Abstract
An in-depth understanding of the genetics and evolution of brain function and behavior requires detailed mapping of gene expression in functional brain circuits across major vertebrate clades. Here we present the Zebra finch Expression Brain Atlas (ZEBrA; www.zebrafinchatlas.org, RRID: SCR_012988), a web-based resource that maps the expression of genes linked to a broad range of functions onto the brain of zebra finches. ZEBrA is a first of its kind gene expression brain atlas for a bird species, and a first for any sauropsid. ZEBrA’s >3,200 high-resolution digital images of in situ hybridized sections for ~650 genes (as of June, 2019) are presented in alignment with an annotated histological atlas and can be browsed down to cellular resolution. An extensive relational database connects expression patterns to information about gene function, mouse expression patterns and phenotypes, and gene involvement in human diseases and communication disorders. By enabling brain-wide gene expression assessments in a bird, ZEBrA provides important substrates for comparative neuroanatomy and molecular brain evolution studies. ZEBrA also provides unique opportunities for linking genetic pathways to vocal learning and motor control circuits, as well as for novel insights into the molecular basis of sex steroids actions, brain dimorphisms, reproductive and social behaviors, sleep function, and adult neurogenesis, among other fundamental themes.
Keywords: molecular signature, gene expression, brain organization, functional circuits, zebra finch, song system, brain atlas
Graphical Abstract
Introduction
A key step towards understanding the genetic basis of brain function and behavior is to map the expression of genes onto functional circuits and cells. The importance of brain gene expression mapping is well illustrated by the Allen Institute’s Mouse Brain Atlas, a resource that has allowed for the examination of >20,000 brain transcripts in a representative mammalian model organism (Hochheiser & Yanowitz, 2007; Lein et al., 2007; Sunkin et al., 2013). This atlas has made numerous contributions to our understanding of the molecular organization of the brain of mammals, including defining molecular signatures of major structures (e.g. cortical layers, hippocampus, amygdala), providing insights into developmental changes in gene expression, and facilitating discoveries linking genetic pathways to circuits that underlie specific functions and/or behaviors (e.g. Acquaah-Mensah & Taylor, 2016; Belgard et al., 2011; Bohland et al., 2010; Hawrylycz et al., 2010; Henry & Hohmann, 2012; Jones et al., 2009; Lein et al., 2017; Li et al., 2017; Sunkin et al., 2013; Thompson et al., 2014; Thompson et al., 2008).
A comparable resource has been missing up until now for birds. Considering that birds have been broadly used as experimental model organisms and have contributed important insights into brain function and behavior, the lack of such a resource constitutes a major gap in neurobiology. For example, birds are increasingly recognized as being capable of sophisticated learned behaviors (e.g. tool use, episodic memory), with corvids (e.g. crows, ravens, jays) and parrots possessing cognitive abilities that rival or even surpass those of mammals, including some non-human primates (e.g. Clayton & Emery, 2015; Gunturkun & Bugnyar, 2016; Jarvis et al., 2005; Pepperberg, 2006). Several avian groups (songbirds, parrots and hummingbirds) have also evolved vocal learning, a trait that enables animals to learn vocalizations by imitation, as extensively studied in zebra finches (Taeniopygia guttata). This trait evolved convergently in these avian vocal learners and in humans (Figure 1a, left, red asterisks), where it provides an important basis for speech and language acquisition, but is largely absent or rudimentary in most non-human mammals, including rodents and non-human primates (Jarvis, 2004, 2019; Petkov & Jarvis, 2012; Wirthlin et al., 2019). Birds also have highly developed visual systems and high visual acuity (e.g. Bennett et al., 1996; Gaffney & Hodos, 2003; Hodos et al., 2003; Martin, 2014; Shimizu et al., 2010), and seminal studies in species like chicken and guinea fowl, and hearing specialists like owls, have been at the forefront of discoveries on central auditory processing and the neurobiological basis of sound localization (Grothe et al., 2004; Hong & Sanchez, 2018; Knudsen & Konishi, 1978; Konishi, 2003; Langner, 1981; Overholt et al., 1992; Scheich & Bonke, 1981; Smith & Rubel, 1979; Theurich et al., 1984; Walton et al., 2017). Many bird species also have complex social behaviors and thus have been excellent models for understanding the neuronal basis of sociality (Goodson & Kabelik, 2009; Kelly & Goodson, 2014). Chicken and quail are also broadly used for development studies, and have contributed numerous insights into early brain patterning, axonal guidance and neurotrophins, among others (e.g. Fraser et al., 1990; Hamburger & Hamilton, 1951; Keynes & Cook, 2018; Keynes & Stern, 1984; Lance-Jones & Landmesser, 1980).
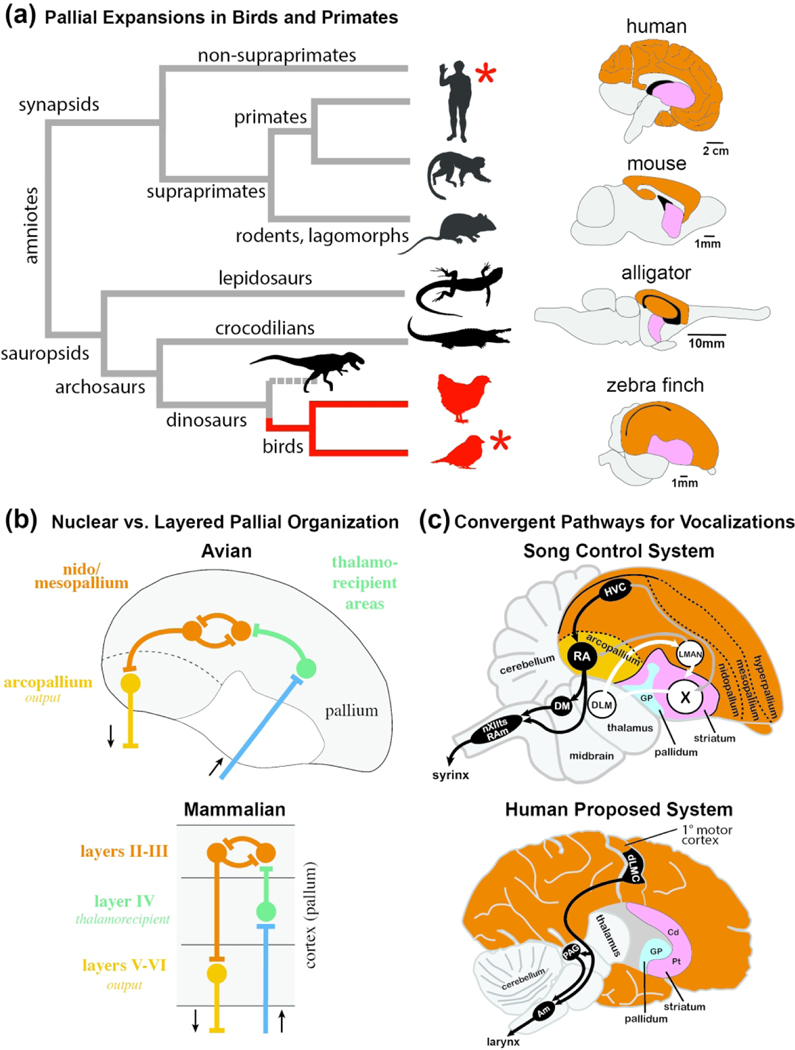
Figure 1. Comparison of brains and circuit organization across vertebrates.
(a) Simplified phylogeny and brain evolution in amniotes. Left: Schematic diagram depicting phylogenetic relationships across major amniote lineages; branches containing bird lineages are highlighted in blue. Only select major extant groups are depicted; dashed line indicates dinosaur lineage extinction. Red asterisks indicate species/groups that possess the trait of vocal learning. Right: Schematic diagrams of representative vertebrate brains highlighting the relative sizes of pallial (orange) and subpallial (pink) areas in representative synapsids and sauropsids. (b) Schematic generic representation of the ‘nuclear’ vs. ‘layered’ hypothesis of pallial organization in avian (top) and mammalian (bottom) brains (after Karten, 1991). Color coding indicates analogous (and possibly homologous) elements between lineages. (c) Schematic diagrams of specialized pathways for birdsong (top) and speech (bottom). Top: Vocal control circuitry of the adult male zebra finch, depicted in the parasagittal plane; some structures have been omitted for clarity. The system consists of a direct motor pathway for song production (in black) that includes connections from HVC to RA, and from RA to brainstem nuclei involved in vocal-motor and respiratory control (nXIIts, RAm), and an anterior forebrain pathway for song learning (in white) containing a cortico-striato-thalamo-cortical loop with projections from pallial LMAN to striatal Area X, from Area X to thalamic DLM, from DLM to LMAN, and from LMAN back to Area X. Additional connections are shown in grey. Bottom: Proposed vocal control circuitry in humans consisting of a direct motor pathway for speech production (in black), and striatal brain regions (Cd, Pt) that have been proposed to be involved in speech learning (in white), adapted from Pfenning et al. 2014. The descending projections from RA and dLMC to brainstem vocal nuclei are thought to be convergently evolved in songbirds and humans. Abbreviations: Am, nucleus ambiguus; Cd, Caudate nucleus of the striatum; dLMC, dorsal part of the laryngeal motor cortex; DLM, medial part of the dorsolateral nucleus of the anterior thalamus; GP, Globus pallidus; HVC, HVC proper name; LMAN, lateral magnocellular nucleus of the anterior nidopallium; nXIIts, tracheosyringeal portion of the nucleus of the hypoglossal nerve (XII); PAG, periaqueductal grey; Pt, Putamen; RA, robust nucleus of the arcopallium; RAm, nucleus retroambiguus medialis; X, Area X.
Notably, while birds possess highly developed cognitive and sensorimotor skills and share important brain features with mammals, their brain organization differs significantly from that of mammals. Birds belong to sauropsids (Figure 1a, left), a large group of amniotes that includes turtles, lizards, snakes, and crocodiles. While the telencephalon of birds and mammals is largely composed of pallium (Figure 1a right, orange area in brain diagrams) (Cobos et al., 2001; Fernandez et al., 1998; Jarvis et al., 2005; Puelles et al., 2000; Redies et al., 2001; Reiner et al., 1998), the avian pallium has expanded considerably relative to the basal ganglia when compared to non-avian sauropsids, similar to how the cortex is expanded in primates compared to more basal mammals (Figure 1a, right; pallium in orange, basal ganglia in pink across species). Intriguingly, bird brains contain more pallial neurons than equivalently-sized mammalian brains, and parrot and corvid brains contain on average twice as many neurons as comparably-sized primate brains (Olkowicz et al., 2016). Birds have a largely non-layered telencephalon (Jarvis et al., 2005; Karten, 2013), and their pallium has a more ‘nuclear’ organization compared to the layered cortex of mammals (Figure 1b). Nonetheless, birds have cortical-like circuitry elements such as distinct thalamo-recipient zones (e.g. auditory field L, visual entopallium) and nuclei that originate descending pallial projections (Mello et al., 1998; Wang et al., 2010; Wild, 1993; Zeier & Karten, 1971). These differences and similarities have raised questions about avian homologs of mammalian pallial structures like neocortex, claustrum, and amygdala (e.g., see Jarvis et al., 2005; Karten, 1997, 2013; Medina et al., 2004; Puelles et al., 1999; Reiner et al., 2004), as well as how the computational needs for sensory, motor, and integrative processing are met by brains with very different layouts (Guy & Staiger, 2017). Whether genetic mechanisms subserving layered vs. non-layered circuits are shared between mammals and birds, and more generally sauropsids, remains unclear despite previous efforts (e.g. Belgard et al., 2013; Dugas-Ford et al., 2012; Jarvis et al., 2013; Montiel et al., 2016; Puelles, 2017; Puelles et al., 2016; Suzuki & Hirata, 2014; Tosches et al., 2018), in part due to limited expression data in an avian species.
Birds also offer excellent opportunities for mapping molecular pathways onto circuits that subserve specific behaviors, largely facilitated by their unique brain features - discrete nuclei with distinct cytoarchitectonics. One of the best examples is the zebra finch, the premier model organism for studying the neurobiology of vocal learning (Fee & Scharff, 2010; Goodson et al., 2009; Mello, 2014; Zann, 1996; Zeigler & Marler, 2004, 2008). This songbird species shares with humans vocal learning features like a critical period, the presence of early babbling-like immature vocalizations, and the ability to modulate vocalizations during sensorimotor learning by reference to auditory feedback (Bolhuis et al., 2010; Doupe & Kuhl, 1999). While some mammals such as bats, marine mammals and elephants (Janik, 2014; Knornschild, 2014; Poole et al., 2005; Prat et al., 2015) may have evolved some of these features, finches are much more amenable to experimentation, and benefit from resources such as a high quality genome assembly and extensive cDNA/EST databases (Korlach et al., 2017; Li et al., 2007; Replogle et al., 2008; Wada et al., 2006; Warren et al., 2010). Most importantly, the finch vocal circuitry has been extensively characterized, and is comprised of: (a) a direct pathway containing discrete cortical pre-motor and motor areas and brainstem vocal and respiratory nuclei (Figure 1c, top, in black) involved in vocal-motor control (Nottebohm & Arnold, 1976; Nottebohm et al., 1982); and (b) an anterior forebrain pathway consisting of a cortico-striato-thalamo-cortical loop (Figure 1c, top, in white) that plays key roles in vocal learning and plasticity (Bottjer & Arnold, 1984; Brainard & Doupe, 2000a, 2000b; Kao et al., 2005; Scharff & Nottebohm, 1991; Sohrabji et al., 1990; Thompson & Johnson, 2007). This system bears remarkable analogies to human vocal areas (Figure 1c, bottom), particularly when considering the descending motor pathway. Knowledge of the organization and physiological properties of finch vocal nuclei has contributed key insights into the roles of cortex and basal ganglia in vocal learning and vocal-motor representation (e.g. Aronov et al., 2008; Dutar et al., 1998; Hahnloser et al., 2002; Kubke et al., 2005; Leonardo & Fee, 2005; Mooney, 2000; Mooney & Prather, 2005; Perkel et al., 2002; Rosen & Mooney, 2003) (see also chapters in Zeigler & Marler, 2004, 2008). Studies in the zebra finch have also been instrumental in elucidating the role of FOXP2, the first gene to be linked to inheritable speech disorders in humans (Burkett et al., 2018; Haesler et al., 2007; Haesler et al., 2004; Teramitsu et al., 2004; Wohlgemuth et al., 2014). Zebra finches have also been widely used to study themes such as the endocrine modulation of reproductive and social behaviors (Goodson & Kabelik, 2009; Kelly & Goodson, 2014), sex steroid actions and brain dimorphisms (Nottebohm & Arnold, 1976; Pinaud et al., 2006; Williams, 1985), the role of sleep in learning (Brawn et al., 2008; Dave & Margoliash, 2000; Hahnloser et al., 2006), how cortical and basal ganglia circuits contribute to motor coding and rhythms (Budzillo et al., 2017; Fee et al., 2004; Gadagkar et al., 2016; Goldberg & Fee, 2010; Hahnloser et al., 2002; Hisey et al., 2018; Leonardo & Fee, 2005; Roberts et al., 2012), adult neurogenesis and neuronal replacement (Alvarez-Buylla et al., 1990), behavioral modulation of gene expression (Drnevich et al., 2012; Hara et al., 2012; Hilliard et al., 2012a; Hilliard et al., 2012b; Li et al., 2007; Stevenson et al., 2012; Thompson et al., 2012; Wada et al., 2006; Whitney et al., 2014), and the role of endocannabinoids (Soderstrom & Johnson, 2003; Soderstrom & Tian, 2004) and ethanol (Olson et al., 2014) in addiction. However, the genetic basis of these functions has remained largely unexplored.
Previous studies have identified specializations of the finch vocal circuitry through focused microarray and in situ hybridization screenings (Li et al., 2007; Lovell et al., 2008; Lovell et al., 2018a; Wada et al., 2004), but there have been no concerted efforts to expand in situ datasets, to study brain-wide gene expression, to present data under a unifying platform, or to generate tools for correlational and comparative analyses. To address these gaps and further our knowledge about the genetic basis of brain function and behavior, we have mapped a large number of brain transcripts in zebra finches and documented the differential expression of genes in vocal nuclei and broader areas. Here we describe the Zebra finch gene Expression Brain Atlas (ZEBrA), an open access website (www.zebrafinchatlas.org, RRID: SCR_012988) that contains >3,200 high-resolution digital in situ hybridization images for ~650 brain transcripts, presented in registration with an annotated histological reference atlas. This atlas is the first of its kind for an avian species, and more generally the first for a sauropsid. To facilitate comparative neuroanatomy, genomics and bioinformatics, ZEBrA includes extensive metadata on gene involvement in speech and language function, neurological and psychiatric diseases and phenotypes, brain expression in mouse, regulatory roles in brain development and function, and links to songbird-related discoveries and to key mammalian databases (Allen Mouse Brain Atlas, OMIM, MGI). Among studies that have already benefited from ZEBrA, we note findings on avian brain organization that give insights into cortical and amygdalar evolution (Jarvis et al., 2013; Mello et al., 2019), as well as the discoveries of convergent specializations of vocal areas shared by vocal learning birds and humans (Pfenning et al., 2014), compensatory changes in response to a lineage-specific loss of a major neuromodulatory gene (Lovell et al., 2015; Lovell et al., 2014), and vocal circuitry specializations related to neuronal excitability regulation (Friedrich et al., 2019; Lovell et al., 2018a). ZEBrA is thus a unique resource that enables exploring brain gene expression patterns in birds, facilitates comparative analyses between birds and other vertebrate lineages, and enables linking molecular pathways to a range of fundamental questions in neurobiology and brain evolution.
Methods
Gene Selection for Portals
The strategy and inclusion criteria used to build the six ZEBrA portals are detailed below. While the criteria are independent across portals, every gene in ZEBrA has been systematically examined to verify if it meets the criteria for inclusion in each portal. As new genes are added to ZEBrA, they are cross-referenced to the primary list of gene entries and/or external databases specific to each portal as described below, so that the contents of each portal/subportal can be updated.
(1). Markers of the Song System.
To build this portal, we examined all relevant in situ hybridization images of adult male zebra finches available in ZEBrA to identify genes that are differentially up- or down-regulated in one or more song nuclei compared to adjacent brain areas, based on visual inspection of images. For some nuclei, we further subcategorized genes that showed sparse cellular expression (e.g. sparse cells in HVC, area X, or RA) and/or were expressed within a restricted subdomain within a nucleus (e.g. paraHVC). For LMAN, a so-called shell region can be distinguished molecularly from a core domain or central nucleus. This LMAN shell has distinct connectivity (Bottjer et al., 2000; Bottjer et al., 1989; Johnson et al., 1995) and has been considered by some a non-vocal motor area (e.g. Feenders et al., 2008), or an auditory area (e.g. Bottjer et al., 2000; Johnson et al., 1995), while other recent studies have suggested a role in vocal learning (Achiro et al., 2017; Bottjer & Altenau, 2010). We use the term LMAN shell as an neuroanatomical description without any implications of function or possible analogy with the shell of the oval nucleus of the anterior nidopallium (NAo), which is considered the parrot equivalent to songbird LMAN. With regards to DLM, a large number of ZEBrA patterns show differential expression in a discrete the dorsal thalamic nucleus that receives projections from striatal Area X, projects to LMAN (Johnson et al., 1995; Luo et al., 2001), and exhibits singing-related up-regulated of DUSP1 (Horita et al., 2012), suggesting a prominent role in vocal learning. This nucleus has been termed DLM in most studies by several labs and has been sometimes referred to as aDLM (after Horita et al., 2012; Kubikova et al., 2010; Wada et al., 2004). Some ZEBrA genes show differential expression in a shell-like region that seems to circle DLM as defined above, and/or in a larger that likely encompasses larger and/or additional nuclei of the dorsal-medial thalamus. It is possible that in some cases this may correspond to the dorsal thalamic region that projects to the LMAN shell (Johnson et al., 1995) and/or that has been labeled DLM to differentiate it from the aDLM (Horita et al., 2012; Kubikova et al., 2010; Wada et al., 2004). A clear definition of this complex region requires efforts beyond the scope of the present paper, but we have taken note of these cases by placing an Unknown Thalamus label over it whenever relevant. A complete list of song nuclei and respective subdomains is presented in SuppInfo:STable1.xlsx. The majority of genes in this portal were identified as candidate markers of song nuclei HVC, RA, or Area X, based on differential microarray screenings (Li et al., 2007; Lovell et al., 2008; Lovell et al., 2018a; Pfenning et al., 2014; Wada et al., 2004). However, every gene in ZEBrA was also evaluated for differential expression in the song system, and has been included in this portal as applicable.
(2). Speech and Language:
To build this portal, we consulted our custom Human Disorder classification tree, which was based on the Online Mendelian Inheritance in Man (OMIM; Dec., 2017) catalog of human genes and genetic disorders. Construction of this custom classification tree is described below under (3) Diseases and Phenotypes. The part of this classification tree that is represented in this portal contains the full set of human genes that were classified as having an association with speech, language, communication, and/or an autism-related disorder. In addition, we consulted the Simons Foundation Autism Research Initiative (SFARI; Q2 Release Oct. 2017) website and identified genes listed in that website at ‘High Confidence’, ‘Strong Candidate’, or ‘Suggestive Evidence’ levels of linkage to autism spectrum disorders. We also included a number of genes that have been highlighted in the literature as being involved (or theoretically involved) in disorders like specific language impairment, dyslexia, childhood dyspraxia, stuttering, and psychiatric disorders with strong language components like schizophrenia. We also included genes that have been linked to speech and language through their shared expression in human and avian vocal circuits (Pfenning et al., 2014). A complete list of speech and language disorders, highlighted genes, and their parent level organizing themes is presented in SuppInfo:STable1.xlsx. The collection of genes in this portal represents an exhaustive effort to list all candidate genes with functions related to speech and language, thus it includes human genes that have not been identified in the genome of zebra finch and/or of birds in general, or for which in situ probes are not currently available.
(3). Diseases and Phenotypes:
To build the Human Diseases subportal, we downloaded OMIM’s Synopsis of the Human Gene Map file (morbidmap.txt; Dec., 2017), which contains a complete listing of genetic disorders and gene associations presented in OMIM. As of December 2017, this corresponded to 11,705 genes, which are associated with a total of 6,668 unique human disorders. We then cross-referenced all genes in ZEBrA with this human gene list. Because OMIM does not provide a hierarchical classification of disorder types, we next constructed a custom Human Disorder classification tree containing 53 unique disorder themes or subthemes that are represented in the ZEBrA database. Specifically, we classified genes in ZEBrA as being associated with ‘Neurological/Psychiatric Disorders’ (n=29 themes), of which we highlight a subset where the associated genes are more highly represented in ZEBrA (n=6 themes: epilepsy, mental retardation, neurodegeneration, ataxia, neuropathy, addiction), or with ‘Other Disorders’ (n=22 themes) which have no obvious neurological and/or psychiatric association. We note that to build this classification each OMIM entry related to a given gene in ZEBrA was determined to belong to one or multiple themes/sub-themes based on the presence of related keywords in the disorder description (e.g. epilepsy, deafness) or an interpretation of the description itself. A complete list diseases, disorders, and their parent level thematic classifications is presented in SuppInfo:STable1.xlsx. To build the Mouse Phenotypes subportal we first downloaded the file: MGI_GenePheno.rpt (Dec. 2018) from the Mouse Genome Informatics (MGI) database. This file contains coded descriptions of specific brain and behavioral phenotypes obtained from gene deletion, knock-down, and/or overexpression studies in experimental mouse models. As of December 2018, this consisted of 3,212 genes related to 1,347 unique phenotypes. Next, we cross-referenced the full set of ZEBrA genes with the genes in this Mouse Phenotype database. Using the MGI Mammalian Phenotype Vocabulary classification system as a starting point, we next constructed a Mouse Phenotype classification tree based on genes that are represented in the ZEBrA database. This classification tree is organized around major phenotypic themes of ‘Abnormal Behavior’ or ‘Brain Morphology Or Physiology’, with a total of 648 phenotypic sub-themes. A complete list of phenotypes and their parent level classifications is presented in SuppInfo:STable1.xlsx. We note that the specific entries in the OMIM and MGI databases for the corresponding genes in ZEBrA can be accessed through the linkouts in this portal.
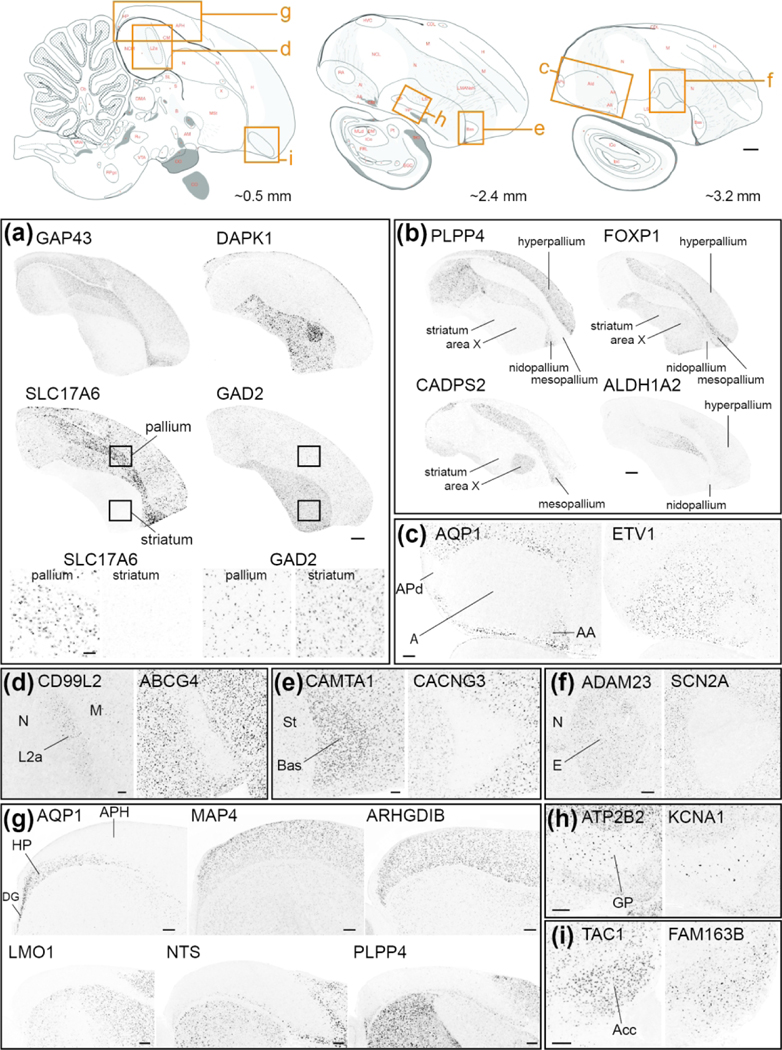
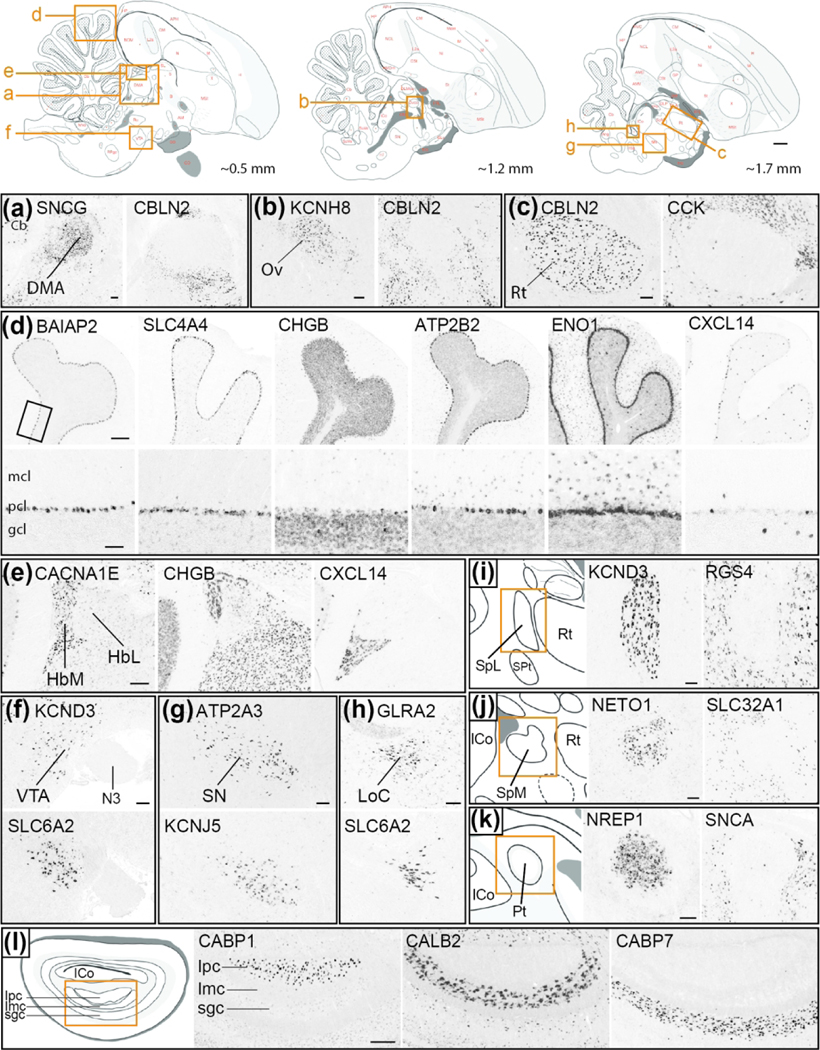
(4). Comparative Neuroanatomy:
To build the Songbird Brain Markers subportal, we systematically examined in situ hybridization images for every gene in ZEBrA, and identified subsets that are differentially up- or down-regulated in various general avian brain areas (other than song control nuclei) compared to adjacent brain areas or subdivisions. A complete list of anatomical structures and their parent level organization within the Songbird Brain Marker subportal is presented in SuppInfo:STable1.xlsx. We note that for n. Ovoidalis (Ov) and for the Entopallium (E) we present markers of both core and shell regions. For the dorsolateral corticoid area (CDL) we present markers of both the entire region and a small and cytoarchitectonically distinct nucleus within the CDL that we have named the core of CDL (CDLco), following the convention used in the chicken stereotaxic atlas (Puelles, 2007). We note that this core nucleus has also been referred to as the dorsal nucleus of the hyperpallium (DNH) by researchers studying night vision and magnetoreception in finches and other birds (e.g. Heyers et al., 2007; Mouritsen et al., 2005; Mouritsen et al., 2016). However, we believe there is currently not conclusive evidence placing this nucleus within the hyperpallium. To build the Mouse Brain Markers subportal, we first constructed a database of molecular markers of mouse brain structures comprised of lists of genes that have been identified by the Allen Mouse Brain Atlas as differential markers of specific cortical layers, basal ganglia, hippocampal and amygdalar subdivisions, as well as a specific thalamic and neuromodulatory cell groups and/or nuclei, among others. For most structures, we retrieved pre-populated list of genes provided by the “Browse by Differential Expression” tool located on the ISH Data page at mouse.brain-map.org. We used default search settings and initially retrieved the 100 top and 100 bottom genes with the largest fold-changes. We next verified the retrieved lists by examination of the Mouse Brain Atlas patterns for genes with fold-change close to 1, and adjusted our cutoff further if genes in this range were identified as non-differential for the structure under consideration. For structures of interest not listed in “ Browse by Differential Expression” tool, we retrieved pre-populated markers sets in the “Fine Structures Search”. In such cases, the lists typically contained less than 50 markers and all markers were retrieved. Next, we cross-listed every gene in ZEBrA against the retrieved mouse brain structure gene lists, and populated the subportal tree accordingly. A complete list of anatomical structures along with their parent level organization within the Mouse Brain Marker subportal is presented in SuppInfo:STable1.xlsx.
(5). Gene Function:
To build this portal, we first adapted the general classification scheme (based on the Gene Families Index of the Human Genome Nomenclature Consortium - HGNC), modifying it to focus on molecular functions of relevance to brain circuits and the regulation of behavior. This includes several aspects of brain development (e.g. regulators of neurite extension, axonal guidance and fasciculation, cell:cell and cell:matrix adhesion, synapse formation, among others) and physiology (neurotransmitter and neuromodulatory receptors, peptides and respective receptors, synaptic function modulators, among others). We also included categories like intracellular signaling and trafficking, gene expression regulation, immune function, myelin-related, and genes that affect neuronal excitability. Other more general categories relate to various aspects of metabolism and gene function. A complete list of gene functions along with their parent level of organization is presented in SuppInfo:STable1.xlsx. We next categorized ZEBrA genes based on this classification scheme. In many cases this was straightforward, as several functional categories are tightly linked to the names of specific genes and/or gene families. In other cases this gene categorization required evaluating Gene Summaries provided by NCBI:Gene, GeneCards.org, and HGNC, or conducting literature searches in NCBI:Pubmed. Genes known to have multiple functions have been listed under multiple categories within this portal.
(6). Songbird Discoveries:
For the ‘Highlighted ZEBrA-Based Discoveries’ subportal, we identified songbird studies that have benefitted from the gene expression patterns available in ZEBrA to derive novel insights into comparative and physiological questions. For the ‘Songbird Studies’ subportal, we identified fundamental themes to which research in zebra finches and other songbirds has contributed novel and basic insights. This is just our own sampling of interesting or important studies in songbirds, and this expanding portal can easily accommodate expansions as more studies are published that reveal functional links for genes in the ZEBrA database, and/or as the ZEBrA database itself expands. Genes within these subportals are arranged first by major theme, and then by a reference in the literature that provides the evidence for the gene inclusion within a given theme. A complete list of themes and their parent level is presented in SuppInfo:STable1.xlsx.
Identification of Zebra Finch Orthologs and Gene Model Curation
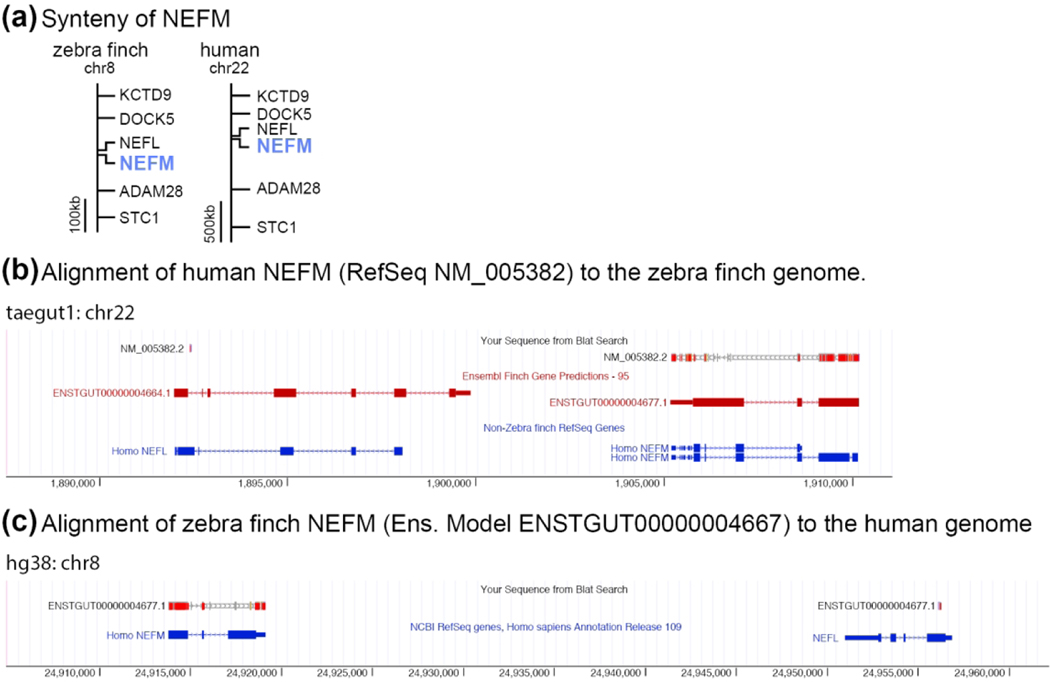
All genes presented in ZEBrA, including those not within a portal, have been individually and systematically vetted to verify their identity and orthology to mammalian genes using a combination of genomic alignments followed by synteny analysis (verification of flanking genes) in available high quality avian genomes (zebra finch, chicken), well assembled/curated mammalian genomes (human, mouse), and when necessary some out-groups (platypus, anole lizard, alligator, xenopus frog). Our curation pipeline consisted of the following steps. We first identified a candidate locus for each gene of interest in the zebra finch genome (taeGut3.2.4) by searching for either an Ensembl (v73.1-v94) or NCBI’s RefSeq gene model with the gene name of interest. We next examined the alignment of the human ortholog to the zebra finch genome by BLAT (UCSC genome browser; Kent, 2002). At this step we examined every locus in the genome with a significant alignment (i.e. BLAT score > 30) to distinguish the putative ortholog from hits to paralogs or related gene family members that may be similar in sequence. From this set of alignments we identified the orthologous locus by verifying which locus in the zebra finch genome with a significant BLAT alignment had the same local synteny (i.e. flanking genes) as the correct locus in the human genome. In cases where synteny was partial due to chromosomal rearrangements or regions where the genome is incomplete, verification was extended to other taxa related to finch and humans (e.g. chicken, mouse, platypus) or to representative species of more ancestral lineages or outgroups to birds and mammals (e.g. alligator, lizard, xenopus, zebra fish, as needed). Finally, we confirmed each locus by reciprocal best BLAT (or BLAST) alignments. The combination of reciprocal best alignments supported by conserved synteny provides the best evidence that a given locus in zebra finch is orthologous to that in humans and mouse. As an example, the medium weight neurofilament (NEFM) gene has nearly identical syntenies on zebra finch chr8 and human chr22 (Figure 2a). However, BLAT alignments of NEFM predictions from finch and human show the highest reciprocal alignment and nucleotide % identity scores to the correct corresponding loci (finch on human BLAT score = 1154, % identity = 87%; human on finch BLAT score = 1085, % identity = 88%; cross-species alignments shown in Figure 2b–c). No similarly high scoring secondary alignments (e.g. paralogs) exist, confirming that the finch and human NEFM loci are orthologous. We note that applying this strategy to this and other molecular studies in finches has resulted in the annotation of a large collection of unannotated Ensembl models, as well as the curation of numerous others that were either incorrectly annotated, or whose orthology could be unambiguously established (Lovell et al., 2013; Lovell et al., 2018b; Lovell & Mello, 2018; Wirthlin et al., 2014). We also note that for cases where a finch locus did not have a predicted model, we provide the chromosomal location where the finch ortholog was identified. For several genes with expression patterns presented in ZEBrA no clear orthologs could be identified in other species. This includes possible novel genes that may be unique to zebra finches or to broader groups like finches, songbirds, or even birds in general. These genes are currently named UNKNOWN in ZEBrA, a terminology that may change as these genes become identified, characterized, and/or named through research efforts.
Figure 2. Gene Orthology Verification.
(a) Schematic depiction showing how conserved synteny establishes the one-to-one homology of zebra finch and human NEFM. (b-c) Screenshots from UCSC’s genome browser showing how NEFM gene predictions from zebra finch (Ensemble model ENSTGUT00000004667) and human (Refseq NM_005382.2) align to the correct loci with the highest reciprocal alignment scores. (b) Human NEFM (NM_005382.2) aligns to a region of the zebra finch genome containing NEFM (ENSTGUT00000004677.1) and NEFL (ENSGUT00000004664.1). The alignment of human NEFM has a higher scoring alignment, and more completely covers (i.e. higher exon coverage) the NEFM locus, than the NEFL locus (BLAT scores = 1085 vs. 25, respectively). (c) Zebra finch NEFM (ENSTGUT00000004677.1) aligns to a region of the human genome that contains NEFM and NEFL. The alignment of zebra finch NEFM model has a higher scoring alignment, and more complete coverage for the NEFM locus than for NEFL locus (BLAT scores = 1154 vs. 65, respectively).
Clone and cDNA template Selection
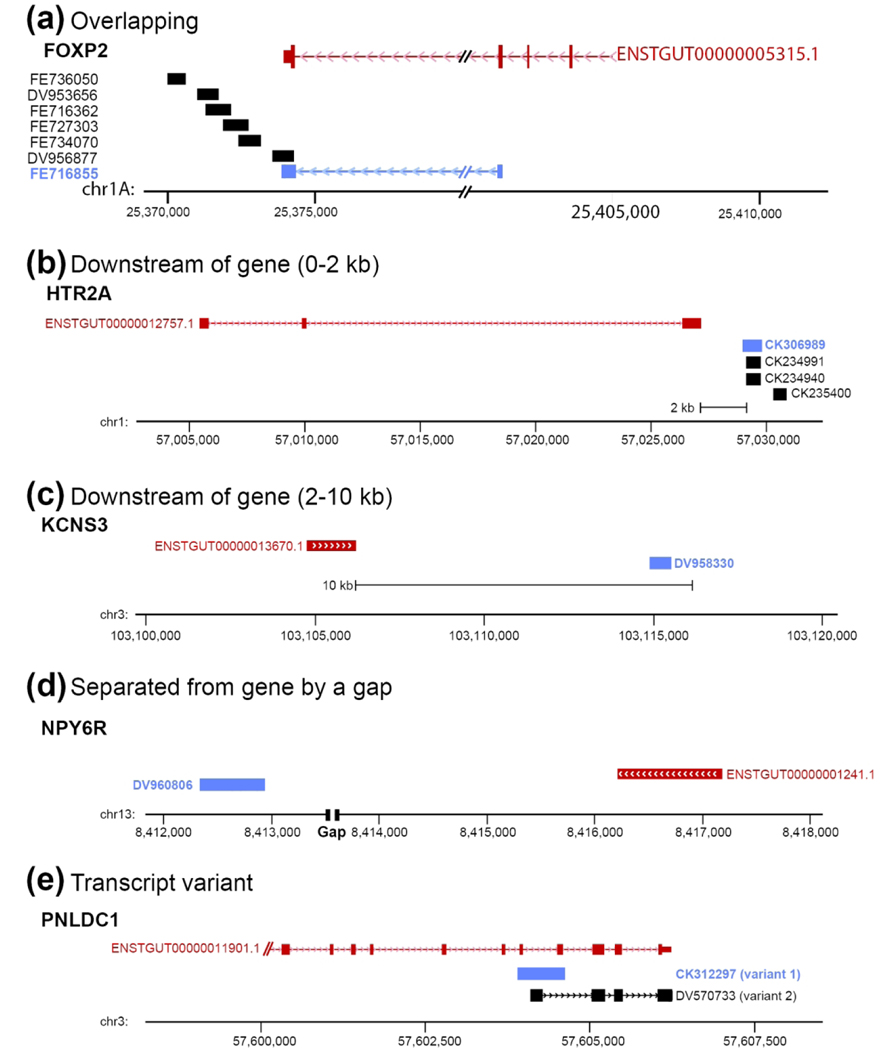
Unless otherwise indicated, all probes were derived from the zebra finch brain cDNA collection generated through the Songbird Neurogenomics Consortium (ESTIMA collection, all ESTs have been deposited in GenBank; details in Replogle et al., 2008). For most genes in ZEBrA we selected a single cDNA clone that is fully or partially overlapping and/or contiguous with the predicted Ensembl gene model based on a series of EST reads, or the corresponding model in chicken (via cross-BLAT alignment). Probes generated from clones of this type are labeled ‘Overlaps gene’ in the Gene Info page (example in Figure 3a). In some cases, we selected clones that were non-overlapping, but found to align within 2 kb of the 3’-end of a gene model. Given the close proximity of these reads to the gene model, evidence of polyadenylation (i.e. poly-A tail), and correct orientation to the +/− DNA strand we are reasonably confident that these cDNAs correspond to the 3’-end of the gene of interest. These clones are labeled ‘Downstream of gene (0–2 kb)’ (e.g. see HTR2A locus in Figure 3b). In other cases, the clones met the other criteria above but were located further downstream, and thus are labeled ‘Downstream of gene (2–10 kb)’ (e.g. see KCNS3 locus in Figure 3c). In some rare cases the only available clone was non-overlapping and separated from the 3’-end of the gene by a gap (or in some instances a few gaps) in the genomic sequence. Since the size of these gaps is not known, we could only tentatively assign these clones to a given gene. Such clones are labeled ‘Separated from gene by a gap’ (e.g. see NPY6R locus in Figure 3d). Clones of the latter two types were only used when no higher confidence alternatives were available. We also identified cases with evidence of alternative splicing, resulting in different transcripts with unique exon arrangements. These are labeled ‘Transcript variant’ (see PNLDC1 locus in Figure 3e). Whenever possible we selected clones consisting primarily of 3’-UTR sequence, since compared to coding sequence these regions are less likely to contain conserved domains present in related members of gene families, although we note that several clones contain some stretches of coding sequence. To confirm clone specificity, we systematically performed BLAT alignments of the selected ESTs to the zebra finch genome. The majority of ESTs aligned unambiguously with high scores to a single locus. In a small number of cases an EST aligned to secondary loci, but the secondary alignment scores were typically <50%, indicating a high likelihood of probe specificity. ZEBrA also includes some probes derived from clones that are not associated with a model in the genome (e.g. novel genes, or known orthologs not predicted by Ensembl or RefSeq). In a few cases where no ESTIMA clones were available, we generated template by RT-PCR cloning.
Figure 3. In Situ Probe Classification.
(a) Example of overlapping probe type: schematic depiction of genomic region for the zebra finch FOXP2 gene (based on UCSC’s genome browser) shows that clone FE716855 (Genbank ID shaded in red) was selected because it overlaps with coding and non-coding (3’-UTR) portions of the FOXP2 Ensembl Gene model (ENSTGUG00000005315.1). The EST for this clone also aligns unambiguously to this locus. Therefore, the Probe Location in the FOXP2 Gene Info page is listed as ‘Overlaps gene’. We note that the FOXP2 gene and related clones are on the minus strand (indicated by arrowheads in the gene model and selected clone alignments). (b-d) Examples of non-overlapping probes according to clone location relative to the gene. In (d), ‘Gap’ refers to a gap present in the genome assembly (taegut1). (e) Example of apparent spliced variants, where only one variant (CK312297) was analyzed. In all panels, Ensembl models are in red, clones selected for in situs are in blue, and orientation of gene and related clones are indicated by the arrowheads in the model and/or alternative variant clone. For each gene in ZEBrA the chromosomal location of the selected clone is listed under Probe Location in the Gene Info Page
Probe preparation and in situ hybridization
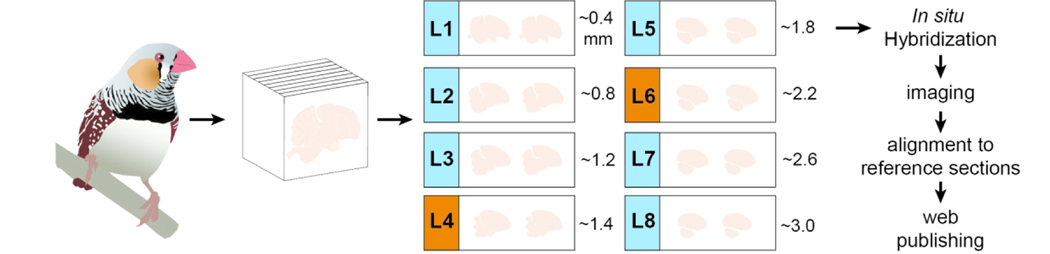
The steps involved in conducting in situ hybridization on zebra finch brain sections, including the preparation of cDNA templates, synthesis of DIG-labeled riboprobes, hybridization and post-hybridization washes, immunohistological detection of DIG probes, chromagen development, and section imaging and processing are described in detail in Carleton et al., 2014 (see also Lovell et al., 2013; Lovell & Mello, 2011). An overview of the basic pipeline we use to generate data for ZEBrA is presented in Figure 4.
Figure 4. In situ pipeline.
Schematic representation of ZEBrA’s in situ hybridization pipeline. Serial cryostat brains sections encompassing most major brain structures are hybridized at close intervals (e.g. L1–8, in blue) for genes that exhibit highly differential patterns, or at select levels (e.g. L4/L6, in vermillion) for non-differential genes, alignment to reference sections from the histological atlas, and ZEBrA web publishing.
Animal and tissue preparation
All birds used in the preparation of ZEBrA were adult male zebra finches (Taeniopygia guttata) obtained from our own breeding colony or purchased from local breeders. All birds were first isolated overnight (12:12-hour light-dark cycle) in custom-built acoustic isolation chambers to reduce non-specific auditory stimulation. On the following morning (~9:00 AM) birds were monitored briefly (~15 min) to confirm they were awake, not singing and not stimulated by song, and then sacrificed by decapitation. Brains were quickly dissected out, split along the midline, and each hemisphere placed in a plastic mold, covered with Tissue-Tek OCT compound (Sakura-Finetek; Torrance, CA), and frozen in a dry ice/isopropanol bath.
ZEBrA in situ Pipeline
Brains were sectioned on a cryostat (10 μm; Leica CM1850), and two consecutive sections mounted per slide to allow for a replication of hybridization in adjacent sections. To minimize the number of zebra finches required for the project, we used both hemispheres interchangeably and did not systematically compare signal across hemispheres. However, the hemisphere used was coded in metadata that is linked to each slide and hybridization experiment, and can be made available to researchers interested in asymmetric brain gene expression.
For every probe we first run an initial hybridization utilizing our standard protocol, on a pair of test slides at 65°C (hybridization and post-hybridization wash temperatures). Following a 3-day incubation in chromogen (BCIP/NBT), slides were washed, coverslipped, and individually evaluated under brightfield to assess both signal strength and background labeling. With some few exceptions, labeling was determined to be strong when the deposition of chromogen was distributed in a characteristic donut-shaped pattern surrounding a central nucleus devoid of signal. Background was deemed low when the tissue overall, including somata and neuropil, as well as the majority of cells within major fiber fields and/or ependymal cells of the lateral ventricle were devoid of labeling. This initial test allowed us to determine best hybridization conditions and provided an initial assessment of brain distribution patterns. If necessary, before each probe was hybridized to a final set of brain sections to be imaged, optimization steps were run to establish hybridization stringency conditions that gave the highest possible signal and lowest background. For instance, if signal was undetectable or deemed too low for imaging in the first test, the hybridization was repeated at 63°C (lower stringency). If the signal in the first test was deemed strong, but the background was also high, the hybridization was repeated at 67° or 69°C to raise the hybridization stringency. In rare cases where a hybridization at 69° C did not reduce background labeling, an additional RNAse H step was applied after the post-hybridization washes. Genes that showed no detectable signal after several attempts to hybridize at lower stringency are presented without images, and labeled “No Signal Detected”. Some notable exceptions to the pipeline above consist of genes that are actually expressed in the aforementioned control structures. These often represent markers of astroglia, oligodendrocytes, and/or ependymal or pial cells, however in such cases the labeling in these control areas resisted and/or became more distinct under higher stringency conditions. In a few other cases strong expression that resisted high stringency conditions occurred in both the somata/neuropil and control structures, representing ubiquitously expressed genes. We also note a few cases where the labeling pattern had a nuclear localization that resisted high stringency conditions.
Genes that exhibited highly differential expression patterns in identified brain nuclei, divisions, and/or cellular fields were subsequently hybridized to a final series. This consisted of sections at 4–8 levels spaced ~0.2 – 4.0 mm from midline (represented schematically in Figure 4, levels L1-L8 in sky blue), encompassing most of the major brain subdivisions, cellular fields, fiber tracts, and nuclei, including the major telencephalic song nuclei (i.e. HVC, RA, LMAN, Area X) and auditory areas (Field L and adjacent parts of the caudal nidopallium and mesopallium). In contrast, genes with relatively non-differential or flat patterns of expression were hybridized to sections at 2–3 levels spaced between ~1.0 and 2.4 mm from midline, but still containing all major telencephalic song nuclei and auditory areas (represented schematically in Figure 4, levels L4 and L6 in vermillion). Since the various steps above were performed in different brains, we typically examined brain expression for each probe in 2–4 birds, providing replication for the expression pattern presented in ZEBrA. We note that the ZEBrA pipeline incorporated a number of quality control measures as well, including: (1) the use of gel electrophoresis to confirm cDNA template quantity and quality, (2) the inclusion of positive control slides hybridized with GAD2, and negative (i.e. no probe) control slides with each hybridization, (3) visual monitoring of the chromogen precipitation reaction, and (4) the custom selection of individual brain sections for imaging. Further variables and issues that are critical for generating high quality in situ hybridization data during high-throughput efforts like ZEBrA are discussed in Carleton et al (2015).
Image Preparation
Completed slides were imaged with a Nanozoomer 2.0HT digital slide scanner (Olympus; Center Valley, PA) in the Mitra lab at Cold Spring Harbor Laboratory and more recently with a Leica Aperio AT2 scanner at the Knight BioLibrary at OHSU. Images were converted from their native Nanozoomer NDPI file into compressed JPEG format. We next used Photoshop CS5 (Adobe; San Jose, CA) to adjust the contrast, brightness, and color balance of each image, and also to correct for any artifacts introduced during slide processing (e.g. scratches, chromagen precipitate). Additional color-balancing was performed across each image series to ensure that background levels were similar across brain sections. Such manipulations were kept minimal and restricted to image regions not containing labeled cells, in order to avoid any misrepresentation of results. Each image was then rotated to approximately match one of the eighteen levels selected from the series of line drawings derived from the histological atlas of the zebra finch brain (Karten et al., 2013). Finally, each image was tiled using Zoomify (Aptos, CA), which converts the large image format into a series of smaller tilesets that can be reconfigured and viewed like a mosaic. Final image series were then uploaded to the ZEBrA webserver at www.zebrafinchatlas.org (RRID: SCR_012988).
Neuroanatomical Marker Annotation
Once posted to the ZEBrA website, each gene series underwent an additional curation step to qualitatively assess whether the gene could be considered a molecular marker for a given structure or region. Using a combination of annotated brain drawings, information derived from other brain atlases (e.g. Pigeon, Karten & Hodos, 1967; Canary, Stokes et al., 1974; Chick, Kuenzel, 1988; Puelles, 2007; Dove, den Boer-Visser, 2004; Zebra finch, Nixdorf-Bergweiler B.E. & Bischof, 2007), and expression data from genes that define known cell populations (e.g. glutamatergic or GABAergic cells), we scored any structure that showed differential mRNA expression as being upregulated (+) or downregulated (−) relative to the surrounding brain tissue. These results were used to populate a Neuroanatomical Marker table subset that is queried when exploring the Markers of the Song System and Bird Brain Markers portals. Each structure that showed differential gene expression was assigned an appropriate anatomical label. These labels can be viewed when the “Labels ON” button is selected in the navigation cluster on the lower right of the In Situ Image Browser.
Results
The ZEBrA database and website
The Zebra finch Expression Brain Atlas (a.k.a. ZEBrA; www.zebrafinchatlas.org, RRID: SCR_012988) is a public access in situ hybridization database that systematically documents the brain distribution of a large collection of genes in an avian species (Figure 5). It consists of high resolution digital images presented in alignment with an annotated histological brain atlas, together with genomics and bioinformatics resources. While an expanding database, ZEBrA currently contains brain expression data for ~650 genes in adult male zebra finches (as of June, 2019), covering a range of gene families and pathways of relevance for brain function, development, and behavior. The data were generated via our in situ pipeline (Figure 4), using a high-throughput non-radioactive protocol optimized for low background and high sensitivity (Carleton et al., 2014). As our main goal is to determine the constitutive expression of genes, the data derive from awake non-singing and not song-stimulated male zebra finches.
Figure 5. ZEBrA: The Zebra finch Expression Brain Atlas.
Image of the homepage of the ZEBrA website (www.zebrafinchatlas.org, RRID: SCR_012988), an online public repository of high-resolution in situ hybridization images for a large collection of genes that are expressed in the brain of the adult male zebra finch.
The Reference Atlas and In Situ Images
The Histological Reference Atlas
The Histological Reference Atlas
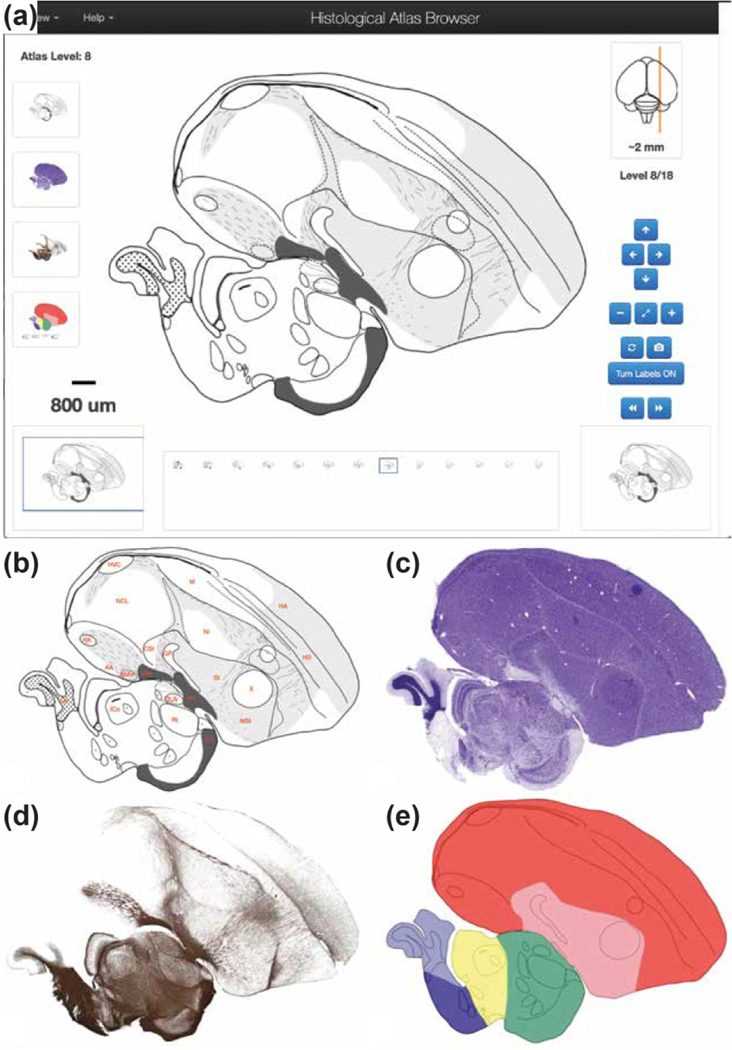
(http://www.zebrafinchatlas.org/gene_display/histological-atlas) consists of images and corresponding drawings from eighteen brain levels covering the main auditory and vocal areas, as well as major subdivisions and structures of the avian forebrain, brainstem, and cerebellum. The images are derived from a set of alternating serial parasagittal sections that were Nissl-stained to visualize cytoarchitectonic features and landmarks, or myelin-stained to visualize fiber tracts (Karten et al., 2013). The complete set of images in ZEBrA can be visualized in the Histological Atlas Browser by clicking on the Nissl image icon located on the left side of the Image Browser or homepage (Figure 6a). The section icon on the lower left indicates the size, position and type of the main image shown in the browser (blue rectangle) relative to the whole section, helped by the scale bar above the section icon. A cluster of clickable and self-explanatory buttons on the lower right of the browser provides options for navigation. When “Turn labels ON” is active, anatomical structure abbreviations or dots (for small structures) appear over the image (e.g., Figure 6b); the full name for each structure is viewed by hovering over that label. When labels are visible, a hyperlink beneath the image provides access to the Nissl image for that level at the repository website for the images (brainarchitecture.org; Karten et al., 2013), where other parasagittal levels for this brain can also be examined. Images of the alternating Nissl- and myelin-stained sections are also included (e.g., Figure 6c–d), and a colorized version indicating major brain divisions and subdivisions (e.g., Figure 6e) can be viewed by clicking the respective icons to the left within the browser window.
Figure 6. The Histological Atlas Browser.
(a) View of the Browser depicting an example of a drawing from one selected level of the Histological Atlas. The full set of parasagittal levels currently available is shown as thumbnails at the bottom of the browser. The thumbnail icons on the left allow users to toggle across each of the four images types. The position of the section being viewed relative to the midline is illustrated by the orange line on the dorsal-view schematic drawing of the brain, in the top right corner of the bowser. The distance (in mm) of the current section from the midline, as well as the section level in the series, are indicated beneath that schematic. The navigation buttons on the lower right provide options for panning (arrows) or zooming (+ and −) an image, resetting the image zoom level (diagonally opposing arrows), toggling across the four image types for a selected level (circular arrows), requesting an image from ZEBrA (camera symbol), turning anatomical labels ON/OFF, and moving to the next or previous image in the series (>> or <<). The Pan function can also be applied by clicking and dragging the main image in any desired direction, and all other navigation functions can be activated by the appropriate keyboard stroke, as described under the “Navigation Help” tab in the upper-left corner of the browser. The size and position of the field of view shown in the browser is indicated by the blue rectangle over the icon on the bottom left, and the image scale is indicated above that icon. (b) View of the same example drawing as in (a), with Turn Labels button activated (ON position). (c-e) Views of the Nissl- and myelin-stained sections, and of the colorized drawing depicting the: pallium (red), sub-pallium (pink), diencephalon (green), midbrain (yellow), pons (dark blue) and cerebellum (light blue).
The In Situ Image Browser
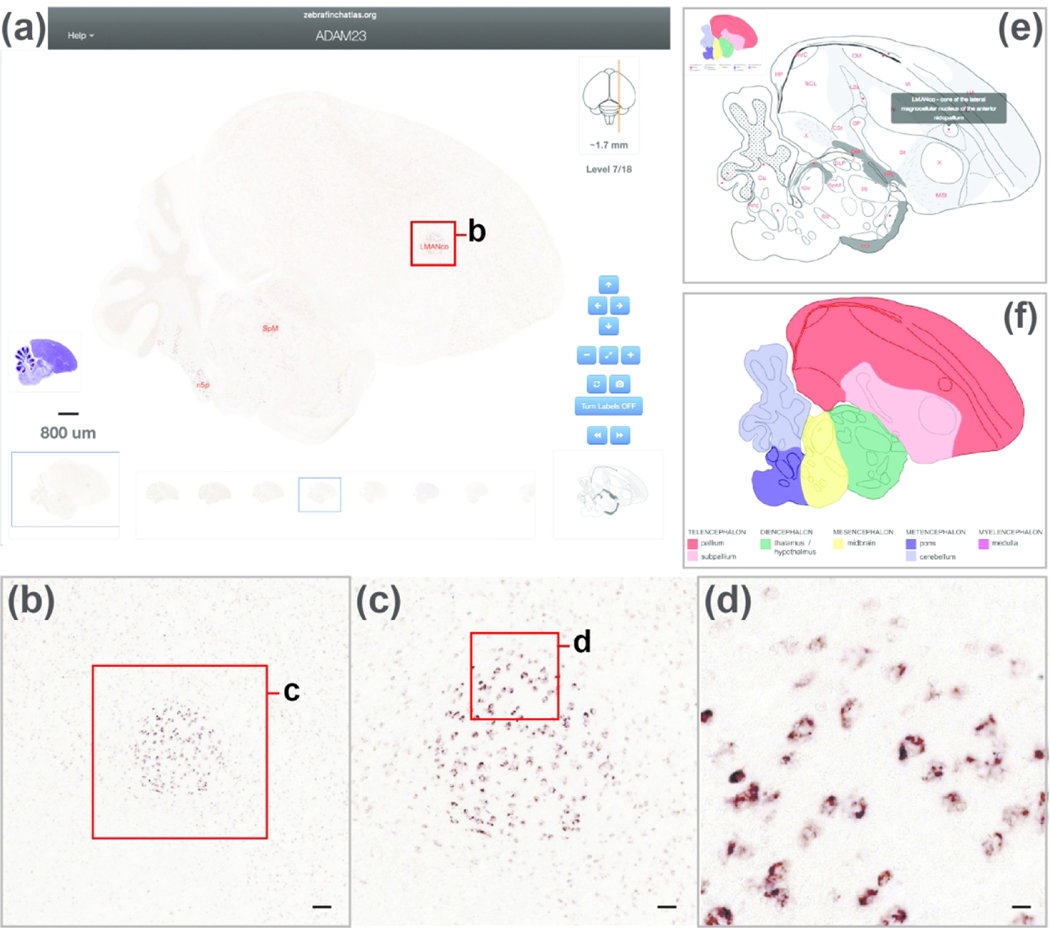
The In Situ Image browser is ZEBrA’s platform for visualizing and navigating high-resolution in situ hybridization images. The navigation features are similar to those of the Histological Atlas Browser, except that drawings cannot be panned or zoomed, and the corresponding Nissl and myelin series are not presented. For example, an in situ image showing ADAM23 reveals that this gene is highly enriched in nucleus LMAN (Figure 7a). This image can be zoomed by double-clicking the image or using the navigation tools on the lower right to allow browsing down to a cellular level of resolution (Figure 7b–d). Each image is presented with a drawing from the Histological Reference Atlas that represents the closest reference level for that image and provides guidance for interpreting gene expression patterns. Toggling between the in situ image and the corresponding reference drawing (e.g. Figure 7e for ADAM23, at the level shown in Figure 7a) aids in data interpretation by displaying which major brain divisions, subdivisions, nuclei, and cytoarchitectonic features such as fiber tracts and laminae are present at the level of the imaged section. An additional colorized schematic providing information about the major brain divisions and subdivision present in the section is presented to the left of the drawing (see inset in Figure 7e, expanded view in Figure 7f). Since the in situ images and the reference drawings are from different brains, the latter cannot provide exact matches for the in situ images, and are meant instead as guides to the approximate brain level and structures in each in situ image. If further anatomical details are needed, the Nissl image icon on the left provides direct access to the full Histological Atlas Browser, which opens as a separate window. Multiple gene series can be opened in dedicated windows, thus by design images from different genes can be viewed simultaneously and in parallel as adjacent windows on the user’s screen.
Figure 7. The in Situ Image Browser.
(a) View of the Browser depicting an in situ image from an example gene (ADAM23), with Turn Labels button activated. The set of icons at the bottom of the browser indicate the levels of the in situ images available for this gene, and the highlighted icon indicates the level of the enlarged image at the center of the browser. The position of the section being viewed relative to the midline is illustrated by the orange line on the dorsal-view schematic drawing of the brain, in the top right corner of the bowser. The distance (in mm) of the current section from the midline, as well as the section level in the series, are indicated beneath that schematic. The navigation buttons on the lower right provide options for panning (arrows) or zooming (+ and −) an image, resetting the image zoom level (diagonally opposing arrows), toggling between the in situ image and the corresponding drawing from the histological reference atlas (circular arrows; note that the atlas drawing cannot be zoomed from this browser), requesting an image from ZEBrA (camera symbol), turning anatomical labels ON/OFF, and moving to the next or previous image in the series (>> or <<). The Pan function can also be applied by clicking and dragging the main image in any desired direction, and all other navigation functions can be activated by the appropriate keyboard stroke, as described under the “Navigation Help” tab in the upper-left corner of the browser. The size and position of the field of view shown at the center of the browser is indicated by the blue rectangle over the icon on the bottom left, and the image scale is indicated above that icon. The icon above the scale provides access to the histological atlas browser, which opens in another window. (b-d) Enlarged views of the areas indicated by the red boxes in (a-c), corresponding to the core region of song nucleus LMAN (LMANco). Scale bars: (b): 100 μm; (c): 50 μm, (d): 25 μm. (e) View of the reference drawing (with labels ON) associated with the in situ image in (a) depicts specific brain structures present at the level of that in situ image. (f) View of the corresponding colorized drawing depicts the position of general brain subdivisions present at that level.
Each gene has been annotated according to its ability to define specific brain regions and nuclei (e.g., ADAM23 is highly enriched in the core region of LMAN, Figure 7a). Only structures for which a given gene is considered a marker (up or down differential expression compared to the surrounding brain areas) are labeled in that gene’s image browser. These marker annotations are also accessible collectively, with the complete anatomical gene marker sets presented in the ‘Markers of the Song System’ portal and/or in the ‘Bird Brain Markers’ sub-portal within the ‘Comparative Neuroanatomy’ portal (discussed below). Overall, the In Situ Image Browser, integrated with the Histological Reference Atlas, allows for detailed explorations of gene expression images documenting molecular specializations of broad avian brain subdivisions, and/or of nuclei that comprise specific avian brain circuits, with emphasis on the vocal control and learning circuitry.
The Gene Info Page
Every gene in ZEBrA is presented along with a Gene Info Page. This page provides probe and genomic information, as well as links to various metadata. Specifically, the ‘Gene/Probe Information’ tab (e.g., FOXP2 Gene Info Page in Figure 8a) includes gene symbols and descriptions, NCBI and Ensembl entries to finch and human orthologs, and the chromosomal location of genes and probes. Aided by these resources, the orthology of each gene in ZEBrA has been confirmed by reciprocal top alignments and synteny verification in comparison with humans, and whenever necessary non-avian out-groups (see Methods for a description of gene orthology verification, and NEFM example in Figure 2). The probe location entry in the ‘Gene/Probe Information’ tab reflects the confidence level for each selected probe. Confidence is highest for clones that overlap the gene model or located within a short distance of that model, but decreases for clones that are far away or separated from the model by a gap (see Methods for full description of probe types in Figure 3). Transcript cases representing apparent splice variants are indicated when relevant. Probes in the low confidence categories were included only when no higher confidence alternatives were present.
Figure 8. The Gene Info Page.
(a) Example of Gene/Probe Information tab (for FOXP2) presents the official gene symbol and related description (HGNC-based), the NCBI entry for the human ortholog, the zebra finch gene entry from Ensembl, the gene’s chromosomal location in the zebra finch genome (UCSC genome browser-based), the GenBank entry and genome location for the clone used to generate the in situ hybridization probe, and information on expression in chick embryos are depicted. A summary of gene function (from NCBI), and hyperlinks to relevant databases are also presented, when available. (b) Example of Gene Attributes tab providing counts and full lists of attributes for a given gene, in this case GRIN2B.
The ‘Gene Attributes’ tab provides counts and lists of all attributes for a given gene (e.g., GRIN2B in Figure 8b). These attributes link genes to their inclusion in the six major thematic portals that are presented in ZEBrA, and provide valuable information about a gene’s association with specific functions, diseases or phenotypes, and marked expression in song circuitry and/or brain areas in both birds and mammals. These data, in turn, facilitate discovering new relationships between gene expression patterns and functional attributes. A description of the content and organization of these portals, and linking of genes to attributes is discussed below. Of note, on the Gene Info page each attribute is associated with a hyperlink that provides access to the full list of genes that share that attribute. This page thus allows systematic explorations of how genes relate to each other by sharing attributes across portals.
Navigating the ZEBrA website and main resources
The ZEBrA website is organized into six thematically organized portals that were designed to facilitate website navigation and user access to images and related resources. These portals are readily accessible from the homepage beneath a short summary of ZEBrA contents (Figure 5). The navigation tools to explore these portals and other features consist of self-explanatory clickable icons and buttons, and when necessary pop-up menus with user instructions. For example, mousing over portal images and clicking the ‘GO’ button provides access to thematic drop-down menus with clickable arrowheads, which in turn provide access to alphabetical gene lists (e.g. Figure 9a). Within a given list, the ‘magnifying glass’ icon provides access to the In Situ Image Browser, the gene name provides access to the ‘Gene info’ Page, the ‘Attributes’ column informs on associations with themes or sub-themes in all portals (e.g. Figure 9b; details below), and the column on the right provides linkouts to databases or references, or informs the direction of differential regulation. Some lists contain multiple entries for the same gene, reflecting multiple entries and linkouts in the respective human or rodent databases.
Figure 9. Structure of gene lists.
(a) View of the organization and layout of a typical list of genes associated with a specific ZEBrA portal theme. In this example, a subset of genes associated with the Neurological/Psychiatric Disorder Epilepsy within the Human Diseases sub-portal is shown. Each gene entry includes a magnifying glass icon under the View In Situ column (which provides access to in situ images in the Image browser), and entries for gene name (which provides access to the Gene Info page), Gene Description, Attributes, and Linkouts (which in this case provide access to an entry in OMIM). (b-c) Views of example pop-up menus showing Attributes associated with the gene RELN that are derived from the Disease and Phenotypes (b) and Speech and Language (c) portals. The association of RELN with the OMIM Attribute “Epilepsy, familial temporal lobe, 7” provides the reasoning behind the classification of this gene under the Human Disease theme of Epilepsy.
Through buttons on the left sidebar of the homepage (Figure 5), or the top navigation bar in each portal, users can access alphabetical gene lists, tools for gene searches by name or description, and the histological reference atlas. All portals can be accessed from within any portal through a drop-down menu on the right side of the navigation bar. The menu bar on the top right provides information on the construction and usage of ZEBrA’s database and website.
ZEBrA portals
The contents of each portal are described below; the current gene numbers in each portal are presented in Table 1. The majority of genes in ZEBrA are part of at least one portal, and are typically presented in multiple portals. A small number of genes are not in any portal but can be accessed through the A to Z index or by gene name search.
Table 1.
Numbers of genes presented in major themes or categories within the six ZEBrA portals.
| # Genes | Portal/Theme/Category |
|---|---|
| Portal 1: Markers of the Song System | |
| 184 | HVC |
| 140 | RA |
| 122 | MMAN / LMAN |
| 61 | Area X |
| 78 | DLM |
| 13 | DM |
| 14 | Nif |
| Portal 2: Speech and Language | |
| 52 | Speech / Language Disorders |
| 57 | Autism (13 OMIM entries; 53 SFARI entries) |
| 41 | FOXP2 targets |
| 36 | Language Readiness Hypothesis |
| 57 | Shared Markers Between Vocal Learners |
| Portal 3: Diseases and Phenotypes | |
| 212 | Human Diseases (326 OMIM entries) |
| 262 | Mouse Phenotypes (511 MGI phenotypes) |
| Portal 4: Comparative Neuroanatomy | |
| 451 | Bird Brain Markers |
| 222 | Mouse Brain Markers (Mouse Brain Atlas entries) |
| Portal 5: Gene Function | |
| 681 | Gene Function |
| Portal 6: Songbird Discoveries | |
| 80 | Highlighted ZEBrA-based Discoveries |
| 641 | Songbird Studies |
Abbreviations: DLM, medial part of the dorsolateral nucleus of the anterior thalamus; DM, medio-dorsal nucleus of the intercollicular complex; HVC, HVC proper name; LMAN, lateral magnocellular nucleus of the anterior nidopallium; MGI: Mouse Genome Informatics; NIf: nucleus interfacialis; OMIM: Online Mendelian Inheritance in Man; RA, robust nucleus of the arcopallium; SFARI, Simons Foundation Autism Research Initiative; X, Area X.
(1). Markers of the Song System:
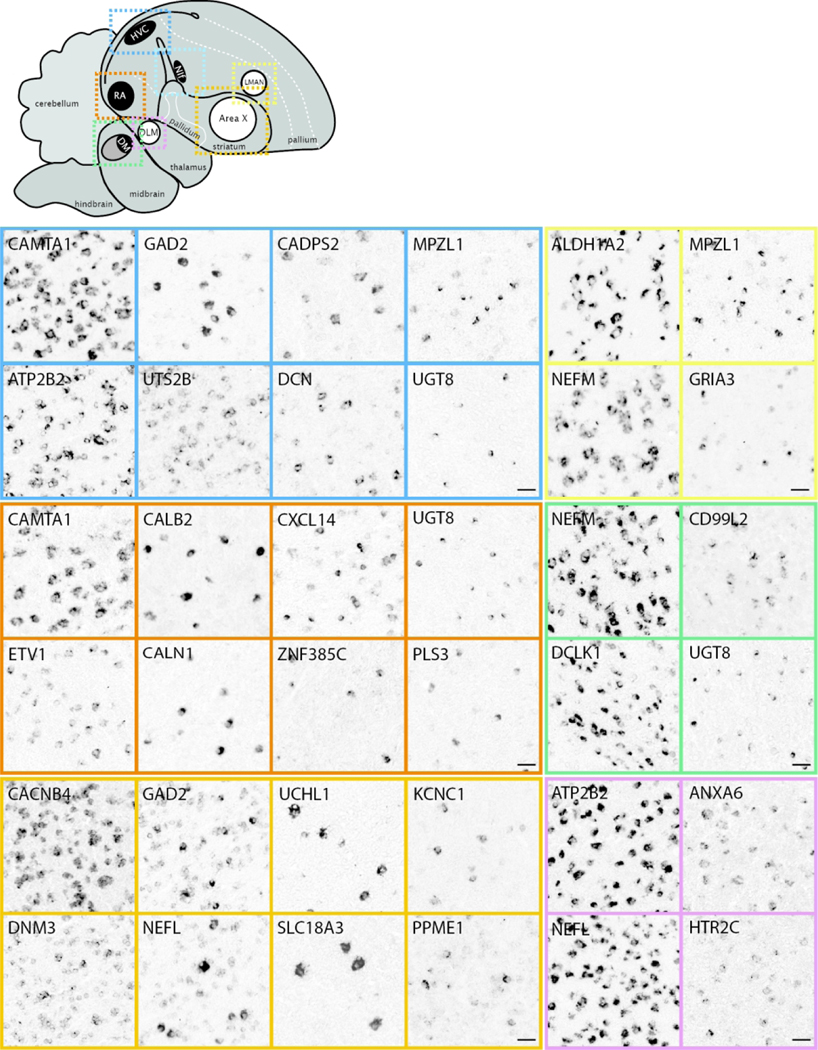
This portal consists of genes that are differentially up-or down-regulated in at least one song control nucleus of adult male zebra finches compared to adjacent brain areas, based on visual assessment of in situ images. Included are markers for most nuclei in both major pathways of the song system (examples in Figure 10). We note the broad diversity of patterns, density of labeling, and contrasts with adjacent areas. Most of these genes were initially identified as candidate song nuclei markers through microarray screenings (Lovell et al., 2008; Lovell et al., 2018a; Pfenning et al., 2014), and in many cases provided stringent and objective criteria for distinguishing true vs. false positives and negatives in these screenings (Lovell et al., 2018a). Other subsets were initially included in other ZEBrA portals but later identified as markers of song system nuclei. Overall, they represent highly validated sets of molecular specializations that are likely linked to the unique anatomical, neurochemical and physiological properties of the song system. Song nuclei markers display broad ranges of labeling patterns at the cellular level (examples in Figure 11). While some markers are strongly expressed in high proportions of cells within various song nuclei (e.g. in Figure 11: CAMTA1 in HVC, CACNB4 in Area X), others label distinctly sparse populations of different sizes and shapes (e.g. in Figure 11: GAD2, CADPS2, UTS2B, and UGT8 in HVC; CALB2 and PLS3 in RA, etc.), suggesting that they might correspond to discrete and distinct cell types within these nuclei. Overall, genes in this portal constitute molecular specializations of song nuclei, but they also serve as markers that can help to define nuclear boundaries, allowing for greater precision in studies of nucleus size, shape, and cell type composition. Furthermore, while the differential expression in song nuclei has been previously reported for some markers (e.g. Hilliard et al., 2012b; Kato & Okanoya, 2010; Kubikova et al., 2010; Lovell et al., 2013; Lovell et al., 2008; Lovell et al., 2018a; Pfenning et al., 2014; Wada et al., 2004; Whitney et al., 2014; Wirthlin et al., 2014; see also Songbird Discoveries portal), ZEBrA provides extensive documentation of the patterns across all song nuclei and throughout the brain.
Figure 10. Markers of the Song System (Portal 1).
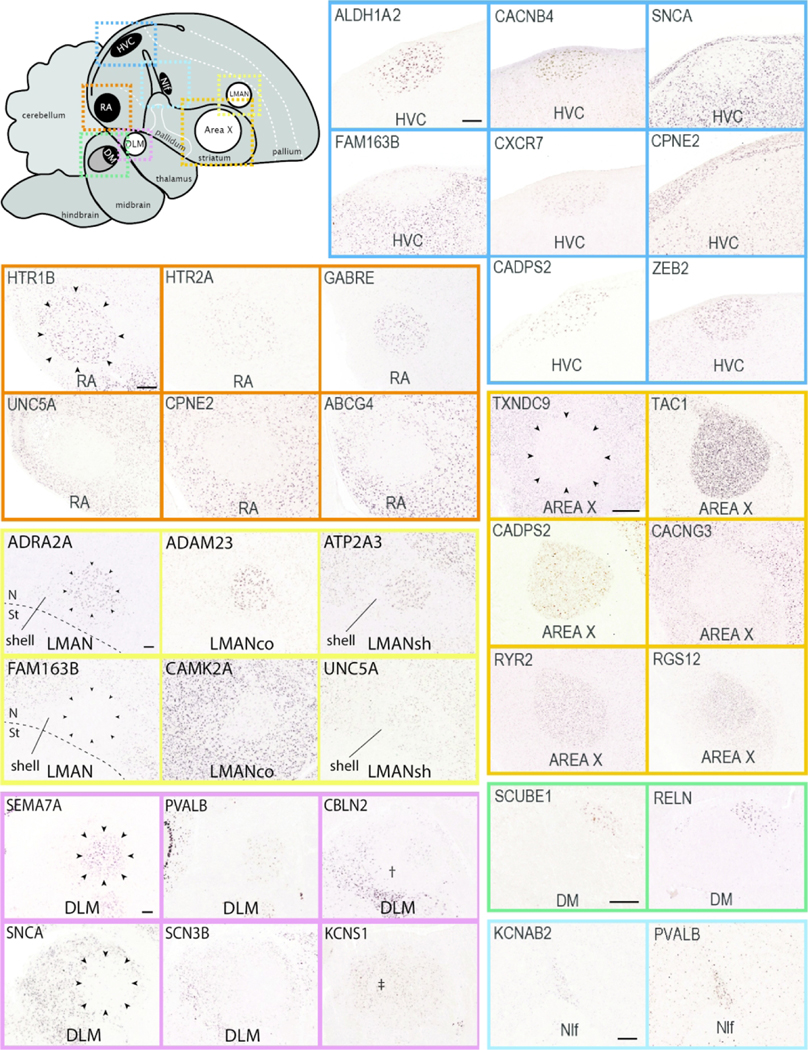
Upper left: Schematic drawing of a parasagittal view of the adult male zebra finch brain depicts major brain subdivisions and the relative location of major song control nuclei; nuclei of the direct vocal-motor pathway are in black, nuclei of the anterior forebrain pathway are in white, colored dashed rectangles indicate the areas shown in the in situ hybridization images. Other panels: Examples of in situ hybridization images of positive or negative markers (up- or down-regulated expression compared to adjacent areas) of zebra finch song nuclei HVC (blue), RA (vermillion), LMAN (yellow), Area X (orange), DLM (reddish purple), DM (green) and NIf (sky blue). We note that various markers of LMAN specifically label the core (LMANco), shell (LMANsh), or both core and shell regions of LMAN. DM: mediodorsal nucleus of the intercollicular complex; NIf: nucleus interfacialis; N: nidopallium; St: Striatum; †: Unknown region of the dorsal thalamus surrounding nucleus DLM; ‡: Unknown region of the dorsal thalamus likely corresponding to the DLM defined by Wada et al., 2004. For other abbreviations, see Figure 1 legend. Scale bars for LMAN, DLM, and Area X = 200 μm; for all other nuclei = 100 μm.
Figure 11. Markers of the Song System: cellular level (Portal 1).
Upper left: Schematic drawing of a parasagittal view of the adult male zebra finch brain depicts major brain subdivisions and the relative location of major song control nuclei; nuclei of the direct vocal-motor pathway are in black, nuclei of the anterior forebrain pathway are in white, colored dashed rectangles indicate the areas shown in the in situ hybridization images. Other panels: Examples of in situ hybridization images of markers that show high density of labeling (left panels for each nucleus) vs sparse labeling (other panels) for song nuclei HVC (blue), RA (vermillion), LMAN (yellow), NIf (sky blue), Area X (orange), and DLM (reddish purple). For abbreviations, see legends of Figures 1 and 11. Scale bars: 50 μm.
(2). Speech and Language:
This portal consists of genes with known or hypothesized involvement in speech and language function and disorders in humans. It provides unique opportunities to determine how genes with known or presumed involvement in speech and language function and disorders are expressed in a circuit that is dedicated to vocal communication and learning. Reflecting its status as a focal gene in human speech research, we highlight FOXP2 (linked to a genetic form of speech dyspraxia in humans and to vocal learning in finches; Haesler et al., 2007; Heston & White, 2015; Lai et al., 2001; Teramitsu et al., 2004; Vargha-Khadem et al., 1998), its main target CNTNAP2 (suspected of involvement in childhood speech apraxia, dyslexia, and autism spectrum disorders; Alarcon et al., 2008; Li et al., 2010; Newbury et al., 2011; Peter et al., 2011; Whitehouse et al., 2011), and candidate FOXP2 targets based on experimental evidence in rodents (Konopka et al., 2009; Spiteri et al., 2007; Vernes et al., 2007). We also present gene lists associated with disorders of speech and language, as well as other selected disorders that cause major disruptions to vocal communication (e.g. deafness, schizophrenia). Genes were selected based on entries in the Online Mendelian Inheritance in Man (OMIM; https://www.omim.org) database, including genes currently linked to human Speech and Language Disorders, as well as experimental or conceptual studies of specific disorders, and listings by the Simons Foundation Autism Research Initiative (SFARI; https://www.sfari.org; Oct. 2017 Release) as ‘High Confidence’, ‘Strong Candidate’, or ‘Suggestive Evidence’ linkages to autism spectrum disorders. This portal also includes genes with suggested involvement in the evolution of speech and language brain circuitry (Murphy & Benitez-Burraco, 2016), as well as shared markers of cortical and striatal vocal areas between humans and vocal learning birds (Pfenning et al., 2014), representing convergent molecular specializations in independently evolved vocal learning systems. Importantly, we indicate genes for which a cDNA clone is not available, which could reflect a temporary limitation of the current expression databases. We also indicate which human disorder genes are not expressed in the song control circuitry and/or are absent in the zebra finch genome.
Many genes in this portal show specialized differential expression within cortical motor nuclei of the direct pathway for song production, and/or striatal nucleus of the anterior forebrain pathway for song learning and plasticity (respectively HVC, RA, and Area X; examples in blue, vermillion, and orange panels in Figure 12), suggesting possible targets for their actions within the vocal circuitry. Notably, some candidate FOXP2 targets (e.g., CACNG3, SLC4A4, and VLDLR (Adam et al., 2016; Konopka et al., 2009; Mendoza & Scharff, 2017; Spiteri et al., 2007; Vernes et al., 2007) are markers of Area X, which is an important brain target of FOXP2 action with regards to vocal learning in finches (Haesler et al., 2007). These genes might thus be mediators of FOXP2 effects in song learning, a compelling hypothesis that has yet to be tested mechanistically. Other candidate FOXP2 targets are differential markers of other song nuclei (e.g. DPP6, SYT4 and others in HVC, CABP1, DCN, and others in RA; Figure 12, in blue and vermillion, respectively), suggesting other possible targets of FOXP2 action. Some genes that have been associated with autism are also differential markers of specific song (e.g., CACNB2 and SEMA5A in HVC, PTCHD1 in Area X; Figure 12, in orange), suggesting their possible relationships to vocal communication pathways. In contrast, genes with involvement in disorders like stuttering (i.e., GNPTG, GNPTAB, AP4E1) have broad brain distributions with no specialized expression in song nuclei (not shown), suggesting either broad functions or mechanisms, or yet unidentified sites of action. We also highlight shared markers of vocal-motor primary cortical areas and striatal nuclei in vocal learning birds and humans (e.g. SLIT1, SYNPR; Figure 12). These convergent molecular specializations of distant vocal learners are thought to reflect important features of vocal learning systems (Pfenning et al., 2014).The patterns in this portal also have a diverse range of expression levels and contrasts with adjacent non-vocal regions, and some genes are expressed in sparse cell populations that might represent distinct cell types within vocal nuclei (e.g. PLS3 in HVC, SYNPR in RA and Area X). Unique to this portal, we list genes for which a cDNA clone is not available as well as genes not present in the zebra finch genome (see Discussion). We also note that while some patterns in this portal (e.g. FOXP1, FOXP2, CNTNAP2) have been previously reported (Condro & White, 2014; Haesler et al., 2004; Panaitof et al., 2010; Teramitsu et al., 2004), ZEBrA documents their expression across all song nuclei and throughout the brain, facilitating comparisons with other patterns of interest to speech and language function, disorders, and related phenotypes.
Figure 12. Speech and Language genes (Portal 2).
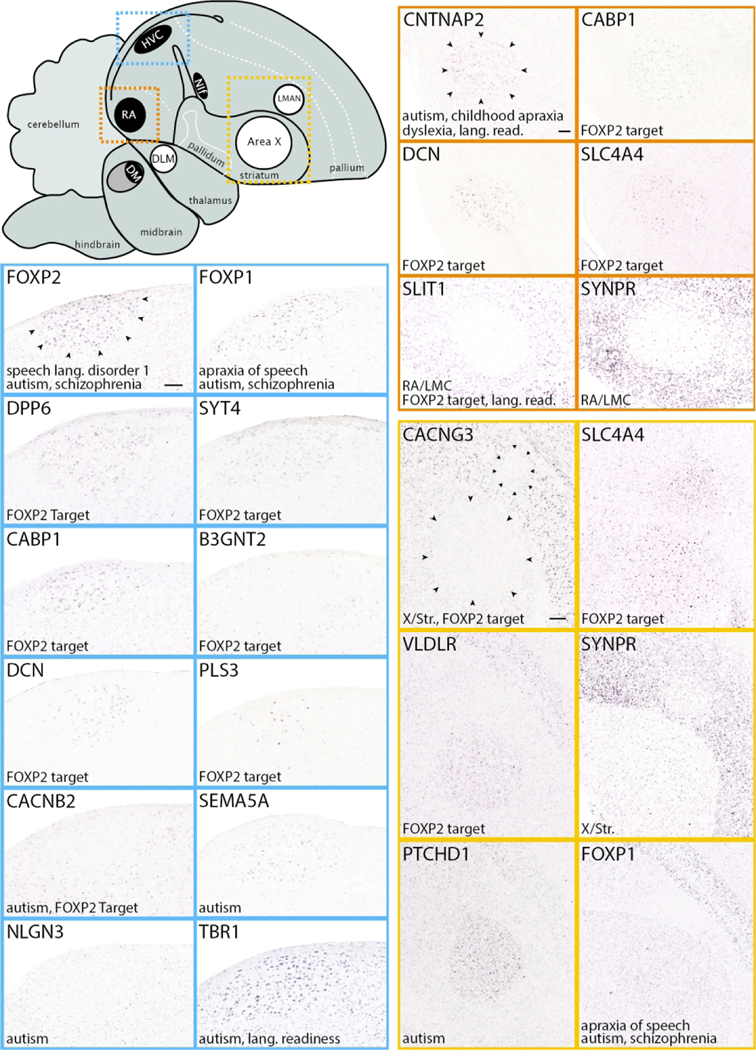
Upper left: Schematic drawing of a parasagittal view of the adult male zebra finch brain depicts major brain subdivisions and the relative location of major song control nuclei; nuclei of the direct vocal-motor pathway are in black, nuclei of the anterior forebrain pathway are in white, colored dashed rectangles indicate the areas shown in the in situ hybridization images. Other panels: Examples of in situ hybridization images of genes linked to Speech and Language function that are positive or negative markers (up- or down-regulated expression compared to adjacent areas) of zebra finch song nuclei HVC (blue), RA (vermillion), and LMAN and/or Area X (orange). Gene attributes related to this portal are indicated in the bottom left of each image; song nuclei borders are indicated by small black arrowheads in the first image for each nucleus. For abbreviations, see legends of Figures 1 and 11. Scale bars for HVC and RA = 200 μm; Area X/LMAN = 400 μm.
(3). Diseases and Phenotypes:
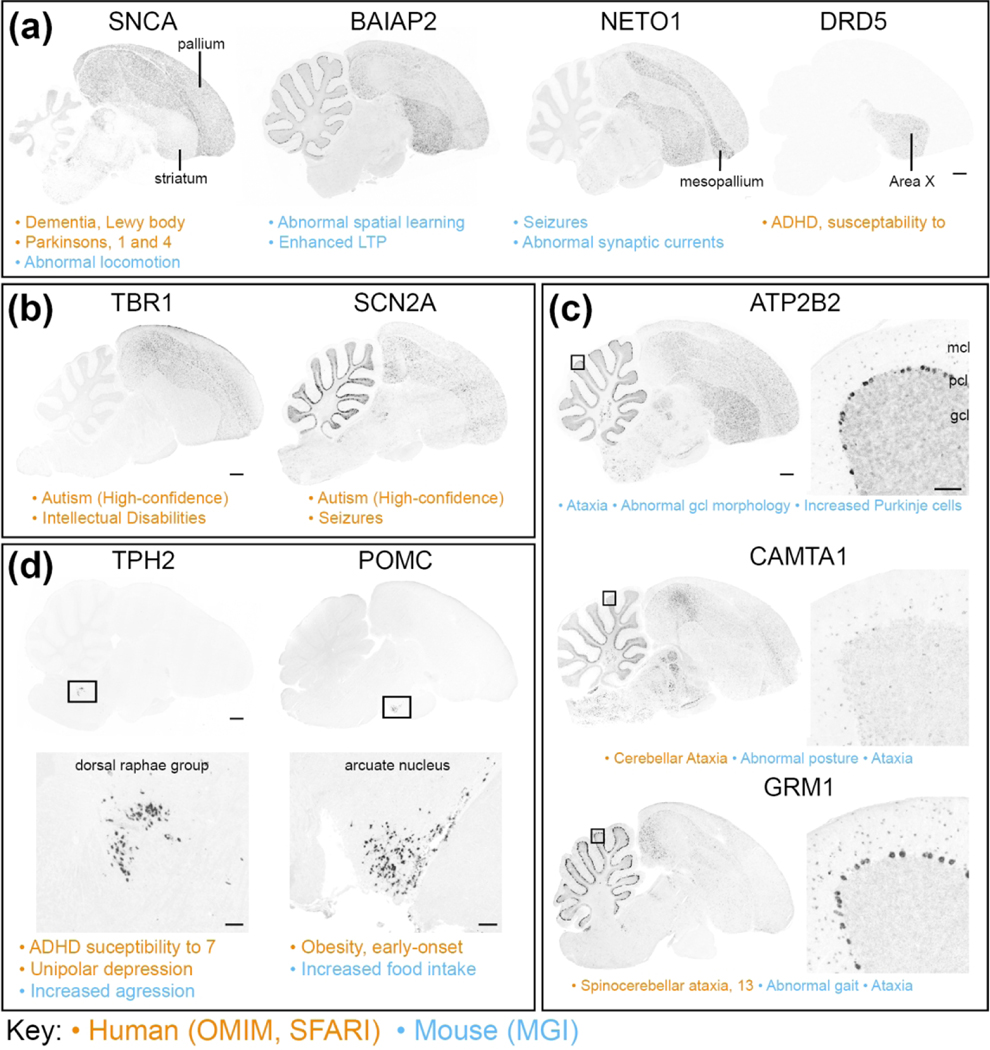
This portal consists of genes with known involvement in brain- and/or behavior-related disorders and phenotypes. Sub-portal 3A (Human Diseases) includes 212 genes with known involvement in at least one of 326 unique human neurological and/or psychiatric disorders, based on OMIM. Gene sets associated with epilepsy and cardiovascular disorders (in the neurological and non-neurological categories, respectively) are the most highly represented, followed by mental retardation and neurodegenerative, metabolic and endocrine disorders. Sub-portal 3B (Mouse Phenotypes) includes 262 genes with established links to at least one of 511 brain and/or behavioral phenotypes in mice, based on the Mouse Genome Informatics (MGI) database (see Table 1). Genes associated with broad phenotypes like seizures and/or disruptions of learning or locomotor function, or with diseases like epilepsy or neurodegenerative disorders, have broad distributions or differential expression in broad telencephalic areas and/or subdivisions, such as the entire pallium, pallial fields, and/or striatum (e.g. Figure 13a), although some show specialized expression in specific nuclei (e.g. SNCA and DRD5 in Area X). Several genes involved in some psychiatric and/or neurological disorders show enriched expression in areas like pallial and/or sub-pallial regions, cerebellar cortical layers, or neuromodulatory brainstem nuclei, in various combinations (examples in various panels of Figure 13). Notably, genes with involvement in autism tend to show broad expression patterns, particularly in the pallium and/or cerebellum (e.g. Figure 13b), although in some cases they also show enriched expression in vocal nuclei (e.g. Figure 10, TBR1 in HVC). Such patterns contrast with cases with more discrete expression, for example the unique cerebellar patterns for genes with involvement in disorders like ataxias (e.g. Figure 13c), or expression in discrete nuclei in the pons for genes associated with disruptions of the serotonergic system (e.g. Figure 13d, left), or in the hypothalamus for genes associated with endocrine disorders (e.g. Figure 13d, right). The complete list of disorders and phenotypes included in this portal and their thematic classifications are presented in SuppInfo:STable1.xlsx.
Figure 13. Diseases and Phenotypes genes (Portal 3).
Examples of in situ hybridization images of genes linked to OMIM- or SFARI-based human neurological and/or psychiatric disorders (sub-portal 3A), or to MGI-based mouse neurological and/or behavioral phenotypes, that show differential expression in various avian brain areas (sub-portal 3B). Gene attributes related to this portal are indicated under each image (vermillion annotations are OMIM- or SFARI-based, blue annotations are MGI-based). (a) Broad expression in telencephalic fields, including pallium, pallial subdivisions, and/or striatum, for genes involved in disorders like seizures or neurodegenerative diseases, or phenotypes disrupting broad learning or locomotor functions. (b) Broad pallial and/or cerebellar expression of genes involved in autism. (c) Differential cerebellar expression for genes involved in cerebellar-related disorders and phenotypes, like ataxias and/or abnormal gait. (d) Discrete expression in nuclei in the pons or hypothalamus for genes involved in disruptions of the serotonergic (left panels) or neuroendocrine (right panels) systems. In (c) and (d), the location of the higher magnification panels in the right or bottom panels are indicated by small rectangles in the left or top panels. Abbreviations for cerebellar cortical layers in (b): gcl, granule cell layer; mcl, molecular layer; pcl, Purkinje cell layer. Scale bars: low-power images = 1 mm; high-power images in (c) and (d) = 200 μm.
(4). Comparative Neuroanatomy:
This portal consists of a comprehensive suite of markers of specific brain areas and subdivisions compared to adjacent areas, providing a better understanding of the molecular brain organization in birds. Sub-portal 4A (Bird Brain Markers) presents in situ data for genes that are differentially expressed (up or down) in various avian brain areas other than song control nuclei compared to adjacent areas or brain subdivisions (examples in Figure 14). For the telencephalon, this includes differential markers of pallium vs. striatum (Figure 14a), pallial subdivisions (e.g. mesopallium, nidopallium, hyperpallium, arcopallium in Figure 14b–c), pallial thalamo-recipient zones (i.e. L2a, basorostralis, and entopallium in Figure 14d–f), the entire hippocampal formation or subdivisions thereof (Figure 14g), or sub-pallial areas like globus pallidus (Figure 14h) or the striatal nucleus accumbens (Figure 14i). Sub-portal 4A also presents numerous markers of thalamic (Figure 15a–c) and hypothalamic (not shown) nuclei, cerebellar cortical layers (Figure 15d), habenular subdivisions (Figure 15e), neuromodulatory cell groups in the midbrain and pontine tegmentum (Figure 15f–h), and specific pre-tectal nuclei (Figure 15i–k) and tectal nuclei or layers (Figure 15l), among others. Close examination reveals a diversity of labeling patterns within many of these structures, with some markers showing very high densities of labeled cells (examples in the left-most panels in Figure 16), whereas other are expressed in sparse cell populations (examples in other panels in Figure 11) that could potentially represent distinct cell types with unique molecular signatures. Notably, some genes are highly expressed in structures that lack neuronal cells such as fiber tracts, ventricular zone, choroid plexus and lining of blood vessels (examples in Figure 16, yellow, black and purple panels) and likely represent markers of non-neuronal cells such as oligodendrocytes, ependymal cells, choroidal cells, and possibly endothelial cells. Sub-portal 4B (Mouse Brain Markers) presents zebra finch brain expression patterns of genes that are markers of specific brain structures in mice, based on lists of markers retrieved from Mouse Brain Atlas; the linkouts provide access to the corresponding gene image set in the Mouse Brain Atlas. This sub-portal is organized so as to facilitate the study of how markers of specific structures in mammals are expressed across avian brain subdivisions, and vice-versa.
Figure 14. Comparative Neuroanatomy genes: telencephalon (Portal 4).
Top row: Schematic drawings of parasagittal sections of the adult male zebra finch brain depict major brain subdivisions and specific nuclei shown in this figure. Red letters and dots indicate structure annotations as they appear in ZEBrA, vermillion rectangles indicate the position of the images shown in other panels, and numbers at bottom right of each diagram provide position (in mm) relative to the midline. Other panels: Examples of in situ hybridization images of genes that show differential expression in various parts of the telencephalon, taken from the Bird Brain Markers (sub-portal 4A). (a) Markers showing differential enrichment in pallium (GAP43, SLC17A6) vs striatum (DAPK1, GAD2); the top and bottom small squares show the position of the left and right insets respectively, depicting pallial vs. striatal differences in cell labeling densities. (b) Markers showing differential enrichment in specific pallial subdivisions, including negative and positive markers of the mesopallium (PLPP4, CADPS2 and FOXP1) and nidopallium (ALDH1A2), in different combinations with St and Area X. (c) General negative/positive markers of the arcopallium (AQP1/ETV1, the latter also a negative marker of subdomains APd and AA). (d-f) Positive/negative markers of pallial thalamo-recipient zones L2a (CD99L2/ABCG4), nucleus basorostralis (CAMTA1/CACNG3) and entopallium (ADAM23/SCN2A). (g) Markers of the entire hippocampal formation (positive: MAP4, ARGHDIB; negative: LMO1, NTS) or parts thereof (AQP1, PLPP4). (h) Markers of the globus pallidus (ATP2B2, KCNA1). (i) Markers of the striatal nucleus accumbens (TAC1, FAM163B). For simplicity, only telencephalic structures are shown in (a) and (b). Abbreviations: A: arcopallium; Acc, nucleus accumbens; APH, parahippocampal area; Bas, nucleus basorostralis; DG, dentate gyrus; E, entopallium; GP, globus pallidus; H, hyperpallium; HA, apical part of the hyperpallium; Hp, hippocampus proper; L2a, subfield L2a with auditory Field L; M, mesopallium; N, nidopallium; St, striatum. For other abbreviations in top row diagrams, please consult the histological atlas in ZEBrA. Scale bars: top row diagrams = 1 mm; (a): low-power photomicrographs = 1mm, high-power insets = 100 μm; (b) = 1 mm; (c) = 400 μm; (d) and (e) = 200 μm; (f) – I = 400 μm.
Figure 15. Comparative Neuroanatomy genes: diencephalon and brainstem (Portal 4).
Top row: Schematic drawings of parasagittal sections of the adult male zebra finch brain depict major brain subdivisions and specific nuclei shown in this figure with the exception of the brain areas shown in panels I-L. Red letters and dots indicate structure annotations as they appear in ZEBrA, vermillion rectangles indicate the position of the images shown in other panels, and numbers at bottom right of each diagram provide position (in mm) relative to the midline. Other panels: Examples of in situ hybridization images of genes that show differential expression in various parts of the diencephalon and brainstem, taken from the Bird Brain Markers (sub-portal 4A). (a-c) Positive/negative markers of thalamic nuclei DMA (SNCG/CBLN2), Ov (KCNH8/CBLN2) and Rt (CBLN2/CCK). (d) Markers of cerebellar cortical layers show distinctly differential expression in pcl (BAIAP2, SLC4A4), gcl (CHGB), pcl/mcl (ATP2B2, ENO1), or in sparse cells in gcl (CXCL14, among other patterns; the bottom panels show high magnification views of interface zone across layers, taken from region depicted by rectangle in top left panel. (e) Markers that delineate the habenula and/or differentiate its medial and lateral subdivisions. (f-h) Markers of VTA (KCND3, SLC6A2), SN (ATP2A3, KCNJ5) and LoC (GLRA2, SLC6A2) in brainstem tegmentum. (i-k) Positive/negative markers of pre-tectal nuclei SpL (KCND1/RGS4), SpM (NETO1/SLC32A1) and Pt (NREP1/SNCA). (l) Markers of ipc (CABP1), imc (CALB2) and sgc (CABP7) in the optic tectum. Abbreviations: Cb, cerebellum; DMA, dorsomedial nucleus of the anterior thalamus; gcl, granule cell layer of Cb; HbL, lateral habenular nucleus; HbM, medial habenular nucleus; Imc, magnocellular part of the isthmic nucleus; Ipc, parvocellular part of the isthmic nucleus; LoC, locus coeruleus; mcl, molecular layer of Cb; N3, root of third cranial nerve; Ov, nucleus ovoidalis; pcl, Purkinje cell layer of Cb; Rt, nucleus rotundus; SN, substantia nigra; sgc, stratum griseum centrale of the optic tectum; SpL, nucleus spiriformis lateralis; SpM, nucleus spiriformis medialis; VTA, ventral tegmental area. For other abbreviations in top row diagrams, please consult the histological atlas in ZEBrA. Scale bars: top row diagrams = 1 mm; (a-c) = 200 μm; (d): low-resolution panels = 200 μm, high-resolution panels = 100 μm; (e-k) = 200 μm; (l) = 400 μm.
Figure 16. Comparative Neuroanatomy genes: cellular level (Portal 4).
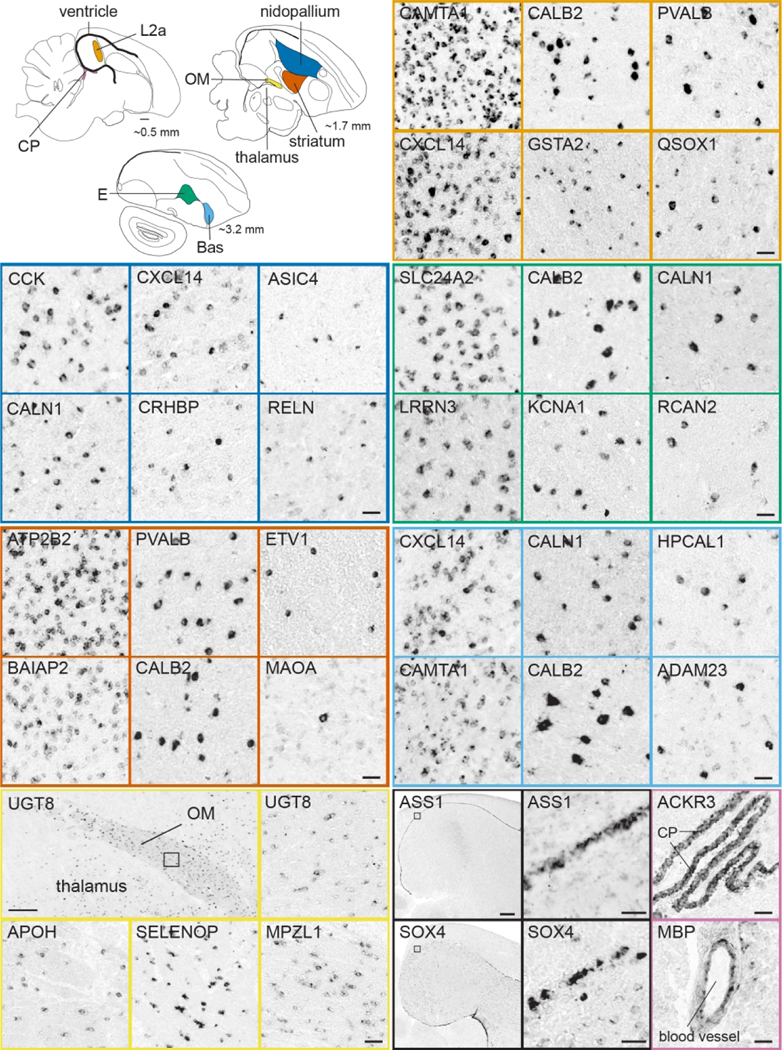
Upper left: Schematic drawings of parasagittal sections of the adult male zebra finch brain depict the locations (colored regions) of the brain regions sampled in the in situ hybridizations images. Other panels: Examples of in situ hybridization images of genes that show high density of labeling (left panels for most brain areas) vs. sparse cell labeling (other panels) in pallium (dark blue), striatum (red), L2a (orange), Bas (light blue) or E (green), as well as markers of OM (yellow), ventricle (black), and choroid plexus and blood vessel endothelium (pinkish purple). Abbreviations: Bas, nucleus basorostralis; CP, choroid plexus; E, entopallium; L2a, subfield L2a within auditory Field L; OM, occipitomesencephalic tract. Scale bars: Top-row drawings = 1 mm; Low-power photomicrographs of UGT8 = 400 μm; ASS1 and SOX4 = 800 μm; all other panels = 50 μm.
(5). Gene Function:
In this portal, genes have been classified according to their association with a gene family, or with major molecular functions and/or pathways, with an emphasis on categories that are most relevant for understanding brain physiology and behavior. The majority of genes in ZEBrA are associated with at least one functional category in this portal. A small subset, however, could not be classified in any of these functions, and thus are not included in this portal. By linking gene function to expression patterns, this portal addresses the need to identify categories of genes that are expressed and/or differentially regulated within specific brain areas and circuits. We note that the functional classification in this portal is not all-encompassing or exhaustive, but rather reflects the current composition of the ZEBrA database.
(6). Songbird Discoveries:
This portal presents gene lists associated with significant contributions in the literature, reflects an attempt to list studies deemed interesting or important, and for which gene patterns are available in ZEBrA. Sub-portal 6A (Highlighted ZEBrA-based Discoveries) highlights discoveries that have benefitted from the analysis of gene expression patterns in ZEBrA. Sub-portal 6B (Songbird Studies) presents sets of genes whose expression or regulation has previously been described in a published study in zebra finches and/or other bird species. Currently we highlight studies of sex dimorphism and the action of sex steroids on brain and behavior, behaviorally-regulated gene expression, the effects of brain plasticity and sleep on learning, the dynamics of adult neurogenesis and neuronal replacement, and gene expression changes during the critical period for vocal learning, among others. We also highlight some key studies that have implicated sets of genes in processes of interest to vocal learning and songbird biology. The specific studies are cited under each theme, and the corresponding gene sets described in those studies are listed, if present in ZEBrA. This portal is not meant to be all-encompassing, but rather to be dynamic, follow expansions in the ZEBrA database, and accommodate additional gene lists and/or future studies describing the roles of genes currently in ZEBrA.
Discussion
ZEBrA documents for the first time and in a systematic manner the expression of a large collection of genes of relevance to brain and behavior in serial brain sections of a bird species. Included are both specialized nuclei within the circuitry for song learning and production, as well as the avian equivalent of cortical and sub-cortical structures, basal forebrain, thalamus, hypothalamus, midbrain, and other brainstem structures. Thus, while providing insights into the molecular properties of the song circuitry, ZEBrA is also a platform to explore how genes linked to brain function and disorders in humans and rodents are expressed in a bird, and to probe the extent of molecular similarities and divergences of brain structures between birds and mammals. These features address important needs of songbird and avian brain researchers, but also inform a broad range of comparative neuroanatomy and physiology questions. We note that an in situ database exists for chicken (GEISHA; Gallus Expression In Situ Hybridization Analysis; http://geisha.arizona.edu/geisha/index.jsp), but it is geared towards embryonic development using whole-mount embryos.
Some technical aspects deserve comments: (1) Behavioral State. The data are from awake non-singing and not song-stimulated birds; thus it reflects the constitutive brain transcriptome, noting that because the birds were awake, some gene regulation by visual stimulation may have occurred in visual areas (e.g. see Feenders et al., 2008; Hara et al., 2009). The data also provides a baseline reference for functional studies involving behavioral states or manipulations known to regulate brain gene expression, such as singing, hearing, sleep, wakefulness, stress, circadian cycle, seasonality, reproductive or hormonal state, among others (Drnevich et al., 2012; Hara et al., 2012; Hilliard et al., 2012a; Hilliard et al., 2012b; Li et al., 2007; Mouritsen et al., 2005; Stevenson et al., 2012; Thompson et al., 2012; Wada et al., 2006; Whitney et al., 2014). All birds in ZEBrA were carefully monitored and controlled for behavioral state, an aspect that has not always been controlled in large scale studies (e.g. behavioral state is not reported in the Mouse Brain Atlas). (2) Gene Annotations. Current annotations of genomic and transcriptomic databases and resources in zebra finches and other birds are far from definitive, as they are fraught with inaccuracies and incompleteness (Lovell et al., 2013; Lovell et al., 2008; Lovell et al., 2018b; Mello et al., 2018; Wirthlin et al., 2014). ZEBrA’s own Gene Curation pipeline (Figure 4) addresses this issue, as all genes have been vetted, using stringent criteria for orthology with other avian and non-avian species including humans. (3) Probes. Due to gaps in genome sequence and scarcity of expression data in birds, there is not sufficient information to fully and unequivocally link all probes to genes of interest. For a limited set of genes, only low confidence probes were available. Such cases are clearly indicated, and will be further assessed as more expression data and higher quality assemblies become available. (4) Brain atlas. A digital histological brain atlas has only recently become available for zebra finches (Karten et al., 2013). The detailed annotations and integration with in situ images for a subset of the atlas levels are important for interpreting the in situ patterns in ZEBrA. However, the annotated atlas is also a stand-alone feature that on its own can be of great use to songbird and avian researchers, independently of the in situ data. (5) Both the atlas and the in situ database are from adult males, a choice justified by the fact that a full-fledged vocal circuitry is absent in adult female zebra finches (e.g. Konishi & Akutagawa, 1985).
Several ongoing and past studies have benefitted from the data in ZEBrA portals, and the large amounts of gene expression images and metadata are open to further explorations and analyses. For example, genes in the Song System Markers portal have helped to identify convergent markers of cortical and striatal vocal areas shared by vocal learning birds and humans (Pfenning et al., 2014), define the transcriptome of song system nuclei (Lovell et al., 2018a), and identify markers related to ion channel families (Friedrich et al., 2019; Lovell et al., 2013). These markers are prime candidates for gene manipulations to gain a mechanistic understanding of the regulation of song production and vocal learning. Furthermore, several markers are shared among song nuclei subsets and could be related to their unique properties, such as the nuclei in the vocal production pathway, the nuclei of the cortico-striatal projection in the anterior pathway for vocal learning and plasticity. Others are also markers of areas outside the song system, possibly reflecting shared specializations between vocal and non-vocal areas. Such relationships are novel and too numerous to be listed, but can be accessed by examining the attributes that Song System Markers share with Avian Brain Markers.
The expression patterns in the Diseases and Phenotypes portal allows for an assessment of how genes with known involvement in brain disorders, physiology and/or behavior based on phenotypes in humans and rodents are expressed in the brain of an experimental avian organism. This in turn could potentially reveal novel and unsuspected insights into the brain substrates underlying such disorders and/or phenotypes. Interestingly, the high representation of gene sets associated with epilepsy and cardiovascular disorders likely reflects the large number of regulators of tissue excitability (e.g. ion channel and neurotransmitter receptor genes, as reported in Lovell et al., 2013; Lovell et al., 2018a). Taken together with the data in Comparative Neuroanatomy, this portal provides cues about possible convergence or divergence of gene function across different vertebrate lineages (birds and mammals) with distinct brain organizations. For example, one can explore whether genes associated with specific brain-related phenotypes in mammals show expression in avian brain areas predicted by those phenotypes (e.g. cerebellar-related phenotypes). Alternatively, the bird expression patterns for some of these genes may not support, or might even contradict the predictions based on mammalian phenotypes, suggesting that these genes have divergent functions in avian vs mammalian lineages. Of particular interest are genes associated with Speech and Language function and disorders. ZEBrA facilitates analyses that are not feasible in humans, since access to appropriate brain samples is very limited, and arguably not possible in rodents and non-human primates, since these more traditional model species lack highly dedicated vocal control and learning cortical circuitry, and have at best limited vocal plasticity abilities (Hammerschmidt et al., 2012; Hammerschmidt et al., 2015; Jurgens, 1998, 2009). Notably, besides revealing possible convergences between humans and vocal learning birds, the data in ZEBrA may also reveal genetic pathways involved uniquely in human speech and language.
Several patterns under Comparative Neuroanatomy define borders not clearly identifiable by cytoarchitectonics or immunostaining, providing precise definitions of brain subdivisions (Jarvis et al., 2013). In other cases they help to determine with accuracy the cell populations that express a given marker, as done to show compensatory changes to the loss of the dopamine transporter (DAT) gene in birds (Lovell et al., 2015). Most importantly, perhaps, these patterns can help to establish molecular signatures of specific areas, subdivisions, and nuclei, or identify subregions not previously appreciated, for example in the arcopallium (Mello et al., 2019) or hippocampus. In many cases, the genes showing regional differential patterns in finch represent markers of specific brain areas or subdivisions in mammals, such as cortical layers or amygdalar subdivisions. While the expression of some shared avian/mammalian markers have been previously reported in some birds (e.g. markers of cortical layers 4 vs 5/6 in rodents; Dugas-Ford et al., 2012; Jarvis et al., 2013) the data in ZEBrA provides more comprehensive brain distribution assessments, thus enabling more systematic comparisons. This is particularly helpful to distinguish markers of single vs. multiple structures, which is critical when utilizing such markers as supportive evidence for molecular convergence of analogous structures or for brain homologies between birds and mammals. As ZEBrA expands, avian brain expression patterns and signatures in comparison with the available data in mammals (e.g. Mouse Brain Atlas) will play increasing roles in defining parallels and/or divergences between these two lineages.
The gene sets under Gene Function reflect a range of pathways of relevance to brain function, but we highlight ion channels or neurotransmitter/ neuromodulatory receptors, as they provide a systematic roadmap to understanding the regulation of electrophysiological properties of song nuclei. This information can help to identify tools for pharmacological manipulation, and thus be key to defining the contributions of specific nuclei to song learning and production. By providing extensive and side-by-side expression data for regional markers and the opportunity to relate the patterns to other gene attributes, the data in ZEBrA complements previous and ongoing descriptions of avian brain gene expression in these families (Friedrich et al., 2019; Lovell et al., 2013; Wada et al., 2004). Other functional categories of potential high impact to songbird researches relate to hormonal, metabolic and cell proliferation pathways, all reflecting topics tightly linked to songbird brain studies, as also highlighted in Songbird Discoveries. More broadly, ZEBrA data also helps to identify molecular pathways that are expressed in non-vocal circuits or in general brain areas and likely to affect broad aspects of brain function and behavior. Of note, the functional data are mostly from studies in non-avian experimental organisms (e.g. drosophila, mice) that are more appropriate for establishing causal links between genes and molecular function. While caution is required, it is reasonable to assume that molecular function is conserved for genes with high sequence identity across lineages, especially in domains critical for function (e.g. enzymatic or allosteric domains, DNA-binding motifs, protein-protein interaction domains).
Since its inception ZEBrA has been a freely available online resource geared towards meeting the needs of the research community. The original high resolution images are freely available upon request, for use in research projects, publications and presentations. We are also open to suggestions on how to improve the features and tools available in the ZEBrA website, as well as on how to expand the database. We also note that the atlas annotations are an ongoing effort, and thus we also welcome suggestions for modifying/correcting current annotations, and/or to label regions or nuclei that are currently unidentified in the histological reference atlas and/or in specific in situ images. Lastly, we welcome suggestions for including past and future studies reporting gene expression patterns in zebra finches to the Songbird Discoveries portal.
Supplementary Material
Acknowledgements:
We wish to thank Tessa Marzulla, Katy Horback, and Vadim Pinsky for their assistance with histological sectioning, in situ hybridizations, and image processing, Michele Hribar for co-authoring a previous version of the ZEBrA website, and NextInteractives and David Weiner for implementing the redesign, branding, and navigation features currently available on ZEBrA.
Funding:
Claudio V. Mello:
National Institutes of Health (R03NS059755, R24GM120464, R24GM092842)
Partha Mitra:
The Crick-Clay Professorship
The Mathers Charitable Foundation
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data Availability
All of the data supporting the findings of this study are presented in this article or in SupportingInfo:SuppTable1.xlsx. High-resolution photomicrographs of in situ hybridization images for most of the genes presented in this study are also available online publicly on ZEBrA (www.zebrafinchatlas.org; RRID: SCR_012988).
References
- Achiro JM, Shen J, & Bottjer SW (2017). Neural activity in cortico-basal ganglia circuits of juvenile songbirds encodes performance during goal-directed learning. Elife, 6. doi: 10.7554/eLife.26973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acquaah-Mensah GK, & Taylor RC (2016). Brain in situ hybridization maps as a source for reverse-engineering transcriptional regulatory networks: Alzheimer’s disease insights. Gene, 586(1), 77–86. doi: 10.1016/j.gene.2016.03.045 [DOI] [PubMed] [Google Scholar]
- Adam I, Mendoza E, Kobalz U, Wohlgemuth S, & Scharff C. (2016). FoxP2 directly regulates the reelin receptor VLDLR developmentally and by singing. Mol Cell Neurosci, 74, 96–105. doi: 10.1016/j.mcn.2016.04.002 [DOI] [PubMed] [Google Scholar]
- Alarcon M, Abrahams BS, Stone JL, Duvall JA, Perederiy JV, Bomar JM, . . . Geschwind DH (2008). Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am J Hum Genet, 82(1), 150–159. doi: 10.1016/j.ajhg.2007.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Buylla A, Kirn JR, & Nottebohm F. (1990). Birth of projection neurons in adult avian brain may be related to perceptual or motor learning. Science, 249(4975), 1444–1446. doi: 10.1126/science.1698312 [DOI] [PubMed] [Google Scholar]
- Aronov D, Andalman AS, & Fee MS (2008). A specialized forebrain circuit for vocal babbling in the juvenile songbird. Science, 320(5876), 630–634. doi: 10.1126/science.1155140 [DOI] [PubMed] [Google Scholar]
- Belgard TG, Marques AC, Oliver PL, Abaan HO, Sirey TM, Hoerder-Suabedissen A, . . . Ponting CP (2011). A transcriptomic atlas of mouse neocortical layers. Neuron, 71(4), 605–616. doi: 10.1016/j.neuron.2011.06.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgard TG, Montiel JF, Wang WZ, Garcia-Moreno F, Margulies EH, Ponting CP, & Molnar Z. (2013). Adult pallium transcriptomes surprise in not reflecting predicted homologies across diverse chicken and mouse pallial sectors. Proc Natl Acad Sci U S A, 110(32), 13150–13155. doi: 10.1073/pnas.1307444110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett ATD, Cuthill IC, Partridge JC, & Maier EJ (1996). Ultraviolet vision and mate choice in zebra finches. Nature, 380, 433–433. [Google Scholar]
- Bohland JW, Bokil H, Pathak SD, Lee CK, Ng L, Lau C, . . . Mitra PP. (2010). Clustering of spatial gene expression patterns in the mouse brain and comparison with classical neuroanatomy. Methods, 50(2), 105–112. doi: 10.1016/j.ymeth.2009.09.001 [DOI] [PubMed] [Google Scholar]
- Bolhuis JJ, Okanoya K, & Scharff C. (2010). Twitter evolution: converging mechanisms in birdsong and human speech. Nat Rev Neurosci, 11(11), 747–759. doi: 10.1038/nrn2931 [DOI] [PubMed] [Google Scholar]
- Bottjer SW, & Altenau B. (2010). Parallel pathways for vocal learning in basal ganglia of songbirds. Nat Neurosci, 13(2), 153–155. doi: 10.1038/nn.2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottjer SW, & Arnold AP (1984). The role of feedback from the vocal organ. I. Maintenance of stereotypical vocalizations by adult zebra finches. J Neurosci, 4(9), 2387–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottjer SW, Brady JD, & Cribbs B. (2000). Connections of a motor cortical region in zebra finches: relation to pathways for vocal learning. J Comp Neurol, 420(2), 244–260. [PubMed] [Google Scholar]
- Bottjer SW, Halsema KA, Brown SA, & Miesner EA (1989). Axonal connections of a forebrain nucleus involved with vocal learning in zebra finches. J Comp Neurol, 279(2), 312–326. doi: 10.1002/cne.902790211 [DOI] [PubMed] [Google Scholar]
- Brainard MS, & Doupe AJ (2000a). Auditory feedback in learning and maintenance of vocal behaviour. Nat Rev Neurosci, 1(1), 31–40. doi: 10.1038/35036205 [DOI] [PubMed] [Google Scholar]
- Brainard MS, & Doupe AJ (2000b). Interruption of a basal ganglia-forebrain circuit prevents plasticity of learned vocalizations. Nature, 404(6779), 762–766. doi: 10.1038/35008083 [DOI] [PubMed] [Google Scholar]
- Brawn TP, Fenn KM, Nusbaum HC, & Margoliash D. (2008). Consolidation of sensorimotor learning during sleep. Learn Mem, 15(11), 815–819. doi: 10.1101/lm.1180908 [DOI] [PubMed] [Google Scholar]
- Budzillo A, Duffy A, Miller KE, Fairhall AL, & Perkel DJ (2017). Dopaminergic modulation of basal ganglia output through coupled excitation-inhibition. Proc Natl Acad Sci U S A, 114(22), 5713–5718. doi: 10.1073/pnas.1611146114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkett ZD, Day NF, Kimball TH, Aamodt CM, Heston JB, Hilliard AT, . . . White SA. (2018). FoxP2 isoforms delineate spatiotemporal transcriptional networks for vocal learning in the zebra finch. Elife, 7. doi: 10.7554/eLife.30649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carleton JB, Lovell PV, McHugh A, Marzulla T, Horback KL, & Mello CV (2014). An optimized protocol for high-throughput in situ hybridization of zebra finch brain. Cold Spring Harb Protoc, 2014(12), 1249–1258. doi: 10.1101/pdb.prot084582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton NS, & Emery NJ (2015). Avian Models for Human Cognitive Neuroscience: A Proposal. Neuron, 86(6), 1330–1342. doi: 10.1016/j.neuron.2015.04.024 [DOI] [PubMed] [Google Scholar]
- Cobos I, Shimamura K, Rubenstein JL, Martinez S, & Puelles L. (2001). Fate map of the avian anterior forebrain at the four-somite stage, based on the analysis of quail-chick chimeras. Dev Biol, 239(1), 46–67. doi: 10.1006/dbio.2001.0423 [DOI] [PubMed] [Google Scholar]
- Condro MC, & White SA (2014). Distribution of language-related Cntnap2 protein in neural circuits critical for vocal learning. J Comp Neurol, 522(1), 169–185. doi: 10.1002/cne.23394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave AS, & Margoliash D. (2000). Song replay during sleep and computational rules for sensorimotor vocal learning. Science, 290(5492), 812–816. doi: 10.1126/science.290.5492.812 [DOI] [PubMed] [Google Scholar]
- den Boer-Visser AM, Brittijn ML, Dubbeldam JL (Ed.) (2004). A stereotaxic atlas of the brain of the collared dove, Streptopelia decaocto: using the old and new nomenclature: Shaker Publishing B.V. [Google Scholar]
- Doupe AJ, & Kuhl PK (1999). Birdsong and human speech: common themes and mechanisms. Annu Rev Neurosci, 22, 567–631. doi: 10.1146/annurev.neuro.22.1.567 [DOI] [PubMed] [Google Scholar]
- Drnevich J, Replogle KL, Lovell P, Hahn TP, Johnson F, Mast TG, . . . Clayton DF (2012). Impact of experience-dependent and -independent factors on gene expression in songbird brain. Proc Natl Acad Sci U S A, 109 Suppl 2, 17245–17252. doi: 10.1073/pnas.1200655109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas-Ford J, Rowell JJ, & Ragsdale CW (2012). Cell-type homologies and the origins of the neocortex. Proc Natl Acad Sci U S A, 109(42), 16974–16979. doi: 10.1073/pnas.1204773109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutar P, Vu HM, & Perkel DJ (1998). Multiple cell types distinguished by physiological, pharmacological, and anatomic properties in nucleus HVc of the adult zebra finch. J Neurophysiol, 80(4), 1828–1838. doi: 10.1152/jn.1998.80.4.1828 [DOI] [PubMed] [Google Scholar]
- Fee MS, Kozhevnikov AA, & Hahnloser RH (2004). Neural mechanisms of vocal sequence generation in the songbird. Ann N Y Acad Sci, 1016, 153–170. doi: 10.1196/annals.1298.022 [DOI] [PubMed] [Google Scholar]
- Fee MS, & Scharff C. (2010). The songbird as a model for the generation and learning of complex sequential behaviors. ILAR J, 51(4), 362–377. doi: 10.1093/ilar.51.4.362 [DOI] [PubMed] [Google Scholar]
- Feenders G, Liedvogel M, Rivas M, Zapka M, Horita H, Hara E, . . . Jarvis ED (2008). Molecular mapping of movement-associated areas in the avian brain: a motor theory for vocal learning origin. PLoS One, 3(3), e1768. doi: 10.1371/journal.pone.0001768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez AS, Pieau C, Reperant J, Boncinelli E, & Wassef M. (1998). Expression of the Emx-1 and Dlx-1 homeobox genes define three molecularly distinct domains in the telencephalon of mouse, chick, turtle and frog embryos: implications for the evolution of telencephalic subdivisions in amniotes. Development, 125(11), 2099–2111. [DOI] [PubMed] [Google Scholar]
- Fraser S, Keynes R, & Lumsden A. (1990). Segmentation in the chick embryo hindbrain is defined by cell lineage restrictions. Nature, 344(6265), 431–435. doi: 10.1038/344431a0 [DOI] [PubMed] [Google Scholar]
- Friedrich SR, Lovell PV, Kaser TM, & Mello CV (2019). Exploring the molecular basis of neuronal excitability in a vocal learner. BMC Genomics, 20(1), 629. doi: 10.1186/s12864-019-5871-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadagkar V, Puzerey PA, Chen R, Baird-Daniel E, Farhang AR, & Goldberg JH (2016). Dopamine neurons encode performance error in singing birds. Science, 354(6317), 1278–1282. doi: 10.1126/science.aah6837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney MF, & Hodos W. (2003). The visual acuity and refractive state of the American kestrel (Falco sparverius). Vision Res, 43(19), 2053–2059. doi: 10.1016/s00426989(03)00304-3 [DOI] [PubMed] [Google Scholar]
- Goldberg JH, & Fee MS (2010). Singing-related neural activity distinguishes four classes of putative striatal neurons in the songbird basal ganglia. J Neurophysiol, 103(4), 2002–2014. doi: 10.1152/jn.01038.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodson JL, & Kabelik D. (2009). Dynamic limbic networks and social diversity in vertebrates: from neural context to neuromodulatory patterning. Front Neuroendocrinol, 30(4), 429–441. doi: 10.1016/j.yfrne.2009.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodson JL, Kabelik D, Kelly AM, Rinaldi J, & Klatt JD (2009). Midbrain dopamine neurons reflect affiliation phenotypes in finches and are tightly coupled to courtship. Proc Natl Acad Sci U S A, 106(21), 8737–8742. doi: 10.1073/pnas.0811821106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grothe B, Carr CE, Casseday JH, Fritzsch B, & Koppl C. (2004). The Evolution of Central Pathways and Their Neural Processing Patterns (Vol. 22). New York, NY: Springer. [Google Scholar]
- Gunturkun O, & Bugnyar T. (2016). Cognition without Cortex. Trends Cogn Sci, 20(4), 291–303. doi: 10.1016/j.tics.2016.02.001 [DOI] [PubMed] [Google Scholar]
- Guy J, & Staiger JF (2017). The Functioning of a Cortex without Layers. Front Neuroanat, 11, 54. doi: 10.3389/fnana.2017.00054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haesler S, Rochefort C, Georgi B, Licznerski P, Osten P, & Scharff C. (2007). Incomplete and inaccurate vocal imitation after knockdown of FoxP2 in songbird basal ganglia nucleus Area X. PLoS Biol, 5(12), e321. doi: 10.1371/journal.pbio.0050321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haesler S, Wada K, Nshdejan A, Morrisey EE, Lints T, Jarvis ED, & Scharff C. (2004). FoxP2 expression in avian vocal learners and non-learners. J Neurosci, 24(13), 3164–3175. doi: 10.1523/JNEUROSCI.4369-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahnloser RH, Kozhevnikov AA, & Fee MS (2002). An ultra-sparse code underlies the generation of neural sequences in a songbird. Nature, 419(6902), 65–70. doi: 10.1038/nature00974 [DOI] [PubMed] [Google Scholar]
- Hahnloser RH, Kozhevnikov AA, & Fee MS (2006). Sleep-related neural activity in a premotor and a basal-ganglia pathway of the songbird. J Neurophysiol, 96(2), 794–812. doi: 10.1152/jn.01064.2005 [DOI] [PubMed] [Google Scholar]
- Hamburger V, & Hamilton HL (1951). A series of normal stages in the development of the chick embryo. J Morphol, 88(1), 49–92. [PubMed] [Google Scholar]
- Hammerschmidt K, Reisinger E, Westekemper K, Ehrenreich L, Strenzke N, & Fischer J. (2012). Mice do not require auditory input for the normal development of their ultrasonic vocalizations. BMC Neurosci, 13, 40. doi: 10.1186/1471-220213-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerschmidt K, Whelan G, Eichele G, & Fischer J. (2015). Mice lacking the cerebral cortex develop normal song: insights into the foundations of vocal learning. Sci Rep, 5, 8808. doi: 10.1038/srep08808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara E, Kubikova L, Hessler NA, & Jarvis ED (2009). Assessing visual requirements for social context-dependent activation of the songbird song system. Proc Biol Sci, 276(1655), 279–289. doi: 10.1098/rspb.2008.1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara E, Rivas MV, Ward JM, Okanoya K, & Jarvis ED (2012). Convergent differential regulation of parvalbumin in the brains of vocal learners. PLoS One, 7(1), e29457. doi: 10.1371/journal.pone.0029457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawrylycz M, Bernard A, Lau C, Sunkin SM, Chakravarty MM, Lein ES, . . . Ng L. (2010). Areal and laminar differentiation in the mouse neocortex using large scale gene expression data. Methods, 50(2), 113–121. doi: 10.1016/j.ymeth.2009.09.005 [DOI] [PubMed] [Google Scholar]
- Henry AM, & Hohmann JG (2012). High-resolution gene expression atlases for adult and developing mouse brain and spinal cord. Mamm Genome, 23(9–10), 539–549. doi: 10.1007/s00335-012-9406-2 [DOI] [PubMed] [Google Scholar]
- Heston JB, & White SA (2015). Behavior-linked FoxP2 regulation enables zebra finch vocal learning. J Neurosci, 35(7), 2885–2894. doi: 10.1523/JNEUROSCI.3715-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyers D, Manns M, Luksch H, Gunturkun O, & Mouritsen H. (2007). A visual pathway links brain structures active during magnetic compass orientation in migratory birds. PLoS One, 2(9), e937. doi: 10.1371/journal.pone.0000937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliard AT, Miller JE, Fraley ER, Horvath S, & White SA (2012a). Molecular microcircuitry underlies functional specification in a basal ganglia circuit dedicated to vocal learning. Neuron, 73(3), 537–552. doi: 10.1016/j.neuron.2012.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliard AT, Miller JE, Horvath S, & White SA (2012b). Distinct neurogenomic states in basal ganglia subregions relate differently to singing behavior in songbirds. PLoS Comput Biol, 8(11), e1002773. doi: 10.1371/journal.pcbi.1002773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisey E, Kearney MG, & Mooney R. (2018). A common neural circuit mechanism for internally guided and externally reinforced forms of motor learning. Nat Neurosci, 21(4), 589–597. doi: 10.1038/s41593-018-0092-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochheiser H, & Yanowitz J. (2007). If I only had a brain: exploring mouse brain images in the Allen Brain Atlas. Biol Cell, 99(7), 403–409. doi: 10.1042/BC20070031 [DOI] [PubMed] [Google Scholar]
- Hodos W, Potocki A, Ghim MM, & Gaffney M. (2003). Temporal modulation of spatial contrast vision in pigeons (Columba livia). Vision Res, 43(7), 761–767. doi: 10.1016/s0042-6989(02)00417-0 [DOI] [PubMed] [Google Scholar]
- Hong H, & Sanchez JT (2018). Need for Speed and Precision: Structural and Functional Specialization in the Cochlear Nucleus of the Avian Auditory System. J Exp Neurosci, 12, 1179069518815628. doi: 10.1177/1179069518815628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horita H, Kobayashi M, Liu WC, Oka K, Jarvis ED, & Wada K. (2012). Specialized motor-driven dusp1 expression in the song systems of multiple lineages of vocal learning birds. PLoS One, 7(8), e42173. doi: 10.1371/journal.pone.0042173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janik VM (2014). Cetacean vocal learning and communication. Curr Opin Neurobiol, 28, 60–65. doi: 10.1016/j.conb.2014.06.010 [DOI] [PubMed] [Google Scholar]
- Jarvis ED (2004). Learned birdsong and the neurobiology of human language. Ann N Y Acad Sci, 1016, 749–777. doi: 10.1196/annals.1298.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis ED (2019). Evolution of vocal learning and spoken language. Science, 366(6461), 50–54. doi: 10.1126/science.aax0287 [DOI] [PubMed] [Google Scholar]
- Jarvis ED, Gunturkun O, Bruce L, Csillag A, Karten H, Kuenzel W, . . . Avian Brain Nomenclature, C. (2005). Avian brains and a new understanding of vertebrate brain evolution. Nat Rev Neurosci, 6(2), 151–159. doi: 10.1038/nrn1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis ED, Yu J, Rivas MV, Horita H, Feenders G, Whitney O, . . . Wada K (2013). Global view of the functional molecular organization of the avian cerebrum: mirror images and functional columns. J Comp Neurol, 521(16), 3614–3665. doi: 10.1002/cne.23404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson F, Sablan MM, & Bottjer SW (1995). Topographic organization of a forebrain pathway involved with vocal learning in zebra finches. J Comp Neurol, 358(2), 260–278. doi: 10.1002/cne.903580208 [DOI] [PubMed] [Google Scholar]
- Jones AR, Overly CC, & Sunkin SM (2009). The Allen Brain Atlas: 5 years and beyond. Nat Rev Neurosci, 10(11), 821–828. doi: 10.1038/nrn2722 [DOI] [PubMed] [Google Scholar]
- Jurgens U. (1998). Neuronal control of mammalian vocalization, with special reference to the squirrel monkey. Naturwissenschaften, 85(8), 376–388. doi: 10.1007/s001140050519 [DOI] [PubMed] [Google Scholar]
- Jurgens U. (2009). The neural control of vocalization in mammals: a review. J Voice, 23(1), 1–10. doi: 10.1016/j.jvoice.2007.07.005 [DOI] [PubMed] [Google Scholar]
- Kao MH, Doupe AJ, & Brainard MS (2005). Contributions of an avian basal ganglia-forebrain circuit to real-time modulation of song. Nature, 433(7026), 638–643. doi: 10.1038/nature03127 [DOI] [PubMed] [Google Scholar]
- Karten HJ (1997). Evolutionary developmental biology meets the brain: the origins of mammalian cortex. Proc Natl Acad Sci U S A, 94(7), 2800–2804. doi: 10.1073/pnas.94.7.2800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karten HJ (2013). Neocortical evolution: neuronal circuits arise independently of lamination. Curr Biol, 23(1), R12–15. doi: 10.1016/j.cub.2012.11.013 [DOI] [PubMed] [Google Scholar]
- Karten HJ, Brzozowska-Prechtl A, Lovell PV, Tang DD, Mello CV, Wang H, & Mitra PP (2013). Digital atlas of the zebra finch (Taeniopygia guttata) brain: a high-resolution photo atlas. J Comp Neurol, 521(16), 3702–3715. doi: 10.1002/cne.23443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karten HJ, & Hodos W. (1967). A stereotaxic atlas of the brain of the pigeon (Columba livia). Baltimore, MD: Johns Hopkins Press. [Google Scholar]
- Kato M, & Okanoya K. (2010). Molecular characterization of the song control nucleus HVC in Bengalese finch brain. Brain Res, 1360, 56–76. doi: 10.1016/j.brainres.2010.09.014 [DOI] [PubMed] [Google Scholar]
- Kelly AM, & Goodson JL (2014). Social functions of individual vasopressin-oxytocin cell groups in vertebrates: what do we really know? Front Neuroendocrinol, 35(4), 512–529. doi: 10.1016/j.yfrne.2014.04.005 [DOI] [PubMed] [Google Scholar]
- Kent WJ (2002). BLAT--the BLAST-like alignment tool. Genome Res, 12(4), 656–664. doi: 10.1101/gr.229202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keynes R, & Cook G. (2018). Segmentation of the chick central and peripheral nervous systems. Int J Dev Biol, 62(1–2-3), 177–182. doi: 10.1387/ijdb.170297rk [DOI] [PubMed] [Google Scholar]
- Keynes RJ, & Stern CD (1984). Segmentation in the vertebrate nervous system. Nature, 310(5980), 786–789. doi: 10.1038/310786a0 [DOI] [PubMed] [Google Scholar]
- Knornschild M. (2014). Vocal production learning in bats. Curr Opin Neurobiol, 28, 80–85. doi: 10.1016/j.conb.2014.06.014 [DOI] [PubMed] [Google Scholar]
- Knudsen EI, & Konishi M. (1978). Center-surround organization of auditory receptive fields in the owl. Science, 202(4369), 778–780. doi: 10.1126/science.715444 [DOI] [PubMed] [Google Scholar]
- Konishi M. (2003). Coding of auditory space. Annu Rev Neurosci, 26, 31–55. doi: 10.1146/annurev.neuro.26.041002.131123 [DOI] [PubMed] [Google Scholar]
- Konishi M, & Akutagawa E. (1985). Neuronal growth, atrophy and death in a sexually dimorphic song nucleus in the zebra finch brain. Nature, 315(6015), 145–147. doi: 10.1038/315145a0 [DOI] [PubMed] [Google Scholar]
- Konopka G, Bomar JM, Winden K, Coppola G, Jonsson ZO, Gao F, . . . Geschwind DH (2009). Human-specific transcriptional regulation of CNS development genes by FOXP2. Nature, 462(7270), 213–217. doi: 10.1038/nature08549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korlach J, Gedman G, Kingan SB, Chin CS, Howard JT, Audet JN, . . . Jarvis ED (2017). De novo PacBio long-read and phased avian genome assemblies correct and add to reference genes generated with intermediate and short reads. Gigascience, 6(10), 1–16. doi: 10.1093/gigascience/gix085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubikova L, Wada K, & Jarvis ED (2010). Dopamine receptors in a songbird brain. J Comp Neurol, 518(6), 741–769. doi: 10.1002/cne.22255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubke MF, Yazaki-Sugiyama Y, Mooney R, & Wild JM (2005). Physiology of neuronal subtypes in the respiratory-vocal integration nucleus retroamigualis of the male zebra finch. J Neurophysiol, 94(4), 2379–2390. doi: 10.1152/jn.00257.2005 [DOI] [PubMed] [Google Scholar]
- Kuenzel WJ, Masson M. (1988). A Stereotaxic Atlas of the Brain of the chick (Gallus domesticus). Baltimore, MD: Johns Hopkins University Press. [Google Scholar]
- Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, & Monaco AP (2001). A forkhead-domain gene is mutated in a severe speech and language disorder. Nature, 413(6855), 519–523. doi: 10.1038/35097076 [DOI] [PubMed] [Google Scholar]
- Lance-Jones C, & Landmesser L. (1980). Motoneurone projection patterns in the chick hind limb following early partial reversals of the spinal cord. J Physiol, 302, 581–602. doi: 10.1113/jphysiol.1980.sp013262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langner G. (1981). Neuronal mechanisms for pitch analysis in the time domain. Exp Brain Res, 44(4), 450–454. doi: 10.1007/bf00238840 [DOI] [PubMed] [Google Scholar]
- Lein ES, Belgard TG, Hawrylycz M, & Molnar Z. (2017). Transcriptomic Perspectives on Neocortical Structure, Development, Evolution, and Disease. Annu Rev Neurosci, 40, 629–652. doi: 10.1146/annurev-neuro-070815-013858 [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, . . . Jones AR (2007). Genome-wide atlas of gene expression in the adult mouse brain. Nature, 445(7124), 168–176. doi: 10.1038/nature05453 [DOI] [PubMed] [Google Scholar]
- Leonardo A, & Fee MS (2005). Ensemble coding of vocal control in birdsong. J Neurosci, 25(3), 652–661. doi: 10.1523/JNEUROSCI.3036-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Hu Z, He Y, Xiong Z, Long Z, Peng Y, . . . Xia K. (2010). Association analysis of CNTNAP2 polymorphisms with autism in the Chinese Han population. Psychiatr Genet, 20(3), 113–117. doi: 10.1097/YPG.0b013e32833a216f [DOI] [PubMed] [Google Scholar]
- Li X, Wang XJ, Tannenhauser J, Podell S, Mukherjee P, Hertel M, . . . Gaasterland T. (2007). Genomic resources for songbird research and their use in characterizing gene expression during brain development. Proc Natl Acad Sci U S A, 104(16), 6834–6839. doi: 10.1073/pnas.0701619104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Chen H, Jiang X, Li X, Lv J, Li M, . . . Liu T. (2017). Transcriptome Architecture of Adult Mouse Brain Revealed by Sparse Coding of Genome-Wide In Situ Hybridization Images. Neuroinformatics, 15(3), 285–295. doi: 10.1007/s12021-017-9333-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell PV, Carleton JB, & Mello CV (2013). Genomics analysis of potassium channel genes in songbirds reveals molecular specializations of brain circuits for the maintenance and production of learned vocalizations. BMC Genomics, 14(1), 470. doi: 10.1186/1471-2164-14-470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell PV, Clayton DF, Replogle KL, & Mello CV (2008). Birdsong “transcriptomics”: neurochemical specializations of the oscine song system. PLoS One, 3(10), e3440. doi: 10.1371/journal.pone.0003440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell PV, Huizinga NA, Friedrich SR, Wirthlin M, & Mello CV (2018a). The constitutive differential transcriptome of a brain circuit for vocal learning. BMC Genomics, 19(1), 231. doi: 10.1186/s12864-018-4578-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell PV, Huizinga NA, Getachew A, Mees B, Friedrich SR, Wirthlin M, & Mello CV (2018b). Curation of microarray oligonucleotides and corresponding ESTs/cDNAs used for gene expression analysis in zebra finches. BMC Res Notes, 11(1), 309. doi: 10.1186/s13104-018-3402-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell PV, Kasimi B, Carleton J, Velho TA, & Mello CV (2015). Living without DAT: Loss and compensation of the dopamine transporter gene in sauropsids (birds and reptiles). Sci Rep, 5, 14093. doi: 10.1038/srep14093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell PV, & Mello CV (2011). Brain expression and song regulation of the cholecystokinin gene in the zebra finch (Taeniopygia guttata). J Comp Neurol, 519(2), 211–237. doi: 10.1002/cne.22513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell PV, & Mello CV (2018). Redefining the convergent brain transcriptome for human speech and birdsong. Paper presented at the Soc. for Neurosci., San Diego, CA. [Google Scholar]
- Lovell PV, Wirthlin M, Wilhelm L, Minx P, Lazar NH, Carbone L, . . . Mello CV (2014). Conserved syntenic clusters of protein coding genes are missing in birds. Genome Biol, 15(12), 565. doi: 10.1186/s13059-014-0565-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Ding L, & Perkel DJ (2001). An avian basal ganglia pathway essential for vocal learning forms a closed topographic loop. J Neurosci, 21(17), 6836–6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GR (2014). The subtlety of simple eyes: the tuning of visual fields to perceptual challenges in birds. Philos Trans R Soc Lond B Biol Sci, 369(1636), 20130040. doi: 10.1098/rstb.2013.0040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina L, Legaz I, Gonzalez G, De Castro F, Rubenstein JL, & Puelles L. (2004). Expression of Dbx1, Neurogenin 2, Semaphorin 5A, Cadherin 8, and Emx1 distinguish ventral and lateral pallial histogenetic divisions in the developing mouse claustroamygdaloid complex. J Comp Neurol, 474(4), 504–523. doi: 10.1002/cne.20141 [DOI] [PubMed] [Google Scholar]
- Mello CV (2014). The zebra finch, Taeniopygia guttata: an avian model for investigating the neurobiological basis of vocal learning. Cold Spring Harb Protoc, 2014(12), 1237–1242. doi: 10.1101/pdb.emo084574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CV, Kaser T, Buckner AA, Wirthlin M, & Lovell PV (2019). Molecular architecture of the zebra finch arcopallium. J Comp Neurol, 527(15), 2512–2556. doi: 10.1002/cne.24688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CV, Lovell PV, & Wirthlin M. (2018). Discovery of Novel Genes and Other Lineage-Specific Features Through Comparative Genomics (Gerlai RT Ed.). Cambridge, MA: Academic Press. [Google Scholar]
- Mello CV, Vates GE, Okuhata S, & Nottebohm F. (1998). Descending auditory pathways in the adult male zebra finch (Taeniopygia guttata). J Comp Neurol, 395(2), 137–160. [PubMed] [Google Scholar]
- Mendoza E, & Scharff C. (2017). Protein-Protein Interaction Among the FoxP Family Members and their Regulation of Two Target Genes, VLDLR and CNTNAP2 in the Zebra Finch Song System. Front Mol Neurosci, 10, 112. doi: 10.3389/fnmol.2017.00112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montiel JF, Vasistha NA, Garcia-Moreno F, & Molnar Z. (2016). From sauropsids to mammals and back: New approaches to comparative cortical development. J Comp Neurol, 524(3), 630–645. doi: 10.1002/cne.23871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney R. (2000). Different subthreshold mechanisms underlie song selectivity in identified HVc neurons of the zebra finch. J Neurosci, 20(14), 5420–5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney R, & Prather JF (2005). The HVC microcircuit: the synaptic basis for interactions between song motor and vocal plasticity pathways. J Neurosci, 25(8), 1952–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouritsen H, Feenders G, Liedvogel M, Wada K, & Jarvis ED (2005). Night-vision brain area in migratory songbirds. Proc Natl Acad Sci U S A, 102(23), 8339–8344. doi:0409575102 [pii] 10.1073/pnas.0409575102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouritsen H, Heyers D, & Gunturkun O. (2016). The Neural Basis of Long-Distance Navigation in Birds. Annu Rev Physiol, 78, 133–154. doi: 10.1146/annurev-physiol-021115-105054 [DOI] [PubMed] [Google Scholar]
- Murphy E, & Benitez-Burraco A. (2016). Bridging the Gap between Genes and Language Deficits in Schizophrenia: An Oscillopathic Approach. Front Hum Neurosci, 10, 422. doi: 10.3389/fnhum.2016.00422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbury DF, Paracchini S, Scerri TS, Winchester L, Addis L, Richardson AJ, . . . Monaco AP (2011). Investigation of dyslexia and SLI risk variants in reading-and language-impaired subjects. Behav Genet, 41(1), 90–104. doi: 10.1007/s10519-010-9424-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixdorf-Bergweiler BE, & Bischof H-J (2007). A stereotaxic atlas of the brain of the zebra finch, Taeniopygia guttata, with special special emphasis on telencephalic visual and song system nuclei in transverse and sagittal sections. . Bethesda: National Library of Medicine, National Center for Biotechnology Information. [Google Scholar]
- Nottebohm F, & Arnold AP (1976). Sexual dimorphism in vocal control areas of the songbird brain. Science, 194(4261), 211–213. doi: 10.1126/science.959852 [DOI] [PubMed] [Google Scholar]
- Nottebohm F, Kelley DB, & Paton JA (1982). Connections of vocal control nuclei in the canary telencephalon. J Comp Neurol, 207(4), 344–357. doi: 10.1002/cne.902070406 [DOI] [PubMed] [Google Scholar]
- Olkowicz S, Kocourek M, Lucan RK, Portes M, Fitch WT, Herculano-Houzel S, & Nemec P. (2016). Birds have primate-like numbers of neurons in the forebrain. Proc Natl Acad Sci U S A, 113(26), 7255–7260. doi: 10.1073/pnas.1517131113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson CR, Owen DC, Ryabinin AE, & Mello CV (2014). Drinking songs: alcohol effects on learned song of zebra finches. PLoS One, 9(12), e115427. doi: 10.1371/journal.pone.0115427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overholt EM, Rubel EW, & Hyson RL (1992). A circuit for coding interaural time differences in the chick brainstem. J Neurosci, 12(5), 1698–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panaitof SC, Abrahams BS, Dong H, Geschwind DH, & White SA (2010). Language-related Cntnap2 gene is differentially expressed in sexually dimorphic song nuclei essential for vocal learning in songbirds. J Comp Neurol, 518(11), 1995–2018. doi: 10.1002/cne.22318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepperberg IM (2006). Grey parrot numerical competence: a review. Anim Cogn, 9(4), 377–391. doi: 10.1007/s10071-006-0034-7 [DOI] [PubMed] [Google Scholar]
- Perkel DJ, Farries MA, Luo M, & Ding L. (2002). Electrophysiological analysis of a songbird basal ganglia circuit essential for vocal plasticity. Brain Res Bull, 57(3–4), 529–532. doi:S0361923001006906 [pii] [DOI] [PubMed] [Google Scholar]
- Peter B, Raskind WH, Matsushita M, Lisowski M, Vu T, Berninger VW, . . . Brkanac Z. (2011). Replication of CNTNAP2 association with nonword repetition and support for FOXP2 association with timed reading and motor activities in a dyslexia family sample. J Neurodev Disord, 3(1), 39–49. doi: 10.1007/s11689-010-9065-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkov CI, & Jarvis ED (2012). Birds, primates, and spoken language origins: behavioral phenotypes and neurobiological substrates. Front Evol Neurosci, 4, 12. doi: 10.3389/fnevo.2012.00012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfenning AR, Hara E, Whitney O, Rivas MV, Wang R, Roulhac PL, . . . Jarvis ED (2014). Convergent transcriptional specializations in the brains of humans and song-learning birds. Science, 346(6215), 1256846. doi: 10.1126/science.1256846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinaud R, Fortes AF, Lovell P, & Mello CV (2006). Calbindin-positive neurons reveal a sexual dimorphism within the songbird analogue of the mammalian auditory cortex. J Neurobiol, 66(2), 182–195. doi: 10.1002/neu.20211 [DOI] [PubMed] [Google Scholar]
- Poole JH, Tyack PL, Stoeger-Horwath AS, & Watwood S. (2005). Animal behaviour: elephants are capable of vocal learning. Nature, 434(7032), 455–456. doi: 10.1038/434455a [DOI] [PubMed] [Google Scholar]
- Prat Y, Taub M, & Yovel Y. (2015). Vocal learning in a social mammal: Demonstrated by isolation and playback experiments in bats. Sci Adv, 1(2), e1500019. doi: 10.1126/sciadv.1500019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puelles L. (2017). Comments on the Updated Tetrapartite Pallium Model in the Mouse and Chick, Featuring a Homologous Claustro-Insular Complex. Brain Behav Evol, 90(2), 171–189. doi: 10.1159/000479782 [DOI] [PubMed] [Google Scholar]
- Puelles L, Ayad A, Alonso A, Sandoval JE, MartInez-de-la-Torre M, Medina L, & Ferran JL (2016). Selective early expression of the orphan nuclear receptor Nr4a2 identifies the claustrum homolog in the avian mesopallium: Impact on sauropsidian/mammalian pallium comparisons. J Comp Neurol, 524(3), 665–703. doi: 10.1002/cne.23902 [DOI] [PubMed] [Google Scholar]
- Puelles L, Kuwana E, Puelles E, Bulfone A, Shimamura K, Keleher J, . . . Rubenstein JL(2000). Pallial and subpallial derivatives in the embryonic chick and mouse telencephalon, traced by the expression of the genes Dlx-2, Emx-1, Nkx-2.1, Pax-6, and Tbr-1. J Comp Neurol, 424(3), 409–438. doi: [DOI] [PubMed] [Google Scholar]
- Puelles L, Kuwana E, Puelles E, & Rubenstein JL (1999). Comparison of the mammalian and avian telencephalon from the perspective of gene expression data. Eur J Morphol, 37(2–3), 139–150. doi: 10.1076/ejom.37.2.139.4756 [DOI] [PubMed] [Google Scholar]
- Puelles L, Paxinos G, Watson C, Martinez S, Martinez-de-la-Torre M. (2007). The Chick Brain in Stereotaxic Coordinates: An Atlas Based on Neuromeres: Elsevier Science & Technology Books. [Google Scholar]
- Redies C, Medina L, & Puelles L. (2001). Cadherin expression by embryonic divisions and derived gray matter structures in the telencephalon of the chicken. J Comp Neurol, 438(3), 253–285. [PubMed] [Google Scholar]
- Reiner A, Medina L, & Veenman CL (1998). Structural and functional evolution of the basal ganglia in vertebrates. Brain Res Brain Res Rev, 28(3), 235–285. doi: 10.1016/s0165-0173(98)00016-2 [DOI] [PubMed] [Google Scholar]
- Reiner A, Perkel DJ, Mello CV, & Jarvis ED (2004). Songbirds and the revised avian brain nomenclature. Ann N Y Acad Sci, 1016, 77–108. doi: 10.1196/annals.1298.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Replogle K, Arnold AP, Ball GF, Band M, Bensch S, Brenowitz EA, . . . Clayton DF (2008). The Songbird Neurogenomics (SoNG) Initiative: community-based tools and strategies for study of brain gene function and evolution. BMC Genomics, 9(1), 131. doi: 10.1186/1471-2164-9-131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts TF, Gobes SM, Murugan M, Olveczky BP, & Mooney R. (2012). Motor circuits are required to encode a sensory model for imitative learning. Nat Neurosci, 15(10), 1454–1459. doi: 10.1038/nn.3206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen MJ, & Mooney R. (2003). Inhibitory and excitatory mechanisms underlying auditory responses to learned vocalizations in the songbird nucleus HVC. Neuron, 39(1), 177–194. doi: 10.1016/s0896-6273(03)00357-x [DOI] [PubMed] [Google Scholar]
- Scharff C, & Nottebohm F. (1991). A comparative study of the behavioral deficits following lesions of various parts of the zebra finch song system: implications for vocal learning. J Neurosci, 11(9), 2896–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheich H, & Bonke BA (1981). Tone-versus FM--induced patterns of excitation and suppression in the 14-C-2-deoxyglucose labeled auditory “cortex” of the guinea fowl. Exp Brain Res, 44(4), 445–449. doi: 10.1007/bf00238839 [DOI] [PubMed] [Google Scholar]
- Shimizu T, Patton TB, & Husband SA (2010). Avian visual behavior and the organization of the telencephalon. Brain Behav Evol, 75(3), 204–217. doi: 10.1159/000314283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DJ, & Rubel EW (1979). Organization and development of brain stem auditory nuclei of the chicken: dendritic gradients in nucleus laminaris. J Comp Neurol, 186(2), 213–239. doi: 10.1002/cne.901860207 [DOI] [PubMed] [Google Scholar]
- Soderstrom K, & Johnson F. (2003). Cannabinoid exposure alters learning of zebra finch vocal patterns. Brain Res Dev Brain Res, 142(2), 215–217. doi: 10.1016/s0165-3806(03)00061-0 [DOI] [PubMed] [Google Scholar]
- Soderstrom K, & Tian Q. (2004). Distinct periods of cannabinoid sensitivity during zebra finch vocal development. Brain Res Dev Brain Res, 153(2), 225–232. doi: 10.1016/j.devbrainres.2004.09.002 [DOI] [PubMed] [Google Scholar]
- Sohrabji F, Nordeen EJ, & Nordeen KW (1990). Selective impairment of song learning following lesions of a forebrain nucleus in the juvenile zebra finch. Behav Neural Biol, 53(1), 51–63. doi: 10.1016/0163-1047(90)90797-a [DOI] [PubMed] [Google Scholar]
- Spiteri E, Konopka G, Coppola G, Bomar J, Oldham M, Ou J, . . . Geschwind DH (2007). Identification of the transcriptional targets of FOXP2, a gene linked to speech and language, in developing human brain. Am J Hum Genet, 81(6), 1144–1157. doi: 10.1086/522237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson TJ, Replogle K, Drnevich J, Clayton DF, & Ball GF (2012). High throughput analysis reveals dissociable gene expression profiles in two independent neural systems involved in the regulation of social behavior. BMC Neurosci, 13, 126. doi: 10.1186/1471-2202-13-126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes TM, Leonard CM, & Nottebohm F. (1974). The telencephalon, diencephalon, and mesencephalon of the canary, Serinus canaria, in stereotaxic coordinates. J Comp Neurol, 156(3), 337–374. doi: 10.1002/cne.901560305 [DOI] [PubMed] [Google Scholar]
- Sunkin SM, Ng L, Lau C, Dolbeare T, Gilbert TL, Thompson CL, . . . Dang C (2013). Allen Brain Atlas: an integrated spatio-temporal portal for exploring the central nervous system. Nucleic Acids Res, 41(Database issue), D996–D1008. doi: 10.1093/nar/gks1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki IK, & Hirata T. (2014). A common developmental plan for neocortical gene-expressing neurons in the pallium of the domestic chicken Gallus gallus domesticus and the Chinese softshell turtle Pelodiscus sinensis. Front Neuroanat, 8, 20. doi: 10.3389/fnana.2014.00020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teramitsu I, Kudo LC, London SE, Geschwind DH, & White SA (2004). Parallel FoxP1 and FoxP2 expression in songbird and human brain predicts functional interaction. J Neurosci, 24(13), 3152–3163. doi: 10.1523/JNEUROSCI.5589-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theurich M, Langner G, & Scheich H. (1984). Infrasound responses in the midbrain of the guinea fowl. Neurosci Lett, 49(1–2), 81–86. doi: 10.1016/0304-3940(84)90140x [DOI] [PubMed] [Google Scholar]
- Thompson CK, Meitzen J, Replogle K, Drnevich J, Lent KL, Wissman AM, . . . Brenowitz EA (2012). Seasonal changes in patterns of gene expression in avian song control brain regions. PLoS One, 7(4), e35119. doi: 10.1371/journal.pone.0035119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CL, Ng L, Menon V, Martinez S, Lee CK, Glattfelder K, . . . Jones AR. (2014). A high-resolution spatiotemporal atlas of gene expression of the developing mouse brain. Neuron, 83(2), 309–323. doi: 10.1016/j.neuron.2014.05.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CL, Pathak SD, Jeromin A, Ng LL, MacPherson CR, Mortrud MT, . . . Lein ES (2008). Genomic anatomy of the hippocampus. Neuron, 60(6), 1010–1021. doi: 10.1016/j.neuron.2008.12.008 [DOI] [PubMed] [Google Scholar]
- Thompson JA, & Johnson F. (2007). HVC microlesions do not destabilize the vocal patterns of adult male zebra finches with prior ablation of LMAN. Dev Neurobiol, 67(2), 205–218. doi: 10.1002/dneu.20287 [DOI] [PubMed] [Google Scholar]
- Tosches MA, Yamawaki TM, Naumann RK, Jacobi AA, Tushev G, & Laurent G. (2018). Evolution of pallium, hippocampus, and cortical cell types revealed by single-cell transcriptomics in reptiles. Science, 360(6391), 881–888. doi: 10.1126/science.aar4237 [DOI] [PubMed] [Google Scholar]
- Vargha-Khadem F, Watkins KE, Price CJ, Ashburner J, Alcock KJ, Connelly A, . . . Passingham RE (1998). Neural basis of an inherited speech and language disorder. Proc Natl Acad Sci U S A, 95(21), 12695–12700. doi: 10.1073/pnas.95.21.12695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernes SC, Spiteri E, Nicod J, Groszer M, Taylor JM, Davies KE, . . . Fisher SE (2007). High-throughput analysis of promoter occupancy reveals direct neural targets of FOXP2, a gene mutated in speech and language disorders. Am J Hum Genet, 81(6), 1232–1250. doi: 10.1086/522238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada K, Howard JT, McConnell P, Whitney O, Lints T, Rivas MV, . . . Jarvis ED (2006). A molecular neuroethological approach for identifying and characterizing a cascade of behaviorally regulated genes. Proc Natl Acad Sci U S A, 103(41), 15212–15217. doi: 10.1073/pnas.0607098103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada K, Sakaguchi H, Jarvis ED, & Hagiwara M. (2004). Differential expression of glutamate receptors in avian neural pathways for learned vocalization. J Comp Neurol, 476(1), 44–64. doi: 10.1002/cne.20201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton PL, Christensen-Dalsgaard J, & Carr CE (2017). Evolution of Sound Source Localization Circuits in the Nonmammalian Vertebrate Brainstem. Brain Behav Evol, 90(2), 131–153. doi: 10.1159/000476028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Brzozowska-Prechtl A, & Karten HJ (2010). Laminar and columnar auditory cortex in avian brain. Proc Natl Acad Sci U S A, 107(28), 12676–12681. doi: 10.1073/pnas.1006645107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren WC, Clayton DF, Ellegren H, Arnold AP, Hillier LW, Kunstner A, . . . Wilson RK (2010). The genome of a songbird. Nature, 464(7289), 757–762. doi: 10.1038/nature08819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse AJ, Bishop DV, Ang QW, Pennell CE, & Fisher SE (2011). CNTNAP2 variants affect early language development in the general population. Genes Brain Behav, 10(4), 451–456. doi: 10.1111/j.1601-183X.2011.00684.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core and region-enriched networks of behaviorally regulated genes and the singing genome, 6215, 346 Cong. Rec. 1256780–1256780 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild JM (1993). Descending projections of the songbird nucleus robustus archistriatalis. J Comp Neurol, 338(2), 225–241. doi: 10.1002/cne.903380207 [DOI] [PubMed] [Google Scholar]
- Williams H. (1985). Sexual dimorphism of auditory activity in the zebra finch song system. Behav Neural Biol, 44(3), 470–484. doi: 10.1016/s0163-1047(85)90904-5 [DOI] [PubMed] [Google Scholar]
- Wirthlin M, Chang EF, Knornschild M, Krubitzer LA, Mello CV, Miller CT, . . . Yartsev MM (2019). A Modular Approach to Vocal Learning: Disentangling the Diversity of a Complex Behavioral Trait. Neuron, 104(1), 87–99. doi: 10.1016/j.neuron.2019.09.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirthlin M, Lovell PV, Jarvis ED, & Mello CV (2014). Comparative genomics reveals molecular features unique to the songbird lineage. BMC Genomics, 15, 1082. doi: 10.1186/1471-2164-15-1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlgemuth S, Adam I, & Scharff C. (2014). FoxP2 in songbirds. Curr Opin Neurobiol, 28, 86–93. doi: 10.1016/j.conb.2014.06.009 [DOI] [PubMed] [Google Scholar]
- Zann RA (1996). The Zebra Finch: a synthesis of field and laboratory studies (illustrated ed.): Oxford University Press. [Google Scholar]
- Zeier H, & Karten HJ (1971). The archistriatum of the pigeon: organization of afferent and efferent connections. Brain Res, 31(2), 313–326. doi: 10.1016/0006-8993(71)90185-5 [DOI] [PubMed] [Google Scholar]
- Zeigler HP, & Marler P. (2004). Behavioral neurobiology of birdsong (Zeigler HP& Marler P. Eds. Vol. 1016). New York: New York Academy of Sciences. [Google Scholar]
- Zeigler HP, & Marler P. (2008). Neuroscience of birdsong (Zeigler HP & Marler P. Eds.). New York, NY: Cambridge University Press. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.