ABSTRACT
Marburg virus (MARV) VP40 protein (mVP40) directs egress and spread of MARV, in part, by recruiting specific host WW domain-containing proteins via its conserved PPxY late (L) domain motif to facilitate efficient virus-cell separation. We reported previously that small-molecule compounds targeting the viral PPxY/host WW domain interaction inhibited VP40-mediated egress and spread. Here, we report on the antiviral potency of novel compound FC-10696, which emerged from extensive structure-activity relationship (SAR) of a previously described series of PPxY inhibitors. We show that FC-10696 inhibits egress of mVP40 virus-like particles (VLPs) and egress of authentic MARV from HeLa cells and primary human macrophages. Moreover, FC-10696 treated-mice displayed delayed onset of weight loss and clinical signs and significantly lower viral loads compared to controls, with 14% of animals surviving 21 days following a lethal MARV challenge. Thus, FC-10696 represents a first-in-class, host-oriented inhibitor effectively targeting late stages of the MARV life cycle.
KEYWORDS: Marburg virus, filovirus, PPxY motif, L domain, antiviral therapeutic, host-oriented, budding, WW domain, Nedd4, virus-host interaction
INTRODUCTION
Marburg virus (MARV) is an emerging pathogen and potential bioterrorism agent that can cause severe hemorrhagic fever in humans and nonhuman primates (1). Currently, there are no approved vaccines or antiviral therapeutics to prevent or treat MARV infections. Development of novel and effective antiviral therapeutics against MARV and other members of the family Filoviridae are urgently needed.
MARV, like many other enveloped RNA viruses, relies on its matrix protein (mVP40) to direct and promote budding of infectious virions. We and others have demonstrated that mVP40 completes the budding process, in part, by using its highly conserved PPxY L domain motif to hijack host proteins/pathways that then facilitate efficient virus-cell separation (2–16). One of the best characterized host interactors is the WW domain-containing E3 ubiquitin ligase Nedd4 (7, 16–18). Notably, we and others have shown that Nedd4 and Nedd4 family members physically interact with viral PPxY motifs via one or more of their WW domains and functionally interact with viral PPxY-containing proteins to enhance or promote budding of virus-like particles (VLPs) and live virus (7, 16–24). The highly conserved physical and functional nature of the PPxY motif in a wide array of RNA viruses makes it an attractive target for the development of antivirals (2, 25–30). Indeed, compounds targeting the PPxY/WW domain virus-host interaction would be predicted to dampen or reduce the ability of the virus to bud or pinch off from infected cells, thus allowing an individual’s immune system more time to combat and clear the virus.
Previously, we described the identification and development of two novel series of small-molecule compounds that significantly inhibited VP40-Nedd4 interaction and PPxY-mediated egress of filovirus VP40 VLPs (26). We went on to show that our lead compounds significantly inhibited budding of a live vesicular stomatitis virus (VSV) recombinant (VSV-M40) that we engineered to express the PPxY motif from Ebola virus (EBOV) VP40 in place of that from the VSV matrix (M) protein (26). Following extensive structure-activity relationship (SAR) and analog testing, we identified lead compound FC-10696, which we have shown is stable in mouse and human liver microsomes and possesses suitable absorption, distribution, metabolism, and excretion (ADME) and pharmacokinetic (PK) properties for intraperitoneal (i.p.) administration and testing in mice. Indeed, here we show that nanomolar concentrations of FC-10696 blocked budding of mVP40 VLPs. Moreover, we demonstrate that similar concentrations of FC-10696 also significantly inhibited egress of live MARV in both HeLa cells and human monocyte-derived macrophages (MDMs) with little or no cytotoxicity at the effective antiviral concentrations. Importantly, we show proof-of-concept in vivo efficacy of FC-10696 in a mouse challenge model of MARV disease. Indeed, FC-10696-treated animals exhibited delayed onset of weight loss, clinical signs, and disease progression compared to control animals, with 14% of animals from the FC-10696 treated group surviving a lethal MARV challenge up to 21 days postinfection. These findings represent the first proof-of-concept efficacy study for a novel host-oriented antiviral that has the potential for broad-spectrum activity against other PPxY-containing viruses, such as EBOV and Lassa virus (LASV), and provides proof of principle for further development of these first-in-class compounds targeting viral L domain/host interactions as effective countermeasures to reduce virus budding and dissemination.
RESULTS
ADME, PK, and antibudding data for compound FC-10696.
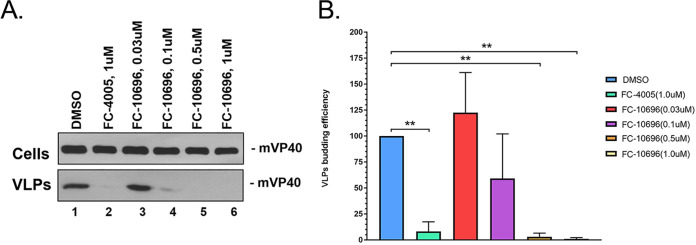
We improved drug disposition properties of a previously described series (26) to generate a novel small molecule, FC-10696, a compound with vastly improved overall ADME and PK properties (Table 1) and with robust antibudding activity against mVP40 VLPs (Table 1 and Fig. 1). Indeed, results from a representative mVP40 VLP budding assay (Fig. 1A) highlight the dose-dependent decrease in mVP40 VLP egress from HEK239T cells treated with increasing concentrations of FC-10696 (Fig. 1, lanes 3 to 6) compared to that from a vehicle (dimethyl sulfoxide [DMSO]) only-treated control (Fig. 1, lane 1). Previously described active compound 1 (FC-4005) at 1.0 μM served as a positive control (Fig. 1, lane 2) (26). As expected, equivalent amounts of mVP40 were detected in cell lysates from all samples (Fig. 1A, top, lanes 1 to 6). Quantitative results from at least three independent mVP40 VLP budding assays in the presence of 0.3 μM and 0.1 μM FC-10696 revealed averages of 91% and 82% inhibition of mVP40 VLP egress, respectively, compared to DMSO alone (Table 1 and Fig. 1B).
TABLE 1.
In vitro Marburg egress, ADME, and PK data
| Parameter | Value for FC-10696 |
|---|---|
| % reduction in Marburg egress assay at concn (μM)a | |
| 1.0 | 99 |
| 0.5 | 94 |
| 0.3 | 91 |
| 0.1 | 86 |
| 0.03 | 0 |
| MW | 357.5 |
| t1/2 (min)b | |
| MLM stability | 76.9 |
| HLM stability | 1670 |
| Mouse PK (i.p. administration)c | |
| Cmax (ng/ml) | 953 |
| AUC (ng · ml/h) | 1,378 |
| t1/2 (s) | 1.5 |
| i.p. bioavailability (%) | 42 |
Percent reduction of Marburg VLPs at the listed compound concentrations in HEK293T cells, with the positive control FC-4005 (26) run at 1 μM (∼90% ± 10%).
MLM, mouse liver microsome; HLM, human liver microsome.
BALB/cJ mice were given one 10-mg/kg dose of drug with plasma collection time points of 0.25, 0.5, 1, 2, and 6 h. Intraperitoneal bioavailability is the ratio of i.p. AUC to i.v. AUC × 100. PK parameters for i.v. administration are not shown.
FIG 1.
FC-10696 inhibits budding of mVP40 VLPs in a dose-dependent manner. HEK293T cells were transfected with mVP40 in the presence of DMSO alone, compound 1, or the indicated concentrations of compound FC-10696. mVP40 was detected in cell lysates and VLPs by Western blotting, and mVP40 levels in VLPs were quantified using NIH ImageJ software. (B) Quantitative results and statistical significance (one-way analysis of variance [ANOVA]) are shown for 3 independent mVP40 VLP budding assays. **, P < 0.01.
We evaluated compound FC-10696 in a single-dose mouse PK experiment with i.p. administration (Table 1). FC-10696 displayed excellent blood levels and good metabolic stability (Table 1). Indeed, FC-10696 was deemed suitable for live virus and mouse efficacy experiments, as it showed good stability to mouse and human liver microsomes and did not inhibit cytochrome P450 3A4 at concentrations up to 33 μM, thus showing low risk for drug/drug interactions. In sum, FC-10696 had overall superior ADME and PK properties, as well as robust antibudding activity in an mVP40 VLP budding assay.
Compound FC-10696 inhibits egress of live MARV in cell culture.
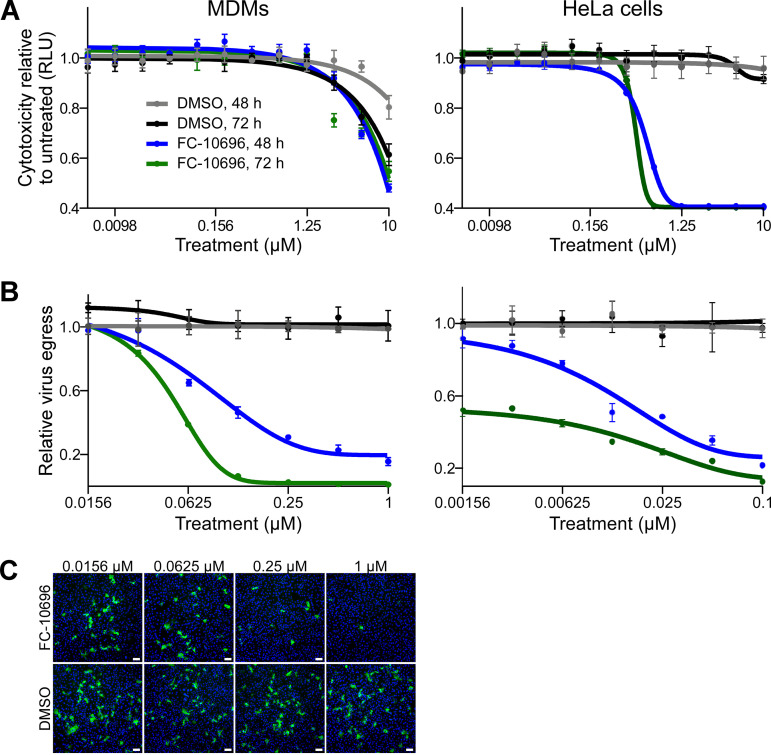
To assess safety of FC-10696, we first assessed its cytotoxicity in a HeLa cell line, routinely used in antifiloviral screens, and human monocyte-derived macrophages (MDMs), the initial targets of filovirus infection in the host (31, 32). Cells treated with 2-fold dilutions of FC-10696 or DMSO as a control were assessed for the number of metabolically active cells after 48 or 72 h. The CC50 (the concentration that reduced the cell viability by 50% compared to the untreated control) showed that the compound was more cytotoxic to HeLa cells than to MDMs (Table 2 and Fig. 2A). Assessment of efficacy revealed that FC-10696 efficiently inhibited live MARV egress in both cell types (Fig. 2B and C). Notably, the half-maximal inhibitory concentration (IC50) in the virion egress assay was in the nanomolar range, and the selectivity index (SI50, calculated as CC50/IC50) was ≥10, signifying antiviral potency and selectivity of this compound against this virus.
TABLE 2.
FC-10696 IC50s and SI50s for live MARV egress at 48 h and 72 ha
| Cell type | 48 h |
72 h |
||||
|---|---|---|---|---|---|---|
| CC50 (μM) | IC50 egress (μM) | SI50 egress | CC50 (μM) | IC50 egress (μM) | SI50 egress | |
| HeLa | 0.49 | 0.042 | 11.7 | 0.39 | 0.003 | 130 |
| Human macrophages | 4.8 | 0.18 | 26.7 | 4.2 | 0.055 | 76.4 |
Cytotoxic properties of FC-10696 were tested in either HeLa cells or human macrophages using a CellTiter-Glo kit 48 or 72 h after treatment to select nontoxic concentration range for antiviral tests. In the virus tests, cells were challenged with MARV at an MOI of 0.01 for 1 h and then incubated with new medium containing serially diluted concentrations of FC-10696. To assess virus egress, cell supernatants were titrated on Vero cells. Infected cells were detected by staining with anti-MARV VLP antibody, and nuclei were stained with Hoechst dye. Samples were subsequently imaged and analyzed. The concentration that reduced cell viability by 50% (CC50) and the half-maximal inhibitory concentration (IC50) for virus egress were determined by nonlinear regression analysis. The selectivity index (SI50) was calculated as CC50/IC50.
FIG 2.
FC-10696 inhibits egress and spread of live MARV from infected human MDMs and HeLa cells. (A) MDMs (left) or HeLa cells (right) were left untreated or treated with FC-10696 at 11 2-fold serially diluted concentrations or DMSO for 48 or 72 h in triplicate. The number of metabolically active cells was determined using a CellTiter-Glo kit. The number of relative light units (RLU) for each concentration is an average with the standard deviation for 3 replicates. (B) MDMs (left) or HeLa cells (right) were challenged with MARV for 1 h, washed, and incubated with new medium containing 7 2-fold serially diluted concentration of the compounds, equal concentrations of DMSO, or no treatment for 48 or 72 h. To assess virus egress from FC-10696-treated MDMs, cell supernatants were titrated on Vero cells for 24 h, stained with anti-VLP antibody and Hoechst dye, and photographed. Nuclei and infected cells were counted using CellProfiler software. The relative infection efficiencies, determined by dividing the number of infected cells by the number of nuclei, are reported relative to the infection efficiency in untreated cells and are averages and standard deviations of 3 replicates. (C) Supernatants of MDMs challenged with MARV and treated as indicated for 72 h were titrated on Vero cells. Samples were treated with anti-VLP antibody to detect infected cells (green) and Hoechst dye to detect nuclei (blue) and imaged using a Nikon imaging system. Bars, 100 μm.
Compound FC-10696 shows efficacy in a mouse challenge model of MARV infection.
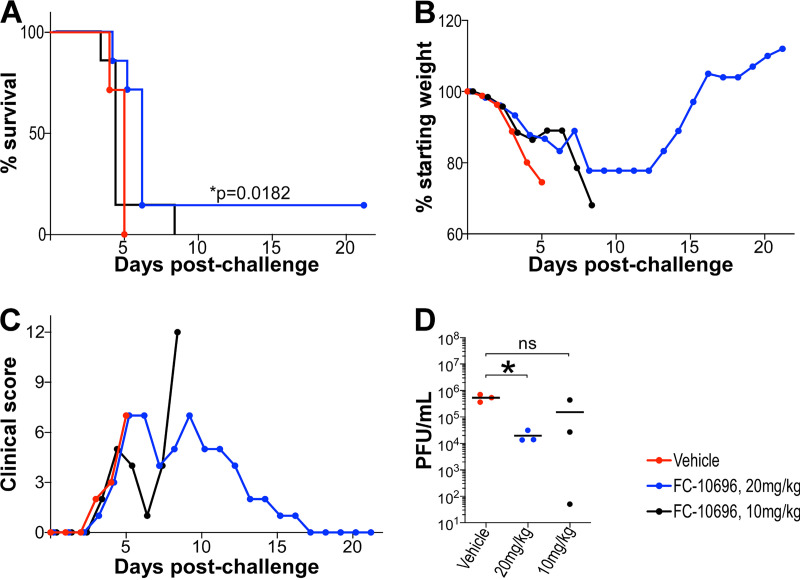
Compounds undergoing preclinical development for antiviral activity need to undergo initial evaluation in a small animal model, typically laboratory mice because of the high degree of similarity between the murine and human immune systems and because the small size of the animals requires smaller amounts of treatment reagents to be injected, thus maximizing reagents that may be in short supply. Because of the limitation of animal biosafety level 4 (ABSL-4) space, the smaller model facilitates proof-of-concept testing, while the inbred nature of the mice also reduces variation in response to treatments and virus, allowing the size of test groups to be limited. To evaluate the antiviral potential of FC-10696 in vivo, BALB/cJ mice challenged intraperitoneally (i.p.) with 1,000 PFU of mouse-adapted MARV were dosed i.p. with a formulation containing the compound twice daily (BID) for 10 consecutive days, starting 6 h postchallenge. BALB/cJ mice are highly susceptible to infection with mouse-adapted MARV, developing disease symptoms and high viremia approximately 3 days postinfection and succumbing to the disease by day 6 (33). We found that treatment with 20 mg/kg of body weight, the highest dose we could prepare, delayed the onset of mortality (P = 0.0182) (Fig. 3A), weight loss (Fig. 3B), and virus load in serum (P = 0.0255) (Fig. 3D). Importantly, the compound was well tolerated in mice at 20 mg/kg, and animals developed only transient treatment-associated toxicity (Fig. 4). Our results demonstrate the proof-of-concept in vivo activity for FC-10696 compound against MARV infection.
FIG 3.
In vivo efficacy of FC-10696 in a mouse model of MARV disease. Three groups of 10 4-week-old female BALB/cJ mice were challenged with 1,000 PFU of mouse-adapted MARV. Intraperitoneal dosing by vehicle or FC-10696 at either 20 or 10 mg/kg started 6 h postchallenge and continued BID for 10 consecutive days. Animals were observed daily for mortality (A), weight loss (B), and clinical signs of disease (C) for 21 days postinfection. The average weight of animals in each group was determined as the ratio of total weight of all live mice to the number of animals. The percent weight change on each day was determined by dividing the average weight on that day by the average weight on day 0. Clinical scores for each group were recorded as a sum of all observations in the group, and if a score of ≥12 was recorded for an individual animal, it was considered terminally ill and euthanized. On day 3 postchallenge, 3 animals/group were euthanized to collect serum for virus load assessment by the neutral red plaque assay (D). The remaining 7 mice/group were used to determine animal survival. Viral burden was analyzed using a Student's t test or one-way ANOVA with Tukey’s test, and survival analysis was performed using a log-rank (Mantel-Cox) test, with a P value of ≤0.05 considered significant in all analyses.
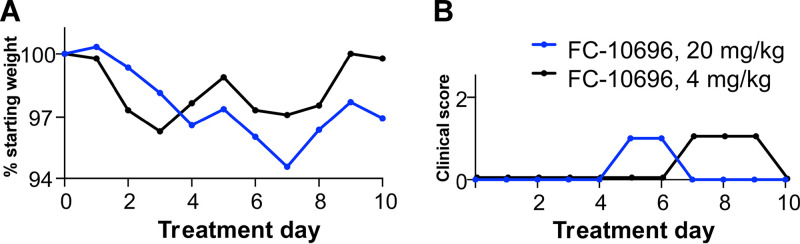
FIG 4.
In vivo toxicity data for FC-10696. FC-10696 was resuspended in a 30% PEG 400–2% DMSO–14% Kleptose HPB parenteral-grade formulation at two different concentrations, 20 and 4 mg/kg, and administered to groups of five 4-week-old female BALB/cJ mice BID via the i.p. route for a period of 10 days. Animals were monitored daily for signs of treatment-associated toxicity: weight loss (A); rough coat, discharge from eyes and nose, diarrhea, and decreased food intake and activity (B); and mortality. The percent weight change in a group on each day was calculated by dividing the average animal weight on that day by the average weight on day 0. Clinical scores for each group were recorded as a sum of all observations in the group.
DISCUSSION
The identification of host-oriented antiviral compounds represents a promising strategy to develop effective, broad-spectrum therapeutics capable of targeting a wide array of emerging pathogens, which may lead to a paradigm shift in the search for new antivirals (25–28, 34–41). MARV and other filoviruses continue to emerge and cause outbreaks of severe hemorrhagic fever, largely originating in Africa but with the potential to spread globally, as observed with the EBOV outbreak in 2014–2015. Here, we report on our continued efforts to develop compounds targeting viral PPxY-mediated host interactions as a novel class of antiviral therapeutics. We demonstrate the antiviral efficacy of the novel compound FC-10696 in an in vitro and in vivo model of MARV infection.
Extensive SAR analysis led to the identification of compound FC-10696, which possesses excellent overall ADME and PK properties. FC-10696 exhibited potent activity at the BSL-2 level in blocking egress of mVP40 VLPs at low-nanomolar concentrations in repeated experiments and in disrupting a PPxY/WW domain-mediated interaction between mVP40 and the human E3 ubiquitin ligase Nedd4, as determined using a bimolecular complementation (BiMC) approach (data not shown). FC-10696 was then moved into the BSL-4 laboratory, where it was assessed for cytotoxicity in both HeLa cells and primary human macrophages, as well as in BALB/cJ mice. After obtaining a satisfactory cytotoxicity profile and CC50s, we went on to demonstrate that FC-10696 significantly inhibited MARV egress from both cell types compared to controls. Most intriguingly, we went on to show that treatment of mice with 20 mg/kg twice daily for 10 days delayed the onset of mortality (P = 0.0182), weight loss, and virus load in serum (P = 0.0255), thus providing strong support for this class of compounds for further development into potent antivirals against filoviruses and possibly other viruses that utilize PPxY L domain motifs for productive infection. These findings represent the first proof-of-concept in vivo activity for our lead host-oriented PPxY inhibitor.
Studies are under way to assess compound FC-10696 and similar analogs for antiviral potency against related PPxY-containing viruses, including EBOV and LASV. Indeed, preliminary findings indicate that FC-10696 can block egress and spread of both eVP40 VLPs and authentic EBOV in vitro, albeit less efficiently than MARV. This may be due to the presence of a second overlapping PTAP L domain motif in the EBOV VP40 protein, which, in addition to the PPxY motif, can also function in promoting efficient egress and spread of EBOV, which is in contrast to the single isolated PPxY motif present in the MARV VP40 protein (24). As such, one could envision the use of a PPxY-mediated inhibitor such as FC-10696 in combination with a PTAP-mediated inhibitor or a viral entry inhibitor, for example as part of an antiviral cocktail strategy targeting multiple stages of the virus life cycle for maximal effect (27, 30, 42–44). Our long-term goal is to develop these compounds alone or in combination primarily for individuals in high-risk situations, including those in the military, health care workers, and first-line responders during an outbreak or epidemic. Additional studies are still needed to precisely identify the mechanism of action and drug interaction site, as well as additional studies to assess cytotoxicity and potential effects on the host in a wider array of cell types.
MATERIALS AND METHODS
Cells, plasmids, and virus strain.
HEK293T, HeLa, and Vero cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum and penicillin (100 U/ml)/streptomycin (100 μg/ml), and the cells were grown at 37°C in a humidified 5% CO2 incubator. Flag-tagged mVP40 plasmid was kindly provided by S. Becker (Institut für Virologie, Marburg, Germany). Monocyte-derived human macrophages (MDMs) were isolated as described previously (45, 46). Peripheral blood was collected from healthy donors according to University of Texas Health-approved IRB protocol 20180013HU. Heparinized blood was overlaid onto a Ficoll-Paque cushion (GE Healthcare, Uppsala, Sweden) to isolate peripheral blood mononuclear cells (PBMCs). PBMCs were cultured in suspension in RPMI medium supplemented with 20% autologous serum for 6 days at 37°C in a humidified 5% CO2 incubator to differentiate monocytes into macrophages. All experiments with live MARV were performed in the BSL-4 laboratory at the Texas Biomedical Research Institute (TBRI; San Antonio, TX). MARV strain Musoke (NCBI accession number NC_001608) was obtained from the virus repository at the Texas Biomedical Research Institute. Mouse-adapted MARV strain Angola (NCBI accession number KM_261523) was generously provided by the National Microbiology Laboratory, Public Health Agency of Canada. Virus stocks were generated and characterized as described previously (33, 47).
ADME and PK data.
Human and mouse liver microsome stability studies and pharmacokinetic studies in mice were performed at Alliance Pharma, Inc. (Malvern, PA). PK parameters from the PK study were calculated by M.S.S.
Human and mouse liver microsome stability.
FC-10696 at a concentration of 0.5 μM was incubated with 0.5 mg/ml of liver microsomes (mouse or human) and an NADPH-regenerating system (cofactor solution) in potassium phosphate buffer (pH 7.4). At 0, 5, 15, 30, and 45 min, an aliquot was taken, and reactions were quenched with an acetonitrile solution containing an internal standard. Midazolam was run as a reference standard. Additionally, controls were measured that do not contain the cofactor solution. Following completion of the experiment, samples were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Results were reported as peak area ratios of each analyte to internal standard. The intrinsic clearance (CLint) was determined from the first-order elimination constant by nonlinear regression.
Pharmacokinetics in mice.
A single-dose study was conducted in adult BALB/cJ male mice weighing 20 to 26 g each. Groups of six animals were administered an intravenous (i.v.) 2-mg/kg dose or an i.p. 10-mg/kg dose of FC-10696, both administered as a soluble 5% DMSO–20% Kleptose aqueous formulation. Plasma samples (3 per time point) were collected from study animals at 5, 15, and 30 min and 1, 2, and 6 h for the i.v. dose and at 15 and 30 min and 1, 2, and 6 h for the i.p. dose. Collected samples were analyzed by LC-MS/MS. PK parameters were calculated using Prism GraphPad.
VLP budding assays.
MARV VP40 VLP budding assays in HEK293T cells were described previously (2). For VLP budding, HEK293T cells were transfected with 0.5 μg of mVP40, and cells were treated with DMSO alone or the indicated concentration of inhibitor (as shown in Fig. 1A) for 24 h posttransfection. The mVP40 protein in cell extracts and VLPs was detected by SDS-PAGE and Western blotting and quantified using NIH ImageJ software. Anti-Flag monoclonal antibody was used to detect Flag-tagged mVP40.
MARV egress assay.
To prepare a stock solution, FC-10696 was resuspended in DMSO at a concentration of 10 mM and stored at <−65°C. HeLa cells or MDMs were plated into wells of a 96-well plate at 2 × 104 or 5 × 104 cells/well, respectively, to determine cytotoxic and antiviral properties of FC-10696. All treatments were performed in triplicate. In cytotoxicity assays, cells were left untreated or treated with the compound at 11 2-fold serially diluted concentrations or DMSO (solvent) for 48 or 72 h. The number of metabolically active cells was determined using a CellTiter-Glo kit. The concentration that reduced the cell viability by 50% compared to the untreated control (CC50) at each time point was determined using nonlinear regression analysis with GraphPad 8 software to select a nontoxic concentration range for antiviral tests.
In virus tests, HeLa cells or MDMs were challenged with MARV at multiplicity of infection (MOI) of 0.01 for 1 h to allow binding, washed, and incubated with new medium containing seven 2-fold serially diluted concentration of the compounds, equal concentrations of DMSO, or no treatment for 48 or 72 h. To assess virus egress, cell supernatants were titrated on Vero cells for 24 h. Infected cells were detected by treatment with MARV VLP antibody (IBT Bioservices, Rockville, MD), and nuclei were visualized by staining with Hoechst dye (Thermo Fisher Scientific, Waltham, MA). Samples were photographed using a Nikon automated system (Nikon, Tokyo, Japan) and analyzed by CellProfiler software (Broad Institute, Cambridge, MA) to quantify virus egress. Infection efficiency in treated samples was determined as a ratio of infected cells and nuclei and reported relative to values for the mock treatment. The half-maximal inhibitory concentration (IC50) for virus egress for each time point was determined by nonlinear regression analysis. The selectivity index (SI50, calculated as CC50/IC50) was used to assess antiviral potential of the compounds.
Animals.
Wild-type 4-week-old female BALB/cJ mice were obtained from The Jackson Laboratory (Bar Harbor, ME). The mouse studies were conducted in strict adherence to the Animal Welfare Act and the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (NIH). The TBRI animal assurance welfare number is D16-00048 (A3082-01) and is on file with the NIH. All mouse procedures were approved by the TBRI Institutional Animal Care and Use Committee (IACUC), which oversees the administration of the IACUC protocols. The mouse studies were performed as outlined in IACUC protocol 1706MU.
In vivo efficacy studies.
To assess toxicity of FC-10696 treatment in mice, the compound was resuspended in a 30% polyethylene glycol 400 (PEG 400)–2% DMSO–14% Kleptose HPB (hydroxypropyl beta-cyclodextrin) parenteral-grade formulation (Roquette, Lestrem, France) at two different concentrations, 20 and 4 mg/kg, and administered to groups of five 4-week-old female BALB/cJ mice twice daily (BID) via the i.p. route for a period of 10 days. Animals were monitored daily for signs of treatment-associated toxicity: weight loss, rough coat, discharge from eyes and nose, diarrhea, decreased food intake and activity, and mortality. The average weight of animals in each group was determined as a ratio of total weight of all live mice to the number of animals. Clinical scores for each group were recorded as a sum of all observations in the group.
To assess the antiviral potential of FC-10696 treatment in a mouse model of MARV disease, three groups of 10 4-week-old female BALB/cJ mice were challenged with 1,000 PFU (as determined on Vero cells) of mouse-adapted MARV by the i.p. route. Intraperitoneal dosing by vehicle or FC-10696 at either 20 or 10 mg/kg started 6 h postchallenge and continued BID for 10 consecutive days. Animals were observed at least twice daily for signs of viral disease (ruffled coat, hunched back, inappetence, weight loss, and decreased movement) and mortality for 21 days postchallenge. The percent weight change in a group on each day was calculated by dividing the average animal weight on that day by the average weight on day 0. Group clinical scores were recorded as the sum of all clinical observations for the group. If a clinical score of ≥12 was recorded for an animal, it was considered terminally ill and euthanized. Three animals from each group were euthanized on day 3 postchallenge to collect blood to determine virus titer by a plaque assay. The remaining 7 mice/group were used to determine animal survival.
Select agent use.
TBRI is a select agent registered entity with Health and Human Services, Centers for Disease Control and Prevention, and U.S. Department of Agriculture, Animal Plant Health Inspection Service, National Select Agent Program. The policies and standard operating procedures for work with select agents at the TBRI comply with applicable federal, state, and municipal regulations and with the guidelines “Biosafety in Microbiological and Biomedical Laboratories” (48) issued by the Centers for Disease Control and Prevention and the National Institutes of Health (NIH). The TBRI Biohazard and Safety Committee (BSC) is responsible for evaluating the ABSL-4 facility, equipment, and staff capabilities to perform work in a safe manner. The protocol BSC19-015 “Pathogenesis and drug efficacy studies of high-containment viruses” detailing the work with replication-competent Marburg viruses described herein was fully approved by the institute’s BSC.
ACKNOWLEDGMENTS
We thank Katie Freeman for developing the formulations for i.v. and i.p. dosing.
Funding was provided in part by National Institutes of Health grants AI138052 and AI138630 to R.N.H., AI129890 to B.D.F., T32-AI070077 to A.S.-M., and an Innovator Award from The Wellcome Trust to M.S.S. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
R.N.H. and B.D.F. are cofounders of Intervir, LLC.
Contributor Information
Olena Shtanko, Email: oshtanko@txbiomed.org.
Ronald N. Harty, Email: rharty@vet.upenn.edu.
REFERENCES
- 1.Feldmann H, Klenk HD. 1996. Marburg and Ebola viruses. Adv Virus Res 47:1–52. 10.1016/s0065-3527(08)60733-2. [DOI] [PubMed] [Google Scholar]
- 2.Han Z, Dash S, Sagum CA, Ruthel G, Jaladanki CK, Berry CT, Schwoerer MP, Harty NM, Freedman BD, Bedford MT, Fan H, Sidhu SS, Sudol M, Shtanko O, Harty RN. 2020. Modular mimicry and engagement of the Hippo pathway by Marburg virus VP40: implications for filovirus biology and budding. PLoS Pathog 16:e1008231. 10.1371/journal.ppat.1008231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harty RN. 2018. Hemorrhagic fever virus budding studies. Methods Mol Biol 1604:209–215. 10.1007/978-1-4939-6981-4_15. [DOI] [PubMed] [Google Scholar]
- 4.Wijesinghe KJ, Urata S, Bhattarai N, Kooijman EE, Gerstman BS, Chapagain PP, Li S, Stahelin RV. 2017. Detection of lipid-induced structural changes of the Marburg virus matrix protein VP40 using hydrogen/deuterium exchange-mass spectrometry. J Biol Chem 292:6108–6122. 10.1074/jbc.M116.758300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liang J, Sagum CA, Bedford MT, Sidhu SS, Sudol M, Han Z, Harty RN. 2017. Chaperone-mediated autophagy protein BAG3 negatively regulates Ebola and Marburg VP40-mediated egress. PLoS Pathog 13:e1006132. 10.1371/journal.ppat.1006132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oda S, Noda T, Wijesinghe KJ, Halfmann P, Bornholdt ZA, Abelson DM, Armbrust T, Stahelin RV, Kawaoka Y, Saphire EO. 2016. Crystal structure of Marburg virus VP40 reveals a broad, basic patch for matrix assembly and a requirement of the N-terminal domain for immunosuppression. J Virol 90:1839–1848. 10.1128/JVI.01597-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Urata S, Yasuda J. 2010. Regulation of Marburg virus (MARV) budding by Nedd4.1: a different WW domain of Nedd4.1 is critical for binding to MARV and Ebola virus VP40. J Gen Virol 91:228–234. 10.1099/vir.0.015495-0. [DOI] [PubMed] [Google Scholar]
- 8.Liu Y, Cocka L, Okumura A, Zhang YA, Sunyer JO, Harty RN. 2010. Conserved motifs within Ebola and Marburg virus VP40 proteins are important for stability, localization, and subsequent budding of virus-like particles. J Virol 84:2294–2303. 10.1128/JVI.02034-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Urata S, Noda T, Kawaoka Y, Morikawa S, Yokosawa H, Yasuda J. 2007. Interaction of Tsg101 with Marburg virus VP40 depends on the PPPY motif, but not the PT/SAP motif as in the case of Ebola virus, and Tsg101 plays a critical role in the budding of Marburg virus-like particles induced by VP40, NP, and GP. J Virol 81:4895–4899. 10.1128/JVI.02829-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kolesnikova L, Bohil AB, Cheney RE, Becker S. 2007. Budding of Marburgvirus is associated with filopodia. Cell Microbiol 9:939–951. 10.1111/j.1462-5822.2006.00842.x. [DOI] [PubMed] [Google Scholar]
- 11.Hartlieb B, Weissenhorn W. 2006. Filovirus assembly and budding. Virology 344:64–70. 10.1016/j.virol.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 12.Swenson DL, Warfield KL, Kuehl K, Larsen T, Hevey MC, Schmaljohn A, Bavari S, Aman MJ. 2004. Generation of Marburg virus-like particles by co-expression of glycoprotein and matrix protein. FEMS Immunol Med Microbiol 40:27–31. 10.1016/S0928-8244(03)00273-6. [DOI] [PubMed] [Google Scholar]
- 13.Kolesnikova L, Berghofer B, Bamberg S, Becker S. 2004. Multivesicular bodies as a platform for formation of the Marburg virus envelope. J Virol 78:12277–12287. 10.1128/JVI.78.22.12277-12287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jasenosky LD, Kawaoka Y. 2004. Filovirus budding. Virus Res 106:181–188. 10.1016/j.virusres.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 15.Kolesnikova L, Bugany H, Klenk HD, Becker S. 2002. VP40, the matrix protein of Marburg virus, is associated with membranes of the late endosomal compartment. J Virol 76:1825–1838. 10.1128/jvi.76.4.1825-1838.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harty RN, Brown ME, Wang G, Huibregtse J, Hayes FP. 2000. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc Natl Acad Sci U S A 97:13871–13876. 10.1073/pnas.250277297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han Z, Sagum CA, Bedford MT, Sidhu SS, Sudol M, Harty RN. 2016. ITCH E3 ubiquitin ligase interacts with Ebola virus VP40 to regulate budding. J Virol 90:9163–9171. 10.1128/JVI.01078-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y, Lee MS, Olson MA, Harty RN. 2011. Bimolecular complementation to visualize filovirus VP40-host complexes in live mammalian cells: toward the identification of budding inhibitors. Adv Virol 2011:341816. 10.1155/2011/341816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han Z, Sagum CA, Takizawa F, Ruthel G, Berry CT, Kong J, Sunyer JO, Freedman BD, Bedford MT, Sidhu SS, Sudol M, Harty RN. 2017. Ubiquitin ligase WWP1 interacts with Ebola virus VP40 to regulate egress. J Virol 91:e00812-17. 10.1128/JVI.00812-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okumura A, Pitha PM, Harty RN. 2008. ISG15 inhibits Ebola VP40 VLP budding in an L-domain-dependent manner by blocking Nedd4 ligase activity. Proc Natl Acad Sci U S A 105:3974–3979. 10.1073/pnas.0710629105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin-Serrano J, Perez-Caballero D, Bieniasz PD. 2004. Context-dependent effects of L domains and ubiquitination on viral budding. J Virol 78:5554–5563. 10.1128/JVI.78.11.5554-5563.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yasuda J, Nakao M, Kawaoka Y, Shida H. 2003. Nedd4 regulates egress of Ebola virus-like particles from host cells. J Virol 77:9987–9992. 10.1128/jvi.77.18.9987-9992.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Timmins J, Schoehn G, Ricard-Blum S, Scianimanico S, Vernet T, Ruigrok RW, Weissenhorn W. 2003. Ebola virus matrix protein VP40 interaction with human cellular factors Tsg101 and Nedd4. J Mol Biol 326:493–502. 10.1016/s0022-2836(02)01406-7. [DOI] [PubMed] [Google Scholar]
- 24.Licata JM, Simpson-Holley M, Wright NT, Han Z, Paragas J, Harty RN. 2003. Overlapping motifs (PTAP and PPEY) within the Ebola virus VP40 protein function independently as late budding domains: involvement of host proteins TSG101 and VPS-4. J Virol 77:1812–1819. 10.1128/jvi.77.3.1812-1819.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shepley-McTaggart A, Fan H, Sudol M, Harty RN. 2020. Viruses go modular. J Biol Chem 295:4604–4616. 10.1074/jbc.REV119.012414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loughran HM, Han Z, Wrobel JE, Decker SE, Ruthel G, Freedman BD, Harty RN, Reitz AB. 2016. Quinoxaline-based inhibitors of Ebola and Marburg VP40 egress. Bioorg Med Chem Lett 26:3429–3435. 10.1016/j.bmcl.2016.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu J, Han Z, Liu Y, Liu W, Lee MS, Olson MA, Ruthel G, Freedman BD, Harty RN. 2014. A host-oriented inhibitor of Junin Argentine hemorrhagic fever virus egress. J Virol 88:4736–4743. 10.1128/JVI.03757-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han Z, Lu J, Liu Y, Davis B, Lee MS, Olson MA, Ruthel G, Freedman BD, Schnell MJ, Wrobel JE, Reitz AB, Harty RN. 2014. Small-molecule probes targeting the viral PPxY-host Nedd4 interface block egress of a broad range of RNA viruses. J Virol 88:7294–7306. 10.1128/JVI.00591-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harty RN. 2009. No exit: targeting the budding process to inhibit filovirus replication. Antiviral Res 81:189–197. 10.1016/j.antiviral.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perez M, Craven RC, de la Torre JC. 2003. The small RING finger protein Z drives arenavirus budding: implications for antiviral strategies. Proc Natl Acad Sci U S A 100:12978–12983. 10.1073/pnas.2133782100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bray M, Geisbert TW. 2005. Ebola virus: the role of macrophages and dendritic cells in the pathogenesis of Ebola hemorrhagic fever. Int J Biochem Cell Biol 37:1560–1566. 10.1016/j.biocel.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 32.Hensley LE, Alves DA, Geisbert JB, Fritz EA, Reed C, Larsen T, Geisbert TW. 2011. Pathogenesis of Marburg hemorrhagic fever in cynomolgus macaques. J Infect Dis 204:S1021–S1031. 10.1093/infdis/jir339. [DOI] [PubMed] [Google Scholar]
- 33.Qiu XG, Wong G, Audet J, Cutts T, Niu YL, Booth S, Kobinger GP. 2014. Establishment and characterization of a lethal mouse model for the Angola strain of Marburg virus. J Virol 88:12703–12714. 10.1128/JVI.01643-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y, Harty RN. 2010. Viral and host proteins that modulate filovirus budding. Future Virol 5:481–491. 10.2217/FVL.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iglesias-Bexiga M, Palencia A, Corbi-Verge C, Martin-Malpartida P, Blanco FJ, Macias MJ, Cobos ES, Luque I. 2019. Binding site plasticity in viral PPxY Late domain recognition by the third WW domain of human NEDD4. Sci Rep 9:15076. 10.1038/s41598-019-50701-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Chassey B, Meyniel-Schicklin L, Vonderscher J, Andre P, Lotteau V. 2014. Virus-host interactomics: new insights and opportunities for antiviral drug discovery. Genome Med 6:115. 10.1186/s13073-014-0115-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heaton SM. 2019. Harnessing host-virus evolution in antiviral therapy and immunotherapy. Clin Transl Immunol 8:e1067. 10.1002/cti2.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sui BQ, Bamba D, Weng K, Ung H, Chang SJ, Van Dyke J, Goldblatt M, Duan R, Kinch MS, Li WB. 2009. The use of random homozygous gene perturbation to identify novel host-oriented targets for influenza. Virology 387:473–481. 10.1016/j.virol.2009.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kinch MS, Yunus AS, Lear C, Mao HW, Chen HS, Fesseha Z, Luo GX, Nelson EA, Li LM, Huang ZH, Murray M, Ellis WY, Hensley L, Christopher-Hennings J, Olinger GG, Goldblatt M. 2009. FGI-104: a broad-spectrum small molecule inhibitor of viral infection. Am J Transl Res 1:87–98. [PMC free article] [PubMed] [Google Scholar]
- 40.Han ZY, Madara JJ, Herbert A, Prugar LI, Ruthel G, Lu JH, Liu YL, Liu WB, Liu XH, Wrobel JE, Reitz AB, Dye JM, Harty RN, Freedman BD. 2015. Calcium regulation of hemorrhagic fever virus budding: mechanistic implications for host-oriented therapeutic intervention. PLoS Pathog 11:e1005220. 10.1371/journal.ppat.1005220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mao HW, Chen HS, Fesseha Z, Chang SJ, Ung-Medoff H, Van Dyke J, Kohli M, Li WB, Goldblatt M, Kinch MS. 2009. Identification of novel host-oriented targets for human immunodeficiency virus type 1 using random homozygous gene perturbation. Virol J 6:154. 10.1186/1743-422X-6-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Palencia A, Martinez JC, Mateo PL, Luque I, Camara-Artigas A. 2006. Structure of human TSG101 UEV domain. Acta Crystallogr D Biol Crystallogr 62:458–464. 10.1107/S0907444906005221. [DOI] [PubMed] [Google Scholar]
- 43.Urata S, Noda T, Kawaoka Y, Yokosawa H, Yasuda J. 2006. Cellular factors required for Lassa virus budding. J Virol 80:4191–4195. 10.1128/JVI.80.8.4191-4195.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Capul AA, de la Torre JC. 2008. A cell-based luciferase assay amenable to high-throughput screening of inhibitors of arenavirus budding. Virology 382:107–114. 10.1016/j.virol.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schlesinger LS. 1993. Macrophage phagocytosis of virulent but not attenuated strains of Mycobacterium tuberculosis is mediated by mannose receptors in addition to complement receptors. J Immunol 150:2920–2930. [PubMed] [Google Scholar]
- 46.Rogers KJ, Shtanko O, Stunz LL, Mallinger LN, Arkee T, Schmidt ME, Bohan D, Brunton B, White JM, Varga SM, Butler NS, Bishop GA, Maury W. 2021. Frontline Science: CD40 signaling restricts RNA virus replication in Mϕs, leading to rapid innate immune control of acute virus infection. J Leukoc Biol 109:309–325. 10.1002/JLB.4HI0420-285RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shtanko O, Sakurai Y, Reyes AN, Noel R, Cintrat JC, Gillet D, Barbier J, Davey RA. 2018. Retro-2 and its dihydroquinazolinone derivatives inhibit filovirus infection. Antiviral Res 149:154–163. 10.1016/j.antiviral.2017.11.016. [DOI] [PubMed] [Google Scholar]
- 48.U.S. Department of Health and Human Services. 2020. Biosafety in microbiological and biomedical laboratories, 6th ed. U.S. Department of Health and Human Services, Washington, DC. [Google Scholar]