LETTER
The emergence of SARS-CoV-2 variants that reduce antibody neutralization and vaccine efficacy is of significant global concern. On 24 March 2021, the Indian SARS-CoV-2 Consortium on Genomics reported that a variant containing a unique combination of two spike receptor-binding domain (RBD) neutralization resistance mutations, L452R and E484Q, made up 15 to 20% of positive cases in the Maharashtra state, which includes Mumbai (https://pib.gov.in/PressReleaseIframePage.aspx?PRID=1707177). Here, we report the rapid recognition of this variant in the San Francisco Bay Area, CA, through investigation of unusual reverse transcriptase real-time PCR (RT-qPCR) curves and confirmation by viral whole-genome sequencing (WGS).
The Stanford Health Care Clinical Virology Laboratory prospectively screened SARS-CoV-2-positive respiratory specimens for three mutations, L452R (HEX), E484K (Cy5), and N501Y (FAM) using a laboratory-developed, multiplex, mutation-specific RT-qPCR (1). This approach allowed high-throughput screening for known variants of concern at the time (B.1.1.7, P.1, B.1.351, and B.1.427/B.1.429) and enabled informed deployment of sequencing resources.
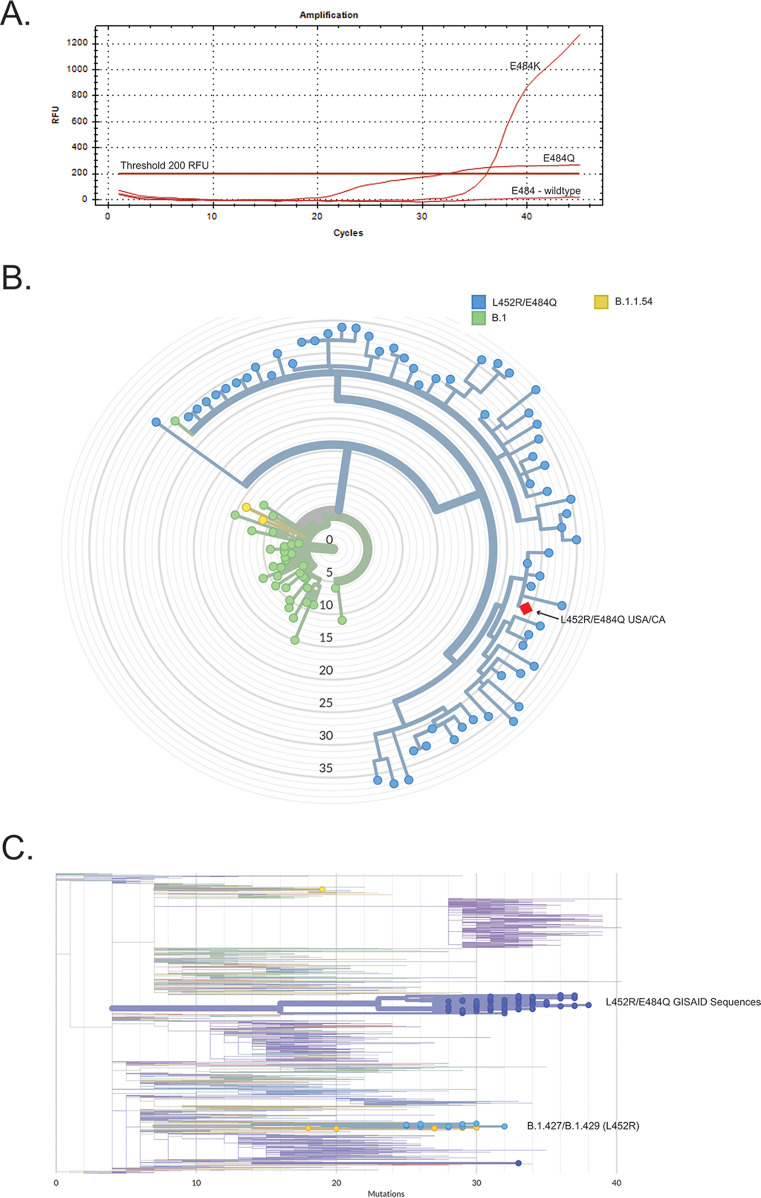
In early March, we observed SARS-CoV-2 samples strongly positive for L452R with unusually shaped, reproducible amplification curves in the E484K Cy5 channel (Fig. 1A). Samples with this pattern of reactivity were identified from five COVID-19 patients. Four of these individuals shared two unrelated household transmission events, one of which involved an individual in their 70s with known exposures in India who presented with moderate symptoms 3 days after their return flight to the United States. The other individuals had mild COVID-19 symptoms; one was infected more than 2 weeks after receiving a second dose of the Pfizer vaccine.
FIG 1.
(A) Unusual real-time, reverse transcription-PCR fluorescence amplification curve in a sample with the E484Q mutation relative to wild-type (E484E) or E484K sequence. The Cy5-labeled E484K probe (CTTGTAATGGTGTTAAAGGTTT) has a single mismatch (indicated with boldface and underlining) with the E484Q template (CTTGTAATGGTGTTCAAGGTTT) resulting in a blunted fluorescence amplification curve. RT-qPCR was performed on the Bio-Rad CFX96 with an annealing temperature of 57°C. (B) Whole-genome phylogenetic tree reveals clustering with other B lineage SARS-CoV-2 containing spike L452R and E484Q mutations. This subtree (150 genomes) was generated using the University of California Santa Cruz (UCSC) Ultrafast Sample placement on Existing tRees (UShER) tool. (C) Whole-genome phylogenetic tree highlighting sequences with the L452R mutation. This tree (1,065 genomes) demonstrates that the L452R/E484Q-containing viruses arose separately from the B.1.427/B.1.429 variants.
SARS-CoV-2 WGS was initially performed on a specimen from one of the mildly symptomatic, unvaccinated individuals. Briefly, viral genome enrichment was conducted using laboratory-developed multiplex RT-PCRs that generate multiple overlapping amplicons ∼1,200 base pairs in length. Fragment libraries were prepared using NEBNext DNA library prep reagents for Illumina (New England BioLabs, Ipswich, MA) and were sequenced on an Illumina MiSeq using single-end 150-cycle sequencing using MiSeq reagent kit v3. Genomes were assembled via a custom assembly and bioinformatics pipeline using NCBI GenBank accession no. NC_045512.2 as reference. We observed 386× mean whole-genome coverage for this sample; 50× coverage was obtained over 92.8% of the genome and 99.95% of spike. The sequence revealed a G/20A clade, B lineage virus containing eight nonsynonymous mutations in the spike protein, G142D, E154K, L452R, E484Q, D614G, P681R, Q1071H, and H1101D (Global Initiative on Sharing All Influenza Data [GISAID] database accession no. EPI_ISL_1379889). This sequence clustered with the other L452R- and E484Q-containing B lineage sequences in GISAID, comprised of 64 sequences primarily from India (59.4%, 38/64) and the United Kingdom (34.4%, 22/64) (Fig. 1B). The nearest neighbor as of 25 March 2021 was hCoV-19/England/CAMC-1322E9F/2021|EPI_ISL_1246284|2021-02-22. The earliest date of collection was a specimen from India, sequence hCoV-19/India/MH-NEERI-NGP-26041/2020|EPI_ISL_1360304|2020-12-05, collected on 12 December 2020. These sequences are distinct from the B.1.427/B.1.429 lineage (Fig. 1C). The four additional cases were subsequently confirmed by whole-genome sequencing and contain the same set of spike mutations (see Table 1 for GISAID accession numbers).
TABLE 1.
SARS-CoV-2 GISAID accession numbers for samples sequenced in this study
| Case no. | Sequence name | GISAID accession no. | Date of collectiona |
|---|---|---|---|
| 1 | hCoV-19/USA/CA-Stanford-12_S42/2021 | EPI_ISL_1379889 | 2021-03-01 |
| 2 | hCoV-19/USA/CA-Stanford-15_S02/2021 | EPI_ISL_1675223 | 2021-03-05 |
| 3 | hCoV-19/USA/CA-Stanford-15_S12/2021 | EPI_ISL_1675224 | 2021-03-12 |
| 4 | hCoV-19/USA/CA-Stanford-15_S27/2021 | EPI_ISL_1675225 | 2021-03-16 |
| 5 | hCoV-19/USA/CA-Stanford-17_S23/2021 | EPI_ISL_1701679 | 2021-03-12 |
Dates given in yr-mo-day format.
The L452R and E484Q mutations are located in the RBD, and viruses harboring these individual mutations have reduced susceptibility to monoclonal antibodies, including bamlanivimab, as well as convalescent plasma (2–8). The combined impact of these mutations on neutralization and vaccine efficacy remains to be determined. This case illustrates the critical nature of assay review and investigation and highlights the rapidity with which potential variants of concern can be transmitted worldwide.
Contributor Information
Benjamin A. Pinsky, Email: bpinsky@stanford.edu.
Michael J. Loeffelholz, Cepheid
REFERENCES
- 1.Wang H, Miller JA, Verghese M, Sibai M, Solis D, Mfuh KO, Jiang B, Iwai N, Mar M, Huang C, Yamamoto F, Sahoo MK, Zehnder J, Pinsky BA. 2021. Multiplex SARS-CoV-2 genotyping PCR for population-level variant screening and epidemiologic surveillance. medRxiv 10.1101/2021.04.20.21255480:2021.04.20.21255480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gottlieb RL, Nirula A, Chen P, Boscia J, Heller B, Morris J, Huhn G, Cardona J, Mocherla B, Stosor V, Shawa I, Kumar P, Adams AC, Van Naarden J, Custer KL, Durante M, Oakley G, Schade AE, Holzer TR, Ebert PJ, Higgs RE, Kallewaard NL, Sabo J, Patel DR, Klekotka P, Shen L, Skovronsky DM. 2021. Effect of bamlanivimab as monotherapy or in combination with etesevimab on viral load in patients with mild to moderate COVID-19: a randomized clinical trial. JAMA 325:632–644. 10.1001/jama.2021.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weisblum Y, Schmidt F, Zhang F, DaSilva J, Poston D, Lorenzi JC, Muecksch F, Rutkowska M, Hoffmann H-H, Michailidis E, Gaebler C, Agudelo M, Cho A, Wang Z, Gazumyan A, Cipolla M, Luchsinger L, Hillyer CD, Caskey M, Robbiani DF, Rice CM, Nussenzweig MC, Hatziioannou T, Bieniasz PD. 2020. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife 9:e61312. 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tortorici MA, Beltramello M, Lempp FA, Pinto D, Dang HV, Rosen LE, McCallum M, Bowen J, Minola A, Jaconi S, Zatta F, De Marco A, Guarino B, Bianchi S, Lauron EJ, Tucker H, Zhou J, Peter A, Havenar-Daughton C, Wojcechowskyj JA, Case JB, Chen RE, Kaiser H, Montiel-Ruiz M, Meury M, Czudnochowski N, Spreafico R, Dillen J, Ng C, Sprugasci N, Culap K, Benigni F, Abdelnabi R, Foo S-YC, Schmid MA, Cameroni E, Riva A, Gabrieli A, Galli M, Pizzuto MS, Neyts J, Diamond MS, Virgin HW, Snell G, Corti D, Fink K, Veesler D. 2020. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science 370:950–957. 10.1126/science.abe3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greaney AJ, Loes AN, Crawford KHD, Starr TN, Malone KD, Chu HY, Bloom JD. 2021. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe 29:463–476.e6. 10.1016/j.chom.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang L, Zhou T, Zhang Y, Yang ES, Schramm CA, Shi W, Pegu A, Oloninyi OK, Ransier A, Darko S, Narpala SR, Hatcher C, Martinez DR, Tsybovsky Y, Phung E, Abiona OM, Cale EM, Chang LA, Corbett KS, DiPiazza AT, Gordon IJ, Leung K, Liu T, Mason RD, Nazzari A, Novik L, Olia AS, Doria-Rose NA, Stephens T, Stringham CD, Talana CA, Teng I-T, Wagner D, Widge AT, Zhang B, Roederer M, Ledgerwood JE, Ruckwardt TJ, Gaudinski MR, Baric RS, Graham BS, McDermott AB, Douek DC, Kwong PD, Mascola JR, Sullivan NJ, Misasi J. 2021. Antibodies with potent and broad neutralizing activity against antigenically diverse and highly transmissible SARS-CoV-2 variants. bioRxiv 10.1101/2021.02.25.432969:2021.02.25.432969. [DOI] [PMC free article] [PubMed]
- 7.Copin R, Baum A, Wloga E, Pascal KE, Giordano S, Fulton BO, Zhou A, Negron N, Lanza K, Chan N, Coppola A, Chiu J, Ni M, Wei Y, Atwal GS, Hernandez AR, Saotome K, Zhou Y, Franklin MC, Hoopper AT, McCarthy S, Hamon S, Hamilton JD, Staples HM, Alfson K, Carrion R, Ali S, Norton T, Somersan-Karakaya S, Sivapalasingam S, Herman GA, Weinreich DM, Lipsich L, Stahl N, Murphy AJ, Yancopoulos GD, Kyratsous CA. 2021. In vitro and in vivo preclinical studies predict REGEN-COV protection against emergence of viral escape in humans. bioRxiv 10.1101/2021.03.10.434834:2021.03.10.434834. [DOI]
- 8.Li Q, Wu J, Nie J, Zhang L, Hao H, Liu S, Zhao C, Zhang Q, Liu H, Nie L, Qin H, Wang M, Lu Q, Li X, Sun Q, Liu J, Zhang L, Li X, Huang W, Wang Y. 2020. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell 182:1284–1294.e9. 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]