Abstract
Prostate cancer is an epithelial malignant tumor of the prostate, and it is one of the malignant tumors with a high incidence of urogenital system in men. The local treatment of prostate cancer is mainly radical resection and radical radiotherapy, but they are not applicable to advanced prostate cancer. Systemic therapy mainly includes targeted therapy and immunotherapy which could cause many complications, and will affect the prognosis and quality of life of patients. It is urgent to find new treatments for prostate cancer. Bioinformatics offers hope for us to find reliable therapeutic targets. Bioinformatics can use the tumor informations in database and analyze them to screen out the best differentially expressed genes. Using the selected differentially expressed genes as targets, a gene interference plasmid was designed, and the constructed plasmid was used for targeted gene therapy. There are some problems about gene therapy that need to be solved, such as how to transfer genes to target cells is also an important challenge. Due to their large molecular weight and hydrophilic nature, they cannot enter cells through passive diffusion mechanisms. Here we synthesized a DNA carrier used surface modified iron based nanoparticles, and used it to load plasmid including ShRNA which can inhibit the expression of oncogene SLC4A4 selected by bioinformatics’ method. After that we use this iron based nanoparticles/plasmid DNA nanocomposite to treat prostate cancer cells in vitro and in vivo. The target gene SLC4A4 we had selected using bioinformatics had a strong effect on the proliferation of prostate cells; Our nanocomposite could inhibit the expression of SLC4A4 effectively, it had strong inhibitory effects on prostate cancer cells both in vivo and in vitro, and can be used as a potential method for prostate cancer treatment.
Keywords: Prostate cancer, bioinformatics, nanoparticles, gene therapy, gene delivery, targeting, cancer inhibition
Introduction
Prostate cancer is the most common malignant tumor in men, with an incidence of about 7.1% of the total number of tumors. It is the second leading cause of cancer death in men after lung cancer and the most common tumor in 105 countries. 1 Most prostate cancer patients have no obvious clinical manifestations, that can only be diagnosed during histological examination, and are often confined to the organs, which has little clinical significance. It is estimated that about 30% of prostate cancer patients over the age of 50 and 60–70% of prostate cancer patients over the age of 80 are asymptomatic patients, they could only be found by prostate specific antigen (PSA) test or during digital rectal exam (DRE) without any other symptoms. 2 The clinical appearance of symptomatic prostate cancer patients is also based on the severity of the disease, and its clinical manifestations are also different. It can range from frequent urination to severe bone pain caused by metastasis. The treatment of localized prostate cancer includes prostate resection, radiation and androgen deprivation therapy (ADT); For advanced prostate cancer ADT is the main treatment method. 3 However, 1.5% man only on active monitoring died from prostate cancer, which did not have difference significantly from death rate (0.9%) after surgery or after radiation (0.7%) for localized prostate cancer. 4 This research shows that surgery and radiation are failed treatments. For advanced prostate cancer, ADT shows related toxicity (decreased bone mineral density, metabolic changes, sexual dysfunction, hot flashes), which leads the decline in quality of life.5–7 So more effective treatment methods should be found.
Gene therapy never failed to fascinate scientists, clinicians and the general public owing to its treatment effects from gene level. This technology has the potential to cure diseases that are treatable but not curable with conventional medications, and to provide treatments for diseases previously classified as untreatable. 8 An effective strategy for clinical gene therapy is to transfer genes in vivo to the target cells or tissue after mitosis, or in vitro to the autologous cells, and then to the patient adoptively. There are several prospective strategies currently being used for targeting cancer using gene therapy including: (a) expressing a gene to induce apoptosis or enhance tumor sensitivity to conventional drug/radiation therapy; (b) inserting a wild type tumor suppressor gene to compensate for its loss/deregulation; (c) blocking the expression of an oncogene by using an antisense (RNA/DNA) approach; and (d) enhancing the immunogenicity of the tumor to stimulate immune cell recognition. 9 For successful gene therapy, it is essential to select the appropriate treatment gene to maximize the efficacy while minimizing the toxicity. For now, the focus is on identifying new genes that are differentially expressed in cancer cells and may modulate the transformed properties. In this respect, bioinformatics is a powerful tool for distinguishing molecular changes in cancer cells. In this case, the ability to perform a comprehensive molecular analysis of the tumor. But how to choose a suitable target gene is a big challenge. Target discovery is the most important problem in the modern drug discovery movement.10–12 Previous studys indicate that the high failure rate in drug development is largely due to poor target selection.13,14 With the development of biological databases, bioinformatics, especially the rise of data mining methods, has changed the way targets are discovered by combining biological ideas with computer tools or statistical methods to extract or filter valuable targets. Text mining and microarray data mining are two main methods of target discovery. 15 Microarray data mining refers to the application of bioinformatics in microarray data analysis to discover the entities and biological pathways that define phenotypes. In typical microarray analysis, the geneome expression profile of pathological tissue and normal tissue should be compared, and we can determine the disease a list of important target genes, or biological pathway in data mining refers to the application of bioinformatics in microarray data analysis method to find the physical and biological pathways phenotype. 16 Researchers had discovered a potential selenium targets in chemoprevention of prostate cancer using microarray data mining. 17 A large number of studies also use this method to find new targets.18–20
Besides, gene fragments are not stable in the blood, and complex cellular and tissue barriers must be overcomed to deliver new genetic information into the target cell to drive proficient expression of a therapeutic molecule. 21 There are two kinds of gene delivery vectors: viral and no-viral vectors. Though viral vectors could deliver DNA more efficiently than non-viral vectors, but it also presents a larger safety defect to patients. Compared with the viral gene delivery vectors, the non-viral vectors have a poor immunogenicity, simple preparation approaches, and may be highly adaptable ability because of their surface modification. 22 Because of its low cost, easy manufacture and modification, magnetic nanoparticles has great application potential in agriculture, medicine and environment. The applications of magnetic nanostructures have been widely demonstrated in the fields of catalysis, biotechnology, biomedicine, magnetic resonance imaging, agriculture, biosensors and environmental pollutant removal. Magnetic nanoparticles as DNA vector have got more and more attentions.23–25 Our previous study also focus on the applications of magnetic nanoparticle on cancer diagnosis and therapy. 26
Here we synthesized a DNA carrier used surface modified iron based nanoparticles, and used it to load plasmid including ShRNA which can inhibit the expression of oncogene SLC4A4 selected by bioinformatics’ method. After that we used this iron based nanoparticle/plasmid DNA nanocomposite to treat prostate cancer cells in vitro and in vivo. The target gene SLC4A4 we had selected using bioinformatics had a strong effect on the proliferation of prostate cells; Our nanocomposite could inhibit the expression of SLC4A4 effectively, it had strong inhibitory effects on prostate cancer cells both in vivo and in vitro, and can be used as a potential method for prostate cancer treatment.
Materials and methods
FeCl3·6H2O, FeSO4·4H2O, Aminopropyltriethoxysilane (APTES), o-di Nitrophenanthrene, acridine orange (AO), ethidium bromide (EB), crystal violet and ammonia were purchased from Shanghai Aladdin Biochemical Technology Co., Ltd.; BALB/C nude mice (male, 6–8 weeks old) were purchased from Beijing Viton Lihua Laboratory Animal Co., Ltd; cell cycle detection kit and apoptosis detection kit were purchased from Shanghai Biyuntian Biotechnology Co., Ltd; the SLC4A4 and β-action antibody were bought from Sigma-Aldrich Co., Ltd.
Targeted gene selection by bioinformatics
Gene expression data (GSE103512, GSE6604, profiling data) were downloaded as raw signals from Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo), interpreted, normalized and log2 scaled using the R software. Exploring of differentially expressed gene sets between normal and cancer stromal profiles in GSE103512 and GSE6604 was via the DAVID online tool (https://david.ncifcrf.gov/).
ShRNA knockdown of SLC4A4
Knockdown of SLC4A4 and a control non-target shRNA was achieved using MISSION shRNA Lentiviral Transduction Particles (SHCLNV-NM, Sigma St. Louis, MO). Total cell populations were generated using puromycin selection on five independent vectors tested for SLC4A4 with 3 giving successful knockdown. Two sequences were successful for C4-2B cells.
Cell culture and the gene silencing effects of SLC4A4 ShRNA
Human prostate cancer cell line LNCap, C4-2B, Du145, PC-3 and prostate prostatic hyperplasia cell line BPH were preserved by our laboratory and were cultured at 37 °C with 5% CO2 in a constant temperature incubator using RPMI-1640 medium (containing 10% fetal bovine serum and 1% penicillin/streptomycin) and DMEM medium (containing 10% fetal bovine serum and 1% penicillin/streptomycin), respectively. The day before transfection, cells in logarithmic growth stage were seeded into 96 and 6-well plates with a density of 1 × 103 and 5 × 105 cells per well respectively and cultured in antibiotic free medium for one day. Change fresh media without serum before cells were tansfected. Add 100 µl of serum-free medium and 1 µg plasmid into the labeled EP tube, and mix it completely. Then add 3 µl of transfection reagent into the diluted 100 µl of plasmid DNA. The transfection reagent DNA plasmid complex EP tube was placed for 15–20 minutes at room temperature. Take out the prepared cells to be transfected, and drop the transfection complex into the cell culture media evenly. Shake the culture dish gently to make the transfection complex fully contact with the cell. Cells were cultured in a 5% CO2 saturated humidity incubator at 37 °C for 24 h. After that, use fluorescence microscope to observe to the transfection efficiency; use Western blot to test the SLC4A4 expression; use MTT to test the cell proliferation; and use flow cytometry to test the cell apoptosis.
Synthesis of iron based nanoparticles
Iron oxide nanoparticles were prepared by a chemical coprecipitation method.26,27 Briefly, Weigh 0.20 g FeCl3 · 6H2O and 0.10 g FeCl2 · 4H2O in 25 mL of distilled water, and stir with nitrogen for 30 min. Place the flask in an 80° C water bath, add 2.5 mL of 25% ammonia water with a syringe under vigorous stirring, and react for 1 h. After the reaction, the temperature was lowered to room temperature, separated with a magnet, and washed with distilled water until neutral. After filtration and vacuum drying, the product was ground into a powder to obtain SPIO particles. 10 ml of (3-aminopropyl) triethoxysilane (APTES) is placed into an aqueous solution of an acid (pH = 4) that acts as a catalyst. The APTES is hydrolyzed, then a condensation reaction takes place to form a silane polymer. In the hydrolysis reaction, alkoxide groups–OC2H5 are replaced by hydroxyl groups (OH) to form reactive silanol groups, which condense with other silanol groups to produce siloxane bonds (Si–O–Si). Alcohole (C2H5OH) and water are produced as by-products. The solution of silane polymer was added to 100 ml of MNP solution in a 250 ml three necked round bottom flask. Under argon protection, the mixture was stirred for 5 h at 60 °C. After cooling at room temperature, the product was washed with deionized water and ethanol for several times respectively, and then dried in vacuum at room temperature. By the above treatment, the surface of nanoparticles was modified with amino-silane named MNP. The products were examined by TEM, Hydrodynamic particle size, Zeta potential and infrared spectroscopy.
Gel retardation assay
To examine the ability of DNA condensation of this nano-medicine, gel electrophoresis was performed on an electrophoresis cell appliance (Liu Yi, DYCP31DN, Beijing, China). Plasmid DNA was added into an appropriate amount of magnetic nano-medicine co-delivery system, and the mass ratio was ranged from 0 to 2. The complex was mixed completely by micropipettor, and then it was transferred into the holes of 1% agarose gels with ethidium bromide (0.1 mg/mL), and ran with Tris–acetate (TAE) buffer at 100 V for 40 min. After that, the gel was placed under a UV transilluminator (BIO-RAD, Gel Doc XRþ, Hercules, CA, USA) to obtain a clear image of DNA bands.
Cell uptake assay
After treating the C4-2B cells with MNP/DNA composite for 6 hours, cells were fixed with 4% paraformaldehyde and stained with Prussian blue staining kit, and then the nanoparticles in cells were observed through a microscope. 48 hours after the cells were added to MNP/DNA composite, cells were fixed for 30 min with 4% paraformaldehyde, and the intensity of green fluorescence of GFP in plasmid in cells was observed through a confocal microscope. The total protein of the cells was extracted, and Western blot experiments were performed to check the expression of SLC4A4 in cells.
The inhibition effects of MNP/DNA composite in vitro
MTT, cell cycle, crystal violet and plate clone formation assay were used to test the inhibition effects of MNP-Sh-SLC4A4 on cell proliferation. C4-2B cells were fully digested with pancreatin, then fresh media was used to suspend the cells, and were seeded in 96 (1 × 103 cells/well), 24-well (7 × 104 cells/well) and 6-well (5 × 105 cells/well) plate respectively. The MNP/DNA nanocomposite was added to the cell culture solution at a mass ratio of 2, the control group was added with an equal volume of PBS. When the cells were cultured for 5 days (24 h in 6-well plate). MTT was added to the 96-well plate of the cells to be tested at the wavelength of 490 nm. Cells in 24-well plate were fixed with methyl alcohol for 30 min, then cells were dyed with 1% gentian violet for 20 min. After washed with water, cells were dried at room temperature, and photos were were taken by a scanner. Cells in 6-well plate were digested and suspended in PBS, then PI was added to cells, FCM was used to test the cells’ cycle situation.
AO/EB staining method was used to test the cells’ death. Immerse the coverslip in 75% alcohol for 10–15 min. Take out the cover glasses submerged in alcohol with tweezers in the super clean bench, and ignite the alcohol on the cover glass with an alcohol lamp to dry it, then put them into 24-well plate. Then, cells were seeded (5 × 105 cells per well) and cultured for 24 h to allow cells to crawl. The MNP/DNA nanocomposite was added to the cell culture solution at a mass ratio of 2, the control group was added with an equal volume of PBS, and incubated with cells for 4 h. After that, change fresh media for cells, and cultured for another 48 h. Discard the media and wash the cells twice with 4 °C PBS. Add 5 µL of 0.01% AO/EB dye solution to the plate (containing 1 ml PBS solution), react at room temperature in the dark for 10–20min. Wash twice with PBS, observe and take pictures under a fluorescence microscope.
Cells’ wound healing experiment was used to test the ability of cell migration and repair capacity. Cells were seeded to 24-well plate at a density of 1 × 105 cells/well to ensure that the cells could grow over the next day in each well. Cells were cultured at 37 °C, 5% CO2 incubator for 24 h. Use a clean pipette head to make a wound line at the bottom of the plate. Then, cells were washed for 3 times with PBS to remove the suspended cells. Fresh media containing MNP/DNA nanocomposite was added to cells at a mass ratio of 2 and incubated for 12 h. After that photos were taken by a microscopy, and five distances were test by Photoshop CC software, and the gained data was analyzed by Graphpad prism 8.0.
The inhibition effects of MNP/DNA composite in vivo
BALB/C mice were used to establish a tumor model. C4-2B cells were digested and washed with PBS; next, they were suspended in serum free media, and were transplanted to mice by subcutaneous injection. While the tumor grew to 5 mm, the therapy with MNP-Sh-SLC4A4 was started twice a week for 3 weeks. Tumor volume was tested while the treatment was under going. When the therapy was finished, mice were sacrificed, and tumor were taken out and photos were taken for them.
In order to provide that the MNP-Sh-SLC4A4 composite could prevent the tumor metastasis in vivo, C4-2B cells were injected by mice tail vein, then MNP/DNA composite was injected by tail vein followed by cells’ injection. And the MNP/DNA injection was continued for another 3 weeks. When the therapy was finished, mice were sacrificed, and their lung and liver were taken out to do the HE stain.
Statistical analysis
Graphpad Prism 8.0 software (SPSS Inc, Chicago, USA) was used to analyze all the data. The Chi-square test was used to compare the differences among each group. Statistical significance was established at the 5% level (P < 0.05) for all statistical analyses.
Results and discussion
The targeted gene SLC4A4 was overexpressed in prostate cancer
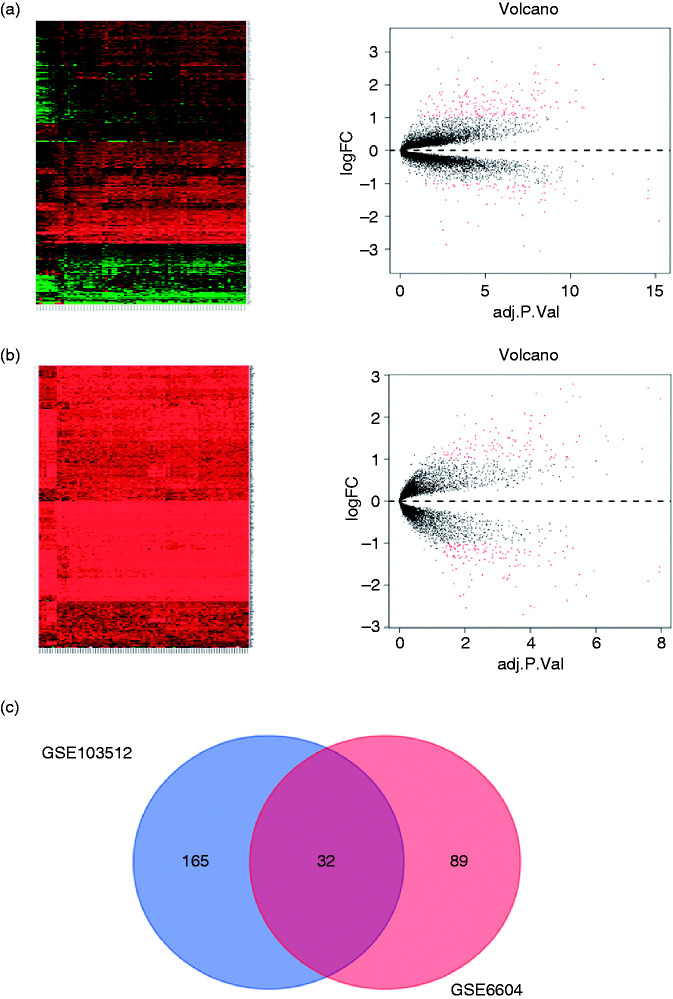
In order to screen the abnormal expressed gene in prostate cancer, we used the GEO database, and found that 165 genes were overexpressed in GSE103512 and 89 genes were overexpressed in GSE6604 (Figure 1(a) and (b)), 32 genes were overexpressed both in GSE103512 and GSE6604 (Figure 1(c)). The targeted gene SLC4A4 was chosen by PCR, after extract total mRNA from prostate cancer cells C4-2B, LNCap, Du-145 and PC-3 compared to prostatic hyperplasia cell BPH, and excluded the genes had been reported in prostate cancer. SLC4A4 is the human gene for sodium bicarbonate cotransporter protein, and it is located on chromosome 4q21 and produces two major transcripts with 5′-end variants resulting from alternative promoter usage. 28 Mutations in SLC4A4 cause severe proximal renal tubular acidosis (pRTA), with a plasma pH of 7.04–7.27 and a plasma bicarbonate concentration of 3–17 mmol/L. 29 Recently, SLC4A4 was reported to be associated with cancer, it could suppress tumor’s proliferation in clear cell renal cell carcinoma. 30 However, previous study also show that SLC4A4 is overexpressed in colon cancer, 31 and use ShRNA to knock down it could inhibit the cell proliferation and metastasis in colon and breast cancer cells. 32 Our results based bioinformatics also get a same verdict.
Figure 1.
The gene expression in two GEO datasets. (a and b) GSE103512 and GSE6604 geneome expression’s hotmap and volcano plot, and (c) their Vemn diagram.
Knockdown SLC4A4 could inhibit prostate cancer cells’ proliferation and promote their apoptosis
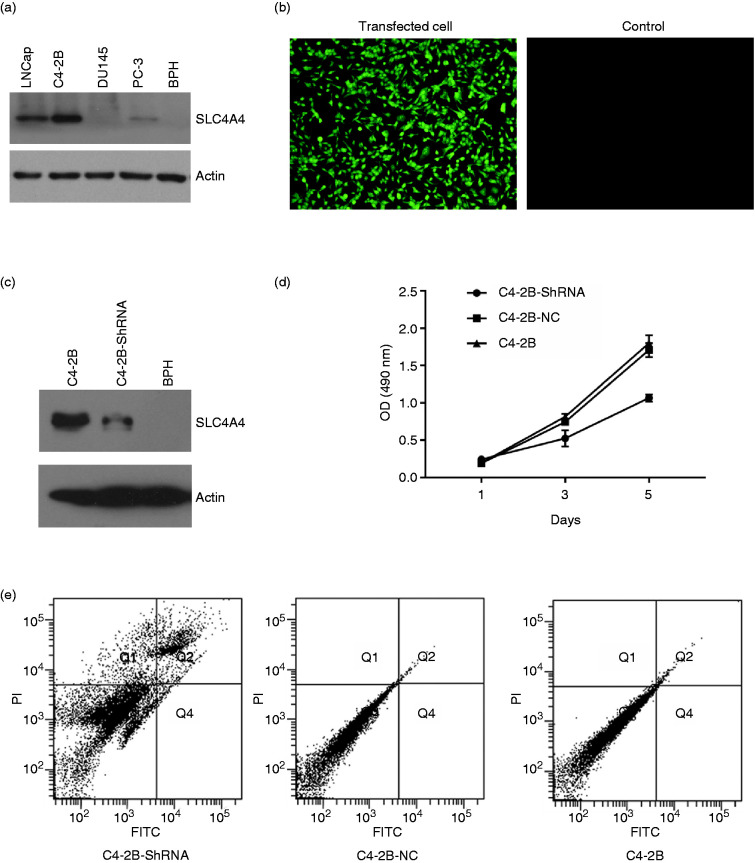
In order to examine the expression of SLC4A4, we firstly extracted the total protein and used Western blot to test it. As it shows in Figure 2(a), SLC4A4 is over expressed in LNCap and C4-2B cell lines, especially in C4-2B cell line. Then we synthesized SLC4A4 ShRNA lentivirus plasmid which containing GFP gene that can express GFP protein and can observed by fluorescence microscopy, and transfected it into C4-2B cells. ShRNA is the abbreviation of Short Hairpin RNA, it could be transfected as plasmid vectors or lentivirus encoding shRNA transcribed by RNA pol III or modified pol II promoters, and integrated to DNA in cells that makes its continuious expression. 33 24 hours after plasmid was transfected, GFP was expressed in C4-2B cells observed by its fluorescence which could prove that the plasmid DNA was sussessfully transfected (Figure 2(b)). Then total protein was extracted and Western blot was used to examine SLC4A4 gene’s expression, the prostatic hyperplasia cell BPH was set as a control. As it shows in Figure 2(c), SLC4A4 gene’s expression is knocked down by its ShRNA obviously. MTT was used to test the effects of MNP/DNA on cell proliferation. As it shows in Figure 2(d), C4-2B cell’s proliferation is depressed obviously after treated by SLC4A4 ShRNA compared to NC plasmid and control. The similar results is shown in Figure 2(e), the cell apoptosis rate of C4-2B increased to 28.6% after treated by SLC4A4 ShRNA, compared to plasmid NC (8.9%) and C4-2B control (9.1%). These results all proved that SLC4A4 is an oncogene in prostate cancer, and play an important role in prostate cancer cells.
Figure 2.
The expression of SLC4A4 and its role in prostate cancer cell lines. (a) The expression of SLC4A4 in LNCap, C4-2B, Du145, PC-3 and prostate prostatic hyperplasia cell line BPH. (b) The photo taken by fluorescence microscopy of C4-2B cells after SLC4A4-ShRNA plasmid was transfected 24 hours later. (c) The expression of SLC4A4 after SLC4A4-ShRNA plasmid was transfected 24 hours later tested by Western blot. D Cell apoptosis of C4-2B cells after SLC4A4-ShRNA plasmid was transfected 24 hours later.
The physicochemical property of MNP
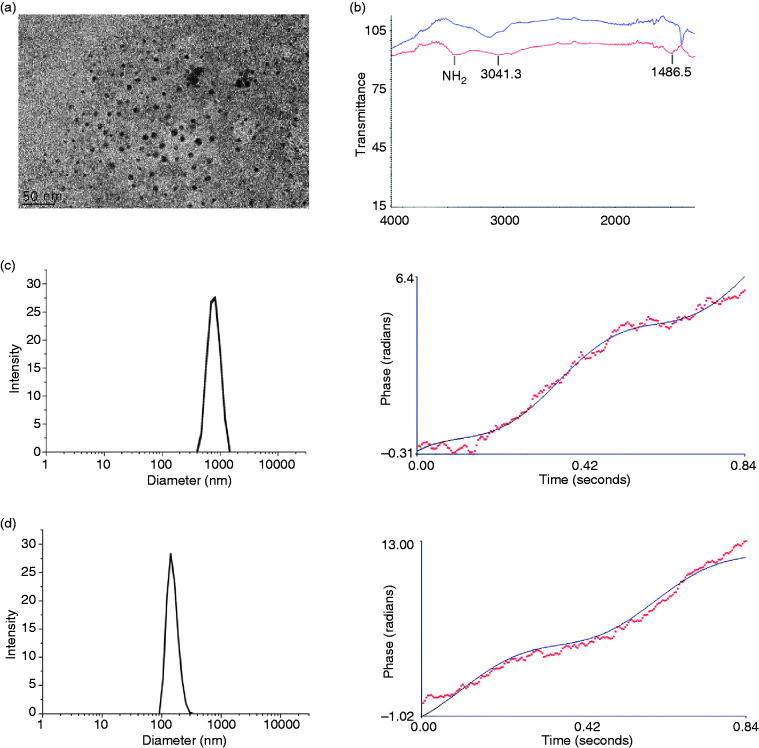
The MNP nanoparticle was synthesized by APTES modified iron oxide nanoparticles. TEM was taken to observe the morphology, as shown in Figure 3(a), the dispersity of our MNP is excellent, and its core diameter is 12.15 ± 2.58 nm. To prove the structure is correct, the infrared spectroscopy was taken. As shown in Figure 3(b), after modified with APTES, there is an amine peak appears at 3500 cm−1 (red) compared with iron oxide nanoparticles (blue). The size and Zeta potential were also test by a dynamic light scattering (DLS). The hydrodynamic diameter is 1350 ± 78 nm of iron oxide nanoparticles, and its Zeta potential is –0.75 ± 0.41 mV (Figure 3(b)). While the hydrodynamic diameter and Zeta potential after modified with APTES is 129.6 ± 2.42 nm and 22.38 ± 1.43 mV, just like the result in Figure 3(c). For gene vectors, their charge should be positive, for the charge of DNA is opposite. Although cationic polymers are identified not safe enough, 34 but their charge will change into neutral after bonding DNA. And because its highly delivery efficiency and protection effects for DNA, flexible properties, facile synthesis, robustness, cationic polymers have become the intense research as non-viral gene delivery systems.35–37 Besides, the MNP also be widely used for gene delivery for its superparamagnetism, low toxicity, biocompatibility, nonimmunogenicity, and high effective surface. 38 Besides, our nanoparticles’ diameter is appropriate. As we know, the pore size of the glomerular basement membrane (GBM) of kidney is approximately 6–10 nm, so the size of nanoparticles should be no less than 10 nm, and nanoparticles should be no more than several hundred nanometers in order to enter and accumulate in tumor’s interstitium through leaky vasculature. 39
Figure 3.
The physicochemical property of MNP. (a) The TEM photo. (b) The infrared spectroscopy of iron oxide (blue) and APTES modified iron oxide nanoparticles (red). (c and d) hydrodynamic diameter and Zeta potential of iron oxide and APTES modified iron oxide nanoparticles respectively.
The cell uptake of MNP
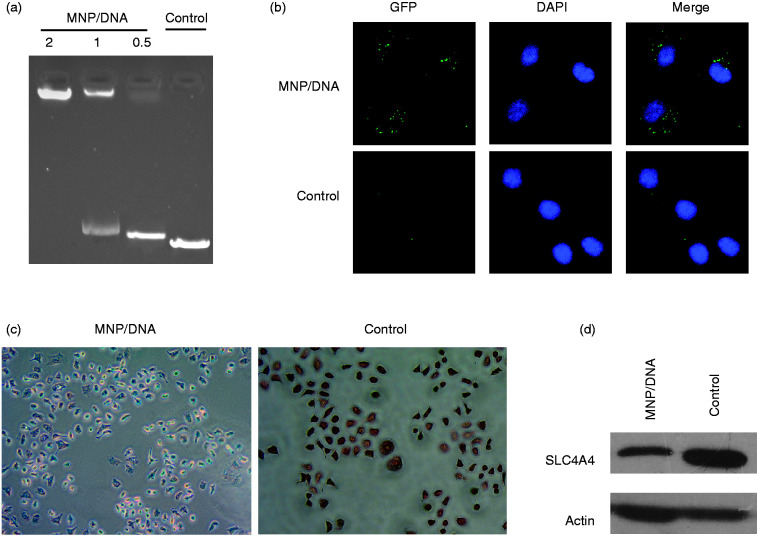
To prove the DNA bonding capacity, the DNA retarding experiment was take. As we see in Figure 4(a), the DNA band is completely retarded while the MNP/DNA mass ratio is 2, and the retarded bond decreases while the the MNP/DNA mass ratio decreases. The control DNA bond is not retarded at all. We use the MNP/DNA nanocomposite to treat C4-2B cells, and a microscope photo was taken. As it shows in Figure 4(b), we could see the GFP fluorescence in cells after treated for 48 hours, while we can’t find any fluorescence in the control group cells (Figure 4(b)). The same thing happens in Prussion blue staining. The iron particles are dyed to blue in cells after treated for 6 hours, while the colour of cells in control group is completely red (Figure 4(c)). The SLC4A4 expression in C4-2B cells is obviously suppressed after treated for 48 hours as well compared with control group (Figure 4(d)). These results prove that our synthesized MNP could bond plasmid DNA and delivery it to tumor cells, after that the plasmid DNA could be released that would play a role in cells’ activity.
Figure 4.
The cell up take of MNP/DNA. (a) The DNA banding capacity of MNP. (b) The confocal microscopy photoes of C4-2B cells after treated with MNP/DNA for 48 h. (c) The Prussion blue staining of C4-2B cells after treated with MNP/DNA for 6 h. (d) The SLC4A4 expression of C4-2B cells after treated with MNP/DNA for 48 h.
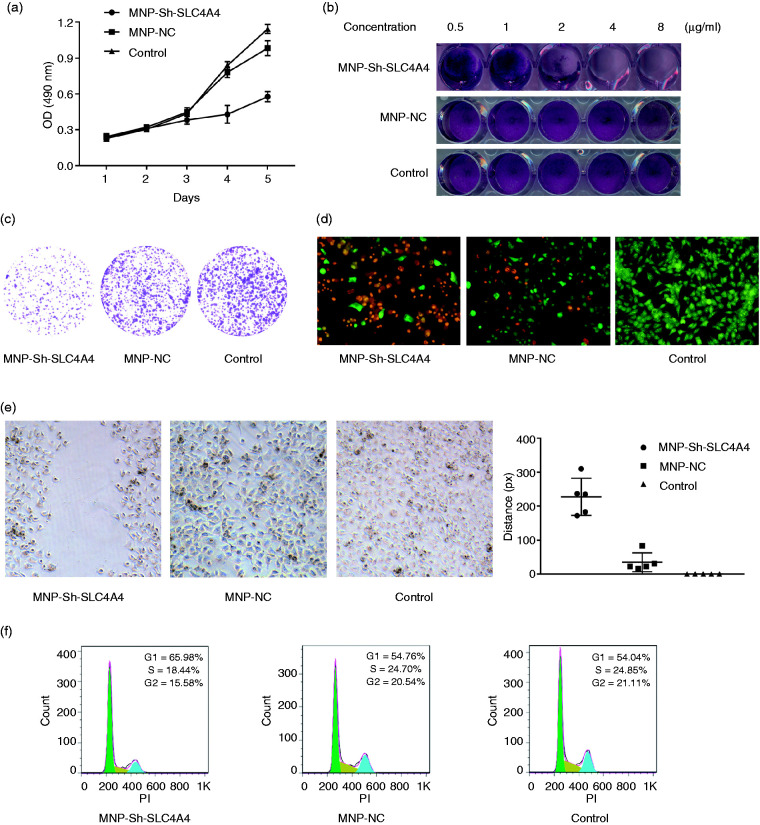
The MNP-Sh-SLC4A4 nanocomposite could inhibit prostate cancer in vitro
To test the inhibiton capacity of MNP/DNA nanocomposite on prostate cancer in vitro. The MTT, cell cycle, crystal violet, plate clone formation, AO/EB staining and cells’ wound healing experiments were taken. As it shows in Figure 5, the proliferation’s inhibition is proved by MTT and crystal violet experiments (Figure 5(a) and (b)). The plate clone formation assay is used to test the affection of MNP/DNA nanocomposite on C4-2B cell line’s clone forming ability. Just like the results in Figure 5(c), the number of clone of C4-2B decreases obviously after treated with MNP/DNA nanocomposite (MNP-SLC4A4-ShRNA) compared to MNP-NC and control group. Cells’ wound healing experiment is used to test the affection of MNP/DNA nanocomposite on C4-2B cell line’s metastasis. As it shows in Figure 5(e), compared to MNP-NC and control group, cells’ wound nearly doesn’t change at all in MNP-Sh-SLC4A4 group. While, the wounds nearly appear in the former two groups. In order to prove the prostate cancer cells were really inhibited, the cell cycle and the dead cells’ staining were taken. From Figure 5(f), we could see that cells’ proliferation is inhibited indeed, the proportion of G2/S cells in decreases obviously in MNP-Sh-SLC4A4 group compared with MNP-NC and control group. The dead cells’ staining was tested by AO/EB staining method. AO and EB both are dyes that can bond to DNA in cells, while AO can penetrate alive cells’ membrane, and EB can only penetrate destroyed cells’ membrane that stands for the cells’ apoptosis and death. The results are showed clearly in Figure 5(d), there are numbers of dead cells (red) in in MNP-Sh-SLC4A4 group, while cells in other two groups are nearly all alive (green).
Figure 5.
The inhibition effects of MNP-Sh-SLC4A4 on prostate cancer cells in vitro. The proliferation inhibition ((a) MTT assay and (b) crystal violet assay). (c) The clone forming ability assay. (d) AO/EB staining assay, the dead cells are stained by EB (red), while the alive cells are stained by AO (green). (e) Cells’ wound healing capacity assay. (f) The cells’ cycle assay. For interpretation of the references to colours in this figure legend, refer to the online version of this article.
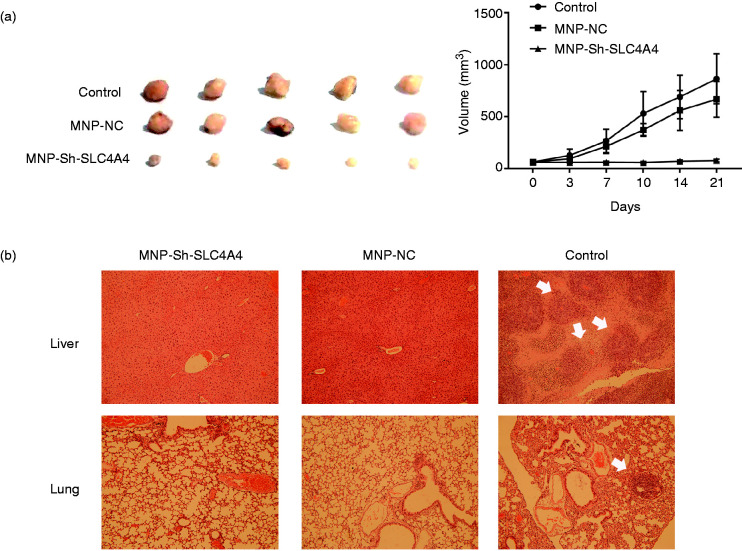
The MNP-Sh-SLC4A4 nanocomposite could inhibit prostate cancer in vivo
In order to provide that the MNP/DNA composite could inhibit the tumor growth in vivo, the animal experiments were taken. As we see in Figure 6(a), tumors’ growth is suppressed obviously after 3 weeks’ treatment by MNP-Sh-SLC4A4 compared to the other two groups, and MNP-NC could also inhibit the tumor growth in a way compared to control group. The tumor growth curve also shows this trend during the therapy process. Interestingly, we found that both the two MNP/DNA composites, MNP-Sh-SLC4A4 and MNP-NC, could prevent the tumor metastasis in vivo. The result is shown in Figure 6(b). The highly malignant prostate cancer cell C4-2B was injected into mice cyculation blood with MNP/DNA composites at the same time, and the MNP/DNA composites injection was continued for another 3 weeks. After treatment we could see that both liver and lung in the control group appear the tumor existence (white arrow). However, the other two groups can not find any. The reason of that maybe because of immunology. Previous studies had used different modified Fe3O4 magnetic nanoparticles for cancer therapy in mice. They found that the MNP could inhibit tumour growth though inducing pro-inflammatory macrophage polarization from M2 to M1 in tumour tissues.40,41 That’s another benefit from this iron based nanoparticle.
Figure 6.
The tumor inhibition in vivo. (a) The tumor volume and growth curve. (b) The HE staining of liver and lung in the tumor metastasis experiment (white arrow shows the tumor foci).
Conclusion
The target gene SLC4A4 we selected by bioinformatics method plays an important role in prostate cancer, it is a potential oncogene in prostate cancer. Based on this, we design a ShRNA to knock it down, the results show that the proliferation of prostate cancer cell line C4-2B is obviously inhibited. Next, we synthesized a magnetic nanoparticle MNP-APTES to delivery the SLC4A4’s ShRNA plasmid to cancer cells. The results show that this MNP/DNA composite could delivery and release plasmid DNA to cancer cells as predicted. And we did a lot of experiments in vitro to prove that the MNP/DNA composite MNP-Sh-SLC4A4 could inhibit the cells’ proliferation, clone forming, metastasis, induce cell death and cell cycle arrest. Moreover, in order to prove that the MNP-Sh-SLC4A4 could inhibit tumor in vivo, we transplanted C4-2B cells to nude mice. After 3 weeks’ treatment, the tumor growth was greatly suppressed. In order to observe the metastasis of prostate cancer cells in vivo, we injected C4-2B cells into mice through tail vein, and the MNP/DNA composite was injected followed by that. Interestingly, we found that both the MNP-Sh-SLC4A4 and MNP-NC could inhibit the metastasis to liver and lung of prostate cancer cells in vivo.
In conclusion, the target gene SLC4A4 we selected by bioinformatics is a potential oncogene in prostate cancer; the nanoparticle MNP-APTES could delivery ShRNA plasmid into cancer cells both in vitro and in vivo. It’s a good method to find a target gene by bioinformatics and use nanotechnology for the gene delivering on cancer therapy.
Acknowledgements
We greatly acknowledge Dr. Xinan Jiang from Department of Urology, The First Affiliated Hospital of Guiyang Medical University for the helping of finding lab to finish this job.
Footnotes
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by funds from the National Natural Science Foundation of China (grant nos 81802543); Guangdong Natural Science Foundation (grant nos 2018A0303130184); and Science and Technology Programme of Guangzhou Municipal Government (grant nos 201707010304, S2013040016969 and 201604010257).
ORCID iD: Hailiang Li https://orcid.org/0000-0001-7759-6699
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018; 68: 394–424. [DOI] [PubMed] [Google Scholar]
- 2.Pentyala S, Lee J, Hsieh K, et al. Prostate cancer: a comprehensive review. Med Oncol 2000; 17: 85–105. [DOI] [PubMed] [Google Scholar]
- 3.Litwin MS, Tan HJ. The diagnosis and treatment of prostate cancer: a review. JAMA 2017; 317: 2532–2542. [DOI] [PubMed] [Google Scholar]
- 4.Hamdy FC, Donovan JL, Lane JA, et al. 10-Year outcomes after monitoring, surgery, or radiotherapy for localized prostate cancer. N Engl J Med 2016; 375: 1415–1424. [DOI] [PubMed] [Google Scholar]
- 5.Wu FJ, Li IH, Chien WC, et al. Androgen deprivation therapy and the risk of iron-deficiency anaemia among patients with prostate cancer: a population-based cohort study. BMJ Open 2020; 10: e34202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scailteux LM, Balusson F, Oger E, et al. Androgen deprivation therapy prescription, blood and bone-density testing in a French population-based study exploring adherence to the French prostate cancer guidelines. Minerva Urol Nefrol. Epub ahead of print 16 April 2020. DOI: 10.23736/S0393-2249.20.03683-8. [DOI] [PubMed]
- 7.Magee DE, Singal RK. Androgen deprivation therapy: indications, methods of utilization, side effects and their management. Can J Urol 2020; 27: 11–16. [PubMed] [Google Scholar]
- 8.Kumar SR, Markusic DM, Biswas M, et al. Clinical development of gene therapy: results and lessons from recent successes. Mol Ther Methods Clin Dev 2016; 3: 16034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Das SK, Menezes ME, Bhatia S, et al. Gene therapies for cancer: strategies, challenges and successes. J Cell Physiol 2015; 230: 259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hicks MA, Hou C, Iranmehr A, et al. Target discovery using biobanks and human genetics. Drug Discov Today 2020; 25: 438–445. [DOI] [PubMed] [Google Scholar]
- 11.Alonso AM, Corvi MM, Diambra L. Gene target discovery with network analysis in Toxoplasma gondii. Sci Rep 2019; 9: 646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldman RC. Target discovery for new antitubercular drugs using a large dataset of growth inhibitors from PubChem. Infect Disord Drug Targets 2020; 20(3): 352–366. [DOI] [PubMed]
- 13.Sams-Dodd F. Target-based drug discovery: is something wrong? Drug Discov Today 2005; 10: 139–147. [DOI] [PubMed] [Google Scholar]
- 14.Butcher SP. Target discovery and validation in the post-genomic era. Neurochem Res 2003; 28: 367–371. [DOI] [PubMed] [Google Scholar]
- 15.Sakharkar MK, Sakharkar KR. Targetability of human disease genes. Curr Drug Discov Technol 2007; 4: 48–58. [DOI] [PubMed] [Google Scholar]
- 16.Yang Y, Adelstein SJ, Kassis AI. Target discovery from data mining approaches. Drug Discov Today 2012; 17Suppl: S16–S23. [DOI] [PubMed] [Google Scholar]
- 17.Zhang H, Dong Y, Zhao H, et al. Microarray data mining for potential selenium targets in chemoprevention of prostate cancer. Cancer Genomics Proteomics 2005; 2: 97–114. [PMC free article] [PubMed] [Google Scholar]
- 18.Hua S, Lei L, Deng L, et al. miR-139-5p inhibits aerobic glycolysis, cell proliferation, migration, and invasion in hepatocellular carcinoma via a reciprocal regulatory interaction with ETS1. Oncogene 2018; 37: 1624–1636. [DOI] [PubMed] [Google Scholar]
- 19.Birgani MT, Hajjari M, Shahrisa A, et al. Long non-coding RNA SNHG6 as a potential biomarker for hepatocellular carcinoma. Pathol Oncol Res 2018; 24(2): 329–337. [DOI] [PubMed]
- 20.Zhu X, Tian X, Yu C, et al. A long non-coding RNA signature to improve prognosis prediction of gastric cancer. Mol Cancer 2016; 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naldini L. Gene therapy returns to centre stage. Nature 2015; 526: 351–360. [DOI] [PubMed] [Google Scholar]
- 22.Majidi S, Zeinali SF, Samiei M, et al. Magnetic nanoparticles: applications in gene delivery and gene therapy. Artif Cells Nanomed Biotechnol 2016; 44: 1186–1193. [DOI] [PubMed] [Google Scholar]
- 23.Luo X, Peng X, Hou J, et al. Folic acid-functionalized polyethylenimine superparamagnetic iron oxide nanoparticles as theranostic agents for magnetic resonance imaging and PD-L1 siRNA delivery for gastric cancer. Int J Nanomed 2017; 12: 5331–5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Assa F, Jafarizadeh-Malmiri H, Ajamein H, et al. Chitosan magnetic nanoparticles for drug delivery systems. Crit Rev Biotechnol 2017; 37: 492–509. [DOI] [PubMed] [Google Scholar]
- 25.Zhao X, Meng Z, Wang Y, et al. Pollen magnetofection for genetic modification with magnetic nanoparticles as gene carriers. Nat Plants 2017; 3: 956–964. [DOI] [PubMed] [Google Scholar]
- 26.Li H, Fu C, Miao X, et al. Multifunctional magnetic co-delivery system coated with polymer mPEG-PLL-FA for nasopharyngeal cancer targeted therapy and MR imaging. J Biomater Appl 2017; 31: 1169–1181. [DOI] [PubMed] [Google Scholar]
- 27.Feng B, Hong RY, Wang LS, et al. Synthesis of Fe3O4/APTES/PEG diacid functionalized magnetic nanoparticles for MR imaging. Colloids Surf A Physicochem Eng Aspects 2008; 328: 52–59. [Google Scholar]
- 28.Demirci FY, Chang MH, Mah TS, et al. Proximal renal tubular acidosis and ocular pathology: a novel missense mutation in the gene (SLC4A4) for sodium bicarbonate cotransporter protein (NBCe1). Mol Vis 2006; 12: 324–330. [PubMed] [Google Scholar]
- 29.Horita S, Simsek E, Simsek T, et al. SLC4A4 compound heterozygous mutations in exon-intron boundary regions presenting with severe proximal renal tubular acidosis and extrarenal symptoms coexisting with Turner's syndrome: a case report. BMC Med Genet 2018; 19: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao W, Wang X, Wang T, et al. MiR-223-3p promotes cell proliferation and metastasis by downregulating SLC4A4 in clear cell renal cell carcinoma. Aging (Albany NY ) 2019; 11: 615–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao ZW, Fan XX, Yang LL, et al. The identification of a common different gene expression signature in patients with colorectal cancer. Math Biosci Eng 2019; 16: 2942–2958. [DOI] [PubMed] [Google Scholar]
- 32.Parks SK, Pouyssegur J. The naþ/HCO3 co-transporter physiology SLC4A4 plays a role in growth and migration of colon and breast cancer cells. J Cell Physiol 2015; 230: 1954–1963. [DOI] [PubMed] [Google Scholar]
- 33.Moore CB, Guthrie EH, Huang MT, et al. Short hairpin RNA (shRNA): design, delivery, and assessment of gene knockdown. Methods Mol Biol 2010; 629: 141–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lv H, Zhang S, Wang B, et al. Toxicity of cationic lipids and cationic polymers in gene delivery. J Control Release 2006; 114: 100–109. [DOI] [PubMed] [Google Scholar]
- 35.Samal SK, Dash M, Van Vlierberghe S, et al. Cationic polymers and their therapeutic potential. Chem Soc Rev 2012; 41: 7147–7194. [DOI] [PubMed] [Google Scholar]
- 36.Wang H, Ding S, Zhang Z, et al. Cationic micelle: a promising nanocarrier for gene delivery with high transfection efficiency. J Gene Med 2019; 21: e3101. [DOI] [PubMed] [Google Scholar]
- 37.Xiao YP, Zhang J, Liu YH, et al. Low molecular weight PEI-based fluorinated polymers for efficient gene delivery. Eur J Med Chem 2019; 162: 602–611. [DOI] [PubMed] [Google Scholar]
- 38.Hw Y MH, Hl L, et al. Potential of magnetic nanoparticles for targeted drug delivery. Nanotechnology, Science and Applications 2012; 5: 73–86. [DOI] [PMC free article] [PubMed]
- 39.Marchetti C, Palaia I, Giorgini M, et al. Targeted drug delivery via folate receptors in recurrent ovarian cancer: a review. Onco Targets Ther 2014; 7: 1223–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zanganeh S, Hutter G, Spitler R, et al. Iron oxide nanoparticles inhibit tumour growth by inducing pro-inflammatory macrophage polarization in tumour tissues. Nat Nanotechnol 2016; 11: 986–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li K, Lu L, Xue C, et al. Polarization of tumor-associated macrophage phenotype via porous hollow iron nanoparticles for tumor immunotherapy in vivo. Nanoscale 2020; 12: 130–144. [DOI] [PubMed] [Google Scholar]