Abstract
Background and Aims
Cannabinoid receptor [CB] activation can attenuate inflammatory bowel disease [IBD] in experimental models and human cohorts. However, the roles of the microbiome, metabolome, and the respective contributions of haematopoietic and non-haematopoietic cells in the anti-colitic effects of cannabinoids have yet to be determined.
Methods
Female C57BL/6 mice were treated with either cannabidiol [CBD], Δ 9-tetrahydrocannabinol [THC], a combination of CBD and THC, or vehicle, in several models of chemically induced colitis. Clinical parameters of colitis were assessed by colonoscopy, histology, flow cytometry, and detection of serum biomarkers; single-cell RNA sequencing and qRT-PCR were used to evaluate the effects of cannabinoids on enterocytes. Immune cell transfer from CB2 knockout mice was used to evaluate the contribution of haematopoietic and non-haematopoietic cells to colitis protection.
Results
We found that THC prevented colitis and that CBD, at the dose tested, provided little benefit to the amelioration of colitis, nor when added synergistically with THC. THC increased colonic barrier integrity by stimulating mucus and tight junction and antimicrobial peptide production, and these effects were specific to the large intestine. THC increased colonic Gram-negative bacteria, but the anti-colitic effects of THC were independent of the microbiome. THC acted both on immune cells via CB2 and on enterocytes, to attenuate colitis.
Conclusions
Our findings demonstrate how cannabinoid receptor activation on both immune cells and colonocytes is critical to prevent colonic inflammation. These studies also suggest how cannabinoid receptor activation can be used as a preventive and therapeutic modality against colitis.
Keywords: Colitis, cannabinoids, microbiome
1. Introduction
The mammalian gastrointestinal [GI] tract provides the architecture for solid and liquid nutrient absorption, while harbouring a diverse and vast array of microbes to maximise catabolic potential for the host. The interplay between absorptive epithelial cells, surveilling immune cells, and commensals calibrate according to age, diet, genetics, geography, immunity, xenobiotics, and numerous other environmental and behavioural variables.1 A perturbation in any of these factors in the GI tract can cause inflammatory bowel diseases [IBD], encompassing Crohn’s disease [CD] and ulcerative colitis [UC]. These chronic idiopathic conditions are a major health concern, with prevalence in North America and Europe reaching 3.5 million people.2 Although the immunomodulatory corticosteroids, anti-tumour necrosis factor-alpha [TNFα] antibodies and 5-aminosalicylic acid [5-ASA] therapies have shown potent remission-inducing ability for IBD patients, they come with risks for infusion reactions and immunosuppression leading to opportunistic infection and malignancy.3 It has been shown conclusively that, besides the pernicious effects of IBD, those afflicted have an increased risk of developing colorectal cancer [CRC] during their lifetime.4,5
The precise pathological mechanisms underlying the development and progression of IBD remain unclear; however certain factors predispose or stave off developing disease. Host genetics can predispose an individual to IBD,6,7 and uncontrolled inflammation via mutation or dysregulated immunogenicity to commensals or dietary antigens precludes the causative pathology in IBD.8 Exogenous factors such as diet and microbial community have been recognised as significant contributors to the pathogenesis and prevention of IBD, such that a ‘Western’ diet will predispose an individual to IBD, whereas a diet high in fibre promotes the production of mucus to protect gut epithelial lining and provide a matrix and substrate where beneficial bacteria can flourish.9–11 Although many studies have investigated the specific microbial clades influencing IBD, no consensus has been reached, and the emerging hypothesis is that perturbations to the collective GI microbiome, deemed dysbiosis, contribute to IBD progression.12 The advances in biologics targeting inflammation, and beneficial effects of diet and exercise on IBD, have stemmed the incidence of IBD in the Westernized world; however, recent epidemiological data suggest an accelerating incidence and prevalence of disease in newly industrialised nations and Asia, highlighting a need for therapies and strategies that address multifaceted mechanisms of disease prevention.2,3,13
Cannabinoids are a class of chemically unique compounds that bind to evolutionarily conserved yet geographically and functionally distinct G protein-coupled receptors: cannabinoid receptors 1 [CB1] and 2 [CB2]. The two most abundant exogenous ligands for the cannabinoid receptors in the plant Cannabis sativa are Δ 9-tetrahydrocannabinol [THC] and cannabidiol [CBD]. Investigation into CB1 and CB2 led to the discovery of the endogenous cannabinoids, N-arachidonoyl-ethanolamine [AEA] and 2-arachidonoylglycerol [2-AG], which are synthesised on demand as bioactive lipids from the precursor arachidonic acid.14–16 Many early studies investigated the psychoactive properties of THC but recent work, along with a plethora of largely anecdotal reports,16,17 have hailed THC, its non-psychoactive cousin CBD, and the endocannabinoids for their therapeutic potential in conditions of autoimmunity, spasticity, nausea, and pain management.17,18 Cannabinoid receptor activation leads to a robust anti-inflammatory response, characterised by reduced antigen-presenting cell [APC] activation,19 a switch from a T helper 1 [Th1] phenotype to a T helper 2 [Th2] phenotype, direct induction of apoptosis in activated T cells,20–22 and induction of immunosuppressive cells such as Tregs and MDSCs.23 In the GI tract, cannabinoids exert a host of anticolitic effects. Recent work from our laboratory has shown CB2 activation to prevent the development of colon cancer in the AOM-DSS model, by: reducing pro-inflammatory cytokines from gut antigen-presenting cells which reduces the chronic release of oncogenic IL-2224; and activation of CB2 in the gut which prompts anti-inflammatory responses that can ameliorate the symptoms of colitis.24–26 Activation of CB1 leads to reduced GI motility, gastric emptying, and increased epithelial integrity.25–27 Further support for the notion that cannabinoids have therapeutic potential for IBD is that knockout mice lacking CB1 or CB2 develop more severe colitis symptoms28; however, the relative contribution of cannabinoid receptor activation on immune cells and enterocytes for colitis prevention has not been investigated.
In contrast to a plethora of in vitro and animal model data, evidence on the efficacy of cannabinoids for treating human IBD is limited to two small trials29,30 that were both negative in meeting their primary outcome of disease remission. In the one study examining inflammatory activity,29 no difference was found between cannabis- and placebo-treated patients with respect to C-reactive protein levels after treatment.
In the current study, the most commonly used cannabinoids, THC and CBD, were used alone or in tandem in several murine models of colitis to investigate how the gut immune cells, intestinal barrier, and gut flora work synergistically after cannabinoid treatment to prevent colitis. We were excited to find that THC prevented inflammation in the colon and attenuated colitis by engaging multiple pathways involving both enterocytes and immune cells.
2. Materials and Methods
2.1. Mice
All mice were housed at the AAALAC-accredited animal facility at the University of South Carolina, School of Medicine [Columbia, SC] under specific pathogen-free conditions and 12-h dark/light cycles in temperature-controlled rooms and given ad libitum access to water and normal chow diet. Female C57BL/6 and BALB/C mice, aged 8–12 weeks, were obtained from Jackson Laboratories [Bar Harbor, ME, USA]. Cnr1-/-, Cnr2-/-, and double knockout Cnr1-/-Cnr2-/- mice are on a C57BL/6 background and were bred and maintained in house. The number of mice for each experimental cohort is described in the figure legends.
2.2. Colitis induction and treatments
For all colitis experiments, unless otherwise indicated, treatments began 3 days before disease induction with CBD [10 mg/kg], THC [10 mg/kg], or a combination of THC+CBD [10 mg/kg, both], or the vehicle control [ethanol]. All treatments were suspended in ethanol and delivered to animals as a ratio of 2:1:18 ethanol:Tween-80:PBS by oral gavage. For the induction of TNBS-induced colitis, BALB/C mice were anesthetised by light isofluorane administration and given an intrarectal administration of 100 μL of 1 mg of TNBS [Millipore, Sigma] dissolved in 50% ethanol. Mice were kept vertical for 30 s after TNBS administration to keep the TNBS in contact with the colonic mucosal surface. DSS colitis was induced by dissolving 2% DSS [MP Biomedicals] in the drinking water and giving mice ad libitum access until the end of the study or humane endpoints were reached. For studies investigating the contribution of CB2 on haematopoietic cells to THC-mediated colitis prevention, 6 to 8 week-old female C57BL/6 WT mice were myelo-ablated via two doses of 600 cGy separated by 3 h, and then immune reconstitution was accomplished by transfer of bone marrow cells from WT [WT → WT] or Cnr2-/-, CB2 knockout [CB2 → WT] mice.
2.3. Assessment of colitis disease parameters
For all colitis models, mice were weighed daily and colon lengths were measured at experimental endpoints. Stool scores were measured according to a modified stool scoring system [Table 1]. Colonoscopy images were taken at indicated time points by anaesthetising the mice and using a high-resolution mouse endoscopic system Karl Storz [Tuttlingen, Germany] Tele Pack Vet X LED endoscope designed for small animals. The severity of colitis was scored using the mouse endoscopy and murine endoscopic index of colitis severity [MEICS] system, detailed in Table 2. To investigate the levels of proteins clinically relevant to the diagnosis of colitis severity, mice were sacrificed and blood was taken via the portal vein, allowed to clot, and serum was taken after centrifugation. Serum samples were subjected to sandwich ELISAs for serum amyloid A [SAA; Abcam], lipocalin-2 [LCN-2; Invitrogen], and myeloperoxidase [MPO; Invitrogen], according to manufacturer’s instructions.
Table 1.
Macroscopic scoring of colitis models.
| Feature | Description | Score |
|---|---|---|
| Stool score | Normal, solid pellets | 0 |
| Loosely-shaped, moist pellets | 1 | |
| Diarrhoea | 2 | |
| Occult blood presenta | 3 → 5 |
a If visible blood was present in stool, +1 was added to the stool score as based on quantification via occult blood test kit.
Table 2.
Mouse endoscopy and murine endoscopic index of colitis severity [MEICS].
| Parameter | Description | Score |
|---|---|---|
| Translucency of the colon mucosa | Transparent | 0 |
| Moderate | 1 | |
| Marked | 2 | |
| Non-transparent | 3 | |
| Vascular pattern | Normal | 0 |
| Moderate | 1 | |
| Marked | 2 | |
| Bleeding | 3 | |
| Fibrin visible | None | 0 |
| Little | 1 | |
| Marked | 2 | |
| Extreme | 3 | |
| Stool consistency | Normal + solid | 0 |
| Still shaped | 1 | |
| Unshaped | 2 | |
| Spread | 3 |
2.4. Gut permeability assay
On Day 2 of TNBS-induced colitis, and Day 9 of DSS-induced colitis, mice had their food and water removed in the evening. The next morning, mice were gavaged with 600 mg/kg of 4kD FITC-dextran [MilliporeSigma] in 100 μL PBS. Food and water were returned, and 4 h later, blood was collected by retro-orbital bleeding and serum was separated, after which FITC-dextran concentrations were determined using a PerkinElmer Life Sciences [Boston, MA] spectrophotometer with excitation wavelength at 480 nm.
2.5. Histology
Proximal colon tissues were excised, rinsed with PBS, and fixed by immersion in 3% paraformaldehyde [PFA] for 24 h. Fixed tissues were embedded in paraffin, sectioned and stained with haematoxylin and eosin. For Periodic Acid-Schiff [PAS] staining, a staining kit purchased from MilliporeSigma was used following manufacturer’s instructions. Colour bright field images and picture montages were taken using a Cytation-5 Imaging Reader [BioTek Instruments, Winooski, VT, USA].
2.6. Tissue processing
Mesenteric lymph nodes [MLNs] and spleens were excised and brought to a single-cell suspension. Spleens were subjected to red blood cell lysis before both spleens and MLNs were passed through a 70-μM filter, spun down, and re-suspended in FACS buffer for flow-cytometric analyses.
To isolate the colonic lamina propria [cLP], colons were excised and luminal contents were removed by gliding curved forceps down the length of the colon; colons were opened longitudinally and mucus was removed by gentle scraping in sterile 1X PBS. Tissue was cut into 0.5-cm pieces and incubated in pre-warmed sterile 1X HBSS [without Ca2+ and mg2+] containing FBS [3% vol/vol], 10 mM EDTA [Cellgro], and 5 mM DL-dithiotreitol [DTT; MilliporeSigma] for 30 min at 37°C while shaking. The intra-epithelial cells [IECs] containing immune cells and enterocytes were recovered by filtering the colon pieces over a 100-μM filter. The supernatant containing the IEC fraction was put on ice for at least 10 min to allow sedimentation of debris, and the IEC fraction was taken from the upper part of the supernatant. Remaining tissue containing the cLP was incubated in pre-warmed 1X HBSS [with Ca2+ and mg2+] solution [15 mL/colon] containing FBS [3% vol/vol], 1% L-glutamine, 1% penicillin–streptomycin, 10 mM HEPES, 0.5 mg/mL collagenase D [Roche], 0.5 mg/mL Dispase [MilliporeSigma], and 0.04 mg/mL DNase I [MilliporeSigma] for 45 min at 37°C while shaking. The supernatant was filtered over a 70-μm cell strainer into ice-cold sterile 1X PBS. cLP cells were passed through a Percoll [GE Healthcare] gradient [40%/80% [v/v] gradient] and spun at 620 x g for 20 min with low acceleration and no brake. Cells at the 40/80 interface were collected and washed twice with supplemented FACS buffer and prepared for flow cytometric analysis.
2.7. Flow cytometry
Relevant tissues were brought to a single-cell suspension, then 1–2 x 106 cells were washed with PBS and then stained with Live/Dead Fixable Aqua Dead Cell Stain Kit [Invitrogen] for 30 min at 4°C. Cells were then washed and incubated with TruStain FcX anti-mouse CD16/32 [Biolegend] to block Fc receptors. Extracellular antigens were stained for 20 min at room temperature in staining buffer. Cells were fixed and permeabilised with BD Cytofix/Cytoperm [for cytokine restimulations] or BD Transcription Factor Buffer Set [for transcription factor staining] per manufacturer’s instructions. Intracellular antigens were stained for 1 h at 4°C in the appropriate 1x Perm/Wash buffer. Cells were washed with staining buffer and passed through a 100-μm nylon mesh before acquisition on a BD FACSCelesta [Becton Dickinson]. Analysis was performed using FlowJo software [FlowJo, BD]. All samples were recorded based on the same live cell threshold per tissue. Compensation was set using UltraComp eBeads [Invitrogen]. Fluorochrome-conjugated antibodies are detailed in Table 3.
Table 3.
List of reagents.
| Fluorescent antibody | Clone | Identifier | Source |
|---|---|---|---|
| APC/Cy7 anti-mouse CD45 | 30-F11 | 103115 | Biolegend |
| Brilliant Violet 785™ anti-mouse CD4 | GK1.5 | 100453 | Biolegend |
| Brilliant Violet 421™ anti-mouse FOXP3 | MF-14 | 126419 | Biolegend |
| Mouse Neuropilin-1 Alexa Fluor® 488-conjugated | 761704 | FAB59941G | RandD Systems |
| Alexa Fluor® 700 anti-mouse/human CD11b | M1/70 | 101222 | Biolegend |
| Alexa Fluor® 700 anti-mouse lineage cocktail with isotype Ctrl | 17A2/RB6-8C5/RA3-6B2/Ter-119/M1/70 | 133313 | Biolegend |
| Brilliant Violet 605™ anti-mouse CD11c | N418 | 117334 | Biolegend |
| PerCP/Cyanine5.5 anti-mouse CD103 | 2E7 | 121416 | Biolegend |
| Brilliant Violet 421™ anti-mouse F4/80 | BM8 | 123137 | Biolegend |
| PE/Dazzle™ 594 anti-mouse CD117 [c-Kit] | 2B8 | 105834 | Biolegend |
| PerCP anti-mouse CD8a | 53–6.7 | 100732 | Biolegend |
| PE anti-mouse FcεRIα | MAR-1 | 134308 | Biolegend |
| PE anti-mouse Ly-6G/Ly-6C [Gr-1] | RB6-8C5 | 108407 | Biolegend |
| Alexa Fluor® 647 anti-mouse I-A/I-E | M5/114.15.2 | 107617 | Biolegend |
| PE mouse anti-mouse RORγt | Q31-378 | 562607 | BD Biosciences |
| Brilliant Violet 605™ anti-T-bet | 4B10 | 644817 | Biolegend |
| PE/Dazzle™ 594 anti-mouse/human Helios | 22F6 | 137232 | Biolegend |
| PE/Dazzle™ 594 anti-mouse CD196 [CCR6] | 29-2L17 | 129822 | Biolegend |
| Brilliant Violet 510™ anti-mouse CD335 [NKp46] | 29A1.4 | 137623 | Biolegend |
| PE/Dazzle™ 594 anti-mouse CD197 [CCR7] | 4B12 | 120122 | Biolegend |
| BV786 rat anti-mouse CD103 | M290 | 564322 | BD Biosciences |
| BB700 rat anti-mouse I-A/I-E | M5/114.15.2 | 746197 | BD Biosciences |
| TruStain FcX™ [anti-mouse CD16/32] | 93 | 101320 | Biolegend |
| or Reagent | Dose/route of administration | Catalogue number | Source |
| Dextran sulphate sodium salt [DSS], colitis grade [36,000–50,000 MW] | Drinking water [2% wt/vol] | 0216011080 | MPBIO |
| Azoxymethane | 10 mg/kg, i.p. | 25843-45-2 | MPBIO |
| Picrylsulphonic acid solution [TNBS] | 1 mg, intrarectal | P2297 | Millipore-Sigma |
| ∆9-tetrahydrocannabinol solution | 10 mg/kg, oral gavage | T2386 | Millipore-Sigma |
| ∆9-THC | 10 mg/kg, oral gavage | 12068 | Cayman Chemical |
| Cannabidiol | 10 mg/kg, oral gavage | 90080 | Cayman Chemical |
| SR 144528 | 10 µM | 5039 | Tocris |
| AM 251 | 10 µM | 1117 | Tocris |
| Recombinant mouse GM-CSF [carrier-free] | 20 ng/mL | 576304 | Biolegend |
| Recombinant mouse IL-4 [carrier-free] | 10 ng/mL | 574304 | Biolegend |
| Recombinant mouse IFN-γ [carrier-free] | 40 ng/mL | 575304 | Biolegend |
| Recombinant human IFN-γ [carrier-free] | 40 ng/mL | 570204 | Biolegend |
| Purified anti-mouse CD3ε [clone 145-2C11] | 1 µg/mL | 100301 | Biolegend |
| Ultra-LEAF™ purified anti-mouse CD28 [clone 37.51] | 4 µg/mL | 102115 | Biolegend |
| Recombinant mouse IL-6 [Animal-Free] | 30 ng/mL | 715202 | Biolegend |
| Recombinant mouse IL-23 [carrier-free] | 20 ng/mL | 589002 | Biolegend |
| Recombinant mouse IL-1β [carrier-free] | 10 ng/mL | 575102 | Biolegend |
| 6-Formylindolo[3,2-b]carbazole [FICZ] | 400 nM | SML1489 | Millipore-Sigma |
| TGF-beta 1, 2, 3 | 5 µg/mL | MAB1835R | RandD Systems |
Wt/vol, weight/volume. i.p., intraperitoneal.
2.8. Cell culture and in vitro treatments
Cells were cultured in a sterile incubator maintained at 37°C and 5% CO2. Caco-2 and LS174T cells were obtained from American Type Culture Collection [ATCC; Manassas, VA], and MC-38 cells were obtained from Kerafast [Boston, MA]. Cells were cultured in Dulbecco’s Modified Eagle Medium [DMEM; Life Sciences] supplemented with 10% fetal bovine serum, 100 U/mL penicillin and 100 U/mL streptomycin, 1% [v/v] non-essential amino acids, and 10 mM HEPES [all Gibco, Paisley, UK]. For experiments involving the addition of compounds, all cell lines were used at a population doubling [PD] between 10 and 20. Cells were seeded at 0.5 x 106 cells/well in a six-well plate. Upon ~80% confluence, medium was removed and replaced with medium containing vehicle with THC, CBD, AM251, SR144528, or a combination [all at 10 μM]. Primary cells were cultured in complete RPMI supplemented with 10% FBS, 100 U/mL penicillin, 100 U/mL streptomycin, 10 mM HEPES [Gibco, Paisley, UK], and 50 μM β-mercaptoethanol [MilliporeSigma, Gillingham, UK] [complete medium].
2.9. RNA extraction and qPCR
After isolation and purification, tissues of interest were snap-frozen in liquid N2, or placed in RNAlater [Qiagen] and transferred to -80°C until ready for processing. Total RNA was isolated using RNeasy kit [Qiagen]. Quality and quantity of RNA was determined by Nanodrop 2000 or Qubit fluorometer [both Invitrogen]. Lithium chloride precipitation was carried out on tissues derived from DSS-treated mice to remove DSS contamination from RNA samples according to Viennois et al.31 Total RNA was used to make cDNA using miScript cDNA synthesis kit from Bio-Rad. qRT-PCR was carried out using SsoAdvanced SYBR green supermix from Bio-Rad. All qRT-PCR experiments were carried out on a CFX96 [or 384] Touch Real-Time PCR Detection System [Bio-Rad], using two-step amplification with a 60°C annealing temperature. Expression levels were normalised to 18S mRNA levels in mouse tissue samples; samples derived from MC38 cells were normalised to a combination of β-actin and 18S expression. Caco-2 samples were normalised to a combination of B2M and RPLPO expression, based on Dowling et al.32 and stable expression between treatment groups. Fold changes were calculated using the 2-ΔΔCT method. Primers for genes of interest are detailed in Table 4.
Table 4.
qRT-PCR primers.
| Gene name | Accession number | Primer | Sequence 5’ → 3’ |
|---|---|---|---|
| mMUC2 | NM_023566 | Forward | CTACCATTACCACCACTAC |
| Reverse | GTCTCTCGATCACCACCATTT | ||
| mMUC5ac | NM O88715 | Forward | CTGTAACACCCAGTGTCCTAAG |
| Reverse | AGGCTGGTAGAAGTAGGTAGAG | ||
| m18S | NR_003278 | Forward | CGTCGTAGTTCCGACCATAAA |
| Reverse | TTTCAGCTTTGCAACCATACTC | ||
| mβ-actin | NM_007393 | Forward | GGCTGTATTCCCCTCCATCG |
| Reverse | CCAGTTGGTAACAATGCCATGT | ||
| mβ-Defensin 1 | NM_007843 | Forward | CACAGGCTTCCTGGGATATAAA |
| Reverse | CGCTCTGGTTGGACAACTTA | ||
| mβ-Defensin 3 | NM_013756 | Forward | TTGAGGAAAGGAGGCAGATG |
| Reverse | CGGGATCTTGGTCTTCTCTATTT | ||
| mZO-1 | NM_009386 | Forward | GCCGCTAAGAGCACAGCAA |
| Reverse | TCCCCACTCTGAAAATGAGGA | ||
| mClaudin18 | NM_001194921 | Forward | TGGGTTTTGTGGTGTCACTG |
| Reverse | GGTAGTTGAATACAGCGGTCAC | ||
| mOccludin | NM_001360536 | Forward | TTGAAAGTCCACCTCCTTACAGA |
| Reverse | CCGGATAAAAAGAGTACGCTGG | ||
| mLyz-1 | NM_013590 | Forward | CCTCCAAGTAACAGGACTTCAG |
| Reverse | CTGACTGACAAGGGAGACTTTG | ||
| mLyz-2 | NM_017372 | Forward | AGTTCTTCAGCCAGGAAGTG |
| Reverse | CCAAGATCAACTGGTCTCCTATAA | ||
| hB2M | NM_004048 | Forward | GAGGCTATCCAGCGTACTCCA |
| Reverse | CGGCAGGCATACTCATCTTTT | ||
| hRPLPO | NM_053275 | Forward | CCATTCTATCATCAACGGGTACAA |
| Reverse | TCAGCAAGTGGGAAGGTGTAATC | ||
| hDEFB1 | NM_005218 | Forward | GGTGGGTCAAAATGTGTGAGT |
| Reverse | GCTGTGGTAGGTCAGGCTTC | ||
| hDEFB103A | NM_001081551.3 | Forward | TGCTCTTCCTGTTTTTGGTGC |
| Reverse | TGCCGATCTGTTCCTCCTTT | ||
| hZO-1 | NM_001301025 | Forward | CGGTCCTCTGAGCCTGTAAG |
| Reverse | GGATCTACATGCGACGACAA | ||
| hMUC2 | NM_002457 | Forward | CACCTGTGCCCTGGAAGGC |
| Reverse | CGGTCACGTGGGGCAGGTTC | ||
| hMUC5AC | NM_017511 | Forward | CGGGTCCACGAGGAGACGGT |
| Reverse | GCTTCTGCAGCCAGGCACGA | ||
| hGAPDH | NM_002046 | Forward | GAAGGTCGGAGTCAACGGATT |
| Reverse | CGCTCCTGGAAGATGGTGAT |
2.10. Short chain fatty acid quantification using GC-MS
At sacrifice, caecal contents were snap-frozen in liquid N2 until ready for processing. Caecal contents were weighed and homogenised in ultrapure water to a concentration of 250 mg/mL; 1:4 volumes of 25% metaphosphoric acid was added to samples for 30 min on ice. Acidified samples were centrifuged at 12 000 x g for 15 min at 4°C before filtering the supernatant over Ultra-free MC columns [MilliporeSigma] using the same spin. An internal standard [IS] of 2-ethylbutyric acid was added to all samples and standards at a concentration of 0.1 mM before addition of methyl tert-butyl ether [MTBE]. Acidified and filtered samples with IS and MTBE were vortexed and spun down at 200 x g for 5 min at room temperature, and the organic layer was recovered and subjected to a HP 5890 gas chromatograph configured with flame-ionisation detectors [GC-FID]. Stabilwax®-DA Column [fused silica] of 30 m × 0.32 mm i.d. coated with 0.50-μm film thickness was used. Helium was supplied as the carrier gas at a flow rate of 15 mL/min. The temperature was programmed to achieve the following run parameters: initial temperature 100°C, hold for 0.5 min, ramp 20°C/min, final temperature 250°C, maintain for 5 min. The injected sample volume for GC analysis was 1 μL splitless and the total run time was 18.0 min. Calibration standards were prepared as aqueous stock solutions using these fatty acids at the given concentration: acetic, propionic, and n-butyric at 400 mM, isovaleric and valeric at 200 mM, isobutyric at 100 mM, and caproic and n-heptanoic at 50 mM [all from MilliporeSigma]. Each standard was injected to identify their retention times. Standard mixture was prepared at several concentrations suitable for the samples. Response factors [RF] were calculated via dividing the peak areas of the responses by the respective concentrations of the standards. To quantify the peak area in terms of concentration, the relative response factor [RRF] was used. The RRF was calculated using the formula RRF = RFStandard/RFIS. The concentration of the samples was calculated using the following equation: Conc. samples = Peak AreaSample x [Conc. IS/Peak AreaIS][1/RRF].
2.11. Measurements of xylulose-5-phosphate and fructose 1,6, bisphosphate by liquid chromatography and mass spectrometry
As described earlier,33 the normal phase chromatographic separation was also used for targeted identification of metabolites. This employed solvents containing water [solvent A], with solvent A modified by the addition of 5 mM a acetate [pH 9.9], and 100% acetonitrile [ACN] solvent B]. The flow rate was 0.2 ml/min with a gradient spanning 80% B to 2% B over a 20-min period followed by 2% B to 80% B for a 5-min period, and followed by 80% B for a 13-min time period. The flow rate was gradually increased during the separation from 0.2 mL/min [0–20 min], 0.3 mL/min [20.1–25 min], 0.35 mL/min [25–30 min], 0.4 mL/min [30–37.99 min], and finally set at 0.2 mL/min [5 min]. Metabolites were separated on a Luna Amino [NH2] column [4 µm, 100A 2.1 x 150 mm, Phenomenex]; 10 µL of suspended samples was injected and analysed using a 6490 triple quadrupole mass spectrometer [Agilent Technologies, Santa Clara, CA] coupled to a HPLC system [Agilent Technologies, Santa Clara, CA] via single reaction monitoring [SRM].
2.12. Antibiotic treatment
BL6 mice were randomised and subjected to antibiotics in their drinking water for at least 3 weeks. Antibiotics included: ampicillin [1 g/L], metronidazole [1 g/L], neomycin [1 g/L], and vancomycin [0.5 g/L]. Fresh antibiotic water replaced this every week. At the end of the antibiotic treatment, stool was collected, DNA was extracted, and PCR using Eubacteria primers was conducted and analysed via agarose gel electrophoresis to determine extinction of bacterial DNA from post-antibiotic treated mice compared with pre-antibiotic mice. Faecal transfer donor mice [3–4 per group] were given at least three administrations of THC [10 mg/kg] or vehicle before being moved to clean cages. After finishing their antibiotic regimen, recipient mice were then randomised again and placed into the old donor mouse cages with their used bedding. Three days after the end of antibiotic treatment, donor mice were placed in clean cages, stool was collected and re-suspended in PBS to 120 mg of faeces/mL of sterile PBS. Stool was homogenised by vortexing and shaking, and spun down at 800 x g for 6 min at room temperature. Supernatant was passed through a 40-μM filter and administered to recipient mice in 200 μL by oral gavage for 3 days before beginning DSS colitis.
2.13. Single cell RNA-seq and analysis
Cell number and viability from cLP samples were measured using a TC20 Automated Cell Counter [BioRad]. Processed samples showed at least 80% viability. Cells were then loaded onto the Chromium Controller [10x Genomics] targeting 1000 cells per lane. The Chromium v2 single-cell 3′ RNA-seq reagent kit [10x Genomics] was used to process samples into single-cell RNA-seq libraries according to the manufacturer’s protocol. Libraries were sequenced with a NextSeq 550 instrument [Illumina] with a depth of 40 k–60 k reads per cell. The 10x Genomics Cell Ranger pipeline [version 2.0] was used to generate FASTQ files, align reads to mm10 genome and summarise read count for each gene in each single cell. Downstream analysis was completed using R scripts and Seurat suite version 3.0 [https://satijalab.org/seurat/]. Data were integrated in Seurat using anchor and integration function. The integrated data were scaled and principal component analysis [PCA] was completed for dimensionality reduction. Clusters were made following PCA analysis by adjusting the granularity resolution to 0.15. We determined the number of principal components [PCs] to use post JackStraw analysis within Seurat to determine PCs with the lowest p-value. Differential expression was determined for each cluster to determine cluster biomarkers, and between the VEH and THC samples using the default Wilcoxon rank sum test. Raw sequencing data are available at the Sequence Read Archive [SRA], accession: PRJNA592156, and processed data files are available at Gene Expression Omnibus [GEO] accession number: GSE155669.
2.14. 16S sequencing
Faecal pellets were collected on indicated days and stored at -80°C. For isolation of mucus-associated bacteria, colons were excised upon sacrifice, luminal contents were removed, and colons were opened longitudinally and gently rinsed in a petri dish with PBS. Mucus was then scraped from the luminal surface of the colons and stored at -80°C. Colon-associated bacteria were harvested by taking a ~1-cm piece of the proximal colon after the mucosal lining was removed and snap-freezing it in liquid N2. Bacterial DNA was extracted using the QIAamp Fast DNA Stool Mini Kit [Qiagen]. Double-stranded DNA was quantified by Qubit Fluorometer [Invitrogen]. Primers for the V4 region of the 16S rRNA bacterial gene were used for amplification, and then samples were individually barcoded to label each sample according to Illumina 16S Sample Preparation Guide [Illumina]. Amplified 16S rDNA was sequenced using Illumina MiSeq. Sequence data were processed using QIIME. Read pairs were quality-filtered and joined to form a complete V4 amplicon sequence. Operational taxonomic units [OTUs] were selected by clustering reads at 99% sequence similarity in relation to the Greengenes reference database using the consensus method implemented in QIIME. Raw sequencing data can be found at the Sequence Read Archive [SRA] under accession number: PRJNA534524.
2.15. Statistical analyses
Data were analysed using GraphPad Prism software with the statistical test and number of experimental repetitions indicated in the respective figure legends. Unless otherwise stated, data are presented as individual dots for each sample/mouse, a line for the mean, and bars indicating standard error of the mean [SEM]. Tests were always two-sided where applicable; p <0.05 was considered significant.
2.16. Study approval
The University of South Carolina Institutional Animal Care and Use Committee approved all experiments.
3. Results
3.1. Cannabinoids ameliorate TNBS-induced colitis and reduce effector cell phenotypes
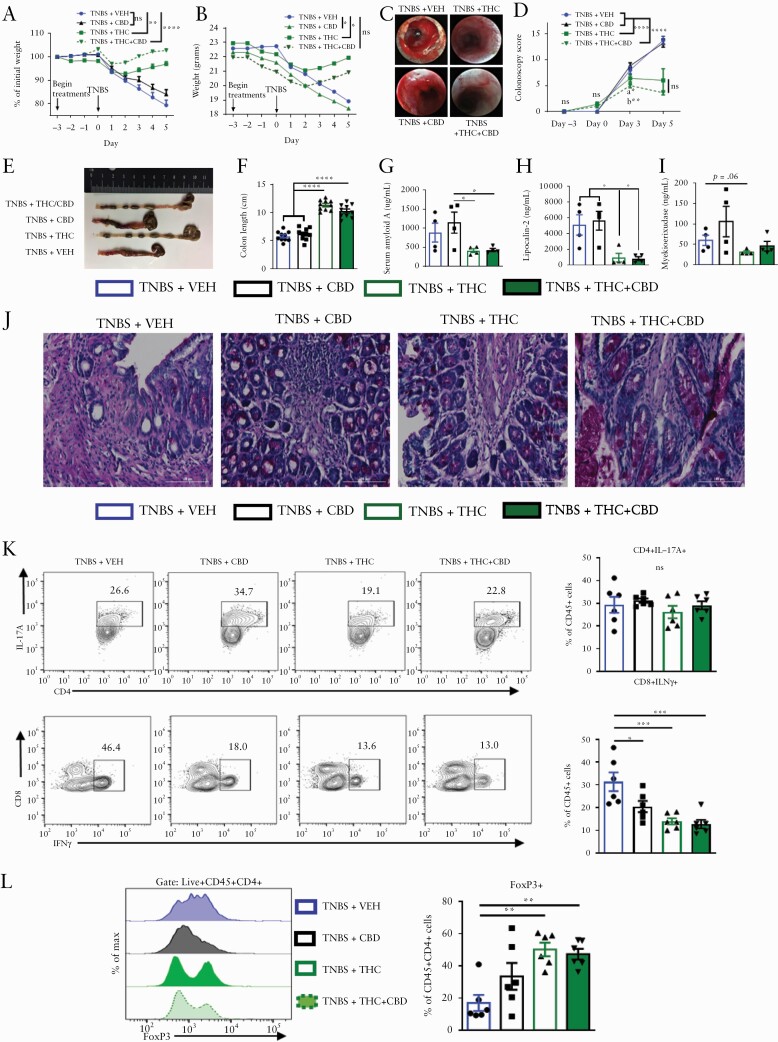
We investigated the beneficial effects of cannabinoids on intestinal inflammation by examining a murine model of acute colitis which mimics the human symptoms of ulcerative colitis. BALB/c mice were injected intrarectally with 100 mg/kg of 2,4,6-trinitrobenzenesulphonic acid [TNBS] in 50% ethanol. To test if cannabinoid use would prevent the onset of the disease, we initiated the treatment 3 days before disease induction. We used four groups of mice: TNBS+vehicle [VEH], TNBS+CBD, TNBS+THC and TNBS+THC+CBD. We used 10 mg/kg of THC or CBD or 10 mg/kg each of THC and CBD in the combination group. We used THC+CBD because these cannabinoids are found together in cannabis and may offer beneficial effects when combined. THC and THC+CBD treatment reduced the body weight loss compared with VEH control, whereas CBD alone failed to reduce weight loss [Figure 1A and B]. Colonoscopy revealed significant inflammation, bleeding, and ulcers in the VEH and CBD groups, which were diminished in the THC and THC+CBD groups [Figure 1C and D; Figure S1A, available as Supplementary data at ECCO-JCC online]. The VEH group had significant colon shortening attenuated with THC or THC+CBD treatment but not with CBD alone [Figure 1E and F]. Inflammatory markers including serum amyloid A [SAA], lipocalin-2 [LCN2], and myeloperoxidase [MPO] were all reduced in the THC or THC+CBD group mice, when compared with vehicle controls, with less striking effects in MPO levels; whereas CBD alone failed to have significant effect [Figure 1G–I]. Periodic acid-Schiff’s [PAS] staining showed that VEH and CBD groups had significant tissue damage, more immune cell infiltration, and less mucus deposition compared with THC and THC+CBD groups [Figure 1J]. To characterise the immune cell populations, the colonic lamina propria [cLP] cells were subjected to flow cytometric analysis. Gating strategies are detailed in [Figure S1B]. There was no change in CD4+ IL-17A secreting Th17 cells [Figure 1K]; however, CBD, THC, and THC+CBD reduced the number of CD8+IFNγ+ cytotoxic T cells [CTLs] compared with the VEH group [Figure 1K]. THC and THC+CBD increased the FoxP3+ T regulatory cells in the cLP compared with VEH and CBD groups [Figure 1L]. Overall, these data demonstrated that although CBD alone at the dose tested [10 mg/kg] was not effective in attenuating TNBS-mediated colitis, THC or a combination of THC+CBD were highly effective. Also, THC alone was as effective as THC+CBD.
Figure 1.
Cannabinoids ameliorate TNBS-induced colitis and reduce effector cell phenotypes. Female BALB/c mice were injected intrarectally with 100 mg/kg TNBS in 50% ethanol. Starting 3 days before disease induction and continuing daily, mice were gavaged with either: vehicle [10% EtOH in PBS+Tween-80], CBD [10 mg/kg], THC [10 mg/kg], or a combination of THC and CBD [10 mg/kg, both], [n = 10]. Mice were sacrificed at 5 days post disease induction, and blood as well as organs of interest were harvested and analysed for colitis-relevant parameters. [A] Percent weight change and [B] actual weight change over the course of disease. [C] Representative colonoscopy images taken on Day 5. [D] Quantification of colitis scores at indicated time points throughout disease course, [n = 5 per group, per time point]. [E] Representative image and [F] length of colons at sacrifice [n = 10]. [G‐I] ELISAs from serum at sacrifice quantifying disease-relevant biomarkers of colitis severity [n = 4–5]. [J] PAS stain of proximal colons from representative mice taken at sacrifice. [K] Representative flow cytometry psuedocolour dot plots [gate: Live,CD45+] displaying effector cell types in the cLP [n = 6]. [L] Offset histograms of FoxP3 expression [gate: Live, CD45+CD4+] in cLP [n = 6]. Each symbol represents an individual mouse. Data are presented as mean ± standard error of the mean [SEM]. NS, not significant; *p <0.05; **p <0.01, ***p <0.001, ****p <0.0001 by two-way ANOVA with Tukey’s multiple comparisons test. Data are from one experiment representative of three independent experiments.
3.2. Cannabinoids prevent DSS-induced colitis and reduce effector cell phenotypes
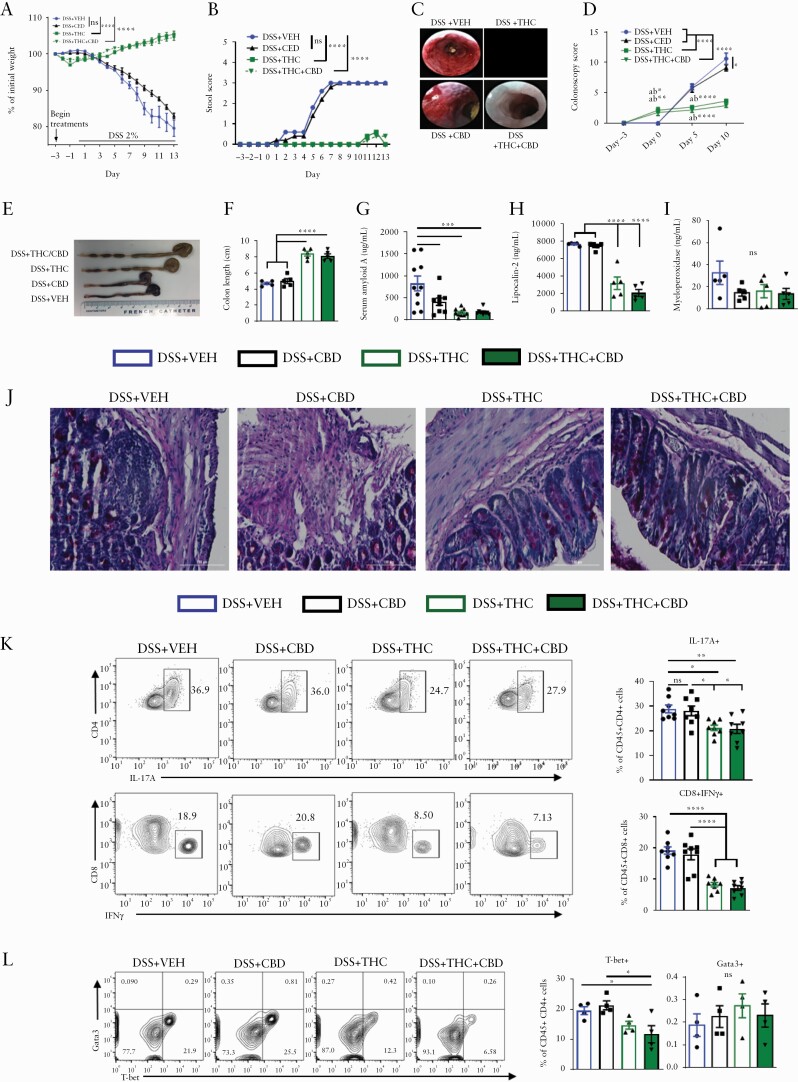
Due to the success of THC and THC+CBD in ameliorating TNBS-induced colitis, we next sought to investigate how cannabinoids may benefit a model of colitis with a different aetiology, dextran sodium sulphate [DSS]-mediated colitis. C57BL/6 mice were treated with VEH or cannabinoids for 3 days before disease induction via administration of 2% DSS in the drinking water. Body weight was monitored daily and stool parameters and colonoscopies were performed periodically throughout the 13-day disease course. THC and THC+CBD were efficacious at preventing weight loss and bloody diarrhoea when compared with VEH controls. but CBD alone was not effective [Figure 2A and B; Figure S2B, available as Supplementary data at ECCO-JCC online]. The colonoscopies revealed less inflammation and a thicker mucus layer in the THC and THC+CBD groups than the VEH and CBD groups throughout the study [Figure 2C and D; Figure S2A]. Rescued colon lengths provided further evidence of the disease-preventive effects of THC and THC+CBD [Figure 2E and F], bolstered by the serum biomarkers SAA and LCN-2 that were also reduced in treatment groups [Figure 2G and H], whereas MPO was not [Figure 2I]. Overall, CBD alone was ineffective except for reducing SAA [Figure 2G]. PAS stains of the proximal colon exhibited prominent immune cell infiltration in the VEH and CBD groups and decreased mucus production from goblet cells reversed in the THC or THC+CBD groups [Figure 2J]. Flow cytometry of cLP effector immune populations revealed a reduction in the THC and THC+CBD groups of inflammatory Th17 cells, CD8+IFNγ+ cells [Figure 2K], and an increase in Tregs [Figure S2C and D]. Additionally, we found a reduction in T-bet+ Th1 cells but no significant change in Gata3+ Th2 population in THC or THC+CBD groups when compared with VEH group [Figure 2L]. Overall, these data are consistent with the findings from the TNBS-induced colitis model in that whereas CBD alone was not effective in attenuating DSS-induced colitis, THC alone or a combination of THC+CBD was highly effective. Also, THC was as effective as THC+CBD, suggesting that CBD, at the dose tested, provided no additional benefit. For this reason, all future studies were pursued with THC only.
Figure 2.
Cannabinoids prevent DSS-induced colitis and reduce effector cell phenotypes. Female C57BL/6 mice were treated with either: vehicle [10% EtOH in PBS+Tween-80], CBD [10 mg/kg], THC [10 mg/kg], or a combination of THC and CBD [10 mg/kg, both] by oral gavage for 3 days before 2% DSS was added to their drinking water. DSS remained in the drinking water until termination of the study 14 days later. Treatments continued daily. Mice were sacrificed at 14 days post disease induction, and blood as well as organs of interest were harvested and analysed for colitis-relevant parameters. [A] Percent weight change and [B] stool score assessed over the course of disease [n = 5]. [C] Representative colonoscopy images taken on Day 10. [D] Quantification of colitis scores at indicated time points throughout disease course [n = 8 per group, per time point]. [E] Representative image and [F] length of colons at sacrifice [n = 5]. [G‐I] ELISAs from serum at sacrifice quantifying disease relevant biomarkers of colitis severity [n = 10, SAA n = 5, LCN-2, MPO]. [J] PAS stain of proximal colons from representative mice taken at sacrifice. [K] Representative flow cytometry psuedocolour dot plots [gate: Live,CD45+] displaying effector cell types in the cLP. [L] Representative flow cytometry contour plots [gate: Live,CD45+CD3+CD4+] of T-bet+ Th1 or Gata3+ Th2 cells in the cLP. Each symbol represents an individual mouse. Data are presented as mean ± standard error of the mean [SEM]. NS, not significant; *p <0.05; **p <0.01, ***p <0.001, ****p <0.0001 by two-way ANOVA with Tukey’s multiple comparisons test. Data are from one experiment representative of four independent experiments.
3.3. THC acts through CB receptors to increase colonic barrier integrity and mucus production, to protect against colitis induction
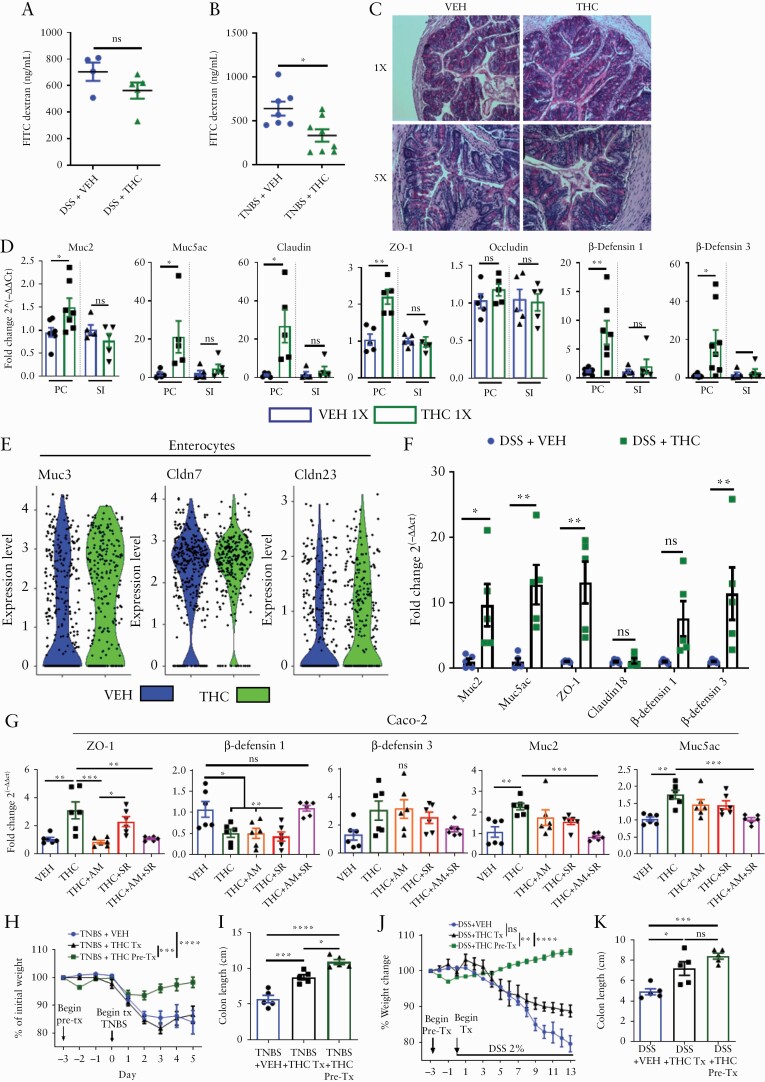
Because barrier integrity plays a critical role in colitis, we investigated the effect of cannabinoids on this function. On the last day of DSS-induced [Day 12] or TNBS-induced colitis [Day 4], mice were gavaged with 4kD FITC-dextran; 4 h later, serum FITC-dextran levels were analysed as a measure of gut permeability. The data showed that THC reduced gut permeability in TNBS-induced colitis and a trending but insignificant reduction in DSS-induced colitis, compared with VEH group [Figure 3A]. To assess mucus deposition in the colonic lumen, mice were given acute [1X] or short-term [5X] VEH or THC, proximal colons were harvested 24 h later, and PAS staining was performed. We observed a striking increase in mucus emanating from the goblet cells in the THC 1X group into the lumen, which although reduced in vibrance in the THC 5X administration group, was still noticeably increased compared with VEH [Figure 3C].
Figure 3.
Cannabinoids use both cannabinoid receptors to specifically increase colonic barrier integrity and mucus production to protect against colitis induction. [A, B] VEH or THC [10 mg/kg] administration by oral gavage began 3 days before disease induction, and at Day 4 of the TNBS colitis [n = 5–10], and Day 13 of the DSS-colitis experiment [n = 5]. Mice were fasted overnight and the next morning were gavaged with 100 µL of 4kD FITC-dextran [600 mg/kg]; 4 h later, blood was collected by retroorbital bleed and serum was analysed for the presence of FITC-dextran as a measure of gut permeability. Data are from one experiment representative of two independent experiments and presented as mean ± ± standard error of the mean [SEM]. NS, not significant; *p <0.05, **p <0.01, ****p <0.0001 by Student’s t test. [C] PAS stain of proximal colon from mice after one or five VEH or THC administrations. [D] qRT-PCR results from proximal colon [PC] or small intestine [SI] 24 h after one administration of VEH or THC [n = 5–7]. [E] scRNA-seq violin plot of indicated mRNA expression in the enterocyte cluster from the colon of mice treated with VEH or THC for 24 h. [F] qRT-PCR results from PC of mice who received DSS [2%] in their drinking water for 13 days. [G] Caco-2 cells were treated with VEH, THC, AM251 [AM], or SR144528 [SR], or a combination where indicated, for 12 h before RNA was collected and qRT-PCR was run on indicated genes [n = 6]. TNBS and DSS colitis models were induced as was done previously but we used three groups. The VEH and THC Pre-Tx [THC, 10 mg/kg, oral gavage] groups received treatments beginning 3 days before colitis initiation, and the THC Tx group began receiving daily treatments the same day as colitis was induced. [H, J] Percent weight change over the disease course, and [I, K] colon lengths at sacrifice [n = 5]. Each symbol represents an individual mouse or well. Data are presented as mean ± ± standard error of the mean [SEM]. NS, not significant; *p <0.05; **p <0.01, ***p <0.001, ****p <0.0001 by two-way ANOVA with Tukey’s multiple comparisons test. In-vivo data are from one experiment representative of two independent experiments.
Next, we looked at mucus and tight-junction protein expression in the proximal colon [PC] and small intestine [SI] in VEH 1X or THC 1X mice, and found that THC increased mRNA expression of gel-forming Muc2 and Muc5ac specifically in the PC [Figure 3D]. Similarly, increases in tight-junction proteins: claudin and zonula occludens-1 [ZO-1], but not occludin, were specifically seen in the PC after acute THC treatment [Figure 3D]. Given that mucin expression and the regulation of antimicrobial peptides, β-defensins, are intertwined,34,35 we examined β-defensin 1 and 3 expression and found it was also increased after short-term THC administration, and specifically in the PC, not in the SI [Figure 3D]. Single-cell RNA sequencing [scRNA-seq] of cells isolated from the colon [Figure S2D, available as Supplementary data at ECCO-JCC online] allowed for discrimination between enterocytes and goblet cells, confirming a THC-mediated increase in enterocyte and goblet cell expression of Cldn 23 and Muc3 and Muc 4 [Figure 3E, Figure S2E].We tested whether the observed expression increases persist in situations of DSS-induced inflammation and found THC increased the colonic expression of Muc2, Muc5ac, ZO-1 and β-defensin 3 [3F]. This effect was also observed in the mouse MC38 adenocarcinoma cell line [Figure S2F]. Treating the human adenocarcinoma cell line, Caco-2 with THC and CB antagonists AM251 [CB1] and/or SR144528 [CB2] revealed that THC-mediated increases in ZO-1 were through CB1, as they were reduced in the THC+AM251 group [Figure 3G]. Contrary to the work in mice, β-defensin 1 expression was reduced by THC and depended on both CB receptors, whereas β-defensin 3 expression was unchanged with treatments [Figure 3G]. Addition of THC to Caco-2 cells increased Muc2 and Muc5ac expression, and this depended on both CB receptors, because only with AM251 and SR144528 were expression levels returned to the level of the vehicle [Figure 3G].
Further evidence that THC requires both CB receptors to increase mucus and defensin production was mirrored by work in Cnr1-/- and Cnr2-/- mice, where THC had no effect [Figure S3A, available as Supplementary data at ECCO-JCC online]. To assess whether THC works better as a preventive ora therapeutic treatment, TNBS and DSS colitis models were initiated as done previously, but with a THC pre-treatment group [Pre-Tx] that received treatment 3 days before disease initiation, and a treatment group [Tx] that received treatment concurrently with disease initiation. Although both methods displayed efficacy in reducing disease parameters, THC pre-treatment was more effective than concurrent treatment at preventing colitis in both TNBS- and DSS-colitis models as evidenced by a decrease in weight loss and colon shortening [Figure 3H–K].
3.4. Haematopoietic cannabinoid receptors mediate THC-mediated increases in TCA cycle and fatty acid metabolites
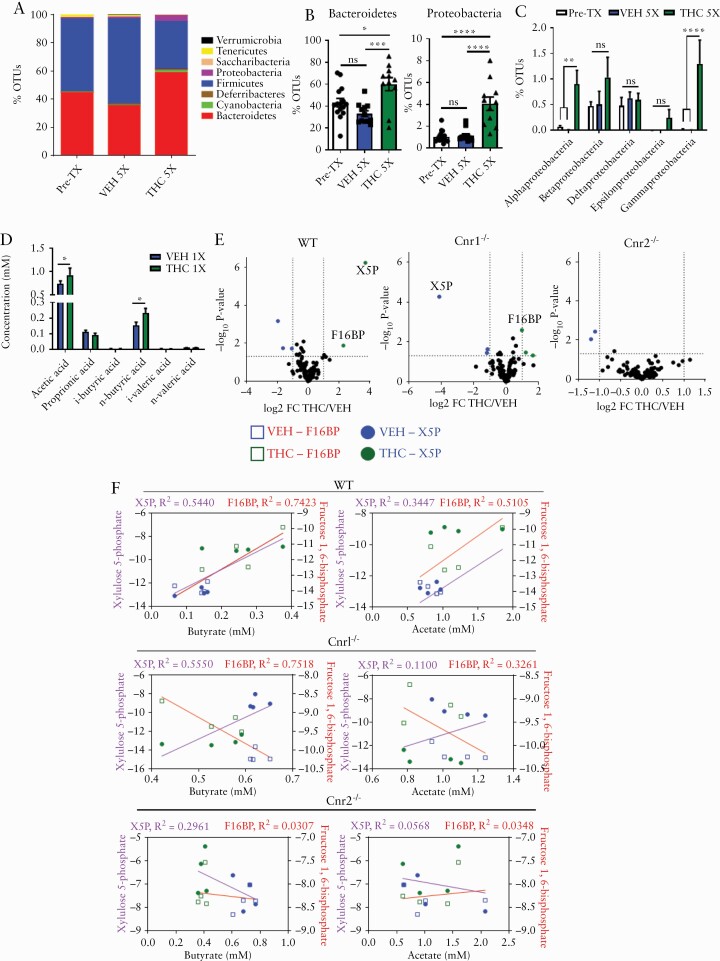
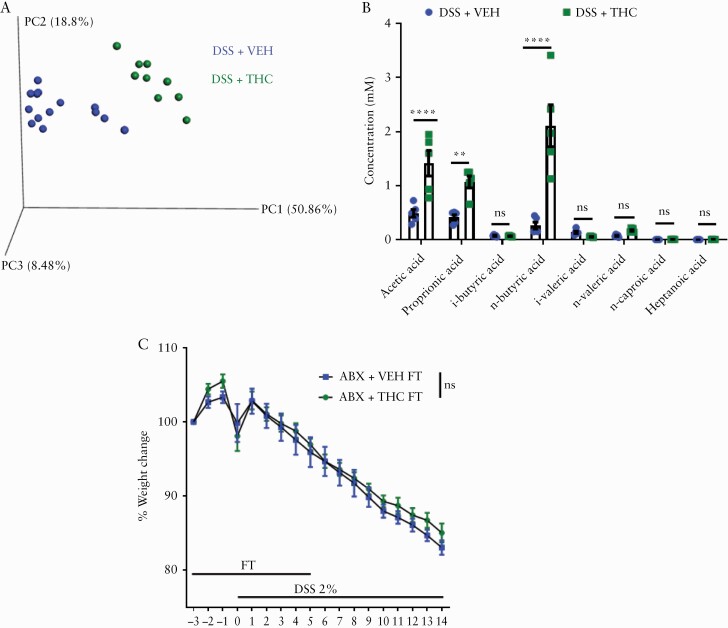
To examine whether the anticolitic effects of THC are due to any changes in the gut flora, we first performed studies assessing how THC affects gut flora in naïve mice. Stool was collected from a pool of naïve mice before [Pre-Tx] and after five administrations of either VEH [VEH 5X] or THC [THC 5X, 10 mg/kg, oral gavage]. Short-term [THC 5X] administration, compared with Pre-Tx mice and VEH 5X mice, showed increases in Gram-negative Bacteroidetes and Proteobacterial phyla [Figure 4A and B]. The specific Proteobacteria altered after THC administration belonged to the classes alpha- and gammaproteobacteria [Figure 4C]. Next, we tested short-chain fatty acids [SCFAs] by collecting caecal contents after 1X and 5X THC or VEH administration, and these data revealed that acetate and butyrate were increased 24 h after a single [1X] administration of THC compared with VEH [Figure 4D]. However, this increase was transient, as there were no differences in caecal SCFA levels after 5X THC administration [Figure S3B].
Figure 4.
Haematopoietic cannabinoid receptors mediate THC-induced increases in TCA cycle and fatty acid metabolites. Stool was collected before any compound administrations [Pre-Tx, n = 15], and after five administrations of VEH [n = 12] or THC [n = 11] by oral gavage. DNA was extracted and subjected to 16S rRNA sequencing. [A] Stacked bar plot displaying % operational taxonomic units [OTUs] of phyla of indicated groups. [B] % OTUs of the phyla bacteroidetes and proteobacteria. [C] % OTUs of the classes of proteobacteria. [D] WT mice were given a single [n = 10] administration of VEH or THC 1 day later and caecal contents were analysed for SCFAs. [E] Volcano plots showing log2 fold change THC/VEH [x-axis] and –log10 of p-value [y-axis] for abundance of TCA cycle metabolites detected in the serum of WT, Cnr1-/-, and Cnr2-/- mice 24 h after a single administration of VEH or THC. Metabolites significantly increased with THC treatment are coloured green, significantly decreased coloured blue. [F] Linear regression analysis of serum TCA cycle metabolites X5P and F16BP [y-axes] plotted against paired caecal butyrate and acetate concentrations [x-axis] collected from mice from indicated genotype 24 h after a single administration of VEH or THC. Circles represent X5P concentrations, and squares represent F16BP concentrations in relation to the SCFA indicated on the x-axis. VEH = blue, THC = green. Each symbol represents an individual mouse. Data are presented as mean ± standard error of the mean [SEM]. NS, not significant; *p <0.05; **p <0.01, ***p <0.001, ****p <0.0001 by two-way ANOVA with Tukey’s multiple comparisons test. 16S sequencing data and SCFA quantification are a combination of two independent experiments.
Because THC increased the SCFAs produced and altered the microbiome toward Gram-negative bacteria possessing increased carbohydrate metabolic potential, we tested if THC would alter the serum metabolic landscape. We studied xylulose 5-phosphate [X5P] and fructose 1,6-bisphosphate [F16BP], which are both intermediates in the oxidative branch of the pentose phosphate pathway [Figure S4A, available as Supplementary data at ECCO-JCC online] and are both markers of a well-fed state, such that X5P can rapidly induce transcription of lipogenesis genes through its associated phosphatase activation of carbohydrate-response element-binding protein [ChREBP].36,37 F16BP is the critical metabolite for signalling glucose availability.38 In this study, we tested the effect of THC in WT mice and in mice deficient in cannabinoid receptors [CBs], to investigate their role. After a single administration of VEH or THC, serum was collected and TCA cycle metabolites were probed in WT mice and those deficient for CB1 [Cnr1-/-] or CB2 [Cnr2-/-]. In WT mice, THC caused an increase in serum X5P and F16BP concentrations compared with VEH-treated mice. In CB1KO mice, X5P was reduced after THC administration and F16BP was increased, albeit at a lower level compared with WT mice. THC did not alter X5P or F16BP in CB2KO mice [Figure 4E]. Also, CB1KO mice given short-term THC did not have similar bacterial changes compared with WT mice given THC, displaying increases in ruminococcaceae and lachnospiraceae; however, CB2KO mice given THC did exhibit an increase in bacteroides and gammaproteobacteria [Figure S3C]. Considering neither single KO mouse could recapitulate the WT metabolic phenotype, and that THC ues both CB receptors to increase mucus production and Gram-negative bacteria, we hypothesised bacterial metabolism of mucus to be the driving force behind increased secretion of high glucose-availability mediators X5P and F16BP in the bloodstream. To clarify the relationship between bacterial fatty acid and TCA cycle metabolites after THC administration, paired serum TCA cycle metabolites and caecal SCFA concentrations from WT, CB1KO, or CB2KO mice given VEH or THC were plotted, to determine if the two were correlated.
In WT mice, X5P and F16BP concentrations correlated strongly with increasing acetate and butyrate production seen after THC administration, whereas in CB1KO mice, there was a positive correlation between butyrate and X5P concentrations and a negative correlation with butyrate and F16BP, but not with acetate. In CB2KO mice, changes in SCFAs did not correlate with metabolite changes [Figure 4F]. These results implicate CB receptors in the modulation of bacterial community development and metabolic activity.
3.5. THC-mediated changes in bacterial composition are inconsequential to DSS progression
Next, we investigated how THC impacts on the microbial balance in the DSS-induced colitis model. We found that the THC group, in which colitis was attenuated, was clustered away from the VEH group, which developed severe colitis [Figure 5A]. Concordantly, the THC group had higher levels of acetate, proprionate, and butyrate in their caecal contents when compared with the VEH group [Figure 5B]. To test whether the changes in bacterial community seen with THC administration were the mechanism of protection against colitis development, we did a faecal transfer [FT] experiment wherein naïve mice were given antibiotics [ABX] in their water for 4 weeks to deplete their microbiota. Then they were taken off ABX water and placed in cages with the used bedding of mice that had received short-term VEH or THC administration which would be the FT donor mice. After 3 days in the used cages with regular drinking water, stool from donor FT mice was collected freshly and gavaged to recipient mice for 3 days before giving DSS, and then for another 5 days after giving DSS. Weight loss was monitored and the data revealed that the faecal bacterial community from THC-treated mice could not protect mice from colitis [Figure 5C].
Figure 5.
Cannabinoid-mediated changes in bacterial composition are inconsequential to DSS progression. [A] PCOA plot displaying the bacterial community clustering from indicated mice on Day 14 of DSS colitis [n = 5]. [B] Caecal SCFA quantification of indicated mice [n = 5]. Mice were given antibiotics [ABX] in their drinking water for 4 weeks before antibiotic-free water was returned, mice were placed in the cages of donor faecal transfer [FT] mice containing their used bedding. Three days after cessation of antibiotics, mice were given daily faecal transfers from stool of indicated donor mice. DSS [2%] was administered in the drinking water after 3 days of faecal transfers; and [C] body weight was recorded throughout the study [n = 7–8 per group]. Data are presented as mean ± standard error of the mean [SEM]. Faecal transfer-DSS experiment was repeated three times. Data presented are from a single representative experiment. NS, not significant. 16S sequencing results and SCFA quantification are from one representative DSS experiment that was repeated four times.
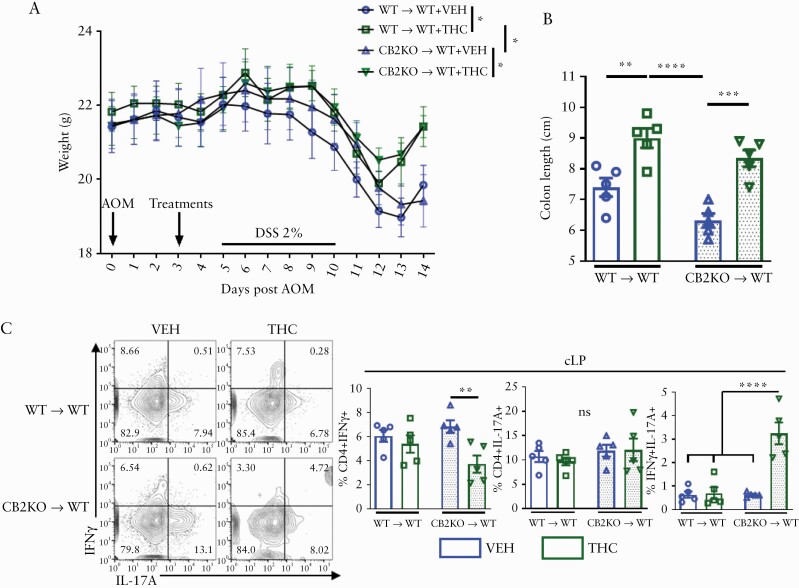
3.6. Haematopoietic and non-haematopoietic cells contribute to THC-mediated colitis protection
To resolve whether CB expression on immune cells or non-haematopoietic cells were responsible for the favourable effects of THC, WT mice were myelo-ablated and reconstituted with BM of CB2KO mice [CB2KO→WT] or WT BM [WT→WT]. Then we induced acute colitis as was done previously23,39 to model the early stages of tumourigenesis before macroscopic tumour development but where the hallmark body weight loss, colon shortening, and inflammation were present. THC compared with VEH was efficacious at preventing body weight loss in the WT mice with CB2KO immune systems [Figure 6A]. Both groups receiving THC treatment had lesser colon shortening than their VEH counterparts [Figure 6B]. In the cLP, there was no difference in the WT→WT group in the inflammatory IFNγ+, IL-17A+ or IFNγ+IL-17A+ secreting Th cells, but in CB2KO→WT mice given THC, there was a decrease in CD4+IFNγ+ single-positive cells, but an increase in IFNγ+IL-17A+ double-positive Th cells [Figure 6C].
Figure 6.
Haematopoietic and non-haematopoietic cells contribute to THC-mediated colitis protection. WT mice were myelo-ablated via two doses of 600 cGy separated by 3 h, then immune reconstitution was accomplished by transfer of bone marrow cells from WT [WT → WT] or Cnr2-/-, CB2 knockout [CB2 → WT] mice. [A] Body weight, and [B] colon lengths from indicated groups [n = 4–5]. [C] Flow cytometric analysis of CD4+ IL-17A+ and IFNγ+ cells in the colonic lamina propria [n = 4–5]. Each symbol represents an individual mouse. Data are from one experiment representative of two independent experiments and presented as mean ± standard error of the mean [SEM]. NS, not significant; *p <0.05; **p <0.01, ***p <0.001, ****p <0.0001 by two-way ANOVA with Tukey’s multiple comparisons test.
4. Discussion
The synergy between immune cells, enterocytes, and symbiotic and pathogenic microbes in the GI tract requires a delicately balanced network that can adapt as new signals are acquired. A disruption in that balance can lead to chronic inflammation. Recent epidemiological evidence suggests this disruption in the form of colitis is on the rise in developing nations, and its current burden in North America and Europe is daunting.1–3 New strategies that can safely tip the gut equilibrium toward host defence, without sparking inflammatory cascades or leaving the host vulnerable to other maladies, will be the most effective strategy for preventing disease. In the current study, we demonstrated the potential for THC in the prevention and amelioration of gut inflammation. Previous reports have found that cannabinoids acting through CB1 can reduce gut motility23,40 and gastric acid secretion,41 and increase barrier integrity.23,25 Studies from our laboratory and others have demonstrated the CB2-dependent anti-inflammatory properties of cannabinoids working directly through immune cells.21,23,24,42 Whereas those studies primarily focused on the effect of cannabinoids on colonic inflammation, our work comprehensively evaluates how THC positively influences host gastrointestinal homeostasis through increased coordination between immune cells, colonocytes and gut flora.
The most frequently consumed cannabinoids come as recreational cannabis, whose primary bioactive components are CBD and THC.16,17 With cannabis legalisation and public consumption of cannabis and CBD extracts increasing, it is important to parse out the scientifically proven beneficial effects of these compounds in order to cut through the noise of increasing anecdotal reports.16,43,44 In the USA, purified forms of CBD come as the FDA-approved drug Epidiolex for epilepsy, and purified THC comes as Dronabinol to treat nausea and vomiting induced during cancer chemotherapy.18,43 A combination of THC and CBD is sold under the trade name Sativex and is prescribed to treat the symptoms of multiple sclerosis.17,18 Our laboratory has also shown that CBD is highly effective against autoimmune hepatitis and the FDA has approved its use to treat this disorder as an orphan drug.45 Numerous reports from animal studies and one prospective placebo-controlled human study tout the effectiveness of cannabinoids for ameliorating colitis and colorectal cancer, but few studies have examined their potential as colitis prophylactics.16, 17, 23–28 Our goal was to understand how cannabinoids can affect disease course, and to best understand the therapeutic potential of these compounds. To accomplish this, we used three models of colitis, looking at gut immunity, barrier function, and host-microbiota interactions, to gain a clear view on the mechanisms through which cannabinoids may help in treating colitis.
In the TNBS and DSS models of colitis, THC and a combination of THC+CBD were effective at preventing the symptoms of colitis, but CBD was not. The lack of efficacy of CBD may be related to the lower dose that we used, and this was because our initial goal was to test if a low dose of CBD when combined with a low dose of THC could exert a synergistic effect. This study provides additional context to previously reported findings that CBD can synergise with THC to reduce TNBS-induced colitis in rats.46 In that study, the colitis induced was far less severe than in this study, the combination of THC and CBD only proved more effective than THC alone at the dose of 5 mg/kg per treatment, and only in terms of MPO activity. Moreover, that study confirms our finding that CBD at 10 mg/kg is ineffective at reducing colitis severity, but that at the 20-mg/kg dose there is efficacy. To that end many reports, including those from our laboratory, demonstrate CBD is effective for treating inflammatory pain43 and liver inflammation.45,47 TNBS and DSS colitis are both induced via luminal delivery of xenobiotics that induce colitis by damage to the epithelial layer and allowing microbes to infiltrate host tissue with DSS, and by disrupting the luminal mucus layer with ethanol and haptenising colonic proteins with TNBS.
In examining tissues from THC-treated mice, we noticed less inflammation than in the VEH group but also more mucus deposition into the luminal layer. We found that THC alone stimulated the production of Muc2 and Muc5ac after a single administration in naïve mice and in inflamed DSS-diseased mice. Besides increasing mucus production, THC also increased the expression of tight-junction proteins ZO-1 and claudin. Importantly, THC-mediated increases in mucus and tight-junction production were specific to the colon. This explains why, despite similarly improved clinical parameters of disease, DSS mice treated with THC had gut leakage comparable to VEH mice, whereas TNBS mice treated with THC did not. DSS delivered in the drinking water also breaks up the epithelial lining of the small intestine and other parts of the GI tract, leading to leakage, but TNBS induced damage is specific to the colon.
Using cannabinoid receptor [CB] knockout mice and MC38 and Caco-2 adenocarcinoma cell lines, we noted that CB1 is responsible for the THC-mediated increases in tight-junction proteins, confirming work by others.25 Novel to this study was that the increase in mucus and β-defensins that occurs after THC administration depends on both CB1 and CB2. The increase in mucus expression was prevalent in mouse tissue, cell lines, and human cell lines, but Caco-2 cells given THC had the opposite trend in β-defensin production when administered THC; this could be attributable to the nature of that cell line or to a non-conserved mechanism. Regardless, data in human subjects are needed to support this finding. It should also be noted that a trend towards increased mucus was seen after THC administration in CB1KO mice, suggesting CB2 could be mediating increases in mucus that were not assessed by qPCR, but were seen to be increased with THC via scRNA-seq in WT mice like Muc3 and Muc4. The observed increases in colonic barrier integrity were possibly important for disease prevention, as mice pre-treated for 3 days with THC developed less severe TNBS and DSS colitis compared with mice whose treatment began concurrently with disease induction. Furthermore, in the model of AOM-DSS colitis, this notion was supported because WT mice that received CB2KO immune reconstitution exhibited reduced colon shortening and weight loss after THC pre-treatment.
Our findings that THC-mediated increases in tight-junction proteins and mucus were seen only in the proximal colon, but not in the small intestine where there is less prolonged contact with microbes, was novel. Mucus once deposited into the lumen polymerises into a polysaccharide gel. This gel is an excellent source of defence from microbes and has been protective in colitis and colon cancer34,35; however, it also acts as fuel for microbes. The increase in mucus production and deposition after THC administration may be why the changes in bacterial composition seen after THC administration favour the Gram-negative phyla of bacteroidetes and proteobacteria, two phyla well known for their voracious appetite for carbohydrates.48 This would also explain why those bacteria, which are commonly thought of as pathogenic with colitis,48,49 were not disease-promoting in our THC-treated mice; those bacteria were not opportunistic pathogens, but there was simply an increase in their preferential food source.
Further support for this notion is the finding that acute administration of THC caused a small increase in acetate and butyrate levels in the caecum which diminished after short-term administration, indicating an initial burst of microbial mucus metabolism which wanes. Release of xylulose 5-phosphate and fructose 1,6-bisphosphate by these microbes into the bloodstream, signalling high glucose availability, reinforces this position. Despite the seemingly beneficial effects of THC on bacterial metabolism, faecal transfer from THC-treated mice was ineffective at suppressing colitis progression. These negative results cannot be used to conclusively state that the gut microbiota do not play a role in THC-mediated effects, because such studies do not accurately mimic direct effects of THC in vivo such as the number of microbiota transferred and lacking a transfer of adherent bacteria.
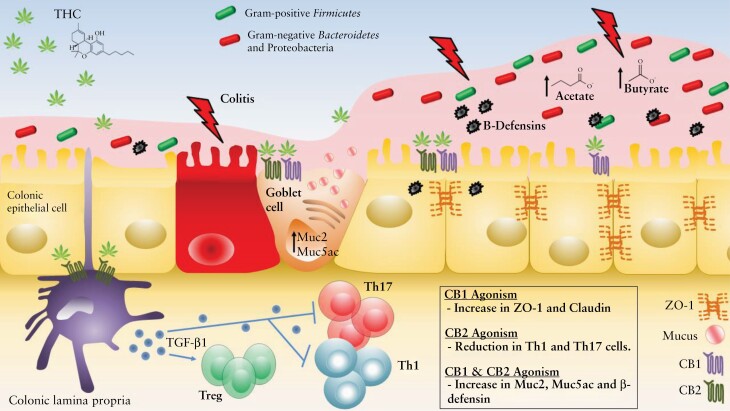
Yet another compensatory mechanism in this complex interplay at the luminal surface is that the increased mucus production is balanced by an increase in defensin production, as seen from both our in vivo animal experiments and in vitro MC38 cells. Taken together, these data indicate a sophisticated network of mechanisms through which THC promotes cooperation and balance in the colonic macro-environment. The decrease in immunogenicity from cLP immune cells23 is counterbalanced by increased barrier integrity, mucus production, and antimicrobial peptide release, which stave off unwanted microbial interference while still allowing for the uptake of their beneficial metabolites [Figure 7]. The presented data provide robust evidence for the multifaceted efficacy of THC in colitis prevention.
Figure 7.
Diagram of the colonic microenvironment and the anti-colitic effects of THC on enterocytes and immune cells.
Supplementary Material
Acknowledgement
We would like to thank Dr Angela Murphy and Dr Jackie Bader for providing us with the MC38 cell line.
Raw sequencing data are available at the Sequence Read Archive [SRA], accession: PRJNA534524 [16S Sequencing data] PRJNA592156 [scRNA-Seq data], and processed data files are available at Gene Expression Omnibus [GEO] accession number: GSE155669.
Funding
These studies were supported in part by National Institutes of Health grants: P01AT003961, P20GM103641, R01AT006888, R01ES030144, R01AI123947 to MN and PN, and American Cancer Society [ACS] Award 127430-RSG-15-105-01-CNE to VP and NP.
Conflict of Interest
The authors have declared that no conflicts of interest exist.
Author Contributions
WB, HRA, MN, and PSN: designed research studies; WB, HRA, PBB, KM, NP, VP, and MW: experimentation and data acquisition; WB, HRA, GC, and KW: data analysis; WB: writing original manuscript draft; WB, MN, and PSN: writing, editing, and revisions; MN and PSN: funding acquisition; MN and PSN: resources; MN and PSN: supervision.
References
- 1. Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med 2016;375:2369–79. [DOI] [PubMed] [Google Scholar]
- 2. Ng SC, Shi HY, Hamidi N, et al. . Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet 2018;390:2769–78. [DOI] [PubMed] [Google Scholar]
- 3. Coward S, Clement F, Benchimol EI, et al. . Past and future burden of inflammatory bowel diseases based on modeling of population-based data. Gastroenterology 2019;156:1345–53.e4. [DOI] [PubMed] [Google Scholar]
- 4. Baker KT, Salk JJ, Brentnall TA, Risques RA. Precancer in ulcerative colitis: the role of the field effect and its clinical implications. Carcinogenesis 2018;39:11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Eaden JA, Abrams KR, Mayberry JF. The risk of colorectal cancer in ulcerative colitis: a meta-analysis. Gut 2001;48:526–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Duerr RH, Taylor KD, Brant SR, et al. . A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science 2006;314:1461–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cho JH. The genetics and immunopathogenesis of inflammatory bowel disease. Nat Rev Immunol 2008;8:458–66. [DOI] [PubMed] [Google Scholar]
- 8. de Souza HS, Fiocchi C. Immunopathogenesis of IBD: current state of the art. Nat Rev Gastroenterol Hepatol 2016;13:13–27. [DOI] [PubMed] [Google Scholar]
- 9. Llewellyn SR, Britton GJ, Contijoch EJ, et al. . Interactions between diet and the intestinal microbiota alter intestinal permeability and colitis severity in mice. Gastroenterology 2018;154:1037–46.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Desai MS, Seekatz AM, Koropatkin NM, et al. . A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell 2016;167:1339–53.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bernstein CN, Shanahan F. Disorders of a modern lifestyle: reconciling the epidemiology of inflammatory bowel diseases. Gut 2008;57:1185–91. [DOI] [PubMed] [Google Scholar]
- 12. Hirata Y, Ihara S, Koike K. Targeting the complex interactions between microbiota, host epithelial and immune cells in inflammatory bowel disease. Pharmacol Res 2016;113:574–84. [DOI] [PubMed] [Google Scholar]
- 13. Kaplan GG, Ng SC. Globalisation of inflammatory bowel disease: perspectives from the evolution of inflammatory bowel disease in the UK and China. Lancet Gastroenterol Hepatol 2016;1:307–16. [DOI] [PubMed] [Google Scholar]
- 14. Devane WA, Hanus L, Breuer A, et al. . Isolation and structure of a brain constituent that binds to the cannabinoid receptor. Science 1992;258:1946–9. [DOI] [PubMed] [Google Scholar]
- 15. Lu HC, Mackie K. An introduction to the endogenous cannabinoid system. Biol Psychiatry 2016;79:516–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Marzo VD. New approaches and challenges to targeting the endocannabinoid system. Nat Rev Drug Discov 2018;17:623–39. [DOI] [PubMed] [Google Scholar]
- 17. Whiting PF, Wolff RF, Deshpande S, et al. . Cannabinoids for medical use: a systematic review and meta-analysis. JAMA 2015;313:2456–73. [DOI] [PubMed] [Google Scholar]
- 18. van Amerongen G, Kanhai K, Baakman AC, et al. . Effects on spasticity and neuropathic pain of an oral formulation of Δ9-tetrahydrocannabinol in patients with progressive multiple sclerosis. Clin Ther 2018;40:1467–82. [DOI] [PubMed] [Google Scholar]
- 19. Roth MD, Castaneda JT, Kiertscher SM. Exposure to Δ9-tetrahydrocannabinol impairs the differentiation of human monocyte-derived dendritic cells and their capacity for T cell activation. J Neuroimmune Pharmacol 2015;10:333–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Eisenstein TK, Meissler JJ. Effects of cannabinoids on T-cell function and resistance to infection. J Neuroimmune Pharmacol 2015;10:204–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Singh UP, Singh NP, Singh B, Price RL, Nagarkatti M, Nagarkatti PS. Cannabinoid receptor-2 [CB2] agonist ameliorates colitis in IL-10[-/-] mice by attenuating the activation of T cells and promoting their apoptosis. Toxicol Appl Pharmacol 2012;258:256–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cabral GA, Rogers TJ, Lichtman AH. Turning over a new leaf: cannabinoid and endocannabinoid modulation of immune function. J Neuroimmune Pharmacol 2015;10:193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hegde VL, Nagarkatti M, Nagarkatti PS. Cannabinoid receptor activation leads to massive mobilization of myeloid-derived suppressor cells with potent immunosuppressive properties. Eur J Immunol 2010;40:3358–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Becker W, Alrafas HR, Wilson K, et al. . Activation of cannabinoid receptor 2 prevents colitis-associated colon cancer through myeloid cell de-activation upstream of IL-22 production. iScience 2020;23:101504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Massa F, Marsicano G, Hermann H, et al. . The endogenous cannabinoid system protects against colonic inflammation. J Clin Invest 2004;113:1202–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Storr MA, Keenan CM, Zhang H, Patel KD, Makriyannis A, Sharkey KA. Activation of the cannabinoid 2 receptor [CB2] protects against experimental colitis. Inflamm Bowel Dis 2009;15:1678–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Alhamoruni A, Lee AC, Wright KL, Larvin M, O’Sullivan SE. Pharmacological effects of cannabinoids on the Caco-2 cell culture model of intestinal permeability. J Pharmacol Exp Ther 2010;335:92–102. [DOI] [PubMed] [Google Scholar]
- 28. Engel MA, Kellermann CA, Burnat G, Hahn EG, Rau T, Konturek PC. Mice lacking cannabinoid CB1-, CB2-receptors or both receptors show increased susceptibility to trinitrobenzene sulfonic acid [TNBS]-induced colitis. J Physiol Pharmacol 2010;61:89–97. [PubMed] [Google Scholar]
- 29. Naftali T, Bar-Lev Schleider L, Dotan I, Lansky EP, Sklerovsky Benjaminov F, Konikoff FM. Cannabis induces a clinical response in patients with Crohn’s disease: a prospective placebo-controlled study. Clin Gastroenterol Hepatol 2013;11:1276–80.e1. [DOI] [PubMed] [Google Scholar]
- 30. Naftali T, Mechulam R, Marii A, et al. . Low-dose cannabidiol is safe but not effective in the treatment for Crohn’s disease, a randomized controlled trial. Dig Dis Sci 2017;62:1615–20. [DOI] [PubMed] [Google Scholar]
- 31. Viennois E, Tahsin A, Merlin D. Purification of total RNA from DSS-treated murine tissue via lithium chloride precipitation. Bio Protoc 2018;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dowling CM, Walsh D, Coffey JC, Kiely PA. The importance of selecting the appropriate reference genes for quantitative real time PCR as illustrated using colon cancer cells and tissue. F1000Res 2016;5:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wangler MF, Chao YH, Bayat V, et al. . Peroxisomal biogenesis is genetically and biochemically linked to carbohydrate metabolism in Drosophila and mouse. PLoS Genet 2017;13:e1006825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kawashima H. Roles of the gel-forming MUC2 mucin and its O-glycosylation in the protection against colitis and colorectal cancer. Biol Pharm Bull 2012;35:1637–41. [DOI] [PubMed] [Google Scholar]
- 35. Cobo ER, Kissoon-Singh V, Moreau F, Chadee K. Colonic MUC2 mucin regulates the expression and antimicrobial activity of β-defensin 2. Mucosal Immunol 2015;8:1360–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kabashima T, Kawaguchi T, Wadzinski BE, Uyeda K. Xylulose 5-phosphate mediates glucose-induced lipogenesis by xylulose 5-phosphate-activated protein phosphatase in rat liver. Proc Natl Acad Sci U S A 2003;100:5107–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Iizuka K, Horikawa Y. ChREBP: a glucose-activated transcription factor involved in the development of metabolic syndrome. Endocr J 2008;55:617–24. [DOI] [PubMed] [Google Scholar]
- 38. Zhang CS, Hawley SA, Zong Y, et al. . Fructose-1,6-bisphosphate and aldolase mediate glucose sensing by AMPK. Nature 2017;548:112–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Malik A, Sharma D, Malireddi RKS, et al. . SYK-CARD9 signaling axis promotes gut fungi-mediated inflammasome activation to restrict colitis and colon cancer. Immunity 2018;49:515–30.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Aviello G, Romano B, Izzo AA. Cannabinoids and gastrointestinal motility: animal and human studies. Eur Rev Med Pharmacol Sci 2008;12[Suppl 1]:81–93. [PubMed] [Google Scholar]
- 41. Kinsey SG, Cole EC. Acute Δ[9]-tetrahydrocannabinol blocks gastric hemorrhages induced by the nonsteroidal anti-inflammatory drug diclofenac sodium in mice. Eur J Pharmacol 2013;715:111–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ke P, Shao BZ, Xu ZQ, et al. . Activation of cannabinoid receptor 2 ameliorates DSS-induced colitis through inhibiting NLRP3 inflammasome in macrophages. PLoS One 2016;11:e0155076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. White CM. A review of human studies assessing Cannabidiol’s [CBD] therapeutic actions and potential. J Clin Pharmacol 2019;59:923–34. [DOI] [PubMed] [Google Scholar]
- 44. Carliner H, Brown QL, Sarvet AL, Hasin DS. Cannabis use, attitudes, and legal status in the U.S.: a review. Prev Med 2017;104:13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hegde VL, Nagarkatti PS, Nagarkatti M. Role of myeloid-derived suppressor cells in amelioration of experimental autoimmune hepatitis following activation of TRPV1 receptors by cannabidiol. PLoS One 2011;6:e18281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jamontt JM, Molleman A, Pertwee RG, Parsons ME. The effects of Delta-tetrahydrocannabinol and cannabidiol alone and in combination on damage, inflammation and in vitro motility disturbances in rat colitis. Br J Pharmacol 2010;160:712–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang Y, Mukhopadhyay P, Cao Z, et al. . Cannabidiol attenuates alcohol-induced liver steatosis, metabolic dysregulation, inflammation and neutrophil-mediated injury. Sci Rep 2017;7:12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Palmela C, Chevarin C, Xu Z, et al. . Adherent-invasive Escherichia coli in inflammatory bowel disease. Gut 2018;67:574–87. [DOI] [PubMed] [Google Scholar]
- 49. Manichanh C, Rigottier-Gois L, Bonnaud E, et al. . Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut 2006;55:205–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.