Abstract
RNA-DNA hybrids form throughout the chromosome during normal growth and under stress conditions. When left unresolved, RNA-DNA hybrids can slow replication fork progression, cause DNA breaks, and increase mutagenesis. To remove hybrids, all organisms use ribonuclease H (RNase H) to specifically degrade the RNA portion. Here we show that, in addition to chromosomally encoded RNase HII and RNase HIII, Bacillus subtilis NCIB 3610 encodes a previously uncharacterized RNase HI protein, RnhP, on the endogenous plasmid pBS32. Like other RNase HI enzymes, RnhP incises Okazaki fragments, ribopatches, and a complementary RNA-DNA hybrid. We show that while chromosomally encoded RNase HIII is required for pBS32 hyper-replication, RnhP compensates for the loss of RNase HIII activity on the chromosome. Consequently, loss of RnhP and RNase HIII impairs bacterial growth. We show that the decreased growth rate can be explained by laggard replication fork progression near the terminus region of the right replichore, resulting in SOS induction and inhibition of cell division. We conclude that all three functional RNase H enzymes are present in B. subtilis NCIB 3610 and that the plasmid-encoded RNase HI contributes to chromosome stability, while the chromosomally encoded RNase HIII is important for chromosome stability and plasmid hyper-replication.
Keywords: Bacillus subtilis, NCIB 310, RNA-DNA hybrid, RNase HI, SOS response
1 |. INTRODUCTION
For all organisms, faithful replication of the chromosome is essential to ensure daughter cells receive an accurate and complete copy of their genetic material. Over the last decade, it has been shown that RNA is often incorporated into genomic DNA, through hybridization to DNA or via covalent linkage (Williams and Kunkel, 2014; Schroeder et al., 2015; Santos-Pereira and Aguilera, 2015). These incorporation events can have severe consequences for cell physiology, leading to replication fork stress, genome instability, and adverse effects on transcription (Prado and Aguilera, 2005; Nick McElhinny et al., 2010a, Nick McElhinny et al., 2010b, Kouzminova et al., 2017; Schroeder et al., 2017). RNA-DNA hybrids form through a variety of processes throughout each phase of bacterial growth, with each type of hybrid impacting genome integrity in a different way.
In exponentially growing cells a common type of RNA-DNA hybrid occurs in the form of Okazaki fragments (Randall et al., 2019). During DNA replication, Okazaki fragments on the lagging strand begin with RNA primers generating an RNA-DNA hybrid with a covalent RNA-DNA junction (Rowen and Kornberg, 1978a; Rowen and Kornberg, 1978b). These RNA primers are later removed and replaced with DNA through the activity of several DNA repair proteins in B. subtilis (Randall et al., 2019). The second type of RNA-DNA hybrid occurs during replication by DNA polymerase, where an rNTP is used in place of the cognate dNTP, resulting in a sugar error (Nick McElhinny et al., 2010b; Yao et al., 2013; Schroeder et al., 2017). Sugar errors tend to be single replicative DNA polymerase errors and have the potential to occur every few thousand base pairs in exponentially growing cells (Nick McElhinny et al., 2010b; Yao et al., 2013). In states of slow growth, it has been proposed that translesion DNA polymerases can use rNTPs in place of scarce dNTPs in a process termed ribopatch repair (Ordonez et al., 2014). Ribopatch repair would generate relatively short polymers of RNA nested in double-stranded DNA to provide a temporary solution for sites in need of repair (Ordonez et al., 2014). RNA polymers covalently joined to DNA can impact genome integrity because the 2′OH of the ribose sugar can facilitate a nucleophilic attack on the 3′ PO4-, resulting in a 2′,3′ cyclic phosphate at the rNMP and a 5′ OH at the adjacent nucleotide (Oivanen et al., 1998). The resulting nick in the phosphodiester backbone is refractory to ligation and unable to function in further DNA synthesis (Das and Shuman, 2013, Das et al., 2014, Schroeder et al., 2017). Failure to heal the end and repair the nick would then result in a double-strand break during the next round of DNA replication.
RNA-DNA hybrids in the form of Okazaki fragments and DNA polymerase errors are similar in that the RNA is covalently linked to DNA through a phosphodiester bond (Randall et al., 2017; Randall et al., 2019). Another prevalent RNA-DNA hybrid forms during transcription when mRNA transcripts are base-paired with the transcribed DNA strand, displacing the coding strand as ssDNA to form an R-loop [for review (Schroeder et al., 2015)]. In the case of R-loops, RNA hybridized to DNA lacks a covalent RNA-DNA junction. Persistent R-loops can impair the progression of replication forks and DNA synthesis, while also decreasing gene expression from the DNA template subsequent to R-loop formation (Lang et al., 2017). Transcription is required during all growth phases, suggesting that R-loop formation could be prevalent during the entire life cycle of a bacterium. Therefore, all cells need to resolve each class of RNA-DNA hybrid that occurs in vivo to maintain genome integrity and efficient gene expression throughout bacterial growth.
To reduce the detrimental consequences that RNA-DNA hybrids impose on genome integrity and transcription, organisms have enzymes dedicated to hybrid resolution (Ohtani et al., 1999b, Cerritelli and Crouch, 2009). The RNase H family of endoribonucleases cleaves the RNA component of RNA-DNA hybrids, resolving all classes of hybrids that occur in vivo (Ohtani et al., 1999b, Cerritelli and Crouch, 2009). RNase H enzymes are highly conserved, with family members present in bacteria, archaea, eukaryotes, and retroviruses, including HIV-1 (Ohtani et al., 2000; Li et al., 2004; Ohtani et al., 2004; Sparks et al., 2012). Bacterial RNase H enzymes are grouped into two general types based on amino acid sequence similarity: type I, which includes RNase HI, and type II, which includes RNase HII and HIII (Kochiwa et al., 2007). RNase HI and RNase HIII enzymes act on both ribopatches (four or more embedded rNMPs) and hybrids lacking a covalent RNA-DNA junction, but are unable to cleave at a single rNMP embedded in DNA (Nowotny et al., 2007; Randall et al., 2017; Randall et al., 2019). Unlike RNase HI and HIII, RNase HII enzymes are adept for cleavage at single embedded rNMPs and ribopatches participating in ribonucleotide excision repair (RER), yet show very poor activity on hybrids that lack a covalent RNA-DNA junction (Sparks et al., 2012; Chon et al., 2013). All three enzymes are active on the RNA primer portions of an Okazaki fragment, suggesting that all three bacterial enzymes could have overlapping functions during lagging strand processing and maturation (Ohtani et al., 1999a; Randall et al., 2017; Randall et al., 2019). In addition to their important contribution to chromosomal replication, evidence suggests that RNase H enzymes function in the regulation of endogenous plasmid replication (Itoh and Tomizawa, 1980) and in regulatory aspects of transcription (Huertas and Aguilera, 2003, Santos-Pereira and Aguilera, 2015).
Comparative sequence analysis of over 300 genomes found that 80% of bacterial genomes contain RNase HI and RNase HII (Kochiwa et al., 2007). Approximately 17% of bacterial genomes lack RNase HI (Kochiwa et al., 2007). The Firmicutes phylum, which includes a group of important Gram-positive pathogens, are among the 17% of bacterial genomes that lack RNase HI and instead encode RNase HII and RNase HIII (Kochiwa et al., 2007). Importantly, Firmicutes were the only group with some representatives that appeared to encode all three RNase H genes (Kochiwa et al., 2007; Randall et al., 2017). One Firmicutes that seemed to encode all three RNase H enzymes is the soil bacterium Bacillus subtilis (Kochiwa et al., 2007; Randall et al., 2017). The RNase HII and HIII enzymes from the lab strain B. subtilis PY79 are active and have been characterized in vitro and in vivo (Ohtani et al., 1999a; Randall et al., 2017; Schroeder et al., 2017; Randall et al., 2019). Functional studies of the putative RNase HI-like proteins from B. subtilis were shown to lack the residues involved in substrate-binding and do not possess nuclease activity in vitro (Kochiwa et al., 2007; Randall et al., 2017). Furthermore, prior work also showed that the simultaneous deletion of genes for both RNase HII (rnhB) and RNase HIII (rnhC) is lethal (Fukushima et al., 2007) or results in a mutator phenotype with the accumulation of compensatory mutations (Randall et al., 2019). These studies suggest that the putative RNase HI-like genes are unable to compensate for the loss of both RNase HII and RNase HIII in vivo. Of the small subset of bacteria that contain putative RNase HI, HII, and HIII there is no experimental evidence to support the coexistence of functional RNase HI and RNase HIII in the same genome (Kochiwa et al., 2007).
B. subtilis NCIB 3610 (referred herein as 3610) is considered a “wild” ancestral strain, which has maintained many of the wild motility and social behaviors associated with B. subtilis strains isolated from the soil (Branda et al., 2001, Kearns and Losick, 2003, Zeigler et al., 2008). In addition to the 4.2 Mbp chromosome, 3610 contains an endogenous 84 Kbp plasmid, pBS32 (Earl et al., 2007; Nye et al., 2017). Plasmid pBS32 encodes 102 genes, many of which include a large contiguous set of genes that appear to encode for a cryptic prophage (Konkol et al., 2013). Other genes on the plasmid encode proteins that control host cell physiology, such as the inhibitor of biofilm formation RapP, the inhibitor of natural competence for DNA uptake ComI, and the cell death promoting sigma factor SigN (Konkol et al., 2013, Parashar et al., 2013, Burton et al., 2019, Omer Bendori et al., 2015). The remaining genes on pBS32 are of unknown function, including zpdC (rnhP), which encodes a putative RNase HI. If zpdC is, indeed, a functional RNase HI enzyme this, to the best of our knowledge, would suggest that B. subtilis 3610 is the first bacterium to encode active RNase HI, HII, and HIII. Moreover, if ZpdC is an active RNase H it is unknown whether ZpdC activity is important for pBS32 plasmid maintenance, integrity of the B. subtilis chromosome, or some other aspect of DNA maintenance.
We show that ZpdC (named herein as RnhP) is capable of cleaving all substrates typical of RNase HI proteins. Deletion of rnhP does not affect pBS32 maintenance, although the deletion of rnhC results in loss of the pBS32 hyper-replication phenotype. We demonstrate that the deletion of rnhP and rnhC results in a 2-fold increase in doubling time, which is attributed to cell filamentation and induction of the SOS response. Together, our data show that both the plasmid-encoded RNase HI (RnhP) and the chromosomally encoded RNase HIII (RnhC) are important for genome maintenance in the ancestral strain of B. subtilis NCIB 3610, demonstrating that bacteria can, indeed, maintain all three RNase H proteins with each enzyme contributing to genome stability.
2 |. RESULTS
2.1 |. RnhP is an active RNase HI enzyme
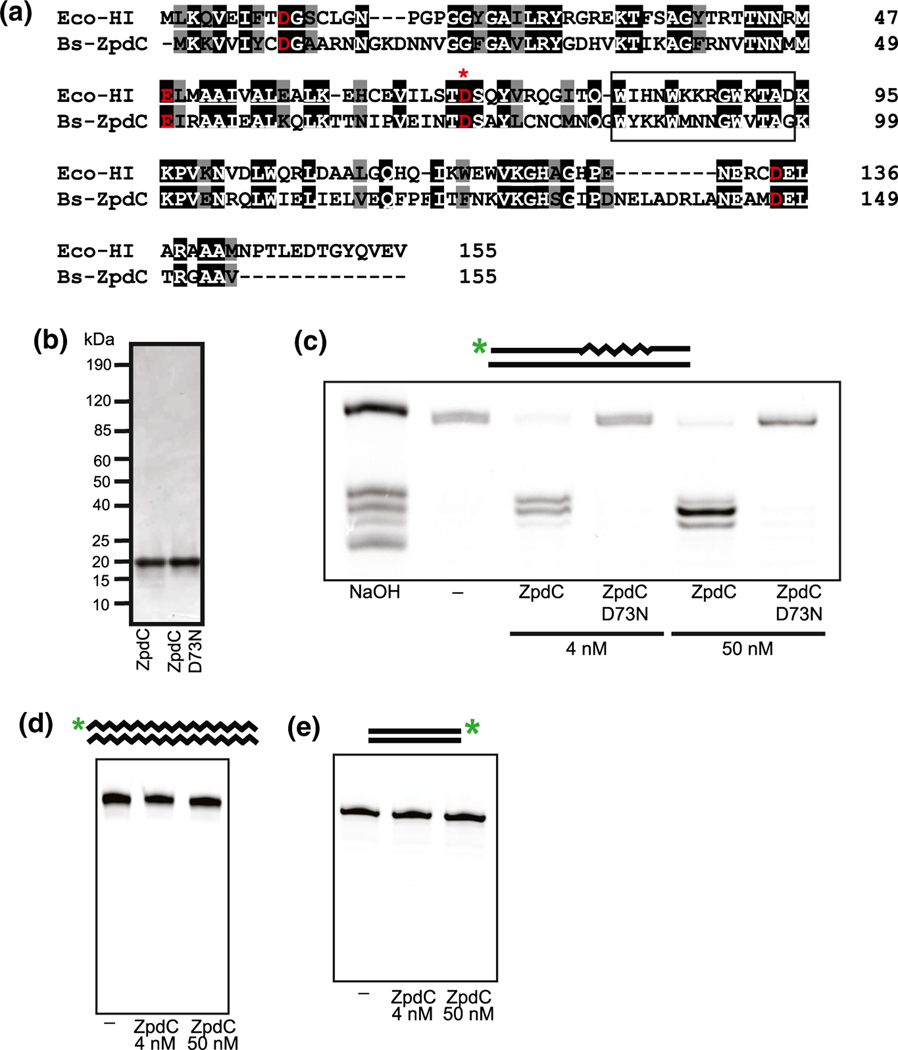
The endogenous 84 Kbp plasmid, pBS32, of the ancestral strain B. subtilis NCIB 3610 contains several uncharacterized genes that encode proteins with sequence homology to bacterial DNA replication and repair proteins (Earl et al., 2007; Earl et al., 2012; Konkol et al., 2013). One such gene, zpdC (rnhP), shares 38.5% primary structure identity and 50.3% primary structure similarity to the RNase HI protein from Escherichia coli (Figure 1a). Importantly, all of the catalytic residues involved in metal coordination are conserved between the two sequences as are the residues within the α-helix 3 basic protrusion handle, which is involved in substrate-binding (Katayanagi et al., 1990), suggesting that ZpdC might have RNase H activity (Figure 1a). As part of our ongoing effort to understand how RNA-DNA hybrids impact genome stability and transcription, we began by purifying ZpdC (RnhP) and a D73N variant, which has been shown to render E. coli RNase HI catalytically inactive (Katayanagi et al., 1990) (Figure 1b).
FIGURE 1.
Plasmid-encoded ZpdC is an active RNase HI protein. (a) Sequence alignment of ZpdC with E. coli RNase HI. Identical and similar residues are indicated in black and gray, respectively. Red indicates conserved catalytic residues. (*) denotes catalytic residue mutated in catalytically inactive variant (D73N). The α-helix 3 basic protrusion handle is boxed. (b) SDS-PAGE stained with Coomassie Brilliant Blue of purified ZpdC and catalytically inactive variant D73N. (c) ZpdC and D73N were incubated with a ribopatch substrate. The 5’ end IR-labeled oligo containing four embedded rNMPs (squiggly lines) within an otherwise DNA oligo (straight lines) was annealed to a complementary DNA oligo (oJR210 and oJR145). A ladder was generated via alkaline hydrolysis of the substrate at the embedded rNMPs (lane one). (d) Incubation of ZpdC with an RNA-RNA substrate. A 5’ end IR-labeled RNA oligo (squiggly line) was annealed to a complementary RNA oligo (oJR227 and oJR166). (e) Incubation of ZpdC with a DNA-DNA substrate. A 3’ end IR-labeled DNA oligo (straight line) was annealed to a complementary DNA oligo (oJR348 and oJR365). For C-E, the reactions were assembled as described in “Experimental Procedures” and products were separated on a 20% denaturing urea-PAGE and subsequently visualized with a LI-COR Odyssey imager
To assay for RNase H activity, we used an oligonucleotide labeled with an IR dye at the 5′ end that contained four embedded rNMPs flanked by DNA on either side. This labeled oligonucleotide was annealed to a complementary DNA strand, creating a double-stranded RNA-DNA chimeric substrate as previously described (Randall et al., 2019). We used this approach because prior work showed that bacterial RNase HI, HII, and HIII are all active on this substrate (Ohtani et al., 1999a; Randall et al., 2017; Randall et al., 2019). We incubated ZpdC (RnhP) and the catalytically inactive variant with this substrate for 10 minutes at 37°C in buffers that mimic in vivo relevant metal concentrations (1 mM Mg2+, 10 μM Mn2+) as described (Randall et al., 2019). The substrate was also exposed to alkaline hydrolysis in a separate reaction to create a ladder corresponding to the positions of each embedded rNMP. The products of the reaction were separated by electrophoresis on a denaturing urea PAGE to measure substrate cleavage. Incubation of the substrate with low (4 nM) and high (50 nM) concentrations of protein results in complete cleavage of the substrate, whereas the catalytically inactive variant did not show any cleavage at either protein concentration (Figure 1c).
To determine if ZpdC (RnhP) has strict RNase H activity, such that it is only capable of cleaving the RNA portion of RNA-DNA hybrids, we tested the ability of ZpdC (RnhP) to cleave double-stranded RNA and DNA substrates. We incubated 4 nM and 50 nM ZpdC (RnhP) with double-stranded RNA and DNA substrates that were end-labeled with IR dyes under the same buffer and incubation conditions described above. The DNA substrate was previously shown to be cleaved by the exonuclease YpcP (Randall et al., 2019) and the RNA substrate was shown to be hydrolyzable by treatment with sodium hydroxide (data not shown). Unlike the RNA-DNA chimera substrate, we did not observe any cleavage of the RNA or DNA substrates when incubated with low or high concentrations of ZpdC (Figure 1d,e). From these data, we conclude that ZpdC is an active and strict RNase H enzyme. Having established that ZpdC is a plasmid-encoded RNase H, we rename zpdC to RNase H from pBS32 (rnhP).
2.2 |. RnhP is active with various metals and at varying temperatures
To determine the parameters of activity for RnhP we assayed for cleavage over a range of temperatures and metal concentrations relevant to B. subtilis growth (Hardwood and Cutting, 1990). RnhP cleaved the RNA-DNA chimeric substrate with four embedded rNMPs when incubated at 25°C, 30°C, and 37°C for 10 minutes with no appreciable difference in activity observed between the three temperatures (Supplementary Figure S1a). We also tested the activity of RnhP on the four embedded rNMP substrate with various metals, holding all other reaction and buffer conditions the same between samples (see Experimental Procedures). RnhP appeared to be most active when incubated with 10 μM Mn2+ as a metal cofactor (Supplementary Figure S1b). RnhP also showed activity when incubated with Mg2+, Zn2+, and Co2+, although RnhP was less active when compared with activity in the presence of Mn2+. We note a reduction in RnhP activity when incubated with 1 mM MgCl2 and 10 μM MnCl2 compared to 10 μM MnCl2 alone, which could be explained by the competition of Mn2+ with Mg2+ for binding to the RnhP active site (Supplementary Figure S1b). We conclude that RnhP is active both under relevant growth temperatures and with several metal cofactors. We note that, compared to the other cofactors tested here, Mn2+ supports the most activity.
2.3 |. RnhP cleaves RNA-DNA covalent chimeras in a different location when compared with B. subtilis RNase HII and RNase HIII
Most bacteria have a functional RNase HII and either RNase HI or RNase HIII (Kochiwa et al., 2007). Further, it has been hypothesized that RNase HI and HIII are mutually exclusive (Kochiwa et al., 2007). RNase HII is classically characterized as having unique activity on a single embedded rNMP within DNA and polymers of rNMPs that are covalently linked to DNA including embedded ribonucleotide polymers (i.e., “ribopatches”) and Okazaki fragments (Gupta et al., 2017). RNase HI and HIII recognize substrates that contain four or more embedded rNMPs that are covalently linked to DNA and RNA-DNA hybrids that interact through hydrogen bonding, such as R-loops (Gupta et al., 2017; Randall et al., 2017; Randall et al., 2019). To empirically determine the cleavage patterns of RNase H enzymes, we purified RnhP, RnhC (RNase HIII), and RnhB (RNase HII) to examine their activities and cleavage patterns on a variety of RNA-DNA hybrid substrates in vitro. The purification of RnhC and RnhB has already been described elsewhere (Randall et al., 2017; Schroeder et al., 2017; Randall et al., 2019).
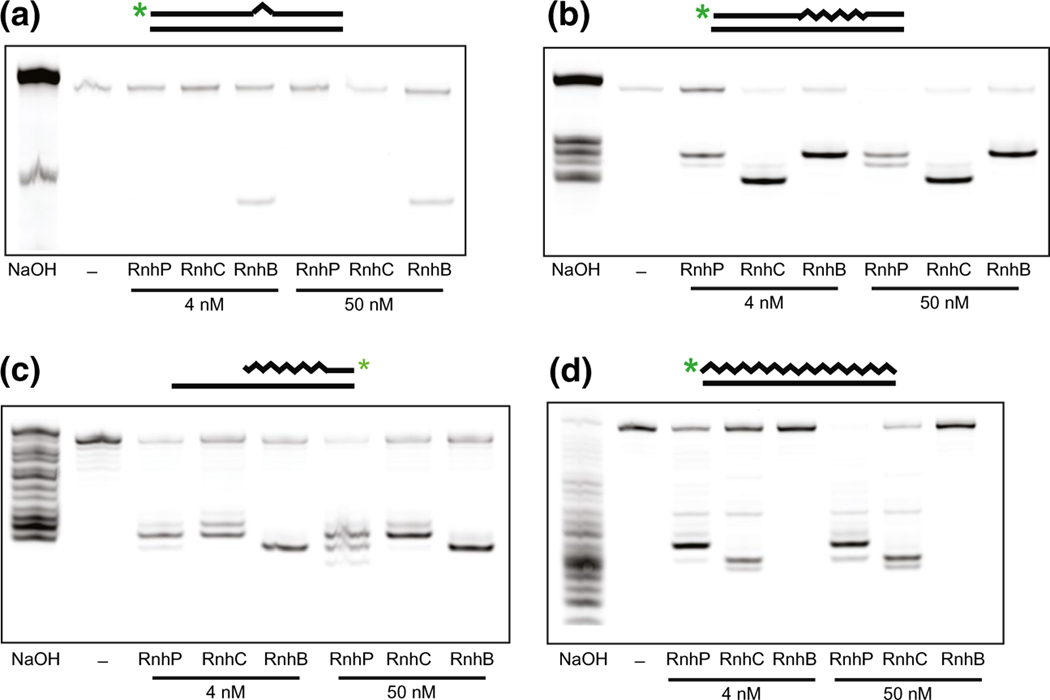
We began by testing the activities of all three enzymes using a substrate labeled with an IR dye at the 5′ end that contained one rNMP flanked by DNA on both sides annealed to a complementary DNA strand (oJR209 and oJR145). Consistent with previously published results (Schroeder et al., 2017), we found that only RnhB (RNase HII) had activity on this substrate under the conditions tested here (see Experimental Procedures) (Figure 2a). We next assayed for activity on the four embedded rNMP substrate. While all three enzymes were capable of incising the substrate at 4 nM and 50 nM protein concentrations, we found that the enzymes differed in their incision patterns. RnhB cleavage yielded a longer product, suggesting cleavage between the third and fourth ribonucleotide from the 5′ IR-dye end label as expected, whereas RnhC cleavage resulted in a shorter product, with cleavage within the middle of the embedded RNA. In contrast to functional redundancy with RnhC, RnhP appeared to cleave the four embedded rNMP substrate more similarly to RnhB (RNase HII), resulting in a longer product than RnhC with cleavage between the third/fourth and second/third rNMPs from the 5′ IR-dye end label (Figure 2b).
FIGURE 2.
RnhP cleaves several RNA-DNA hybrid substrates. For each reaction, the indicated substrate was incubated separately with RnhP, RnhB (RNase HII), or RnhC (RNase HIII) in reaction buffer for 10 min at 37°C (see Experimental Procedures). For each substrate, a ladder was created via alkaline hydrolysis of the substrate at the rNMPs (lane one). The products were separated on a 20% denaturing urea-PAGE and subsequently visualized with a LI-COR Odyssey imager. (a) Incubation of RnhP, RnhB, and RnhC with a single rNMP substrate. A 5’ end IR-labeled oligo containing one embedded rNMP (triangle) within an otherwise DNA oligo (straight lines) was annealed to a complementary DNA oligo (oJR209 and oJR145). (b) Incubation of RnhP, RnhB, and RnhC with a ribopatch substrate. A 5’ end IR-labeled oligo containing four embedded rNMPs (squiggly lines) within an otherwise DNA oligo (straight lines) was annealed to a complementary DNA oligo (oJR210 and oJR145). (c) Incubation of RnhP, RnhB, and RnhC with an Okazaki fragment-like substrate. A 3’ IR-dye end-labeled oligo with rNMPs at the 5’ end covalently linked to a stretch of DNA was hybridized to an oligo that was complementary at the 5’ end of the molecule but was significantly longer to generate a 3’ overhang (oJR339 and oJR340). (d) Incubation of RnhP, RnhB, and RnhC with a complementary RNA-DNA hybrid substrate. A 5’ end IR-labeled RNA oligo (squiggly line) was annealed to a complementary DNA oligo (straight lines) to create an RNA-DNA hybrid (oJR227 and oJR145)
Both the single and quadruple embedded rNMP substrates are intended to represent misincorporation events that occur when DNA polymerases erroneously add rNTPs during replication or ribopatch repair, accounting for as many as 2,000 rNMPs incorporated into a bacterial genome per round of replication (Yao et al., 2013). Significantly more rNMPs (~23,000) are expected to be incorporated into the genome in the form of Okazaki fragments during lagging strand synthesis (Schroeder et al., 2015). In B. subtilis, these primers are removed and replaced with dNMPs through the combined action of RNase HIII, DNA polymerase I, and YpcP with RNase HII also contributing to RNA removal (Randall et al., 2019). To test how RnhP activity compared to RnhC (RNase HIII) and RnhB (RNase HII) on an Okazaki fragment, we constructed an oligo with rNMPs at the 5′ end covalently linked to a stretch of DNA with a 3′ IR-dye end label. This oligo was hybridized to another oligo that was complementary to the 5′ end of the RNA-containing oligo but was significantly longer to generate a 3′ overhang (oJR339 and oJR340). We incubated this substrate, as previously described (Randall et al., 2019), with RnhB, RnhC, and RnhP to measure activity. Consistent with previous work, we observe Okazaki fragment substrate cleavage for both RnhB and RnhC (Randall et al., 2019). Furthermore, we show that RnhP has activity on the model Okazaki fragment substrate, with multiple cleavage sites and some incision sites overlapping with RnhC (Figure 2c).
2.4 |. RnhP cleaves RNA-DNA hybrids differently than RNase HIII
A defining feature of the single rNMP substrate, the polymer of four embedded rNMPs, and the Okazaki fragment substrate is that each has an RNA-DNA covalent junction. In contrast, an R-loop represents a different type of RNA-DNA hybrid, which is produced during transcription when RNA hybridizes with complementary DNA in the template strand, displacing the DNA coding strand (Asai and Kogoma, 1994, Santos-Pereira and Aguilera, 2015). This substrate differs in that it does not contain a covalent RNA-DNA linkage. To determine if RnhP, like other RNase HI enzymes, is capable of cleaving substrates without a covalent RNA-DNA linkage we began by testing activity on a substrate labeled at the 5′ end with an IR dye that contains an all RNA strand hybridized to a complementary DNA strand (oJR227 and oJR145). Consistent with previously published results, we observe cleavage when the complementary RNA-DNA hybridized substrate was incubated with RNase HIII but not RNase HII (Randall et al., 2017; Randall et al., 2019). We then tested RnhP and demonstrate that RnhP does, indeed, have activity on an RNA-DNA hybridized substrate lacking an RNA-DNA covalent junction. However, the site of incision differs between RnhP and RNase HIII (RnhC) (Figure 2d). This result, considered with the results described above, shows that RnhP recognizes the same substrates as a canonical RNase HI (Gupta et al., 2017) and that RnhP often cleaves at a different location relative to cleavage observed by RNase HIII.
2.5 |. RNase HIII is required for plasmid hyper-replication
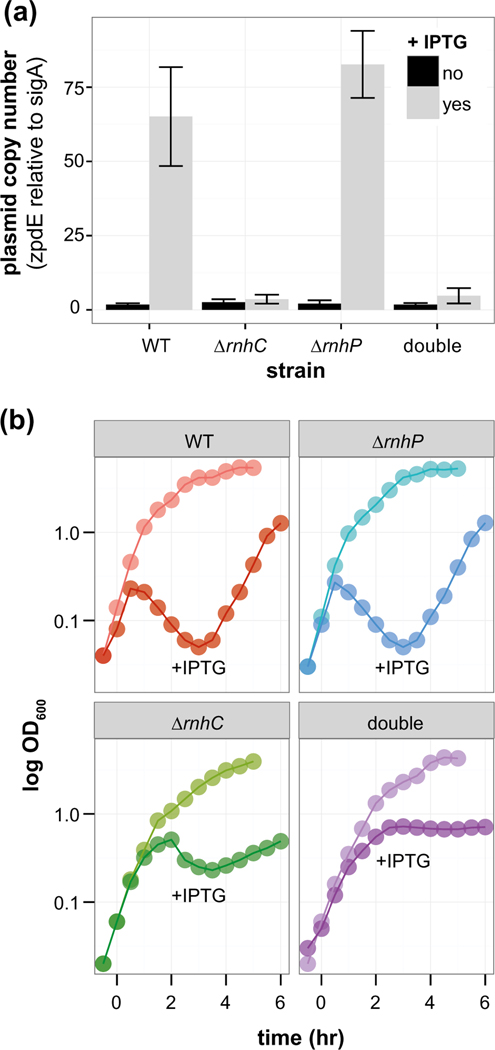
We initially hypothesized that RnhP would be important for the replication of pBS32 in 3610 based on the notion that rnhP is plasmid-borne. It was previously reported that overexpression of the plasmid-specific sigma factor, SigN (ZpdN), causes pBS32 to hyper-replicate and promoted cell lysis (Burton et al., 2019). Plasmid copy number was measured by quantitative PCR during normal growth and following hyper-replication by expressing SigN from an IPTG inducible promoter (Burton et al., 2019). We found that pBS32 was maintained at a low copy number similar to WT in ΔrnhC, ΔrnhP, and the double mutant (ΔrnhP rnhC::erm) when the expression of SigN was not induced (Figure 3a). Induction of SigN caused the plasmid to hyper-replicate in WT and ΔrnhP cells, but not in ΔrnhC cells (Figure 3a). Further, cell viability was assessed in all strains induced with SigN by measuring optical density and scoring CFUs every 30 min up to four hours post-induction (Figure 3, Supplementary Figure S2a). Induction of SigN in the WT and ΔrnhP strains caused a similar loss of cell viability, suggesting that RnhP is not required for the pBS32-mediated cell death phenotype (Figure 3b, Supplementary Figure S2a). In contrast, the strain with an rnhC deletion showed a slight drop and then plateau in OD, while CFUs were reduced less drastically than the WT or ΔrnhP mutant but recovered much slower (Figure 3b, Supplementary Figure S2a). In the double mutant, OD reached a plateau while CFUs dropped and did not recover over a four-hour time course experiment (Figure 3b, Supplementary Figure S2a). Moreover, we found that the double mutant cells displayed a severely filamentous phenotype throughout the course of the experiment (Supplementary Figure S2b-e). With these data, we conclude that chromosomally encoded RNase HIII is important for plasmid hyper-replication and recovery, while the loss of rnhP alone has no effect on pBS32 maintenance or hyper-replication.
FIGURE 3.
RnhC, not RnhP, is required for plasmid hyper-replication. (a) The average plasmid copy number for IPTG inducible sigN strains in WT, ΔrnhC, ΔrnhP, and ΔrnhP rnhC::erm backgrounds with (light gray bars) and without (dark gray bars) IPTG. The plasmid copy number was assessed via qPCR ratio of the plasmid-encoded zpdE gene to the chromosomally encoded housekeeping sigma factor sigA. Error bars represent the standard deviation or range in the case of WT and rnhC. (b) OD600 (y-axis) of sigN inducible strains in WT, ΔrnhC, ΔrnhP, and ΔrnhP rnhC::erm backgrounds over time (x-axis). Representative curves for uninduced and IPTG induced strains are indicated. IPTG was added at time 0
2.6 |. Cells lacking both RnhP and RnhC activity have a reduced growth rate
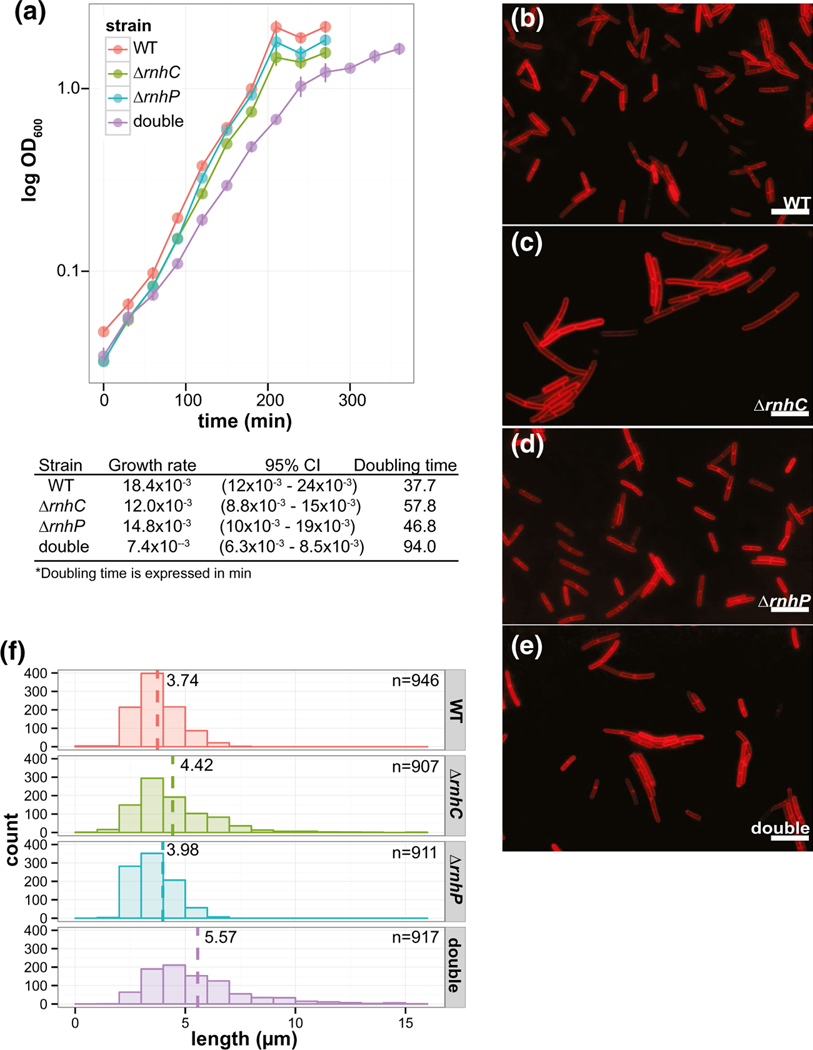
Our results thus far demonstrate that RnhP has activity on RNADNA hybrids with four or more ribonucleotides in vitro and that RnhP does not contribute to pBS32 maintenance or hyper-replication. Therefore, we asked if RnhP contributes to chromosomal maintenance in 3610. If so, it would suggest that 3610 has a fitness advantage when maintaining active RNase HI (RnhP), HII, and HIII enzymes for the purpose of resolving the variety of RNA-DNA hybrids that form on the chromosome. It has been shown that in the absence of RNase HIII, but not RNase HII, there is a decrease in growth rate in B. subtilis PY79 (Randall et al., 2017; Schroeder et al., 2017; Randall et al., 2019). To test whether RnhP activity could compensate for the decrease in growth observed in the absence of RNase HIII, we performed growth curves for 3610 in liquid LB media for the WT, ΔrnhC, ΔrnhP, and ΔrnhP rnhC::erm strains. While the doubling times for the ΔrnhC and ΔrnhP single deletion strains (57.8 and 46.8 min, respectively) appeared to be slower than WT (37.7 min), there was no statistically significant difference in growth rate between WT, ΔrnhC, or ΔrnhP strains based on the growth model used here (Figure 4a, see Experimental Procedures). However, upon loss of both rnhP and rnhC the growth rate was significantly slower than WT (37.7 min) and slower than either of the single deletion strains, with a doubling time over two times greater than WT at 94 min (Figure 4a). As described in greater detail later in the results, we show that ectopic expression of rnhP in a ΔrnhC background of the lab strain PY79 rescues ΔrnhC growth defects to WT levels (Figure 7b, described below). With these results, we conclude that RnhP can compensate for RNase HIII (rnhC) and that these enzymes have overlapping functions in 3610.
FIGURE 4.
Loss of RnhP and RnhC results in decreased cell growth and increased cell length during exponential growth. (a) Growth curves for WT, ΔrnhC, ΔrnhP, and ΔrnhP rnhC::erm in LB media with shaking at 30°C. The growth curves were fit to a Gompertz growth model and the estimated growth rates and corresponding doubling times are indicated with 95% confidence intervals. (b-e) Representative images for cell length of WT, ΔrnhC, ΔrnhP, and ΔrnhP rnhC::erm, respectively. Cells were grown in LB media with shaking at 30°C to mid-exponential growth and treated with a membrane strain for subsequent imaging. (f) The distributions of cell lengths plotted for each strain. The dashed line for each strain indicates the average cell length. The number of cells scored is indicated
FIGURE 7.
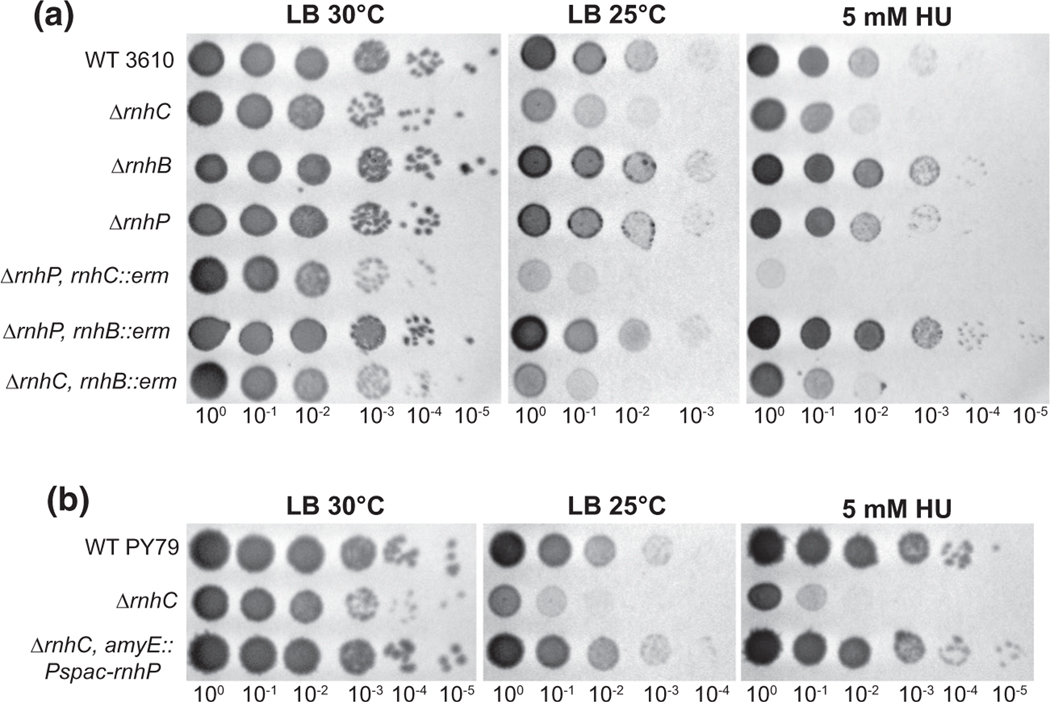
RnhP contributes to the mitigation of cell stress caused by DNA damage. (a) Single and pairwise deletion strains in NCIB 3610 were serially diluted 10-fold and spotted onto LB agar media at 30°C, 25°C, and with 5 mM hydroxyurea added to the plates and incubated at 30°C. Plates were imaged after overnight incubation. (b) PY79 and derived strains were serially diluted 10-fold and spotted onto LB agar media at 30°C, 25°C, and at 30°C with 5 mM hydroxyurea added to the plates. Plates were imaged after overnight incubation
2.7 |. Cells lacking rnhP and rnhC genes filament relative to WT
To test if the differences in growth rate were caused by an inhibition of cell division we assayed cell length in exponentially growing cultures of WT, ΔrnhC, ΔrnhP, and ΔrnhP rnhC::erm strains. Cell membranes were imaged with a lipophilic fluorescent dye and cell length was measured as described (Experimental Procedures). Consistent with the slight decrease in growth rate observed in both ΔrnhC and ΔrnhP single deletions at 30°C, the average cell lengths of ~4.4 μm and ~4.0 μm for each strain, respectively, was longer than that of the WT strain measuring at ~3.7 μm (Figure 4c–e). A slight tail on the distribution of cell lengths can be observed for each single deletion strain, representing a subpopulation of cells that are slow to complete septation, resulting in a portion of longer cells (Figure 4f). The distribution of cell lengths for the ΔrnhP rnhC::erm double mutant has a more pronounced tail and an average cell length greater than WT or the single deletions alone at ~5.6 μm (Figure 4f). These results support the conclusion that single deletions of rnhC or rnhP are well tolerated by the cell. We suggest that one gene can compensate for the loss of the other. However, the double deletion results in a severe growth defect that is, at least in part, the product of improper cell division, suggesting genome integrity may be compromised in the double mutant during normal growth in the absence of exogenous DNA damage.
2.8 |. Cells lacking rnhP and rnhC activity are induced for the SOS response
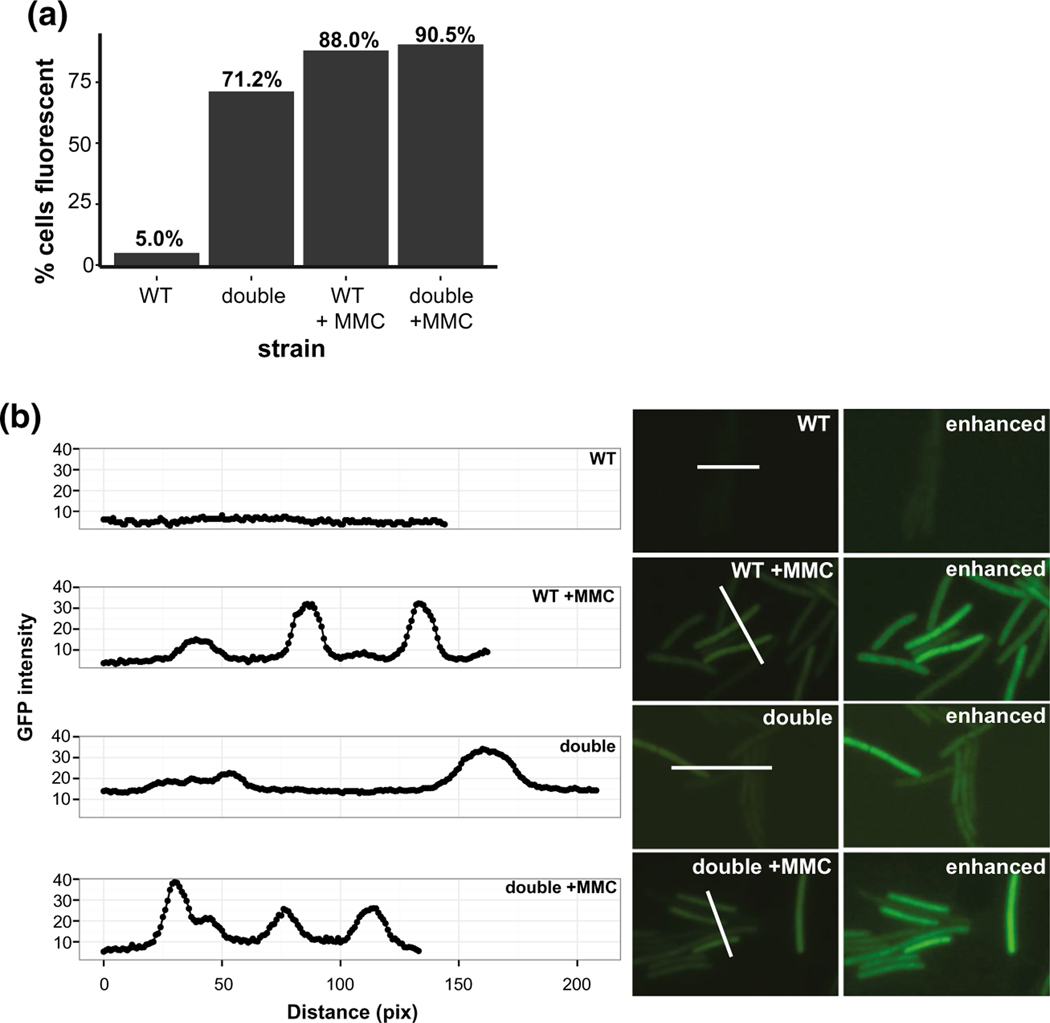
During periods of DNA damage, cell division is inhibited by the cell division inhibitor, YneA, to allow for the chromosome to be properly repaired and replicated before cell division resumes (Kawai et al., 2003; Mo and Burkholder, 2010; Burby et al., 2018). The YneA-enforced DNA damage checkpoint ensures that daughter cells receive a complete copy of the chromosome after replication is complete (Kawai et al., 2003; Mo and Burkholder, 2010; Burby et al., 2018). It has been shown previously that multiple deletions of RNase H and RNase H-like genes in B. subtilis lacking the pBS32 plasmid show SOS induction (Fukushima et al., 2007). Further, given the defects in growth and cell filamentation of the double deletion strain studied here, we asked if cells lacking rnhP and rnhC are SOS induced. We used the SOS reporter construct tagC::tagC-gfp, which like yneA is highly upregulated during the SOS response, as a single cell proxy for SOS induction (Britton et al., 2007; Simmons et al., 2009). In exponentially growing WT cells (OD600 = 0.5–0.7) at 30°C in LB media, we found that ~5.0% of cells expressed the SOS reporter, while ~88.0% of WT cells expressed the reporter when the DNA damage response was induced following the addition of mitomycin C (MMC) (Figures 5a,b and S3a,c). In contrast to the WT cells, ~71.2% of the double deletion cells expressed the SOS reporter under normal growth conditions, which increased to ~ 91.3% upon treatment with MMC (Figure 5a,b, Figure S3b,d).
FIGURE 5.
Loss of RnhP and RnhC activates the SOS response under normal growth conditions. (a) Scoring of cells expressing the tagC::tagC-gfp reporter in WT and ΔrnhP rnhC::erm with and without mitomycin C treatment. The strain backgrounds and treatment status are indicated on the x-axis and the percent of cells expressing the reporter is indicated on the y-axis. The percent of fluorescent cells for each strain is indicated above the bar. For WT, WT with MMC treatment, ΔrnhP rnhC::erm, and ΔrnhP rnhC::erm with MMC treatment reporters 799, 744, 799, and 767 cells were scored per strain, respectively. (b) Single image GFP intensities for tagC::tagC-gfp reporter strains in WT and ΔrnhP rnhC::erm backgrounds plus treatment with mitomycin C. The GFP intensity per pixel was quantified for each strain and plotted. The white line used to quantify pixels for GFP intensity is indicated in each image. An enhanced image for each strain is also shown for comparison. The GFP intensity per pixel was quantified for each strain and plotted to demonstrate background fluorescence in (WT) relative to the fluorescence intensity observed in cells inducing SOS as measured by TagC-GFP fluorescence. The red bar in the upper right panel represents 4.5 μm
Therefore, we show that cells lacking rnhP and rnhC experience a ~14-fold increase in SOS induction during normal growth conditions, which may explain the slow growth and cell elongation results described above (Figure 4). These results further show that 3610 is able to mitigate the deleterious effects of RNA-DNA hybrids when either RnhP or RNase HIII (rnhC) is active. When both genes are nonfunctional, the consequences to genome integrity cause the majority of cells to induce the SOS response delaying cell division and impairing proliferation.
2.9 |. Cells lacking rnhP and rnhC exhibit replication stress near the terminus region
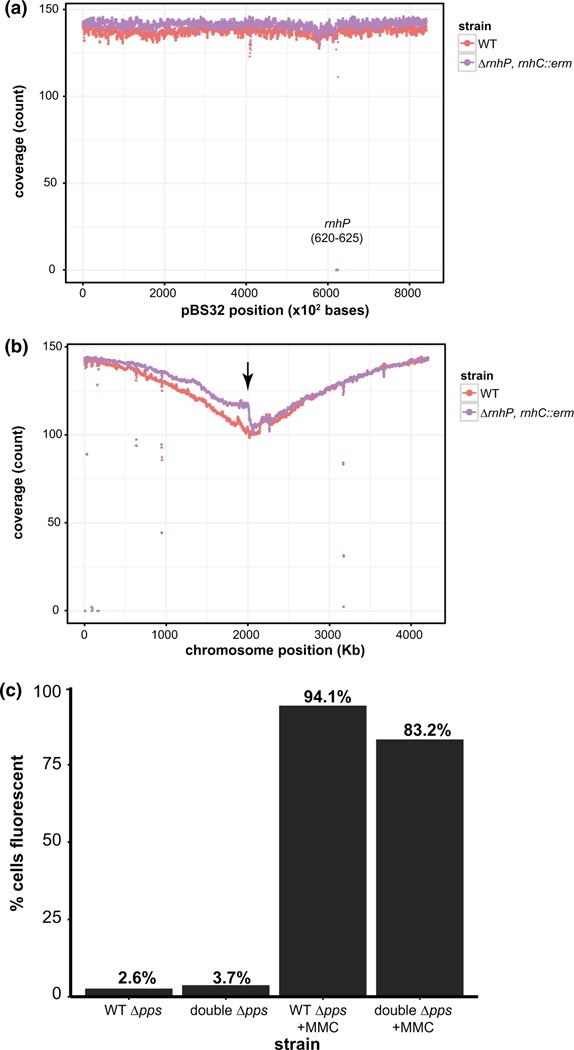
Having established that loss of rnhP and rnhC results in SOS induction for most cells during normal growth, we investigated the genome-wide replication status of the WT and double deletion strains in exponential phase cultures in an effort to understand the underlying cause. We isolated DNA from each strain in triplicate for single-end DNA-sequencing to determine chromosome and plasmid replication status. The resulting reads were aligned to the NCIB 3610 reference chromosome (Nye et al., 2017) and plasmid separately and the average coverage was plotted over the length of the reference. There was little to no difference in sequencing coverage between the WT and double deletion strain over the length of pBS32 (Figure 6a). We found a severe drop in sequencing coverage around 60 Kbp for pBS32 in the double deletion strain, which corresponds to the location of the deleted rnhP gene (62,030–62,497). When visualizing the sequencing coverage map for the chromosome and comparing the WT and double deletion strains, we noticed an abrupt drop in sequencing reads in the terminus region for the right replichore. This result shows that replication fork progression is slowed in this region for the double deletion strain (Figure 6b, below arrow).
FIGURE 6.
Loss of RnhP and RnhC results in laggard replication progression near the terminus. (a) Average plasmid coverage of exponentially growing WT and ΔrnhP rnhC::erm cells. The average count of sequencing coverage at each base (y-axis) for three independent replicates for reads aligned to the pBS32 reference over 100 bp regions are plotted in 10 bp sliding windows over the length of the plasmid on the x-axis. The plots for the WT and ΔrnhP rnhC::erm strains are indicated. (b) Average genome coverage of exponentially growing WT and ΔrnhP rnhC::erm cells. Average count of sequencing coverage at each base (y-axis) for three independent replicates for reads aligned to the NCIB 3610 reference (Nye et al., 2017) chromosome over 10 Kb regions is plotted in 1Kb sliding windows over the length of the chromosome (x-axis). The first origin proximal base in the reference genome represents position 1. The median value across each window was plotted. The plots for the WT and ΔrnhP rnhC::erm are indicated. The ppsA-E locus (1,960,230–1,997,989) is indicated by a black arrow. (c) Shown is a graph reporting the percent of cells with TagC-GFP expression (SOS) in the Δpps (TMN 141) or the ΔrnhPrnhC::erm Δpps (TMN142) strains. The number of cells scored for the analysis are as follows: TMN 141 untreated (n = 806), TMN 142 untreated (n = 852), TMN141 MMC treated (n = 909), and TMN 142 MMC treated (n = 850). The percent of cells expressing the SOS reporter is indicated above each bar
2.10 |. Deletion of the pps operon restores the replication defects in the terminus region and mitigates the SOS response in cells lacking rnhP and rnhC
The abrupt drop in sequencing reads near the terminus of the right replichore prompted us to examine the genes in this region more carefully. We began by identifying genes located immediately before the 2000 Kbp locus (Figure 6b). The dacC gene (1,998,372‒1,999,847), which encodes a carboxypeptidase, is a non-essential average length (1,476 nt) gene that is transcribed in the head-on direction with replication fork progression. Immediately upstream of the dacC gene is the pps operon (1,960,230–1,997,989) which encodes plipastatin synthase. The pps operon consists of five very long genes, ppsA (7,685 nt), ppsB (7,682 nt), ppsC (7,667 nt), ppsD (10,811 nt), and ppsE (3,839 nt), that are all transcribed in the head-on direction relative to replication (Nye et al, 2017). Due to the length and the orientation of these genes, it seems possible that R-loops could accumulate in this region and impair fork movement. We hypothesize that R-loops in the pps operon could be resolved in WT but not in the double deletion strain due to the lack of RnhP and RnhC activity. As previously discussed, R-loops in head-on oriented genes are well known to impair replication fork progression (Prado and Aguilera, 2005; Wang et al., 2007; Kim et al., 2007; Srivatsan et al., 2010; Lang et al., 2017), which could explain the observed drop in sequencing coverage at the terminus region. To test whether the pps operon was contributing to the replication defects near the terminus region of the right replichore, we created a clean deletion of the 37.7 Kbp pps operon (see supporting “Experimental Procedures”) in WT and ΔrnhP rnhC::erm backgrounds followed by harvesting genomic DNA from WT, ΔrnhP rnhC::erm, and the ΔrnhP rnhC::erm ΔppsA-E strains for paired-end sequencing. We note that the much higher sequencing coverage causes a less drastic appearance of the observed effects. We do, however, again observe replication defects in the terminus region of the right replichore (Figure S4a) that were present in all three biological replicates tested (Figure S4b,c). Further, we found that deletion of the pps operon in the ΔrnhP rnhC::erm background ameliorated the drop in sequencing coverage in the terminus region of the right replichore to WT levels (Figure S4d,e).With these results, we suggest that conflicts at the pps operon result in the replication defects observed in the double deletion strain.
Impaired replication fork movement in the double deletion strain due to replication-transcription conflict at the pps operon would also explain the SOS induction we observe with the tagC-gfp reporter. If replication-transcription conflicts within the pps operon were responsible for the SOS induction observed in the double deletion strain, then the deletion of the pps operon in the double RNase H mutant background should mitigate the constitutive SOS induction observed during normal cell growth in cells lacking rnhC and rnhP. We tested this idea by deleting the pps operon in the SOS reporter backgrounds for WT and the double rnhC and rnhP deletion (Figures 5 and S3). We note that upon the deletion of the pps operon, we were unable to visualize the cell membrane stained by FM4–64 with high enough quality to use the membrane stain as a proxy for cell boundaries. Therefore, when imaging cells with the pps deletion we used DIC images paired with their corresponding GFP images (Figure S5). It is possible that per image, the number of cells with the SOS response turned on were under-represented because when quantifying DIC images, we cannot visualize all closed septa. However, with that unavoidable caveat, when scoring WT and the double RNase H mutant bearing the pps deletion, cell scoring remained consistent (“Experimental Procedures”) (Figures 6c and Figure S5).
In contrast to the 71.2% of cells that expressed the SOS reporter during normal growth (Figure 5a) in the double RNase H deletion background, only 3.7% of double deletion cells expressed the SOS reporter under normal growth conditions in the absence of the pps operon. As a control, we challenged the ΔrnhPrnhC::erm Δpps cells with mitomycin C and find that 83.1% of cells were SOS induced (Figures 6c and S5). Taken together, we conclude that in the absence of rnhP and rnhC replication forks become laggard in the terminus after encountering R-loops or the transcription machinery in the head-on 37.7 Kbp pps operon that cannot be resolved in the absence of RnhC and RnhP.
2.11 |. Cells lacking both rnhP and rnhC show increased sensitivity to cellular stress
Having established that defects in both plasmid-encoded rnhP and chromosomally encoded rnhC genes results in decreased growth rate and inhibition of cell division, we asked how cells respond to various stressors in the absence of one or both of these RNase H enzymes. In B. subtilis PY79, ΔrnhC cells are sensitive to a myriad of cellular stresses, including cold shock, osmotic stress, and treatment with genotoxic agents (Lang et al., 2017; Randall et al., 2017; Randall et al., 2019). To test how the RNase H genes contribute to genotoxic stress responses, we tested the susceptibility of the single deletions, ΔrnhB, ΔrnhC, and ΔrnhP, as well as pairwise deletion strains, ΔrnhC rnhB::erm, ΔrnhP rnhB::erm, and ΔrnhP rnhC::erm, in 3610 to various stressors. We began by testing sensitivity to cold stress (growth at 25°C), which has been hypothesized to contribute to the stability of Okazaki fragments, and growth on sublethal concentrations of hydroxyurea (HU) (Davies et al., 2009; Randall et al., 2019). In contrast to WT B. subtilis PY79, deletion of the rnhC gene in 3610 resulted in only a modest sensitivity relative to WT in response to cold shock or growth in the presence of HU (compare second row Figure 7a,b). For cold shock and HU treatment conditions, the ΔrnhP rnhC::erm double mutant displayed ~100 and ~1,000-fold increases in sensitivity relative to WT cells, respectively (Figure 7a).
Given that RnhP can compensate for the loss of RnhC activity in the ancestral strain NCIB 3610, we asked if the expression of rnhP could rescue the cold and HU sensitivities observed in the ΔrnhC strain for B. subtilis PY79, which lacks pBS32 and thus a native rnhP gene. We created a strain that expresses rnhP from an IPTG inducible promoter from an ectopic chromosomal locus in the ΔrnhC background for PY79 and tested susceptibility to cold and HU stress. In support of our results from 3610, we find that ectopic expression of rnhP in a PY79 ΔrnhC background completely restored the cold and HU sensitivities to WT survival in the absence of IPTG, with > 100- and > 1,000-fold growth relative to the ΔrnhC strain for cold stress and HU, respectively (Figure 7b). With these results we conclude that uninduced expression of rnhP compensates for loss of RNase HIII when challenged with cold stress or HU demonstrating overlapping functions of rnhC and rnhP genes in B. subtilis.
3 |. DISCUSSION
RNase H enzymes are biologically universal and required for cleavage of the RNA moiety in an RNA-DNA hybrid (Ohtani et al., 1999b; Cerritelli and Crouch, 2009). RNase H genes are present in the genomes of bacteria, archaea, eukaryotes, and retroviruses (Ohtani et al., 2000; Li et al., 2004; Ohtani et al., 2004; Sparks et al., 2012). Eukaryotes show less diversity in the RNase H genes they encode. Almost all eukaryotes contain RNase HI and RNase HII (Ohtani et al., 1999b, Cerritelli and Crouch, 2009). Plants, including Arabidopsis, contain multiple RNase HI homologs because different RNase HIs are targeted to the nucleus, mitochondria, and chloroplast (Yang et al., 2017; Kucinski et al., 2020).
In the genomes of prokaryotes, RNase H enzymes show striking diversity between organisms (Ohtani et al., 1999b, Cerritelli and Crouch, 2009). Phylogenetic studies have shown that all prokaryotic genomes analyzed contain at least one RNase H with most genomes containing two RNase H genes (Kochiwa et al., 2007). In general, most bacteria contain RNase HI and HII, while a subset contains RNase HII and RNase HIII (Kochiwa et al., 2007). As RNase HI and HIII are active on the same class of substrates and because a prokaryotic genome had not been identified to encode functional RNase HI and RNase HIII, it had been proposed that RNase HI and HIII are mutually exclusive (Kochiwa et al., 2007). We show that rnhP (RNase HI) and rnhC (RNase HIII) contribute to genome maintenance in 3610 demonstrating that 3610 contains functional RNase HI, RNase HII, and RNase HIII enzymes. RNase HIII is chromosomally encoded, while rnhP is plasmid-encoded. We suggest that rnhP was acquired through horizontal gene transfer and has resided on the nonessential plasmid pBS32. Our experiments in vivo show that 3610 grows well with an ΔrnhC allele; however, 3610 grows poorly and experiences constitutive SOS induction when rnhP and rnhC are deficient, indicating that either RNase HI or RNase HIII is important for normal growth and resolution of RNA-DNA hybrids that form in vivo. Therefore, although rnhP is plasmid-borne, RNase HI activity from this gene product is important for genome maintenance in 3610 and, to our knowledge, this is the first organism described where functional RNase HI and RNase HIII have been shown to coexist.
As discussed above, we initially hypothesized that RnhP would be required for pBS32 maintenance or hyper-replication. In contrast, we find that RNase HIII (rnhC) was required for plasmid hyper-replication, while neither rnhP nor rnhC was important for normal plasmid maintenance. We find that while the rnhP deletion alone does not confer a phenotype, the ΔrnhC does confer slight growth interference to DNA damage from cold stress and hydroxyurea (HU), suggesting that RNase HIII is the more important enzyme in vivo. The double deletion of ΔrnhP rnhC::erm shows ~100-fold and ~1,000-fold growth interference from cold stress and HU treatment, respectively. If we compare the results of ΔrnhP rnhC::erm on HU for 3610 to the phenotype for ΔrnhC from B. subtilis strain PY79 we find the same extent of growth interference. Therefore, the comparison of phenotypes between 3610 and PY79 shows that ΔrnhP rnhC::erm in 3610 largely phenocopies the single ΔrnhC deletion for PY79 in the presence of HU and for cold sensitivity. Finally, we show that the PY79 ΔrnhC phenotype is rescued with ectopic expression of rnhP, further demonstrating functional overlap between RnhP and RNase HIII (rnhC) in B. subtilis.
Biochemical characterization shows that RnhP is an RNase H with specificity for substrates with four or more embedded ribonucleotides. RnhP is not active on a dsDNA or dsRNA substrate. Further, RnhP does not cleave a substrate with a single ribonucleotide nested in duplex DNA. Therefore, the biochemical characterization of RnhP shows that it is a strict RNase H with a preference for Mn2+. Our prior work characterizing RNase HIII showed that this enzyme was most active with Mg2+ on the canonical substrates for RNase HIII (Randall et al., 2017). One explanation for the coexistence of RNase HIII and RnhP is that metals could be scarce for wild Bacillus during growth in the soil. We speculate that RNase HIII is most active when magnesium concentrations are sufficient to support activity. During conditions when magnesium concentrations are lower and manganese concentrations are sufficient, RnhP could be more active providing RNase H activity and a fitness advantage for cells carrying both rnhC and rnhP. Given our studies with the double RNase H mutant, all experiments point to a model where the growth of 3610 is well supported with either RNase HIII or RnhP. It is the double deletion that grows poorly and is constitutively induced for the DNA damage response. We suggest that RNase HIII activity predominates and RnhP activity can be used to supplement RNase HIII during specific growth conditions or when the burden of RNA-DNA hybrid resolution overwhelms the capacity of RNase HIII.
Another important finding from this work is that the 37.7 Kbp plipastatin synthase operon (pps) transcribed head-on with the direction of DNA replication causes laggard replication fork progression in ΔrnhP rnhC::erm cells. We show that the laggard fork movement coincides with SOS induction in 3610 cells lacking RnhC (RNase HIII) and RnhP (RNase HI). Further, we show that deletion of the pps operon rescues the high percentage of SOS induction observed during normal growth in cells deficient for RnhC and RnhP. It is well established that head-on transcription can impair replication fork progression (Wang et al., 2007; Kim et al., 2007; Srivatsan et al., 2010; Lang et al., 2017). We speculate that R-loops are prevalent in the pps operon during normal growth and that it is an R-loop-mediated conflict with the replisome that results in SOS induction in the majority of cells lacking RnhC and RnhP. This result provides a native locus in B. subtilis that is a strong candidate for persistent R-loop formation that requires resolution by RNase HI or RNase HIII during normal growth conditions. It will be interesting to determine if there are other regions in the B. subtilis chromosome that are prone to R-loop formation causing impaired replication fork progression.
Prior phylogenetic work shows that only a small subset of bacteria in the phylum Firmicutes, including B. subtilis and Lactobacillus, contains genes for all three RNase H proteins (Kochiwa et al., 2007). Sequence comparisons showed that the predicted RNase HI genes in organisms with RNase HIII lack the catalytic residues and the substrate-binding α-helix 3 basic protrusion handle found in active RNase HI enzymes (Kochiwa et al., 2007). Moreover, prior functional studies of the chromosomally encoded and predicted RNase HI genes from B. subtilis, including YpdQ and YpeP, failed to detect RNase H activity, further supporting the argument that RNase HI and RNase HIII activities do not coexist (Randall et al., 2017). One possible limitation of prior phylogenetic studies would be if this work only interrogated core genomes. Our finding that RnhP has a different metal preference and cleavage site selection relative to RNase HIII could also provide a biochemical difference that allows for these genes to coexist as both contribute to RNA-DNA hybrid resolution. We speculate that functional RNase HI and RNase HIII are unlikely to coexist as chromosomally encoded genes. We wish to speculate that other bacteria will be identified to have RNase HI and RNase HIII with one gene encoded as part of the accessory genome and the other as part of the core genome. This would allow for acquisition, transfer, and loss of one RNase H gene and maintenance of both when a fitness advantage is conferred. As more genome sequences become available, it will be interesting to learn if other bacteria encode functional RNase HI and RNase HIII and how these genes contribute to growth and genome integrity.
4 |. EXPERIMENTAL PROCEDURES
4.1 |. General bacteriology
Unless otherwise specified, the antibiotic concentrations used in this study are as follows: 0.5 μg/ml of erythromycin, 100 μg/ml of spectinomycin, 5 mM hydroxyurea, and 20 ng/μl of mitomycin C. Strains were grown in Lysogeny Broth (LB) at 30°C. All strains, plasmids, and oligonucleotides used are listed in Tables S1, S2, and S3 of the Supplemental Material.
4.2 |. RNase H alignments
Global alignments were performed on the GenBank protein sequences for ZpdC (AGQ21310.1) from Bacillus subtilis NCIB 3610 and RnhA (NP_414750) from Escherichia coli MG1655 using the pairwise sequence alignment tool from Clustal Omega (https://www.ebi.ac.uk/Tools/psa/).
4.3 |. Spot plates
Designated strains were streaked from frozen stocks and grown overnight at 30°C on LB agar plates. Plates were washed in LB liquid media and used to inoculate 2 ml of cultures to an OD600 of 0.05. Cultures were grown in a 30°C rolling rack to an OD600 0.9–1.5. Cultures were diluted to OD600 of 0.5 in phosphate-buffered saline (PBS) pH 7.4 followed by 10-fold serial dilutions in 1x PBS. The dilution series was then spotted onto LB plates plus the indicated antibiotic or incubated at the indicated temperatures.
4.4 |. Growth rate analysis
Designated strains were streaked onto LB agar plates from frozen stocks and grown overnight at 30°C. Plates were washed in LB liquid media and inoculated at an OD600 of 0.05 into 25 mL of pre-warmed LB liquid media. The cultures were grown in a shaking water bath at 200 RPM at 30°C. The OD600 measurement for each culture was recorded every 30 minutes. Biological replicates were performed in triplicate on three separate days for each strain and the average growth measurement with corresponding standard errors were plotted. A modified Gompertz growth model in the form was fit to the replicates for each strain to obtain estimated growth rates (Zwietering et al., 1990). The parameters A, μm, and λ represent the time (t) when the growth rate equals zero (asymptote), the maximum growth rate, and the lag time, respectively (Zwietering et al., 1990). The estimated growth rate (μm) from the Gompertz model was then used to calculate doubling time estimates as ln(2)/μm for each strain (Zwietering et al., 1990; Randall et al., 2019). Additional growth rate parameters are shown in Table S4.
4.5 |. Genomic DNA purifications
Designated strains were streaked from frozen stocks onto LB agar plates and grown overnight at 30°C. Plates were washed in LB liquid media and used to inoculate 10 mL of LB liquid media at an OD600 of 0.05. The strains were grown in triplicate over three separate days prior to harvesting chromosomal DNA. At an OD600 of 0.5–0.7, the cells were pelleted via centrifugation, washed in 1 mL of re-suspension buffer (50 mM Tris–HCl pH 8 with 5% glycerol), and mixed in a final volume of 150 μl of re-suspension buffer. For cell lysis, Triton 100 was added to 1% (v/v), 10 μl of 10 mg/ml of RNase A, and lysozyme from the MasterPure™ Gram-positive DNA purification kit (Lucigen) were added and used as described. Subsequent lysis and purification steps were performed as described in the MasterPure™ Gram-positive DNA purification kit (Lucigen) protocol per the manufacturer’s instructions with the exception of the RNase treatment step, which was omitted because RNase treatment was performed during cell lysis.
4.6 |. DNA sequencing and chromosome coverage analysis
Library preparation and DNA single-end sequencing were performed by the University of Michigan DNA Sequencing Core. Sequencing reads were aligned using bwa (v 0.7.8-r455) to the NCIB 3610 chromosome reference (CP020102.1) and pBS32 (CP020103.1) reference (Li and Durbin, 2009, Nye et al., 2017). The resulting bam files were subsequently sorted and filtered using Samtools (v 0.1.18) for quality values greater than 30 (Li et al., 2009). PCR duplicates were removed using Picard tools (https://github.com/broadinstitute/picard ). The filtered bam files were used to calculate the genome coverage at each base for each replicate using genomeCoverageBed from bedtools (v 2.29.1) (Quinlan and Hall, 2010). The coverage at each base was averaged for the three replicates. The median coverage over 10kb windows was plotted every 1kb throughout the length of the chromosome and median coverage over 100 bp windows was plotted every 10 bp throughout the length of the plasmid using the packages ggplot2 and zoo in R (v 3.1.3) (Wickham, 2016, Zeileis and Grothendieck, 2005).
4.7 |. RnhP (D73N)
To create a catalytically inactive RnhP variant we mutated the aspartic acid residue (GAT) responsible for metal ion coordination at position 73 to asparagine (AAT) using overlapping PCR. The 5′ block was created using B. subtilis NCIB 3610 genomic DNA as a template with primers oTN58 and oTN61. Similarly, the 3′ block was created using primers oTN59 with oTN60. A PCR cleanup was performed and the purified products were combined using Gibson assembly (Gibson et al., 2009) to create pE-SUMOrnhPD73N. The generated plasmid was subsequently used to transform competent MC1061 cells and plated on 25 μg/ml of kanamycin. Resulting colonies were screened by PCR using primers oTN58 and oTN59 for the pE-SUMOrnhPD73N vector and the insert sequences were verified as correct using Sanger sequencing through the University of Michigan Core sequencing facility.
4.8 |. Protein Purification
Recombinant proteins were purified from E. coli BL21DE3 cells containing pE-SUMOrnhP and pE-SUMOrnhPD73N as described (Schroeder et al., 2017; Randall et al., 2019). Briefly, Cultures were grown in 2 liters of LB with 25 μg/ml of kanamycin at 37°C shaking to an OD600 of 0.7. Overexpression was induced by adding IPTG to 0.5 mM followed by growth for 3 additional hours. Cells were then pelleted by centrifugation and stored at −80°C. Once thawed, the pellet was resuspended in lysis buffer [50 mM Tris–HCl pH 8, 300 mM NaCl, 10% sucrose, 10 mM imidazole, 1x protease inhibitors (Roche 11,873,580,001)] and cells were sonicated using a Fisherbrand™ Q500 Sonicator at an amplitude of 70% for cycles of 10 seconds on and 20 seconds off for a total of 5 minutes on ice. Cell debris was cleared and pelleted by centrifugation. Supernatant was then applied to a 4 mL of Ni2+-NTA agarose gravity-flow column. The column was washed with wash buffer (50 mM Tris-HCl pH 8, 25 mM imidazole, 2 M NaCl, 5% glycerol) and eluted with elution buffer (50 mM Tris–HCl pH 8, 400 mM imidazole, 150 mM NaCl, 5% glycerol). Following elution, 1 mM DTT and SUMO protease were added to the eluate and incubated for 2 hours at room temperature. The SUMO protease treated sample was dialyzed into storage buffer (50 mM Tris–HCl pH 8, 150 mM NaCl, 5% glycerol) overnight at 4°C. The product was fractionated by application to a 4 mL of Ni2+-NTA gravity-flow column to separate the recombinant protein from the SUMO tag. SDS-PAGE was performed to confirm the SUMO tag was removed. The sample was then dialyzed into cation exchange start buffer (20 mM Tris–HCl pH 7.5, 5% glycerol, 1 mM DTT) overnight at 4°C. The dialyzed sample was purified using a HiTrap SP HP column (GE: 17–1152-01) with an elution gradient of 50–500 mM NaCl at a flow rate of 1 ml/min over 90 minutes. SDS-PAGE was performed and fractions containing pure protein were pooled. The RnhP (D73N) protein was concentrated, glycerol was added to 25%, aliquoted, flash-frozen in liquid nitrogen, and stored at −80°C. The RnhP protein eluted slightly earlier from the S-column and required further purification with size exclusion chromatography. The concentrated protein was applied to a HiPrep 16/60 Sephacryl S200 HR column (GE: 17–1166-01) at a flow rate of 0.6 ml/min with sizing column buffer (20 mM Tris–HCl pH 7.5, 200 mM NaCl, 1 mM DTT) and eluted in one peak. SDS-PAGE was again performed and fractions containing only pure protein were pooled, concentrated, glycerol was added to 25%, aliquoted, flash-frozen, and stored at −80°C.
4.9 |. RNase H activity assays
The end infrared (IR) dye-labeled substrates for the 1-rNMP, 4-rNMP, and all RNA substrates were created by mixing oJR209, oJR210, and oJR227, respectively, with oJR145 in a 1:2 μM ratio diluted in Buffer A (20 mM Tris–HCl pH8, 50 nM NaCl, 1 mM DTT) (Randall et al., 2017; Randall et al., 2019). The Okazaki fragment substrate was assembled by mixing oJR339 with oJR340 in a 1:2 μM ratio in Buffer A. The RNA-RNA and DNA-DNA hybrids were created by mixing oJR227/oJR166 and oJR348/oJR365, respectively, in 1:2 μM ratios in Buffer A. The oligos were annealed by heating at 98ᵒC for 1 min followed by cooling on the bench top to room temperature. Reactions totaling 10 μL in volume included 100 nM substrate and 4 or 50 nM protein as indicated (diluted from stock concentrations in Buffer A) in the in vivo metal concentration buffer (20 mM Tris–HCl pH8, 50 nM NaCl, 1 mM MgCl2, 10 μM MnCl2, and 1 mM DTT) (Randall et al., 2017; Randall et al., 2019). For NaOH treated samples, 200 mM of NaOH was added to 500 nM substrate. Reactions were allowed to proceed for 10 min at 37°C unless otherwise indicated. For all reactions except the RNA-RNA hybrid, 10 μl of stop buffer (95% formamide, 20 mM EDTA, 0.01% bromophenol blue) was added after 10 min, and reactions were placed at 98ᵒC for 5 min and subsequently snapcooled on ice. A denaturing 8M urea 20% polyacrylamide gel was prepared by pre-electrophoresing the gel at 250V for 30 min in TBE buffer. The gel was subsequently loaded with 4 μL of each reaction and electrophoresed at 250V for 1.5 hrs. For the RNA-RNA hybrid, 10 μL of RNA hybrid stop buffer (66% formamide, 9% formaldehyde, 17.5 mM EDTA, and 0.65x MOPS Buffer (10x MOPS buffer: 200 mM MOPS, 50 mM sodium acetate, 10 mM EDTA) was added after 10 min and the reactions were placed at 55ᵒC for 15 minutes. A denaturing 8M urea 20% polyacrylamide gel was prepared by pre-electrophoresing the gel at 100V for 30 min in 0.5× MOPS buffer. The gel was subsequently loaded with 4 μl of each reaction and electrophoresed at 150V for 2 hrs. For all gels, the products were visualized with a LI-COR Odyssey imager.
4.10 |. Plasmid growth analysis following induction
Strains grown overnight in LB at 22°C were subcultured into 50 ml of fresh LB to an OD600 of 0.1 and cultured at 37°C. OD600 was measured every 30 min until OD600 reached between 0.07 and 0.12. IPTG was added to a final concentration of 1 mM, and OD600 was measured every 30 min for a total of 4 hrs post-induction. Simultaneously, 100 μl of multiple 10-fold serial dilutions of each sample culture was plated on LB plates containing spectinomycin and incubated overnight at 37°C. The following day, plates containing individual colony forming units (CFUs) were counted to determine CFU/ml.
4.11 |. Plasmid copy number following induction
Strains grown overnight in LB at 22°C were subcultured into 50 ml of fresh LB to an OD600 of 0.1 and cultured at 37°C until OD600 reached between 0.07 and 0.12. IPTG was added to a final concentration of 1 mM and strains continued to grow for an additional 60 min. Four OD600 units of each sample were pelleted, and genomic and plasmid DNA was isolated from cells by Qiagen DNeasy Blood & Tissue Kit (Cat #69504). The concentration of isolated DNA was quantified by Nanodrop, samples were standardized to a DNA concentration of 10 ng/μl, and diluted 10- and 100-fold to 1 ng/μl and 0.1 ng/μl, respectively. Quantitative PCR was performed with all three dilutions to determine the plasmid copy number as previously described (Skulj et al., 2008). Primers 3106/3107 (sigA) were used to measure chromosomal DNA, and primers 6527/6528 (zpdE) were used to measure pBS32 DNA.
4.12 |. Live-cell microscopy
Each strain imaged was streaked from frozen stocks onto LB agar plates and grown at 30°C for 16 hrs. Plates were then washed with LB and 25 ml cultures were inoculated to an initial OD600 of approximately 0.05. Cultures were then placed in a water bath at 30°C with shaking at 212 RPM until reaching an OD600 of 0.6–0.9. Once the desired OD600 was reached, the cultures were filtered to concentrate the cells, and subsequently washed three times with 1x PBS. Cells were then pelleted via centrifugation and resuspended in 300 μL 1x PBS. The membrane was stained using 0.25 ng/μL of FM4–64. About 200 μL of cells were then placed onto a microscope slide with a 1% agarose pad as described (Simmons et al., 2008). After each slide was prepared, the slides were placed under an Olympus BX61 microscope equipped with a Hamamatsu camera (Lenhart et al., 2013). The microscope was focused under exposure of 20–30 ms. Then the microscope was switched from DIC to RFP setting in order to observe the membrane stain of FM4–64 with an exposure of 300 ms. Once the microscope was properly focused, an image was recorded. After a combined total of approximately 900 cells were imaged for each strain, images were adjusted for brightness, contrast, and gamma using the cellSens software (Olympus). The length of the cells was measured using the polyline tool of cellSens (Olympus). In order for a cell to be considered scorable, the cell membrane had to be clearly imaged from pole to pole. For cells undergoing division, incomplete septa were scored as one cell and complete septa were scored as two cells. For cells scored with the Δpps operon deletion all preparation was the same except individual cells were scored by visualizing invaginating septa through DIC imaging. We expect that this method could underestimate the number of cells scored since all septa and complete septa cannot be completely visualized through this procedure. We suggest that in some cases a cell scored as one could have actually been two cells. However, as described in the Results section, MMC addition turns on the SOS response allowing for a clear comparison between the untreated and treated conditions. Further, given the example micrographs shown in Figure S4, the scoring of DIC images and their corresponding GFP images was straightforward and consistent between the pps operon deletion strains.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Kevin Chang and Dr. Peter Freddolino for their guidance and assistance with the DNA sequencing analysis. This study was supported by the National Institutes of Health grant R35 GM131783 to DBK, National Science Foundation grant MCB 1714539, and in part by R35 GM131772 to LAS. AMB was supported by a National Institutes of Health post-doctoral fellowship GM123635 and TMN was supported by a National Science Foundation predoctoral fellowship (#DEG 1256260).
Funding information
Division of Graduate Education, Grant/Award Number: DEG 1256260; Division of Molecular and Cellular Biosciences, Grant/Award Number: MCB 1714539; National Institute of General Medical Sciences, Grant/Award Number: GM123635, GM131772 and R35 GM131783; National Institutes of Health, Grant/Award Number: GM123635; National Science Foundation, Grant/Award Number: MCB 1714539 and DEG 1256260
Footnotes
DATA AVAILABILITY STATEMENT
All data underlying each figure are available upon request without restrictions. The raw and processed files for the DNA sequencing data presented in Figures 6 and S4 have been deposited in the Gene Expression Omnibus by NCBI (Edgar et al., 2002) and are available through the SuperSeries accession number GSE15 4586 (https://www.ncbi.nlm.nih.gov/geo) ([Dataset] et al., 2020).
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section.
CONFLICT OF INTEREST
The authors have no conflict of interest to declare.
REFERENCES
- Asai T. and Kogoma T. (1994) D-loops and R-loops: alternative mechanisms for the initiation of chromosome replication in Escherichia coli. Journal of Bacteriology, 176, 1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda SS, Gonzalez-Pastor JE, Ben-Yehuda S, Losick R. and Kolter R. (2001) Fruiting body formation by Bacillus subtilis. Proceedings of the National Academy of Sciences USA, 98, 11621–11626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton RA, Kuster-Schock E, Auchtung TA and Grossman AD (2007) SOS induction in a subpopulation of structural maintenance of chromosome (Smc) mutant cells in Bacillus subtilis. Journal of Bacteriology, 189, 4359–4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burby PE, Simmons ZW, Schroeder JW and Simmons LA (2018) Discovery of a dual protease mechanism that promotes DNA damage checkpoint recovery. PLoS Genetics, 14, e1007512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton AT, DeLoughery A, Li GW, and Kearns DB (2019) Transcriptional regulation and mechanism of SigN (ZpdN), a pBS32-encoded sigma factor in Bacillus subtilis. mBio, 10(5), e01899–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerritelli SM and Crouch RJ (2009) Ribonuclease H: the enzymes in eukaryotes. FEBS Journal, 276, 1494–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chon H, Sparks JL, Rychlik M, Nowotny M, Burgers PM, Crouch RJ, et al. (2013) RNase H2 roles in genome integrity revealed by unlinking its activities. Nucleic Acids Research, 41, 3130–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [Dataset], Nye TM, Kearns DB, and Simmons LA (2020) DNA sequencing coverage in wild type and RNase H deficient Bacillus subtilis cells. GEO. [Google Scholar]
- Das U, Chauleau M, Ordonez H. and Shuman S. (2014) Impact of DNA3’pp5’G capping on repair reactions at DNA 3’ ends. Proceedings of the National Academy of Sciences USA, 111, 11317–11322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das U. and Shuman S. (2013) Mechanism of RNA 2’,3’-cyclic phosphate end healing by T4 polynucleotide kinase-phosphatase. Nucleic Acids Research, 41, 355–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies BW, Kohanski MA, Simmons LA, Winkler JA, Collins JJ and Walker GC (2009) Hydroxyurea induces hydroxyl radical-mediated cell death in Escherichia coli. Molecular Cell, 36, 845–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl AM, Eppinger M, Fricke WF, Rosovitz MJ, Rasko DA, Daugherty S, et al. (2012) Whole-genome sequences of Bacillus subtilis and close relatives. Journal of Bacteriology, 194, 2378–2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl AM, Losick R. and Kolter R. (2007) Bacillus subtilis genome diversity. Journal of Bacteriology, 189, 1163–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M. and Lash AE (2002) Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Research, 30, 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima S, Itaya M, Kato H, Ogasawara N. and Yoshikawa H. (2007) Reassessment of the in vivo functions of DNA polymerase I and RNase H in bacterial cell growth. Journal of Bacteriology, 189, 8575–8583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd and Smith HO (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Methods, 6, 343–345. [DOI] [PubMed] [Google Scholar]
- Gupta R, Chatterjee D, Glickman MS and Shuman S. (2017) Division of labor among Mycobacterium smegmatis RNase H enzymes: RNase H1 activity of RnhA or RnhC is essential for growth whereas RnhB and RnhA guard against killing by hydrogen peroxide in stationary phase. Nucleic Acids Research, 45, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwood CR and Cutting SM (1990) Molecular Biological Methods for Bacillus. Chichester: John Wiley & Sons, pp. 5–14. [Google Scholar]
- Huertas P. and Aguilera A. (2003) Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Molecular Cell, 12, 711–721. [DOI] [PubMed] [Google Scholar]
- Itoh T. and Tomizawa J. (1980) Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H. Proceedings of the National Academy of Sciences USA, 77, 2450–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayanagi K, Miyagawa M, Matsushima M, Ishikawa M, Kanaya S, Ikehara M, et al. (1990) Three-dimensional structure of ribonuclease H from E. coli. Nature, 347, 306–309. [DOI] [PubMed] [Google Scholar]
- Kawai Y, Moriya S. and Ogasawara N. (2003) Identification of a protein, YneA, responsible for cell division suppression during the SOS response in Bacillus subtilis. Molecular Microbiology, 47, 1113–1122. [DOI] [PubMed] [Google Scholar]
- Kearns DB and Losick R. (2003) Swarming motility in undomesticated Bacillus subtilis. Molecular Microbiology, 49, 581–590. [DOI] [PubMed] [Google Scholar]
- Kim N, Abdulovic AL, Gealy R, Lippert MJ and Jinks-Robertson S. (2007) Transcription-associated mutagenesis in yeast is directly proportional to the level of gene expression and influenced by the direction of DNA replication. DNA Repair (Amst), 6, 1285–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochiwa H, Tomita M. and Kanai A. (2007) Evolution of ribonuclease H genes in prokaryotes to avoid inheritance of redundant genes. BMC Evolutionary Biology, 7, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkol MA, Blair KM and Kearns DB (2013) Plasmid-encoded ComI inhibits competence in the ancestral 3610 strain of Bacillus subtilis. Journal of Bacteriology, 195, 4085–4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzminova EA, Kadyrov FF and Kuzminov A. (2017) RNase HII Saves rnhA Mutant Escherichia coli from R-Loop-associated chromosomal fragmentation. Journal of Molecular Biology, 429, 2873–2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucinski J, Chamera S, Kmera A, Rowley MJ, Fujii S, Khurana P, et al. (2020) Evolutionary history and activity of RNase H1-Like proteins in arabidopsis thaliana. Plant and Cell Physiology, 61, 1107–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang KS, Hall AN, Merrikh CN, Ragheb M, Tabakh H, Pollock AJ, et al. (2017) Replication-transcription conflicts generate R-loops that orchestrate bacterial stress survival and pathogenesis. Cell, 170(787–799), e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenhart JS, Sharma A, Hingorani MM and Simmons LA (2013) DnaN clamp zones provide a platform for spatiotemporal coupling of mismatch detection to DNA replication. Molecular Microbiology, 87, 553–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. and Durbin R. (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics, 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. (2009) The sequence alignment/map format and SAMtools. Bioinformatics, 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li TK, Barbieri CM, Lin HC, Rabson AB, Yang G, Fan Y, et al. (2004) Drug targeting of HIV-1 RNA.DNA hybrid structures: thermodynamics of recognition and impact on reverse transcriptase-mediated ribonuclease H activity and viral replication. Biochemistry, 43, 9732–9742. [DOI] [PubMed] [Google Scholar]
- Mo AH and Burkholder WF (2010) YneA, an SOS-induced inhibitor of cell division in Bacillus subtilis, is regulated posttranslationally and requires the transmembrane region for activity. Journal of Bacteriology, 192, 3159–3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Kumar D, Clark AB, Watt DL, Watts BE, Lundstrom EB, et al. (2010a) Genome instability due to ribonucleotide incorporation into DNA. Nature Chemical Biology, 6, 774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Watts BE, Kumar D, Watt DL, Lundstrom EB, Burgers PM, et al. (2010b) Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proceedings of the National Academy of Sciences of the USA, 107, 4949–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowotny M, Gaidamakov SA, Ghirlando R, Cerritelli SM, Crouch RJ and Yang W. (2007) Structure of human RNase H1 complexed with an RNA/DNA hybrid: insight into HIV reverse transcription. Molecular Cell, 28, 264–276. [DOI] [PubMed] [Google Scholar]
- Nye TM, Schroeder JW, Kearns DB and Simmons LA (2017) Complete genome sequence of undomesticated Bacillus subtilis strain NCIB 3610. Genome Announc, 5, pii: e00364–00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani N, Haruki M, Morikawa M, Crouch RJ, Itaya M. and Kanaya S. (1999a) Identification of the genes encoding Mn2+-dependent RNase HII and Mg2+-dependent RNase HIII from Bacillus subtilis: classification of RNases H into three families. Biochemistry, 38, 605–618. [DOI] [PubMed] [Google Scholar]
- Ohtani N, Haruki M, Morikawa M. and Kanaya S. (1999b) Molecular diversities of RNases H. Journal of Bioscience and Bioengineering, 88, 12–19. [DOI] [PubMed] [Google Scholar]
- Ohtani N, Haruki M, Muroya A, Morikawa M. and Kanaya S. (2000) Characterization of ribonuclease HII from Escherichia coli overproduced in a soluble form. Journal of Biochemistry, 127, 895–899. [DOI] [PubMed] [Google Scholar]
- Ohtani N, Yanagawa H, Tomita M. and Itaya M. (2004) Cleavage of double-stranded RNA by RNase HI from a thermoacidophilic archaeon, Sulfolobus tokodaii 7. Nucleic Acids Research, 32, 5809–5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oivanen M, Kuusela S. and Lonnberg H. (1998) Kinetics and mechanisms for the cleavage and isomerization of the phosphodiester bonds of RNA by bronsted acids and bases. Chemical Reviews, 98, 961–990. [DOI] [PubMed] [Google Scholar]
- Omer Bendori S, Pollak S, Hizi D. and Eldar A. (2015) The RapP-PhrP quorum-sensing system of Bacillus subtilis strain NCIB3610 affects biofilm formation through multiple targets, due to an atypical signal-insensitive allele of RapP. Journal of Bacteriology, 197, 592–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordonez H, Uson ML and Shuman S. (2014) Characterization of three mycobacterial DinB (DNA polymerase IV) paralogs highlights DinB2 as naturally adept at ribonucleotide incorporation. Nucleic Acids Research, 42, 11056–11070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parashar V, Konkol MA, Kearns DB and Neiditch MB (2013) A plasmid-encoded phosphatase regulates Bacillus subtilis biofilm architecture, sporulation, and genetic competence. Journal of Bacteriology, 195, 2437–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado F. and Aguilera A. (2005) Impairment of replication fork progression mediates RNA polII transcription-associated recombination. EMBO Journal, 24, 1267–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR and Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics, 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall JR, Hirst WG and Simmons LA (2017) Substrate specificity for bacterial RNase HII and HIII is influenced by metal availability. Journal of Bacteriology, 200, pii: e00401–00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall JR, Nye TM, Wozniak KJ and Simmons LA (2019) RNase hiii is important for Okazaki fragment processing in Bacillus subtilis. Journal of Bacteriology, 201, pii: e00686–00618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowen L. and Kornberg A. (1978a) Primase, the dnaG protein of Escherichia coli. An enzyme which starts DNA chains. Journal of Biological Chemistry, 253, 758–764. [PubMed] [Google Scholar]
- Rowen L. and Kornberg A. (1978b) A ribo-deoxyribonucleotide primer synthesized by primase. Journal of Biological Chemistry, 253, 770–774. [PubMed] [Google Scholar]
- Santos-Pereira JM and Aguilera A. (2015) R loops: new modulators of genome dynamics and function. Nature Reviews Genetics, 16, 583–597. [DOI] [PubMed] [Google Scholar]
- Schroeder JW, Randall JR, Hirst WG, O’Donnell ME and Simmons LA (2017) Mutagenic cost of ribonucleotides in bacterial DNA. Proceedings of the National Academy of Sciences USA, 114, 11733–11738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder JW, Randall JR, Matthews LA and Simmons LA (2015) Ribonucleotides in bacterial DNA. Critical Reviews in Biochemistry and Molecular Biology, 50, 181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons LA, Davies BW, Grossman AD and Walker GC (2008) Beta clamp directs localization of mismatch repair in Bacillus subtilis. Molecular Cell, 29, 291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons LA, Goranov AI, Kobayashi H, Davies BW, Yuan DS, Grossman AD, et al. (2009) Comparison of responses to double-strand breaks between Escherichia coli and Bacillus subtilis reveals different requirements for SOS induction. Journal of Bacteriology, 191, 1152–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skulj M, Okrslar V, Jalen S, Jevsevar S, Slanc P, Strukelj B, et al. (2008) Improved determination of plasmid copy number using quantitative real-time PCR for monitoring fermentation processes. Microbial Cell Factories, 7, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks JL, Chon H, Cerritelli SM, Kunkel TA, Johansson E, Crouch RJ, et al. (2012) RNase H2-initiated ribonucleotide excision repair. Molecular Cell, 47, 980–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivatsan A, Tehranchi A, MacAlpine DM and Wang JD (2010) Coorientation of replication and transcription preserves genome integrity. PLoS Genetics, 6, e1000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JD, Berkmen MB and Grossman AD (2007) Genome-wide coorientation of replication and transcription reduces adverse effects on replication in Bacillus subtilis. Proceedings of the National Academy of Sciences USA, 104, 5608–5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. (2016) ggplot2: Elegant Graphics for Data Analysis. Measurement: Interdisciplinary Research and Perspectives, 3rd edition. New York: Springer-Verlag, Vol. 17, 160–167. [Google Scholar]
- Williams JS and Kunkel TA (2014) Ribonucleotides in DNA: origins, repair and consequences. DNA Repair (Amst), 19, 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Hou Q, Cheng L, Xu W, Hong Y, Li S. and et al. (2017) RNase H1 cooperates with DNA gyrases to restrict R-loops and maintain genome integrity in arabidopsis chloroplasts. The Plant Cell, 29, 2478–2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao NY, Schroeder JW, Yurieva O, Simmons LA and O’Donnell ME (2013) Cost of rNTP/dNTP pool imbalance at the replication fork. Proceedings of the National Academy of Sciences of the USA, 110, 12942–12947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeigler DR, Pragai Z, Rodriguez S, Chevreux B, Muffler A, Albert T, et al. (2008) The origins of 168, W23, and other Bacillus subtilis legacy strains. Journal of Bacteriology, 190, 6983–6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeileis A. and Grothendieck G. (2005) zoo: S3 infrastructure for regular and irregular time series. Journal of Statistical Software, 14, 1–27. [Google Scholar]
- Zwietering MH, Jongenburger I, Rombouts FM and van ‘t Riet K. (1990) Modeling of the bacterial growth curve. Applied and Environment Microbiology, 56, 1875–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.