ABSTRACT
Ciprofloxacin is one of the most widely used antibiotics for treating Pseudomonas aeruginosa infections. However, P. aeruginosa acquires mutations that confer ciprofloxacin resistance, making treatment more difficult. Resistance is multifactorial, with mutations in multiple genes influencing the resistance phenotype. However, the contributions of individual mutations and mutation combinations to the amounts of ciprofloxacin that P. aeruginosa can tolerate are not well understood. Engineering P. aeruginosa strain PAO1 to contain mutations in any one of the resistance-associated genes gyrA, nfxB, rnfC, parC, and parE showed that only gyrA mutations increased the MIC for ciprofloxacin. Mutations in parC and parE increased the MIC of a gyrA mutant, making the bacteria ciprofloxacin resistant. Mutations in nfxB and rnfC increased the MIC, conferring resistance, only if both were mutated in a gyrA background. Mutations in all of gyrA, nfxB, rnfC, and parC/E further increased the MIC. These findings reveal an epistatic network of gene-gene interactions in ciprofloxacin resistance. We used this information to predict ciprofloxacin resistance/susceptibility for 274 isolates of P. aeruginosa from their genome sequences. Antibiotic susceptibility profiles were predicted correctly for 84% of the isolates. The majority of isolates for which prediction was unsuccessful were ciprofloxacin resistant, demonstrating the involvement of additional as yet unidentified genes and mutations in resistance. Our data show that gene-gene interactions can play an important role in antibiotic resistance and can be successfully incorporated into models predicting resistance phenotype.
KEYWORDS: Pseudomonas aeruginosa, ciprofloxacin resistance, epistatic interactions, antibiotic resistance prediction, ciprofloxacin, gene-gene interactions
INTRODUCTION
The frequency of antibiotic resistance is rapidly increasing, making infections more difficult to treat. One of the most problematic bacteria in this regard is Pseudomonas aeruginosa, a member of the so-called ESKAPE pathogens (1). P. aeruginosa is an opportunistic pathogen responsible for a large number and broad range of infections, ranging from acute infections that can lead to death within 2 to 3 days to chronic infections that can last several decades (2, 3). Treatment strategies for bacterial infections are commonly based on measurement of antibiotic resistance (4), but this can be time-consuming and, for acute infections, can cost valuable time before appropriate treatment can begin. Inappropriate treatment of acute infections can significantly increase patient mortality (5, 6).
Analysis of genome sequences is emerging as a potential approach to more rapidly predict the antibiotic resistance of bacteria and hence allow effective treatment at an early stage of infection (4, 7–10). The possibility of using this approach with P. aeruginosa has been explored, and shows promise (11–13), but a number of challenges must be overcome for it to be implemented effectively. This approach depends on a strong understanding of the genetic basis of resistance which, in P. aeruginosa, is multifactorial and complex (14–17).
The fluoroquinolone antibiotic ciprofloxacin is one of the most commonly prescribed antibiotics used to treat P. aeruginosa infections but its extensive use is leading to the emergence of resistant bacteria (18, 19). Analysis of clinical isolates and of experimentally evolved ciprofloxacin-resistant mutants shows that resistance commonly arises through mutations in the target-encoding genes gyrA, gyrB, parC, and parE, and in the nfxB gene that regulates the mexCD efflux pump (16, 20–23). A probable ferredoxin gene, rnfC (PA3491), has also been implicated in resistance (24). Plasmid-mediated ciprofloxacin resistance is rare in P. aeruginosa (25–27), although a plasmid-borne gene crpP that may contribute to resistance has been reported (28, 29). Ciprofloxacin resistance-associated alleles can also be present in isolates that are susceptible to this antibiotic, highlighting the incomplete understanding of the relationship between genotype and resistance phenotype (13, 16). A possible explanation for a lack of correlation between genotype and phenotype is that gene-gene interactions need to be taken into account, something that has not been explored to date (9). Gene-gene interactions occur when the effect of an allele on resistance is dependent on the alleles present in other resistance-associated genes (30, 31). Resistance mutations can also have different effects in different genetic backgrounds, indicating epistatic interactions with other unidentified genome components (15, 30). A second potential complication is that different mutations are likely to have different effects on the amounts of ciprofloxacin that can be tolerated by P. aeruginosa. One approach to quantifying the contributions of different mutations to resistance is to engineer mutants that are isogenic with a wild-type strain and then determine the effects of the mutations on the antibiotic MIC (15, 22).
The purpose of this study was to quantify resistance conferred by individual mutations and by combinations of mutations and then use the resulting data to predict ciprofloxacin susceptibility/resistance of clinical isolates of P. aeruginosa from genome analysis. Our findings demonstrate that gene-gene interactions play an important role in ciprofloxacin resistance in P. aeruginosa and that understanding the contributions of different genes can provide an effective framework for predicting the susceptibility of clinical isolates to ciprofloxacin.
RESULTS
Subsets of mutations confer ciprofloxacin resistance in experimentally evolved mutants.
P. aeruginosa strain PAO1 is sensitive to ciprofloxacin, with an MIC of 0.0625 mg/liter. Previously, 13 mutants of strain PAO1 were experimentally evolved to become ciprofloxacin resistant (MIC above 2 mg/liter) by exposing the bacteria in a step-wise manner to sequentially increasing concentrations of ciprofloxacin (24). The ciprofloxacin MIC for mutants was up to 128 mg/liter. Each mutant had mutations in between 4 and 8 genes. To characterize the effects of subsets of these mutations, bacteria from that study that had undergone fewer selection steps and could only tolerate smaller amounts of ciprofloxacin were analyzed by whole-genome sequencing and by MIC testing. Three independent mutants that had the same combination of gyrA, nfxB, and rnfC mutations all had MICs of 4 mg/liter (Table S1 in the supplemental material). Four independent mutants with the combination of gyrA, nfxB, rnfC, and parC mutations all had MICs of 32 mg/liter (Table S1). These data indicate that subsets of mutations can cause the MIC to be above the clinical breakpoint for resistance (2 mg/liter; CLSI, clsi.org), as well as demonstrating the reproducibility of the quantitative contribution of mutation combinations to resistance. They also show that subsets of mutations cause the MIC to be between that of the parental strain PAO1 and the maximally resistant mutants that contain up to 8 mutations, emphasizing the multifactorial nature of resistance.
gyrA mutations increase the MIC of ciprofloxacin for P. aeruginosa.
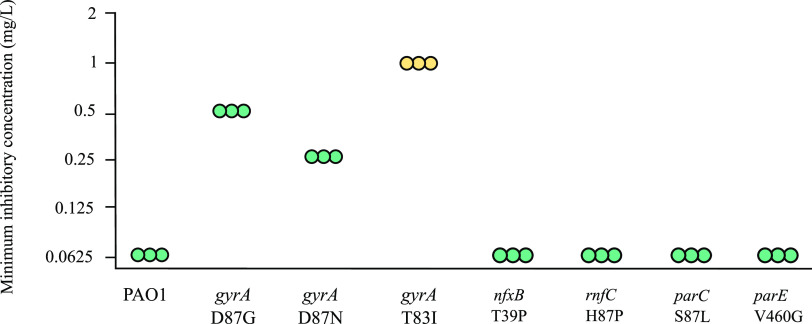
The mutants described above contained combinations of mutations that collectively enable growth in the presence of high concentrations of ciprofloxacin. To quantify the effects of individual mutations, we engineered mutants of P. aeruginosa containing only specific mutations of interest. We first investigated mutations in gyrA that are frequently associated with ciprofloxacin resistance. Three different gyrA mutations, T83I, D87N and D87G, are frequently observed in experimentally evolved mutants of PAO1 and also in clinical isolates of P. aeruginosa (20). Each of these mutations was engineered into P. aeruginosa PAO1 and MICs were determined (Fig. 1). The MICs of the resulting mutants were between 4- and 16-fold higher than that of the parental strain PAO1 (Fig. 1), confirming that gyrA mutations increase the ciprofloxacin MIC and showing that different mutations have different effects.
FIG 1.
Effects of single mutations on the amount of ciprofloxacin required to inhibit growth of P. aeruginosa PAO1. MICs of ciprofloxacin for parental strain PAO1 and isogenic mutants are shown. Each point represents a biological replicate. The breakpoint for ciprofloxacin susceptibility is ≤0.5 mg/liter and intermediate resistance is 1 mg/liter (clsi.org). MICs for the gyrA T83I mutant are in yellow (intermediate resistance), and MICs of strains carrying the other mutations are in turquoise (susceptible).
Clinically, P. aeruginosa with MICs for ciprofloxacin of 0.5 mg/liter are considered susceptible; those with an MIC of 1 mg/liter are considered to have intermediate resistance; and those with an MIC of 2 mg/liter or more, to be resistant (clsi.org). The gyrA T83I mutation conferred intermediate resistance to ciprofloxacin and mutants containing gyrA D87N or D87G remained susceptible to ciprofloxacin.
Mutations in other genes do not increase the ciprofloxacin MIC in the absence of a gyrA mutation.
As well as mutations in gyrA, experimentally evolved ciprofloxacin-resistant mutants commonly have mutations in other genes, including nfxB, parC, parE (20), and rnfC (24). Mutations in each of these genes were engineered into strain PAO1. None of the mutations altered the MIC (Fig. 1). This result showed that these mutations do not by themselves increase the MIC of ciprofloxacin for strain PAO1.
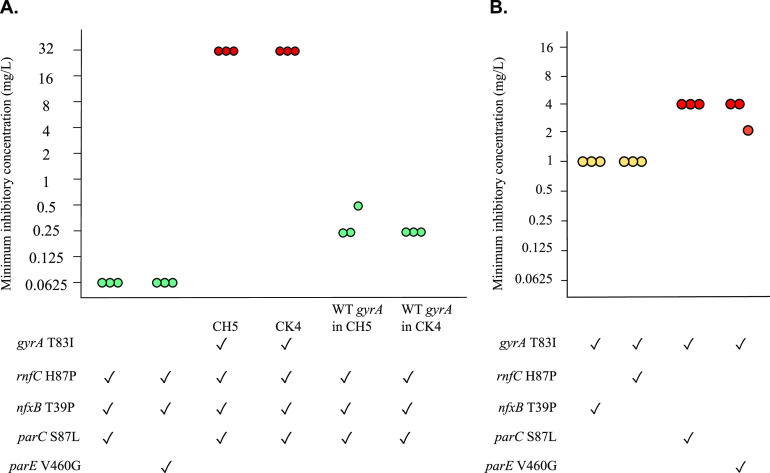
Mutants were constructed to test the possibility that combinations of mutations would increase the amount of ciprofloxacin tolerated by the mutants. Bacteria containing three mutations, in rnfC, nfxB, and parC or parE, had the same MIC as PAO1 (Fig. 2A). These data show that in the absence of a gyrA mutation, mutations in these genes do not increase the ciprofloxacin MIC even when present together.
FIG 2.
A gyrA mutation is required for ciprofloxacin resistance. (A) Effects of combinations of mutations with and without a gyrA mutation. Mutants of strain PAO1 were engineered to contain combinations of rnfC, nfxB, parC, and parE mutations in the absence of a gyrA mutation, and also by replacing a gyrA mutation with the wild-type allele in experimentally evolved mutants CH5 and CK4. Genotypes of mutants are shown, where ✓ denotes the presence of mutation. (B) Effects of mutations in combination with a gyrA mutation. Mutants were engineered to contain the mutations shown. Each point represents one replicate of MIC testing. Turquoise, yellow and red denote susceptible (≤0.5 mg/liter), intermediate (1 mg/liter), and resistant range (≥2 mg/liter) of MICs, respectively.
In a complementary approach, we took advantage of two experimentally evolved mutants of PAO1, CH5 and CK4, which have mutations in all 4 genes gyrA, nfxB, rnfC, and parC. The gyrA T83I mutation was replaced with the wild-type allele in each of these mutants, giving rise to strains containing only the nfxB, rnfC, and parC mutations. The absence of the gyrA mutation led to a reduction in resistance by 128-fold relative to the parent mutant, with an MIC of 0.25 mg/liter (Fig. 2A). Whole-genome sequencing confirmed the presence of only the intended mutations in these bacteria. This result confirmed that when the gyrA wild-type allele was present, none of the other tested mutations led to resistance either individually or in combination. Intriguingly, the MIC of these mutants was reproducibly 4-fold higher than for bacteria engineered to contain combinations of mutations, despite the genome sequences being identical.
parC and parE mutations confer ciprofloxacin resistance when combined with gyrA T83I.
We next tested the possibility that mutations in parC, parE, nfxB, or rnfC might increase the amount of ciprofloxacin tolerated by P. aeruginosa when combined with mutated gyrA. The MICs of double mutants containing gyrA T83I and one other mutation are shown in Fig. 2B. Mutations in parC and parE made the bacteria resistant to ciprofloxacin, increasing the MIC by 4-fold relative to the gyrA mutant. The presence of nfxB and rnfC mutations did not increase the MIC. As parC and parE mutations did not increase the ciprofloxacin MIC for bacteria with wild-type gyrA, the increase in MIC that they caused in the gyrA mutant indicates that gene-gene interactions are important in determining amounts of ciprofloxacin that can be tolerated by P. aeruginosa.
nfxB and rnfC mutations combine to increase the amount of ciprofloxacin that can be tolerated by P. aeruginosa PAO1.
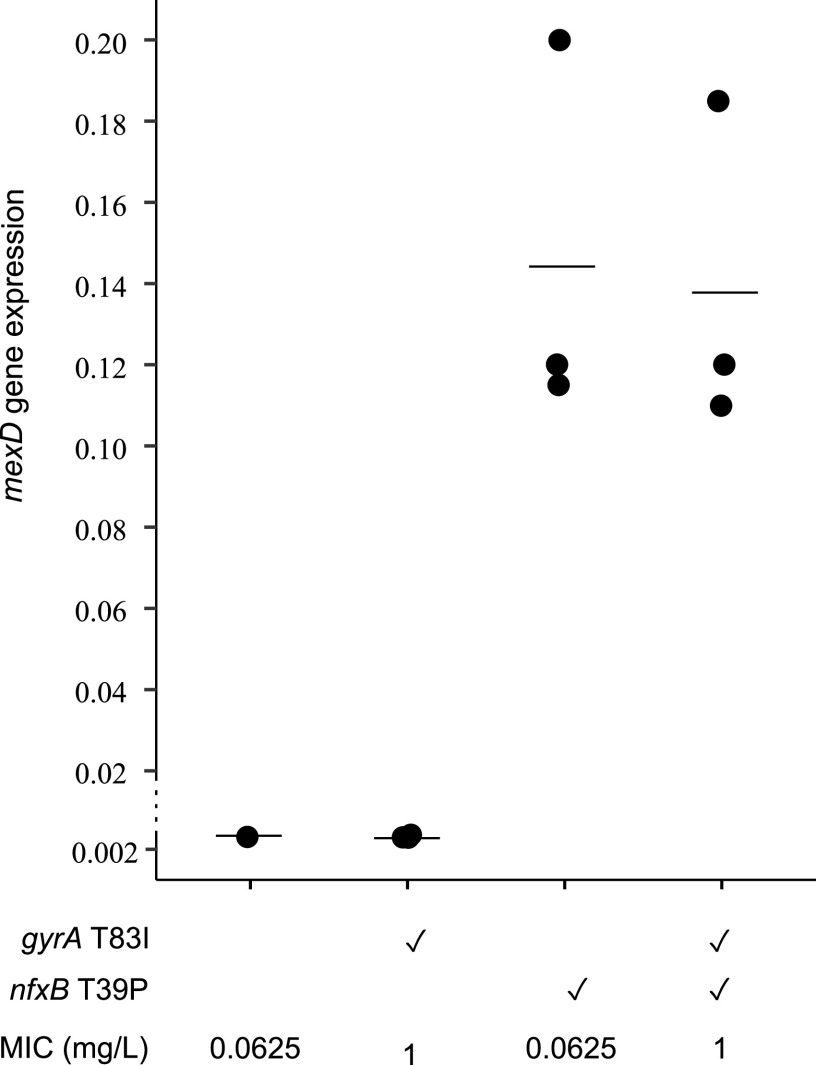
Mutations nfxB T39P and rnfC H87P were present in experimentally evolved ciprofloxacin resistant mutants of strain PAO1 at a high frequency, indicating that they contribute to resistance (24). However, in contrast to previous studies where mutations in nfxB have been associated with ciprofloxacin resistance in laboratory-evolved mutants of P. aeruginosa strain PA14 (15, 22), the presence of the nfxB mutation did not increase the MIC above that of PAO1 containing the gyrA T83I mutation (Fig. 2B). These findings led to the question of how much the mutation effects the function of NfxB. NfxB represses the expression of genes encoding the MexCD-OprJ efflux pump, which is associated with ciprofloxacin resistance (32–34). To evaluate the effect of the nfxB T39P mutation on expression of the efflux pump, quantitative reverse transcriptase PCR (RT-qPCR) of the mexD gene was carried out. The results showed that the nfxB T39P mutation increased mexD expression by approximately 50-fold (Fig. 3; Tables S2 and S3). This indicated that the nfxB T39P mutation resulted in significant derepression of the mexCD-oprJ efflux pump, but this increase was not sufficient to increase the ciprofloxacin MIC.
FIG 3.
Effects of mutation in nfxB on expression of mexD. Expression of mexD was quantified by RT-qPCR. Bars represent the average value of three biological replicates, which are shown as dots. Each dot is the average of 2 technical replicates. Individual values are listed in Table S2. The ✓ denotes the presence of mutations. P values were calculated from one-way ANOVA followed by Tukey’s test (post hoc t test) and showed significant (P < 0.01) difference in mexD expression between NfxB+ and nfxB mutant bacteria (Table S3).
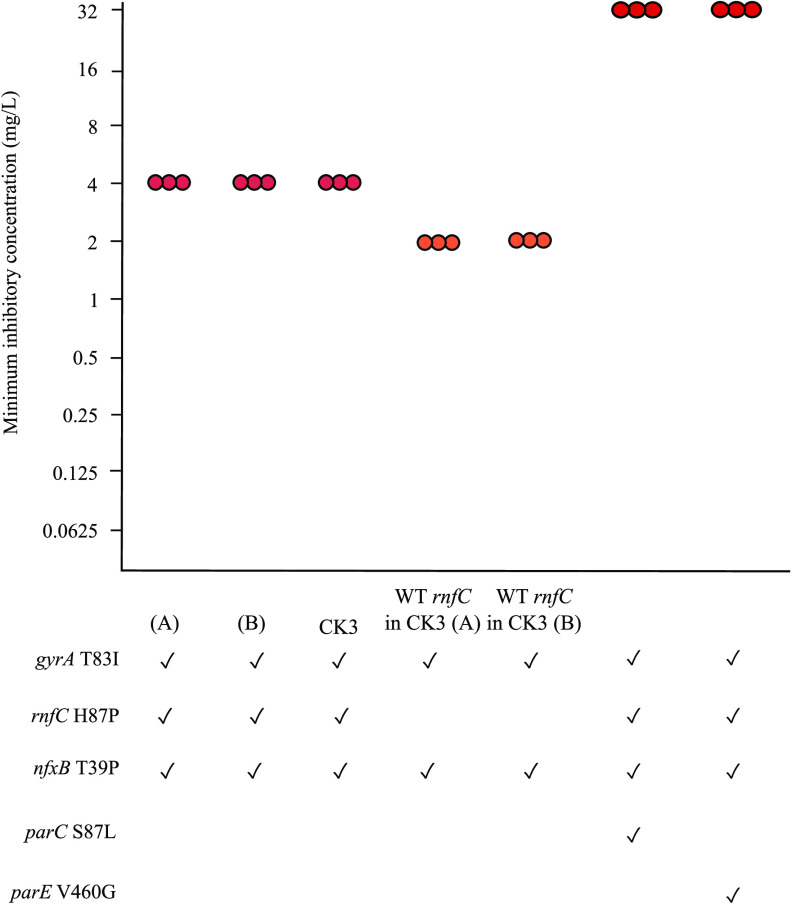
The co-occurrence of the nfxB T39P and rnfC H87P mutations in experimentally evolved mutants (Table S1) suggested that these mutations may interact to increase the amount of ciprofloxacin that can be tolerated by the bacteria. To test this possibility, a mutant containing gyrA T83I, nfxB T39P, and rnfC H87P was constructed. This mutant was resistant to ciprofloxacin with an MIC of 4 mg/liter (Fig. 4), the same as experimentally evolved bacteria with this combination of mutations (CK3, Fig. 4) and 4-fold higher than double mutants containing gyrA T83I and nfxB T39P or gyrA T83I and rnfC H87P (Fig. 2B). These findings indicated that nfxB and rnfC can increase the ciprofloxacin MIC when present together, but not separately, a further example of gene-gene interactions in ciprofloxacin resistance.
FIG 4.
Effects of rnfC H87P on ciprofloxacin MIC. P. aeruginosa PAO1 was engineered to contain mutations in gyrA, rnfC, nfxB, and parC or parE. CK3 is an experimentally evolved mutant of strain PAO1 containing gyrA, rnfC, and nfxB mutations. The wild-type rnfC allele was engineered into CK3 with two independent derivatives being analyzed. Genotypes of mutants are shown, where ✓ denotes the presence of mutation. (A) and (B) are independently engineered biological replicates. Each point represents one replicate of MIC testing. Orange and red denote MICs of 2 mg/liter and >2 mg/liter, respectively.
As rnfC has not been previously associated with antibiotic resistance, its involvement was examined further. In the experimentally evolved CK3 mutant that has mutations gyrA T83I, nfxB T39P, and rnfC H87P, the rnfC mutation was replaced with the wild-type allele. The resulting strain had an MIC of 2 mg/liter, 2-fold lower than the parent strain (Fig. 4). MIC results were reproducible when mutant construction and MIC testing were repeated (Fig. 4). These data show that rnfC contributes to ciprofloxacin resistance in PAO1 when in combination with the nfxB T39P mutation. The MIC of these strains was reproducibly 2-fold higher than that of the engineered mutant in which the gyrA and nfxB mutations were introduced separately (Fig. 2B), despite the bacteria having the same genotype, and the reason for this difference is not clear.
A combination of four mutations further increases ability to tolerate to ciprofloxacin.
To determine whether the amount of ciprofloxacin that can be tolerated by P. aeruginosa is further increased by combinations of more mutations, quadruple mutants were constructed by engineering parC and parE mutations into a gyrA nfxB rnfC triple mutant. The resulting quadruple mutants had an 8-fold higher MIC (32 mg/liter) than the parental mutant, and also higher MICs than gyrA parC and gyrA parE double mutants (Fig. 4). The MICs of both quadruple mutants were the same as experimentally evolved mutants carrying the same mutations (Table S1). These results show that mutations in multiple resistance-associated genes can act synergistically to enable P. aeruginosa to grow despite the presence of high concentrations of ciprofloxacin.
Combinations of resistance mutations can reduce growth rate in antibiotic-free medium.
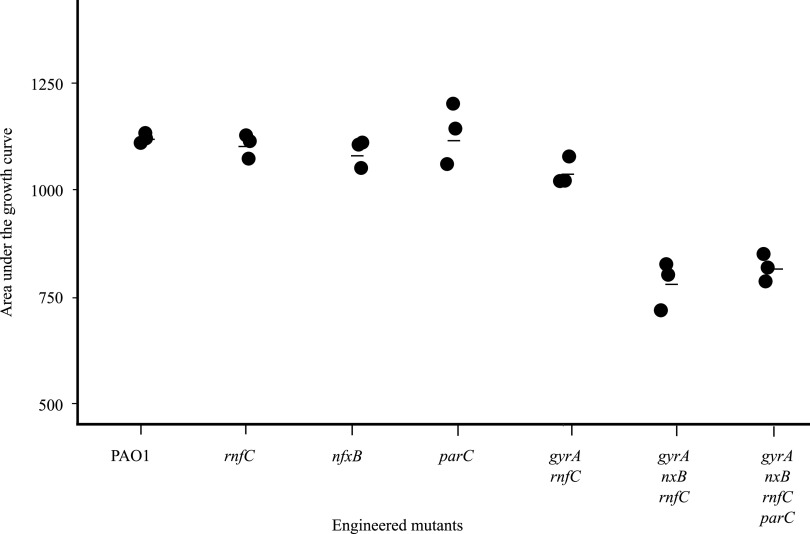
Growth of resistant mutants in the absence of antibiotic is an important consideration in determining how well resistant bacterial populations would be maintained when antibiotic exposure is interrupted (35). Generally, resistance-associated mutations result in changes in proteins that are involved in important biological functions and so may affect the growth of bacteria in the absence of antibiotic (35). The growth of mutants in antibiotic-free medium was quantified (Fig. 5; Fig. S1 and Table S4). None of the mutations affected the growth rate when present individually. However, a significant reduction in growth was observed in bacteria containing mutations in each of nfxB, rnfC, and gyrA (Fig. 5) (36).
FIG 5.
Effects of mutations on growth in antibiotic-free medium. Culture growth, represented as area under the curve, is shown. Each point represents a biological replicate. Average values (Table S4) are represented by black bars. Tukey’s test (post hoc t test) showed that the difference of AUC between PAO1 and strains containing three or four mutations was significant (P value 0.0005) and the difference between PAO1 and mutants containing one or two mutations was not significant.
Prediction of ciprofloxacin resistance from genome sequences.
We next tested whether analysis of variants in genes associated with ciprofloxacin resistance would predict the MICs of clinical isolates of P. aeruginosa. Ciprofloxacin resistance-associated variants used for prediction are listed in Table 1. Sequence variants were chosen for inclusion based on meeting at least two of three criteria: (i) the gene had an established association with antibiotic resistance, (ii) effects of the variant on MIC had been quantified, and (iii) the sequence variant was predicted to have a significant effect on protein function. The gene rnfC was not included in the resistance prediction because its association with resistance, beyond strain PAO1, has not been established. Conversely, variants in gyrB were included because, although they did not arise in our earlier studies with strain PAO1, they were present in 2 out of 5 mutants of strain PA14 that had been evolved to become ciprofloxacin resistant (Table S5) and gyrB mutations are known to contribute to ciprofloxacin resistance (15, 20). The effects of gyrB gene variants on resistance have been quantified previously (15, 20). The plasmid-encoded CrpP protein has also been associated with increased ciprofloxacin resistance in P. aeruginosa (29) and so was included in the analysis.
TABLE 1.
List of amino acid variations searched in clinical isolates of P. aeruginosa to predict the ciprofloxacin resistance
| Protein | Amino acid variation | Reference for presence in clinical isolates or experimentally evolved mutants | Reference for quantification of resistanceb | PROVEAN valuea |
|---|---|---|---|---|
| G81C | 24 | - | −8.7 | |
| G81D | 23 | - | −6.8 | |
| GyrA | T83I | This study, 15, 24 | This study, 15 | −3.7 |
| Y86N | 23 | - | −8.7 | |
| D87N | This study, 15, 24 | This study, 15, 22 | −4.6 | |
| D87G | 23 | This study, 22 | −6.6 | |
| Q106L | 23 | - | −6.8 | |
| E153K | 21 | - | −3.8 | |
| S612L | 24 | - | −5.3 | |
| GyrB | S466Y | This study, 15 | 15 | −5.1 |
| S466F | This study, 15, 16 | 15 | −5.1 | |
| P749S | This study | - | −5.9 | |
| NfxB | P22Q | This study | - | −6.7 |
| A30V | This study | - | −3.9 | |
| A38G | This study | - | −3.8 | |
| T39P | 24 | This study, 22 | −5.6 | |
| R42C | This study, 76 | - | −7.5 | |
| E75G | This study | - | −3.8 | |
| R82L | This study, 77 | - | −4.5 | |
| H87R | 76 | - | −4.8 | |
| ParC | S87L | This study, 24 | This study | −5.9 |
| S87W | 15 | 15 | −6.9 | |
| ParE | M437I | 15 | - | −2.9 |
| P438S | This study, 23 | - | −7.9 | |
| S457C | This study, 23 | - | −4.9 | |
| V460G | 24 | This study | −6.9 | |
| A473V | This study, 78 | - | −2.4 | |
| Plasmid mediated protein | CrpP | This study, 29 | - | - |
PROVEAN analysis was carried out as described in reference 24. Variants with PROVEAN values equal to or below −2.5 are likely to be function-altering variants.
The symbol - indicates not tested.
Criteria to predict the MICs and resistance phenotypes of clinical isolates of P. aeruginosa (Table 2) were developed from the MICs of engineered mutants of strain PAO1 (Table S1). They were then applied to 274 clinical isolates for which genome sequences were available (Table S6). Protein BLAST was carried out on the proteins of interest to identify amino acid variations from strain PAO1. Amino acid variations and ciprofloxacin MICs of each isolate are listed in Table S7. MIC values were available for 200 of the isolates and were determined for the remaining 74 (Table S7). Predicted and actual MICs were then compared to evaluate the accuracies of these criteria for prediction. Ciprofloxacin MICs determined experimentally and predicted from genotypes are termed actual and predicted MICs, respectively. A comparison of predicted and actual MICs (Table 3) showed that 225/274 (82%) isolates had correctly predicted MICs or within one doubling dilution of the actual MIC, whereas 18% had a ≥4-fold MIC difference between actual and predicted MICs (Table 3), indicating a high predictive power for our approach.
TABLE 2.
Criteria used to predict ciprofloxacin MICs and resistance/susceptibility phenotypes for clinical isolates
| Gene variantsa | Corresponding PAO1 mutants | Predicted MIC (predicted ciprofloxacin resistance phenotype)b |
|---|---|---|
| No analyzed variants in gyrA, parC, nfxB, gyrB, or parE | - | Less than 1 (S) |
| Wild-type gyrA and variant parC, nfxB, gyrB, parE, or have crpP genec | parC S87L | Less than 1 (S) |
| parE V460G | ||
| nfxB T39P | ||
| gyrB d | ||
| gyrA D87N | gyrA D87N | 0.25 (S) |
| gyrAT83I | gyrA T83I | 1 (I) |
| gyrAT83I + parC | gyrAT83I + parCS87L | 4 (R) |
| gyrAT83I + parE | gyrAT83I + parEV460G | 2 (R) |
| gyrAT83I + nfxB | gyrAT83I + nfxBT39P | 1 (I) |
| gyrAT83I + nfxB + parE/parC | gyrAT83I + nfxB T39P + parCS87L | 2–4 (R) |
TABLE 3.
Antibiotic resistance prediction in isolates of P. aeruginosa using genomes
| Resistance alleles | Predicted MICa | Total no. of isolates | MICactual equal to, or with one doubling of MICpredicted |
MICactual greater than MICpredictedb |
MICactual less than MICpredictedb |
|||
|---|---|---|---|---|---|---|---|---|
| No. of isolates | MIC range in mg/liter | No. of isolates | MIC range in mg/liter | No. of isolates | MIC range in mg/liter | |||
| Clinical isolates with MIC predicted to be less than or equal to 1 mg/liter | ||||||||
| No analyzed resistance alleles present | 0.05 to 0.5 | 136 | 112 (99 + 13) | 0.05–1 | 24 | 2–4 | 0 | NA |
| WT gyrA in combination with resistance alleles in other genes nfxB/parC/parE/gyrB and crpP | 0.05 to 0.5 | 63 | 57 (43 + 14) | 0.125–1 | 6 | 2–3 | 0 | NA |
| Only gyrAT83I | MIC = 1 | 28 | 17 | 0.5–2 | 6 | 4–12 | 5 | 0.05–0.25 |
| Clinical isolates with MIC predicted to be greater than 1 mg/liter | ||||||||
| Both gyrAT83I and parCS87L | MIC ≥ 4 | 33 | 31 | ≥4 | 1 | 24 | 1 | 0.125 |
| gyrAT83I and parE | MIC = 2 | 5 | 4 | 2–4 | 0 | NA | 1 | 0.5 |
| gyrAT83I + nfxB + parE/parC | MIC ≥ 2 | 9 | 5 | 2–8 | 1 | 12 | 3 | 0.05–1 |
| Total | 274 | 226 | 0.05–8 | 38 | 2–24 | 10 | 0.05–1 | |
Predictions were based on the resistance alleles in genes gyrA, gyrB, parC, parE and nfxB (Table 1) and MICs of mutants carrying those alleles (Table S1).
A value of ± 4-fold or more was considered a significant difference between actual and predicted MICs. NA, not applicable.
For clinical purposes, bacteria are classified as being susceptible (sensitive or intermediate resistance) or resistant (clsi.org). Using these definitions, 84% of isolates were correctly predicted to be susceptible or resistant to ciprofloxacin, with 97% (187/192) of susceptible isolates and 50% of the resistant isolates (41/82) being predicted correctly (Table 4). The largest group of isolates that were incorrectly predicted was 24 resistant isolates that were predicted as susceptible (false negatives) because they did not have any of the analyzed resistance-associated variants but had MICs greater than 1 mg/liter (Table 3, Table S7).
TABLE 4.
Predicted ciprofloxacin resistance/susceptibility in isolates of P. aeruginosa
| Predicted antibiotic susceptibility profilea | Actual antibiotic susceptibility profileb |
|
|---|---|---|
| S/I (n = 192) | R (n = 82) | |
| S/I | 187 | 41 |
| R | 5 | 41 |
Predictions were based on the alleles in genes gyrA, gyrB, parC, parE, and nfxB (Table 1) and MICs of mutants carrying those alleles (Table 2).
CLSI breakpoints were used to categorize isolates; S, susceptible (MIC ≤ 0.5 mg/liter); I, intermediate (MIC = 1 mg/liter); R, resistant (MIC ≥ 2 mg/liter); n, number of isolates.
The approach described above incorporated information on gene-gene interactions into the prediction framework. For comparison, we carried out the same analysis without taking into account gene-gene interactions. In this case, isolates were predicted to be resistant if they had any of the ciprofloxacin resistance-associated alleles (Table 2), without considering the effects of other alleles. Isolates containing no ciprofloxacin resistance-associated alleles were predicted to be susceptible. Using this approach, resistance/susceptibility phenotypes of 63% of clinical isolates were predicted correctly, significantly less than the 84% correctly predicted when taking combinations of alleles into account. The majority of the incorrectly predicted isolates were ciprofloxacin sensitive, having been predicted to be resistant.
DISCUSSION
Antibiotic resistance in P. aeruginosa, as in other species, is multifactorial with sequence variants in multiple genes determining the amounts of antibiotic that can be tolerated by the bacteria. In the case of ciprofloxacin, mutations in gyrA, gyrB, parC, parE, and nfxB are all associated with resistance (20). However, the effects of individual mutations and of combinations of mutations on the amount of ciprofloxacin tolerated by P. aeruginosa have not been well quantified. Here, we show that mutation of gyrA is required for the increased ciprofloxacin MIC in P. aeruginosa PAO1 and that mutations in other genes act in combination to further increase the ability of the bacteria to grow in the presence of ciprofloxacin, conferring resistance. We also show that analysis of appropriate combinations of genetic variants is a strong predictor of ciprofloxacin resistance/susceptibility in isolates of P. aeruginosa. We also confirm rnfC as a contributor to ciprofloxacin resistance in strain PAO1.
The MICs of strain PAO1 derivatives and of clinical isolates showed that the gyrA T83I allele is a key determinant of ciprofloxacin resistance in P. aeruginosa, consistent with earlier studies (15). Mutations other than gyrA T83I did not increase the MIC of P. aeruginosa PAO1 individually or in combination when gyrA was wild type (Fig. 1 and 2A). Similarly, clinical isolates containing other genetic variants, but not gyrA T83I, were predominantly (57/63) susceptible to ciprofloxacin with 43 being fully sensitive (Table 3, Table S7). Mutations in gyrA reduce the affinity of ciprofloxacin for GyrA, its primary target (37). Mutations in parC and parE that encode DNA topoisomerase, a secondary target of ciprofloxacin, would not prevent inhibition of GyrA by ciprofloxacin, explaining why mutations in these genes do not by themselves increase the MIC. Mutations in nfxB that cause upregulation of the mexCD efflux pump are also evidently insufficient to overcome the inhibitory effect of ciprofloxacin on GyrA.
Quantification of the effects of combinations of mutations showed that gene-gene interactions contribute to ciprofloxacin resistance in P. aeruginosa. For example, although mutations in parC or parE did not by themselves increase the ciprofloxacin MIC above that of PAO1, they did increase the amount of ciprofloxacin that could be tolerated by a gyrA mutant (Fig. 2B), reflecting reduced inhibition of ParCE protein variants by ciprofloxacin. Similarly, a mutation in nfxB increased the MIC only in bacteria with a gyrA mutation, presumably because increased efflux associated with loss of functional NfxB does not reduce the intracellular concentration of ciprofloxacin sufficiently to prevent inhibition of wild-type DNA gyrase. The nfxB mutation only increased the MIC when a mutation was also present in rnfC, but the biochemical function of the RnfC protein is not known.
Unexpectedly, the MICs of evolved mutants were in some cases different from bacteria that had been engineered to have the same mutations (Fig. 2 and 4). Whole-genome sequencing confirmed that the same mutations were present in evolved and engineered bacteria and so the reason for the difference in MICs is not clear. It may be that the evolved bacteria contain one or more additional mutations that were not detected when the sequence reads were aligned with the parental PAO1 genome. Alternatively, there is some evidence that epigenetic factors can play a role in antibiotic resistance in other bacterial species (38, 39) and the same may apply to ciprofloxacin resistance in P. aeruginosa.
Although the nfxB T39P mutation did not by itself increase ciprofloxacin resistance in our study, the same mutation in P. aeruginosa strain PA14 increased the MIC for ciprofloxacin by 8-fold (22), supporting the proposal that genetic background impacts the quantitative contribution of mutations to ciprofloxacin resistance (15). Mutations in nfxB cause increased expression of mexC-mexD-oprJ efflux pump genes (Fig. 3) (34). Further, nfxB variants arise at high frequency in experimental evolution studies selecting ciprofloxacin-resistant P. aeruginosa (22, 24, 40) but are much less frequent in clinical isolates (Table S7) (41–43). The lower frequency of nfxB variants in clinical isolates is likely due to a fitness cost associated with nfxB genetic variations. Although a nfxB mutation did not by itself affect growth, mutants containing combinations of nfxB and other mutations had reduced growth (Fig. 5), consistent with earlier studies (22, 35, 44). Gene-gene interactions also affect fitness in ciprofloxacin-resistant E. coli (36). It is notable that gene-gene interactions when multiple mutations are present result in increased resistance (Fig. 2 and 4; Table S1) but reduced growth (Fig. 5) in the absence of ciprofloxacin.
The enhanced ability of strain PAO1 to tolerate ciprofloxacin when other mutations were present in combination with gyrA T83I demonstrated the importance of gene-gene interactions in ciprofloxacin resistance. Emphasizing this point, clinical isolates with gyrA T83I and other ciprofloxacin resistance-associated mutations also had (on average) higher MICs than isolates with the gyrA T83I variant alone. The correct prediction of MIC in a high proportion of clinical isolates containing multiple resistance-associated variants (Table 3) suggested that the variants analyzed are good predictors of ciprofloxacin resistance. An alternative approach to predict ciprofloxacin resistance from P. aeruginosa genome sequences, involving the use of machine learning without consideration of candidate genes or gene-gene interactions, also had a high (90%) predictive value (13). It is likely that combining experimental and machine learning-based approaches will maximize the ability to predict antibiotic susceptibility/resistance from genome sequences. Quantitatively predicting the effectiveness of antibiotics has the potential to improve chemotherapeutic dosages and antibiotic-resistance management (9). Isolates able to survive high concentrations of antibiotic may not be eradicated with dosages that eliminate P. aeruginosa with lower MICs (9).
In P. aeruginosa, resistance to many other antibiotics is also multifactorial, with levels of antibiotic resistance being determined by sequence variants of several genes, as well as the presence or absence of horizontally acquired resistance genes (45, 46). However, the role of gene-gene interactions in determining levels of antibiotic resistance has not been investigated, except in the case of colistin resistance (47). Our results indicated that gene-gene interactions play a major role in determining levels of ciprofloxacin resistance, and may well do so for other antibiotics from different classes. Resistance is also multifactorial for a range of antibiotics in other species, including both Gram-negative and Gram-positive species (48–50), although there has been little or no investigation of the importance of gene-gene interactions. It may well be that such interactions play a role in determining levels of resistance to many antibiotics, in many species, with widespread implications for prediction of antibiotic resistance/susceptibility from genome sequences.
Overall, our genome-based methodology correctly predicted 98% of susceptible isolates and 50% of resistant isolates. The relatively large number of false negatives (i.e., resistant strains predicted to be ciprofloxacin susceptible) was primarily due to 24 isolates that did not have resistance-associated variants in gyrA or any of the other analyzed genes but were nonetheless resistant to ciprofloxacin (Table 3). These isolates did not have any other sequence variants in the genes listed in Table 1, indicating that other genes contribute to the resistance phenotype. All but 2 of these isolates had MICs of 2 mg/liter, only just reaching the threshold for resistance. Other studies (15, 16, 51–55) have also reported ciprofloxacin-resistant isolates that did not have gyrA variants. Resistance variants in such isolates could be due to sequence variants in gyrA, gyrB, parC, or parE that were not included in our prediction framework (Table 2), or changes in efflux pump genes that confer a resistance phenotype (56–59). A number of other genes can affect ciprofloxacin susceptibility (14) and variants of these genes, or currently unidentified genes acquired by horizontal gene transfer, may also influence susceptibility of clinical isolates. Identification of resistance mechanisms that do not involve any of the genes or variants included in this study will be an important step in optimizing the genome-based prediction of ciprofloxacin susceptibility in P. aeruginosa.
In conclusion, gene-gene interactions determine the amount of ciprofloxacin that can be tolerated by P. aeruginosa. The gyrA T83I mutation is needed for an increased ciprofloxacin MIC in strain PAO1, with other mutations increasing the amount of antibiotic that can be tolerated only if the gyrA T83I mutation is present. Analysis of gene variants, taking gene-gene interactions into account, successfully predicted resistance phenotypes in the majority of clinical isolates but showed that in some isolates the genetic basis of resistance is not yet fully understood. Our findings have important implications for understanding the genetic basis of antibiotic resistance in bacteria, as well as for using genomic data to predict resistance phenotypes.
MATERIALS AND METHODS
Strains used and growth of bacteria.
Experimentally evolved ciprofloxacin-resistant mutant derivatives of P. aeruginosa reference strain PAO1 (24) and PA14 (this study) were obtained using the antibiotic gradient agar plate method (60). Bacteria were grown in LB medium at 37°C with shaking (200 rpm).
Bacterial growth analysis.
Growth analysis of PAO1 mutants was carried out using the method previously described (24). Briefly, mutants were grown overnight in LB medium and diluted to 1.5 × 106 CFU/ml. Cultures were diluted and 200 μl portions were dispensed into wells in a 96-well tissue culture plate (JETbiofil). Plates were incubated in a BMG FLUOstar Omega microplate reader at 37°C/200 rpm for 18 h. The optical density at 600 nm (OD600) was measured every 30 min as a measure of growth. The blank-corrected OD600 data were used to calculate the area under the growth curve (AUC) using GrowthCurver package version 0.2.1 (61) in software RStudio. The logistic area under the curve (AUC) was used as the parameter for quantifying bacterial growth.
Determination of antibiotic MIC.
MIC testing of engineered and experimentally evolved mutants was conducted using the protocol given in Wiegand et al. (62) and Wardell et al. (24). Briefly, overnight cultures in L-broth were diluted to 106 CFU/ml and spotted onto DifcoTM Muller-Hinton (MH) agar plates containing ciprofloxacin (Mylan, New Zealand Ltd.) in doubling dilutions. A control MH agar plate without antibiotic supplementations was also inoculated with bacterial culture. Plates were dried and incubated overnight at 37°C. Results were recorded the next day.
Whole-genome sequencing.
Genomic DNA was extracted using the MoBio UltraClean microbial DNA isolation kit from overnight-grown cultures of bacteria. Genome sequencing was carried out by New Zealand Genomics Limited using Illumina HiSeq2000 and Illumina MiSeq.
Allelic exchange.
Mutations were engineered in laboratory reference strain PAO1 using a two-step allelic exchange method (63). Genomic DNA extracted from cultures of experimentally evolved mutants containing the mutation of interest was used as a PCR template. The primers used are listed in Table S8 in the supplemental material. Each PCR product incorporated approximately 1,000 bp of DNA upstream and downstream of the target mutation. Each amplified fragment was cloned into plasmid pEX18Tc (64). Recombinant plasmids were extracted using a plasmid isolation kit (Macherey-Nagel, Germany) and sequenced using universal M13 primers and primers specific to the mutation-containing DNA region (Table S8) to confirm that only the intended mutation was present. A list of the recombinant plasmids constructed in this study is given in Table S9. Mutations were engineered into the PAO1 genome through homologous recombination as described by (65). PCR sequencing was carried out to screen the mutant candidates, using primers specific to the mutation-containing DNA region (Table S8). Mutants with combinations of mutations were constructed with the same procedure. Whole-genome sequencing of selected engineered mutants was carried out to make sure the mutants had only the intended mutations.
RT-qPCR.
RNA was extracted from bacterial cultures grown in LB medium to an OD600 of 0.8 (exponential growth phase) using an RNA extraction kit (RNeasy minikit, Qiagen) according to the manufacturer’s instructions. Gene-specific primers mexD, and reference gene primers clpX and oprL (66), are in Table S8. Primer efficiencies were calculated using dilutions of genomic DNA and were between 1.8 and 2 for all primer pairs. For RT-qPCR, RNA was reverse transcribed to cDNA. RT-qPCR was carried out using a LightCycler 480 SYBR Green I Master kit (Roche) as described previously (67). Two technical replicate reactions were carried out for each sample. Melt curve analysis was carried out and PCR products were detected by agarose gel electrophoresis to confirm single amplification products from each reaction. LightCycler 480 software was used to calculate the crossing points, target/reference ratios, and melting temperatures. All RT-qPCR experiments were performed on three biological replicates.
Bioinformatics and statistical analysis.
Mutations were identified through comparison of genome sequences to P. aeruginosa reference strain PAO1 (PAO1-Otago) or PA14 using BreSeq (68) as described previously (24). One-way ANOVA and Tukey’s HSD test were carried out and P values were calculated to evaluate the multiple comparisons. Dot plots were created using R Studio package ggplot2 (69).
Analysis of clinical isolate genomes.
The 237 P. aeruginosa genomes from the International Pseudomonas Consortium Database (IPCD) (70), available at http://ipcd.ibis.ulaval.ca, and 37 from International Health Management Associates (IHMA) analyzed in this study are listed in Table S6. These isolates were chosen based on the availability of MIC data and genotypic diversity. To annotate the genomes, Prokka v1.10 (71) was used and a protein blast database of annotated genomes was prepared. Protein BLAST of sequences was carried out to identify amino acid sequence variants between each genome and that of the reference sequence PAO1. BLAST tabular output format 6 was produced. BLAST results were filtered by setting the minimum percent identity at 95% and minimum coverage at 90%. Sequences were retrieved from BLAST results and MUSCLE (72) was used to align the protein sequences from all 274 isolates. Alignments were visualized in Jalview (version 2) (73) to identify the amino acid variants.
Criteria to predict ciprofloxacin MICs of isolates from genome sequences were designed based on MIC data of engineered mutants. The effects of variants on protein function were predicted using PROVEAN version 1.1 (74) using the method described in Wardell et al. (24). The Software Resistance Gene Identifier (RGI) (75) was used to search the plasmid-borne gene crpP in P. aeruginosa isolates using assembled genomes.
ACKNOWLEDGMENTS
This research was supported in part by a research grant from the New Zealand Health Research Council (17/372) (to I.L.L.). A.R. was supported by a New Zealand International Doctoral Research Scholarship and a travel award from the Marjorie McCallum fund.
We are grateful to all members of the International Pseudomonas Consortium Database (IPCD) who donated strains utilized in this study.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Pendleton JN, Gorman SP, Gilmore BF. 2013. Clinical relevance of the ESKAPE pathogens. Expert Rev Anti Infect Ther 11:297–308. 10.1586/eri.13.12. [DOI] [PubMed] [Google Scholar]
- 2.Driscoll JA, Brody SL, Kollef MH. 2007. The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs 67:351–368. 10.2165/00003495-200767030-00003. [DOI] [PubMed] [Google Scholar]
- 3.Bassetti M, Vena A, Croxatto A, Righi E, Guery B. 2018. How to manage Pseudomonas aeruginosa infections. Drugs Context 7:212527. 10.7573/dic.212527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Belkum A, Burnham CD, Rossen JWA, Mallard F, Rochas O, DunneWM, Jr.. 2020. Innovative and rapid antimicrobial susceptibility testing systems. Nat Rev Microbiol 18:299–311. 10.1038/s41579-020-0327-x. [DOI] [PubMed] [Google Scholar]
- 5.Coulter S, Roberts JA, Hajkowicz K, Halton K. 2017. The use of bloodstream infection mortality to measure the impact of antimicrobial stewardship interventions: assessing the evidence. Infect Dis Rep 9:6849–6849. 10.4081/idr.2017.6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonine NG, Berger A, Altincatal A, Wang R, Bhagnani T, Gillard P, Lodise T. 2019. Impact of delayed appropriate antibiotic therapy on patient outcomes by antibiotic resistance status from serious Gram-negative bacterial infections. Am J Med Sci 357:103–110. 10.1016/j.amjms.2018.11.009. [DOI] [PubMed] [Google Scholar]
- 7.Tamma PD, Fan Y, Bergman Y, Pertea G, Kazmi AQ, Lewis S, Carroll KC, Schatz MC, Timp W, Simner PJ. 2018. Applying rapid whole-genome sequencing to predict phenotypic antimicrobial susceptibility testing results among carbapenem-resistant Klebsiella pneumoniae clinical isolates. Antimicrob Agents Chemother 63:e01923-18. 10.1128/AAC.01923-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freschi L, Vincent AT, Jeukens J, Emond-Rheault J-G, Kukavica-Ibrulj I, Dupont M-J, Charette SJ, Boyle B, Levesque RC. 2019. The Pseudomonas aeruginosa pan-genome provides new insights on its population structure, horizontal gene transfer, and pathogenicity. Genome Biol Evol 11:109–120. 10.1093/gbe/evy259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su M, Satola SW, Read TD. 2018. Genome-based prediction of bacterial antibiotic resistance. J Clin Microbiol 57:e01405-18. 10.1128/JCM.01405-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shelburne SA, Kim J, Munita JM, Sahasrabhojane P, Shields RK, Press EG, Li X, Arias CA, Cantarel B, Jiang Y, Kim MS, Aitken SL, Greenberg DE. 2017. Whole-genome sequencing accurately identifies resistance to extended-spectrum β-lactams for major Gram-negative bacterial pathogens. Clin Infect Dis 65:738–745. 10.1093/cid/cix417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drouin A, Giguère S, Déraspe M, Marchand M, Tyers M, Loo VG, Bourgault AM, Laviolette F, Corbeil J. 2016. Predictive computational phenotyping and biomarker discovery using reference-free genome comparisons. BMC Genomics 17:754. 10.1186/s12864-016-2889-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeukens J, Freschi L, Kukavica-Ibrulj I, Emond-Rheault JG, Tucker NP, Levesque RC. 2019. Genomics of antibiotic-resistance prediction in Pseudomonas aeruginosa. Ann N Y Acad Sci 1435:5–17. 10.1111/nyas.13358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khaledi A, Weimann A, Schniederjans M, Asgari E, Kuo T-H, Oliver A, Cabot G, Kola A, Gastmeier P, Hogardt M, Jonas D, Mofrad MR, Bremges A, McHardy AC, Häussler S. 2020. Predicting antimicrobial resistance in Pseudomonas aeruginosa with machine learning-enabled molecular diagnostics. EMBO Mol Med 12:e10264. 10.15252/emmm.201910264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Breidenstein EBM, Khaira BK, Wiegand I, Overhage J, Hancock REW. 2008. Complex ciprofloxacin resistome revealed by screening a Pseudomonas aeruginosa mutant library for altered susceptibility. Antimicrob Agents Chemother 52:4486–4491. 10.1128/AAC.00222-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bruchmann S, Dötsch A, Nouri B, Chaberny IF, Häussler S. 2013. Quantitative contributions of target alteration and decreased drug accumulation to Pseudomonas aeruginosa fluoroquinolone resistance. Antimicrob Agents Chemother 57:1361–1368. 10.1128/AAC.01581-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kos VN, Déraspe M, McLaughlin RE, Whiteaker JD, Roy PH, Alm RA, Corbeil J, Gardner H. 2015. The resistome of Pseudomonas aeruginosa in relationship to phenotypic susceptibility. Antimicrob Agents Chemother 59:427–436. 10.1128/AAC.03954-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeukens J, Kukavica-Ibrulj I, Emond-Rheault JG, Freschi L, Levesque RC. 2017. Comparative genomics of a drug-resistant Pseudomonas aeruginosa panel and the challenges of antimicrobial resistance prediction from genomes. FEMS Microbiol Lett 364. 10.1093/femsle/fnx161. [DOI] [PubMed] [Google Scholar]
- 18.Sader HS, Farrell DJ, Flamm RK, Jones RN. 2014. Antimicrobial susceptibility of Gram-negative organisms isolated from patients hospitalized in intensive care units in United States and European hospitals (2009–2011). Diagn Microbiol Infect Dis 78:443–448. 10.1016/j.diagmicrobio.2013.11.025. [DOI] [PubMed] [Google Scholar]
- 19.Shortridge D, Gales AC, Streit JM, Huband MD, Tsakris A, Jones RN. 2019. Geographic and temporal patterns of antimicrobial resistance in Pseudomonas aeruginosa over 20 years from the SENTRY antimicrobial surveillance program, 1997–2016. Open Forum Infect Dis 6:S63–S68. 10.1093/ofid/ofy343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rehman A, Patrick WM, Lamont IL. 2019. Mechanisms of ciprofloxacin resistance in Pseudomonas aeruginosa: new approaches to an old problem. J Med Microbiol 68:1–10. 10.1099/jmm.0.000873. [DOI] [PubMed] [Google Scholar]
- 21.Cabot G, Zamorano L, Moyà B, Juan C, Navas A, Blázquez J, Oliver A. 2016. Evolution of Pseudomonas aeruginosa antimicrobial resistance and fitness under low and high mutation rates. Antimicrob Agents Chemother 60:1767–1778. 10.1128/AAC.02676-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Melnyk AH, McCloskey N, Hinz AJ, Dettman J, Kassen R. 2017. Evolution of cost-free resistance under fluctuating drug selection in Pseudomonas aeruginosa. mSphere 2:e00158-17. 10.1128/mSphere.00158-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.López-Causapé C, Cabot G, Del Barrio-Tofiño E, Oliver A. 2018. The versatile mutational resistome of Pseudomonas aeruginosa. Front Microbiol 9:685–685. 10.3389/fmicb.2018.00685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wardell SJT, Rehman A, Martin LW, Winstanley C, Patrick WM, Lamont IL. 2019. A large-scale whole-genome comparison shows that experimental evolution in response to antibiotics predicts changes in naturally evolved clinical Pseudomonas aeruginosa. Antimicrob Agents Chemother 63:e01619–19. 10.1128/AAC.01619-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogbolu DO, Daini OA, Ogunledun A, Alli AO, Webber MA. 2011. High levels of multidrug resistance in clinical isolates of Gram-negative pathogens from Nigeria. Int J Antimicrob Agents 37:62–66. 10.1016/j.ijantimicag.2010.08.019. [DOI] [PubMed] [Google Scholar]
- 26.Liu J, Yang L, Chen D, Peters BM, Li L, Li B, Xu Z, Shirtliff ME. 2018. Complete sequence of pBM413, a novel multidrug resistance megaplasmid carrying qnrVC6 and blaIMP-45 from Pseudomonas aeruginosa. Int J Antimicrob Agents 51:145–150. 10.1016/j.ijantimicag.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 27.Wang F, Wu K, Sun J, Wang Q, Chen Q, Yu S, Rui Y. 2012. Novel ISCR1-linked resistance genes found in multidrug-resistant Gram-negative bacteria in southern China. Int J Antimicrob Agents 40:404–408. 10.1016/j.ijantimicag.2012.06.016. [DOI] [PubMed] [Google Scholar]
- 28.Chávez-Jacobo VM, García Merinos JP, López Y, Meza-Carmen V, Ramírez-Díaz MI. 2020. Identification of essential residues for ciprofloxacin resistance of ciprofloxacin-modifying enzyme (CrpP) of pUM505. Microbiology (Reading) 166:367–374. 10.1099/mic.0.000889. [DOI] [PubMed] [Google Scholar]
- 29.Chávez-Jacobo VM, Hernández-Ramírez KC, Romo-Rodríguez P, Pérez-Gallardo RV, Campos-García J, Gutiérrez-Corona JF, García-Merinos JP, Meza-Carmen V, Silva-Sánchez J, Ramírez-Díaz MI. 2018. CrpP is a novel ciprofloxacin-modifying enzyme encoded by the Pseudomonas aeruginosa pUM505 plasmid. Antimicrob Agents Chemother 62:e02629-17. 10.1128/AAC.02629-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hughes D, Andersson DI. 2017. Environmental and genetic modulation of the phenotypic expression of antibiotic resistance. FEMS Microbiol Rev 41:374–391. 10.1093/femsre/fux004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sommer MOA, Munck C, Toft-Kehler RV, Andersson DI. 2017. Prediction of antibiotic resistance: time for a new preclinical paradigm? Nat Rev Microbiol 15:689–696. 10.1038/nrmicro.2017.75. [DOI] [PubMed] [Google Scholar]
- 32.Poole K, Gotoh N, Tsujimoto H, Zhao Q, Wada A, Yamasaki T, Neshat S, Yamagishi J, Li XZ, Nishino T. 1996. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol Microbiol 21:713–724. 10.1046/j.1365-2958.1996.281397.x. [DOI] [PubMed] [Google Scholar]
- 33.Shiba T, Ishiguro K, Takemoto N, Koibuchi H, Sugimoto K. 1995. Purification and characterization of the Pseudomonas aeruginosa NfxB protein, the negative regulator of the nfxB gene. J Bacteriol 177:5872–5877. 10.1128/jb.177.20.5872-5877.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Purssell A, Poole K. 2013. Functional characterization of the NfxB repressor of the mexCD-oprJ multidrug efflux operon of Pseudomonas aeruginosa. Microbiology (Reading) 159:2058–2073. 10.1099/mic.0.069286-0. [DOI] [PubMed] [Google Scholar]
- 35.Andersson DI, Hughes D. 2010. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat Rev Microbiol 8:260–271. 10.1038/nrmicro2319. [DOI] [PubMed] [Google Scholar]
- 36.Marcusson LL, Frimodt-Møller N, Hughes D. 2009. Interplay in the selection of fluoroquinolone resistance and bacterial fitness. PLoS Pathog 5:e1000541. 10.1371/journal.ppat.1000541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jacoby GA. 2005. Mechanisms of resistance to quinolones. Clin Infect Dis 41 Suppl 2:S120–S126. 10.1086/428052. [DOI] [PubMed] [Google Scholar]
- 38.Ghosh D, Veeraraghavan B, Elangovan R, Vivekanandan P. 2019. Antibiotic resistance and epigenetics: more to it than meets the eye. Antimicrob Agents Chemother 64:e02225-19. 10.1128/AAC.02225-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Riber L, Hansen LH. 2021. Epigenetic memories: the hidden drivers of bacterial persistence? Trends Microbiol 29:190–194. 10.1016/j.tim.2020.12.005. [DOI] [PubMed] [Google Scholar]
- 40.Wong A, Rodrigue N, Kassen R. 2012. Genomics of adaptation during experimental evolution of the opportunistic pathogen Pseudomonas aeruginosa. PLoS Genet 8:e1002928. 10.1371/journal.pgen.1002928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jeannot K, Elsen S, Köhler T, Attree I, van Delden C, Plésiat P. 2008. Resistance and virulence of Pseudomonas aeruginosa clinical strains overproducing the MexCD-OprJ efflux pump. Antimicrob Agents Chemother 52:2455–2462. 10.1128/AAC.01107-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Henrichfreise B, Wiegand I, Pfister W, Wiedemann B. 2007. Resistance mechanisms of multiresistant Pseudomonas aeruginosa strains from Germany and correlation with hypermutation. Antimicrob Agents Chemother 51:4062–4070. 10.1128/AAC.00148-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kiser TH, Obritsch MD, Jung R, MacLaren R, Fish DN. 2010. Efflux pump contribution to multidrug resistance in clinical isolates of Pseudomonas aeruginosa. Pharmacotherapy 30:632–638. 10.1592/phco.30.7.632. [DOI] [PubMed] [Google Scholar]
- 44.Sánchez P, Linares JF, Ruiz-Díez B, Campanario E, Navas A, Baquero F, Martínez JL. 2002. Fitness of in vitro selected Pseudomonas aeruginosa nalB and nfxB multidrug resistant mutants. J Antimicrob Chemother 50:657–664. 10.1093/jac/dkf185. [DOI] [PubMed] [Google Scholar]
- 45.Poole K. 2011. Pseudomonas aeruginosa: resistance to the max. Front Microbiol 2:65. 10.3389/fmicb.2011.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dötsch A, Becker T, Pommerenke C, Magnowska Z, Jänsch L, Häussler S. 2009. Genomewide identification of genetic determinants of antimicrobial drug resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 53:2522–2531. 10.1128/AAC.00035-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jochumsen N, Marvig RL, Damkiær S, Jensen RL, Paulander W, Molin S, Jelsbak L, Folkesson A. 2016. The evolution of antimicrobial peptide resistance in Pseudomonas aeruginosa is shaped by strong epistatic interactions. Nat Commun 7:13002. 10.1038/ncomms13002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Redgrave LS, Sutton SB, Webber MA, Piddock LJV. 2014. Fluoroquinolone resistance: mechanisms, impact on bacteria, and role in evolutionary success. Trends Microbiol 22:438–445. 10.1016/j.tim.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 49.Eichenberger EM, Thaden JT. 2019. Epidemiology and mechanisms of resistance of extensively drug resistant Gram-negative bacteria. Antibiotics (Basel) 8:37. 10.3390/antibiotics8020037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karaman R, Jubeh B, Breijyeh Z. 2020. Resistance of Gram-positive bacteria to current antibacterial agents and overcoming approaches. Molecules 25:2888. 10.3390/molecules25122888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salma R, Dabboussi F, Kassaa I, Hamze M, Dabboussi F, Hamze M, Khudary R. 2013. gyrA and parC mutations in quinolone-resistant clinical isolates of Pseudomonas aeruginosa from Nini Hospital in north Lebanon. J Infect Chemother 19:77–81. 10.1007/s10156-012-0455-y. [DOI] [PubMed] [Google Scholar]
- 52.Wang Y-T, Lee M-F, Peng C-F. 2014. Mutations in the quinolone resistance-determining regions associated with ciprofloxacin resistance in Pseudomonas aeruginosa isolates from Southern Taiwan. Biomark Genom Med 6:79–83. 10.1016/j.bgm.2014.03.003. [DOI] [Google Scholar]
- 53.Yang X, Xing B, Liang C, Ye Z, Zhang Y. 2015. Prevalence and fluoroquinolone resistance of Pseudomonas aeruginosa in a hospital of South China. Int J Clin Exp Med 8:1386–1390. [PMC free article] [PubMed] [Google Scholar]
- 54.Nguyen KV, Nguyen TV, Nguyen HTT, Le DV. 2018. Mutations in the gyrA, parC, and mexR genes provide functional insights into the fluoroquinolone-resistant Pseudomonas aeruginosa isolated in Vietnam. Infect Drug Resist 11:275–282. 10.2147/IDR.S147581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Telling K, Laht M, Brauer A, Remm M, Kisand V, Maimets M, Tenson T, Lutsar I. 2018. Multidrug resistant Pseudomonas aeruginosa in Estonian hospitals. BMC Infect Dis 18:513. 10.1186/s12879-018-3421-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reinhardt A, Köhler T, Wood P, Rohner P, Dumas J-L, Ricou B, van Delden C. 2007. Development and persistence of antimicrobial resistance in Pseudomonas aeruginosa: a longitudinal observation in mechanically ventilated patients. Antimicrob Agents Chemother 51:1341–1350. 10.1128/AAC.01278-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goli HR, Nahaei MR, Rezaee MA, Hasani A, Samadi Kafil H, Aghazadeh M, Sheikhalizadeh V. 2016. Contribution of mexAB-oprM and mexXY (-oprA) efflux operons in antibiotic resistance of clinical Pseudomonas aeruginosa isolates in Tabriz, Iran. Infect Genet Evol 45:75–82. 10.1016/j.meegid.2016.08.022. [DOI] [PubMed] [Google Scholar]
- 58.Llanes C, Köhler T, Patry I, Dehecq B, van Delden C, Plésiat P. 2011. Role of the MexEF-OprN efflux system in low-level resistance of Pseudomonas aeruginosa to ciprofloxacin. Antimicrob Agents Chemother 55:5676–5684. 10.1128/AAC.00101-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morita Y, Tomida J, Kawamura Y. 2015. Efflux-mediated fluoroquinolone resistance in the multidrug-resistant Pseudomonas aeruginosa clinical isolate PA7: identification of a novel MexS variant involved in upregulation of the mexEF-oprN multidrug efflux operon. Front Microbiol 6:8. 10.3389/fmicb.2015.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bryson V, Szybalski W. 1952. Microbial selection. Science 116:45–51. 10.1126/science.116.3003.45. [DOI] [PubMed] [Google Scholar]
- 61.Sprouffske K, Wagner A. 2016. Growthcurver: an R package for obtaining interpretable metrics from microbial growth curves. BMC Bioinformatics 17:172. 10.1186/s12859-016-1016-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wiegand I, Hilpert K, Hancock REW. 2008. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc 3:163–175. 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 63.Hmelo LR, Borlee BR, Almblad H, Love ME, Randall TE, Tseng BS, Lin C, Irie Y, Storek KM, Yang JJ, Siehnel RJ, Howell PL, Singh PK, Tolker-Nielsen T, Parsek MR, Schweizer HP, Harrison JJ. 2015. Precision-engineering the Pseudomonas aeruginosa genome with two-step allelic exchange. Nat Protoc 10:1820–1841. 10.1038/nprot.2015.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. 10.1016/S0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 65.Thoma S, Schobert M. 2009. An improved Escherichia coli donor strain for diparental mating. FEMS Microbiol Lett 294:127–132. 10.1111/j.1574-6968.2009.01556.x. [DOI] [PubMed] [Google Scholar]
- 66.Konings AF, Martin LW, Sharples KJ, Roddam LF, Latham R, Reid DW, Lamont IL. 2013. Pseudomonas aeruginosa uses multiple pathways to acquire iron during chronic infection in cystic fibrosis lungs. Infect Immun 81:2697–2704. 10.1128/IAI.00418-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martin LW, Robson CL, Watts AM, Gray AR, Wainwright CE, Bell SC, Ramsay KA, Kidd TJ, Reid DW, Brockway B, Lamont IL. 2018. Expression of Pseudomonas aeruginosa antibiotic resistance genes varies greatly during infections in cystic fibrosis patients. Antimicrob Agents Chemother 62:e01789-18. 10.1128/AAC.01789-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Deatherage DE, Barrick JE. 2014. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol Biol 1151:165–188. 10.1007/978-1-4939-0554-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wickham H. 2016. ggplot2 elegant graphics for data analysis. Springer-Verlag, New York, NY. [Google Scholar]
- 70.Freschi L, Jeukens J, Kukavica-Ibrulj I, Boyle B, Dupont MJ, Laroche J, Larose S, Maaroufi H, Fothergill JL, Moore M, Winsor GL, Aaron SD, Barbeau J, Bell SC, Burns JL, Camara M, Cantin A, Charette SJ, Dewar K, Déziel É, Grimwood K, Hancock RE, Harrison JJ, Heeb S, Jelsbak L, Jia B, Kenna DT, Kidd TJ, Klockgether J, Lam JS, Lamont IL, Lewenza S, Loman N, Malouin F, Manos J, McArthur AG, McKeown J, Milot J, Naghra H, Nguyen D, Pereira SK, Perron GG, Pirnay JP, Rainey PB, Rousseau S, Santos PM, Stephenson A, Taylor V, Turton JF, Waglechner N. 2015. Clinical utilization of genomics data produced by the international Pseudomonas aeruginosa consortium. Front Microbiol 6:1036. 10.3389/fmicb.2015.01036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 72.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. 2009. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Choi Y, Chan AP. 2015. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 31:2745–2747. 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, Tsang KK, Lago BA, Dave BM, Pereira S, Sharma AN, Doshi S, Courtot M, Lo R, Williams LE, Frye JG, Elsayegh T, Sardar D, Westman EL, Pawlowski AC, Johnson TA, Brinkman FS, Wright GD, McArthur AG. 2017. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 45:D566–d573. 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Monti MR, Morero NR, Miguel V, Argaraña CE. 2013. nfxB as a novel target for analysis of mutation spectra in Pseudomonas aeruginosa. PLoS One 8:e66236. 10.1371/journal.pone.0066236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jalal S, Ciofu O, Hoiby N, Gotoh N, Wretlind B. 2000. Molecular mechanisms of fluoroquinolone resistance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Antimicrob Agents Chemother 44:710–712. 10.1128/aac.44.3.710-712.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pasca MR, Dalla Valle C, De Jesus Lopes Ribeiro AL, Buroni S, Papaleo MC, Bazzini S, Udine C, Incandela ML, Daffara S, Fani R, Riccardi G, Marone P. 2012. Evaluation of fluoroquinolone resistance mechanisms in Pseudomonas aeruginosa multidrug resistance clinical isolates. Microb Drug Resist 18:23–32. 10.1089/mdr.2011.0019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental figure and tables. Download AAC.02696-20-s0001.pdf, PDF file, 1.0 MB (989.9KB, pdf)