Abstract
Wolbachia are maternally transmitted, intracellular bacteria that can often selfishly spread through arthropod populations via cytoplasmic incompatibility (CI). CI manifests as embryonic death when males expressing prophage WO genes cifA and cifB mate with uninfected females or females harboring an incompatible Wolbachia strain. Females with a compatible cifA-expressing strain rescue CI. Thus, cif-mediated CI confers a relative fitness advantage to females transmitting Wolbachia. However, whether cif sequence variation underpins incompatibilities between Wolbachia strains and variation in CI penetrance remains unknown. Here, we engineer Drosophila melanogaster to transgenically express cognate and non-cognate cif homologs and assess their CI and rescue capability. Cognate expression revealed that cifA;B native to D. melanogaster causes strong CI, and cognate cifA;B homologs from two other Drosophila-associated Wolbachia cause weak transgenic CI, including the first demonstration of phylogenetic type 2 cifA;B CI. Intriguingly, non-cognate expression of cifA and cifB alleles from different strains revealed that cifA homologs generally contribute to strong transgenic CI and interchangeable rescue despite their evolutionary divergence, and cifB genetic divergence contributes to weak or no transgenic CI. Finally, we find that a type 1 cifA can rescue CI caused by a genetically divergent type 2 cifA;B in a manner consistent with unidirectional incompatibility. By genetically dissecting individual CI functions for type 1 and 2 cifA and cifB, this work illuminates new relationships between cif genotype and CI phenotype. We discuss the relevance of these findings to CI’s genetic basis, phenotypic variation patterns, and mechanism.
Keywords: cytoplasmic incompatibility, Wolbachia, phage WO, Drosophila melanogaster, Two-by-One model
Introduction
Wolbachia are intracellular bacteria that occur in 40–65% of arthropod species (Hilgenboecker et al. 2008; Zug and Hammerstein 2012; Weinert et al. 2015; Charlesworth et al. 2019). While often horizontally transmitted between species (Boyle et al. 1993; Huigens et al. 2004; Gerth et al. 2013; Tolley et al. 2019; Scholz et al. 2020), vertical transmission from mother to offspring predominates within species (Turelli and Hoffmann 1991; Narita et al. 2009). Wolbachia can frequently increase their rate of spread in host populations through the matriline by causing cytoplasmic incompatibility (CI). CI results in embryonic death of uninfected embryos after fertilization by Wolbachia-modified sperm (Yen and Barr 1973; Shropshire et al. 2020b). Embryos with compatible Wolbachia are rescued from CI-induced lethality, yielding a relative fitness advantage to Wolbachia-infected females that transmit the bacteria to their offspring (Hoffmann et al. 1990; Turelli 1994; Turelli and Hoffmann 1995). CI frequently manifests between arthropods infected with different Wolbachia strains. Strains may be reciprocally incompatible (bidirectional CI), or only one of the two strains can rescue the other’s sperm modification (unidirectional CI). CI-inducing Wolbachia have garnered attention for their role in suppressing vector populations (Lees et al. 2015; Nikolouli et al. 2018; Crawford et al. 2020), curbing the transmission of pathogenic RNA viruses (O’Neill 2018; Moretti et al. 2018; Gong et al. 2020), and reproductive isolation and incipient speciation (Bordenstein et al. 2001; Jaenike et al. 2006; Brucker and Bordenstein 2012; Shropshire and Bordenstein 2016).
Two adjacent genes in the eukaryotic association module of Wolbachia’s prophage WO cause CI when expressed in males (cifA and cifB) (Bordenstein and Bordenstein 2016; Beckmann et al. 2017; LePage et al. 2017; Chen et al. 2019; Shropshire and Bordenstein 2019), and one of the same genes rescues CI when expressed in females (cifA) (Shropshire et al. 2018; Chen et al. 2019; Shropshire and Bordenstein 2019). These results established the Two-by-One genetic model of CI (Figure 1A) (Shropshire and Bordenstein 2019), but its generality across cif homologs remains to be evaluated. Singly expressing a small set of cifA variants that have only annotated domains or cifB variants that exhibit in vitro deubiquitilase and nuclease activities also does not cause rescuable embryonic death (Beckmann et al. 2017; LePage et al. 2017; Chen et al. 2019; Shropshire and Bordenstein 2019). Cif proteins segregate into at least five phylogenetic clades (types 1–5) (LePage et al. 2017; Lindsey et al. 2018; Bing et al. 2020; Martinez et al. 2020), and distant Cif-like homologs are found in Orientia and Rickettsia bacteria, which are not known to cause CI (Gillespie et al. 2018). To date, the genetic basis for CI (Figure 1A) has been tested using cif transgenes from the types 1 and 4 clades in wMel Wolbachia of Drosophila melanogaster and wPip Wolbachia of Culex pipiens mosquitoes (Beckmann et al. 2017; Chen et al. 2019; Shropshire and Bordenstein 2019). Thus, a considerable amount of phylogenetic diversity remains untested.
Figure 1.
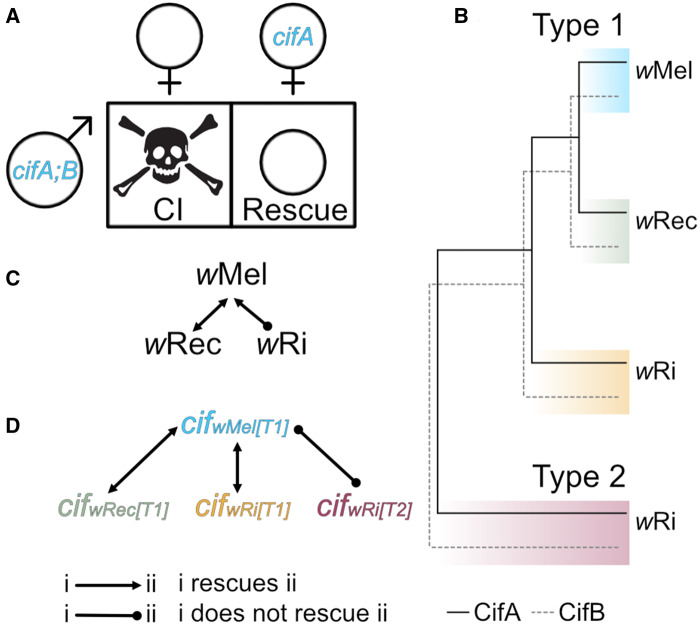
Two-by-One model, Cif phylogeny, and predicted relationships between wMel, wRec, and wRi strains and cif gene variants. (A) The Two-by-One genetic basis of CI: cifA;B causes CI that can be rescued by females expressing cifA (Shropshire and Bordenstein 2019). (B) Schematic representation of the evolutionary relationships between CifA and CifB proteins from wMel, wRec, and wRi (LePage et al. 2017). (C) Incompatibilities between wMel, wRec, and wRi Wolbachia strains. Unidirectional CI between wMel and wRi is based on crossing experiments after the transinfection of wMel into D. simulans (Poinsot et al. 1998). Compatibility between wMel and wRec is based on the prediction that strains with closely related cif gene sequences are compatible. (D) Predicted incompatibility relationships between cif homologs from each of the three strains, based on sequence relationship. Lines between strains/genes indicate compatibility relationships. If the line ends in an arrowhead, then the strain/gene(s) at the beginning of the arrow can rescue CI caused by the strain/gene(s) the arrow points toward. If the line ends in a circle, then rescue is not expected. Skull art is modified from vecteezy.com with permissions.
Moreover, while the genetic basis of CI between infected and uninfected insects is resolved for some strains, the genetic basis of unidirectional or bidirectional CI between insects harboring different Wolbachia strains remains largely unknown. Phylogenetic and sequence analyses of cif genes from incompatible Wolbachia strains in Drosophila or Culex reveal that incompatible Wolbachia strains differ in genetic relationship and/or copy number (Bonneau et al. 2018, 2019; LePage et al. 2017), supporting cif variation as the basis of strain incompatibilities. Moreover, since cifA is involved in both CI induction and rescue, a single-step evolutionary model for bidirectional CI has been proposed where a single mutation in cifA leads to incompatibility between the ancestral and derived variants while retaining compatibility with the emergent variant and requiring cifB only as an accessory function (Shropshire et al. 2018). However, these hypotheses have not been empirically tested.
In this study, we first test cif homologs from wMel of D. melanogaster, wRec of D. recens, and wRi of D. simulans for CI and rescue when transgenically expressed in uninfected D. melanogaster. We previously determined that wMel genes adhere to the Two-by-One model (Shropshire and Bordenstein 2019), but the genetic bases of wRec and wRi CI remain unknown. wRec and wRi are strong CI inducers that cause high degrees of embryonic death (Turelli and Hoffmann 1991; Werren and Jaenike 1995; Shoemaker et al. 1999). Both encode phylogenetic type 1 cif genes similar to wMel, and wRi also encodes a type 2 cif pair that is highly diverged from wMel (Figure 1B) (LePage et al. 2017). Like wMel, we predict that wRec and wRi CI have a Two-by-One genetic basis. This is the first time type 2 cif genes have been functionally interrogated.
Next, we test the crossing relationships between divergent cif homologs to investigate the basis of interstrain incompatibilities. wRi and wMel Wolbachia are unidirectionally incompatible in a common D. simulans host background. In other words, wRi can rescue wMel-induced CI, but wMel cannot rescue wRi-induced CI (Figure 1C) (Poinsot et al. 1998). Thus, we hypothesize that wRi can rescue wMel-induced CI because it has type 1 cif genes comparable to wMel, and wMel cannot rescue wRi because it does not have genes capable of rescuing wRi’s type 2 genes (Figure 1D) (LePage et al. 2017). Moreover, since wRec’s type 1 cif genes are closely related to wMel genes, we predict them to be compatible upon transgenic expression (Figure 1C and D). We discuss our results in the context of the genetic basis of CI in these strains, the causes of CI strength variation and strain incompatibilities, and CI’s molecular basis.
Materials and Methods
Fly lines and maintenance
The following Upstream Activation Sequence (UAS) transgenic constructs were generated for this study: cifAwRec[T1], cifBwRec[T1],cifAwRi[T1],cifBwRi[T1;N], cifBwRi[T1;C], cifBwRi[T1;poly], cifAwRi[T2], and cifBwRi[T2]. Each transgene was codon-optimized for expression in D. melanogaster and synthesized by GenScript (Hong Kong, China). Valine start codons were replaced with methionine. Wild-type and codon-optimized gene sequences are reported in Supplementary Table S2. Each gene was cloned into the pTIGER plasmid at GenScript. pTIGER is a pUASp-based vector designed for germline expression and was previously used to generate cifAwMel[T1] and cifBwMel[T1] transgenes also used in this study (LePage et al. 2017). pTIGER enables phiC31 integration into the D. melanogaster genome, contains a UAS promoter region intended for GAL4/UAS expression, and has a red-eye marker for screening. D. melanogaster embryo injections were conducted by Best Gene (Chino Hills, California) using phiC31 integrase to place cifA and cifB homologs into the Attp40 (chromosome 2) and Attp2 (chromosome 3) insert sites, respectively. Transformants were screened via eye color, and homozygous transgenic lines were generated for all lines. All lines were negative for Wolbachia based on PCR using Wolb_F and Wolb_R3 primers (Casiraghi et al. 2005). Dual expressing UAS transgenic lines were generated via standard genetic crossing schemes.
In addition, the following D. melanogaster stocks were used in this study: infected and uninfected y1w* (BDSC 1495), uninfected nos-GAL4:VP16 (BDSC 4937), and uninfected UAS transgenic lines homozygous for cifAwMel[T1], cifBwMel[T1], and cifA;BwMel[T1] (LePage et al. 2017). Genotypes and infection states were regularly confirmed for transgene expressing fly lines using primers listed in Supplementary Table S3. D. melanogaster stocks were maintained at 12:12 light:dark at 25°C on 50 ml of a standard media. Stocks for virgin collections were stored at 18°C overnight to slow eclosion rate, and virgin flies were kept at room temperature.
Hatch rate assays
To test for CI, hatch rate assays were conducted as previously described (LePage et al. 2017; Shropshire et al. 2018). Briefly, virgin nos-GAL4:VP16 adult females were aged 9–11 days post-eclosion, to control for the paternal grandmother age effect (Layton et al. 2019), and mated with UAS transgenic or y1w* males. GAL4-UAS males and females were paired in 8-oz round bottom Drosophila bottles (Genesee Scientific) affixed with a grape-juice agar plate smeared with yeast affixed to the opening with tape. To control the impact of male age and the younger brother effect on CI level (Reynolds and Hoffmann 2002; Yamada et al. 2007), only young early emerging males (0–48 h) were used in crossings. Conversely, 5–7-day-old females were used since they are highly fecund. The flies and bottles were stored at 25°C for 24 h at which time the plates were replaced with freshly smeared plates and again stored for 24 h. Plates were then removed, and the number of embryos on each plate was counted and stored at 25°C. After 30 h, the remaining unhatched embryos were counted. The percentage of embryos that hatched into larvae was calculated by dividing the number of hatched embryos by the initial embryo count and multiplying by 100.
Embryonic cytology
Flies were collected, aged, and crossed as described for hatch rate assays. However, 60 females and 12 males were included in each bottle with a grape-juice agar plate attached. Flies were siblings of those in hatch rate assays. Embryos laid in the first 24 h were discarded due to low egg-laying. During the second day, embryos were aged 1–2 h and then dechorionated, washed, and fixed in methanol as previously described (LePage et al. 2017; Shropshire et al. 2018). Embryos were stained with propidium iodide and imaged (LePage et al. 2017; Shropshire et al. 2018). Scoring of cytological defects was conducted using previously defined characteristics (LePage et al. 2017).
Sequence analyses
Sequence similarity between Cif proteins was determined using pairwise MUSCLE alignments of protein sequences using default settings. Glimmer 3 was used to identify open reading frames in cifBwRi[T1] after the early stop codon that truncates the gene. These analyses were conducted in Geneious Prime.
Statistical analyses
All statistical analyses were conducted in GraphPad Prism 8. Hatch rate statistical comparisons were made using Kruskal–Wallis followed by a Dunn’s multiple comparison test. Samples with fewer than 25 embryos laid were removed from hatch rate analyses as previously described (LePage et al. 2017). Hatch rates in main text figures display the combination of two replicate experiments, which were analyzed simultaneously, and those in the supplement display only single experiments (N = 8–58 per cross after exclusion). Replicate data were statistically comparable in all cases. Cytological abnormalities were compared using a pairwise Fisher’s exact test followed by a Bonferroni–Dunn correction test (N = 43–167 embryos per cross). Figure esthetics were edited in Affinity Designer 1.7 (Serif Europe, Nottingham, UK). All P-values are reported in Supplementary Table S1, and the exact sample sizing information is available in Supplementary File S1.
Data availability
All data are made publicly available in the supplement of this manuscript. Fly lines not otherwise available in the Bloomington Drosophila Stock Center are available upon request.
Supplemental material is available at figshare DOI: https://doi.org/10.25386/genetics.13215503.
Results
To distinguish between different cifA and cifB genetic variants, we use a gene nomenclature that identifies the Wolbachia strain in subscript and the cif phylogenetic type associated with the variant in brackets (Shropshire et al. 2019), following published standards (The Journal of Bacteriology 2018). For instance, cif genes of the wMel strain belong to the type 1 clade and are referred to as cifAwMel[T1] and cifBwMel[T1]. We used the GAL4-UAS system (Duffy 2002) to drive the germline expression of cif transgenes in D. melanogaster, and all transgenes are expressed in uninfected flies using the nos-GAL4:VP16 driver that causes strong cifwMel[T1] CI and rescue (Shropshire and Bordenstein 2019). We measure CI as the percentage of embryos that hatch into larvae relative to a compatible control in which cifA;BwMel[T1] CI from males is rescued by cifAwMel[T1] females (LePage et al. 2017; Shropshire et al. 2018; Shropshire and Bordenstein 2019). This cross is included in all experiments and will hereafter be referred to as the “compatible control”. All protein annotations are derived from prior works (Lindsey et al. 2018).
Do phylogenetic type 1 cif genes from wRec transgenically induce and rescue CI?
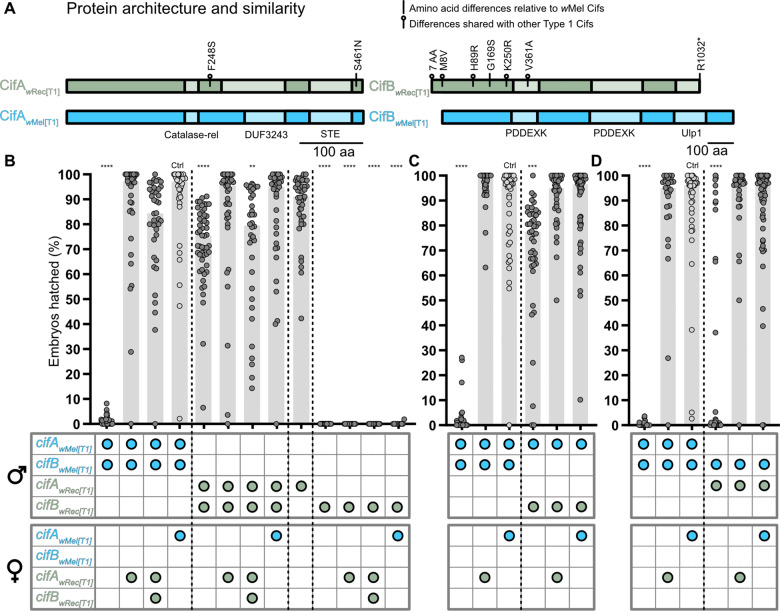
Relative to CifwMel[T1] proteins, CifAwRec[T1] has two amino acid substitutions in unannotated regions: one prior to CifA’s putative DUF3243 and another after the annotated STE domain (Figure 2A). CifBwRec[T1] has 13 amino acid changes that include a seven amino acid extension on the N-terminus, four substitutions in the N-terminal unannotated region, a single substitution in the first putative PD-(D/E)XK-like nuclease domain (hereafter PDDEXK), and a stop codon that truncates amino acids at residues 1032–1173 on the C-terminus of the protein (Figure 2A). wRec causes strong CI in D. recens (Shoemaker et al. 1999; Werren and Jaenike 1995), the wRec genome lacks other cif genes (Metcalf et al. 2014), and these variants are highly similar to cifwMel[T1] genes (Figure 2A). Thus, we predicted that cifA;BwRec[T1] expression in uninfected males will cause CI, transgenic cifAwRec[T1] expression in uninfected females will rescue CI, and CI induced by cifwRec[T1] transgenes will be compatible with cifwMel[T1] transgenes.
Figure 2.
CifwRec[T1] protein similarity and results of transgenic crosses including CifwRec[T1] proteins. (A) Protein architecture of CifwMel[T1] and CifwRec[T1] (Lindsey et al. 2018). Substitutions inside schematics represent sequence identity relative to CifwMel[T1]. Substitutions marked with a circle above the protein schema are shared between CifwRec[T1] and CifwRi[T1]. R1032* represents an arginine to stop codon mutation. Hatch rate analyses testing (B) cifAwRec[T1], cifBwRec[T1], and cifA;BwRec[T1] for CI and rescue (N = 12–51 where each dot represents a clutch of embryos from a single mating pair), (C) cifAwMel[T1];cifBwRec[T1] for CI (N = 36–55), and (D) cifAwRec[T1];cifBwMel[T1] for CI (N = 27–58). Horizontal bars represent median embryonic hatching from single pair matings. Genotypes for each cross are illustrated below the bars where the genes expressed in each sex are represented by colored circles. Blue circles represent cifwMel[T1] genes, and green circles represent cifwRec[T1] genes. Each hatch rate contains the combined data of two replicate experiments, each containing all crosses shown. Asterisks above bars represent significant differences relative to a control transgenic rescue cross (denoted Ctrl) with an α = 0.05. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Exact P-values are provided in Supplementary Table S1.
Consistent with prior reports in D. melanogaster, cifA;BwMel[T1] males induce strong CI that is rescued by the compatible control cross with cifAwMel[T1] females (Figure 2B) (Shropshire and Bordenstein 2019). cifA;BwRec[T1] males also cause a small but statistically significant reduction in hatching (Mdn = 75.4% hatching; P < 0.0001; Figure 2B) that is rescued by cifAwRec[T1] females but not by cifA;BwRec[T1] females (Mdn = 79.6% hatching; P = 0.0054). Results therefore suggest that cifAwRec[T1] is a rescue gene, weak cifA;BwRec[T1] CI is rescuable, and cifBwRec[T1] may reduce cifAwRec[T1] rescue capacity in embryos. Since neither cifAwMel[T1] nor cifBwMel[T1] induces CI alone (LePage et al. 2017; Shropshire and Bordenstein 2019), we predicted neither cifAwRec[T1] nor cifBwRec[T1] would reduce hatching. Indeed, cifAwRec[T1] males did not reduce hatching (P > 0.99). However, cifBwRec[T1] males caused complete embryonic death (Mdn = 0% hatching; P < 0.0001; Figure 2B) that was not rescued by cifAwRec[T1] (Mdn = 0% hatching), cifA;BwRec[T1] (Mdn = 0% hatching), cifAwMel[T1] (Mdn = 0% hatching), or wMel-infected (Mdn = 0% hatching) females (Figure 2B and Supplementary Figure S1). Embryos fertilized by cifBwRec[T1] males had an unusually high percentage of early mitotic failures and single puncta indicative of unfertilized embryos or embryos undergoing mitotic failure in the first division (Supplementary Figure S2). However, unlike cifA;BwMel[T1] males, there were no later stage regional mitotic failures or chromatin bridging phenotypes, and the cifBwRec[T1] defects were unrescuable (Supplementary Figure S2) (LePage et al. 2017). Taken together, these results indicate that cifBwRec[T1] alone causes an atypical embryonic lethality relative to cifA;BwMel[T1]-induced CI.
Next, we tested crossing relationships between cifwMel[T1] and cifwRec[T1] transgenic males and females. Weak cifA;BwRec[T1]-induced CI was reduced when mated with cifAwMel[T1] females (P > 0.99) relative to the compatible control. Similarly, cifA;BwMel[T1] CI was reduced when mated with cifAwRec[T1] (P > 0.99) or cifA;BwRec[T1] (P = 0.10) females (Figure 2B). However, cifA;BwRec[T1] females only partially rescue cifA;BwMel[T1] CI, and since cifA;BwRec[T1] females do not rescue cifA;BwRec[T1] CI (Figure 2B), a firm conclusion cannot be made on whether cifA;BwRec[T1] females can rescue cifA;BwMel[T1] CI. Together, these data indicate that cifAwMel[T1] and cifAwRec[T1], which differ by two amino acid substitutions in the putative DUF3243 and STE domains (Figure 2A), rescue the other strain’s transgenic CI. This is perhaps unsurprising since prior mutagenesis assays suggest conserved sites in DUF3243 and STE domains are not related to rescue (Shropshire et al. 2020a).
Finally, since cifA;BwRec[T1] males induce weak CI relative to cifA;BwMel[T1] males, we hypothesized that cifAwRec[T1] or cifBwRec[T1] sequence variation underpins CI level variation. We tested this hypothesis by engineering and expressing non-cognate combinations of cifwRec[T1] and cifwMel[T1] transgenes. We report that cifAwMel[T1];cifBwRec[T1] males cause a weak but statistically significant reduction in hatching relative to the compatible control (Mdn = 77.6% hatching; P = 0.0008; Figure 2C), and this hatch rate reduction was comparable to that of cognate cifA;BwRec[T1] (Mdn = 75.4% hatching; Figure 2B) and likewise rescued when crossed to cifAwMel[T1] (P > 0.99) or cifAwRec[T1] (P > 0.99) females (Figure 2C). In contrast, cifAwRec[T1];cifBwMel[T1] males caused strong CI (Mdn = 0% hatching; P < 0.0001) that was also rescued by cifAwRec[T1] (Mdn = 97.1% hatching; P > 0.99) or cifAwMel[T1] (Mdn = 95.9% hatching; P > 0.99) females (Figure 2D). These data demonstrate that the two closely related cifA sequences are interchangeable and fully capable of CI and rescue and that sequence variation in cifB is crucially responsible for weak cifA;BwRec[T1] transgenic CI in D. melanogaster.
Do phylogenetic type 1 cif genes from wRi transgenically induce and rescue CI?
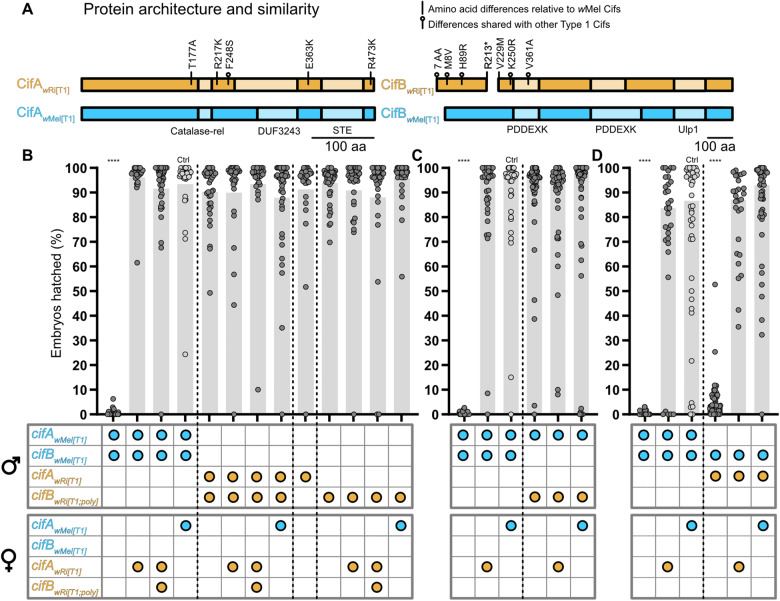
wRi has three cif gene pairs: two identical type 1 pairs and one type 2 pair (LePage et al. 2017; Lindsey et al. 2018). We first focus attention on the cifwRi[T1] genes, their protein sequence differences, and CI phenotype variation. Relative to CifAwMel[T1], CifAwRi[T1] protein has five amino acid substitutions in unannotated regions flanking the predicted domains (Figure 3A). One of these CifA substitutions is also present in CifAwRec[T1]. CifBwRi[T1] has an in-frame stop codon introduced at residue 213 in the 1173-amino-acid-long protein (Figure 3A), and Glimmer 3 predicts that another protein coding sequence begins 16 amino acids later at residue 229. Thus, cifBwRi[T1] may yield two proteins: an N-terminal 212 amino acid protein and a C-terminal 945 amino acid protein. We refer to the gene sequences yielding the N-terminal and C-terminal peptides as cifBwRi[T1;N] and cifBwRi[T1;C], respectively. Relative to CifBwMel[T1], CifBwRi[T1;N] has two amino acid substitutions, a seven amino acid N-terminal extension, and an early stop codon. In this region, CifBwRec[T1] has the same sequence variations, excluding the early stop codon in addition to one extra substitution. CifBwRi[T1;C] has three substitutions relative to CifBwMel[T1]: one in the first PDDEXK domain, a valine to methionine substitution marking the translation start site, and one in the unannotated region prior to the first PDDEXK domain (Figure 3A). In this C-terminal region, CifBwRec[T1] shares all but the valine to methionine substitution. To investigate the genetic basis of wRi CI, we generated cifAwRi[T1], cifBwRi[T1;N], and cifBwRi[T1;C] transgenes. We additionally created a polycistronic cifBwRi[T1] transgene that expressed both the N-terminal and C-terminal peptides from a single transcript using a T2A sequence between the two proteins (Donnelly et al. 2001a,b). We refer to this polycistronic transgenic construct as cifBwRi[T1;poly].
Figure 3.
CifwRi[T1] protein similarity and results of transgenic crosses including CifwRi[T1] proteins. (A) Protein architecture of CifwMel[T1] and CifwRi[T1] (Lindsey et al. 2018). Substitutions inside schematics represent sequence identity relative to CifwMel[T1]. Substitutions marked with a circle above the protein schema are shared between CifwRec[T1] and CifwRi[T1]. R213* represents an arginine to stop codon mutation. Hatch rate analyses testing (B) cifAwRi[T1], cifBwRi[T1;poly], and cifA;BwRi[T1;poly] for CI and rescue (N = 26–44 where each dot represents a clutch of embryos from a single mating pair), (C) cifAwMel[T1];cifBwRi[T1;poly] for CI (N = 32–56), and (D) cifAwRi[T1];cifBwMel[T1] for CI (N = 27–47). Horizontal bars represent median embryonic hatching from single pair matings. Genotypes for each cross are illustrated below the bars where the genes expressed in each sex are represented by colored circles. Blue circles represent cifwMel[T1] genes and orange circles represent cifwRi[T1] genes. All flies were uninfected with Wolbachia. Each hatch rate contains the combined data of two replicate experiments, each containing all crosses shown. Asterisks above bars represent significant differences relative to a control transgenic rescue cross (denoted Ctrl) with an α = 0.05. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Exact P-values are provided in Supplementary Table S1.
We first tested cifA;BwRi[T1;poly] males for their ability to induce CI and found that they did not reduce hatching (P = 0.55) (Figure 3B). Males dually expressing cifAwRi[T1] with either cifBwRi[T1;N] (P = 0.55; Supplementary Figure S3A) or cifBwRi[T1;C] (P = 0.32; Supplementary Figure S4A) also failed to reduce hatching, suggesting that dual expression of cifwRi[T1] transgenes cannot recapitulate CI. In addition, singly expressing cifAwRi[T1] (P > 0.99) or cifBwRi[T1;poly] (P > 0.99) males does not cause CI (Figure 3B). Next, to test if cifwRi[T1] genes can rescue strong cifwMel[T1] CI, we crossed cifA;BwMel[T1] males with cifAwRi[T1] (P > 0.99) and cifA;BwRi[T1;poly] (P > 0.99) females, both of which yielded hatching levels comparable to cifAwMel[T1] rescue (Figure 3B). These results indicate that cifAwRi[T1] is a rescue gene, and cifwRi[T1] transgenes do not cause CI when singly or dually expressed as cognate partners in D. melanogaster.
To further evaluate if cifwRi[T1] transgenes are capable of CI and whether variation in cifA or cifB may underpin the lack of CI above, we engineered and dually expressed non-cognate pairs of cifwRi[T1] genes with cifwMel[T1] genes. cifAwMel[T1];cifBwRi[T1;poly] males did not yield a reduction in hatching compared to the compatible cross (P > 0.99; Figure 3C). Similarly, males dually expressing cifAwMel[T1] and either cifBwRi[T1;N] (P > 0.99; Supplementary Figure S3B) or cifBwRi[T1;C] (P > 0.99; Supplementary Figure S4B) did not reduce hatching. However, cifAwRi[T1];cifBwMel[T1] males caused near-complete embryonic death (Mdn = 0% hatching; P < 0.0001) that could be rescued by cifAwRi[T1] and cifAwMel[T1] females (Figure 3D). These findings suggest that cifAwRi[T1] contributes to both rescue and CI induction, but cifBwRi[T1] transgenes fail to contribute to CI.
Do the phylogenetic type 2 cif genes from wRi transgenically induce and rescue CI?
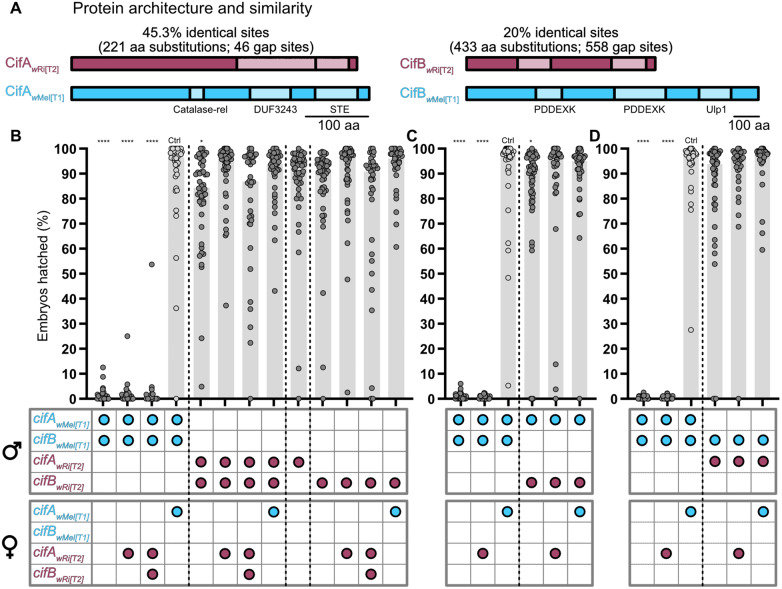
Pairwise alignments of CifwMel[T1] and CifwRi[T2] proteins (488 and 1239 amino acids for CifA and CifB, respectively) reveal major divergence. First, CifAwMel[T1] and CifAwRi[T2] differ by 267 sites (45.3% identical sites), with 221 amino acid substitutions and 46 gap sites in the alignment (Figure 4A and Supplementary Figure S5). CifAwRi[T2] has substitutions in all six of the sites that vary in CifAwRec[T1] and CifAwRi[T1], and two of the CifAwRi[T2] substitutions are shared with both proteins, and a third is shared with CifAwRi[T1]. Second, CifBwMel[T1] and CifBwRi[T2] differ by 991 sites (20% identical sites), with 433 substitutions and 558 gap sites in the alignment (Figure 4A and Supplementary Figure S5). In addition, CifBwRi[T2] has substitutions in four of the six sites that vary in CifBwRi[T1] and CifBwRec[T1], but the specific amino acids are unique to CifBwRi[T2] (Supplementary Figure S5). Moreover, while the sequence lengths of the two CifA variants are comparable, CifBwRi[T2] does not have the C-terminal Ulp1 domain that, for other distant type 1 Cif variants, acts in vitro as a deubiquitinase (Beckmann et al. 2017). It also has an eight-amino-acid N-terminal extension (Supplementary Figure S5), of which four amino acids are shared in the N-terminal extensions of CifBwRec[T1] and CifBwRi[T1].
Figure 4.
CifwRi[T2] protein similarity and results of transgenic crosses including CifwRi[T2] proteins. (A) Protein architecture of CifwMel[T1] and CifwRi[T2] (Lindsey et al. 2018). In an alignment of CifAwMel[T1] and CifAwRi[T2] (488 aa), there are 221 identical sites, 221 aa substitutions, and 46 gap sites. In an alignment of CifBwMel[T1] and CifBwRi[T2] (1239 aa), there are 248 identical sites, 433 aa substitutions, and 558 gap sites. Specific details on the kinds and locations of sequence variations are illustrated in Supplementary Figure S5. Hatch rate analyses testing (B) cifAwRi[T2], cifBwRi[T2], and cifA;BwRi[T2] for CI and rescue (N = 35–55 where each dot represents a clutch of embryos from a single mating pair), (C) cifAwMel[T1];cifBwRi[T2] for CI (N = 39–56), and (D) cifAwRi[T2];cifBwMel[T1] for CI (N = 31–45). Horizontal bars represent median embryonic hatching from single pair matings. Genotypes for each cross are illustrated below the bars where the genes expressed in each sex are represented by colored circles. Blue circles represent cifwMel[T1] genes and purple circles represent cifwRi[T2] genes. All flies were uninfected with Wolbachia. Each hatch rate contains the combined data of two replicate experiments, each containing all crosses shown. Asterisks above bars represent significant differences relative to a control transgenic rescue cross (denoted Ctrl) with an α = 0.05. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Exact P-values are provided in Supplementary Table S1.
First, we test if cifwRi[T2] transgenes cause and rescue CI in D. melanogaster. cifA;BwRi[T2] males caused a weak but statistically significant hatch rate reduction (Mdn = 84.4% hatching; P = 0.01; Figure 4B) that was rescued upon crossing with cifAwRi[T2] females (P > 0.99; Figure 4B). Similar to results with cifA;BwRec[T1] females above (Figure 2B), crossing cifA;BwRi[T2] males with cifA;BwRi[T2] females only slightly improved hatching such that it was no longer statistically different from the compatible control (Mdn = 86.9% hatching; P = 0.15); however, the median hatch rate was comparable when cifA;BwRi[T2] males were mated to uninfected females (Mdn = 84.4% hatching; Figure 4B). Thus, similar to cifwRec[T1], it cannot be concluded that cifA;BwRi[T2] females are rescue-capable yet, but cifAwRi[T2] females clearly rescue cifA;BwRi[T2] CI as expected under the Two-by-One Model. In parallel, we showed that neither cifAwRi[T2] (P = 0.84) nor cifBwRi[T2] (P = 0.13) males alone reduce hatching, as expected (Figure 4B). These data suggest that cifA;BwRi[T2] males can cause very weak CI that can be rescued by cifAwRi[T2] females.
Next, we aimed to determine if the considerable intertype divergence between cifAwRi[T2] and cifAwMel[T1] underpins incompatibility between the strains (Figures 1C and 3A). Embryo death was observed when cifA;BwMel[T1] males mated with cifAwRi[T2] (Mdn = 0%; P < 0.0001) or cifA;BwRi[T2] (Mdn = 0%; P < 0.0001) females (Figure 4B), suggesting incompatibility between the gene variants. Reciprocally, embryonic hatching increased to compatible levels when cifA;BwRi[T2] males mated with cifAwMel[T1] females (P > 0.99) (Figure 4B). Together, these data suggest unidirectional CI between cifwMel[T1] and cifwRi[T2] such that cifAwMel[T1] can rescue CI caused by both variants, but cifAwRi[T2] can only rescue its own lethality. This is the first empirical finding that type 1 and 2 cif genes are partially compatible and thus likely share similar CI mechanisms.
Finally, as previously done for cifwRec[T1] and cifwRi[T1], we combinatorially tested if non-cognate expression of cifwRi[T2] and cifwMel[T1] genes underpins the genetic basis of CI level variation. First, cifAwMel[T1];cifBwRi[T2] males yielded a small but significant hatch rate reduction (Mdn = 92.0% hatching; P = 0.01), relative to the compatible control. Second, cifAwRi[T2] (P > 0.99) and cifAwMel[T1] (P = 0.40) females rescued the weak hatch rate reduction (Figure 4C). Finally, cifAwRi[T2];cifBwMel[T1] males had a similar, but statistically insignificant, impact on hatching (P = 0.07) relative to cifAwMel[T1];cifBwRi[T2] males (Figure 4D). Thus, dual expression of both non-cognate pairs yields a small reduction in hatching, and weak cifAwMel[T1];cifBwRi[T2] CI was rescuable. Contrary to non-cognate expression of cifAwRec[T1] or cifAwRi[T1] with cifBwMel[T1], neither non-cognate pairing of cifwRi[T2] and cifwMel[T1] yielded strong CI. These data again suggest that divergent cif types can work together to cause a weak CI-like phenotype.
Discussion
The Two-by-One genetic model of CI states that cifA;B males cause CI, and cifA females rescue that CI (Shropshire and Bordenstein 2019). However, it remains unknown if this model can be widely generalized across cif variants. Likewise, it is unknown whether cif variation alone explains incompatibilities between Wolbachia strains and CI level variation. Here, we use transgenic tools in D. melanogaster to test if cif homologs from wRec and wRi contribute to CI and rescue, whether cif genetic variation relates to strain incompatibility (Charlat et al. 2001; LePage et al. 2017; Shropshire et al. 2018; Bonneau et al. 2018, 2019), and if cif sequence variation determines transgenic CI levels.
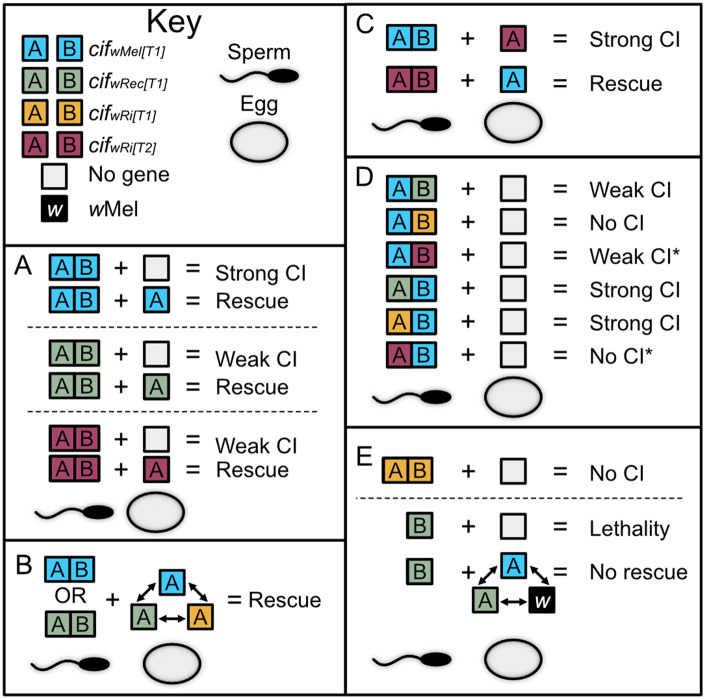
We report four key findings (Figure 5): (i) Evidence is consistent with a Two-by-One genetic basis for rescuable CI, but only weak CI is caused by cifwRec[T1] and cifwRi[T2] homologs (Figure 5A). (ii) Both type 1 cifA homologs rescue strong cifA:BwMel[T1] CI (Figure 5B), supporting the hypothesis that closely related cif genes are compatible (Charlat et al. 2001; LePage et al. 2017; Shropshire et al. 2018; Bonneau et al. 2018). (iii) Type 2 cifAwRi[T2] homologs cannot rescue cifA;BwMel[T1] CI, but the type 1 cifAwMel[T1] can rescue weak cifA;BwRi[T2] CI (Figure 5C), suggesting that different cif types may mechanistically work together, and genetic distance may contribute to unidirectional CI instead of the simple expectation of bidirectional CI. (iv) Type 1 cifB genetic variants determine CI level variation when paired with cifAwMel[T1] whereas both type 1 cifA homologs contribute to strong CI when paired with cifBwMel[T1] (Figure 5D). We also report two results contrary to our initial predictions: cifBwRec[T1] males yield unrescuable sperm infertility or embryonic death, and cifBwRi[T1] does not induce transgenic CI alone or with any cifA variant (Figure 5E). Below we interpret these findings in the context of the cif genotype–phenotype relationship for CI level variation, incompatibility relationships between Wolbachia strains, cif genotype by host genotype interactions, and CI mechanisms.
Figure 5.
Summary of findings. (A) cifwRec[T1] and cifwRi[T2] induce CI phenotypes in a manner consistent with the Two-by-One genetic model of CI previously established with cifwMel[T1] genes (Shropshire and Bordenstein 2019). (B) CI induced by type 1 cif pairs can be interchangeably rescued by cifAwMel[T1], cifAwRec[T1], and cifAwRi[T1] transgene expressing females. (C) Unidirectional CI is caused between cifwRi[T2] and cifwMel[T1] transgenes such that cifAwMel[T1] can rescue type 2 transgenic CI but cifAwRi[T2] fails to rescue type 1 transgenic CI. (D) Dual non-cognate expression of type 1 homologs reveals that cifB homologs cause weak or no CI while cifA homologs can contribute to strong transgenic CI. Non-cognate pairs that cause CI can be rescued by cifA-expressing females. Dual non-cognate expression of type 1 and 2 cif homologs reveals that despite amino acid and domain divergence, they may functionally work together to induce weak or marginal CI. * denotes significant or nearly significant levels of very weak CI. (E) cifwRi[T1] do not contribute to CI and cifBwRec causes complete embryonic death that cannot be rescued by cifAwRec, cifAwMel, or wMel-infected females.
The genetic basis of wRec (type 1) and wRi (type 2) CI and rescue
wRec and wRi induce strong CI in their native hosts (Turelli and Hoffmann 1991; Werren and Jaenike 1995; Shoemaker et al. 1999), leading to the prediction that their corresponding cif genes could yield strong transgenic CI in D. melanogaster. However, a small but significant and repeatable CI was observed when cifA;BwRec[T1] and cifA;BwRi[T2] were expressed in uninfected D. melanogaster males, and that CI was rescued by females expressing their cognate cifA or cifAwMel[T1]. Thus, we conclude that these gene pairs function in accordance with the Two-by-One genetic model of CI (Shropshire and Bordenstein 2019). Moreover, this is the first report of a CI-like phenotype caused by the phylogenetic type 2 cif genes. However, it is important to emphasize that a firm conclusion about the full genetic determinants of CI and rescue for these gene pairs is inhibited by the weakened CI levels. Unlike cifA;BwRec[T1] and cifA;BwRi[T2], dual expression of cifA;BwRi[T1] failed to cause CI. We propose three non-exclusive hypotheses for why weak CI is induced by cifwRec[T1] and cifwRi[T2] transgenes, and we discuss interpretations for why cifA;BwRi[T1] males fail to cause CI, and why cifBwRec[T1] alone causes embryonic death.
First, strong transgenic CI can be impacted by the method of transgenic expression. Indeed, the first report of transgenic wMel CI with cifA;BwMel[T1] expression in males revealed incomplete CI (LePage et al. 2017), and later optimization of the expression driver was necessary to cause consistently strong transgenic CI (Shropshire and Bordenstein 2019). Here, we used the expression system optimized for transgenic expression of wMel cif genes (Shropshire and Bordenstein 2019), and thus, it is plausible that the level or location of expression optimal for wMel-induced CI is not the same as for these other gene products. Second, other genes may be necessary to cause strong CI alongside cifwRec[T1] and cifwRi[T2] genes. These may include additional cif gene copies or other Wolbachia and prophage WO genes. For instance, wRi contains both type 1 and type 2 cif genes (LePage et al. 2017), and all Wolbachia strains known to carry type 2 cifs also harbor genes from other cif types (LePage et al. 2017; Lindsey et al. 2018). Thus, co-expression of both cif types may be necessary to cause strong CI, or additional genes predicted to interact with eukaryotes may modulate CI (Wu et al. 2004; Yamada et al. 2011; Bordenstein and Bordenstein 2016). Third, several transinfection and introgression studies show that host genotype affects CI levels (Poinsot et al. 1998; Bordenstein et al. 2003). The proximal basis of this affect remains unknown, but it is predicted to be related to Wolbachia titers and cif expression profiles (Shropshire et al. 2020b). For instance, wMel is considered as a weak CI inducer, but strict control of several variables that covary with Wolbachia titers and cif expression enables strong CI (Reynolds and Hoffmann, 2002, Yamada et al. 2011, Layton et al. 2019). Moreover, strong wMel transgenic CI is possible (Shropshire et al. 2019), thus suggesting that a weak CI strain can cause strong transgenic CI. However, while titer and cif gene expression likely control CI strength within a system, it is plausible that Cif amino acid sequence also corresponds with a change in efficiency when binding to D. melanogaster targets in a heterologous expression system.
In summary, while these data are currently in line with the Two-by-One genetic model of CI, optimization of the transgenic expression system in D. melanogaster (Duffy 2002) will be necessary to confirm that these genes can recapitulate strong CI and rescue. If optimization fails to improve the pentrance, then other proteins may modulate the phenotypic potency of CI and be required for strong CI. Alternatively, homologous proteins may not efficiently bind to targets in other hosts, preventing strong CI under heterologous expression. Notably, since non-cognate expression of cifA homologs with cifBwMel[T1] yielded strong CI, it is clear that cifA sequence variation is not responsible for weakened CI. This is perhaps unsurprising given that mutagenesis assays of CifwMel[T1] proteins reveal that CI expression is more likely to be impacted by mutations in CifB than in CifA (Shropshire et al. 2020a). Thus, the aforementioned effects of suboptimal expression, need for additional genes, or inefficient binding to D. melanogaster targets could be related to the expression of cifB homologs.
Similarly, cifA;BwRi[T1] males do not cause CI, but notably non-cognate dual expression with cifwMel[T1] genes revealed that cifAwRi[T1], but not cifBwRi[T1], contributes to strong CI. This result is perhaps expected since cifBwRi[T1] has an early in-frame stop codon relative to cifBwMel[T1] that contributes to its annotation in the wRi genome as a nonfunctional pseudogene. Despite this, we hypothesized that cifBwRi[T1] would contribute to CI since wRi’s expression of both type 1 and 2 cif genes aligns with the patterns of unidirectional CI between wRi and wMel (Figure 1D). We provide four hypotheses to explain the absence of CI under cifA;BwRi[T1] expression. First, cifBwRi[T1] is a pseudogene and is not capable of contributing to CI. Second, since wRi harbors two identical type 1 gene pairs and a type 2 gene pair (LePage et al. 2017), both type pairs may be required for complete CI expression. Third, the early stop codon in cifBwRi[T1] may not prevent translation of the full-length protein since some stop codons slow translation instead of halting it (Wangen and Green 2020). Thus, a full-length CifBwRi[T1] protein may be generated despite the internal stop codon, and we did not test that here. Finally, to co-express the N-terminal and C-terminal CifBwRi[T1] proteins, we introduced a sequence between the two proteins that causes translational slippage and multi-protein translation from a single transcript (Donnelly et al. 2001a,b). This method yields a C-terminal sequence extension to the first protein that may alter protein function. In summary, these data currently support pseudogenization of the cifBwRi[T1] gene, but transgenic optimization and co-expression with other cif proteins will be necessary to fully rule out alternative explanations.
Contrary to initial predictions, cifBwRec[T1] transgenic males cause sperm infertility and/or embryonic death when mated with uninfected females. At its surface, cifBwRec[T1] alone may be interpreted to cause CI. However, this lethality is not rescued by cifAwRec[T1], cifBwMel[T1], or wMel females, and it associates with unusual cytological defects relative to wMel transgenic CI. As bona fide CI is defined by male embryonic lethality, a standard set of cytological defects, and the ability to rescue them, we do not interpret cifBwRec[T1] lethality as CI. However, it is plausible that a wRec-infected fly may rescue cifBwRec[T1] lethality. Since testing this requires difficult transinfection of wRec into D. melanogaster, we did not test this hypothesis. Conversely, cifA:BwRec[T1] males also weakly reduce hatching that is rescuable by cifAwRec[T1] and cifAwMel[T1]. Thus, these data suggest that while cifBwRec[T1] alone may cause an unusual lethality, a CI-like phenotype is only achieved when CifA and CifB proteins are dually expressed in males. We discuss our mechanistic interpretations of the results below (see “CI mechanism” section).
Incompatibility relationships
wMel and wRi Wolbachia are unidirectionally incompatible when wMel Wolbachia are transinfected into D. simulans (Poinsot et al. 1998) (Figure 1C). Specifically, wRi rescues wMel CI, but wMel does not rescue wRi CI. We hypothesized that cif sequence and copy number variation controls these incompatibility relationships (LePage et al. 2017). Since wRi has both type 1 and 2 cif genes, we expected cifAwRi[T1] to rescue cifA;BwMel[T1]-induced CI because the cifA variants are closely related, and cifAwMel[T1] would not rescue cifA;BwRi[T2]-induced CI because cifAwMel[T1] is highly divergent from the type 2 gene pair (LePage et al. 2017) (Figure 1D). In addition, wRec and wMel have only type 1 genes with a few amino acid changes, leading to the prediction that they are compatible (Figure 1C and D). We tested three key predictions of this cif genotype—CI phenotype hypothesis: (i) cifAwRi[T1] rescues transgenic cifwMel[T1] CI , (ii) cifAwRi[T1] cannot rescue transgenic cifwMel[T1] CI, and (iii) cifAwMel[T1] cannot rescue transgenic cifwRi[T2] CI.
As predicted, type 1 cifAwRec[T1] and cifAwRi[T1] can each rescue transgenic cifA;BwMel[T1] CI. In addition, cifAwRi[T2] cannot rescue cifA;BwMel[T1] CI, despite being able to rescue cifA;BwRi[T2] CI. These data align with expectations that only closely related cif homologs are compatible (Figure 1D). However, we also hypothesized that cifAwMel[T1] does not rescue cifA;BwRi[T2] CI, but rescue occurred at the same levels for both cifAwMel[T1] or cifAwRi[T2] females, suggesting that both cifA variants were capable of rescuing transgenic cifA;BwRi[T2] CI. These results imply a unidirectional incompatibility between type 1 and type 2 genes where type 1 genes cannot be rescued by type 2 genes, but the reciprocal cross is compatible. Not only are these results contrary to our expectations, but they also fail to sufficiently explain the reported unidirectional CI between wMel and wRi since rescue occurs in the opposite direction than we report here (Poinsot et al. 1998). We propose two possible explanations for these results.
First, host genotype may impact incompatibility relationships. Two studies evaluated the CI relationships between wMel and wRi, revealing unidirectional CI when wMel is transinfected into D. simulans (Poinsot et al. 1998) and no incompatibility when wMel-infected D. melanogaster is crossed with wRi-infected D. simulans (Gazla and Carracedo 2009). Similarly, two Wolbachia from the Nasonia longicornis parasitoid wasp switched from being unidirectionally to bidirectionally incompatible when moved into the same genetic background (Raychoudhury and Werren 2012). Thus, there is support for host control of Wolbachia reproductive parasitism and incompatibility relationships. It is unknown what kind of incompatibility relationships might occur if both wMel and wRi are in a D. melanogaster host background. However, our transgenic cif expression data suggest that wMel can rescue wRi, but not vice versa. Thus, we hypothesize that rescue, in particular, is impacted by host genotype such that cifA expressed natively (e.g. wMel in D. melanogaster or wRi in D. simulans) has expanded rescue capability as compared to expression in introduced strains. This hypothesis can be tested through transinfection of wRi into a D. melanogaster background or through transgenic expression of cifwMel[T1], cifwRi[T1], and cifwRi[T2] in D. simulans. Second, it remains possible that there are dynamic interactions between Cifs such that multiple phylogenetic types interact with one another to impact the phenotypic output. For instance, since wRi naturally maintains both type 1 and 2 cif genes (LePage et al. 2017; Lindsey et al. 2018), expression of both may be required to induce the reported compatibility relationships between wMel and wRi (Poinsot et al. 1998). This hypothesis can be tested through the dual expression of both types 1 and 2 gene pairs and crossing to cifwMel expressing flies.
CI mechanism
The cellular and molecular bases of CifA and CifB in CI remain an active area of investigation. To date, in vitro assays determined that CifBwMel[T1] and CifBwPip[T1] act in part as deubiquitinases, CifBwPip[T4] acts in part as a nuclease, cognate CifA;B pairs of wMel and wPip can bind, and both CifA and CifB interact with host proteins when transgenically expressed in D. melanogaster (Beckmann et al. 2017, 2019c; Chen et al. 2019). There are two mechanistic models for CI that are currently debated: host modification (HM) and toxin antidote (TA) (Hurst 1991; Poinsot et al. 2003; Shropshire et al. 2019; Beckmann et al. 2019a,b). HM models posit that CifA;B proteins cause CI by modifying host factors during spermatogenesis, and those modifications are transferred to the embryo. Rescue occurs when CifA in females reverses those sperm modifications in the embryo (Shropshire et al. 2018, 2019). Conversely, TA models suggest that CifB is transferred to the embryo via the sperm and kills the embryo unless its lethality is rescued through binding to CifA in the embryo (Beckmann et al. 2019a; Shropshire et al. 2019). Notably, there is no evidence of paternal transfer of Cif toxin(s), and it remains unclear whether CifA-B binding is related to CI or rescue (Shropshire et al. 2019). Thus, current data are insufficient to support one model over the other. Here, we place three findings above into the context of CI’s mechanistic basis: (i) CifB sequence variation impacts CI level variation, (ii) closely related type 1 CifA can be interchanged for both CI and rescue, and (iii) CifBwRec[T1] induces complete embryonic death when singly expressed.
A key finding of this study is that cifBwRec[T1] and cifBwRi[T1] sequence variation impacts cifA;B-induced CI level when transgenically expressed in D. melanogaster. We propose two mechanistic explanations. First, foreign CifB homologs in a new host may be less efficient or unable to bind host proteins or to CifA. Proteomic analyses of synthesized CifwPip[T1] proteins bound to a column and washed with D. melanogaster lysate revealed that CifBwPip[T1], CifAwPip[T1], or CifA;BwPip[T1] proteins bind between 15 and 60 fly proteins (Beckmann et al. 2019c). The sheer number of potential CifB-binding partners may contribute to the large impact of cifBwRec[T1] and cifBwRi[T1] sequence variation on CI levels. Alternatively, cifBwRec[T1] and cifBwRi[T1] sequence variation may contribute to variation in its tissue localization, subcellular localization, or ability to diffuse between cellular components. CI levels have been correlated with the number of Wolbachia-infected spermatocytes and spermatids during spermatogenesis in wRi-infected D. simulans (Clark and Karr 2002; Veneti et al. 2003; Clark et al. 2003), but even uninfected spermatocytes often result in modified sperm that can cause CI (Riparbelli et al. 2007), suggesting that CifA and/or CifB are diffusible between spermatocytes or during earlier stages of spermatogenesis. Binding and localization studies would elucidate these hypotheses.
While cifBwRec[T1] and cifBwRi[T1] sequence variation clearly impacts the CI level in transgenic D. melanogaster, type 1 cifAwReci[T1] and cifAwRi[T1] homologs were notably interchangeable and contribute to both strong CI and rescue. These data importantly suggest that while cifBwRec[T1] and cifBwRi[T1] sequence variation may be specifically attuned to a distinct host background, transgenic CifA is less subject to variation in host background. For instance, it is plausible that while CifB is interacting with rapidly evolving host targets in an arms race, CifA interacts with a set of conserved targets. One prediction of this hypothesis would be that CifA would be under purifying selection to retain compatibility with conserved host targets. Indeed, comparative sequence analyses reveal not only that type 1 CifAs are under strong purifying selection (Shropshire et al. 2018), but also that CifA sequence length is highly conserved across the phylogenetic types (LePage et al. 2017; Lindsey et al. 2018) and less prone to pseudogenization than CifB (Martinez et al. 2020). Thus, a type of HM model could be proposed whereby CifB acts simply as an “accessory” to bind CifA and unlock its access to conserved host processes not otherwise accessible in the absence of CifB. In addition, theory suggests that hosts will evolve resistance to CI while maintaining rescue (Turelli 1994), and many of the same predictions above would also apply in this scenario. For instance, if CifA’s targets in rescue and CI are similar, then one would predict the conservation of those targets to maintain rescue, while also maintaining CifA’s ability to contribute to CI. However, variation in CifB’s targets would only inhibit CI induction; thus, selection may favor variation in CifB targets to develop resistance against CI.
Finally, cifBwRec[T1] males cause complete infertility and/or embryonic death, but these defects are not rescuable and associate with unusual cytological defects. As such, cifBwRec[T1]-induced effects are not consistent with our expectations for CI induction. We propose two hypotheses for these results. First, CifB may cause CI in the absence of CifA. Singly expressing cifB homologs in yeast causes temperature-sensitive lethality that can be reduced when dually expressed with cognate cifA (Beckmann et al. 2017, 2019a,b; Chen et al. 2019). However, aside from singly expressing cifBwRec[T1] in this study, only cifBwPip[T4] males cause weak embryonic lethality (Chen et al. 2019), but there is also no evidence that cifBwPip[T4]-induced lethality can be rescued; moreover, more embryonic death is induced when cifBwPip[T4] is dually expressed with cifAwPip[T4] (Chen et al. 2019). Thus, cifBwRec[T1]-induced lethality varies from these historical results because cifBwRec[T1] yields near-complete embryonic death that is weakened and becomes rescuable only when dually expressed with cifAwRec[T1]. It is plausible that the reduced embryonic death from cifA;BwRec[T1] relative to cifBwRec[T1] alone is explained by cifA protection of a cifB-mediated sperm toxicity. However, it then becomes unclear why embryonic death increases in all other cases of dual transgene expression in insects and why cifBwRec[T1] is the only cifB homolog to cause near-complete embryonic death. Second, cifBwRec[T1] embryonic lethality may be a transgenic, off-target artifact. CifA’s binding to CifB (Beckmann et al. 2017) may be required for proper function, such as localizing CifB to its cellular target or priming its activity (Shropshire et al. 2019). Thus, in the absence of CifAwRec[T1], CifBwRec[T1] may result in off-target enzymatic activity and/or disruption of crucial host processes unrelated to CI induction, thus leading to a sterility independent of CI. This may explain why CifBwRec[T1] defects cannot be rescued. However, why would CifBwRec[T1] cause artifactual embryonic death when singly expressing other CifB homologs does not? CifBwRec[T1] has a unique C-terminal truncation beyond the putative deubiquinase domain. Numerous insecticidal toxins and bacterial protoxins have C-terminal self-inhibitors that prevent enzymatic activity, including latrotoxins (Rohou et al. 2007), Cry toxins (Peña-Cardeña et al. 2018), and botulinum neurotoxins (Mizanur et al. 2013). As such, some CifB proteins may contain C-terminal self-inhibitors that prevent their action in males. If CifBwRec[T1] lacks this self-inhibitor, then its activity would not require cleavage. When expressed by Wolbachia, this toxicity may not be observed if the expression profile is tightly regulated or if other proteins are expressed that suppress CifBwRec[T1] function. Support of these hypotheses will require the characterization of CifB’s C-terminus and the functional role of CifA-B binding. In summary, cifBwRec[T1]-induced effects are of interest, but significant caution is warranted as this lethality is not rescuable, which is a requirement for bona fide CI.
Summary
Here, we set out to investigate the hypothesis that cif sequence variation directly relates to CI phenotypic variation by evaluating cognate combinations of the cif genes and their incompatibility relationships. Moreover, we engineered non-cognate gene sets to test CI capacity and links between cif sequence variation and variation in CI level. In summary, we determined for the first time that type 1 cif homologs from wRec and type 2 cif homologs from wRi cause weak CI when transgenically expressed in D. melanogaster, variation in cifB contributes to CI level variability, divergent cifA fails to rescue transgenic cifA;BwMel[T1] CI, and type 1 cifA homologs are interchangeable for inducing both strong CI and rescue. We discuss these results in the context of CI’s Two-by-One genetic basis in wRec and wRi, incompatibility relationships, and CI mechanism. The work expands upon our understanding of the genetics of CI and incompatibilities between Wolbachia strains, and they establish novel hypotheses regarding the cif mechanism, CI level variation, and the relationship between CI phenotypes and host genetics.
Acknowledgments
We thank members of the Bordenstein Lab for feedback throughout this study and two anonymous reviewers for excellent feedback on the manuscript.
J.D.S. and S.R.B. designed research, analyzed data, and wrote the paper. J.D.S. and R.R. performed research.
Funding
This work was supported by National Institutes of Health awards R01 AI132581 and R01 AI143725 to S.R.B., the Vanderbilt Microbiome Initiative, and National Science Foundation Graduate Research Fellowship DGE-144519 and National Science Foundation Postdoctoral Research Fellowship in Biology DBI-2010210 to J.D.S. Any opinions, findings, conclusions, or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Institutes of Health or the National Science Foundation.
Conflicts of interest
J.D.S. and S.R.B. are listed as inventors on a pending patent relevant to this work. S.R.B. is a coinventor on two other pending patents related to controlling arthropods.
Literature cited
- Beckmann J, Bonneau M, Chen H, Hochstrasser M, Poinsot D, et al. 2019a. The toxin–antidote model of cytoplasmic incompatibility: genetics and evolutionary implications. Trends Genet. 35:175–185. 10.1016/j.tig.2018.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann J, Bonneau M, Chen H, Hochstrasser M, Poinsot D, et al. 2019b. Caution does not preclude predictive and testable models of cytoplasmic incompatibility: a reply to Shropshire. Trends Genet. 35:399–400. 10.1016/j.tig.2019.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann J, Ronau JA, Hochstrasser M.. 2017. A Wolbachia deubiquitylating enzyme induces cytoplasmic incompatibility. Nat Microbiol. 2:17007. 10.1038/nmicrobiol.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann J, Sharma GD, Mendez L, Chen H, Hochstrasser M.. 2019c. The Wolbachia cytoplasmic incompatibility enzyme CidB targets nuclear import and protamine-histone exchange factors. eLife. 8:e50026. 10.7554/eLife.50026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bing X-L, Zhao D-S, Sun J-T, Zhang K-J, Hong X-Y.. 2020. Genomic analysis of Wolbachia from Laodelphax striatellus (Delphacidae, Hemiptera) reveals insights into its “Jekyll and Hyde” mode of infection pattern. Genome Biol Evol. 10.1093/gbe/evaa006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneau M, Atyame C, Beji M, Justy F, Cohen-Gonsaud M, et al. 2018. Culex pipiens crossing type diversity is governed by an amplified and polymorphic operon of Wolbachia. Nat Commun. 9. 10.1038/s41467-017-02749-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneau M, Caputo B, Ligier A, Caparros R, Unal S, et al. 2019. Variation in Wolbachia cidB gene, but not cidA, is associated with cytoplasmic incompatibility mod phenotype diversity in Culex pipiens. Mol Ecol. 28:4725–4736. 10.1111/mec.15252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordenstein SR, Bordenstein SR.. 2016. Eukaryotic association module in phage WO genomes from Wolbachia. Nat Commun. 7:13155. 10.1038/ncomms13155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordenstein SR, O'Hara FP, Werren JH.. 2001. Wolbachia-induced incompatibility precedes other hybrid incompatibilities in Nasonia. Nature. 409:707–710. 10.1038/35055543. [DOI] [PubMed] [Google Scholar]
- Bordenstein SR, Uy JJ, Werren JH.. 2003. Host genotype determines cytoplasmic incompatibility type in the haplodiploid genus Nasonia. Genetics. 164:223–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle L, O'Neill S, Robertson H, Karr T.. 1993. Interspecific and intraspecific horizontal transfer of Wolbachia in Drosophila. Science. 260:1796–1799. 10.1126/science.8511587. [DOI] [PubMed] [Google Scholar]
- Brucker RM, Bordenstein SR.. 2012. Speciation by symbiosis. Trends Ecol Evol. 27:443–451. 10.1016/j.tree.2012.03.011. [DOI] [PubMed] [Google Scholar]
- Casiraghi M, Bordenstein SR, Baldo L, Lo N, Beninati T, et al. 2005. Phylogeny of Wolbachia pipientis based on gltA, groEL and ftsZ gene sequences: clustering of arthropod and nematode symbionts in the F supergroup, and evidence for further diversity in the Wolbachia tree. Microbiol Sgm. 151:4015–4022. 10.1099/mic.0.28313-0. [DOI] [PubMed] [Google Scholar]
- Charlat S, Calmet C, Merçot H.. 2001. On the mod resc model and the evolution of Wolbachia compatibility types. Genetics. 159:1415–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth J, Weinert LA, Araujo EV, Welch JJ.. 2019. Wolbachia, Cardinium and climate: an analysis of global data. Biol Lett. 15:20190273. 10.1098/rsbl.2019.0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Ronau JA, Beckmann J, Hochstrasser M.. 2019. A Wolbachia nuclease and its binding partner provide a distinct mechanism for cytoplasmic incompatibility. Proc Natl Acad Sci USA. 116:22314–22321. 10.1073/pnas.1914571116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark ME, Karr TL.. 2002. Distribution of Wolbachia within Drosophila reproductive tissue: implications for the expression of cytoplasmic incompatibility. Integr Comp Biol. 42:332–339. 10.1093/icb/42.2.332. [DOI] [PubMed] [Google Scholar]
- Clark ME, Veneti Z, Bourtzis K, Karr TL.. 2003. Wolbachia distribution and cytoplasmic incompatibility during sperm development: the cyst as the basic cellular unit of CI expression. Mech Dev. 120:185–198. 10.1016/S0925-4773(02)00424-0. [DOI] [PubMed] [Google Scholar]
- Crawford JE, Clarke DW, Criswell V, Desnoyer M, Cornel D, et al. 2020. Efficient production of male Wolbachia-infected Aedes aegypti mosquitoes enables large-scale suppression of wild populations. Nat Biotechnol. 38:482–492., 10.1038/s41587-020-0471-x. [DOI] [PubMed] [Google Scholar]
- Donnelly ML, Hughes LE, Luke G, Mendoza H, ten Dam E, et al. 2001a. The “cleavage” activities of foot-and-mouth disease virus 2A site-directed mutants and naturally occurring “2A-like” sequences. J Gen Virol. 82:1027–1041. 10.1099/0022-1317-82-5-1027. [DOI] [PubMed] [Google Scholar]
- Donnelly ML, Luke G, Mehrotra A, Li X, Hughes LE, et al. 2001b. Analysis of the aphthovirus 2A/2B polyprotein “cleavage” mechanism indicates not a proteolytic reaction, but a novel translational effect: a putative ribosomal “skip”. J Gen Virol. 82:1013–1025. 10.1099/0022-1317-82-5-1013. [DOI] [PubMed] [Google Scholar]
- Duffy JB. 2002. GAL4 system in Drosophila: a fly geneticist’s Swiss army knife. Genesis. 34:1–15. 10.1002/gene.10150. [DOI] [PubMed] [Google Scholar]
- Gazla IN, Carracedo MC.. 2009. Effect of intracellular Wolbachia on interspecific crosses between Drosophila melanogaster and Drosophila simulans. Genet Mol Res. 8:861–869. 10.4238/vol8-3gmr595. [DOI] [PubMed] [Google Scholar]
- Gerth M, Rothe J, Bleidorn C.. 2013. Tracing horizontal Wolbachia movements among bees (Anthophila): a combined approach using multilocus sequence typing data and host phylogeny. Mol Ecol. 22:6149–6162. 10.1111/mec.12549. [DOI] [PubMed] [Google Scholar]
- Gillespie JJ, Driscoll TP, Verhoeve VI, Rahman MS, Macaluso KR, et al. 2018. A tangled web: origins of reproductive parasitism. Genome Biol Evol. 10:2292–2309. 10.1093/gbe/evy159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J-T, Li Y, Li T-P, Liang Y, Hu L, et al. 2020. Stable introduction of plant-virus-inhibiting Wolbachia into planthoppers for rice protection. Curr Biol. 10.1016/j.cub.2020.09.033. [DOI] [PubMed] [Google Scholar]
- Hilgenboecker K, Hammerstein P, Schlattmann P, Telschow A, Werren JH.. 2008. How many species are infected with Wolbachia?—a statistical analysis of current data. FEMS Microbiol Lett. 281:215–220. 10.1111/j.1574-6968.2008.01110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A, Turelli M, Harshman L.. 1990. Factors affecting the distribution of cytoplasmic incompatibility in Drosophila simulans. Genetics. 126:933–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huigens ME, de Almeida RP, Boons PAH, Luck RF, Stouthamer R.. 2004. Natural interspecific and intraspecific horizontal transfer of parthenogenesis-inducing Wolbachia in Trichogramma wasps. Proc R Soc Lond B. 271:509–515. 10.1098/rspb.2003.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst LD. 1991. The evolution of cytoplasmic incompatibility or when spite can be successful. J Theor Biol. 148:269–277. 10.1016/s0022-5193(05)80344-3. [DOI] [PubMed] [Google Scholar]
- Jaenike J, Dyer KA, Cornish C, Minhas MS.. 2006. Asymmetrical reinforcement and Wolbachia infection in Drosophila. PLoS Biol. 4:e325. 10.1371/journal.pbio.0040325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layton EM, On J, Perlmutter JI, Bordenstein SR, Shropshire JD.. 2019. Paternal grandmother age affects the strength of Wolbachia-induced cytoplasmic incompatibility in Drosophila melanogaster. mBio. 10. 10.1128/mBio.01879-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees RS, Gilles JR, Hendrichs J, Vreysen MJ, Bourtzis K.. 2015. Back to the future: the sterile insect technique against mosquito disease vectors. Curr Opin Insect Sci. 10:156–162. 10.1016/j.cois.2015.05.011. [DOI] [PubMed] [Google Scholar]
- LePage DP, Metcalf JA, Bordenstein SR, On J, Perlmutter JI, et al. 2017. Prophage WO genes recapitulate and enhance Wolbachia-induced cytoplasmic incompatibility. Nature. 543:243–247., 10.1038/nature21391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey A, Rice DW, Bordenstein SR, Brooks AW, Bordenstein SR, et al. 2018. Evolutionary genetics of cytoplasmic incompatibility genes cifA and cifB in prophage WO of Wolbachia. Genome Biol Evol. 10:434–451., 10.1093/gbe/evy012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Klasson L, Welch JJ, Jiggins FM.. 2020. Life and death of selfish genes: comparative genomics reveals the dynamic evolution of cytoplasmic incompatibility. Mol. Biol. Evol. 10.1093/molbev/msaa209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf JA, Jo M, Bordenstein SR, Jaenike J, Bordenstein SR.. 2014. Recent genome reduction of Wolbachia in Drosophila recens targets phage WO and narrows candidates for reproductive parasitism. PeerJ. 2:e529. 10.7717/peerj.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizanur RM, Frasca V, Swaminathan S, Bavari S, Webb R, et al. 2013. The C terminus of the catalytic domain of type A botulinum neurotoxin may facilitate product release from the active site. J Biol Chem. 288:24223–24233., 10.1074/jbc.M113.451286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti R, Marzo GA, Lampazzi E, Calvitti M.. 2018. Cytoplasmic incompatibility management to support incompatible insect technique against Aedes albopictus. Parasit Vectors. 11:649. 10.1186/s13071-018-3208-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita S, Shimajiri Y, Nomura M.. 2009. Strong cytoplasmic incompatibility and high vertical transmission rate can explain the high frequencies of Wolbachia infection in Japanese populations of Colias erate poliographus (Lepidoptera: Pieridae). Bull Entomol Res. 99:385–391. 10.1017/S0007485308006469. [DOI] [PubMed] [Google Scholar]
- Nikolouli K, Colinet H, Renault D, Enriquez T, Mouton L, et al. 2018. Sterile insect technique and Wolbachia symbiosis as potential tools for the control of the invasive species Drosophila suzukii. J Pest Sci. 91:489–503., https://doi.org/10/gc55cr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill SL. 2018. The use of Wolbachia by the World Mosquito Program to interrupt transmission of Aedes aegypti transmitted viruses. Adv. Exp. Med. Biol. 1062:355–360. 10.1007/978-981-10-8727-1_24. [DOI] [PubMed] [Google Scholar]
- Peña-Cardeña A, Grande R, Sánchez J, Tabashnik BE, Bravo A, et al. 2018. The C-terminal protoxin region of Bacillus thuringiensis Cry1Ab toxin has a functional role in binding to GPI-anchored receptors in the insect midgut. J Biol Chem. 293:20263–20272. 10.1074/jbc.RA118.005101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poinsot D, Bourtzis K, Markakis G, Savakis C, Mercot H.. 1998. Wolbachia transfer from Drosophila melanogaster into D. simulans: host effect and cytoplasmic incompatibility relationships. Genetics. 150:227–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poinsot D, Charlat S, Mercot H.. 2003. On the mechanism of Wolbachia-induced cytoplasmic incompatibility: confronting the models with the facts. Bioessays. 25:259–265. 10.1002/bies.10234. [DOI] [PubMed] [Google Scholar]
- Raychoudhury R, Werren JH.. 2012. Host genotype changes bidirectional to unidirectional cytoplasmic incompatibility in Nasonia longicornis. Heredity (Edinb). 108:105–114. 10.1038/hdy.2011.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds KT, Hoffmann AA.. 2002. Male age, host effects and the weak expression or non-expression of cytoplasmic incompatibility in Drosophila strains infected by maternally transmitted Wolbachia. Genet Res. 80:79–87. [DOI] [PubMed] [Google Scholar]
- Riparbelli MG, Giordano R, Callaini G.. 2007. Effects of Wolbachia on sperm maturation and architecture in Drosophila simulans Riverside. Mech Dev. 124:699–714. 10.1016/j.mod.2007.07.001. [DOI] [PubMed] [Google Scholar]
- Rohou A, Nield J, Ushkaryov YA.. 2007. Insecticidal toxins from black widow spider venom. Toxicon. 49:531–549. 10.1016/j.toxicon.2006.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz M, Albanese D, Tuohy K, Donati C, Segata N, et al. 2020. Large scale genome reconstructions illuminate Wolbachia evolution. Nat Commun. 11:5235. 10.1038/s41467-020-19016-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker DD, Katju V, Jaenike J.. 1999. Wolbachia and the evolution of reproductive isolation between Drosophila recens and Drosophila subquinaria. Evolution. 53:1157–1164. 10.1111/j.1558-5646.1999.tb04529.x. [DOI] [PubMed] [Google Scholar]
- Shropshire JD, Bordenstein SR.. 2019. Two-by-one model of cytoplasmic incompatibility: synthetic recapitulation by transgenic expression of cifA and cifB in Drosophila. PLoS Genet. 15:e1008221. 10.1371/journal.pgen.1008221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shropshire JD, Bordenstein SR.. 2016. Speciation by symbiosis: the microbiome and behavior. mBio. 7:e01785–15. 10.1128/mBio.01785-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shropshire JD, Kalra M, Bordenstein SR.. 2020a. Evolution-guided mutagenesis of the cytoplasmic incompatibility proteins: identifying CifA’s complex functional repertoire and new essential regions in CifB. PLOS Pathog. 16:e1008794. 10.1371/journal.ppat.1008794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shropshire JD, Leigh B, Bordenstein SR.. 2020b. Symbiont-mediated cytoplasmic incompatibility: what have we learned in 50 years? eLife. 9:e61989. 10.7554/eLife.61989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shropshire JD, Leigh B, Bordenstein SR, Duplouy A, Riegler M, et al. 2019. Models and nomenclature for cytoplasmic incompatibility: caution over premature conclusions—a response to Beckmann. Trends Genet. 35:397–399. 10.1016/j.tig.2019.03.004 [DOI] [PubMed] [Google Scholar]
- Shropshire JD, On J, Layton EM, Zhou H, Bordenstein SR.. 2018. One prophage WO gene rescues cytoplasmic incompatibility in Drosophila melanogaster. Proc Natl Acad Sci Usa. 115:4987–4991. 10.1073/pnas.1800650115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Journal of Bacteriology. 2018. Instructions to authors. (Accessed: 2020 November 8). https://jb.asm.org/content/nomenclature.
- Tolley SJA, Nonacs P, Sapountzis P.. 2019. Wolbachia horizontal transmission events in ants: what do we know and what can we learn? Front Microbiol. 10:296. 10.3389/fmicb.2019.00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli M. 1994. Evolution of incompatibility-inducing microbes and their hosts. Evolution. 48:1500–1513. 10.1111/j.1558-5646.1994.tb02192.x [DOI] [PubMed] [Google Scholar]
- Turelli M, Hoffmann AA.. 1991. Rapid spread of an inherited incompatibility factor in California Drosophila. Nature. 353:440–442. 10.1038/353440a0 [DOI] [PubMed] [Google Scholar]
- Turelli M, Hoffmann AA.. 1995. Cytoplasmic incompatibility in Drosophila simulans: dynamics and parameter estimates from natural populations. Genetics. 140:1319–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veneti Z, Clark ME, Zabalou S, Karr TL, Savakis C, et al. 2003. Cytoplasmic incompatibility and sperm cyst infection in different Drosophila-Wolbachia associations. Genetics. 164:545–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wangen JR, Green R.. 2020. Stop codon context influences genome-wide stimulation of termination codon readthrough by aminoglycosides. eLife. 9:e52611. 10.7554/eLife.52611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert LA, Araujo-Jnr EV, Ahmed MZ, Welch JJ.. 2015. The incidence of bacterial endosymbionts in terrestrial arthropods. Proc R Soc B. 282. 10.1098/rspb.2015.0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren JH, Jaenike J.. 1995. Wolbachia and cytoplasmic incompatibility in mycophagous Drosophila and their relatives. Heredity (Edinb). 75:320–326. [DOI] [PubMed] [Google Scholar]
- Wu M, Sun LV, Vamathevan J, Riegler M, Deboy R, et al. 2004. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2:e69–341. 10.1371/journal.pbio.0020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada R, Floate KD, Riegler M, O'Neill SL.. 2007. Male development time influences the strength of Wolbachia-induced cytoplasmic incompatibility expression in Drosophila melanogaster. Genetics. 177:801–808. 10.1534/genetics.106.068486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada R, Iturbe-Ormaetxe I, Brownlie JC, O’Neill SL.. 2011. Functional test of the influence of Wolbachia genes on cytoplasmic incompatibility expression in Drosophila melanogaster. Insect Mol Biol. 20:75–85. 10.1111/j.1365-2583.2010.01042.x. [DOI] [PubMed] [Google Scholar]
- Yen JH, Barr AR.. 1973. The etiological agent of cytoplasmic incompatibility in Culex pipiens. J Invertebr Pathol. 22:242–250. 10.1016/0022-2011(73)90141-9. [DOI] [PubMed] [Google Scholar]
- Zug R, Hammerstein P.. 2012. Still a host of hosts for Wolbachia: analysis of recent data suggests that 40% of terrestrial arthropod species are infected. PLoS One. 7:e38544. 10.1371/journal.pone.0038544. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are made publicly available in the supplement of this manuscript. Fly lines not otherwise available in the Bloomington Drosophila Stock Center are available upon request.
Supplemental material is available at figshare DOI: https://doi.org/10.25386/genetics.13215503.