ABSTRACT
Accurate SARS-CoV-2 serological assays are critical for COVID-19 serosurveillance. However, previous studies have indicated possible cross-reactivity of these assays, including in areas where malaria is endemic. We tested 213 well-characterized prepandemic samples from Nigeria using two SARS-CoV-2 serological assays, Abbott Architect IgG and Euroimmun NCP IgG assay, both targeting SARS-CoV-2 nucleocapsid protein. To assess antibody binding strength, an avidity assay was performed on these samples and on plasma from SARS-CoV-2 PCR-positive persons. Thirteen (6.1%) of 212 samples run on the Abbott assay and 38 (17.8%) of 213 run on the Euroimmun assay were positive. Anti-Plasmodium IgG levels were significantly higher among false positives for both Abbott and Euroimmun; no association was found with active Plasmodium falciparum infection. An avidity assay using various concentrations of urea wash in the Euroimmun assay reduced loosely bound IgG: of 37 positive/borderline prepandemic samples, 46%, 86%, 89%, and 97% became negative using 2 M, 4 M, 5 M, and 8 M urea washes, respectively. The wash slightly reduced avidity of antibodies from SARS-CoV-2 patients within 28 days of PCR confirmation; thereafter, avidity increased for all urea concentrations except 8 M. This validation found moderate to substantial cross-reactivity on two SARS-CoV-2 serological assays using samples from a setting where malaria is endemic. A simple urea wash appeared to alleviate issues of cross-reactivity.
KEYWORDS: SARS-CoV-2, cross-reactivity, malaria, serology
INTRODUCTION
The COVID-19 pandemic has led to more than 100 million confirmed cases and more than 2.2 million deaths from COVID-19 globally as of early February 2021 (1). However, with mild or asymptomatic disease presentations (2) and access to SARS-CoV-2 molecular and antigen testing still limited in many places, cumulative infections may be underestimated. Serological assays that detect antibodies can be useful for understanding the true extent of SARS-CoV-2 exposure in a population (3, 4). A multitude of rapid and laboratory-based SARS-CoV-2 serological assays have been developed since the beginning of the pandemic: as of early February 2021, 65 SARS-CoV-2 serological tests have received emergency use authorization (EUA) from the United States Food and Drug Administration (5).
In addition to manufacturer validation results, results from independent validations of SARS-CoV-2 immunoassay performance are becoming increasingly available (6–9). An important concern in development of SARS-CoV-2 serologic assays is to ensure that measured antibody responses are specific to SARS-CoV-2 infection in the human host. High specificity becomes even more relevant when seropositivity levels are low in a population (10–12), as even small declines in test specificity can lead to large proportions of false-positive serological tests.
Most independent validations of SARS-CoV-2 serological assays have used samples from Chinese, European, or North American COVID-19 cases and negative (typically pre-2020) controls (7, 13–15). A concern for certain geographical areas is cross-reactivity to endemic pathogens that were not included in validation studies. Previous serological studies for Zika (16), dengue (17), and HIV (18) have shown false-positive results from persons exposed to malaria parasites, although the mechanisms for these false-positive test results have not been fully elucidated.
A recent study found false-positive SARS-CoV-2 serology tests with four commercially available IgG enzyme-linked immunosorbent assay (ELISA) kits in samples from Nigeria and Ghana but not in samples from Madagascar, Germany, Columbia, or Lao People's Democratic Republic (19). Data from Benin showed that approximately 25% of 60 samples from patients with acute malaria in 2019 had positive SARS-CoV-2 serological results (20).
An urgent need exists for specific SARS-CoV-2 serologic assays appropriate for a wide variety of settings; the accuracy of such assays in the context of other endemic infectious diseases needs to be carefully assessed. Here, we present results from laboratory testing of two commercially available SARS-CoV-2 serological assays. These assays were performed on a well-characterized panel of Nigerian samples collected in 2018 as well as on samples from SARS-CoV-2 PCR-positive patients from 2020. The prevalence of false-positive serological test results was investigated to determine any association with malaria infection and antibody levels. Strength of IgG binding from false-positive and true-positive test results was examined.
MATERIALS AND METHODS
Specimens tested.
Deidentified samples from Nigeria’s national biorepository at the National Reference Laboratory (NRL) that were initially collected as part of the 2018 Nigeria HIV/AIDS Indicator and Impact Survey (NAIIS) (21) were tested for SARS-CoV-2 antibodies. Whole blood was collected from participants and, for those consenting, stored as plasma at NRL at −80°C. Through the Nigeria Multidisease Serologic Surveillance of Stored Specimens (NMS4) project (22), these samples had been tested for the presence of malaria antigens and IgG against a variety of pathogens endemic to Nigeria (22, 23). The multiplex bead assay (MBA) for IgG against a panel of infectious and vaccine-preventable diseases was performed on the MAGPIX platform as described previously (23–25), with a serum dilution of approximately 1:400. The multiplex malaria antigen detection assay was also performed on the MAGPIX platform as described previously (26, 27) at a whole-blood dilution of 1:40. All assays were performed at the NRL (Nigeria Centre for Disease Control, NCDC, Abuja, Nigeria).
For SARS-CoV-2 serology, we sampled 107 children <15 years old and 106 adults >15 years old (Table 1). Approximately half of the samples were intentionally selected based on histidine-rich protein 2 (HRP2) antigen positivity, indicating current or recent infection with Plasmodium falciparum. Of HRP2 antigen positives, one-third had low-positive, one-third medium-positive, and one-third high-positive malaria antigen values based on antigenemia tertiles.
TABLE 1.
Characteristics of individuals and samples from the 2018 Nigeria HIV/AIDS Indicator and Impact Survey (n = 213)
| Parameter | Value | % |
|---|---|---|
| Age (yr) | ||
| Median [IQR] | 14 [10, 23] | |
| <5 | 5 | 2.4 |
| 5–9 | 42 | 19.7 |
| 10–14 | 60 | 28.2 |
| 15–19 | 30 | 14.1 |
| 20–24 | 24 | 11.3 |
| 25–29 | 14 | 6.6 |
| 30–34 | 11 | 5.2 |
| 35–39 | 10 | 4.7 |
| 40–44 | 15 | 7.0 |
| 45–60 | 2 | 0.9 |
| Sex | ||
| Female | 127 | 60.1 |
| Malaria | ||
| Positivea | 107 | 50.2 |
Based on HRP2 antigen positivity from a bead-based immunoassay.
We also tested plasma samples from 32 SARS-CoV-2 PCR-positive patients in Lagos that had been collected at various time points since PCR confirmation.
Laboratory methods for SARS-CoV-2 serological assays.
Two commercially available SARS-CoV-2 IgG assays were assessed using the prepandemic samples. The Euroimmun anti-SARS-CoV-2 NCP ELISA (IgG [Euroimmun Medizinische Labordiagnostika, Lübeck, Germany]) assay detects IgG antibodies against SARS-CoV-2 nucleocapsid (NCP) protein. Using the automated Abbott Architect plus i2000sr analyzer (Abbott, IL, USA) and SARS-CoV-2 IgG kit is a method for detecting IgG antibodies against the SARS-CoV-2 NCP. Both tests were performed according to the manufacturer’s recommendations and using an avidity assay with a urea wash (see the supplemental material).
Samples were initially run at the Center for Human Virology and Genomics (CHVG; at the Nigerian Institute of Medical Research [NIMR] in Lagos, Nigeria). To examine interlaboratory variations, the Euroimmun and Abbott tests were also run on additional sample aliquots at the NRL.
Statistical analyses.
Log-transformed antibody and malaria antigen values were compared among true-negative and false-positive prepandemic samples using the Wilcoxon rank-sum test for nonnormally distributed data. Given the exploratory nature of the analyses and the relatively large number of comparisons (n = 78), we used the Benjamini-Hochberg adjustment and a false discovery rate of 10% to define which comparisons were statistically significant (28). Agreement among test results from the two laboratories was measured with kappa statistics for categorical test outcomes (e.g., positive, negative, borderline, and signed-rank nonparametric tests to compare optical density ratios on the same test samples). Significant differences among avidity index (AI) means were determined by two-sided t tests using unequal variances. Sensitivity and specificity confidence limits were calculated using binomial exact formulas. Stata 16.0 (College Station, Texas) and Microsoft Excel were used for analyses.
Ethical approval.
Written informed consent for future testing of collected blood samples was provided by participants during NAIIS data collection. Written consent was obtained from SARS-CoV-2-positive patients for plasma collection and storage for future testing. This cross-reactivity evaluation was approved by the National Health Research Ethics Committee of Nigeria (NHREC) (protocol number NHREC/01/01/2007-31/08/2020) and by the U.S. Centers for Disease Control and Prevention.
RESULTS
Of 213 prepandemic samples from the 2018 NAIIS, the median age was 14 years (interquartile range [IQR], 10 years, 23 years), and 127 (60.1%) were from females (Table 1). In total, 107 (50.2%) were positive for P. falciparum HRP2 antigen, indicating current/recent malaria infection, 139 (65.3% were seropositive for glurp, 162 (76.1%) were seropositive for pfama1, and 193 (90.6%) were seropositive for pfmsp1. All 213 samples were tested with the Euroimmun assay and 212 with the Abbott assay (one sample had insufficient volume for the Abbott assay). Twenty Euroimmun results were borderline after the first run and were repeated. Two Abbott tests had invalid results after the first run and were repeated.
Test specificity.
For the prepandemic samples, the Abbott test had two (0.9%) invalid results after two test runs, 197 (92.9%) negative, and 13 (6.1%) positive results (Table 2). The Euroimmun had seven (3.3%) borderline results after repeating, 168 (78.9%) negative results, and 38 (17.8%) positive results. All but two of the 13 positive Abbott results were also positive on Euroimmun, while the remaining two were negative (Table 3). Excluding invalid and borderline results, specificity was 81.6% (95% confidence interval [CI], 75.6%, 86.3%) for Euroimmun and 93.8% for Abbott (95% CI, 89.6%, 96.4%) assays. Using a sequential algorithm (both tests negative), specificity was 94.6% (95% CI, 90.5%, 97.0%).
TABLE 2.
Results of two SARS-CoV-2 serological assays on selected samples from the 2018 Nigeria HIV/AIDS Indicator and Impact Survey
| Result | Value [no. (%)] for: |
|
|---|---|---|
| Abbott (n = 212a) | Euroimmun (n = 213) | |
| Invalidb | 2 (0.9) | NA |
| Borderlinec | NAd | 7 (3.3) |
| Negative | 197 (92.9) | 168 (78.9) |
| Positive | 13 (6.1) | 38 (17.8) |
One sample had insufficient volume to be tested with the Abbott assay.
Two samples had invalid results after two runs with the Abbott assay.
Twenty samples were borderline initially on Euroimmun, and seven remained borderline after a repeat test.
NA, not applicable.
TABLE 3.
Combinations of Abbott and Euroimmun SARS-CoV-2 assay results on selected samples from the 2018 Nigeria HIV/AIDS Indicator and Impact Survey
| Abbott | Euroimmun | N | % |
|---|---|---|---|
| − | − | 165 | 77.8 |
| − | + | 25 | 11.8 |
| + | + | 11 | 5.2 |
| − | Borderlinea | 7 | 3.3 |
| + | − | 2 | 0.9 |
| Invalidb | + | 1 | 0.5 |
| Invalidb | − | 1 | 0.5 |
| Total | 212 | 100 | |
Borderline assay result defined as a second borderline response after a first borderline value according to Euroimmun.
Invalid assay result defined by Abbott analyzer.
Interlaboratory result agreement.
For prepandemic samples, there was moderate to strong agreement (29) between the Euroimmun and the Abbott assay results from tests run at NIMR and at NRL, with kappa statistics of 0.6220 for Euroimmun (0.7655) if borderline results were excluded and 0.8621 for Abbott (see Tables S1 and S2 in the supplemental material). Tests run at NIMR had, on average, higher optical density (OD) ratios for both Euroimmun and Abbott than tests run at NRL (P value for sign-rank test, <0.001) for both.
Relationships between positive SARS-CoV-2 serological tests and levels of malaria and other antibodies in prepandemic samples.
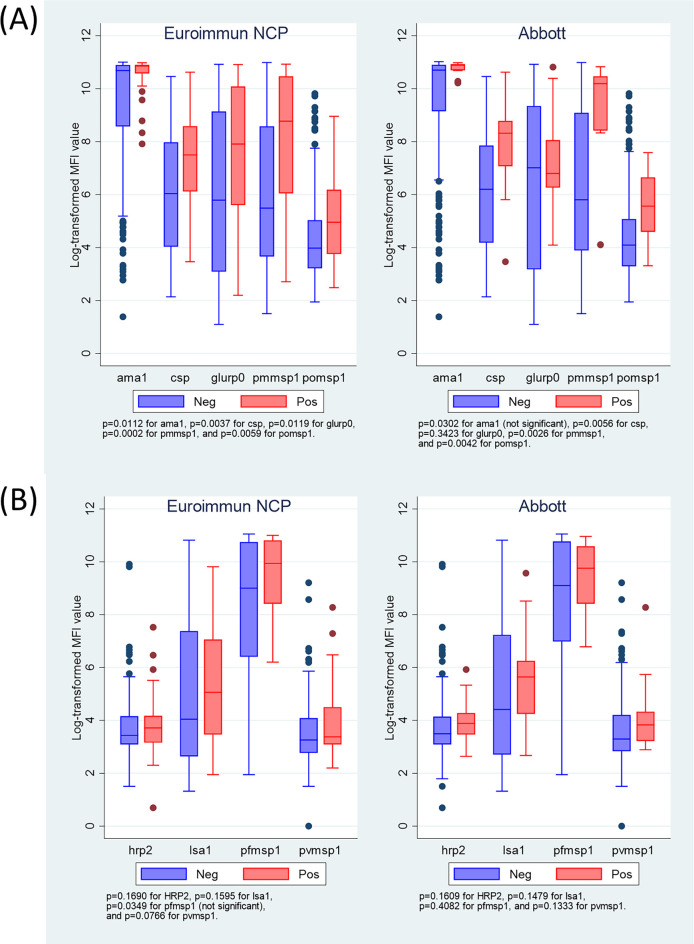
Levels of malaria antibodies were significantly higher for prepandemic samples with positive SARS-CoV-2 antibody test results for five of nine malaria IgG targets: PfCSP, glurp (Euroimmun only), Pfama1 (Euroimmun only), pmmsp1, and pomsp-1 (Fig. 1 and Table S3). There was no significant association with pfmsp1, pvmsp1, hrp2, or lsa1 malaria IgG antibodies. In assessing active malaria infection, no significant association was observed with the presence or levels of any of the four malaria antigen targets (Fig. S1).
FIG 1.
Levels of anti-Plasmodium IgG antibodies by SARS-CoV-2 antibody test result for Euroimmun (n = 168 for negative, n = 38 for positive) and Abbott (n = 197 negative, n = 13 positive) for prepandemic samples (2018 Nigeria HIV/AIDS Indicator and Impact Survey). Plots display five anti-malaria IgG antibodies significantly associated with SARS-CoV-2 IgG positivity (A) and four not significantly associated (B). Boxes shows interquartile range (IQR), lines displaying medians, and whiskers extending 1.5× above and below IQR. Markers display values outside 1.5× IQR. NCP, nucleocapsid protein; MFI, median fluorescent intensity.
For either Abbott or Euroimmun, but not for both, positive SARS-CoV-2 serological results had significantly higher antibodies for several other pathogens included in the NMS4 multiplex bead-based assay, including lymphatic filariasis (Abbott assay), onchocerciasis (Abbott), syphilis/yaws (Euroimmun), cysticercosis (Abbott), and taeniasis (Abbott) (Table S3).
Avidity assay for prepandemic samples with SARS-CoV-2 IgG assay.
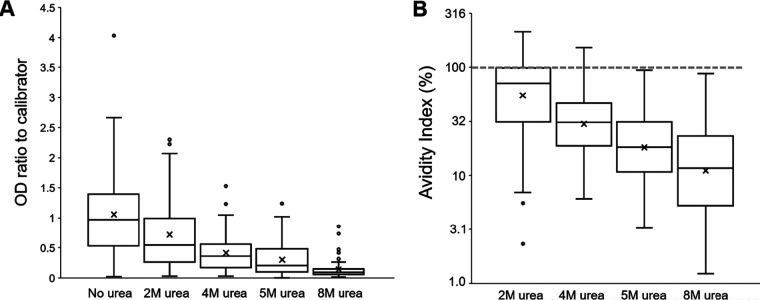
Forty prepandemic samples (32 positive, 5 borderline, and 3 negative by Euroimmun assay) were run using four concentrations of urea wash (Fig. 2A). The three negative samples remained negative, and the five borderline samples became negative at all four urea concentrations. Of the 32 positive samples, 11, 3, 1, and 0 remained positive and 9, 2, 3, and 1 became borderline using the 2 M, 4 M, 5 M, and 8 M washes, respectively (Fig. S2). Of these initial 32 positives, 12 (38%), 27 (84%), 28 (88%), and 31 (97%) became negative using the 2 M, 4 M, 5 M, and 8 M washes, respectively. For all prepandemic samples, the OD ratio to calibrator (Fig. 2A and Fig. S3A) and avidity index (AI) (Fig. 2B) steadily decreased with increasing urea concentrations. Although the 2 M urea wash had only a slight effect on the amount of retained anti-NCP IgG (median AI, 71.5%), the more stringent 4 M (median AI, 31.0%), 5 M (median AI, 18.1), and 8 M (median AI, 11.7%) removed the vast majority of cross-binding IgG antibodies in prepandemic samples.
FIG 2.
Test results with or without urea-based avidity assays at 2 M, 4 M, 5 M, and 8 M with the Euroimmun NCP assay for 40 prepandemic samples (2018 Nigeria HIV/AIDS Indicator and Impact Survey). The panel includes samples with positive (n = 32), borderline (n = 5), and negative (n = 3) calls. (A) Boxplots display OD-to-calibrator ratios for all samples at each wash. (B) Avidity index for all samples at different molarities of urea wash. Gray hash line displays an avidity index of 100%, which would represent no loss of IgG signal. For panels A and B, boxes show interquartile range (IQR), lines display medians, X symbol show means, and whiskers extend 1.5× above and below IQR. Markers display values outside 1.5× IQR.
Avidity assay for samples from SARS-CoV-2 PCR-positive persons with SARS-CoV-2 IgG assay.
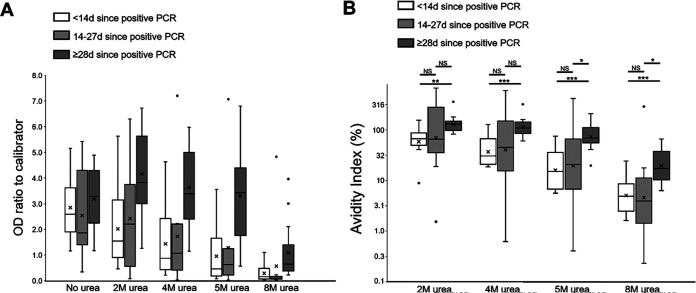
Using 32 samples from patients testing positive for SARS-CoV-2 by PCR, OD ratios decreased at higher urea concentrations, but this was dependent on time since PCR positivity (Fig. 3 and Fig. S3B). Persons with samples collected <28 days after a PCR positive test showed a decrease in OD ratio with increasing urea concentrations (Fig. 3A). However, samples collected ≥28 days after a positive PCR largely retained the OD ratio through the 2, 4, and 5 M urea washes before substantially dropping off at the 8 M wash (Fig. 3A). This was reflected in the strength of IgG binding, with significant differences in AIs for all urea wash concentrations for samples of <14 days versus ≥28 days post-PCR positivity (Fig. 3B). Using the more stringent 5 M and 8 M urea washes, samples collected 14 to 27 days post-PCR positivity had AIs significantly lower than those collected at ≥28 days. For samples collected ≥28 days post-PCR positivity, median AIs were largely similar at urea concentrations of ≤5 M (2 M, 129.6%; 4 M, 109.2%; 5 M, 89.9%; 8 M, 21.2%) but dropped quickly for samples collected 14 to 28 days post-PCR positivity (2 M, 65.4%; 4 M, 38.4%; 5 M, 20.6%; 8 M, 5.1%). Positive associations were observed between time since PCR positivity and AI at all urea concentrations, but correlations were not strong (Fig. S4).
FIG 3.
Test results with or without urea-based avidity assays at 2 M, 4 M, 5 M, and 8 M with the Euroimmun NCP assay for 32 samples from persons with previous SARS-CoV-2-positive PCR. Each plot has three categories indicating persons testing SARS-CoV-2 PCR positive less than 14 days prior to sample collection, between 14 and 27 days prior, and 28 days or greater. (A) Boxplots display OD/calibrator ratios for all samples at each wash. (B) Avidity index for all samples at each molarity of urea wash, with statistically significant differences indicated. NS, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001. For panels A and B, boxes show interquartile ranges (IQR), lines display medians, X symbol show means, and whiskers extend 1.5× above and below IQR. Markers display values outside 1.5× IQR.
Level of anti-NCP IgG versus strength of binding.
For both the prepandemic and the SARS-CoV-2 PCR-positive sample sets, a general negative trend was observed between total amount of anti-NCP IgG detected (by OD ratio to calibrator) and AI for different urea wash concentrations (Fig. S5), but these trends showed high variability. The OD/calibrator ratio was significantly higher for all urea washes for SARS-CoV-2 PCR positives versus prepandemic samples; differences in AIs between these sample sets were only seen for 4 and 5 M urea washes (Fig. S6A and B).
Sensitivity and specificity of Euroimmun NCP assay with different concentrations of urea.
Combining the prepandemic and the SARS-CoV-2 PCR-positive panels to examine effects of various concentrations of a urea wash step on test performance, sensitivity decreased with increasing concentrations of urea (to 12.5%) at 8 M, while specificity increased (to 100%) at 8 M (Table 4). Samples collected ≥14 days post-PCR did not show as sharp declines in sensitivity; samples collected ≥28 days post-PCR retained 100% sensitivity up to 5 M, at which point sensitivity dropped to 83.3% (Table 4).
TABLE 4.
Sensitivity and specificity estimates when including urea wash for Euroimmun assayc
| Wash | Sensitivity, % (95% CI) |
Specificity, % (95% CI) (n = 207b) | ||
|---|---|---|---|---|
| All SARS-CoV-2+ (n = 32) | ≥14 days SARS-CoV-2+ (n = 22) | ≥28 days SARS-CoV-2+ (n = 12) | ||
| No wash | 96.9 (89.1, 100a) | 95.5 (72.2, 99.9) | 100.0 (73.5, 100a) | 84.1 (78.4, 88.9) |
| 2 M urea | 78.1 (60.0, 90.7) | 81.8 (59.7, 94.8) | 100.0 (73.5, 100a) | 94.4 (90.3, 97.2) |
| 4 M urea | 62.5 (43.7, 78.9) | 72.7 (49.8, 89.3) | 100.0 (73.5, 100a) | 98.5 (95.7, 99.7) |
| 5 M urea | 53.1 (34.7, 70.9) | 63.6 (40.7, 82.8) | 83.3 (51.6, 97.9) | 99.5 (97.3, 100.0) |
| 8 M urea | 12.5 (3.5, 29.0) | 18.2 (5.2, 40.3) | 33.3 (9.9, 65.1) | 100.0 (98.2, 100*) |
One-sided, 97.5% confidence interval.
Those false-positive and borderline samples not able to run with all urea wash concentrations were subtracted from the numerator and denominator; samples with persistent borderline results were excluded from analysis.
Sensitivity was calculated from the SARS-CoV-2 PCR+ panel, and specificity was calculated from prepandemic samples.
DISCUSSION
Our results from this setting where malaria is highly endemic showed a high level of false-positive results with the Euroimmun NCP SARS-CoV-2 serological assay (17.8%) and lower levels with the Abbott Architect assay (6.1%), both yielding specificity levels below the WHO-recommended 97% for SARS-CoV-2 serological assays (30). Although active malaria infection was not associated with reduced specificity of these two assays, levels of anti-Plasmodium IgGs against multiple malaria antigen targets were significantly higher in false-positive samples than true negatives. The IgGs leading to false-positive serological results were found to be weakly bound to the SARS-CoV-2 antigens, and most were removed with low concentrations of the protein denaturant urea. No significant correlation was seen between the level of cross-binding IgG and the strength of IgG binding, suggesting that these IgGs that are binding SARS-CoV-2 antigens are not due to a true affinity maturation process. Importantly, a relatively simple urea wash step during the Euroimmun assay improved assay specificity.
The 93.8% specificity we found from this Nigerian sample set with Abbott Architect is lower than estimates from previous evaluations, including 99.6% reported by the manufacturer using a panel of pre-COVID-19 samples and samples from patients with other respiratory illnesses (total n = 1,070) (31) and 99.6% by an independent evaluation using 1,099 prepandemic samples (32). The specificity of 81.6% on the Euroimmun NCP assay we found was substantially lower than the manufacturer-reported specificity of 99.8% using pre-COVID-19 panels from Germany, the United States, and China, including some samples positive for influenza and Epstein-Barr virus and rheumatoid factor-positive samples (n = 1,140) (33).
Our study is the first to demonstrate an association between SARS-CoV-2 antibody cross-reactivity and existing malaria antibodies. Previous specificity experiments with SARS-CoV-2 serological assays have typically included samples from non-malaria-endemic areas positive for autoimmune diseases, other human coronaviruses, Epstein-Barr virus, cytomegalovirus, and other respiratory pathogens, and most have shown low to no cross-reactivity (13, 34, 35).
However, a recent study found much higher levels of cross-reactivity to SARS-CoV-2 (primarily to the NCP among prepandemic samples from Tanzania and Zambia compared to those from the United States); given that the cross-reactive samples also showed strong reactivity against other human coronaviruses, the authors concluded that exposure to other coronaviruses may induce cross-reactive antibodies against SARS-CoV-2 in sub-Saharan Africa (36). However, seasonal coronaviruses are not unique to the African continent, and the lower specificity of SARS-CoV-2 serological assays with African samples may have been due to other factors as well.
Findings from previous studies using samples from Benin, Nigeria, and Ghana have led to speculation that malaria contributes to cross-binding antibodies or other humoral factors (19, 20). An additional study in the country of Gabon, where malaria is highly endemic, found that 32 of 135 (23.7%) samples from 2014 were positive for SARS-CoV-2 antibodies using an NCP antigen serological assay, although the authors acknowledged that the cause of the cross-reactivity could not be isolated with certainty (37). Our current study found anti-Plasmodium IgG levels to be significantly higher in samples with false-positive SARS-CoV-2 results than in those with negative results. This was found for 5 of 9 Plasmodium antigen targets in our malaria panel and encompassed three malaria parasite species: P. falciparum, P. malariae, and P. ovale. This significant association held true for 3 of 5 of these targets (PfCSP, PmMSP1, and PoMSP1 for both Euroimmun NCP and Abbot assays). Although levels of several NTD antibodies were higher in the samples with false-positive Abbott SARS-CoV-2 results, levels of only one NTD antibody, to syphilis/yaws, were significantly higher in samples with Euroimmun NCP false-positive results, and no NTD antibodies were higher for both tests; thus, NTDs is a less likely contributor to SARS-CoV-2 cross-reactivity than malaria. In addition, previous immunological studies support that malaria antibodies cross-reactive with other pathogens could arise from polyclonal and atypical B cell populations promoted during malaria infection (38, 39).
Our current study evaluated the strength of IgG cross-binding to SARS-CoV-2 antigens that elicit these false-positive results. Using relatively low concentrations of 2 M, 4 M, and 5 M the protein denaturant urea (typically 6 M or 8 M) removes loosely bound IgG (40, 41), and most borderline or false-positive prepandemic samples were recategorized as negative. A more stringent 8 M concentration was found to also substantially reduce binding of actual SARS-CoV-2 antibodies, leading to many false negatives. Given sensitivity-specificity trade-offs with increasing urea concentration, the 4 M wash appeared to yield promising results, especially for samples taken ≥28 days post-PCR confirmation. The finding of strong IgG binding 28 days postexposure suggests that reliable results can be obtained for population serostudies for SARS-CoV-2 IgG that do not enroll many individuals with recent COVID-19. These findings are consistent with previous studies using avidity assays for SARS-CoV-2 serology, with clear increases in IgG avidity as time from exposure increases (42, 43).
An avidity assay is not specific for malaria IgG; any weak binding of IgG would be removed by this process. Regardless of the exact mechanisms contributing to cross-reactivity on SARS-CoV-2 serological assays, the relatively simple urea wash step holds the potential to mitigate this problem of false positives on NCP-based assays. This might be especially important for samples from sub-Saharan Africa or other areas where malaria is endemic.
A major limitation of our study is that the prepandemic samples had not been tested for the presence of antibodies to other human coronaviruses. Evidence suggesting some cross-reactivity of SARS-CoV-2 serological tests with malaria was found, but it cannot be ruled out that the primary cause of cross-reactivity is exposure to other human coronaviruses, which may be more prevalent in sub-Saharan Africa versus other parts of the world. Another limitation of our study was the modest interlaboratory agreement for the Euroimmun test results, possibly due to different laboratory equipment for this open-system assay. It is important to note that the variability was primarily with low-positive, borderline, or negative samples that have lower ODs and, thus, are more sensitive to change with minor OD variation; additionally, the Euroimmun result is a ratio of two OD values and, therefore, has more potential for variability than a raw signal. The agreement between the Abbott test results was strong, even though NIMR used the Abbott Architect Plus i2000sr analyzer while NRL used the Abbott Architect i1000sr analyzer; chemiluminescent assays are known to perform more reliably than ELISA spectrophotometers.
Our study indicated substantial cross-reactivity to two commercial, SARS-CoV-2 IgG serological assays targeting the NCP antigen using Nigerian plasma samples from 2018. Cross-reactive samples had significantly higher levels of malaria antibodies, although it is unclear whether this is directly responsible for false-positive results. Use of a simple urea wash appeared to substantially reduce cross-reactivity and should be considered when testing samples from regions where malaria is endemic using SARS-CoV-2 ELISA platforms.
ACKNOWLEDGMENTS
We acknowledge the contribution of valuable specimens by the NAIIS Group, including the Republic of Nigeria Federal Ministry of Health (FMoH), National Agency for the Control of AIDS (NACA), National Population Commission (NPopC), National Bureau of Statistics (NBS), U.S. Centers for Disease Control and Prevention (CDC), The Global Funds to Fight AIDS, Tuberculosis, and Malaria, University of Maryland Baltimore (UMB), ICF International, African Field Epidemiology Network (AFENET), University of Washington (UW), the Joint United Nations Program on HIV and AIDS (UNAIDS), World Health Organization (WHO), and United Nations Children’s Fund (UNICEF). NAIIS was supported by the President’s Emergency Plan for AIDS Relief (PEPFAR) through the CDC under the cooperative agreement U2GGH002108 to UMB and by the Global Funds to Fight AIDS, Tuberculosis, and Malaria through NACA, under the contract number NGA-H-NACA to UMB.
The findings and conclusions of this report are those of the authors and do not necessarily represent the official positions of the NAIIS Group or the Centers for Disease Control and Prevention. Use of trade names and commercial sources is for identification only and does not constitute endorsement by the Centers for Disease Control and Prevention or the U.S. Department of Health and Human Services.
We have no conflicts of interest to declare.
This work was supported by the Nigerian Institute of Medical Research and by the Centers for Disease Control and Prevention. Funding support for Y.H., M.X.R., and I.A. comes from the Bill and Melinda Gates Foundation and the National Institute for Health Research (RP-PG-0217-20009).
Footnotes
Supplemental material is available online only.
Contributor Information
Laura C. Steinhardt, Email: LSteinhardt@cdc.gov.
Michael J. Loeffelholz, Cepheid
REFERENCES
- 1.Johns Hopkins University Center for Systems Science and Engineering. 2020. COVID-19 Dashboard on Johns Hopkins University of Medicine. https://coronavirus.jhu.edu/map.html. Accessed 11 November 2020.
- 2.Meyerowitz EA, Richterman A, Bogoch II, Low N, Cevik M. 7 December 2020. Towards an accurate and systematic characterisation of persistently asymptomatic infection with SARS-CoV-2. Lancet Infect Dis 10.1016/S1473-3099(2030837-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization. 2020. Population-based age-stratified seroepidemiological investigation protocol for coronavirus 2019 (COVID-19 infection. WHO, Geneva, Switzerland. [Google Scholar]
- 4.Rostami A, Sepidarkish M, Leeflang MMG, Riahi SM, Nourollahpour Shiadeh M, Esfandyari S, Mokdad AH, Hotez PJ, Gasser RB. 24 October 2020. SARS-CoV-2 seroprevalence worldwide: a systematic review and meta-analysis. Clin Microbiol Infect 10.1016/j.cmi.2020.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.U.S. Food & Drug Administration. 2020. EUA authorized serology test performance. https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/eua-authorized-serology-test-performance. Accessed 11 January 2020.
- 6.Bond K, Nicholson S, Lim SM, Karapanagiotidis T, Williams E, Johnson D, Hoang T, Sia C, Purcell D, Mordant F, Lewin SR, Catton M, Subbarao K, Howden BP, Williamson DA. 2020. Evaluation of serological tests for SARS-CoV-2: implications for serology testing in a low-prevalence setting. J Infect Dis 222:1280–1288. 10.1093/infdis/jiaa467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lassaunière R, Frische A, Harboe ZB, Nielsen AC, Fomsgaard A, Krogfelt KA, Jørgensen CS. 2020. Evaluation of nine commercial SARS-CoV-2 immunoassays. medRxiv 10.1101/2020.04.09.20056325. [DOI] [Google Scholar]
- 8.Mairesse A, Favresse J, Eucher C, Elsen M, Tre-Hardy M, Haventith C, Gruson D, Dogne JM, Douxfils J, Gobbels P. 2020. High clinical performance and quantitative assessment of antibody kinetics using a dual recognition assay for the detection of SARS-CoV-2 IgM and IgG antibodies. Clin Biochem 10.1016/j.clinbiochem.2020.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tre-Hardy M, Wilmet A, Beukinga I, Favresse J, Dogne JM, Douxfils J, Blairon L. 2021. Analytical and clinical validation of an ELISA for specific SARS-CoV-2 IgG, IgA, and IgM antibodies. J Med Virol 93:803–811. 10.1002/jmv.26303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Havers FP, Reed C, Lim T, Montgomery JM, Klena JD, Hall AJ, Fry AM, Cannon DL, Chiang CF, Gibbons A, Krapiunaya I, Morales-Betoulle M, Roguski K, Rasheed MAU, Freeman B, Lester S, Mills L, Carroll DS, Owen SM, Johnson JA, Semenova V, Blackmore C, Blog D, Chai SJ, Dunn A, Hand J, Jain S, Lindquist S, Lynfield R, Pritchard S, Sokol T, Sosa L, Turabelidze G, Watkins SM, Wiesman J, Williams RW, Yendell S, Schiffer J, Thornburg NJ. 2020. Seroprevalence of Antibodies to SARS-CoV-2 in 10 Sites in the United States, March 23-May 12, 2020. JAMA Intern Med 180:1576. 10.1001/jamainternmed.2020.4130. [DOI] [PubMed] [Google Scholar]
- 11.Pollan M, Perez-Gomez B, Pastor-Barriuso R, Oteo J, Hernan MA, Perez-Olmeda M, Sanmartin JL, Fernandez-Garcia A, Cruz I, Fernandez de Larrea N, Molina M, Rodriguez-Cabrera F, Martin M, Merino-Amador P, Leon Paniagua J, Munoz-Montalvo JF, Blanco F, Yotti R, Group E-CS . 2020. Prevalence of SARS-CoV-2 in Spain (ENE-COVID: a nationwide, population-based seroepidemiological study. Lancet 10.1016/S0140-6736(2031483-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stringhini S, Wisniak A, Piumatti G, Azman AS, Lauer SA, Baysson H, De Ridder D, Petrovic D, Schrempft S, Marcus K, Yerly S, Arm Vernez I, Keiser O, Hurst S, Posfay-Barbe KM, Trono D, Pittet D, Getaz L, Chappuis F, Eckerle I, Vuilleumier N, Meyer B, Flahault A, Kaiser L, Guessous I. 2020. Seroprevalence of anti-SARS-CoV-2 IgG antibodies in Geneva, Switzerland (SEROCoV-POP: a population-based study. Lancet 396:313–319. 10.1016/S0140-6736(2031304-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beavis KG, Matushek SM, Abeleda APF, Bethel C, Hunt C, Gillen S, Moran A, Tesic V. 2020. Evaluation of the EUROIMMUN anti-SARS-CoV-2 ELISA assay for detection of IgA and IgG antibodies. J Clin Virol 129:104468. 10.1016/j.jcv.2020.104468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ejazi SA, Ghosh S, Ali N. 2021. Antibody detection assays for COVID-19 diagnosis: an early overview. Immunol Cell Biol 99:21–33. 10.1111/imcb.12397. [DOI] [PubMed] [Google Scholar]
- 15.Whitman JD, Hiatt J, Mowery CT, Shy BR, Yu R, Yamamoto TN, Rathore U, Goldgof GM, Whitty C, Woo JM, Gallman AE, Miller TE, Levine AG, Nguyen DN, Bapat SP, Balcerek J, Bylsma SA, Lyons AM, Li S, Wong AW, Gillis-Buck EM, Steinhart ZB, Lee Y, Apathy R, Lipke MJ, Smith JA, Zheng T, Boothby IC, Isaza E, Chan J, Acenas DD, Jr, Lee J, Macrae TA, Kyaw TS, Wu D, Ng DL, Gu W, York VA, Eskandarian HA, Callaway PC, Warrier L, Moreno ME, Levan J, Torres L, Farrington LA, Loudermilk RP, Koshal K, Zorn KC, Garcia-Beltran WF, Yang D, et al. 2020. Evaluation of SARS-CoV-2 serology assays reveals a range of test performance. Nat Biotechnol 38:1174–1183. 10.1038/s41587-020-0659-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwarz NG, Mertens E, Winter D, Maiga-Ascofare O, Dekker D, Jansen S, Tappe D, Randriamampionona N, May J, Rakotozandrindrainy R, Schmidt-Chanasit J. 2017. No serological evidence for Zika virus infection and low specificity for anti-Zika virus ELISA in malaria positive individuals among pregnant women from Madagascar in 2010. PLoS One 12:e0176708. 10.1371/journal.pone.0176708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hunsperger EA, Yoksan S, Buchy P, Nguyen VC, Sekaran SD, Enria DA, Pelegrino JL, Vazquez S, Artsob H, Drebot M, Gubler DJ, Halstead SB, Guzman MG, Margolis HS, Nathanson CM, Rizzo Lic NR, Bessoff KE, Kliks S, Peeling RW. 2009. Evaluation of commercially available anti-dengue virus immunoglobulin M tests. Emerg Infect Dis 15:436–440. 10.3201/eid1503.080923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gasasira AF, Dorsey G, Kamya MR, Havlir D, Kiggundu M, Rosenthal PJ, Charlebois ED. 2006. False-positive results of enzyme immunoassays for human immunodeficiency virus in patients with uncomplicated malaria. J Clin Microbiol 44:3021–3024. 10.1128/JCM.02207-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emmerich P, Murawski C, Ehmen C, von Possel R, Pekarek N, Oestereich L, Duraffour S, Pahlmann M, Struck N, Eibach D, Krumkamp R, Amuasi J, Maiga-Ascofare O, Rakotozandrindrainy R, Asogun D, Ighodalo Y, Kann S, May J, Tannich E, Deschermeier C. 2021. Limited specificity of commercially available SARS-CoV-2 IgG ELISAs in serum samples of African origin. Trop Med Int Health 10.1111/tmi.13569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yadouleton A, Sander AL, Moreira-Soto A, Tchibozo C, Hounkanrin G, Badou Y, Fischer C, Krause N, Akogbeto P, de Oliveira Filho EF, Dossou A, Brunink S, Aissi MAJ, Djingarey MH, Hounkpatin B, Nagel M, Drexler JF. 2021. Limited specificity of serologic tests for SARS-CoV-2 antibody detection. Emerg Infect Dis 27:233–237. 10.3201/eid2701.203281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Federal Ministry of Health. 2019. Nigeria HIV/AIDS indicator and impact survey (NAIIS. Federal Ministry of Health, Abuja, Nigeria. [Google Scholar]
- 22.Martin D. 2020. Symposium 31: using laboratory methods to increase data available for public health decisions: the Nigeria multi-disease serologic surveillance using stored specimens (NMS4 experience). American Society of Tropical Medicine and Hygiene, Arlington, VA. [Google Scholar]
- 23.Plucinski MM, Candrinho B, Chambe G, Muchanga J, Muguande O, Matsinhe G, Mathe G, Rogier E, Doyle T, Zulliger R, Colborn J, Saifodine A, Lammie P, Priest JW. 2018. Multiplex serology for impact evaluation of bed net distribution on burden of lymphatic filariasis and four species of human malaria in northern Mozambique. PLoS Negl Trop Dis 12:e0006278. 10.1371/journal.pntd.0006278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Njenga SM, Kanyi HM, Arnold BF, Matendechero SH, Onsongo JK, Won KY, Priest JW. 2020. Integrated cross-sectional multiplex serosurveillance of IgG antibody responses to parasitic diseases and vaccines in coastal Kenya. Am J Trop Med Hyg 102:164–176. 10.4269/ajtmh.19-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Priest JW, Jenks MH, Moss DM, Mao B, Buth S, Wannemuehler K, Soeung SC, Lucchi NW, Udhayakumar V, Gregory CJ, Huy R, Muth S, Lammie PJ. 2016. Integration of multiplex bead assays for parasitic diseases into a national, population-based serosurvey of women 15–39 years of age in Cambodia. PLoS Negl Trop Dis 10:e0004699. 10.1371/journal.pntd.0004699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plucinski MM, Herman C, Jones S, Dimbu R, Fortes F, Ljolje D, Lucchi N, Murphy SC, Smith NT, Cruz KR, Seilie AM, Halsey ES, Udhayakumar V, Aidoo M, Rogier E. 2019. Screening for Pfhrp2/3-deleted Plasmodium falciparum, non-falciparum, and low-density malaria infections by a multiplex antigen assay. J Infect Dis 219:437–447. 10.1093/infdis/jiy525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rogier E, Nace D, Ljolje D, Lucchi NW, Udhayakumar V, Aidoo M. 2020. Capture and detection of Plasmodium vivax lactate dehydrogenase in a bead-based multiplex immunoassay. Am J Trop Med Hyg 102:1064–1067. 10.4269/ajtmh.19-0772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glickman ME, Rao SR, Schultz MR. 2014. False discovery rate control is a recommended alternative to Bonferroni-type adjustments in health studies. J Clin Epidemiol 67:850–857. 10.1016/j.jclinepi.2014.03.012. [DOI] [PubMed] [Google Scholar]
- 29.McHugh ML. 2012. Interrater reliability: the kappa statistic. Biochem Med 22:276–282. 10.11613/BM.2012.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.World Health Organization. 2020. Target product profiles for priority diagnostics to support response to the COVID-19 pandemic v.1.0. WHO, Geneva, Switzerland. [Google Scholar]
- 31.Abbott. 2020. SARS-CoV-2 IgG for use with ARCHITECT package insert. Abbott, Chicago, IL. [Google Scholar]
- 32.Patel EU, Bloch EM, Clarke W, Hsieh YH, Boon D, Eby Y, Fernandez RE, Baker OR, Keruly M, Kirby CS, Klock E, Littlefield K, Miller J, Schmidt HA, Sullivan P, Piwowar-Manning E, Shrestha R, Redd AD, Rothman RE, Sullivan D, Shoham S, Casadevall A, Quinn TC, Pekosz A, Tobian AAR, Laeyendecker O. 2020. Comparative performance of five commercially available serologic assays to detect antibodies to SARS-CoV-2 and identify individuals with high neutralizing titers. J Clin Microbiol 59:e02257-20. 10.1128/JCM.02257-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.EUROIMMUN Medizinische Labordiagnostika. 2020. Anti-SARS-CoV-2-NCP ELISA (IgG Instruction for use. EUROIMMUN Medizinische Labordiagnostika, Lubeck, Germany. [Google Scholar]
- 34.Okba NMA, Muller MA, Li W, Wang C, GeurtsvanKessel CH, Corman VM, Lamers MM, Sikkema RS, de Bruin E, Chandler FD, Yazdanpanah Y, Le Hingrat Q, Descamps D, Houhou-Fidouh N, Reusken C, Bosch BJ, Drosten C, Koopmans MPG, Haagmans BL. 2020. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease patients. Emerg Infect Dis 26:1478–1488. 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peterhoff D, Gluck V, Vogel M, Schuster P, Schutz A, Neubert P, Albert V, Frisch S, Kiessling M, Pervan P, Werner M, Ritter N, Babl L, Deichner M, Hanses F, Lubnow M, Muller T, Lunz D, Hitzenbichler F, Audebert F, Hahnel V, Offner R, Muller M, Schmid S, Burkhardt R, Gluck T, Koller M, Niller HH, Graf B, Salzberger B, Wenzel JJ, Jantsch J, Gessner A, Schmidt B, Wagner R. 2021. A highly specific and sensitive serological assay detects SARS-CoV-2 antibody levels in COVID-19 patients that correlate with neutralization. Infection 49:75–82. 10.1007/s15010-020-01503-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tso FY, Lidenge SJ, Pena PB, Clegg AA, Ngowi JR, Mwaiselage J, Ngalamika O, Julius P, West JT, Wood C. 2020. High prevalence of pre-existing serological cross-reactivity against SARS-CoV-2 in sub-Sahara Africa. Int J Infect Dis 10.1016/j.ijid.2020.10.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mveang Nzoghe A, Essone PN, Leboueny M, Maloupazoa Siawaya AC, Bongho EC, Mvoundza Ndjindji O, Avome Houechenou RM, Agnandji ST, Djoba Siawaya JF. 2021. Evidence and implications of pre-existing humoral cross-reactive immunity to SARS-CoV-2. Immun Inflamm Dis 9:128–133. 10.1002/iid3.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ly A, Hansen DS. 2019. Development of B cell memory in malaria. Front Immunol 10:559. 10.3389/fimmu.2019.00559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Silveira ELV, Dominguez MR, Soares IS. 2018. To B or not to B: understanding B cell responses in the development of malaria infection. Front Immunol 9:2961. 10.3389/fimmu.2018.02961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olsson J, Johansson J, Honkala E, Blomqvist B, Kok E, Weidung B, Lovheim H, Elgh F. 2019. Urea dilution of serum for reproducible anti-HSV1 IgG avidity index. BMC Infect Dis 19:164. 10.1186/s12879-019-3769-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taylor DW, Bobbili N, Kayatani A, Tassi Yunga S, Kidima W, Leke RFG. 2020. Measuring antibody avidity to Plasmodium falciparum merozoite antigens using a multiplex immunoassay approach. Malar J 19:171. 10.1186/s12936-020-03243-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benner SE, Patel EU, Laeyendecker O, Pekosz A, Littlefield K, Eby Y, Fernandez RE, Miller J, Kirby CS, Keruly M, Klock E, Baker OR, Schmidt HA, Shrestha R, Burgess I, Bonny TS, Clarke W, Caturegli P, Sullivan D, Shoham S, Quinn TC, Bloch EM, Casadevall A, Tobian AAR, Redd AD. 2020. SARS-CoV-2 antibody avidity responses in COVID-19 patients and convalescent plasma donors. J Infect Dis 222:1974–1984. 10.1093/infdis/jiaa581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu T, Hsiung J, Zhao S, Kost J, Sreedhar D, Hanson CV, Olson K, Keare D, Chang ST, Bliden KP, Gurbel PA, Tantry US, Roche J, Press C, Boggs J, Rodriguez-Soto JP, Montoya JG, Tang M, Dai H. 2020. Quantification of antibody avidities and accurate detection of SARS-CoV-2 antibodies in serum and saliva on plasmonic substrates. Nat Biomed Eng 4:1188–1196. 10.1038/s41551-020-00642-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental methods, Tables S1 to S3, and Fig. S1 to S6. Download JCM.00514-21-s0001.pdf, PDF file, 934 KB (933.9KB, pdf)