A transcriptional module amplifies the gibberellin and brassinosteroid signal to accelerate endosperm rupture to promote seed germination in Arabidopsis.
Keywords: Arabidopsis thaliana, BEE2, bHLH, brassinosteroids (BRs), GASA6, gibberellins (GAs), HBI1, seed germination
Abstract
Seed germination is regulated by multiple phytohormones, including gibberellins (GAs) and brassinosteroids (BRs); however, the molecular mechanism underlying GA and BR co-induced seed germination is not well elucidated. We demonstrated that BRs induce seed germination through promoting testa and endosperm rupture in Arabidopsis. BRs promote cell elongation, rather than cell division, at the hypocotyl–radicle transition region of the embryonic axis during endosperm rupture. Two key basic helix–loop–helix transcription factors in the BR signaling pathway, HBI1 and BEE2, are involved in the regulation of endosperm rupture. Expression of HBI1 and BEE2 was induced in response to BR and GA treatment. In addition, HBI1- or BEE2-overexpressing Arabidopsis plants are less sensitive to the BR biosynthesis inhibitor, brassinazole, and the GA biosynthesis inhibitor, paclobutrazol. HBI1 and BEE2 promote endosperm rupture and seed germination by directly regulating the GA-Stimulated Arabidopsis 6 (GASA6) gene. Expression of GASA6 was altered in Arabidopsis overexpressing HBI1, BEE2, or SRDX-repressor forms of the two transcription factors. In addition, HBI1 interacts with BEE2 to synergistically activate GASA6 expression. Our findings define a new role for GASA6 in GA and BR signaling and reveal a regulatory module that controls GA and BR co-induced seed germination in Arabidopsis.
Introduction
In plants, the freshly formed seeds maintain dormancy until the proper time of germination. Seed germination is a critical process in the plant life cycle and relies on networks of interconnected signal transduction pathways that integrate multiple hormonal and environmental signals (Bewley, 1997; Koornneef et al., 2002; Bentsink and Koornneef, 2008; Seo et al., 2009; Shu et al., 2016; Topham et al., 2017). Gibberellins (GAs) and brassinosteroids (BRs) promote seed germination, while abscisic acid (ABA) represses it (Finch-Savage and Leubner-Metzger, 2006; Bentsink and Koornneef, 2008; Weitbrecht et al., 2011; Gallego-Bartolome et al., 2012; Tuan et al., 2018; Kim et al., 2019). A high GA/BR and low ABA level is a favorable condition for seed germination.
Multiple ABA-responsive transcription factors (TFs), including ABSCISIC ACID INSENSITIVE5 (ABI5), play key roles in inhibition of seed germination (Finkelstein and Lynch, 2000; Carles et al., 2002; Lopez-Molina et al., 2002; Skubacz et al., 2016). ABI5 binds to the ABA-responsive element (ABRE) in the promoters of the genes encoding late embryogenesis abundant (LEA) proteins, such as EARLY METHIONINE-LABELED 1 (EM1) and EM6, to repress their expression (Carles et al., 2002). ABI5 also plays roles in integrating external signals and the crosstalk between several growth hormones, including GA and BR (Skubacz et al., 2016).
GA promotes seed germination by directly inducing the expression of the genes involved in cell division and elongation or derepression of gene expression by degrading DELLA proteins, negative regulators of GA signaling. Degradation of DELLA proteins, particularly RGL2, also reduces ABA biosynthesis and promotes seed germination (Piskurewicz et al., 2008). The antagonistic interaction between GA and ABA in controlling seed germination has been extensively studied (Bewley, 1997; Olszewski et al., 2002; Finch-Savage and Leubner-Metzger, 2006; Piskurewicz et al., 2008; Weitbrecht et al., 2011; Liu et al., 2016; Liu and Hou, 2018).
BR promotes seed germination by controlling the inhibitory effect of ABA on seed germination (Hu and Yu, 2014; Zhao et al., 2019). Extensive physiological, biochemical, and genetic studies, mainly using Arabidopsis, have led to the identification and functional characterization of the components of BR signal transduction (Li et al., 2001, 2002; Nam and Li, 2002; Tang et al., 2011). BR signaling begins with the perception of the hormone ligand by the plasma membrane-associated receptor complex consisting of BRASSINOSTEROID-INSENSITIVE1 (BRI1) and BRI-ASSOCIATED KINASE1 (BAK1). The activated BRI1–BAK1 receptor complex phosphorylates BR-SIGNALING KINASE 1 (BSK1) and CONSTITUTIVE DIFFERENTIAL GROWTH 1 (CDG1), which further phosphorylates the PP1 type phosphatase BRI1 SUPPRESSOR 1 (BSU1). BSU1, along with PROTEIN PHOSPHATSE 2A (PP2A), dephosphorylates and inactivates the glycogen synthase kinase3-like kinase BRASSINOSTEROID INSENSITIVE2 (BIN2). Inactivation of BIN2 promotes the accumulation of positive regulators of BR signaling, BRASINAZONE-RESISTANT 1 (BZR1) and BRI1-EMS-SUPPRESSOR 1 (BES1), which directly control the transcription of BR-responsive genes to regulate plant developmental events (Li et al., 2001; Kim et al., 2011; Planas-Riverola et al., 2019). Overexpression of BZR1 diminishes the inhibitory effect of ABA in transgenic Arabidopsis plants (Tsugama et al., 2013). In the absence of BR, BIN2 phosphorylates BZR1 and BES1 to repress their DNA binding capacity (He et al., 2002; Wang et al., 2002; Yin et al., 2002; Ryu et al., 2007). BES1 physically interacts with ABI5 to hinder its DNA binding capacity, attenuating the ABA-mediated suppression of seed germination by lowering the expression of ABI5 targets (Zhao et al., 2019). BIN2 is a repressor of BR signal, but it promotes the ABA responses. During seed germination, BIN2 physically interacts with ABI5 to phosphorylate and stabilize AB15 in the presence of ABA. BIN2 and ABI5 mutually modulate the ABA-induced inhibition of seed germination. However, BRs antagonize the BIN2–ABI5 cascade and promote seed germination (Hu and Yu, 2014), indicating a complex hormonal crosstalk during seed germination.
Both BR and GA promote cell expansion and seed germination. The physical interaction of DELLA and BZR1 seems to be the molecular basis for the BR–GA crosstalk (Gallego-Bartolome et al., 2012; Ross and Quittenden, 2016; Ross et al., 2016). BR can rescue the germination phenotypes of the GA biosynthetic mutant, ga1-3, and the GA-insensitive mutant, sleepy1 (Steber and McCourt, 2001), suggesting that BR-induced seed germination does not totally depend on GA response, but rather the hormones working in parallel. Seeds of both the BR biosynthetic mutant det2-1 and the BR-insensitive mutant bril1-1 are able to germinate without BR and are hypersensitive to ABA. In rice, seed germination and seedling growth are significantly affected by the BR biosynthetic inhibitor brassinazole (BRZ) which is completely recovered by treatment with GA (Li et al., 2020). These observations indicate that BRs play an auxiliary role in the GA-promoted regulation of seed germination and can reverse the inhibitory effect of ABA (Steber and McCourt, 2001). A recent study using iTRAQ (isobaric tag for relative and absolute quantification) proteomic analysis has revealed that GAs and BRs coordinately regulate rice seed germination and embryo development by modulating the expression of several common targets (Li et al., 2018). However, the molecular mechanisms underlying GA- and BR-induced seed germination have not been thoroughly investigated.
The interacting transcriptional module, DELLA/BZR1/PHYTOCHROME INTERACTING FACTOR4 (PIF4), integrates GA, BR, and light signals to mediate cell elongation (Bai et al., 2012b; Li et al., 2012; Oh et al., 2012). Under low GA conditions, DELLA interacts with BZR1 and PIF4 to inhibit their DNA binding activity, thus inhibiting cell growth and elongation. The promotion of cell elongation by BZR1–PIF4 requires a tripartite helix–loop–helix/basic helix–loop–helix (HLH/bHLH) module that consists of PACLOBUTRAZOL-RESISTANT (PRE), ILI1 BINDING bHLH PROTEIN1 (IBH1), and HOMOLOG OF BEE2 INTERACTING WITH IBH1 (HBI1) (Bai et al., 2012a; Fan et al., 2014; Zheng et al., 2019). HBI1 is also a positive regulator of BR signaling and functionally redundant with another bHLH TF, BEE2 (BRASSINOSTEROID ENHANCED EXPRESSION2) (Malinovsky et al., 2014). Genetic evidence suggests that HBI1 plays a pivotal role in GA-induced cell elongation. A total of 177 direct targets of HBI1 have been identified by chromatin immunoprecipitation sequencing (ChIP-Seq) and RNA sequencing (RNA-Seq), several of which encode cell wall-related proteins, such as expansins (EXP2) and GA-stimulated Arabidopsis (GASA) family proteins (GASA4 and GASA6) (Rubinovich and Weiss, 2010; Bai et al., 2012a; Fan et al., 2014; Yan et al., 2014; Zhong et al., 2015). In Arabidopsis, the GASA family is represented by 14 members, of which GASA4 and GASA6 are positive regulators of GA response ( Zhang and Wang, 2008; Rubinovich and Weiss, 2010; Zhong et al., 2015). Our previous study suggests that GASA6 regulates seed germination by serving as an integrator for the GA, ABA, and glucose (Glc) signaling cascades (Zhong et al., 2015).
In this study, we demonstrate that, similarly to GA, BRs also promote seed germination by accelerating endosperm rupture through promoting cell elongation at the hypocotyl–radicle transition region. In addition, we provide genetic and molecular evidence that two GA- and BR-responsive bHLH TFs, HBI1 and BEE2, directly bind to E box elements in the GASA6 promoter to regulate its expression. We illustrate a mechanism in which a bHLH TF complex mediates GA/BR-induced seed germination through activation of GASA6.
Materials and methods
Plant material and growth conditions
All mutant and transgenic lines were in the Arabidopsis thaliana accession Col-0. Seeds were surface-sterilized and sown on plates with half-strength basal Murashige and Skoog (MS) medium (Sigma-Aldrich, USA) containing 0.8% (w/v) agar (MBCHEM, China). Plants were grown in a climate-controlled room (22 °C, photoperiod of 16 h light/8 h dark, light intensity of ~100 µmol m−2 s−1, and relative humidity of 70%). HBI1-OE (overexpressing) and BEE2-OE lines were kindly provided by Dr Cyril Zipfel (Sainsbury laboratory, Norwich, UK), and HBI1-SRDX and BEE2-SRDX lines were kindly provided by Dr Masaru Ohme-Takagi (Bioproduction research institute, Tsukuba, Japan). HBI1-OE/gasa6 and BEE2-OE/gasa6 were generated by a genetic cross between gasa6 (SALK_072904) and HBI1-OE or BEE2-OE, and homozygous lines were verified by PCR using the primers listed in Supplementary Table S1.
Germination assay and hypocotyl length assay
For each germination assay, three independently grown seed batches of the wild type (WT), HBI1-OE, or BEE2-OE were compared. To ensure synchronous germination, seeds were imbibed at 4 °C for 3 d, then moved to a growth chamber with a 16 h/8 h light/dark cycle at 22 °C. The experiments were performed on half-strength MS medium supplemented with 1 µM 2,4-epibrassinolide (BR), 1 µM BRZ (Sigma-Aldrich, USA), 100 µM gibberellin (GA3), 1 µM paclobutrazol (PAC) (Sigma-Aldrich, USA), or 100 µM ABA (Sigma-Aldrich, USA). At least 80 seeds were imbibed for each treatment and examined for testa and endosperm rupture under a SMZ1500 stereomicroscope (Nikon, Japan), and photographed with a high-resolution digital camera (COOLPIX4500, Nikon, Japan). Germination rate was determined by calculating the percentage of testa and endosperm rupture in the control and different treatments. In the hypocotyl length assay, seeds were incubated for 36 h to attain 100% germination because BR-treated seeds germinate faster than those of the WT. After germination, testas were stripped and 50–70 embryos were photographed with a BX51 camera (Olympus, Japan). Hypocotyl length was measured using the Image J software (https://imagej.nih.gov/ij/index.html). The SPSS software (http://www.spss.com/) was used for statistical analysis throughout this study.
Measurement of embryonic axis epidermal cells
To ensure the synchronous and full germination of both untreated and treated seeds, we incubated the seeds for 36 h at room temperature before taking the measurements. Seeds were collected at 36 h and fixed in 50% (v/v) methanol and 10% (v/v) acetic acid overnight at 4 °C. Embryos were dissected from testas and stained as described previously (Sliwinska et al., 2009), and subsequently photographed with an LSM510 Meta confocal laser-scanning microscope (Zeiss, Germany). Photographs were enlarged electronically for measurement of cell length and width with Image J software.
Gene expression analysis
Quantitative real-time PCR (qRT-PCR) was performed as previously described (Zhong et al. 2015). Briefly, total RNA was extracted from 2-week-old seedlings or seeds using the total RNA isolation Kit (Promega, USA) according to the manufacturer’s instruction. About 800 ng of total RNA for each sample was reverse transcribed using the PrimeScript RT Reagent Kit with gDNA Eraser (TAKARA, Japan). All PCRs were performed using SYBR Premix Ex Taq Mix (TAKARA, Japan) in triplicate and repeated at least three times. The transcript levels were measured by the comparative cycle threshold (Ct) method (bulletin no. 2; Applied Biosystems, http://www.appliedbiosystems.com). Ubiquitin1 (UBQ1) (Jiang et al., 2012) and Tubulin 3 (TUB3) (Patra et al., 2013) were used as internal controls. Primers used for qRT-PCR are listed in Supplementary Table S1. β-Glucuronidase (GUS) assay was performed as previously described (Zhong et al., 2015). Briefly, T3 transgenic lines carrying different truncated GASA6 promoters fused with the GUS reporter gene were analyzed for GUS histochemical staining. Samples were incubated in the GUS staining solution [1 mg ml–1 5-bromo-4-chloro-3-indolyl glucuronide (X-Gluc) dissolved in 50 mM Na-phosphate buffer] at 37 °C overnight, then bleached using 70% (v/v) ethanol. All the samples were photographed under an Olympus BX Microscope (Olympus, Japan).
Yeast two-hybrid assay
The cDNA encoding either the full length or fragments of the desired proteins were fused to pGADT7 [activation domain (AD)] or pGBKT7 [DNA-binding domain (BD)]. The AD and BD fusion plasmids were paired in different combinations and co-transformed into Saccharomyces cerevisiae strain AH109 (Clontech, USA). Transformed colonies were then selected on synthetic dropout (SD) medium lacking leucine and tryptophan (–Leu –Trp). Interactions were determined by growth of the colonies on SD medium lacking histidine, leucine, and tryptophan (–His –Leu –Trp), and containing 5 mM (for HBI1) 3-amino-1,2,4-triazole (3-AT). Primers used for plasmid construction for yeast two-hybrid assay are listed in Supplementary Table S1.
Bimolecular fluorescence complementation (BiFC) assay
Full-length HBI1, BEE2, or IBH1 cDNAs were cloned into the pSAT6-nYFPC1 or pSAT6-cYFPC1 vectors, which contained either the N- or the C-terminal half of yellow fluorescent protein (YFP). The resulting constructs were paired in different combinations and co-transformed into the A. thaliana mesophyll protoplasts as described previously (Yoo et al., 2007). The YFP signals were observed with an LSM510 Meta confocal laser-scanning microscope (Zeiss, Germany). Primers used for plasmid construction for the BiFC assay are listed in Supplementary Table S1.
Protoplast transient assay
Different fragments of the GASA6 (1.4, 1.2, 1.1, or 0.9 kb) promoter were each cloned into the pGreen II 0800-LUC vector (Hellens et al., 2005) to generate reporter constructs. Full-length HBI1, BEE2, or IBH1 cDNAs were cloned into the pBlueScript vector with the Cauliflower mosaic virus (CaMV) 35S promoter and rbcS terminator to generate effector constructs. Each reporter construct, together with either 35S::HBI1, 35S::BEE2, or 35S::IBH1, was co-transformed into the mesophyll protoplasts of A. thaliana for transcriptional activity assay.
Single or double mutants of the GASA6 (1.4 kb) promoter were generated with the MutanBEST Kit (TAKARA, Japan) and subsequently cloned into the pGreen II 0800- LUC vector. Firefly and Renilla luciferase activities were assayed with the microplate luminometer (Turner Biosystems, USA) and the Dual-Luciferase Reporter Assay reagents (Promega, USA). Primers used for plasmid construction for protoplast assay are listed in Supplementary Table S1.
ChIP-PCR assay
ChIP assays were performed as previously described (Hou et al., 2014). Briefly, 5-day-old seedlings of 35S::HBI1-YFP-HA or 35S::BEE2-YFP-HA were fixed on ice for 45 min in 1% formaldehyde under vacuum. Fixed tissues were homogenized, and chromatin was isolated and sonicated to generate DNA fragments with an average size of 500 bp. The solubilized chromatins were immunoprecipitated by Protein A+G magnetic beads (Magna, USA) with anti-HA (Sigma-Aldrich, USA), and the co-immunoprecipitated DNAs were subsequently recovered and analyzed by qPCR with the SYBR Premix Ex Taq Mix (TAKARA, Japan). The relative fold enrichment was calculated by normalizing the amount of target DNA fragments against the respective input DNA samples and then against the amount of PP2A genomic fragments. Primers used for ChIP-PCR are listed in Supplementary Table S1.
Recombinant protein production in bacteria, and EMSA
To produce recombinant HBI1 and BEE2 proteins in bacteria, the corresponding ORFs were cloned into the pGEX4T1 vector (GE Healthcare Biosciences, USA). The resulting plasmids were transformed into BL21 cells containing pRIL (Agilent, USA). Protein expression was induced by adding 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG) to the cell cultures at A600 ~1.0 and incubated for 3 h at 37 °C. The cells were harvested and lysed using CelLytic B (Sigma-Aldrich, USA). The glutathione S-transferase (GST) fusion proteins were bound to glutathione–Sepharose 4B columns (Amersham, USA) and then eluted by 10 mM reduced glutathione in 50 mM Tris–HCl buffer (pH 8.0) (Patra et al., 2018).
For EMSA, two (GASA1 37 bp and GASA2 30 bp) probes were synthesized, with or without 5′ biotin labeling (Integrated DNA Technology, USA). The probes were designed to include the potential binding sites, as indicated by the ChIP experiment and protoplast transient assay. Oligos were annealed to produce double-stranded probes for EMSA using nuclease-free duplex buffer (Integrated DNA Technology, USA). The DNA-binding reactions were carried out in 10 mM Tris, pH 7.5, 50 mM KCl, 5 mM MgCl2, 1 mM DTT, 2.5% glycerol, 0.5% NP-40, and 50 ng of poly(dI–dC) in a final volume of 20 µl. Purified proteins were incubated with 25 fmol DNA probe at room temperature for 45 min. For the competition experiment, cold probes were added in an excess molar ratio (1000 times). The DNA–protein complexes were resolved by electrophoresis on 6% non-denaturing polyacrylamide gels and then transferred to BiodyneB modified membrane (0.45 mm; Pierce, USA). The band shifts were detected by a chemiluminescent nucleic acid detection module (Pierce, USA) and exposed to X-ray films.
Results
BRs promote seed germination by accelerating cell expansion
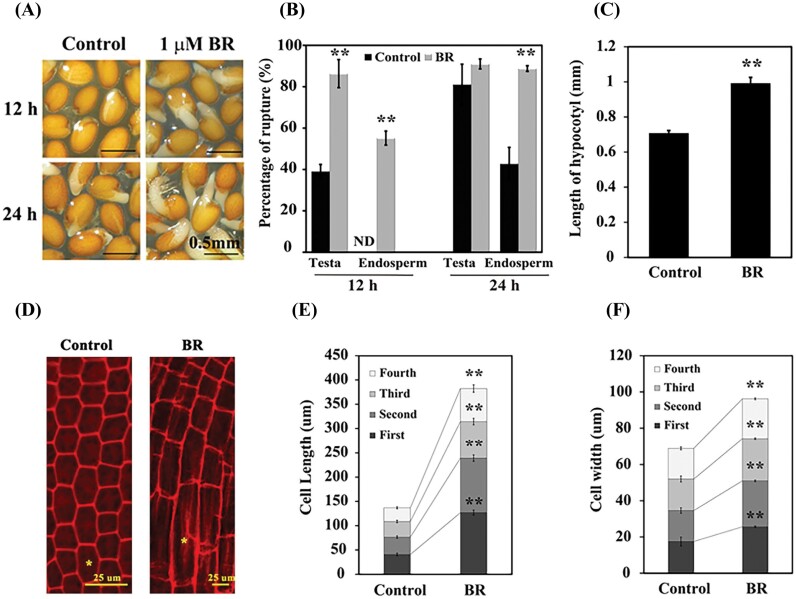
The completion of seed germination requires two sequential steps: shortly after imbibition, the testa and endosperm rupture consecutively, followed by radicle emergence (Bentsink and Koornneef, 2008; Weitbrecht et al., 2011). We analyzed the Arabidopsis seed germination in the presence of 2,4-epibrassinolide (one of the biologically active brassinosteroids, hereafter referred to as BR). Seeds were germinated on half-strength MS medium alone or supplemented with 1 µM BR. At 12 h, ~40% of testa rupture was observed in control seeds, compared with 86% in BR-treated seeds. Endosperm rupture was significantly higher (55%) in BR-treated seeds compared with the control (undetectable). Similar patterns of endosperm rupture were also observed at 24 h (Fig. 1A, B), suggesting that BR promotes seed germination by accelerating testa and endosperm rupture.
Fig. 1.
BR accelerates seed germination by promoting cell expansion. (A) Germination phenotypes of wild-type (WT) Arabidopsis (Col) seeds treated with 1 µM BR for 12 h and 24 h. (B) Percentages of testa and endosperm rupture in (A). (C) Hypocotyl length of WT seeds treated with 1 µM BR at 36 h. (D) Images of embryonic axis cells of WT seeds treated with 1 µM BR at 36 h; the yellow asterisk indicates the first cell. (E and F) Cell length and width, respectively, of the embryonic axis of WT seeds in (D). Seeds germinated on half-strength basal MS were used as control. ND, no detection. The black asterisks indicate significant differences compared with control (one-way ANOVA was used to analyze the significant differences). Three biological replicates were used for analysis. *P<0.05; **P<0.01.
Hypocotyl elongation assays were conducted under similar conditions to those of germination assays. BR significantly promoted the hypocotyl length by ~30% compared with the control (Fig. 1C). The sizes of the four cells in the hypocotyl–radicle transition region were measured to determine whether the effects of BR on embryo axis growth were caused by cell division or cell elongation. The cell lengths were greater in the presence of BR compared with the control (Fig. 1D, E). The effects of BR on the four cells in the transition region seem to be gradual, as in the presence of BR the lengths of the cell closest to the radicle (the first cells) increased 86 µm in contrast to the fourth cells which increased 40 µm (Fig. 1D, E). A similar effect of BR was observed on cell width (Fig. 1D, F). These observations imply that BR accelerates endosperm rupture by promoting cell elongation in the hypocotyl–radicle transition region of the embryo.
HBI1 and BEE2 mediate endosperm rupture mainly via enhancing BR and GA responses
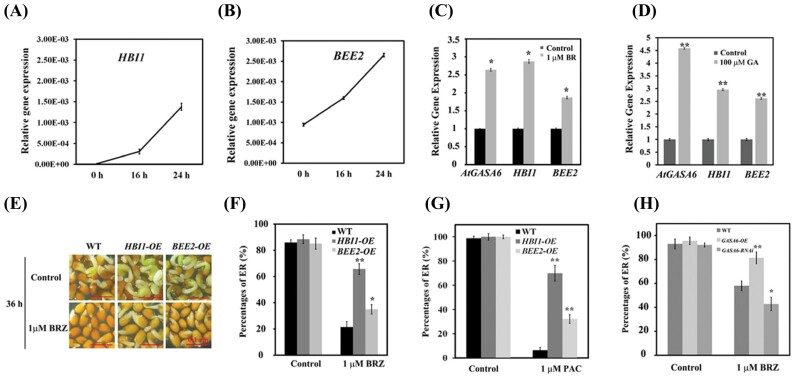
HBI1 and its closest homolog BEE2 are known to act downstream of BR and GA signaling pathways to promote cell elongation (Bai et al., 2012a). However, their roles in seed germination have not been investigated. We performed qRT-PCR to examine the expression of HBI1 and BEE2 during Arabidopsis seed germination as well as their response to BR and GA treatments. The seeds were stratified for 3 d, and expression of HBI1 and BEE2 was measured at 0, 16, and 24 h of light exposure. The expression of both HBI1 and BEE2 increased gradually during seed germination. Transcript abundance of BEE2 was higher than that of HBI1 in germinating seeds (Fig. 2A, B; Supplementary Fig. S1). Consistent with previous observations, the expression of both regulators was significantly induced following BR or GA treatment (Fig. 2C, D; Supplementary Fig. S2A, B). Next, we determined whether ABA regulates expression of HBI1 and BEE2 to influence endosperm rupture. We found that the expression of HBI1 and BEE2 was significantly increased in the abi5 mutant compared with that in the WT (Supplementary Fig. S3). Additionally, ABA repressed the expression of HBI1 in WT seeds while BEE2 expression did not change significantly (Supplementary Fig. S4). Next, we compared endosperm rupture in seeds of HBI1 and BEE2 overexpression lines (HBI1-OE and BEE2-OE) with that of the control (WT) in the presence of the BR biosynthesis inhibitor, BRZ. Compared with the WT, the HBI1-OE and BEE2-OE seeds were less sensitive to BRZ, as evidenced by significantly higher percentages of endosperm rupture in the BRZ-supplemented medium (Fig. 2E, F). In addition, we compared the percentage of edosperm rupture of HBI1-OE and BEE2-OE with that of the WT in the presence of the GA biosynthesis inhibitor, PAC. Under the control condition, HBI1-OE showed no difference in seed germination compared with the WT, but in the presence of PAC, HBI1-OE showed an ~65% increase of seed germination (Fig. 2G). Similarly, the percentage of endosperm rupture of BEE2-OE was significantly higher than that of the WT in the presence of PAC (Fig. 2G). In addition, overexpression of HBI1 (HBI1-OE) diminished the negative effect of ABA on seed germination, as evident by the higher percentage of endosperm rupture compared with that of WT and BEE2-OE seeds when treated with ABA (Supplementary Fig. S5A). These findings suggest that overexpression of HBI1 enhances the BR and GA responses in regulating endosperm rupture, and HBI1 is capable of counteracting the inhibitory effect of ABA during seed germination.
Fig. 2.
HBI1 and BEE2 are involved in BR-mediated endosperm rupture. Relative transcript levels of GASA6, HBI1, and BEE2 in response to BR and GA3 as measured using quantitative RT-PCR (qRT-PCR). The expression of HBI1 (A) and BEE2 (B) during the course of seed germination; 0 h was marked as the time point when seeds were exposed to light after 3 d of stratification. Two-week-old seedlings were treated with 1 µM BR (C) or 100 µM GA3 (D) for 2 h. The transcript levels were normalized to UBQ1. Data represent the mean ±SE (n=3). One-way ANOVA was used to analyze any significant difference. All experiments were repeated at least twice with similar results. (E) Images of germination phenotypes of WT (Col), HBI1-OE, or BEE2-OE seeds treated with 1 µM BRZ for 36 h; scale bar=0.1 cm. (F) Percentages of endosperm rupture (ER) in WT, HBI1-OE, or BEE2-OE seeds in (E). (G) Percentages of ER of WT, HBI1-OE, or BEE2-OE seeds treated with 1 µM PAC for 54 h. (H) Percentage of ER in WT, GASA6-OE and GASA6-RNAi seeds treated with 1 µM BRZ at 48 h. Seeds germinated on half-strength basal MS was used as the control. The asterisks indicate significant differences compared with control or the WT (one-way ANOVA was used to analyze significant differences). Three biological replicates were used for analysis. *P<0.05; **P<0.01.
HBI1 and BEE2 promote seed germination, probably via GASA6
HBI1 is a potential regulator of genes encoding many cell wall-related proteins, such as expansins and GASAs (Fan et al., 2014). A previous study has shown that GASA6 acts as a positive regulator in GA-, ABA-, and Glc-mediated seed germination (Zhong et al., 2015). However, the involvement of GASA6 in BR signaling is not well studied. We performed qRT-PCR to determine the effect of BR on GASA6 expression. Similar to HBI1 and BEE2, GASA6 expression was significantly activated by BR and GA (Fig. 2C, D; Supplementary Fig. S2A, B). Also similar to HBI1, expression of GASA6 was induced in abi5 seeds and reduced in WT seeds in the presence of ABA (Supplementary Figs S3A, B, S4A, B). In addition, the seed germination efficiencies of GASA6-overexpressing lines (GASA6-OE) and RNAi lines (GASA6-RNAi) were evaluated in the presence of BRZ. In the control conditions, no significant change in seed germination was observed for either GASA6-OE or RNAi lines (Fig. 2H). However, in the presence of 1 µM BRZ, seeds of GASA6-OE showed increased germination compared with the WT, whereas seeds of GASA6-RNAi displayed decreased germination (Fig. 2H). Similarly, in the presence of ABA, GASA6-OE seeds showed a significantly improved germination rate compared with the WT and GASA6-RNAi (Supplementary Fig. S5B). These results suggest that HBI1 and GASA6 act coordinately to promote BR- and GA-mediated seed germination and attenuate the negative effect of ABA during seed germination.
HBI1 and BEE2 directly regulate GASA6 expression in Arabidopsis
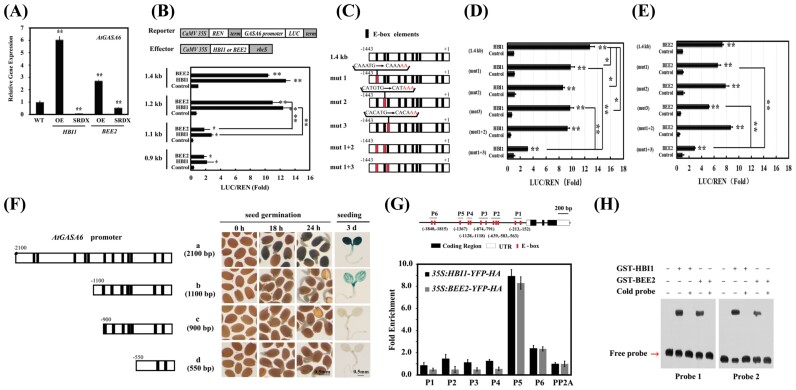
In silico analysis identified a total of 12 bHLH TF-binding motifs (E-box elements) in the GASA6 promoter (Supplementary Fig. S6). In addition, the co-expression analysis using the ATTED-II network drawer revealed that both HBI1 and BEE2 are co-expressed with GASA6 (Supplementary Figs S7–S9). To test whether HBI1 and BEE2 regulate GASA6 expression through binding to the E-box elements, we first determined GASA6 expression in the HBI1-OE and BEE2-OE lines, as well as in the HBI1-SRDX and BEE2-SRDX lines. As HBI1 and BEE2 are functionally redundant, it is assumed that the single knockout mutants will not show an obvious phenotype (Malinovsky et al., 2014). Therefore, we used the HBI1-SRDX and BEE2-SRDX lines, in which the dominant repressor form of HBI1 or BEE2 specifically suppresses the target genes, thus preventing the possible interference of functional redundancy (Hiratsu et al., 2003; Ikeda et al., 2012). The results showed that GASA6 expression was significantly higher in HBI1-OE or BEE2-OE lines than that in the WT but was repressed markedly in the HBI1-SRDX or BEE2-SRDX lines (Fig. 3A; Supplementary Fig. S10), indicating that HBI1 and BEE2 regulate the expression of GASA6 in Arabidopsis.
To identify the binding sites of HBI1 and BEE2 in the GASA6 promoter, we performed transient expression assays using Arabidopsis protoplasts. Truncated fragments of the GASA6 promoter, cloned into the pGreenII 0800-LUC vector (Hellens et al., 2005), served as reporters. HBI1 or BEE2, expressed under the control of the CaMV 35S promoter, were used as effectors. The reporters and effectors were co-transformed into Arabidopsis mesophyll protoplasts in different combinations. HBI1 or BEE2 significantly induced the activities of the 1.4 kb and 1.2 kb GASA6 promoters (upstream of the ATG start codon). However, their effects were dramatically reduced on the 1.1 kb and 0.9 kb promoter (Fig. 3B). These observations indicate that the 100 bp region between 1.2 kb and 1.1 kb of the GASA6 promoter is likely to be critical for the HBI1- and BEE2-induced expression. We identified three E-box elements (CAAATG, CATGTG, and CACATG) between 1.4 kb and 1.1 kb. To clarify the importance of these three E-box elements in HBI1/BEE2-controlled expression, point mutants were generated individually in the three E-boxes and used in the Arabidopsis transient expression assays. As shown in Fig. 3C, the last two nucleotides (TG) in all three E-box elements were replaced with AA to generate mutant promoters, mut 1, 2, 3, and their combination, mut 1 + 2 and mut 1 + 3 (Fig. 3C). The transient expression assays showed that, compared with the activation of the WT promoter, the activation of the three single mutant promoters by HBI1 was significantly reduced, whereas only mut 3 affected the activation by BEE2. However, the double mutation, mut1 + 3, significantly reduced the activation by either HBI1 or BEE2 (P<0.001) (Fig. 3D, E), indicating that two or more distantly located E-boxes are required for the regulation of GASA6 by the two factors.
Fig. 3.
HBI1 and BEE2 regulate GASA6 expression by binding to the E-box-like elements in vivo and in vitro. (A) Transcript levels of GASA6 in HBI1- or BEE2-OE and SRDX lines measured using quantitative RT-PCR (qRT-PCR). Data were normalized to UBQ1. (B) Transactivation of the full-length and truncated GASA6 promoter–reporters by HBI1 or BEE2 in Arabidopsis protoplasts. Various constructs used in transient expression assays are shown in the upper panel. (C) Schematic diagram of the 1.4 kb GASA6 promoter with all E-boxes (black) and mutated E-boxes (red) used in (D) and (E). Transactivation of the GASA6 promoter and its mutants by HBI1 (D) or BEE2 (E) in Arabidopsis protoplasts. (F) Schematic diagram of different fragments of the GASA6 promoter (left); the numbers indicate the promoter length. Analysis of GUS activities in different pGASA6::GUS lines (right). Scale bar=0.5 mm. (G) Schematic diagram of the GASA6 promoter. P1 to P6 indicate fragments used for chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR) amplification. ChIP-qPCR analysis of HBI1-HA or BEE2-HA binding to the GASA6 promoter upon precipitation with anti-HA antibody. Five-day-old 35S::HBI1-YFP-HA, 35S::BEE2-YFP-HA, or WT seedlings were used in ChIP-qPCR. Fold enrichments indicate the enrichment of HBI1-HA or BEE2-HA binding to the GASA6 promoter compared with that of the WT. Data represent the mean ±SE of three replicates. (H) EMSA of HBI1 or BEE2 after incubation with biotin-labeled DNA probes containing the E-box sequences of the GASA6 promoter (Probe1 and Probe2). In competition experiments to demonstrate the specific binding of proteins to the probes, non-labeled probes (cold probes) were added in 1000-fold excess of the labeled probes. Values represent the mean ±SE of at least four biological replicates. Except when specifically indicated, asterisks indicate significant differences compared with control (one-way ANOVA was used to analyze the significant differences). *P<0.05; **P<0.01.
To further validate this, we performed 5′ end deletion analysis of the GASA6 promoter using transgenic plants expressing various truncated promoter fragments fused to the GUS reporter gene. Analysis of the transgenic plants revealed that the region between –2100 bp and –1100 bp, where multiple E-boxes reside, is potentially important for GASA6 expression during seed germination and seedling growth (Fig. 3F).
Next, we performed ChIP-PCR assay using transgenic plants expressing HBI1-YFP-HA or BEE2-YFP-HA to measure enrichment of the E-boxes in the GASA6 promoter. The WT plants served as control. HBI1 strongly bound to the E-box-containing regions of P5 (–1367 bp upstream of ATG) in the GASA6 promoter (Fig. 3G). A similar DNA enrichment pattern by the BEE2-YFP-HA protein was also detected in the GASA6 promoter (Fig. 3G), suggesting that HBI1 and BEE2 co-target the GASA6 promoter, probably via direct binding to the E-box elements. However, the E-box elements (CACATG and CATGTG) in the 100 bp region are different from the one (CAAATG) in the P5 fragment, suggesting that the mechanisms of HBI1 or BEE2 regulating GASA6 expression are somewhat intricate. Direct binding of HBI1 or BEE2 to the E-box elements in the GASA6 promoter was further verified by EMSA, where two DNA probes, with or without 5′ biotin labeling, were synthesized based on the E-box elements of the GASA6 promoter (Probe 1, caatttttaatcacaaatgctattttattggacgacc; Probe 2, GTCTCCCATGTGAGTGCACATGGAGTTATG). The results showed that both HBI1 and BEE2 individually bind to the E-box elements, resulting in a mobility shift. The specificity of the DNA–protein interaction was confirmed by the competition experiment, in which the addition of excess (1000×) non-labeled probe eliminated the interaction between labeled probe and HBI1 or BEE2 (Fig. 3H).
BEE2 and HBI1 form a heterodimer and synergistically regulate expression of GASA6
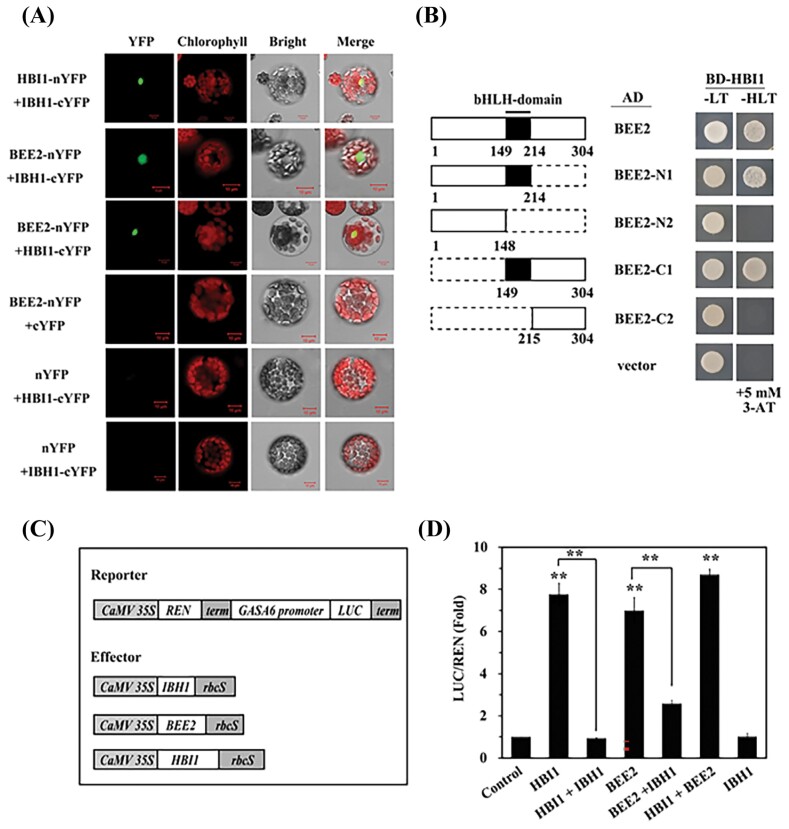
It has been well demonstrated that bHLH factors homo- or heterodimerize through the bHLH domain (Heim et al., 2003; Toledo-Ortiz et al., 2003). To determine whether HBI1 and BEE2 form a homo- or heterodimer, we first performed BiFC in Arabidopsis mesophyll protoplasts. Reciprocal fusions of BEE2, IBH1, and HBI1 with the N- or C-terminal half of YFP (nYFP and cYFP, respectively) were generated and co-transformed into the protoplasts in combinations. IBH1, a known interactor of BEE2 and HBI1, served as a positive control. Strong YFP fluorescence signals were observed in the nucleus when HBI1–nYFP or BEE2–nYFP was co-transformed with IBH1–cYFP and BEE2–nYFP was co-transformed with HBI1–cYFP, indicating specific interactions of HBI1–IBH1, BEE2–IBH1, and BEE2–HBI1 (Fig. 4A). Next, we performed yeast two-hybrid assays to confirm that the HBI1–BEE2 interaction is mediated by the bHLH domains. BEE2 was truncated into four fragments, with or without the bHLH domain, fused with the GAL4 AD, and co-expressed with HBI1 fused with the GAL4 BD (Fig. 4B. Only the bHLH domain-containing BEE2 fragments interacted with HBI1, confirming the requirement of the bHLH domain in dimerization.
Fig. 4.
HBI1 and BEE2 modulate the expression of GASA6 by forming a homodimer or heterodimer in vivo. (A) Bimolecular fluorescent complementation (BiFC) assay shows that HBI1 and BEE2 interact with each other and IBH1 to form heterodimers in Arabidopsis mesophyll protoplasts. YFP, signal of yellow fluorescence protein; Chlorophyll, autofluorescence of chloroplasts; Bright, protoplasts in light view; Merge, merge of YFP, chlorophyll, and light view. (B) Diagram of the BEE2 domain structures and various deletions (left); yeast two-hybrid assays show the heterodimer of HBI1, and the interaction domain. Protein–protein interactions were detected by yeast growth on triple (–His–Leu–Trp) dropout selection medium, with 5 mM 3-amino-1,2,4-triazole (3-AT) (right). (C) Schematic diagram of various constructs used in transient expression assays (D). (D) Transactivation of the GASA6 promoter (1.4 kb) by HBI1, BEE2, and IBH1 in Arabidopsis mesophyll protoplasts. The GASA6 promoter fused to the LUC reporter was co-transformed with effectors or empty vector (control) into mesophyll protoplasts. Fold of LUC/REN indicates the expression level of GASA6 activation by various effectors. Values represent the mean ±SE of four biological replicates. Except when specifically indicated, asterisks indicate significant differences compared with control (one-way ANOVA was used to analyze the significant differences). **P<0.01.
To determine the biological significance of the dimerization among HBI1, BEE2, and IBH1, we performed transient expression assays in the protoplasts with different combinations of these effector proteins (Fig. 4C) on the GASA6 promoter (pGASA6-luc). As shown in Fig. 4D, activation of the GASA6 promoter was significantly higher when HBI1 and BEE2 were co-expressed (HBI1+BEE2) compared with HBI1 or BEE2 expressed alone (Fig. 4D). On the other hand, the addition of IBH1 significantly attenuated the activity of HBI1 or BEE2 (Fig. 4D), possibly by forming a non-DNA-binding complex with HBI1 or BEE2 as described previously (Ikeda et al., 2012; Zhiponova et al., 2014)
GASA6 acts downstream of HBI1 and BEE2 to promote cell elongation
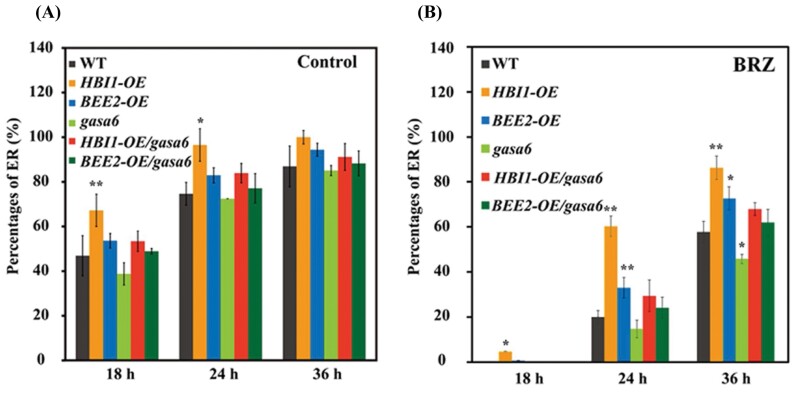
To investigate the relationship between HBI1 or BEE2 and GASA6 at the genetic level, HBI1-OE/gasa6 and BEE2/gasa6 plants were generated by making a genetic cross between HBI1-OE or BEE2-OE transgenic plants and the homozygous GASA6 T-DNA insertion mutant (gasa6). HBI1-OE or BEE2-OE lines exhibited a BRZ-resistant phenotype. Although no significant difference was observed between the WT and gasa6 under the control condition (Fig. 5A), the endosperm rupture percentage of gasa6 was markedly reduced compared with the WT under 1 µM BRZ treatment (Fig. 5B). Furthermore, HBI1-OE or BEE2-OE in the gasa6 background showed increased sensitivity to BRZ (~60%; Fig. 5B), compared with that without BRZ treatment (~90%; Fig. 5A), suggesting that, in the BR signaling cascade, GASA6 acts downstream of HBI1 and BEE2 to promote cell elongation.
Fig. 5.
GASA6 acts downstream of HBI1 and BEE2 to promote seed germination. Endosperm rupture (ER) percentage of WT, HBI1-OE, BEE2-OE, gasa6, HBI1-OE/gasa6 and BEE2-OE/gasa6 seeds after 18, 24, and 36 h of germination (A) control (half-strength basal MS medium) and (B) half-strength basal MS medium with 1 mM BRZ. The black asterisks indicate significant differences compared with the WT (one-way ANOVA was used to analyze the significant differences). *P<0.05; **P<0.01.
Discussion
BR and GA are principal plant growth regulators that function redundantly to control many important physiological functions, including seed germinations and cell elongation (Steber and McCourt, 2001; Hu and Yu, 2014; Li et al., 2018; Zhao et al., 2019). Physical interactions between BZR1/BES1 and DELLAs mediate the crosstalk between BRs and GAs during cell elongation in Arabidopsis (Bai et al., 2012b; Gallego-Bartolome et al., 2012; Li et al., 2012); however, the molecular mechanism underlying GA–BR crosstalk during seed germination is not well studied. In this study, we demonstrated that BR accelerates endosperm rupture by enhancing the growth of the hypocotyl–radicle transition region of the embryo (Fig. 1), similar to what has been observed previously in GASA6 overexpression (Zhong et al., 2015). The elongation of the embryonic axis in a completely germinated Arabidopsis seed is a result of cell elongation rather than cell division (Sliwinska et al., 2009). As expected, the length and width of the cells in the hypocotyl–radicle transition region are significantly increased in the presence of BR (Fig. 1D–F), suggesting that BR affects cell elongation and width during embryonic axis elongation before endosperm rupture in Arabidopsis. Although the underlying mechanism requires further investigation, our findings provide a new insight into the coherent events at BR-promoted cell elongation during seed germination.
BZR1 and BES1 contribute to regulation of seed germination (Ryu et al., 2014; Zhao et al., 2019). The gain-of-function mutant bes1-D exhibits reduced sensitivity to ABA during seed germination, a phenotype not observed in the bzr1-D mutant, suggesting that BES1, but not BZR1, is the major contributor to BR-mediated suppression of ABA signaling during seed germination (Ryu et al., 2014). In addition, BES1 physically interacts with ABI5 to attenuate the ABA-mediated suppression of seed germination by lowering the expression of ABI5 targets (Zhao et al., 2019). Similar to that of GASA6 (Zhong et al., 2015), we found that HBI1 and BEE2 expression increased gradually during seed germination (Fig. 2A, B; Supplementary Fig. S1) and increased significantly in abi5 mutant seeds (Supplementary Fig. S3). ABA repressed the expression of HBI1 in WT seeds, while BEE2 expression remained unchanged (Supplementary Fig. S4). We thus hypothesized that HBI1 and BEE2 are involved in regulation of seed germination. Supporting this notion is that the endosperm rupture of the HBI1-OE or BEE2-OE lines showed decreased sensitivity to the BR biosynthesis inhibitor BRZ (Fig. 2 E, 2F). Seeds of HBI1-OE did not show the negative effect of ABA on germination (Supplementary Fig. S5), suggesting that, similar to BZR1 and BES1, HBI1, and possibly BEE2, breaks ABA-induced dormancy and promotes GA–BR-indued seed germination. Our results indicate that HBI1 and BEE2 are involved in BR-mediated seed germination by promoting endosperm rupture through controlling cell elongation.
It has been demonstrated that the tripartite HLH/bHLH module, PRE–IBH1–HBI1, regulates cell elongation in response to GA and BRs (Bai et al., 2012a; Fan et al., 2014). Consistent with the role of HBI1 in promoting cell elongation (Bai et al., 2012a), BEE2, the closest homolog of HBI1, plays redundant roles in cell elongation (Carretero-Paulet et al., 2010). The germination phenotypes of the BEE2-OE and HBI1-OE lines (Fig. 2F, G) support the individual role of HBI1 and BEE2. To identify downstream targets of HBI1 and BEE2, we performed co-expression network analysis and found that GASA6, known to integrate GA, ABA, and Glc signaling to regulate seed germination (Zhong et al., 2015), is co-expressed with HBI1 (Supplementary Fig. S8) and BEE2 (Supplementary Fig. S9). In addition, similar to HBI1 and BEE2, GASA6 expression is also activated by BR and GA (Fig. 2C, D; Supplementary Fig. S2). Both HBI1 and BEE2 function in the GA- and BR-mediated seed germination processes (Fig. 2E–G). Our findings suggest that the inhibitory effect on seed germination by BRZ is overcome by GASA6 overexpression but enhanced by GASA6-RNAi (Fig. 2H). We showed that HBI1 and BEE2 promote seed germination by directly regulating the expression of GASA6. The transcript levels of GASA6 were significantly altered in the overexpression and repression lines of HBI1 and BEE2 (Fig. 3A; Supplementary Fig. S10). In addition, the region between 2.1 kb and 0.9 kb of the GASA6 promoter is important for GASA6 expression during seed germination and seedling growth (Fig. 3F). ChIP-qPCR, EMSA, and protoplast transactivation assays suggested that HBI1 and BEE2 activate the GASA6 promoter mainly through binding the region between 1.4 kb and 1.1 kb (Fig. 3). Furthermore, mutations in the potential bHLH-binding motifs in the GASA6 promoter significantly affected the promoter activity in Arabidopsis protoplasts (Fig. 3D, E), indicating that the two E-box motifs (CACATG and CATGTG) in the GASA6 promoter are crucial for the activation by HBI1 and BEE2. HBI1 and BEE2 overexpression in the gasa6 mutant increased the sensitivity to BRZ (Fig. 5). Collectively, these results indicated that GASA6 is one of the downstream targets of HBI1 and BEE2 in the regulation of BR–GA-regulated seed germination.
Combinatorial transcriptional regulation is a hallmark of eukaryotic gene expression. Tight regulatory control is achieved by the highly dynamic nature of transcriptional activators and repressors. Heterodimeric TFs increase options of gene expression control. bHLH TFs are known to form homo- and heterodimers to regulate the expression of target genes (Toledo-Ortiz et al., 2003). Here, we demonstrated the interaction between HBI1, BEE2, and IBH1 in both yeast cells and Arabidopsis mesophyll protoplasts (Fig. 4A, B). In addition, BEE2 is found to interact with HBI1 to synergistically activate GASA6 (Fig. 4D). On the other hand, IBH1 antagonizes the function of HBI1 and BEE2 in activating GASA6 expression, possibly by forming non-DNA-binding complexes, HBI1–IBH1 or BEE2–IBH1 (Fig. 4D). Accumulating evidence suggests that interactions between activators and repressors fine-tune plant growth, development, and metabolic outcomes. In barley, the GA pathway is controlled by the interaction of two transcriptional activators and two repressors (Zou et al., 2008). In Catharanthus roseus, GBF1 and GBF2 interact with and antagonize transcriptional activities of MYC2 on the pathway gene promoters (Sui et al., 2018). Similarly, the subgroup IIId bHLH TF RMT1 competes with MYC2 and antagonizes its activity (Patra et al., 2018). In Arabidopsis, TCP4 interacts with AP2/ERF WRINKLED1 to attenuate its transcriptional activity to fine-tune seed oil accumulation (Kong et al., 2020). We showed that the HBI1–BEE2–IBH1 module is critical in regulation of BR–GA-induced seed germination.
BR and GA pathways are well characterized for triggering expression of downstream genes, such as GASA6. Less known is how the combined effects of BRs and GA regulate the gene expression. Prior to this study, the transcriptional hub that amplifies the BR–GA signal to GASA6 was elusive. Our findings reveal a new role for GASA6 in BR signaling and uncover an additional molecular mechanism of GA–BR-induced seed germination in Arabidopsis. HBI1 and BEE2 promote endosperm rupture and seed germination by directly activating the expression of GASA6. Moreover, further dissections of the protein–protein and protein–DNA interactions associated with the regulatory network advance our understanding of GA–BR-induced cell elongation during endosperm rupture.
Supplementary data
The following supplementary data are available at JXB online.
Fig. S1. Relative gene expression of HBI1 and BEE2 during seed germination.
Fig. S2. Relative gene expression of HBI1, BEE2, and GASA6 in response to BR and GA.
Fig. S3. Relative expression of GASA6, HBI1, and BEE2 in abi5 mutant seeds.
Fig. S4. Relative gene expression of HBI1, BEE2, and GASA6 in response to ABA.
Fig. S5. Effect of ABA on endosperm rupture of WT, HBI1-OE, or BEE2-OE seeds.
Fig. S6. Cis-motif analysis of the AtGASA6 promoter.
Fig. S7. The co-expression network of GASA6 in Arabidopsis as analyzed by the ATTED-II network drawer.
Fig. S8. The co-expression network of HBI1 in Arabidopsis as analyzed by the ATTED-II network drawer.
Fig. S9. The co-expression network of BEE2 in Arabidopsis as analyzed by the ATTED-II network drawer.
Fig. S10. Relative gene expression of GASA6 in HBI1- or BEE2-OE and SRDX lines.
Table S1. List of gene-specific primer sequences.
Acknowledgements
Our sincere thanks go to Dr Sitakanta Pattanaik (University of Kentucky) for his thoughtful suggestions and critical review of this manuscript. We thank Dr Cyril Zipfel (Sainsbury Laboratory, UK) for kindly providing the overexpression lines of HBI1 and BEE2, and Dr Masaru Ohme-Takagi (Institute for Environmental Science and Technology of Saitama University, Japan) for kindly providing the SRDX lines of HBI1 and BEE2. This work was funded by the National Natural Science Foundation of China (grant no. 31700282 to CZ, and 90917011 and 31372099 to XW). This work is also supported partially by the Harold R. Burton Endowed Professorship to LY and by the National Science Foundation under Cooperative Agreement no. 1355438 to LY.
Author contributions
CZ, BP, LY, and XWL: study design; CZ, BP, YT, and XL: performing the experiments; CZ, B.P, LY, and XW: data analysis; CZ, BP, XW, and LY: writing.
Conflict of interest
The authors declare no conflicts of interest.
Data availability
Sequence data from this study can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: HBI1 (At2g18300), BEE2 (At4g36540), IBH1 (At2g43060), GASA6 (At1g74670), UBQ1 (At3g52590), PP2A (At1g69960), and TUB3 (At5g 62700). All data supporting the findings of this study are available within the paper and within its supplementary data published online.
References
- Bai MY, Fan M, Oh E, Wang ZY. 2012a. A triple helix–loop–helix/basic helix–loop–helix cascade controls cell elongation downstream of multiple hormonal and environmental signaling pathways in Arabidopsis. The Plant Cell 24, 4917–4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai MY, Shang JX, Oh E, Fan M, Bai Y, Zentella R, Sun TP, Wang ZY. 2012b. Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nature Cell Biology 14, 810–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink L, Koornneef M. 2008. Seed dormancy and germination. The Arabidopsis Book 6, e0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley JD. 1997. Seed germination and dormancy. The Plant Cell 9, 1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carles C, Bies-Etheve N, Aspart L, Léon-Kloosterziel KM, Koornneef M, Echeverria M, Delseny M. 2002. Regulation of Arabidopsis thaliana Em genes: role of ABI5. The Plant Journal 30, 373–383. [DOI] [PubMed] [Google Scholar]
- Carretero-Paulet L, Galstyan A, Roig-Villanova I, Martínez-García JF, Bilbao-Castro JR, Robertson DL. 2010. Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiology 153, 1398–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan M, Bai MY, Kim JG, et al. . 2014. The bHLH transcription factor HBI1 mediates the trade-off between growth and pathogen-associated molecular pattern-triggered immunity in Arabidopsis. The Plant Cell 26, 828–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch-Savage WE, Leubner-Metzger G. 2006. Seed dormancy and the control of germination. New Phytologist 171, 501–523. [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ. 2000. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. The Plant Cell 12, 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Bartolomé J, Minguet EG, Grau-Enguix F, Abbas M, Locascio A, Thomas SG, Alabadí D, Blázquez MA. 2012. Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proceedings of the National Academy of Sciences, USA 109, 13446–13451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JX, Gendron JM, Yang Y, Li J, Wang ZY. 2002. The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proceedings of the National Academy of Sciences, USA 99, 10185–10190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC. 2003. The basic helix–loop–helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Molecular Biology and Evolution 20, 735–747. [DOI] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA. 2005. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M. 2003. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. The Plant Journal 34, 733–739. [DOI] [PubMed] [Google Scholar]
- Hou X, Zhou J, Liu C, Liu L, Shen L, Yu H. 2014. Nuclear factor Y-mediated H3K27me3 demethylation of the SOC1 locus orchestrates flowering responses of Arabidopsis. Nature Communications 5, 4601. [DOI] [PubMed] [Google Scholar]
- Hu Y, Yu D. 2014. BRASSINOSTEROID INSENSITIVE2 interacts with ABSCISIC ACID INSENSITIVE5 to mediate the antagonism of brassinosteroids to abscisic acid during seed germination in Arabidopsis. The Plant Cell 26, 4394–4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Fujiwara S, Mitsuda N, Ohme-Takagi M. 2012. A triantagonistic basic helix–loop–helix system regulates cell elongation in Arabidopsis. The Plant Cell 24, 4483–4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Li H, Wang T, Peng C, Wang H, Wu H, Wang X. 2012. Gibberellin indirectly promotes chloroplast biogenesis as a means to maintain the chloroplast population of expanded cells. The Plant Journal 72, 768–780. [DOI] [PubMed] [Google Scholar]
- Kim TW, Guan S, Burlingame AL, Wang ZY. 2011. The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Molecular Cell 43, 561–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Warpeha KM, Huber SC. 2019. The brassinosteroid receptor kinase, BRI1, plays a role in seed germination and the release of dormancy by cold stratification. Journal of Plant Physiology 241, 153031. [DOI] [PubMed] [Google Scholar]
- Kong Q, Singh SK, Mantyla JJ, Pattanaik S, Guo L, Yuan L, Benning C, Ma W. 2020. TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL FACTOR4 interacts with WRINKLED1 to mediate seed oil biosynthesis. Plant Physiology 184, 658–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Bentsink L, Hilhorst H. 2002. Seed dormancy and germination. Current Opinion in Plant Biology 5, 33–36. [DOI] [PubMed] [Google Scholar]
- Li J, Nam KH, Vafeados D, Chory J. 2001. BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiology 127, 14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC. 2002. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110, 213–222. [DOI] [PubMed] [Google Scholar]
- Li QF, Wang C, Jiang L, Li S, Sun SS, He JX. 2012. An interaction between BZR1 and DELLAs mediates direct signaling crosstalk between brassinosteroids and gibberellins in Arabidopsis. Science Signaling 5, ra72. [DOI] [PubMed] [Google Scholar]
- Li QF, Wang JD, Xiong M, Wei K, Zhou P, Huang LC, Zhang CQ, Fan XL, Liu QQ. 2018. iTRAQ-based analysis of proteins co-regulated by brassinosteroids and gibberellins in rice embryos during seed germination. International Journal of Molecular Sciences 19, 3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QF, Zhou Y, Xiong M, Ren XY, Han L, Wang JD, Zhang CQ, Fan XL, Liu QQ. 2020. Gibberellin recovers seed germination in rice with impaired brassinosteroid signalling. Plant Science 293, 110435. [DOI] [PubMed] [Google Scholar]
- Liu X, Hou X. 2018. Antagonistic regulation of ABA and GA in metabolism and signaling pathways. Frontiers in Plant Science 9, 251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Hu P, Huang M, Tang Y, Li Y, Li L, Hou X. 2016. The NF–YC–RGL2 module integrates GA and ABA signalling to regulate seed germination in Arabidopsis. Nature Communications 7, 12768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, McLachlin DT, Chait BT, Chua NH. 2002. ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. The Plant Journal 32, 317–328. [DOI] [PubMed] [Google Scholar]
- Malinovsky FG, Batoux M, Schwessinger B, Youn JH, Stransfeld L, Win J, Kim SK, Zipfel C. 2014. Antagonistic regulation of growth and immunity by the Arabidopsis basic helix–loop–helix transcription factor homolog of brassinosteroid enhanced expression2 interacting with increased leaf inclination1 binding bHLH1. Plant Physiology 164, 1443–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam KH, Li J. 2002. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110, 203–212. [DOI] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Wang ZY. 2012. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nature Cell Biology 14, 802–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski N, Sun TP, Gubler F. 2002. Gibberellin signaling: biosynthesis, catabolism, and response pathways. The Plant Cell 14 Suppl, S61–S80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra B, Pattanaik S, Schluttenhofer C, Yuan L. 2018. A network of jasmonate-responsive bHLH factors modulate monoterpenoid indole alkaloid biosynthesis in Catharanthus roseus. New Phytologist 217, 1566–1581. [DOI] [PubMed] [Google Scholar]
- Patra B, Pattanaik S, Yuan L. 2013. Ubiquitin protein ligase 3 mediates the proteasomal degradation of GLABROUS 3 and ENHANCER OF GLABROUS 3, regulators of trichome development and flavonoid biosynthesis in Arabidopsis. The Plant Journal 74, 435–447. [DOI] [PubMed] [Google Scholar]
- Piskurewicz U, Jikumaru Y, Kinoshita N, Nambara E, Kamiya Y, Lopez-Molina L. 2008. The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. The Plant Cell 20, 2729–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas-Riverola A, Gupta A, Betegon-Putze I, Bosch N, Ibanes M, Cano-Delgado AI. 2019. Brassinosteroid signaling in plant development and adaptation to stress. Development 146, dev151894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JJ, Miraghazadeh A, Beckett AH, Quittenden LJ, McAdam EL. 2016. Interactions between gibberellins and other hormones. Annual Plant Reviews 49, 229–252. [Google Scholar]
- Ross JJ, Quittenden LJ. 2016. Interactions between brassinosteroids and gibberellins: synthesis or signaling? The Plant Cell 28, 829–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinovich L, Weiss D. 2010. The Arabidopsis cysteine-rich protein GASA4 promotes GA responses and exhibits redox activity in bacteria and in planta. The Plant Journal 64, 1018–1027. [DOI] [PubMed] [Google Scholar]
- Ryu H, Cho H, Bae W, Hwang I. 2014. Control of early seedling development by BES1/TPL/HDA19-mediated epigenetic regulation of ABI3. Nature Communications 5, 4138. [DOI] [PubMed] [Google Scholar]
- Ryu H, Kim K, Cho H, Park J, Choe S, Hwang I. 2007. Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. The Plant Cell 19, 2749–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M, Nambara E, Choi G, Yamaguchi S. 2009. Interaction of light and hormone signals in germinating seeds. Plant Molecular Biology 69, 463–472. [DOI] [PubMed] [Google Scholar]
- Shu K, Liu XD, Xie Q, He ZH. 2016. Two faces of one seed: hormonal regulation of dormancy and germination. Molecular plant 9, 34–45. [DOI] [PubMed] [Google Scholar]
- Skubacz A, Daszkowska-Golec A, Szarejko I. 2016. The role and regulation of ABI5 (ABA-Insensitive 5) in plant development, abiotic stress responses and phytohormone crosstalk. Frontiers in Plant Science 7, 1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliwinska E, Bassel GW, Bewley JD. 2009. Germination of Arabidopsis thaliana seeds is not completed as a result of elongation of the radicle but of the adjacent transition zone and lower hypocotyl. Journal of Experimental Botany 60, 3587–3594. [DOI] [PubMed] [Google Scholar]
- Steber CM, McCourt P. 2001. A role for brassinosteroids in germination in Arabidopsis. Plant Physiology 125, 763–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui X, Singh SK, Patra B, Schluttenhofer C, Guo W, Pattanaik S, Yuan L. 2018. Cross-family transcription factor interaction between MYC2 and GBFs modulates terpenoid indole alkaloid biosynthesis. Journal of Experimental Botany 69, 4267–4281. [DOI] [PubMed] [Google Scholar]
- Tang W, Yuan M, Wang R.et al. . 2011. PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nature Cell Biology 13, 124–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Huq E, Quail PH. 2003. The Arabidopsis basic/helix–loop–helix transcription factor family. The Plant Cell 15, 1749–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topham AT, Taylor RE, Yan D, Nambara E, Johnston IG, Bassel GW. 2017. Temperature variability is integrated by a spatially embedded decision-making center to break dormancy in Arabidopsis seeds. Proceedings of the National Academy of Sciences, USA 114, 6629–6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugama D, Liu S, Takano T. 2013. Arabidopsis heterotrimeric G protein beta subunit, AGB1, regulates brassinosteroid signalling independently of BZR1. Journal of Experimental Botany 64, 3213–3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuan PA, Kumar R, Rehal PK, Toora PK, Ayele BT. 2018. Molecular mechanisms underlying abscisic acid/gibberellin balance in the control of seed dormancy and germination in cereals. Frontiers in Plant Science 9, 668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Nakano T, Gendron J, et al. . 2002. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Developmental Cell 2, 505–513. [DOI] [PubMed] [Google Scholar]
- Weitbrecht K, Müller K, Leubner-Metzger G. 2011. First off the mark: early seed germination. Journal of Experimental Botany 62, 3289–3309. [DOI] [PubMed] [Google Scholar]
- Yan A, Wu M, Yan L, Hu R, Ali I, Gan Y. 2014. AtEXP2 is involved in seed germination and abiotic stress response in Arabidopsis. PLoS One 9, e85208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J. 2002. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109, 181–191. [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. 2007. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature Protocols 2, 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zhang SC, Wang XJ. 2008. Expression pattern of GASA, downstream genes of DELLA, in Arabidopsis. Chinese Science Bulletin 53, 3839–3846. [Google Scholar]
- Zhao X, Dou L, Gong Z, Wang X, Mao T. 2019. BES1 hinders ABSCISIC ACID INSENSITIVE5 and promotes seed germination in Arabidopsis. New Phytologist 221, 908–918. [DOI] [PubMed] [Google Scholar]
- Zheng K, Wang Y, Wang S. 2019. The non-DNA binding bHLH transcription factor Paclobutrazol Resistances are involved in the regulation of ABA and salt responses in Arabidopsis. Plant Physiology and Biochemistry 139, 239–245. [DOI] [PubMed] [Google Scholar]
- Zhiponova MK, Morohashi K, Vanhoutte I, Machemer-Noonan K, Revalska M, Van Montagu M, Grotewold E, Russinova E. 2014. Helix–loop–helix/basic helix–loop–helix transcription factor network represses cell elongation in Arabidopsis through an apparent incoherent feed-forward loop. Proceedings of the National Academy of Sciences, USA 111, 2824–2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong C, Xu H, Ye S, Wang S, Li L, Zhang S, Wang X. 2015. Gibberellic acid-stimulated Arabidopsis6 serves as an integrator of gibberellin, abscisic acid, and glucose signaling during seed germination in Arabidopsis. Plant Physiology 169, 2288–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou X, Neuman D, Shen QJ. 2008. Interactions of two transcriptional repressors and two transcriptional activators in modulating gibberellin signaling in aleurone cells. Plant Physiology 148, 176–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data from this study can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: HBI1 (At2g18300), BEE2 (At4g36540), IBH1 (At2g43060), GASA6 (At1g74670), UBQ1 (At3g52590), PP2A (At1g69960), and TUB3 (At5g 62700). All data supporting the findings of this study are available within the paper and within its supplementary data published online.