Abstract
Rationale:
The workup and longitudinal monitoring for subjects presenting with pulmonary nodules is a pressing clinical problem. A blood-based biomarker panel potentially has utility for identifying subjects at higher risk for harboring a malignant nodule for whom additional work-up would be indicated or subjects at reduced risk for whom imaging based follow-up would be indicated.
Objectives:
To assess whether a previously described four-protein biomarker panel, previously reported to improve assessment of lung cancer risk compared to a smoking-based lung cancer risk model, can provide discrimination between benign and malignant indeterminate pulmonary nodules.
Methods:
A previously validated multiplex enzyme linked immunoassay was performed on matched case and control samples from each cohort.
Measurements:
The biomarker panel was tested in two case-control cohorts of patients presenting with indeterminate pulmonary nodules at the University of Pittsburgh Medical Center and The University of Texas Southwestern.
Main Results:
In both cohorts, the biomarker panel resulted in improved prediction of lung cancer risk over a model based on nodule size alone. Of particular note, the addition of the marker panel to nodule size greatly improved sensitivity at a high specificity in both cohorts.
Conclusions:
A four-marker biomarker panel, previously validated to improve lung cancer risk prediction, also shows utility in distinguishing benign from malignant indeterminate pulmonary nodules. Its performance in improving sensitivity at a high specificity indicates potential utility of the marker panel in assessing likelihood of malignancy in otherwise indeterminate nodules.
Keywords: lung neoplasms, early detection of cancer, enzyme-linked immunosorbent assay, incidental findings
Introduction
Lung nodules are a common finding on chest CT scans. While most are incidentally discovered, some are discovered through low-dose CT (LDCT)-based lung cancer screening programs. A relatively high percentage of subjects in the general population (~1.9%) have a chest CT done per year, with incidental nodules discovered in some 24–31%.1 similar to the incidence of nodules reported in the National Lung Screening Trial (24%).2 The risk that a nodule is a cancer largely revolves around its size, with nodules greater than 20 mm often referred for work-up. Recommendations for smaller nodules are to follow them with additional imaging, for instance PET/CT, or short follow up repeated CT scans.3,4 Such an approach carries a risk of missing an early-stage lung cancer. Thus, lung nodules represent a significant diagnostic conundrum, with many institutions establishing clinics devoted solely to diagnosing and following nodules discovered incidentally or through screening.
A 4-protein marker panel (4MP) was subjected to blinded validation using plasmas collected up to one year before diagnosis of lung cancer, with significant performance in distinguishing cases from controls.5 The panel includes the proprotein form of surfactant protein B (pro-SFTPB) and three other markers with known utility in diagnosing lung cancer: cancer antigen 125 (CA125), cytokeratin-19 fragment (CYFRA 21–1), and carcinoembryonic antigen (CEA). This panel, integrated with a lung cancer risk model that included smoking history and patient age, improved the area under the curve (AUC) of the receiver operating characteristic (ROC) curve from 0.73 (95% CI 0.64−0.82) for the smoking model alone to 0.83 (95% CI 0.76−0.90). At a specificity of 83%, the integrated risk model showed a sensitivity of 63% (95% CI, 0.49–0.76).
Given the performance of the panel in the pre-diagnostic setting, we assessed the performance of the panel in distinguishing benign from malignant nodules using plasma samples from two cohorts of subjects presenting with pulmonary nodules. Our findings indicate improved sensitivity and specificity in predicting cancer compared to a model based on nodule size alone.
Materials and Methods
Sources of Study Populations
The Cooper lung nodule and cancer proteomics and genomics research registry (IRB # 03072024) was approved by the University of Pittsburgh IRB. The protocol enrolled patients with a confirmed benign lung nodule or diagnosed lung cancer from the Medical Oncology, Thoracic Surgery, and Pulmonary Medicine Clinics. The protocol authorized blood collections for research at periodic intervals, including before and at the time of diagnosis, after surgery, and at the time of lung cancer recurrence. Since 2004, this protocol enrolled 666 patients, with 521 of them eventually diagnosed with lung cancer and the remaining 145 with pulmonary benign nodules. All of them donated blood samples for research use.
The Pittsburgh Lung Screening Study (PLuSS) Cohort was approved by the University of Pittsburgh IRB (IRB# 011171). PLuSS is a community-based research cohort that recruited 3,642 smokers (current or former) during 2002–2006.6 Each PLuSS participant completed a questionnaire, underwent spirometry for pulmonary function testing (PFT), received a chest low-dose CT exam, and provided a blood sample. All of the 3,642 participants received a baseline low-dose CT scan, and 3,423 participants received a follow-up low-dose CT scan one-year later. Beginning in 2006, we re-enrolled original participants of PLuSS with the highest lung cancer risk (referenced as PLuSS X). Overall 970 individuals were enrolled into the PLuSS X, who received biennial low-dose CT scans, spirometry and blood draws during 2006–2015. The PluSS X and Cooper registry used the same protocol for blood sample collection, processing and storage (Figure S1A).
Blood samples were also obtained through a similar prospective protocol at the University of Texas at Southwestern (IRB# STU 072010–082, approved 8/17/2010 and terminated 6/8/2018). 193 total patients presenting for evaluation of pulmonary nodules at one of four affiliated hospitals (Parkland Hospital, Simmons Cancer Center Seay Clinic, St. Paul’s Clinic, and the Dallas Veterans Administration Medical Center) were enrolled into this protocol. Samples met PRoBE requirements for optimal biomarker collection.7 Seven patients failed screening, for a total initial enrollment of 186. Of these, 33 were known lung cancer patients, 62 were eventually diagnosed with lung cancer, 40 were negative in a lung cancer workup, and 5 were diagnosed with another cancer, with the rest with no eventual diagnosis (Figure S1B).
Design of UPMC and UTSW Case-Control Study
The UPMC cohort included 100 patients with early stage lung cancer. The median maximum nodule size on diagnostic CT scan was 20 mm (ranging from 7.5 to 38 mm) at initial diagnosis. For each case, we selected one control subject with a similar nodule size (maximum nodule size: 6.0 to 39.0 mm). The selected control was matched to index case by smoking status at the time of blood draw and gender. If a perfect match could not be identified, we dropped the gender as a matching criterion. Due to a small pool of nodule controls available from the Cooper registry, we also selected nodule controls from the PLuSS X participants. Control subjects were defined as having benign nodules if the nodule did not show radiographic growth for a minimum of two years, or were biopsy-proven negative for cancer. Despite attempts, perfect matching in nodule size between case and control cohorts was not achieved across the cohort. For the present study, we selected a plasma sample that was collected within 6 months prior to CT scan that revealed a pulmonary nodule of 6–39 mm. All 200 samples were pulled from the biorepository and sent to a test lab. The case-control status of the biospecimens were blinded to the team for biomarker test. A similar approach was performed with the UTSW cohort, with selection of 32 cases and control matched on age and gender. Control subjects were defined as those having negative cancer diagnosis after tissue sampling, improvement on radiographic follow-up, or alternative clinical diagnosis such as pulmonary fibrosis or pneumonia. These subjects were followed for 2 or more years with radiographic surveillance to ensure they were true negatives for cancer. Plasma specimens were collected at the time of study enrollment. Due to the size of the cohort, there were significant differences in smoking history and nodule size between lung cancer cases and controls (Figure S1).
Biomarker validation adhered to guidelines outlined by the Institute of Medicine (IOM) and the REMARK criteria.8,9 Briefly, samples were drawn under a standard operating procedure for venipuncture and aliquoted in a clinical research laboratory adhering to Clinical Laboratory Improvement Amendment (CLIA) guidelines. The 4MP was already validated in a lung cancer screening population and here was tested, with the same coefficients, in two blinded cohorts of a new intended use population of patients with indeterminate pulmonary nodules. Sensitivities and specificities are reported on this population, building on previous analytical validation on our previous study.
Luminex Bead Based Assay
Human pro-SFTPB, CEA, CYFRA21–1 and CA125 protein markers were quantified using Luminex bead-based immunoassay and the measured fluorescence intensities was measured with a MAGPIX instrument (Luminex Corporation, Austin TX). Pro-SFTPB Luminex assay was developed in-house as a sandwich ELISA using Mouse monoclonal antibodies against the N-terminus of pro-SFTPB. CEA and CA125 were assayed using a multiplex assay from EMD Millipore Corp. CYFRA21–1 was assayed using a single-plex kit from R&D Systems (Minneapolis, MN, USA). Plasma samples were thawed at 4°C and centrifuged at 1200g for 10 mins at 4°C before plating and testing. Samples were diluted 40X for pro-SFTPB, 6X for CEA/CA125 and 2X for CYFRA21–1. Samples were plated and analyzed in a blinded fashion. Each assay plate contained 7 calibration standards and a blank sample in duplicates. Quality controls include spike-in QCs and low/high plasma controls. The inter-plate and intra-plate coefficients of variation were 3% and 3.6% for pro-SFTPB, 3.19% and 10.4% for CEA, 1.33% and 4.4% for CA125 and 5.01% and 13.9% for CYFRA21–1 respectively.
Statistical Analyses
The ROC curve estimates are empirically based. 95% confidence intervals and standard errors of the AUC estimates as well those referring to the sensitivity (specificity) at a given specificity (sensitivity) are derived using the bootstrap scheme presented in the Appendix. To derive the ROC curves and corresponding AUC estimates for various fixed values of the covariates of interest, such the packyear and the nodule size, we considered a Cox based modeling technique. The details of this method, named HCNS, can be found in Bantis et al.10,11 This estimates the baseline cumulative hazard of a marker and then projects it through a Cox model for the desired covariate level. This is done separately for the control and the case group. Using these cumulative hazard estimates we derive the corresponding estimates of the cumulative distributions for both groups. These, in turn, allow us to derive the ROC for the covariate profile Z, by . We illustrate both the empirical estimates as well as the corresponding spline-based estimates given by the HCNS method. All corresponding p-values and confidence intervals are derived with the use of the bootstrap (see Appendix). Risks were calculated based on a logistic regression model, using simultaneously both covariates of interest, for instance the pack-year and the nodule size. Such a model induces a risk surface illustrated in Figure S2 on which we also overlay the raw data and provide the plot from different angles for better visualization. Such a surface allows us to graphically illustrate how steep the risk change is, as we increase either the pack-year or the nodule size value. Numerical results regarding these risks are presented in the results section. The logit link function was used throughout and standard GLM theory is used to fit these logistic regression models.
Results
Performance of the 4MP in the Pittsburgh nodule cohort
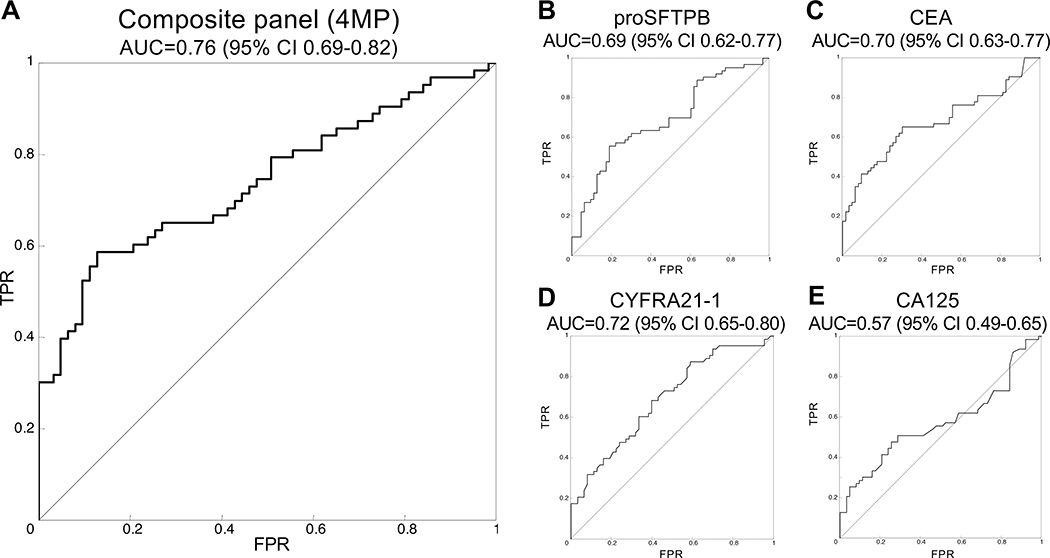
The study design consisted of 200 subjects with pulmonary nodules that were referred to the pulmonary clinic at the University of Pittsburgh Medical Center. The cohort consisted of 100 subjects who were subsequently diagnosed with lung cancer and 100 control patients with benign nodules that were matched for gender, age, and smoking history (Table 1, Table S1). The mean and distributions of age, gender, smoking status, and pack-years of smoking were comparable between cases and controls. The mean (±SD) maximum nodule size was significantly larger for cases (21±7.8 mm) than that of controls (11.6±5.8, p<0.001). Assays of plasmas for the 4MP was performed in a blinded fashion using the same standard operating procedure and fixed coefficients utilized in the pre-diagnostic study.5 The 4MP showed an area under the curve (AUC) of 0.76, with 95% CI 0.69–0.82 (Figure 1A). Three of the four independent markers also showed significant discrimination (Figure 1B–E).
Table 1 –
Baseline characteristics of study subjects from the University of Pittsburgh Medical Center (UPMC)
| All | With cancer | Without cancer | p-value | |
|---|---|---|---|---|
| Age | 67.7 ± 8.7 | 68.5 (Std = 9.2) | 67.0 (Std = 8.18) | 0.2191 |
| Gender | 0.1543 | |||
| Female | 88 | 49 | 39 | |
| Male | 112 | 51 | 61 | |
| Smoking status | 1.000 | |||
| Current | 74 | 37 | 37 | |
| Former | 126 | 63 | 63 | |
| Never | 0 | |||
| Pack years | 48.4 (Std = 23.1) | 47.8 (Std = 24.9) | 49.1 (Std = 21.2) | 0.6926 |
| Nodule size (mm) | 16.55 (Std = 8.47) | 21.5 (Std = 7.81) | 11.6 (Std = 5.81) | <0.001 |
Figure 1: Performance of the 4MP in the Pittsburgh nodule cohort.
A) The 4MP shows an AUC of 0.76 (95% CI 0.69–0.82). Three of the markers performed moderately well, including pro-SFTPB (B) with an area under the curve (AUC) of the receiver operative characteristic (ROC) of 0.69 (95% CI 0.62–0.77), CEA (C) at 0.70 (95% CI 0.63–0.77), and CYFRA21–1 (D) at 0.72 (95% CI 0.65–0.80). CA125 (E) did not show statistical significance with an AUC of 0.57 (95% CI 0.49–0.65, Table 1E).
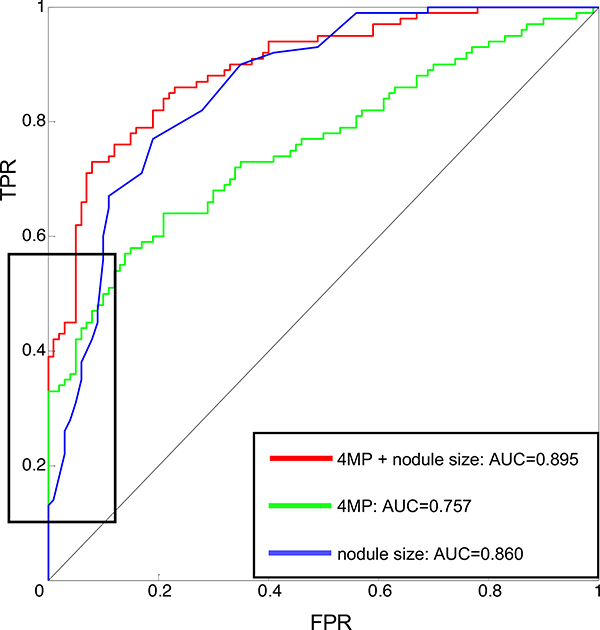
A Cox model accounting for age, gender, smoking history, and nodule size showed significant interaction between the 4MP and nodule size but none of the other variables. There were no significant differences in either the individual markers or the full 4MP between current and former smokers (Table S2, Figure S3–4). Pro-SFTPB and CYFRA21–1 exhibited significantly higher performance in larger nodules (Figure 2A–E). Compared to nodule size alone, the 4MP increased AUC from 0.86 to 0.90 (p-value of the comparison 0.033). Moreover, adding the 4MP to nodule size markedly increased sensitivity at a high specificity. At 99% specificity, the sensitivity of nodule size alone was 14%, which increased to 42% in combination with the 4MP. At 95% specificity, sensitivity increased from 31% to 62%, and at 90% specificity, sensitivity increased from 60% to 73% (Figure 3).
Figure 2: The 4MP in the Pittsburgh cohort by nodule size.
A) A Cox model accounting for age, gender, smoking history, and nodule size showed significant interaction between the 4MP and nodule size but none of the other variables. Compared to nodule size alone, the 4MP moderately improved performance, increasing AUC from 0.86 to 0.90 (p-value of the comparison 0.033). Pro-SFTPB (B) and CYFRA21–1 (D) drove this interaction, showing significantly higher performance in larger nodules. CEA and CA125 (C, E) did not show significant interaction with nodule size.
Figure 3: Performance of the 4MP combined with a nodule size-based risk model.
The 4MP improves performance of a risk model for lung cancer based on nodule size, increasing AUC from 0.86 to 0.89. The black box indicates that this performance improvement is pronounced on the left side of the ROC, indicating an increase in sensitivity at a high specificity.
Additional validation in an independent cohort
Given the findings in the first validation cohort, we sought to further validate the 4MP in an independent cohort of 60 patients with nodules from the University of Texas Southwestern (UTSW, Table 2, Table S2). The cohort consisted of 30 subjects subsequently diagnosed with lung cancer and 30 subjects with benign nodules matched for age and gender. This cohort had a lower pack-year history and included subjects with smaller nodules. Of the 60 subjects, 27 had nodules ≤ 6 mm.
Table 2 –
Baseline characteristics of study subjects from the University of Texas at Southwestern
| All | With cancer | Without cancer | p-value | |
|---|---|---|---|---|
| Age | 59.3 (Std = 10.7) | 59.9 (Std = 9.60) | 58.59 (Std = 11.8) | 0.63 |
| Gender | ||||
| Female | 30 | 15 | 15 | |
| Male | 34 | 17 | 17 | |
| Smoking status | ||||
| Current | 39 | 22 | 32 | |
| Former | 20 | 10 | 13 | |
| Never | 5 | 0 | 11 | |
| Pack years | 30.1 (Std = 33.3) | 44.1 (Std = 40.2) | 16.6 (Std = 16.4) | 0.0011 |
| Nodule size (mm) | 14.0 (Std = 15.4) | 19.2 (Std = 18.8) | 8.40 (Std = 7.59) | 0.0052 |
The 4MP performed well in identifying cases of lung cancer in this cohort (Figure 4A), with an AUC of 0.87 (95% CI 0.79–0.96). Given the higher number of smaller nodules, nodule size poorly predicted cancer risk, with an AUC of the ROC of 0.54 (95% CI 0.37–0.70). Addition of nodule size to the 4MP did not further improve the 4MP performance, with an AUC of 0.86 (95% CI 0.76–0.96). Again, the 4MP significantly improved performance at a 95% specificity, increasing sensitivity from 4% to 26%. (Figure S5). The performance of the full panel in the subset of 27 subjects with nodule size ≤ 6 mm was particularly striking (Figure 4B). In these patients, with 15 controls and 12 cases, the combination of nodule size and the 4MP showed an AUC of 0.95 with 95% CI 0.85–1.00.
Figure 4: Performance of the composite 4MP in the Southwestern nodule cohort.
A) The 4MP shows an AUC of 0.87 (95% CI 0.79–0.96), consistent with its performance in the Pittsburgh nodule cohort. Individual marker performance ranged from CYFRA21–1 at an AUC of 0.63 (95% CI 0.49–0.78), CEA at 0.72 (95% CI 0.59–0.86), to pro-SFTPB at 0.76 (95% CI 0.63–0.88) and CEA at 0.80 (95% CI 0.69–0.91). B) In a subset of nodules < 6 mm, the 4MP markedly improved performance of the nodule-size risk model. While nodule size alone predicted cancer with an AUC of 0.57 (95% CI 0.35–0.79), addition of the 4MP increased this to 0.95 (95% CI 0.85–1.000).
Discussion
Large numbers of pulmonary nodules are discovered incidentally or through low-dose CT-based screening, and represent a significant clinical dilemma. Most nodules are radiographically followed, with the Fleischner society recommending 24 months of follow-up to ensure a nodule is not cancerous. This requires continued coordination of treatment teams and utilization of medical resources. Small nodules represent a low risk that are followed with long intervals between imaging, leaving providers and patients anxious about interval development. The high rate of false-positive scan produced by CT screening is often cited as an impediment to implementation, as these nodules require structured, longitudinal follow-up that is a major contributor to the workload of a screening trial.
Here, we show that a biomarker panel previously reported to improve a smoking-based risk model for lung cancer screening also has utility in distinguishing benign from malignant pulmonary nodules. This panel improves the performance of nodule size alone in predicting the risk of cancer in a large cohort of heavy smokers recruited from the University of Pittsburgh pulmonary clinics. The panel was influenced by nodule size, performing better in larger nodules, but retained performance even in small nodules. The performance of the 4MP was not significantly altered by smoking history. Notably, the panel improved sensitivity from 14% to 42% at 99% specificity. This points to a potential clinical role in identifying nodules at higher risk. Marker-positive nodules could then be followed or biopsied more aggressively, with a high potential for earlier identification and treatment of disease. These findings were largely confirmed in a second, smaller cohort of lighter smokers from the University of Texas Southwestern. Notably, in this cohort, which included plasma samples from subjects with small nodules, the 4MP showed very large improvements over the nodule-size based clinical risk model alone. The fact that we used two different validation cohorts from different institutions with a range of nodule sizes is a strength of our study. Of interest, this second cohort contained 12 cases and 15 controls with nodule size ≤ 6 mm. In this small subset, the 4MP performed particularly well, with an AUC of the ROC of 0.95. While this obviously requires validation in a larger group, it again carries strong clinical ramifications.
This study was limited by relatively small size of the cohorts and limited demographic and radiographic data available from the subjects. This precluded the use of standard malignancy risk calculators. For instance, the Brock University calculator utilizes family history, presence of emphysema, nodule quality (solid, part-solid, or nonsolid), lobar location, nodule count, and spiculation.12 The Mayo Clinic model uses spiculation, upper lobe location, and history of extrathoracic cancer.13 These data were not available for either cohort. Another weakness of our study is the fact that the nodule size in the cancer cases is larger than in the benign nodules, and that smoking history was unable to be fully matched between cases and controls. Future studies will require integration of these and other established and emerging markers of increased risk, for instance occupational exposure, interstitial disease, and radiomic nodule features such as kurtosis, sphericity, and lung densitometry.14–16
Nodules under 6 mm are a common CT finding with a low risk of cancer. The 2017 Fleischner Society guidelines recommends only optional follow up CT scan at 12 months in high-risk patients. A biomarker that could identify patients at higher risk again has a strong potential to identify cancers that could be missed while they are still at early, curable stage. A negative biomarker score would be an effective reassurance that a nodule represented a benign process not requiring further imaging. While a prospective trial is necessary to definitively demonstrate the utility of this panel, these results give a promising foundation for such a trial. As such, these findings may represent a key step toward clinical utility as outlined by the American Thoracic Society guidelines.17 Finally, this panel, comprised of 4 protein markers, was measured on a multiplex immunoassay system, and additionally has the potential to provide a cost-effective means to secondarily stratify indeterminate nodules in these clinical scenarios. This could facilitate additional CT-screening, in resource-limited settings, or as a basis to broaden CT screening to lower risk groups.
Supplementary Material
Figure S1: Flowchart of subject recruitment. A) The UPMC cohort of malignant and benign lung nodules was assembled from a lung cancer screening program at Cooper Lung Cancer Center, supplemented by benign nodules identified from the PLuSS-X lung cancer screening trial. B) The UTSW cohort of malignant and benign lung nodules was assembled from subjects recruited from four pulmonary clinics.
Figure S2: 3D visualization of the 4MP with nodule size and smoking pack-years. A) A 3-dimensional projection of the 4MP logistic regression model using nodule size and probability of cancer. The same projection is displayed from above and 3 directions. B) A similar 3-dimensional projection of the model utilizing pack-years and nodule size and probability of cancer. A lack of slope on the pack-years axis indicates that it lends only small contribution towards cancer prediction.
Figure S3: Performance of the 4MP in the Pittsburgh cohort by smoking status. The 4MP and its individual components did not show significant differences between former (A) and current (B) smokers in the Pittsburgh cohort.
Figure S4: The 4MP in the Pittsburgh cohort by smoking pack-years. In the Pittsburgh cohort, the 4MP (A) or its individual components (B-E) did not show significant changes based on pack-years of smoking. Curves are plotted based on the Cox model.
Figure S5: The 4MP in the UTSW cohort combined with a nodule size-based lung cancer risk model. In the UTSW cohort, the 4MP combined with nodule size improved performance over nodule size alone, showing an AUC of 0.86 (95% CI 0.76–0.96) compared to nodule size alone which had an ROC of 0.54 (95% CI 0.37–0.70). Again noted was an improvement in sensitivity at a high specificity (black outline).
Acknowledgements
This study was supported by the Department of Defense Congressionally Mandated Lung Cancer Research Program W81XWH-10-1-0632. The authors would like to thank Ehsan Irajizad for review of the manuscript, and Jessica Saltarski and Joan Schiller for help with the UTSW protocol.
Appendix
Appendix 1:
Inferences for the AUCs.
Step 1: Sample with replacement n1 individuals from the controls and n2 from the cases where n1 and n2 are the total sample sizes for the controls and cases respectively.
Step 2: Based on the samples drawn in Step 1, use the empirical ROC estimate to derive the empirical based AUC estimate for this current bootstrap sample denoted by with b=1,2,..., B.
Step 3: Repeat Steps 1–3 B=1000 times to collect 1000 values of estimated AUCs.
Step 4: The SE of the original AUC estimate is given by:
For assessing the significance of the AUC estimate we consider under the null hypothesis that:
based on which we calculate the reported p-values.
Appendix 2:
Comparing AUCs.
To compare two competing AUCs we considered the following bootstrap scheme. The same bootstrap scheme is analogously extended for the comparison of the sensitivity (specificity) at a given level of specificity (sensitivity).
Step 1: Sample with replacement n1 controls and n2 cases where n1 and n2 are the total sample sizes for the controls and cases respectively.
Step 2: Based on Step 1 calculate the areas under both curves, denoted by and where b=1,2,..., B. We have used 1000 bootstrap samples i.e. B=1000.
Step 3: Repeat Steps 1–3 1000 times to collect 1000 values of estimated AUCs.
Step 4: The SE of the original AUC estimates for our data set are given by:
and
and
Finally, for the comparison of and we note that under the null hypothesis it holds that:
based on which we calculate the reported p-values.
For the AUCs, related SEs, and the corresponding comparisons that refer to the HCNS method which is based on an underlying Cox model, the above bootstrap schemes are analogously extended. That is, for each bootstrap sample the HCNS method is re-applied and the Cox model re-fitted (see also Bantis et al. 2012, 2013)10,11.
References
- 1.Gould MK, Tang T, Liu IL, et al. Recent Trends in the Identification of Incidental Pulmonary Nodules. Am J Respir Crit Care Med 2015;192(10):1208–1214. [DOI] [PubMed] [Google Scholar]
- 2.National Lung Screening Trial Research T, Aberle DR, Adams AM, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 2011;365(5):395–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.MacMahon H, Naidich DP, Goo JM, et al. Guidelines for Management of Incidental Pulmonary Nodules Detected on CT Images: From the Fleischner Society 2017. Radiology. 2017;284(1):228–243. [DOI] [PubMed] [Google Scholar]
- 4.American College of Radiology. Lung CT Screening Reporting and Data System (Lung-RADS). American College of Radiology. http://www.acr.org/Quality-Safety/Resources/LungRADS. Published 2014. Accessed. [Google Scholar]
- 5.Integrative Analysis of Lung Cancer E, Risk Consortium for Early Detection of Lung C, Guida F, et al. Assessment of Lung Cancer Risk on the Basis of a Biomarker Panel of Circulating Proteins. JAMA Oncol. 2018;4(10):e182078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson DO, Weissfeld JL, Fuhrman CR, et al. The Pittsburgh Lung Screening Study (PLuSS): outcomes within 3 years of a first computed tomography scan. Am J Respir Crit Care Med 2008;178(9):956–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pepe MS, Feng Z, Janes H, Bossuyt PM, Potter JD. Pivotal evaluation of the accuracy of a biomarker used for classification or prediction: standards for study design. J Natl Cancer Inst 2008;100(20):1432–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Micheel CMNSJ, Omenn GS In: Micheel CM, Nass SJ, Omenn GS, eds. Evolution of Translational Omics: Lessons Learned and the Path Forward. Washington (DC)2012. [PubMed] [Google Scholar]
- 9.Bossuyt PM, Reitsma JB, Bruns DE, et al. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. BMJ. 2015;351:h5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bantis LE, Tsimikas JV, Georgiou SD. Survival estimation through the cumulative hazard function with monotone natural cubic splines. Lifetime Data Anal 2012;18(3):364–396. [DOI] [PubMed] [Google Scholar]
- 11.Bantis LE, Tsimikas JV, Georgiou SD. Smooth ROC curves and surfaces for markers subject to a limit of detection using monotone natural cubic splines. Biom J 2013;55(5):719–740. [DOI] [PubMed] [Google Scholar]
- 12.McWilliams A, Tammemagi MC, Mayo JR, et al. Probability of cancer in pulmonary nodules detected on first screening CT. N Engl J Med 2013;369(10):910–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swensen SJ, Silverstein MD, Ilstrup DM, Schleck CD, Edell ES. The probability of malignancy in solitary pulmonary nodules. Application to small radiologically indeterminate nodules. Arch Intern Med 1997;157(8):849–855. [PubMed] [Google Scholar]
- 14.Dilger SK, Uthoff J, Judisch A, et al. Improved pulmonary nodule classification utilizing quantitative lung parenchyma features. J Med Imaging (Bellingham). 2015;2(4):041004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Field RW, Withers BL. Occupational and environmental causes of lung cancer. Clin Chest Med 2012;33(4):681–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naccache JM, Gibiot Q, Monnet I, et al. Lung cancer and interstitial lung disease: a literature review. J Thorac Dis 2018;10(6):3829–3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mazzone PJ, Sears CR, Arenberg DA, et al. Evaluating Molecular Biomarkers for the Early Detection of Lung Cancer: When Is a Biomarker Ready for Clinical Use? An Official American Thoracic Society Policy Statement. Am J Respir Crit Care Med 2017;196(7):e15–e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Flowchart of subject recruitment. A) The UPMC cohort of malignant and benign lung nodules was assembled from a lung cancer screening program at Cooper Lung Cancer Center, supplemented by benign nodules identified from the PLuSS-X lung cancer screening trial. B) The UTSW cohort of malignant and benign lung nodules was assembled from subjects recruited from four pulmonary clinics.
Figure S2: 3D visualization of the 4MP with nodule size and smoking pack-years. A) A 3-dimensional projection of the 4MP logistic regression model using nodule size and probability of cancer. The same projection is displayed from above and 3 directions. B) A similar 3-dimensional projection of the model utilizing pack-years and nodule size and probability of cancer. A lack of slope on the pack-years axis indicates that it lends only small contribution towards cancer prediction.
Figure S3: Performance of the 4MP in the Pittsburgh cohort by smoking status. The 4MP and its individual components did not show significant differences between former (A) and current (B) smokers in the Pittsburgh cohort.
Figure S4: The 4MP in the Pittsburgh cohort by smoking pack-years. In the Pittsburgh cohort, the 4MP (A) or its individual components (B-E) did not show significant changes based on pack-years of smoking. Curves are plotted based on the Cox model.
Figure S5: The 4MP in the UTSW cohort combined with a nodule size-based lung cancer risk model. In the UTSW cohort, the 4MP combined with nodule size improved performance over nodule size alone, showing an AUC of 0.86 (95% CI 0.76–0.96) compared to nodule size alone which had an ROC of 0.54 (95% CI 0.37–0.70). Again noted was an improvement in sensitivity at a high specificity (black outline).