ABSTRACT
Organs stop growing to achieve a characteristic size and shape in scale with the body of an animal. Likewise, regenerating organs sense injury extents to instruct appropriate replacement growth. Fish fins exemplify both phenomena through their tremendous diversity of form and remarkably robust regeneration. The classic zebrafish mutant longfint2 develops and regenerates dramatically elongated fins and underlying ray skeleton. We show longfint2 chromosome 2 overexpresses the ether-a-go-go-related voltage-gated potassium channel kcnh2a. Genetic disruption of kcnh2a in cis rescues longfint2, indicating longfint2 is a regulatory kcnh2a allele. We find longfint2 fin overgrowth originates from prolonged outgrowth periods by showing Kcnh2a chemical inhibition during late stage regeneration fully suppresses overgrowth. Cell transplantations demonstrate longfint2-ectopic kcnh2a acts tissue autonomously within the fin intra-ray mesenchymal lineage. Temporal inhibition of the Ca2+-dependent phosphatase calcineurin indicates it likewise entirely acts late in regeneration to attenuate fin outgrowth. Epistasis experiments suggest longfint2-expressed Kcnh2a inhibits calcineurin output to supersede growth cessation signals. We conclude ion signaling within the growth-determining mesenchyme lineage controls fin size by tuning outgrowth periods rather than altering positional information or cell-level growth potency.
KEY WORDS: Appendage development, Calcineurin, Fin regeneration, Ion signaling, Organ size, Zebrafish
Summary: Ectopic kcnh2a underlies the classic longfin zebrafish mutant and links ion signaling within a growth-determining mesenchyme lineage to fin size control by tuning outgrowth periods rather than altering positional information.
INTRODUCTION
Animal organs develop to and maintain a species-specific size and shape in scale with the overall body of an individual. Remarkably, regenerating organs can sense injury extents to restore their original proportions. Fish fins neatly demonstrate both organ size establishment and regeneration mysteries. Fins display tremendous morphological diversity to optimize swimming, predator avoidance and courtship, while also contributing to the aesthetic appeal of fishes (Nelson et al., 2016). Furthermore, most fins, including those of the zebrafish model organism, regenerate to their original form, providing a striking example of organ size control and scaling.
Zebrafish fin skeletons comprise segmented cylindrical bony rays that define the overall fin form. Each ray segment embodies two opposing hemi-cylindrical bones produced by tightly associated osteoblasts. Vasculature, sensory axons and fibroblasts reside within rays that are themselves encased by a stratified epidermis. During larval development, fins initiate rapid asynchronous allometric growth that culminates in their mature forms, including the familiar bi-lobed shape of the well-studied caudal fin (Goldsmith et al., 2006). Adult fins switch to isometric growth, slowly expanding in scale with the rest of the body throughout the life of an animal. Adult fin regeneration engages the appropriate extent of rapid allometric growth to restore a properly proportioned appendage (Goldsmith et al., 2006; Iovine and Johnson, 2000).
The zebrafish fin regeneration model affords a tractable platform to determine fin size control mechanisms. No matter where a fin is amputated, the regenerated fin restores its original size and shape (Chen and Poss, 2017; Sehring and Weidinger, 2020). Fin amputation triggers a wound epithelium formed by migrating epidermal cells. De-differentiation of mature cells to lineage-restricted progenitors then generates a regenerative blastema for each ray (Knopf et al., 2011; Singh et al., 2012; Sousa et al., 2011; Stewart and Stankunas, 2012; Tu and Johnson, 2011). Heterogeneous blastemas organize by cell lineage and state to enable progressive regeneration (reviewed by Wehner and Weidinger, 2015). De-differentiated, proliferative osteoblasts (pObs) migrate to blastema peripheries and hierarchically arrange along the distal-to-proximal axis, with the most progenitor ‘state’ cells distally concentrated (Stewart et al., 2014). Similarly, fibroblasts residing between hemi-rays de-differentiate, form the major blastema mesenchyme population radially interior to pObs and then contribute to regenerated intra-ray tissue (Tornini et al., 2016). An outgrowth phase follows that integrates spatially segregated proliferation (distal) and differentiation (proximal) activities for the progressive restoration of replacement tissue.
Developmental signaling pathways promote tissue interactions and cell behaviors to replace lost cells and re-form mature patterned tissue (Wehner and Weidinger, 2015). A distal blastema ‘organizing center’ produces essential Wnt signals that coordinate other pathways, including FGFs and BMPs (Wehner et al., 2014). FGFs likely are the primary mitogens (Lee et al., 2005, 2009; Poss et al., 2000; Shibata et al., 2016), whereas BMP signaling is implicated in osteoblast differentiation (Quint et al., 2002; Smith et al., 2006; Stewart et al., 2014). Yet the cell behaviors and their mechanistic control, which ensure cessation of regrowth once the proper fin size is reached, are unresolved. Prevailing models for fin size restoration suggest that positional identities of cells at an injury site establish appropriate outgrowth-determining blastema pre-patterns (Lee et al., 2005; Rabinowitz et al., 2017; Tornini et al., 2016). The nature of such cellular memories, how they establish blastema positional information and how those pre-patterns are converted into outgrowth extents all remain largely mysterious.
Genetic studies of adult viable zebrafish mutants with abnormally sized fins provide an entry for molecular insights into fin growth control (Van Eeden et al., 1996). These studies universally implicate ion signaling as a major determinant of fin size. Gain-of-function mutations in the K+ channel kcnk5b in another longfin (alf) develop and regenerate long fins (Perathoner et al., 2014). Furthermore, fin overgrowth in the schleier mutant is caused by loss of kcc4a/slc12a7a, a K+ Cl− co-transporter (Lanni et al., 2019). Viral insertion-driven ectopic (kcnj13) or transgenic (kcnj1b, kcnj10a, kcnk9c) overexpression of additional K+ channels also cause overgrowth (Silic et al., 2020). Finally, loss of gap junction protein alpha 1b (gja1, also known as connexin 43) in shortfin mutants suggests involvement of intercellular ion exchange (Iovine and Johnson, 2000). This literature, together with studies in other animals, points to central roles of ion signaling, or ‘bioelectricity’, in organ size establishment (McLaughlin and Levin, 2018). However, how ion signaling dynamics directly alter cell states and behaviors to instruct and/or enable scaled growth is poorly understood.
Cooperating factors that promote and effect ion signaling dynamics for fin size control are unresolved. As one key insight, small molecule inhibition of the Ca2+-dependent phosphatase calcineurin causes dramatic fin overgrowth (Kujawski et al., 2014). As a decoder of the ubiquitous cytosolic Ca2+ second messenger, calcineurin links ion signaling and protein effector phosphorylation (Crabtree, 1999). Calcineurin may act directly upstream of Kcnk5b (alf) to temper outgrowth and restore fin proportions (Daane et al., 2018; Yi et al., 2021). In mammals, diverse stimuli activate calcineurin by elevating intracellular Ca2+, including antigen engagement of the T-cell receptor complex (Rao, 2009), depolarization of cardiomyocytes (Parra and Rothermel, 2017) and neuronal ion channel activity (Baumgärtel and Mansuy, 2012). Calcineurin dephosphorylates multiple proteins, including ion channels and transcription factors, to promote context-dependent cell behaviors. Characterizing the upstream regulators and downstream effectors of calcineurin in the fin is a promising path towards revealing organ size control mechanisms.
The classic dominant longfint2 (loft2) mutant also develops and regenerates exceptionally long fins, producing a ‘schleierform’, or flowing veil-like appearance (Elias, 1984; Van Eeden et al., 1996) (Fig. 1A,B). Notably, the widely used stock strain Tüpfel-Longfin (TL) contains compound loft2/t2; leopardt1/t1 (leo) mutations, the respective fin and pigmentation phenotypes of which allow visual identification of mixed genotypes (Haffter et al., 1996). Developing fins of loft2 fish do not have a greater maximal growth rate (Iovine and Johnson, 2000). Rather, loft2 fins fail to return to isometric growth as animals reach maturity (Goldsmith et al., 2006; Iovine and Johnson, 2000). Unlike kcnk5b (alf) and kcc4a/slc12a7a (schlier) mutants or calcineurin-inhibited animals, loft2 overgrown fins lack other defects, such as tissue hyper-vascularization and elongated ray segments (Kujawski et al., 2014; Lanni et al., 2019; McMillan et al., 2018; Perathoner et al., 2014; Silic et al., 2020). The last phenotype likely reflects the independent disruption of joint formation rather than overgrowth, as ray segment number and fin size are not correlated; evx1 mutants, which are devoid of all fin joints, have normal fin sizes (Schulte et al., 2011; Ton and Iovine, 2013). Regardless, loft2 provides a uniquely clean model to explore fin size control.
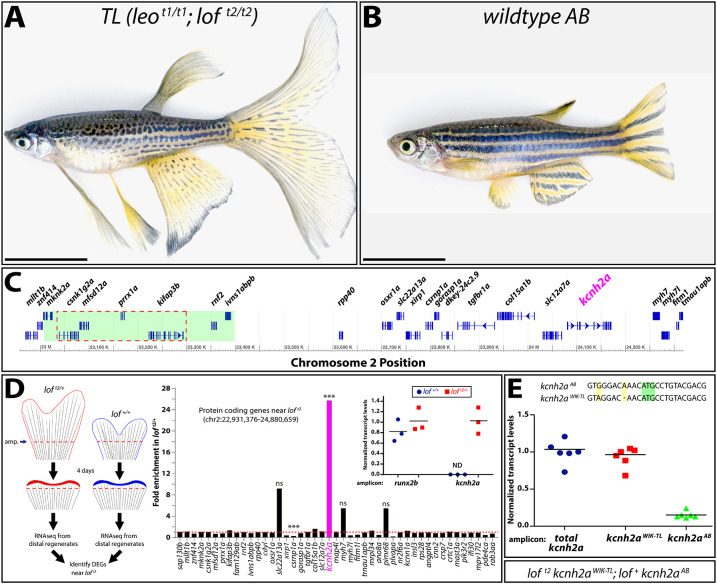
Fig. 1.
The loft2 mutation causes cis-ectopic expression of kcnh2a. (A,B) Bright-field images of adult TL (Tüpfel longfin: leot1/t1; loft2/t2) and wild-type AB zebrafish. Scale bars: 1 cm. (C) Schematic diagram of 1.5 Mb region of zebrafish chromosome 2 (chr2). The putative location of the loft2 mutation is outlined with a dashed red line and the region deleted in the suppressed lofjg1 is highlighted by a green box, as determined by Iovine and Johnson (2002). (D) Left: schematic of RNA-Seq experimental design to identify genes mis-expressed in regenerated loft2. Right: RNA-Seq data showing gene expression at chr2:22,931,376-24,880,659 from loft2/+ relative to clutchmate lof+/+ controls. The asterisks indicate the two differentially expressed genes (***P<10−4). ns, not significant. Inset: expression of kcnh2a in loft2/+ (in red) and lof+/+ (in blue) clutchmates determined by RT-qPCR using 4 dpa fin cDNA. Data were normalized to rpl8 reference expression levels and are presented as fold change relative to loft2/+. runx2b expression levels are shown for comparative purposes and were not significantly changed between the two genotypes. Expression of kcnh2a was below limits of detection in lof+/+ fish (indeterminate, ND). Each point represents a cohort of three animals. (E) RT-qPCR on 4 dpa caudal fin cDNA from loft2 kcnh2aWIK-TL; lof+ kcnh2aAB fish to detect chromosome-specific expression of kcnh2a. Sequences of non-coding kcnh2a polymorphisms that specifically amplify either kcnh2aWIK-TL, which is located on the loft2 mutant chr2 (red squares), or kcnh2aAB, which is located on AB chr2 (green triangles). Data were normalized to total kcnh2a levels (blue circles) determined using primers that amplify both alleles indiscriminately. Each data point represents a result from an individual fish. RT-qPCR statistical analyses used one-way ANOVA with Tukey's multiple comparisons tests.
Further motivated by the historical significance and prominence of loft2, we sought to identify the cause of its eponymous phenotype. We show the dominant loft2 fin overgrowth phenotype is caused by cis-ectopic expression of kcnh2a, a K+ channel-encoding gene mapping near the lof locus. During regeneration, the activity of ectopic Kcnh2a, an ortholog of cardiac arrhythmia-associated ether-a-go-go channels, promotes fin overgrowth exclusively by extending the outgrowth period. Labeled loft2 genetic chimeras demonstrate that Kcnh2a acts tissue autonomously within size-determining intra-ray mesenchymal lineage cells. Finally, we provide evidence Kcnh2a disrupts a Ca2+/calcineurin pathway that gradually terminates allometric fin outgrowth. Our results suggest readily tunable ion signaling alters organ size by modulating growth periods rather than establishing growth-defining positional information or directly impacting cell cycling rates.
RESULTS
cis-overexpression of kcnh2a in loft2
The dominant loft2 allele that specifically causes dramatic fin overgrowth maps to an essential ∼250 kb region (Fig. 1A-C) of chromosome 2 (chr2) (Iovine and Johnson, 2002). Therefore, loft2 likely results from a dominant-negative or gain-of-function mutation rather than from haploinsufficiency. One explanation is that loft2 alters a transcriptional regulatory element causing overexpression or ectopic expression of a gene(s) within or near the lof region (Fig. 1C). We explored this possibility using mRNA sequencing (RNA-seq) to identify differentially expressed genes (DEGs) from 4 days post-amputation (dpa) caudal fin regenerates (Fig. 1D). We reasoned 4 dpa regenerates would highlight primary transcriptional changes because loft2 and wild-type fin sizes were indistinguishable at this stage of regeneration (Fig. S1, discussed below). We identified 39 increased and 111 decreased DEGs (±2-fold change) in loft2 fins (Table S1). Confining the DEG analysis to the loft2 region and surrounding genes (Fig. 1C), a single transcript ∼1 Mb from loft2, potassium voltage-gated channel, subfamily H (eag-related), member 2a (kcnh2a), was greatly elevated in loft2/+ animals (+25.8-fold) (Fig. 1D). One gene, csrnp1a, showed decreased transcript levels (−3.9-fold). Quantitative RT-PCR (RT-qPCR) confirmed kcnh2a was expressed uniquely in loft2/+ fin regenerates, at levels similar to the runx2b osteoblast-lineage transcription factor (Fig. 1D, inset).
To determine whether loft2 affects kcnh2a expression in cis or trans, we leveraged sequence polymorphisms (Butler et al., 2015) in the 5′ UTR that distinguish kcnh2a from TL and WIK (kcnh2aWIK-TL) versus our wild-type AB (kcnh2aAB) strains (Fig. 1E). We used allele-specific primers and RT-qPCR to observe that kcnh2aWIK-TL transcripts accounted for nearly all kcnh2a expression in loft2 kcnh2aWIK-TL; lof+ kcnh2aAB regenerating caudal fins (Fig. 1E; P<0.0001). The apparent 10% residual contribution of kcnh2aAB to total kcnh2a expression likely reflects partial primer cross-hybridization. We conclude that most, if not all, kcnh2a expression in loft2 regenerating fins arises from cis effects of the loft2 mutation.
loft2 is a regulatory neomorph allele of kcnh2a
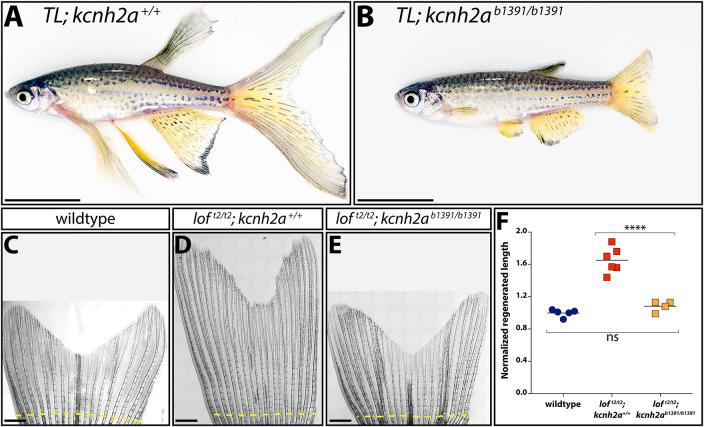
We used CRISPR/Cas9 to mutate kcnh2a in the TL (leot1/t1; loft2/t2) background to determine whether overexpressed kcnh2a causes loft2 fin overgrowth. We outcrossed founders to wild-type fish and identified numerous obligatory loft2/+ F1 animals with normal sized fins carrying germline-transmitted kcnh2a loss-of-function alleles. One of these, kcnh2ab1391, contained an 8 bp deletion causing a frameshift at codon 3 of the predicted polypeptide. Homozygous loft2/t2; kcnh2ab1391/b1391 fish were phenotypically normal with fin sizes indistinguishable from wild-type clutchmates (Fig. 2A,B; P<0.0001 between loft2/t2 and either wild-type or loft2/t2; kcnh2ab1391/b1391 fish; no difference between wild-type and loft2/t2; kcnh2ab1391/b1391 genotypes). As loft2 kcnh2ab1391; lof+ kcnh2a+ and loft2 kcnh2ab1391; loft2 kcnh2a+ fish developed normal and long fins, respectively, the kcnh2ab1391 allele uniquely suppresses loft2 in cis. Regenerative outgrowth also was identical between wild-type and loft2/t2; kcnh2ab1391/b1391 caudal fins (Fig. 2C-F). Finally, loft2-linked kcnh2a had no exon or intron/exon boundary non-synonymous mutations or unappreciated SNPs. We conclude cis-ectopic expression of kcnh2a on the loft2 chr2 causes the dominant loft2 fin overgrowth phenotype. Therefore, loft2 likely is a regulatory kcnh2a neomorphic allele.
Fig. 2.
Fin overgrowth in loft2 requires kcnh2a. (A,B) Bright-field images of clutchmate TL; kcnh2a+/+ (A) and TL; kcnh2ab1391/b1391 (B) adult zebrafish. Scale bars: 1 cm. (C-E) Representative images of regenerated caudal fins at 27 days post-amputation (dpa) from (C) wild-type control, (D) loft2/t2; kcnh2a+/+ and (E) loft2/t2; kcnh2ab1391/b1391 fish. The dashed yellow line indicates the site of fin resection. Scale bars: 1 mm. (F) Quantification of fin ray outgrowth of wild-type (blue circles), loft2/t2; kcnh2+/+ (red squares) and loft2/t2; kcnh2ab1391/b1391 (orange squares) fish at 27 dpa. Each data point represents the normalized length of ray 3 from an individual animal of the indicated genotype. ****P<0.005 (one-way ANOVA with Tukey's multiple comparisons tests). ns, not significant.
Kcnh2a actively promotes fin overgrowth by slowing outgrowth termination
We measured regenerating fin outgrowth of wild-type and clutchmate loft2 fish over a 30-day period to explore temporal effects of ectopic kcnh2a on fin overgrowth. For each genotype, regenerated lengths over time fit well to a logistic growth curve, consistent with a lag for blastema establishment prior to a prolonged outgrowth phase. Wild-type and loft2 fish had indistinguishable maximal growth rates reached between 3 and 5 dpa (Fig 3A,B and Fig. S1). Growth rates then gradually declined, approximating an exponential decay equation. However, the growth rate declined slower in loft2 fish, progressively increasing the fin length difference compared with wild-type animals. Wild-type fins largely ceased regenerative outgrowth by 21 dpa, whereas loft2 fins continued to extend through the 30 dpa period. This prolonged outgrowth dynamic matches that of loft2 fin development (Iovine and Johnson, 2000). We conclude fin overgrowth in loft2 fish is due to an extended outgrowth period beginning around 6 dpa rather than elevated maximum growth.
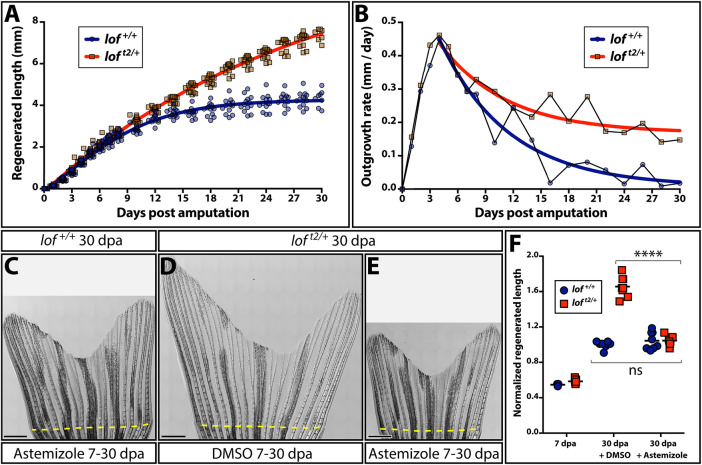
Fig. 3.
Kcnh2a actively prolongs the fin outgrowth period in loft2. (A) Plots comparing the growth (A) and growth rate (B) of regenerating loft2/+ (blue) and clutchmate lof+/+ (red) caudal fins. Curves show actual data fit to (A) a logistic equation reflecting establishment and then progressively slowing outgrowth phases, and (B) an exponential decay equation using time points after the peak growth rate is reached. All data points (≥12 fish per time) are shown in A and mean values are used in B. (C-E) Stitched DIC images showing clutchmate lof+/+ astemizole-treated (C; 500 nM), DMSO-treated loft2/+ (D) and loft2/+, astemizole-treated (E; 500 nM) fish at 30 days post-caudal fin amputation (dpa). The dashed yellow line indicates the amputation plane. Scale bars: 1 mm. (F) Quantification of normalized caudal fin regenerative outgrowth from control and astemizole-treated animals. Data points represent individual lof+/+ (blue circles) and loft2/+ (red squares) animals prior to treatment (7 dpa) and post-treatment (30 dpa) with DMSO or astemizole (500 nM). Graph shows mean lengths of the third ventral regenerated ray normalized to DMSO-treated controls at 30 dpa. Each point is an individual animal. ****P<0.0001 for DMSO treated from 7-30 dpa compared with wild-type and loft2/+ fin lengths; no significant differences between genotypes at 7 dpa or when comparing 7-30 dpa astemizole-treated wild-type and loft2/+ regenerative outgrowth. Statistical tests used one-way ANOVA with Tukey's multiple comparisons tests.
Kcnh2a is related to ether-a-go-go voltage-gated K+ channels (Vandenberg et al., 2012), the channel activity of which is blocked by the small molecule astemizole (Sanguinetti and Tristani-Firouzi, 2006; Suessbrich et al., 1996; Zhou et al., 1999). We treated loft2/+ fish with astemizole from 7-30 dpa to determine whether ectopic Kcnh2a actively extends fin outgrowth period. This inhibitor regimen fully suppressed excessive loft2/+ fin outgrowth to wild-type levels (Fig. 3C-F; P<0.0001). Furthermore, starting astemizole treatment at 6 dpa was as effective as administration throughout regeneration (Fig. S2). Therefore, although loft2/+ fins express kcnh2a relatively early during regeneration (4 dpa, Fig. 1), its functional impact on growth at these times is negligible. Furthermore, consistent with wild-type-sized kcnh2ab1391/b1391 caudal fins, Kcnh2a activity normally does not contribute to regenerative outgrowth. We conclude ectopic Kcnh2a exclusively causes regenerative fin overgrowth by actively preventing growth termination during late-stage outgrowth (∼7 dpa and onward).
Ectopic kcnh2a acts in the intra-ray mesenchyme lineage to promote fin overgrowth in loft2
Kcnh2a could function fin autonomously or systemically (e.g. via circulating endocrine factors) to prolong loft2 fin outgrowth periods. We used F0 CRISPR/Cas9 targeting to induce mosaic kcnh2a mutations to discriminate between these scenarios. The tissue-autonomous model predicts fin size heterogeneity depending on the extent of kcnh2a targeting across rays of a given fin and between different fins. In contrast, the systemic hypothesis anticipates kcnh2a loss-of-function mutations always would equally suppress loft2/+ fin overgrowth. We injected embryos from a wild-type×loft2/+ cross with Cas9 and a kcnh2a-targeting gRNA, and scored reared adults for fin overgrowth. Roughly half of uninjected control animals displayed long fins, as anticipated (51:40, wild type:overgrown). In contrast, 21.8% of kcnh2a crispants showed partial regionalized fin overgrowth (17 of 78, of which half would be loft2/+ fish). A representative partially overgrown caudal fin is shown in Fig. 4A. Another striking example displayed one pectoral fin rescued to wild-type length with the other exhibiting pronounced overgrowth indicative of loft2 (Fig. 4B). Therefore, ectopic Kcnh2a likely functions within fin tissue rather than systemically to disrupt the cessation of fin outgrowth.
Fig. 4.
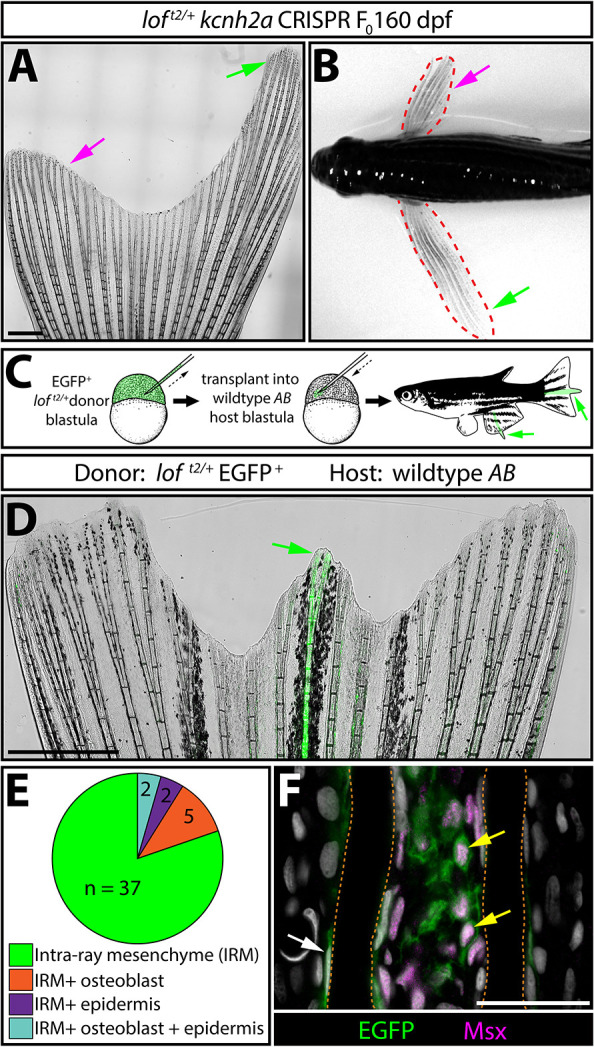
kcnh2a acts fin autonomously and within the intra-ray mesenchyme lineage to promote overgrowth. (A) Representative whole-mount image of a F0 kcnh2a CRISPR loft2/+ adult caudal fin at 160 days post-fertilization. The magenta arrow indicates suppressed overgrowth in otherwise long fins (green arrow). Scale bar: 1 mm. (B) Pectoral fin asymmetry in a kcnh2a F0 CRISPR loft2/+ animal. The magenta arrow indicates a wild-type-sized fin indicative of phenotypic suppression. The green arrow indicates the overgrown contralateral fin expected of the loft2/+ genotype. (C) Upper panel schematic highlighting the nature of the blastula stage transplant experiment. EGFP+ loft2/+ blastula cells were transplanted into wild-type AB embryos. Reared adults with partial or complete fin overgrowth were scored for cell type(s) with EGFP expression. (D) A representative example of a caudal fin displaying overgrown EGFP+ fin rays. The green arrow indicates overgrown EGFP+ fin rays. Scale bar: 1 mm. (E) The pie chart indicates the EGFP+ lineage(s) present in 39 overgrown regions across 30 total fins with extended rays. (F) Caudal fin section of an overgrown chimeric fin ray immunostained with EGFP and Msx antibodies. Yellow arrows indicate EGFP+/Msx+ intra-ray mesenchymal cells. The white arrow highlights an EGFP+ osteoblast. Fin rays are outlined with a dashed orange line. Hoechst-stained nuclei are in gray. Scale bar: 50 µm.
We generated chimeras by blastula cell transplantations (Kimmel et al., 1990) to examine loft2 tissue autonomy further. We introduced EGFP-labeled loft2/+ blastula-stage cells into AB wild-type host embryos and raised them to adulthood (Fig. 4C). 13% of transplants (39 fins with overgrowth from 28 of 214 screened chimeras) exhibited EGFP+ fin tissue with notable overgrowth indicative of chimerism (Fig. 4D and Fig. S3). Several fins had strong EGFP+ overgrown rays flanked by intermediate length rays with minimal EGFP expression. We attribute these neighbor effects to diffusible factors emanating from the primary overgrown ray, to anatomical influences on growth between adjacent rays or to scarce EGFP+ cells sufficient to cause lesser overgrowth. In a few cases, chimeric animals displayed asymmetrically sized pectoral fins, as observed with kcnh2a CRISPR-targeted F0 loft2 mosaic fish (Fig. S3). All overgrown chimeric rays contained EGFP+ intra-ray mesenchymal cells (n=30 overgrown fins containing only EGFP+ mesenchyme; Fig. 4E), although some also displayed EGFP-expressing osteoblasts and/or epidermis (n=5 EGFP+ mesenchyme/osteoblast; n=2 EGFP+ mesenchyme/epidermis; n=2 EGFP+ mesenchyme/osteoblast/epidermis Fig. 4E). We immunostained sections from representative overgrown fins for the mesenchymal marker Msx (Akimenko et al., 1995) to confirm overgrown chimeric fins always included EGFP+ loft2/+-derived intra-ray mesenchyme (Fig. 4F and Fig. S3). In contrast, chimeras harboring only EGFP+ loft2/+ epidermal cells were of normal length (n=3, Fig. S4). We conclude ectopic Kcnh2a functions in intra-ray mesenchyme and/or blastema cells derived from this lineage to disrupt outgrowth-slowing mechanisms.
We used RNAscope (Wang et al., 2012) to localize kcnh2a mRNA in 4 dpa TL caudal fin sections. As predicted from our transplant studies, kcnh2a was expressed in TL regenerating intra-ray mesenchyme, including at lower levels in growth-promoting distal blastema cells of shared lineage (Tornini et al., 2017; Wehner et al., 2014) (Fig. 5A). We did not detect kcnh2a in AB control fins, as expected from RNA-Seq and qRT-PCR (Fig. 5B). We next combined kcnh2a RNAscope with immunostaining for the intra-ray mesenchyme lineage markers Msx (Akimenko et al., 1995) or tph1b:mCherry (Tornini et al., 2016). kcnh2a was expressed in Msx+ or tph1b:mCherry+ cells in proximal regenerating tissue (Fig. 5C-F) with lower levels in tph1b:mCherry+ distal blastema cells (Fig. S5), reinforcing that ectopic Kcnh2a acts in this lineage to promote fin overgrowth.
Fig. 5.
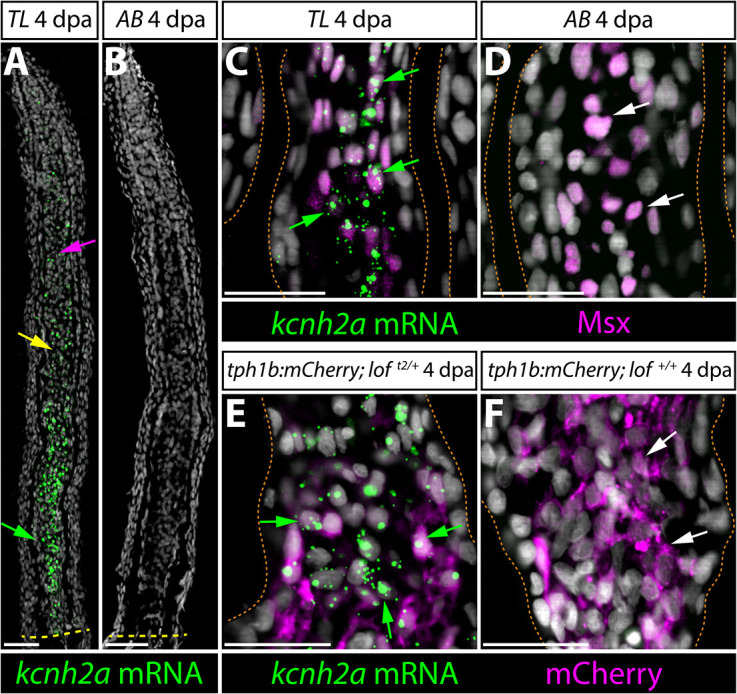
kcnh2a is expressed ectopically in loft2 intra-ray mesenchyme lineage cells during fin regeneration. (A,B) kcnh2a mRNA localization (in green) detected by RNAscope in longitudinal caudal fin sections from 4 days post-amputation (dpa) TL (loft2/t2, A) and AB (B) animals. The green arrow indicates proximal intra-ray cells expressing high levels of kcnh2a, which is also detected in medial mesenchyme (yellow arrow) and distal cells (magenta arrow) of TL fish. The dashed yellow line indicates the site of amputation. Hoechst stained nuclei are in gray. Scale bars: 50 µm. (C,D) Double kcnh2a RNAscope (green) and Msx immunostaining (magenta) of 4 dpa caudal fin sections from TL (C) and AB (D) fish. (E,F) Combination kcnh2a RNAscope (green) and mCherry immunostaining (magenta) of 4 dpa fin sections from (E) tph1b:mCherry; loft2/+ and (F) tph1b:mCherry; lof+/+ fish. For C-F, green arrows highlight Msx+ or tph1b:mCherry+ cells with overlapping kcnh2a mRNA in proximal regenerating lof tissue. White arrows indicate Msx+ or tph1b:mCherry+ nuclei in corresponding regions from control fins lacking kcnh2a expression. Fin rays are outlined with a dashed orange line. Hoechst-stained nuclei are in gray. Scale bars: 50 µm.
Intra-ray mesenchymal cell proliferation rates monitored by EdU incorporation were similar in 4 dpa wild-type and loft2/+ regenerating fins (Fig. S6). Cell cycling was distally concentrated in both genotypes. The fraction of proliferating cells was significantly higher in 10 dpa loft2/+ fin regenerates, being only modestly reduced from 4 dpa (Fig. S6). EdU-incorporating intra-ray cells remained largely confined to the distal blastema of loft2/+ fin regenerates in spite of ectopic kcnh2a throughout ray mesenchyme. Furthermore, the wnt5a growth factor remained distal-enriched in 10 dpa loft2/+ regenerating fins (Stoick-Cooper et al., 2007; Stewart et al., 2014; Fig. S6). Therefore, matching our outgrowth rate measurements, an extended outgrowth period leading to accumulated cell proliferation rather than an increased maximum cycling rate causes loft2 fin overgrowth. Furthermore, kcnh2a prolongs outgrowth without being autonomously sufficient to drive cell cycling and/or a growth factor-producing distal blastema cell state.
Calcineurin promotes outgrowth cessation to help establish fin size
Inhibiting the Ca2+/calmodulin-dependent phosphatase calcineurin with the immunosuppressants FK506 or cyclosporin leads to pronounced fin overgrowth (Daane et al., 2018; Kujawski et al., 2014). Maximal growth rates in calcineurin-inhibited regenerating caudal fins are the same as wild type (Daane et al., 2018; Kujawski et al., 2014), implying that, like loft2/+, such fins do not properly terminate growth. We treated fish with FK506 at various time points post-caudal fin amputation to test this hypothesis. Optimization experiments demonstrated that daily 4 h immersion from 1-21 dpa in water containing 500 nM FK506 was sufficient to overgrow fins to the same degree as loft2/t2 with no apparent adverse effects on animal health (Fig. S7). We treated regenerating animals from either 1-5, 6-21 or 1-21 dpa following this acute drug delivery regimen and measured regeneration extents at 21 dpa. Treating animals with FK506 from 1-5 dpa had no effect on fin outgrowth but still prevented joint formation (Fig. 6A,B), a known additional phenotype upon calcineurin inhibition (Kujawski et al., 2014). In contrast, FK506 treatment from 6-21 or 1-21 dpa caused the same extent of pronounced fin overgrowth (Fig. 6C,E). Thus, like ectopic Kcnh2a in loft2 mutants, calcineurin inhibition appears uniquely to cause fin overgrowth by disrupting growth cessation.
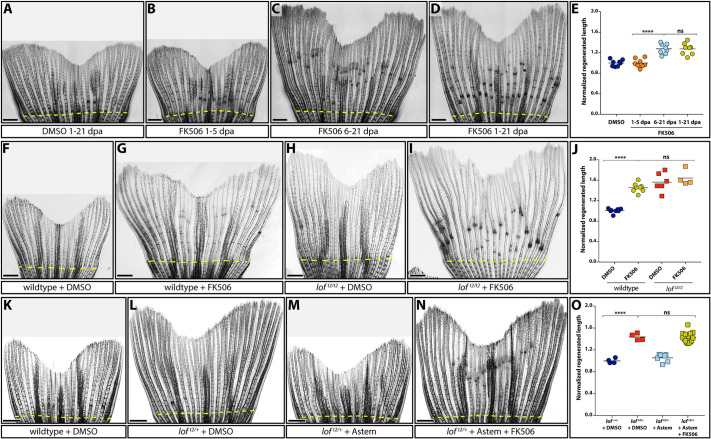
Fig. 6.
Kcnh2a-disrupted calcineurin signaling gradually terminates fin outgrowth. (A-D) Calcineurin gradually ends the fin outgrowth period. Stitched DIC images showing wild-type 21 day post amputation (dpa) caudal fins treated with DMSO (A) or 500 nM FK506 from either 1-5 (B), 6-21 (C) or 1-21 (D) dpa. (E) Quantification of the experiments presented in A-D. Data are the average regenerated lengths of the third ray normalized to DMSO-treated controls at 21 dpa. Each point is a single wild-type animal treated with either DMSO 1-21 dpa (dark blue circles) or FK506 for the indicated times (orange circles, 1-5 dpa; light-blue circles, 6-21 dpa; gold circles, 1-21 dpa). (F-I) Ectopic Kcnh2a and FK506 treatment do not cooperate during overgrowth. DIC-imaged regenerated caudal fins from wild-type (F,G) and loft2/t2 (H,I) animals treated with DMSO (F,H) or 500 nM FK506 (G,I) daily from 1-23 dpa. (J) Graph showing relative regenerate lengths of ray 3 at 23 dpa from wild-type fish treated with DMSO or FK506 (blue or gold circles, respectively), and loft2/t2 fish treated with DMSO or FK506 (red or orange squares, respectively). Each data point is from an individual fish. (K-N) Kcnh2a activity is not required for FK506-induced overgrowth. Images showing wild-type (K) and loft2/+ clutchmate (L-N) caudal fin regenerates at 21 dpa. Animals were treated from 7-21 dpa with DMSO vehicle (K,L), 500 nM astemizole (M) or 500 nM astemizole+500 nM FK506 (N). (O) Plot showing relative regenerative fin growth of ray 3 from the samples described in K-N. Data points represent individual animals: DMSO-treated lof+/+ or loft2/+ (dark-blue circles and red squares, respectively), astemizole-treated loft2/+ (light-blue squares), and loft2/+ treated with both FK506 and astemizole (gold squares). Data are normalized to DMSO-treated wild-type samples. Dashed yellow lines indicate amputation sites. Each data point represents an individual animal. ****P<0.001; ns, not significant. Scale bars: 1 mm. One-way ANOVA with Tukey's multiple comparisons tests tested differences between regenerated fin lengths.
Kcnh2a in loft2 likely disrupts calcineurin signaling to cause fin overgrowth
We hypothesized that ectopic Kcnh2a disrupts calcineurin signaling, given that loft2 and FK506 experiments highlight similar growth dynamic effects and temporal functions during fin regeneration. If so, loft2 should not enhance FK506-induced fin regenerative overgrowth. Accordingly, we observed no difference in caudal fin overgrowth in wild-type versus loft2/t2 fish treated with FK506 from 7 to 21 dpa (Fig. 6F-J). The loft2/t2 genetic background did not alter FK506 bio-availability because joint formation remained dramatically disrupted (Fig. 6H,I). Additionally, treating loft2/+ with both astemizole and FK506 from 7-21 dpa still produced FK506-induced fin overgrowth (Fig. 6K-O; P<0.001). Therefore, Kcnh2a activity, unlike Kcnk5b/alf (Daane et al., 2018), is not downstream of calcineurin. Furthermore, loft2-ectopic Kcnh2a throughout fin development, and even the first week of fin regeneration, does not impact how an amputated fin responds to calcineurin inhibition. We conclude ectopic Kcnh2a likely disrupts calcineurin signaling, which otherwise impacts fin size by gradually terminating outgrowth.
DISCUSSION
loft2 is a neomorph allele of the ether-a-go-go-related kcnh2a potassium channel
loft2 was one of the first zebrafish mutant lines used scientifically (hence, its ‘t2’ – Tübingen 2 – designation), having been originally isolated by tropical fish hobbyists (Elias, 1984; Haffter et al., 1996; Van Eeden et al., 1996). loft2 remains widely used because its highly specific fin overgrowth provides a convenient phenotypic marker to discriminate zebrafish of mixed genotypes. We demonstrate that loft2 is a regulatory neomorphic allele of kcnh2a (kcnh2alof) that causes its ectopic expression in fin mesenchyme, resolving the basis of its remarkable and long-appreciated phenotype. The genetic lesion is likely a chromosomal inversion linking kcnh2a to a displaced enhancer, explaining why kcnh2a lies just outside the originally mapped lof region (Daane et al., 2021 preprint; Iovine and Johnson, 2000).
kcnh2a encodes an ether-a-go-go (EAG)-related voltage-gated K+ channel. EAG/KCNH channels support membrane repolarization in various excitable cells, including neurons and myocytes (Vandenberg et al., 2012). The human ortholog KCNH2 produces IKr, the rapid component of the cardiac delayed rectifier current (Curran et al., 1995; Noble and Tsien, 1969; Sanguinetti et al., 1995). The voltage-dependent gating properties of KCNH2, namely slow opening and closing but fast inactivation, uniquely enables it to repolarize cardiac tissue and terminate action potentials (Bohnen et al., 2017; Vandenberg et al., 2012). KCNH2 mutations are a frequent cause of inherited arrhythmias known as long QT syndrome, whereby patients exhibit prolonged cardiac action potentials (Bohnen et al., 2017; Curran et al., 1995; Sanguinetti et al., 1995). KCNH2 is also a notorious pharmaceutical ‘off-target’, leading to withdrawal of many drugs due to arrhythmia side effects. We find homozygous loss of kcnh2a has no overt effects on zebrafish development to adulthood, including fin length, although we have not assessed cardiac function. Ectopic expression of kcnh2a in loft2 may be confined to fin mesenchyme, explaining the highly specific phenotype and otherwise healthy fish. However, we have not explored whether kcnh2a is misexpressed in other loft2 tissues, where it could adversely impact cardiac or other excitable cell functions. If so, loft2 likely should be avoided as a ‘wild-type’ strain.
Size establishment by modulation of outgrowth periods by intra-ray mesenchymal cells
Our study of fin overgrowth in loft2 advances a mechanistic view of fin growth control centered on growth cessation rather than acceleration or potency. Ectopic Kcnh2a in loft2 does not alter initial or maximal growth rates during caudal fin regeneration, similar to loft2 fin development growth dynamics (Iovine and Johnson, 2000). Rather, Kcnh2a expression dampens growth rate deceleration, effectively extending the allometric growth period. Kcnh2a specifically may enhance later outgrowth phases because mitogenic drive is saturated during the initial regenerative growth response (3-6 dpa), overshadowing the growth-promoting effects of Kcnh2a. Alternatively, upstream signals activating Kcnh2a or the ion signaling pathways disrupted by Kcnh2a may be inactive early in regeneration.
Our CRISPR and transplant chimera studies indicate ectopic Kcnh2a within fins and their intra-ray mesenchyme lineage is necessary and sufficient for loft2 fin overgrowth. Concordantly, loft2 fish ectopically express kcnh2a in blastema fin mesenchyme but not other cell types, with transcripts nearly undetectable in wild-type regenerating caudal fins. alf transplant experiments suggest hypermorphic Kcnk5b also acts within the mesenchyme lineage to cause fin overgrowth (Perathoner et al., 2014). Further evidence indicating this population is growth-determining include lineage-tracing experiments showing intra-ray mesenchyme contributes to the growth-promoting distal blastema (Tornini et al., 2016). Finally, we recently proposed a model explaining slowing fin outgrowth by the progressive depletion of these distal blastema cells by biased differentiation versus self-renewal (Stewart et al., 2019 preprint). Therefore, Kcnh2a may disrupt ion signaling that normally promotes mesenchyme lineage cell transitions from a growth-promoting to differentiated state.
Alternatively, ectopic Kcnh2a may prolong or enhance the production of pro-growth signaling molecules during the slowing outgrowth phase of fin regeneration. In support, Wnts and FGFs are produced by a mesenchyme-derived ‘organizing center’ or ‘niche’ cells at the distal blastema (Stewart et al., 2019 preprint; Tornini et al., 2016; Wehner et al., 2014), and the Kcn5b (alf) K+ channel cell-autonomously promotes growth factor production (Yi et al., 2021). We observed kcnh2a expression throughout regenerating mesenchyme, highest near the amputation site but still detectable in medial and distal blastema. However, we did not observe elevated proliferation or ectopic wnt5a, a representative distal growth factor, in proximal kcnh2a-expressing loft2 mesenchyme. Nevertheless, ectopic Kcnh2a could prolong growth factor production by acting directly in distal blastema/niche cells or in more proximal mesenchyme by disrupting negative feedback to the distal cells.
Calcineurin as an ion signaling node for growth cessation
Mutations in kcnk5b (a K+ channel; Perathoner et al., 2014), kcc4a/slc12a7A (a K+ Cl− co-transporter; Lanni et al., 2019), or over/ectopic expression of the K+ channels kcnj13, kcnj1b, kcnj10a and kcnk9c (Silic et al., 2020), and now kcnh2a (this study) all cause fin overgrowth. Each model may disrupt a common ‘ion signaling’ pathway featuring the fin outgrowth-restraining Ca2+-dependent phosphatase calcineurin (Daane et al., 2018; Harris et al., 2020; Kujawski et al., 2014; Lanni et al., 2019; Yi et al., 2021). Here, we found fin overgrowth in regenerating loft2 fins was not enhanced by the calcineurin inhibitor FK506, suggesting ectopic Kcnh2a also inhibits calcineurin signaling. Ectopic Kcnh2a in loft2 could derail calcineurin output either upstream or downstream of calcineurin itself. Supporting the former, the phosphatase activity of calcineurin is modulated by sustained elevated cytosolic Ca2+ (Klee et al., 1998; Rao, 2009; Timmerman et al., 1996) and KCNH2 effectively shortens Ca2+ fluxes during the cardiac conduction cycle (Bohnen et al., 2017; Vandenberg et al., 2012). Furthermore, recent work indicates reduced calcineurin activity in loft2 fins (Cao et al., 2021). Alternatively, ectopic Kcnh2a could short-circuit calcineurin-promoted ion signaling dynamics mediated by Kcnk5b inhibition (Daane et al., 2018; Yi et al., 2021). These possibilities could be distinguished by determining whether expressing constitutively active calcineurin in the intra-ray mesenchyme lineage suppresses loft2 fin overgrowth.
Ion signaling and interpretation of size-instructing positional information
Bioelectricity is widely linked to organ size control and regeneration (McLaughlin and Levin, 2018). Bioelectric fields are suggested to pre-pattern undifferentiated tissue (including the fin blastema) to establish positional information instructing the correct amount of growth. However, loft2 does not seem to change positional information established at the outset of fin regeneration because inhibiting ectopic Kcnh2a during only the late outgrowth phase restores a normal sized fin. Likewise, calcineurin need be inhibited only late during regeneration to maximally overgrow fins. Therefore, elevated Kcnh2a and calcineurin inhibition appear to disrupt growth deceleration mechanisms tuned to interpret, rather than set, positional information and thereby help establish (and re-establish) correct proportions. Regeneration has the additional challenge that outgrowth has to ‘read’ some form of memory within cells or in higher tissue-level organization to direct the correct amount of outgrowth while also re-establishing said memories. At least proximally, loft2 overgrown caudal fins do not carry abnormal positional memory as fins amputated here regenerate normally when ectopic Kcnh2a is inhibited. Similarly, clonal analyses show calcineurin inhibition does not alter blastema pre-patterning (Tornini et al., 2016) and re-amputation of previously FK506-treated animals results in normal fin size (Daane et al., 2018). The nature of fin positional information and memory is unresolved but, as mentioned, likely reflects properties of intra-ray fibroblasts and derived blastema mesenchyme cells and/or their population sizes (Stewart et al., 2019 preprint).
Identifying and characterizing ectopic kcnh2a as the cause of the classic loft2 zebrafish allele provides a framework to consider bioelectric control of organ size and shape through ion-mediated intracellular signaling. Subtle changes in calcium dynamics that tune signaling output and then growth period durations could produce profound changes in organ scale, while retaining overall form and function. More local expression changes in ion signaling components (therefore, acting as effectors of ‘positional information’) could then readily alter organ proportions that support evolutionary innovations and phenotypic diversity. Extending this concept, intersecting systemic gene regulatory signals, including hormones, could underlie sexual dimorphic traits or environmental effects on organ morphology arising after embryonic development. Swordtail fish provide a compelling example by their male-specific, dramatically elongated rays (the ‘sword’) on the ventral edge of the caudal fin. Strikingly, the swordtail phenotype was recently linked to the kcnh2a-related gene kcnh8 (Schartl et al., 2021). How fin growth periods are highly sensitive to alterations in K+ channels and Ca2+/calcineurin is unclear. Key future steps include characterizing membrane potentials and putative depolarizing signals, and determining how Ca2+ signaling alters states and/or activities of intra-ray mesenchymal cells during fin outgrowth.
MATERIALS AND METHODS
Zebrafish strains and maintenance
Zebrafish were housed in the University of Oregon Aquatic Animal Care Services facility at 28-29°C. The University of Oregon Institutional Animal Care and Use Committee oversaw animal use. Wild-type AB (University of Oregon Aquatic Animal Care Services), TL (Haffter et al., 1996), Tg(tph1b:mCherry)ens700Tg (Tornini et al., 2016), Tg(sp7:EGFP)b1212Tg (DeLaurier et al., 2010) and Tg(Xla.Eef1a1-actb2:LOXP-LOX5171-FRT-F3-EGFP,mCherry)vu295aTg (Boniface et al., 2009), abbreviated herein as Tg(eab:FlEx), zebrafish lines were used.
CRISPR/Cas9-generation of kcnh2a mutants
A guide RNA (gRNA) encompassing the putative kcnh2a start codon (in bold) 5′ GACAACATGCCTGTACGACG 3′ was synthesized in vitro and co-injected with recombinant Cas9 protein (500 ng/µl; Thermo Fisher Scientific) into one-cell stage TL embryos. F0 fish were outcrossed and those F1 clutches harboring mutant alleles identified by PCR sequencing reared to adulthood. Genotyping of F1 adults identified a predicted nonsense allele, kcnh2ab1391, that deletes 8 bp (underlined) at the gRNA target site (GACAACATGCCTGTACGACG) to produce a frameshift at codon 3 of the predicted polypeptide and remove a Hpy99I restriction endonuclease site. kcnh2ab1391 was genotyped by amplifying genomic DNA using primers kcnh2a_P1 and kcnh2a_P2, and digesting PCR products with Hpy99I.
To distinguish between a tissue autonomous versus systemic function for kcnh2a in loft2/+ fin outgrowth, CRISPR/Cas9 mutagenesis was carried out on one-cell stage embryos from a loft2/+×lof+/+ cross. These animals were raised to adulthood and examined for fin ray length heterogeneity within fins and fin size disparities between paired fins.
Sequencing of kcnh2a in longfint2
kcnh2a-coding exons were PCR amplified from TL genomic DNA (primers listed below), ligated to a vector (pCRII, Thermo Fisher Scientific) and sequenced using T7 and SP6 primers. Additional sequencing primers were used to ensure complete coverage of the clone.
Adult whole zebrafish and fin imaging
Whole animal images were captured using a homemade light box made from a fenestrated styrofoam container, a halogen bicycle lamp and a consumer Fujifilm X-A1 camera with Fujinon 28 mm 1.4R lens. High-resolution fin images were obtained from tricaine-euthanized adult fish placed in water or mounted in 0.75% low melting agarose. Stitched differential interference contrast (DIC) images were then captured using a motorized Nikon Eclipse Ti widefield microscope with a 10× objective and NIS-Elements software.
RNA-Seq
Caudal fins of adult loft2/+ and lof+/+ Tg(sp7:EGFP) clutchmates were amputated. At 4 dpa, regenerated tissue beyond the GFP-marked differentiated osteoblast domain was homogenized in TRIzol reagent (Thermo Fisher Scientific). Tissue pooled from four animals constituted matched replicate samples. RNA was isolated following the manufacturer's instructions with minor alterations. The RNA was precipitated overnight at −80°C and then pelleted at 21,000 g for 30 min at 4°C. Pellets were washed twice with 70% ethanol, dried for 10 min at room temperature and resuspended in RNase-free water (Thermo Fisher Scientific). RNA-Seq libraries were prepared from 1 µg of isolated RNA using a Kapa Biosystems Stranded mRNA-Seq kit. Bar-coded libraries were pooled and sequenced using a HiSeq 4000 (Illumina), and reads were aligned to the zebrafish genome (GRCz11) using TopHat2 (Kim et al., 2013). Aligned reads were scored using HTseq (Anders et al., 2015) with edgeR (Robinson et al., 2010) used to identify differentially expressed transcripts.
RT-qPCR
RNA was isolated from whole caudal fin regenerates of adult loft2/+ and lof+/+ clutchmates at 96 hpa. Each replicate represented a pooled sample of three fin regenerates. RNA was isolated as described in the preceding section. cDNA was synthesized from 500 ng of total RNA using oligo(dT)18 primer and Maxima H Minus RT (Thermo Fisher Scientific) following the manufacturer's protocol. Primers kcnh2a_P1 and kcnh2a_P2 were used to determine kcnh2a expression. Primers for runx2b and rpl8 were used as a control and a ubiquitously expressed reference gene, respectively.
The kcnh2aAB polymorphism was identified serendipitously in University of Oregon Aquatic Animal Care Services facility stock AB fish and has been observed previously in other zebrafish strains (Butler et al., 2015). To demonstrate monoallelic expression of kcnh2a on mutant loft2 chromosome 2, we generated loft2 kcnh2aWIK-TL; lof+ kcnh2aAB fish and prepared cDNA from 4 dpa caudal fins as described above. Quantitative PCR used the primer combinations: kcnh2a_P1 and kcnh2a_P2, kcnh2a_P1 and kcnh2aWIK-TL, and kcnh2a_P1 and kcnh2aAB. For all studies, qPCR was quantified by the ΔΔCt method using rpl13 mRNA levels for normalization as described (Stewart et al., 2014). RT-qPCR statistical analyses used a repeated measures one-way ANOVA with Tukey's multiple comparisons test.
Small molecule treatments
For astemizole treatments, fins of loft2/+ and clutchmate animals were amputated and allowed to regenerate for the indicated times. When appropriate, astemizole (5 mM stock solution in DMSO) was added directly to fish water to a final concentration of 500 nM; control fish received an equal volume of DMSO. Fish were fed and changed to fresh drug- or DMSO-containing water every 48 h. The unusually long half-life of astemizole [>7 days in mammals (Paton and Webster, 1985)] precluded testing how exclusively early treatments affect outgrowth. For FK506 experiments, fins were amputated and animals treated with indicated concentrations of FK506 (typically, 500 nM; stock solution in DMSO) in static water for 4 h every day for the indicated periods and then returned to normal water flow conditions. For Kcnh2a/calcineurin epistasis experiments, we combined treatments with astemizole and FK506 from 7-21 dpa for 4 h in static water containing 500 nM astemizole±500 nM FK506, and then returned animals to system water. At the conclusion of the experiment, each caudal fin was imaged on a Leica M205 FA stereomicroscope and lengths of the third ray from the amputation site to the tip of the fin were measured (detailed below). For each experimental group, representative animals were imaged by DIC stitched imaging as described previously.
Fin measurements
To quantify fin growth after regeneration, the length of the third ray was measured from the amputation site to the tip of the fin using stereomicroscope images and FIJI software (NIH). Growth measurements from individuals comprising each cohort were normalized to the mean of the control group. A one-way ANOVA with Tukey's multiple comparisons tests tested differences between regenerated fin lengths. Logistic curves were fit to the fin regeneration length over time data to account for a slowly outgrowing establishment phase, followed by rapid acceleration and then slow deceleration during the outgrowth phase. An exponential decay curve was fit to the outgrowth rate over time data, starting from the peak growth rate at 4 dpa. Prism 7 was used for statistical analyses, curve fitting and graph plotting (GraphPad Software).
Immunostaining and EdU staining
Fin tissue was paraffin wax embedded and sectioned as described previously (Stewart et al., 2014). Slides were antigen retrieved for 5 min in 1 mM EDTA and 0.1% Tween-20 using a pressure cooker followed by blocking in 10% milk in phosphate-buffered saline containing 0.1% Tween-20. Antibodies were applied overnight in blocking buffer at 4°C. Individual antigens were visualized using Alexa dye-conjugated secondary antibodies (Thermo Fisher Scientific). Stained sections were imaged using Olympus FV1000 or Zeiss LSM880 laser scanning confocal microscopes.
For 5-ethynyl-2′-deoxyuridine (EdU) incorporation studies, caudal fins from tph1b:mCherry and clutchmate tph1b:mCherry; loft2/+ fish were collected at 4 or 10 dpa following a 4 h pulse with 1 mg/kg EdU (Thermo Fisher Scientific) delivered by intra-peritoneal injection. Fins were processed for immunostaining as described above. Only the ventral lobe was collected for long-finned fish. EdU detection was carried out according to the manufacturer's recommendations (Thermo Fisher Scientific). Slides were imaged on a Zeiss LSM880 confocal microscope. EdU+ mesenchymal cells were quantified using the Imaris software package Spots function on a masked channel excluding non-blastema cells. The statistical analysis used a two-way ANOVA with Tukey's multiple comparison tests.
RNAscope combined with EdU or immunostaining
RNAscope probes to detect kcnh2a or wnt5a mRNA were designed and synthesized by ACD Bio. RNAscope was performed using the Multiplex Fluorescent kit (ACD Bio) according to the manufacturer's recommendations for paraffin wax embedded sections with minor modifications. RNAscope followed by EdU was performed for dual imaging. Combined immunostaining using Msx (Developmental Studies Hybridoma Bank; 1:20 dilution of hybridoma supernatant) or mCherry (Sicgen; 1:100) antibodies was carried out after RNAscope, as directed by the manufacturer (ACD Bio). Nuclei were visualized by Hoechst staining (Thermo Fisher Scientific). Imaging used Olympus FV1000 or Zeiss LSM880 laser scanning confocal microscopes.
Generation of chimeric animals
Blastula stage cell transplantations were performed as described previously (Kimmel et al., 1990). Briefly, 20-50 mesoderm-targeted single cells from high-stage loft2/+; Tg(eab:FlEx) donors were transplanted into AB host high-stage embryos (Kimmel et al., 1995). Chimeric animals were reared to adulthood and fins screened for EGFP expression and overgrowth. Those with apparent fin overgrowth were imaged on a Leica M205 FA stereomicroscope. Stitched high-resolution images of select overgrown fins were also collected. Epifluorescent images of overgrown fins were examined to determine lineages, including EGFP+ cells, as described previously (Stewart and Stankunas, 2012). Lineage-labeling of select samples was confirmed by immunostaining fin sections using EGFP (Aves Labs; 1:1000), Msx (Developmental Studies Hybridoma Bank; 1:20), sp7 (A-13; Santa Cruz Biotechnology; 20 ng/ml) and p63 (PA5-36069; Thermo Fisher Scientific; 1:100) antibodies as described earlier.
Primers
The following primers were used: kcnh2a_P1, 5′ GGATTTGCGCCCTCCGAACACTAAC 3′; kcnh2a_P2, 5′ GTGCGCGCATCCTCGAGCTCAG 3′; kcnh2a_P3, 5′ ACTGGAGACTTTGAAGAGACGC 3′; kcnh2a_P4, 5′ GAATGATGGTGTCCAGAAAGGT 3′; kcnh2aAB, 5′ GTACAGGCATGTTTGTCCC 3′; kcnh2aWIK-TL, 5′ GTACAGGCATGTTGTCCT 3′; kcnh2a_exon_1-2_for, 5′ CCCCAACCATCCTCTCACTGCCTCTCC 3′; kcnh2a_exon_1-2_rev, 5′ CAGGGCCCCTAAGCTGTCCTGG 3′; kcnh2a_exon_3_for, 5′ CTGGTGATCGCAGACCTTAACACAGACCTC 3′; kcnh2a_exon_3_rev, 5′ GAATTGACCCCCTCCCTGATCGTCAACG 3′; kcnh2a_exon_4_for, 5′ CTGGTGAACTGCTGCAAGTAAGTTACTGTAGATTC 3′; kcnh2a_exon_4_rev, 5′ CTGTATGACCACTGTATGACCAGGTGTACC 3′; kcnh2a_exon_5_for, 5′ GTACAGAGGTGGTGGTCCTCCAGGAACGTGG 3′; kcnh2a_exon_5_rev, 5′ GTCTGTGGTGAATACACCATGTGTGCCTTTTAAAT 3′; kcnh2a_exon_6_for, 5′ CAGCAGCATTTACTTCAATCACGATGATCAC 3′; kcnh2a_exon_6_rev, 5′ GGTTTGGAGCCACCTGAGTGTGAG 3′; kcnh2a_exon_7_for, 5′ GCACCCTAGCAACTGTAATAACTGCTGC 3′; kcnh2a_exon_7_rev, 5′ GAATAAGCACTTAGCAGCACTAACCTCCG 3′; kcnh2a_exon_8-9_for, 5′ GTTCAACCACTGGGTGTCAAACTTACACACT 3′; kcnh2a_exon_8-9_rev, 5′ GGGTGGGGAGTGTCTGGGTAAGGG 3′; kcnh2a_exon_10-12_for, 5′ GGCATGCAGCGCCACCTGCTGTTGATTTC 3′; kcnh2a_exon_10-12_rev, 5′ GGTACGGCAACTATTCTCCCAATCAACATAC 3′; kcnh2a_exon_13-14_for, 5′ GAGGTTAGAATGACTTGAATGAGTATAATCAGTG 3′; kcnh2a_exon_13-14_rev, 5′ GACCCAGCCGAGGCTTGAACCA 3′; kcnh2a_exon_15_for, 5′ GAGGGTGTGTGATATCAATAGCATGGGG 3′; kcnh2a_exon_15_rev, 5′ GCGTTCACACAGCACAGCGTAACCAG 3′; runx2b_forward, 5′ AGCTTCACCCTGACGATTACA 3′; runx2b_reverse, 5′ CCAGTTCACTGAGACGGTCA 3′; rpl8_forward, 5′ CCGAGACCAAGAAATCCAGA 3′; and rpl8_reverse, 5′ GAGGCCAGCAGTTTCTCTTG 3′.
Supplementary Material
Acknowledgements
We thank the University of Oregon Aquatic Animal Care Services for animal husbandry; A. Lasseigne and A. Miller for assisting with blastula transplantations; J. Chehab for sequence analysis; H. Markovic for assistance with inhibitor studies; the Stankunas lab for discussions; C. Kimmel, D. Grimes and V. Lewis for manuscript feedback; and K. Poss for providing the Tg(tph1b:mCherry) line.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: S.S., K.S.; Methodology: S.S., H.K.L., G.A.Y., A.L.H., K.S.; Formal analysis: S.S., H.K.L., G.A.Y., A.L.H., K.S.; Investigation: S.S., H.K.L., G.A.Y., A.L.H., A.E.R., J.A.B., K.S.; Data curation: S.S., G.A.Y., K.S.; Writing - original draft: S.S., K.S.; Writing - review & editing: S.S., H.K.L., G.A.Y., A.L.H., K.S.; Visualization: S.S., H.K.L., G.A.Y., A.L.H., K.S.; Supervision: S.S., K.S.; Project administration: S.S., K.S.; Funding acquisition: S.S., K.S.
Funding
G.A.Y. was supported by a National Institutes of Health National Research Service Award graduate fellowship (F31AR071283). H.K.L. received funding from the University of Oregon Developmental Biology Training Program (T32HD007348). A.E.R. was supported by the University of Oregon Genetics Training Program (T32GM007413) and by a National Institutes of Health National Research Service Award fellowship (F31GM139343). The National Institutes of Health provided research funding (R01GM127761 and R03AR067522). Deposited in PMC for release after 12 months.
Data availability
RNA-Seq data have been deposited in GEO under accession number GSE137352.
Peer review history
The peer review history is available online at https://journals.biologists.com/dev/article-lookup/doi/10.1242/dev.199384
References
- Akimenko, M. A., Johnson, S. L., Westerfield, M. and Ekker, M. (1995). Differential induction of four msx homeobox genes during fin development and regeneration in zebrafish. Development 121, 347-357. [DOI] [PubMed] [Google Scholar]
- Anders, S., Pyl, P. T. and Huber, W. (2015). HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166-169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgärtel, K. and Mansuy, I. M. (2012). Neural functions of calcineurin in synaptic plasticity and memory. Learn. Mem. 19, 375-384. 10.1101/lm.027201.112 [DOI] [PubMed] [Google Scholar]
- Bohnen, M. S., Peng, G., Robey, S. H., Terrenoire, C., Iyer, V., Sampson, K. J. and Kass, R. S. (2017). Molecular pathophysiology of congenital long QT syndrome. Physiol. Rev. 97, 89-134. 10.1152/physrev.00008.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boniface, E. J., Lu, J., Victoroff, T., Zhu, M. and Chen, W. (2009). FlEx-based transgenic reporter lines for visualization of Cre and Flp activity in live zebrafish. Genesis 47, 484-491. 10.1002/dvg.20526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler, M. G., Iben, J. R., Marsden, K. C., Epstein, J. A., Granato, M. and Weinstein, B. M. (2015). SNPfisher: tools for probing genetic variation in laboratory-reared zebrafish. Development 142, 1542-1552. 10.1242/dev.118786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, Z., Meng, Y., Gong, F., Xu, Z., Liu, F., Fang, M., Zou, L., Liao, X., Wang, X., Luo, L.et al. (2021). Calcineurin controls proximodistal blastema polarity in zebrafish fin regeneration. Proc. Natl. Acad. Sci. USA 118, e2009539118. 10.1073/pnas.2009539118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C.-H. and Poss, K. D. (2017). Regeneration genetics. Annu. Rev. Genet. 51, 63-82. 10.1146/annurev-genet-120116-024554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabtree, G. R. (1999). Generic signals and specific outcomes: signaling through Ca2+, calcineurin, and NF-AT. Cell 96, 611-614. 10.1016/S0092-8674(00)80571-1 [DOI] [PubMed] [Google Scholar]
- Curran, M. E., Splawski, I., Timothy, K. W., Vincent, G. M., Green, E. D. and Keating, M. T. (1995). A molecular basis for cardiac arrhythmia: HERG mutations cause long QT syndrome. Cell 80, 795-803. 10.1016/0092-8674(95)90358-5 [DOI] [PubMed] [Google Scholar]
- Daane, J. M., Lanni, J., Rothenberg, I., Seebohm, G., Higdon, C. W., Johnson, S. L. and Harris, M. P. (2018). Bioelectric-calcineurin signaling module regulates allometric growth and size of the zebrafish fin. Sci. Rep. 8, 10391. 10.1038/s41598-018-28450-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daane, J. M., Blum, N., Lanni, J., Boldt, H., Iovine, M. K., Higdon, C. W., Johnson, S. L., Lovejoy, N. R. and Harris, M. P. (2021). Novel regulators of growth identified in the evolution of fin proportion in flying fish. bioRxiv 434157. [Google Scholar]
- DeLaurier, A., Eames, B. F., Blanco Sánchez, B., Peng, G., He, X., Swartz, M. E., Ullmann, B., Westerfield, M. and Kimmel, C. B. (2010). Zebrafish sp7:EGFP: A transgenic for studying otic vesicle formation, skeletogenesis, and bone regeneration. Genesis 48, 505-511. 10.1002/dvg.20639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias, J. (1984). Brachydanio frankei-Schleierform. Aquaria 34, 81-85. [Google Scholar]
- Goldsmith, M. I., Iovine, M. K., O'Reilly-Pol, T. and Johnson, S. L. (2006). A developmental transition in growth control during zebrafish caudal fin development. Dev. Biol. 296, 450-457. 10.1016/j.ydbio.2006.06.010 [DOI] [PubMed] [Google Scholar]
- Haffter, P., Odenthal, J., Mullins, M. C., Lin, S., Farrell, M. J., Vogelsang, E., Haas, F., Brand, M., Van Eeden, F. J. M., Furutani-Seiki, M.et al. (1996). Mutations affecting pigmentation and shape of the adult zebrafish. Dev. Genes Evol. 206, 260-276. 10.1007/s004270050051 [DOI] [PubMed] [Google Scholar]
- Harris, M. P., Daane, J. M. and Lanni, J. (2020). Through veiled mirrors: fish fins giving insight into size regulation. Wiley Interdiscip. Rev. Dev. Biol. 99, e381. 10.1002/wdev.381 [DOI] [PubMed] [Google Scholar]
- Iovine, M. K. and Johnson, S. L. (2000). Genetic analysis of isometric growth control mechanisms in the zebrafish caudal fin. Genetics 155, 1321-1329. 10.1093/genetics/155.3.1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovine, M. K. and Johnson, S. L. (2002). A genetic, deletion, physical, and human homology map of the long fin region on zebrafish linkage group 2. Genomics 79, 756-759. 10.1006/geno.2002.6769 [DOI] [PubMed] [Google Scholar]
- Kim, D., Pertea, G., Trapnell, C., Pimentel, H., Kelley, R. and Salzberg, S. L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36. 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel, C. B., Warga, R. M. and Schilling, T. F. (1990). Origin and organization of the zebrafish fate map. Development 108, 581-594. [DOI] [PubMed] [Google Scholar]
- Kimmel, C. B., Ballard, W. W., Kimmel, S. R., Ullmann, B. and Schilling, T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253-310. 10.1002/aja.1002030302 [DOI] [PubMed] [Google Scholar]
- Klee, C. B., Ren, H. and Wang, X. (1998). Regulation of the calmodulin-stimulated protein phosphatase, calcineurin. J. Biol. Chem. 273, 13367-13370. 10.1074/jbc.273.22.13367 [DOI] [PubMed] [Google Scholar]
- Knopf, F., Hammond, C., Chekuru, A., Kurth, T., Hans, S., Weber, C. W., Mahatma, G., Fisher, S., Brand, M., Schulte-Merker, S.et al. (2011). Bone regenerates via dedifferentiation of osteoblasts in the Zebrafish Fin. Dev. Cell 20, 713-724. 10.1016/j.devcel.2011.04.014 [DOI] [PubMed] [Google Scholar]
- Kujawski, S., Lin, W., Kitte, F., Börmel, M., Fuchs, S., Arulmozhivarman, G., Vogt, S., Theil, D., Zhang, Y. and Antos, C. L. (2014). Calcineurin regulates coordinated outgrowth of zebrafish regenerating fins. Dev. Cell 28, 573-587. 10.1016/j.devcel.2014.01.019 [DOI] [PubMed] [Google Scholar]
- Lanni, J. S., Peal, D., Ekstrom, L., Chen, H., Stanclift, C., Bowen, M. E., Mercado, A., Gamba, G., Kahle, K. T. and Harris, M. P. (2019). Integrated K+ channel and K+Cl- cotransporter functions are required for the coordination of size and proportion during development. Dev. Biol. 456, 164-178. 10.1016/j.ydbio.2019.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y., Grill, S., Sanchez, A., Murphy-Ryan, M. and Poss, K. D. (2005). Fgf signaling instructs position-dependent growth rate during zebrafish fin regeneration. Development 132, 5173-5183. 10.1242/dev.02101 [DOI] [PubMed] [Google Scholar]
- Lee, Y., Hami, D., De Val, S., Kagermeier-Schenk, B., Wills, A. A., Black, B. L., Weidinger, G. and Poss, K. D. (2009). Maintenance of blastemal proliferation by functionally diverse epidermis in regenerating zebrafish fins. Dev. Biol. 331, 270-280. 10.1016/j.ydbio.2009.05.545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin, K. A. and Levin, M. (2018). Bioelectric signaling in regeneration: mechanisms of ionic controls of growth and form. Dev. Biol. 433, 177-189. 10.1016/j.ydbio.2017.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan, S. C., Zhang, J., Phan, H.-E., Jeradi, S., Probst, L., Hammerschmidt, M. and Akimenko, M.-A. (2018). A regulatory pathway involving retinoic acid and calcineurin demarcates and maintains joint cells and osteoblasts in regenerating fin. Development 145, dev161158. 10.1242/dev.161158 [DOI] [PubMed] [Google Scholar]
- Nelson, J. S., Grande, T. C. and Wilson, M. (2016). Fishes of the World, 5th Edn. John Wiley & Sons. [Google Scholar]
- Noble, D. and Tsien, R. W. (1969). Outward membrane currents activated in the plateau range of potentials in cardiac Purkinje fibres. J. Physiol. 200, 205-231. 10.1113/jphysiol.1969.sp008689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra, V. and Rothermel, B. A. (2017). Calcineurin signaling in the heart: the importance of time and place. J. Mol. Cell. Cardiol. 103, 121-136. 10.1016/j.yjmcc.2016.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paton, D. M. and Webster, D. R. (1985). Clinical pharmacokinetics of H1-receptor antagonists (the antihistamines). Clin. Pharmacokinet. 10, 477-497. 10.2165/00003088-198510060-00002 [DOI] [PubMed] [Google Scholar]
- Perathoner, S., Daane, J. M., Henrion, U., Seebohm, G., Higdon, C. W., Johnson, S. L., Nüsslein-Volhard, C. and Harris, M. P. (2014). Bioelectric signaling regulates size in zebrafish fins. PLoS Genet. 10, e1004080. 10.1371/journal.pgen.1004080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss, K. D., Shen, J., Nechiporuk, A., McMahon, G., Thisse, B., Thisse, C. and Keating, M. T. (2000). Roles for Fgf signaling during zebrafish fin regeneration. Dev. Biol. 222, 347-358. 10.1006/dbio.2000.9722 [DOI] [PubMed] [Google Scholar]
- Quint, E., Smith, A., Avaron, F., Laforest, L., Miles, J., Gaffield, W. and Akimenko, M.-A. (2002). Bone patterning is altered in the regenerating zebrafish caudal fin after ectopic expression of sonic hedgehog and bmp2b or exposure to cyclopamine. Proc. Natl. Acad. Sci. USA 99, 8713-8718. 10.1073/pnas.122571799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz, J. S., Robitaille, A. M., Wang, Y., Ray, C. A., Thummel, R., Gu, H., Djukovic, D., Raftery, D., Berndt, J. D. and Moon, R. T. (2017). Transcriptomic, proteomic, and metabolomic landscape of positional memory in the caudal fin of zebrafish. Proc. Natl. Acad. Sci. USA 114, E717-E726. 10.1073/pnas.1620755114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, A. (2009). Signaling to gene expression: calcium, calcineurin and NFAT. Nat. Immunol. 10, 3-5. 10.1038/ni0109-3 [DOI] [PubMed] [Google Scholar]
- Robinson, M. D., McCarthy, D. J. and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139-140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanguinetti, M. C. and Tristani-Firouzi, M. (2006). hERG potassium channels and cardiac arrhythmia. Nature 440, 463-469. 10.1038/nature04710 [DOI] [PubMed] [Google Scholar]
- Sanguinetti, M. C., Jiang, C., Curran, M. E. and Keating, M. T. (1995). A mechanistic link between an inherited and an acquired cardiac arrhythmia: HERG encodes the IKr potassium channel. Cell 81, 299-307. 10.1016/0092-8674(95)90340-2 [DOI] [PubMed] [Google Scholar]
- Schartl, M., Kneitz, S., Ormanns, J., Schmidt, C., Anderson, J. L., Amores, A., Catchen, J., Wilson, C., Geiger, D., Du, K.et al. (2021). The developmental and genetic architecture of the sexually selected male ornament of swordtails. Curr. Biol. 12, 911-922.e4. https://doi.org/10.1016/j.cub.2020.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte, C. J., Allen, C., England, S. J., Juárez-Morales, J. L. and Lewis, K. E. (2011). Evx1 is required for joint formation in zebrafish fin dermoskeleton. Dev. Dyn. 240, 1240-1248. 10.1002/dvdy.22534 [DOI] [PubMed] [Google Scholar]
- Sehring, I. M. and Weidinger, G. (2020). Recent advancements in understanding fin regeneration in zebrafish. Wiley Interdiscip. Rev. Dev. Biol. 9, e367. 10.1002/wdev.367 [DOI] [PubMed] [Google Scholar]
- Shibata, E., Yokota, Y., Horita, N., Kudo, A., Abe, G., Kawakami, K. and Kawakami, A. (2016). Fgf signalling controls diverse aspects of fin regeneration. Development 143, 2920-2929. 10.1242/dev.140699 [DOI] [PubMed] [Google Scholar]
- Silic, M. R., Wu, Q., Kim, B. H., Golling, G., Chen, K. H., Freitas, R., Chubykin, A. A., Mittal, S. K. and Zhang, G. (2020). Potassium channel-associated bioelectricity of the dermomyotome determines Fin patterning in Zebrafish. Genetics 215, 1067-1084. 10.1534/genetics.120.303390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, S. P., Holdway, J. E. and Poss, K. D. (2012). Regeneration of amputated zebrafish fin rays from de novo osteoblasts. Dev. Cell 22, 879-886. 10.1016/j.devcel.2012.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, A., Avaron, F., Guay, D., Padhi, B. K. and Akimenko, M. A. (2006). Inhibition of BMP signaling during zebrafish fin regeneration disrupts fin growth and scleroblast differentiation and function. Dev. Biol. 299, 438-454. 10.1016/j.ydbio.2006.08.016 [DOI] [PubMed] [Google Scholar]
- Sousa, S., Afonso, N., Bensimon-Brito, A., Fonseca, M., Simões, M., Leon, J., Roehl, H., Cancela, M. L. and Jacinto, A. (2011). Differentiated skeletal cells contribute to blastema formation during zebrafish fin regeneration. Development 138, 3897-3905. 10.1242/dev.064717 [DOI] [PubMed] [Google Scholar]
- Stewart, S. and Stankunas, K. (2012). Limited dedifferentiation provides replacement tissue during zebrafish fin regeneration. Dev. Biol. 365, 339-349. 10.1016/j.ydbio.2012.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, S., Gomez, A. W., Armstrong, B. E., Henner, A. and Stankunas, K. (2014). Sequential and opposing activities of Wnt and BMP coordinate zebrafish bone regeneration. Cell Rep. 6, 482-498. 10.1016/j.celrep.2014.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, S., Yette, G. A., Le Bleu, H. K., Henner, A. L., Braunstein, J. A., Chehab, J. W., Harms, M. J. and Stankunas, K. (2019). Skeletal geometry and niche transitions restore organ size and shape during zebrafish fin regeneration. bioRxiv 606970. [Google Scholar]
- Stoick-Cooper, C. L., Weidinger, G., Riehle, K. J., Hubbert, C., Major, M. B., Fausto, N. and Moon, R. T. (2007). Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development 134, 479-489. 10.1242/dev.001123 [DOI] [PubMed] [Google Scholar]
- Suessbrich, H., Waldegger, S., Lang, F. and Busch, A. E. (1996). Blockade of HERG channels expressed in Xenopus oocytes by the histamine receptor antagonists terfenadine and astemizole. FEBS Lett. 385, 77-80. 10.1016/0014-5793(96)00355-9 [DOI] [PubMed] [Google Scholar]
- Timmerman, L. A., Clipstone, N. A., Ho, S. N., Northrop, J. P. and Crabtree, G. R. (1996). Rapid shuttling of NF-AT in discrimination of Ca2+ signals and immunosuppression. Nature 383, 837-840. 10.1038/383837a0 [DOI] [PubMed] [Google Scholar]
- Ton, Q. V. and Iovine, M. K. (2013). Identification of an evx1-dependent joint-formation pathway during FIN regeneration. PLoS ONE 8, e81240. 10.1371/journal.pone.0081240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornini, V. A., Puliafito, A., Slota, L. A., Thompson, J. D., Nachtrab, G., Kaushik, A.-L., Kapsimali, M., Primo, L., Di Talia, S. and Poss, K. D. (2016). Live monitoring of blastemal cell contributions during appendage regeneration. Curr. Biol. 26, 2981-2991. 10.1016/j.cub.2016.08.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornini, V. A., Thompson, J. D., Allen, R. L. and Poss, K. D. (2017). Live fate-mapping of joint-associated fibroblasts visualizes expansion of cell contributions during zebrafish fin regeneration. Development 144, 2889-2895. 10.1242/dev.155655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu, S. and Johnson, S. L. (2011). Fate restriction in the growing and regenerating zebrafish fin. Dev. Cell 20, 725-732. 10.1016/j.devcel.2011.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Eeden, F. J., Granato, M., Schach, U., Brand, M., Furutani-Seiki, M., Haffter, P., Hammerschmidt, M., Heisenberg, C. P., Jiang, Y. J., Kane, D. A.et al. (1996). Genetic analysis of fin formation in the zebrafish, Danio rerio. Development 123, 255-262. [DOI] [PubMed] [Google Scholar]
- Vandenberg, J. I., Perry, M. D., Perrin, M. J., Mann, S. A., Ke, Y. and Hill, A. P. (2012). hERG K+ channels: structure, function, and clinical significance. Physiol. Rev. 92, 1393-1478. 10.1152/physrev.00036.2011 [DOI] [PubMed] [Google Scholar]
- Wang, F., Flanagan, J., Su, N., Wang, L.-C., Bui, S., Nielson, A., Wu, X., Vo, H.-T., Ma, X.-J. and Luo, Y. (2012). RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn. 14, 22-29. 10.1016/j.jmoldx.2011.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehner, D. and Weidinger, G. (2015). Signaling networks organizing regenerative growth of the zebrafish fin. Trends Genet. 31, 336-343. 10.1016/j.tig.2015.03.012 [DOI] [PubMed] [Google Scholar]
- Wehner, D., Cizelsky, W., Vasudevaro, M. D., Özhan, G., Haase, C., Kagermeier-Schenk, B., Röder, A., Dorsky, R. I., Moro, E., Argenton, F.et al. (2014). Wnt/β-catenin signaling defines organizing centers that orchestrate growth and differentiation of the regenerating zebrafish caudal fin. Cell Rep. 6, 467-481. 10.1016/j.celrep.2013.12.036 [DOI] [PubMed] [Google Scholar]
- Yi, C., Spitters, T. W., Al-Far, E. A.-D. A., Wang, S., Xiong, T., Cai, S., Yan, X., Guan, K., Wagner, M., El-Armouche, A.et al. (2021). A calcineurin-mediated scaling mechanism that controls a K+-leak channel to regulate morphogen and growth factor transcription. eLife 10, e60691. 10.7554/eLife.60691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Z., Vorperian, V. R., Gong, Q., Zhang, S. and January, C. T. (1999). Block of HERG potassium channels by the antihistamine astemizole and its metabolites desmethylastemizole and norastemizole. J. Cardiovasc. Electrophysiol. 10, 836-843. 10.1111/j.1540-8167.1999.tb00264.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.