Abstract
Background:
Heparanase (HPSE) is the only known mammalian enzyme that can degrade heparan sulfate. Heparan sulfate proteoglycans are essential components of the glycocalyx, and maintain physiological barriers between the blood and endothelial cells. HPSE increases during sepsis, which contributes to injurious glyocalyx degradation, loss of endothelial barrier function, and mortality.
Objectives:
As platelets are one of the most abundant cellular sources of HPSE, we sought to determine whether HPSE expression and activity increases in human platelets during clinical sepsis. We also examined associations between platelet HPSE expression and clinical outcomes.
Patients/Methods:
Expression and activity of HPSE was determined in platelets isolated from septic patients (n = 59) and, for comparison, sex-matched healthy donors (n = 46) using complementary transcriptomic, proteomic, and functional enzymatic assays. Septic patients were followed for the primary outcome of mortality, and clinical data were captured prospectively for septic patients.
Results:
The mRNA expression of HPSE was significantly increased in platelets isolated from septic patients. Ribosomal footprint profiling, followed by [S35] methionine labeling assays, demonstrated that HPSE mRNA translation and HPSE protein synthesis were significantly upregulated in platelets during sepsis. While both the pro- and active forms of HPSE protein increased in platelets during sepsis, only the active form of HPSE protein significantly correlated with sepsis-associated mortality. Consistent with transcriptomic and proteomic upregulation, HPSE enzymatic activity was also increased in platelets during sepsis.
Conclusions:
During clinical sepsis HPSE, translation, and enzymatic activity are increased in platelets. Increased expression of the active form of HPSE protein is associated with sepsis-associated mortality.
Keywords: blood platelets, heparanase, inflammation, sepsis, translation
1 |. INTRODUCTION
The luminal surface of endothelial cells is lined with a protective layer consisting of membrane-bound proteoglycans (PGs), glycoproteins, and glycosaminoglycans (GAGs), which play key roles in health and disease. This complex network of glycoconjugates compose the glycocalyx, a cell surface layer that contributes to vascular homeostasis by regulating vessel permeability, nitric oxide production, coagulation, and adhesion of circulating cells (including platelets and leukocytes).1 The backbone of the glycocalyx is made up of PGs, which contain one or more GAG chains, and the sulfated, negatively charged heparan sulfate (HS) polymer accounts for more than 50% of all GAGs within the glycocalyx.2 However, dramatic remodeling of the vessel surface occurs in response to systemic inflammation, such as is the case during sepsis. Degradation or “shedding” of glycocalyx components results in the loss of protective functions and leads to enhanced vascular permeability, tissue edema, leukocyte adhesion, and platelet aggregation.3– 5 Degradation products of the glycocalyx such as HS and hyaluronan are also increasingly recognized as highly potent damage-associated molecular patterns (DAMPs), which can function as significant contributors to critical illnesses and may also serve as diagnostic and therapeutic targets in septic patients.6,7 Dysfunction of the glycocalyx is thought to have an early role in clinical sepsis progression and leads to the lethal coagulopathy associated with acute kidney injury, respiratory failure, and hepatic dysfunction observed during the course of critical illness.5 Pre-clinical models support these clinical observations as therapeutic strategies to protect or restore the glycocalyx improve survival in mouse models of sepsis.2,8
Heparanase (HPSE) is the only known mammalian endo- β- glucuronidase capable of degrading HS9 and cleaves the GAG polymer within highly sulfated regions.10– 12 HPSE is synthesized by cells as an inactive, glycosylated 65 kDa pro- enzyme13 that interacts with heparin sulfate proteoglycans (HSPGs) at the cell surface following secretion.14 HPSE pro-enzyme may either remain at the cell surface or become internalized via receptor- mediated endocytosis, where it is targeted to the lysosome and processed into 50 kDa and 8 kDa subunits by cathepsin L.15– 17 These two subunits non-covalently associate to form the active HPSE heterodimer, which can be trafficked back to the cell surface or secreted into the extracellular space. HPSE also has been shown to possess non-enzymatic functions, including cell signaling, adhesion, and differentiation.18,19 HPSE- mediated degradation of the extracellular matrix is known to promote cell migration and release of HS-bound cytokines and growth factors associated with inflammation,20 angiogenesis,21 and tumor metastasis.12 Soluble HS fragments generated by HPSE activity act as endogenous danger signals capable of amplifying the pro-inflammatory response via Toll-like receptor 4 both in vitro and in murine models of sepsis.22–24
HPSE is elevated in many inflammatory diseases characterized by vascular remodeling, including cancer,11,25,26 diabetes,27 and sepsis.28,29 Circulating levels of HS fragments, and corresponding HPSE activity, are increased in plasma collected from patients with clinical sepsis,7,30 and contribute to disease pathogenesis and progression in pre-clinical studies. Murine models of polymicrobial sepsis demonstrate that HPSE enhances glycocalyx shedding and increases sepsis-associated mortality8 and that HPSE-mediated glycocalyx degradation exposes endothelial surface adhesion molecules including intracellular adhesion molecule 1 and vascular cell adhesion molecule 1, and promotes neutrophil adhesion to the pulmonary endothelial surface.30 HPSE also mediates septic renal dysfunction in mice by augmenting renal inflammatory gene expression independently of neutrophil recruitment,29 suggesting that the mechanisms by which HPSE activity mediates organ injury in sepsis can vary. Importantly, HPSE inhibition in these studies leads to preservation of HS within the glycocalyx and attenuated sepsis-induced injury, supporting observations that HPSE directly contributes to disease progression.
Although platelets are anucleate, it is now clear that the platelet transcriptome and proteome are dynamic and become altered during the course of disease.31– 33 During sepsis, platelet transcriptome, translational activity, and functional responses are altered and, in some cases, are associated with increased mortality.34– 37 Sepsis survivors have an increased risk of thromboembolic events,38 and overexpression of HPSE in transgenic mice is associated with increased platelet adhesion, spreading on fibrinogen, surface P-selectin expression, and enhanced thrombosis in a FeCl3 model of carotid artery injury.39 While many studies of HPSE in sepsis have focused on endothelial expression of HPSE, platelets are recognized as one of the most abundant cellular sources of HPSE25,40,41 yet the expression and function of platelet HPSE during sepsis remains largely unstudied.
Given the potential of platelet-derived HPSE to enhance to platelet reactivity, glycocalyx degradation, and sepsis severity, in this report we apply complementary techniques of RNA-seq, ribosome footprint profiling, metabolic labeling, and functional enzymatic assays to examine whether sepsis is associated with changes in platelet HPSE and clinical outcomes.
2 |. METHODS
2.1 |. Participant recruitment
During the course of enrolling septic patients, we amended our protocol to match updated definitions of sepsis.42 When we first began our study, we recruited septic patients who had either a diagnosis of sepsis (any cause) or septic shock defined as a clinical construct of sepsis with persisting hypotension requiring vasopressors to maintain mean arterial pressure ≥ 65 mmHg and having a serum lactate level >2 mmol/L (18 mg/dl) despite adequate volume resuscitation. We later amended our protocol to conform with updated definitions of sepsis. For this amended protocol, septic patients were eligible if they had a suspected or confirmed infection and a defined sequential organ failure assessment (SOFA) score ≥2. Septic shock was defined as having a lactate of >2 mmol/L (18 mg/dl) despite adequate volume resuscitation and/or the use of vasopressors. All septic patients were enrolled within 72 h of intensive care unit (ICU) admission. Table 1 indicates patient characteristics and Figure S1 in supporting information indicates known causes of sepsis within our population by site of infection (Figure S1A) and identified pathogen (Figure S1B).
TABLE 1.
Characteristics of septic patients and healthy donors. Septic patients were prospectively enrolled within 72 h of ICU admission
| Healthy (N = 46) | Septic (N = 59) | p- value | |
|---|---|---|---|
| Male sex, n (%) | 24 (52.2%) | 30 (50.9%) | .9790 |
| Age, years | 41.6 ± 6.1 | 54.5 ± 2.3 | .0002 |
| Platelet count, K/μl | — | 211.9 ± 10.6 | - |
| White blood cell count, K/μl | — | 14.7 ± 1.0 | - |
| Mortality, n (%) | — | 5 (8.5%) | - |
| Hemoglobin, mg/dl | — | 10.9 ± 0.3 | - |
| Hypotension requiring Vasopressors, n (%) | — | 33 (55.9%) | - |
| Ventilated, n (%) | — | 21 (35.6%) | - |
| APACHE II | — | 17.5 ± 4.0* | - |
| SOFA | — | 5.4 ± 0.9* | - |
Abbreivations: APACHE, Acute Physiologic Assessment and Chronic Health Evaluation; ICU, intensive care unit; SOFA, sequential organ failure assessment.
APACHE II scores were missing for 1 septic patient and SOFA scores were missing for 3 septic patients. Data is expressed as the average ± the standard deviation, unless otherwise specified.
Healthy male and female consenting donors were eligible if they were above the age of 18, medication- free, and without any acute or chronic illnesses. Healthy donors were matched to septic patients on the variable of sex. Local institutional review boards approved all of the studies, and each participant or a legally authorized representative provided written, informed consent.
For selected transcriptomic comparisons of HPSE expression in platelets, we also studied three other acutely infected cohorts (and matched healthy donors) in addition to septic patients: (1) patients with primary influenza A/H1 N1, (2) patients with dengue, and (3) patients with malaria. Influenza A/H1 N1 patients were recruited during the 2009 influenza pandemic in Salt Lake City, Utah. Influenza A/H1 N1 was diagnosed on an appropriate respiratory specimen by real- time polymerase chain reaction (RT-P CR). All influenza A/H1 N1 patients were studied within 72 h of ICU admission. Age- , sex- , and race-matched healthy donors from the Salt Lake City area were recruited for transcriptomic comparisons, as previously reported.36
Dengue patients were defined as patients with acute, primary, or secondary dengue infection, and were recruited from the Instituto Nacional de Infectologia Evandro Chagas, Fundação Oswaldo Cruz in Rio de Janeiro, Brazil. Dengue infection was confirmed serologically and/or molecularly using ELISA for immunoglobulin M and G quantification (PanBio) and RT-PCR, respectively. Age- , sex- , and race- matched healthy donors from the same area in Brazil were recruited for transcriptomic comparisons, as previously reported.36
Malaria patients infected by Plasmodium vivax (P. vivax) were recruited at the Fundacao de Medicina Tropical - Dr. Heitor Vieira Dourado (FMT-HVD), a tertiary care center for infectious diseases in Manaus, Amazonas State, Brazil. P. vivax was diagnosed by quantitative PCR (qPCR) analysis, according to recommended diagnostic guidelines.43,44 Age- , sex- , and race- matched healthy donors from the same area in Brazil were recruited for transcriptomic comparisons.
2.2 |. Platelet isolation
Whole blood was drawn into 3.2% acid- citrate-dextrose (ACD) tubes and centrifuged at 100 g for 20 min at room temperature (RT) to obtain platelet rich plasma (PRP); 100 nM prostaglandin E1 (PGE1; Cayman Chemicals) was added to the PRP. The PRP was then centrifuged again at 500 g for 20 min at RT. The platelet pellet was then resuspended in phosphate-buffered saline (PBS) containing 2 mM EDTA, 0.5% human albumin, and 100 nM PGE1. Anti-CD45 beads were added for 20 min at RT to bind leukocytes and then the negative fraction (e.g., platelets) was isolated with an Automax Pro (Miltenyi) for sepsis and H1 N1 patients or EasySep (StemCell) for dengue and malaria patients according to the manufacturer’s recommendations. Isolated platelets were then centrifuged at 500 g for 20 min at RT and resuspended in warmed (37°C) Medium 199 (M199 Lonza, Biologics). This method results in a highly purified population of platelets, generally with fewer than 3 leukocytes per 107 platelets (>99.9% purity).45
2.3 |. Platelet transcriptomics
For RNA-seq studies, 1 × 109 platelets were lysed in TRIzol and then DNase treated. Total RNA was isolated and an Agilent bioanalyzer was used to quantify the amount and quality of RNA. RNA-seq libraries were prepared with TruSeq V2 with oligo- dT selection (Illumina). Reads were aligned (Novoalign) to the reference genome at the time of these studies and a pseudotranscriptome containing splice junctions. The Deseq2 analysis package was used to assign reads to composite transcripts (one per gene) and quantitate fragments per kilobase of transcript per million mapped reads (FPKMs) as previously described.45 Post-RNA-seq analysis of an index leukocyte transcript (PTPRC; CD45) confirmed that the samples were highly purified platelet preparations. We detected equivalent PTPRC levels between control and sepsis samples (p > .05) that were >5000 times lower in FPKM than the index platelet transcript PF4. Differential expression and significance were determined according to the Deseq2 algorithm, which controls for multiple comparisons using a false discovery rate (FDR) according to the Benjamini-Hochberg procedure.46 Significant variance in expressed transcripts were prespecified as those transcripts with an FDR < 0.05 and a log2 fold change ≥1.5 in sepsis, compared to non-septic conditions. The raw RNA-seq data for sepsis, influenza A/H1 N1, and dengue patients were published and made publicly available by our group previously.35,36
For qRT-PCR experiments, 5 × 108 platelets from septic patients and healthy donors were isolated and lysed in Trizol. RNA was isolated and measured using a Nanodrop 2000 (ThermoFisher Scientific). The RNA was converted into cDNA using SuperScript II. cDNA was analyzed on an iCycler from BioRad. Samples were analyzed for HPSE and tubulin expression (tubulin served as a control). The human HPSE forward primer is CTTGGGAACTAGGCAATGAACC and the reverse primer is CCTTCAGGAAGCTCTTCAGCAT.
2.4 |. Ribosome footprinting
For ribosome footprint profiling experiments, 1 × 109 platelets were treated with cycloheximide (100 mg/ml) for 1 min at RT to preserve ribosomes as natively attached to mRNAs.35 The platelets were lysed in Mammalian Lysis Buffer (ARTseq, Epicentre). Total RNA and ribosome-protected read RNAs (RPRs) were prepared per the manufacturer’s instructions (ARTseq-R ibosome Profiling Kit, Epicentre). Sequencing libraries (25 pM) were chemically denatured and applied to an Illumina HiSeq v4 single-read flow cell using an Illumina cBot. Hybridized molecules were clonally amplified and annealed to sequencing primers with reagents from an Illumina HiSeq SR Cluster Kit v4-cBot (GD- 401– 4001). Following transfer of the flow cell to an Illumina HiSeq 2500 instrument (HCSv2.2.38 and RTA v1.18.61), a 50-cycle single-read sequence run was performed using HiSeq SBS Kit v4 sequencing reagents (FC- 401– 4002). Ribosome profiling (eukaryote) library preparation was completed using the NEBNext Multiplex Small RNA Library Prep Set.
2.5 |. ELISA
5 × 108 platelets were isolated from septic patients and healthy donors, suspended in 500 μl M199, and stimulated with a total of 0.01 units/mL of thrombin (Sigma) for 1 h at 37°C. Platelets were centrifuged at 12,000 g for 2 min at RT to separate the cell pellet from the supernatant. Cell lysates and supernatants were collected. Cell lysates were washed with PBS and resuspended in 1X PBS. Platelets were lysed via snap freezing the samples three times in liquid nitrogen. Phosphatase and protease inhibitors (ThermoFisher) were added to prevent protein degradation. Total protein levels of HPSE in lysed platelets and supernatants were measured via ELISA (LSBio), according to the manufacturer’s recommendations.
2.6 |. De novo protein synthesis
Platelets were isolated from septic patients and healthy donors in parallel as described above. In addition, platelets were leukocyte and red blood cell depleted (Glycophorin A beads). 6 × 108 platelets, at a concentration of 2 × 108/ml, were resuspended in warmed (37°C) methionine- free M199 media and then incubated with S[35] methionine at 37°C for 8 h. Next, platelets were centrifuged down through centrifugation at 12,000 g for 2 min at RT and carefully washed three times with 1X PBS. The cell lysates were collected and lysed via snap freezing the samples three times in liquid nitrogen. HPSE was captured and bound using the antibody in ELISA as described above. HPSE from the platelet lysate was removed from the plate using a combination of TRIzol and 1x Laemmli (0.1% 2-Mercaptoethanol, 0.0005% Bromophenol Blue, 10% Glycerol, 2% SDS, and 63 mM Tris- HCl pH 6.8). Scintillation counts were then collected using a LS 6500 multi- purpose scintillation counter from Beckman Coulter.
2.7 |. Western blot
5 × 108 platelets from septic patients and healthy donors were lysed in 1x Laemmli (1% 2-Mercaptoethanol, 0.0005% Bromophenol Blue, 10% Glycerol, 2% SDS, and 63 mM Tris-HCl pH 6.8). Platelets were boiled at 100°C for 10 min, and 1/10 of the sample loaded onto a 10% SDS gel. Protein was transferred onto a nitrocellulose membrane and then blocked using blocking buffer (LiCor) for 60 min at RT with swirling movement. The membrane was incubated with primary antibodies against HPSE (Santa Cruz [since discontinued]) and β- actin (Santa Cruz) overnight at 4°C with swirling movement. Membranes were then washed three times with filtered 1X TBS with 1% Tween- 20. Secondary antibodies (LiCor) were incubated with the membrane for 60 min at RT at a 1/10,000 dilution. Membranes were then washed three times with filtered 1X TBS with 1% Tween- 20 and imaged utilizing the Odyssey LiCor machine. Exposures in displayed gels were selected for optimal visualization as the active form of HPSE is expressed more robustly in human platelets than the pro- form of HPSE and we were unable to find a single exposure that allowed clear visualization of both forms of HPSE protein at the same time. All quantifications and analyses of HPSE protein expression (both the pro- and active forms) between septic patients and healthy controls were done using the equivalent intensities. For quantification, measured densitometry of HPSE bands was first normalized to β- actin (loading control) and then to the average expression of HPSE in healthy subjects.
2.8 |. Heparanase activity assay
Platelets from septic and healthy individuals were isolated and 1 × 109 platelets were centrifuged (12,000 g for 2 min at RT) and washed with 1X PBS. Platelets were lysed in 100 μl of PBS containing phosphate and protease inhibitors (ThermoFisher). Cells were lysed through snap freezing three times in liquid nitrogen. Total protein in the platelet lysates was measured using a Bradford assay. HPSE activity measurements were performed using a fluorescence resonance energy transfer (FRET)-based assay as described previously.47 Polymeric heparin, which contains GlcA—GlcNS6S glycosidic bonds sensitive to HPSE, was labeled using carbodiimide-based linking chemistry to generate a FRET-enabled heparin analog modified with donor-acceptor pairs. Briefly, a 200 μl solution containing either platelet lysate (10 μl or a total of 1 × 108 platelets) or recombinant human HPSE (1 μg/ml; R and D Biotechnology) and the FRET-substrate (66 μM) in assay buffer (20 mM sodium acetate buffer, pH 5.0 containing 100 mM NaCl or 100 mM sodium acetate buffer, pH 4.5) were shaken overnight at 37°C in the dark. The HPSE-specific inhibitor OGT2115 (1 μM; Tocris) was used to measure the specific activity of HPSE. Hydrolysis of the FRET-enabled heparan by HPSE was monitored in 96-well plates on a fluorescence microplate reader FlexStation III (Molecular Devices) and measured at 500 nm (λEX = 340 nm). The observed change in fluorescence was normalized to initial fluorescence to calculate HPSE activity.
2.9 |. Statistical analyses
Summary statistics were used to describe the study cohort, and clinical variables are expressed as the mean ± standard error deviation or as a number and percentage. Parametric 2- tailed Student’s t tests or analysis of variance was used for continuous variables, and the χ2 and Fisher’s exact test were used for categorical variables. Statistical analyses were performed using GraphPad Prism version 7 software (GraphPad Software). A two-tailed P < .05 was considered statistically significant. In clinical assays with smaller sample sizes, data are shown as median (interquartile range [IQR]).
3 |. RESULTS
3.1 |. HPSE mRNA expression is increased in platelets isolated from septic patients
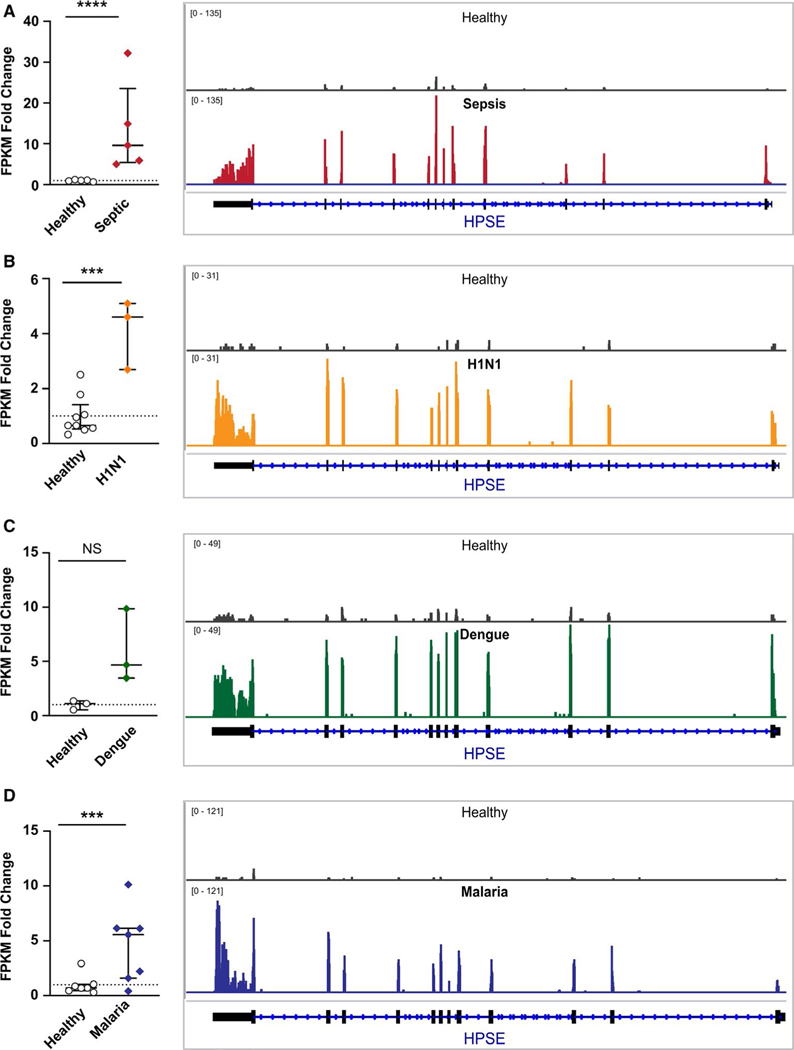
As a key modulator of the glycocalyx, HPSE plays a role in the severity of many illnesses.2,5,8 Therefore, we first examined and compared the expression of HPSE mRNA using RNA-sequencing of platelets isolated from four cohorts of patients with acute infectious diseases (sepsis, influenza A/H1 N1, dengue, and P. vivax malaria). HPSE was one of two genes identified that increased in platelets from all four conditions, with the other gene being DNA Mismatch Repair Protein MLH3. While HPSE mRNA expression was increased in each infectious setting compared to sex-matched healthy donors (Figure 1A-D, Table S1 in supporting information), the greatest change was observed in sepsis (~13-fold increase vs. healthy donors, Figure 1A). We thus chose to focus in further detail on the expression and activity of HPSE in platelets during sepsis.
FIGURE 1.
RNA-seq identified that expression of heparanase (HPSE) mRNA is increased in platelets during clinical infectious diseases. Platelets from healthy donors or patients with sepsis (A), influenza A/H1 N1 (B), dengue virus (C), or P. vivax malaria (D) were isolated and HPSE mRNA expression analyzed by RNA-seq. The mRNA expression of HPSE was measured as fragments per kilobase of transcript per million mapped reads (FPKM). Data as shown as the fold-change in patients compared to healthy donors (dotted line). A representative IGVB image of HPSE mRNA expression is shown on the right next to each bar graph, with greater height on the y-axis indicating greater relative mRNA expression. An adjusted p- value (e.g., false discovery rate [FDR]) was utilized for statistical significance according to the Benjamini-Hochberg procedure46 (***≤.0005, ****<.0001)
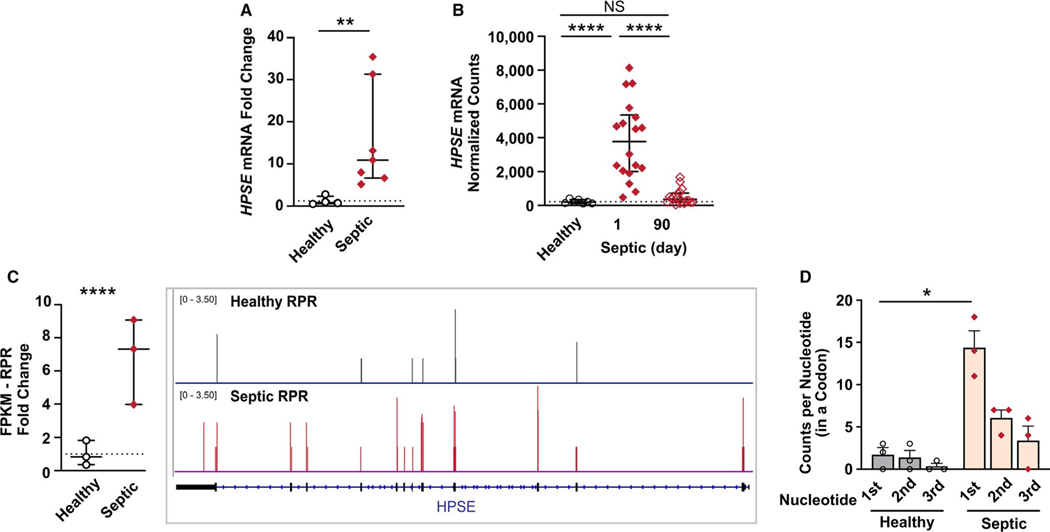
We next examined HPSE expression in an independent cohort of septic patients and sex-matched healthy donors. Using qRT-PCR, we identified approximately a 16-fold increase in HPSE mRNA expression in isolated platelets from septic patients compared to healthy donors (Figure 2A). Previous work demonstrates that changes in platelet gene expression during sepsis may correlate with disease course.35,48 Moreover, acute, sepsis-induced changes in the platelet transcriptome may normalize after patients recover.35 Using a cohort of septic patients followed longitudinally and prospectively, we examined the expression of platelet HPSE mRNA expression acutely during sepsis and in the same patients following recovery (approximately 90 days after their hospitalization for sepsis). While HPSE mRNA expression increased acutely during sepsis (day 1), HPSE mRNA expression in platelets returned to levels generally similar to healthy sex-matched donors ~90 days later (Figure 2B).
FIGURE 2.
Heparanase (HPSE) mRNA expression and ribosomal protected RNAs are significantly and dynamically upregulated in platelets during clinical sepsis. A, Quantitative real-time polymerase chain reaction (qRT-PCR) for HPSE mRNA was performed in platelets isolated from an independent cohort of septic patients or matched healthy donors. The fold change in HPSE expression in septic patients, compared to the average HPSE expression in healthy donors (denoted with a dotted line) was plotted. Statistical significance was measured using an unpaired non-parametric Mann-Whitney statistical analysis (*≤.05; average HPSE expression in healthy subjects is shown with a dotted line). B, RNA-seq analysis was performed on platelets isolated from the septic patients assessed within 72 h of intensive care unit admission for sepsis (Day 1) and then, in the same patients, approximately 90 days later during recovery. Statistical significance was measured using a one-way analysis of variance (***≤.0001; average HPSE expression in healthy subjects is shown with a dotted line). C, Ribosomal protected regions (RPR), an indicator of RNAs undergoing active translation, were increased in platelets from septic patients. The fold-change of HPSE RPR (measured in fragments per kilobase of transcript per million mapped reads [FPKM]) in septic patients, related to average levels in healthy donors (denoted with a dotted line) was plotted. An adjusted P-value was used to determine statistical significance (****<.0001). D, Periodicity of HPSE was used to determine where the ribosome was present on the codon. 1st represents the first nucleic acid in a codon, 2nd the second, and 3rd the third (*<0.05)
3.2 |. HPSE mRNA translation and de novo protein synthesis are increased in platelets during sepsis
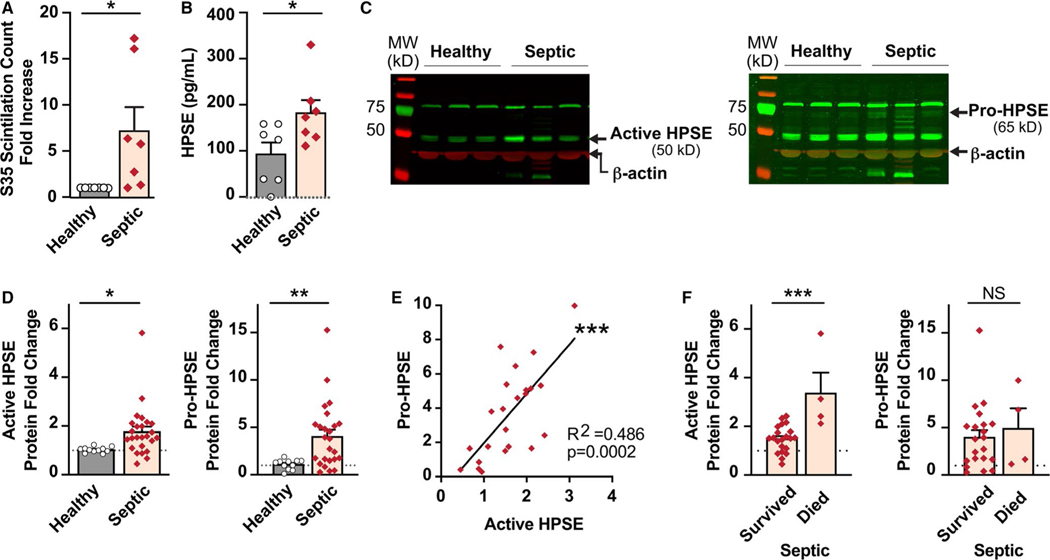
During sepsis, platelets are hyperreactive49 and have increased translational activity.35 As we had identified that HPSE mRNA expression was increased in platelets during sepsis, we next sought to determine if HPSE mRNA translation and de novo protein synthesis were also upregulated. We first examined a previously published data set that sequenced ribosomal protected RNAs in platelets from septic patients.35 We observed significant enrichment in ribosomal protected exons of HPSE mRNA in platelets from septic patients (Figure 2C). HPSE exhibited the expected periodicity with enrichment of the first nucleic acid in a codon, further suggesting that HPSE mRNA was being translated (Figure 2D). To confirm that HPSE mRNA was translated into new protein, we performed [S35] methionine incorporation assays. De novo protein synthesis of HPSE was significantly increased in platelets from septic patients (Figure 3A).
FIGURE 3.
Heparanase (HPSE) mRNA translation and protein synthesis is increased in platelets during sepsis. A, S[35] methionine incorporation assays measured levels of de novo HPSE protein synthesis. Platelets from healthy donors and septic patients were incubated with S[35] for 8 h. HPSE protein was collected and measured for scintillation counts. The fold change of scintillation counts in platelets from septic donors over healthy donors was plotted. B, HPSE protein levels in platelet lysates were detected via ELISA. C, Western blot analyses of HPSE protein allowed for differentiation of the active and pro- forms of HPSE. The active form is detected around 50 kDa (left) while the pro- form is detected around 65 kDa (right). The active form of HPSE is more robustly expressed in platelets than the pro- form of HPSE. Therefore, the right panel shows the same gel with higher intensity. D, Densitometry of the western blot bands was performed and normalized to β-actin expression. E, Correlation plot of the pro- and active forms of HPSE protein expression in platelets from septic patients. F, Active HPSE protein expression was significantly increased in septic patients who did not survive. There were no significant differences in the pro- form of HPSE between septic survivors and non-survivors. (*<.05, **<.005, ***<.0005; average HPSE expression in healthy subjects is shown with a dotted line)
By ELISA, we confirmed that HPSE total protein (including both pro- and active forms of HPSE) was significantly increased in platelets from septic patients (Figure 3B). As ELISA does not differentiate between the pro- and active form of HPSE, we next used western blotting to determine the proportion of HPSE present in active and latent forms. In platelets from septic patients, both the pro- and active forms of HPSE were significantly increased (Figure 3C–D). Increased expression of the pro- and active forms of HPSE in platelets from septic patients were significantly correlated (Figure 3E).
We next sought to determine whether there were any clinical outcomes or features of sepsis associated with increased HPSE protein expression in platelets. In univariate analyses, septic patients who died had modest, but significant, increased levels of the active form of HPSE protein expression compared to septic patients who survived (Figure 3F). In contrast, the pro-form of HPSE protein did not differ between sepsis survivors and non-survivors (Figure 3F). Active HPSE protein expression did not correlate significantly with other variables tested, including platelet counts, patient age or sex, inflammatory or thrombotic markers, or indices of illness severity (Table S2 and Figure S3 in supporting information). While platelets are often activated during clinical sepsis, ex vivo stimulation of platelets did not affect HPSE protein expression in platelets from either septic patients or healthy donors (Figure S2A in supporting information). We also did not see any evidence that HPSE protein was secreted by platelets (Figure S2B, and data not shown), suggesting that HPSE may be retained at the platelet surface upon activation.
3.3 |. HPSE enzymatic activity is increased in platelets during sepsis
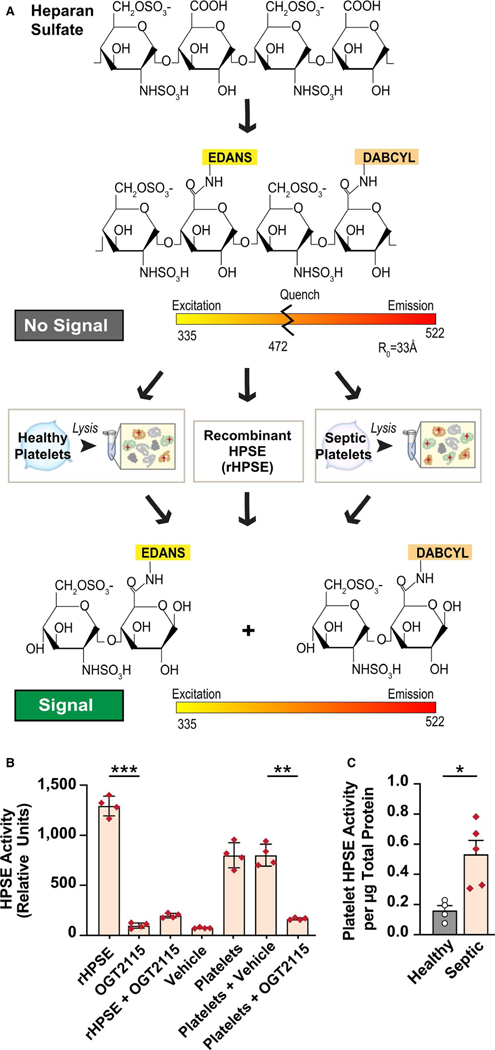
As HPSE expression, translation, and protein synthesis are all increased in platelets during sepsis (Figures 1– 3), we next measured the activity of HPSE in platelets. To determine whether platelet-derived HPSE was active, we labeled heparin, which contains GlcA—GlcNS6S glycosidic bonds sensitive to HPSE, with a fluorescent label and a quencher (schematic shown in Figure 4A). To confirm specificity, we measured HPSE activity in the presence or absence of OGT2115, a known HPSE inhibitor. In control experiments, OGT2115 or vehicle alone demonstrated no detectable activity (Figure 4B) and in the presence of OGT2115, HPSE activity was maximally inhibited as expected (Figure 4B). We next measured HPSE activity in platelets isolated from septic patients and determined that HPSE enzymatic activity was significantly increased in platelets from septic patients compared to healthy controls (Figure 4C), consistent with the observed increase in the active form of HPSE in platelets from these patients (Figure 3).
FIGURE 4.
Heparanase (HPSE) enzymatic activity is increased in platelets from septic patients. HPSE activity was measured through measurement of heparan sulfate degradation using a fluorescence resonance energy transfer-based assay. A, A schematic representation of the HPSE activity assay. We used either platelet lysates or recombinant HPSE (rHPSE, as a positive control) to measure HPSE activity. B, To confirm specificity, HPSE activity was measured in platelet lysates from healthy donors with and without OGT2115 (a specific HPSE inhibitor). C, HPSE activity was measured in platelets from septic patients and healthy donors. Activity was calculated from the readings with subtraction of the background. Statistical analysis was performed using a one-way analysis of variance for (B) and an unpaired student T- test for panel (C; *<.05, **<.005)
4 |. DISCUSSION
During sepsis, platelet gene expression (e.g., transcriptome and proteome) and functional responses are altered.34– 37 Platelets are one of the most abundant cellular sources of HPSE,25,40,41 and HPSE is the only human enzyme known to directly cleave heparan sulfate from HSPGs.9 Whether clinical sepsis is associated with increases in platelet HPSE expression and enzymatic activity has never before been reported, to the best of our knowledge.
Through complementary transcriptomic, proteomic, and biochemical approaches, we identified that HPSE expression is dynamically increased in platelets during clinical sepsis. Moreover, while HPSE mRNA expression is increased in other acute viral and parasitic infectious diseases, changes were greatest in sepsis. Additionally, while both pro- and active forms of HPSE protein were increased in platelets during sepsis, only levels of the active form of HPSE significantly correlated with sepsis-associated mortality. Importantly, we did not find any association between active HPSE expression and other variables that can be associated with sepsis mortality. Nevertheless, we recognize that our study cohort was relatively small and we could not control for all potential confounding variables. Additionally, we acknowledge that our healthy donors and septic patients were not age matched. However, as changes in HPSE expression during sepsis were not associated with age, we do not believe this substantially impacts our findings. We also determined that increased HPSE mRNA and protein expression is accompanied by greater HPSE activity in platelets during sepsis. These data support and extend other clinical and experimental observations demonstrating that while anucleate, platelet gene expression is dynamically altered during sepsis, contributing to alterations in platelet functional responses with other circulating blood cells and tissue beds.35–37
HPSE is a well-conserved enzyme with functions contributing to systemic inflammatory disease at the endothelial surface. Our data raise several interesting implications for the role of platelet-derived HPSE in sepsis. In blood flow, platelets distributed toward the endothelium are in intimate contact with the glycocalyx in circulation where they are poised to recognize components of the extracellular matrix. Enzymatic cleavage of HS chains leaves the proteoglycan core protein susceptible to enzymatic degradation by proteases such as thrombin,50,51 and also reveals cell adhesion molecules and cytokine receptors for binding and activation, thereby enhancing inflammation and coagulation.14,30,52 In endotoxemic mice, HPSE-mediated degradation of the GC increases the availability of cell-adhesion molecules and enhances neutrophil adhesion to the vessel surface, and inhibition of HPSE prevents glycocalyx loss and neutrophil adhesion.30
Unbiased proteomic analysis of vascular surface molecules altered during the course of murine sepsis indicate that platelet degranulation and extracellular matrix remodeling are the top pair of proteomic changes observed in the vasculature.53 Recent reports have demonstrated that HS chains have an unexpected role as regulators of the platelet receptor G6b-B, which functions to attenuate platelet activation and play key roles in platelet production.54 Interestingly, overexpression of HPSE in mice promotes megakaryopoiesis,55 and it is plausible that increased HPSE expression by megakaryocytes could play a direct role in generation of hyperreactive platelets. Apart from the contribution of serglycin to alpha-granule development, PGs remain understudied in the context of platelets and it is plausible that G6b-B interactions with HSPGs at the platelet surface function to restrain inappropriate platelet activation. We speculate that platelet HPSE may contribute to sepsis pathophysiology by directly mediating degradation and shedding of heparan sulfate from vascular surfaces, and may also contribute to control of platelet activation by regulating G6b-B interactions with HS.
The strengths of our study include the clinical nature of our investigations, our interrogation of HPSE mRNA and protein expression, and the use of complementary techniques to examine HPSE transcriptional and translational events in platelets during sepsis. We also employed a specific, and relevant, functional assay to confirm that increased HPSE expression in sepsis was accompanied by greater HPSE activity. In a subset of patients, we were able to study HPSE expression in septic patients acutely during their hospitalization and during recovery. This repeated, longitudinal sampling—which demonstrated that acute increases in HPSE during sepsis normalized in patients after recovery—suggests that upregulation of HPSE was due to sepsis and not due to differences in comorbid conditions or medications between septic patients and healthy donors. This dynamic change in HPSE expression in platelets also parallels findings we recently described for ITGA2B, a subunit of integrin aIIbb3.35 We recognize that given the clinical nature of our studies, we could not mechanistically establish the regulation of HPSE in vivo during sepsis. While the role of endothelial HPSE in regulating the pulmonary endothelial glycocalyx is better defined, our studies shed light on the emerging role of platelets as novel regulators of the glycocalyx in disease.
5 |. CONCLUSIONS
In conclusion, these studies provide new evidence demonstrating that HPSE expression and functional activity is increased dynamically in platelets during clinical sepsis and its expression is associated with adverse outcomes.
Supplementary Material
Essentials.
The expression of heparanase is elevated in platelets during sepsis.
Heparanase protein synthesis is increased in platelets during clinical sepsis.
Heparanase protein activity is increased in platelets during clinical sepsis.
The expression of heparanase protein in platelets is significantly higher in sepsis non-survivors.
ACKNOWLEDGMENTS
This work was supported by the National Heart, Lung, and Blood Institute (HL145237 to Andrew Weyrich, HL142804 to Matthew Rondina and Andrew Weyrich, HL130541 and HL126547 to Matthew Rondina, HL135265 to Aaron Petrey, and HL044525 to Guy Zimmerman). This work was also supported by the National Institute on Aging (AG048022 and AG059877 to Matthew Rondina); the National Institute of General Medical Sciences (GM103806 to Jesse Rowley); and the National Institute of Neurological Disorders and Stroke (U10NS086606 to Robert Campbell). This work was further supported by the German Research Foundation (Research Fellowship KR 4945/1-1 to Krystin Krauel). This work was also supported in part by Merit Review Award Number I01 CX001696 to Matthew Rondina from the U.S. Department of Veterans Affairs Clinical Sciences R&D (CSRD). This material is the result of work supported with resources and the use of facilities at the George E. Wahlen VA Medical Center, Salt Lake City, Utah. The contents do not represent the views of the U.S. Department of Veterans Affairs or the United States Government.
Funding information
National Institute of Neurological Disorders and Stroke, Grant/Award Number: U10NS086606; Deutsche Forschungsgemeinschaft, Grant/Award Number: KR 4945/1-1; National Heart, Lung, and Blood Institute, Grant/ Award Number: HL126547, HL044525, HL130541, HL135265, HL142804 and HL145237; Department of Veterans Affairs Clinical Sciences R&D (CSRD), Grant/Award Number: I01 CX001696; National Institute on Aging, Grant/Award Number: AG048022 and AG059877; National Institute of General Medical Sciences, Grant/Award Number: GM103806
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section.
CONFLICTS OF INTEREST
The authors declare no conflicts or competing financial interests.
REFERENCES
- 1.Sieve I, Munster-Kuhnel AK, Hilfiker-Kleiner D. Regulation and function of endothelial glycocalyx layer in vascular diseases. Vascul Pharmacol. 2018;100:26–33. [DOI] [PubMed] [Google Scholar]
- 2.Reitsma S, Slaaf DW, Vink H, van Zandvoort M, oude Egbrink M. The endothelial glycocalyx: composition, functions, and visualization. Pflügers Arch. 2007;454:345–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nelson A, Berkestedt I, Schmidtchen A, Ljunggren L, Bodelsson M. Increased levels of glycosaminoglecans during septic shock: relation to mortality and the antibacterial actions of plasma. Shock. 2008;30(6):623–627. [DOI] [PubMed] [Google Scholar]
- 4.Martin L, Koczera P, Zechendorf E, Schuerholz T. The endothelial glycocalyx: new diagnostic and therapeutic approaches in sepsis. Biomed Res Int. 2016;2016:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chelazzi C, Villa G, Mancinelli P, De Gaudio AR, Adembri C. Glycocalyx and sepsis-induced alterations in vascular permeability. Crit Care. 2015;19(1):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uchimido R, Schmidt EP, Shapiro NI. The glycocalyx: a novel diagnostic and therapeutic target in sepsis. Crit Care. 2019;23(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmidt EP, Li G, Li L, et al. The circulating glycosaminoglycan signature of respiratory failure in critically ill adults. J Biol Chem. 2014;289(12):8194–8202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song J, Zullo JA, Liveris D, Dragovich M, Zhang XF, Goligorsky MS. Therapeutic restoration of endothelial glycocalyx in sepsis. J Pharmacol Exp Ther. 2017;361:115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vreys V, David G. Mammalian heparanase: what is the message? J Cell Mol Med. 2007;11(3):427–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peterson S, Liu J. Unraveling the specificity of heparanase utilizing synthetic substrates. J Biol Chem. 2010;285(19):14504–14513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vlodavsky I, Friedmann Y, Elkin M, et al. Mammalian heparanase: Gene cloning, expression and function in tumor progression and metastasis. Nat Med. 1999;5(7):793–802. [DOI] [PubMed] [Google Scholar]
- 12.Hulett MD, Freeman C, Hamdorf BJ, Baker RT, Harris MJ, Parish CR. Cloning of mammalian heparanase, an important enzyme in tumor invasion and metastasis. Nat Med. 1999;5(7):803–809. [DOI] [PubMed] [Google Scholar]
- 13.Nadav L, Eldor A, Yacoby-Zeevi O, et al. Activation, processing and trafficking of extracellular heparanase by primary human fibroblasts. J Cell Sci. 2002;115:2179–2187. [DOI] [PubMed] [Google Scholar]
- 14.Levy- Adam F, Feld S, Suss-Toby E, Vlodavsky I, Ilan N. Heparanase facilitates cell adhesion and spreading by clustering of cell surface heparan sulfate proteoglycans. PLoS ONE. 2008;3(6):e2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gingis-Velitski S, Zetser A, Kaplan V, et al. Heparanase uptake is mediated by cell membrane heparan sulfate proteoglycans. J Biol Chem. 2004;279:44084–44092. [DOI] [PubMed] [Google Scholar]
- 16.Zetser A, Levy- Adam F, Kaplan V, et al. Processing and activation of latent heparanase occurs in lysosomes. J Cell Sci. 2004;117:2249–2258. [DOI] [PubMed] [Google Scholar]
- 17.Abboud-Jarrous G, Atzmon R, Peretz T, et al. Cathepsin L is responsible for processing and activation of proheparanase through multiple cleavages of a linker segment. J Biol Chem. 2008;283:18167–18176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masola V, Bellin G, Gambaro G, Onisto M. Heparanase: A Multitasking protein involved in extracellular matrix (ECM) remodeling and intracellular events. Cells. 2018;7(236):192–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roy M, Marchetti D. Cell surface heparan sulfate released by heparanase promotes melanoma cell migration and angiogenesis. J Cell Biochem. 2009;106(2):200–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldberg R, Meirovitz A, Hirshoren N, et al. Versatile role of heparanase in inflammation. Matrix Biol. 2013;32(5):234–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elkin M, Ilan N, Ishai-Michaeli R, et al. Heparanase as mediator of angiogenesis: mode of action. FASEB J. 2001;15(9):1661–1663. [DOI] [PubMed] [Google Scholar]
- 22.Johnson GB, Brunn GJ, Platt JL. Cutting edge: an endogenous pathway to systemic inflammatory response syndrome (SIRS)-like reactions through Toll-like receptor 4. J Immunol. 2004;172(1):20–24. [DOI] [PubMed] [Google Scholar]
- 23.Johnson GB, Brunn GJ, Kodaira Y, Platt JL. Receptor-mediated monitoring of tissue well-being via detection of soluble heparan sulfate by Toll-like receptor 4. J Immunol. 2002;168(10):5233–5239. [DOI] [PubMed] [Google Scholar]
- 24.Goodall KJ, Poon IK, Phipps S, Hulett MD. Soluble heparan sulfate fragments generated by heparanase trigger the release of pro-inflammatory cytokines through TLR-4. PLoS ONE. 2014;9(10):e109596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eldor A, Bar-Ner N, Yahalom J, Fuks Z, Vlodavsky I. Role of heparanase in platelet and tumor cell interactions with the subendothelial extracellular matrix. Semin Thromb and Hemost. 1987;13(4):475–478. [DOI] [PubMed] [Google Scholar]
- 26.Mohan C, Hari S, Preetham HD, et al. Targeting heparanase in cancer: inhibition by synthetic, chemically modified, and natural compounds. iScience. 2019;15:360–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shafat I, Ilan N, Zoabi S, Vlodavsky I, Nakhoul F. Heparanase levels are elevated in the urine and plasma of type 2 diabetes patients and associate with blood glucose levels. PLoS ONE. 2011;6(2):e17312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen S, He Y, Hu Z, et al. Heparanase mediates intestinal inflammation and injury in a mouse model of sepsis. J Histochem Cytochem. 2017;65(4):241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lygzios M, Yang Y, Altmann CJ, et al. Heparanase mediates renal dysfunction during early sepsis in mice. Physiol Rep. 2013;1(6):e00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmidt EP, Yang Y, Janssen WJ, et al. The pulmonary endothelial glycocalyx regulates neutrophil adhesion and lung injury during experimental sepsis. Nat Med. 2012;18(8):1217–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ple H, Maltais M, Corduan A, Rousseau G, Madore F, Provost P. Alteration of the platelet transcriptome in chronic kidney disease. Thromb Haemost. 2012;108(4):605–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lood C, Amisten S, Gullstrand B, et al. Platelet transcriptional profile and protein expression in patients with systemic lupus erythematosus: up-regulation of the type I interferon system is strongly associated with vascular disease. Blood. 2010;116(11):1951–1957. [DOI] [PubMed] [Google Scholar]
- 33.Eicher JD, Wakabayashi Y, Vitseva O, et al. Characterization of the platelet transcriptome by RNA sequencing in patients with acute myocardial infarction. Platelets. 2016;27(3):230–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ren Y, Wang J, Xia J, et al. The Alterations of mouse plasma proteins during septic development. J Proteome Res. 2007;6:2812–2821. [DOI] [PubMed] [Google Scholar]
- 35.Middleton E, Rowley JW, Campbell RA, et al. Sepsis alters the transcriptional and translational landscape of human and murine platelets. Blood. 2019;134(12):911–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campbell R, Schwertz H, Hottz ED, et al. Human megakaryocytes possess intrinsic antiviral immunity through regulated induction of IFITM3. Blood. 2019;133(19):2013–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Freishtat R, Natale J, Benton AS, et al. Sepsis alters the megakaryocyte-platelet transcriptional axis resulting in granzyme b-mediated lymphotoxicity. Am J Respir Crit Care Med. 2009;179(6):467–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaplan D, Casper TC, Elliot CG, et al. VTE incidence and risk factors in patients with severe sepsis and septic shock. Chest. 2015;148(5):1224–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cui H, Tan Y, Osterholm C, et al. Heparanase expression upregulates platelet adhesion activity and thrombogenicity. Oncotarget. 2016;7(26):39486–39496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Freeman C, Parish CR. Human platelet heparanase: purification, characterization, and catalytic activity. Biochem J. 1998;330:1341–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ishai-Michaeli R, Eldor A, Vlodavsky I. Heparanase activity expressed by platelets, neutrophils, and lymphoma cells releases active fibroblast growth factor from extracellular matrix. Cell Regulation. 1990;1:833–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gul F, Arslantas MK, Cinel I, Kumar A. Changing definitions of sepsis. Turk J Anaesthesiol Reanim. 2017;45(3):129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Mobrison F, Angei C, Staal K, Kaiser K, Picot S. Simultaneous identification of the four human Plasmodium species and quantification of plasmodium DNA Load in human blood by real-time polymerase chain reaction. Trans R Soc Trop Med Hyg. 2003;97:387–390. [DOI] [PubMed] [Google Scholar]
- 44.Rosanas-Urgell A, Mueller D, Betuela I, et al. Comparison of diagnostic methods for the detection and quantification of the four sympatric Plasmodium species in field samples from Papua New Guinea. Malar J. 2010;9(361). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Campbell R, Franks Z, Bhatnagar A, et al. Granzyme A in human platelets regulates the synthesis of proinflammatory cytokines by monocytes in aging. J Immunol. 2018;200(1):295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Love M, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sistla J, Morla S, Alabbas AB, et al. Polymeric flyorescent heparin as one-step FRET substrate of human heparanase. Carbohyd Polym. 2019;205:385–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rondina M, Schwertz H, Harris ES, et al. The septic milieu triggers expression of spliced tissue factor mRNA in human platelets. J Thromb Haemost. 2011;9(4):748–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Assinger A, Schrottmaier WC, Salzmann M, Rayes J. Platelets in sepsis: an update on experimental models and clinical data. Front Immunol. 2019;10:1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramani VC, Pruett PS, Thompson CA, DeLucas LD, Sanderson RD. Heparan sulfate chains of syndecan- 1 regulate ectodomain shedding. J Biol Chem. 2012;287(13):9952–9961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Manon-Jensen T, Multhaupt HA, Couchman JR. Mapping of matrix metalloproteinase cleavage sites on syndecan-1 and syndecan-4 ectodomains. FEBS J. 2013;280(10):2320–2331. [DOI] [PubMed] [Google Scholar]
- 52.Constantinescu AA, Vink H, Spaan JA. Endothelial cell glycocalyx modulates immobilization of leukocytes at the endothelial surface. Arterioscler Thromb Vasc Biol. 2003;23(9):1541–1547. [DOI] [PubMed] [Google Scholar]
- 53.Toledo AG, Golden G, Campos AR, et al. Proteomic atlas of organ vasculopathies triggered by Staphylococcus aureus sepsis. Nat Commun. 2019;10(1):4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vogtle T, Sharma S, Mori J, et al. Heparan sulfates are critical regulators of the inhibitory megakaryocyte- platelet receptor G6b-B. Elife. 2019;8:e46840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tan Y, Cui H, Wan LM, et al. Overexpression of heparanase in mice promoted megakaryopoiesis. Glycobiology. 2018;28(5):269–275. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.