ABSTRACT
Eis promoter mutations can confer reduced Mycobacterium tuberculosis kanamycin susceptibility. GenoType MTBDRsl, a widely used assay evaluating this region, wrongly classified 17/410 isolates as eis promoter wild type. Six out of seventeen isolates harbored mutations known to confer kanamycin resistance, and the remainder harbored either novel eis promoter mutations (7/11) or disputed mutations (4/11). GenoType MTBDRsl can miss established and new variants that cause reduced susceptibility. These data highlight the importance of reflex phenotypic kanamycin testing.
KEYWORDS: Mycobacterium tuberculosis, extensive drug resistance, second-line injectables
INTRODUCTION
The drugs amikacin (AMK), kanamycin (KAN), and capreomycin (CAP) have been part of the recommended second-line antituberculosis treatment since the 1970s. The most common genetic resistance marker for these drugs is a single-nucleotide variant (SNV) at position 1401 of the rRNA 16S encoding gene, rrs (1, 2). An alternative mechanism conferring (low-level) resistance to KAN includes SNVs in the promoter region of eis (Rv2416c) (Fig. S1 in the supplemental material) (3). Amikacin is often used as a surrogate for KAN phenotypic drug-susceptibility testing (pDST) based on the assumption of complete cross-resistance. Similarly, if the strain was susceptible to AMK, KAN susceptibility was assumed, and low-level KAN resistance was potentially overlooked. Until 2017, eis promoter mutations were not routinely tested for in South Africa, leading to undetected resistance and less effective treatment.
This study investigated the presence, type, and detection of eis promoter mutations in clinical Mycobacterium tuberculosis isolates collected in South Africa using the line probe assay GenoType MTBDRsl VER 2.0 (MTBDRsl; Hain Lifescience, Germany), Sanger sequencing, and whole-genome sequencing (WGS).
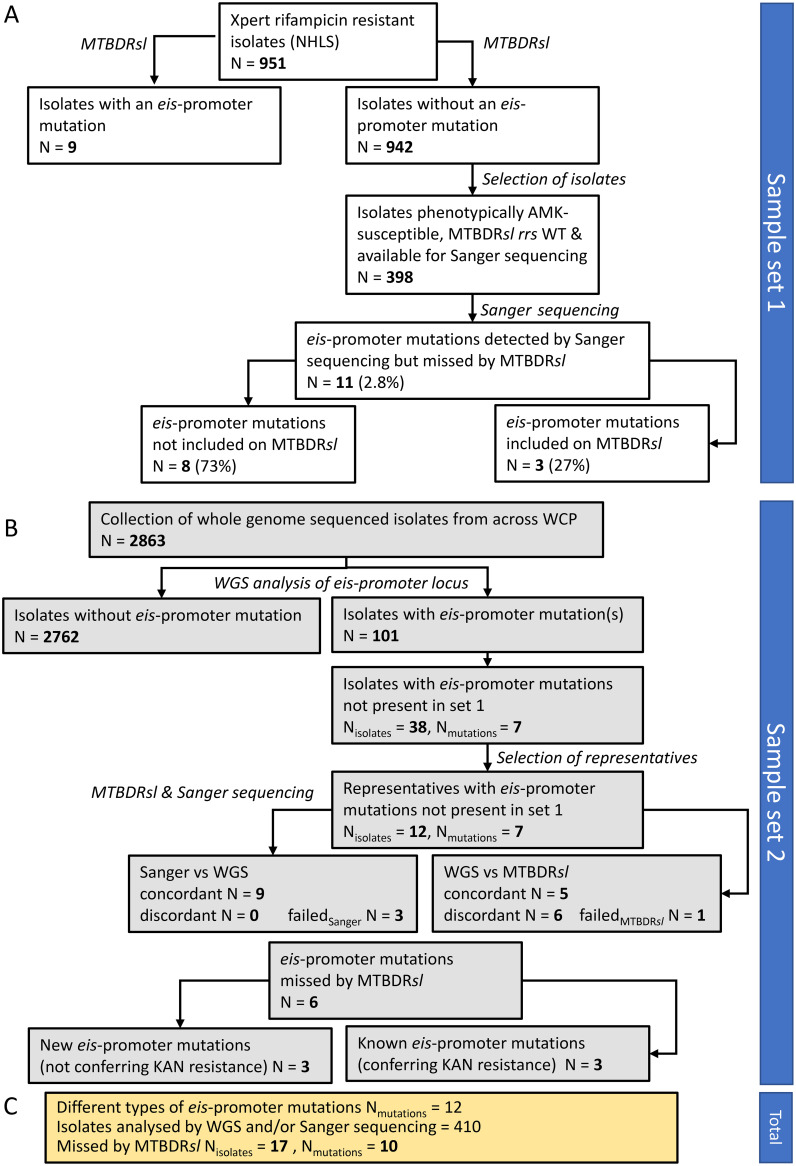
Two unique sample sets were analyzed. Sample set 1 consisted of 951 M. tuberculosis isolates from Xpert MTB/RIF (Cepheid) rifampin (RIF)-resistant specimens from South Africa that were collected between June 2016 and June 2017 as part of routine diagnostics by the National Health Laboratory Services, Cape Town. These isolates were analyzed using the GenoType MTBDRplus assay (detecting resistance against RIF and/or isoniazid [INH]) and MTBDRsl (4). To determine the number of eis promoter mutations missed by MTBDRsl, isolates that were phenotypically susceptible to AMK, wild type (WT) for eis promoter and rrs by MTBDRsl, and available in the Stellenbosch University biobank (n = 398) were Sanger sequenced (i.e., the region covering 222 bp upstream of the transcriptional start site of the eis gene, subsequently referred to as “eis promoter region”; Fig. S1). Sample set 2 consisted of a convenience sample of 2,863 whole-genome sequences of clinical M. tuberculosis isolates derived from sputum samples collected between 1993 and 2018 and sequenced as part of different research projects (5–9). These sequences were screened in silico for eis promoter mutations (genome positions 2715332 to 2715582 of M. tuberculosis H37Rv; GenBank accession no. AL123456). Of those isolates with eis promoter mutations, representatives for each (combination of) mutation(s) were selected for further analyses with targeted Sanger sequencing, MTBDRsl, and pDST. An overview of the study workflow for both sample sets is given in Fig. 1.
FIG 1.
Workflow diagram. (A) Workflow and number of isolates included in each step for sample set 1. (B) Workflow and number of isolates included in each step for sample set 2. (C) Total number of eis promoter mutations detected and missed by routine MTBDRsl across both sample sets. NHLS, National Health Laboratory Services; WCP, Western Cape Province; WGS, whole-genome sequencing.
For isolates of sample set 1, PCR amplification—and subsequent Sanger sequencing—was conducted on thermal lysates, whereas purified DNA was used for sample set 2. Briefly, the PCR mixture contained the following final concentrations: 1× HotStartTaq Plus master mix (Qiagen, San Diego, CA, USA), 500 nM each primer (forward, 5′-CCATGGGACCGGTACTTGCT-3′; reverse, 5′-ACTTCACCAGGCACCGTCAA-3′), and 1× SYTO 9 green fluorescent nucleic acid stain (Thermo Fisher Scientific). As a template, 1 μl of thermal lysate (sample set 1) or purified DNA (sample set 2) was added to the reaction mixture. Amplification of the eis promoter region of the selected isolates was carried out using a CFX96TM real-time system C1000 Touch thermal cycler (Bio-Rad) running the following thermocycling protocol: initial denaturation at 95°C for 5 min, followed by 40 cycles of 95°C for 1 min, annealing at 62°C for 1 min, and elongation at 72°C for 1 min, followed by a final elongation at 72°C for 10 min. Successful amplification was confirmed by a high-resolution melt from 80°C to 95°C with an increment of 0.5°C, each increment temperature held for 5 s. Successfully amplified PCR products were sent to the Central DNA Sequencing Facilities of Stellenbosch University for targeted Sanger sequencing using the forward PCR primer. The MTBDRsl assay was conducted according to the manufacturer’s protocol using the same DNA used for WGS. The assay defines specific banding patterns (i.e., presence or absence of WT and MUT bands) for the following most common eis promoter mutations: −37 G > T, −14 T > C, −12 T > C, −10 G > A, and −2 A > C. In this study, these mutations were therefore defined as “detectable by MTBDRsl.” However, only the mutation −14 T > C is explicitly detected by a MUT probe (4). Other known eis promoter mutations (Fig. S1) may also cause one of the WT bands to fail but appear not to have been validated by the manufacturer. In this study, these mutations were therefore defined as “not included in MTBDRsl.” Phenotypic DST was performed on all isolates using solid Löwenstein-Jensen medium according to the 1% proportion method at clinical breakpoints of 0.2 μg/ml for INH, 40.0 μg/ml for RIF, 30 μg/ml for AMK, and 2 μg/ml for ofloxacin (10, 11). MICs for KAN were subsequently determined for isolates with an eis promoter mutation missed by the MTBDRsl (sample set 1) and for representatives of each additional (combination of) eis promoter mutation(s) (sample set 2). These MICs were done using 2-fold serial dilutions ranging from 10.0 μg/ml to 1.25 μg/ml using the Bactec MGIT 960 system with the TB eXiST module of the EpiCentre software (12). Susceptibility to KAN was determined using the 1% proportion method based on a clinical breakpoint of 2.5 μg/ml. For WGS, each isolate was recultured from culture stocks, and DNA was extracted as previously described (13). Whole-genome sequencing libraries were prepared according to the manufacturer’s protocol (Illumina, Inc, San Diego, CA), and sequenced on an Illumina HiSeq or Illumina NextGen Seq platform. The resulting sequencing reads were mapped to the M. tuberculosis H37Rv reference strain (GenBank accession no. AL123456). Variant calling and annotation were conducted using a within-house pipeline as previously described (6). The genotypic drug resistance profile of each isolate was determined using markers defined by Miotto et al. and Coll et al. (14, 15). Raw sequencing reads of the isolates listed in Tables 1 and 2 have been deposited at the European Nucleotide Archive (ENA accession no. PRJEB41458). Additional details for all methods are described in Supplemental File 1.
TABLE 1.
Genotypic drug susceptibility testing results of selected representative isolatesb
| Patient | Isolate no. | Collection yr | WGS eis promoter mutation (% of reads) | WGS rrs 1401 mutation (% of reads) | Sanger sequencing eis promoter | MTBDRsl result eis promoter | MTBDRsl eis result interpretation | MTBDRsl result rrs | MTBDRsl rrs result interpretation | WGS vs MTBDRsl eis promoter | Lineage |
|---|---|---|---|---|---|---|---|---|---|---|---|
| P3 | WGS-3 | 2009 | −6 G > A (17) | WT | −6 G > K | All WT bands present, no MUT band | WT | WT1 and WT2 present, MUT1 present | Heteroresistance, rrs 1401 A > G and WT | Discrepant (not detectable by sl) | 2.2 |
| P14 | WGS-22 | 2007 | −32 C > T (91) | WT | −32 C > Y | All WT bands present, no MUT band | WT | WT1 and WT2 present, no MUT band | WT | Discrepant (not detectable by sl) | 4.1.1.3 |
| WGS-22 | 2007 | NA | NA | No eis promoter mutation detected (additional Sanger sequencing of rrs locus found rrs 1401A > G) | All WT bands present, no MUT band | WT | WT1 missing, MUT1 present | rrs 1401 A > G | NA | NA | |
| P17 | WGS-26 | 2008 | −10 G > A (63) and −15 C > G (36) | WT | −10 G > R and −15 C > S | All WT bands present, no MUT band | WT | WT1 and WT2 present, no MUT band | WT | Discrepant (detectable by sl) | 2.2 |
| P19 | WGS-30 | 2009 | −14 C > T (17) | rrs 1401 A > G (21) | Failed | Failed | NA | Failed | NA | NA | 2.2 |
| P27 | WGS-48 | 2014 | −14 C > T (67) | WT | −14 C > T | WT2 missing, MUT1 present | eis promoter mutation −14 | WT1 and WT2 present, no MUT band | WT | Concordant | 4.1.1.3 |
| P29 | WGS-50 | 2010 | −37 G > T (22) | rrs 1401 A > G (35) | −37 G > K | All WT bands present, no MUT band | WT | WT1 and WT2 present, MUT1 present | Heteroresistance, rrs 1401 A > G and WT | Discrepant (detectable by sl) | 2.2 |
| P31 | WGS-54 | 2010 | −12 C > T (18) and −14 C > T (9)a and −37 G > T (60) | WT | −12 C > Y and −14 C > Ya and −37 G > K | WT3 present; WT1 and WT2 weakly present, MUT present | Heteroresistance, eis promoter mutations −12 or −10, −14, −37 and WT mixed (not interpretable without additional information) | WT1 and WT2 present, no MUT band | WT | Concordant | 4.9 |
| P34 | WGS-58 | 2012 | −14 C > T (11) and −37 G > T (11) | rrs 1401 A > G (53) | −14 C > Ya and −37 G > Ka | All WT bands present and MUT band present | Heteroresistance, eis promoter mutation −14 and WT | WT1 and WT2 present, MUT1 present | Heteroresistance, rrs 1401 A > G and WT | Concordant | 2.2 |
| P40 | WGS-71 | 2012 | −14 C > T (45) | rrs 1401 A > G (8) | −14 C > Y | All WT bands present and MUT band present | Heteroresistance, eis promoter mutation −14 and WT | WT1 and WT2 present, MUT1 present | Heteroresistance, rrs 1401 A > G and WT | Concordant | 2.2 |
| P40 | WGS-72 | 2012 | −14 C > T (39)and −10 G > A (12) | rrs 1401 A > G (41) | Failed | All WT bands present and MUT band present | Heteroresistance, eis promoter mutation −14 and WT | WT1 (weak) and WT2 present, MUT1 present | Heteroresistance, rrs 1401 A > G and WT | Concordant | 2.2 |
| P47 | WGS-79 | 2015 | −8 C > A (93) | WT | Failed | All WT bands present and MUT band present | WT | WT1 and WT2 present, no MUT band | WT | Discrepant (not detectable by sl) | 2.2 |
| P65 | WGS-97 | 2015 | −104 G > A (62) | WT | −104 G > R | All WT bands present, no MUT band | WT | WT1 and WT2 present, no MUT band | WT | Discrepant (not detectable by sl) | 4.9 |
| Summary | 7/12 WT, 5/12 rrs 1401 A > G and WT | 9/12 confirmed WGS, 3/12 failed | 6/12 WT, 1/12 eis promoter −14 MUT, 3/12 eis promoter −14 MUT and WT, 1/12 combination of eis promoter mutations, 1/12 NA | 5/12 WT, 5/12 rrs 1401 and WT, 1/12 1× WT, 1× rrs 1401 (P14), 1/12 NA | 5/12 concordant, 4/12 discrepant (not detectable by sl), 2/12 discrepant (detectable by sl), 1/12 no result | 8/12 lineage 2.2, 2/12 lineage 4.9, 2/12 lineage 4.1.1.3 | |||||
Difficult to distinguish from background noise.
Changes in rrs or eis promoter are indicated as nucleotide changes using the IUPAC nucleotide code. WGS, whole-genome sequencing; sl, MTBDRsl assay; WT, wild type; MUT, mutant; NA, not applicable.
TABLE 2.
Phenotypic drug susceptibility testing results of selected representative isolatesb
| Patient | Isolate No. | Collection yr | Amikacin pDST result (routine diagnostics) | gDST vs pDST (routine diagnostics) of AMK resistance | Amikacin pDST result (repeat by SU) | Kanamycin pDST result (SU) | Kanamycin MIC (μg/ml) |
|---|---|---|---|---|---|---|---|
| P3 | WGS-3 | 2009 | S | Discrepant | ND | Failed to regrow | NA |
| P14 | WGS-22 | 2007 | R | Discrepant | R | S | 1.25 and intermediate growth at all measured concentrations |
| WGS-22 | 2007 | Ra | Concordanta | ND | R | >10 | |
| P17 | WGS-26 | 2008 | R | Discrepant | S | R | >10 |
| P19 | WGS-30 | 2009 | R | Concordant | R | R | >10 |
| P27 | WGS-48 | 2014 | S | Concordant | ND | Failed to regrow | NA |
| P29 | WGS-50 | 2010 | R | Concordant | ND | R | >10 |
| P31 | WGS-54 | 2010 | R | Discrepant | ND | R | 10 |
| P34 | WGS-58 | 2012 | R | Concordant | R | R | >10 |
| P40 | WGS-71 | 2012 | R | Concordant | ND | R | >10 |
| P40 | WGS-72 | 2012 | S | Concordant | ND | R | >10 |
| P47 | WGS-79 | 2015 | R | Discrepant | ND | R | >10 |
| P65 | WGS-97 | 2015 | S | Concordant | S | S | 2.5 |
| Summary | 8/12 R, 4/12 S | 7/12 concordant, 5/12 discrepant | 7/12 ND, 4/12 confirmed DST at diagnosis, 1/12 discrepant to diagnosis | 8/12 R, 2/12 S, 2/12 failed | 7/12 > 10, 1/12 10, 1/12 2.5, 1/12 1.25 and intermediate growth, 2/12 failed | ||
Result from the original diagnostic isolate.
Changes in rrs or eis promoter are indicated as nucleotide changes using the IUPAC nucleotide code. SU, Stellenbosch University; WGS, whole-genome sequencing; gDST, genotypic drug susceptibility testing; pDST, phenotypic drug susceptibility testing; AMK, amikacin; R, resistant; S, susceptible; ND, not done; NA, not applicable.
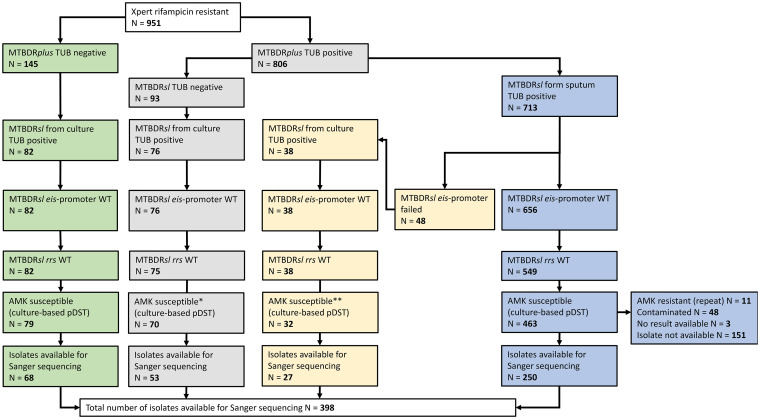
In sample set 1, eis promoter mutations were detected in 9/951 (0.95%) isolates by MTBDRsl. These isolates were phenotypically AMK susceptible with no rrs 1401 mutation (Table 3). From the 951 isolates, 398 were phenotypically AMK susceptible, eis promoter, and rrs WT, based on the MTBDRsl, and available in the biobank (Fig. 2). Sanger sequencing revealed that 11/398 (2.8%) isolates classified as eis promoter and rrs WT by routine diagnostics harbored at least one eis promoter mutation (Table 4). Three of those 11 carried the known KAN resistance markers 12 C > T and −10 G > A and should have been detected by the MTBDRsl. As Sanger sequencing revealed no heteroresistance for these isolates, it is unlikely that MTBDRsl missed this mutation because of the detection limit. Two of the three were phenotypically resistant to KAN (Table 4). The failure to detect these mutations therefore falsely classified the isolates as KAN susceptible, impacting the patient’s treatment options. The third isolate was phenotypically susceptible to KAN despite carrying an eis promoter mutation, −12 C > T. Previous studies also reported variable KAN pDST results for this mutation, including KAN susceptibility (2, 16–19). The eis promoter mutations of the remaining eight isolates could potentially have been detected through failing WT bands but were missed by the MTBDRsl. These mutations are either considered not to confer KAN resistance (n = 4; eis promoter mutation, −10 G > C) or undescribed (n = 4; eis promoter mutations, −50 T > C and −100 C > T) (Table 4). The latter are unlikely to affect the transcription of the eis gene, as they are located upstream of the usual promoter area. Since none of these mutations elevated the KAN MIC, patient treatment should not have been affected despite undetected mutations.
TABLE 3.
eis promoter mutations detected in sample set 1 by the MTBDRsl assay as part of routine diagnosticsa
| Patient | Isolate | MTBDRsl result eis promoter (banding pattern) | eis promoter mutation | AMK pDST result |
|---|---|---|---|---|
| Pa-1 | NHLS-1 | WT2 and MUT1 missing | −10 G > A or −12 C > T | S |
| Pa-2 | NHLS-2 | WT2 and MUT1 missing | −10 G > A or −12 C > T | S |
| Pa-3 | NHLS-3 | WT2 and MUT1 missing | −10 G > A or −12 C > T | S |
| Pa-4 | NHLS-4 | WT2 and MUT1 missing | −10 G > A or −12 C > T | S |
| Pa-5 | NHLS-5 | WT1-3 and MUT1 present | WT and −14 C > T mixed | S |
| Pa-6 | NHLS-6 | WT1-3 and MUT1 present | WT and −14 C > T mixed | S |
| Pa-7 | NHLS-7 | WT1-3 and MUT1 present | WT and −14 C > T mixed | S |
| Pa-8 | NHLS-8 | WT1-3 and MUT1 present | WT and −14 C > T mixed | S |
| Pa-9 | NHLS-9 | WT1-3 and MUT1 present | WT and −14 C > T mixed | S |
WT, wild type; pDST, phenotypic drug susceptibility testing; AMK, amikacin; S, susceptible; MUT, mutation.
FIG 2.
Flowchart describing the sample selection for sample set 1. WT, wild type; TUB, tuberculosis control band of the assay; AMK, amikacin; pDST, phenotypic drug susceptibility testing. *, the remaining 5 cultures were contaminated and pDST could therefore not be performed; **, the remaining 6 cultures were contaminated, and pDST could therefore not be performed.
TABLE 4.
eis promoter mutations and kanamycin MICs of isolates diagnosed as eis promoter wild type by the MTBDRsl assayb
| Patient | Isolate | eis promoter mutation | Detectable by MTBDRsla | AMK pDST result | KAN pDST result | KAN MIC (ug/ml) |
|---|---|---|---|---|---|---|
| Pa-10 | NHLS-10 | −10 G > C | No | S | S | 2.5 |
| Pa-11 | NHLS-11 | −12 C > T | Yes | S | S | 2.5 |
| Pa-12 | NHLS-12 | −100 C > T | No | S | S | 2.5 |
| Pa-13 | NHLS-13 | −10 G > C | No | S | S | 2.5 |
| Pa-14 | NHLS-14 | −10 G > C | No | S | S | 2.5 |
| Pa-15 | NHLS-15 | −50 T > C | No | S | S | 2.5 |
| Pa-16 | NHLS-16 | −100 C > T | No | S | S | 2.5 |
| Pa-17 | NHLS-17 | −12 C > T | Yes | S | R | 5 |
| Pa-18 | NHLS-18 | −10 G > C | No | S | Failed regrowth | Failed regrowth |
| Pa-19 | NHLS-19 | −10 G > A | Yes | S | R | 10 |
| Pa-20 | NHLS-20 | −100 C > Y | No | S | Failed regrowth | Failed regrowth |
“Detectable by MTBDRsl” refers to those mutations for which the MTBDRsl provides specific banding patterns (see text).
MICs are reported as the lowest concentration tested at which no growth was observed; however, the MIC can be lower than the reported number. S, susceptible; R, resistant; AMK, amikacin; KAN, kanamycin.
The screening of 2,863 WGS of clinical M. tuberculosis isolates (sample set 2) identified 101 isolates from 69 patients that carried at least one mutation in the eis promoter region (Tables S1 and S2). Seven mutations (−6 G > A, −8 C > A, −14 C > T, −15 G > A, −32 C > T, −37 G > T, −104 G > A) were not present in sample set 1. The mutations −6 G > A, −32 C > T, and −104 G > A were previously undescribed. More in-depth analyses of 12 representative isolates revealed that 6 (50%) were wrongly classified as eis promoter WT by the MTBDRsl, 4 with eis promoter mutations not included in the MTBDRsl and 2 isolates with mutations detectable by MTBDRsl (−37 G > T; −10 G > A and−15 C > G) (Tables 1 and 2). The reasons for the failure of detecting these mutations remain unclear. However, the assay failed to detect the −10 G > A mutation when in combination with −15 C > G in all four isolates with that eis promoter combination (Table S2), even when the majority of the WGS reads belonged to the M. tuberculosis subpopulation with the −10 G > A mutation (i.e., 63% of reads versus 36%; Tables 1 and 2). It is therefore unlikely that the mutant subpopulation was missed due to the detection limit of the assay. As all other isolates with different combinations of eis promoter mutations were correctly identified as mutant, the presence of more than one SNV in the same isolate does not generally seem to affect the assay’s performance. For one isolate with three eis promoter mutations (−12 C > T, −14 C > T, and −37 G > T), MTBDRsl correctly identified all mutations, but the result would not have been properly interpretable without the additional information of WGS and Sanger sequencing.
The phenotypic and genotypic results were partially discrepant (Tables 1 and 2): three of six isolates misclassified as WT carried an eis promoter mutation known to confer low-level KAN resistance (−8 C > A, −37 G > T, and −10 G > A) and were thereby falsely classified as KAN susceptible. At the time these isolates were collected, the routine diagnostic algorithm did not yet include MTBDRsl but only pDST. All three isolates were phenotypically AMK resistant, which, following the national treatment guidelines, would have led to the exclusion of KAN from the treatment regimen for those patients. An isolate with eis promoter mutation −32 C > T was phenotypically KAN susceptible, yet intermediate growth (<1%) was observed at all drug concentrations measured (1.25, 2.5, 5.0, and 10.0 μg/ml). The latter is usually an indication of heteroresistance with an underlying resistant M. tuberculosis subpopulation. However, the eis promoter mutant subpopulation was found to be the dominant subpopulation by both WGS (eis promoter mutation, −32 C > T in 91% of reads) and Sanger sequencing, indicating that the −32 C > T mutation may not be the reason for the intermediate growth under KAN pressure. For this isolate, additional pDST under KAN pressure was conducted, and subsequent MTBDRsl and Sanger sequencing revealed the rrs 1401 mutation but not the −32 C > T eis promoter mutation as being present in this subpopulation. Phenotypic DST for this isolate showed high KAN resistance (MIC > 10 μg/ml). This subpopulation had been present in a concentration below the detection limit of the pDST (1%) in the original culture but is clinically relevant, as treatment with KAN could have failed due to high-level KAN resistance (20).
In addition to the eis promoter mutations, the presence of the rrs 1401 mutation was investigated (Tables 1 and 2). Phenotypic DST revealed AMK resistance in 8/12 isolates at diagnosis, but for only 4/8, the genotypic marker rrs 1401 was detected by MTBDRsl and/or WGS. For two isolates with no rrs 1401 mutation, pDST was repeated, confirming the phenotypic resistance for one isolate, whereas the other was phenotypically susceptible, matching the genotypic results. Genotypic and phenotypic results correlated for 3/4 isolates that were typed AMK susceptible at diagnosis, but for 1, MTBDRsl detected heteroresistance (i.e., WT and rrs 1401 present). WGS, however, did not detect the rrs 1401 mutation, suggesting a false-positive MTBDRsl result (Tables 1 and 2).
This study used comprehensive data sets that nevertheless bare limitations (for a more comprehensive discussion of the limitations, see Supplemental File 1). Not all isolates of set 1 were available for Sanger sequencing; the proportion of missed eis promoter mutations could therefore be higher. Despite analyzing data collected over 25 years, no conclusions about the prevalence of eis promoter mutations across that period can be drawn, as sample set 2 was a convenience sample from several studies. All WGS isolates were screened for eis promoter mutations, but only representatives were further analyzed. However, in combination, our data provide insights on the type and frequency of eis promoter mutations present in South Africa and reflect the complexity of antibiotic resistance in M. tuberculosis. Our results indicate the most reliable option for comprehensive individual DST to be a combination of genotypic methods, including (targeted) WGS and the phenotypic analysis of consecutively collected isolates of a patient. This reduces the limitations of current diagnostic algorithms and allows adaptation to newly emerging resistance markers (5, 21) but remains an unaffordable option for low- and middle-income countries where most tuberculosis (TB) cases occur. With more and less expensive WGS-based tools becoming available, targeted use of this strategy for severe cases could nevertheless be implemented (22).
The prevalence of eis promoter mutations detected in routine surveillance data and the proportion of missed low-level KAN resistance were low in this setting but nevertheless represent a potential cause of treatment failure. WHO released new tuberculosis treatment guidelines in 2019, no longer recommending the use of KAN (23). However, some eis promoter mutations (e.g., −14 C > T) also cause low-level resistance to AMK, which remains part of the WHO-recommended treatment guidelines. More importantly, though, many countries may not be able to timely implement the new treatment recommendations and will continue using AMK or KAN (23, 24). It therefore remains important to continue monitoring the prevalence of eis promoter mutations in circulating M. tuberculosis to preserve as many treatment options as possible.
Ethics.
This study was designed and carried out in accordance with relevant guidelines and regulations. It was reviewed and approved by the Health Research Ethics Committee of Stellenbosch University (HREC) and the Western Cape Province Department of Health.
ACKNOWLEDGMENTS
The presented work was supported by funding from the South African Medical Research Council (R.M.W.), the Swiss National Science Foundation (S.D.L.; grant number P2BSP3_165379), and by funding provided through the Flemish Fund for Scientific Research (S.D.L. and M.V.; grant number FWO G0F8316N).
S.P. acknowledges funding from the Stellenbosch University Faculty of Health Sciences, the National Research Foundation, and the Harry Crossley Foundation. Y.F.V.D.H. was supported by the National Institutes of Health (grant number K08-AI106420). G.T. acknowledges funding from the EDCTP2 program supported by the European Union (grant number SF1401, Optimal Diagnosis), and the Faculty of Medicine and Health Sciences, Stellenbosch University. G.T. received funding from Hain Lifesciences for a separate project.
Hain Lifesciences donated consumables used to generate some of the data analyzed; however, Hain Lifesciences had no say in the results or interpretation.
We declare no conflict of interest.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Sirgel FA, Tait M, Warren RM, Streicher EM, Böttger EC, van Helden PD, Gey van Pittius NC, Coetzee G, Hoosain EY, Chabula-Nxiweni M, Hayes C, Victor TC, Trollip A. 2012. Mutations in the rrs A1401G gene and phenotypic resistance to amikacin and capreomycin in Mycobacterium tuberculosis. Microb Drug Resist 18:193–197. 10.1089/mdr.2011.0063. [DOI] [PubMed] [Google Scholar]
- 2.Kambli P, Ajbani K, Nikam C, Sadani M, Shetty A, Udwadia Z, Georghiou SB, Rodwell TC, Catanzaro A, Rodrigues C. 2016. Correlating rrs and eis promoter mutations in clinical isolates of Mycobacterium tuberculosis with phenotypic susceptibility levels to the second-line injectables. Int J Mycobacteriology 5:1–6. 10.1016/j.ijmyco.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sandgren A, Strong M, Muthukrishnan P, Weiner BK, Church GM, Murray MB. 2009. Tuberculosis drug resistance mutation database. PLoS Med 6:e2. 10.1371/journal.pmed.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hain Lifescience. 2016. Instructions for use for GenoType MTBDRsl VER 2.0 package insert. Hain Lifescience, Nehren, Germany. [Google Scholar]
- 5.de Vos M, Ley SD, Wiggins KB, Derendinger B, Dippenaar A, Grobbelaar M, Reuter A, Dolby T, Burns S, Schito M, Engelthaler DM, Metcalfe J, Theron G, van Rie A, Posey J, Warren R, Cox H. 2019. Bedaquiline microheteroresistance after cessation of tuberculosis treatment. N Engl J Med 380:2178–2180. 10.1056/NEJMc1815121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Black PA, de Vos M, Louw GE, van der Merwe RG, Dippenaar A, Streicher EM, Abdallah AM, Sampson SL, Victor TC, Dolby T, Simpson JA, van Helden PD, Warren RM, Pain A. 2015. Whole genome sequencing reveals genomic heterogeneity and antibiotic purification in Mycobacterium tuberculosis isolates. BMC Genomics 16:857. 10.1186/s12864-015-2067-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dheda K, Limberis JD, Pietersen E, Phelan J, Esmail A, Lesosky M, Fennelly KP, Te Riele J, Mastrapa B, Streicher EM, Dolby T, Abdallah AM, Ben-Rached F, Simpson J, Smith L, Gumbo T, van Helden P, Sirgel FA, McNerney R, Theron G, Pain A, Clark TG, Warren RM. 2017. Outcomes, infectiousness, and transmission dynamics of patients with extensively drug-resistant tuberculosis and home-discharged patients with programmatically incurable tuberculosis: a prospective cohort study. Lancet Respir Med 5:269–281. 10.1016/S2213-2600(16)30433-7. [DOI] [PubMed] [Google Scholar]
- 8.Ezewudo M, Borens A, Chiner-Oms Á, Miotto P, Chindelevitch L, Starks AM, Hanna D, Liwski R, Zignol M, Gilpin C, Niemann S, Kohl TA, Warren RM, Crook D, Gagneux S, Hoffner S, Rodrigues C, Comas I, Engelthaler DM, Alland D, Rigouts L, Lange C, Dheda K, Hasan R, McNerney R, Cirillo DM, Schito M, Rodwell TC, Posey J. 2018. Integrating standardized whole genome sequence analysis with a global Mycobacterium tuberculosis antibiotic resistance knowledgebase. Sci Rep 8:15382. 10.1038/s41598-018-33731-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dippenaar A, De Vos M, Marx FM, Adroub SA, van Helden PD, Pain A, Sampson SL, Warren RM. 2019. Whole genome sequencing provides additional insights into recurrent tuberculosis classified as endogenous reactivation by IS6110 DNA fingerprinting. Infect Genet Evol 75:103948. 10.1016/j.meegid.2019.103948. [DOI] [PubMed] [Google Scholar]
- 10.Canetti G, Fox W, Khomenko A, Mahler HT, Menon NK, Mitchison DA, Rist N, Smelev NA. 1969. Advances in techniques of testing mycobacterial drug sensitivity, and the use of sensitivity tests in tuberculosis control programmes. Bull World Health Organ 41:21–43. [PMC free article] [PubMed] [Google Scholar]
- 11.World Health Organization. 2018. Technical report on critical concentrations for drug susceptibility testing of medicines used in the treatment of drug-resistant tuberculosis. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 12.Springer B, Lucke K, Calligaris-Maibach R, Ritter C, Böttger EC. 2009. Quantitative drug susceptibility testing of Mycobacterium tuberculosis by use of MGIT 960 and EpiCenter instrumentation. J Clin Microbiol 47:1773–1780. 10.1128/JCM.02501-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warren R, de Kock M, Engelke E, Myburgh R, Gey van Pittius N, Victor T, van Helden P. 2006. Safe Mycobacterium tuberculosis DNA extraction method that does not compromise integrity. J Clin Microbiol 44:254–256. 10.1128/JCM.44.1.254-256.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miotto P, Tessema B, Tagliani E, Chindelevitch L, Starks AM, Emerson C, Hanna D, Kim PS, Liwski R, Zignol M, Gilpin C, Niemann S, Denkinger CM, Fleming J, Warren RM, Crook D, Posey J, Gagneux S, Hoffner S, Rodrigues C, Comas I, Engelthaler DM, Murray M, Alland D, Rigouts L, Lange C, Dheda K, Hasan R, Ranganathan UDK, McNerney R, Ezewudo M, Cirillo DM, Schito M, Köser CU, Rodwell TC. 2017. A standardised method for interpreting the association between mutations and phenotypic drug resistance in Mycobacterium tuberculosis. Eur Respir J 50:1701354. 10.1183/13993003.01354-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coll F, Phelan J, Hill-Cawthorne GA, Nair MB, Mallard K, Ali S, Abdallah AM, Alghamdi S, Alsomali M, Ahmed AO, Portelli S, Oppong Y, Alves A, Bessa TB, Campino S, Caws M, Chatterjee A, Crampin AC, Dheda K, Furnham N, Glynn JR, Grandjean L, Minh Ha D, Hasan R, Hasan Z, Hibberd ML, Joloba M, Jones-López EC, Matsumoto T, Miranda A, Moore DJ, Mocillo N, Panaiotov S, Parkhill J, Penha C, Perdigão J, Portugal I, Rchiad Z, Robledo J, Sheen P, Shesha NT, Sirgel FA, Sola C, Oliveira Sousa E, Streicher EM, Helden PV, Viveiros M, Warren RM, McNerney R, Pain A, et al. 2018. Genome-wide analysis of multi- and extensively drug-resistant Mycobacterium tuberculosis. Nat Genet 50:307–316. 10.1038/s41588-017-0029-0. [DOI] [PubMed] [Google Scholar]
- 16.Pholwat S, Stroup S, Heysell S, Ogarkov O, Zhdanova S, Ramakrishnan G, Houpt E. 2016. eis promoter C14G and C15G mutations do not confer kanamycin resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 60:7522–7523. 10.1128/AAC.01775-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gikalo MB, Nosova EY, Krylova LY, Moroz AM. 2012. The role of eis mutations in the development of kanamycin resistance in Mycobacterium tuberculosis isolates from the Moscow region. J Antimicrob Chemother 67:2107–2109. 10.1093/jac/dks178. [DOI] [PubMed] [Google Scholar]
- 18.Zaunbrecher MA, Sikes RD, Metchock B, Shinnick TM, Posey JE. 2009. Overexpression of the chromosomally encoded aminoglycoside acetyltransferase eis confers kanamycin resistance in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 106:20004–20009. 10.1073/pnas.0907925106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Magnet S, Blanchard JS. 2005. Molecular insights into aminoglycoside action and resistance. Chem Rev 105:477–498. 10.1021/cr0301088. [DOI] [PubMed] [Google Scholar]
- 20.Engelthaler DM, Streicher EM, Kelley EJ, Allender CJ, Wiggins K, Jimenez D, Lemmer D, Vittinghoff E, Theron G, Sirgel FA, Warren RM, Metcalfe JZ. 2019. Minority Mycobacterium tuberculosis genotypic populations as an indicator of subsequent phenotypic resistance. Am J Respir Cell Mol Biol 61:789–791. 10.1165/rcmb.2019-0178LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nimmo C, Millard J, van Dorp L, Brien K, Moodley S, Wolf A, Grant AD, Padayatchi N, Pym AS, Balloux F, O'Donnell M. 2020. Population-level emergence of bedaquiline and clofazimine resistance-associated variants among patients with drug-resistant tuberculosis in southern Africa: a phenotypic and phylogenetic analysis. Lancet Microbe 1:e165–e174. 10.1016/S2666-5247(20)30031-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.World Health Organization. 2020. WHO operational handbook on tuberculosis, module 4: treatment - drug-resistant tuberculosis treatment. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 23.World Health Organization. 2019. WHO consolidated guidelines on drug-resistant tuberculosis treatment. World Health Organization, Geneva, Switzerland. [PubMed] [Google Scholar]
- 24.Gygli SM, Borrell S, Trauner A, Gagneux S. 2017. Antimicrobial resistance in Mycobacterium tuberculosis: mechanistic and evolutionary perspectives. FEMS Microbiol Rev 41:354–373. 10.1093/femsre/fux011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental text, figure, and legends. Download AAC.02502-20-s0001.pdf, PDF file, 0.9 MB (893.9KB, pdf)
Supplemental Table S1. Download AAC.02502-20-s0002.xlsx, XLSX file, 0.01 MB (11.3KB, xlsx)
Supplemental Table S2. Download AAC.02502-20-s0003.xlsx, XLSX file, 0.01 MB (14.6KB, xlsx)