Abstract
Prior to updating global influenza-associated mortality estimates, the World Health Organization convened a consultation in July 2017 to understand differences in methodology and implications for results of 3 influenza mortality projects from the US Centers for Disease Control and Prevention (CDC), the Netherlands Institute for Health Service Research’s Global Pandemic Mortality Project II (GLaMOR), and the Institute for Health Metrics and Evaluation (IHME). The expert panel reviewed estimates and discussed differences in data sources, analysis, and modeling assumptions. We performed a comparison analysis of the estimates. Influenza-associated respiratory death counts were comparable between CDC and GLaMOR; the IHME estimate was considerably lower. The greatest country-specific influenza-associated fold differences in mortality rate between CDC and IHME estimates and between GLaMOR and IHME estimates were among countries in Southeast Asia and the Eastern Mediterranean region. The data envelope used for the calculation was one of the major differences (CDC and GLaMOR: all respiratory deaths; IHME: lower-respiratory infection deaths). With the assumption that there is only one cause of death for each death, IHME estimates a fraction of the full influenza-associated respiratory mortality that is measured by the other 2 groups. Wide variability of parameters was observed. Continued coordination between groups could assist with better understanding of methodological differences and new approaches to estimating influenza deaths globally.
Keywords: comparison, estimates, global, influenza, mortality
Seasonal influenza epidemics contribute significantly to annual global mortality (1). The availability of influenza-associated burden estimates, especially in resource-limited settings (2), can support evidence-based policy decisions by guiding funding priorities for prevention and control, such as influenza vaccine introduction or expansion of existing immunization programs (3), or availability and use of antiviral medications (4).
National estimates of influenza-associated deaths may use different outcomes. Some studies use ecological models beginning with all respiratory-coded deaths to calculate influenza-associated excess deaths by subtracting out deaths that are unlikely to be associated with an influenza virus infection (i.e., chronic respiratory deaths included in the baseline death calculation). Other groups might use a multiplier approach, beginning with lower-respiratory infection (LRI) deaths, and assign a proportion of those to influenza.
Previous global influenza-associated mortality estimates from the World Health Organization (WHO) were widely referenced, despite lack of documentation on the methodology and data sources (5). In parallel, the increased number of countries with high-quality influenza viral surveillance data and national disease mortality registries has highlighted a need to update global estimates and document methods.
Prior to updating global influenza-associated mortality estimates, WHO convened a consultation to compare the 3 recent global influenza-mortality projects from: 1) the US Centers for Disease Control and Prevention (CDC), 2) the Netherlands Institute for Health Service Research, and 3) the Institute for Health Metrics and Evaluation (IHME). These groups used slightly different outcomes measures to arrive at global estimates.
CDC led a collaboration of more than 47 countries to estimate global seasonal influenza-associated deaths, focusing on respiratory mortality (6). The Netherlands Institute for Health Service Research, in collaboration with the US National Institutes of Health and CDC, conducted the Global Pandemic Mortality Project II (GLaMOR), funded by WHO (7), to estimate global seasonal influenza-associated respiratory mortality. IHME leads the Global Burden of Disease Study (GBD), funded by Bill and Melinda Gates Foundation. Estimates of LRI and etiology-specific LRI mortality (influenza) from GBD 2016 were used in this comparison (8).
We describe the different approaches to estimate global influenza mortality, focusing on data sources, analysis, and model assumptions, and compare the differences between estimates. We compare methods and provide additional details regarding methodology to understand factors that might explain differences in these estimates and to make recommendations for future global burden efforts.
Table 1 summarizes the approach taken by each research group: CDC, GLaMOR, and IHME.
Table 1.
Description of 3 Institutional Models to Estimate Influenza Respiratory Mortality, Multiple Countries
| Research Group | Envelope | Sources of Data | Age Groups, years | Indicators | Extrapolation Methodology |
|---|---|---|---|---|---|
| CDCa | All respiratory causes of death | EMRs from 33 countries or territories. Annual respiratory deaths from 14 countries. WHO GHE RI deaths, 2015. United Nations and US Census Population Estimates 2015.b Study period: 1999–2015. |
<65, 65–74, ≥75, all | WHO GHE RI deaths (2015) as a proxy to account for differences in MMR | Bayesian hierarchical model with multiplier: • Estimate country-specific influenza-associated respiratory EMR for 33 countries using time series log-linear regression models with vital death records and influenza surveillance data. • Extrapolate estimates to countries without data. Countries divided into 3 ADs using WHO GHE RI mortality rates. • Country-specific MRR generated from WHO GHE RI deaths • Random average country-specific mortality rate from a contributing country multiplied by country/specific MMR to calculate country-specific mortality rate distributions |
| GLaMORc | All respiratory causes of death | EMRs from 33 countries or territories. Study period: 2002–2011. |
<65, ≥65, all | 10 country-specific indicators: WHO Region; age group all-cause mortality rates; physician density; obesity; population density; major infectious diseases; gross national income per capita; rural population; population age structure; latitude |
Multiple imputation method for extrapolation: • Two-step approach with a data creation step (stage 1) followed by a hierarchical regression modeling step to project burden for all countries (stage 2). • For data creation, used statistical correlations between country-specific indicators and contributing country mortality estimates to create distribution of possible mortality values. |
| IHMEd | LRI deaths | LRI mortality data from 10,312 site-year vital registration, 915 site-year verbal autopsy, and 928 site-year surveillance data, between 1980 and 2016. | 23 age groups: 0–6 days, 7–27 days, 28–364 days, 1–4 years, and every 5 years up to 99 |
13 covariates involved in the modeling process of LRI mortality: pneumococcal conjugate vaccine coverage; indoor air pollution; LRI summary exposure variable; mean BMI; smoking prevalence; DTP3 vaccine coverage; health-care access and quality index; education per capita; LDI per capita; sociodemographic index; alcohol liters per capita; outdoor air pollution (PM2.5); water and sanitation summary exposure value | Counterfactual approach to attribute influenza Estimate of LRI deaths. Attribution of specific pathogens (influenza) by: • Systematic review to estimate prevalence of influenza virus in patients with LRI; • Calculation of PAF using proportion influenza positive and OR of LRI given the presence of influenza virus; • Attributable fraction adjusted by the viral: bacterial pneumonia CFR ratio. |
Abbreviations: AD, analytical divisions; BMI, body mass index; CDC, US Centers for Disease Control and Prevention; CFR, case-fatality rate; DPT3, diphtheria-tetanus-pertussis; EMR, excess mortality estimates; GHE, Global Health Estimates; GLaMOR, Global Pandemic Mortality Project; IHME, Institute for Health Metrics and Evaluation; LDI, lag-distributed income; LRI, lower respiratory tract infection; MMR, mortality risk between countries; MRR, mortality rate ratio; OR, odds ratio; PAF, population attributable fraction; PM2.5, particulate matter with an aerodynamic diameter less than or equal to 2.5 μm; RI, respiratory tract infection; URI, respiratory tract infection; WHO, World Health Organization.
Iuliano et al. (6).
UN World Population Prospects (26).
Paget et al. (7).
Global Burden of Disease 2016 LRI Collaborators. (8).
CDC used a multistep approach to extrapolate country-specific (185 countries/territories), regional, and global seasonal influenza-associated excess deaths, under the assumption that influenza burden scales with respiratory infection death rates (6). Deaths were estimated for 1999–2015 and for 3 age groups (<65 years, 65–74 years, and ≥75 years). First, they estimated influenza-associated excess mortality rates (EMRs) for 33 contributing countries or territories by applying time-series linear regression models to weekly or monthly national vital statistics and viral surveillance data. Prior to extrapolation, Bayesian hierarchical models were used to calculate the mean influenza-associated EMR for multiple years for each contributing country. These mean EMRs were applied to countries without data to estimate influenza-associated mortality. For extrapolation, countries were divided into analytical divisions (ADs) using age-specific 2015 WHO Global Health Estimates (GHE) respiratory infection mortality rates. To adjust for differences in the risk of respiratory infection death between countries, country-specific respiratory mortality rate ratios were calculated by comparing GHE respiratory infection mortality rates from countries that do not have EMR estimates with those that have estimates. Extrapolation was conducted within each age-specific AD, and uncertainty was quantified via 5,000 Markov chain Monte Carlo simulations for each country. For the 33 contributing countries, extrapolated estimates were replaced by the mean influenza-associated EMR provided by the country.
The GLaMOR model used a 2-stage approach to estimate global and region-specific influenza-associated respiratory mortality, using a multiple imputation method for extrapolation(9).They calculated seasonal influenza-associated death estimates for 2 age groups (<65 years and ≥65 years). In stage 1, they used age-specific seasonal influenza-associated EMRs from countries or subnational regions. Thirty-three countries contributed data, of which 30 overlapped with CDC contributing countries. In stage 2, GLaMOR extrapolated EMRs to 196 countries using a multiple imputation method that involved a data-creation step followed by a hierarchical regression modeling step. Ten broad country-level indicators of development, demographic factors, or health were used to match countries in the data-creation step. In the GBD 2016, IHME estimated the number of LRI deaths directly attributable to influenza, using a 2-step modeling approach. First, they estimated overall LRI mortality according to 23 age groups (0–6 days, 7–27 days, 28–364 days, 1–4 years, and then in 5-year increments up to 99 years), sex, location (195 countries), and year for 1980–2016 (8, 10). LRI mortality was modeled in a Cause of Death Ensemble model (11). Next, they estimated the etiology-specific LRI mortality using a counterfactual approach to calculate the population attributable fraction (PAF) for influenza, or the proportion of LRI that could be averted if exposure to influenza was eliminated. PAF for influenza was estimated by:
where Prop is the proportion of influenza-positive LRI cases and OR is the odds ratio of LRI given influenza detection (12). Additionally, the influenza mortality PAF was adjusted for the age-specific relative case-fatality rate of viral to bacterial pneumonia episodes.
METHODS
Country- and age-specific influenza-associated respiratory mortality rates were provided by CDC, GLaMOR, and IHME for 3 age groups (<65 years, ≥65 years, and all ages) and for the 183 countries common between the groups for this comparison. CDC and GLaMOR used 2015 United Nations World Population Prospects, and IHME used their own population estimates for 2015 (Web Figure 1, available at https://doi.org/10.1093/aje/kwaa196). CDC provided death counts and rates, as well as 95% credible intervals and the 2.5th and the 97.5th intervals from the posterior Markov chain Monte Carlo distribution. GLaMOR provided the mean death counts, rates, and the range (lowest to highest annual estimate) across estimated years. IHME provided death counts, rates, and the 95% uncertainty interval. Given the different interval estimates provided, we compared the point estimates. Analyses were completed for 183 common countries and 3 age groups.
We compared influenza mortality rate estimates between CDC and IHME, CDC and GLaMOR, and IHME and GLaMOR by calculating the fold difference or rate ratios between country-and age-specific mortality rates. We obtained the coefficient of variation within a WHO Region for the country- and age-specific mortality rates using the ratio of the standard deviation to the mean. Then we compared between-country variability within the WHO Regions for CDC, IHME, and GLaMOR. We calculated pairwise Spearman’s ranked correlations by age group and WHO Region.
RESULTS
Magnitude of difference in influenza mortality estimates
Overall, global influenza-associated respiratory deaths and rates (per 100,000 population) overlapped between CDC (409,111, 95% credible interval, 291,243, 645,832; 5.6, 95% credible interval, 4.0, 8.8) and GLaMOR (389,213, range, 293,980–518,230; 5.9, range, 4.0–8.0) methods. In contrast, both CDC and GLaMOR influenza-associated death estimates were higher than IHME influenza-attributed LRI deaths (58,193, 95% uncertainty interval, 43,953, 74,175; 0.8,95%uncertaintyinterval,0.6,1.0),a difference of nearly 350,000 deaths (Table 2).
Table 2.
Global Influenza-Associated Mortality Estimates According to Age Group, Multiple Countries, 1980 –2016a–c
| Age Group | CDCa | GLaMORb | IHMEc | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Median | 95% Crl | Rates Per 100,000 | 95% Crl | Median | Ranged | Rates Per 100,000 | Ranged | Median | 95% Ul | Rates Per 100,000 | 95% Ul | |
| All ages | 409,111 | 291,243, 645,832 | 5.6 | 4.0, 8.8 | 389,213 | 293,980–518,230 | 5.9 | 4.0–8.0 | 58,193 | 43,953,74,175 | 0.8 | 0.6, 1.0 |
| <65 years | 175,303 | 67,255, 342,576 | 2.6 | 1.0, 5.1 | 128,512 | 91,764–184,980 | 2.1 | 1.6–3.1 | 30,041e | 13,019, 60,297 | 0.4e | 0.2, 0.9 |
| >65 years | 244,012e | 188,261, 330,694 | 40.2e | 31.0, 54.5 | 260,701 | 187,430–333,250 | 53.7 | 35.6–71.7 | 27,884e | 13,718,49,871 | 4.6e | 2.3, 8.3 |
Abbreviations:CDC, US Centers for Disease Control and Prevention;CrI, credible interval;GLaMOR, Global Pandemic Mortality Project;IHME, Institute for Health Metrics and Evaluation; UI, uncertainty interval.
Iuliano et al. (6).
Paget et al. (7).
Global Burden of Disease 2016 LRI Collaborators (8).
Range, minimum and maximum value.
Estimates for these age groups were not included in publications.
Among those <65 years of age, CDC and GLaMOR estimated the highest mortality rates for African region countries, followed by the Eastern Mediterranean, Southeast Asia, and European region countries (Web Figure 2A and 2B). In comparison, the highest IHME influenza-attributed LRI mortality rates for this age group were observed in the European and Eastern Mediterranean countries, then African countries (Web Figure 2C). For those aged <65 years and all countries, the median mortality rate was 1.6/100,000 population for CDC and GLAMOR and 0.5/100,000 population for IHME. Among those aged ≥65 years, CDC and GLaMOR estimates were higher, particularly among American, African, and Southeast Asian countries (Web Figures 3A and 3B). The median mortality rate for all countries in the ≥65-years age group was lowest for IHME (6.2/100,000), compared with CDC (46.0/100,000) and GLaMOR (40.4/100,000) (Web Figure 3C). The median all age mortality rates were 1.0/100,000 for IHME, compared with 5.4/100,000 and 4.2/100,000 for CDC and GLaMOR, respectively (Figure 1). Scatter plots comparing influenza-associated mortality rates show the highest differences in the African region for CDC and IMHE as well as the CDC and GLaMOR models. The comparison of rates between IHME and GLaMOR is consistent across WHO Regions (Web Figure 4).
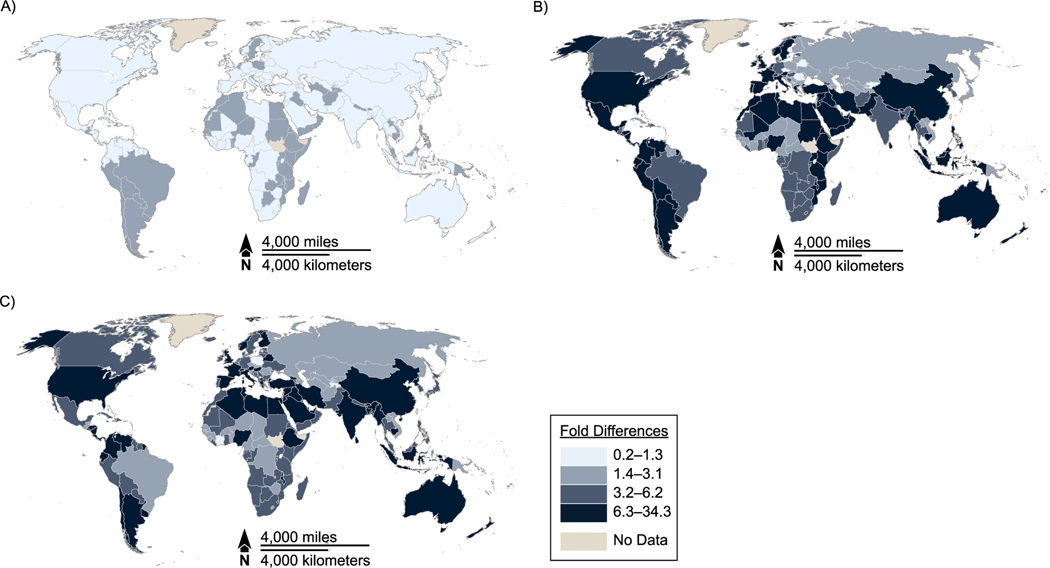
Figure 1.
Country-specific influenza respiratory mortality rates by quartile, all ages, multiple countries. A) US Centers for Disease Control and Prevention (CDC); B) Global Pandemic Mortality Project (GLaMOR); C) Institute for Health Metrics and Evaluation (IHME). Mortality rate per 100,000 population. Study periods: CDC, 1999–2015; GLaMOR, 2002–2011; IHME, 1980–2016. Rates for 183 countries included. Lighter colors indicate a lower mortality rate.
Overall, fold differences comparing influenza mortality rate estimates were lowest between CDC and GLaMOR (Figure 2A; Web Figures 5A and 6A). Across all age groups, country-specific influenza rate fold differences were the highest in the Southeast Asia and Eastern Mediterranean regions between CDC and IHME (Figure 2B; Web Figures 5B and 6B) and GLaMOR and IHME (Figure 2C; Web Figures 5C and 6C). Among those aged ≥65 years, the influenza-associated mortality-rate mean fold differences observed between CDC and IHME and GLaMOR and IHME were similar, at 7.6 and 8.1, respectively (Web Figures 6B and 6C).
Figure 2.
Fold differences comparing pairwise influenza mortality rate estimates by quartile, all ages, multiple countries. A) US Centers for Disease Control and Prevention (CDC) versus Global Pandemic Mortality Project (GLaMOR); B) CDC versus Institute for Health Metrics and Evaluation (IHME); C) GLaMOR versus IHME. Study periods: CDC, 1999–2015; GLaMOR, 2002–2011; IHME, 1980–2016. Rates for 183 countries included. Lighter colors indicate a low fold difference.
Across methods, the coefficients of variation for the influenza-associated mortality estimates were similar (Table 3), except among those aged <65 years in the European region, where IHME had a higher coefficient of variation (3.5) compared with CDC (2.8) and GLaMOR (2.0). Among those aged ≥65 years, IHME had the smallest (2.4) coefficient of variation in the Western Pacific region. Comparing the groups, the mortality rate coefficient of variation within WHO Regions was consistent between CDC, IHME, and GLaMOR, with the largest coefficient of variation observed in the Western Pacific and the lowest coefficient of variation in the African region.
Table 3.
Coefficients of Variation for the Country-Specific Influenza-Associated Mortality Estimates According to World Health Organization Region and Age Group, Multiple Countries, 1980–2016a–c
| WHO Region | Aged <65 Years | Aged ≥65 Years | All Ages | ||||||
|---|---|---|---|---|---|---|---|---|---|
| CDCa | GLaMORb | IHMEc | CDCa | GLaMORb | IHMEc | CDCa | GLaMORb | IHMEc | |
| AFRO | 1.51 | 1.60 | 1.24 | 1.48 | 1.54 | 1.25 | 1.49 | 1.57 | 1.23 |
| AMRO | 1.76 | 1.60 | 2.27 | 2.18 | 2.37 | 2.25 | 2.00 | 2.12 | 2.20 |
| EMRO | 1.82 | 1.53 | 1.77 | 1.27 | 1.64 | 1.55 | 1.53 | 1.58 | 1.66 |
| EURO | 2.79 | 2.00 | 3.52 | 1.64 | 1.62 | 1.62 | 1.71 | 1.65 | 2.54 |
| SEARO | 1.90 | 2.16 | 2.28 | 2.42 | 2.05 | 2.57 | 2.11 | 2.10 | 2.41 |
| WPRO | 2.65 | 3.39 | 2.21 | 3.75 | 3.61 | 2.43 | 3.56 | 3.56 | 2.27 |
Abbreviations: AFRO, African Region; AMRO, Region of the Americas; CDC, US Centers for Disease Control and Prevention; EMRO, Eastern Mediterranean Region; EURO, European Region; GLaMOR, Global Pandemic Mortality Project; IHME, Institute for Health Metrics and Evaluation; SEARO, Southeast Asia Region; WHO, World Health Organization; WPRO, Western Pacific Region.
Iuliano et al. (6).
Paget et al. (7).
Global Burden of Disease 2016 LRI Collaborators (8).
Across age groups, higher correlations for WHO Regions were observed between CDC and IHME estimates, except in the African region, compared with correlations between GLaMOR and IHME estimates (Table 4).
Table 4.
Pairwise Spearman’s Ranked Correlations of Influenza-Associated Mortality Estimates According to World Health Organization Region and Age Group, Multiple Countries, 1980–2016a–c
| WHO Region | Aged <65 Years | Aged ≥65 Years | All Ages | ||||||
|---|---|---|---|---|---|---|---|---|---|
| CDC–IHME | CDC–GLaMOR | GLaMOR–IHME | CDC–IHME | CDC–GLaMOR | GLaMOR–IHME | CDC–IHME | CDC–GLaMOR | GLaMOR–IHME | |
| AFRO | −0.12 | 0.09 | 0.48 | 0.01 | −0.03 | 0.42 | 0.15 | 0.48 | 0.40 |
| AMRO | 0.62 | 0.56 | 0.57 | 0.64 | 0.66 | 0.60 | 0.65 | 0.62 | 0.52 |
| EMRO | 0.85 | 0.72 | 0.82 | 0.67 | 0.29 | 0.53 | 0.83 | 0.73 | 0.61 |
| EURO | 0.73 | 0.78 | 0.60 | 0.37 | 0.30 | −0.01 | 0.14 | 0.45 | −0.15 |
| SEARO | 0.32 | 0.74 | 0.71 | 0.80 | −0.47 | −0.35 | 0.72 | 0.13 | 0.52 |
| WPRO | 0.67 | 0.84 | 0.76 | 0.61 | 0.60 | 0.40 | 0.38 | 0.44 | 0.28 |
Abbreviations: AFRO, African Region; AMRO, Region of the Americas; CDC, US Centers for Disease Control and Prevention; EMRO, Eastern Mediterranean Region; EURO, European Region; GLaMOR, Global Pandemic Mortality Project; IHME, Institute for Health Metrics and Evaluation; SEARO, Southeast Asia Region; WHO, World Health Organization; WPRO, Western Pacific Region.
CDC, Iuliano et al. (6).
GLaMOR, Paget et al. (7).
Global Burden of Disease 2016 LRI Collaborators (8).
Potential reasons for differences between global influenza mortality estimates
Data sources and processing.
Different data sources and inclusion criteria were used by the groups. Thirty-three countries contributed EMR data to the CDC by providing either weekly or monthly vital records of mortality data for all respiratory causes of death (International Classification of Diseases, Ninth Revision: 460–519; International Classification of Diseases, Tenth Revision: J00–J99), according to 3 age groups, that were analyzed by CDC. Alternatively, countries provided age- and country-specific influenza-associated respiratory mortality rates analyzed by the collaborator. Viral surveillance data was used by 26 countries. Countries without sufficient viral surveillance data used a Serfling approach (13) and defined their influenza epidemic period using pneumonia and influenza mortality data or the limited viral surveillance data available. Rate estimates were calculated or provided for at least 4 years/seasons, excluding the 2009 pandemic period. Mortality rate ratios were calculated to adjust for differences in the risk of death between countries using the 2015 WHO GHE respiratory infection mortality rates, which included estimates for both lower-respiratory (International Classification of Diseases, Tenth Revision: J09–J22, P23, U04) and upper-respiratory (International Classification of Diseases, Tenth Revision: J00–J06) infection mortality. Population estimates were obtained from the United Nations World Population Prospects and from the US Census Bureau mid-year population estimates for 2015.
In stage 1 of the GLaMOR approach, weekly respiratory deaths and viral surveillance data were used to calculate annual EMRs. Data from 33 countries were included; EMRs from 30 of these were also used in the CDC model. Data were available for 2002–2011, excluding the 2009 pandemic year from the analysis. EMR estimates from India and Kenya were not included because death codes were based on verbal autopsy assessment of respiratory cause of death; however, these countries were included in the sensitivity analysis. Ten country indicators (Table 1) were used in stage-2 analysis to account for differences between countries.
In the IHME approach, a total of 801,600 LRI mortality data points from 161 countries were used in the modeling, including all available data from vital registration systems, verbal autopsy, and surveillance systems. Nonspecific causes of death, such as sepsis or heart failure, were reallocated to specific causes of death (8), including LRI, according to regression models designed to assign specific causes (14). The PAF was used to attribute LRI deaths to influenza and was calculated using the proportion of LRI cases positive for influenza, which was estimated by modeling data from a systematic literature review. There were 649 data points that informed this model, representing all age groups, 102 geographic locations, and years 1990–2016. Of these, 349 (53.8%) were from inpatient populations, and 392 (60.4%) used polymerase chain reaction diagnostic techniques (10). The value used to capture the odds of LRI given influenza detection came from one source (15). The PAF was further adjusted for the relative frequency of nonfatal-versus-fatal LRI episodes using a scalar from DisMod-MR 2.1 proportion models (16), and for the case-fatality rate of viral to bacterial pneumonia episodes by age. The data sources for the case-fatality adjustment factor were representative of Austria, Brazil, Mexico, and the United States (8).
Comparison of data sources and processing.
The first major difference between the groups is the underlying data sources used to estimate influenza mortality burden. While CDC and GLaMOR estimated EMRs using vital records mortality data for all respiratory deaths and extrapolating these data to countries, IHME used GBD LRI deaths and applied a proportion to obtain influenza deaths in a country. IHME calculated the maximum number of LRI deaths and attributed some to influenza. The IMHE model assumes that 1 death can only be counted once and categorically attributes deaths to a single underlying cause; thus, influenza-related LRI deaths in patients with other underlying causes were assigned to another category of death. With this approach, the sum of deaths does not exceed the overall global mortality, and this resulted in a significantly lower number of LRI deaths to attribute to influenza compared with the other groups.
Another difference between IHME and the other groups might be the use of the fraction of influenza-related respiratory deaths occurring outside the LRI envelope. In particular, deaths occurring after complications of severe influenza infection, such as secondary bacterial infection or the exacerbation of underlying medical conditions, including chronic obstructive pulmonary disease (17, 18), were considered in CDC and GLaMOR approaches. However, the IHME method would have assigned these deaths to a different cause. Considering that 3.2 million people die from chronic obstructive pulmonary disease every year (19) and that people ill with influenza and underlying chronic respiratory conditions have a higher risk of severe outcomes (20), if the IHME method were adjusted to use broader mortality categories or allow deaths to be assigned to more than 1 cause, the influenza-related estimates would likely increase.
Additionally, the quality of source data for extrapolation models might contribute to differences. Both CDC and GLaMOR required high-quality vital records and viral surveillance data to estimate influenza-associated excess mortality from a small subset of countries, which were used to extrapolate global deaths. Both groups lacked EMRs from the Eastern Mediterranean region and had few EMRs from the African and Southeast Asia regions. Moreover, GLaMOR excluded 2 countries with verbal autopsy data, limiting the use of data from these countries to the sensitivity analysis. In contrast, IHME was more flexible on data inclusion, allowing verbal autopsy, vital registration, and surveillance data from 161 countries for modeling LRI deaths. The different approaches highlight the importance of balancing quality and geographical distribution of data. For example, in influenza-associated mortality across sub-Saharan countries in the CDC and GLaMOR models, the exclusion of Kenya from GLaMOR analysis might have underestimated the burden in the region or perhaps overestimated burden by including Kenya in the CDC model. More flexible inclusion criteria might increase the number of contributing countries but might also increase risk of introducing biases in the models (21) due to incomplete data or biased representation of the cause-specific mortality from representative national samples compared with inclusive national data. Determining the appropriate balance between additional data points (particularly from low- and middle-income countries), the risk of bias the model, and the consideration of complete data versus imputed data requires further investigation.
In addition, IHME’s calculation of the PAF might benefit from more representative data according to age and geography. Specifically, the odds ratio used in the PAF calculation for all age groups was obtained from the Shi et al. (15) meta-analysis, which focused on children aged <5 years. Also, the case-fatality rate ratio for viral/bacterial pneumonia, used as an adjustment factor for the death PAF, was calculated from data for 4 countries, mainly from the Americas, and applied to all countries. Limitations in data in calculating the PAF could lead to an under- or overestimation of deaths in certain age groups and regions and might increase the uncertainty of the influenza mortality estimate.
Modeling assumptions, statistical methods, and addressing uncertainty.
Additional potential reasons for differences in the influenza mortality estimates between the groups might be the diverse modeling assumptions and methods to manage statistical challenges in extrapolation.
CDC categorized countries into ADs using the 2015 WHO GHE respiratory infection mortality rates, which were ranked from lowest to highest, and countries were distributed across 3 ADs, resulting in a total of 9 age-specific ADs. Countries with variation in respiratory infection mortality between age groups could be classified into different ADs for the age groups. The number of EMR-contributing countries within each AD ranged from 6 to 18 depending on categorization into the AD and available data. To validate the ADs, authors performed different sensitivity analyses. When removing ADs, the range in mortality estimates became larger, supporting their assumption that ADs generate more conservative estimates and that estimating influenza deaths between countries with similar LRI mortality was appropriate.
In the GLaMOR approach, a multiple imputation model was applied to each year in 2002–2011 (excluding 2009). Mortality estimates provided by countries were used in the imputation for each year. If a contributing country did not have complete data for 2002–2011, the missing years were not filled in to ensure a complete group of contributing countries for each year. The stage-2 analysis was performed for those years where data from 19 or more stage-1 countries were available. Results of the sensitivity analysis, which included stage-1 input for India and Kenya, showed an important variation of the estimates in the <65-years age group for those years where data from these 2 countries were included (7).
In the IHME approach, a smoothing approach was used to calculate the proportion of LRI episodes that tested positive for influenza by 5-year increments starting in 1990. This approach does not capture single-year heterogeneity in influenza circulation and might not be reliable in the event of large influenza outbreaks or implementation of preventative interventions. However, a significant epidemic can be included as an external factor after completing modeling, specifically for the 2009 influenza A(H1N1) pandemic (22).
Each group had a different approach regarding the use of covariates in their models (Table 1). CDC used GHE respiratory infection mortality estimates as a proxy to account for differences in mortality risk between countries and used these ratios to extrapolate influenza-associated excess mortality rates from countries with available estimates to those without. GLaMOR selected10country-specificindicatorsto generate influenza-associated respiratory mortality for their stage-2 analysis, including non–influenza-related indicators (e.g., WHO Region). IHME’s method used 13 covariates to model LRI mortality in the Cause of Death Ensemble model (11), which is based on an algorithm that selects the most plausible biological relationship with LRI mortality (10).
Each group described the uncertainty of their estimates using different approaches. To describe the uncertainty in the age-specific influenza-associated respiratory death estimates for each country, CDC calculated 95% credible intervals from the posterior distribution generated from 5,000 extrapolation iterations, using different values for unknown parameters obtained from a Bayesian framework. IHME presented 95% uncertainty intervals, calculated from 1,000 draws of the parameters in the PAF equation including LRI mortality, modeled proportions, odds ratios, and case-fatality rate scalars. GLaMOR presented the variability in mortality over time as a range (minimum and maximum) of the annual influenza-associated mortality estimates rather than a statistical interval estimate.
Comparison of modeling assumptions, statistical methods, and addressing uncertainty.
Differences in modeling strategy also contributed to the variation in the estimates between the groups. This is particularly notable between CDC and GLaMOR models, where despite the use of similar EMR data, the modeling approaches generated different results. Further, including data from India and Kenya in the GLaMOR model might have increased their estimates and potentially made their maximum values more similar to the credible intervals presented by CDC. In addition, imputing missing season/year rate estimates for contributing countries could have reduced the year-to-year fluctuation of EMRs observed in the sensitivity analyses in the GLaMOR project (7).
The use of smoothing processes in IHME models limits the ability to evaluate yearly variation of influenza burden. Similarly, the CDC estimates represent a mean mortality rate over the time period of 1999–2015.The GLaMOR approach, which imputed EMRs for each influenza season, allows for better evaluation of annual fluctuations in influenza burden because of changes in virus circulation.
The use of covariates in modeling is common to improve estimate precision; however, covariates might also introduce bias when imputing or extrapolating to generate estimates. Covariate selection is especially important in absence of high-quality input data (21) for models. While the groups considered several covariates, none of the covariates used were common among the models, potentially contributing further to the differences in the estimates.
DISCUSSION
Although time-series models are commonly used to estimate influenza-associated mortality, these estimates could lead to under- or overestimates of influenza mortality (21, 23). Further, it is not feasible for all countries to use these methods due to lack of reliable data. Methods to estimate influenza-associated mortality that can be used by more countries are not available (24), though needed. Efforts by CDC, GLaMOR, and IHME help to fill this gap in influenza burden knowledge by calculating country-specific, regional, and global influenza-associated mortality estimates. By contributing to this comparison, these groups assist in developing a better understanding of differences in estimates and reasons behind those differences.
As demonstrated by these groups, global influenza mortality burden can be conceptualized and presented using different methods. While CDC and GLaMOR mainly focused on defining the number of influenza respiratory deaths, IHME measured only influenza-attributable deaths within LRI mortality. By modeling respiratory excess mortality, CDC and GLaMOR estimated those deaths that might be associated with influenza, while IHME through a counterfactual approach estimated LRI deaths caused by influenza. As expected given the similarity of the approach and use of all respiratory deaths, the CDC and GLaMOR models generated similar overall estimates. Given that IHME started from a more restrictive mortality outcome (LRI deaths), their estimates were 4 to 5 times lower than the others. However, reassuringly, ranking the estimates across regions and age groups showed similar variability in some cases, suggesting true differences in disease burden rather than modeling artifacts.
Complete transparency of the modeling approach and openness to refine future estimates has been a fundamental component of this joint collaboration. The commitment to further improve modeling approaches by IHME were documented in the recent GBD 2017 publication (18). A revision of the ratio of mortality in viral compared with bacterial causes of LRI death resulted in a 2-fold increase of LRI deaths attributable to influenza (25).
Accurate quantification of influenza mortality is challenging because several factors can contribute to differences in modeled estimates, including source data quality, parameter and covariate choice, and modeling assumptions. The variability of several parameters across the groups made it difficult to identify which aspects of the models drove the observed differences. A standard approach to assess data quality and suitability for global modeling and the use of common covariates might reduce differences across the estimates, thus resulting in the remaining differences being related to the actual model selected. Continued collaboration between modeling groups and sharing of data could improve geographic representation in future global models and improve validation and enhancement of methods.
The review of these global influenza mortality efforts provided WHO with necessary information for a consensus on the newly adopted WHO influenza-associated respiratory mortality estimate (4). However, the true burden of influenza is expected to be higher when taking into consideration influenza-associated deaths that exacerbate non-respiratory conditions, such as cardiovascular diseases. To achieve greater understanding of the true burden of influenza mortality, a continued collaboration between groups could promote further improvements in methodology, greater appreciation for strengths and weaknesses of the different approaches, and possibly convergence over time in global mortality estimates.
Supplementary Material
ACKNOWLEDGMENTS
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the World Health Organization or the Centers for Disease Control and Prevention.
Abbreviations:
- AD
analytical division
- CDC
US Centers for Disease Control and Prevention
- EMR
excess mortality rate
- GBD
Global Burden of Disease Study
- GHE
Global Health Estimates
- GLaMOR
Global Pandemic Mortality Project II
- IHME
Institute for Health Metrics and Evaluation
- LRI
lower-respiratory infections
- PAF
population attributable fraction
- WHO
World Health Organization
Footnotes
Conflict of interest: none declared.
REFERENCES
- 1.Simonsen L. The global impact of influenza on morbidity and mortality. Vaccine. 1999;17(suppl 1):S3–S10. [DOI] [PubMed] [Google Scholar]
- 2.de Francisco Shapovalova N, Donadel M, Jit M, et al. A systematic review of the social and economic burden of influenza in low- and middle-income countries. Vaccine. 2015;33(48):6537–6544. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization. Manual for Estimating the Economic Burden of Seasonal Influenza. https://apps.who.int/iris/bitstream/handle/10665/250085/WHO-IVB-16.04-eng.pdf. Accessed November 26, 2019.
- 4.Duque J, McMorrow ML, Cohen AL. Influenza vaccines and influenza antiviral drugs in Africa: are they available and do guidelines for their use exist? BMC Public Health. 2014; 14:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization. News Release. “Up to 650 000 people die of respiratory diseases linked to seasonal flu each year.” https://www.who.int/en/news-room/detail/14-12-2017-up-to-650-000-people-die-of-respiratory-diseases-linked-to-seasonal-flu-each-year. Accessed November 26, 2019.
- 6.Iuliano AD, Roguski KM, Chang HH, et al. Estimates of global seasonal influenza-associated respiratory mortality: a modelling study. Lancet. 2018;391(10127):1285–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paget J, Spreeuwenberg P, Charu V, et al. Global mortality associated with seasonal influenza epidemics: new burden estimates and predictors from the GLaMOR project. J Glob Health. 2019;9(2):020421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.GBD 2016 Lower Respiratory Infections Collaborators. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory infections in 195 countries, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect Dis. 2018;18(11):1191–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simonsen L, Spreeuwenberg P, Lustig R, et al. Global mortality estimates for the 2009 influenza pandemic from the GLaMOR project: a modeling study. PLoS Med. 2013; 10(11):e1001558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.GBD 2016 Causes of Death Collaborators. Global, regional, and national age-sex specific mortality for 264 causes of death, 1980–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet. 2017;390(10100): 1151–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foreman KJ, Lozano R, Lopez AD, et al. Modeling causes of death: an integrated approach using CODEm. Popul Health Metr. 2012;10:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miettinen OS. Proportion of disease caused or prevented by a given exposure, trait or intervention. Am J Epidemiol. 1974; 99(5):325–332. [DOI] [PubMed] [Google Scholar]
- 13.Serfling RE. Methods for current statistical analysis of excess pneumonia-influenza deaths. Public Health Rep. 1963;78(6): 494–506. [PMC free article] [PubMed] [Google Scholar]
- 14.Naghavi M, Makela S, Foreman K, et al. Algorithms for enhancing public health utility of national causes-of-death data. Popul Health Metr. 2010;8:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi T, McLean K, Campbell H, et al. Aetiological role of common respiratory viruses in acute lower respiratory infections in children under five years: a systematic review and meta-analysis. J Glob Health. 2015;5(1): 010408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flaxman AD, Vos C, Murray CJL, eds. An Integrative Metaregression Framework for Descriptive Epidemiology. Seattle, WA. University of Washington Press. 2015. [Google Scholar]
- 17.Taubenberger JK, Morens DM. The pathology of influenza virus infections. Annu Rev Pathol. 2008;3:499–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Papi A, Bellettato CM, Braccioni F, et al. Infections and airway inflammation in chronic obstructive pulmonary disease severe exacerbations. Am J Respir Crit Care Med. 2006;173(10):1114–1121. [DOI] [PubMed] [Google Scholar]
- 19.GBD 2017 Causes of Death Collaborators. Global, regional, and national age-sex-specific mortality for 282 causes of death in 195 countries and territories, 1980–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2018;392(10159):1736–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coleman B, Fadel S, Fitzpatrick T, et al. Risk factors for-serious outcomes associated with influenza illness in high-versus low- and middle- income countries: systematic literature review and meta-analysis. Influenza Other Respi Viruses. 2018;12(1):22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kovacs SD, Mullholland K, Bosch J, et al. Deconstructing the differences: a comparison of GBD 2010 and CHERG’s approach to estimating the mortality burden of diarrhea, pneumonia, and their etiologies. BMC Infect Dis. 2015;15:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.GBD 2015 Mortality and Causes of Death Collaborators. Global, regional, and national life expectancy, all-cause and cause-specific mortality for 249 causes of death, 1980–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016;388(10053):1459–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li L, Wong JY, Wu P, et al. Heterogeneity in estimates of the impact of influenza on population mortality: a systematic review. Am J Epidemiol. 2018;187(2):378–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicoll A, Ciancio BC, Lopez Chavarrias V, et al. Influenza-related deaths—available methods for estimating numbers and detecting patterns for seasonal and pandemic influenza in Europe. Euro Surveill. 2012;17(18): pii20162. [DOI] [PubMed] [Google Scholar]
- 25.GBD 2017 Influenza Collaborators. Mortality, morbidity, and hospitalisations due to influenza lower respiratory tract infections, 2017: an analysis for the Global Burden of Disease Study 2017. Lancet Respir Med. 2019;7(1): 69–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.United Nations Department of Economic and Social Affairs. World Population Prospects; 2015. https://population.un.org/wpp/Download/Archive/Standard/. Accessed June 21, 2020. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.