ABSTRACT
We evaluated the quantitative DiaSorin Liaison severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antigen test in symptomatic and asymptomatic individuals consulting their general practitioners (GPs) during a period of stable intense virus circulation (213/100,000 habitants per day). Leftover reverse transcription-PCR (RT-PCR) positive (n = 204) and negative (n = 210) nasopharyngeal samples were randomly selected among fresh routine samples collected from patients consulting their GPs. Samples were tested on Liaison XL according to the manufacturer’s instructions. Equivocal results were considered negative. The overall sensitivity and specificity of the Liaison antigen test compared to RT-PCR were 65.7% (95% confidence interval [CI], 58.9% to 71.9%) and 100% (CI, 97.8% to 100%). Sensitivity in samples with viral loads of ≥105, ≥104, and ≥103 copies/ml were 100% (CI, 96.3% to 100.0%), 96.5% (CI, 91.8% to 98.7%), and 87.4% (CI, 81.3% to 91.5%), respectively. All samples with ≤103 copies/ml were antigen negative. The ratio of antigen concentration to viral load in samples with ≥103 copies/ml was comparable in symptomatic and asymptomatic individuals (P = 0.58). The proportion of RT-PCR-positive participants with a high viral load (≥105 copies/ml) was not significantly higher in symptomatic than in asymptomatic participants (63.9% [CI, 54.9% to 72.0%] versus 51.9% [CI, 41.1% to 62.6%]; P = 0.11), but the proportion of participants with a low viral load (<103 copies/ml) was significantly higher in asymptomatic than in symptomatic RT-PCR-positive participants (35.4% [CI, 25.8% to 46.4%] versus 14.3% [CI, 9.0% to 21.8%]; P < 0.01). Sensitivity and specificity in samples with a viral load of ≥104 copies/ml were 96.5% and 100%. The correlation of antigen concentration with viral load was comparable in symptomatic and asymptomatic individuals.
KEYWORDS: COVID-19 testing, SARS-CoV-2, diagnosis, sensitivity and specificity, ambulatory care, antigen
INTRODUCTION
Rapid testing of symptomatic individuals remains an important diagnostic challenge during the current coronavirus disease 2019 (COVID-19) pandemic (1). Reverse transcription-PCR (RT-PCR) is considered the gold standard for accurate diagnosis of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) with a theoretical turnaround time (TAT) of only a few hours (1, 2). However, the real TAT can easily be more than 24 h due to limited laboratory capacity, high sample volumes, and recurrent worldwide shortages of reagents and consumables. In September 2020, the World Health Organization (WHO) published a guidance document on the potential role of antigen-detecting rapid diagnostic tests (Ag-RDT) to diminish pressure on molecular laboratories and reduce delays in diagnosis (1). While Ag-RDT are substantially less sensitive than RT-PCR, they could offer the possibility of rapid, inexpensive, and early detection of most infectious COVID-19 cases. An evaluation of the potential economic benefit of a repeated screening program with Ag-RDT showed that the fiscal, macroeconomic, and health benefits far exceed the cost (3).
Similar to rapid antibody assays for SARS-CoV-2, the quality of Ag-RDT varies, and few published data are available for many assays (4, 5). Studies with some of the widely used Ag-RDT, such as the Panbio COVID-19-Ag rapid test (distributed by Abbott) and SD Biosensor Standard Q-Covid-19 test (distributed by Roche), have demonstrated that Ag-RDT can identify symptomatic individuals with a cycle threshold (CT) value of ≤25 but that sensitivity decreases after 7 days (6–8). Unfortunately, direct comparison of the performance of Ag-RDT and RT-PCR is hampered by the use of test-specific swabs and buffers in many Ag-RDT (including the Panbio and SD Biosensor Ag-RDT) and the increasing use of virus-neutralizing media for RT-PCR. If the Ag-RDT and RT-PCR are not performed using the same sample, it is not possible to distinguish false-negative and false-positive results caused by lack of sensitivity or specificity of the Ag-RDT from discordant results due to sampling.
Ag-RDT are easy to use and have a short TAT (typically less than 30 min) but are difficult to scale up. Most Ag-RDT are lateral-flow assays which require visual readout and lack proper internal quality control, making them more prone to errors. The introduction of high-throughput automated antigen assays with daily quality control could help address these limitations. The DiaSorin Liaison SARS-CoV-2 antigen test is one of the first automated quantitative SARS-CoV-2 antigen assays to obtain a CE-IVD (Conformité Européenne for in vitro diagnostics) mark (26 October 2020). There are currently only a few peer-reviewed studies which evaluated the performance of quantitative SARS-CoV-2 antigen assays and no peer-reviewed studies which evaluated a quantitative antigen assay in an outpatient setting (9–13).
The first aim of this study was to evaluate the performance of the Liaison antigen test compared to RT-PCR in symptomatic and asymptomatic individuals consulting their general practitioners (GPs) during a period of stable intense virus circulation (213 per 100,000 habitants per day). The second aim was to compare the relation between the antigen concentration and viral load in symptomatic and asymptomatic individuals.
MATERIALS AND METHODS
Study design.
This study was approved by the local ethics committee (S65002). Between 23 November and 18 December 2020, 206 leftover RT-PCR-positive and 210 RT-PCR-negative fresh nasopharyngeal samples were randomly selected among routine samples collected from patients consulting their GPs. Samples were collected in 3 ml universal transport medium (UTM) or virus stabilization tubes (VST). After collection, samples were sent to the laboratory of the Medical Centre for GPs (Medisch Centrum voor Huisartsen, Leuven, Belgium). Only one swab was collected per patient. Two of the selected samples were excluded from the study once the clinical information was collected, based on the predefined exclusion criterion (collected more than 10 days after symptom onset).
RT-PCR.
RT-PCR was performed upon arrival of the sample in the laboratory using the Viasure SARS-CoV-2 (N1 + N2) real-time PCR detection kit (Certest Biotec SL, Zaragoza, Spain) for the BD MAX system (Beckton Dickinson [BD], Franklin Lakes, NJ, USA). This assay targets both the N1 and the N2 regions of the nucleocapsid phosphoprotein gene. The test is considered positive if the cycle threshold (CT) value for the N1 and/or the N2 gene region is <40. Viral load (copies/ml) based on CT values was calculated after calibration using a prequantified viral stock prepared by the Belgian National Reference Center for Respiratory Pathogens and distributed by the laboratory of Sciensano (the Belgian Institute for Health). The correlation of CT values and viral load is presented in Table S1 in the supplemental material. Calculations are based on the mean viral load of the N1 and the N2 gene regions.
Antigen testing.
Samples selected for antigen testing were stored for a maximum of 12 h between 2 and 8°C, after which they were frozen before processing according to the manufacturer’s instructions. First, 1 ml of the UTM or the VST contents was pipetted into a tube containing 1 ml sample inactivation buffer (DiaSorin, Saluggia, Italy). Next, the tubes were kept at room temperature for at least 120 min before testing for virus inactivation. If samples could not be tested immediately after inactivation, they were stored according to the manufacturer’s instructions (up to 5 days between 2 and 8°C or at −20°C for up to 1 month). Antigen concentrations were determined using the DiaSorin Liaison SARS-CoV-2 antigen test for the platform Liaison XL. Results are expressed as 50% tissue culture infectious doses (TCID50) per milliliter. Results of ≥200.00 TCID50/ml are considered positive, results between ≥100.00 and <200.00 TCID50/ml are equivocal, and results of <100.00 TCID50/ml are negative. The limit of detection and upper limit of quantitation are 22.00 and 100,000.00 TCID50/ml, respectively.
See the supplemental material for more detailed information on data collection and analysis. To calculate performance characteristics, equivocal results were treated as negative. Thirteen patients were excluded for subanalyses based on symptoms and test indication because this information was not available.
RESULTS
Study population.
The median age of the 414 participants was 37 years (range, 2 to 101), with a male/female ratio of 40/60. Based on the clinical information, 223 participants were symptomatic and 178 were asymptomatic (Table S2). Of 151 patients for whom the date of symptom onset was available, 132 presented within the first 3 days, 14 between days 4 and 7, and 5 between days 8 and 10. Of the 178 asymptomatic individuals, 74 were tested in context of a high-risk contact. Indications in asymptomatic participants without a high-risk contact included travel, preoperative screening, or admission to a residential care facility. No clinical information was available for 13 participants.
Sensitivity and specificity.
The study included 204 RT-PCR positive samples and 210 RT-PCR negative samples (Table 1). With the Liaison antigen test, 134 were positive, 4 were equivocal, and 276 were negative. The four equivocal samples were RT-PCR positive, with CT values ranging between 23.8 and 26.2 for both the N1 and the N2 gene regions. Overall sensitivity and specificity (with equivocal results considered negative) of the Liaison antigen test compared to RT-PCR were 65.7% (95% confidence interval [CI], 58.9% to 71.9%) and 100% (CI, 97.8% to 100%), respectively. There were no false-positive results with Liaison in this study. Of the 210 RT-PCR-negative samples, 208 had a Liaison result below the limit of detection (LOD) of 22.00 TCID50/ml. The two remaining samples had concentrations of 72.92 TCID50/ml and 38.47 TCID50/ml.
TABLE 1.
Comparison of the results of the Liaison antigen test with those of RT-PCRa
| Liaison antigen test result | No. of samples with RT-PCR result |
Total | |
|---|---|---|---|
| Positive | Negative | ||
| Positive | 134 | 0 | 134 |
| Equivocal | 4 | 0 | 4 |
| Negative | 66 | 210 | 276 |
| Total | 204 | 210 | 414 |
Sensitivity when equivocal results were considered negative, 65.7% (CI, 58.9% to 71.9%); sensitivity when equivocal results were considered positive, 67.7% (CI, 60.9% to 73.8%); specificity, 100% (CI, 97.8% to 100%).
When viral load (copies/ml) was calculated based on CT values, sensitivity was 100% (CI, 96.3% to 100.0%) for samples with a viral load of ≥105 copies/ml (Table 2). The sensitivity in samples with viral loads of ≥104 copies/ml and ≥103 copies/ml decreased to 96.5% (CI, 91.8% to 98.7%) and 86.8% (CI, 80.6% to 91.3%), respectively. All samples with ≤103 copies/ml were negative with Liaison. When sensitivity was calculated according to the CT value of the N1 or N2 gene region (whichever was the lowest), the sensitivity was 96.4% (CI, 91.8% to 98.8%) for a CT value of <25.0 and 100% (CI, 96.5% to 100.0%) for a CT value of <23.0.
TABLE 2.
Sensitivity of the Liaison antigen test compared to RT-PCR (equivocal results were considered negative)
| Measurement used for sensitivity | No. of samples | % sensitivity (CI) |
|---|---|---|
| No. of copies/ml | ||
| ≥106 | 92 | 100 (95.2–100.0) |
| ≥105 | 121 | 100 (96.3–100.0) |
| ≥104 | 142 | 93.7 (88.2–96.8) |
| ≥103 | 159 | 84.3 (77.8–89.2) |
| CT for N1 or N2a | ||
| <20.0 | 103 | 100 (95.7–100.0) |
| <25.0 | 141 | 94.3 (89.0–97.3) |
| <30.0 | 161 | 83.2 (76.7–88.3) |
| <35.0 | 188 | 71.3 (64.4–77.3) |
CT value of the N1 or the N2 region, whichever was lower.
Relation between antigen concentration and RT-PCR results.
CT values of the N1 and N2 gene region were significantly lower in Liaison-positive than Liaison-negative samples (Fig. 1A and B) (P < 0.01). There was a good correlation of CT values for the N1 and N2 gene regions (r = 0.99; P < 0.01). CT values of the N1 and the N2 gene region ranged from 10.2 to 26.2 and from 10.0 to 26.2, respectively, in the samples which tested positive or equivocal with Liaison (n = 138). In the group of RT-PCR-positive but Liaison-negative samples (n = 66), CT values of the N1 and the N2 gene regions varied from 23.2 to 39.0 and from 23.70 to 39.70, respectively. Viral load (copies/ml) was significantly higher in Liaison-positive than in Liaison-negative samples (P < 0.01).
FIG 1.
RT-PCR results and viral load according to Liaison antigen test results. (A to C) RT-PCR results for the N1 (A) and N2 (B) gene regions and viral loads (C) in samples that were positive and negative in the Liaison antigen test. (D) Correlation of the CT values for the N1 and N2 gene regions in RT-PCR-positive samples.
Relation between antigen concentration and viral load.
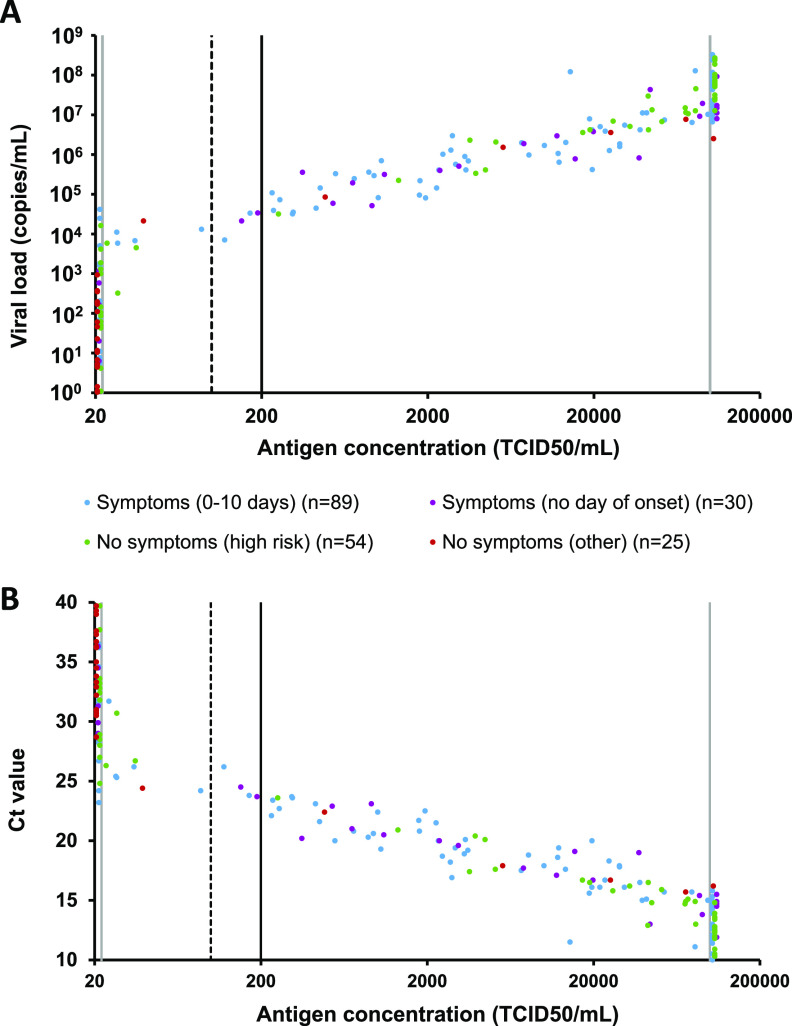
The relation between antigen concentration and viral load in 119 symptomatic and 79 asymptomatic individuals is shown in Fig. 2. Antigen concentration and viral load correlated in the 97 samples with a viral load of ≥103 copies/ml and an antigen concentration between 20 and 100,000 TCID50/ml (r = 0.41; P < 0.01). The ratio of antigen concentration to viral load in Liaison-positive samples was comparable in the 71 symptomatic and 26 asymptomatic participants (P = 0.58) (Fig. S1). The proportion of symptomatic individuals with a high viral load, defined as ≥105 copies/ml, was not significantly higher than in asymptomatic individuals (Fig. 3A) (63.9% [CI, 54.9% to 72.0%] versus 51.9% [CI, 41.1% to 62.6%], respectively; P = 0.11), but the proportion of asymptomatic individuals with a low viral load, defined as <103 copies/ml, was significantly higher than that of symptomatic individuals (Fig. 3) (35.4% [CI, 25.8% to 46.4%] versus 14.3% [CI, 9.0% to 21.8%], respectively; P < 0.01). This explains the higher overall sensitivity of the Liaison antigen test in symptomatic patients (72.3% [CI, 63.6% to 79.6%]) compared to asymptomatic participants (54.4% [CI, 43.5% to 65.0%]) (Fig. 3B) (P < 0.01). If only patients presenting within the first 3 or 7 days were included, the sensitivity was 75.9% (CI, 65.4% to 84.1%) and 74.7% (CI, 64.6% to 82.7%) (both P < 0.01 versus asymptomatic participants).
FIG 2.
Quantitative Liaison antigen test results in symptomatic and asymptomatic patients compared to viral load (A) and lowest CT value of the N1 or N2 gene region (B). Vertical lines represent the limit of detection (22 TCID50/ml), cutoff for equivocal result (100 TCID50/ml), cutoff for positive result (200 TCID50/ml), and upper limit of quantification (100,000 TCID50/ml).
FIG 3.
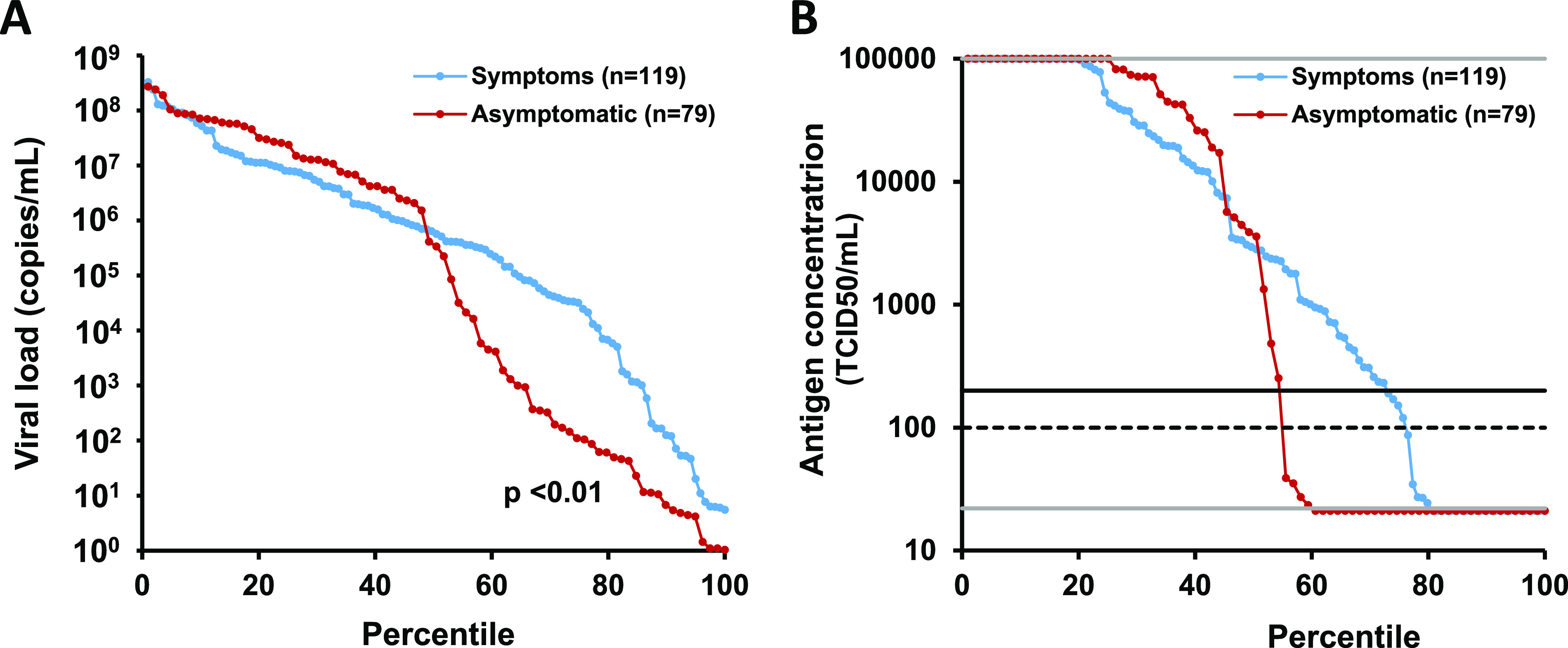
Distribution of results for viral load (A) and quantitative Liaison antigen test results (B) in 119 symptomatic and 79 asymptomatic RT-PCR positive patients. Results are ordered from high to low. Horizontal lines represent the limit of detection (22 TCID50/ml), cutoff for equivocal result (100 TCID50/ml), cutoff for positive result (200 TCID50/ml), and upper limit of quantification (100,000 TCID50/ml).
DISCUSSION
We evaluated the quantitative high-throughput Liaison antigen test in comparison to RT-PCR in symptomatic and asymptomatic outpatients during a stable period of intense virus circulation. The sensitivity in symptomatic and asymptomatic individuals with a high viral load (≥105 copies/ml) was 100%, and there were no false-positive results. The proportion of participants with a low viral load (<103 copies/ml) was significantly higher in asymptomatic than in symptomatic participants, explaining the lower overall sensitivity in asymptomatic compared to symptomatic participants.
Ag-RDT are generally characterized by a relatively low sensitivity and a good specificity compared to RT-PCR (6). A recent Cochrane meta-analysis reported an average sensitivity and specificity of 59.4% (CI, 50.7% to 67.5%) and 99.6% (CI, 97.4% to 99.9%) in 3 studies with nasopharyngeal swabs (from symptomatic and asymptomatic individuals) (5). The overall sensitivity of 67.7% and specificity of 100% of the Liaison antigen test in our study were comparable to these results. An ongoing Dutch evaluation of the Liaison antigen test reported a sensitivity of 84.5% (CI, 76.7% to 90.0%) and specificity of 99.2% (CI, 98.5% to 99.6%) in 116 RT-PCR-positive and 948 RT-PCR-negative samples from mildly symptomatic patients (14). We found a sensitivity of 75.6% in 119 symptomatic patients using randomly selected samples.
The quantitative Liaison results correlated with viral load in RT-PCR-positive patients. All patients with a viral load of ≥105 copies/ml were antigen positive. Previous studies have shown that individuals with a viral load of >106 copies/ml in respiratory samples are highly infectious and that samples with a viral load of <105 copies/ml typically lead to negative viral culture (7, 15, 18, 24–26). However, correlation between viral load, viral culture, and infectivity is complex and there is not yet a consensus in the literature regarding a cutoff for infectiousness. Taken together, these results support the use of the Liaison antigen test for rapid identification of highly infectious individuals and efficient epidemiological control. Despite the excellent performance with a sensitivity of 86.8% in randomly selected fresh samples with a viral load of ≥103 copies/ml, the Liaison antigen test did not meet the WHO acceptance criterion for sensitivity of SARS-CoV-2 Ag-RDT. The WHO acceptance criteria require a sensitivity of ≥80% and a specificity of ≥97% compared to RT-PCR (1). The failure to meet the sensitivity criterion is most likely due to the fact that our group of randomly selected RT-PCR-positive samples included a significant number of patients with a low viral load (<103 copies/ml). We believe that an acceptance criterion for antigen tests should take into account the viral load of samples (e.g., calculated in samples with a viral load of ≥103 copies/ml), since these tests are intended for rapid identification of infectious individuals.
The correlation of antigen concentrations with viral load in samples of symptomatic and asymptomatic individuals with a viral load of ≥103 copies/ml indicates that RNA shedding and antigen production occur in parallel. A recent systematic review reported similar viral loads at the start of infection in symptomatic and asymptomatic individuals (16). The fact that the proportion of asymptomatic RT-PCR-positive individuals with a high viral load (>105 copies/ml) was not significantly lower than in symptomatic patients is in line with other studies (17–19). The higher proportion of asymptomatic individuals with a viral load <103 copies/ml could be explained by earlier decrease in viral load after infection but similar median duration of viral shedding in asymptomatic individuals compared to symptomatic individuals (16, 20). The lower overall sensitivity in asymptomatic participants in our study is therefore not unexpected. We can, however, not rule out that asymptomatic individuals with a low viral load were tested in the presymptomatic window. A recent study with the Abbott BinaxNOW Ag-RDT reported a sensitivity of 35.8% in asymptomatic persons compared to 64.2% in symptomatic persons (21).
The Liaison antigen test was initially launched with a cutoff of 200 TCID50/ml. The company DiaSorin subsequently proposed an additional equivocal category for results between 100 and 200 TCID50/ml. Lowering the cutoff to 100 TCID50/ml had no effect on specificity in our study. The cutoff could in fact be lowered to the LOD of 22 TCID50/ml in our study while maintaining a specificity of 99.0% (96.4% to 100%). This would have resulted in 9 additional positive results in patients with a viral load of <105 copies/ml and would have increased overall sensitivity to 72.1% (CI, 65.5% to 77.8%). Of note, any modification to an in vitro diagnostic assay should be validated by the laboratory, and the impact of changing the cutoff after European In Vitro Diagnostic Regulation 2017/746 goes into effect in May 2022 is not yet clear (22).
The strength of our study is the fact that we used randomly selected RT-PCR-positive and RT-PCR-negative samples from symptomatic and asymptomatic participants consulting their GPs rather than samples collected in a hospital setting. Outpatients typically present within the first days after symptom onset, as was the case in our study, while the median time between symptom onset and hospital admission is estimated to be around 7 days (23). A limitation of this study is the relatively small sample size (n = 414). This was partially offset by randomly selecting a cohort of RT-PCR-positive and a cohort of RT-PCR-negative samples, with each a little over 200 samples. Due to this approach, the positivity rate in this study is not a reflection of the prevalence. Another limitation of our study is the use of 3 ml medium for sample collection (to allow simultaneous RT-PCR and antigen quantification) in combination with a 1-in-2 dilution prior to inactivation. Directly introducing the swab in a smaller volume of inactivation buffer as is customary for Ag-RDT (e.g., 300 μl for Panbio) could increase the sensitivity in samples with a viral load between 103 and 105 copies/ml and improve turnaround time.
In summary, the performance of the Liaison antigen test compared to RT-PCR was good, with a sensitivity in symptomatic and asymptomatic individuals with a high viral load (≥105 copies/ml) of 100% and a specificity of 100%. The Liaison antigen assay can help diminish pressure on molecular laboratories and reduce delays in diagnosis. Further studies are needed to determine if the sensitivity can be increased by directly introducing the swab in a smaller volume of inactivation buffer.
ACKNOWLEDGMENTS
P. Vermeersch is a senior clinical investigator of the FWO-Vlaanderen.
We thank the company DiaSorin for providing the reagents free of charge.
The research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
P.V. devised the study, collected data, and drafted the manuscript; S.L. and C.I. collected data and drafted the manuscript; K.D. and C.I. collected data. All authors critically reviewed the data and the manuscript.
Footnotes
Supplemental material is available online only.
Contributor Information
Pieter Vermeersch, Email: pieter.vermeersch@uzleuven.be.
Randall Hayden, St. Jude Children's Research Hospital.
REFERENCES
- 1.World Health Organization. 2020. SARS-CoV-2 antigen-detecting rapid diagnostic tests: an implementation guide. https://www.who.int/publications/i/item/9789240017740
- 2.Young S, Taylor SN, Cammarata CL, Roger-Dalbert C, Montano A, Griego-Fullbright C, Burgard C, Fernandez C, Eckert K, Andrews JC, Ren H, Allen J, Ackerman R, Cooper CK. 2020. Clinical evaluation of BD VeritorTM SARS-CoV-2 point-of-care test performance compared to PCR-based testing and versus the Sofia 2 SARS antigen point-of-care test. J Clin Microbiol 17:e02338-20. 10.1128/JCM.02338-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atkeson A, Droste M, Stock JH, Mina MJ. 2020. Economic benefits of COVID-19 screening tests. medRxiv 10.1101/2020.10.22.20217984. [DOI] [Google Scholar]
- 4.Van Elslande J, Houben E, Depypere M, Brackenier A, Desmet S, André E, Van Ranst M, Lagrou K, Vermeersch P. 2020. Diagnostic performance of seven rapid IgG/IgM antibody tests and the Euroimmun IgA/IgG ELISA in COVID-19 patients. Clin Microbiol Infect 26:1082–1087. 10.1016/j.cmi.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dinnes J, Deeks JJ, Adriano A, Berhane S, Davenport C, Dittrich S, Emperador D, Takwoingi Y, Cunningham J, Beese S, Dretzke J, Ferrante di Ruffano L, Harris IM, Price MJ, Taylor-Phillips S, Hooft L, Leeflang MM, Spijker R, Van den Bruel A, Cochrane COIVD-19 Diagnostic Test Accuracy Group . 2020. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst Rev 26:CD013705. 10.1002/14651858.CD013705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Linares M, Pérez-Tanoira R, Carrero A, Romanyk J, Pérez-Garcia F, Gomez-Herruz P, Arroyo T, Cuadros J. 2020. Panbio antigen rapid test is reliable to diagnose SARS-CoV-2 infection in the first 7 days after the onset of symptoms. J Clin Virol 133:104659. 10.1016/j.jcv.2020.104659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alemany A, Baro B, Ouchi D, Rodo P, Ubals M, Corbacho-Monné M, et al. 2021. Analytical and clinical performance of the panbio COVID-19 antigen-detecting rapid diagnostic test. J Infect 10.1016/j.jinf.2020.12.033. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Möckel M, Corman VM, Stegemann MS, Hofmann J, Stein A, Jones TC, Gastmeier P, Seybold J, Offermann R, Bachmann U, Lindner T, Bauer W, Drosten C, Rosen A, Somasundaram R. 2021. SARS-CoV-2 antigen rapid immunoassay for diagnosis of COVID-19 in the emergency department. Biomarkers 26:213–220. 10.1080/1354750X.2021.1876769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirotsu Y, Maejima M, Shibusawa M, Nagakubo Y, Hosaka K, Amemiya K, Sueki H, Hayakawa M, Mochizuki H, Tsutsui T, Kakizaki Y, Miyashita Y, Yagi S, Kojima S, Omata M. 2020. Comparison of automated SARS-CoV-2 antigen test for COVID-19 infection with quantitative RT-PCR using 313 nasopharyngeal swabs, including from seven serially followed patients. Int J Infect Dis 99:397–402. 10.1016/j.ijid.2020.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirotsu Y, Maejima M, Shibusawa M, Amemiya K, Nagakubo Y, Hosaka K, Sueki H, Hayakawa M, Mochizuki H, Tsutsui T, Kakizaki Y, Miyashita Y, Omata M. 2021. Prospective study of 1,308 nasopharyngeal swabs from 1,033 patients using the LUMIPULSE SARS-CoV-2 antigen test: domparison with RT-qPCR. Int J Infect Dis 10.1016/j.ijid.2021.02.005. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aoki K, Nagasawa T, Ishii Y, Yagi S, Okuma S, Kashiwagi K, Maeda T, Miyazaki T, Yoshizawa S, Tateda K. 2020. Clinical validation of quantitative SARS-CoV-2 antigen assays to estimate SARS-CoV-2 viral loads in nasopharyngeal swabs. J Infect Chemother 10.1016/j.jiac.2020.11.021. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobayashi R, Murai R, Asanuma K, Fujiya Y, Takahashi S. 2021. Evaluating a novel, highly sensitive, and quantitative reagent for detecting SARS-CoV-2 antigen. J Infect Chemother 27:800–807. 10.1016/j.jiac.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pollock NR, Savage TJ, Wardell H, Lee RA, Mathew A, Stengelin M, Sigal GB. 2021. Correlation of SARS-CoV-2 nucleocapsid antigen and RNA concentrations in nasopharyngeal samples from children and adults using an ultrasensitive and quantitative antigen assay. J Clin Microbiol 59:e03077-20. 10.1128/JCM.03077-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han W, Benschop K, Meijer A, Reusken C. 2021. Status validatie SARS-CoV-2 antigeen sneltesten. Rijksinstituut voor Volksgezondheid en Milieu. https://www.rivm.nl/documenten/status-validatie-sars-cov-2-antigeen-sneltesten
- 15.van Kampen JJA, van de Vijver DAMC, Fraaij PLA, Haagmans BL, Lamers MM, Okba N, van den Akker JPC, Endeman H, Gommers DAMPJ, Cornelissen JJ, Hoek RAS, van der Eerden MM, Hesselink DA, Metselaar HJ, Verbon A, de Steenwinkel JEM, Aron GI, van Gorp ECM, van Boheemen S, Voermans JC, Boucher CAB, Molenkamp R, Koopmans MPG, Geurtsvankessel C, van der Eijk AA. 2021. Duration and key determinants of infectious virus shedding in hospitalized patients with coronavirus disease-2019 (COVID-19). Nat Commun 12:8–13. 10.1038/s41467-020-20568-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cevik M, Tate M, Lloyd O, Maraolo AE, Schafers J, Ho A. 2021. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe 2:E13–E22. 10.1016/S2666-5247(20)30172-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ra SH, Lim JS, Kim GU, Kim MJ, Jung J, Kim SH. 2021. Upper respiratory viral load in asymptomatic individuals and mildly symptomatic patients with SARS-CoV-2 infection. Thorax 76:61–63. 10.1136/thoraxjnl-2020-215042. [DOI] [PubMed] [Google Scholar]
- 18.Walsh KA, Jordan K, Clyne B, Rohde D, Drummond L, Byrne P, Ahern S, Carty PG, O'Brien KK, O'Murchu E, O'Neill M, Smith SM, Ryan M, Harrington P. 2020. SARS-CoV-2 detection, viral load and infectivity over the course of an infection. J Infect 81:357–371. 10.1016/j.jinf.2020.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee S, Kim T, Lee E, Lee C, Kim H, Rhee H, Park SY, Son H-J, Yu S, Park JW, Choo EJ, Park S, Loeb M, Kim TH. 2020. Clinical course and molecular viral shedding among asymptomatic and symptomatic patients with SARS-CoV-2 infection in a community treatment center in the Republic of Korea. JAMA Intern Med 180:1447–1452. 10.1001/jamainternmed.2020.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li W, Su Y-Y, Zhi S-S, Huang J, Zhuang C-L, Bai W-Z, Wan Y, Meng X-R, Zhang L, Zhou Y-B, Luo Y-Y, Ge S-X, Chen Y-K, Ma Y. 2020. Virus shedding dynamics in asymptomatic and mildly symptomatic patients infected with SARS-CoV-2. Clin Microbiol Infect 26:1556.e1–1556.e6. 10.1016/j.cmi.2020.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prince-Guerra JL, Almendares O, Nolen LD, Gunn JKL, Dale AP, Buono SA, Deutsch-Feldman M, Suppiah S, Hao L, Zeng Y, Stevens VA, Knipe K, Pompey J, Atherstone C, Bui DP, Powell T, Tamin A, Harcourt JL, Shewmaker PL, Medrzycki M, Wong P, Jain S, Tejada-Strop A, Rogers S, Emery B, Wang H, Petway M, Bohannon C, Folster JM, MacNeil A, Salerno R, Kuhnert-Tallman W, Tate JE, Thornburg NJ, Kirking HL, Sheiban K, Kudrna J, Cullen T, Komatsu KK, Villanueva JM, Rose DA, Neatherlin JC, Anderson M, Rota PA, Honein MA, Bower WA. 2021. Evaluation of Abbott BinaxNOW rapid antigen test for SARS-CoV-2 infection at two community-based testing sites—Pima County, Arizona, November 3–17, 2020. MMWR Morb Mortal Wkly Rep 70:100–105. 10.15585/mmwr.mm7003e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vermeersch P, Van Aelst T, Dequeker EM. 2021. The new IVD Regulation 2017/746: a case study at a large university hospital laboratory in Belgium demonstrates the need for clarification on the degrees of freedom laboratories have to use lab-developed tests to improve patient care. Clin Chem Lab Med 59:101–106. 10.1515/cclm-2020-0804. [DOI] [PubMed] [Google Scholar]
- 23.Van Elslande J, Decru B, Jonckheere S, Van Wijngaerden E, Houben E, Vandecandelaere P, Indevuyst C, Depypere M, Desmet S, André E, Van Ranst M, Lagrou K, Vermeersch P. 2020. Antibody response against SARS-CoV-2 spike protein and nucleoprotein evaluated by four automated immunoassays and three ELISAs. Clin Microbiol Infect 26:1557.E1–1557.E7. 10.1016/j.cmi.2020.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singanayagam A, Patel M, Charlett A, Bernal JL, Saliba V, Ellis J, Ladhani S, Zambon M, Gopal R. 2020. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Eurosurveillance 25:1–5. https://www.eurosurveillance.org/content/10.2807/1560-7917.ES.2020.25.32.2001483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.La Scola B, Le Bideau M, Andreani J, Hoang VT, Grimaldier C, Colson P, Gautret P, Raoult D. 2020. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis 39:1059–1061. 10.1007/s10096-020-03913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, Niemeyer D, Jones TC, Vollmar P, Rothe C, Hoelscher M, Bleicker T, Brünink S, Schneider J, Ehmann R, Zwirglmaier K, Drosten C, Wendtner C. 2020. Virological assessment of hospitalized patients with COVID-2019. Nature 581:465–469. 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Materials and Methods, Tables S1 and S2, and Fig. S1. Download JCM.00374-21-s0001.pdf, PDF file, 228 KB (227.4KB, pdf)