Abstract
Optogenetics utilizes photosensitive proteins to manipulate the localization and interaction of molecules in living cells. Because light can be rapidly switched and conveniently confined to the sub-micrometer scale, optogenetics allows for controlling cellular events with an unprecedented resolution in time and space. The past decade has witnessed an enormous progress in the field of optogenetics within the biological sciences. The ever-increasing amount of optogenetic tools, however, can overwhelm the selection of appropriate optogenetic strategies. Considering that each optogenetic tool may have a distinct mode of action, a comparative analysis of the current optogenetic toolbox can promote the further use of optogenetics, especially by researchers new to this field. This review provides such a compilation that highlights the spatiotemporal accuracy of current optogenetic systems. Recent advances of optogenetics in live cells and animal models are summarized, the emerging work that interlinks optogenetics with other research fields is presented, and exciting clinical and industrial efforts to employ optogenetic strategy toward disease intervention are reported.
Keywords: cross-disciplinary interface, gene regulation, optogenetics, organelle manipulation, signal transduction
1. Introduction
Optogenetics has been rapidly evolving for the past decade. Blessed by the precision and convenience of light manipulation, optogenetics empowers precise control of molecular activities at an unprecedented resolution in time and space. However, even with emerging online resources that aim to provide comprehensive, annotated databases for the ever-evolving field of optogenetics,[1] as well as the effort to streamline and standardize “customized” optogenetic tools,[2] it remains overwhelming to select an optogenetic device with appropriate photokinetics, dynamic range, and mode of action for a specific application. This challenge inspires us to carry out a comparative analysis of recent work in optogenetics to gain insights into the improved use of this emerging biotechnology.
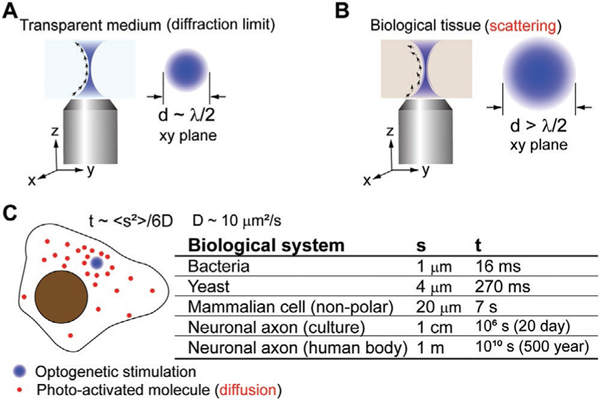
The sub-micrometer spatial resolution empowers optogenetics to probe molecular activities at the single-cell or subcellular level. However, this resolution could be compromised by tissue scattering or diffusion of photoactivated molecules (Figure 1). In a transparent medium, lens-based optical microscopy could focus a coherent light beam (e.g., laser) into a tiny spot, whose dimension is comparable to the size of the wavelength of the light. The diameter of the smallest beam waist is about half the size of the wavelength, which is referred to as the diffraction limit. Thus, for visible light, the theoretical diffraction limit is between 200 and 400 nm, much smaller than the size of a single cell (Figure 1A). However, in biological tissues that significantly scatter and absorb visible light, the spatial resolution could be compromised. In multicellular organisms, light absorption limits the penetration depth. The scattering of the light by the opaque biological tissues would expand the volume of light stimulation and reduce the spatial resolution. The scattering from the biological tissue can be thought of as a random variation of the coherent beam’s wavevector, disrupting the coherence and results in an enlargement of the focal volume (Figure 1B).
Figure 1.
A) Light diffraction limits the size of a coherent beam to approximately half of the excitation wavelength in lens-based optical microscopy. B) In opaque biological tissues, light scattering varies the coherent beam’s wavevector randomly and causes an expansion of the effective focal volume. C) molecular diffusion in cells further compromises the spatial resolution of the optogenetic stimulation. A back-of-envelop estimation of the traversing time of a small protein across a cell is presented. The typical value of a small protein’s diffusion coefficient in the cytoplasm is selected based on experimental measurement.[3]
In cultured cells where light absorption and scattering are not as significant, another factor could compromise the spatial resolution of optogenetic stimulation. After photostimulation, even within a diffraction-limited focal volume, diffusion of the photoexcited molecules (the experimentally measured diffusion coefficient can be found in reference [3]) could expand the region of activity (Figure 1C). To address this issue, one can use a mechanism to deactivate molecules that diffuse away from the focal spot of excitation light. For example, certain optogenetic systems can be switched off with a different color of light; therefore a patterned deactivation light can be designed with a spatially light modulator. Alternatively, in polarized cell types such as neurons, traversing across the whole cell takes a significantly longer time based on diffusion. Thus, it is easier to interpret phenotypes from a spatially localized optogenetic stimulation with a subcellular resolution. For instance, neuronal regeneration in live organisms could be guided by local optogenetic activation of Rac1,[4] Raf, and AKT[5] protein activities.
Because these spatial regulation issues are common to all optogenetic systems, we will not explicitly discuss the spatial resolution of individual ones covered in this work. On the other hand, biological events span a broad spectrum of temporal scale – conformational changes of macromolecules occur within micro- to milliseconds, molecular transport ranges from seconds to minutes, intracellular signal transduction typically takes minutes, gene expression lasts hours, and behavior changes happen within days or longer term. How can one take advantage of the temporal accuracy of optogenetics to study biological processes spanning these distinct timescales?
Here, we first introduce newly discovered or improved optogenetic tools, followed by discussing their categorized applications in live cells and animals. We then propose a procedure for optogenetic system design and validation, point out the cross-disciplinary research interlinking optogenetics with other research fields, and end by presenting the clinical and industrial efforts in pushing optogenetics toward disease intervention. Considering the fast growth of work in this field, we only focus on recent work from the past three years and encourage interested readers to refer to other excellent reviews on optogenetic research in delineating signal transduction[6] regulating embryonic development,[7] as well as their use in specific model systems.[8] For the same reason, channelrhodopsin and its derivatives, genetically encoded voltage indicators (GEVIs), genetically encoded calcium indicators (GECIs), and ion pump based optogenetics will not be discussed here.
2. Recent Advances in the Development of the Optogenetic Toolbox
Besides the tremendous growth of optogenetic applications using previously established photoactivatable proteins, new photoactivatable proteins have been discovered, and existing ones have been improved. In this section, we briefly highlight the progress in tool development.
2.1. New Photoactivatable Proteins
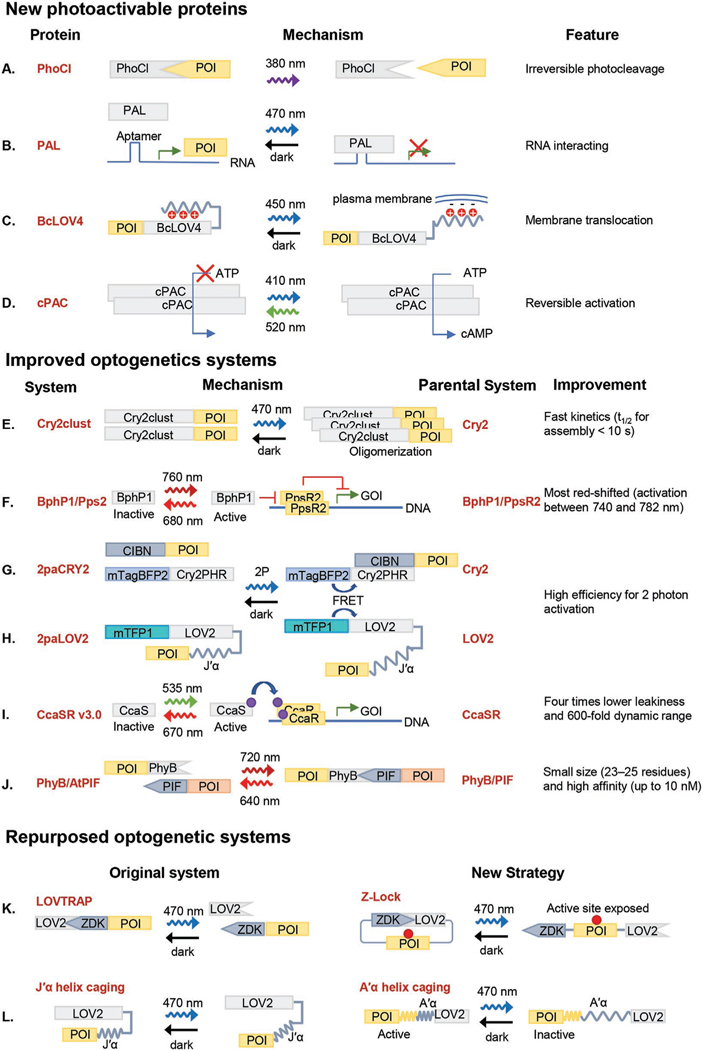
Most current photoactivatable proteins respond to light stimulation by undergoing allosteric change (e.g., the light-oxygen-voltage sensing domain, LOV) or intermolecular association such as UV resistance Locus 8 (UVR8), phytochrome (PhyB) and phytochrome interacting factor (PIF), cryptochrome and cryptochrome-interacting basic-helix–loop–helix (CIB), and light-induced dimerize (iLID). In contrast, light-inducible dissociation protein pairs are rare except for the recently developed LOV2 trap and release of protein (LOVTRAP) system, where blue light induces the dissociation between LOV2 and the small protein zDark.[9] A recent addition to this category is the photocleavable protein (PhoCl),[10] which is engineered from photoconvertible fluorescent protein mMaple. Violet light (380 nm) induces β-elimination, which cleaves a small peptide fragment from the backbone (Figure 2A). The finalized PhoCl system has a broad absorbance from 400 to 500 nm, and the dissociation rate is ≈500 s. Many proteins, such as Cre recombinase, Gal4 transcription factor, Pannexin-1 ion channel, could be controlled by PhoCl.[10] Compared with the reversible interaction of LOVTRAP, photocleavage of PhoCl is irreversible, and therefore, can be useful in applications with a low bearing threshold for phototoxicity.
Figure 2.
Recent advancements of optogenetic systems. A–D) Recently developed new photoactivatable protein systems. E–J) Improvement of existing optogenetic systems. K–L) Existing optogenetic systems with repurposed functions. References of each photoactivatable protein are listed as follows: A) PhoCl,[10] B) PAL,[11] C) BcLOV4,[12] D) cPAC,[14] E) CRY2clust,[16] F) BphP1/Pps2,[19] G,H) FRAPA,[23] I) CcaSR v3.0,[26] J) PhyB/AtPIF,[29] K) Z-Lock,[31] and L) usage of the A’α helix of LOV.[33]
Instead of employing protein–protein interaction in these optogenetic modalities, a recently identified bacterial LOV receptor whose architecture comprising Per-ARNT-Sim (PAS), AmiR and NasR transcription antitermination regulators, and LOV domain, dubbed as PAL, interacts with RNA. Aided by systematic evolution of ligands by exponential enrichment (SELEX) screening, the authors engineered short (fewer than 20 nucleotides in size) RNA aptamers that bind to PAL with around 20 × 10−9 m affinity in blue light and weaker than 1 × 10−6 m in darkness (Figure 2B).[11] These RNA aptamers were then embedded into the 5′-untranslated region of reporter genes, whose translation could then be inhibited by blue light in prokaryotes and eukaryotes.
Another new class of LOV variants contains a dikarya fungal LOVs associated with the regulator of G-protein signaling (RGS), which is located to the N-terminal of LOV. In addition, there is a C-terminal domain of unidentified function (DUF) with mixed α-helix/β-sheet. One of these variants, Botrytis cinerea BcLOV4, is dynamically and reversibly associated with the plasma membrane by binding to the anionic phospholipids (Figure 2C). Membrane association and undocking kinetics are fast (τon = 1.11 s; τoff = 89.1 s). Structural determination reveals that lipid-binding results from light-induced exposure of a polybasic amphipathic helix at the LOV-DUF linker inhibited by the RGS domain in the dark. [12] Membrane translocation of BcLOV4-Rac1 leads to actin polymerization and lamellipodia formation in HEK cells.[13]
Photoactivated adenylyl cyclases (PACs) have been used to manipulate cellular processes through light-dependent cAMP production. Flavin-based PACs have been successfully utilized in mammalian cells, but these PACs can only be turned off by spontaneous thermal decay. A cyanobacteriochrome-based photoswitchable adenylyl cyclase (cPAC) that can be reversibly activated (blue light) and inactivated (green light) overcame this drawback (Figure 2D).[14] This bidirectional control enables cPAC to reversibly control cAMP levels in live cells at a timescale of seconds to minutes. In Escherichia coli, this system yields ≈2.5 times more production under blue light illumination than cells exposed to green light. By changing the cyanobacteriochrome (CBCR) GAF domain, the cPAC system can be modified to absorb near IR light, increasing its versatility.
2.2. Improvement of the Photophysical Properties of Existing Photoactivatable Proteins
The photophysical properties of existing photoactivatable protein have been continuously improved, including reduced leakiness in the dark, broader responsive spectrum, increased affinity, and enhanced activation efficiency. Here we highlight several examples in this category.
Light-dependent homo-oligomerization is a common optogenetic strategy. For instance, cryptochrome2 (CRY2) from Arabidopsis thaliana oligomerizes under blue light.[15] An enhanced module termed “CRY2clust” was created by tagging a newly identified short peptide (ARDPPDLDN) (Figure 2E).[16] CRY2clust shows much faster kinetics (cluster assembly half-time t1/2 < 10 s; cluster disassembly half-time t1/2 ≈ 225 s) compared to the previously developed oligomerization tools CRY2[17] or CRY2olig.[18] CRY2clust also has a higher sensitivity, as clustering efficiency does not depend on CRY2cluster fusion proteins’ expression level.
Optogenetic tools with redshifted wavelength have attracted considerable attention because of the efficacy of deep-tissue applications. The bacteriophytochrome photoreceptor 1 (BphP1) is a near-infrared (NIR) sensing protein that can bind and inhibit the transcriptional repressor PpsR2.[19] By engineering a PpsR2-responding promoter in Escherichia coli, Ong et al. developed a bacterial sensor whose transcription can be controlled by near-infrared light.[19] This system shows a strong response to an activation wavelength of 740–782 nm, deactivation wavelength of 636–677 nm within a half-maximal activation time of 27 min (Figure 2F). Interestingly, the fusion of bacteriophytochrome (Bph) and bacterial adenylate cyclase (AC) generates a near-infrared light-activated AC, termed IlaM,[20] which has been optimized for expression and functionality in mammalian cells compared to the previous NIR PAC system.[21] Notably, by integrating the blue-light-sensitive AsLOV domain and the red light-sensitive BphP1/PpsR2 system with intrabodies, Redchuck et al. achieved multidirectional subcellular targeting of endogenous proteins.[22]
Two-photon optogenetic stimulation could also increase the penetration depth but remains challenging in vivo because of the small absorption cross-section for photoactivatable proteins in the red-infrared light region. This challenge has been indirectly addressed by Förster resonance energy transfer (FRET)-assisted photoactivation, or FRAPA, to activate photo activatable proteins. Kinjo et al. developed 2paCRY2 by selecting mTagBFP2 and flavin adenine dinucleotide (FAD) as the donor and acceptor FRET pair, respectively. mTagBFP2 effectively absorbs 840 nm and activates cryptochrome by energy transfer to its cofactor, FAD (Figure 2G). This system enabled extracellular signal-regulated kinase (ERK) activation in the mouse auricular epidermis. Notably, the FRAPA system also allowed two-photon activation of the LOV domain by using mTFP1 (a cyan fluorescent protein) as the donor to activate flavin mononucleotide, the cofactor of LOV (Figure 2H).[23]
Multiplexing optogenetics is another attractive feature to take full advantage of the visible spectrum for optogenetic applications. Commonly used optogenetic proteins include the blue light-sensitive LOV, cryptochrome, and the red light-sensitive phytochromes. A new addition to the family of optogenetic tools is the green light-sensitive photoactivable proteins. CcaSR is a two-component system activated by the green light and deactivated by the red light.[24] The system consists of cyanobacteriochrome sensor histidine kinase, CcaS, and response regulator, CcaR. Upon illumination, CcaR will be phosphorylated by CcaS and bind to promotors to activate gene transcription. The recent improvement, including the CcaSR v 2.0 system (with optimized output promoter)[25] and the CcaSR v 3.0 system (deletion of two PAS domains within CcaS), produced lower leakiness and higher dynamic range (Figure 2I).[26] The CcaSR system has been successfully used to control gene transcription in Bacillus subtilis.[27] Another green light-sensitive system is the cobalamin (vitamin B12) binding domains (CBDs) of bacterial CarH transcription factors, which undergo green light-dependent dissociation. In the dark, CarH is a dimer of dimer; upon green light irradiation, photocleavage of the 5′-deoxyadenosylcobalamin (AdoCbl) cofactor causes dissociation.[28] These new tools make it possible to employ multiplexed optogenetic stimulations with different colors of light.
Searching for the minimally required structure for optogenetic stimulation would benefit the broader usage of optogenetic tools. Recent work has developed a series of A. thaliana PIFs (AtPIF) (Figure 2J).[29] The identified small-size (23–25 residues) and high-affinity (up to 10 × 10−9 m) variants improve their usage in mammalian cells.
Eliminating the need for exogenous cofactors would facilitate the use of optogenetics in more diverse model systems. For example, PhyB-PIF interaction requires phycocyanobilin (PCB) as a cofactor, which is not produced in mammalian cells. To efficiently synthesize PCB in mammalian cells, vectors that coexpress heme oxygenase1 (HO1) and ferredoxin oxidoreductase (PcyA) with Ferredoxin and Ferredoxin-NADP+ reductase was generated.[30] Moreover, the PCB concentration can be further increased by depletion of biliverdin reductase A. This genetically encoded PCB synthesis system would help generate transgenic animals that allow for PhyB-PIF based optogenetics without PCB injection.
2.3. Repurposing Existing Optogenetic Systems
Considering the module-like optogenetic proteins, one could create new functionalities by repurposing optogenetic tools. Here we briefly introduce some examples in this category.
In contrast to the light-mediated association, LOVTRAP enables photoinducible protein dissociation between the LOV domain and the Zdk epitope.[9] By fusion of the LOV and Zdk domains to the termini of the target protein, LOVTRAP has been repurposed for photocaging, which was termed as the Z-lock system.[31] Light-mediated dissociation of LOV and Zdk exposes the active site and turns on protein activity (Figure 2K). Optimization of the Z-lock system can be performed by computer-assisted protein design. As demonstrated in the control of cofilin and αTAT, the Z-lock system works for target proteins with a single active site.
The allosteric change of the Jα helix in LOV2 has been commonly used to cage specific proteins fused to the C-terminus of LOV2. However, the N-terminal A’α helix is also destabilized during photoactivation.[32] Recently, the caging effect of the A’α helix for optogenetic control has been demonstrated in controlling protein tyrosine phosphatase 1B (PTP1B), where the A’α helix of LOV2 was fused to the C-terminal α7 helix of PTP1B.[33] A light-dependent unwinding of the A’α helix successfully destabilizes the α7 helix and further disrupts the activate site of PTP1B (Figure 2L).
New functions can also be achieved by combinatorial use of optogenetic modules. For instance, to design a generalizable platform for optogenetic protein control, the generalizable light modulated protein stabilization system (GLIMPSe)[34] was developed by combining the blue light-responsive tobacco etch virus (TEV) protease fused to the light-inducible nuclear export system (LEXY)[35] and the caging effect of LOV.[36] Different families of proteins, such as kinases and phosphatases, can be controlled by the same GLIMPSe platform.
Besides the commonly used photoinduced association of gene activation domains to the promoter, controlling the transcription factor’s accessibility is another way to modulate gene activity. By sequestering the transcription factor on the plasma membrane, one can limit its accessibility to the nucleus. Using calcium- and light-gated switch to cleave the membrane-sequestered transcription factor, Lee et al. and Wang et al. have developed Cal-Light[37] and fast light- and activity-regulated expression (FLARE),[36] respectively. Both systems have combined the modality of photo-uncaging (of the TEV cleavage site) and protein translocation (of the protease) to achieve gene transcription control in vivo. These tools enable the dissection of neural circuits underlying complex animal behaviors.
3. Spatiotemporal Optical Control of Intracellular Signal Transduction
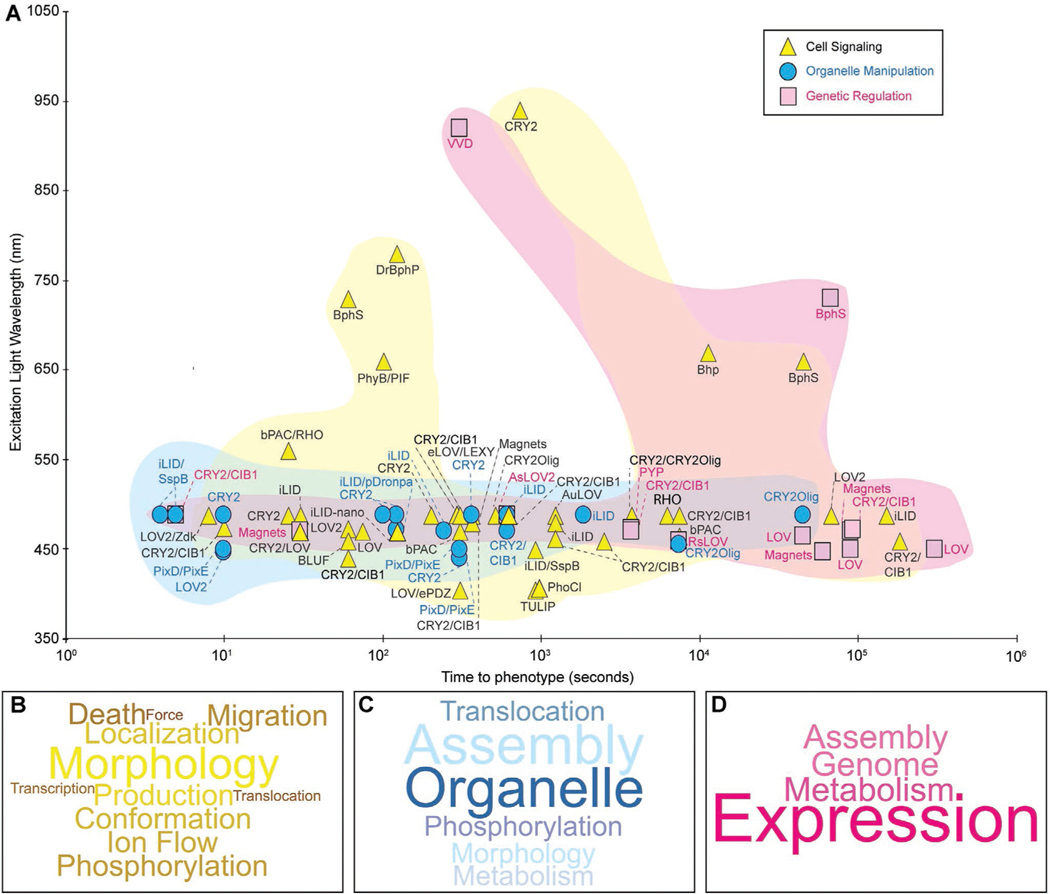
This section highlights recent updates in the optical control of intracellular signal transduction, which transmits extracellular signals to the cell interior. We will follow an “outside-in” route of signal transduction and group our discussion based on the subcellular localization of signaling molecules, i.e., from the membrane-bound receptor, cytosolic signaling molecules, organelles, to gene regulation in the nucleus. To better comprehend the recent progress of optogenetic control of molecular activities, we created a graph mapping the excitation wavelength and the typical time scale from the selected work (Figure 3). We also compiled a list to highlight the experimental conditions, including the wavelength, power density, model organisms (Table 1), and temporal kinetics of the phenotypes reported in these work. (Table 2). Note that the temporal kinetics is not the direct association or dissociation kinetics for specific photoactivatable proteins per se, but rather the time scale during which photoactivatable proteins elicit functional outcomes.
Figure 3.
Applications of optogenetics in steering molecular activity in cells and organisms. A) A scattered plot depicting selected work of optogenetic control of molecular activities in the past three years. Strategies are grouped into three categories: cell signaling (yellow),[20,34,38–49,51a,52–59,61,67,69–76,79–83,66] organelle manipulation (cyan),[84–87,89–100] and genetic regulation (magenta).[101,102,105–107,109–118] Excitation wavelength refers to the light used to activate the optogenetic proteins, which are individually labeled. The time to phenotype was chosen based on reported timescales outlined in Table 2. Word clouds indicating the frequency of each outcome per category: signaling B), organelle manipulation C), and genetic regulation D). The outcomes are also summarized in Table 2. The word clouds were generated in Wordcloud.com.
Table 1.
Experimental parameters in selected optogenetic research cited in this work. The color code of the table matches those in Figure 3 and corresponds to the selected topics in the main text, i.e., optogenetic control of signal transduction (yellow), organelle manipulation (cyan), and gene regulation (pink). In cases where power density is not reported, the total power is filled.
| Optogenetic protein(s) | λ[nm] | P [mW cm−2a)] | Model organisms | Ref. |
|---|---|---|---|---|
| AuLOV | 488 | 0.3–5.0 | PCI 2 (Rattus norvegicus) | [38] |
| DrBphP | 780 | 0.3–3 | PC6–3 (Rattus norvegicus), mouse | [39] |
| LOV2pep/PDZ | 447 | 0.3 | PC6–3 | [39] |
| iLI D/tdrano | 488 | 0.4–9700 | PCI 2 | [40] |
| CRY2PHR/CIBN | 460 | 0.4–5.0 | HEK293T (Homo sapiens), Xenopus laevis embryos | [41] |
| CRY2olig | 488 | 10–1500 | Mouse embryonic fibroblast, mouse lateral amygdala | [43] |
| LOV2/Zdk | 470 | 0.016 | Jurkat (Homo sapiens) | [44] |
| CRY2/CRY20lig | 488 | 0.6 | Drosophila | [47] |
| MagnetsHigh | 488 | 4–49 | HEK293T, NIH3T3 (Mus musculus), HeLa (Homo sapiens) | [49] |
| eLOV/LEXY | 488 | 0.5 | HEK293T, PCI 2 | [34] |
| LOV | 470 | 0.6–6.4 | Cerebellar granule neuron (Rattus norvegicus), HEK293T | [52] |
| CRY2/iLID | 470 | 4 | HeLa, HEK293, COS-7 (Cercopithecus aethiops) | [54] |
| CRY2 | 473 | 0.1–50 | HeLa, HEK293T | [55] |
| CRY2/CIBN | 475 | 737 | HEK293T | [57] |
| LOV2 | 473 | 3–100 | Mouse, hippocampal slices, HeLa, HEK293 | [58] |
| bPAC | 470 | 7 | HEK293, Caenorhabditis elegans motor neurons | [61] |
| bPAC | 470 | 18 | MIN6 and /βTC48 /β-cells (Mus musculus) | [66] |
| EB1 | 460 | 0.1–2.0 | Escherichia coli (Bacteria) | [69] |
| BphS | 660 | 1.33 | Escherichia coli | [71] |
| TULIP | 405 | a)<l μW | NIH3T3 | [72] |
| Rhodopsin | 475 | 0.3 | Drosophila | [46] |
| iLID-nano | 470 | 1 | Mouse kidney fibroblast | [75] |
| PhoCI | 405 | 2–150 | MDCK (Canisfamitiaris), MCF7 (Homo sapiens) | [76] |
| LOV2 | 488 | a)1.2–5.0 mW | Drosophila | [80] |
| iLI D/SspB/Zdk/LOV2 | 480 | 1–2.5 | Yersinia enterocolitica (Bacteria) | [81] |
| iLI D/pdDronpal | 472 | 5300 | U20S (Homo sapiens), COS-7 | [84a] |
| iLID | 488 | 0.5 | HeLa, HEK293T, ETNA (in-house derived, Mus musculus) | [84b] |
| iLID | 488 | a)25 mW | HCT116 (Homo sapiens), HeLa | [85] |
| iLI D/WD52C | 470 | 0.02–20 | HeLa, U20S, COS-7 | [86] |
| CRY2/CIBN | 470 | a)200 mW | MIN6 (Mus musculus), INS-1E (Rattus norvegicus) | [87] |
| CRY2olig | 488 | a)180 nW | N1H3T3, Xenopus laevis oocytes | [90] |
| CRY2PHR | 440 | a)0.02–1.5 μW | HEK293T | [91] |
| iLI D/SspB | 488 | a)<0.1 μW | NIH3T3, HEK293T | [93] |
| PixD/PixE | 450/488 | b)1.9–2.2 | Saccharomyces cerevisiae | [95] |
| iLI D/SspB | 488 | 8.4e6 | HeLa, HEK293, U20S, Lenti-X293T (Homo sapiens) | [96] |
| CRY2PHR | 488 | 4.5e3–2.5e9 | U20S, iPSC neurons (Homo sapiens) | [97] |
| CRY2olig | 456 | 0.69 | Zebrafish | [99] |
| PYP | 473 | a)0.2–1.5 mW | HEK293T, primary hippocampal neurons, mouse | [101] |
| Magnets | 448 | 0.2–20 | HEK293T, mouse liver | [105] |
| VVD | 920 | a)5–25 mW | Mouse brain | [106] |
| MagHigh | 470 | 40–1000 | HEK293T, mouse brain | [107] |
| BphS | 730 | c)2.0–3.3 | HEK293, iPSC, mouse | [110] |
| CRY2/CIB1 | 488 | 49–70 | HeLa, NIH3T3, hippocampal culture (Rattus norvegicus) | [114] |
| LOV | 450 | 2.0 | Saccharomyces cerevisiae | [115] |
| LOV | 450 | 0.1–2 | Mesenchymal stem cell (Rattus norvegicus) | [116] |
| LOV | 465 | 0.8 | HEK293T, Neuro-2a (Mus musculus) | [117] |
| RsLOV | 460 | 0.059–3 | Escherichia coli | [118] |
| CRY2/CIBN | 470 | 1.5–5 | ES (Mus musculus) | [113] |
When the power density is not reported explicitly, the entry refers to the total power;
Converted from the reported value of 73–82 μmole (m2 s)−1 at 450 nm;
Converted from the reported value of 77.5–129 μmole (m2 s)−1 at 465 nm.
Table 2.
Compilation of optogenetic control of molecular activity and cellular functions cited in this work. The color code of the table matches those in Figure 2 and corresponds to the selected topics in the main text, i.e., optogenetic control of signal transduction (yellow), organelle manipulation (cyan), and gene regulation (pink). Each phenotype has been assigned to a category, and the resource is highlighted as figure numbers in the original reference.
| Protein (s) | λ[nm] | t[s] | Phenotype | Category | Resource | Ref. |
|---|---|---|---|---|---|---|
| AuLOV | 460 | 1200 | BiFC split Venus protein reconstitution | Conformation | Figure 2D | [38] |
| AuLOV | 460 | 600 | Protein phosphorylation | Phosphorylation | Figure 4A | [38] |
| DrBphP | 780 | 120 | Protein translocation | Localization | Figure 4D | [39] |
| DrBphP | 780 | 20 | Calcium signaling | Ion flow | Figure 5B | [39] |
| iLID | 488 | 144 000 | PCI2 neurite outgrowth | Morphology | Figure 3C | [40] |
| CRY2/CIB1 | 460 | 172 800 | cFos luciferase assay | Transcription | Figure 2D | [41] |
| CRY2/CIB1 | 460 | 54 000 | Tail-like structure formation | Morphology | Figure 3B | [41] |
| CRY2/CIB1 | 440 | 60 | Protein translocation | Localization | Figure 2B | [42] |
| CRY2olig | 488 | 500 | Cluster formation | Conformation | Figure 1C | [43] |
| LOV2/Zdk | 488 | 8 | DAG production | Production | Figure 2C | [44] |
| PhyB/PIF | 660 | 100 | Calcium influx | Ion flow | Figure 5B/6B | [45] |
| CRY/CRY2olig | 488 | 3600 | Ventral furrow formation | Morphology | Figure 4C | [47] |
| Magnets | 488 | 360 | Optobody binding | Conformation | Figure 1C | [49] |
| CRY2 | 488 | 198 | AM PAR membrane translocation | Localization | Figure 2C | [48] |
| eLOV/LEXY | 488 | 300 | Protein nuclear export | Localization | Figure 3A | [34] |
| LOV | 470 | 73 | Protein photoswitching | Conformation | Figure 3A | [52] |
| CRY2/CIB1 | 401/491 | 360 | GEF recruitment | Localization | Figure 1C | [53] |
| CRY2 LOV | 470 | 28 | Calcium signaling | Ion flow | Figure 1E | [54] |
| CRY2 | 488 | 25 | Calcium flux | Ion flow | Figure 1D | [55] |
| CRY2 | 940 | 300 | Nuclear translocation | Localization | Figure 4B | [56] |
| CRY2/CIB1 | 475 | 50 | Calcium oscillation suppression | Ion flow | Figure 3B/C | [57] |
| LOV2 | 473 | 60 | Inhibition of structural LTP | Morphology | Figure 4A | [58] |
| CRY2/CIB1 | 488 | 600 | PA sensor accumulation | Production | Figure S3 | [59] |
| iLID | 488 | 30 | PIP2 depletion | Production | Figure 2C | [60] |
| bPAC | 470 | 3–8 | Crawling speed increase | Morphology | Figure 1B | [61] |
| Bph | 660 | 10 800 | cAMP-de pen dent protein expression | Production | Figure 5C | [20] |
| bPAC | 470 | 300 | cAMP increase | Production | Figure 1D | [66] |
| bPAC | 465 | 7200 | Blood glucose levels decrease | Production | Figure 3D | [67] |
| BphS | 610–730 | 60 | Aerotaxis | Migration | Figure 3A | [70] |
| BLUF | 460 | 60 | Swim zone increase | Migration | Figure 4A | [69] |
| BphS | 660 | 129 600 | Tryptophan production | Production | Figure 5C | [71] |
| TULIP | 405 | 900 | Actin accumulation | Localization | Figure 1E | [72] |
| LOV/ePDZ | 405 | 300 | Cell junction contraction | Morphology | Figure 1D | [73] |
| CRY2/CIB1 | 460 | 1200 | Cell migration | Migration | Figure 3A | [74] |
| Rhodopsm | 488 | 6000 | Cell protrusion | Morphology | Figure 4 S5B | [46] |
| iLID-nano | 470 | 120 | Force generation | Force | Figure 5A | [75] |
| PhoCI | 405 | 1800 | Photocleavage of PC-cadherin | Conformation | Figure 2 A | [76] |
| CRY2/CIB1 | 488 | 93 600 | Cell apoptosis | Death | Figure 2B | [79] |
| LOV2 | 488 | 64 800 | Neuronal cell degeneration | Death | Figure S8C | [80] |
| iLID/Zdk | 480 | 1200 | Protein secretion inhibition | Translocation | Figure S8 | [81] |
| CRY2/CIB1 | 488 | 300 | Damping of cortical contraction | Morphology | Figure S6B | [82] |
| CRY2/CIB1 | 463 | 1200 | ERK phosphorylation | Phosphorylation | Figure 3B | [83] |
| i LI D/SspB | 450 | 900 | ERK-KTR translocation | Phosphorylation | Figure S6C | [51a] |
| iLID/pdDronpal | 472 | 120 | Membrane curvature change | Morphology | Figure 3D | [84a] |
| iLID | 488 | 1800 | Mitophagy | Organelle | Figure 2A | [84b] |
| iLID | 488 | 390 | Mitotic spindle pulling | Organelle | Figure 1B | [85] |
| iLID/WD52C | 470 | 240 | Endosome recruitment | Organelle | Figure 4H | [86] |
| CRY2/CIB1 | 470 | 900 | Mitochondria recruitment | Organelle | Figure 1J | [87] |
| LOV2 | 488 | 10 | Myosin VI tethering to peroxisomes | Organelle | Figure 4A | [89] |
| CRY2 | 488 | 180 | Change in nucleolus material property | Organelle | Figure 1B | [90] |
| CRY2 | 440 | 300 | IDP condensation | Assembly | Figure 1C | [91] |
| CRY2 | 488 | 360 | 53BP1 cluster formation | Assembly | Figure 4A | [92] |
| iLID/SspB | 488 | 4 | Droplet condensation | Assembly | C-E | [93] |
| PixD/PixE | 450 | 10 | Protein droplet disassembly | Assembly | Figure 3D | [94] |
| CRY2 | 450 | 600 | RTK signaling activation | Phosphorylation | Figure S6C | [94] |
| CRY2 | 488 | 300 | Assembly of metabolic enzyme | Assembly | Figure S2AB | [95] |
| PixD/PixE | 488 | 300 | Disassembly of metabolic enzyme | Assembly | Figure S2C | [95] |
| iLID/SspB | 488 | 5 | Intracellular droplet condensation | Assembly | Figure 1C | [96] |
| CRY2 | 488 | 15 | OptoGranule formation | Assembly | Figure 1B | [97] |
| CRY2olig | 488 | 43 200 | Inclusion formation | Assembly | Figure 1C | [98] |
| CRY2olig | 456 | 7200 | TDP-43 nuclear export | Translocation | Figure 2D/E | [99] |
| CRY2 | 488 | 6000 | Oxygen consumption | Metabolism | Figure S6B/8B | [100] |
| PYP | 473 | 7200 | CREB expression | Expression | Figure 1E | [101] |
| CRY2/CIB1 | 470 | 3600 | ZLD-dependent gene expression | Expression | Figure 1B/1E | [102] |
| Magnets | 448 | 43 200 | Inducible gene expression | Expression | Figure 2C | [105] |
| WD | 900 | 360 | Inducible gene expression | Expression | Figure S2 | [106] |
| Magnets | 473 | 30 | PA-Flp activation | Assembly | Figure S3 | [107] |
| Magnets | 470 | 86 400 | mRNA level change | Expression | Figure 3A | [109] |
| BphS | 730 | 21 600 | ASCL1 mRNA expression | Expression | Figure 3B | [110] |
| CRY2/CIB1 | 488 | 60 | RNA trapping | Assembly | Figure 1F | [114] |
| LOV | 450 | 288 000 | Glucose consumption | Metabolism | Figure 3C/D | [115] |
| LOV | 450 | 86 400 | BMP2 and Lhx8 expression | Expression | Figure 1C/D | [116] |
| LOV | 465 | 43 200 | TFEB expression | Expression | Figure 3A/B | [117] |
| RsLOV | 460 | 7200 | RNA expression | Expression | Figure 3B/C | [118] |
| Magnets | 470 | 86 400 | Gene editing | Genome | Figure 2C | [111] |
| AsLOV2 | a)488 | 600 | H2B deubiquitination | Epigenetics | Figure 5A | [112] |
| CRY2/CIB1 | 470 | 86 400 | Looping of genome | Genome | Figure 4A/B | [113] |
The excitation wavelength is retrieved from cited reference in the original publication.
3.1. Signaling Cascades
3.1.1. Receptors
Intracellular signal transduction often starts with binding of a ligand to a membrane-bound receptor, which then activates the receptor, followed by the recruitment of adaptor proteins and activation of the downstream signaling pathways. Here, we introduce recent strategies to turn on several receptors, including receptor tyrosine kinases (RTKs), T-cell receptors (TCRs), Wnt receptor, Notch receptor, glutamate receptor, and antibodies.
RTK:
RTKs are single-transmembrane receptors whose activation requires receptor dimerization. By fusing the photoactivatable dimer, the LOV domain of aureochrome 1 from Vaucheria frigida or AuLOV, to the intracellular domain of TrkA, Khamo et al. induced the activation of tropomyosin receptor kinase A (TrkA) in the absence of nerve growth factor.[38] The system allows for delineating the synergistic contribution of the residues Y490 and Y785 to the activation of the downstream ERK signaling pathway.[38] Using a fusion protein of Deinococcus radiodurans bacterial phytochrome, DrBphP, and the cytoplasmic domains of Trks, Leopold et al. developed Dr-TrkA and Dr-TrkB, whose activation is induced by 780 nm (or darkness) and inactivation is triggered by 650 nm light.[39] Interestingly, activation of RTK can be achieved by recruiting the cytosolic domain of RTK from the cytoplasm to the plasma membrane.[40] Such a design has been shown to exhibit amenable dark activity for the fibroblast growth factor receptor (FGFR), TrkA, TrkB, and TrkC, which is critical for their use in developing Xenopus laevis embryos.[41]
Membrane-recruitment of the cytosolic domain of receptors was also used to activate plexin, a membrane-bound receptor for semaphorin, an axonal growth cone guidance molecule to deflect axons from repulsive environments.[42] Optogenetic activation of Plexin-B1 at the leading edge of migrating osteoblasts induces local retraction at the illumination site and protrusions at distal regions. Also, activation of Ephrin type-B receptor 2 (EphB2) can be achieved by oligomerization of the receptor’s intracellular domain. Optical activation of EphB2 in mice’s lateral amygdala during fear conditioning specifically enhances long-term auditory fear memory.[43]
TCR:
T cells are faithful patrollers that protect self- from foreign pathogens by recognizing the small differences in ligand-receptor binding half-lives, resulting in either “go” or “no-go” action for targeted cell death. A kinetic proofreading mechanism has been suggested to account for the T cell receptor’s capacity to differentiate the foreign peptide-major histocompatibility complex (pMHC) from self-pMHCs. In this model, the low-abundance, high-affinity foreign pMHC activates T cells, whereas high-abundance, low-affinity self pMHCs do not. A common strategy to probe the kinetic proofreading in T cells is to use altered peptides sequence to change the binding half-lives, but the altered ligand sequence could lead to changes of both binding half-lives and stability, confounding the interpretation of T-cell activation. To address this challenge, Tischer and Weiner developed an optogenetic chimeric antigen receptor (CAR) that tune the binding half-lives independent of bond stability.[44] This system uses LOVTRAP, in which Zdk is fused to CAR, and purified LOV2 serves as the ligand. The authors demonstrated that binding half-lives, rather than receptor occupancy, dominates CAR signaling.[44] Supporting the same idea, Yousefi et al. developed an opto-ligand–TCR system with the PhyB-PIF system.[45] In this system, PIF was fused to TCR, which can be activated when its binding partner PhyB is in the active state under red light stimulation. The results of both work suggest that kinetic proofreading takes place in the TCR.[44,45]
Wnt Receptor:
The Wnt signaling pathway is crucial for axis formation during embryonic development. Depending on the type of ligand, either canonical or noncanonical Wnt pathways can be activated. The canonical Wnt pathway involves the activation of low-density lipoprotein receptor-related protein 6 (LRP6, a membrane-bound coreceptor of Wnt5 ligand), whereas the noncanonical Wnt pathway involves activation of Frizzled 7 (Fz7) to regulate planar cell polarity (PCP). Interestingly, by oligomerizing the cytosolic domain of LPR6, Bugaj et al. achieved optical control of the canonical Wnt signaling in culture cells.[15] To control the noncanonical Wnt signaling pathway, Capek et al. substituted the intracellular domain of the light-sensitive rhodopsin with the corresponding domain of Fz7. Activation of opto-Fz7 caused the formation of the mesenchymal cell protrusion and directed migration of prechordal plate (ppl) progenitors in fz7 mutant zebrafish embryos.[46]
Notch Receptor:
In contrast to the dimerization of receptors, the induction of ligand oligomerization is sufficient to cause the clustering of some membrane receptors. For instance, the opto-Delta system induces ligand (Delta) activation to inhibit the Notch receptor in the transgenic Drosophila. In this system, both alleles of the Notch ligand, Delta, were fused with a gene encoding CRY2PHR for blue light-mediated oligomerization.[47]
Glutamate Receptor:
Glutamate is an excitatory neurotransmitter that plays a major role in learning and memory. Upon neuronal activity, glutamate receptors are enriched in the stimulated postsynaptic spine, enhancing the capacity of neurons to respond to further stimulation (synaptic plasticity). Thus, one way to control the neurotransmitter receptor is to modulate its membrane occupancy instead of changing receptor conformation. Sinnen et al. developed a system to fine-tune the molecular abundance of specific molecules at the postsynaptic density (PSD) protein.[48] In this system, CIB1 is fused with the α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor, and CRY2 is fused with PSD anchors. A single burst of 50 ms blue light exposure recruits AMPA receptors to the PSD within 3 min in a spine.[48] Additional AMPA receptors activate synapses with few receptors but have little influence on already established synapses, which indicates that other remodeling events are required to strengthen those synapses.
Antibodies:
Inspired by the neutralizing feature of antibodies, Yu et al. developed the optobody using split antibody fragments fused with blue-light responsive heterodimerizing protein partners.[49] Optobodies against β2 adrenergic receptor successfully suppresses the endogenous target protein activity.[49] Light-inducible recombination of antibody fragment provides a mechanism to regulate intracellular antibody activity and manipulate the activity of endogenous proteins.
3.1.2. Optical Control of Cytosolic Signaling Molecules
Following receptor activation, cytosolic signaling molecules will typically be recruited in the receptor’s vicinity, activated, and diffused to other subcellular localization to trigger the downstream signaling pathways. A key advantage of optogenetics resides in its ability to interrogate signal transduction at the intermediate signaling nodes within a signaling cascade. This modality allows dynamic signaling perturbation and delineation of cellular and developmental outcomes.[50]
Protein Signaling Transducer:
One of the most frequent targets of optogenetic signaling manipulation is the direct control of protein signaling transducer. To date, optogenetic activation of the Ras/Raf/MEK/ERK signaling axis has been achieved at both cellular and organismal levels.[23,50d,51] To enable bidirectional control of the ERK signaling pathway, Mondal et al. developed a post-translational knock-in system to enhance the intracellular concentration of mitogen-activated protein kinase phosphatase 3.[34] Additionally, optogenetic inhibition of the p38 mitogen-activated protein kinase (p38MAPK) and c-Jun N-terminal kinase (JNK) has been achieved with LOV-mediated photocaging.[52]
Notably, other than the Raf/MEK/ERK cascade, Ral GTPase, RalB, also functions downstream of Ras. Optogenetic activation of RalB has been achieved by membrane recruitment of its corresponding Ral guanine exchange factor (GEF). By coexpression of RalGEF-CRY2-mCherry and CIBN-GFP-CaaX, Zago et al. demonstrated that blue light activates Ral protein and causes cell protrusions independent of Rac1, a small GTPase that regulates actin dynamics.[53]
Calcium Ion:
The calcium ion is involved in various intracellular signaling processes. Using the prototypical Ca2+ response activated Ca2+ (CRAC) system, Ma et al. extended the current genetically encoded Ca2+ actuator, GECA, to delineate protein oligomerization, conformational changes, and protein-target interactions.[54] Two general optogenetic strategies can be employed to recapitulate Stromal interaction molecule (STIM)-mediated signaling events: CRY2 oligomer-induced clustering of STIM1 mimics its Ca2+ depletion-induced oligomerization state, or by replacing the autoinhibitory domain (CC1) with LOV domain to enable light-inducible removal of the autoinhibition. Using these synthetic GECAs, the authors delineated important events for Ca2+ entry into cells.[54] An improved version of optoSTIM1 has enabled noninvasive Ca2+ modulation in mouse brains[55] and single T cells.[56]
To achieve calcium spike inhibition, Hannanta-Anan et al. used an opto-split approach for the regulator of G-protein signaling2 (RGS2) protein by integrating the CRY2-CIBN system. Light-induced heterodimerization reconstitutes the activity of RGS2, which inhibits the Gαq-mediated calcium spike.[57] Calcium-calmodulin kinase II (CaMKII), a calcium-activated protein, is required to induce long-term potentiation (LTP) and the associated structural plasticity of dendritic spines. To accurately control CaMKII activity duration, Murakoshi et al. equipped LOV2 to cage a CaMKII inhibitory peptide to develop photoactivatable autocamtide inhibitory peptide 2, which dynamically inhibited CaMKII functionality upon irradiation.[58]
Lipid Signaling Transducers:
To regulate lipid molecules, Tei and Baskin developed optoPLD with CRY2 and CIBN. OptoPLD engages organelle-specific recruitment of phospholipase D and the subsequent generation of phosphatidic acid.[59] Another type of lipid signaling transducer is the phospholipid molecules, which mediate protein trafficking through cargo transport. For example, phosphatidylinositol-4,5-bisphosphate (PI(4,5)P2) regulates cargo exocytosis during synaptic transmission and hormone secretion. To demonstrate the effects of PI(4,5)P2 on exocytosis, Ji et al. developed an optogenetic system to regulate PI(4,5)P2 by recruiting 5′-phosphatase to the plasma membrane by iLID. Results showed that local PI(4,5) P2 abundancy correlates with vesicle-PM docking for secretory pathway in INS-1 cells.[60]
3.1.3. Immediate Second Messengers
Unlike other commonly used optogenetic tools, some light-sensitive proteins allow for the production or degradation of small molecule-second massagers such as cyclic adenosine monophosphate (cAMP) and cyclic guanosine monophosphate (cGMP). Signaling via second messengers occurs at various time scales ranging from seconds to hours. Induction of expression of enzymes involved in the synthesis or degradation of second messengers could modulate slow processes, but lacks the time resolution to interrogate the fast, seconds-to-minutes time scale. Here we highlight recent work that uses light to produce or degrade second messengers to address this challenge.
cAMP Signaling Pathway:
The cAMP signaling plays an essential role in synaptic transmission. To better understand the mechanism underlying cAMP signaling, Steuer Costa et al. used the Beggiatoa-photoactivated adenylyl cyclase (bPAC) to produce cAMP in Caenorhabditis elegans motor neurons. Several seconds of illumination is sufficient to increase the cAMP level, enhance synaptic vesicle fusion, evoke the dense-core vesicle (DCV) release of neuropeptides, and induce the animal behavior.[61] By optimizing an AuLOV domain and fusing with the PAC, Hepp et al. enhanced the dynamic range of the PAC activity that reduces the dark activity even under a stronger promoter. The dose-dependent production of cAMP in the range of 20–60 pmol mg−1 (dry weight)[62] was higher than a typical cAMP concentration in the wild-type Saccharomyces cerevisiae, 6–12 pmol mg−1 dry weight,[63] but within the range of an engineered yeast strain, which reached about 100 pmol mg−1 dry weight.[64]
To facilitate the use of light-sensitive adenylyl cyclase (AC) in vivo, Ryu engineered NIR activatable AC (NIRW-AC).[21] To further increase the sensitivity, Fomicheva et al. developed the second-generation of NIRW-AC, which induces cAMP-dependent gene expression in mammalian cells and the ventral posteromedial nucleus and nucleus reticularis of mouse thalamus.[20] Because cAMP directly activates the hyperpolarized cyclic nucleotide (HCN)-dependent ion channel,[65] the NIRW-AC was used to inhibit the spindle oscillations during the light stages of nonrapid eye movement sleep in mice, which also depends on the current of the HCN-dependent ion channel.[20] Because cAMP also triggers signaling cascades by insulin secretion in β-cell, photostimulation was applied to boost insulin secretion from β-cells expressing the PAC gene from Beggiatoa (bPAC).[66] By engineering MIN6 cells stably expressing bPAC, Zhang et al. could further induce higher plasma insulin concentrations and concurrent lower blood glucose concentrations in mice within 30 min post light activation.[67]
Cyclic di-GMP (c-di-GMP)-Dependent Signaling Pathways:
Cyclic di-GMP-dependent signaling pathways control many bacterial physiologies and behaviors. Early work has developed several light-activated systems to control the synthesis of cyclic di-GMP.[68] However, the complementary optogenetic phosphodiesterases (PDEs) that specifically hydrolyze cyclic di-GMP was not available. Ryu et al. characterized a light-activated c-diGMP phosphodiesterase named BldP that consists of c-di-GMP phosphodiesterase domain, EAL, and blue light sensory domain, BLUF.[69] Combining this optogenetic PDE with the red/near-infrared-light-regulated diguanylate cyclase (DGC) that the same laboratory has developed,[68a] the authors demonstrated bidirectional regulation of cyclic di-CGMP bacterial cells.[70] Additionally, light-mediated c-di-GMP signaling can facilitate the biotransformation of indole to tryptophan by E. coli biofilms.[71]
3.1.4. Cytoskeleton and Cell Motility
Small GTPase regulates cytoskeletal dynamics during cell motility, shape, attachment, and junctions. For example, RhoA regulates actin polymerization and myosin light chain phosphorylation. Oakes et al. fused tandom PDZ domains to the DH domain of the RhoA-specific GEF, LARG, and anchored the binding partner LOVpep at the plasma membrane. Within 15 min of illumination, LARG is recruited to the plasma membrane and activates RhoA,[72] which simulated local recruitment of actin, myosin, and zyxin while increasing traction forces through stress fibers. Using a similar system, Cavenaugh et al. found that short RhoA activation led to reversible junction length changes, while intense or prolonged RhoA activation drove irreversible junction shortening. Additionally, episodic RhoA activation with periods of quiescence induced greater irreversible changes than a sustained pulse of the same strength.[73]
RhoA has additional roles in regulating cell mobility. The beginning of cell migration often requires the breaking of cell symmetry. In events with no external polarity cues, spontaneous symmetry breaking can occur via the cell cytoskeleton through several proposed mechanisms. Hennig et al. developed a single-cell 1D migration assay to determine the role of RhoA dynamics on spontaneous symmetry breaking events. This assay mimics the in vivo fibrillary environment through high-resolution force measurements, quantitative microscopy, and an optogenetics system. Blue light-inducible opto-GEF activation results in the localization of ArhGEF11, the upstream regulator of cell rear retraction, to the membrane and subsequent RhoA activation, which resulted in an immediate, local increase of traction force similar to that seen in spontaneous symmetry breaking.[74]
Cell migration and polarization can also be controlled by noncanonical Wnt-Frizzled signaling. While the role of this signaling in cell polarization is relatively well-defined, previous work indicated an additional permissive function since overexpression of noncanonical Wnt signaling ligands rescues defective mesenchymal cell polarization. To better understand the permissive nature, a light-responsive frizzled 7 receptor was constructed (opto-fz7).[46] Upon activation of opto-Fz7, the formation of mesenchymal cell protrusions and directed migration was observed within 20 min in both cultured cells and zebrafish embryos.[46]
To quantitatively determine the optogenetic systems’ mechanical stability, Yu et al. used a magnetic tweezer setup to measure the force that would dissociate the iLID–nanoprotein complex.[75] It was found that iLID–nano can withstand forces up to 10 pN for tens of seconds. The system’s mechanical stability suggests that the iLID–nano module can be employed to modulate mechanotransduction processes that involve similar force ranges.[75] The cell cytoskeleton transforms a physical signal into a biochemical signal and regulates mechanotransduction in cells. This occurs through the transmembrane protein E-cadherin, which forms contact with neighboring cells’ E-cadherin extracellular domains. The tension created in this interaction produces contractions within the cell through actomyosin. To study this mechanical stress, PhoCl was fused between the N and C terminals of E-cadherin and named PC-cadherin. In response to 405 nm light stimulation, the PC-cadherin cleaves into two fragments, resulting in an inhibition of mechanotransduction at the intercellular junctions of epithelial cells.[76]
3.1.5. Programmed Cell Death
Apoptosis, a type of programmed cell death, plays a key role during tissue development and can be initiated through intrinsic or extrinsic pathways.[77] The intrinsic pathway senses cell stress and uses proapoptotic signaling to perturb mitochondrial membranes and release cytochrome c. In contrast, the extrinsic pathway senses signals from other cells and often involves activation of death receptors such as Fas receptor and tumor necrosis factor (TNF) receptors. To date, both types of apoptotic pathways can be controlled by light. An optoBax system[78] uses CRY2/CIBN to recruit the proapoptotic protein Bax to the mitochondrial membrane and activates the intrinsic apoptosis pathway. The extrinsic apoptotic pathway can be regulated by the dimerization of Fas and its adapter protein fas-associated protein with death domain (FADD).[79]
Caspase 3 is the executioner protein during apoptosis. To control Caspase 3 activity, Smart et al. inserted the LOV2 into the inter-subunit linker of caspase. Upon illumination, the conformational change of the Jα helix allows the active site of caspase to fold into an active conformation.[80] In vitro assay of the system showed that Caspase-LOV has similar activity to wild-type caspase-3. Combining with tissue-specific expression systems, Caspase-LOV successfully causes cell death in both whole flies and specific tissues.
Interestingly, by hijacking the type III secretion system (T3SS) from bacteria, Lindner et al. demonstrated the light-regulated protein translocation from bacteria to eukaryotic host. When a pro-apoptotic protein, the truncated human BH3 interacting-domain death agonist (tBID), was injected into the host eukaryotic cell, active host apoptosis was observed.[81]
3.1.6. Cell Cycle
The cell cycle requires tightly regulated signal transduction for cell fate determination. The cell cycle consists of a growth phase (G1), DNA synthesis (S), a second growth phase while preparing for mitosis (G2), mitosis (M), and the resting state (G0). Each phase is divided by “checkpoints” that allow progression toward mitosis or rest. Here, we summarize recent progress in the optogenetic control of the cell cycle.
During early embryogenesis, spatially coordinated and synchronized cleavage division is orchestrated by cell cycle oscillators and cytoskeleton dynamics. However, there is a lack of understanding of how spatially confined biochemical signals work along with the embryo’s physical properties to create the collective dynamics necessary for nuclear spreading. To better understand nuclear spreading in Drosophila embryos, Deneke et al. applied optogenetic RhoGEF to induce cortical actomyosin-mediated membrane contraction. It was found that the nuclei of embryos exposed to uniform light were unable to spread along the anterior-posterior axis as they should during interphase. The results indicate that the actomyosin gradients are necessary to generate contractile forces to generate cytoplasmic flow and nuclear spreading during G1, S, and G2 phases.[82]
To investigate how the Ras-Son of Sevenless (SOS)-ERK signaling dynamics affect the cell cycle, Goglia et al. developed a high-throughput screening assay to search for drugs that change the ERK dynamics. By testing more than 400 kinase inhibitors in primary mouse keratinocytes, they found that drugs that altered ERK’s dynamics also affected cell proliferation. OptoSOS-induced change in ERK activity duration was sufficient to explain the proliferation observed in the screen, but the proliferation was not directly proportional to the dose of ERK activation.[51a] To determine the mechanism for the ERK dynamics on the cell cycle. De et al. used optogenetic tools (pmCIBN, CRY2-Raf) and determined that prolonged ERK activation led to cdc25 phosphorylation, promoting the accumulation of pro-mitotic factors such as polo-like kinase 1 (PLK1).[83] PLK1 then lowered the threshold needed for the cell passing through the G2 checkpoint.
3.2. Organelle Manipulation
Intracellular communication is often powered by dynamic distributions of organelles and constant change of organelle compositions. This dynamic nature makes it challenging to correlate the localization and chemical make-up of organelles with their functions. Optogenetics, featured by its precise control of organelle shape, localization, and interactions,[84] could provide new insights into the molecular machinery of cell communication.
3.2.1. Generation of Membrane Curvature
The plasma membrane encapsulates the intracellular contents and serves as a physical barrier for the cell. Instead of merely accommodating proteins on the cell surface, the plasma membrane has been recognized as a crucial organelle that plays a role in various cellular processes during material transport (e.g., endocytosis and exocytosis) and actin dynamics. Indeed, even the shape of the membrane curvature affects protein functions and intracellular signaling, but few methods can precisely manipulate the membrane shape. Using the Bin/Amphiphysin/Rvs (BAR) domain, Jones et al. established opto-FBAR and opto-IBAR, which could use light to generate membrane invagination (positive curvature) and filopodia (negative curvature), respectively.[84a] The idea is that the oligomerization of the monomer bar domain with different shape and charge on the plasma membrane can cause membrane deformation. For example, the F-BAR (extended Fes-CIP4 homology (EFC)/FCH-BAR) proteins are banana-shaped with positive charges on the concave side, whereas the I-BAR (IRSp53-MIM homology domain I-BAR/inverse-BAR) proteins are cigar-shaped with positive charges on the convex side. The distinct shape and charge distribution enable these BAR domain monomers to bend the plasma membrane toward their respective curvature preference side and generate distinct membrane curvature.
3.2.2. Regulation of Vesicle Transport
Motor protein-mediated vesicle delivery system is the primary material-transporting mechanism in live cells. Cargo localization and properties can be modified by motor proteins such as dynein, kinesin, and myosin. To elucidate the role of dynein, one of the retrograde motor proteins, in mitotic spindle-pulling activity, Okumura et al. developed an optogenetic system by anchoring iLID to the membrane and fusing nuclear mitotic apparatus (NuMA) protein with Nano for cortical recruitment of dynein–dynactin complex.[85] Localization of NuMA to the mitotic cell cortex is sufficient for dynein–dynactin recruitment, and the spindle-pulling NuMA protein level reaches three times higher than that of endogenous NuMA in metaphase.[85]
Kinesin is an anterograde motor protein that travels to the plus end of the microtubule. An improved optogenetic kinesin module was developed by replacing the neck coil of kinesin with the small LOV domain of the Vivid (VVD) photoreceptor. Without light, the kinesin remains an inactive monomer. Upon blue light stimulation, VVD homodimerization activates the kinesin. By fusing the SspB domain to the N-terminal of the opto-kinesin and the iLID domain to the cargo binding domain (e.g., Rab11 or Rab5 to mark endosomes), opto-kinesin allows for light-inducible anterograde cargo movement.[86] Interestingly, anterograde recruitment of mitochondria to the cell cortex discourages the glucose spiked cytosolic calcium concentration and insulin secretion in beta cells.[87]
Notably, both dyneins and kinesins use microtubules as the cytoskeletal track. The growth of microtubule plus ends requires interaction with functionally diverse microtubule plusend-tracking proteins (+TIPs) mediated by end-binding proteins (EBs). To study the spatiotemporal influences of +TIP complexes, van Haren et al. constructed a photoinactivated EB1 variant (π-EB1) by inserting LOVTRAP between the microtubule-binding and +TIP binding domains of EB1. Reversible attenuation of microtubule growth can be achieved by alternating the dark-light conditions. In the dark, π-EB1 replaces endogenous EB1 function, while light exposure caused dissociation of the two domains and led to +TIP complex disassembly.[88]
In addition to microtubule-based motor proteins, myosin is an actin-based motor protein regulating protein trafficking and cell contraction. Among the members of myosin, myosin VI can travel to the directed end of the actin filament. Utilizing the unique feature of myosin VI, a spatially sensitive optogenetics system was engineered by caging Disabled2 (Dab2), a myosin VI cargo protein with LOV2.[89]
3.2.3. Phase Transition
Optical induction of oligomerization of specific proteins can cause in-cell phase transition of protein/nucleic acid complex. Nucleophosmin (NPM1) is a nucleolar protein at the outer layer of the nucleolus associated with processed rRNA. when fused with CRY2olig, NPM1 undergoes a liquid-to-gel-like transformation upon blue light stimulation. This phase transition alters NPM1’s diffusion and motility dynamics, which subsequently inhibits the rRNA processing rate.[90] Another phase-transition event occurs during the DNA damage-induced assembly of tumor suppressor p53-binding protein 1 (53BP1) protein around DNA lesions. Using CRISPR/Cas9 to tag the endogenous protein 53BP1 with CRY2oligo,[91] Kilic et al. developed an optogenetic system that allows light-inducible 53BP1 assembly, mimicking its response to DNA damage.[92] In addition, Shin et al. characterized the role of nucleolus liquid-liquid phase separation of intrinsically disordered proteins in chromatin landscape reconstructions. A CRISPR-Cas9-based optogenetic system, CasDrop, was applied to control liquid condensation of intrinsically disordered proteins (IDR) at specific genome loci. The CasDrop system revealed that the intrinsically disordered region composed of condensates prefers areas with low chromatin density. Light-induced condensation leads to the mechanical exclusion of nontargeting chromatin.[93] Other than nucleus protein phase separation, light-stimulated phase separation enables long-term membrane plasticity,[94] control of metabolic influx at the cytoplasm,[95] and establishment of local and global intracellular phase diagrams.[96]
Because protein aggregation often occurs in pathology, light-inducible protein phase separation could mimic the pathological conditions without introducing a constitutive effect that causes lethality. For example, stress granules (SGs) are phase-separated RNA-protein complexes typically shown in amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). To determine the effects of SG dynamics on pathology, Zhang et al. constructed the light-inducible SG system (OptoGranules) to induce the multimerization of Ras GTPase-activating protein-binding protein 1 (G3BP1), a required protein for stress granule assembly. SGs formed upon G3BP1 oligomerization and consequently induced cell death, phenocopying the pathology of ALS and FTD. Persistent SGs were found to be less cytotoxic than intermittent SGs formed by pulsed activation.[97] On the other hand, oligomerization of transactivation response element DNA-binding protein 43 kDa (optoTDP43) with CRY2olig causes stress granule (SG)-an independent agglomeration of TDP-43, which was alleviated by RNA oligonucleotides treatment composed of TDP-43 target sequences.[98] The optical induction of TDP-43 aggregation in the zebrafish neuromuscular system results in the exacerbation of locomotion behavior.[99]
Amyloid-β plaque is a prominent factor that accelerates the progression of Alzheimer’s disease (AD). Blue-light-mediated oligomerization of Amyloid-β peptide was developed by the fusion of the peptide with CRY2. Although cytotoxicity of Amyloid-β was confirmed in both soluble and light-induced solid phases, metabolic damage occurs only in the solid phase upon light stimulation.[100]
3.3. Genetic Regulation
Control of gene expression has been demonstrated in early work by recruiting transcription activation domains to reporter gene promoters. Over the past few years, optogenetic control for gene expression has been significantly improved with more specificity, endogenous gene targeting, and live animal applications. Here, we focus on the new or improved modality for optogenetic control of gene expression.
3.3.1. Targeting Endogenous Gene Expression
The homodimer of cyclic AMP response element-binding protein (CREB) activates the transcription of its target gene. Heterodimerization of wild-type CREB with a dominant-negative inhibitor of CREB (A-CREB) represses its activity. This heterodimer can be disrupted when A-CREB is caged by the photoactive yellow protein (PYP) under blue light stimulation, resulting in a recovery of wild-type CREB activity.[101]
Direct transcription inhibition can be achieved by oligomerization of a transcription factor. For example, the Drosophila transcription factor Zelda is a master regulator of genome activation. When Zelda is fused with CRY2 (CRY2-Zelda), blue light-mediated CRY2 clustering causes inactivation of Zelda protein transcription.[102] Using this dynamic light-dependent Zelda activity regulation tool, it was revealed that Zelda’s continued activity is required to activate zygomatic gene expression and early embryonic development.
3.3.2. Improving Optogenetic Control of DNA Recombinase
Cre recombinase is derived from a bacteriophage that can carry out site-specific gene recombination. Since the early development of photoactivatable Cre (PA-Cre)[103] and PA-Cre 2.0,[104] PA-Cre 3.0 was developed via optimization of the protein dimerizers (CRY2-CIB1, Magnets), promoter, and 2A self-cleavage peptide.[105] The improved PA-Cre 3.0 system significantly reduces the dark basal activity in vivo. A large pool of optogenetic recombinases has been developed, including Cre, Dre, and Flp (RecVs). The VVD-based RecVs allows for genomic modifications when excited by blue light and longer-wavelength (900 nm) light under two-photon excitation microscopy.[106] Another recent work also used photoactivatable Flp recombinase (PA-Flp) to achieve highly light-sensitive property in deep mouse brain regions.[107]
3.3.3. Genomic and Epigenetic Control
Optogenetic CRISPR–Cas9-based photoactivatable transcription system (CPTS) was first developed via light-induced recruitment of p65 to the nuclease-dead Cas9, dCas9.[108] An improved CPTS system was developed[109] with a split dCas9, which could be reconstituted by the nMag/pMag protein pair. This new system achieved 1200-fold upregulation of the target gene, compared with the previous nine-fold increase, although both systems use blue light for stimulation. To extend the stimulation spectrum to redshifted wavelength, which could increase the penetration depth and reduce the phototoxicity, Shao et al. developed a far-red light (FRL)-activated CRISPR-dCas9 effector (FACE) system.[110] FACE system consists of BphS, an FRL-activated c-di-GMP synthase, and a transcription factor BldD, which is fused with p65 and VP64 for transcription activation. The authors demonstrated that genes involved in muscle mass and regeneration including laminin subunit alpha1 (Lama1) or follistatin (Fst), as well as those involved in neuronal differentiation of inducible pluripotent stem cells (iPSCs) such as neurogenin 2 (NEUROG2) could be targeted and activated by FACE in live mice.
Using a split protein strategy for Cpf1, a sister protein of Cas9 with better target sequence specificity, The Sato group has fused the N-terminal and C-terminal of Cpf1 to the Magnet protein pair. Blue light reconstitutes full-length Cpf1 activity with minimal dark activity.[111] Besides genomic editing, optogenetics has also been used to study histone H2B monoubiquitination (H2Bub1), an essential component for the trans-histone regulation of H3K4 and H3K79 methylation. To measure H2B ubiquitination and deubiquitination kinetics in vivo, a rapid and reversible optogenetic tool, the light-inducible nuclear exporter (LINX), was used to control nuclear localization of H2Bub1 E3 ligase, Bre1. From the Bre1-LINX system, deubiquitination mediated repressed proteolysis was observed within 10 min upon light illumination.[112]
Besides transcription factors, genome architecture also regulates transcription. Recent work shows a promising way to control the 3D arrangement of genomic structure. The light-activated dynamic looping (LADL) system comprises two different genomic anchoring proteins, dCas9-CIBN fusion proteins, and free CRY2 proteins. Blue light pulse promoted interaction between CRY2 and CIBN, as well as CRY2-CRY2 homologous interaction to bring two different genomic anchor sites near each other’s proximity. By redirecting the stretch enhancer (SE) away from its endogenous Klf4 target gene to the Zfp462 promoter, the authors observed de novo formation of the Zfp462-SE loop, which correlates with a modest increase in Zfp462 expression.[113]
3.3.4. Controlling Gene Expression at the Translational Level
As previously mentioned, the light-activated PAL:RNA interaction can regulate gene expression at the RNA level.[11] Translational inhibition can also be achieved by clustering, and therefore, trapping mRNA in an inactive state. A four-component optogenetic module (multimeric protein (MP)-CIBN, CRY2-GFP nanobody, MCP–GFP, MS2-binding site-tagged (MBS)-mRNA) was made for manipulating the localization and translation of specific mRNA by trapping the mRNA inside the light-induced cluster.[114] Functionally, this sequestration reduced mRNAs’ accessibility to ribosomes, thus markedly and rapidly attenuating protein synthesis. A spatiotemporally resolved analysis indicats that sequestration of endogenous β-actin mRNA attenuated cell motility by regulating focal adhesion dynamics.
3.3.5. Control Diverse Cellular Activities by Gene Expression
New optogenetic circuits have been engineered in S. cerevisiae to enable light-controlled fermentation. This strategy allows for a new mode of bioreactor operation with light-tuned enzyme expression.[115] Another application is the tuning of cell fate to favor specific functions, e.g., tissue regeneration, as evidenced by the work showing that optical activation of bone morphogenetic protein 2 (BMP2) and LIM homeobox 8 (Lhx8) expression could turn mesenchymal cell fate toward bone regeneration in rats.[116] It has also been observed that Transcription factor EB (TFEB)-controlled expression could help clear p-Tau from neurons via autophagy. This capacity allows for the clearance of p-Tau from AD patient-derived human iPSC-neurons.[117] Lastly, by regulating the expression of FtsZ and CheZ genes in bacterial cells, the eLightOn system allows for the control of cell division and swimming.[118]
4. Generation and Validation of Optogenetic Actuators
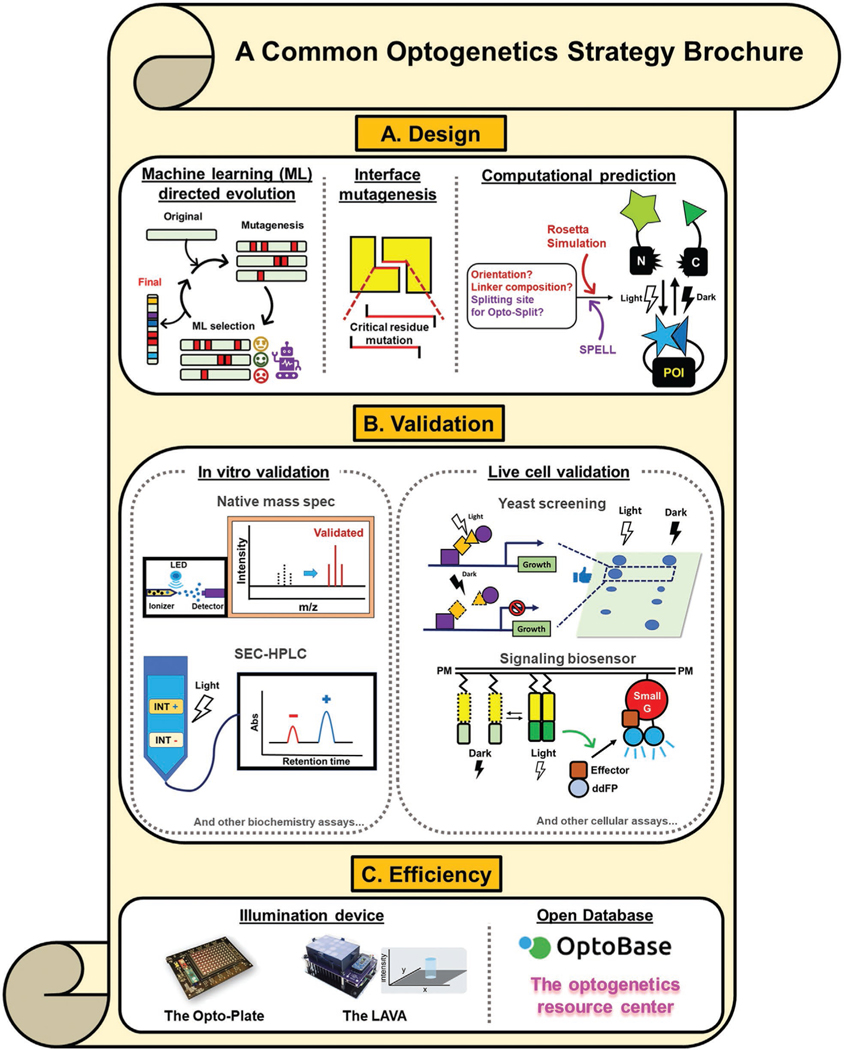
To make optogenetics more approachable, we highlight some commonly employed strategies in the design and validation of optogenetic actuators (Figure 4).
Figure 4.
Collection of commonly used optogenetic strategies. a) Optically active protein design and engineering approaches. POI: Protein of interest. b) Live cell or in vitro validation modules. INT+: Positive interaction; INT−, negative interaction; ddFP, dimerization-dependent fluorescence protein c) Efficient resources for conducting an experiment or collecting data. References of each strategy are listed as follows: machine learning directed evolution,[119,120] interface directed mutagenesis,[122] computational prediction,[31,123,124] native mass spectrometry,[128] SEC-HPLC,[129] yeast screening,[122,130] signaling biosensor,[131] optoplate,[132] LAVA plate,[133] optobase,[1a] and the optogenetic resource center.[1] The image of the optoplate is reproduced with permission.[51b] Copyright 2018, The American Association for the Advancement of Science. The image of the LAVA plate is reproduced with permission.[133] Copyright 2020, Elsevier.
4.1. Design of the System
Optogenetic system design aims to identify a specific protein sequence that performs a specific function through protein engineering. Besides rational design and directed evolution, recent work resorts to machine learning-directed evolution to optimize protein function. Whereas directed evolution discards information from unimproved protein sequences, the machine-learning methods include this information to guide and expand the properties that can be optimized by intelligently selecting new variants from the unbiased screen and reaching better fitness.[119] The machine-learning engineering method has improved the functionality of channelrhodopsin (ChR) variants, which eventually yielded simultaneous high-photocurrent ChRs with high light sensitivity.[120]
The advancement of omics research has provided another resource for identifying, designing, and comparing optogenetic proteins. By genomic and transcriptomic mining of diverse organisms, Glantz et al. reported the bioinformatic identification of over 6700 candidate LOV domains through motif analysis. Also, conserved upstream and downstream effector domains can be identified, and their functions can be annotated across archaea, bacteria, fungi, protists, and land plants.[121]
Directed mutagenesis at the protein interface has been a useful method to produce functionally enhanced photoactivatable protein pairs. For example, by varying the protein interface between circularly permuted photoactive yellow protein (cPYP) and binder of PYP dark state (BoPD), distinct photoswitchable variants with altered affinity, kinetics, and apo-state binding could be recovered.[122] Several computational prediction programs were developed to predict the photophysical properties of engineered protein variants. For instance, Rosetta uses energy and distance constraints to predict favorable linker composition and fusion protein orientation in silico.[31] The Rosetta simulation for three different light-activatable fusion proteins was validated in a separate study.[123] In addition, Dagliyan et al. developed an automated approach to design effective split proteins regulated by a ligand or by light (SPELL).[124] The scoring algorithm derived from the computational design approach ensured dynamic reassembly of protein fractions with low background activity in living cells. The schemes of each engineering strategies are illustrated in Figure 4A.
Besides the computational efforts to help with protein engineering, Harrigan et al. developed a closed-loop optogenetic compensation (CLOC) strategy to explore the feedback regulator dynamics in signaling networks.[125] The system can monitor the real-time pathway output and automatically adjust the light input. Typical genetic complementation experiment tests like gene knockouts or rescue studies were able to determine feedback regulation. However, these approaches cannot reveal information about the regulation dynamic of the signaling network. Application of CLOC to feedback-deletion pathways could identify the dynamics of feedback demands.[125] Also, several other in silico optogenetic systems were developed for the dynamic control of transcription intensity[126] and reconfiguration of gene circuits with logic gates.[127]
4.2. Validation
Functional validation of optogenetic systems is typically carried out either based on cell harvests or live–cell readouts (Figure 4B). The conventional cell harvest-based assay, such as native gel electrophoresis, provides a quick readout of protein interaction but often suffers from suboptimal detectability of reagents such as antibodies. A recent study used native mass spectrometry to quantify protein changes upon photoactivation by installing the stimulation light source between the sample capillary and the ion detector. The light-mediated protein conformational change could be validated by either m/z scoring for protein size or ion mobility for structural dynamics.[128] Additionally, the size exclusion chromatography (SEC) and dynamic light scattering facilitate the measurement of the physical interaction and useful photophysical parameters.[129]
Live-cell readout, such as yeast cell growth, could be repurposed to validate interactions between photoactivatable proteins, particularly dimerizers.[122,130] An alternative way is to use biosensors that respond to the activation of photoactivatable proteins to confirm the system’s functionality. For instance, the dimer dependent red fluorescence protein (ddFP) was engineered only to emit fluorescence when the synthetic G protein receptor is appropriately activated by light.[131]
4.3. Improving the Efficiency of Optogenetic Operation
Here, we highlight strategies that improved the efficiency of optogenetics (Figure 4C). Optimization of experimental procedure often requires fine-tuning several experimental parameters such as light power, illumination duration, frequency, or light wavelengths. To facilitate parallel operation, a high-throughput OptoPlate has been developed recently. The OptoPlate uses 3D printing to fabricate an economical device (less than $600) that enables 96-well or 384-well scaled experiments with independent control in each well.[132] Similar engineered illumination devices for optogenetic photostimulation and light activation at variable amplitude (LAVA) has been developed. Time-varying and spatially localized light patterns provide a low-cost and user-friendly method for high-throughput optogenetic control of cell signaling.[133]
To streamline the production of optogenetic systems, Tichy et al. proposed a generalizable genetic engineering strategy that permits fusing a protein of interest to multiple types of the light-sensitive domain (LSD) to generate tens of working constructs by a single cloning step.[2a] Tissue-specific viral vectors[134] and organelle-specific optogenetic tool library[2b] could be conveniently created.
Last but not least, there are online resources, such as Optobase, which provide a comprehensively annotated publication database for photoactivatable protein-based optogenetics (optobase.org).[1a] Also, the optogenetic resource center hosted by the Deisseroth lab (web.stanford.edu/group/dlab/optogenetics) offers technical know-how such as optogenetic viral vector preparation and delivery protocol, optogenetic hardware, along with other assorted information.[1b] Although the design and validation of optogenetic applications solely depend on the specific purpose of research, we expect that information listed in this section could be useful for current and new optogenetic users. We also hope this information could help establish an “optogenetics handbook” that further catalyzes efficiency and significance for optogenetics research.
5. Interlinking Optogenetics with Other Research Fields
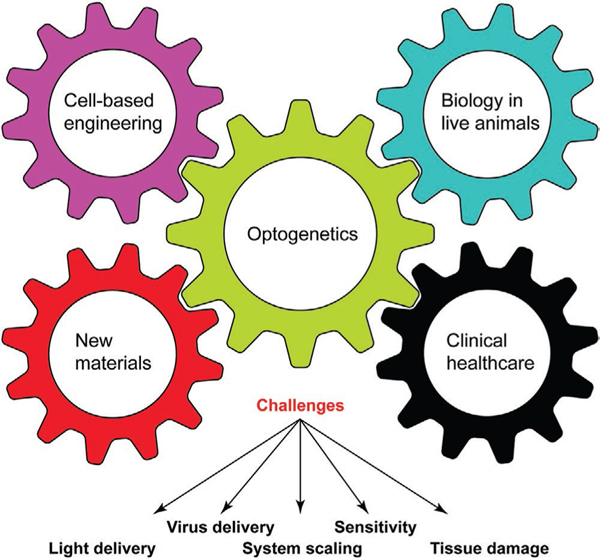
Parallel to the continuous improvement of the optogenetic toolbox, the photoactuators and their derivatives start to provide opportunities to advance other fields (Figure 5).
Figure 5.
The cross-disciplinary interplay between optogenetics and other fields. Optogenetics could be applied to facilitate cell-based engineering (e.g., tissue engineering) or generate new biomaterials. By overcoming technical challenges, such as light and virus delivery, sensitivity, toxicity, and scaling up to large animals, optogenetic tools would be increasingly applied in the live animals. Advanced technologies, such as the two-photon optogenetic stimulation and light upconversion, will continue pushing optogenetic technology toward clinical settings in the healthcare sector.
5.1. Engineering Devices for Live-Animal Optogenetics
The successful application of optogenetics in live animals needs to overcome several technical barriers, such as efficient light delivery into deep tissue and wireless optogenetic devices. The emerging implantable wireless micro-LEDs powered by radio-frequency scavengers[135] removed the spatial constraints from wired fiber optics. Since the initial success of the design, multiple lines of research have employed miniature LEDs for live-animal optogenetics studies.[136]
Another strategy to improve the light penetration depth is to use nanometer-sized upconversion nanoparticles (UCNP)[137] to produce visible light (for optogenetic stimulation) from near-infrared light (800–980 nm), which has a much deeper penetration depth in biological tissues. Substantial progress has been made in the past three years by using the UCNP in optogenetic applications.[138]
5.2. Biomaterials
The mechanical properties of the extracellular microenvironment determine many cellular behaviors such as cell migration and differentiation. Synthetic hydrogels are widely used as biomimetic in vitro model systems to study cell–extracellular environment interactions, but these hydrogels’ mechanical properties could not be easily controlled. To address this issue, Wu et al. developed a hydrogel platform with tunable mechanical properties based on photo-induced switch of the cross-linker fluorescent protein, Dronpa145N, between the monomeric and tetrameric state. This hydrogel allows for reversible control of stiffness, which can be used to direct cell migration.[139] The stiffness,[140] local density,[141] and durability[142] of the hydrogel can be fine-tuned by the light-inducible conformational changes of photoactivatable proteins.
Hydrogel has been commonly used as a reservoir of chemicals and proteins. However, the current setting lacks precise control over releasing the cargo at the right place and time. For releasing nanoscale cargo, PhoCl was utilized to release the epidermal growth factor to promote 3D anisotropic cellular proliferation in the cultured cell.[143] When biotinylated PhyB was conjugated to NeutrAvidin-containing fibrin hydrogel, PIF-fusion proteins can be trapped in (upon 660 nm light exposure) or released from (upon 740 nm light exposure) the hydrogel for multiple cycles.[144] Unlike other protein-embedding approaches to release cargo to the media, Sankaran et al. developed hydrogel with bacteria embedding approach to synthesize antimicrobial and antitumoral drug, deoxyviolacein, and deliver the chemical to the target cells.[145] Aside from the practical applications of the light-responsive hydrogel, these hydrogels showed delicate sensitivity[146] and great shape and volume morphing potentials.[147] These studies highlight the opportunities of leveraging optogenetics to fabricate hydrogels with versatile functionality.[142]
Optogenetic stimulation of cell–cell interaction could regulate biofilm formation, which involves quorum sensing, a chemical-based intercellular communication between cells. A light-controlled quorum quenching system uses near-infrared light-activated diguanylate cyclase (DGC; c-di-GMP synthase) and blue light-activated phosphodiesterase (PDE; c-di-GMP hydrolase) to fine-tune the c-di-GMP level in E. coli. Activation of this system can successfully mitigate biofouling on water purification membranes by expressing a quorum quenching enzyme.[148] Further studies have demonstrated precise bacterial adhesion,[149] cell patterning,[150] and living biofilm formation.[148] Notably, bacteria could be engineered to recognize red, green, and blue light stimulation by changing gene expression,[151] allowing for full spectral programming and multiplexed optogenetic control.[152]
5.3. Tissue Engineering
Tissue engineering integrates the concept of engineering into biological sciences to recapitulate synthetic tissue and organs. Technologies such as 3D bioprinting[153] significantly accelerate synthetic tissue production but still suffer from slow printing and low cell viability. Optogenetic control of cell fate provides an attractive approach where single cells and their interactions with other cells or extracellular matrix can be manipulated with minimal toxicity. The importance of cell–matrix interaction has prevailed in various human body locations, such as between soft tissue and epithelial tissue. Tissue engineering has been adorned by hydrogel to mimic the human soft tissue embedding epithelial cells.[154] The light-responsive hydrogel can become an undisputed partner for fine-tuning the extracellular matrix to reflect a dynamic physiological environment[155] and initiate cell signaling.[139,156]
Besides cell–matrix interaction, fine-tuning of cell–cell interactions are of equal importance in tissue engineering. Blue light-mediated self-sorting of cells was achieved by engineering cells with heterodimer protein pairs (iLID/Nano and nMagHigh/pMagHigh).[157] To enable the multiplexing potential of cell–cell interactions, Yuz et al. developed a cell module that can independently control two different cell types with the blue and red light.[158] Computer-assisted automated optogenetics can also benefit the field of tissue engineering. A representative study showed that computational optogenetics could determine the intensity of light input precisely based on the readout from tens of cell patches.[126a]
5.4. Manipulation of Molecular Activity in Cell-Free Systems
Although optogenetics has been primarily utilized in live cells or animals, it starts to shine a light on the cell-free systems. A cell-free system is a powerful tool that can provide insights into biological processes by reconstituting functional modules in a user-defined environment. One example is the study of motor protein–cytoskeleton interaction. The microtubule gliding assay is commonly used to study the collective behavior of active and self-organizing motor proteins. Herein, Tas et al. used light to recruit molecular motors to control microtubule gliding activity in the synthetic cell. The ePDZ domain was tagged to kinesin, and the LOVpep was immobilized on the surface. Blue light triggers ePDZ-LOVpep binding, which recruits the microtubule to the surface for directional movement. Maximum association between microtubule and motor proteins occurs 6 s after blue light exposure, and dissociation occurs in the dark with a half-life of 13 s.[159] Apart from the microtubule dynamics, photoactivatable cell-free systems can be applied to cell motility,[160] pattern formation,[161] and cell interaction.[162]
6. Outlook: The Road to the Clinic
In the past decade, a large amount of effort has been used to develop versatile tools to control molecular activities in living cells. As more researchers consider optogenetics as an emerging strategy to investigate biological questions, we expect a steady progress that will continue pushing optogenetics toward in vivo and clinical applications, which is parallel to the fast pace of gene therapy.
Indeed, besides the insights that optogenetics offers into basic sciences and drug discovery,[163] there is a growing number of translational research applications such as in pain management,[164] strokes,[165] epilepsy,[166] behavior,[167] heart diseases,[168] motor functions,[169] memory,[170] and psychiatric diseases, including depression, anxiety, addiction, schizophrenia, and autism, to name a few.[171] However, there are a few bumps on the road to the clinic for optogenetics, mainly light delivery, virus delivery, scaling from rodent brains, sensitivity, and minimizing tissue damage.[172]
Light delivery is one of the main challenges that are often faced by visible light-based optogenetics. In some cases, such as in the eye or on the skin’s surface, light penetration is not a problem. However, subdermal light delivery can be more problematic. There have been many recent technological advances to overcome this barrier, such as subdermal magnetic coil antennas that are connected to injectable LED,[173] injectable light delivery devices without batteries,[136] deep tissue activatable nanoparticles,[174] implantable optoelectronic probes,[175] and hybrid-drive combining optogenetics, pharmacology and electrophysiology (HOPE) implants.[176] As we mentioned, redshifting of excitation wavelength and light upconversion are promising approaches to mitigate the challenge of limited penetration depth. We continue holding great hope for the use of upconversion nanoparticles—a recent work showed exciting results that photoreceptor-binding upconversion nanoparticles, when injected in the eyes of mice, empowers the mice to see infrared light without compromising their ability to see visible light.[177]
On the other hand, viral gene therapy is a field experiencing growth and attention pre-dating optogenetics. Therefore, the issue of virus delivery is not a new problem. The US FDA has approved 17 viral therapies as of 2019 (fda.gov), such as therapies for spinal muscular atrophy (Zolgensma, AveXis, Inc.), large B-cell lymphoma (Yescarta, Kite Pharma, Inc), prostate cancer (Provenga, Dendreon Corp.), cartilage defects (MACI, Vericel Corp.), and retinal dystrophy (Luxturna, Spark). More detailed information has been discussed in recent reviews.[178] Because the encoding gene’s size determines the titer of the virus (e.g., the typical packaging capacity for lentivirus and the adeno-associated virus is 9 and 4.7 kb, respectively), it would be beneficial to develop single-transcript optogenetic systems using small-size photosensitive modules.
With extraordinary examples of optogenetics in mice, such as smartphone-controlled glucose homeostasis in diabetic mice,[179] the next step becomes to scale the system to a human-sized patient. Since a mouse brain is 2500 times smaller than a human brain,[180] many researchers have first turned to nonhuman primates as model systems.[181] We encourage interested readers to check out these excellent reviews for a more detailed discussion of optogenetics in nonhuman primates.[170,172,182]
Even with these bottlenecks, some optogenetic systems have made the leap from translational science to clinical trials, such as retinopathy treatment.[183] In this approach, a viral plasmid is delivered directly to the eye containing a microbial opsin for vision restoration. There are currently three clinical trials in phases 1 and 2 treating retinopathy with optogenetics. The first trial of RST-001 started in 2015 for advanced retinitis pigmentosa and is sponsored by Allergen (NCT2556736) with an anticipated primary completion date of 2020. Gensight Biologics began a trial in 2018 (NCT03326336) treating nonsyndromic retinitis pigmentosa with optogenetic system GS030-DP combined with the biomimetic goggle. The anticipated primary completion date is also in 2020. Most recently, Bionic Sight LLC began a trial in 2020 (NCT04278131) for the treatment of retinitis pigmentosa with BS01, and primary completion is scheduled for 2022. We anticipate that as the technology for virus and light delivery grows, more non-neuronal optogenetic systems will be entering the clinic applications.
In summary, there has been rapid progress in the field of optogenetics in the past few years. The unprecedented spatiotemporal accuracy that light offers to control intracellular signaling and cell behavior will be likely to continue drawing more researchers in this field. The portfolio of optogenetic toolbox also benefits from the constant interplay across different research fields such as material sciences and engineering. We are eager to learn new, exciting biology uncovered by this emerging technology in the near future.
Acknowledgements
T.-J.O., H.F., S.S.S. contributed equally to this work. H.F. thanks for the support of the Charles F. Kade Fellowship from the Department of Biochemistry at UIUC. S.S.S. thanks for the support of the Westcott Bioscience Fellowship from the Department of Biochemistry at UIUC. K.Z. thanks for the support from NIH grants R01GM132438 and R01MH124827, and the School of Molecular and Cellular Biology at UIUC.
Biography

Teak-Jung Oh graduated from Southern Illinois University at Carbondale (SIUC) in 2018 with a Bachelor of Science in Physiology and a Master of Science in Biochemistry and Molecular Biology. He is currently a Ph.D. candidate under the guidance of Dr. Kai Zhang at the University of Illinois at Urbana–Champaign, where he focused on engineering optogenetics platform to control intracellular cell signaling pathways.

Huaxun Fan received his Bachelor of Science from Zhejiang University in 2017 and worked there as a research assistant for one year. He started his graduate school at the University of Illinois at Urbana–Champaign in 2018. Working in Dr. Kai Zhang’s laboratory in the Biochemistry Department, his research focuses on developing and implementing optogenetic systems to study cell signaling.

Kai Zhang received his Bachelor of Science from the University of Science and Technology of China (USTC) in 2002 and Ph.D. in Chemistry from the University of California, Berkeley, in 2008. Following a postdoctoral study at Stanford University, Dr. Zhang joined the Biochemistry Department of the University of Illinois at Urbana–Champaign as an assistant professor in 2014. The Zhang laboratory develops new biotechnologies, including optogenetics and single-molecule fluorescence microscopy, to investigate how growth-factor-mediated signal transduction regulates cell fate determination during embryonic development and diseases such as neurological disorders and cancers.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- [1] a).Kolar K, Knobloch C, Stork H, Znidaric M, Weber W, ACS Synth. Biol 2018, 7, 1825; [DOI] [PubMed] [Google Scholar]; b) Deisseroth K, Optogenetics Resource Center https://web.stanford.edu/group/dlab/optogenetics/ (accessed: December 2020). [Google Scholar]
- [2] a).Tichy AM, Gerrard EJ, Legrand JMD, Hobbs RM, Janovjak H, J. Mol. Biol 2019, 431, 3046; [DOI] [PubMed] [Google Scholar]; b) Muhlhauser WWD, Weber W, Radziwill G, ACS Synth. Biol 2019, 8, 1679. [DOI] [PubMed] [Google Scholar]
- [3].Kumar M, Mommer MS, Sourjik V, Biophys. J 2010, 98, 552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Harris JM, Wang AY, Boulanger-Weill J, Santoriello C, Foianini S, Lichtman JW, Zon LI, Arlotta P, Dev. Cell 2020, 53, 577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wang Q, Fan H, Li F, Skeeters SS, Krishnamurthy VV, Song Y, Zhang K, Elife 2020, 9, e57395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6] a).Kwon E, Heo WD, Biochem. Biophys. Res. Commun 2020, 527, 331; [DOI] [PubMed] [Google Scholar]; b) Wu DZ, Lackner RM, Aonbangkhen C, Lampson MA, Chenoweth DM, Methods Enzymol. 2019, 624, 25; [DOI] [PubMed] [Google Scholar]; c) Wittmann T, Dema A, van Haren J, Curr. Opin. Cell Biol 2020, 66, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7] a).Rogers KW, Muller P, Curr. Top. Dev. Biol 2020, 137, 37; [DOI] [PubMed] [Google Scholar]; b) Krueger D, Izquierdo E, Viswanathan R, Hartmann J, Pallares Cartes C, Renzis S. De, Development 2019, 146, dev175067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8] a).Marsafari M, Ma J, Koffas M, Xu P, Curr. Opin. Biotechnol 2020, 64, 175; [DOI] [PubMed] [Google Scholar]; b) Varady A, Distel M, Front. Cell Dev. Biol 2020, 8, 418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wang H, Vilela M, Winkler A, Tarnawski M, Schlichting I, Yumerefendi H, Kuhlman B, Liu R, Danuser G, Hahn KM, Nat. Methods 2016, 13, 755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zhang W, Lohman AW, Zhuravlova Y, Lu X, Wiens MD, Hoi H, Yaganoglu S, Mohr MA, Kitova EN, Klassen JS, Pantazis P, Thompson RJ, Campbell RE, Nat. Methods 2017, 14, 391. [DOI] [PubMed] [Google Scholar]
- [11].Weber AM, Kaiser J, Ziegler T, Pilsl S, Renzl C, Sixt L, Pietruschka G, Moniot S, Kakoti A, Juraschitz M, Schrottke S, Lledo Bryant L, Steegborn C, Bittl R, Mayer G, Moglich A, Nat. Chem. Biol 2019, 15, 1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Glantz ST, Berlew EE, Jaber Z, Schuster BS, Gardner KH, Chow BY, Proc. Natl. Acad. Sci. USA 2018, 115, E7720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Berlew EE, Kuznetsov IA, Yamada K, Bugaj LJ, Chow BY, Photochem. Photobiol. Sci 2020, 19, 353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Blain-Hartung M, Rockwell NC, Moreno MV, Martin SS, Gan F, Bryant DA, Lagarias JC, J. Biol. Chem 2018, 293, 8473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Bugaj LJ, Choksi AT, Mesuda CK, Kane RS, Schaffer DV, Nat. Methods 2013, 10, 249. [DOI] [PubMed] [Google Scholar]
- [16].Park H, Kim NY, Lee S, Kim N, Kim J, Heo WD, Nat. Commun 2017, 8, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Che DL, Duan L, Zhang K, Cui B, ACS Synth. Biol 2015, 4, 1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Taslimi A, Vrana JD, Chen D, Borinskaya S, Mayer BJ, Kennedy MJ, Tucker CL, Nat. Commun 2014, 5, 4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ong NT, Olson EJ, Tabor JJ, ACS Synth. Biol 2018, 7, 240. [DOI] [PubMed] [Google Scholar]
- [20].Fomicheva A, Zhou C, Sun QQ, Gomelsky M, ACS Synth. Biol 2019, 8, 1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ryu MH, Kang IH, Nelson MD, Jensen TM, Lyuksyutova AI, Siltberg-Liberles J, Raizen DM, Gomelsky M, Proc. Natl. Acad. Sci. USA 2014, 111, 10167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Redchuk TA, Karasev MM, Verkhusha PV, Donnelly SK, Hulsemann, Virtanen J, Moore HM, Vartiainen MK, Hodgson L, Verkhusha VV, Nat. Commun 2020, 11, 605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kinjo T, Terai K, Horita S, Nomura N, Sumiyama K, Togashi K, Iwata S, Matsuda M, Nat. Methods 2019, 16, 1029. [DOI] [PubMed] [Google Scholar]
- [24].Levskaya A, Chevalier AA, Tabor JJ, Simpson ZB, Lavery LA, Levy M, Davidson EA, Scouras A, Ellington AD, Marcotte EM, Voigt CA, Nature 2005, 438, 441. [DOI] [PubMed] [Google Scholar]
- [25].Schmidl SR, Sheth RU, Wu A, Tabor JJ, ACS Synth. Biol 2014, 3, 820. [DOI] [PubMed] [Google Scholar]
- [26].Ong NT, Tabor JJ, ChemBioChem 2018, 19, 1255. [DOI] [PubMed] [Google Scholar]
- [27].Castillo-Hair SM, Baerman EA, Fujita M, Igoshin OA, Tabor JJ, Nat. Commun 2019, 10, 3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kainrath S, Stadler M, Reichhart E, Distel M, Janovjak H, Angew. Chem., Int. Ed. Engl 2017, 56, 4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Golonka D, Fischbach P, Jena SG, Kleeberg JRW, Essen LO, Toettcher JE, Zurbriggen MD, Moglich A, Commun. Biol 2019, 2, 448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Uda Y, Goto Y, Oda S, Kohchi T, Matsuda M, Aoki K, Proc. Natl. Acad. Sci. USA 2017, 114, 11962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Stone OJ, Pankow N, Liu B, Sharma VP, Eddy RJ, Wang H, Putz AT, Teets FD, Kuhlman B, Condeelis JS, Hahn KM, Nat. Chem. Biol 2019, 15, 1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zayner JP, Antoniou C, Sosnick TR, J. Mol. Biol 2012, 419, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hongdusit A, Zwart PH, Sankaran B, Fox JM, Nat. Commun 2020, 11, 788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Mondal P, Krishnamurthy VV, Sharum SR, Haack N, Zhou H, Cheng J, Yang J, Zhang K, ACS Synth. Biol 2019, 8, 2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Niopek D, Wehler P, Roensch J, Eils R, Di Ventura B, Nat. Commun 2016, 7, 10624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Wang W, Wildes CP, Pattarabanjird T, Sanchez MI, Glober GF, Matthews GA, Tye KM, Ting AY, Nat. Biotechnol 2017, 35, 864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lee D, Hyun JH, Jung K, Hannan P, Kwon HB, Nat. Biotechnol 2017, 35, 858. [DOI] [PubMed] [Google Scholar]
- [38].Khamo JS, Krishnamurthy VV, Chen Q, Diao J, Zhang K, Cell Chem. Biol 2019, 26, 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Leopold AV, Chernov KG, Shemetov AA, Verkhusha VV, Nat. Commun 2019, 10, 1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Hope JM, Liu A, Calvin GJ, Cui B, J. Mol. Biol 2020, 13, 3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Krishnamurthy VV, Fu J, Oh T, Khamo J, Yang J, Zhang K, J. Mol. Biol 2020, 432, 3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Deb Roy A, Yin T, Choudhary S, Rodionov V, Pilbeam CC, Wu YI, Nat. Commun 2017, 8, 15831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Alapin JM, Dines M, Vassiliev M, Tamir T, Ram A, Locke C, Yu J, Lamprecht R, Cell Rep. 2018, 23, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Tischer DK, Weiner OD, Elife 2019, 8, e42498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Yousefi OS, Gunther M, Horner M, Chalupsky J, Wess M, Brandl SM, Smith RW, Fleck C, Kunkel T, Zurbriggen MD, Hofer M, Weber W, Schamel WW, Elife 2019, 8, e42475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Capek D, Smutny M, Tichy AM, Morri M, Janovjak H, Heisenberg CP, Elife 2019, 8, e42093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Viswanathan R, Necakov A, Trylinski M, Harish RK, Krueger D, Esposito E, Schweisguth F, Neveu P, De Renzis S, EMBO Rep. 2019, 20, e47999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Sinnen BL, Bowen AB, Forte JS, Hiester BG, Crosby KC, Gibson ES, Dell’Acqua ML, Kennedy MJ, Neuron 2017, 93, 646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Yu D, Lee H, Hong J, Jung H, Jo Y, Oh BH, Park BO, Heo WD, Nat. Methods 2019, 16, 1095. [DOI] [PubMed] [Google Scholar]
- [50] a).Keenan SE, Blythe SA, Marmion RA, Djabrayan NJ, Wieschaus EF, Shvartsman SY, Dev. Cell 2020, 52, 794; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Eritano AS, Bromley CL, Bolea Albero A, Schutz L, Wen FL, Takeda M, Fukaya T, Sami MM, Shibata T, Lemke S, Wang YC, Dev. Cell 2020, 53, 212; [DOI] [PubMed] [Google Scholar]; c) Kaur P, Saunders TE, Tolwinski NS, Sci. Rep 2017, 7, 16636; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Johnson HE, Toettcher JE, Dev. Cell 2019, 48, 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51] a).Goglia AG, Wilson MZ, Jena SG, Silbert J, Basta LP, Devenport D, Toettcher JE, Cell Syst. 2020, 10, 240; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Bugaj LJ, Sabnis AJ, Mitchell A, Garbarino JE, Toettcher JE, Bivona TG, Lim WA, Science 2018, 361, eaao3048; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Patel AL, Yeung E, McGuire SE, Wu AY, Toettcher JE, Burdine RD, Shvartsman SY, Proc. Natl. Acad. Sci. USA 2019, 116, 25756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Melero-Fernandez de Mera RM, Li LL, Popinigis A, Cisek K, Tuittila M, Yadav L, Serva A, Courtney MJ, Nat. Commun 2017, 8, 15017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Zago G, Veith I, Singh MK, Fuhrmann L, De Beco S, Remorino A, Takaoka S, Palmeri M, Berger F, Brandon N, El Marjou A, Vincent-Salomon A, Camonis J, Coppey M, Parrini MC, Elife 2018, 7, e40474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Ma G, He L, Liu S, Xie J, Huang Z, Jing J, Lee YT, Wang R, Luo H, Han W, Huang Y, Zhou Y, Nat. Commun 2020, 11, 1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Kim S, Kyung T, Chung JH, Kim N, Keum S, Lee J, Park H, Kim HM, Lee S, Shin HS, o Heo WD, Nat. Commun 2020, 11, 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Bohineust A, Garcia Z, Corre B, Lemaitre F, Bousso P, Nat. Commun 2020, 11, 1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Hannanta-Anan P, Chow BY, ACS Synth. Biol 2018, 7, 1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Murakoshi H, Shin ME, Parra-Bueno P, Szatmari EM, Shibata ACE, Yasuda R, Neuron 2017, 94, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Tei R, Baskin JM, J. Cell Biol 2020, 219, e201907013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Ji C, Fan F, Lou X, Cell Rep. 2017, 20, 1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Steuer Costa W, Yu SC, Liewald JF, Gottschalk A, Curr. Biol 2017, 27, 495. [DOI] [PubMed] [Google Scholar]
- [62].Hepp S, Trauth J, Hasenjäger S, Bezold F, Essen L, Taxis C, JMB 2020, 432, 1880. [DOI] [PubMed] [Google Scholar]
- [63].Muller D, Exler S, Aguilera-Vazquez L, Guerrero-Martin E, Reuss M, Yeast 2003, 20, 351. [DOI] [PubMed] [Google Scholar]
- [64].Dong J, Fu XM, Wang PF, Dong SS, Li X, Xiao DG, Zhang CY, J. Food Biochem 2019, 43, e12846. [DOI] [PubMed] [Google Scholar]
- [65].Flynn GE, Zagotta WN, Proc. Natl. Acad. Sci. USA 2018, 115, E8086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Zhang F, Tzanakakis ES, Sci. Rep 2017, 7, 9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Zhang F, Tzanakakis ES, ACS Synth. Biol 2019, 8, 2248. [DOI] [PubMed] [Google Scholar]
- [68] a).Ryu MH, Gomelsky M, ACS Synth. Biol 2014, 3, 802; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Misra R, Hirshfeld A, Sheves M, Chem. Sci 2019, 10, 7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Ryu MH, Fomicheva A, Moskvin OV, Gomelsky M, J. Bacteriol 2017, 199, e00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].O’Neal L, Ryu MH, Gomelsky M, Alexandre G, J. Bacteriol 2017, 199, e00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Hu Y, Liu X, Ren ATM, Gu JD, Cao B, ChemSusChem 2019, 12, 5142. [DOI] [PubMed] [Google Scholar]
- [72].Oakes PW, Wagner E, Brand CA, Probst D, Linke M, Schwarz US, Glotzer M, Gardel ML, Nat. Commun 2017, 8, 15817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Cavanaugh KE, Staddon MF, Munro E, Banerjee S, Gardel ML, Dev. Cell 2020, 52, 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Hennig PMK, Wang I, Valon L, DeBeco S, Coppey M, Miroshnikova YA, Albiges-Rizo C, Favard C, Voituriez R, Balland M, Sci. Adv 2020, 6, eaau5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Yu M, Le S, Barnett S, Guo Z, Zhong X, Kanchanawong P, Yan J, Phys. Rev. X 2020, 10, 021001. [Google Scholar]
- [76].Endo M, Iwawaki T, Yoshimura H, Ozawa T, ACS Chem. Biol 2019, 14, 2206. [DOI] [PubMed] [Google Scholar]
- [77].Fulda S, Debatin KM, Oncogene 2006, 25, 4798. [DOI] [PubMed] [Google Scholar]
- [78] a).Hughes RM, Freeman DJ, Lamb KN, Pollet RM, Smith WJ, Lawrence DS, Angew. Chem., Int. Ed. Engl 2015, 54, 12064; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Godwin WC, Hoffmann GF, Gray TJ, Hughes RM, J. Biol. Chem 2019, 294, 16918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Zhang M, Lin X, Zhang J, Su L, Ma M, Ea VL, Liu X, Wang L, Chang J, Li X, Zhang X, Oncogene 2020, 39, 2118. [DOI] [PubMed] [Google Scholar]
- [80].Smart AD, Pache RA, Thomsen ND, Kortemme T, Davis GW, Wells JA, Proc. Natl. Acad. Sci. USA 2017, 114, E8174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Lindner F, Milne-Davies B, Langenfeld K, Stiewe T, Diepold A, Nat. Commun 2020, 11, 2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Deneke VE, Puliafito A, Krueger D, Narla AV, De Simone A, Primo L, Vergassola M, De Renzis S, Di Talia S, Cell 2019, 177, 925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].De S, Campbell C, Venkitaraman AR, Esposito A, Cell Rep. 2020, 30, 2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84] a).Jones TT, Liu A, Cui B, ACS Synth. Biol 2020, 9, 893; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) D’Acunzo P, Strappazzon F, Caruana I, Meneghetti G, Di Rita A, Simula L, Weber G, Del Bufalo F, Dalla Valle L, Campello S, Locatelli F, Cecconi F, Nat. Commun 2019, 10, 1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Okumura M, Natsume T, Kanemaki MT, Kiyomitsu T, Elife 2018, 7, e36559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Nijenhuis W, van Grinsven MMP, Kapitein LC, J. Cell Biol 2020, 219, e201907149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Griesche N, Sanchez G, Hermans C, Idevall-Hagren O, J. Cell Sci 2019, 132, jcs228544. [DOI] [PubMed] [Google Scholar]
- [88].van Haren J, Charafeddine RA, Ettinger A, Wang H, Hahn KM, Wittmann T, Nat. Cell Biol 2018, 20, 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].French AR, Sosnick TR, Rock RS, Proc. Natl. Acad. Sci. USA 2017, 114, E1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Zhu L, Richardson TM, Wacheul L, Wei MT, Feric M, Whitney G, Lafontaine DLJ, Brangwynne CP, Proc. Natl. Acad. Sci. USA 2019, 116, 17330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Shin Y, Berry J, Pannucci N, Haataja MP, Toettcher JE, Brangwynne CP, Cell 2017, 168, 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Kilic S, Lezaja A, Gatti M, Bianco E, Michelena J, Imhof R, Altmeyer M, EMBO J. 2019, 38, 101379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Shin Y, Chang YC, Lee DSW, Berry J, Sanders DW, Ronceray P, Wingreen NS, Haataja M, Brangwynne CP, Cell 2018, 175, 1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Dine E, Gil AA, Uribe G, Brangwynne CP, Toettcher JE, Cell Syst. 2018, 6, 655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Zhao EM, Suek N, Wilson MZ, Dine E, Pannucci NL, Gitai Z, Avalos JL, Toettcher JE, Nat. Chem. Biol 2019, 15, 589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Bracha MTWD, Wei M, Zhu L, Kurian M, Avalos JL, Toettcher JE, Brangwynne CP, Cell 2018, 6, 1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Zhang P, Fan B, Yang P, Temirov J, Messing J, Kim HJ, Taylor JP, Elife 2019, 8, e39578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Mann JR, Gleixner AM, Mauna JC, Gomes E, DeChellis-Marks MR, Needham PG, Copley KE, Hurtle B, Portz B, Pyles NJ, Guo L, Calder CB, Wills ZP, Pandey UB, Kofler JK, Brodsky JL, Thathiah A, Shorter J, Donnelly CJ, Neuron 2019, 102, 321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Asakawa K, Handa H, Kawakami K, Nat. Commun 2020, 11, 1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Lim CH, Kaur P, Teo E, Lam VYM, Zhu F, Kibat C, Gruber J, Mathuru AS, Tolwinski NS, Elife 2020, 9, e52589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Park A, Jacob AD, Walters BJ, Park S, Rashid AJ, Jung JH, Lau J, Woolley GA, Frankland PW, Josselyn SA, Neuropsychopharmacology 2020, 45, 916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].McDaniel SL, Gibson TJ, Schulz KN, Fernandez Garcia M, Nevil M, Jain SU, Lewis PW, Zaret KS, Harrison MM, Mol. Cell 2019, 74, 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, Tucker CL, Nat. Methods 2010, 7, 973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104] a).Taslimi A, Zoltowski B, Miranda JG, Pathak GP, Hughes RM, Tucker CL, Nat. Chem. Biol 2016, 12, 425; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Meador K, Wysoczynski CL, Norris AJ, Aoto J, Bruchas MR, Tucker CL, Nucleic Acids Res. 2019, 47, e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Morikawa K, Furuhashi K, de Sena-Tomas C, Garcia-Garcia AL, Bekdash R, Klein AD, Gallerani N, Yamamoto HE, Park SE, Collins GS, Kawano F, Sato M, Lin CS, Targoff KL, Au E, Salling MC, Yazawa M, Nat. Commun 2020, 11, 2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Yao S, Yuan P, Ouellette B, Zhou T, Mortrud M, Balaram P, Chatterjee S, Wang Y, Daigle TL, Tasic B, Kuang X, Gong H, Luo Q, Zeng S, Curtright A, Dhaka A, Kahan A, Gradinaru V, Chrapkiewicz R, Schnitzer M, Zeng H, Cetin A, Nat. Methods 2020, 17, 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Jung H, Kim SW, Kim M, Hong J, Yu D, Kim JH, Lee Y, Kim S, Woo D, Shin HS, Park BO, Heo WD, Nat. Commun 2019, 10, 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Nihongaki Y, Yamamoto S, Suzuki H, Sato M, Chem. Biol 2015, 22, 169. [DOI] [PubMed] [Google Scholar]
- [109].Nihongaki Y, Furuhata Y, Otabe T, Hasegawa S, Yoshimoto K, Sato M, Nat. Methods 2017, 14, 963. [DOI] [PubMed] [Google Scholar]
- [110].Shao J, Wang M, Yu G, Zhu S, Yu Y, Heng BC, Wu J, Ye H, Proc. Natl. Acad. Sci. USA 2018, 115, E6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Nihongaki Y, Otabe T, Ueda Y, Sato M, Nat. Chem. Biol 2019, 15, 882. [DOI] [PubMed] [Google Scholar]
- [112].Meriesh HA, Lerner AM, Chandrasekharan MB, Strahl BD, J. Biol. Chem 2020, 295, 6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Kim JH, Rege M, Valeri J, Dunagin MC, Metzger A, Titus KR, Gilgenast TG, Gong W, Beagan JA, Raj A, Phillips-Cremins JE, Nat. Methods 2019, 16, 633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Kim NY, Lee S, Yu J, Kim N, Won SS, Park H, Heo WD, Nat. Cell Biol 2020, 22, 341. [DOI] [PubMed] [Google Scholar]
- [115].Zhao EM, Zhang Y, Mehl J, Park H, Lalwani MA, Toettcher JE, Avalos JL, Nature 2018, 555, 683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Wang W, Huang D, Ren J, Li R, Feng Z, Guan C, Bao B, Cai B, Ling J, Zhou C, Theranostics 2019, 9, 8196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Binder JL, Chander P, Deretic V, Weick JP, Bhaskar K, PLoS One 2020, 15, e0230026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Li X, Zhang C, Xu X, Miao J, Yao J, Liu R, Zhao Y, Chen X, Yang Y, Nucleic Acids Res. 2020, 48, e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Yang KK, Wu Z, Arnold FH, Nat. Methods 2019, 16, 687. [DOI] [PubMed] [Google Scholar]
- [120].Bedbrook CN, Yang KK, Robinson JE, Mackey ED, Gradinaru V, Arnold FH, Nat. Methods 2019, 16, 1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Glantz ST, Carpenter EJ, Melkonian M, Gardner KH, Boyden ES, Wong GK, Chow BY, Proc. Natl. Acad. Sci. USA 2016, 113, E1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Woloschuk RM, Reed PMM, McDonald S, Uppalapati M, Woolley GA, J. Mol. Biol 2020, 432, 3113. [DOI] [PubMed] [Google Scholar]
- [123].Teets FD, Watanabe T, Hahn KM, Kuhlman B, J. Mol. Biol 2020, 432, 805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Dagliyan O, Krokhotin A, Ozkan-Dagliyan I, Deiters A, Der CJ, Hahn KM, Dokholyan NV, Nat. Commun 2018, 9, 4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Harrigan P, Madhani HD, El-Samad H, Cell 2018, 175, 877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126] a).Perkins ML, Benzinger D, Arcak M, Khammash M, Nat. Commun 2020, 11, 1355; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Benzinger D, Khammash M, Nat. Commun 2018, 9, 3521; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Rullan M, Benzinger D, Schmidt GW, Milias-Argeitis A, Khammash M, Mol. Cell 2018, 70, 745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Mahajan T, Rai K, PLoS One 2018, 13, e0183242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Camacho IS, Theisen A, Johannissen LO, Diaz-Ramos LA, Christie JM, Jenkins GI, Bellina B, Barran P, Jones AR, Proc. Natl. Acad. Sci. USA 2019, 116, 1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].a) Tateyama S, Kobayashi I, Hisatomi O, Biochemistry 2018, 57, 6615; [DOI] [PubMed] [Google Scholar]; b) Korepanova A, Matayoshi ED, Curr. Protoc. Protein Sci 2012, 68, 29.5.1. [DOI] [PubMed] [Google Scholar]
- [130].Lu H, Mazumder M, Jaikaran ASI, Kumar A, Leis EK, Xu X, Altmann M, Cochrane A, Woolley GA, ACS Synth. Biol 2019, 8, 744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Kim J, Lee S, Jung K, Oh WC, Kim N, Son S, Jo Y, Kwon HB, Heo WD, Nat. Commun 2019, 10, 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Bugaj LJ, Lim WA, Nat. Protoc 2019, 14, 2205. [DOI] [PubMed] [Google Scholar]
- [133].Repina NA, McClave T, Johnson HJ, Bao X, Kane RS, Schaffer DV, Cell Rep. 2020, 31, 107737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Juttner J, Szabo A, Gross-Scherf B, Morikawa RK, Rompani SB, Hantz P, Szikra T, Esposti F, Cowan CS, Bharioke A, Patino-Alvarez CP, Keles O, Kusnyerik A, Azoulay T, Hartl D, Krebs AR, Schubeler D, Hajdu RI, Lukats A, Nemeth J, Nagy ZZ, Wu KC, Wu RH, Xiang L, Fang XL, Jin ZB, Goldblum D, Hasler PW, Scholl HPN, Krol J, Roska B, Nat. Neurosci 2019, 22, 1345. [DOI] [PubMed] [Google Scholar]
- [135] a).Montgomery KL, Yeh AJ, Ho JS, Tsao V, Iyer SM, Grosenick L, Ferenczi EA, Tanabe Y, Deisseroth K, Delp SL, Poon AS, Nat. Methods 2015, 12, 969; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kim T, McCall JG, Jung YH, Huang X, Siuda ER, Li Y, Song J, Song YM, Pao HA, Kim R, Lu C, Lee SD, Song I, Shin G, Al-Hasani R, Kim S, Tan MP, Huang Y, Omenetto FG, Rogers JA, Bruchas M. l. R., Science 2013, 340, 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136] a).Matarese BFE, Feyen PLC, de Mello JC, Benfenati F, Front. Bioeng. Biotechnol 2019, 7, 278; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Mao D, Li N, Xiong Z, Sun Y, Xu G, iScience 2019, 21, 403; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Qazi R, Gomez AM, Castro DC, Zou Z, Sim JY, Xiong Y, Abdo J, Kim CY, Anderson A, Lohner F, Byun SH, Chul Lee B, Jang KI, Xiao J, Bruchas MR, Jeong JW, Nat. Biomed. Eng 2019, 3, 655; [DOI] [PubMed] [Google Scholar]; d) Zhang Y, Castro DC, Han Y, Wu Y, Guo H, Weng Z, Xue Y, Ausra J, Wang X, Li R, Wu G, Vazquez-Guardado A, Xie Y, Xie Z, Ostojich D, Peng D, Sun R, Wang B, Yu Y, Leshock JP, Qu S, Su CJ, Shen W, Hang T, Banks A, Huang Y, Radulovic J, Gutruf P, Bruchas MR, Rogers JA, Proc. Natl. Acad. Sci. USA 2019, 116, 21427; [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Wu X, Zhu X, Chong P, Liu J, Andre LN, Ong KS, Brinson K Jr., Mahdi AI, Li J, Fenno LE, Wang H, Hong G, Proc. Natl. Acad. Sci. USA 2019, 116, 26332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Zhang Y, Huang L, Li Z, Ma G, Zhou Y, Han G, ACS Nano 2016, 10, 3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138] a).Zheng B, Wang H, Pan H, Liang C, Ji W, Zhao L, Chen H, Gong X, Wu X, Chang J, ACS Nano 2017, 11, 11898; [DOI] [PubMed] [Google Scholar]; b) Kawano F, Okazaki R, Yazawa MSM, Nat. Chem. Biol 2016, 12, 1059; [DOI] [PubMed] [Google Scholar]; c) Sasaki Y, Oshikawa M, Bharmoria P, Kouno H, Hayashi-Takagi A, Sato M, Ajioka I, Yanai N, Kimizuka N, Angew. Chem., Int. Ed. Engl 2019, 58, 17827; [DOI] [PubMed] [Google Scholar]; d) Rao P, Wang L, Cheng Y, Wang X, Li H, Zheng G, Li Z, Jiang C, Zhou Q, Huang C, Biomed. Opt. Express 2020, 11, 1401; [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Wang Y, Xie K, Yue H, Chen X, Luo X, Liao Q, Liu M, Wang F, Shi P, Nanoscale 2020, 12, 2406; [DOI] [PubMed] [Google Scholar]; f) Pan H, Wang H, Yu J, Huang X, Hao Y, Zhang C, Ji W, Yang M, Gong X, Wu X, Chang J, Biomaterials 2019, 199, 22. [DOI] [PubMed] [Google Scholar]
- [139].Wu X, Huang W, Wu W-H, Xue B, Xiang D, Li Y, Qin M, Sun F, Wang W, Zhang W-B, Cao Y, Nano Res. 2018, 11, 5556. [Google Scholar]
- [140].Xiang D, Wu X, Cao W, Xue B, Qin M, Cao Y, Wang W, Front. Chem 2020, 8, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Liu L, Shadish JA, Arakawa CK, Shi K, Davis J, DeForest CA, Adv. Biosyst 2018, 2, 1800240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Hammer JA, Ruta A, West JL, Ann. Biomed. Eng 2019, 7, 1885. [DOI] [PubMed] [Google Scholar]
- [143].Shadish JA, Strange AC, DeForest CA, J. Am. Chem. Soc 2019, 141, 15619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Beyer HM, Thomas OS, Riegel N, Zurbriggen MD, Weber W, Horner M, Acta Biomater. 2018, 79, 276. [DOI] [PubMed] [Google Scholar]
- [145].Sankaran S, Becker J, Wittmann C, Del Campo A, Small 2019, 15, 1804717. [DOI] [PubMed] [Google Scholar]
- [146].Beyer HM, Engesser R, Horner M, Koschmieder J, Beyer P, Timmer J, Zurbriggen MD, Weber W, Adv. Mater 2018, 30, 1800472. [DOI] [PubMed] [Google Scholar]
- [147].Rivera-Tarazona LK, Bhat VD, Kim H, Campbell ZT, Ware TH, Sci. Adv 2020, 6, eaax8582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Mukherjee M, Hu Y, Tan CH, Rice SA, Cao B, Sci. Adv 2018, 4, eaau1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Chen F, Wegner SV, ACS Synth. Biol 2017, 6, 2170. [DOI] [PubMed] [Google Scholar]
- [150].Jin X, Riedel-Kruse IH, Proc. Natl. Acad. Sci. USA 2018, 115, 3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Fernandez-Rodriguez J, Moser F, Song M, Voigt CA, Nat. Chem. Biol 2017, 13, 706. [DOI] [PubMed] [Google Scholar]
- [152].Olson EJ, Tzouanas CN, Tabor JJ, Mol. Syst. Biol 2017, 13, 926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Dzobo K, Thomford NE, Senthebane DA, Shipanga H, Rowe A, Dandara C, Pillay M, Motaung K, Stem Cells Int. 2018, 2018, 2495848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Toda H, Yamamoto M, Uyama H, Tabata Y, Acta Biomater. 2016, 29, 215. [DOI] [PubMed] [Google Scholar]
- [155].Horner M, Raute K, Hummel B, Madl J, Creusen G, Thomas OS, Christen EH, Hotz N, Gubeli RJ, Engesser R, Rebmann B, Lauer J, Rolauffs B, Timmer J, Schamel WWA, Pruszak J, Romer W, Zurbriggen MD, Friedrich C, Walther A, Minguet S, Sawarkar R, Weber W, Adv. Mater 2019, 31, 1806727. [DOI] [PubMed] [Google Scholar]
- [156].Baaske J, Muhlhauser WWD, Yousefi OS, Zanner S, Radziwill G, Horner M, Schamel WWA, Weber W, Commun. Biol 2019, 2, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Mueller M, Rasoulinejad S, Garg S, Wegner SV, Nano Lett. 2020, 20, 2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Yuz SG, Ricken J, Wegner SV, Adv. Sci 2018, 5, 1800446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Tas RP, Chen CY, Katrukha EA, Vleugel M, Kok M, Dogterom M, Akhmanova A, Kapitein LC, Nano Lett. 2018, 18, 7524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Bartelt SM, Steinkuhler J, Dimova R, Wegner SV, Nano Lett. 2018, 18, 7268. [DOI] [PubMed] [Google Scholar]
- [161].Jia H, Kai L, Heymann M, Garcia-Soriano DA, Hartel T, Schwille P, Nano Lett. 2018, 18, 7133. [DOI] [PubMed] [Google Scholar]
- [162].Chervyachkova E, Wegner SV, ACS Synth. Biol 2018, 7, 1817. [DOI] [PubMed] [Google Scholar]
- [163].Song C, Knopfel T, Nat. Rev. Drug Discovery 2016, 15, 97. [DOI] [PubMed] [Google Scholar]
- [164].Bonin RP, Wang F, Desrochers-Couture M, Ga Secka A, Boulanger ME, Cote DC, De Koninck Y, Mol. Pain 2016, 12, 10.1177/1744806916629051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Azad TD, Veeravagu A, Steinberg GK, Neurosurg. Focus 2016, 40, E2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Tonnesen J, Kokaia M, Clin. Sci 2017, 131, 1605. [DOI] [PubMed] [Google Scholar]
- [167].Chunshan Deng HY, i Dai J, Neuroscientist 2017, 24, 526. [DOI] [PubMed] [Google Scholar]
- [168] a).Boyle PM, Karathanos TV, Trayanova NA, JACC Clin. Electrophysiol 2018, 4, 155; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Ambrosi CM, Klimas A, Yu J, Entcheva E, Prog. Biophys. Mol. Biol 2014, 115, 294; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Pianca N, Zaglia T, Mongillo M, Biochem. Biophys. Res. Commun 2017, 482, 515. [DOI] [PubMed] [Google Scholar]
- [169].Bryson JB, Machado CB, Lieberam I, Greensmith L, Curr. Opin. Biotechnol 2016, 40, 75. [DOI] [PubMed] [Google Scholar]
- [170].Hardt O, Nadel L, Neurosci. Lett 2018, 680, 54. [DOI] [PubMed] [Google Scholar]
- [171].McDevitt RA, Reed SJ, Britt JP, Neuropsychiatr. Dis. Treat 2014, 10, 1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Galvan A, Stauffer WR, Acker L, El-Shamayleh Y, Inoue KI, Ohayon S, Schmid MC, Neurosci J. 2017, 37, 10894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Shin G, Gomez AM, Al-Hasani R, Jeong YR, Kim J, Xie Z, Banks A, Lee SM, Han SY, Yoo CJ, Lee JL, Lee SH, Kurniawan J, Tureb J, Guo Z, Yoon J, Park SI, Bang SY, Nam Y, Walicki MC, Samineni VK, Mickle AD, Lee K, Heo SY, McCall JG, Pan T, Wang L, Feng X, Kim TI, Kim JK, Li Y, Huang Y, Gereau R, Ha JS, Bruchas MR, Rogers JA, Neuron 2017, 93, 509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174] a).Yu N, Huang L, Zhou Y, Xue T, Chen Z, Han G, Adv. Healthcare Mater 2019, 8, 1801132; [DOI] [PubMed] [Google Scholar]; b) All AH, Zeng X, Teh DBL, Yi Z, Prasad A, Ishizuka T, Thakor N, Hiromu Y, Liu X, Adv. Mater 2019, 31, 1803474. [DOI] [PubMed] [Google Scholar]
- [175].Iseri E, Kuzum D, J. Neural Eng 2017, 14, 031001. [DOI] [PubMed] [Google Scholar]
- [176].Delcasso S, Denagamage S, Britton Z, Graybiel AM, Front. Neural Circuits 2018, 12, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Ma Y, Bao J, Zhang Y, Li Z, Zhou X, Wan C, Huang L, Zhao Y, Han G, Xue T, Cell 2019, 177, 243. [DOI] [PubMed] [Google Scholar]
- [178] a).Naso MF, Tomkowicz B, Perry WL 3rd, Strohl WR, BioDrugs 2017, 31, 317; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wang D, Tai PWL, Gao G, Nat. Rev. Drug Discovery 2019, 18, 358; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Colella P, Ronzitti G, Mingozzi F, Mol. Ther.–Methods Clin. Dev 2018, 8, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Shao J, Xue S, Yu YYG, Yang X, Bai Y, Zhu S, Yang L, Yin J, Wang Y, Liao S, Guo S, Xie M, Fussenegger M, Ye H, Sci. Transl. Med 2017, 9, eaal2298. [DOI] [PubMed] [Google Scholar]
- [180].Towne C, Thompson KR, Curr. Protoc. Pharmacol 2016, 75, 11.19.1. [DOI] [PubMed] [Google Scholar]
- [181].Gerits A, Vanduffel W, Trends Genet. 2013, 29, 403. [DOI] [PubMed] [Google Scholar]
- [182] a).Galvan A, Caiola MJ, Albaugh DL, J. Neural Transm 2018, 125, 547; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Alikaya A, Rack-Wildner M, Stauffer WR, J. Neural Transm 2018, 125, 565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Duebel J, Marazova K, Sahel JA, Curr. Opin. Ophthalmol 2015, 26, 226. [DOI] [PMC free article] [PubMed] [Google Scholar]