Abstract
Gene Set Enrichment Analysis (GSEA) is used to identify differentially expressed gene sets that are enriched for annotated biological functions. The existing GSEA R code is not in the form of a flexible package with analysis and plotting customization options, and the results produced are not generated in the form of R objects. In this study, we introduce the GSEAplot R package with novel functionality for saving relevant information from the analysis to the current R workspace, and we introduce the ability to customize plots and databases. The GSEAplot package provides a novel utility that facilitates the implementation of GSEA R-based in genomics analysis pipelines.
Keywords: functional enrichment analysis, gene set analysis, R package
1. Introduction
Gene expression analyses aim to provide insights into biological mechanisms. Gene Set Enrichment Analysis (GSEA) allows for evaluating gene expression data by considering groups of genes that are collectively differentially expressed. The analysis of gene sets, in contrast to analyses of individual genes, augments statistical power and increases the likelihood of detecting biological pathways associated with differentially expressed genes (Subramanian et al., 2005).
The primary open source GSEA software is available in the form of a Java application and an R script. There also exist multiple implementations of GSEA in R package form. For instance, the phenoTest and clusterProfiler R packages perform GSEA (Yu et al., 2012; Planet, 2020). However, the existing packages do not support customized gene set libraries, analysis, and plotting. Although individual researchers have generated customized GSEA plots (Calabrese et al., 2016; Normand et al., 2018), there does not exist an R package to facilitate the generation of such plots. We developed the GSEAplot package to address these limitations by introducing novel features to the analysis: making relevant data accessible in the R environment and providing customization options for analysis and plotting.
2. Methods
GSEAplot was formulated using the publicly available GSEA R code (https://software.broadinstitute.org/cancer/software/gsea/wiki/index.php/R-GSEA_Readme). The original version of GSEA saves plot and report files automatically. The GSEAplot package contains modified GSEA source code to allow the user direct access to relevant data within the R environment. This feature is new to our package and was not included in previous implementations. GSEAplots() is our new wrapper function introduced in the package to perform GSEA and output the necessary information for data analysis and plot generation in R. The primary outputs include the following: gene.set.reference.matrix provides a table of the gene sets being studied along with corresponding gene symbols, and gene.set.leading is a list containing the leading edge sets [i.e., sets of genes with differential expression that is most related to a change in phenotype; Subramanian et al. (2005)]. Our package offers the novel option to include new gene sets in existing gene set libraries, as well as functionality for formulating novel gene set libraries. We included common gene set libraries, such as the molecular signatures hallmark database (Liberzon et al., 2015), in our package. We also included transcription factor targets and ligand–receptor pair gene lists in our package (Eward, 2016; Kadoki et al., 2017). We used GSEAplots to perform GSEA using adipose tissue gene expression data from male and female participants of the GTEx Consortium (GTEx Consortium et al., 2017). The data set consisted of 581 subcutaneous adipose tissue RNA-seq samples (33% female) that were processed as described elsewhere (Anderson et al., 2020).
3. Results and Conclusions
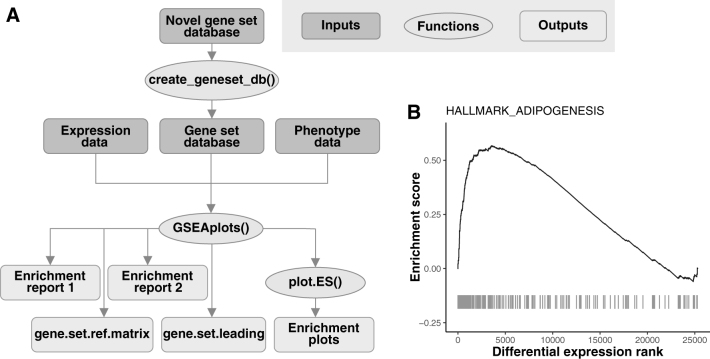
The GSEAplot R package is accompanied by thorough documentation and a vignette available on github (https://github.com/kelsiereinaltt/GSEAplot/). Figure 1A details the inputs and outputs of GSEAplot and highlights novel attributes of our package. Figure 1B demonstrates an example plot created by the package. All aspects of the plots are modifiable, and the plot data are available to the user for further analysis/plotting. The GSEAplot package provides a user-friendly implementation of GSEA in R. This package saves relevant data to the user's working directory, and includes outputs of interest for further analysis and plotting. In particular, the user can customize the appearance of enrichment plots and customize gene sets for analysis. Therefore, this package can streamline the application GSEA in genomics data analysis.
FIG. 1.
The GSEAplot R package includes novel functions for customizing gene set enrichment analysis. (A) The GSEAplot flowchart includes data inputs, essential functions, and outputs. (B) The customized enrichment plot shows the running enrichment score with respect to genes ranked by signal-to-noise ratio, where genes with elevated expression in females have lower differential expression ranks. The peak on the left indicates that genes related to adipogenesis have elevated expression in females relative to males (false discovery rate = 0). The tick marks denote adipogenesis-related genes. GSEA, Gene Set Enrichment Analysis.
Author Disclosure Statement
No competing financial interests exist.
Funding Information
This study has been supported by American Heart Association Postdoctoral Fellowship #18POST33990082 (W.D.A.), NIH T32 DK007646 (W.D.A.), NIH T32 HL007284 (W.D.A.), NIH/NIDDK R01 DK118287 (M.C.), American Diabetes Association 1-19-IBS-105 (M.C,), and NIH/NIDDK R01 DK118243 (M.C.).
References
- Anderson, W.D., Soh, J.Y., Innis, S.E., et al. . 2020. Sex differences in human adipose tissue gene expression and genetic regulation involve adipogenesis. Genome Res. 30, 1379–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese, G., Mesner, L.D., Foley, P.L., et al. . 2016. Network Analysis Implicates Alpha-Synuclein (Snca) in the regulation of ovariectomy-induced bone loss. Sci. Rep. 6, 29475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eward, K. 2016. Available at: https://github.com/slowkow/tftargets. Accessed April6, 2021
- GTEx Consortium, Laboratory, Data Analysis & Coordinating Center (LDACC)—Analysis Working Group, Statistical Methods groups—Analysis Working Group, Enhancing GTEx (eGTEx) groups, NIH Common Fund, NIH/NCI, NIH/NHGRI, NIH/NIMH, NIH/NIDA, Biospecimen Collection Source Site—NDRI, et al. . 2017. Genetic effects on gene expression across human tissues. Nature 550, 204–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoki, M., Patil, A., Thaiss, C.C., et al. . 2017. Organism-level analysis of vaccination reveals networks of protection across tissues. Cell 171, 398–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon, A., Birger, C., Thorvaldsdóttir, H., et al. . 2015. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normand, R., Du, W., Briller, M., et al. . 2018. Found In Translation: A machine learning model for mouse-to-human inference. Nat. Methods 15, 1067–1073 [DOI] [PubMed] [Google Scholar]
- Planet, E. 2020. phenoTest: Tools to Test Association Between Gene Expression and Phenotype in a Way That Is Efficient, Structured, Fast and Scalable. R Package Version 1.36.0. Available at: https://bioconductor.org/packages/release/bioc/html/phenoTest.html. Accessed April6, 2021
- Subramanian, A., Tamayo, P., Mootha, V.K., et al. . 2005. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, G., Wang, L., Han, Y., et al. . 2012. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287 [DOI] [PMC free article] [PubMed] [Google Scholar]