Abstract
Mouse mast cell proteases (mMCP)-1 and −2 are specifically expressed in mucosal mast cells (MCs). However, the transcriptional regulation mechanism of the Mcpt1 and Mcpt2 genes induced in mucosal MCs is largely unknown. In the current study, we found that TGF-β stimulation drastically induced upregulation of Mcpt1 and Mcpt2 mRNA in mouse bone marrow–derived MCs (BMMCs). TGF-β–induced expression of Mcpt1 and Mcpt2 was markedly suppressed by transfection with small interfering RNA targeting Smad2 or Smad4 and moderately reduced by Smad3 small interfering RNA. We next examined the roles of the hematopoietic cell–specific transcription factors GATA1 and GATA2 in the expression of Mcpt1 and Mcpt2 and demonstrated that knockdown of GATA1 and GATA2 reduced the mRNA levels of Mcpt1 and Mcpt2 in BMMCs. The recruitment of GATA2 and acetylation of histone H4 of the highly conserved GATA–Smad motifs, which were localized in the distal regions of the Mcpt1 and Mcpt2 genes, were markedly increased by TGF-β stimulation, whereas the level of GATA2 binding to the proximal GATA motif was not affected by TGF-β. A reporter assay showed that TGF-β stimulation upregulated GATA2-mediated transactivation activity in a GATA–Smad motif-dependent manner. We also observed that GATA2 and Smad4 interacted in TGF-β–stimulated BMMCs via immunoprecipitation and Western blotting analysis. Taken together, these results demonstrate that TGF-β induced mMCP-1 and −2 expression by accelerating the recruitment of GATA2 to the proximal regions of the Mcpt1 and Mcpt2 genes in mucosal MCs.
Mast cells (MCs) express high affinity IgE receptor (FcεRI) on their surface. Cross-linking of IgE-binding FcεRI by Ag induces MC degranulation accompanied by the release of histamine, effector proteases, and eicosanoids as an early phase reaction and induces the production of cytokines in MCs as a late-phase reaction. MCs are derived from hematopoietic stem cells in the bone marrow and are released into the peripheral blood as MC progenitors. The immature MCs maturate in peripheral tissues, including the intestinal mucosa and dermis, to obtain tissue-specific characteristics. MCs are phenotypically divided into two subpopulations, mucosal MCs (MMCs) and connective tissue MCs (CTMCs), on the basis of protease expression profiles. In mice, MMCs found on mucosal surfaces, such as the gut, abundantly express mouse MC proteases (mMCP)-1 and −2, whereas CTMCs detected in the dermis express mMCP-4 and −5 (1–4). Various cytokines and growth factors, such as SCF, TGF-β, IL-3, IL-9, and IL-10, are involved in the development of MMCs or CTMCs, and subtype-specific expression of mMCPs (5–10). However, the transcriptional regulation mechanism of the Mcpt genes induced by stimulation signaling is largely unknown.
Several transcription factors critical for the development and cell-type specific gene expression in MCs have been identified. MITF is a well-known transcription factor that is essential for the expression of MC-specific genes, including c-kit and mMCPs (11–13). We previously demonstrated that the hematopoietic cell–specific transcription factors GATA1, GATA2, and PU.1 coordinately regulate the expression of allergy-related molecules in MCs, including FcεRI-α, and -β, c-Kit, ST2/IL1RL1, and Syk (14–18). Although any of these transcription factors relating to MC-specific gene expression may be involved in the stimulation signaling-induced expression of mMCPs, the detailed mechanism remains unclear.
These observations prompted us to analyze the effect of stimulation signaling on the function of transcription factors involved in the expression of mMCPs. In the current study, we examined the involvement of GATA1, GATA2, and MITF in transactivation of the genes encoding mMCP-1 and mMCP-2 and found that GATA1 and GATA2 are involved in TGF-β–mediated transactivation of mMCP-1 and 2. We also showed that the TGF-β–Smad axis accelerates GATA2 recruitment toward cis-enhancing elements conserved in distal region of the Mcpt1 and Mcpt2 genes in MCs.
Materials and Methods
Mice and cells
Bone marrow–derived MCs (BMMCs) were generated from bone marrow cells of C57BL/6 mice purchased from Japan SLC (Hamamatsu, Japan) by cultivation in RPMI 1640 (Sigma-Aldrich, St. Louis, MO) containing 10% heat-inactivated FCS (Biowest), 100 U/ml penicillin, 100 μg/ml streptomycin, 50 μM 2-ME, 100 μM MEM nonessential amino acid solution, and 5 ng/ml murine IL-3 (BioLegend) at 37°C for more than 5 wk. FACS was used to confirm that the frequency of BMMCs expressing both FcεRI and c-kit was over 95%. All animal experiments were performed in accordance with the approved guidelines of the Institutional Review Board of Tokyo University of Science, Tokyo, Japan. The Animal Care and Use Committees of Tokyo University of Science specifically approved this study. The human embryonic kidney cell line HEK293T was maintained in DMEM (Sigma-Aldrich) supplemented with 10% FCS. Recombinant mouse TGF-β1 (no. 5231; Cell Signaling Technology) was added to the culture medium at a 1 or 10 ng/ml concentration to stimulate cells.
Quantitative analysis of mRNA
An RNAeasy kit (QIAGEN, Hilden, Germany) and a ReverTra Ace qPCR RT kit (TOYOBO, Osaka, Japan) were used for preparation of total RNA and synthesis of cDNA, respectively. The mRNA levels were determined by real-time PCR using a Step-One Real-Time PCR system (Applied Biosystems) with TaqMan Gene Expression Assays (Applied Biosystems), no. Mm00484678_m1 for mouse GATA1, no. Mm00492300_m1 for mouse GATA2, no. 4352339E for rodent GAPDH, and a THUNDERBIRD probe qPCR Mix (TOYOBO). For measurement of mouse Mcpt1, Mcpt2, Mitf, Smad2, Smad3, and Smad4, the following primers were used with THUNDERBIRD SYBR qPCR Mix (TOYOBO): Mcpt1 forward, 5′-AAGTTCCACAAAGTTAAAAACAGCATAC-3′, reverse, 5′-GTGAATCCCCATAAGATACAATACCAT-3′, Mcpt2 forward, 5′-AAAGTTTCAGTACCTTTCGGG-3′, reverse, 5′-CATCCACATCAGAATTCAACTCT-3′, Mitf forward, 5′-GGAACAGCAACGAGCTAAGG-3′, reverse, 5′-TGATGATCCGATTCACCAGA-3′, Smad2 forward, 5′-GTGTCACCATACCAAGCACTTGC-3′, reverse, 5′-CCTGTTGTGTCCCACTGATCTACC-3′, Smad3 forward, 5′-ACCAAGTGCATTACCATCC-3′, reverse, 5′-CAGTAGATAACGTGAGGGAGCCC-3′, Smad4 forward, 5′-CATCCTGGACATTACTGGCCA-3′, reverse, 5′-CCTACCTGAACGTCCATTTCAA-3′.
ELISA
Mouse MCPT-1 Uncoated ELISA Kit (no. 88–7503-22; Invitrogen) was used to determine the concentration of mMCP-1 protein.
Introduction of small interfering RNAs
The following small interfering RNAs (siRNAs) were purchased from Invitrogen (Carlsbad, CA); mouse Gata1 (MSS236579), Gata2 (MSS204585), Smad2 (MSS206405), Smad3 (MSS206420), and Smad4 (MSS206437) and control siRNAs (Stealth RNAi Negative Universal Control Lo GC, Med GC, and Hi GC [no. 12935–200, −300, and −400]). BMMCs (2 × 106 or 2 × 105) were transfected with 10 or 1 μl of 20 μmol/L siRNA with a Neon 100 μl kit or a Neon 10 μl kit using a Neon transfection system (Invitrogen) set at Program no. 5.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assays were performed using a ChIP Assay Kit (Upstate, Lake Placid, NY) with anti-GATA2 Ab (H-116, sc-9008), anti–acetyl histone H4 Ab (06–866), or control rabbit IgG (02–6102; Invitrogen) as previously described (19). The amount of chromosomal DNA was determined by quantitative PCR. The following synthesized oligonucleotides were used as primers for PCR: the Mcpt1 promoter −3300/−3230 (forward primer; 5′-CTGGACTGATGTTGAGAACTGATAGAC-3′, and reverse primer; 5′-CCGGGATGCTGATCCTATGA-3′), −127/−58 (forward primer; 5′-CCACCAGTGGTCAGAGTATGAGAA-3′, and reverse primer; 5′-GAACCACAGATCTGGCTTGGA-3′), the Mcpt2 promoter −3492/−3422 (forward primer; 5′-CCCAACAGCTTACCAGATATAAGAAA-3′, and reverse primer; 5′-TCAGCCCAGCCTCATCAG-3′) −61/+11 (forward primer; 5′-CACAGACTCAACACCACCAGAGA-3′, and reverse primer; 5′-TCTGGTTTGGACAAGCTCTACTTTC-3′).
Immunoprecipitation and Western blotting of transfectants expressing exogenous GATA2 and Smads
The expression plasmids pCMV-Myc-N-mSmad2, pCMV-Myc-N-mSmad4, and pCMV-HA-N-mSmad4 were generated by insertion of Smad2 cDNA and Smad4 cDNA amplified from mouse RNAs via PCR into multicloning sites of pCMV-Myc-N (Clontech) and pCMV-HA-N (Clontech), respectively. For the expression of Flag-tagged GATA2, pCR3.1-Flag-GATA2, which was termed pCR3.1-GATA2 in our previous study (16), was used. HEK293T cells were transfected with 5 μg of each plasmid DNA (pCMV-Myc-N-mSmad2, pCMV-HA-N-mSmad4, and pCR3.1-Flag-GATA2) using the calcium phosphate method. At 24 h after transfection, 10 ng/ml TGF-β was added into the culture medium, and cells were incubated for an additional 2 h. To perform immunoprecipitation, cell lysate was mixed with anti-Flag Ab (M2, F1804; Sigma-Aldrich). After incubation for 3 h at 4°C with rotation, protein A/G agarose was added to the lysate, which was incubated for an additional 1 h.
Western blot analyses were performed as previously described (19) with the following Abs: anti-Smad2/3 (FL-425, sc-8332; Santa Cruz Biotechnology), anti-HA (631207; Clontech Laboratories), anti–c-Myc (631206; Clontech Laboratories), and anti-GATA2 (same as used in ChIP assays). A densitometric analysis was performed using ImageJ (https://imagej.nih.gov/ij/).
Luciferase assay
Reporter plasmids carrying the luciferase gene under the control of the Mcpt1 promoter (−299/+32) with or without an enhancer region of the Mcpt1 gene (−3440/−3218) were generated by insertion of DNA fragments amplified by PCR from the C57BL/6 genomic DNA into the multicloning site of pGL4.10[luc2] (Promega, Madison, WI). To delete 28 bp of element (−3280/−3253) from an enhancer region of a reporter plasmid, site-directed mutagenesis was performed using a PrimeSTAR Mutagenesis Basal Kit (Takara Bio) with the following primers (5′-GAGAACTGTATCATAGGATCAGCAT-3′, joining −3287/−3281 and −3252/−3235 and 5′-TATGATACAGTTCTCAACATCAGTCC-3′, joining −3245/−3252 and −3281/−3298). To create a simple cis-reporter luciferase system, tandem repeated three copies of −3281/−3252 and its mutants were inserted into the luciferase plasmid carrying the Mcpt1 minimum promoter. The following synthesized oligonucleotides and complementary oligonucleotides were used to generate plasmids including tandem repeated elements: wild-type repeat; 5′-KpnI-(TGATAGACACATTATCAGACAGACAGATAG)×3-SacI-3′, Smad Mut repeat; 5′-KpnI-(TGATgGgaACATTATCgGgagGgaAGATAG)×3-SacI-3′, GATA–Smad Mut repeat; 5′-KpnI-(TGcTgGgaACATTAgCgGgagGgaAGcTAG)×3-SacI-3′ (mutated nucleotides are shown as small letters). Cotransfection of HEK293T cells with reporter plasmid and expression plasmids, pCR3.1-GATA1 (20), pCR3.1-GATA2 (16), pCMV-HA-N-mSmad4, and determination of promoter activity were performed as previously described (21).
Statistical analysis
The p values were determined by a two-tailed Student t test.
Results
The role of Smad molecules in the expression of mMCP-1 and −2 in MCs
Previous studies have shown that TGF-β upregulates the expression of the MC proteases mMCP-1 and −2 in MCs (5, 6). To clarify the mechanism by which TGF-β signaling regulates the expression of mMCP-1 and −2, we analyzed the change in Mcpt1 and Mcpt2 mRNA levels following TGF-β stimulation in MCs. As shown in Fig. 1A, the addition of TGF-β into culture medium rapidly induced upregulation of the Mcpt1 mRNA level, briefly, 3-fold at 2 h, 10-fold at 5 h, and over 100-fold at 12 h after stimulation, and the increase continued for at least 36 h. This rapid and marked upregulation was also observed in the expression of Mcpt2 mRNA. When TGF-β–treated BMMCs were stimulated with A23187 (Ca2+ ionophore), mMCP-1 protein was released into culture supernatants, whereas mMCP-1 protein was not detected in those of TGF-β–untreated BMMCs (Fig. 1B), suggesting that the TGF-β–induced upregulation of Mcpt1 transcription was reflected to mMCP-1 protein production. To investigate the role of Smads, well-known intracellular signaling molecules downstream of TGF-β receptor, in expression of Mcpt1 and Mcpt2 mRNAs, Smad knockdown MCs was analyzed (Fig. 1C). The introduction of siRNA for Smad2, Smad3, or Smad4 significantly reduced the amount of target mRNA in MCs. Under these experimental conditions, we measured the mRNA levels of Mcpt1 and Mcpt2 in MCs 8 h after TGF-β stimulation and found that TGF-β–induced upregulation of Mcpt1 and Mcpt2 transcripts were obviously suppressed in Smad2 or Smad4 knockdown MCs and moderately reduced in Smad3 knockdown MCs (Fig. 1D). These results suggest that TGF-β induces the expression of Mcpt1 and Mcpt2 genes primarily through Smad2 and Smad4 and in part through Smad3.
FIGURE 1.
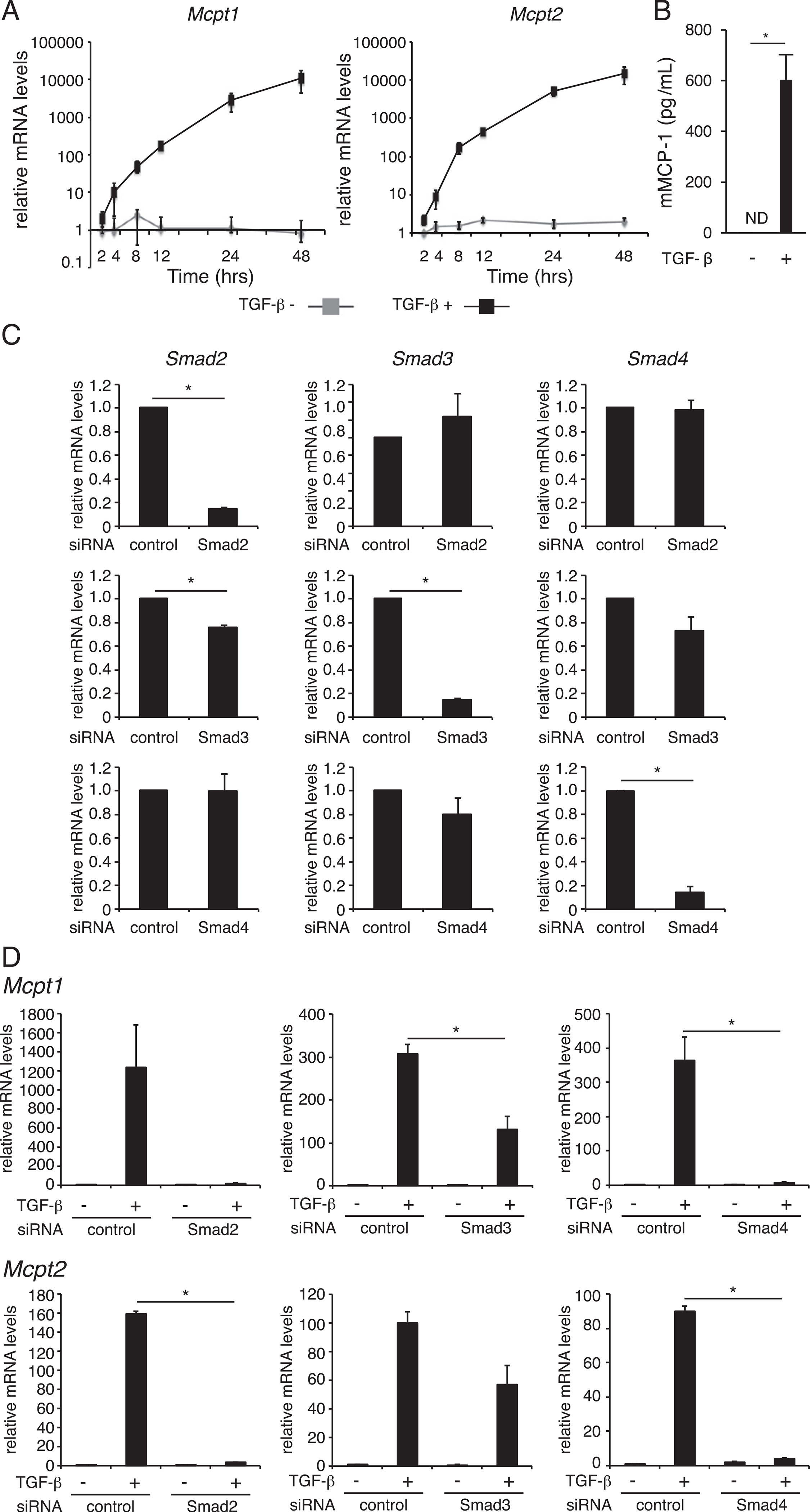
Effect of TGF-β signaling on the expression of Mcpt1 and Mcpt2 in BMMCs. (A) BMMCs were treated with 1 ng/ml TGF-β for the indicated times. The mRNA expression levels of Mcpt1 and Mcpt2 were assessed by quantitative RT-PCR. (B) mMCP-1 protein concentrations in culture media of BMMCs. BMMCs were cultured in the presence of 1 ng/ml TGF-β for 72 h. The culture media of BMMCs (1.0 × 106 cells/500 μl), which were stimulated with 1 m-M A23187 for 1 h, were harvested to determine mMCP-1 protein concentrations by an ELISA. (C) The mRNA levels of Smad2, Smad3, and Smad4 in siRNA-transfected BMMCs at 48 h after siRNA transfection. (D) The mRNA levels of Mcpt1 and Mcpt2 in siRNA-transfected BMMCs at 8 h after addition of TGF-β. The expression of each mRNA was normalized to that of GAPDH mRNA by calculation of the cycle threshold values. The data are presented as the mean ± SD of three independent experiments performed with triplicate samples (A, C, and D). A typical result of two independent experiments performed with triplicate samples is shown (B). *p < 0.05.
The involvement of GATA1 and GATA2 in mMCP-1 and −2 expression
Although the expression of mMCP-1 and −2 is restricted to MCs, especially mucosal type MCs, the transcription factors regulating the cell-type specific expression of the Mcpt1 and Mcpt2 genes are largely unknown. We previously found that GATA1 and/or GATA2 are critical for regulation of MC-expressed genes, including FCERIA (14, 21), MS4A2 (15, 21), IL1RL1 (17), and Kit (16). First, to investigate whether these GATA molecules are involved in the expression of the MMC-specific genes Mcpt1 and Mcpt2, we evaluated the role of GATA1 or GATA2 in transcription of Mcpt1 and Mcpt2 genes. Introduction of siRNAs for Gata1 and Gata2 significantly reduced the Gata1 and Gata2 mRNA levels, respectively, in BMMCs (Fig. 2A). Under these conditions, the mRNA levels of Mcpt1 and Mcpt2 were significantly decreased, whereas knockdown of GATA1 or GATA2 did not affect the mRNA level of another MC-specific transcription factor, Mitf (Fig. 2A). These results indicate that GATA1 and GATA2 are involved in the expression of Mcpt1 and Mcpt2 genes independently of MITF.
FIGURE 2.
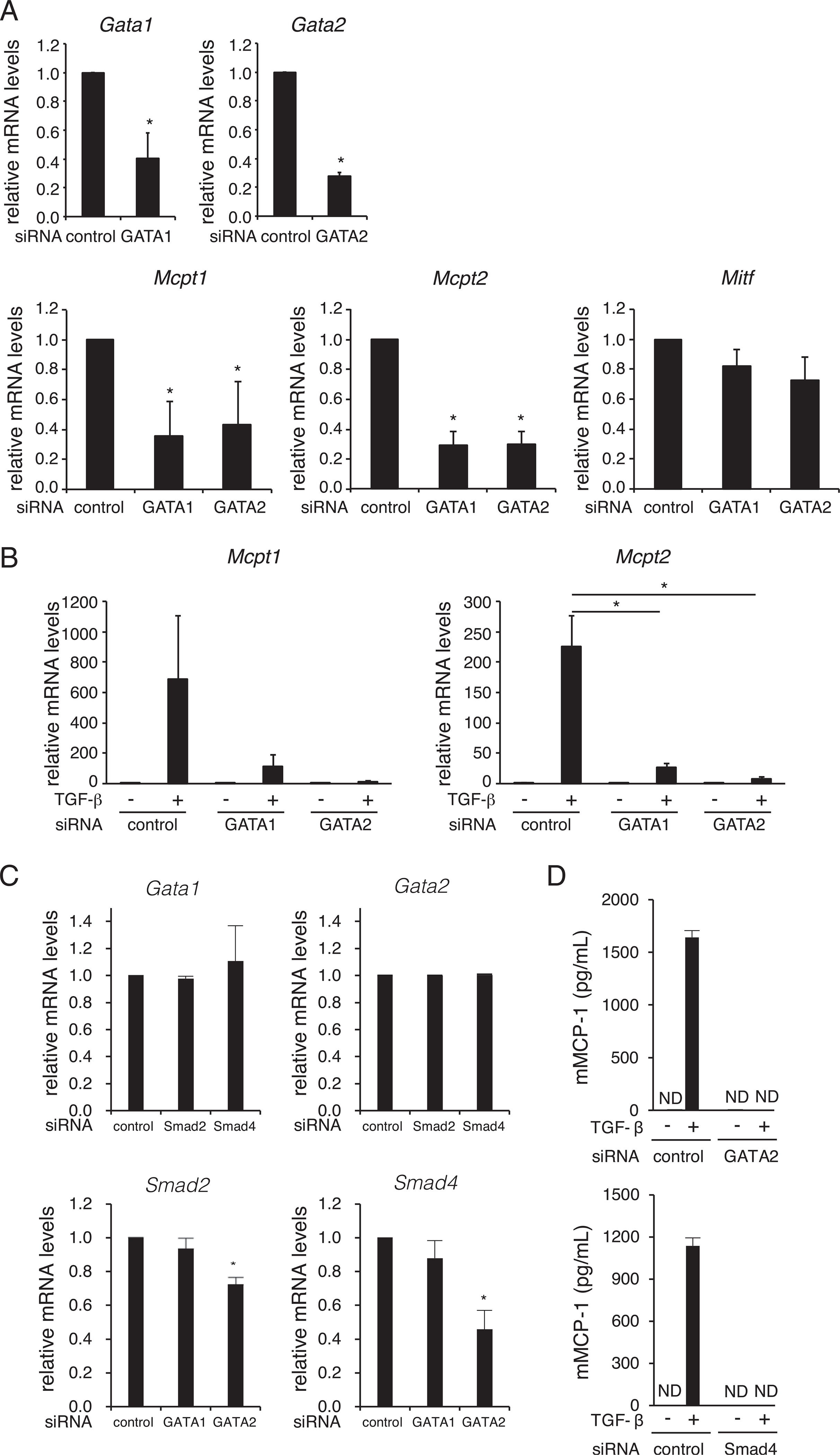
Effects of GATA1 and GATA2 knockdown on the expression of Mcpt1 and Mcpt2 in BMMCs. (A) The mRNA levels of transcription factors (Gata1, Gata2, and Mitf) and mMCPs (Mcpt1 and Mcpt1) in siRNA-transfected BMMCs *p < 0.05. (B) The mRNA levels of Mcpt1 and Mcpt2 in siRNA-introduced and TGF-β–treated BMMCs. *p < 0.05. (C) The mRNA levels of Gata1 and Gata2 in Smad siRNA-transfected BMMCs (top), and the mRNA levels of Smad2 and 4, in GATA siRNA-transfected BMMCs (bottom). *p < 0.05. The expression of each mRNA was evaluated relative to that of GAPDH (A-C). The data are shown as the mean ± SD of three (A and B) and two (C) independent experiments performed with triplicate samples. (D) mMCP-1 protein concentrations in culture media of siRNA-transfected BMMCs. TGF-β (1 ng/ml) was added to culture media of BMMCs (1.0 × 106 cells/500 μl) at 48 h after siRNA transfection. After additional 72 h incubation, BMMCs were stimulated with A23187. The concentrations of mMCP-1 in culture supernatants at 1 h after A23187-stimulation were determined by an ELISA. The data are shown as the mean ± SD of triplicate samples. Similar results were obtained in two independent experiments. ND, not detected.
Next, we examined the effect of GATA1 and GATA2 knockdown on TGF-β–induced upregulation of Mcpt1 and Mcpt2 mRNA expression. As shown in Fig. 2B, the TGB-β–induced upregulation of Mcpt1 mRNA was reduced to ~15 and 5% that of control siRNA-introduced cells by knockdown of GATA1 and GATA2, respectively. The knockdown of GATA1 and GATA2 exhibited a further striking effect on TGF-β–induced upregulation of Mcpt2 mRNA. These results demonstrate that GATA1 and GATA2 are involved in both the basal expression and TGF-β–induced expression of mMCP-1 and mMCP-2 in MCs.
The siRNA experiments showed that knockdown of Smad2 and Smad4 (Fig. 1D) and of GATA1 and GATA2 (Fig. 2B) dramatically suppressed TGF-β–induced transactivation of Mcpt1 and Mcpt2 in a similar yield. To clarify whether Smads and GATAs regulate expression of each other, we determined mRNA levels of Gata1 and Gata2 in BMMCs when siRNAs for Smad2 or Smad4 were introduced (Fig. 2C, top), and vice versa (Fig. 2C, bottom). We found that the mRNA levels of Gata1 and Gata2 in Smad2 or Smad4 siRNA-tranfected BMMCs were similar to those of control BMMCs (Fig. 2C, top). These results suggest that the suppression of Smad2 and Smad4 reduced the expression of Mcpt1 and Mcpt2 without affecting the expression levels of GATA1 and GATA2. We also found that the knockdown of GATA1 did not affect the mRNA levels of Smad2 and Smad4 in BMMCs (Fig. 2C, bottom). Although the amounts of Smad2 and Smad4 mRNAs were significantly decreased in Gata2 siRNA-transfected BMMCs, the reduction levels of Smad2 and Smad4 mRNAs were moderate. These results demonstrate that GATA1 and GATA2 are involved in TGF-β–induced transactivation of the Mcpt1 and Mcpt2 genes in a manner distinguishable from regulation of the Smad genes.
The above-mentioned results suggest that knockdown of Smads and GATAs reduced TGF-β–induced upregulation of Mcpt1 and Mcpt2 mRNAs in BMMCs. To evaluate the effect of the knockdown on mMCP-1 protein production, we measured mMCP-1 in the culture supernatants of siRNA-transfected BMMCs. As shown in Fig. 2D, mMCP-1 protein was not detected in the culture supernatants of BMMCs in which Gata2 siRNA or Smad4 siRNA was introduced, whereas control BMMCs released a significant amount of mMCP-1 following the A23187 stimulation. These results indicate that the knockdown of GATA2 or Smad4 completely suppressed the Ca2+ ionophore-induced release of mMCP-1 protein from TGF-β–treated BMMCs.
GATA2-binding and histone acetylation of Mcpt1 and Mcpt2 genes modulated by TGF-β–stimulation
GATA family transcription factors bind to chromosomal DNA via the “GATA” sequence as a core motif. As shown in Fig. 3A, several GATA sequences were located just upstream of the transcription start site of the Mcpt1 and Mcpt2 genes. Furthermore, we found tandem repeats of a closely located GATA-motif and Smad-binding sequence set over 3 kb upstream of the transcription start site of the Mcpt1 and Mcpt2 genes. To evaluate the effect of TGF-β on recruitment of GATA molecules and the histone acetylation degree around the Mcpt1 and Mcpt2 genes, we performed ChIP assays using BMMCs cultivated with or without TGF-β. A ChIP assay using anti-GATA2 Ab showed that the amount of GATA2 binding to the GATA-Smad region in the Mcpt1 gene was dramatically upregulated by TGF-β stimulation, whereas TGF-β stimulation did not exhibit an apparent effect on the binding degree of GATA2 to the proximal promoter region (Fig. 3B). This observation that the TGF-β–dependent increase in GATA2 binding to the GATA–Smad region but not to the proximal promoter was also detected in the Mcpt2 gene. In addition, TGF-β increased the acetylation degree of histone H4 around the GATA–Smad sites in the Mcpt1 and Mcpt2 genes (Fig. 3C). When siRNA for Smad4 or Gata2 was introduced into BMMCs, the TGF-β–induced increase in acetyl-H4 was significantly suppressed (Fig. 3D). The effect of TGF-β on the levels of GATA2 binding was not observed in the cis-enhancing element of the Il1rl1 gene, which was transactivated by GATA2 but not affected by TGF-β stimulation (Fig. 3E). Taken together, we conclude from these results that TGF-β stimulation accelerates the recruitment of GATA2 to the distal region containing GATA–Smad motifs in the Mcpt1 and Mcpt2 genes, resulting in the enhancement of histone acetylation of the Mcpt1 and Mcpt2 genes. In the current study, we cannot clarify the relationship between recruitment of GATA1 and TGF-β stimulation because significant binding of GATA1 to the Mcpt1 and Mcpt 2 gens was not detected in the ChIP assays using anti-GATA1 Ab.
FIGURE 3.
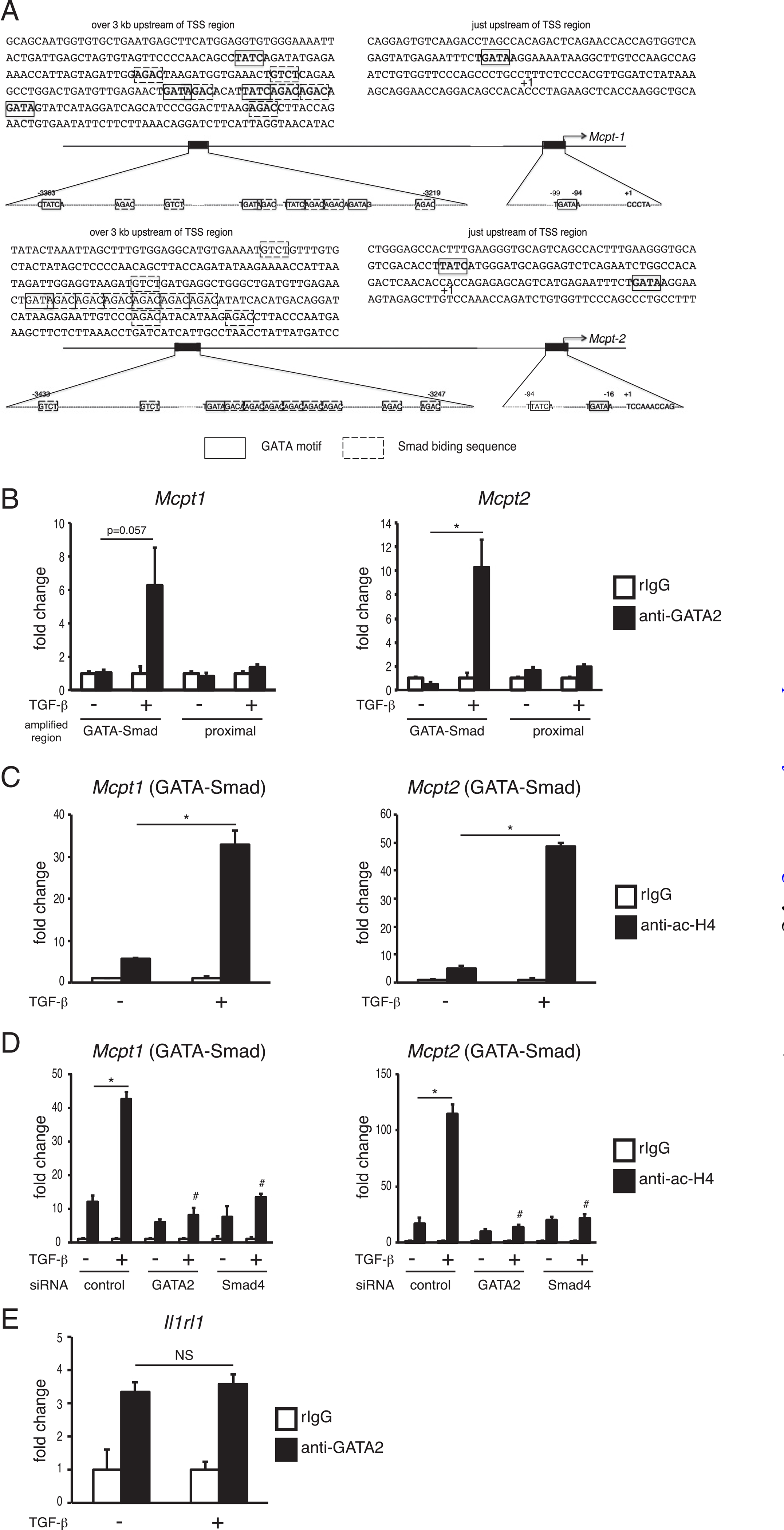
The levels of GATA2 binding and histone acetylation on the Mcpt1 and Mcpt2 genes in TGF-β–stimulated BMMCs. (A) Schematic diagrams of the Mcpt1 and Mcpt2 genes. (B-E) ChIP assay data. ChIP assays were performed with control rabbit IgG (rIgG) and anti-GATA2 Ab (B and E) or anti–acetyl histone H4 Ab (C and D). After 2 h of TGF-β treatment, cells were harvested and subjected to ChIP assays. The amount of chromosomal DNA immunoprecipitated with anti-acH4 Ab or anti-GATA2 Ab is shown as the ratio to that immunoprecipitated with rIgG. The results are expressed as the mean ± SD of triplicate samples, and similar results were obtained in another experiments (B-E). *p < 0.05, #p < 0.05 versus TGF-β–stimulated control.
TGF-β accelerates the association between GATA2 and Smads
It was confirmed that the mRNA levels of Gata1 and Gata2 in TGF-β–stimulated MCs were comparable to those in nonstimulated MCs (Fig. 4A), suggesting that enhancement of GATA2-binding to the distal GATA–Smad motifs is regulated by a posttranslational mechanism. First, to clarify whether GATA2 and Smad4 physically interact in living cells, Flag–GATA2 and Myc–Smad4 were exogenously expressed in HEK293T cells. Immunoblotting by anti-Myc Ab showed that a substantial amount of Myc–Smad4 was present in immunoprecipitates of anti-Flag Ab but not in those of control IgG (Fig. 4B). This result suggests that GATA2 physically (directly or indirectly) interacted with Smad4 in cells. However, the effect of TGF-β signaling on the association between GATA2 and Smad4 was not observed. HEK293T cells were cotransfected with expression plasmids for Flag–GATA2, Myc–Smad2, and HA–Smad4. We confirmed that expression level of HA–Smad4 was not in a large excess compared with Myc–Smad4 (data not shown). As shown in Fig. 4C, the amount of HA-tagged Smad4 (marked with an arrowhead in immunoblotting) immunoprecipitated with anti-Flag Ab was significantly increased by TGF-β treatment under this experimental condition. This result indicates that TGF-β stimulation induces and/or enhances formation of a complex that includes GATA2 and Smad4.
FIGURE 4.
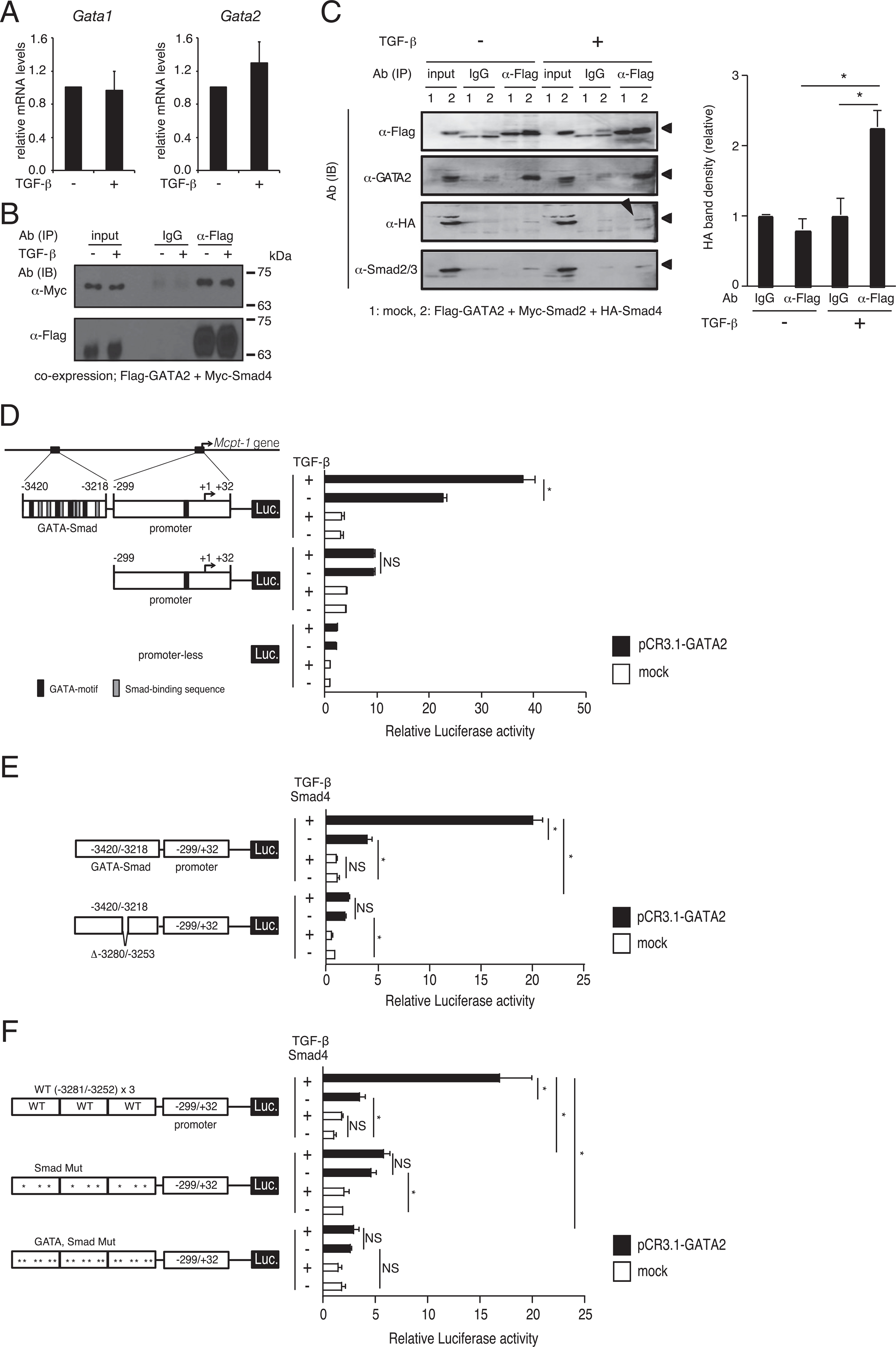
TGF-β induced GATA2/Smad4 complex formation and enhanced Mcpt1 transactivation through the GATA-Smad region. (A) The mRNA levels of GATA1 and GATA2 in TGF-β–treated BMMCs. The data are shown as the mean ± SD of three independent experiments performed with triplicate samples. (B) Association between exogenous GATA2 and Smad4. HEK293T cells cotransfected with expression plasmids for Flag–GATA2 and Myc–Smad4 were stimulated by TGF-β at 24 h after transfection and were harvested after 2 h incubation in the presence or absence of TGF-β. The cell lysates were subjected to immunoprecipitation (IP) and immunoblotting (IB). Aliquot lysates were loaded to independent gels; one for staining by anti-Myc and another for anti-Flag. A typical result of three independent experiments was shown. (C) Western blotting profiles of immunoprecipitated samples (left) and band intensity determined in three independent experiments (right). HEK293T cells were cotransfected with Flag-tagged GATA2 and Myc-tagged Smad2 and HA-tagged Smad4 expression plasmids. After 24 h, cells were treated with TGF-β for an additional 2 h and then subjected to IP and Western blotting assays using the indicated Abs. A single transferred membrane was reprobed. Band densities are shown as ratio to that of control IgG without TGF-β stimulation (n = 3; three independent experiments performed with a single sample). (D) Transcriptional activity driven by the promoter (−299/+32) or the GATA–Smad (−3420/−3218) + promoter (−299/+32) of the Mcpt1 gene was determined by a luciferase assay. TGF-β (10 ng/ml) was added to the culture medium of 293T transfectants at 4 h after transfection, and the cells were harvested after an additional 24 h incubation. (E) The effects of wild-type and 28 bp-deleted GATA-Smad regions on GATA2 and TGF-β signaling-mediated transcriptional activity. After 4 h incubation in culture media without FCS, transfectants were stimulated by 10 ng/ml TGF-β (E and F). (F) Transcriptional activity of tandem repeats of the minimal elements and its mutants. Luciferase activity was normalized to that of β-galactocidase (D–F). The data are expressed as mean ± SD of triplicate samples and are shown as fold change relative to promoterless, untreated cells (D–F). Similar results were obtained in two independent experiments (D–F). *p < 0.05.
TGF-β enhances transactivation activity of GATA2 through the GATA–Smad motifs
To evaluate the effect of TGF-β on the transactivation activity of GATA2, we performed a reporter assay using HEK293T cells. As shown in Fig. 4D, luciferase activity in cells in which a reporter plasmid carrying the proximal promoter region of the Mcpt1 gene was significantly increased by coexpression of GATA2 but was not affected by TGF-β stimulation. In contrast, luciferase activity driven by the GATA–Smad region-inserted promoter was synergistically upregulated by TGF-β stimulation and cotransfection with GATA2. Interestingly, GATA1 did not exhibit a synergistic effect on the GATA–Smad region (data not shown). To further strengthen the TGF-β signaling, we modified a reporter assay in two points: 1) a Smad4-expressing plasmid (pCMV-HA-N-Smad4) was cotransfected, and 2) all transfected cells were incubated in the FCS-free medium for 4 h before the addition of rTGF-β for starvation of bovine TGF-β. Under these experimental conditions, luciferase activity driven by the GATA–Smad region-inserted promoter was enhanced by 4-fold by coexpression of GATA2 and was synergistically upregulated by up to 20-fold by cooperation of GATA2 coexpression and TGF-β stimulation, whereas Smad4/TGF-β stimulation alone did not affect the GATA–Smad sequence-mediated luciferase activity (Fig. 4E). Deletion of 28 bp ( −3280/−3253), to which three GATA motifs and three Smad-binding sequences are localized, markedly reduced the effects of GATA2 coexpression (2-fold of mock), and the synergistic effect of GATA2 coexpression and Smad4/TGF-β stimulation was not observed (Fig. 4E). These results suggest that the 28 bp sequence may be critical for the cis-enhancing activity. To clarify the role of the minimal elements of GATA and Smad-binding motifs in the cis-enhancing activity, we generated a reporter plasmid carrying three copies of tandem repeats of these minimal elements. As shown in Fig. 4F, luciferase activity driven by this WT×3 reporter plasmid was significantly increased by GATA2 coexpression and was further dramatically upregulated by the combination of GATA2 coexpression and Smad4/TGF-β stimulation. When all Smad sequences were mutated, the synergistic effect was not observed, whereas GATA2 coexpression significantly increased the luciferase activity. Mutation of all GATA motifs and Smad sequences resulted in the lack of responsiveness to both GATA2 and TGF-β. These results suggest that the transactivation activity of GATA2 was increased by TGF-β stimulation when the Smad-binding motif was closely located to the cis-enhancing GATA motif.
Discussion
The mMCP family members, which are primarily produced in MCs and basophils, play important roles in IgE-mediated immunoresponses, including the pathology of allergy-related diseases and the host defense against parasite infection. These proteases are stored as granule compounds in the steady-state and are rapidly secreted from activated cells. In addition to their importance in immunological events, the expression profile of mMCPs is a unique issue. Briefly, mMCPs are useful as a hallmark to distinguish the MC subtypes and basophils because the expression of several mMCPs is restricted to MMCs, CTMCs, or basophils. The characteristic specific transactivation of these promoters makes inducible ablation of CTMCs and basophils possible, as follows. Mcpt5-Cre; iDTR mice (22), which express the Cre recombinase under the control of the Mcpt5 promoter and carry a floxed diphtheria toxin receptor transgene, are used for inducible ablation of CTMCs (23). Similarly, the basophil-specific conditional knockout system has been developed by using the Mcpt8 promoter specificity (24). In the current study, we focused on the regulatory mechanism of the Mcpt1 and Mcpt2 genes that are expressed in an MMC-specific manner to understand the function of transcription factors involved in the development of MMCs.
Cultivation of BMMCs in the presence of TGF-β increased the frequency of mMCP-1–positive cells and the amount of mMCP-1 accumulated in culture medium (5) (and Fig. 1B in the current study). The expression of mMCP-1 and −2 was reduced in the intestinal MMCs of mice lacking the integrin αvβ6, which is essential for activation of TGF-β (7). These observations indicate that TGF-β upregulates the expression of mMCP-1 in MCs in vitro and in vivo. Although the transcriptional activity of the Mcpt1 gene may be enhanced by TGF-β stimulation because the Mcpt1 mRNA level was increased in BMMCs maintained in the presence of TGF-β for 7 d (5), the molecular mechanism of transcriptional regulation of the Mcpt1 gene by TGF-β has not been revealed. The siRNA experiments showed that any knockdown of Smad2, 3, or 4 or GATA1 or 2 reduced the TGF-β–induced upregulation of Mcpt1 and Mcpt2 mRNA levels in BMMCs (Figs. 1, 2). We also found tandem GATA and Smad sequences at ~3 kb upstream of the transcription start site in both the Mcpt1 and Mcpt2 genes, to which GATA2 was recruited following TGF-β stimulation (Fig. 3), resulting in increased transcription activity (Fig. 4). A Western blotting analysis suggested that formation of the complex containing GATA2 and Smads (at least Smad4) may occur through TGF-β stimulation (Fig. 4). The TGF-β–Smad axis induces the development of cells following the context of each cell lineage. TGF-β signaling is involved in cell-type specific gene expression by accelerating the binding of master transcription factors to the target sites through Smad3 activation in various cells: Oct4 in stem cells, Myod1 in myotubes, and PU.1 in pro-B cells (25). Analysis of the DNA motifs, including the binding sequences of a master transcription factor and closely located Smad-binding sites, would help in understanding TGF-β–mediated development and/or differentiation of cells. For GATA family members, the Th2 master regulator GATA3 is involved in TGF-β–dependent expression of the Th2 cytokines IL-5 and IL-10 via interaction with Smad3 (26). Although GATA2 is an essential transcription factor for development and cell-type gene expression of MCs, the relationship between GATA2 and TGF-β signaling in MCs has been largely unknown. Under our experimental conditions, knockdown of Smad2 or Smad4 rather than Smad3 drastically suppressed the TGF-β–dependent increase in Mcpt1 and Mcpt2 mRNAs, likely suggesting that Smad2 and Smad4 play more prominent roles than Smad3. However, the current study cannot exclude the possibility that Smad3 functions as an efficient modulator of GATA2-mediated transactivation of the Mcpt1 and Mcpt2 genes because Smad2 siRNA slightly but significantly reduced the Smad3 mRNA level (Fig. 1B). As for transcriptional regulation of the Tpsabl gene encoding mMCP-7, a CTMC-specific protease, Smad3 and Smad4 but not Smad2 are required for the TGF-β–induced transactivation in MCs (27, 28). Further detailed analyses will be required to clarify the importance of Smad3 in TGF-β–induced expression of mMCP-1 and −2 in MCs.
The Mcpt1 and Mcpt2 genes are interesting targets not only for understanding the transcription factor network but also for development of MMC-specific gene targeted mice. We will conduct further studies regarding gene regulation of the Mcpt1 and Mcpt2.
Acknowledgments
We are grateful to the members of Laboratory of Molecular and Cellular Immunology, Department of Biological Science and Technology, Tokyo University of Science for constructive discussions and technical support.
This work was supported by the Funding Program for Next Generation World-Leading Researchers from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT) (LS111 to C.N.), a Grant-in-Aid for Challenging Exploratory Research (to C.N.), a Grant-in-Aid for Young Scientists (to K.K. and T.Y.), a Grant-in-Aid for Scientific Research (C) (to K.K. and T.Y.), the MEXT-Supported Program for the Strategic Research Foundation at Private Universities (Translational Research Center, Tokyo University of Science), the Tokyo Biochemical Research Foundation (to C.N.), the Tojuro Iijima Foundation for Food Science and Technology (to C.N. and T.Y.), the Takeda Science Foundation (to C.N.), the Tokyo University of Science Grant for President’s Research Promotion (to C.N.), and the Nipponham Foundation for the Future of Food and Foundation for Dietary Scientific Research (to K.K.). K.K. was supported by Research Fellowships of the Japanese Society for the Promotion of Science (JSPS) for Young Scientists (JSPS Research Fellowships for Young Scientists 3241, from 2016 to 2018).
Abbreviations used in this article:
- BMMC
bone marrow–derived MC
- ChIP
chromatin immunoprecipitation
- CTMC
connective tissue MC
- MC
mast cell
- MMC
mucosal MC
- mMCP
mouse MC protease
- siRNA
small interfering RNA
Footnotes
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Gurish MF, and Austen KF. 2012. Developmental origin and functional specialization of mast cell subsets. Immunity 37: 25–33. [DOI] [PubMed] [Google Scholar]
- 2.Pejler G, Abrink M, Ringvall M, and Wernersson S. 2007. Mast cell proteases. Adv. Immunol. 95: 167–255. [DOI] [PubMed] [Google Scholar]
- 3.Galli SJ, Tsai M, Marichal T, Tchougounova E, Reber LL, and Pejler G. 2015. Approaches for analyzing the roles of mast cells and their proteases in vivo. Adv. Immunol. 126: 45–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wernersson S, and Pejler G. 2014. Mast cell secretory granules: armed for battle. Nat. Rev. Immunol. 14: 478–494. [DOI] [PubMed] [Google Scholar]
- 5.Miller HR, Wright SH, Knight PA, and Thornton EM. 1999. A novel function for transforming growth factor-beta1: upregulation of the expression and the IgE-independent extracellular release of a mucosal mast cell granule-specific beta-chymase, mouse mast cell protease-1. Blood 93: 3473–3486. [PubMed] [Google Scholar]
- 6.Wright SH, Brown J, Knight PA, Thornton EM, Kilshaw PJ, and Miller HR. 2002. Transforming growth factor-beta1 mediates coexpression of the integrin subunit alphaE and the chymase mouse mast cell protease-1 during the early differentiation of bone marrow-derived mucosal mast cell homologues. Clin. Exp. Allergy 32: 315–324. [DOI] [PubMed] [Google Scholar]
- 7.Knight PA, Brown JK, Wright SH, Thornton EM, Pate JA, and Miller HR. 2007. Aberrant mucosal mast cell protease expression in the enteric epithelium of nematode-infected mice lacking the integrin alphavbeta6, a transforming growth factor-beta1 activator. Am. J. Pathol. 171: 1237–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown JK, Donaldson DS, Wright SH, and Miller HR. 2003. Mucosal mast cells and nematode infection: strain-specific differences in mast cell precursor frequency revisited. J. Helminthol. 77: 155–161. [DOI] [PubMed] [Google Scholar]
- 9.Ghildyal N, Friend DS, Nicodemus CF, Austen KF, and Stevens RL. 1993. Reversible expression of mouse mast cell protease 2 mRNA and protein in cultured mast cells exposed to IL-10. J. Immunol. 151: 3206–3214. [PubMed] [Google Scholar]
- 10.Yamazaki S,Nakano N, Honjo A, Hara M, Maeda K, Nishiyama C, Kitaura J, Ohtsuka Y, Okumura K, Ogawa H, and Shimizu T. 2015. The transcription factor Ehf is involved in TGF-β-induced suppression of FcεRI and c-kit expression and FcεRI-mediated activation in mast cells. J. Immunol. 195: 3427–3435. [DOI] [PubMed] [Google Scholar]
- 11.Kitamura Y, Morii E, Jippo T, and Ito A. 2002. Effect of MITF on mast cell differentiation. Mol. Immunol. 38: 1173–1176. [DOI] [PubMed] [Google Scholar]
- 12.Kitamura Y, Morii E, Jippo T, and Ito A. 2002. Regulation of mast cell phenotype by MITF. Int. Arch. Allergy Immunol. 127: 106–109. [DOI] [PubMed] [Google Scholar]
- 13.Morii E, Oboki K, Ishihara K, Jippo T, Hirano T, and Kitamura Y. 2004. Roles of MITF for development of mast cells in mice: effects on both precursors and tissue environments. Blood 104: 1656–1661. [DOI] [PubMed] [Google Scholar]
- 14.Nishiyama C, Hasegawa M, Nishiyama M, Takahashi K, Akizawa Y, Yokota T, Okumura K, Ogawa H, and Ra C. 2002. Regulation of human Fc epsilon RI alpha-chain gene expression by multiple transcription factors. J. Immunol. 168: 4546–4552. [DOI] [PubMed] [Google Scholar]
- 15.Maeda K, Nishiyama C, Tokura T, Nakano H, Kanada S, Nishiyama M, Okumura K, and Ogawa H. 2006. FOG-1 represses GATA-1-dependent FcepsilonRI beta-chain transcription: transcriptional mechanism of mastcell-specific gene expression in mice. Blood 108: 262–269. [DOI] [PubMed] [Google Scholar]
- 16.Maeda K, Nishiyama C, Ogawa H, and Okumura K. 2010. GATA2 and Sp1 positively regulate the c-kit promoter in mast cells. J. Immunol. 185: 4252–4260. [DOI] [PubMed] [Google Scholar]
- 17.Baba Y, Maeda K, Yashiro T, Inage E, Kasakura K, Suzuki R, Niyonsaba F, Hara M, Tanabe A, Ogawa H, et al. 2012. GATA2 is a critical transactivator for the human IL1RL1/ST2 promoter in mast cells/basophils: opposing roles for GATA2 and GATA1 in human IL1RL1/ST2 gene expression. J. Biol. Chem. 287: 32689–32696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oda Y, Kasakura K, Fujigaki I, Kageyama A, Okumura K, Ogawa H, Yashiro T, and Nishiyama C. 2018. The effect of PU.1 knockdown on gene expression and function of mast cells. Sci. Rep. 8: 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kitamura N, Yokoyama H, Yashiro T, Nakano N, Nishiyama M, Kanada S, Fukai T, Hara M, Ikeda S, Ogawa H, Okumura K, and Nishiyama C. 2012. Role of PU.1 in MHC class II expression through transcriptional regulation of class II transactivator pI in dendritic cells. J. Allergy Clin. Immunol. 129: 814–824.e6. [DOI] [PubMed] [Google Scholar]
- 20.Nishiyama C, Yokota T, Okumura K, and Ra C. 1999. The transcription factors Elf-1 and GATA-1 bind to cell-specific enhancer elements of human high-affinity IgE receptor alpha-chain gene. J. Immunol. 163: 623–630. [PubMed] [Google Scholar]
- 21.Inage E, Kasakura K, Yashiro T, Suzuki R, Baba Y, Nakano N, Hara M, Tanabe A, Oboki K, Matsumoto K, et al. 2014. Critical Roles for PU.1, GATA1, and GATA2 in the expression of human FcεRI on mast cells: PU.1 and GATA1 transactivate FCER1A, and GATA2 transactivates FCER1A and MS4A2. J. Immunol. 192: 3936–3946. [DOI] [PubMed] [Google Scholar]
- 22.Dudeck A, Dudeck J, Scholten J, Petzold A, Surianarayanan S, Köhler A, Peschke K, Vöhringer D, Waskow C, Krieg T, et al. 2011. Mast cells are key promoters of contact allergy that mediate the adjuvant effects of haptens. Immunity 34: 973–984. [DOI] [PubMed] [Google Scholar]
- 23.Scholten J, Hartmann K, Gerbaulet A, Krieg T, Muller W, Testa G, and Roers A. 2008. Mast cell-specific Cre/loxP-mediated recombination in vivo. Transgenic Res. 17: 307–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wada T, Ishiwata K, Koseki H, Ishikura T, Ugajin T, Ohnuma N, Obata K, Ishikawa R, Yoshikawa S, Mukai K, et al. 2010. Selective ablation of basophils in mice reveals their nonredundant role in acquired immunity against ticks. J. Clin. Invest. 120: 2867–2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mullen AC, Orlando DA, Newman JJ, Lovén J, Kumar RM, Bilodeau S, Reddy J, Guenther MG, DeKoter RP, and Young RA. 2011. Master transcription factors determine cell-type-specific responses to TGF-β signaling. Cell 147: 565–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blokzijl A, ten Dijke P, and Ibáñez CF. 2002. Physical and functional interaction between GATA-3 and Smad3 allows TGF-βeta regulation of GATA target genes. Curr. Biol. 12: 35–45. [DOI] [PubMed] [Google Scholar]
- 27.Funaba M, Ikeda T, Murakami M, Ogawa K, Nishino Y, Tsuchida K, Sugino H, and Abe M. 2006. Transcriptional regulation of mouse mast cell protease-7 by TGF-beta. Biochim. Biophys. Acta 1759: 166–170. [DOI] [PubMed] [Google Scholar]
- 28.Funaba M, Ikeda T, Murakami M, Ogawa K, Tsuchida K, Sugino H, and Abe M. 2003. Transcriptional activation of mouse mast cell Protease-7 by activin and transforming growth factor-beta is inhibited by microphthalmia-associated transcription factor. J. Biol. Chem. 278: 52032–52041. [DOI] [PubMed] [Google Scholar]


