ABSTRACT
The increasing emergence of carbapenemase-producing Klebsiella pneumoniae (CPK) isolates is a global health alarm. Rapid methods that require minimum sample preparation and rapid data analysis are urgently required. Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) has recently been used by clinical laboratories for identification of antibiotic-resistant bacteria; however, discrepancies have arisen regarding biological and technical issues. The aim of this study was to standardize an operating procedure and data analysis for identification of CPK by MALDI-TOF MS. To evaluate this approach, a series of 162 K. pneumoniae isolates (112 CPK and 50 non-CPK) were processed in the MALDI BioTyper system (Bruker Daltonik, Germany) following a standard operating procedure. The study was conducted in two stages; the first is denominated the “reproducibility stage” and the second “CPK identification.” The first stage was designed to evaluate the biological and technical variation associated with the entire analysis of CPK and the second stage to assess the final accuracy of MALDI-TOF MS for the identification of CPK. Therefore, we present an improved MALDI-TOF MS data analysis pipeline using neural network analysis implemented in Clover MS Data Analysis Software (Clover Biosoft, Spain) that is designed to reduce variability, guarantee interlaboratory reproducibility, and maximize the information selected from the bacterial proteome. Using the random forest (RF) algorithm, 100% of CPK isolates were correctly identified when all the peaks in the spectra were selected as input features and total ion current (TIC) normalization was applied. Thus, we have demonstrated that real-time direct tracking of CPK is possible using MALDI-TOF MS.
KEYWORDS: carbapenemases, Klebsiella pneumoniae, MALDI-TOF
INTRODUCTION
The increasing emergence of carbapenemase-producing Klebsiella pneumoniae (CPK) is recognized as a global health alarm by different organizations, such as the ECDC, CDC, and WHO (1–4). In addition, carbapenemases can confer resistance to almost all available beta-lactam antibiotics, which are the antibiotics most commonly used to treat infections caused by Enterobacterales (5). Antimicrobial resistance detection and bacterial typing are usually based on widely approved molecular techniques (6). However, they remain time consuming, labor intensive, and expensive. Methods involving minimum sample preparation and providing rapid results are therefore urgently needed.
Matrix-assisted laser desorption–ionization mass spectrometry (MALDI-TOF MS) is a proteomic technique used for identification of microorganisms by analysis of ribosomal proteins of between 2 and 20 kDa from whole cells, indicating the high diversity of these proteins among different species of bacteria (7–9). Mass spectra consist of hundreds or thousands of peaks of mass-to-charge (m/z) ratios per bacterial isolate, intensity levels, and areas under the curve for each mass peak. This is translated into thousands of data points with a specific statistical distribution. Neural network tools, such as machine learning and deep learning, are powerful classification strategies that have been used in health sciences, such as cancer genomics (10, 11). As advancements in high-throughput technologies lead to the production of large amounts of data, the classification features are expanding the use to other omics areas, such as proteomics in clinical microbiology.
Due to the popularity and the good performance of MALDI-TOF MS, the primary purpose of the technique has been redirected from microorganism identification to discrimination of subgroups within the same species of pathogens (12–15). Various different mass spectrometers, with the corresponding experimental procedures and databases, have been successfully evaluated for identifying resistance mechanisms by means of the protein profiles of K. pneumoniae (16), Staphylococcus aureus (17), and Enterococcus faecium (18, 19). This MALDI-TOF MS application has also served for typing a large number of bacterial species, such as Pseudomonas aeruginosa (20, 21), K. pneumoniae (22), Escherichia coli (23), and Clostridium difficile (24). Several MALDI-TOF MS-based procedures and data analyses have been published; however, inconsistencies and discrepancies in the identification of antibiotic-resistant bacteria have arisen regarding biological and technical reproducibility and data analysis that limit the overall applicability of MALDI-TOF MS as a first-line typing methodology (25–29).
Biological variation, defined as the physiological fluctuation of the constituents of living organisms around a homeostatic point, has two components: intraindividual (within-subject) and interindividual (between-subject) variation. The variation in protein expression in bacteria is reflected by the numbers and intensities of average peaks. This source of variability may be significant in relation to replicating the results of different studies.
Technical variation occurs due to the imprecision of the different steps and conditions throughout the entire analytical process, ranging from sample preparation to instrumental conditions of the equipment used (MALDI-TOF MS). Sample preparation is often a critical point because it consists of several steps with a high level of associated imprecision. MALDI-TOF MS has multiple sources of imprecision (instrumental conditions), including the laser power, distance to the sample, cleanliness of the source, and detector capacity. To reduce the technical variation, computational procedures like normalization of the resulting spectra are typically used.
In this study, we evaluated the biological and technical variation associated with the detection of CPK isolates by MALDI-TOF MS to design a standard operating procedure and a MALDI-TOF MS data analysis pipeline for direct tracking of CPK isolates that could be implemented in routine screening in clinical microbiology laboratories at no additional cost and with no requirement for specific laboratory expertise. Besides this, we recommend good practice guidelines for MALDI-TOF MS identification of antibiotic-resistant bacteria and protein biomarker performance.
MATERIALS AND METHODS
Bacterial isolates.
The study included a representative collection of 162 unduplicated clinical isolates. One hundred twelve of the 162 were CPK isolates, and the remaining 50 isolates were not carbapenemase producers. Regarding the CPK isolates, 93 were collected during a nationwide survey of carbapenemase-producing Enterobacterales (CPE) from 15 participating hospitals throughout Spain. The call was promoted by the Spanish Society of Infectious Diseases and Clinical Microbiology (SEIMC) and by the Spanish Network for Research in Infectious Diseases (REIPI). Another 19 isolates used in the study form part of our own collection (30).
Isolates were screened for carbapenemase production according to the screening cutoff values recommended by European Committee on Antimicrobial Susceptibility Testing (EUCAST), that is, having a meropenem or ertapenem MIC of >0.125 mg/liter. The isolates belonging to the Spanish national survey were characterized by whole-genome sequencing, and the isolates from our laboratory collection were characterized by PCR.
Genomic characterization of the isolates.
The 93 clinical isolates belonging to the national survey on CPE used in the study were analyzed by whole-genome sequencing (WGS). Total genomic DNA was obtained using a genomic DNA buffer set with a genomic-tip 20/G (Qiagen). Purified genomic DNA from all isolates was sequenced in parallel using short-read (Illumina MiSeq benchtop; Illumina) (31) and long-read (MinION; Oxford Nanopore Technologies) approaches (32–34). The resultant long and short reads from each isolate were assembled using the Unicycler version 0.4.6 hybrid assembler. The contigs were visualized using Bandage software (32). The assemblies obtained were finally annotated using Prokka version 1.13 (33). Analysis of the total antimicrobial resistance gene content of the isolates was carried out in silico using Resfinder software version 3.2 (34) and the Comprehensive Antibiotic Resistance Database (CARD) (33). Multilocus sequence typing (MLST) was performed in silico from assembled whole-genome sequencing data using available online databases (https://cge.cbs.dtu.dk/services/MLST/).
The 19 isolates belonging to our laboratory collection were characterized by routinely used genomic techniques. A PCR assay was performed to detect the genes coding for the carbapenemases OXA-48 and KPC. DNA was extracted using the boiling method (10 min at 95°C). Specific oligonucleotides were used to amplify the different genes (OXA-48 Fw, GCGTGGTTAAGGATGAACAC; OXA-48 Rv, CATCAAGTTCAACCCAACCG; KPC Fw, CGTCTAGTTCTGCTGTCTTG; and KPC Rv, CTTGTCATCCTTGTTAGGCG) (35). The presence of the different carbapenemase genes was confirmed by sequencing the PCR products. MLST of K. pneumoniae was conducted according to the reference protocol (https://bigsdb.web.pasteur.fr/klebsiella/klebsiella.html), under the following conditions: initial denaturation at 94°C for 2 min; 35 cycles of 20 s at 94°C, 30 s at 50°C, and 30 s at 72°C; and final elongation for 5 min at 72°C. Nucleotide sequences were compared with existing entries in the MLST database (https://bigsdb.web.pasteur.fr/cgi-bin/bigsdb/bigsdb.pl?db=pubmlst_klebsiella_seqdef) for generation of allelic numbers and assignment of STs.
Study design.
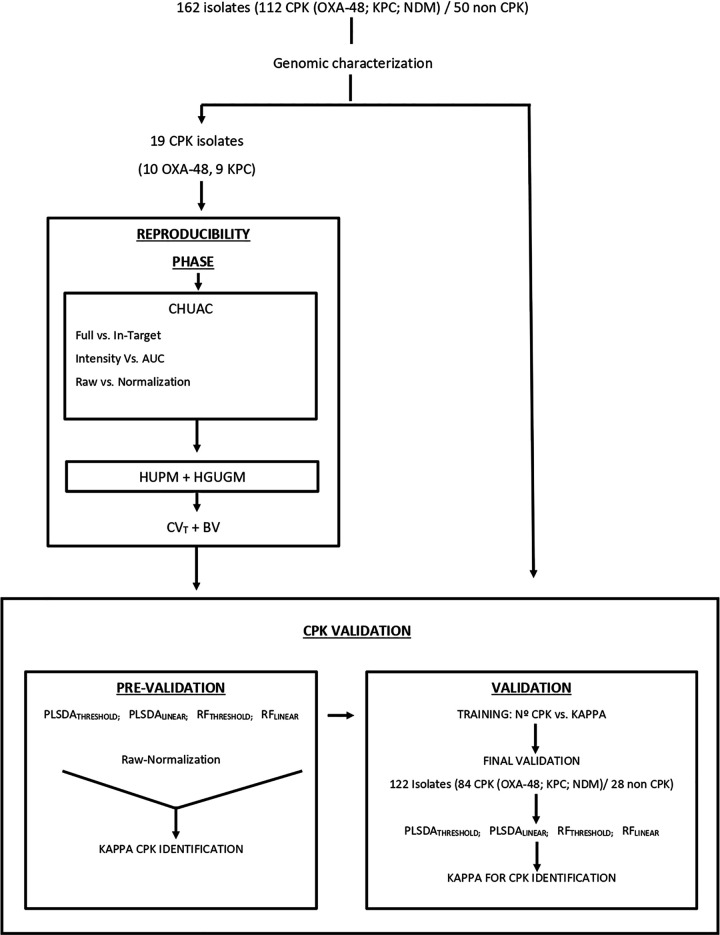
The first part of the study, the reproducibility stage (Fig. 1), was designed to evaluate the biological and technical variation associated with the entire analysis of CPK isolates by MALDI-TOF MS. Initially, imprecision associated with two different extraction methods (“full” versus “in-target”) and two different integration peak methods (“intensity” versus “area under the curve” [AUC]) were evaluated in the Complejo Hospitalario Universitario de A Coruña (CHUAC). The least imprecise methods were chosen for completion of this stage in the Hospital General Universitario Gregorio Marañón (HGUGM) in Madrid and the Hospital Universitario Puerta del Mar (HUPM) in Cádiz.
FIG 1.
The different steps involved in formulation of the study, the reproducibility phase and the CPK identification, are in turn divided into prevalidation and validation steps. CPK, carbapenemase-producing Klebsiella pneumoniae; AUC, area under the curve; CHUAC, Complejo Hospitalario Universitario A Coruña; HUPM, Hospital Universitario Puerta del Mar; HGUGM, Hospital General Universitario Gregorio Marañón, CV, coefficient of variation; BV, biological variation; PLSDA, partial least squares discriminant analysis; RF, random forest; MLINEAR and MTHRESHOLD, methods of generating mass spectra.
The second part of the study, CPK identification, was carried out in several stages. In the first, the prevalidation stage, a brief study was carried out to determine the state-of-the-art use of MALDI-TOF MS to identify CPK. The accuracy of identification was also related to the technical imprecision obtained in the previous reproducibility stage to check for any possible correlation. It was decided that, if the results of the prevalidation stage were very good (kappa > 80%) (36), a more extensive validation study would be performed to assess the final accuracy of MALDI-TOF MS for the identification of CPK. The complete analytical scheme is shown in Fig. 2.
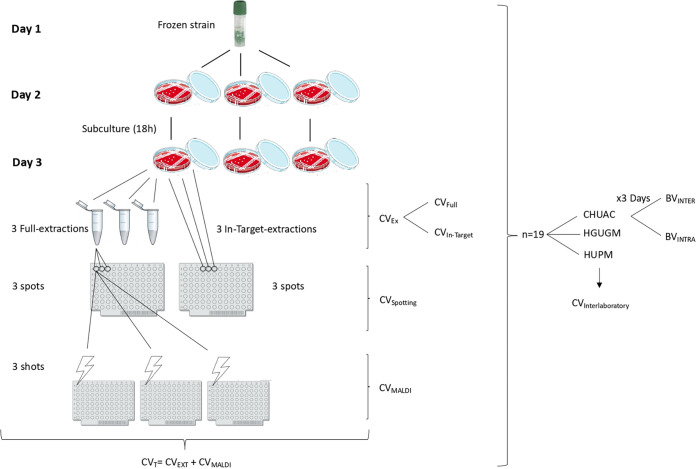
FIG 2.
Standard operating procedure for evaluation of the biological and technical variability in the identification of CPK isolates followed by the Complejo Hospitalario Universitario de A Coruña (CHUAC), Hospital General Universitario Gregorio Marañón (HGUGM), and Hospital Universitario Puerta del Mar (HUPM). The HGUGM and HUPM only evaluated the methods associated with least imprecision to complete the reproducibility phase. The CVs associated with each step of the procedure are illustrated in the figure. CV, level of imprecision associated with the extraction (CVEXT) or the MALDI-TOF MS process (CVMALDI) and total or technical imprecision (CVT); BV, biological variation of isolates; BVINTER, interindividual BV; BVINTRA, intraindividual BV.
(i) Reproducibility stage.
In the reproducibility stage, the mass peak at 5,380 Da was chosen as the reference peak, since it represents 50S ribosomal protein L34, a highly conserved protein in K. pneumoniae, which thus appears in all spectra (Uniprot identification number A0A0V9I726). Imprecision was expressed by the coefficient of variation (%CV), so that the higher the CV, the greater the imprecision. We then identified possible sources of imprecision throughout the analytical process and grouped these sources into “extraction,” “spotting,” and “MALDI.” Thus, “CVEXT” represents imprecision during protein extraction from CPK isolates. Two of the most widely used extraction methods were initially evaluated in the CHUAC: the full extraction method (CVFULL) and the in-target extraction method (CVIN-TARGET). “CVSPOTTING” represents the imprecision associated with the formation, homogenization, and spotting of the sample-matrix mixture on the plate. As the in-target extraction method is performed on the same plate, it does not have a CVSPOTTING-associated imprecision, unlike the full method; that is to say, in the in-target extraction protocol, we cannot differentiate the extraction from the spotting process. Finally, “CVMALDI” represents imprecision associated with conditions inherent in the MALDI-TOF MS equipment. In the CHUAC, MALDI peak integration was conducted in two different ways, by intensity and area under the curve. All isolates were analyzed in triplicate in each step, “extraction,” “spotting,” and “MALDI.” All CVs were calculated for both raw and normalized data. In addition, in the CHUAC, isolates were analyzed on three different days in order to calculate biological variation (BV). Finally, the least imprecise methods (full versus in-target and intensity versus AUC) were chosen to complete the reproducibility stage in the HGUGM and HUPM. The intraindividual biological variation (BVINTRA) was determined as the mean of the CVs obtained for each isolate over 3 different days, while the interindividual biological variation (BVINTER) was determined as the mean of the CVs of all the individuals on 3 days. The testing of the isolates for the analysis of the biological variation was performed after 36 h, and always after thawing and resubculturing. This workflow was adopted for every analysis reported herein (Fig. 2). Mathematical formulas for calculating all CVs are included in Table S1 in the supplemental material.
(ii) CPK identification stage.
The CPK identification part of the study was carried out to assess the accuracy of MALDI-TOF MS for the identification of CPK (blaOXA-48, blaKPC, and blaNDM)-producing isolates. Therefore, the mass spectrum of each isolate was generated by two methods (MTHRESHOLD and MLINEAR), both for raw and normalized data. The mass spectra were then analyzed with two of the most-used multivariate analytical tools based on neural networks, i.e., partial least squares discriminant analysis (PLSDA) and random forest (RF) analysis, implemented in the Clover MS Data Analysis Software (Clover Biosoft, Spain). The software analyzes the similarities and differences between the mass peaks in the spectra and assigns a relative weight in the algorithm to each mass peak in the spectrum for final classification. Both neural network tools are supervised, that is, the algorithm is trained on “historical” data and thus “learns” to assign the appropriate output label to the new sample, i.e., it predicts the result based on proper training data. The PLSDA method is a variant of the partial least squares regression method, where the relationship between the intensity values of the spectrum and the input categories (OXA-48, NDM, KPC, and non-CPK) is modeled by linear regression. Otherwise, the RF classifier builds a model based on multiple decision trees that differentiate the input categories associated with each spectrum (37). Finally, kappa values were obtained for the correct identification of the CPK isolates and, more specifically, the correct identification of blaOXA-48, blaKPC, and blaNDM carbapenemases. Non-carbapenemase-producing isolates were introduced as a category in the analysis, so κ values were also obtained for these isolates. In summary, we evaluated the accuracy of CPK identification for eight possible combinations of data tools, PLSDA-MTHRESHOLD-RAW, PLSDA-MTHRESHOLD-NORM, PLSDA-MLINEAR-RAW, PLSDA-MLINEAR-NORM, RF-MTHRESHOLD-RAW, RF-MTHRESHOLD-NORM, RF-MLINEAR-RAW, and RF-MLINEAR-NORM.
(a) Prevalidation stage. Mass spectra from the reproducibility stage were processed as explained above. Thus, the kappa statistics for the eight tool combinations were obtained in each laboratory, as well as the total kappa for CPK identification. With the results obtained, we had an initial view of the state of the art of this technology for CPK identification, and if the results were very good (kappa > 80%) (36), a more extensive validation study would be performed. The possible correlation between the eight tools’ combination kappa values and their CVs calculated in the reproducibility stage was also studied.
(b) Validation stage. CPK isolates that were not used in the prevalidation stage were processed in CHUAC. Isolates were analyzed by the same operator, with in-target extraction, the peak intensity method, and the same MALDI-TOF MS instrument. A training step was developed for the eight tool combinations to allow the software to learn to identify CPK correctly. We determined how many samples were required to obtain a kappa peak for each of the eight groups. When the training step was completed, we performed the final validation step, in which the accuracy (kappa) of the eight combinations in CKP identification was obtained.
MALDI-TOF MS spectrum acquisition.
Bacterial isolates were stored at −80°C using glass cryopearls in a small vial (Deltalab, Barcelona, Spain). On the first day, isolates were thawed on a blood agar plate by removing one of the pearls from the tube with a sterile loop and then rolling it on the plate. After 18 h of incubation, isolates were subcultured for another 18 h on a blood agar plate for analysis under standard conditions. Incubation was performed in an aerobic atmosphere at 37°C (Fig. 2). All isolates were of the same age to control for senescence-associated changes in the mass peak spectrum. Isolates were submitted to a modified Hodge test to check for the presence of the carbapenemase enzyme (38). No discordant results were found in relation to the phenotypic and genotypic annotation.
The same operator performed the analysis of all isolates in each laboratory to reduce the associated variability. The operators were given a precise protocol (Fig. 1) for conducting the whole procedure to avoid externally introduced experimental imprecision. Bacterial proteomes were analyzed by in-target and full protein extraction (39). Briefly, the in-target extraction consists of a protein extraction made directly in the MALDI target by spotting 1 μl of formic acid once the sample is dried and then adding the IVD HCCA (α-cyano-4-hydroxycinnamic acid)-portioned matrix (Bruker Daltonics, Germany). The full protein extraction consists of performing a full ethanol/formic acid extraction of the isolate in a tube and then spotting the extract in the MALDI target plate. Protocols are provided in Table S2.
All participating laboratories used the MALDI Biotyper (Bruker Daltonics, Germany). MALDI-TOF MS spectra were acquired in a microflex LT/SH smart instrument with the FlexControl software version 3.4 in a linear positive ion mode within a mass range of 2 to 20 kDa. A total of 240 satisfactory laser shots were acquired in 40 shot steps for each spectrum using the spiral small movement. External calibration was performed using bacterial test standard (BTS; Bruker Daltonics, Germany) prior to each run. Species had to be confirmed in comparison with the mass spectrum library using the MALDI Biotyper Compass software (version 4.1.100, Bruker Daltonics).
Spectra were processed with the Clover MS Data Analysis Software (Clover Biosoft, Granada, Spain). The first step involved preprocessing all spectra by applying noise reduction with the Savitzky-Golay filter (smoothing filter with window length of 11 and polynomial order 3) and then subtracting the baseline by using the Top-Hat filter (baseline-removing filter with a factor of 0.02). Nine replicates (3 spots with 3 spectra each) of each isolate were processed. For the data from the three laboratories, an average spectrum for each isolate was obtained by a two-step process of alignment. The first step consisted of aligning the spectra for all spots and then obtaining a unique average spectrum. The second step involved repeating this process but aligning the spectra obtained for each spot to produce the final average spectrum. Once this process was completed, only one average spectrum remained for each isolate. The main goal of performing an alignment-plus-average spectrum process was to minimize the variability between replicates of the same isolate. Once one spectrum per isolate was available in the software platform, all the spectra were again aligned in order to increase the accuracy of the next steps. All the alignment processes were performed by considering the most representative peaks of each sample included in the set to be aligned. These peaks were then used to form a reference peak list. Each spectrum peak was shifted within a linear tolerance of 2,000 ppm to correspond to this list.
Up to this point, all of the procedures explained are common to both acquisition methods, MTHRESHOLD and MLINEAR, and it is in the subsequent steps where the methods differ.
For generating a peak matrix in the MTHRESHOLD method, a threshold value (0.01) was established so that all peaks with at least 1% of the maximum intensity of the spectrum were considered. All the resultant peaks were merged into a common list within a linear tolerance of 2,000 ppm. The MLINEAR method did not perform the peak finding step, and the mass lists for each spectrum were merged into a common list. Peak matrices were later normalized by the total ion current (TIC) method. This method consists of accumulating all the values of the resulting spectrum (for MTHRESHOLD, the values are the peaks obtained, and for MLINEAR, the values are the raw intensities which compose the spectrum) and then dividing each intensity value by the resulting accumulated value.
Bioinformatics tools for identification of CPK isolates.
After the spectrum acquisition, we obtained four different matrices: MTHRESHOLD-RAW, MTHRESHOLD-NORM, MLINEAR-RAW, and MLINEAR-NORM. Two supervised machine learning algorithms were then applied to these matrices: PLSDA and RF. The main purpose of these analyses was to study how each of the eight different combinations of tools discriminated the different categories (OXA-48, KPC, NDM, and non-CPK). The kappa statistic value was used to explain the efficacy of the combined results.
Statistical analysis.
Statistical analysis was performed using GraphPad Prism 8.0 software. The normalized distribution was tested with the D’Agostino-Pearson test. Continuous variables are presented as median values and interquartile ranges (IQRs) (25th to 75th percentiles), and categorical data are displayed as counts and percentages. Analysis of differences between 2 groups was performed using the Wilcoxon matched-pairs signed-rank test. A two-sided P value of <0.05 was considered statistically significant for all calculations. The kappa value was the score of a k-fold (k = 10) cross-validation analysis, obtained as the relationship between the number of samples correctly classified and the total number of spectra under study (40).
Data availability.
The BioProject accession number for strain genomes is PRJEB39112. It is anticipated that this accession number will be released by early 2022; until that time, the strain genome data will be available from the corresponding author upon request.
RESULTS
Bacterial isolates.
The 112 CPK isolates represent a not-geographically related sampling, coming from 15 different hospitals throughout the territory, in addition to 16 isolates of unknown origin. Among them, 11 different sequence types (STs) were detected: ST-11 (n = 27), ST-512 (n = 27), ST-15 (n = 12), ST-147 (n = 13), ST-307 (n = 12), ST-1961 (n = 8), ST-101 (n = 6), ST-273 (n = 3), ST-437 (n = 2), ST-395 (n = 1), and ST-405 (n = 1). A total of 47 of the 112 isolates carried the blaKPC gene, 58 harbored the blaOXA-48 gene, and 7 carried the blaNDM gene. Carbapenemase-nonproducing K. pneumoniae isolates were not submitted to detailed genomic analysis. Data are summarized in Table S3.
Nineteen of the 112 representative and randomly selected CPK isolates were used in the reproducibility stage, including 9 KPC-producing isolates (4 belonging to ST11 and to 5 ST512) and 10 OXA-48-producing isolates (5 belonging to ST11 and 5 to ST15) (Table S3). These isolates were examined in triplicate in the three laboratories (CHUAC, HGUGM, and HUPM).
Reproducibility stage. (i) Intensity versus AUC.
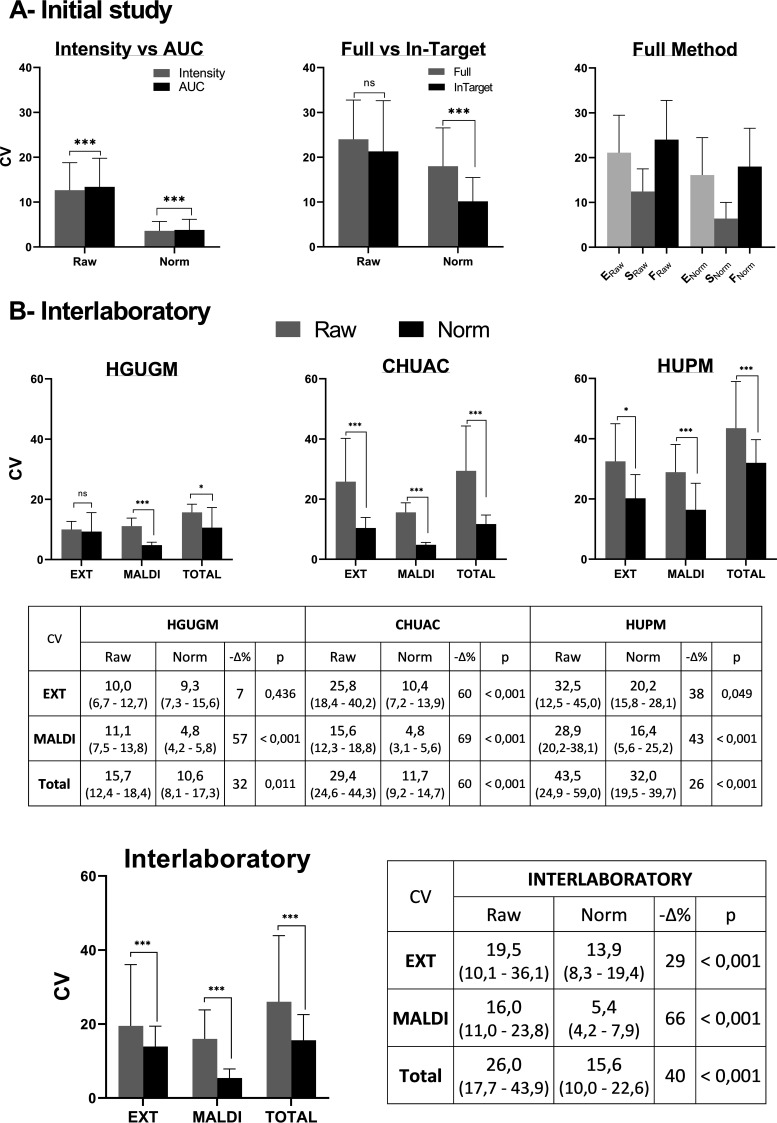
In the CHUAC, the peak at m/z 5,380 was integrated in two different ways, area under the curve (AUC) and intensity. AUC integration was slightly more imprecise (although clearly significant [P < 0.0001]) for both raw and normalized data (Fig. 3A). The imprecision for raw data was a CVAUC value of 13.4 (8.2 to 19.8) and a CVINTENSITY value of 12.7 (8.1 to 18.8). The imprecision for normalized data was a CVAUC value of 3.8 (2.3 to 6.2) and a CVINTENSITY value of 3.6 (2.3 to 5.7). Throughout the study, all peaks from the MALDI spectra were subsequently integrated using the intensity mode.
FIG 3.
Reproducibility stage. (A) Initial study. Coefficients of variation (CVs) obtained in the CHUAC to compare intensity versus AUC methods of peak acquisition and full versus in-target methods of extraction. Norm, normalized data; E, extraction; S, spotting; F, full extraction. (B) Interlaboratory CVs obtained in 3 hospitals for K. pneumoniae isolates analyzed by MALDI-TOF MS (top) and considering the results from the 3 laboratories together (bottom). EXT, extraction step (in-target method); MALDI, MALDI-TOF MS process; TOTAL, the entire process (EXT+MALDI); HGUGM, Hospital General Universitario Gregorio Marañon; CHUAC, Complexo Hospitalario Universitario A Coruña; HUPM, Hospital Universitario Puerta del Mar; interlaboratory, represents overall CV from the 3 hospitals involved; −Δ%, % decrease in kappa of normalized versus raw data. All data are presented as median values (bars) and interquartile ranges (error bars). All adjusted P values were obtained by Wilcoxon matched-pairs signed-rank test. ns, P ≥ 0.05; *, P < 0.05; **, P < 0.005; ***, P < 0.001.
(ii) Full versus in-target.
Comparison of the full versus in-target extraction methods revealed that the full method was slightly more imprecise than the in-target method. For raw data, the CVFULL value was 24.0 (17.5 to 32.8) and the CVIN-TARGET value was 21.3 (13.2 to 32.6), with no significant difference between them (P = 0.3004) (Fig. 3A). For normalized data, the CVFULL value was 18.0 (14.3 to 26.6) and the CVIN-TARGET value was 10.2 (7.0 to 15.5), with a significant difference between the full and in-target extraction values (P < 0.0001). Throughout the study, all isolates were subsequently extracted using the in-target extraction method.
For further examination of these results, we must first remember that the total imprecision associated with an extraction method was calculated as the sum of the imprecision from protein extraction plus that from the spotting process. As the in-target extraction method is performed on the same plate, it does not have an associated CVSPOTTING imprecision, unlike the full method. To evaluate the imprecision associated with the full method, we represent the two components (extraction and spotting) separately. As observed from the results in Fig. 3, the variability in the protein extraction component (E) is almost double that in the spotting (S) process.
(iii) Interlaboratory variability.
Once the CHUAC study was completed, the same 19 isolates were processed in the HGUGM and HUPM using the in-target extraction method, and the peak at m/z 5,380 was integrated as intensity. Thus, in each laboratory, we obtained the levels of imprecision associated with the extraction (CVEXT), with the MALDI-TOF MS process (CVMALDI), and with the total or technical imprecision (CVT). The results are shown in Fig. 3B, both for raw and normalized spectra, including significant differences. Intra- and interlaboratory levels of imprecision were also calculated.
The lowest total levels of imprecision for both raw and normalized spectra were obtained in the HGUGM, and the highest levels in the HUPM. In all three laboratories, there was a significant reduction in imprecision after normalization of the data (except for extraction in the HGUGM, which was already low initially). The total interlaboratory imprecision values obtained were a CVT value of 26.0 (17.7 to 43.9) for raw spectra and a CVT value of 15.6 (10.0 to 22.6) for normalized spectra. Therefore, a significant reduction in the total imprecision of 40% (P < 0.0001) was achieved. For normalized spectra, the total imprecision due to extraction (CVEXT = 3.9 [8.3 to 19.4]) was twice that due to the MALDI reading (CVMALDI = 5.4 [4.2 to 7.9]).
(iv) Biological variation.
Biological variation (BV) was calculated in the CHUAC for the 19 isolates processed during three different days. The intraindividual variation, BVINTRA, was 28 (19 to 42), and the interindividual variation, BVINTER, was 43 (30 to 58).
CPK identification. (i) Prevalidation.
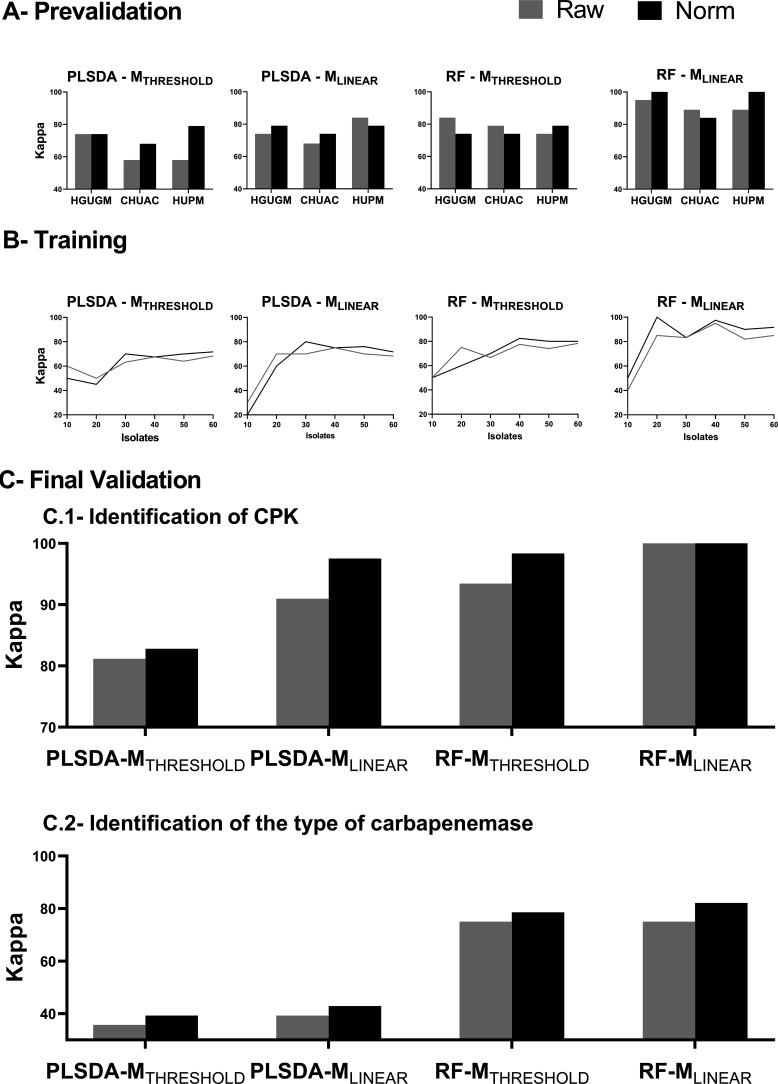
The results obtained in the PLSDA-MTHRESHOLD group showed a clear improvement for correct CPK identification (Δκ = +18%) after normalization of the mass spectra, except in the HGUGM, where the imprecision was the same for raw and normalized spectra (it was very low in both groups) (Fig. 4A). Consequently, for the PLSDA-MTHRESHOLD group, technical imprecision (CV) appears to be inversely related to the accuracy of CPK identification (kappa). For the other groups, no clear association between imprecision and CPK identification accuracy was observed. The kappa results were good (kappa > 65) for all eight tool combinations and very good for the RF-MLINEAR group, which achieved a mean kappa of 95% for correct CPK identification (Fig. 4). The kappa results were remarkably similar among the three hospitals for all tool combinations, and the MALDI-TOF MS replicability between laboratories for CPK identification was confirmed. These promising results led us to continue with the next validation stages.
FIG 4.
CPK identification. (A) Prevalidation. Kappa values for CPK identification (KPC versus OXA-48) obtained in three hospitals for four data processing bioinformatic tool combinations (PLSDA-MTHRESHOLD, PLSDA-MLINEAR, RF-MTHRESHOLD, and RF-MLINEAR) for both raw and normalized spectra. PLSDA, partial least square discriminant analysis; RF, random forest; THRESHOLD, threshold mode of spectrum acquisition; LINEAR, linear mode of spectrum acquisition; Norm, normalized; HGUGM, Hospital General Universitario Gregorio Marañon; CHUAC, Complexo Hospitalario Universitario A Coruña; HUPM, Hospital Universitario Puerta del Mar. (B) Training. Kappa values obtained in the training set versus the number of isolates used for both raw and normalized spectra. (C) Final validation. 1. Identification of CPK isolates. Kappa values obtained with a group of 122 isolates (84 CPK and 38 non-CPK) processed at the CHUAC by MALDI-TOF MS and four bioinformatic tool combinations. 2. Identification of the type of carbapenemase. Kappa values obtained for the 84 CPK isolates when identification of the type of carbapenemase (OXA-48, KPC, and NDM) was performed.
(ii) Validation stage.
The training sets used were the spectra obtained in the CHUAC from the 110 isolates. The kappa values for the eight tool combinations were obtained using an increasing number of isolates. Thus, the training set began with 10 isolates and reached up to 60 isolates, increasing in groups of 10. The isolates used for the training sets were randomly selected by the software. The maximum kappa value (in which correct CPK identification was maximal) was achieved with 40 samples for the RF algorithm, applying either the MTHRESHOLD or the MLINEAR method. When analyzing the PLSDA algorithm, the maximum kappa was obtained with 50 isolates using the MLINEAR and with 60 isolates using the MTHRESHOLD method (Fig. 4B). The graphs showed that once a maximum was reached, the curves flattened out or increased very slowly. In general, the accuracy of CPK identification was slightly higher for normalized than for raw spectra, especially as the kappa values increased. The need of fewer isolates in the MLINEAR than in the MTHRESHOLD method and in the RF than in the PLSDA algorithm for achieving the maximum kappa reveals that the method with higher classifying capabilities is the RF-MLINEAR. To unify criteria, the same number of isolates was chosen in the training set for all methods. The criterion applied was using the smallest number of isolates to achieve the maximum value for the kappa. Thus, a sampling size of 40 isolates was chosen for the definitive final validation stage. The maximum kappa value was achieved for the RF-MLINEAR method after normalization, yielding 97.5% accuracy. In this case, the RF feature importance, that is, the description of the mass peaks in the spectrum that are most relevant in the formulation of the model, is described in Fig. S1. However, no specific mass peaks or biomarkers in the spectrum that were specific to a type of carbapenemase were identified, but the entire mass peak data set was taken into account in the final algorithm in which each mass peak has its relative importance (16).
The final validation stage was carried out with the remaining 122 isolates, which in turn consisted of 84 CPK and 38 non-CPK isolates. Among the CPK isolates, 45 of the 84 were OXA-48, 36 of the 84 were KPC, and 3 of the 84 were NDM (Table 1). Thus, with a greater number of samples than in the previous stages, the relationship between tool combinations and imprecision depended similarly on both the type of spectrum acquisition (threshold versus linear) and the machine learning tool used for the analysis (PLSDA versus RF) (Fig. 4C). Therefore, we observed that for all the methods of analysis, there was a considerable increase in the kappa value for normalized versus raw spectra. In contrast, for RF-MLINEAR, there was no such dependence, with this tool combination yielding similar kappa values regardless of the normalization of the spectra, especially in the identification of CPK.
TABLE 1.
Results of the optimized model for CPK identification by MALDI-TOF
| Characterization | No. of isolates characterized as CPK or as bearing indicated type of carbapenemase using: |
% agreementc | |||
|---|---|---|---|---|---|
| Genomic analysisa | MALDI-TOF MS using the RF-MLINEAR algorithm for data analysisb |
||||
| CPK | Non-CPK | ||||
| CPK | 84 | 84 | 0 | 100 | |
| Non-CPK | 38 | 0 | 38 | 100 | |
| Total | 122 | 100 | |||
| KPC | NDM | OXA-48 | |||
| KPC | 36 | 25 | 0 | 11 | 69 |
| NDM | 3 | 0 | 2 | 1 | 66 |
| OXA-48 | 45 | 0 | 3 | 42 | 93 |
| Total | 84 | 82 | |||
| Overall accuracy | 122 | 88 | |||
Number of isolates included for validation in each group.
Results for the identification of carbapenemase-producing K. pneumoniae (CPK) or of the type of carbapenemase, obtained by MALDI-TOF MS using the RF-MLINEAR algorithm for data analysis.
Percentage of agreement between the genomic analysis obtained by whole-genome sequencing and the proteomic analysis obtained by MALDI-TOF MS.
The final validation was divided into two subsequent stages. The first was the identification of CPK (Fig. 4C.1). As in previous stages, RF-MLINEAR yielded the highest overall kappa value, reaching correct identification of 100% (84/84) of CPK isolates and also reaching 100% (38/38) specificity, meaning that the method is able to identify a CPK isolate among all K. pneumoniae isolates with maximum reliability (Table 1). Regarding the identification of the type of carbapenemase (Fig. 4C.2), again, the RF-MLINEAR method yielded the highest overall kappa value, reaching correct identification of 82% (69/84) of the isolates; thus very good results were also obtained in the final validation stage. The highest rate of correct identifications was accomplished for the OXA-48 isolates, with 93% (42/45) identified correctly. For KPC and NDM isolates, the rates of correct identifications were very similar, 69% (25/36) and 66% (2/3), respectively, thus obtaining good results. However, the number of NDM isolates was too low to reach significance (Table S4).
DISCUSSION
Analysis of the operating procedure and subsequent data management are critical for reliable recovery of information from mass spectrum profiles obtained by MALDI-TOF MS. In this study, we analyzed the biological and technical reproducibility of the operating procedure, we evaluated how this variance can affect the final results of the analysis, and finally, we introduced an improved MALDI-TOF MS data analysis pipeline for the identification of CPK. Accordingly, we suggest some solutions for a number of current bottlenecks in processing MALDI-TOF MS data and provide technical know-how regarding CPK identification.
Regarding the analysis of biological variation, it is known that for a method to be adequate in terms of technical specifications, the imprecision associated with the technique must be half the intraindividual biological variation (BVINTRA) (41, 42). In the present study, this held true when the spectra were normalized, as the total interlaboratory imprecision was a CVT value of 15.6%, which is approximately half of the BVINTRA value of 28%. However, raw spectra with a CVT value of 26% did not fit the above criterion, and normalization of MALDI-TOF MS spectra is therefore greatly encouraged.
In addition to BV, several experimental and instrumental influences introduce systematic and random variations into the mass spectra obtained by MALDI-TOF MS. Oberle et al. studied an ESBL-producing-E. coli outbreak by MALDI-TOF MS and reported that the technical reproducibility was the most critical part of the process and should be further investigated (43). The performance of MALDI-TOF MS could be evaluated by the modifications in growth conditions, the type of extraction method, the use of various matrixes, and/or a change in the mass range or the post-data treatment of the mass peaks in the spectra. The type of medium and other culture conditions have some effects but do not affect the overall ability of MALDI-TOF MS for identification of bacterial species, as previously demonstrated (25). To simplify the process, we chose HCCA, which is the most widely used matrix in the routine identification of microorganisms by MALDI-TOF, and a mass range from 2 to 20 kDa, so the software parameters would be the same as those already used in the routine identification. When evaluating the type of extraction, the in-target method proved more precise than the full extraction method. This finding favors the application of the CPK identification, as it can be performed in real-time in practice, allowing add-on information, as only one spectrum is used for bacterial identification and detection of antibiotic resistance for an isolate. When evaluating the full extraction method, the extraction step is the most critical part in the process, as the variability is almost twice that in the spotting step, and extra attention and care must be taken with the sample when choosing this procedure. When replicating the analysis in different laboratories, the variability in the process differed significantly between laboratories. The CV values obtained correlated with the relative experience of the centers: the greater the experience of the center, the lower the CV value obtained, with the HGUGM and CHUAC being the most experienced and the HUPM the least experienced. In general, normalizing the spectra obtained almost halves (40%) the associated imprecision. Therefore, normalization is crucial for reducing the variability of the results at all stages of the analytical process. The spectra associated with the highest levels of imprecision will be the most improved by the normalization process. The integration of the mass peaks is significantly better and contributes to less imprecision when the intensity of the mass peaks is used rather than the AUC.
In the prevalidation step for CPK identification using neural networks tools, we found that for the PLSDA-MTHRESHOLD group, technical imprecision (CV) appeared to be inversely related to the accuracy of CPK identification (kappa). However, for the other groups, no such clear association between imprecision and CPK identification accuracy was observed. This finding led us to conclude that when using specific and definite numbers of peaks and classical statistics, the imprecision significantly affected the results of the analysis, but when using neural network tools, this imprecision was minimized. In addition, comparison of the accuracy of the different methods of analysis among the three laboratories showed that the levels of accuracy were very similar for the different types of analysis, although the associated imprecision was highly dependent on the laboratory concerned. This again supports the idea that normalization helps in reducing variability and so improves the interlaboratory transferability, previously identified as a factor precluding the use of MALDI-TOF MS for detection of antibiotic resistance and for typing purposes (25–29).
In the training of neural network tools, normalization reduces the number of samples available for achieving the maximum accuracy but also improves the accuracy (higher kappa values). Furthermore, in the final validation stage, we observed that the addition of the MLINEAR method with the RF algorithm reduces the imprecision the maximum amount, being less dependent on the normalization procedure and increasing the kappa values. Thus, we conclude that using the whole spectra for the analysis, rather than specific peaks, reduces the imprecision and has less impact on the results of the analysis. Focusing the analysis on specific peaks has proved to be less accurate, as it is strongly influenced by the variability in the processing. In addition, peak shifts and variable intensities can depend on the solubility or on missense mutations with changes in the amino acid sequence.
Being aware of the error and the variability of the procedure is fundamental for implementation of good practice guidelines and improving the accuracy of the results obtained. Thus, we provide a summary of our study findings that could lay the groundwork for future studies regarding antimicrobial resistance detection and bacterial typing using MALDI-TOF MS (44), as follows.
Good practice guidelines for MALDI-TOF MS identification of antibiotic-resistant isolates.
1. Include large and well-characterized collections of nonepidemiologically related isolates to build a training set.
2. Integrate mass spectra with the intensity of the mass peaks.
3. The in-target extraction method can be used not only to build the model but also for clinical validation and for use in clinical practice (need for preliminary study).
4. If using the full extraction method as the operating procedure, pay special attention to the extraction step, which is strongly influenced by variability.
5. Highly trained personnel operating MALDI-TOF MS generally provide the most accurate results.
6. When using specific biomarkers, ensure the validity of the extraction method and train individuals in the operating procedure, as the results are very imprecise.
7. For generating spectra, use methods that use the maximum number of peaks possible, as these methods are less influenced by the variability in the processing steps.
8. The use of neural network analysis to detect antimicrobial resistance patterns is highly recommended, as the analysis is less influenced by the expression of certain peptides or proteins, variability is reduced, and transferability is guaranteed.
9. Normalization helps to reduce the variability and is therefore strongly encouraged, especially when definite biomarkers are used.
MALDI-TOF MS proved valuable for assessing potential different groups of resistance using high-quality data focused on the existence or the lack of definite spectral mass peaks to recognize antibiotic-resistant bacteria (16–21). However, unlike most other studies, we did not focus on target peaks but based the classification on neural network analysis. This type of analysis is especially useful for identifying patterns or trends that are hidden in the data in automatic learning, as interpreting the amount and complexity of the information generated by visual inspection of large data sets is often not possible. RF is a supervised machine learning technique based on decision trees. Its main advantage is that, for similar training performances, it obtains a better generalization yield than other supervised machine learning algorithms. In our case, an improved MALDI-TOF MS data analysis pipeline was designed with a succession of steps that implied noise reduction with the Savitzky-Golay filter, baseline subtraction by using the Top-Hat filter, a two-step process of alignment, selection of all peaks contained in the bacterial spectra, integration of mass spectra with the intensity of the mass peaks, TIC normalization, and an RF algorithm for analysis that provided a rate of 100% correct CPK identifications. The whole procedure, presented herein, is designed to reduce variability, guarantee interlaboratory reproducibility, and maximize the information obtained from the bacterial proteome. In contrast to approaches based on genomic assays, this approach is much less expensive, faster, and simpler. Thus, we think the ideal workflow for this MALDI-TOF application would be as an initial screening test for identification of CPK isolates in a clinical laboratory, as direct tracking can be performed at the same time as bacterial identification, using the same hands-on procedure and the same spectrum. The only difference would be the later processing by machine learning algorithms that would only take minutes to apply, once the software is designed with the optimized parameters (Clover MS Data Analysis Software; Clover Biosoft, Spain). The processing is fully automated, so no particular skills beyond the basic use of MALDI-TOF MS are needed, allowing full integration into the routine laboratory workflow.
The identification of the type of carbapenemase contributes to the prescription of targeted treatments against specific classes of carbapenemases, such as the new β-lactam–β-lactamase inhibitors imipenem/relebactam and ceftazidime/avibactam, which are particularly valuable in treating KPC- and OXA-48-producing isolates, respectively (45). The analysis yielded 82% correct identifications and had special success in identifying the OXA-48 type of carbapenemase, with 93% agreement between the proteomic and the genomic analysis. Further introduction of KPC and NDM isolates into the training set would enhance the rates of accuracy for these groups of carbapenemases. Replacement of molecular techniques by MALDI-TOF MS at this stage is still far from consideration. However, the determination of the type of carbapenemase in a specific epidemiological setting, such as in areas with a high prevalence of OXA-48-producing K. pneumoniae isolates, could be very helpful until molecular results are available.
Validation in a further collection of K. pneumoniae isolates is necessary before the method can be fully integrated into the normal workflow in clinical microbiological laboratories. It would also be interesting to expand the application to the identification of carbapenemases in other Enterobacterales. However, we have demonstrated that direct tracking of CPK isolates using MALDI-TOF MS is possible within minutes, reproducible with technical know-how available for all users, and significantly less expensive than genomic-based technologies. In a potential outbreak situation, MALDI-TOF MS-based identification of CPK isolates might provide the first evidence to initiate intensive health care and infection control measures, modifying the clinical outcome of these infections.
ACKNOWLEDGMENTS
This work was supported by funding awarded to M.O. through the Competitive Research Call 2019 of the Spanish Society of Infectious Diseases and Clinical Microbiology (SEIMC). The research was also funded by the Fondo de Investigación Sanitaria through grants no. PI15/00860 and PI18/00860 to G.B., integrated in the Plan Nacional de I+D and funded by the Instituto de Salud Carlos III (ISCIII) and by the Spanish Network of Research in Infectious Diseases (REIPI), grant no. RD16/0016/0006, integrated in the National Plan for Scientific Research, Development and Technological Innovation 2013-2016 and funded by the ISCIII, General Subdirection of Assessment and Promotion of the Research, European Regional Development Fund (FEDER) A Way of Making Europe. M.O. was financially supported by the Juan Rodés program (ISCIII-SERGAS, JR18/00006) and E.G. by the research call 2019 of the SEIMC.
Clover Biosoft received funding from the European Union’s Horizon H2020 research and innovation program under grant agreement no. 868365.
Footnotes
Supplemental material is available online only.
Contributor Information
Marina Oviaño, Email: marina.oviano.garcia@sergas.es.
Nathan A. Ledeboer, Medical College of Wisconsin
REFERENCES
- 1.European Center for Disease Prevention and Control. 2020. The European Surveillance System Antimicrobial Resistance (AMR) reporting protocol 2020. Available at: https://www.ecdc.europa.eu/sites/default/files/documents/EARS-Net-reporting-protocol-2020.pdf.
- 2.Centers for Disease Control and Prevention. 2019. Antibiotic resistance threats in the United States, 2019. CDC, Atlanta, GA. [Google Scholar]
- 3.WHO. 2017. WHO publishes list of bacteria for which new antibiotics are urgently needed. Saudi Med J 38:444–446. [Google Scholar]
- 4.European Centre for Disease Prevention and Control. 2019. Surveillance of antimicrobial resistance in Europe 2018. ECDC, Stockholm, Sweden. Available at: https://www.ecdc.europa.eu/sites/default/files/documents/surveillance-antimicrobial-resistance-Europe-2018.pdf. [Google Scholar]
- 5.Rodriguez-Baño J, Gutiérrez-Gutiérrez B, Machuca I, Pascual A. 2018. Treatment of infections caused by extended-spectrum-beta-lactamase-, AmpC-, and carbapenemase-producing Enterobacteriaceae. Clin Microbiol Rev 31:e00079-17. 10.1128/CMR.00079-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Magiorakos AP, Burns K, Rodríguez Baño J, Borg M, Daikos G, Dumpis U, Lucet JC, Moro ML, Tacconelli E, Simonsen GS, Szilágyi E, Voss A, Weber JT. 2017. Infection prevention and control measures and tools for the prevention of entry of carbapenem-resistant Enterobacteriaceae into healthcare settings: guidance from the European Centre for Disease Prevention and Control. Antimicrob Resist Infect Control 6:113. 10.1186/s13756-017-0259-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bizzini A, Greub G. 2010. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry, a revolution in clinical microbial identification. Clin Microbiol Infect 16:1614–1619. 10.1111/j.1469-0691.2010.03311.x. [DOI] [PubMed] [Google Scholar]
- 8.Clark AE, Kaleta EJ, Arora A, Wolk DM. 2013. Matrix-assisted laser desorption ionization-time of flight mass spectrometry: a fundamental shift in the routine practice of clinical microbiology. Clin Microbiol Rev 26:547–603. 10.1128/CMR.00072-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suarez S, Ferroni A, Lotz A, Jolley KA, Guérin P, Leto J, Dauphin B, Jamet A, Maiden MC, Nassif X, Armengaud J. 2013. Ribosomal proteins as biomarkers for bacterial identification by mass spectrometry in the clinical microbiology laboratory. J Microbiol Methods 94:390–396. 10.1016/j.mimet.2013.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang S, Nianguang CAI, Penzuti Pacheco P, Narrandes S, Wang Y, Xu W. 2018. Applications of support vector machine (SVM) learning in cancer genomics. Cancer Genomics Proteomics 15:41–51. 10.21873/cgp.20063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bibault JE, Giraud P, Burgun A. 2016. Big Data and machine learning in radiation oncology: state of the art and future prospects. Cancer Lett 382:110–117. 10.1016/j.canlet.2016.05.033. [DOI] [PubMed] [Google Scholar]
- 12.Flores-Treviño S, Garza-González E, Mendoza-Olazarán S, Morfín-Otero R, Camacho-Ortiz A, Rodríguez-Noriega E, Martínez-Meléndez A, Bocanegra-Ibarias P. 2019. Screening of biomarkers of drug resistance or virulence in ESCAPE pathogens by MALDI-TOF mass spectrometry. Sci Rep 9:18945. 10.1038/s41598-019-55430-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Figueroa-Espinosa R, Costa A, Cejas D, Barrios R, Vay C, Radice M, Gutkind G, Di Conza J. 2019. MALDI-TOF MS based procedure to detect KPC-2 directly from positive blood culture bottles and colonies. J Microbiol Methods 159:120–127. 10.1016/j.mimet.2019.02.020. [DOI] [PubMed] [Google Scholar]
- 14.Pliakos EE, Andreatos N, Shehadeh F, Ziakas PD, Mylonakis E. 2018. The cost-effectiveness of rapid diagnostic testing for the diagnosis of bloodstream infections with or without antimicrobial stewardship. Clin Microbiol Rev 31:e00095-17. 10.1128/CMR.00095-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodríguez-Sánchez B, Cercenado E, Coste AT, Greub G. 2019. Review of the impact of MALDI-TOF MS in public health and hospital hygiene, 2018. Eurosurveillance 24:1800193. 10.2807/1560-7917.ES.2019.24.4.1800193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lau AF, Wang H, Weingarten RA, Drake SK, Suffredini AF, Garfield MK, Chen Y, Gucek M, Youn JH, Stock F, Tso H, DeLeo J, Cimino JJ, Frank KM, Dekker JP. 2014. A rapid matrix-assisted laser desorption ionization-time of flight mass spectrometry-based method for single-plasmid tracking in an outbreak of carbapenem-resistant Enterobacteriaceae. J Clin Microbiol 52:2804–2812. 10.1128/JCM.00694-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Josten M, Dischinger J, Szekat C, Reif M, Al-Sabti N, Sahl HG, Parcina M, Bekeredjian-Ding I, Bierbaum G. 2014. Identification of agr-positive methicillin-resistant Staphylococcus aureus harbouring the class A mec complex by MALDI-TOF mass spectrometry. Int J Med Microbiol 304:1018–1023. 10.1016/j.ijmm.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 18.Griffin PM, Price GR, Schooneveldt JM, Schlebusch S, Tilse MH, Urbanski T, Hamilton B, Venter D. 2012. Use of matrix-assisted laser desorption ionization-time of flight mass spectrometry to identify vancomycin-resistant enterococci and investigate the epidemiology of an outbreak. J Clin Microbiol 50:2918–2931. 10.1128/JCM.01000-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holzknecht BJ, Dargis R, Pedersen M, Pinholt M, Christensen JJ, Danish Enterococcal Study Group . 2018. Typing of vancomycin-resistant enterococci with MALDI-TOF mass spectrometry in a nosocomial outbreak setting. Clin Microbiol Infect 24:1104.e1–1104.e4. 10.1016/j.cmi.2018.03.020. [DOI] [PubMed] [Google Scholar]
- 20.Cabrolier N, Sauget M, Bertrand X, Hocquet D. 2015. Matrix-assisted laser desorption ionization-time of flight mass spectrometry identifies Pseudomonas aeruginosa high-risk clones. J Clin Microbiol 53:1395–1398. 10.1128/JCM.00210-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mulet X, García R, Gayá M, Oliver A. 2019. O-antigen serotyping and MALDI-TOF, potentially useful tools for optimizing semi-empiric antipseudomonal treatments through the early detection of high-risk clones. Eur J Clin Microbiol Infect Dis 38:541–544. 10.1007/s10096-018-03457-z. [DOI] [PubMed] [Google Scholar]
- 22.Rodrigues C, Novais Sousa C, Ramos H, Coque TM, Cantón R, Lopes JA, Peixe L. 2017. Elucidating constraints for differentiation of major human Klebsiella pneumoniae clones using MALDI-TOF MS. Eur J Clin Microbiol Infect Dis 36:379–386. 10.1007/s10096-016-2812-8. [DOI] [PubMed] [Google Scholar]
- 23.Egli A, Tschudin-Sutter S, Oberle M, Goldenberger D, Frei R, Widmer AF. 2015. Matrix-assisted laser desorption/ionization time of flight mass-spectrometry (MALDI-TOF MS) based typing of extended-spectrum β-lactamase producing E. coli—a novel tool for real-time outbreak investigation. PLoS One 10:e0120624. 10.1371/journal.pone.0120624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rizzardi K, Åkerlund T. 2015. High molecular weight typing with MALDI-TOF MS—a novel method for rapid typing of Clostridium difficile. PLoS One 10:e0122457. 10.1371/journal.pone.0122457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lasch P, Fleige C, Stämmler M, Layer F, Nübel U, Witte W, Werner G. 2014. Insufficient discriminatory power of MALDI-TOF mass spectrometry for typing of Enterococcus faecium and Staphylococcus aureus isolates. J Microbiol Methods 100:58–69. 10.1016/j.mimet.2014.02.015. [DOI] [PubMed] [Google Scholar]
- 26.O’Connor JA, Corcoran GD, O’Reilly B, O’Mahony J, Lucey B. 2020. Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) for investigation of Mycobacterium tuberculosis complex outbreaks: a type dream? J Clin Microbiol 58:e02077-19. 10.1128/JCM.02077-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sauget M, Valot B, Bertrand X, Hocquet D. 2017. Can MALDI-TOF mass spectrometry reasonably type bacteria? Trends Microbiol 25:447–455. 10.1016/j.tim.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 28.Brackmann M, Leib SL, Tonolla M, Schürch N, Wittwer M. 2020. Antimicrobial resistance classification using MALDI-TOF-MS is not that easy: lessons from vancomycin-resistant Enterococcus faecium. Clin Microbiol Infect 26:391–393. 10.1016/j.cmi.2019.10.027. [DOI] [PubMed] [Google Scholar]
- 29.Spinali S, Van Belkum A, Goering RV, Girard V, Welker M, Van Nuenen M, Pincus DH, Arsac M, Durand G. 2015. Microbial typing by matrix-assisted laser desorption ionization-time of flight mass spectrometry: do we need guidance for data interpretation? J Clin Microbiol 53:760–765. 10.1128/JCM.01635-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vázquez-Ucha JC, Seoane-Estévez A, Rodiño-Janeiro BK, González-Bardanca M, Conde-Pérez K, Martínez-Guitián M, Alvarez-Fraga L, Arca-Suárez J, Lasarte-Monterrubio C, Gut M, Gut I, Álvarez-Tejado M, Oviaño M, Beceiro A, Bou G. 3 March 2021. Activity of imipenem/relebactam against a Spanish nationwide collection of carbapenemase-producing Enterobacterales. J Antimicrob Chemother 10.1093/jac/dkab043. [DOI] [PubMed] [Google Scholar]
- 31.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31:3350–3352. 10.1093/bioinformatics/btv383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 34.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nordmann P, Naas T, Poirel L. 2011. Global spread of carbapenemase producing Enterobacteriaceae. Emerg Infect Dis 17:1791–1798. 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Altman DG. 1991. Practical statistics for medical research. Chapman and Hall, London, UK. [Google Scholar]
- 37.Shi L, Westerhuis JA, Rosén J, Landberg R, Brunius C. 2019. Variable selection and validation in multivariate modelling. Bioinformatics 35:972–980. 10.1093/bioinformatics/bty710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vasoo S, Cunningham SA, Kohner PC, Simner PJ, Mandrekar JN, Lolans K, Hayden MK, Patel R. 2013. Comparison of a novel, rapid chromogenic biochemical assay, the Carba NP test, with the modified Hodge test for detection of carbapenemase-producing Gram-negative bacilli. J Clin Microbiol 51:3097–3101. 10.1128/JCM.00965-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsuda N, Matsuda M, Notake S, Yokokawa H, Kawamura Y, Hiramatsu K, Kikuchi K. 2012. Evaluation of a simple protein extraction method for species identification of clinically relevant staphylococci by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol 50:3862–3866. 10.1128/JCM.01512-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuhn M, Johnson K. 2018. Applied predictive modeling. Springer-Verlag, New York, NY. [Google Scholar]
- 41.Fraser CG. 2001. Biological variation: from principles to practice. AACC Press, Washington, DC. [Google Scholar]
- 42.Ricós C, Álvarez V, Minchinela J, Fernández-Calle P, Perich C, Boned B, González E, Simón M, Díaz-Garzón J, García-Lario JV, Cava F, Fernández-Fernández P, Corte Z, Biosca C. 2017. Biologic variation approach to daily laboratory. Clin Lab Med 37:47–56. 10.1016/j.cll.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 43.Oberle M, Wohlwend N, Jonas D, Maurer FP, Jost G, Tschudin-Sutter S, Vranckx K, Egli A. 2016. The technical and biological reproducibility of matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) based typing: employment of bioinformatics in a multicenter study. PLoS One 11:e0164260. 10.1371/journal.pone.0164260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oviaño M, Bou G. 2019. Matrix-assisted laser desorption ionization–time of flight mass spectrometry for the rapid detection of antimicrobial resistance mechanisms and beyond. Clin Microbiol Rev 32:e00037-18. 10.1128/CMR.00037-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haidar G, Clancy CJ, Chen L, Samanta P, Shields RK, Kreiswirth BN, Nguyen MH. 2017. Identifying spectra of activity and therapeutic niches for ceftazidime-avibactam and imipenem-relebactam against carbapenem-resistant Enterobacteriaceae. Antimicrob Agents Chemother 61:e00642-17. 10.1128/AAC.00642-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
. Download JCM.00800-21-s0001.pdf, PDF file, 207 KB (206KB, pdf)
Data Availability Statement
The BioProject accession number for strain genomes is PRJEB39112. It is anticipated that this accession number will be released by early 2022; until that time, the strain genome data will be available from the corresponding author upon request.