Abstract
BACKGROUND
Abnormal tuftelin 1 (TUFT1) has been reported in multiple cancers and exhibits oncogenic roles in tumor progression. However, limited data are available on the relationship between TUFT1 and hepatocellular carcinoma (HCC), and the exact biological mechanism of TUFT1 is still poorly understood in HCC.
AIM
To investigate TUFT1 expression in HCC and how interfering TUFT1 transcription affects HCC growth.
METHODS
TUFT1 in HCC and non-HCC tissues based on databases of the Cancer Genome Atlas and Oncomine were analyzed, and TUFT1 in human HCC tissues on microarray were detected by immunohistochemistry for clinicopathological features, overall survival, and disease-free survival. HCC cells were transfected with constructed vectors of TUFT1 that interfere or over-express TUFT1 for analyzing the biological behaviors of HCC cells. Proliferation, invasion, migration, and apoptosis of cells were detected by cell counting kit-8, scratch assay, transwell tests, and flow cytometry and confirmed by Western blotting, respectively.
RESULTS
Abnormal TUFT1 levels in databases expressed in HCC at messenger RNA (mRNA) level and HCC tissues were mainly located in cytoplasm and membrane. The level of TUFT1 expression in the HCC group was significantly higher (χ2 = 18.563, P < 0.001) than that in the non-cancerous group, closely related to clinical staging, size, vascular invasion of tumor, hepatitis B e-antigen positive, and ascites (P < 0.01) of HCC patients, and negatively to HCC patients’ overall survival and disease-free survival (P < 0.001). After interfering with TUFT1 transcription at mRNA level in the MHCC-97H cells by the specific TUFT1-short hairpin RNA, cell proliferation, invasion, and metastasis were significantly inhibited with increasing apoptosis rate. In contrast, proliferation, invasion, and migration were significantly enhanced after over-expression of TUFT1 mRNA in Hep3B cells in vitro.
CONCLUSION
Oncogenic TUFT1 was associated with the progression of HCC and could be a potential molecular-target for inhibiting HCC growth.
Keywords: Hepatocellular carcinoma, Tuftelin 1, Prognosis, Molecular-target, Growth
Core Tip: Tuftelin1 (TUFT1) has been reported to be regulated by hypoxia and involved in the Hedgehog signaling pathway, with over-expression in hepatocellular carcinoma (HCC) tissues or cell lines. Abnormal TUFT1 level was significantly related to tumor size, vascular invasion, positive hepatitis B e-antigen, advanced tumor-node-metastasis stage of HCC, patients with ascites, and shorter overall survival and disease-free survival. Interfering TUFT1 transcription could markedly suppress the growth and metastasis of high TUFT1 MHCC-97H cell lines in vitro through accelerating apoptosis. Moreover, increasing TUFT1 expression might promote the growth and metastasis of low TUFT1 Hep3B cell lines in vitro. The data suggested that TUFT1 is involved in HCC progression via the mechanism of inhibiting apoptosis and might serve as a potential therapeutic target for inhibiting HCC growth.
INTRODUCTION
Hepatocellular carcinoma (HCC) is usually lethal because of late diagnosis and early metastasis[1]. HCC is still one of the most common cancers with increasing incidence in the inshore area of the Yangtze River[2]. Aberrant up-regulated or down-regulated expression of many genes was identified during malignant transformation of hepatocytes in patients with chronic inflammation or damage by hepatitis B viruses (HBV), hepatitis C viruses (HCV), toxic intake, liver cirrhosis, and non-alcoholic fatty liver disease[3,4]. Given the limited effective therapies available for HCC patients with advanced stages, surgical resection and radiofrequency ablation are still the most common curative treatments for early stage patients, but the prognosis as overall survival (OS) or disease free survival (DFS) rate is very poor[5,6]. Therefore, the pathogenesis and feasible therapeutic treatments of HCC need urgent investigation[7-9]. Recently, tuftelin 1 (TUFT1) has been reported to be elevated during multiple cancer types, and the abnormal expression of TUFT1 has attracted medical attention[10,11]. TUFT1 was first identified and sequenced from a bovine ameloblast-enriched complementary DNA library with a highly conserved gene localized in chromosome 1q21-31 that contains 13 exons encoding an acidic, phosphorylated glycoprotein of 390 amino acids and plays a critical role in the development and mineralization of enamel[12].
TUFT1 belongs to the enamel associated teeth proteins and is thought to play a role in enamel mineralization. Also, TUFT1 is expressed in embryonic stem cells, neuronal cells, and non-mineralizing tissues like eyes, brain, adrenal gland, lung, liver, kidneys, testis, and tumor tissues[13-15]. TUFT1 can modulate the Rab GTPase-regulated process to activate the mammalian target of rapamycin complex 1 signaling that is activated in many cancers[16] and associated with poor prognosis in breast or pancreatic cancer and thyroid carcinoma[10,11,14]. In addition, TUFT1 is correlated with the metastasis and epithelial-mesenchymal transition[10], which could be regulated by hypoxia that enhanced TUFT1 by down-regulating miR-671-5p via interaction with 3'-ultranslated region of TUFT1 messenger RNA (mRNA) rather than by directly promoting hypoxia inducible factor-1α (HIF-1α) binding to TUFT1 promoter, and the Hedgehog signaling pathway[17-19]. Abnormal TUFT1 has been reported in multiple cancers and exhibits oncogenic roles in tumor progression[17,20]. However, limited data are available on the relationship between TUFT1 and HCC, and the exact biological mechanism of TUFT1 in HCC is still poorly understood. In this present study, the landscape of TUFT1 expression in human HCC tissues was investigated, and its molecular mechanism was confirmed through intervening or over-expressing TUFT1 activation in vitro.
MATERIALS AND METHODS
Bioinformatics analysis
Differential expressions of TUFT1 mRNA between HCC and normal liver tissues were collected according to the bioinformatics data from the Gene Expression Profiling Interactive Analysis (GEPIA) public Website[21] (http://GEPIA.cancer-pku.cn/detail.php) and the Oncomine database (https://www.Oncomine.org/resource).
Liver tissues and clinical data
This study was approved by the Ethics Committee permission (TDFY2018-025) at the Affiliated Hospital of Nantong University, China from Jan 2009 to Dec 2014, and the prior written informed consent was obtained from HCC patients according to the Helsinki Declaration of World Medical Association. A total 132 pairs of self-controlled HCC tissues (HCC group) and their para-cancerous tissues (2 cm to cancer, Para-C group) were collected from patients with HCC after post-operation, frozen in liquid nitrogen, and kept at -85 °C until used. According to their medical records, the patients were 111 males and 21 females with 21-79-years-old (average 60.04 ± 15.8 years). Serum alpha-fetoprotein (AFP) concentrations were 104 cases with ≥ 20 ng/mL and 28 cases with < 20 ng/mL. There were 120 cases with positive HBV surface antigen and 45 cases with positive hepatitis B e-antigen (HBeAg). There were 106 cases with tumor size ≥ 3.0 cm, 58 cases with vascular invasion, and 32 cases with ascites.
According to the 2019 edition of the diagnosis and treatment of primary liver cancer, there were three groups: Well (I), medium (II), and poor (III) differentiation. HCC patients were divided into I-IV stages, with 59 cases at I-II, and 73 cases at III-IV staging on the tumor-node-metastasis (TNM) classification of the International Union Against Cancer. All patients underwent histopathology and did not receive radiation or chemotherapy prior to surgery. HCC patients had regular follow-up from operation to death until Jan 2020. Diagnosis of HCC was confirmed with the criteria set by the Chinese National Collaborative Cancer Research Group[22].
Tissue microarray
Tissue microarrays (TMA) were constructed by the Departments of Pathology, Affiliated Hospital of Nantong University, China, containing formalin-fixed and paraffin-embedded specimens from 132 HCC tissues and their non-cancerous tissues. Tissue cores (1.5 mm) from the representative areas were constructed into the TMA slices.
Immunohistochemistry
Immunohistochemistry staining was determined for assessing TUFT1 expression by GTVisionTM III Detection System/Mo&Rb (GK600710) according to the manufacturer′s protocol. TMA slices were dewaxed and rehydrated, underwent antigen retrieval, and were blocked endogenous peroxidase and antibody nonspecific binding with 3% H2O2 and 10% goat serum, respectively. Slices were incubated with the primary anti-human TUFT1 antibody (1: 250, Abcam, Cambridge, United Kingdom) overnight at 4 °C. The slices after incubated with horseradish peroxidase (HRP)-conjugated goat anti-rabbit immunoglobulin G (Everest Biotech, Oxford, United Kingdom) were stained with diamino-benzidine, and counterstained with hematoxylin, cleared in xylene, and covered. The levels of TUFT1 expression were calculated with the Image-Pro Plus 6.0 software (Media Cybernetics, Rockville, MD, United States). Blinded evaluations of TUFT1 immunostaining and independent observation by two independent pathologists were carried out simultaneously, and the score was calculated by the sum of staining intensities and the rates of 100 positive cells. TUFT1 level in HCC tissues was divided into low and high expression. The expressing intensities of TUFT1 were divided into four categories: 0, negative; 1, weakly positive; 2, moderate positive; and 3, strongly positive.
Cell culture
All human HCC (Hep3B, MHCC-97L, MHCC-97H, BEL-7404, HepG2, HCCLM3, and SMMC-7721) cells and LO2 cell line were purchased from the Zhongqiao Xinzhou Science and Technology Co. (Shanghai, China). HCC cells were divided into three groups: Control, negative control (sh-NC or NC), and intervening (sh-TUFT1 or over-expressing) groups. HCC (MHCC-97L, MHCC-97H, BEL-7404, HepG2, and HCCLM3) cells in high glucose Dulbecco’s modified Eagle’s Medium with 10% fetal bovine serum (FBS, Gibco Laboratories, Gaithersburg, MD, United States), SMMC-7721 and LO2 cell lines in Roswell Park Memorial Institute-1640 medium with 10% FBS, and Hep3B cells in Minimal Essential Medium with 15% FBS were cultured containing 100 U/mL of penicillin/streptomycin at 37 °C with 5% of CO2. Cell density to about 85% was used to next cell passage.
Plasmid coding short hairpin RNA against TUFT1
Specific short hairpin RNA (shRNA) targeting sites of TUFT1 sequence (GenBank: AH009496.2)[23] were designed and synthesized (Guangzhou Cyagen Biotechnology Co., Ltd., China) and used to construct lentivirus LV-U6 > Scramble-shRNA-PGK > enhanced green fluorescent protein (EGFP)/T2A/Puro vector. Inserted interfering sequences in plasmids were TUFT1-shRNA1: 5’-AGAAGCTCCGG GAGGATATAACTCGAGTTATATCCTCCCGGAGCTCT-3’, TUFT1-shRNA2: 5’-TGA GGTGGACACCTGTATAAACTCGAGTTTATACAGGTGTCCACCTCA-3’, and TUFT1 -shRNA3: 5’-GATTCACGAGAAGAATATTAACTCGAGTTAATATTCTTCTCGTGAA TC-3’, respectively. TUFT1-negative plasmid was constructed similarly with a shRNA sequence that does not suppress TUFT1 expression, and the Scramble vector (Cyagen, Santa Clara, CA, United States) as a control. All inserted sequences were confirmed by sequencing.
Expressing TUFT1 plasmid
The exon sequence of TUFT1 gene (GenBank: AH009496.2)[23] was inserted into the Scramble vector (GenePharma, Shanghai, China) to construct pEX-4 (pGCMV/MCS/ T2A/EGFP/ Neo) vector. Negative or control plasmids were equal to above vectors.
Cell transfection
Human MHCC-97H cell line in high glucose Dulbecco’s modified Eagle’s Medium with 10% of FBS or Hep3B cell line in MEM with 15% of FBS was cultured at 37 °C with 5% CO2. When Hep3B cells up to optimal confluence over 70% were transfected at time, the cells of blank, negative control, and TUFT1 over-expressing groups were transfected with corresponding plasmids using Lipofectamine 3000 (Invitrogen, USA) according to the manufacturer’s instructions. MHCC-97H cells were also divided into blank, negative control, and sh-TUFT1 groups. They were transfected with corresponding plasmids using polybrene (Cyagen) according to the manufacturer’s instructions.
Western blotting
Western blot analysis was performed as described previously[24]. Briefly, 50 µg of purified protein from transfected cells was separated on electrophoresis of 10% sodium dodecyl sulfate-polyacrylamide gel, transferred to polyvinylidene fluoride membranes (Millipore, Burlington, MA, United States), blocked in 5% of nonfat dry milk in Tris-buffered saline (pH 7.5, 100 mmol/L NaCl, 50 mmol/L Tris, 0.1% Tween-20), and incubated with anti-human TUFT1 antibodies (Abcam) overnight at 4 °C, followed by HRP conjugated immunoglobulin G (Everest Biotech, Oxford, United Kingdom). Alterations of proteins were analyzed by Quantity-one software (Bio-Rad, Laboratories, Hercules, CA, United States) with Immobilon ECL Chemiluminescence HRP Substrate (Millipore) and quantified using Gel-Pro Analyzer software.
CCK-8 assay
Cell counting kit (CCK-8) was employed to evaluate HCC cell proliferation. There were three groups in the experiment: Blank, negative control, sh-TUFT1 intervention or TUFT1 over-expressing groups. Cells from different treatment groups were counted, adjusted to concentration 4 × 104/mL, and seeded in a 96-well plate with 100 μL/per well. Cells were seeded in triplicate for each group. The 96-well plate was placed in the incubator (37 °C and 5% CO2), and cells were cultured till the appropriate time. Then, 10 μL CCK-8 solution was added in each well, and the culture plate was incubated for 1-4 h. The absorbance (A) at 450 nm was detected using a plate reader.
Detection of apoptosis
Cell culture and experiment groups were set up as described above. Cells were cultured in a 6-well plate at about 70% confluence. Cells were aspirated and placed in a centrifuge tube, centrifuged at 1000 rpm for 5 min, rinsed with phosphate-buffered saline twice, and centrifuged again at 1000 rpm for 5 min. Cells (1 × 106) with 5 μL of Annexin V Alexa Fluor 647 staining solution were mixed in 100 μL binding buffer, collected, and placed in dark at room temperature for 10 min. Then, 400 μL binding buffer and 10 μL propidium iodide were mixed and placed in the dark at room temperature for 10 min. Fluorescent microscope observation or flow cytometry was conducted within 1 h. FL2 channel was used to detect the yellow fluorescence of propidium iodide with excitation wavelength of 488 nm. FL4 channel was used to detect the red fluorescence of Alexa Fluor 647 with emission wavelength of 640 nm. Cells without apoptotic induction were used as control for fluorescence compensation to remove spectral overlap and determine the position of cross-over.
Cell invasion
Transwell assays were performed to examine the invasion of cells transfected with small interfering RNA that targeted TUFT1 or a scrambled negative control. Cell invasion was defined using Transwell polycarbonate membrane inserts (Millipore). Cells (1 × 105) in serum-free medium were placed into the upper chamber of an insert coated with Matrigel (TheWell Bioscience Inc., North Brunswick, NJ, United States). Medium containing 10% FBS was added to the lower chamber to stimulate invasion, and the cells remaining on the upper membrane were removed with cotton wool after cultivation for 48-72 h. Cells that had invaded through the membrane were stained with methanol and 0.1% crystal violet, imaged, and counted using an IX71 inverted microscope (Olympus, Tokyo, Japan). Each assay was repeatedly conducted three times.
Scratch migration assay
To measure the logarithmic growth period, cells were seeded on plates for scratch analysis. When the cell growth density was about 90%, the scratch migration assay was performed to test cell migration. Using the ruler perpendicular to the ix-hole plate, with the 200 μL spear head along the ruler, a scratch was made, keeping strength as far as possible. A picture was taken, and the cells were allowed to continue to grow; a picture was taken every 24 h. The images were then compared, and the data analyzed.
Statistical analysis
Statistical analysis was performed by GraphPad Prism 8.0 software (GraphPad Co. Ltd., San Diego, CA, Unites States) and SPSS statistical package Version 25.0 (SPSS Inc., Armonk, NY, United States). The Student’s t test, a rank-sum test, χ2 test, and Fisher's exact probability test were used for categorical variables[25]. Cumulative survival curves of OS and DFS were calculated using the Kaplan–Meier method and compared by the log-rank test. The Cox proportional hazards model was used to determine independent factors of recurrence based on the variables selected by univariate analysis. Estimated 95% confidence intervals (CIs) with hazard ratio (HR) were combined to evaluate the prognostic and clinicopathologic values according to the Tierney method[26]. All the significant predictors of recurrence in the univariate analysis were analyzed in a logistic regression model to show an independent value at the multivariate analysis. A two-tailed value of P < 0.05 was considered statistically significant.
RESULTS
TUFT1 in bioinformatics database
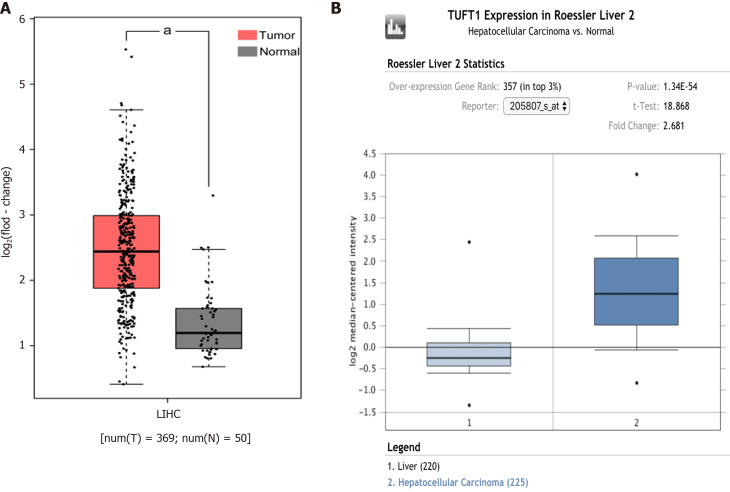
The comparative analysis of TUFT1 mRNA box diagrams between HCC (n = 594) and normal liver (n = 270) tissues from the bioinformatics websites are shown in Figure 1. In the Cancer Genome Atlas database (Figure 1A) from the GEPIA public website, the levels of TUFT1 mRNA expression in the HCC group were significantly higher (P < 0.05) than those in the normal group. Based on the Oncomine database (Figure 1B), the levels of TUFT1 mRNA expression in the HCC group were also significantly higher (P < 0.05) than those in the normal group. The data from online databases indicated that hepatic TUFT1 should be over-expressing status in HCC progression.
Figure 1.
Comparative analysis of tuftelin 1 messenger RNA between hepatocellular carcinoma and normal livers based on bioinformatics databases. A: The expression of tuftelin 1 (TUFT1) messenger RNA in the cancerous tissues of hepatocellular carcinoma (HCC) patients (n = 369, red, aP < 0.05) and normal liver tissues (n = 50, grey) from the Cancer Genome Atlas database; B: The expression of TUFT1 messenger RNA in the cancerous tissues of HCC patients (n = 225, dark blue, aP < 0.05) and normal liver tissues (n = 220, light blue) from the Oncomine database. The horizontal line represents the median of the two groups. Vertical bars indicate the range of data. LIHC: Liver hepatocellular carcinoma.
TUFT1 expression in Human HCC tissues
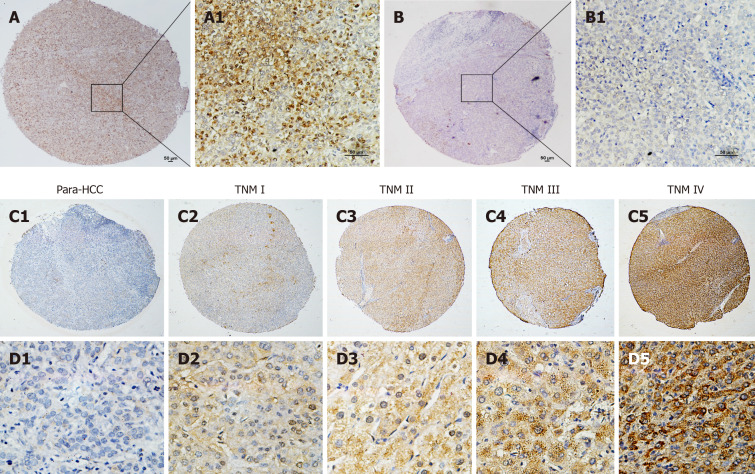
The immunohistochemistry analysis of hepatic TUFT1 expression in 132 cases of HCC and their non- HCC tissue microarray with clinical staging and prognosis is shown in Figure 2. The cellular distribution of hepatic TUFT1 expression was mainly located in cytoplasm and cell membrane. The intensity of TUFT1 staining was stronger in the HCC group (Figure 2A and A1) than that in the non-HCC group (Figure 2B and B1). The positive rate of TUFT1 in the HCC group (87.1%, 115/132) was significantly higher (χ2 = 18.563, P < 0.001) than that in the non-HCC group (64.4%, 85/132). Comparative analysis of TUFT1 expressions in the HCC or non-HCC tissues is summarized in Table 1. The significant difference (Z = 4.911, P < 0.001) of TUFT1 staining scores was found between HCC and non-HCC tissues by Z test, with higher expression in 50.0% HCC tissues (2-3 scores, 66/132), and low or no expression in 76.5% their non-HCC tissues (0-1 scores, 101/132). Regarding the relationship between TUFT1 and HCC staging (Figure 2C and D), there was lower/no TUFT1 staining in non-HCC tissues (Figure 2C1 and D1) with brown staining of TUFT1 gradually increasing from stage I to IV (Figure 2C2-5 or Figure 2D2-5).
Figure 2.
Tuftelin 1 expression with clinical staging of hepatocellular carcinoma. A, A1: Hepatocellular carcinoma (HCC) tissue with strongly positive staining; B, B1: Adjacent non-HCC tissue with negative staining, original magnifications of × 40 (scale bar: 50 μm) in A and B, and × 200 (scale bar: 50 μm) in A1 and B1; C, D: Tuftelin 1 (TUFT1) expression in different HCC staging; C1, D1: Low TUFT1 expression in the non-HCC tissues; C2-C5: The brown staining of TUFT1 gradually increases in the HCC tissues from stage I to IV (original magnification × 40); D2-D5: The staining of TUFT1 in the non-HCC tissues from stage I to IV (original magnification × 400). TNM: Tumor-node-metastasis.
Table 1.
Comparative analysis of hepatic tuftelin 1 immunohistochemical staining and expressing intensity between hepatocellular carcinoma and their para-cancerous tissues
|
Group
|
n
|
TUFT1
|
χ
2
value
|
P
value
|
TUFT1 score
|
Z value
|
P
value
|
||||
|
Neg.
|
Pos.
|
0
|
1
|
2
|
3
|
||||||
| HCC | 132 | 17 | 115 | 18.563 | < 0.001 | 17 | 49 | 55 | 11 | 4.911 | < 0.001 |
| Non-HCC | 132 | 47 | 85 | 47 | 54 | 23 | 8 | ||||
HCC: Hepatocellular carcinoma; Neg: Negative; Non-HCC: Non-cancerous tissue; Pos: Positive; TUFT1: Tuftelin 1.
Figure 5.
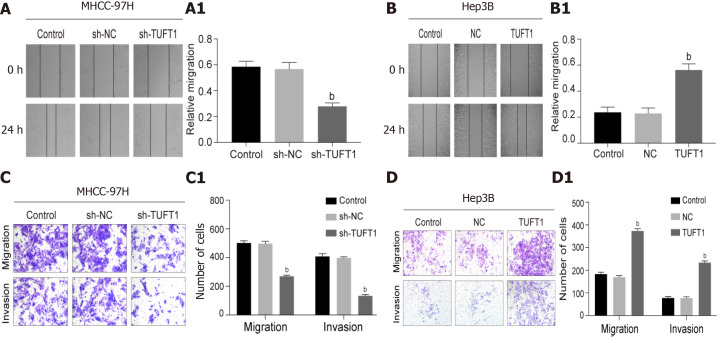
Effects of tuftelin 1 on healing, invasion, and migration of hepatocellular carcinoma cells. A: The scratch test of the MHCC-97H cells, the cell healing ability was decreasing after the high tuftelin 1 (TUFT1) cells were transfected with the TUFT1-shRNA3 plasmid, and A1, the relative cell healing ability was significantly inhibited compared with the control or negative sh-RNA for tuftelin 1 mRNA (sh-NC) cells (bP < 0.001); B: The scratch test of the Hep3B cells, the cell healing ability was increased after the cells were transfected with the over-expressing TUFT1 plasmid; B1, the relative cell healing ability was significantly improved compared with the control or blank control (NC) cells (bP < 0.001); C: The migration and invasion abilities of the MHCC-97H cells were analyzed by the transwell test; C1: The histograms of the cell migration and invasion abilities among the different groups (bP < 0.001): D: The migration and invasion abilities of the Hep3B cells were analyzed by the transwell test; D1: The histograms of the cell migration and invasion abilities among the different groups (bP < 0.001). Data are presented as mean ± standard error of the mean of at least three independent experiments. sh-RNA: sh-RNA3 for tuftelin 1 mRNA.
Clinicopathologic features and prognostic value of TUFT1
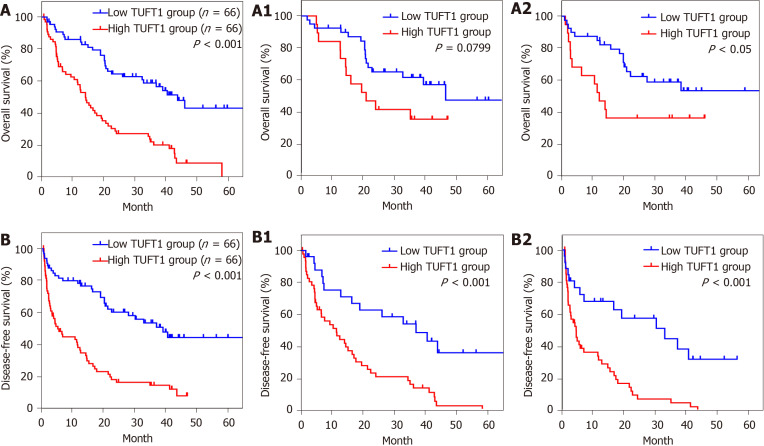
The clinicopathological features of TUFT1 expression in 132 HCC patients are shown in Table 2. The higher TUFT1 level was associated with tumor size, vascular invasion, HBeAg, advanced TNM stage of HCC, and ascites of patients. However, no significant difference was found between TUFT1 and age, sex, AFP level, HBV surface antigen, and Edmondson-Steiner grading. Analysis of univariate or multivariate Cox regression of TUFT1 in HCC tissues (Table 3) was significantly correlated with HBeAg, tumor size, vascular invasion, ascites, TNM stage, and AFP level with OS of patients, and further confirmed that TUFT1, tumor size, vascular invasion, ascites, and TNM stage were independent predictive factors for OS. On the contrary, TUFT1 and TNM stage should be independent prognostic factors for DFS of HCC. The OS or DFS of HCC with high or low TUFT1 was analyzed at early or advanced HCC (Figure 3A or Figure 3B). Compared to low TUFT1 group, the cases with high TUFT1 expression had significantly decreased OS and DFS, suggesting that TUFT1 should be a promoter or prognostic marker for HCC.
Table 2.
Relationship between tuftelin 1 and clinicopathological characteristics in hepatocellular carcinoma patients
|
Group
|
n
|
TUFT1 expression
|
Pearson χ2
|
P
value
|
|
|
Low
|
High
|
||||
| AFP (ng/mL) | 2.901 | 0.089 | |||
| < 20 | 28 | 18 | 10 | ||
| ≥ 20 | 104 | 48 | 56 | ||
| HBsAg | 3.300 | 0.069 | |||
| Neg. | 12 | 9 | 3 | ||
| Pos. | 120 | 57 | 63 | ||
| HBeAg | 4.080 | 0.043 | |||
| Neg. | 87 | 49 | 38 | ||
| Pos. | 45 | 17 | 28 | ||
| Tumor size (cm) | 9.388 | 0.002 | |||
| < 3 | 26 | 20 | 6 | ||
| ≥ 3 | 106 | 46 | 60 | ||
| Differentiation degree | 3.013 | 0.083 | |||
| Well-Med. | 19 | 13 | 6 | ||
| Poor | 113 | 53 | 60 | ||
| Vascular invasion | 14.885 | < 0.001 | |||
| With | 58 | 18 | 40 | ||
| Without | 74 | 48 | 26 | ||
| Ascites | 5.940 | 0.015 | |||
| With | 32 | 10 | 22 | ||
| Without | 100 | 56 | 44 | ||
| TNM staging | 13.516 | < 0.001 | |||
| I-II | 59 | 40 | 19 | ||
| III-IV | 73 | 26 | 47 | ||
AFP: Alpha-fetoprotein; HBeAg: Hepatitis B virus e antigen; HBsAg: Hepatitis B virus surface antigen; HCC: Hepatocellular carcinoma; Neg: Negative expression; Pos: Positive expression; TUFT1: Tuftelin 1.
Table 3.
Univariate and multivariate analysis of tuftelin 1 in the prognosis of hepatocellular carcinoma
|
Group
|
Univariate analysis
|
Multivariate analysis
|
||||
|
HR
|
95%CI
|
P
value
|
HR
|
95%CI
|
P
value
|
|
| TUFT1 expression, high vs low | 3.026 | 1.927-4.752 | < 0.001 | 1.743 | 1.044-2.910 | 0.034 |
| AFP (ng/mL), ≥ 20 vs < 20 | 1.866 | 1.066-3.266 | 0.027 | 1.482 | 0.835-2.630 | 0.178 |
| HBsAg, pos. vs neg. | 1.495 | 0.652-3.431 | 0.339 | |||
| HBeAg, pos. vs neg. | 1.670 | 1.084-2.574 | 0.019 | 1.070 | 0.659-1.739 | 0.784 |
| Tumor size (cm), ≥ 3 vs < 3 | 3.680 | 1.837-7.370 | < 0.001 | 2.235 | 1.043-4.485 | 0.039 |
| Differentiation degree, Poor vs well-med. | 1.642 | 0.869-3.100 | 0.123 | |||
| Vascular invasion, with vs without | 3.297 | 2.133-5.094 | < 0.001 | 2.018 | 1.186-3.434 | 0.010 |
| Ascites, with vs without | 2.592 | 1.648-4.076 | < 0.001 | 1.929 | 1.177-3.161 | 0.009 |
| TNM staging, III-IV vs I-II | 2.411 | 1.532-3.796 | < 0.001 | 1.110 | 0.614-2.005 | 0.731 |
AFP: Alpha-fetoprotein; HBeAg: hepatitis B virus e antigen; HBsAg: hepatitis B virus surface antigen; HR: Hazard ratio; Neg: Negative expression; Pos: Positive expression; TNM: Tumor-node-metastasis.
Figure 3.
Relationship between tuftelin 1 expression and prognosis of hepatocellular carcinoma. A: Kaplan-Meier analysis was performed to compare the overall survival (OS) between hepatocellular carcinoma (HCC) with high (n = 66) and low (n = 66) TUFT1; A1: The high (n = 19) and low (n = 40) tuftelin 1 (TUFT1) with OS at early HCC; A2: The high (n = 47) and low (n = 26) TUFT1 with OS at advanced HCC; B: Kaplan-Meier analysis was performed to compare the disease-free survival (DFS) between HCC with high (n = 66) and low (n = 66) TUFT1; B1: The high (n = 19) and low (n = 40) TUFT1 with DFS at early HCC; B2: The high (n = 47) and low (n = 26) TUFT1 with DFS at advanced HCC.
Effects of interfering TUFT1 transcription on HCC cells
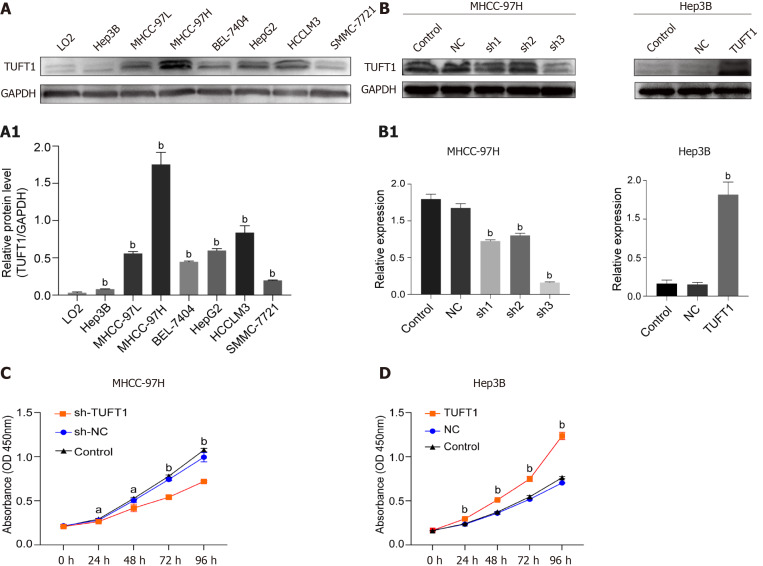
Screening TUFU1 and interfering TUFT1 mRNA transcription on effects of HCC cell lines are shown in Figure 4. Differential expressions of TUFT1 among different HCC cell lines were screened (Figure 4A). The relative ratio from TUFT1 to glyceraldehyde 3-phosphate dehydrogenase showed Hep3B cells with lowest and MHCC-97H cells with strongest level of TUFT1 expression (P < 0.001, Figure 4A1). Based on the status of TUFT1 expression in the MHCC-97H or Hep3B cells, the MHCC-97H cells were chosen to be used for interfering TUFT1 mRNA transcription with shRNA1-3 (Figure 4B left), and the most significantly inhibiting effect was shRNA3 (sh-3) plasmid for TUFT1 (Figure 4B1 left). The Hep3B cells were selected to over-express TUFT1 with constructed pEX-4 (pGCMV/MCS/T2A/EGFP/ Neo) plasmid (Figure 4B right) and showed markedly increasing TUFT1 expression (Figure 4B1 right). Using a different style of TUFT1 interference, the TUFT1-shRNA3 significantly suppressed proliferation of MHCC-97H cells (P < 0.001, Figure 4C), and TUFT1 activation markedly increased proliferation of Hep3B cells (P < 0.001, Figure 4D). The results confirmed that TUFT1 could promote HCC cell proliferation.
Figure 4.
Expression, interfering of tuftelin 1 with proliferation of hepatocellular carcinoma cells. A: Expressions of tuftelin 1 (TUFT1) among different hepatocellular carcinoma (HCC) cell lines were detected by Western blotting, and the expressing levels of all HCC cells were significantly higher than that of the control LO2 cells (bP < 0.001); A1: The relative ratio from TUFT1 to glyceraldehyde 3-phosphate dehydrogenase with the MHCC-97H cells showed the strongest TUFT1 expression, and the Hep3B cells showed the lowest TUFT1 expression; B: The MHCC-97H cells and interfering with TUFT1 short hairpin RNA (sh-RNA)1-3 (left); the TUFT1 was over-expressed after the Hep3B cells transfected with the constructed pEX-4 (pGCMV/MCS/T2A/EGFP/Neo) plasmid (Right); B1: The TUFT1 expression was significantly inhibited by the TUFT1-shRNA transfection (bP < 0.001, left); the markedly increasing TUFT1 Level was compared with the control cells (bP < 0.001, right); C: The TUFT1 activation interfering by TUFT1- shRNA3 significantly inhibited the proliferation of MHCC-97H cells (aP < 0.05, bP < 0.001); D: the over-expression of TUFT1 significantly enhanced the proliferation of the Hep3B cells (bP < 0.001). Data are presented as mean ± standard error of the mean of at least three independent experiments. NC: Blank control; sh-NC: Negative sh-RNA for tuftelin 1 mRNA.
Effects of TUFT1 on healing, invasion, and migration of HCC cells
According to the above results, the effects of higher or lower level of TUFT1 on the healing, invasion, and migration of HCC cells were further observed. The effects of different TUFT1 levels on the healing, invasion, and migration of the MHCC-97H or Hep3B cells are shown in Figure 5. The healing abilities of the MHCC-97H cells transfected with the TUFT1-shRNA3 plasmid were averaged down to 50% in the sh-NC group (Figure 5A), and the healing abilities in the sh-RNA3 group were significantly inhibited compared with the sh-NC group (P < 0.001, Figure 5A1). However, the healing abilities of the Hep3B cells transfected with the over-expressing TUFT1 plasmid were averaged up to 200% in the sh-NC group (Figure 5B); and the healing abilities of the Hep3B cells were significantly improved compared with the sh-NC group (P < 0.001, Figure 5B1). Compared with the sh-NC group, the migration or invasion abilities of the MHCC-97H cells were significantly suppressed (Figure 5C); and the migration or invasion abilities of the MHCC-97H cells were down to 50% or 30% less than those in the sh-NC group (P < 0.001, Figure 5C1). However, the migration or invasion abilities of the Hep3B cells were significantly increased compared with the sh-NC group (Figure 5D); and the migration and invasion abilities of the Hep3B cells were up to 200% or 300% more than those in the sh-NC group [P < 0.001, (Figure 5D1)].
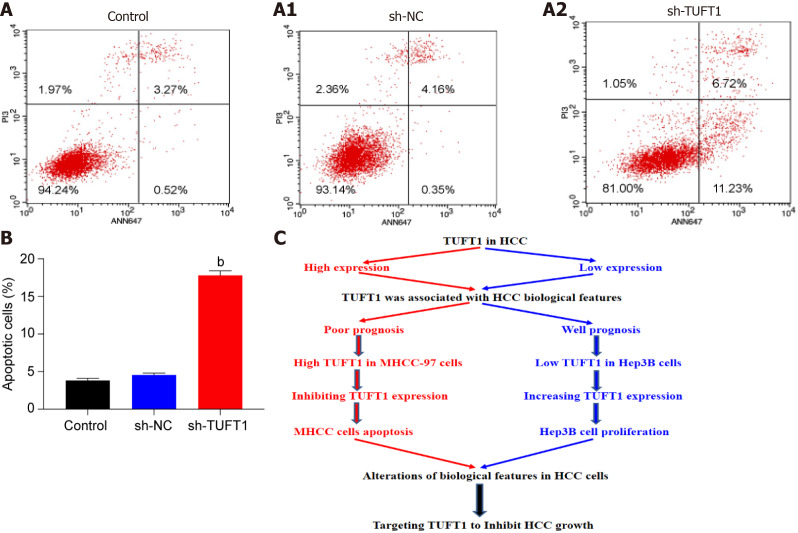
Anti-apoptotic mechanism of TUFT1
After the MHCC-97H cells were transfected with the most effective TUFT1-shRNA3 plasmid, the relationship between TUFT1 expression and apoptosis of MHCC-97H cells was determined (Figure 6). After the MHCC-97H cells were transfected with the more specific TUFT1-shRNA3 plasmid, the numbers of the apoptotic cells in the sh-TUFT1 group (Figure 6A2) were markedly increased than those in the control (Figure 6A) or sh-NC groups (Figure 6A1). The ratio of the apoptotic cells in the sh-TUFT1 group was significantly higher (P < 0.001, Figure 6B) than those in the control or sh-NC groups.
Figure 6.
Possible mechanism of tuftelin 1 expression in hepatocellular carcinoma cells progression. A: The MHCC-97H cells were transfected with the tuftelin 1 (TUFT1)-short hairpin RNA (sh-RNA)3 plasmid, and the apoptotic cells in the sh-TUFT1 group (A2) were increased more than those in the blank control (NC) (A) or negative sh-RNA for tuftelin 1 mRNA (sh-NC) group (A1); B: The histograms of the apoptotic cells among the different groups, the rate of the MHCC-97H cell apoptosis in the sh-TUFT1 group was significantly higher (bP < 0.001) than those in the NC or sh-NC group. Data are presented as mean ± standard error of the mean of at least three independent experiments; C: Speculation of possible mechanisms of TUFT1 promotion of the progression of hepatocellular carcinoma (HCC). Aberrant TUFT1 activation promoted the proliferation and metastasis via anti-apoptosis of HCC cells.
Speculations based on the above data and the possible mechanism of TUFT1 promoting the progression of HCC are shown in Figure 6C. Aberrant up-regulating or down-regulating expressions of TUFT1 was associated with the biological features of HCC. Increased hepatic TUFT1 expression promoted the proliferation, growth, and metastasis of HCC by inhibiting tumor cell apoptosis.
DISCUSSION
Carcinogenesis of hepatocytes is a multi-factor, multi-step, and complex process with major risk factors including chronic infections with HBV or hepatitis C virus, exposure to dietary toxin, hereditary liver disease, and non-alcoholic fatty liver disease of any etiology[2,27]. Despite the urgent need for molecular targeted anti-HCC agents, the therapeutic strategies are difficult to be identified. Accumulating data have demonstrated that many kinds of abnormal signaling pathways are associated with HCC by interacting with highly complex genetic aberrations such as abnormal methylation, histone modification, and chromosome remodeling[28]. Recently, TUFT1 has been shown to exist in many tissues and has been identified as a useful biomarker associated with poor prognosis in several cancers[29-31]. However, only limited studied were available on the relationship between TUFT1 and HCC. In this study, the alterations of TUFT1 expression in HCC tissues and if the oncogenic TUFT1 might promote HCC growth and metastasis via an anti-apoptotic mechanism were investigated.
TUFT1 was originally discovered and mostly studied in the tooth, where it plays an important role in the initial stages of the mineralization of ectodermal enamel[12,29]. Even though TUFT1 is widely distributed and expressed in different embryonic and adult tissues, TUFT1 has been shown to be associated with cellular adaptation to hypoxia and recently even with cell differentiation[32]. TUFT1 activation is related to the progression of tumors, and abnormal TUFT1 has been reported in breast, lung, pancreatic, and thyroid carcinomas[10,11,13]. TUFT1 expression was significantly higher in the HCC tissues than in non-HCC tissues, with invasion and metastasis of tumors[14]. In this study, the comparative analysis of TUFT1 in HCC and non-HCC tissues showed high TUFT1 expression related to tumor size, vascular invasion, advanced TNM stage of HCC, and HBV replication or ascites formation of HCC patients, suggesting that abnormal TUFT1 activation could play oncogenic roles in the progression of HCC.
TUFT1 expression in tissue hypoxia was associated with activations of signaling pathways and poor outcome of patients[18,19]. Previous studies have indicated high HIF-1α level in the hypoxic microenvironment of HCC[33,34] because of tissue hypoxia inducing HIF-1α expression and binding to TUFT1 promoter, thereby promoting tumor progression via the activation of the Rac1/β-catenin, mitogen activated protein kinase, calcium/phosphatidylinositol 3 kinase/AKT, Akt- mammalian target of rapamycin/glycogen synthase kinase 3β, and HIF1-SNAIL signaling pathway[11,14,18]. In this study, TUFT1 level was shown to be positively correlated with tumor size, histological grade, and lymph node metastasis rate, and elevated TUFT1 levels were related to unfavorable clinicopathologic characteristics and poor survival. Furthermore, the univariate and multivariate analysis of clinical data indicated that TUFT1 could be an independent prognostic marker for HCC.
The levels of inhibiting or expressing TUFT1 might alter the biological features of HCC cells[12,23]. In this study, evidence from different cell line studies in vitro suggested that the higher or lower TUFT1 level could affect the biological behaviors of HCC cells. The MHCC-97H cells with high TUFT1 expression were transfected with the specific interfering shRNA3 plasmid, and the proliferation, healing, invasion, and migration abilities of the cells were significantly inhibited less than those in the sh-NC group. However, the Hep3B cells with low TUFT1 expression were transfected with the over-expressing TUFT1 plasmid, and those abilities of the cells were significant increased compared to those in the sh-NC group. These results have confirmed that TUFT1 could promote the growth and metastasis of HCC cells in vitro via an anti-apoptosis mechanism, suggesting that TUFT1 might function as a potential therapeutic target for inhibiting HCC growth.
CONCLUSION
In conclusion, high TUFT1 levels based on human specimen studies were associated with HCC progression. Further inhibiting TUFT1 in vitro could significantly alleviate the proliferation and metastasis of HCC cells. Moreover, TUFT1 over-expression could promote the proliferation and metastasis of HCC cells in vitro. Furthermore, TUFT1 inhibition led to significant changes of cell apoptosis rate. More studies in future are needed to clarify the molecular mechanisms underlying the TUFT1 up-regulation contribution to the progression of HCC. However, high expression of TUFT1 could be considered as a novel effective molecular target for inhibiting HCC growth.
ARTICLE HIGHLIGHTS
Research background
Tuftelin 1 (TUFT1) has been reported to be elevated in multiple cancer types, and its abnormal expression has attracted medical attention as a novel oncogenic biomarker. However, the relationship between TUFT1 and hepatocellular carcinoma (HCC) remains to be identified. In this study, the levels of TUFT1 in HCC tissues were investigated and the roles of TUFT1 expression were explored in vitro by intervening in TUFT1 activation of HCC cell lines.
Research motivation
TUFT1 was first identified and sequenced from a bovine ameloblast-enriched complementary DNA library with a highly conserved gene localized in chromosome 1q21-31 that contained 13 exons, encoding an acidic, phosphorylated glycoprotein of 390 amino acids, and it plays a critical role in the development and mineralization of enamel. Recently, TUFT1 has been reported to be elevated in multiple cancers, and abnormal TUFT1 has attracted medical attention for HCC. However, limited data is available on the relationship between TUFT1 and HCC, and the exact biological mechanism of TUFT1 is still poorly understood in HCC.
Research objectives
The pathogenesis and feasible therapeutic treatments of HCC need urgent investigation. Abnormal TUFT1 has been reported in multiple cancers, and it exhibits oncogenic roles in tumor progression. In this study, the landscape of TUFT1 expression in human HCC tissues was investigated, and its mechanism was confirmed through intervening or over-expressing TUFT1 activation in vitro.
Research methods
TUFT1 on databases of the Cancer Genome Atlas and Oncomine or in HCC tissues were analyzed for clinicopathological features, overall survival, and disease-free survival. High and low expressing HCC cell lines were screened among different HCC cell lines and transfected with constructed vectors that interfere or over-express TUFT1 to analyze biological behaviors. Proliferation, invasion, migration, and apoptosis of cells were detected by CCK-8, scratch assay, transwell tests, and flow cytometry, respectively, and confirmed by Western blotting.
Research results
The research results confirmed that TUFT1 was over-expressed in HCC tissues, and its expression was significantly related to tumor size, vascular invasion, positive hepatitis B e-antigen, advanced tumor-node-metastasis staging, ascites, shorter overall survival, and disease-free survival of HCC patients. Novel findings were that interfering with TUFT1 gene transcription could markedly suppress the proliferation and metastasis of the higher TUFT1 MHCC-97H cell lines through an apoptosis mechanism. Moreover, over-expressing TUFT1 promoted the growth and metastasis of the lower TUFT1 Hep3B cell lines in vitro.
Research conclusions
Based on human specimen studies, high TUFT1 levels were associated with HCC progression, inhibiting TUFT1 affected proliferation and metastasis, over-expressing TUFT1 promoted proliferation and metastasis, and interfering activation of TUFT1 led to significant increase in cell apoptosis. TUFT1 could be a novel useful target for HCC effective therapy.
Research perspectives
Basic and clinical studies have confirmed that the alterations of oncogenic TUFT1 expression might promote HCC growth and metastasis via an anti-apoptotic mechanism, and abnormal TUFT1 level could be a new prognostic marker or potential molecular target for HCC therapy. More studies in the future are needed to clarify the molecular mechanisms underlying TUFT1 up-regulation contribution to the progression of HCC.
ACKNOWLEDGEMENTS
All authors thank Professor Zhi-Zhen Dong of Nantong University for comments on earlier drafts of the manuscript.
Footnotes
Institutional review board statement: This study was approved by the Ethics Committee of the Affiliated Hospital of Nantong University (TDFY2018-025), China.
Conflict-of-interest statement: No benefits in any form have been received or will be received from a commercial party related directly or indirectly to the subject of this article.
Manuscript source: Unsolicited manuscript
Corresponding Author's Membership in Professional Societies: Chinese Society of Gastroenterology, No. 1006110078.
Peer-review started: December 14, 2020
First decision: December 27, 2020
Article in press: May 19, 2021
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Kanda T, Zapater P S-Editor: Wang JL L-Editor: Filipodia P-Editor: Wang LL
Contributor Information
Meng-Na Wu, Research Center of Clinical Medicine, The Affiliated Hospital of Nantong University, Nantong 226001, Jiangsu Province, China.
Wen-Jie Zheng, Research Center of Clinical Medicine, The Affiliated Hospital of Nantong University, Nantong 226001, Jiangsu Province, China.
Wen-Xin Ye, Department of Medical Immunology, Medical School of Nantong University, Nantong 226001, Jiangsu Province, China.
Li Wang, Department of Medical Informatics, Medical School of Nantong University, Nantong 226001, Jiangsu Province, China.
Ying Chen, Department of Oncology, The Affiliated Second Hospital of Nantong University, Nantong 226001, Jiangsu Province, China.
Jie Yang, Department of Molecular Biology, Life Science School of Nantong University, Nantong 226009, Jiangsu Province, China.
Deng-Fu Yao, Research Center of Clinical Medicine, The Affiliated Hospital of Nantong University, Nantong 226001, Jiangsu Province, China. yaodf@ahnmc.com.
Min Yao, Department of Medical Immunology, Medical School of Nantong University, Nantong 226001, Jiangsu Province, China.
Data sharing statement
No additional data are available.
References
- 1.Singal AG, Lampertico P, Nahon P. Epidemiology and surveillance for hepatocellular carcinoma: New trends. J Hepatol. 2020;72:250–261. doi: 10.1016/j.jhep.2019.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen JG, Zhu J, Zhang YH, Zhang YX, Yao DF, Chen YS, Lu JH, Ding LL, Chen HZ, Zhu CY, Yang LP, Zhu YR, Qiang FL. Cancer survival in Qidong between 1972 and 2011: A population-based analysis. Mol Clin Oncol. 2017;6:944–954. doi: 10.3892/mco.2017.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petruzziello A. Epidemiology of Hepatitis B Virus (HBV) and Hepatitis C Virus (HCV) Related Hepatocellular Carcinoma. Open Virol J. 2018;12:26–32. doi: 10.2174/1874357901812010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Younossi Z, Tacke F, Arrese M, Chander Sharma B, Mostafa I, Bugianesi E, Wai-Sun Wong V, Yilmaz Y, George J, Fan J, Vos MB. Global Perspectives on Nonalcoholic Fatty Liver Disease and Nonalcoholic Steatohepatitis. Hepatology. 2019;69:2672–2682. doi: 10.1002/hep.30251. [DOI] [PubMed] [Google Scholar]
- 5.Chen S, Cao Q, Wen W, Wang H. Targeted therapy for hepatocellular carcinoma: Challenges and opportunities. Cancer Lett. 2019;460:1–9. doi: 10.1016/j.canlet.2019.114428. [DOI] [PubMed] [Google Scholar]
- 6.Tang A, Hallouch O, Chernyak V, Kamaya A, Sirlin CB. Epidemiology of hepatocellular carcinoma: target population for surveillance and diagnosis. Abdom Radiol (NY) 2018;43:13–25. doi: 10.1007/s00261-017-1209-1. [DOI] [PubMed] [Google Scholar]
- 7.Chung W, Kim M, de la Monte S, Longato L, Carlson R, Slagle BL, Dong X, Wands JR. Activation of signal transduction pathways during hepatic oncogenesis. Cancer Lett. 2016;370:1–9. doi: 10.1016/j.canlet.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin YH, Wu MH, Yeh CT, Lin KH. Long Non-Coding RNAs as Mediators of Tumor Microenvironment and Liver Cancer Cell Communication. Int J Mol Sci. 2018;19 doi: 10.3390/ijms19123742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qin Y, Liao P. Hepatitis B virus vaccine breakthrough infection: surveillance of S gene mutants of HBV. Acta Virol. 2018;62:115–121. doi: 10.4149/av_2018_210. [DOI] [PubMed] [Google Scholar]
- 10.Zhou B, Zhan H, Tin L, Liu S, Xu J, Dong Y, Li X, Wu L, Guo W. TUFT1 regulates metastasis of pancreatic cancer through HIF1-Snail pathway induced epithelial-mesenchymal transition. Cancer Lett. 2016;382:11–20. doi: 10.1016/j.canlet.2016.08.017. [DOI] [PubMed] [Google Scholar]
- 11.Liu W, Chen G, Sun L, Zhang Y, Han J, Dai Y, He J, Shi S, Chen B. TUFT1 Promotes Triple Negative Breast Cancer Metastasis, Stemness, and Chemoresistance by Up-Regulating the Rac1/β-Catenin Pathway. Front Oncol. 2019;9:617. doi: 10.3389/fonc.2019.00617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delgado S, Deutsch D, Sire JY. Evolutionary Analysis of the Mammalian Tuftelin Sequence Reveals Features of Functional Importance. J Mol Evol. 2017;84:214–224. doi: 10.1007/s00239-017-9789-5. [DOI] [PubMed] [Google Scholar]
- 13.Liu W, Zhang L, Jin Z, Zhao M, Li Z, Chen G, Sun L, Chen B. TUFT1 is expressed in breast cancer and involved in cancer cell proliferation and survival. Oncotarget. 2017;8:74962–74974. doi: 10.18632/oncotarget.20472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu H, Zhu J, Mao Z, Zhang G, Hu X, Chen F. Tuft1 promotes thyroid carcinoma cell invasion and proliferation and suppresses apoptosis through the Akt-mTOR/GSK3β signaling pathway. Am J Transl Res. 2018;10:4376–4384. [PMC free article] [PubMed] [Google Scholar]
- 15.Shilo D, Cohen G, Blumenfeld A, Goren K, Hanhan S, Sharon S, Haze A, Deutsch D, Lazarovici P. Tuftelin Is Required for NGF-Induced Differentiation of PC12 Cells. J Mol Neurosci. 2019;68:135–143. doi: 10.1007/s12031-019-01292-1. [DOI] [PubMed] [Google Scholar]
- 16.Kawasaki N, Isogaya K, Dan S, Yamori T, Takano H, Yao R, Morishita Y, Taguchi L, Morikawa M, Heldin CH, Noda T, Ehata S, Miyazono K, Koinuma D. TUFT1 interacts with RABGAP1 and regulates mTORC1 signaling. Cell Discov. 2018;4:1. doi: 10.1038/s41421-017-0001-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma C, Nie ZK, Guo HM, Kong Y. MiR-671-5p plays a promising role in restraining osteosarcoma cell characteristics through targeting TUFT1. J Biochem Mol Toxicol. 2020;34:e22490. doi: 10.1002/jbt.22490. [DOI] [PubMed] [Google Scholar]
- 18.Dou C, Zhou Z, Xu Q, Liu Z, Zeng Y, Wang Y, Li Q, Wang L, Yang W, Liu Q, Tu K. Hypoxia-induced TUFT1 promotes the growth and metastasis of hepatocellular carcinoma by activating the Ca2+/PI3K/AKT pathway. Oncogene. 2019;38:1239–1255. doi: 10.1038/s41388-018-0505-8. [DOI] [PubMed] [Google Scholar]
- 19.Qin Y, Liu HJ, Li M, Zhai DH, Tang YH, Yang L, Qiao KL, Yang JH, Zhong WL, Zhang Q, Liu YR, Yang G, Sun T, Yang C. Salidroside improves the hypoxic tumor microenvironment and reverses the drug resistance of platinum drugs via HIF-1α signaling pathway. EBioMedicine. 2018;38:25–36. doi: 10.1016/j.ebiom.2018.10.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li ZJ, Li JS, Li DY, Pu T. [Effect of TUTF1 expression on the proliferation, apoptosis and prognosis of hepatocellular carcinoma cells] Zhonghua Gan Zang Bing Za Zhi. 2019;27:879–884. doi: 10.3760/cma.j.issn.1007-3418.2019.11.011. [DOI] [PubMed] [Google Scholar]
- 21.Tang Z, Li C, Kang B, Gao G, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie DY, Ren ZG, Zhou J, Fan J, Gao Q. 2019 Chinese clinical guidelines for the management of hepatocellular carcinoma: updates and insights. Hepatobiliary Surg Nutr. 2020;9:452–463. doi: 10.21037/hbsn-20-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mao Z, Shay B, Hekmati M, Fermon E, Taylor A, Dafni L, Heikinheimo K, Lustmann J, Fisher LW, Young MF, Deutsch D. The human tuftelin gene: cloning and characterization. Gene. 2001;279:181–196. doi: 10.1016/s0378-1119(01)00749-1. [DOI] [PubMed] [Google Scholar]
- 24.Fang M, Yao M, Yang J, Zheng WJ, Wang L, Yao DF. Abnormal CD44 activation of hepatocytes with nonalcoholic fatty accumulation in rat hepatocarcinogenesis. World J Gastrointest Oncol. 2020;12:66–76. doi: 10.4251/wjgo.v12.i1.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qian J, Yao D, Dong Z, Wu W, Qiu L, Yao N, Li S, Bian Y, Wang Z, Shi G. Characteristics of hepatic igf-ii expression and monitored levels of circulating igf-ii mRNA in metastasis of hepatocellular carcinoma. Am J Clin Pathol. 2010;134:799–806. doi: 10.1309/AJCPTFDSE2V3LCZP. [DOI] [PubMed] [Google Scholar]
- 26.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 28.Kanda T, Goto T, Hirotsu Y, Moriyama M, Omata M. Molecular Mechanisms Driving Progression of Liver Cirrhosis towards Hepatocellular Carcinoma in Chronic Hepatitis B and C Infections: A Review. Int J Mol Sci. 2019;20 doi: 10.3390/ijms20061358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Januszyk P, Januszyk K, Wierzbik-Strońska M, Boroń D, Grabarek B. Analysis of the differences in the Expression of mRNAs and miRNAs Asso-ciated with Drug Resistance in Endometrial Cancer Cells Treated with Salinomycin. Curr Pharm Biotechnol. 2020 doi: 10.2174/1389201021666200629151008. [DOI] [PubMed] [Google Scholar]
- 30.Dou C, Sun L, Wang L, Cheng J, Wu W, Zhang C, Xu Q, Tu K, Liu J. Bromodomain-containing protein 9 promotes the growth and metastasis of human hepatocellular carcinoma by activating the TUFT1/AKT pathway. Cell Death Dis. 2020;11:730. doi: 10.1038/s41419-020-02943-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bobek J, Oralova V, Kratochvilova A, Zvackova I, Lesot H, Matalova E. Tuftelin and HIFs expression in osteogenesis. Histochem Cell Biol. 2019;152:355–363. doi: 10.1007/s00418-019-01813-4. [DOI] [PubMed] [Google Scholar]
- 32.Shilo D, Blumenfeld A, Haze A, Sharon S, Goren K, Hanhan S, Gruenbaum-Cohen Y, Ornoy A, Deutsch D. Tuftelin's involvement in embryonic development. J Exp Zool B Mol Dev Evol. 2019;332:125–135. doi: 10.1002/jez.b.22855. [DOI] [PubMed] [Google Scholar]
- 33.Dong ZZ, Yao M, Wang L, Wu W, Gu X, Yao DF. Hypoxia-inducible factor-1alpha: molecular-targeted therapy for hepatocellular carcinoma. Mini Rev Med Chem. 2013;13:1295–1304. doi: 10.2174/1389557511313090004. [DOI] [PubMed] [Google Scholar]
- 34.Li S, Yao D, Wang L, Wu W, Qiu L, Yao M, Yao N, Zhang H, Yu D, Ni Q. Expression characteristics of hypoxia-inducible factor-1α and its clinical values in diagnosis and prognosis of hepatocellular carcinoma. Hepat Mon. 2011;11:821–828. doi: 10.5812/kowsar.1735143X.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No additional data are available.