ABSTRACT
KPC-82 is a KPC-2 variant identified in a carbapenem-nonsusceptible Citrobacter koseri that confers high-level resistance to ceftazidime-avibactam. Genomic analysis revealed that blaKPC-82 is carried by a chromosomally integrated Tn4401 transposon (disrupting porin gene phoE) and evolved by a 6-nucleotide tandem repeat duplication causing a two-amino-acid insertion (Ser-Asp) within the Ala267-Ser275 loop. Similar to related KPC variants, KPC-82 showed decreased carbapenemase activity when expressed in a heterologous background and remained susceptible to carbapenem/β-lactamase inhibitor combinations.
KEYWORDS: CRE, Citrobacter koseri, KPC, carbapenems, ceftazidime-avibactam
INTRODUCTION
Carbapenem-resistant Enterobacteriaceae (CRE) are a significant threat to modern medicine. In particular, isolates producing carbapenem-hydrolyzing β-lactamase enzymes (carbapenemases) are increasingly prevalent and a cause for further concern given their ability to spread, the severity of infections, and the lack of effective therapeutics (1). Though colistin and tigecycline have been used as first-line treatment, newer antimicrobials with better safety profiles and potent activity against CRE are increasingly being employed as preferable therapeutic options (2).
Among them, ceftazidime-avibactam (CZA) is a β-lactam/β-lactamase inhibitor combination recently introduced into clinical practice (2). It has proven active against serine β-lactamases, including Klebsiella pneumoniae carbapenemases (KPC), which otherwise confer resistance to most β-lactams and β-lactam/β-lactamase inhibitor combinations (1). Despite limited clinical use worldwide, acquired resistance has been reported in multiple independent occurrences and by several mechanisms in both patients with or without a history of CZA therapy (3–10). Most frequently, resistance is caused by KPC variants exhibiting amino acid substitutions, insertions, or deletions in one of 4 loops (loop Leu102 to Ser106, Ω-loop Arg164 to Asp179, or loops Cys238 to Thr243 and Ala267 to Ser275) (11). At the time of writing (April 2021), 82 blaKPC alleles have been deposited in GenBank, including 20 conferring CZA resistance. In this report, we use genomic and molecular genetic approaches to characterize KPC-82, a KPC-2 variant that confers CZA resistance.
Citrobacter koseri MRSN 755319 was cultured from the blood of a patient in a U.S. hospital in 2020. The patient had been hospitalized for several months after suffering a gunshot wound to the abdomen. During this time, the patient had frequent infections caused by multidrug-resistant (MDR) bacteria, including a recurrent respiratory infection due to a carbapenem-susceptible Klebsiella aerogenes (days 159, 197, and 231) as well as a bloodstream infection caused by a carbapenem-resistant (CR), blaKPC-2-carrying Serratia marcescens (MRSN 696556, day 109), that ultimately resolved after ∼4 weeks of treatment with CZA (Fig. 1A). Two and a half months after CZA was discontinued, the patient developed another infection, and blood cultures yielded C. koseri (MRSN 755319, day 250). The isolate was carbapenem resistant (Table 1), and the blaKPC gene was detected using the Cepheid Xpert Carba-R assay. On day 252, the patient was prescribed tigecycline and CZA, which was substituted on day 260 with meropenem-vaborbactam (MVB) following extended antibiotic susceptibility testing (AST) that indicated the isolate was nonsusceptible to CZA (MIC, 128 μg/ml) but susceptible to MVB (MIC, 0.125 μg/ml).
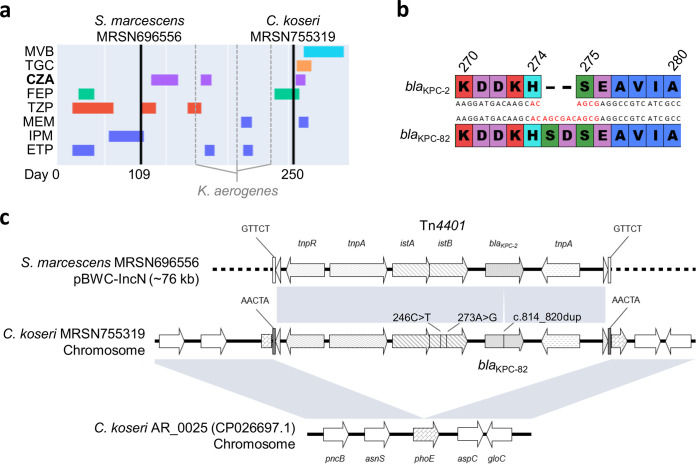
FIG 1.
Identification and characterization of KPC-82. (a) Patient treatment course and sample collection. (b) Nucleotide and amino acid alignment (Ambler numbering and Clustal color scheme) of KPC-2 and KPC-82. (c) Alignment of the plasmid-borne KPC-2 carrying Tn4401 (MRSN 696556) with the chromosomally integrated KPC-82 carrying Tn4401 (MRSN 755319) and its corresponding insertion site in reference genome C. koseri AR_0025.
TABLE 1.
MICs of β-lactams for isolates S. marcescens MRSN 696556, C. koseri MRSN 755319, and recombinant strains E. coli TOP10 with or without KPC-82 or KPC-2
| β-lactam(s)a | MIC (μg/ml) of: |
||||
|---|---|---|---|---|---|
| S. marcescensb MRSN 696556 (KPC-2) | C. koserib MRSN 755319 (KPC-82) | E. colic TOP10 (KPC-82) | E. colic TOP10 (KPC-2) | E. colic TOP10 (pBCSK) | |
| Ampicillin | >16 | >16 | >16 | >16 | ≤8 |
| Ampicillin-sublactam | >16 | >16 | >16 | >16 | ≤4 |
| Piperacillin-tazobactam | >64 | >128 | 32 | >128 | ≤8 |
| Ceftriaxone | >64 | >32 | 32 | >32 | ≤0.5 |
| Cefepime | >16 | >32 | 16 | 32 | ≤4 |
| Ceftazidime | 16 | >16 | >16 | >16 | 4 |
| Ceftazidime-avibactam | 8 | 128 | 64 | 0.5 | 0.25 |
| Ceftolozane-tazobactam | NDd | >16 | >16 | >16 | ≤1 |
| Aztreonam | ND | >16 | >16 | >16 | ≤1 |
| Ertapenem | >8 | 4 | 0.5 | 4 | ≤0.25 |
| Imipenem | >4 | 4 | 1 | >8 | ≤0.5 |
| Meropenem | >4 | 2 | ≤0.5 | >8 | ≤0.5 |
| Meropenem-vaborbactam | 0.125 | 0.125 | ND | ND | ND |
Tazobactam and avibactam were added at a fixed concentration of 4 μg/ml.
Performed in duplicate using a Vitek 2 in the MRSN College of American Pathologists (CAP)-accredited laboratory.
Performed in two biological duplicates (distinct transformants confirmed by Sanger sequencing) using a Gram-negative GN4F AST plate (Thermo Fisher).
ND, not determined.
As part of routine surveillance of MDR organisms, isolates S. marcescens 696556 and C. koseri 755319 were forwarded to the Multidrug-Resistant Organism Repository and Surveillance Network (MRSN). Whole-genome sequencing was performed on an Illumina MiSeq sequencer (Illumina, Inc., San Diego, CA), and genomes were processed as previously described (12). For S. marcescens 696556, long-read sequencing was performed using a MinION sequencer (Oxford Nanopore Technologies). Base-calling was performed using Guppy (configuration r9.4.1_450bps_hac) and filtered using Filtlong (https://github.com/rrwick/Filtlong), and hybrid assembly was performed using Unicycler (13).
Genome analysis revealed that CR and CZA-susceptible (Table 1) S. marcescens MRSN 696556 carried the blaKPC-2 allele. In contrast, CR and CZA-nonsusceptible C. koseri MRSN 755319 carried a mutated blaKPC-2 allele (hereby named blaKPC-82; GenBank accession no. MW485086) and no other acquired β-lactamase. The mutated allele was identical to blaKPC-2 with the exception of a 6-nucleotide (ACAGCG) tandem repeat (TR) insertion causing a two-amino-acid insertion (Ser-Asp) between positions 274 and 275 (Ambler numbering) in the KPC protein (Fig. 1B). TR insertions within the KPC Ala267 to Ser275 loop have been reported previously (11), including KPC-50, a KPC-3 variant with a three-amino-acid insertion (Glu-Ala-Val) at this exact position (3).
To investigate whether the two-amino-acid insertion identified within KPC-82 was responsible for the phenotypic resistance to CZA, the blaKPC-82 gene was cloned into vector pBCSK (Stratagene, La Jolla, CA) and expressed in E. coli TOP10. AST showed that KPC-82 conferred resistance to all β-lactams, including ceftazidime, as well as high-level resistance to CZA (Table 1). Importantly, and similar to KPC-50 (3), E. coli expressing blaKPC-82 remained susceptible to the carbapenems (ertapenem, imipenem, and meropenem).
Further investigations into the genetic context of blaKPC-82 in MRSN 755319 revealed that it was carried by an ∼10-kb Tn4401-like transposon that inserted into the chromosome and disrupted the gene coding for the outer membrane protein PhoE (Fig. 1C and D). Porin loss, such as OprD in P. aeruginosa (14) and OmpK36 in K. pneumoniae (15), has been widely implicated in β-lactam and carbapenem resistance in other bacterial species. Notably, PhoE downregulation has been hypothesized as a possible reason for carbapenem resistance in K. pneumoniae (16), suggesting that its inactivation in MRSN 755319 could cause the otherwise unexplained low-level carbapenem resistance (Table 1).
Interestingly, in S. marcescens MRSN 696556 from the same patient, the blaKPC-2 allele was also carried by a nearly identical Tn4401 (only 2 synonymous mutations in istB in addition to the TR insertion in blaKPC). However, unlike MRSN 755319 but similar to previous reports (17), this transposon was not chromosomally located and was instead carried by an ∼76-kb IncN plasmid named pBWC01 (Fig. 1C and D). The backbone of pBWC01 was absent in MRSN 755319, but both S. marcescens and C. koseri isolates carried an identical ∼4-kb Col440i-type plasmid (Fig. 1C). Similar Col440i cryptic plasmids have been identified in a variety of Enterobacteriaceae and have been documented to coconjugate with a larger IncN KPC-carrying plasmid (including across genus, in vitro) (18). Altogether, and despite missing intermediate isolates, a hypothesis for the emergence of blaKPC-82 would be that (i) both plasmids cotransferred from Serratia to Citrobacter within the host, and (ii) Tn4401 inserted into the chromosome of Citrobacter while the remaining of pBWC01 was lost. In this proposed chain of events, whether blaKPC-82 evolved from blaKPC-2 in Serratia, as a result of CZA exposure, or once acquired by Citrobacter MRSN 755319 still remains unresolved.
In summary, a novel KPC-type enzyme conferring resistance to CZA was identified from a multidrug-resistant C. koseri. Similar to other KPC mutants conferring resistance to CZA, KPC-82 showed decreased carbapenemase activity and remained susceptible to carbapenem/β-lactamase inhibitor combinations, including meropenem-vaborbactam, which successfully cleared the infection in this patient.
Data availability.
Genomes of S. marcescens MRSN 696556 and C. koseri MRSN 755319 have been deposited at NCBI (BioProject accession no. PRJNA692233).
ACKNOWLEDGMENTS
This study was funded by the U.S. Army Medical Command and the Defense Medical Research and Development Program. The manuscript has been reviewed by the Walter Reed Army Institute of Research. There is no objection to its presentation. The opinions or assertions contained herein are the private views of the authors and are not to be construed as official or reflecting the views of the Department of the Army or the Department of Defense.
A.I. was supported through Physician Scientist Incubator Program at the University of Pittsburgh sponsored by the Burrows Wellcome Fund. Y.D. was supported by research grants from the National Institutes of Health (R01AI104895, R21AI151362, and R21AI035522).
Contributor Information
Francois Lebreton, Email: francois.lebreton.ctr@mail.mil.
Jason W. Bennett, Email: jason.w.bennett.mil@mail.mil.
REFERENCES
- 1.Iovleva A, Doi Y. 2017. Carbapenem-resistant Enterobacteriaceae. Clin Lab Med 37:303–315. 10.1016/j.cll.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doi Y. 2019. Treatment options for carbapenem-resistant Gram-negative bacterial infections. Clin Infect Dis 69:S565–S575. 10.1093/cid/ciz830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poirel L, Vuillemin X, Juhas M, Masseron A, Bechtel-Grosch U, Tiziani S, Mancini S, Nordmann P. 2020. KPC-50 confers resistance to ceftazidime-avibactam associated with reduced carbapenemase activity. Antimicrob Agents Chemother 64:e00321-20. 10.1128/AAC.00321-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Humphries RM, Yang S, Hemarajata P, Ward KW, Hindler JA, Miller SA, Gregson A. 2015. First report of ceftazidime-avibactam resistance in a KPC-3-expressing Klebsiella pneumoniae isolate. Antimicrob Agents Chemother 59:6605–6607. 10.1128/AAC.01165-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shields RK, Chen L, Cheng S, Chavda KD, Press EG, Snyder A, Pandey R, Doi Y, Kreiswirth BN, Nguyen MH, Clancy CJ. 2017. Emergence of ceftazidime-avibactam resistance due to plasmid-borne blaKPC-3 Mutations during treatment of carbapenem-resistant Klebsiella pneumoniae infections. Antimicrob Agents Chemother 61:e02097-16. 10.1128/AAC.02097-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hemarajata P, Humphries RM. 2019. Ceftazidime/avibactam resistance associated with L169P mutation in the omega loop of KPC-2. J Antimicrob Chemother 74:1241–1243. 10.1093/jac/dkz026. [DOI] [PubMed] [Google Scholar]
- 7.Räisänen K, Koivula I, Ilmavirta H, Puranen S, Kallonen T, Lyytikäinen O, Jalava J. 2019. Emergence of ceftazidime-avibactam-resistant Klebsiella pneumoniae during treatment, Finland, December 2018. Euro Surveill 24:1900256. 10.2807/1560-7917.ES.2019.24.19.1900256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giddins MJ, Macesic N, Annavajhala MK, Stump S, Khan S, McConville TH, Mehta M, Gomez-Simmonds A, Uhlemann A-C. 2017. Successive emergence of ceftazidime-avibactam resistance through distinct genomic adaptations in blaKPC-2-harboring Klebsiella pneumoniae sequence type 307 isolates. Antimicrob Agents Chemother 62:e02101-17. 10.1128/AAC.02101-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaibani P, Ambretti S, Campoli C, Viale P, Re MC. 2020. Genomic characterization of a Klebsiella pneumoniae ST1519 resistant to ceftazidime/avibactam carrying a novel KPC variant (KPC-36). Int J Antimicrob Agents 55:105816. 10.1016/j.ijantimicag.2019.09.020. [DOI] [PubMed] [Google Scholar]
- 10.Sun L, Chen W, Li H, Li L, Zou X, Zhao J, Lu B, Li B, Wang C, Li H, Liu Y, Cao B. 2020. Phenotypic and genotypic analysis of KPC-51 and KPC-52, two novel KPC-2 variants conferring resistance to ceftazidime/avibactam in the KPC-producing Klebsiella pneumoniae ST11 clone background. J Antimicrob Chemother 75:3072–3074. 10.1093/jac/dkaa241. [DOI] [PubMed] [Google Scholar]
- 11.Hobson CA, Bonacorsi S, Jacquier H, Choudhury A, Magnan M, Cointe A, Bercot B, Tenaillon O, Birgy A. 2020. KPC beta-lactamases are permissive to insertions and deletions conferring substrate spectrum modifications and resistance to ceftazidime-avibactam. Antimicrob Agents Chemother 64:e01175-20. 10.1128/AAC.01175-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galac MR, Snesrud E, Lebreton F, Stam J, Julius M, Ong AC, Maybank R, Jones AR, Kwak YI, Hinkle K, Waterman PE, Lesho EP, Bennett JW, Mc Gann P. 2020. A diverse panel of clinical Acinetobacter baumannii for research and development. Antimicrob Agents Chemother 64:e00840-20. 10.1128/AAC.00840-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H, Luo YF, Williams BJ, Blackwell TS, Xie CM. 2012. Structure and function of OprD protein in Pseudomonas aeruginosa: from antibiotic resistance to novel therapies. Int J Med Microbiol 302:63–68. 10.1016/j.ijmm.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.García-Sureda L, Doménech-Sánchez A, Barbier M, Juan C, Gascó J, Albertí S. 2011. OmpK26, a novel porin associated with carbapenem resistance in Klebsiella pneumoniae. Antimicrob Agents Chemother 55:4742–4747. 10.1128/AAC.00309-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaczmarek FM, Dib-Hajj F, Shang W, Gootz TD. 2006. High-level carbapenem resistance in a Klebsiella pneumoniae clinical isolate is due to the combination of bla(ACT-1) beta-lactamase production, porin OmpK35/36 insertional inactivation, and down-regulation of the phosphate transport porin PhoE. Antimicrob Agents Chemother 50:3396–3406. 10.1128/AAC.00285-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Partridge SR. 2014. Tn4401 carrying blaKPC is inserted within another insertion in pKpQIL and related plasmids. J Clin Microbiol 52:4448–4449. 10.1128/JCM.02426-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barry KE, Wailan AM, Sheppard AE, Crook D, Vegesana K, Stoesser N, Parikh HI, Sebra R, Mathers AJ. 2019. Don't overlook the little guy: an evaluation of the frequency of small plasmids co-conjugating with larger carbapenemase gene containing plasmids. Plasmid 103:1–8. 10.1016/j.plasmid.2019.03.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Genomes of S. marcescens MRSN 696556 and C. koseri MRSN 755319 have been deposited at NCBI (BioProject accession no. PRJNA692233).