Abstract

Minicells are nanosized membrane vesicles produced by bacteria. Minicells are chromosome-free but contain cellular biosynthetic and metabolic machinery, and they are robust due to the protection provided by the bacterial cell envelope, which makes them potentially highly attractive in biomedical applications. However, the applicability of minicells and other nanoparticle-based delivery systems is limited by their inefficient accumulation at the target. Here we engineered the minicell-producing Escherichia coli strain to overexpress flagellar genes, which enables the generation of motile minicells. We subsequently performed an experimental and theoretical analysis of the minicell motility and their responses to gradients of chemoeffectors. Despite important differences between the motility of minicells and normal bacterial cells, minicells were able to bias their movement in chemical gradients and to accumulate toward the sources of chemoattractants. Such motile and chemotactic minicells may thus be applicable for an active effector delivery and specific targeting of tissues and cells according to their metabolic profiles.
Keywords: chemotaxis, motility, minicell, nanoparticle, drug delivery
Introduction
Nanoparticles are highly promising as containers for targeted drug delivery in such biomedical applications as tumor therapy, and a large spectrum of different nanoparticle designs has been developed over the recent years.1−3 Nevertheless, the efficiency of drug delivery by nanoparticles remained relatively low,3 with a particular challenge being to enrich nanoparticles within the targeted tissues. One type of nanosized delivery vehicles is bacterial minicells, ∼0.5 μm spheres surrounded by a cell envelope, which are spontaneously generated through an aberrant division of bacteria close to cell poles. The production of minicells is particularly frequent in bacterial min mutants that have lost control of the cell-division site placement.4,5 Minicells carry no chromosomes and are therefore nonliving, but they can contain plasmid DNA and other cellular components, including metabolic enzymes and cellular machineries that are required for energy generation and for transcription and translation.5,6 Hence, minicells are metabolically and biosynthetically active, meaning that—similar to the intact bacteria—they can be utilized as specific biosensors7 and engineered to express a wide range of toxins, cytokines, tumor antigens, and apoptosis-inducing factors under the control of specific external stimuli.8 Because of the protection provided by the bacterial cell wall and membranes, minicells are highly robust and do not spontaneously release their content. Their small size enables minicells to penetrate fenestrated blood vessels and to accumulate at tumor sites, where they can be subsequently endocytosed and release their content within the target cells.9 Minicells were engineered to target cancer cells via bispecific antibodies and equipped with various payloads including chemotherapeutic drugs or inhibitory RNAs9−14 as well as with a secretion system for antigen injection into the host cells.15
Intact bacteria can also be utilized as drug delivery vehicles, with the bacterial ability to swim in liquid media providing a particular advantage for efficient delivery. The bacterial swimming motion is typically mediated by the rotation of several flagellar filaments that bundle together to propel the cell,16 and it can be biased in chemical gradients by the chemotaxis signaling pathway. This pathway perceives temporal changes in chemical stimulation as cells swim in the gradient, and it signals to flagellar motors to modulate the frequency of cell reorientation, thereby increasing the duration of cell runs in a favorable direction.17,18 This mechanism of gradient sensing by temporal comparisons of ligand concentration along the swimming path is necessary because of the small size of bacteria, and it is physically limited by a gradual reorientation of the cell body due to Brownian diffusion or—at high cell densities—to the emergent collective motion.19−21 Because of this importance of rotational diffusion, the processivity of swimming and thus efficiency of chemotaxis can increase with bacterial cell length.22,23
Bacterial chemosensory systems were reported to perceive chemical signals released by the host epithelium24−26 and in tumor microenvironments,27,28 indicating that chemotaxis could be applicable for specific tumor targeting. Furthermore, flagellar motility can promote an attachment to epithelial cells29 and tissue penetration.30,31 Motile bacteria can also be specifically loaded with cargo nanoparticles carrying customized therapeutics,32 and such bacteriabots are chemotactic as long as the cargo does not strongly reduce their swimming speed.22 However, despite these potential advantages for autonomous active delivery, the in vivo application of intact bacteria remains severely limited by biosafety concerns.
Here we report a system that combines advantages provided by small and chromosome-less minicells with the chemotactic capability of motile bacteria. We engineered an Escherichia coli strain that produces minicells with an inducible expression of the flagellar system and investigated their motility and chemotaxis. We demonstrate that, at higher levels of flagellar gene expression, these minicells are well-motile, despite their small size and hence small number of flagella and faster rotational diffusion. Moreover, although minicells were previously shown to contain functional chemosensory complexes,33,34 it was unclear whether their swimming could be fast and processive enough to enable a proper functioning of the bacterial chemotaxis strategy. We show that, despite these potential limitations, the chemotactic efficiency of minicells is comparable to that of regular bacteria, and we develop an analytical model of the minicell motility that can largely account for our experimental observations. This proof-of-concept implementation of motility and chemotaxis in minicells makes it possible to further increase the efficiency of minicell-based drug delivery as well as its specific targeting relying on chemical gradients emanating from particular microenvironments such as tumors.
Results and Discussion
Engineering E. coli for an Inducible Production of Flagellated Minicells
To generate minicells, we used a derivative of the E. coli strain MG1655 that carries a deletion of the minCDE operon encoding the division-site positioning system. This deletion results in frequent cell divisions at cell poles, pinching off multiple minicells.5 We further introduced an A115V amino acid replacement in the actin-like protein mreB, which decreases the E. coli width and therefore leads to the production of minicells with even smaller diameter.34 Whereas cells of the wild-type strain MG1655 are ∼2.3 μm long and ∼1 μm wide when grown in tryptone broth (TB) (Figure S1A,B), the minCDE mreBA115V strain is elongated and produces spherical minicells of 440 ± 49 nm diameter (Figure 1A–-E). We observed that our minCDE mreBA115V strain also acquired a spontaneous (apparently adaptative) deletion that inactivated the operon encoding flhDC, the upstream master regulator of the flagellar regulatory network (Figure S2A), thus effectively shutting down the expression of all flagellar genes. Consequently, both minicell-producing mother cells (Figure 1A) and minicells (Figure 1B) were not flagellated. In order to tune the levels of flagellar and chemotaxis proteins, we engineered this strain to express the flhDC operon from a plasmid under an arabinose-inducible promoter. The induction of flhDC expression in minicell-producing mother cells indeed led to the increased activity of the flagellin (fliC) promoter (Figure S2B,C) that is representative for the expression of flagellar and chemotaxis genes (Figure S2A).35,36 Consistently, the activation of flagellar gene expression led to the appearance of flagellar filaments in both mother cells (Figure 1C and Figure S3A,B) and in minicells (Figure 1D,F,G and Figure S3C,D). On average, flagellar filaments in minicells were ∼7.5 μm in length (Figure 1G and Figure S4A), similar to the length of flagellar filaments in the parental MG1655 cells (Figure S1C). The filament length remained constant over the whole range of the flhDC expression levels, above the initial activation threshold (Figure 1G and Figure S4A), whereas the number of flagellar motors increased with induction up to a maximum of approximately two motors per minicell (Figure 1F and Figure S4B).
Figure 1.
Flagellation of minicells dependent on the induction of flhDC expression. (A–D) Negative staining electron microscopy images of E. coli minicell-producing strain (A, C) and of purified minicells (B, D) for cultures of the minicell-producing strain without the flhDC expression construct (A, B) or upon induction of the flhDC expression with 0.01% arabinose (C, D). Scale bars are 1 μm. (E) Distribution of the minicell size measured in the electron microscopy images as in (D). (F–G) Dependence of flagellar number (F) and flagellar length (G) in minicells, measured in the electron microscopy images, on PfliC promoter activity in the minicell-producing culture. Colors indicate different levels of flhDC induction (Figure S2B,C). Solid lines are hyperbolic fits to the data for visualization. Error bars indicate the standard deviation for 10–25 flagella measured at each condition.
Motility and Chemotaxis of Flagellated Minicells
To investigate whether flagellated minicells generate enough energy to power the rotation of flagellar motors, and whether a rotation of one to two flagellar filaments produces a sufficient force to processively propel the minicell, we next compared the motion of nonflagellated and flagellated minicells. Consistent with them lacking an active propulsion system, trajectories of nonflagellated minicells were clearly Brownian (Figure 2A). In contrast, trajectories of flagellated minicells showed significantly persistent swimming (Figure 2B and Figure S5), comparably to the trajectories of the parental MG1655 cells (Figure S5D). Cell tracking confirmed that, as expected for diffusive behavior, the mean squared displacement (MSD) increases linearly as a function of time for nonflagellated minicells (Figure 2D) and that the distribution of their displacements is Gaussian (Figure S5A). For flagellated minicells and MG1655 cells, the MSD grows quadratically with time (Figure 2D), which is characteristic for a ballistic motion. Distributions of displacements were also similar for flagellated minicells and MG1655 cells (Figure S5A), with minor differences being likely explained by a slightly higher fraction of minicells that were nonmotile, ∼10–30% (independent of flhDC induction) compared to 5–10% for MG1655, as determined by microscopy analysis (see Materials and Methods). This subpopulation of nonmotile minicells might arise from cell or flagella damage during the culture preparation or from the residual heterogeneity of the flhDC induction (Figure S2C). The average velocity of swimming minicells saturated below 15 μm/s (Figure 2C), which is significantly lower than the velocity of MG1655 cells (24 μm/s). In contrast, the duration of runs was longer for the swimming minicells (Figure 2E). Finally, consistent with their small size, swimming minicells were reoriented more rapidly due to the rotational diffusion, being thus less able to maintain their swimming direction than normal E. coli cells (Figure 2F).
Figure 2.
Motility of flagellated minicells. (A, B) Trajectories of minicells without flagellar filaments (A) strain without the flhDC expression construct) or with flagellar filaments upon induction of flhDC expression with 0.01% arabinose (B). (C) Dependence of the average velocity of minicells on flagellar gene expression. Error bars indicate the standard deviation for three independent measurements. (D–F) Characterization of the cell motion for minicells carrying either an empty vector or the flhDC expression plasmid induced with indicated concentrations of arabinose. Data for MG1655 cells, measured previously,47 are shown for comparison. (D) The mean squared displacement MSD(t) as a function of the lag time t, shown on a logarithmic scale. Black bars indicate the power laws MSD(t) ∝ t (corresponding to diffusive behavior) and MSD(t) ∝ t2 (corresponding to ballistic motion) as labeled. (E) Mean durations of the minicell runs. Statistical significance was evaluated using an unpaired Student t-test. (F) The time autocorrelation function of the direction of swimming ⟨ui(t)ui(0)⟩ as a function of the lag time t. Multitime scale decay, expected given the various processes contributing to minicell reorientation, was fitted as a stretched exponential (solid line) exp(−(λt)β), yielding the typical decay time λ. The stretching exponent is β = 0.7 for the minicells, and β = 1 for normal MG1655 cells. (D–F) Error bars are the standard error of the mean on four (motile cells) or two (empty plasmid) independent data sets.
To further test whether, despite
these differences in their swimming
behavior, motile minicells are capable of performing chemotaxis, we
probed the motility of minicells in gradients of α-methyl-d,l-aspartate (MeAsp), a nonmetabolizable analogue
of aspartate and potent chemoattractant for E. coli. We first used a previously described microfluidic device (Figure 3A inset) that allows
measurements of the chemotactic drift of a bacterial population in
a steady linear chemical gradient.35,37 A significant
population drift up the MeAsp gradient could be observed, with the
drift velocity of the minicell population growing with flagellar gene
expression (Figure 3A). This drift velocity apparently increased linearly as a function
of the average number of flagellar motors of the minicells (Figure 3B). Thus, although
a single flagellum might already be sufficient to propel a minicell
at nearly maximum speed, the efficiency of the chemotaxis is apparently
higher for minicells that are propelled by two flagella. This higher
efficiency might stem from physical effects of the number of flagella
on the tumbling rate and processivity of swimming and/or from an increased
expression of chemotaxis proteins at higher levels of flhDC induction (Figure S2A and Supporting Information). Notably, even at the
highest induction of flagellar genes the chemotactic drift of the
minicells (
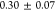 μm/s)
remained lower than the one
of the parent MG1655 cells (
μm/s)
remained lower than the one
of the parent MG1655 cells (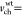
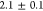 μm/s).
μm/s).
Figure 3.

Chemotaxis of minicells. (A, B) Drift velocity of minicells in a linear gradient of MeAsp (0–100 μM) formed in the microfluidic device shown in inset, as a function of the flagellar gene expression (A) and the flagellar number (B). Error bars for the drift velocity values indicate standard deviation for three independent measurements. Values for the flagellar numbers are taken from Figure 1F. (C, D) Schematic representation of the microfluidic chip used for the minicell chemotaxis-mediated accumulation analysis (C) and microscopic image (top view) of the observation channel used to determine chemotactic accumulation (D). The observation channel connects a well containing the chemoattractant MeAsp (2 mM) (right side; source) with another well containing the minicell suspension (left side; sink). See Figure S6 for the exact design and dimensions of the microfluidic chip. (E–H) Accumulation of sfGFP-expressing minicells in the observation channel in the presence or absence of MeAsp. Numbers of minicells under indicated conditions (E) and representative images showing the accumulation of motile minicells in the presence (F) or absence (G) of MeAsp in the source chamber, as well as nonflagellated minicells (H) in a chamber with a MeAsp gradient as control. Error bars in (E) indicate the standard deviation for three independent measurements.
We further confirmed that chemotactic minicells can efficiently accumulate toward the sources of chemoattractants, by using another microfluidic device where MeAsp is continuously released at the end of the channel38−40 (Figure 3C,D and Figure S6). This device mimics natural situations where chemoeffectors are released by a source, such as tumor tissue. Consistent with their ability to perform chemotaxis, minicells showed an increased migration into the observation channel, thus accumulating toward the source of attractant (Figure 3E,F). No accumulation was observed in the absence of MeAsp in the microfluidic chamber (Figure 3E,G) or for nonmotile minicells (Figure 3E,H).
Modeling of Motility and Chemotaxis of Minicells
In
order to better understand physical limitations on the swimming and
chemotaxis of minicells, we used a common model for the chemotactic
drift of E. coli (Supporting Information).20,41−43 The chemotactic
drift is described in this model as a function of biochemical properties
of the signaling pathway as well as of the physical parameters that
characterize cell swimming. We assumed that the functioning of the
chemotaxis pathway in minicells is similar to that of the normal cells.33 Two important physical parameters, which are
affected by the cell dimensions, are the rotational diffusion coefficient
for swimming cells Dr and the tumble persistence
time τT (Figure 4). According to our data (Figure 2F), minicells are less able to keep swimming
in a given direction, with the rotational diffusion coefficient of
the minicells being 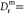
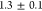 s–1 and therefore much
larger compared with
s–1 and therefore much
larger compared with 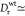
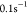 for normal cells.19 This increase is
well-accounted for by a simple model of rotational
diffusion that considers the rotation of the cell body and the flagellum
(Supporting Information).44,45 The tumble persistence time measures the time it takes for a cell
to randomize its direction of motion via tumbling. It is expected
to be
for normal cells.19 This increase is
well-accounted for by a simple model of rotational
diffusion that considers the rotation of the cell body and the flagellum
(Supporting Information).44,45 The tumble persistence time measures the time it takes for a cell
to randomize its direction of motion via tumbling. It is expected
to be 
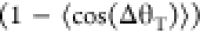 , where τ0 is the mean
run duration, and ΔθT is the angular change
in direction during a tumble. For minicells, the angular change ΔθT is expected to be larger compared to normal cells because
of their smaller body, whereas the run duration τ0 is expected to increase because of their smaller number of flagella.
Indeed, the run duration of minicells was
, where τ0 is the mean
run duration, and ΔθT is the angular change
in direction during a tumble. For minicells, the angular change ΔθT is expected to be larger compared to normal cells because
of their smaller body, whereas the run duration τ0 is expected to increase because of their smaller number of flagella.
Indeed, the run duration of minicells was 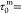
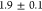 s, compared
with
s, compared
with 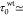
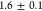 s for normal cells (Figure 2E). This moderate increase could be well-accounted
for by an effective veto model for bacterial tumbling46 (Supporting Information). We
also assumed a complete randomization of the swimming direction of
minicells during a tumble (⟨cos(ΔθT)⟩
= 0), in contrast to only a partial reorientation for longer normal
cells (⟨cos(ΔθT)⟩ ≃ 1/2)
as reported previously.19 Assuming the
biochemical properties of the chemotaxis pathway are unchanged in
the minicells, we predict a chemotactic velocity for the minicells
s for normal cells (Figure 2E). This moderate increase could be well-accounted
for by an effective veto model for bacterial tumbling46 (Supporting Information). We
also assumed a complete randomization of the swimming direction of
minicells during a tumble (⟨cos(ΔθT)⟩
= 0), in contrast to only a partial reorientation for longer normal
cells (⟨cos(ΔθT)⟩ ≃ 1/2)
as reported previously.19 Assuming the
biochemical properties of the chemotaxis pathway are unchanged in
the minicells, we predict a chemotactic velocity for the minicells 
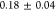 μm/s.
This estimate is in very good
agreement with the experimentally observed value (Figure 3B), confirming that the difference
in chemotactic ability between normal and minicells is primarily due
to the difference in their physical properties, especially their increased
rotational diffusion coefficient. The model also highlights that,
besides cell propulsion, flagella play another essential role in ensuring
that minicells are capable of chemotaxis, namely, by reducing rotational
diffusion and therefore stabilizing the direction of the minicell
motion. Nevertheless, the model seems to underestimate their chemotaxis
efficiency, which might be either because of the oversimplified model
assumptions or due to the slightly different signaling parameters
of minicells, such as higher concentrations of chemotaxis proteins
or faster signaling due to shorter distances between the chemosensory
complexes and flagellar motors.
μm/s.
This estimate is in very good
agreement with the experimentally observed value (Figure 3B), confirming that the difference
in chemotactic ability between normal and minicells is primarily due
to the difference in their physical properties, especially their increased
rotational diffusion coefficient. The model also highlights that,
besides cell propulsion, flagella play another essential role in ensuring
that minicells are capable of chemotaxis, namely, by reducing rotational
diffusion and therefore stabilizing the direction of the minicell
motion. Nevertheless, the model seems to underestimate their chemotaxis
efficiency, which might be either because of the oversimplified model
assumptions or due to the slightly different signaling parameters
of minicells, such as higher concentrations of chemotaxis proteins
or faster signaling due to shorter distances between the chemosensory
complexes and flagellar motors.
Figure 4.
Schematics illustrating motility and swimming parameters of wild-type cells and minicells. (A, B) Trajectory of a wild-type cell (A) and a minicell (B), with Dr representing the rotational diffusion coefficient and τ0 representing the mean run duration. ΔθT represents the angular change in direction during a tumble.
Concluding Remarks
Concluding, we observed that E. coli minicells that were engineered to have high levels of a flagellar gene expression are motile. The swimming pattern of these minicells was different from that of the parental E. coli cells, with minicells exhibiting a lower swimming velocity and directional persistence but increased run duration. These differences were consistent with the mathematical model describing minicell motility, and they could be accounted for by their small size and thus faster rotational diffusion as well as by the smaller number of flagella per minicell. Despite the potential major impact of these factors on the bacterial chemotaxis strategy, minicells were capable of following chemical gradients and accumulating toward sources of chemoattractants. Given increasing evidence that chemotactic bacteria can follow local chemical gradients to accumulate toward specific sites within their animal hosts,24−26 motility and chemotaxis could therefore be used to largely enhance the efficiency and specificity of the delivery of various drugs and protein and nucleic acid effectors that can be carried by minicells.
Materials and Methods
Strains and Plasmid Construction
The E. coli strain MG1655 was used as the wild-type for all experiments. A minCDE deletion was conducted using λ red recombination relying on pKD46.48 The kanamycin resistance cassette was removed using pCP20.49 pKOV was used to generate mreBA115V point mutation.50 Green fluorescent protein (GFP) promoter reporter for fliC (pAM109)35,36 was constructed based on pUA66.51 pBAD18 or pBAD24 vectors carrying flhDC genes were used to express FlhDC.52 sfGFP was expressed using a pTrc99a-backbone-based vector.
Minicell Production and Purification
For the minicell production, overnight cultures were inoculated into TB supplemented with kanamycin and grown at 30 °C with shaking (180 rpm) for 8 h. When necessary, different concentrations of arabinose (0%, 0.001%, 0.01%, 0.1%) were added in the culture to induce FlhDC expression, and 50 μM isopropyl β-d-1-thiogalactopyranoside (IPTG) was used to induce the sfGFP expression. For minicell purification, parental cells were first removed from the cell culture by centrifugation at 10 000g for 20 min, and minicells were subsequently harvested by centrifugation at 40 000g for 20 min.
Analysis of Swimming Velocity and Chemotaxis
The average swimming and chemotactic drift velocity of minicells were measured as described previously.35,36,53 Swimming velocity and chemotactic drift velocity of purified minicells were measured by recording the cell motion in a poly(dimethylsiloxane) (PDMS) microchamber using phase-contrast microscopy (Nikon TI Eclipse, 10× objective with numerical aperture (NA) = 0.3, CMOS camera EoSens 4CXP). The cell motion was analyzed as described previously both via Fourier-based algorithms,54,55 for measuring the swimming velocity and chemotactic drift, and cell tracking53 for measuring mean squared displacements, run durations, and swimming persistence. A suspension of 100 μM MeAsp in a tethering buffer (6.15 mM K2HPO4, 3.85 mM KH2PO4, 100 μM ethylenediaminetetraacetic acid (EDTA), 1 μM l-methionine, 10 mM lactic acid, pH 7.0) was used to generate a chemical gradient in PDMS chambers. All data were analyzed using ImageJ (https://imagej.nih.gov/ij/) with custom-written plugins.
The chemotactic accumulation of minicells in response to releasing gradients of MeAsp was measured with a microfluidic device described previously,38−40 with a slight modification. For the microfluidic devices preparation, 0.3% agarose was added to fill the whole microfluidic chamber. Ten microliters of tethering buffer each was then added into both source and sink sides. Afterward, purified minicells were added into the sink pore and allowed to diffuse into the observation channel for 2 h. A solution of 2 mM MeAsp was then added to the source pore and allowed to gradually diffuse through the agarose gel into the observation channel. The minicell density in the observation channel was monitored over time, starting immediately after compound addition, using Nikon Ti-E inverted fluorescence microscope with a 20× objective lens and Lumencor SOLA-SEII equipped with Andor Zyla sCMOS camera.
Analysis of Cell-Tracking Data
The mean squared displacement
MSD(t) = ⟨(ri(t + t0) – ri(t0))2⟩i,t0, with ri(t0) = (xi(t0),yi(t0)) the two-dimensional (2D) position of particle i at time t0, was computed as a function
of the lag time t, averaging over particles i and initial times t0. The
MSD was displayed until a lag time of t = 6 s, corresponding
to one-tenth of the duration of the movies (61 s, 2500 frames) and
above which statistics gets poor (less than 10 independent time steps
per average). For the quantification of the tumbling rate and swimming
persistence, trajectories were sorted into swimmer and nonswimmer
based on their radius of gyration 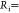
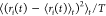 as previously described,53 and these properties were characterized on the
swimmer
trajectories only. Quantification of tumbling rate was also performed
as described previously.53 For measuring
the swimming persistence, the instantaneous swimming direction was
defined as the 2D instantaneous velocity measured on a 10 frames (0.25
s) wide sliding window normalized to its norm (ui(t) = vi(t)/|vi(t)|). The time autocorrelation
of ui(t) was computed by averaging over initial times and swimming cells,
as previously described,53 and these properties were characterized on the
swimmer
trajectories only. Quantification of tumbling rate was also performed
as described previously.53 For measuring
the swimming persistence, the instantaneous swimming direction was
defined as the 2D instantaneous velocity measured on a 10 frames (0.25
s) wide sliding window normalized to its norm (ui(t) = vi(t)/|vi(t)|). The time autocorrelation
of ui(t) was computed by averaging over initial times and swimming cells, 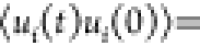
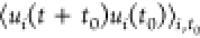 .
.
Quantification of Flagellar Length and Number of Minicells
Purified minicells were suspended in a tethering buffer with 10% glycerol and frozen at −80 °C before analysis by electon microscopy. For the sample preparation, 5 μL of the minicell suspension was applied onto hydrophilized carbon-coated copper grids (400 mesh). After a brief wash with filtered water, bacteria were stained with 2% uranyl acetate. All samples were analyzed using a JEOL JEM-2100 transmission electron microscope with an acceleration voltage of 120 kV. For the image acquisition, an F214 FastScan CCD camera (TVIPS; Gauting) was used. The flagellar number and length of minicells were quantified manually with ImageJ.
Promoter Activity Analysis
The activity of the gfp reporter of the fliC promoter was assayed using a BD LSRFortessa SORP cell analyzer (BD Biosciences) as described previously.35,36
Acknowledgments
We thank T. Heimerl, E. Kaganovitch, and N. Krink for their help with experiments.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acssynbio.1c00012.
Mathematical models for minicell motility, supporting figures (PDF)
Author Present Address
§ (B.N.) College of Resources and Environmental Science, National Academy of Agriculture Green Development, China Agricultural University, Beijing, 100193, China.
Author Contributions
∥ (B.N. and R.C.) These authors contributed equally. B.N., R.C., and V.S. designed the research. B.N. performed the experiments. B.N. and R.C. analyzed the data. R.C. performed the mathematical modeling. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
This work was supported by the Max Planck Society. R.C. acknowledges support from the Deutsche Forschungsgemeinschaft, Grant No. CO 1813/2–1.
The authors declare no competing financial interest.
Supplementary Material
References
- Wang A. Z.; Langer R.; Farokhzad O. C. (2012) Nanoparticle delivery of cancer drugs. Annu. Rev. Med. 63, 185–198. 10.1146/annurev-med-040210-162544. [DOI] [PubMed] [Google Scholar]
- Vader P.; Mol E. A.; Pasterkamp G.; Schiffelers R. M. (2016) Extracellular vesicles for drug delivery. Adv. Drug Delivery Rev. 106, 148–156. 10.1016/j.addr.2016.02.006. [DOI] [PubMed] [Google Scholar]
- Wilhelm S.; Tavares A. J.; Dai Q.; Ohta S.; Audet J.; Dvorak H. F.; Chan W. C. (2016) Analysis of nanoparticle delivery to tumours. Nat. Rev. Mater. 1, 1–12. 10.1038/natrevmats.2016.14. [DOI] [Google Scholar]
- de Boer P. A.; Crossley R. E.; Rothfield L. I. (1989) A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell 56, 641–649. 10.1016/0092-8674(89)90586-2. [DOI] [PubMed] [Google Scholar]
- Farley M. M.; Hu B.; Margolin W.; Liu J. (2016) Minicells, Back in Fashion. J. Bacteriol. 198, 1186–1195. 10.1128/JB.00901-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer A. C.; Curtiss R. 3rd. (1975) Production, properties and utility of bacterial minicells. Curr. Top. Microbiol. Immunol. 69, 1–84. 10.1007/978-3-642-50112-8_1. [DOI] [PubMed] [Google Scholar]
- Rampley C. P.; Davison P. A.; Qian P.; Preston G. M.; Hunter C. N.; Thompson I. P.; Wu L. J.; Huang W. E. (2017) Development of SimCells as a novel chassis for functional biosensors. Sci. Rep. 7, 1–10. 10.1038/s41598-017-07391-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes N. S. (2010) Engineering the perfect (bacterial) cancer therapy. Nat. Rev. Cancer 10, 785–794. 10.1038/nrc2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDiarmid J. A.; Brahmbhatt H. (2011) Minicells: versatile vectors for targeted drug or si/shRNA cancer therapy. Curr. Opin. Biotechnol. 22, 909–916. 10.1016/j.copbio.2011.04.008. [DOI] [PubMed] [Google Scholar]
- MacDiarmid J. A.; Mugridge N. B.; Weiss J. C.; Phillips L.; Burn A. L.; Paulin R. P.; Haasdyk J. E.; Dickson K. A.; Brahmbhatt V. N.; Pattison S. T.; James A. C.; Al Bakri G.; Straw R. C.; Stillman B.; Graham R. M.; Brahmbhatt H. (2007) Bacterially derived 400 nm particles for encapsulation and cancer cell targeting of chemotherapeutics. Cancer Cell 11, 431–445. 10.1016/j.ccr.2007.03.012. [DOI] [PubMed] [Google Scholar]
- MacDiarmid J. A.; Amaro-Mugridge N. B.; Madrid-Weiss J.; Sedliarou I.; Wetzel S.; Kochar K.; Brahmbhatt V. N.; Phillips L.; Pattison S. T.; Petti C.; Stillman B.; Graham R. M.; Brahmbhatt H. (2009) Sequential treatment of drug-resistant tumors with targeted minicells containing siRNA or a cytotoxic drug. Nat. Biotechnol. 27, 643–651. 10.1038/nbt.1547. [DOI] [PubMed] [Google Scholar]
- van Zandwijk N.; Pavlakis N.; Kao S. C.; Linton A.; Boyer M. J.; Clarke S.; Huynh Y.; Chrzanowska A.; Fulham M. J.; Bailey D. L.; Cooper W. A.; Kritharides L.; Ridley L.; Pattison S. T.; MacDiarmid J.; Brahmbhatt H.; Reid G. (2017) Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: a first-in-man, phase 1, open-label, dose-escalation study. Lancet Oncol. 18, 1386–1396. 10.1016/S1470-2045(17)30621-6. [DOI] [PubMed] [Google Scholar]
- Tsuji S.; Chen X.; Hancock B.; Hernandez V.; Visentin B.; Reil K.; Sabbadini R.; Giacalone M.; Godbey W. T. (2016) Preclinical evaluation of VAX-IP, a novel bacterial minicell-based biopharmaceutical for nonmuscle invasive bladder cancer. Mol. Ther Oncolytics 3, 16004. 10.1038/mto.2016.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M.; Khan M. W.; Grenier S.; Tsuji S.; Giacalone M. A.; McGuire K. L. (2016) Abstract B017: Bacterial minicells decrease tumor development and modulate immunity in a mouse mouse model of colon cancer. Cancer Immunol Res. 4, B017–B017. 10.1158/2326-6066.IMM2016-B017. [DOI] [Google Scholar]
- Carleton H. A.; Lara-Tejero M.; Liu X.; Galan J. E. (2013) Engineering the type III secretion system in non-replicating bacterial minicells for antigen delivery. Nat. Commun. 4, 1590. 10.1038/ncomms2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner L.; Ryu W. S.; Berg H. C. (2000) Real-time imaging of fluorescent flagellar filaments. J. Bacteriol. 182, 2793–2801. 10.1128/JB.182.10.2793-2801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colin R.; Sourjik V. (2017) Emergent properties of bacterial chemotaxis pathway. Curr. Opin. Microbiol. 39, 24–33. 10.1016/j.mib.2017.07.004. [DOI] [PubMed] [Google Scholar]
- Berg H. C.; Brown D. A. (1972) Chemotaxis in Escherichia coli analysed by three-dimensional tracking. Nature 239, 500–504. 10.1038/239500a0. [DOI] [PubMed] [Google Scholar]
- Berg H. C. (1983) Random Walks in Biology ,Princeton University Press, Princeton, NJ. [Google Scholar]
- Colin R.; Drescher K.; Sourjik V. (2019) Chemotactic behavior of Escherichia coli at high cell density. Nat. Commun. 10, 5329. 10.1038/s41467-019-13179-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg H. C.; Purcell E. M. (1977) Physics of Chemoreception. Biophys. J. 20, 193–219. 10.1016/S0006-3495(77)85544-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer O., Mostaghaci B., Colin R., Hurtgen D., Kraus D., Sitti M., and Sourjik V.. Motility and chemotaxis of bacteria-driven microswimmers fabricated using antigen 43-mediated biotin display, Sci. Rep. 2018, 8. 10.1038/s41598-018-28102-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guadayol O.; Thornton K. L.; Humphries S. (2017) Cell morphology governs directional control in swimming bacteria. Sci. Rep. 7, 2061. 10.1038/s41598-017-01565-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes J. G.; Sourjik V. (2018) Chemotaxis of Escherichia coli to major hormones and polyamines present in human gut. ISME J. 12, 2736–2747. 10.1038/s41396-018-0227-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J. Y.; Sweeney E. G.; Sigal M.; Zhang H. C.; Remington S. J.; Cantrell M. A.; Kuo C. J.; Guillemin K.; Amieva M. R. (2015) Chemodetection and destruction of host urea allows Helicobacter pylori to locate the epithelium. Cell Host Microbe 18, 147–156. 10.1016/j.chom.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sule N., Pasupuleti S., Kohli N., Menon R., Dangott L. J., Manson M. D., and Jayaraman A.. The norepinephrine metabolite 3,4-dihydroxymandelic acid is produced by the commensal microbiota and promotes chemotaxis and virulence gene expression in enterohemorrhagic Escherichia coli, Infect. Immun. 2017, 85. 10.1128/IAI.00431-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasinskas R. W.; Forbes N. S. (2007) Salmonella typhimurium lacking ribose chemoreceptors localize in tumor quiescence and induce apoptosis. Cancer Res. 67, 3201–3209. 10.1158/0008-5472.CAN-06-2618. [DOI] [PubMed] [Google Scholar]
- Kasinskas R. W.; Forbes N. S. (2006) Salmonella typhimurium specifically chemotax and proliferate in heterogeneous tumor tissue in vitro. Biotechnol. Bioeng. 94, 710–721. 10.1002/bit.20883. [DOI] [PubMed] [Google Scholar]
- Furter M.; Sellin M. E.; Hansson G. C.; Hardt W. D. (2019) Mucus Architecture and Near-Surface Swimming Affect Distinct Salmonella Typhimurium Infection Patterns along the Murine Intestinal Tract. Cell Rep. 27, 2665–2678. 10.1016/j.celrep.2019.04.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt C. K.; Medina-Sanchez M.; Edmondson R. J.; Schmidt O. G. (2020) Engineering microrobots for targeted cancer therapies from a medical perspective. Nat. Commun. 11, 5618. 10.1038/s41467-020-19322-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno V. M.; Álvarez E.; Izquierdo-Barba I.; Baeza A.; Serrano-López J.; Vallet-Regí M. (2020) Bacteria as Nanoparticles Carrier for Enhancing Penetration in a Tumoral Matrix Model. Adv. Mater. Interfaces 7, 1901942. 10.1002/admi.201901942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseinidoust Z.; Mostaghaci B.; Yasa O.; Park B. W.; Singh A. V.; Sitti M. (2016) Bioengineered and biohybrid bacteria-based systems for drug delivery. Adv. Drug Delivery Rev. 106, 27–44. 10.1016/j.addr.2016.09.007. [DOI] [PubMed] [Google Scholar]
- Liu J.; Hu B.; Morado D. R.; Jani S.; Manson M. D.; Margolin W. (2012) Molecular architecture of chemoreceptor arrays revealed by cryoelectron tomography of Escherichia coli minicells. Proc. Natl. Acad. Sci. U. S. A. 109, E1481–1488. 10.1073/pnas.1200781109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt A.; Cassidy C. K.; Ames P.; Bacia-Verloop M.; Baulard M.; Huard K.; Luthey-Schulten Z.; Desfosses A.; Stansfeld P. J.; Margolin W.; Parkinson J. S.; Gutsche I. (2020) Complete structure of the chemosensory array core signalling unit in an E. coli minicell strain. Nat. Commun. 11, 743. 10.1038/s41467-020-14350-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni B.; Ghosh B.; Paldy F. S.; Colin R.; Heimerl T.; Sourjik V. (2017) Evolutionary Remodeling of Bacterial Motility Checkpoint Control. Cell Rep. 18, 866–877. 10.1016/j.celrep.2016.12.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni B.; Colin R.; Link H.; Endres R. G.; Sourjik V. (2020) Growth-rate dependent resource investment in bacterial motile behavior quantitatively follows potential benefit of chemotaxis. Proc. Natl. Acad. Sci. U. S. A. 117, 595–601. 10.1073/pnas.1910849117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colin R.; Zhang R.; Wilson L. G. (2014) Fast, high-throughput measurement of collective behaviour in a bacterial population. J. R. Soc., Interface 11, 20140486. 10.1098/rsif.2014.0486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi S.; Jin F.; Sourjik V. (2018) Inverted signaling by bacterial chemotaxis receptors. Nat. Commun. 9, 2927. 10.1038/s41467-018-05335-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi S. Y.; Yu D. Q.; Si G. W.; Luo C. X.; Li T. Q.; Ouyang Q.; Jakovljevic V.; Sourjik V.; Tu Y. H.; Lai L. H. (2013) Discovery of novel chemoeffectors and rational design of Escherichia coli chemoreceptor specificity. Proc. Natl. Acad. Sci. U. S. A. 110, 16814–16819. 10.1073/pnas.1306811110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si G. W.; Yang W.; Bi S. Y.; Luo C. X.; Ouyang Q. (2012) A parallel diffusion-based microfluidic device for bacterial chemotaxis analysis. Lab Chip 12, 1389–1394. 10.1039/c2lc21219f. [DOI] [PubMed] [Google Scholar]
- Celani A.; Shimizu T. S.; Vergassola M. (2011) Molecular and Functional Aspects of Bacterial Chemotaxis. J. Stat. Phys. 144, 219–240. 10.1007/s10955-011-0251-6. [DOI] [Google Scholar]
- Dufour Y. S.; Fu X.; Hernandez-Nunez L.; Emonet T. (2014) Limits of Feedback Control in Bacterial Chemotaxis. PLoS Comput. Biol. 10, e1003694 10.1371/journal.pcbi.1003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Y. (2013) Quantitative modeling of bacterial chemotaxis: signal amplification and accurate adaptation. Annu. Rev. Biophys. 42, 337–359. 10.1146/annurev-biophys-083012-130358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locsei J. T.; Pedley T. J. (2009) Run and Tumble Chemotaxis in a Shear Flow: The Effect of Temporal Comparisons, Persistence, Rotational Diffusion, and Cell Shape. Bull. Math. Biol. 71, 1089–1116. 10.1007/s11538-009-9395-9. [DOI] [PubMed] [Google Scholar]
- Tirado M. M.; Martinez C. L.; Delatorre J. G. (1984) Comparison of Theories for the Translational and Rotational Diffusion-Coefficients of Rod-Like Macromolecules - Application to Short DNA Fragments. J. Chem. Phys. 81, 2047–2052. 10.1063/1.447827. [DOI] [Google Scholar]
- Mears P. J.; Koirala S.; Rao C. V.; Golding I.; Chemla Y. R. (2014) Escherichia coli swimming is robust against variations in flagellar number. eLife 3, e01916 10.7554/eLife.01916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micali G.; Colin R.; Sourjik V.; Endres R. (2017) Drift and behavior of E. coli cells. Biophys. J. 113, 2321–2325. 10.1016/j.bpj.2017.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko K. A.; Wanner B. L. (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97, 6640–6645. 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov P. P.; Wackernagel W. (1995) Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158, 9–14. 10.1016/0378-1119(95)00193-A. [DOI] [PubMed] [Google Scholar]
- Link A. J.; Phillips D.; Church G. M. (1997) Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179, 6228–6237. 10.1128/JB.179.20.6228-6237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaslaver A.; Bren A.; Ronen M.; Itzkovitz S.; Kikoin I.; Shavit S.; Liebermeister W.; Surette M. G.; Alon U. (2006) A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat. Methods 3, 623–628. 10.1038/nmeth895. [DOI] [PubMed] [Google Scholar]
- Sourjik V.; Vaknin A.; Shimizu T. S.; Berg H. C. (2007) In vivo measurement by FRET of pathway activity in bacterial chemotaxis. Methods Enzymol. 423, 365–391. 10.1016/S0076-6879(07)23017-4. [DOI] [PubMed] [Google Scholar]
- Suchanek V. M.; Esteban-Lopez M.; Colin R.; Besharova O.; Fritz K.; Sourjik V. (2020) Chemotaxis and cyclic-di-GMP signalling control surface attachment of Escherichia coli. Mol. Microbiol. 113, 728–739. 10.1111/mmi.14438. [DOI] [PubMed] [Google Scholar]
- Colin R.; Zhang R.; Wilson L. G. (2014) Fast, high-throughput measurement of collective behaviour in a bacterial population. J. R. Soc., Interface 11, 20140486. 10.1098/rsif.2014.0486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson L. G.; Martinez V. A.; Schwarz-Linek J.; Tailleur J.; Bryant G.; Pusey P. N.; Poon W. C. (2011) Differential dynamic microscopy of bacterial motility. Phys. Rev. Lett. 106, 018101. 10.1103/PhysRevLett.106.018101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.





