efficiently recycle plant-bound nutrients from above-ground structures back to roots may be an essential trait ensuring its success. Indeed, nutrient islands surrounding perennial plants are a common feature of dryland plant communities (Schlesinger et al., 1995).
We used a multi-pronged approach to test our hypotheses. We determined the endophyte contribution to decomposition of freshly senesced standing litter with a field study of mass loss rates from sterile and non-sterile tillers over a 6-month period, predicting that if tillers contain endophytes that are latent saprophytes, sterilized tillers would decompose more slowly. We also assessed whether endophytes isolated from S. sabulicola were efficient decomposers in vitro and compared results of this experiment with the field study. Finally, we used next-generation sequencing (NGS) methods to estimate the overlap between living plant endophyte and year-old litter saprophyte communities. If plants support a diverse endophyte community because these fungi are an important inoculum source that initiates efficient standing litter decomposition immediately following senescence, we would expect a large proportion of the endophyte community to remain present and active in the decomposing surface litter.
Materials and Methods
Sites and Endophyte Sampling
Living tiller and dead tiller (litter) S. sabulicola samples originated from two regions, which span the moisture gradient of the NSS (Figure 1). NRM sources predominate at the two western sites: Rooibank (RB: S 23.186566° E 14.640453°) and Gobabeb (GB: S 23.588148° E 15.052270°) (Evans et al., 2020; Logan et al., 2021). In contrast, rainfall is the predominant moisture source at the two eastern sites: Euphorbia Hill (EH: S 23.694694°, E 15.593583°) and Far East Dune (FE: S 23.765639°, E 15.714639°). At Gobabeb, mean annual moisture derived from NRM is three times greater than mean annual rainfall (∼52 mm vs. ∼19 mm respectively) (Lancaster et al., 1984; Eckardt et al., 2013) whereas at the Far East Dune, mean annual rainfall is ∼65 mm and NRM is ten times less frequent than at Gobabeb (Evans et al., 2020; Logan et al., 2021).
Tiller samples were collected in January 2014 from asymptomatic living plants, with no visible fungal infection (western region: n = 5 (GB:2, RB:3); eastern region: n = 3 (FE:2, EH:1). Each sample consisted of at least ten tillers (4–5 mm diameter stems) from a single grass hummock (1–3 m diameter) located at least 20 m apart from other sample positions at the site. Tillers were cut to 20 cm lengths with a sterile razorblade and stored at 4°C in sterile Whirlpak plastic bags until fungal or DNA isolation was performed.
Characterization of S. sabulicola Fungal Endophyte Community
Fungal Isolation
Endophytes were isolated according to a protocol adapted from Kumar and Kaushik (2013) and (Arnold et al., 2007). Twelve 15 mm-tiller segments from each sample were surface-sterilized, by submersion for 2 min each in 0.5% sodium hypochlorite followed by 70% ethanol, followed by a sterile water rinse. The effectiveness of the sterilization procedure was confirmed by imprinting on nutrient media (Schulz et al., 1998), prior to plating the tiller segment onto 2% MEA nutrient medium supplemented with antibiotic streptomycin (30 ug/ml). Subcultures from the growing edge of each emerging colony were taken prior to sporulation, transferred to 2% MEA nutrient plates, and incubated at 25°C in a dark growth chamber. Pure isolates were generated by subsequent rounds of sub-culturing. We made replicate stock cultures from each of these purified isolates that were stored at 4°C for immediate use for DNA extraction and the decomposition assay, and for long-term storage at −80°C in the KJCC permanent culture collection at Grinnell College (KJCC#1166-1236).
Identification, Characterization, and Culture-Dependent Diversity Assessment
Isolates were initially grouped into morphologically distinct operational taxonomic units (OTUs) and identified to genus and species using available taxonomic resources. DNA was then extracted from all unique OTUs and multiple isolates of common OTUs (60 total) according to the E.Z.N.A.®, Fungal DNA Mini Kit Protocol for fresh or frozen specimens (OMEGA Bio-Tek 2011). DNA was quantified using fluorometry and portions of the ITS region and the 18S rDNA gene were amplified via polymerase chain reaction (PCR) with Platinum®, PCR SuperMix (Life Technologies) and primers ITS1 + ITS4 (ITS region) and NS5 + NS6 (18 S rDNA gene) respectively. PCR products were purified using the QIAquick® PCR Purification Kit Protocol and sequenced on an ABI Prism 3730XL sequencing instrument using the ABI prism BigDyeTM terminator cycle sequence kit (Applied Biosystems, Foster City, CA, United States). The SeqMan sequence alignment and analysis software (Lasergene; DNASTAR, Madison, WI, United States) was used for sequence editing and alignment. Sequences were identified by BLAST to Genbank TYPE entries1 (Ko et al., 2011) and CBS2 databases, and positive identification was determined as coverage of >80% and identity match of >98.0% (Raja et al., 2017). Sequence data for all identified isolates is archived in GenBank (accessions MF038136-MF038191). We performed two-sample t-tests (Minitab Version 19) to determine whether measures of abundance (mean isolation frequency, mean number of isolates/sample) and richness of isolates at the western sites differed from those at the east sites. We used the Welch’s test (not assuming equal variances) to account for possible effects of differences in sample size between groups.
Metagenomic Comparative Assessment of Whole Communities in Living and Senesced Host Material
We used NGS to compare endophytic communities in living tillers and saprophytic communities in dead surface litter (6 plants, 3 samples per plant) collected at Gobabeb and Far East Dune sites In November 2014. DNA was extracted from subsets of the eight S. sabulicola tiller samples used in the culture-dependent studies described above. Isolations were done on tillers that were surface-sterilized as above, using a MoBio Powersoil DNA extraction kit. DNA was stored at −80°C until further analysis.
For saprophytic communities, we wanted to identify taxa not only present on dead tillers, but also actively decomposing litter (i.e., functioning as saprophytes). To do this, we incubated samples with Bromodeoxyuridine (BrdU) and separated active DNA using immunocapture. BrdU is an analog of thymidine, and therefore can be used to tag proliferating, active cells (Borneman, 1999; Goldfarb et al., 2011; McMahon et al., 2011). We incubated the 18 litter samples at 25°C in sterile falcon tubes for 36 h, while growing microorganisms incorporated BrdU molecules into replicating DNA. We chose 36 h because it captures the period when much of the growth and activity of microbes after rewetting is known to occur (Iovieno and Baath, 2008; Placella et al., 2012; Göransson et al., 2013), but short enough to avoid significant fungal turnover or use of BrdU as a substrate by other microbes. DNA was extracted immediately after BrDU incubations using MoBio PowerSoil DNA Isolation Kit (MoBio Laboratories, Solano, CA, United States). After extraction, antibody immunocapture was performed to isolate BrdU-labeled DNA from inactive DNA (McMahon et al., 2011), resulting in 18 DNA samples of active DNA and 18 samples of ‘total’ DNA from the litter samples. We evaluated the efficacy of the BrdU immunocapture by visualizing DNA and amplified PCR product on agarose gels, and by comparing bands to a control sample incubated with only water, without BrdU.
Fungi from both the dead litter (n = 18 active and 18 total) and living tillers (n = 8) samples were characterized by amplifying the ITS1 region using primers ITS1-F/ITS2 and sequencing on the Illumina MiSeq platform (Michigan State University Genomics Core Facility). Sequences were processed using the USEARCH pipeline (v.8.1). We identified representative sequences for each OTU with referenced (UNITE) based chimera checking, and made taxonomic assignments using utax (v. 9.2) classifier against UNITE 7.1. The OTU table was constructed using USEARCH with taxonomic assignments from RDP (Kõljalg et al., 2013). Sequencing resulted in an average of 31,032 sequences per sample, before filtering. We included three blanks that followed the same extraction protocol as the samples but that instead used sterile water. The 32 species reported in the blanks were removed from the 44 experimental samples. Filtering included exclusion of samples with sequence numbers < 1000, taxa that were not present in more than two samples of the whole dataset (endophyte & saprophyte), and taxa with less than 5 reads across endophyte samples (n = 8) and 25 reads across decomposer samples (n = 36). All sequences are available in NCBI under the BioProject ID of PRJNA726949.
To examine endophyte diversity, we rarefied samples to the lowest sequence per sample (∼18,000) and calculated mean richness per site. We were interested in the dual presence of OTUs in dead litter (acting as saprophytes) and surface-sterilized live stems (acting as endophytes), so we summed the sequence counts for all saprophyte samples within sites and quantified the occurrence of endophytes in the aggregated saprophytes. We also examined the relative abundance of these taxa in endophyte versus saprophyte samples, testing differences using the same statistical procedures as above.
In vitro Decomposition Assays
Newly senesced standing Stipagrostis sabulicola tiller litter was collected from plants at the Gobabeb site in June 2014 for the decomposition assay. The flowering stalks of these tillers were still in place indicating recent senescence. Yellow tillers (4–5 mm diameter) with no signs of coloration, cuticle damage, or fungal infection, were cut with a sterile razor blade in the late afternoon when relative humidity was 10–20%, air-dried, and kept in sterile Whirlpak bags at room temperature until used.
The decomposition assay was based on a protocol developed by Osono and Takeda, 2006. Yellow tiller litter was cut to 10 mm lengths, halved laterally, and dried at 60°C overnight. Sets of three segments comprised each sample and were weighed, placed in microcentrifuge tubes and sterilized by dry autoclaving at 120°C for 20 min. Autoclaving did not affect the mass of the segments: dry mass of three randomly chosen samples before and after autoclaving did not differ (T-stat = 1.0, p = 0.423). Using sterile technique, the three sterile segments comprising each sample were placed approximately 10 mm apart and 15 mm from a designated central inoculation site on Petri dishes containing water agar. Subcultures (2 × 2 × 2 mm) from the growing margin of 66 endophyte isolates were placed on the central inoculation point of each Petri dish (n = 10). Controls were similarly prepared but without inoculation. The assay and control plates were incubated in a growth chamber for 28 days with a diel cycle of thermal and light regimes that simulated average summer ambient conditions at ∼ 30 cm above the sand surface on S. sabulicola tillers at Gobabeb (Ebner et al., 2011): 10 h - 15°C, dark; 2 h - 20°C light, 1 h - 25°C light; 1 h - 30°C, light; 4 h - 35°C, light, 2 h - 30°C, light; 2 h - 25°C, light; and 2 h - 20°C, light. At the conclusion of the incubation period, all tiller samples were brushed free of superficial mycelial growth using a dry task wiper (Kimberly-ClarkTM Professional Kimtech ScienceTM KimwipesTM), oven-dried at 60°C, and weighed to determine final dry mass.
Mean percent mass loss was computed for each fungal isolate as a measure of decomposition ability (Osono and Takeda, 2006). We determined that an isolate was able to decompose S. sabulicola tillers if mean percent mass loss was significantly greater (p < 0.05) than that of the un-inoculated controls, using 1-way 2-sample t-tests. Mean normalized decomposition ability (NDA) was then computed for each isolate by subtracting the mean percent mass loss of un-inoculated controls from percent mass loss of each isolate sample. Mean NDA for each isolate most accurately represents the percent mass loss attributable solely to the fungal treatments, by accounting for mass loss that occurred from leaching into the water agar medium or by photodegradation. A t-test was used to determine whether the mean percentage of saprophytic isolates (i.e., with positive NDA) differed between the western and eastern regions of the dune field.
Laccase and peroxidase enzyme assays (Viswanath et al., 2008) were conducted on all endophytic isolates. We predicted that as primary colonizers, these isolates would exhibit minimal lignin-degrading enzyme activity. We also performed a post hoc 2-sample t-test to determine whether there was a significant difference in the NDA of isolates based on presence or absences of lignin-degrading enzymes.
Endophyte-Mediated Decomposition in situ
We deployed sterilized and un-sterilized newly senesced tillers in wooden racks 40 cm above the ground at Gobabeb for 6 months, to determine whether endophytes play a measurable role in standing tiller decomposition immediately following senescence (Figure 1C). Ambient moisture and temperature conditions for 15 June through 15 December 2016 were determined from data downloaded from the Gobabeb Met SASSCAL meteorological station3. Twenty tillers from Gobabeb (see decomposition assay above for details) were cut to standard 9 cm lengths, half were dry-autoclaved to eliminate the endogenous fungal community, and half were not. Each tiller was weighed using an analytical balance (to 0.1 mg) and loaded randomly into two wooden racks. After 6 months of exposure, the mass of each tiller was determined as above, and differences in percent mean mass loss between the endophyte and sterilized treatments were compared using two-sample t-tests.
Field mass loss resulting from the endophyte community was also compared with the range of NDA obtained for the isolates by converting both to hourly rates. The total number of hours of fungal activity during the field experiment was conservatively estimated by summing the number of hours during the 6-month period when relative humidity was greater than 90%, and lasted for at least 6 h (55 events, 457 h). Previous investigations have shown that saprophyte communities of S. sabulicola are metabolically active under these conditions (Jacobson et al., 2015; Evans et al., 2020). The total number of hours of fungal activity during the in vitro experiments was 672 h (28 days × 24 h), because the isolates were able to grow at all temperatures used in the diel cycle (Jacobson et al., 2015) and moisture was never a limiting factor.
Results
Richness and Abundance of Endophytes Associated With Living and Senesced S. sabulicola Tillers
We obtained 71 endophyte isolates from 96 S. sabulicola tiller segments, corresponding to an isolation frequency across both regions of 74%. All but three of the 71 isolates were identified at the order level or lower using sequence data. These three had no morphologically identifiable features and were excluded from subsequent analyses. The remaining 68 isolates comprised 20 different taxa (Figure 2 and Table 1), of which four were identified at the generic level but could not be resolved to species. The sequences of these four taxa were aligned with congeneric taxa from the data set and were distinct, so were included in subsequent analyses.
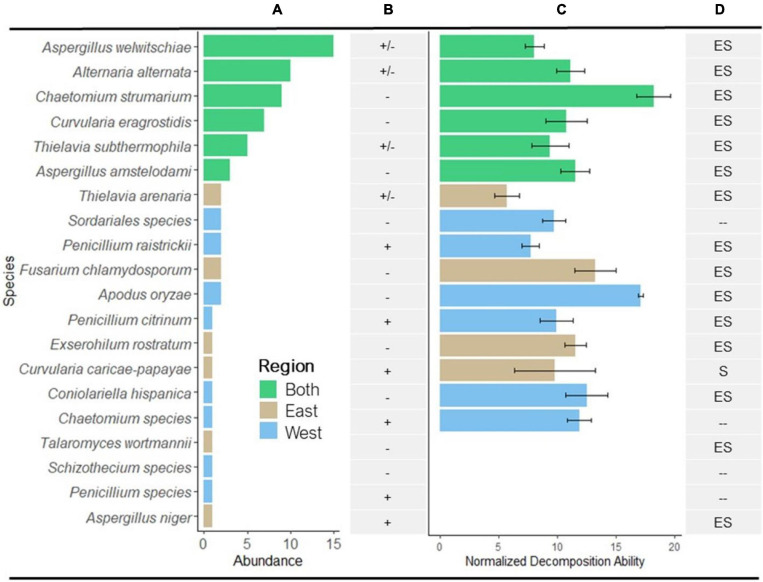
FIGURE 2.
Abundance and decomposition ability of isolated fungi. (A) Distribution of the 20 identified species from eastern and western regions of the Namib Sand Sea (see Table 1 for number of isolates/species). Bar colors correspond to geographical regions. (B) Presence or absence of lignin-degrading enzymes laccase and peroxidase; (+), enzyme presence in all isolates; (–), no enzyme present; (±), enzyme present in some isolates and not present in others. (C) Mean normalized decomposition ability (NDA; mean % mass loss) of isolates that were able to decompose sterile S. sabulicola tillers over 28 days in the Namib diel cycle. Values were normalized relative to mass loss in un-inoculated tillers (Isolate sample size as in A, error bars = 1 × SE; n = 10 for each isolate). (D) Summary of known nutritional modes of endophytic species isolated from S. sabulicola: E, endophyte; S, saprophyte; –, no data as taxa was not identified to species (see Table 1 for references).
TABLE 1.
Endophytic taxa isolated from Stipagrostis sabulicola.
| Taxon (order, family, genus, and species) | Coverage/similarity (%) | Laccase and peroxidase assay | Endophyte references | Saprophyte references |
| Eurotiales | ||||
| Aspergillaceae | ||||
| Aspergillus amstelodami Thom and Church | 99/99 | No (3/3) | Sanchel Marquez et al., 2008 | Pinciroli, 2013 |
| Aspergillus niger Tiegh | 92/99 | Yes (1/1) | Sanchel Marquez et al., 2008 | Jacobson et al., 2015 |
| Aspergillus welwitschiae (Bres.) Henn. | 92/99 | Yes (12/14) | Liu et al., 2019 | Hong et al., 2013 |
| Penicillium citrinum Thom | 100/100 | Yes (1/1) | Khan et al., 2008 | Pinciroli, 2013 |
| Penicillium raistrickii G. Sm. | 100/100 | Yes (2/2) | Busby et al., 2016 | Pandey and Palni, 1998 |
| Penicillium species | No match | Yes (1/1) | NA | NA |
| Trichocomaceae | ||||
| Talaromyces wortmannii (Kloeker) C.R. Benj. | 100/98 | No (1/1) | Bara et al., 2013 | Yilmaz et al., 2014 |
| Hypocreales Nectriaceae | ||||
| Fusarium chlamydosporum Wollenw and Reinking | 100/100 | No (2/2) | Ananda and Sridhar, 2002 | Jacobson et al., 2015 |
| Sordariales | ||||
| Sordariales species | No match | No (2/2) | NA | NA |
| Chaetomiaceae | ||||
| Chaetomium strumarium (Rai, Tew, and Muki) Cann. | 98/99 | No (10/10) | Uma Anitha and Mythili, 2017 | Jacobson et al., 2015 |
| Chaetomium species | No match | Yes (1/1) | NA | NA |
| Thielavia arenaria Mouch. | 98/99 | Yes (1/2) | Maciá-Vicente et al., 2008 | Jacobson et al., 2015 |
| Thielavia subthermophila Mouch. | 99/99 | Yes (2/5) | Mouchacca, 2007 | Jacobson et al., 2015 |
| Lasiosphaeriaceae | ||||
| Apodus oryzae Carolis and Arx | No TYPE; 99/99 | No (2/2) | Angelini et al., 2012 | Geydan et al., 2012 |
| Schizothecium species | No match | No (2/2) | NA | NA |
| Pleosporales Pleosporaceae | ||||
| Alternaria alternata E.G. Simmons | 98/99 | Yes (6/10) | Kumar and Kaushik, 2013 | Jacobson et al., 2015 |
| Curvularia caricae-papayae H.P. Srivast. and Bilgrami | 99/100 | Yes (1/1) | Not known | Not known |
| Curvularia eragrostidis (Henn.) J.A. Mey. | 98/99 | No (6/6) | Kaushik et al., 2014 | Granados-Montero et al., 2018 |
| Xylariales Xylariaceae | ||||
| Exserohilum rostratum (Drechsler) K.J. Leonard and Suggs | 99/98 | No (1/1) | Costa et al., 2012 | Gomaa et al., 2015 |
| Coniolariella hispanica Checa, Arenal, and J.D. Rogers | 100/99 | No (1/1) | Checa et al., 2008 | Hao et al., 2017 |
NCBI Blast results % coverage and % Similarity are all TYPE search results unless otherwise indicated. Results of the laccase and peroxidase enzyme assays: Yes vs. No indicates whether isolates tested positive or negative in enzyme assay (with bracketed fraction indicating the proportion of the isolates tested). Summary of known nutritional modes for each taxon.
Of the 20 identified taxa, the six most abundant species were found in both regions, collectively comprising 72% of all identified isolates: Aspergillus welwitschiae (15 isolates), Alternaria alternata (10), Chaetomium strumarium (9), Curvularia eragrostidis (7), Thielavia subthermophila (5), and Aspergillus amstelodami (3) (Figure 2A). Of the remaining 14 species (19 isolates), comprising 28% of the isolates, 12% (6 species, 8 isolates) were found only in the eastern region and 16% (8 species, 11 isolates) were found only in the western region. We observed no difference in mean richness and abundance between the eastern and western regions of the NSS (p = 0.921 and p = 0.414, respectively) (Table 2).
TABLE 2.
Comparison of the endophyte communities in living tillers in the eastern and western regions of the Namib Sand Sea. Mean abundance, richness and decomposition metrics (± SD) of fungal endophyte communities of S. sabulicola obtained by (1) isolate-based sampling and (2) next-generation sequencing.
| 1. Isolate-based sampling | ||||
| Region |
||||
| West | East | T-stat | p value | |
| Mean % isolation frequency (isolates/segment) | 63 (9.5) | 78 (5.2) | 1.58 | 0.364 |
| Mean isolates/site | 19.5 (2.1) | 14.5 (4.9) | 1.31 | 0.414 |
| Mean richness/site | 9 (5.6) | 8.5 (0.7) | 0.12 | 0.921 |
| % NDA + isolates/site1 | 64 (8) | 45 (12) | 0.75 | 0.590 |
| Mean % mass loss | 12.64 (4.78) | 10.37 (3.91) | 1.70 | 0.098 |
| 2. Sequence-based sampling | ||||
|
Region |
||||
| West | East | T-stat | p value | |
| Mean OTU richness/site – all samples | 37.6 (6.61) | 38.8 (7.4) | 0.19 | 0.854 |
| Mean OTU richness/site – abundant2 samples only | 22 (6.55) | 27.2 (6.61) | 1.082 | 0.335 |
| Mean% active saprophytic richness in living tillers3 | 57.35 (8.14) | 67.18 (9.87) | 1.46 | 0.242 |
| Mean% active saprophytic abundance in living tillers4 | 55.5 (21.4) | 83.4 (23.6) | 1.67 | 0.193 |
1NDA + isolates are those capable of decomposing dead S. sabulicola tillers. 2Abundant defined as sum of occurrence across all 8 samples >20. 3OTUs present in living tillers that were also in active litter. 4Reads of OTUs present in living tillers that were also in active litter.
Corroborating the results from the isolate-based approach, we observed no significant differences in richness between the eastern and western regions using sequence-based community profiling (Table 2). NGS sequencing identified 168 endophyte taxa associated with living tillers. This is much greater than the richness values obtained from the isolate-based community assessment, which was expected because previous studies have shown that very large numbers of isolate samples are needed to yield comparable richness assessments to high-throughput sequencing (O’Brien et al., 2005; Arnold et al., 2007; Higgins et al., 2011). Despite the differences in richness estimates from the two methods, both revealed communities with similar structure, comprised of a small number of abundant taxa and a large number of rare taxa: the 50 most abundant taxa (30% of taxa) comprised 96% of the read abundance (isolates: top 30% comprise 72%). A comparison of only the most abundant OTUs across the two regions confirm the insignificant results from the isolate-based study.
Furthermore, both the mean proportion of active saprophytic taxa (latent saprophytes) and their abundance in the living tillers were not significantly different in the east and west (p = 0.242, Table 2). Saprophytic function of latent saprophytes borne within the host plant does not differ at the sites. Notably, in both regions active saprophytic taxa make up a large proportion of endophytic communities, comprising 57–67% of the taxa that account for 56–83% of endophyte abundance.
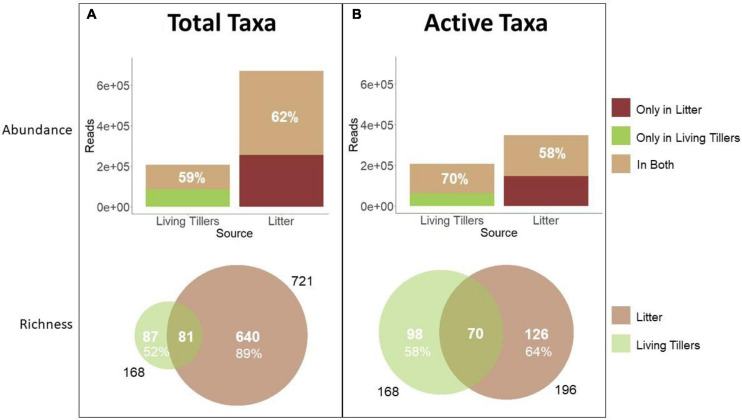
Total litter saprophyte community richness (721 taxa) was much greater than the living tiller endophyte community (168 taxa), in part because of differences in sampling effort (n = 18 vs. 8) (Figure 3). However, even at the sample level, mean richness was significantly greater in the total litter (119 ± 14) than in the living tillers (38 ± 3) (Table 3; p < 0.001). Likewise total read abundance of all litter samples was larger because of sampling effort differences, but even at the sample level, mean read abundance was 1.5× greater in the year-old litter samples (Table 3; p = 0.04). Other studies have similarly shown that saprophytic communities are more diverse than endophyte communities (Guerreiro et al., 2018). Overall, 81 latent saprophyte taxa (shared OTUs present in both living tillers and year-old gray litter) comprised 48% (81/168) of the endophyte (living tiller-derived) community richness, but only 11% (81/721) of the total saprophyte (litter-derived) community richness (Figure 3A). Despite these differences in overall richness, these shared taxa comprised roughly 3/5 of the read abundance in both endophyte and saprophyte communities (59 and 62%, respectively; Figure 3A), which did not differ significantly at the sample level (Table 3; shared richness: p = 0.172, shared abundance: p = 0.567). Thus, while the latent saprophyte taxa are proportionally abundant in the endophyte community prior to senescence, this relatively small number of latent saprophytic taxa (81/720) comprise a disproportionately larger fraction of the read abundance in the total litter (62%).
FIGURE 3.
Abundance and richness of saprophytic fungi in living and senescent tillers. Comparison between the endophyte community and (A) the total litter community or (B) the active litter community. Sequence read abundance and OTU richness of latent saprophytes in living tillers (olive; n = 8 samples) and of saprophytes in dead litter (brown; n = 18 samples) is based on NGS data.
TABLE 3.
Next-Generation Sequencing (NGS) data comparing living tillers and litter communities on a per sample basis.
| Total litter | ||||||
| Mean | SE | N | T-value | p-value | ||
| Richness | Living tillers | 38 | 3 | 8 | 5.84 | < 0.001 |
| Total litter | 119 | 14 | 18 | |||
| Abundance | Living tillers | 25,878 | 4,166 | 8 | 2.28 | 0.04 |
| Total litter | 37,188 | 2,685 | 18 | |||
| Shared richness | Living tillers | 22 | 1 | 8 | 1.41 | 0.172 |
| Total litter | 18 | 2 | 18 | |||
| Shared abundance | Living tillers | 19,628 | 4,540 | 8 | 0.58 | 0.567 |
| Total litter | 23,079 | 3,779 | 18 | |||
|
Active litter | ||||||
| Mean | SE | N | T-value | p-value | ||
| Richness | Living tillers | 38 | 3 | 8 | 0.54 | 0.588 |
| Active litter | 35 | 2 | 18 | |||
| Abundance | Living tillers | 25,878 | 4,166 | 8 | 1.21 | 0.243 |
| Active litter | 19,367 | 3,385 | 18 | |||
| Shared richness | Living tillers | 23 | 1 | 8 | 0.23 | 0.817 |
| Active litter | 24 | 3 | 18 | |||
| Shared abundance | Living tillers | 18,049 | 4,509 | 8 | 1.37 | 0.200 |
| Active litter | 11,092 | 2,307 | 18 | |||
While this result hints at the functional importance of the latent saprophytes in the litter community, we used BrDU analysis to explicitly quantify taxon activity. We first identified the active litter community after 36 h of wetting and then determined the proportion of latent saprophytes that were amongst these active taxa. Roughly 27% (196/721) of all the litter OTUs were active in the litter (Figure 3B), and a mean of 35 (± 2) OTUs were active in each litter sample (Table 3). Active latent saprophytes (70 shared OTUs) comprised 42% of the endophyte community richness, and 70% of the read abundance in living tillers prior to senescence (Figure 3B). The latent saprophytes are similarly abundant (58%) in the active litter samples a year post-senescence; and read abundance of living tillers and active litter on a per sample basis were not significantly different (Table 3; p = 0.200). Clearly, latent saprophytes comprise an important part of the functional saprophytic community in the litter more than a year post-senescence.
Thus, both NGS methodologies yielded similar outcomes: more than half (59–70%) of the S. sabulicola endophyte community is comprised of latent saprophytes, and these dual-niche fungi still comprise a similarly large proportion (58–62%) of the litter community more than a year after senescence.
Decomposition Potential of S. sabulicola Endophyte Isolates
A literature survey revealed that all but one of the identified endophytic species have been previously reported as both endophytes and as saprophytes (Figure 2D and Table 1).
Furthermore, of the 20 taxa evaluated, 80% were capable of decomposing tillers during the 28-day in vitro decomposition assay (mean NDA = 11.14%; SD = 3.20, SE = 0.80) (Figure 2C). There was no relationship between mean NDA and relative abundance, although 4/6 of the most abundant species had mean NDA values greater than the overall mean. The range of mean NDA for isolates capable of decomposing litter material was 6.50–17.88%, or 0.97–2.66 × 10–2%/hour.
There was no significant difference in the proportion of isolates from the west (64%) and east (45%) that could decompose S. sabulicola tillers during the 28-day experiment, nor in the mean % mass loss of these isolates from the two locations (Table 2). Six saprophytic species occurred in both the western and eastern regions, four were unique to the east, and six were unique to the west (Figure 2C). This relatively even distribution of cosmopolitan and region-specific saprophytic endophytes (latent saprophytes) across the NSS suggests that endophytes capable of saprotrophy are equally prevalent in both regions. While the taxonomic composition of the endophyte communities may differ across regions, the overall saprophytic ability of the endophyte community is consistent across the study area.
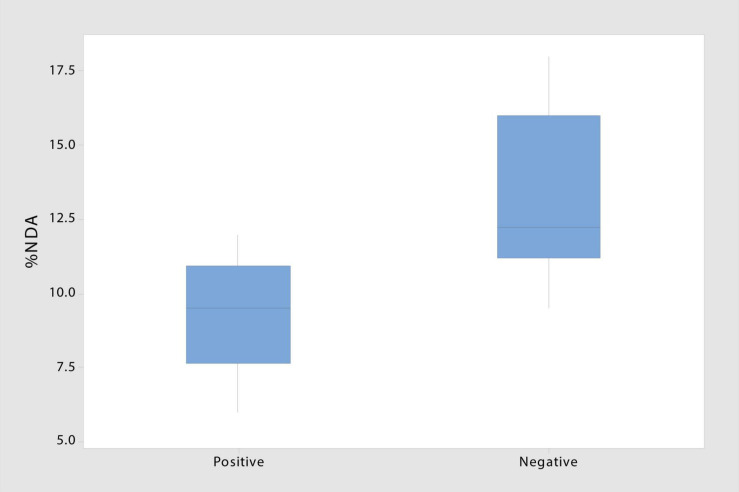
The presence of laccase and peroxidase enzymes varied greatly across taxonomic groups and even within species (Figure 2B and Table 1). Two of the four taxa with insignificant NDA values tested negative for both enzymes, suggesting that these species (T. wortmannii and Schizothecium spp.) may lack saprophytic abilities and function strictly as endophytes. Of the 16 saprophytic taxa tested, half tested positive for lignin-degrading enzymes, but notably, isolates with no enzyme activity had significantly higher NDA (p = 0.009) than those that tested positive (Figure 4). While this result requires further study, a plausible interpretation is that high NDA (lignin-degrading enzymes absent) isolates are those that perform well as early colonizers competing for simpler cellulose moieties; whereas low NDA (enzymes present) isolates are specialized for later stages of decomposition when recalcitrant lignin becomes available (Frankland, 1976; Deacon, 2006). It is also plausible that the two taxa (Penicillium sp. and Aspergillus niger) that tested positive for lignin-degrading enzymes but had insignificant NDA values were simply not efficient enough to register in our brief decomposition assay. Furthermore, A. niger is known to be a particularly efficient degrader of tannins, and because tannins bind with proteins, Van Diepeningen et al. (2004) proposed that this fungus may play a unique role modulating nitrogen availability in litter communities. It thus appears that most of the endophytic taxa of S. sabulicola that we isolated are decomposers, and that this latent-saprophytic community is comprised of taxa showing specialization for different stages of the decomposition process.
FIGURE 4.
Decomposition ability of taxa testing negative for laccase and peroxidase enzymes involved in lignin degradation is significantly higher than that of taxa testing positive (T-stat = 3.09; p = 0.009) (n = 8).
In situ Decomposition Ability of the S. sabulicola Endophyte Community
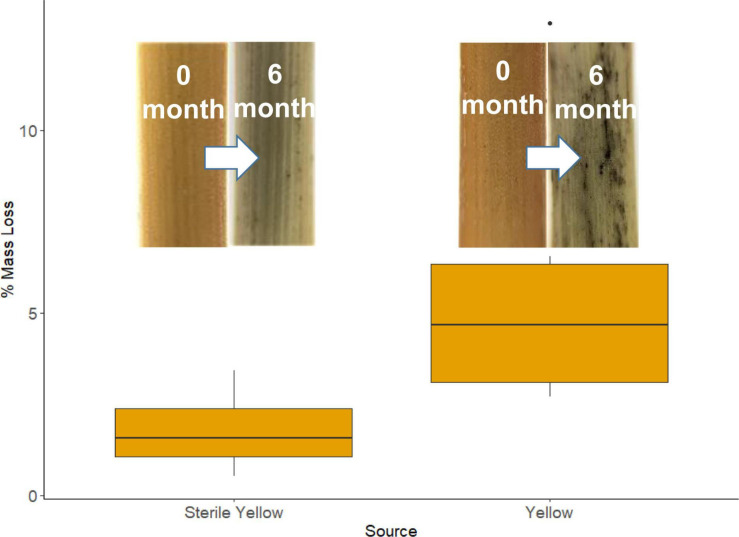
During the 183-day period when tillers were in the field, there was no rainfall at the study site, but there were 55 nights that met our “wet” criteria, amounting to 457 h during which fungi could be metabolically active (Jacobson et al., 2015). The mean length of each event was 8.5 h (SE = 0.3). We observed significantly greater mass loss from unsterilized tillers containing endophytes compared to sterilized tillers: 5.00% (SE = 0.74) vs. 1.78% (SE = 0.31) (Figure 5; p = 0.002). The tiller mass loss rate of 1.2 × 102%/hour from the unsterilized material provided a whole-community estimate of mass loss from recently senesced tillers which falls well within the range of mass loss rate for individual taxa (0.97–2.66 × 10–2%/hour) obtained via the in vitro decomposition assays. While a possible explanation for the smaller mass loss from the sterile material is that the sterilization process made the substrates unpalatable, it should be noted that dry-autoclaved litter was readily decomposed by fungal isolates in the in vitro decomposition trials, suggesting that sterilization did not alter the physical or chemical properties in a fashion that prevented fungi using them as substrates.
FIGURE 5.
Mean percent mass loss from recently senesced sterile and non-sterile yellow S. sabulicola tillers. Inserts are of lower portions of sterile yellow tillers at 0 and 6 months, and unsterilized yellow tillers at 0 and 6 months.
Discussion
Our multi-pronged study yields strong evidence supporting the hypothesis that dryland plants tolerate complex endophyte communities in living tillers because these fungi initiate decomposition of standing tillers immediately post-senescence. Both NGS- and isolate-based sampling methods revealed that endophyte communities are abundant and species-rich in S. sabulicola stems throughout the NSS (Tables 2, 3 and Figures 2A, 3). This community plays a large role in actively decomposing litter post-senescence: litter with intact endophyte communities decomposed twice as fast over a 6-month period as sterile litter colonized solely by airborne fungal propagules (Figure 5). Furthermore, all but one of the taxa we isolated as endophytes are also known saprophytes of plant litter (Figure 2D and Table 1). This overlap in potential function is well known from endophyte studies in a variety of habitats (Promputtha et al., 2007; Su et al., 2010; Purahong and Hyde, 2011; Sun et al., 2011; Sánchez Márquez et al., 2012), but no studies have confirmed that specific endophyte isolates are functional saprophytes (Porras-Alfaro and Bayman, 2011; Vázquez de Aldana et al., 2013). Our decomposition assay conclusively showed that 80% of these endophyte taxa were efficient decomposers of their host tillers (Figure 2C), which is compelling evidence that the endophyte community is composed of “dual-niche” fungal genotypes that function as both saprophytes and endophytes in their host plants.
The decomposition assay also revealed inter- and intra-specific differences in degradation ability among endophytes (6.5–18%) (Figure 2C). This difference emerged even over our short assay period, which is just a mere snapshot of their ability to decompose newly senescent litter. An analysis of functional ability throughout the multi-year decomposition process was beyond the scope of this study, but we did find that isolates lacking lignin-degradation capabilities were more efficient decomposers than those that had laccase and peroxidase function (Figure 4). This suggests that the latent saprophytic communities that occupy living plants are functionally diverse, exhibiting trade-offs between strategies of efficient cellulose decomposition immediately post-senescence versus specialization for more recalcitrant lignin substrates, likely later in the decomposition process (Dix and Webster, 1995). Remarkably, once corrected for hours of wetness when fungi would be active, the decomposition rates measured for the community in the field fell well within the range of those measured for individual species in our short lab-based assay.
We also found that the latent saprophyte community remains an important part of the litter saprophyte community at least a year after senescence. Active taxa comprised roughly comprised roughly 40% of both the living tillers (42%) and the litter (36%) community richness (Figure 3B). Not surprisingly, these 70 taxa comprise a large share of the endophyte abundance in living tillers (70% of reads), but remarkably, also comprise almost 3/5 of the abundance in year-old gray litter (58%), despite opportunities for the senesced litter to be colonized by the 126 active litter saprophyte taxa present in the environment which do not colonize living tillers. Other recent studies have similarly found that plants harbor endophyte communities that include latent saprophytes (Baldrian et al., 2012; Szink et al., 2016; Guerreiro et al., 2018). In particular, Guerreiro et al. (2018) found that endophytes comprised a very large portion of the reads and about a half of the richness in year-old beech leaf litter.
Contrary to our hypothesis, endophyte community richness and abundance did not differ across the abiotic stress gradient (Table 2). This suggests that, unlike mycorrhizal fungi (Jacobson, 1997), the benefits of possessing a functionally diverse latent-saprophyte endophytic community outweigh the costs to the living plants. Furthermore, the functional diversity of eastern and western communities did not differ in the proportion of decomposition-capable isolates, the mean percent mass loss of all isolates, or the richness and abundance proportions of the endophyte community that were also in active litter. These asymptomatic latent saprophytes appear to be of sufficient functional importance that plants support them in their living tissues regardless of the abiotic stress they are experiencing. Our results corroborate a recent study that found differences in community composition across the gradient, but functional performance (litter decomposition rates) did not differ, nor did fungal diversity and richness amongst these surface litter communities (Logan et al., 2021).
Based on our study, we propose that the need to efficiently recycle nutrients is why diverse foliar endophyte communities are present in plants in extreme environments (e.g., Rosa et al., 2009; Massimo et al., 2015; González-Menéndez et al., 2018). While individual species may confer advantages to the living plant, our data strongly supports the hypothesis that plants tolerate diverse communities of latent saprophytes throughout their lifetime because of the role that the community plays at senescence. By hosting latent saprophytes as endophytic communities, living tillers are pre-inoculated with a functionally diverse saprophytic community that can immediately commence decomposition in response to frequent NRM and rain events that moisten standing litter (Jacobson et al., 2015; Evans et al., 2020; Logan et al., 2021). Such a strategy may be particularly important to dryland perennial plants because inorganic soil nitrogen and phosphorus are often limiting (Ronca et al., 2015), and therefore plant organic matter is a critical nutrient source. Nutrient islands beneath perennial plants are a well-documented dryland phenomenon, where N & P levels may be 3–10 times higher than in the soil matrix between plants (Klopatek and Stock, 1994; Abrams et al., 1997; André et al., 1997; Throop et al., 2020). Plant morphological traits, such as the caespitose stem structure of bunch grasses (see Figure 1A), provide the structural support for these islands to the benefit of perennial plants. Likewise, we propose that the ability of perennial plants to accept/recruit and maintain specific latent saprophytes in their living tissue, in preparation for senescence, is an essential trait that contributes to efficient mineralization and retention in these nutrient islands.
Foliar endophyte communities in perennial plants in more mesic habitats are also composed of complex assemblages of latent saprophytes, and a host of fungi whose functional attributes are not yet known (Arnold, 2007; Porras-Alfaro and Bayman, 2011). Given the large proportion of latent saprophytes in beech leaves that are present a year after senescence (Guerreiro et al., 2018), endophytes may play a similar recycling role in the root zone of forest trees. It may even be important in mesic nutrient-rich prairies where space in the root zone is at a premium and thus nutrient retention by individual plants is important. Indeed caespitose grasses, such as Andropogon gerardii (big blue stem) and Sorghastrum nutans (Indian grass), but not rhizomatous grasses, have “nutrient islands” of enriched N & P in their root zones (Derner and Briske, 2001). It is worth noting that fungal communities on standing big bluestem litter are as responsive to NRM events (dew, fog and high humidity) as those in the NSS, and that “wet events” (including rainfall) occur 77% of the time in temperate prairie habitats (Evans et al., 2020). Likewise, fungi on standing litter of little bluestem species in a longleaf pine savanna ecosystem account for 55–65% of carbon loss (Lodato et al., 2021), and all dominant taxa are known endophytes. The ability to tolerate communities of asymptomatic latent saprophytes as endophytes in living tissue, so that they can initiate in situ nutrient cycling immediately upon senescence, may well be an essential trait of any perennial plant facing nutrient limitations.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
AW and KJ conceived of the study and wrote the manuscript. KJ, PJ, MS, JL, and SE conducted field work. AW, AD, KJ, JL, and SE did lab work and data analysis. All authors edited the manuscript and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Roland Vogt for meteorological data, Cassandra Miller for field assistance, and Gobabeb Research and Training Centre for logistical support. We greatly appreciate the opportunity to work in the Namib-Naukluft Park, courtesy of the Namibian Ministry of Environment and Tourism (Permit Number 2116/2016). SE thanks Will West for preliminary analysis of sequence data and Lukas Bell-Dereske for assistance developing fungal bioinformatic pipelines.
Funding. Funding for field- and lab-work was provided to AW, KJ, and PJ by Grinnell College. SE was supported by Michigan State University, the Kellogg Biological Station, and NSF FSML grant (1722621). JL was supported by an NSF GRFP.
References
- Abrams M. M., Jacobson P. J., Jacobson K. M., Seely M. K. (1997). Survey of soil chemical properties across a landscape in the Namib Desert. J. Arid Environ. 35:139. 10.1006/jare.1995.0139 [DOI] [Google Scholar]
- Allen M. F. (1991). The ecology of Mycorrhizae. New York: Cambridge University Press. [Google Scholar]
- Ananda K., Sridhar K. R. (2002). Diversity of endophytic fungi in the roots of mangrove species on the west coast of India. Can. J. Microbiol. 48:10. 10.1139/w02-080 [DOI] [PubMed] [Google Scholar]
- André H. M., Noti M.-I., Jacobson K. M. (1997). The soil microarthropods of the Namib Desert: A patchy mosaic. J. Afr. Zool. 111 499–517. [Google Scholar]
- Angelini P., Rubini A., Gigante D., Reale L., Pagiotti R., Venanzoni R. (2012). The endophytic fungal communities associated with the leaves and roots of the common reed (Phragmites australis) in Lake Trasimeno (Perugia, Italy) in declining and healthy stands. Fungal. Ecol. 5 683–693. 10.1016/j.funeco.2012.03.001 [DOI] [Google Scholar]
- Arnold A. E. (2007). Understanding the diversity of foliar endophytic fungi: progress, challenges, and frontiers. Fungal. Biol. Rev. 21 51–66. 10.1016/j.fbr.2007.05.003 [DOI] [Google Scholar]
- Arnold A. E., Henk D. A., Eells R. L., Lutzoni F., Vilgalys R. (2007). Diversity and Phylogenetic Affinities of Foliar Fungal Endophytes in Loblolly Pine Inferred by Culturing and Environmental PCR. Mycologia 99 185–206. 10.1080/15572536.2007.11832578 [DOI] [PubMed] [Google Scholar]
- Baldrian P., Kolařík M., Stursová M., Kopecký J., Valášková V., Větrovský T., et al. (2012). Active and total microbial communities in forest soil are largely different and highly stratified during decomposition. ISME J. 6 248–258. 10.1038/ismej.2011.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bara R., Aly A. H., Pretsch A., Wray V., Wang B., Proksch P., et al. (2013). Antibiotically active metabolites from Talaromyces wortmannii, an endophyte of Aloe vera. J. Antibiot. 66 491–493. 10.1038/ja.2013.28 [DOI] [PubMed] [Google Scholar]
- Borneman J. (1999). Culture-Independent Identification of Microorganisms That Respond to Specified Stimuli. Appl. Environ. Microbiol. 65 3398–3400. 10.1128/AEM.65.8.3398-3400.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busby P. E., Peay K. G., Newcombe G. (2016). Common foliar fungi of Populus trichocarpa modify Melampsora rust disease severity. N. Phytol. 209 1681–1692. 10.1111/nph.13742 [DOI] [PubMed] [Google Scholar]
- Checa J., Arenal F., Blanco N., Rogers J. D. (2008). Coniolariella hispanica sp. nov. and other additions to Coniolariella. Mycol. Res. 112 795–801. 10.1016/j.mycres.2008.01.014 [DOI] [PubMed] [Google Scholar]
- Costa I. P. M. W., Assuncao M. M. C., Lima T. E. F., Oliveira R. J. V., Cavalcanti M. A. Q. (2012). Checklist of endophytic fungi from tropical regions. Mycotaxon 119:494. [Google Scholar]
- Deacon J. W. (2006). Fungal Biology. Vol. 4. Malden: Blackwell. [Google Scholar]
- Derner J. D., Briske D. D. (2001). Below-ground carbon and nitrogen accumulation in perennial grasses: a comparison of caespitose and rhizomatous growth forms. Plant Soil 237 117–127. [Google Scholar]
- Dirks I., Navon Y., Kanas D., Dumbur R., Grünzweig J. M. (2010). Atmospheric water vapor as driver of litter decomposition in Mediterranean shrubland and grassland during rainless seasons. Global. Change Biol. 16 2799–2812. 10.1111/j.1365-2486.2010.02172.x [DOI] [Google Scholar]
- Dix N. J., Webster J. (1995). Fungal Ecology. Netherland: Springer. [Google Scholar]
- Ebner M., Miranda T., Roth-Nebelsick A. (2011). Efficient fog harvesting by Stipagrostis sabulicola (Namib dune bushman grass). J. Arid Environ. 75 524–531. 10.1016/j.jaridenv.2011.01.004 [DOI] [Google Scholar]
- Eckardt F. D., Soderberg K., Coop L. J., Muller A. A., Vickery K. J., Grandin R. D., et al. (2013). The nature of moisture at Gobabeb, in the central Namib Desert. J. Arid Environ. 93 7–19. 10.1016/j.jaridenv.2012.01.011 [DOI] [Google Scholar]
- Evans S., Todd-Brown K. E. O., Jacobson K., Jacobson P. (2020). Non-rainfall Moisture: A Key Driver of Microbial Respiration from Standing Litter in Arid, Semiarid, and Mesic Grasslands. Ecosystems 23 1154–1169. 10.1007/s10021-019-00461-y [DOI] [Google Scholar]
- Frankland J. C. (1976). Decomposition of bracken litter. Biol. J. Linn. Soc. 73 133–143. 10.1111/j.1095-8339.1976.tb02018.x [DOI] [Google Scholar]
- Geydan T. D., Debets A. J. M., Verkley G. J. M., Van Diepeningen A. D. (2012). Correlated evolution of senescence and ephemeral substrate use in the Sordariomycetes. Mol. Ecol. 21 2816–2828. 10.1111/j.1365-294X.2012.05569.x [DOI] [PubMed] [Google Scholar]
- Goldfarb K. C., Karaoz U., Hanson C. A., Santee C. A., Bradford M. A., Treseder K. K., et al. (2011). Differential Growth Responses of Soil Bacterial Taxa to Carbon Substrates of Varying Chemical Recalcitrance. Front. Microbio. 2:94. 10.3389/fmicb.2011.00094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomaa M., Hifney A. F., Fawzy M. A., Issa A. A., Abdel-Gawad K. M. (2015). Biodegradation of Palisada perforata (Rhodophyceae) and Sargassum sp. (Phaeophyceae) biomass by crude enzyme preparations from algicolous fungi. J. Appl. Phycol. 27 2395–2404. 10.1007/s10811-014-0517-x [DOI] [Google Scholar]
- González-Menéndez V., Crespo G., de Pedro N., Diaz C., Martín J., Serrano R., et al. (2018). Fungal endophytes from arid areas of Andalusia: high potential sources for antifungal and antitumoral agents. Sci. Rep. 8:9729. 10.1038/s41598-018-28192-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göransson H., Godbold D. L., Jones D. L., Rousk J. (2013). Bacterial growth and respiration responses upon rewetting dry forest soils: Impact of drought-legacy. Soil Biol. Biochem. 57 477–486. 10.1016/j.soilbio.2012.08.031 [DOI] [Google Scholar]
- Granados-Montero M., Minter D. W., Castañeda Ruiz R. F. (2018). A checklist of asexual fungi from Costa Rica. Mycotaxon 133 365–365. 10.5248/133.365 [DOI] [Google Scholar]
- Guerreiro M. A., Brachmann A., Begerow D., Peršoh D. (2018). Transient leaf endophytes are the most active fungi in 1-year-old beech leaf litter. Fungal Div. 89 237–251. 10.1007/s13225-017-0390-4 [DOI] [Google Scholar]
- Hao L., Haiyun Q., Qiwu Z., Zhenhui R., Ruixu X., Baoyu Z., et al. (2017). Endogenous fungi isolated from three locoweed species from rangeland in western China. Afr. J. Microbiol. Res. 11 155–170. 10.5897/AJMR2016.8392 [DOI] [Google Scholar]
- Higgins K. L., Coley P. D., Kursar T. A., Arnold A. E. (2011). Culturing and direct PCR suggest prevalent host generalism among diverse fungal endophytes of tropical forest grasses. Mycologia 103 247–260. 10.3852/09-158 [DOI] [PubMed] [Google Scholar]
- Hong S.-B., Lee M., Kim D.-H., Varga J., Frisvad J. C., Perrone G., et al. (2013). Aspergillus luchensis, an industrially important black Aspergillus in East Asia. PLoS One 8:e63769. 10.1371/journal.pone.0063769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovieno P., Baath E. (2008). Effect of drying and rewetting on bacterial growth rates in soil: Rewetting and bacterial growth in soil. FEMS Microbiol. Ecol. 65 400–407. 10.1111/j.1574-6941.2008.00524.x [DOI] [PubMed] [Google Scholar]
- Jacobson K., van Diepeningen A., Evans S., Fritts R., Gemmel P., Marsho C., et al. (2015). Non-Rainfall Moisture Activates Fungal Decomposition of Surface Litter in the Namib Sand Sea. PLoS One 10:e0126977. 10.1371/journal.pone.0126977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson K. M. (1997). Moisture and substrate stability determine VA-mycorrhizal fungal community distribution and structure in an arid grassland. J. Arid Environ. 35 59–75. 10.1006/jare.1995.0140 [DOI] [Google Scholar]
- Kaushik N., Murali T., Sahal D., Suryanarayanan T. (2014). A search for antiplasmodial metabolites among fungal endophytes of terrestrial and marine plants of southern India. Acta Parasitol. 59 745–57. 10.2478/s11686-014-0307-2 [DOI] [PubMed] [Google Scholar]
- Khan S., Hamayun M., Yoon H., Kim H.-Y., Suh S.-J., Hwang S.-K., et al. (2008). Plant growth promotion and Penicillium citrinum. BMC Microbiol. 8:231. 10.1186/1471-2180-8-231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopatek J., Stock W. (1994). Partitioning of nutrients in Acanthosicyos horrdus, a keystone endemic species in the Namib Desert. J. Arid Environ. 26 233–240. 10.1006/jare.1994.1026 [DOI] [Google Scholar]
- Ko T. W., Stephenson S. L., Bahkali A. H., Hyde K. D. (2011). From morphology to molecular biology: can we use sequence data to identify fungal endophytes? Fungal. Div. 50 113–120. 10.1007/s13225-011-0130-0 [DOI] [Google Scholar]
- Kõljalg U., Nilsson R. H., Abarenkov K., Tedersoo L., Taylor A. F. S., Bahram M., et al. (2013). Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 22 5271–5277. 10.1111/mec.12481 [DOI] [PubMed] [Google Scholar]
- Kuehn K., Gessner M. O., Wetzel R. G., Suberkropp K. (1999). Decomposition and CO 2 Evolution from Standing Litter of the Emergent Macrophyte Erianthus giganteus. Microbial. Ecol. 38 50–57. 10.1007/s002489900154 [DOI] [PubMed] [Google Scholar]
- Kumar S., Kaushik N. (2013). Endophytic Fungi Isolated from Oil-Seed Crop Jatropha curcas Produces Oil and Exhibit Antifungal Activity. PLoS One 8:e56202. 10.1371/journal.pone.0056202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster J., Lancaster N., Seely M. K. (1984). Climate of the central Namib Desert. Madoqua 14 5–61. [Google Scholar]
- Liu Y., Ding Z., Peng D., Liu S., Kong L., Peng H., et al. (2019). Evaluation of the biocontrol potential of Aspergillus welwitschiae against the root-knot nematode Meloidogyne graminicola in rice (Oryza sativa L.). J. Integr. Agricult. 18 2561–2570. 10.1016/S2095-3119(19)62610-9 [DOI] [Google Scholar]
- Lodato M. B., Boyette J. S., Smilo R. A., Jackson C. R., Halvorson H. M., Kuehn K. A. (2021). Functional importance and diversity of fungi during standing grass litter decomposition. Oecologia 195 499–512. 10.1007/s00442-020-04838-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan J. R., Jacobson K. M., Jacobson P. J., Evans S. E. (2021). Fungal Communities on Standing Litter Are Structured by Moisture Type and Constrain Decomposition in a Hyper-Arid Grassland. Front. Microbiol. 12:596517. 10.3389/fmicb.2021.596517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciá-Vicente J. G., Jansson H.-B., Abdullah S. K., Descals E., Salinas J., Lopez-Llorca L. V. (2008). Fungal root endophytes from natural vegetation in Mediterranean environments with special reference to Fusarium spp: Fungal root endophytes in stressed environments. FEMS Microbiol. Ecol. 64 90–105. 10.1111/j.1574-6941.2007.00443.x [DOI] [PubMed] [Google Scholar]
- Massimo N. C., Nandi Devan M. M., Arendt K. R., Wilch M. H., Riddle J. M., Furr S. H., et al. (2015). Fungal Endophytes in Aboveground Tissues of Desert Plants: Infrequent in Culture, but Highly Diverse and Distinctive Symbionts. Microb. Ecol. 70 61–76. 10.1007/s00248-014-0563-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon S. K., Wallenstein M. D., Schimel J. P. (2011). A cross-seasonal comparison of active and total bacterial community composition in Arctic tundra soil using bromodeoxyuridine labeling. Soil Biol. Biochem. 43 287–295. 10.1016/j.soilbio.2010.10.013 [DOI] [Google Scholar]
- Mouchacca J. (2007). Heat tolerant fungi and applied research: Addition to the previously treated group of strictly thermotolerant species. World J. Microbiol. Biotechnol. 23 1755–1770. 10.1007/s11274-007-9426-3 [DOI] [PubMed] [Google Scholar]
- O’Brien H. E., Parrent J. L., Jackson J. A., Moncalvo J.-M., Vilgalys R. (2005). Fungal Community Analysis by Large-Scale Sequencing of Environmental Samples. AEM 71 5544–5550. 10.1128/AEM.71.9.5544-5550.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osono T. (2006). Role of phyllosphere fungi of forest trees in the development of decomposer fungal communities and decomposition processes of leaf litter. Can. J. Microbiol. 52 701–716. 10.1139/w06-023 [DOI] [PubMed] [Google Scholar]
- Osono T., Takeda H. (2006). Fungal decomposition of Abies needle and Betula leaf litter. Mycologia 98 172–179. 10.1080/15572536.2006.11832689 [DOI] [PubMed] [Google Scholar]
- Pandey A., Palni L. M. S. (1998). Isolation of Pseudomonas corrugata from Sikkim Himalaya. World J. Microbiol. Biotechnol. 14 411–413. 10.1023/A:1008825514148 [DOI] [Google Scholar]
- Pinciroli M. (2013). Mycobiota evolution during storage of paddy, brown and milled rice in different genotypes. Summa Phytopathol. 39 157–161. 10.1590/s0100-54052013000300002 [DOI] [Google Scholar]
- Placella S. A., Brodie E. L., Firestone M. K. (2012). Rainfall-induced carbon dioxide pulses result from sequential resuscitation of phylogenetically clustered microbial groups. Proc. Natl. Acad. Sci. U. S. A. 109 10931–10936. 10.1073/pnas.1204306109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porras-Alfaro A., Bayman P. (2011). Hidden Fungi, Emergent Properties: Endophytes and Microbiomes. Annu. Rev. Phytopathol. 49 291–315. 10.1146/annurev-phyto-080508-081831 [DOI] [PubMed] [Google Scholar]
- Promputtha I., Lumyong S., Dhanasekaran V., McKenzie E. H. C., Hyde K. D., Jeewon R. (2007). A Phylogenetic Evaluation of Whether Endophytes Become Saprotrophs at Host Senescence. Microb. Ecol. 53 579–590. 10.1007/s00248-006-9117-x [DOI] [PubMed] [Google Scholar]
- Purahong W., Hyde K. D. (2011). Effects of fungal endophytes on grass and non-grass litter decomposition rates. Fungal. Div. 47 1–7. 10.1007/s13225-010-0083-8 [DOI] [Google Scholar]
- Raja H. A., Miller A. N., Pearce C. J., Oberlies N. H. (2017). Fungal Identification Using Molecular Tools: A Primer for the Natural Products Research Community. J. Nat. Prod. 80 756–770. 10.1021/acs.jnatprod.6b01085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronca S., Ramond J.-B., Jones B. E., Seely M., Cowan D. A. (2015). Namib Desert dune/interdune transects exhibit habitat-specific edaphic bacterial communities. Front. Microbiol. 6:845. 10.3389/fmicb.2015.00845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa L. H., Vaz A. B. M., Caligiorne R. B., Campolina S., Rosa C. A. (2009). Endophytic fungi associated with the Antarctic grass Deschampsia antarctica Desv. (Poaceae). Polar Biol. 32 161–167. 10.1007/s00300-008-0515-z [DOI] [Google Scholar]
- Sanchel Marquez S., Bills G. F., Zabalgogeazcoa I. (2008). Diversity and structure of the fungal endophytic assemblages from two sympatric coastal grasses. Fungal Div. 2:33. [Google Scholar]
- Sánchez Márquez S., Bills G. F., Herrero N., Zabalgogeazcoa I. (2012). Non-systemic fungal endophytes of grasses. Fungal Ecol. 5 289–297. 10.1016/j.funeco.2010.12.001 [DOI] [Google Scholar]
- Schlesinger W. H., Raikes J. A., Hartley A. E., Cross A. F. (1995). On the Spatial Pattern of Soil Nutrients in Desert Ecosystems: Ecological Archives E077-002. Ecology 77 364–374. 10.2307/2265615 [DOI] [Google Scholar]
- Schulz B., Guske S., Dammann U., Boyle C. (1998). Endophyte-Host Interactions. II. Defining Symbiosis of the Endophyte-Host Interaction. Symbioisis 25 213–227. [Google Scholar]
- Su Y.-Y., Guo L.-D., Hyde K. D. (2010). Response of endophytic fungi of Stipa grandis to experimental plant function group removal in Inner Mongolia steppe, China. Fungal Div. 43 93–101. 10.1007/s13225-010-0040-6 [DOI] [Google Scholar]
- Sun X., Guo L.-D., Hyde K. D. (2011). Community composition of endophytic fungi in Acer truncatum and their role in decomposition. Fungal Div. 47 85–95. 10.1007/s13225-010-0086-5 [DOI] [Google Scholar]
- Sun Y., Wang Q., Lu W., Okane I., Kakishima M. (2012). Endophytic fungal community in stems and leaves of plants from desert areas in China. Mycol. Progr. 11 790–790. 10.1007/s11557-011-0790-x [DOI] [Google Scholar]
- Szink I., Davis E. L., Ricks K. D., Koide R. T. (2016). New evidence for broad trophic status of leaf endophytic fungi of Quercus gambelii. Fungal Ecol. 22 2–9. 10.1016/j.funeco.2016.04.003 [DOI] [Google Scholar]
- Throop H. L., Seely M. K., Marufu V. J. Summer Drylands Program Participants. (2020). Multiple scales of spatial heterogeneity control soil respiration responses to precipitation across a dryland rainfall gradient. Plant Soil 453 423–443. 10.1007/s11104-020-04614-0 [DOI] [Google Scholar]
- Uma Anitha K. P. G., Mythili S. (2017). Antioxidant and hepatoprotective potentials of novel endophytic fungus Achaetomium sp., from Euphorbia hirta. Asian Pac. J. Tropical Med. 10 588–593. 10.1016/j.apjtm.2017.06.008 [DOI] [PubMed] [Google Scholar]
- Van Diepeningen A. D., Debets A. J. M., Varga J., Van Der Gaag M., Swart K., Hoekstra R. F. (2004). Efficient degradation of tannic acid by black Aspergillus species. Mycol. Res. 108 919–925. 10.1017/s0953756204000747 [DOI] [PubMed] [Google Scholar]
- Vázquez de Aldana B. R., Bills G., Zabalgogeazcoa I. (2013). Are endophytes an important link between airborne spores and allergen exposure? Fungal Div. 60 33–42. 10.1007/s13225-013-0223-z [DOI] [Google Scholar]
- Viswanath B., Chandra M. S., Pallavi H., Reddy B. R. (2008). Screening and assessment of laccase producing fungi isolated from different environmental samples. African J. Biotech. 7, 1129–1133. [Google Scholar]
- Wang J., Liu L., Wang X., Yang S., Zhang B., Li P., et al. (2017). High night−time humidity and dissolved organic carbon content support rapid decomposition of standing litter in a semi−arid landscape. Funct. Ecol. 31 1659–1668. 10.1111/1365-2435.12854 [DOI] [Google Scholar]
- Yilmaz N., Visagie C. M., Houbraken J., Frisvad J. C., Samson R. A. (2014). Polyphasic taxonomy of the genus Talaromyces. Stud. Mycol. 78 175–341. 10.1016/j.simyco.2014.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabalgogeazcoa I., Gundel P. E., Helander M., Saikkonen K. (2013). Non-systemic fungal endophytes in Festuca rubra plants infected by Epichloë festucae in subarctic habitats. Fungal. Div. 60 25–32. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.