Abstract
Background:
CHD7 pathogenic variants are identified in more than 90% of infants and children with CHARGE syndrome. Approximately 10% of cases have no known genetic cause identified.
Methods:
We report a male child with clinical features of CHARGE syndrome and non-diagnostic genetic testing that included chromosomal microarray, CHD7 sequencing and deletion/duplication analysis, SEMA3E sequencing, and trio Exome and Whole Genome Sequencing. We used a comprehensive clinical assessment, genome-wide methylation analysis (GMA), reanalysis of Whole Genome Sequencing (WGS) data, and CHD7 RNA studies to discover a novel variant that causes CHD7 haploinsufficiency.
Results:
The 7-year-old Hispanic male proband has typical phenotypic features of CHARGE syndrome. GMA revealed a CHD7-associated epigenetic signature. Reanalysis of the WGS data with focused bioinformatic analysis of CHD7 detected a novel, de novo 15 base pair deletion in intron 4 of CHD7, (c.2239–20_2239–6delGTCTTGGGTTTTTGT (NM_017780.3)). Using proband RNA, we confirmed that this novel deletion causes CHD7 haploinsufficiency by disrupting the canonical 3’ splice site and introducing a premature stop codon.
Conclusion:
Integrated genomic, epigenomic, and transcriptome analyses discovered a novel CHD7 variant that causes CHARGE syndrome.
Keywords: CHARGE syndrome, CHD7, epigenetics, genome sequencing, exome sequencing
Background
CHARGE syndrome (MIM 214800) is an autosomal dominant condition characterized by multiple congenital anomalies. CHARGE syndrome (Coloboma of the iris, retina, and/or optic disk; congenital Heart defects, choanal Atresia, Retardation of growth and development, Genital hypoplasia, and characteristic outer and inner Ear anomalies and deafness) may also include cranial nerve dysfunction, characteristic dysmorphic facies, tracheoesophageal fistula, and orofacial cleft (Lalani, Hefner, Belmont, & Davenport, 1993). Clinical diagnosis is based on the Blake criteria, which were later adjusted by Amiel et al and Verloes (Amiel et al., 2001; Blake et al., 1998; Lalani et al., 1993; Verloes, 2005). Initially described as an association, CHARGE syndrome is now known to result from haploinsufficiency of CHD7 in most cases. Variants in SEMA3E have also been observed in 2 patients with CHARGE syndrome (Lalani et al., 2004).
CHD7 (Chromodomain Helicase DNA-Binding Protein 7) is located on chromosome 8q12.2 and has 37 exons that encode a 2,997-amino acid protein. CHD7 belongs to the superfamily of chromodomain helicases and is involved in transcription regulation via chromatin remodeling (Basson & van Ravenswaaij-Arts, 2015). Most CHD7 pathogenic variants are de novo, nonsense or frameshift, and cause haploinsufficiency as the mechanism of disease (Basson & van Ravenswaaij-Arts, 2015; Lalani et al., 1993). A causative variant in CHD7 is found in more than 90% of cases that fully meet diagnostic criteria for CHARGE syndrome,(Bergman et al., 2011) and in about 65%−70% of definitive and suspected cases combined (Aramaki et al., 2006; Jongmans et al., 2006; Lalani et al., 1993). Approximately 5%−10% of cases have no confirmed pathogenic variant.
We report a male child patient with clinical features of CHARGE syndrome but without a pathogenic CHD7 variant discovered after extensive clinical genetic testing. We integrated genome-wide methylation analysis, whole genome sequencing, and transcript analysis to discover this child’s novel CHD7 pathogenic variant.
Case Report
The 7-year-old Hispanic male proband presented with global developmental delay and multiple congenital anomalies typical of CHARGE syndrome that included bilateral optic nerve coloboma, strabismus, nystagmus, bilateral hearing loss, repaired persistent ductus arteriosus, bicuspid aortic valve, micropenis, and undescended testicles. He is the third child of a nonconsanguineous couple of Mexican descent. Prenatal ultrasounds were notable for a possible renal collecting system anomaly, abnormal genitalia, and enlarged mega cisterna magna. The proband was born at term via vaginal delivery complicated by breech presentation and was admitted to the neonatal intensive care unit for evaluation of his multiple anomalies. His persistent ductus arteriosus required surgical ligation in the newborn period. He also has a bicuspid aortic valve with coronary fistula but has no cardiac symptoms. He had amblyopia, nystagmus, and strabismus and underwent strabismus surgery at age 6 years. His micropenis was treated with intramuscular testosterone with appropriate response. Bilateral cryptorchidism was surgically corrected at 9 months. A left ureteropelvic junction obstruction resulted in thinned renal parenchyma and no cortical function and required left nephrectomy. Neuropsychological testing at age 6 years revealed low intellectual functioning (Fluid-Crystallized Index of 66 on the Kaufman Assessment Battery for Children, 2nd Ed., Normative Update) and attention deficits but normal adaptive functioning. He has not had developmental regression. He receives special education and occupational, physical, and speech therapies. He also has bilateral sensorineural hearing loss for which he uses hearing aids and has required three sets of tympanostomy tubes due to bilateral otitis media with effusion.
A brain MRI at 2 days of life showed midline cerebellar hypoplasia. A computed tomography scan of his temporal bones at age 7 years demonstrated abnormalities consistent with CHARGE syndrome (i.e., complete aplasia of the semicircular canals with hypoplasia of the vestibules, congenital hypoplasia of the basal portion of the occipital bone, and hypoplasia of the left internal auditory canal).
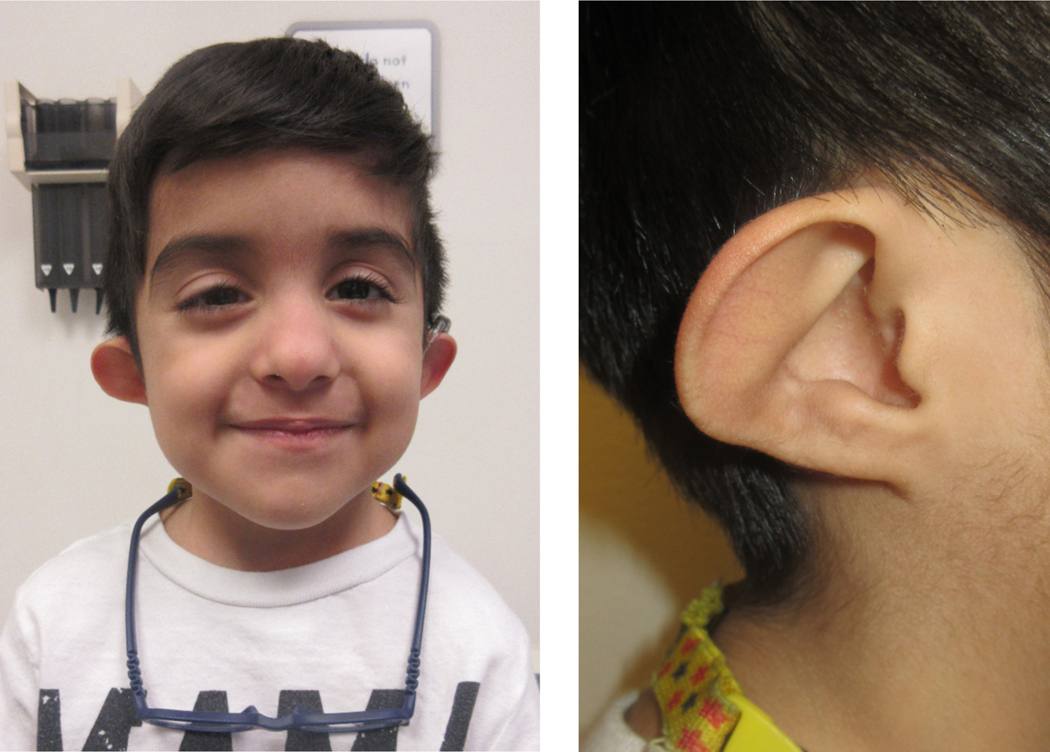
His physical examination at age 7 years was notable for short stature (Z-score: −3.40), dolichocephaly, low anterior hairline, squared face, low-set CHARGE-like ears (thin dysplastic helices, absent earlobes, prominent antihelix which was discontinuous with the antitragus), downslanting palpebral fissures, right exophoria, long eyelashes, intermittent horizontal nystagmus, prominent nose, and fifth digit clinodactyly (Figure 1).
Figure 1.
Photograph of the patient. Note low anterior hairline, squared face, low-set CHARGE-like ears, downslanting palpebral fissures, prominent nose.
CHD7 sequencing with deletion/duplication analysis, SEMA3E sequencing, chromosomal microarray, a cerebellar/pontocerebellar gene panel, trio exome sequencing, and trio whole genome sequencing were non-diagnostic. Due to his CHARGE syndrome phenotype, we undertook integrated genomic, epigenomic, and transcript analyses.
Methods
Enrollment in the Undiagnosed Diseases Network (UDN)
After review and approval by the Institutional Review Board of Washington University School of Medicine in St. Louis, we obtained informed consent from the proband’s parents for enrollment in the UDN. The proband underwent a full evaluation by multiple pediatric subspecialists, and we obtained peripheral blood samples for DNA and RNA extraction as well as a skin punch biopsy for establishment of a fibroblast cell line.
Genome-Wide Methylation Analysis (GMA)
A peripheral blood specimen was sent to the Greenwood Genetic Center and genomic DNA was isolated using standard techniques. Following bisulfite conversion with the EZ DNA Methylation Kit (Zymo Research, Tustin, CA), methylation data was generated using the Illumina Infinium Methylation EPIC BeadChip array according to manufacturer protocols. Methylation data was analyzed at London Health Sciences Centre Molecular Diagnostic Laboratory using a clinically validated machine-learning algorithm as previously described (Aref-Eshghi et al., 2018).
Whole Genome Sequencing (WGS)
Trio clinical WGS (Baylor Clinical Genomics Laboratory/UDN Sequencing Core at Baylor College of Medicine) provided unaligned BAM files which we realigned with BWA (v0.7.15) and called variants with GATK (v3.5.0). We annotated variants with ANNOVAR (v2019–10-24,)(Wang, Li, & Hakonarson, 2010) and selected variants with <1% minor allele frequency in public databases (GnomAD v2.0.1 and v3.0). We assessed variants in coding regions and splice regions (+/−12 bp) for predicted pathogenicity using CADD, M-CAP, REVEL, and dbscSNV scores from the dbNSFP database within ANNOVAR.
RNA analysis
We extracted RNA from peripheral blood (PaxGene RNA tubes and blood RNA kit (Qiagen, Germantown, MD)) from the proband and synthesized cDNA using SuperScript III (Invitrogen, Carlsbad, CA). To assess RNA splicing, we designed PCR primers that specifically amplify cDNA that includes exons 3 through 8 of CHD7, spanning several splice junctions. We separated and gel purified individual cDNAs using agarose gel electrophoresis and confirmed sequence of individual cDNAs with Sanger sequencing to evaluate the splicing effects of the CHD7 variant.
Results
GMA revealed a methylation pattern consistent with a CHD7-associated epigenetic signature (Figure 2, panel A). Although initial analysis of the WGS data by the Baylor Sequencing Center was non-diagnostic, research reanalysis of the data with focused analysis of CHD7 detected a de novo 15 bp deletion in intron 4 of CHD7 (c.2239–20_2239–6delGTCTTGGGTTTTTGT (NM_017780.3)). This variant was confirmed by the Baylor clinical laboratory via Sanger sequencing and was not detected in the samples from the proband’s unaffected parents. In silico splicing analysis (Alamut Visual (Interactive Biosoftware, Rouen, France)) predicted that this variant would disrupt the canonical 3’ splice site of exon 5 of CHD7. Analysis of proband CHD7 RNA (Supp. Figure 1) confirmed that this deletion causes disruption of the canonical 3’ splice site of exon 5, activates an alternative upstream splice site, adds 321 bp to the cDNA after exon 4, and introduces a stop codon 63 codons into the new sequence (Figure 2, panel B). This abnormal transcript would likely be subject to nonsense-mediated decay. These results confirmed that the proband has CHARGE syndrome due to a novel, de novo CHD7 pathogenic variant.
Figure 2.
Methylation analysis and transcriptional consequence of our patient’s CHD7 variant. (A) Heatmap comparing the genome-wide methylation patterns of controls (green) Vs patients with confirmed CHARGE syndrome (red). The current patient is indicated with a blue arrow. (B) Effect of our patient’s variant in CHD7 splicing. The upper section shows the localization of the intronic deletion (red circle). The middle section details the effects of the deletion as predicted by Alamut and confirmed by RNA studies. The lower section is a schematic representation of the resulting alternative transcript with retained intron 4 sequence and generation of a premature stop codon (red pentagon)
Discussion
The combination of strong phenotypic evidence of CHARGE syndrome and non-diagnostic genetic testing prompted further characterization of this patient’s epigenomic, genomic, and transcript signatures. Using GMA, we confirmed a CHARGE syndrome methylation signature (Aref-Eshghi et al., 2018). Research reanalysis of the WGS data with a focused analysis of CHD7 discovered a novel intronic variant which was computationally predicted to result in aberrant RNA splicing. Lack of recognition of this variant in initial analysis may have resulted from its location outside a pre-determined splice region, annotation of this deletion from its start position (−20), and computational difficulty in predicting pathogenicity of insertion/deletions. In this case, the CHD7 deletion variant would not be scored by splicing prediction algorithms such as dbscSNV or SpliceAI and could be missed. Assessment of the proband’s CHD7 transcripts confirmed the pathogenicity of the discovered CHD7 deletion variant. Our case emphasizes the importance of reanalyzing sequence data and integrating epigenomic, genomic, and transcript evaluation for genetic syndrome diagnosis.
Supplementary Material
Acknowledgments
This work was supported by the National Human Genome Research Institute (U01 HG010215, FSC) and the Children’s Discovery Institute (FSC, JAW, DJW). We thank the patient and his family for their participation in this study.
Data Accessibility
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Footnotes
Conflict of Interests
The authors have no conflict of interests to disclose
References
- Amiel J, Attieé-Bitach T, Marianowski R, Cormier-Daire V, Abadie V, Bonnet D, . . . Lyonnet S. (2001). Temporal bone anomaly proposed as a major criteria for diagnosis of CHARGE syndrome. Am J Med Genet, 99(2), 124–127. doi: [DOI] [PubMed] [Google Scholar]
- Aramaki M, Udaka T, Kosaki R, Makita Y, Okamoto N, Yoshihashi H, . . . Kosaki K. (2006). Phenotypic spectrum of CHARGE syndrome with CHD7 mutations. J Pediatr, 148(3), 410–414. doi: 10.1016/j.jpeds.2005.10.044 [DOI] [PubMed] [Google Scholar]
- Aref-Eshghi E, Rodenhiser DI, Schenkel LC, Lin H, Skinner C, Ainsworth P, . . . Sadikovic B. (2018). Genomic DNA Methylation Signatures Enable Concurrent Diagnosis and Clinical Genetic Variant Classification in Neurodevelopmental Syndromes. Am J Hum Genet, 102(1), 156–174. doi: 10.1016/j.ajhg.2017.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basson MA, & van Ravenswaaij-Arts C. (2015). Functional Insights into Chromatin Remodelling from Studies on CHARGE Syndrome. Trends in genetics : TIG, 31(10), 600–611. doi: 10.1016/j.tig.2015.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman JE, Janssen N, Hoefsloot LH, Jongmans MC, Hofstra RM, & van Ravenswaaij-Arts CM (2011). CHD7 mutations and CHARGE syndrome: the clinical implications of an expanding phenotype. J Med Genet, 48(5), 334–342. doi: 10.1136/jmg.2010.087106 [DOI] [PubMed] [Google Scholar]
- Blake KD, Davenport SL, Hall BD, Hefner MA, Pagon RA, Williams MS, . . . Graham JM Jr. (1998). CHARGE association: an update and review for the primary pediatrician. Clin Pediatr (Phila), 37(3), 159–173. doi: 10.1177/000992289803700302 [DOI] [PubMed] [Google Scholar]
- Jongmans MCJ, Admiraal RJ, van der Donk KP, Vissers LELM, Baas AF, Kapusta L, . . . van Ravenswaaij CMA (2006). CHARGE syndrome: the phenotypic spectrum of mutations in the CHD7 gene. J Med Genet, 43(4), 306–314. doi: 10.1136/jmg.2005.036061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalani SR, Hefner MA, Belmont JW, & Davenport SLH (1993). CHARGE Syndrome. In Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, & Amemiya A. (Eds.), GeneReviews(®). Seattle (WA): University of Washington, Seattle [Google Scholar]
- Copyright © 1993–2020, University of Washington, Seattle. GeneReviews is a registered trademark of the University of Washington, Seattle. All rights reserved.
- Lalani SR, Safiullah AM, Molinari LM, Fernbach SD, Martin DM, & Belmont JW (2004). SEMA3E mutation in a patient with CHARGE syndrome. J Med Genet, 41(7), e94–e94. doi: 10.1136/jmg.2003.017640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verloes A. (2005). Updated diagnostic criteria for CHARGE syndrome: a proposal. Am J Med Genet A, 133a(3), 306–308. doi: 10.1002/ajmg.a.30559 [DOI] [PubMed] [Google Scholar]
- Wang K, Li M, & Hakonarson H. (2010). ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res, 38(16), e164. doi: 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.