Abstract
The role of the RNAi/Dicer/Ago system in degrading RNA viruses has been elusive in mammals in the past, which has prompted authors to think that interferon (IFN) synthesis is essential in this clade, relegating the RNAi defense strategy against viral infection as an accessory function. However, recent publications highlight the existence of abundant viral small interference and micro RNAs (VsiRNAs and VmiRNAs) in both cell-line and whole organism based experiments, indicating a contribution of these molecules in host responses and/or viral replication. We explore the theoretical possibility that RNAi triggered by SARS-CoV-2 might degrade some host transcripts in the opposite direction, although this hypothesis seems counterintuitive. The SARS-CoV-2 genome was therefore computationally searched for exact intrapairing within the viral RNA and exact hybrid pairing with the human transcriptome over a minimum of 20 bases in length. Minimal segments of 20-base lengths of SARS-CoV-2 RNA were found based on the theoretical matching with existing complementary strands in the human host transcriptome. Few human genes potentially annealing with SARS-CoV-2 RNA, including mitochondrial deubiquitinase USP30, the subunit of ubiquitin protein ligase complex FBXO21 and two long noncoding RNAs, were retrieved. The hypothesis that viral-originated RNAi might mediate degradation of host transcriptome messages was corroborated by published high throughput sequencing of RNA from infected tissues and cultured cells, clinical observation and phylogenetic comparative analysis, indicating a strong specificity of these SARS-CoV-2 hybrid pairing sequences for human genomes.
Keywords: SARS-CoV-2, dsRNA, Dicer, RNA cleavage, Host transcriptome, Viral genome
SARS-CoV-2; dsRNA; Dicer; RNA cleavage; Host transcriptome; Viral genome.
1. Introduction
Two quarantine-scale epidemics have already recently occurred, caused by two coronaviruses: severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV) (Marra et al., 2003; Van Boheemen et al., 2012). The third recent new viral outbreak causing a quickly propagating pandemic is provoked by a new coronavirus, named SARS-CoV-2 that exhibits terrifying uncontrolled spread. Metagenomic sequencing along with phylogenetic analysis of the virus from a sample of bronchoalveolar lavage fluid from an infected patient has been performed at the early days of epidemic (Wu et al., 2020; Zhou et al., 2020). The high homology of the predicted protein of the spike protein receptor-binding domain (RBD) with that of the precedent SARS-CoV confirms that human angiotensin-converting enzyme 2 (ACE2) acts as a receptor for binding to an entry into human host cells (Wrapp et al., 2020; Letko et al., 2020). In mammalian infected host cells, the role of RNAi machinery in degrading viruses has been controversial in the past. In fungi, plants and invertebrates, viral dsRNAs are trapped and cleaved by the Dicer/Ago machinery into small interfering RNAs (siRNAs) that degrade the targeted viral messenger RNAs within the RNA-induced silencing complex (RISC) (Aliyari and Ding, 2009; Bartel, 2018; Fukunaga, 2012). In mammals/humans, the detection of virus-derived small RNAs has been mainly undetectable in the past upon RNA virus infection (Umbach and Cullen, 2009). However, more recently a short catalog of pre miRNAs has been found in the genome of DNA and RNA viruses (tenOever, 2013). The proof for miRNA qualification in virus resides in their experimentally determined presence associated to Ago2 along with the structural secondary structure of their precursor molecules (hairpin forms). According to these criteria, in the Rhesus lymphocryptovirus (closely related to EBV) 68 miRNAs were found (Riley et al., 2010, 2012). In herpesvirus infection, the molecular machinerie includes the processing of viral miRNA precursors, which contributes to virus latency by modifying the infected host cell biology and allowing the virus survival and persistence (Grey, 2015). Viral dsRNA might have other function like to bind dsRNA dependent kinase PKR and inhibit its activity (tenOever, 2013). Upon complexing with dsRNA segments, PKR phosphorylates the translation initiation factor eIF2 whose the consequence is to stop the host translation program at the beginning of viral infection (tenOever, 2013). Although in plants, nematodes and arthropods, siRNAs and miRNAs have been evolutionary selected in response to viral infections, chordates have privileged sophisticated immune mechanisms based on cellular and humoral immune responses. Thus the siRNAs and miRNAs appeared like an accessory second layer of weaponry to fight many families of viruses whereas viral sRNAs have been detected in Ago2 complex for only few viruses (Grey, 2015). Up to date, a general acceptance is that an intertwined RNA metabolism take place in infected cells in which viruses use opportunistically various strategies and tricks to manipulate host enzymes in order to produce useful small RNAs. Those alternative innovative pathways might sign the evolutionary success for the viral adaptation to their host (tenOever, 2013; Kazimierz et al., 2015). On the other side and more importantly, small viral RNAs (svRNAs) of about 22–27 nt in length have been found in tissues infected with viruses like Influenza A. and their accumulation was the result of RNA-dependent RNA polymerase activity (Perez et al., 2010). Authors have proposed that svRNAs orchestrate the viral switch from transcription to replication although the refined mechanism is still elusive (Perez et al., 2010; Tahamtan et al., 2016). This observation was found independent of virus subtypes and the host species. Those svRNAs were not related to siRNAs or miRNAs from viral precursor and not related either to Drosha/Dicer/Ago endonucleases (Perez et al., 2010).
The unique Dicer in humans processes long double-stranded RNA (dsRNA) and hairpin dsRNA into siRNAs and microRNAs, respectively, but its effective role as a producer of antiviral RNAis is still debated with divergent observations depending on virus strains and host cells/tissues. However, mammalian viruses were successfully eliminated, and replication was inhibited in cultured host cells via synthetic siRNAs (Umbach and Cullen, 2009). In mammals, powerful and non-sequence-specific synthesis of interferon (IFN) appears to be the major molecular weapon to fight RNA viruses (Sadler and Williams, 2008). This effector response might have overwhelmed the initial RNAi role, which is inferred by the rare detection of virus-derived small RNAs that were observed in tissues infected with only few virus families (Sadler and Williams, 2008; Koshiba, 2013; Berkhout, 2018).
The question of whether RNA virus infection of mammalian cells can trigger an effective and powerful antiviral RNAi weapon remains a hot topic, as no siRNAs of viral origin were initially found in mammalian infected cells owing to potent RNAi suppressor proteins, in contrast with observations in plants and invertebrates (Koshiba, 2013; Berkhout, 2018). The absence of virus-specific siRNAs in mammalian cells observed in some experimental set-ups might be also explained by the inactivity of host endonucleases type III to dsRNAs offered by the virus genomes. The question that viral infection in mammals leads to in vivo production of VsiRNAs with an impact on Dicer processing has been debated and still under investigation in many laboratories.
However, some new reports reveal that antiviral response in mammals are mediated by the RNAi mechanism with strong similarities to the siRNA-directed response against virus in plants and invertebrates (Han et al., 2020). Recent studies have shown that infection of mammalian cells by RNA viruses of 4 families (Nodaviridae, Orthomyxoviridae, Picornaviridae, Flaviviridae) triggers Dicer activation which processes the viral dsRNA replicative intermediates appearing transiently during the replication process. This was observed to be concomitant to abundant production of virus-derived siRNAs (Maillard et al., 2019; Guo et al., 2019). The avoidance strategy, evolutionary selected in Nodamura virus (NoV; Nodaviridae), Influenza A virus (IAV; Orthomyxoviridae), human enterovirus 71 (HEV71; Picornaviridae), and dengue virus-2 (DENV2;Flaviviridae), was to encode a viral suppressor of RNAi (VSR), designated protein B2, NS1, 3A, and 2A, respectively (Maillard et al., 2013; Li et al., 2013, 2016; Qiu et al., 2017, 2020). For instance authors have shown that the Nodamura virus, a mosquito-transmissible positive-strand RNA virus (NoV), is linked to an abundant VsiRNAs production that turned out to be very efficient to induce and guide specific RNA slicing by Ago2. This slicing was shown also to be inhibited by the viral suppressor of RNAi, the protein B2 blocking both VsiRNAs production and VsiRNA-RISC complex (Han et al., 2020). Genetic studies reported by authors show that NoV RNA replication is inhibited by the RNAi pathway dependent of Dicer and Argonaute 2 slicing activity in mouse fibroblasts and enhanced by the B2 viral RNAi suppressor in RNAi-competent cells (Han et al., 2020). Silenced B2 gene in virus leads to defective infection and absence of pathogenicity (Han et al., 2020). The production and loading of abundant VsiRNAs on Argo 2 is observed in adult mice with an intact IFN system in at least few viruses. In those cases, the IFN response does not trigger an inhibitory effect of VsiRNAs production or VsiRNA-RISC competent slicing activity, but seems to act as a parallel response. Authors conclude that VSR-B2 protein in adult mice suppressed the production of VsiRNAs and the RNA slicing by VsiRNA-RISC but has no observable effect on the endogenous miRNAs status or the induction of the IFN dependent genes (Han et al., 2020).
In SARS-CoV-2, the transcriptome is highly complex due to a high number of discontinuous transcription events. In addition to the 9 subgenomic RNAs, SARS-CoV-2 present unknown ORFs encoding transcripts orchestrated by various insertion, deletion, and/or frameshift in the genome (Kim et al., 2020; Sola et al., 2015). Authors have also found at least 41 RNA modification sites on viral RNAs, the most frequent motif being AAGAA. Modified RNAs have shorter poly(A) tails than unmodified RNAs, which suggest that the RNA modifications might affect their stability and therefore contribute to the avoidance of host immunity (Kim et al., 2020). On the other side, it is debated whether some accessory genes might be embedded in genome with potential expression induced by rapid reactions of SARS CoV-2 infected host cells, which could constitute an evolutionary success to respond to new challenges. Moreover, the new ORFs may also lead to accessory proteins with advantageous natural selection to dodge immune defense of its hosts. Finally, each coronaviral RNA contains the common 50 ‘‘leader’’ sequence of 70 nt fused to the ‘‘body’’ sequence (Kim et al., 2020). Recently, we performed a computational search of the SARS-CoV-2 full RNA sequence to explore whether viral short sequences presenting perfect matches with reverse complementary strands from human transcriptome might exist. We also examined the possibility of intrapairing within the full SARS-CoV-2 RNA at a minimum of 20-base lengths. Comparative studies were conducted with SARS-CoV, MERS-CoV and two nonvirulent coronaviruses (HCoV-229E and HCoV-OC43) along with three SARS-CoV-2 variants. The purpose of this search was guided by the hypothesis that hybrid duplex RNA (one strand from the human transcriptome, the other from SARS-CoV-2) could be theoretically formed and consequently might lead to degradation of RNA targets not only in SARS-CoV-2 but also in opposite direction in the human transcriptome. In this study, the pairing length for computational searching was set minimally to 20 bases, which leads to 24-mers considering the 2 nt overhang in both 3′ extremities for Dicer cleavage specificity. Exact duplexes at, and/or above, 20 nt length for intra annealing of SARS-CoV-2 genome in which high density of potential hairpin structures exists, were not found, although numerous possibilities of imperfect pairing or very short pairing lengths have been found. Inversely, the computational search of hybrid pairing RNAs retrieved a few human genes related to ubiquitin metabolism and two long noncoding RNAs (lncRNAs). This observation argues in favor of the hypothesis that RISC/Ago might be fueled by diverse sources of RNAi, byproducts of the action of Dicer originating from hybrid and asymmetric RNA duplexes, with one strand coming from the host and the other from the virus. The new SARS-CoV-2 variants (UK, Brazil, South Africa) were also analyzed for comparison with the original strain (Wuhan).
2. Results
2.1. List of human transcripts theoretically duplexed with SARS-CoV-2 RNA
Short segments of SARS-CoV-2 RNAs that are potential sources of siRNA by hybridization with human RNAs were computationally found, identifying the transcripts of 7 coding human genes that are complementary to a segment of SARS-CoV-2 RNA. The results are summarized in Table 1, and the full list of paired transcript isoforms is shown in Supplemental Table S1. Among the human genes, we noticed the presence of DNAJC13 (regulates endosomal membrane trafficking (Yoshida et al., 2018), FBXO21 (an F-box protein that is one of the four subunits of the ubiquitin protein ligase complex (Yu et al., 2016)), FLRT2 (fibronectin leucine-rich transmembrane protein), ELP4 (a histone acetyltransferase, a subunit associated with RNA polymerase type II), USP31 (a ubiquitin-specific peptidase that has been described to activate transcription factor NF-κB, which stimulates interferon synthesis (Tzimas et al., 2006)) and USP30 (a mitochondrial ubiquitin-specific peptidase (Cunningham et al., 2015). The segment of USP30 RNA having a reverse complement in SARS-CoV-2 RNA was found exclusively in humans, which highlights the unexpected and intriguing specificity of this coronavirus. The viral pairing sequences originate from 3 putative open reading frames (orf): orf1a/b, Spike and orf3b. In a recent study, Chan et al. showed that there are no remarkable differences between orf1a/b in SARS-CoV-2 and that in SARS-CoV, the major distinction being located in orf3b, Spike and orf8 (Chan et al., 2020). The authors note that orf3a encodes a completely novel short protein that may have a role in viral pathogenicity. Orf3a also includes a 20 nt sequence that potentially targets ubiquitin-specific peptidase 30 (USP30). The complementary sequence of a segment of USP30 within SARS-CoV-2 was found only in this coronavirus strain, whereas the corresponding USP30 segment was not found in other mammalian species.
Table 1.
List of hybrid duplex RNAs potentially formed with one strand from the human transcriptome and the other from SARS-CoV-2. The table shows the position of the sequence in the RNA virus (start position and ORF name), the DNA sequence encoding the human RNA and the corresponding features in humans. The numbers in the column ‘Human target’ are specified using the format x(y), where x and y are the numbers of known and predicted transcripts, respectively.
| Pos. in virus |
Human sequence forming hybrid duplex RNA |
Length | Human target | |
|---|---|---|---|---|
| Start | ORF | |||
| 3778 | orf1a/b | AGACAGCTAAGTAGACATTT | 20 | 2(1) mRNAs of DNAJC13 |
| 4395 | orf1a/b | TTCTGCATGTGCAAGCATTT | 20 | 3 mRNAs of ELP4 |
| 7803 | orf1a/b | AGAGAGAGAATGTCTTTCAT | 20 | 2(3) mRNAs of FBXO21 |
| 10971 | orf1a/b | TGTTCTTTTCACTGCACTTT | 20 | 1 ncRNA of LOC105377468 |
| 11755 | orf1a/b | CATCTATGCTATTCTTGGGT | 20 | 4 mRNAs of FLRT2 |
| 16896 | orf1a/b | GCACAAAATAATCACCAACA | 20 | 1(4) mRNAs of USP31 |
| 21820 | S | AGCAAAATAAACACCATCATTA | 22 | 1 ncRNA of LOC105372614 |
| 23034 | S | GAAACCATATGATTGTAAAG | 20 | 1 mRNA of ZNF443 |
| 25698 | orf3a | AAAGATAGAGAAAAGGGGCT | 20 | 2(13) mRNAs of USP30 |
2.2. List of human transcripts theoretically duplexing with RNA of other coronaviruses
The same type of analysis was conducted with other Coronaviridae strains for comparison: SARS-CoV (having caused the precedent China pandemic), MERS-CoV and two nonvirulent strains (HCoV-229E and HCoV-OC43). The results are shown in Tables 2, 3, and 4 and Supplemental Table S1. We noticed that each strain presented a unique set of sequences of 20-base lengths matching in reverse complement with human transcripts, and these unique sets targeted no common human genes. Remarkably, SARS-CoV mostly targets ncRNAs (Table 2 and Supplemental Table S1). MERS targets a histone methyltransferase (KMT2C) and a miscRNA included in this gene; a small GTPase of the Rho subfamily (CDC42); a ubiquitin protein ligase (ARH1); a calcium-activated chloride channel (ANO9); a voltage- and calcium-sensitive potassium channel (KCNMA1); and an adhesion glycoprotein (THBS3) (Table 3 and Supplemental Table S1). Regarding the nonvirulent coronaviruses (HCoV-229E and HCoV-OC43), few human genes are potentially targeted by their viral RNA sequences; among them are a few noncoding RNAs for which a role in defense against viral infection is not documented. The few coding genes do not appear to qualify for Gene Ontology (GO) annotation of cellular signaling susceptible to alteration by virus infection (see the NCBI database for information).
Table 2.
List of hybrid duplex RNAs potentially formed with one strand from the human transcriptome and the other from SARS-CoV. The table shows the start position of the sequence in the RNA virus, the DNA sequence encoding the human RNA and the corresponding features in humans. The numbers in the column ‘Human target’ are specified using the format x(y), where x and y are the numbers of known and predicted transcripts, respectively.
| Start pos. in virus |
Human sequence forming hybrid duplex RNA |
Length | Human target |
|---|---|---|---|
| 15256 | AGAGGCCATTATCCTAAGCA | 20 | 1 lncRNA of NUTM2B-AS1 |
| 19102 | AAACAAACAAACACCATCAG | 20 | 0(3) ncRNAs of LOC101927871 |
| 20547 | TCAACATGTCCATCCTTACA | 20 | 1 lncRNA of LRP1-AS |
| 20792 | CAGAGCCAGCACCAAAGTGA | 20 | 0(2) mRNAs of LOC102724965 |
| 27270 | AAGAGAATAATTTTCATGTT | 20 | 0(5) lncRNAs of LINC02272 |
Table 3.
List of hybrid duplex RNAs potentially formed with one strand from the human transcriptome and the other from MERS-CoV. The table shows the start position of the sequence in the RNA virus, the DNA sequence encoding the human RNA and the corresponding features in humans. The numbers in the column ‘Human target’ are specified using the format x(y), where x and y are the numbers of known and predicted transcripts, respectively.
| Start pos. in virus | Human sequence forming hybrid duplex RNA |
Length | Human target |
|---|---|---|---|
| 9467 | CAGCAGCAACAACAGCAATT | 20 | 1(27) mRNAs, 0(1) miscRNA of KMT2C |
| 15455 | TGCAGTGGTGGCATCTCCGCT | 21 | 2(0) mRNAs of CDC42 |
| 21230 | TTAATAGTACCCAAGTAATT | 20 | 1 mRNA of ARIH1 |
| 22581 | TCAACACCTTCAGCCTGTTC | 20 | 2(5) mRNAs, 0(10) miscRNAs of ANO9 |
| 23098 | AAGCCAGCCACCACCTTCAA | 20 | 6(12) mRNAs of KCNMA1 |
| 25583 | TTTGAGGATGCAGTGACCAA | 20 | 0(2) mRNAs, 1 ncRNA of THBS3 |
Table 4.
List of hybrid duplex RNAs potentially formed with one strand from the human transcriptome and the other from HCoV-229E and HCoV-OC43. The first 5 lines (∗1888 to 25074) refer to HCoV-229E, and the last 6 lines (°849 to 23025) refer to HCoV-OC43. The table shows the start position of the sequence in the RNA virus, the DNA sequence encoding the human RNA and the corresponding features in humans. The numbers in the column ‘Human target’ are specified using the format x(y), where x and y are the numbers of known and predicted transcripts, respectively.
| Start pos. in virus | Human sequence forming hybrid duplex RNA |
Length | Human target |
|---|---|---|---|
| ∗1888 | GTTCAACACGTGAAGATTTA | 20 | 1 ncRNAs of GTF2IP1 |
| ∗1888 | GTTCAACACGTGAAGATTTA | 20 | 1 ncRNAs of GTF2IP4 |
| ∗9372 | TATTATAGAAAAATTATGCA | 20 | 1(6) mRNAs of GCNA |
| ∗16936 | TTGGCATTACAGGCATGAGC | 20 | 2 mRNAs of SRGAP1 0(2) mRNAs of COX11 |
| ∗25074 | AACTGAAGAATAACAATGAATA | 22 | 0(1) ncRNA of LOC107986455 |
| °849 | TCTTCAACTTTAAAATCATA | 20 | 1 lncRNA of LINC01829 |
| °5404 | CAAATCAACTTGACTAAACA | 20 | 0(1) ncRNA of LOC107986018 |
| °12038 | CCTCATCAAGATTTTTCTTA | 20 | 0(1) mRNAs of SSH1 |
| °13797 | TTCAACAAAATCATACCAAT | 20 | 0(1) ncRNA of LOC105373593 |
| °21968 | TTGATGGTGATTTTATAATT | 20 | 1 mRNA of PTP4A1 |
| °23025 | CACCAGTGTCTTTATTAAAA | 20 | 0(1) ncRNA of LOC107985904 |
2.3. List of human transcripts theoretically duplexing with SARS-CoV-2 variants
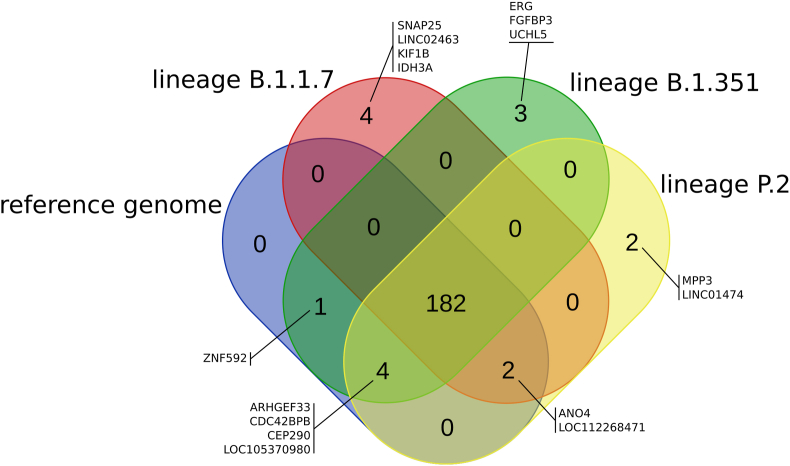
To get an insight into how SARS-CoV-2 mutations are impacting hybridization with human RNAs, we performed the same analysis on three different SARS-CoV-2 variants: B.1.1.7, B.1.351 and P.2., commonly named UK, south african and brazilian variants. Considering a perfect pairing with a minimum of 20 bases length, the results obtained are exactly the same for the three variants and are identical to those obtained with the SARS-CoV-2 genome reference (see Table 1). When one mismatch was introduced in the 20 bases length of hybrid dsRNAs, few genes appeared specifically in one or the other variants: LINC01474 (LncRNA) and MPP3 (member of MAGUKs family that are membrane-associated guanylate kinase), for the brazilian variant; ERG (transcription factor), FGFBP3 (fibroblast growth factor binding protein) and UCHL5 (Ubiquitin C-Terminal Hydrolase) for the south African variant; IDH3A (Isocitrate dehydrogenase), KIF1B (ATPase activity and microtubule motor activity), LINC02463 (long non coding RNA) and SNAP25 (Synaptosome Associated Protein) for the UK variant. In parallel, few genes were absent in one or the other variants for the same one mismatch protocol: ZNF592 (zinc finger protein) for the brazilian and UK variants; ANO4 (intracellular calcium activated chloride channel activity and phospholipid scramblase activity) and LOC112268471 (non coding RNA) for the south African variant; ARHGEF33 (Rho guanyl-nucleotide exchange factor activity), CDC42BPB (serine/threonine protein kinase family), CEP290 (centrosomal component) and LOC105370980 (uncharacterized protein) for the UK variant. (See Figure 1 for an overview, Supplemental Table S2 for the full list of paired transcript isoforms and Supplemental Table S3 for the list of targeted genes).
Figure 1.
Diagram of overlapping targeted human genes between SARS-CoV-2 variants. One mismatch was tolerated in the hybrid ARN duplex. Assuming that the host machinerie works still minimally to produce Ago2 compatible dsRNA, we compare the genes targeted by SARS-CoV-2 between 4 sequenced variants: lineage B.1.1.7 (UK variant), lineage B.1.351 south african avariant), lineage P.2 (brazilian variant) and the original strain (Wuhan). The names of the genes that are targeted by only some of the variants are specified in the figure.
2.4. Theoretical hybrid human/SARS-CoV-2 dsRNA is related to the ubiquitin pathway and mitochondria
Upon RNA viral infection, host RNA helicases (RIG-I and MDA-5) bind virus-derived RNAs (Takeuchi and Akira, 2009; Iwasaki and Medzhitov, 2010; Fujita, 2009). The complexes then translocate at the outer mitochondrial membrane by binding with mitochondrial antiviral signaling proteins (MAVS) (Sun et al., 2006). The bound MAVS acting as scaffold protein recruits downstream effectors to form a “MAVS signalosome”, of which the major function resides in drastically inducing the stimulation of the NF-κB and IRF-3 factors, a molecular scenario well documented in viral infection of mammalian host cells (Sun et al., 2006). Mitochondria appear to constitute a hub of communication that implicates a cascade of effector molecules recruited for antiviral defense (Sun et al., 2006). Regarding the human genes targeted by SARS-CoV-2, we observed the presence of USP30, a deubiquitinase specific to mitochondria, and FBXO21, which is a subunit in a ubiquitin protein ligase complex. Remarkably, the FBXO21 ubiquitin ligase complex has been reported to be required for the antiviral innate response (Yu et al., 2016). The host ubiquitin system is known to be crucial in innate immunity, and host E3-ubiquitin ligases have clear antiviral functions (Rajsbaum et al., 2018). Putatively, dsRNAs, hybrid forms with one strand coming from the human transcriptome and the other from viral RNA, might have an unfortunate role in directing the Dicer/ago/RISC defense system against its own host.
2.5. Phylogenetic comparison of the computationally retrieved 20-base lengths of RNA sequences among coronavirus strains and between mammalian species
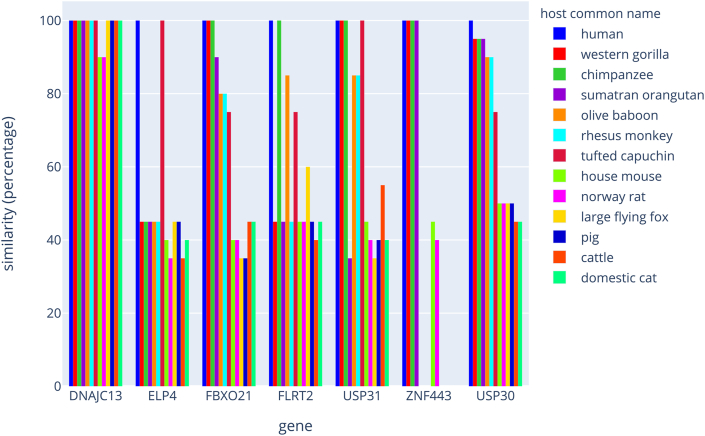
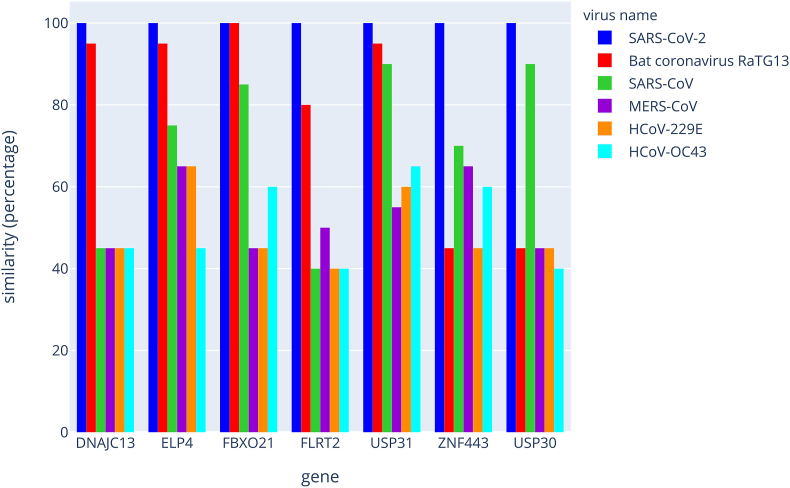
We conducted a comparison of the RNA segments within the orthologous genes of numerous vertebrate species, with a particular emphasis placed on the class of primates. The results are shown in Figure 2 and Supplemental Tables S4–S10. In summary, one 20-base sequence (FBXO21) presents 100% homology among humans, gorillas and chimpanzees. One other sequence (USP30) is unique to humans, whereas another sequence (USP31) was found in humans, gorillas, chimpanzees and capuchin monkeys. All these sequences were found to be phylogenetically distant in other mammalian species. Overall, the combinatorial arrangement of the 7 RNA segments in humans was unmatched with another primate species, which highlights that a pattern of gene targets might be unique for humans. The same type of analysis was conducted between strains of coronaviruses (see Figure 3 and Supplemental Tables S11–S17). As such, the conservation of the 20-base sequences is weak, with significant divergences. We noted that SARS-CoV-2 and the bat coronavirus RaTG13 present 4 sequences that are identical or with one base change, whereas the other coronaviruses known to cause the same clinical symptoms and physiopathology in humans as SARS-CoV-2 are highly divergent for the 7 sequences.
Figure 2.
Phylogenetic comparison of the human strand of hybrid dsRNAs with those of other mammalian species The graph represents the comparison of the sequences within the orthologous genes among a few mammalian species. The results are plotted as a percentage of similarities with the human sequence.
Figure 3.
Phylogenetic comparison of the SARS-CoV-2 strand of hybrid dsRNAs with those of other coronaviruses The graph represents the comparison of the 20-base sequences among a few coronavirus strains. The results are plotted as a percentage of similarities with the SARS-CoV-2 sequence.
2.6. Assessing the existence of VmiRNAs and VsiRNA encoded in SARS-Cov-2 genome
Viral miRNAs have been found based on qualifying secondary structure features of pre-miRNAs and their functions have been assumed to target host transcripts involved in many cell biology fields like apoptosis, and immunity (Kincaid and Sullivan, 2012). The virus miRNAs have been described to orchestrate opportunistic mechanisms to degrade host mRNAs and hijack host cellular miRNAs to benefit the viral replication efficiency (Trobaugh et al., 2014, 2016). Moreover, recent studies indicate the virus-encoded miRNAs, host-encoded miRNAs, and their respective targets together constitute a highly sophisticated intertwined regulatory networks which dictate the fate of host death, survival or tolerance in relation to the degree of pathogenicity of the virus. Hence, it is crucial to comprehensively address the role of these novel regulatory pathways during SARS-CoV-2 infection in order to unravel possible innovative events of RNA metabolism emerged from the non pathogen common coronaviruses. To this regard, a study has proposed to computationally predict the putative SARS-CoV-2 miRNA and if they exist their putative targets in the human genome (Liu et al., 2020). Authors conclude that a virus-encoded miRNA MR147-3p could enhance the expression of a protease named TMPRSS2 of which the reported function was to strengthen SARS-CoV-2 infection (Liu et al., 2020). To qualify as VmiRNA or VsiRNA, the physical presence of these molecules associated to Ago2 would have to be documented although we cannot exclude innovative pathways to process them and novel functions independent of Dicer/Ago2. The numerous negative results of co precipitation VsRNA/Ago2 with a large spectrum of viruses might be also due to technical limitations and sensitivity of the pull down procedures. In our study we conducted a wide scale genome survey to detect intra pairing sequence within the SARS-CoV-2 genome presenting the canonical features of Dicer substrates. Based on 20 bases minimum length perfect pairing sequences, the computational analysis did not retrieve a RNA motif with a complementary sequence that complies with canonical Dicer slicing competence (see Table 5). When one or two mismatches were introduced in the matching sequence, very rare candidates fulfilled the criteria although they might not be valid substrate for Dicer. Numerous hairpin structures are present in the genome of SARS-CoV-2 like in many RNA virus families. However, these structures have been scarcely described as host endonuclease type III substrates (Drosha/Dicer). Upon infection, the genomic mRNA is translated with the host cytosol thus mixing the host and viral transcriptomes in the same compartment accessible to Dicer/ago2. The replication has been described to occur in internal membranous invaginations of the endoplasmic reticulum, which likely isolates physically the replication machinerie from host endonuclease type III and thus avoids dsRNA detection by Drosha/Dicer (Paul and Bartenschlager, 2013). The complexity of architecture of SARS-Cov-2 genome in 3D that associates protein partners along with the partial isolation of the viral machinerie within a membranous organelle might be a barrier to Dicer accessibility and activity. However Dicer/ago should be exposed to viral RNA degradome in the cytosol allowing the opportunism of virus to hijack host enzymes for innovative and unexpected functions. Few reported experimental datasets on the original SARS-CoV that emerged more than 10 years ago, argue in favor of these hypotheses (Morales et al., 2017). Deep sequencing of small RNAs from the lungs of infected mice with SARS-CoV was carried out by experimenters that discovered three 18–22 nt small viral RNAs (svRNAs) encoded from the nsp3 genomic regions of SARS-CoV. This biogenesis was Pol III dependent and cell type or host species independent. This viral small RNA biogenesis contributes to the success of the infectious process in lung and to the production of pro-inflammatory cytokines (Morales et al., 2017). This leads to the paradigm that Sars-CoV encode few ncRNAs transcribed by Pol III to an extreme high number of copies in infected cells that are not related to the parasitism of the canonical of host endonuclease type III enzymes and therefore independent of the ordered cascade Drosha/Dicer/Ago. To prove that svRNAs were not processed by the cell miRNA enzymes (Drosha and Dicer), authors have transfected a plasmid in a cultured cell host in which was inserted a 200–500 nt sequences flanking svRNAs of the SARS-CoV genome as potential miRNA precursors. No viral small RNAs from plasmids was detected indicating the host dsRNA generating enzymes did not process this precursor, which argues for a major dependence of viral enzyme equipment to synthetize those fragments. Another set of experiments have been designed in order to establish whether vsRNAs in SARS-CoV infection depend or not on canonical pathway of miRNA biogenesis: authors have infected the RNase III nucleases, Drosha and Dicer deficient cells after transient expression of mACE2 and the results of vsRNAs turned out to be similar with or without the host RNase III nucleases, suggesting a non canonical process (Morales et al., 2017).
Table 5.
Count of potential internal pairing in SARS-CoV-2 RNA. The table shows the distribution of the number of potential internal pairing in SARS-CoV-2 by length. The calculation was performed considering an exact matching and one or two mismatches for lengths of 5 bases or more.
| length of potential internal pairing | number of potential internal pairing |
||
|---|---|---|---|
| No mismatch | One mismatch | Two mismatches | |
| 28 | 1 | ||
| 23 | 1 | ||
| 21 | 1 | ||
| 20 | 1 | 7 | |
| 19 | 1 | 7 | |
| 18 | 24 | ||
| 17 | 8 | 41 | |
| 16 | 2 | 6 | 183 |
| 15 | 1 | 34 | 478 |
| 14 | 3 | 100 | 1519 |
| 13 | 8 | 343 | 4594 |
| 12 | 36 | 1141 | 14014 |
| 11 | 141 | 3789 | 42100 |
| 10 | 537 | 12829 | 120466 |
| 9 | 1784 | 41682 | 335027 |
| 8 | 6628 | 138361 | 880569 |
| 7 | 24071 | 384414 | 2074296 |
| 6 | 89038 | 951538 | 3893583 |
| 5 | 329507 | 1804325 | 4940827 |
2.7. Viral RNA internal pairing stretches might interfere weakly with hybrid dsRNA annealing
The full RNA of SARS-CoV-2, like other coronaviruses, have secondary structures that are involved in protein association involved in regulatory functions. Translation initiation, elongation, and termination of translation are guided by higher order mRNA secondary structures located predominantly in non-coding regions (Kiening et al., 2019). After cleavage by mono strand ribonucleases, the secondary structures of RNA like hairpin is undone or impaired by weak constant of association of pairing segments (high Kd). For our studies we assumed that the secondary structures interfere very little with our analysis due to the fact that the RNA degradome constituted of byproducts of the original viral RNA are likely the major resources to feed the Dicer/Ago system and the dsRNA dependent kinase R known to inhibit translation by phosphorylating an elongation factor (Tycowski et al., 2015). However we aimed to assess the extent to which the complex mixture of viral RNA could interfere with the hypothetical formation of hybrid dsRNA. In order to address this issue we have proceeded to a computational search for internal pairing sequences over at least 5 bases length. Results are presented in Table 5. We observed that sequences that hybridize above 20 bases length are absent and when one or two mismatches are introduced in the searching protocol the candidates remain rare. In parallel we have proceeded to a computational search of overlapping sequences for different length of at least 5 bases long, operating a competition for the potential hybridization with our identified >20 nt segments of human transcripts The results are shown in Table 6. We see that a massive amount of potential base pairing occurs for lengths of 5 bases and that the numbers decline exponentially for longer lengths. The pairing of 5–10 bases lengths leads to very weak double strand association with a dissociation constant at equilibrium (Kd) very high. Consequently, the competition against our hybrid dsRNA appears very limited if existent. The other massive source of internal viral RNA pairing comes from the replication. The transient and intermediate viral dsRNA during the replication process is theoretically prone to Dicer dependent degradation but the isolation of replication in invaginated compartment from endoplasmic reticulum along with the co presence of VSR proteins would likely block this process (Paul and Bartenschlager, 2013).
Table 6.
Distribution of potential viral sequences overlapping with those that potentially hybridize with the human transcriptome over 20 bases length. Each row in the table represents a sequences in SARS-CoV-2 that could hybridize with the human transcriptome. The first three columns contain the start, the length and the potential human target of the sequence, respectively. The remaining columns show the distribution of the number of potential internal pairings that overlap with the sequence.
| sequence in SARS-CoV-2 |
Human target | Number of overlapping sequences |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| start | length | 5 bases | 6 bases | 7 bases | 8 bases | 9 bases | 10 bases | 11 bases | |
| 3778 | 20 | DNAJC13 | 334 | 98 | 13 | 6 | |||
| 4395 | 20 | ELP4 | 328 | 65 | 11 | 4 | 1 | ||
| 7803 | 20 | FBXO21 | 380 | 79 | 30 | 6 | 2 | 1 | |
| 10971 | 20 | LOC105377468 | 464 | 96 | 33 | 12 | 3 | 1 | |
| 11755 | 20 | FLRT2 | 395 | 117 | 24 | 11 | 2 | ||
| 16896 | 20 | USP31 | 430 | 110 | 23 | 5 | |||
| 21820 | 22 | LOC105372614 | 441 | 105 | 30 | 10 | 2 | ||
| 23034 | 20 | ZNF443 | 412 | 99 | 32 | 7 | 1 | ||
| 25698 | 20 | USP30 | 332 | 83 | 12 | 7 | 1 | ||
2.8. Dicer flexibility and computational mismatch analysis within the 20 nt hybrid DsRNAs
Dicer can trap dsRNAs pairing over a 19-base length with at least a 2 nt overhang at the 3′ ends and can then directly transfer the modified/unmodified siRNA to the Ago2 site (Snead and Rossi, 2012; Snead et al., 2013; Sun et al., 2008). This mechanism has been further demonstrated with fluorogenic dsRNA (Bernard et al., 2015). Some paired 19-mers with a dTdT overhang at the 3′ end bind Dicer with high affinity; these sequences are not cleaved by Dicer, are fully transferred to Ago and trigger high-efficiency gene-silencing activity (Snead and Rossi, 2012; Snead et al., 2013). Dicer also binds single RNAs at a specific site with high affinity, the function of which is to accelerate hybridization with a partner from the milieu of RNA metabolism (Kurzynska-Kokorniak et al., 2016; Wojnicka et al., 2020). This leads to the paradigm that hDicer acts as an annealer and presents a RNA molecule for searching a complementary segment in the RNA degradome, thus facilitating pairing between a small RNA and a complementary longer RNA in asymmetric mode. Authors have also shown that the pairing length can be as short as 16 nt, with a several-base overhang at the 3′ end to fully bind to Dicer and subsequently activate Ago in the RISC context (Snead and Rossi, 2012; Snead et al., 2013; Sun et al., 2008). In the past, RNA duplex with mismatches have been tested for their efficiency to promote RNA target degradation by Ago2. In some examples a single mismatch in a 19-bases length sequence can prevent and/or fragilize duplex formation by creating thermodynamic asymmetry (Parrish et al., 2000; Theotokis et al., 2017). In other examples these heteroduplexes can still promote RNAi activity on mRNA targets, but with moderately to drastically reduced effectiveness when they are compared to perfect RNA pairing. The introduction of mismatches that lower Tm or to add modified bases that increase Tm within the siRNA duplex was tested for Dicer compatibility. The conclusions were that the non complementary bases must be localized at the 3′-end of the passenger strand (sense strand) and not into the 5′-end of the guide strand (anti sense strand) to conserve the annealing and cleavage of to the RNA target (Amarzguioui et al., 2003). Finally, Dicer displays extremely stringent conditions as a single base pair mismatch over crucial location in the pairing length turned out to be enough to block its endonuclease activity (Amarzguioui et al., 2003). Ago2 in RISC complex is bound to 3′ and 5′ overhang dsRNA and the guide strand has a “seed” sequence 2–8 at 5'end that is crucial to engage in target recognition/annealing and subsequent cleavage (MacRae et al., 2006, 2007). Interestingly, hDicer can produce RNA species shorter or longer than characteristic ~22-nt RNAs in a very flexible manner depending on the protein partners and protein association (Lima et al., 2009; Chakravarthy et al., 2010). A piece of Dicer without PAZ and helicase domains that resemble to the basic model endonuclease type III of E coli., generates a major product of 15 nt fragment from dsRNA substrate. This shorter fragment was always seen with the full length hDicer combining the major 22 nt RNAs and a minor 15 nt RNA (Sinha et al., 2018; MacRae et al., 2006, 2007; Chakravarthy et al., 2010). As another example, the interaction of hDicer with a with 5-lipooxygenase observed during inflammation produce fragment of 55 nt and 12 nt length from a pre miRNA substrate (Dincbas-Renqvist et al., 2009). This indicate that cleavage of the hDicer by endogenous proteases that separate PAZ/helicase domains and the endonuclease/dsRBD domains would generate complex pattern of Dicer products with different sizes, preserving their loading on and triggering cleavage capacity of Ago2. Therefore, the canonical 22 nt siRNA might be more heterogenous than expected. This Dicer flexibility in cleavage properties was well documented by authors that use extensive catalogue de substrates along with the amazing fact that mono strand RNAs display strong affinity to the same order than dsRNA (Lima et al., 2009). Moreover, few seminal studies have shown that dsRNA bound to active Ago2 can be also independent of Dicer cleavage and loading (Cheloufi et al., 2010; Cifuentes et al., 2010). The complexity of viral biology resides partly in the overwhelming concentrations of viral RNA in host cell along with their degradome. These massive amount of viral RNAs likely capture the host enzymes activities for viral needs, leading likely to unsuspected metabolic pathways of RNA metabolism. Based on this complex outlook, we performed a computational search of hybrid pairing (human/virus) on minimal 20 nt length in introducing randomly one mismatch in the dsRNAs with the assumption that these species might still work in vivo regarding the Dicer/Ago2 flexibility. The analysis gave 39 pairing zones within 9 genes for perfect matches in hybrid dsRNAs (SARS-CoV-2 and human) and 836 pairing zones within 189 genes when a mismatch was tolerated (Figure 1 and Supplemental Table S2–S3). In this later case, 2 new genes were identified in Brazilian variant, 3 for the south African variant and 4 new ones for the UK variant (see the gene function in the above paragraph: List of human transcripts theoretically duplexing with SARS-CoV-2 variants). The final validation will be to show the existence of these small RNA molecules bound to Ago2 in pull down analysis. This type of biochemical validation could document the degree of tolerance for mismatch(es) in dsRNA in a context where the “law of mass action” and the associated constants(Kd, Ka) change the enzymes activity in favor of the highly abundant viral substrates.
2.9. Experimental evidences for a validation of the RNAi hypothesis
In order to validate the theoretical scenario of RNAi directed against the host, we re analyzed the data from a recent publication which proposes the short- and long-range RNA-RNA interactome of SARS-CoV-2 (Ziv et al., 2020). In this study, RNA:RNA pairs were crosslinked and secondarily linked to a biotin by means of a “click” chemistry reaction. After fractionation and isolation of the dsRNA, the crosslink was reversed, the dsRNA were ligated by the ends and finally the RNAs were sequenced. The protocol was carried out with the full viral genome and the fragmental subgenomic mRNAs (sgmRNAs) used to transcribe viral proteins. This study uncovered a large network of RNA-RNA pairing and demonstrated that viral genome and subgenomic mRNAs as well adapt multiple conformations more complex and numerous than expected. This study proves also that cross pairing between the host messengers and viral RNAs is significant and constitute a robust fraction of found pairing (Ziv et al., 2020). Coronaviruses mRNAs, so-called subgenomic mRNAs, are processed by discontinuous genomic RNA reading due to the recognition of specific motifs on the genome acting as transcriptional regulating sequences (Mateos-Gomez et al., 2013; Wu and Brian, 2010). This contrasts with the replication process that involves the complete reading for the full-length genome. The synchronization of the dual processing: discontinuous transcription with “nested” sgmRNAs of different sizes and genome replication is still under scrutiny with unknown aspects (Madhugiri et al., 2016; Mateos-Gomez et al., 2013; Wu and Brian, 2010). In the lists provided by these authors (Ziv et al., 2020), we have calculated that the interactions host/SARS-CoV-2 are 3% of the total obtained with the isolated full viral genome material whereas this ratio increases up to 10% if we consider separately the sgmRNAs component. The same measure gives 12% for the host/MERS hybrid RNA:RNA with only the isolated full genome only being available. Table 7 summarizes the quantitative data on these experimentally isolated hybrid host/virus RNA:RNA for both the MERS and Covid-19 viruses. These data are validated by 4 independent assays on each virus (Table 7). Moreover, our analysis did not show the presence of our human sequences susceptible to be Dicer/ago substrate after annealing with viral RNA complementary strand. The analysis of these lists of proved viral RNA:RNA lead us to observe that exact pairing above 20 bases length are inexistent and the long stretches of internal pairing are interrupted by mismatches along the sequences. This likely would impair and/or prohibit the efficiency of Dicer, an enzyme that is fully active with exact matching of dsRNA. We assume that the cleavage off triggered by Dicer and subsequent transfer to Ago2 would have subtracted and removed the dsRNA fragments from the isolated viral genomic material. The perfect matches of hybrid dsRNAs at minimum 20 bases length would provoke their excision and therefore their absence in the viral RNA pull down. Intramolecular base-pairing within the same viral genome is highly predominant as expected. However the intermolecular base-pairing between the viral and host genomes is likely a solid component for which little is known at the full genome scale for both host and virus (Ziv et al., 2020). Even though the full demonstration is still missing, at this stage we cannot rule out the possibility that an RNAi process as part of the host immune system is directed against the host via Dicer and hybrid dsRNA. Therefore the data sets from deep sequencing on the crosslinked paired RNA-RNA, are consistent with our bio informatics analysis of hybrid RNA:RNA as potential substrate of Dicer/ago and consolidate the fact that immune pathway can be turned against its host during Covid-19 infection. Finally the VSR proteins appear to leave intact the endogenous production of miRNA, which lead to the hypothesis that the Dicer/Ago could dodge VSR proteins inhibitory effect, likely because these later are compartmentalized in organelle like structure in which replication occurs.
Table 7.
Experimental isolation of hybrid dsRNAs host/virus from published datasets. Recently published datasets of the sequences corresponding to experimentally isolated RNA:RNA paired segments from cells infected by SARS-CoV-2 and MERS-CoV (Ziv et al., 2020) were re –analyzed to quantify the host/virus dsRNA fragments compared to internal pairing. This publication reports the wide scale genome catalog of dsRNAs from which one strand has at least a viral origin.
| sample | virus/virus interactions |
virus/host interactions |
other |
total count |
|||
|---|---|---|---|---|---|---|---|
| count | % of total | count | % of total | count | % of total | ||
|
a) RNA-RNA interactions spanning the entirety of SARS-CoV-2 gRNA | |||||||
| GSM4676629 | 315174 | 85% | 23849 | 6% | 29684 | 8% | 368707 |
| GSM4676630 | 1383051 | 95% | 31519 | 2% | 43506 | 3% | 1458076 |
| GSM4676631 | 330792 | 83% | 30435 | 8% | 39324 | 10% | 400551 |
| GSM4676632 | 1594186 | 92% | 45475 | 3% | 88199 | 5% | 1727860 |
| Total |
3623203 |
92% |
131278 |
3% |
200713 |
5% |
3955194 |
|
b) RNA-RNA interactions spanning the entirety of SARS-CoV-2 sgmRNA | |||||||
| GSM4676637 | 2421822 | 73% | 522778 | 16% | 372774 | 11% | 3317374 |
| GSM4676638 | 9676252 | 85% | 817233 | 7% | 934193 | 8% | 11427678 |
| GSM4676639 | 2835137 | 70% | 669965 | 17% | 531431 | 13% | 4036533 |
| GSM4676640 | 9648232 | 81% | 970620 | 8% | 1279706 | 11% | 11898558 |
| Total |
24581443 |
80% |
2980596 |
10% |
3118104 |
10% |
30680143 |
|
c) RNA-RNA interactions spanning the entirety of MERS-CoV gRNA | |||||||
| GSM4676625 | 577463 | 66% | 137638 | 16% | 161706 | 18% | 876807 |
| GSM4676626 | 1399538 | 70% | 239390 | 12% | 367489 | 18% | 2006417 |
| GSM4676627 | 440294 | 68% | 89809 | 14% | 122030 | 19% | 652133 |
| GSM4676628 | 767696 | 74% | 98421 | 10% | 166175 | 16% | 1032292 |
| Total | 3184991 | 70% | 565258 | 12% | 817400 | 18% | 4567649 |
2.10. Experimental evidence of the RNAi hypothesis based on host cell/tissue transcriptomes upon SARS-Cov-2 infection
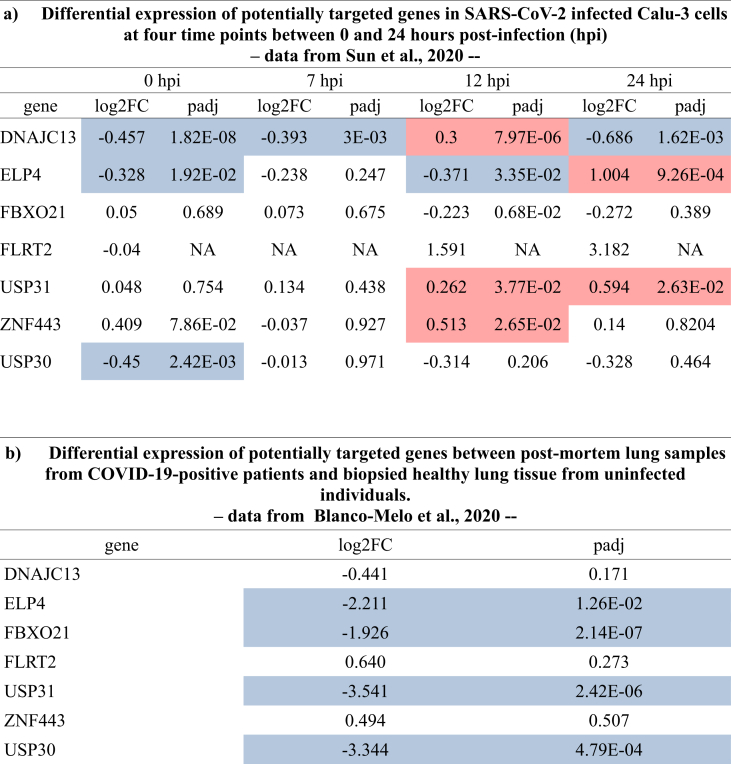
We re-analyzed data sets from two recently published studies referring to modification of host cell or tissue transcriptomes induced by SARS-CoV-2 infection (Sun et al., 2020; Blanco-Melo et al., 2020). In the first study, authors have compared mock-infected and SARS-CoV-2-infected Calu-3 cells at four different time points: 0, 7, 12, and 24 h post-infection (hpi). The time-series data simultaneously obtained on SARS-CoV-2 and host transcriptomes showed an early intensive response of host genes. The comparative transcriptome of the host cells in response to SARS-CoV-2, SARS-CoV and MERS-CoV leads to drastic differences (Sun et al., 2020). Upon infection, SARS-CoV-2 induced hundreds of up-regulated host genes mostly related to cytokine virus-specific host antiviral responses. The early rapid host responses in SARS-CoV-2 cases underscored a remarkably up-regulated gene expression of a specific protease, named TPRMSS2, soon after infection and reported responsible of concomitant fusion of infected cells (Sun et al., 2020). In this context, we observed, in the early stages following infection, an overall under-expression of our host gene candidates potentially targeted by SARS-CoV-2 (Figure 4). At the very early stage (0 hpi) DNAJC13, ELP4 and USP30 are found to be significantly under-expressed, while the other genes are not differentially expressed. At 7 hpi, DNAJC13 is still under-expressed, while there is no significant difference in expression for the other genes. At 12 and 24 hpi, the result is less clear-cut: some of the genes we have pinpointed for engaging in potential hybrid dsRNA are over-expressed, others are under-expressed, while more differentially expressed genes in these late stages compared to early stages were observed. The second study (Blanco-Melo et al., 2020) refers to host RNAs quantitatively modified in patient tissues after SARS-CoV-2 infection. The results of this study shows significant down-regulations of our retrieved candidates genes potentially targeted by SARS-CoV-2. Of the 7 coding genes, 4 are, indeed, under-expressed in COVID-19 patients with a factor of 4 to nearly 12 (log2FC from -1,926 to -3,344). This significant down regulation of the human host transcript candidates susceptible to hybrid pairing with viral RNA was obtained with direct extracts of infected patient tissues. This clear cut collapse in the transcripts levels of our candidate genes obtained with patient tissues contrasts with the heterogeneity of levels when the analysis is carried out with cell cultures. Moreover strong differences in transcriptomic metrics were observed from one cellular type to another in infected cell culture protocol, which suggests that transformed cells with their own phenotypic alterations specify their interaction with viruses.
Figure 4.
Differential expression of potentially targeted genes after SARS-CoV-2 infection. The table shows the differential expression of genes encoding mRNAs listed in table 1. Under-expressions (negative log2FC and adjusted pvalue < 0.05) are highlighted in blue while over-expressions (positive log2FC associated with an adjusted pvalue < 0.05) are highlighted in red. a) comparison between mock-infected and SARS-CoV-2-infected Calu-3 cells at 0, 7, 12 and 24 hours post-infection (hpi); data from Sun et al., 2020 b) comparision of post-mortem lung samples from COVID-19-positive patients and biopsied healthy lung tissue from uninfected individuals; data from Lanco-Melo et al., 2020.
3. Discussion
The existence of small non-coding RNAs originated from SARS-CoV was demonstrated by deep sequencing of RNAs from the lungs of infected mice in which the length of 18–22 nt small viral RNAs (svRNAs) were found (Morales et al., 2017). The Biogenesis of CoV svRNAs are generated by RNase III and their apparition are correlated with extensive phase of replication. Viruses presenting a nuclear step like retroviruses encode ncRNAs in the form of miRNAs that appear to be implicated in establishing installation of infection (Tycowski et al., 2015; tenOever, 2013). In addition to virus-encoded miRNAs, other small viral ncRNAs generated by cytoplasmic RNA viruses were detected although their roles have not been fully investigated up to date. However, based on few examples, the viral ncRNAs are produced by various alternative modes orchestrated by the entangled metabolic process involving both cellular and viral factors (Perez et al., 2010; Weng et al., 2014). On the other side, there are several targets for human miRNAs on RNAs of SARS-CoV-2 that have been found in the 5′ and 3’ untranslated regions (Perez et al., 2010; Weng et al., 2014). It was recently proposed that the ncRNA catalogue of host cells by mediating SARS-CoV-2 RNA degradome further dictates or guide the susceptibility and efficiency of infection success depending on tissue specificity (Henzinger et al., 2020). Conversely, virus infection also changes the host ncRNA expression patterns (Henzinger et al., 2020). Authors have recently shown that human siRNAs and miRNAs inhibit SARS CoV-2 spike protein synthesis in dose dependent levels in both HEK293 cells and a primary human airway tracheal cell line (Gallicano et al., 2020). Our analysis propose that the entangled outcome of RNA metabolism in both host and SARS-CoV-2 might constitute a more complicated intertwined networks leading to host defense mechanisms that might act against itself. Our observations lead to the following conclusions: i) segments presenting intraRNA pairing above 20-base lengths within the 5 coronaviruses RNAs are rare, although perfect pairing on shorter length and/or longer pairing length with mismatches are frequent; ii) the list of hybrid duplex coronaviruses/human RNAs was significantly more important in SARS-CoV-2 than in the other strains; and iii) regarding SARS-CoV-2, amazingly, hybrid duplexing sequences were found in three important human genes: mitochondrial ubiquitin-specific peptidase 30 (USP30), a subunit of the ubiquitin ligase complex (FBXO21) and, finally, ubiquitin-specific peptidase 31 (USP31). SARS-CoV-2 might affect mitochondria via ubiquitination alteration, which could be a plausible pathway of interference to explain the dramatic deteriorating conditions of many patients affected by this virus. SARS-CoV-2 might have a “stolen” piece of genetic information from Homo sapiens and/or humanoids acquired along the co-evolutionary history over millions years, or alternatively, the SARS-CoV-2 genome might have randomly drifted, allowing the appearance of matching pieces of sequences with the human genome. All these genetic innovations could have occurred with ultimately catastrophic results, directing the human immunity defense system against itself. This hypothesis argues in favor of the idea that the immune defense Dicer/Ago/RISC system could work against its host during viral infection. RNA viruses over million years of coevolution not only have dodged the immune system of hosts but also, in some cases, could trigger a “self-destruction” or “programmed suicide” of infected mammalian host cells. A recent report documented that interferon molecules (types I, II and III) are expressed at low levels or not at all in cells and tissues infected by SARS-CoV-2, whereas in contrast, high levels of chemokines and interleukin type 6, a pro inflammatory cytokine involved in innate immunity, were observed (Blanco-Melo et al., 2020). The authors observed that the ongoing infection downregulated a large panel of genes, among which are some of our genes making hybrid dsRNA. This puzzling transcriptomic picture regarding the classic immune responses to viral infection suggests a scenario in which SARS-CoV-2 induces molecular pathways in infected cells that became derailed from expected patterns.
Many reports have described the functional role of hybrid dsRNA host/virus regarding few human non coding RNAs. For instance, host-virus RNA-RNA interactions has been described between hepatitis C virus RNA and human microRNA miR-122 (Jopling et al., 2005), Zika virus RNA and human miR-21 (Ziv et al., 2018). As another example, the HIV-1 replication was found to implicate human tRNAs (Mak and Kleiman, 1997). However the wide scale genome analysis of the potential hybrid dsRNA host/virus has not been carried out in the past at our knowledge for any virus and host. The recent publication on MERS and Sars-CoV-2 RNA pairing shows clearly the physical isolation of hybrid dsRNAs and demonstrate that these later are generated massively during the process of infection (Ziv et al., 2020). Moreover, these hybrid dsRNAs are likely produced mostly with the viral sgmRNAs (subgenomic coding fragments) and the degradation products of viral genome. These two pools of viral RNAs lack internal secondary structures that protect against inter pairing process. Finally if a substantial number of viral intra pairing occurs, the absence of Dicer compatible substrates in these lists argues in favor of the fact that transient viral dsRNA appearing during replication along with the multiple hairpin structure in RNA viruses might be Dicer targets. However, the organelle like architecture in which replication takes place with the co presence of VSR proteins highlights the compartmentalization of RNA viruses biology: replication protected inside a membrane invaginated compartment from endoplasmic reticulum and a subgenome, RNA degradome component mixed with host transcriptome in cytosol. The hybrid dsRNAs would be excised by Dicer and transferred to Ago(s) and would be subtracted from the pull down of the viral genome physically isolated (Ziv et al., 2020). Whereas the viral internal hairpin secondary structures seem not Dicer substrate, the degradation products and the sgmRNAs without secondary structures unambiguously offer numerous host/virus dsRNAs associations to qualify for Dicer activity.
Recently, authors reported that they retrieved the full catalog of human proteins that bind with strong affinity to the 26 tagged SARS-CoV-2 proteins by double hybrid technology (Gordon et al., 2020). The datasets indicate that 42 human proteins are strong candidates for high-affinity binding with viral partners (corrected p values from 1.41E-29 to 0.00578) and that 280 other human proteins show weak statistical evidence for interaction (Gordon et al., 2020). Among this first group (n = 42), 13 are mitochondrial proteins, each interacting with different viral proteins, and 6 are centrosomal proteins interacting with a unique viral partner. Mitochondrial proteins are also heavily represented in the second group (n = 280). In the full dataset of interacting human proteins, no protein linked to the Dicer/ago pathways was found, which suggests that the RNAi process is not altered by SARS-CoV-2 viral infection (Gordon et al., 2020). Overall, this information suggests that the physio pathological landscape provoked by SARS-CoV-2 infection might be multifactorial, including inactivation of major host proteins by high-affinity binding with a viral partner along with drastic degradation of host messages by RNAi machinery. On the other side, the numerous dsRNA could trigger the activation of the dsRNA dependent protein kinase R (PKR) that has a known role in inhibition of protein synthesis (Clemens, 1997; Clemens and Elia, 1997; García et al., 2007). This enzyme is induced by interferon and has a significant and well documented function in fighting viral infection. PKR phosphorylates the eukaryotic translation initiation factor eIF2α. The addition of phosphate group inhibits cellular mRNA translation, thereby preventing viral and host protein synthesis as well (Clemens, 1997; Clemens and Elia, 1997; García et al., 2007). Thus, production of dsRNA that can come from Dicer action during viral infection can also lead to PKR activation. As its role does not seem specialized stricto sensus for a category of proteins, this molecular weapon could down load the viral charges and provoke apoptosis in host cells as well. All together these elements highlight the prospect that the cellular and molecular weaponry against virus could trigger in parallel disastrous auto lysis in host cells due to the very relative specificity of pathogen targeting by the immune system.
4. Material and methods
4.1. Viral sequences
We obtained the sequences of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), previously provisionally named 2019-nCoV; Middle East respiratory syndrome-related coronavirus (MERS-CoV); SARS coronavirus (SARS-CoV); human coronavirus 229E (HCoV-229E); and human coronavirus OC43 (HCoV-OC43) from NCBI GenBank (RefSeq accession: NC_045512.2, NC_019843, NC_004718.3, NC_002645.1 and NC_006213.1, respectively) (Wu et al., 2020; Van Boheemen et al., 2012; Marra et al., 2003; Thiel et al., 2001; St-Jean et al., 2004). We also included in our study representative genomes of three variants spreading in the UK (lineage B.1.1.7, accession: MW487270.1), South Africa (lineage B.1.351, accession: MW580574.1) and Brazil (lineage P.2, accession: MW411871.1) (Rambaut et al., 2020; Tegally et al., 2020; Voloch et al., 2021).
4.2. Human transcriptome
RNA products annotated on the Genome Reference Consortium Human Build 38 patch release 13 (GRCh38.p13) were downloaded from the NCBI FTP site (RefSeq accession: GCF_ 00000 1405.39).
4.3. Identification of potential double-stranded RNA fragments
Our previously published analysis, along with other published works, confirmed that the catalog of siRNAs is far more complex and extensive and that it encompasses larger sets of the transcriptome than previously thought (Pasquier et al., 2020; Ghildiyal et al., 2008; Czech et al., 2008; Li et al., 2012). A study has shown that the extensive presence of dsRNAs in Drosophila and Caenorhabditis elegans obtained by high-throughput sequencing involves many categories of RNA, including mRNAs, in which miRNA and lncRNA populations appear as a minority component (Li et al., 2012). Following the same approach used by Pasquier et al., each virus sequence was cut into short overlapping sequences of 15 bases. These 15-base sequences were extracted at intervals of 6 bases to ensure that any sequence of at least 20 bases belonging to the complete virus sequence at least one of the 15-base sequences. The potential pairing was computed by aligning short 15-base sequences on RNA sequences with the STAR RNA-seq aligner (Dobin et al., 2013) and by retaining only perfect matches for which the short sequence was reverse complemented. We then extended the matches to obtain the maximum alignment length. We subsequently applied a second post processing step to eliminate duplicate alignments and to remove sequences aligned over less than 20 bases. This procedure has been applied to compute the potential pairing between RNA viruses and human transcripts and to identify the possible internal pairing within RNA virus sequences. Briefly, regarding SARS-CoV-2, the pairing segment was observed in all the isoforms (2 known and 13 predicted) of the human transcripts of USP30. The pairings were identified at the junction between exons 17 and 18 of DNAJC13, in exon 10 of ELP4, in exon 12 of FBXO21, in exon 2 of FLRT2, in exon 13 of USP30, in exon 16 of USP31 and in exon 4 of ZNF443. The 20-base sequence originating from orf3a of SARS-CoV-2 has the potential to target the sequence AAAGATAGAGAAAAGGGGCT, which is shared by 15 predicted and/or confirmed transcripts of USP30. On the other hand, regarding the USP30 gene, the data gathered by the “1000 Genomes Project” show that this 20-base sequence is present in all Homo sapiens, with the exception of 0.49 % of Han Chinese individuals in Beijing that have a mutation A/T in the 2nd position within this matching sequence (Siva, 2008). Finally, DNAJC13 is an endosome-related protein and is believed to regulate endosomal membrane trafficking. Mutations in DNAJC13 provoke Parkinson's disease and neurodegeneration (Yoshida et al., 2018). In parallel, the SARS-CoV-2 fragments that pair with USP31, FLRT2, and FBXO21 were found in only 5, 4 and 5 isoforms (predicted or experimentally found), respectively. Regarding the MERS virus, the collection of hybrid dsRNA seems more disparate, targeting 2 ion channels and a GTPase of the Rho subfamily. The analysis of nonvirulent viruses (HCoV-229E and HCoV-OC43) did not reveal targeting of essential genes except the coding gene: SSH1 (protein tyrosine phosphatase involved in actin filament dynamics in cellular lamellipodia formation), PTP4A1 (protein tyrosine phosphatase involved in progression G1-S mitosis), GCNA (protein with an acidic domain involved in genome stability, for which mutants accumulate DNA-protein crosslinks resulting in chromosome instability), COX11 (a component of the mitochondrial respiratory chain that catalyzes the electron transfer from reduced cytochrome c to oxygen) and SRGAP1 (GTPase activator involved in neuronal migration) (see the NCBI database for information).
4.4. Identification of potential intra pairing double-stranded RNA sequences in Coronaviridae
In parallel, we also investigated whether the structure of the 5 tested coronavirus RNAs permits the formation of intermolecular double-stranded structures. We computed the potential internal pairing minimally over 20-base lengths and identified only one candidate in SARS-CoV, involving a sequence of 20 bases starting at position 5745 and located in orf1.
Further analysis, considering potential internal pairing in SARS-CoV-2 RNA sequence with a length as low as 5 bases was performed. As expected, the number of potential internal pairing increases as the minimum required length is decreased; it reaches 659006 for potential internal pairing of size 5 (Table 5). Given that the size of the SARS-CoV-2 genome is 29903 bases, this means that each base can potentially be part of an average of 22 internal matches of 5 bases in length, or about 440 for a sequence of 20 bases. Performing the same processing for B.1.1.7, B.1.351 and P.2 variants leads to similar results (Supplemental Table S18) The search for the number of potential internal pairings included in sequences that potentially hybridize with the human transcriptome is in line with the expected average number (Table 6).
4.5. Assessment of the scope of hybrid duplexing against other species or viruses
To assess whether the sequences identified in SARS-CoV-2 could impact other species, we used BLAST to calculate the similarities between each human sequence and the following 12 other species: Gorilla gorilla, Pan troglodytes, Pongo abelii, Papio anubis, Macaca mulatta, Sapajus apella, Mus musculus, Rattus norvegicus, Pteropus vampyrus, Sus scrofa, Bos taurus and Felis catus. The potential of hybrid pairing depends on the species and candidate mRNAs in the host (Figure 2 and Supplemental Tables S4 to S10). USP30 features only human-specific hybrid pairing. Conversely, using BLAST, we also calculated similarities of sequences originating from SARS-CoV-2 with the following viruses: bat coronavirus RaTG13, SARS-CoV, MERS-CoV, HCoV-229E and HCoV-OC43. The results are presented in Figure 3 and Supplemental Tables S11 to S17.
4.6. Assessment of the existence of base-pairing between RNA molecules of viral origin and the host transcriptome
To confirm the presence of host-virus RNA-RNA interactions and quantify the occurrence of this phenomenon, we relied on data generated by Ziv et al., in their study “The short-and long-range RNA-RNA Interactome of SARS-CoV-2” published in Molecular Cell (2020). The authors experimentally mapped the in vivo RNA-RNA interactome of the full-length SARS-CoV-2 genome and subgenomic mRNAs. Their base-pairing prediction data are available as supplementary data associated with the raw sequencing data deposited on GEO with accession number GSE154662. The chimeric reads are listed in tabulated files with their annotation to the respective genome.
We processed these files with a python script to compute the number of identified interactions and the distribution of base pairings between those occurring within the same RNA molecule and those involving different RNA molecules. We performed the processing for RNA-RNA interactions spanning the entirety of SARS-CoV-2 genomic RNA (gRNA) and subgenomic mRNAs (sgmRNAs). We also performed the same calculations for RNA-RNA interactions spanning the entirety of MERS-CoV gRNA. The results are presented in Table 7.
4.7. Assessment of the impact of SARS-CoV-2 infection on host transcriptome
Calu-3 cell line: FASTQ files related to the study of Sun et al. (2020) were downloaded from the China National Genomics Data Center (https://bigd.big.ac.cn/) with the accession number PRJCA002617. The dataset contains the sequencing of triplicate samples of mock-infected and SARS-CoV-2-infected Calu-3 cells at four different time points: 0, 7, 12, and 24 h post-infection (hpi). COVID19 patient samples: FASTQ files related to the study of Blanco-Melo et al. (2020) were downloaded from the NCBI Gene Expression Omnibus (GEO) server under the accession number GSE147507. The dataset contains the sequencing of uninfected human lung biopsies derived from one male (age 72) and one female (age 60) and additionally, lung samples derived from a single male COVID19 deceased patient (age 74) processed in technical replicates.
We used Salmon version 1.4.0 (Patro et al., 2017) to quantify abundance and effective transcript lengths for all samples. We quantified against the NCBI Build 38 patch release 13 (GRCh38.p13) reference human genome and associated RefSeq Transcripts. We used tximport (Soneson et al., 2015) to import counts, abundances and effective transcript lengths into R. Gene-level statistical analysis was then performed using DESeq2 (Love at al., 2014). The differential expressions of genes potentially targeted by SARS-CoV-2 results are presented in Figure 4.
Declarations
Author contribution statement
Claude Pasquier: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Alain Robichon: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This work was supported by the ANR grant “Methylclonome” ANR-12-BSV6-006-01 to Alain Robichon and Claude Pasquier. This work was also supported by the French National Research Agency (ANR) through the LABEX SIGNALIFE program (reference # ANR-11-LABX-0028-01).
Data availability statement
Data included in article/supp. material/referenced in article.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Aliyari R., Ding S.W. RNA-based viral immunity initiated by the Dicer family of host immune receptors. Immunol. Rev. 2009;227:176. doi: 10.1111/j.1600-065X.2008.00722.x. 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarzguioui M., Holen T., Babaie E., Prydz H. Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res. 2003;31:589–595. doi: 10.1093/nar/gkg147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. Metazoan microRNAs. Cell. 2018;173:20–51. doi: 10.1016/j.cell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkhout B. RNAi-mediated antiviral immunity in mammals. Curr. Opin. Virol. 2018;32:9–14. doi: 10.1016/j.coviro.2018.07.008. [DOI] [PubMed] [Google Scholar]
- Bernard M.A., Wang L., Tachado S.D. Dicer-Argonaute 2 complex in continuous fluorogenic assays of RNA interference enzymes. PloS One. 2015;10 doi: 10.1371/journal.pone.0120614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D., Wang T.T., Schwartz R.E., Lim J.K., Albrecht R.A., tenOever B.R. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045. doi: 10.1016/j.cell.2020.04.026. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthy S., Sternberg S.H., Kellenberger C.A., Doudna J.A. Substrate-specific kinetics of dicer-catalyzed RNA processing. J. Mol. Biol. 2010;404:392–402. doi: 10.1016/j.jmb.2010.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.W., Kok K.H., Zhu Z., Chu H., To K.K., Yuan S., Yuen K.J. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microb. Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheloufi S., Dos Santos C.O., Chong M.M., Hannon G.J. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes D., Xue H., Taylor D.W., Patnode H., Mishima Y., Cheloufi S., Ma E., Mane S., Hannon G.J., Lawson N.D., Wolfe S.A., Giraldez A.J. A novel miRNA processing pathway independent of Dicer requires Argonaute 2 catalytic activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens M.J. PKR: a protein kinase regulated by double-stranded RNA. Int. J. Biochem. Cell Biol. 1997;29:945–949. doi: 10.1016/s1357-2725(96)00169-0. [DOI] [PubMed] [Google Scholar]
- Clemens M.J., Elia A. The double-stranded RNA-dependent protein kinase PKR: structure and function. J. Interferon Cytokine Res. 1997;17:503–524. doi: 10.1089/jir.1997.17.503. [DOI] [PubMed] [Google Scholar]
- Cunningham C.N., Baughman J.M., Phu L., Tea J.S., Yu C., Coons M., Kirkpatrick D.S., Bingol B., Corn J.E. USP30 and parkin homeostatically regulate atypical ubiquitin chains on mitochondria. Nat. Cell Biol. 2015;17:160–169. doi: 10.1038/ncb3097. [DOI] [PubMed] [Google Scholar]
- Czech B., Malone C.D., Zhou R., Stark A., Schlingeheyde C., Dus M., Perrimon N., Kellis M., Wohlschlegel J.A., Sachidanandam T., Hannon G.J., Brennecke J. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:98–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dincbas-Renqvist V., Pepin G., Rakonjac M., Plante I., Ouellet D.L., Hermansson A., Goulet I., Doucet J., Samuelsson B., Radmark O. Human Dicer C-terminus functions as a 5-lipoxygenase binding domain. Biochim. Biophys. Acta. 2009;1789:99–108. doi: 10.1016/j.bbagrm.2008.10.002. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batu P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y.T. RNA recognition and signal transduction by RIG-I-like receptors. Immunol. Rev. 2009;227:54–65. doi: 10.1111/j.1600-065X.2008.00727.x. [DOI] [PubMed] [Google Scholar]
- Fukunaga R., Han B.W., Hung J.H., Xu J., Weng Z., Zamore P.D. Dicer partner proteins tune the length of mature miRNAs in flies and mammals. Cell. 2012;151:533–546. doi: 10.1016/j.cell.2012.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallicano G.I., Casey J.L., Fu J., Mahapatra S. Molecular targeting of vulnerable RNA sequences in SARS CoV-2: identifying clinical feasibility. Gene Therapy. 2020:1–8. doi: 10.1038/s41434-020-00210-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García M.A., Meurs E.F., Esteban M. The dsRNA protein kinase PKR: virus and cell control. Biochimie. 2007;89:799–811. doi: 10.1016/j.biochi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Ghildiyal M., Seitz H., Horwich M.D., Li C., Du T., Lee S., Xu J., Kittler E.L., Zapp M.L., Weng Z., Zamore P.D. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008;320:1077–1081. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M.…Krogan N.J. SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grey F. Role of microRNAs in herpesvirus latency and persistence. J. Gen. Virol. 2015;96(Pt 4):739–751. doi: 10.1099/vir.0.070862-0. [DOI] [PubMed] [Google Scholar]
- Guo Z., Li Y., Ding S.W. Small RNA-based antimicrobial immunity. Nat. Rev. Immunol. 2019;19:31–44. doi: 10.1038/s41577-018-0071-x. [DOI] [PubMed] [Google Scholar]
- Han Q., Chen G., Wang J., Jee D., Li W.X., Lai E.C., Ding S.W. Mechanism and function of antiviral RNA interference in mice. mBio. 2020;11 doi: 10.1128/mBio.03278-19. 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henzinger H., Barth D.A., Klec C., Pichle M. Non-coding RNAs and SARS-related coronaviruses. Viruses. 2020;12:1374. doi: 10.3390/v12121374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A., Medzhitov R. Regulation of adaptive immunity by the innate immune system. Science. 2010;327:291–295. doi: 10.1126/science.1183021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling C.L., Yi M., Ancaster L.A.M., Lemon S.M., Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- Kazimierz T. Tycowski, Yang Eric Guo, Nara Lee, Walter N. Moss, Tenaya K. Vallery, Mingyi Xie, Joan A.Steitz. Viral noncoding RNAs: more surprises. Genes Dev. 2015;29(6):567–584. doi: 10.1101/gad.259077.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiening M., Ochsenreiter R., Hellinger H.J., Rattei T., Hofacker I., Frishman D. Conserved secondary structures in viral mRNAs. Viruses. 2019;11:401–423. doi: 10.3390/v11050401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Lee J.Y., Yang J.S., Kim J.W., Narry Kim V., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181:914–921. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kincaid R.P., Sullivan C.S. Virus-encoded microRNAs: an overview and a look to the future. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1003018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshiba T. Mitochondrial mediated antiviral immunity. Biochim. Biophys. Acta. 2013;1833:225–232. doi: 10.1016/j.bbamcr.2012.03.005. [DOI] [PubMed] [Google Scholar]
- Kurzynska-Kokorniak A., Pokornowska M., Koralewska N., Hoffmann W., Bienkowska-Szewczyk K., Figlerowicz M. Revealing a new activity of the human Dicer DUF283 domain in vitro. Sci. Rep. 2016 doi: 10.1038/srep23989. article 23989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Zheng Q., Ryvkin P., Dragomir I., Desai Y., Aiyer S., Valladares O., Yang J., Bambina S., Sabin L.R., Murray J.L., Lamitina T., Raj A., Cherry S., Wang L.S., Gregory B.D. Global analysis of RNA secondary structure in two metazoans. Cell Rep. 2012;1:69–82. doi: 10.1016/j.celrep.2011.10.002. [DOI] [PubMed] [Google Scholar]
- Li Y., Lu J., Han Y., Fan X., Ding S.W. RNA interference functions as an antiviral immunity mechanism in mammals. Science. 2013;342:231–234. doi: 10.1126/science.1241911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Basavappa M., Lu J., Dong S., Cronkite D.A., Prior J.T., Reinecker H.-C., Hertzog P., Han Y., Li W.-X., Cheloufi S., Karginov F.V., Ding S.-W., Jeffrey K.L. Induction and suppression of antiviral RNA interference by influenza A virus in mammalian cells. Nat. Microbiol. 2016;2:16250. doi: 10.1038/nmicrobiol.2016.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima W.F., Murray H., Nichols J.G., Wu H., Hong Sun H., Prakash T.P., Berdeja A.R., Hans J., Gaus H.J., Stanley T., Crooke S.T. Human Dicer binds short single-strand and double-strand RNA with high affinity and interacts with different regions of the nucleic acids. J. Biol. Chem. 2009;284:2535–2548. doi: 10.1074/jbc.M803748200. [DOI] [PubMed] [Google Scholar]
- Liu Z., Wang J., Xu Y., Guo M., Mi K., Xu R., Pei Y., Zhang Q., Luan X., Hu Z., Liu X. arXiv preprint; 2020. Implications of the Virus-Encoded miRNA and Host miRNA in the Pathogenicity of SARS-CoV-2. arXiv:2004.04874. [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):1–21. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae I.J., Zhou K., Li F., Repic A., Brooks A.N., Cande W.Z., Adams P.D., Doudna J.A. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- MacRae I.J., Zhou K., Doudna J.A. Structural determinants of RNA recognition and cleavage by Dicer. Nat. Struct. Mol. Biol. 2007;14:934–940. doi: 10.1038/nsmb1293. [DOI] [PubMed] [Google Scholar]
- Madhugiri R., Fricke M., Marz M., Ziebuhr J. Coronavirus cis-acting RNA elements. Adv. Virus Res. 2016;96:127–163. doi: 10.1016/bs.aivir.2016.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard P.V., Ciaudo C., Marchais A., Li Y., Jay F., Ding S.W., Voinnet O. Antiviral RNA interference in mammalian cells. Science. 2013;342:235–238. doi: 10.1126/science.1241930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard P.V., Veen A.G., Poirier E.Z., Reis e Sousa C. Slicing and dicing viruses: antiviral RNA interference in mammals. EMBO J. 2019;38 doi: 10.15252/embj.2018100941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak J., Kleiman L. Primer tRNAs for reverse transcription. J. Virol. 1997;71:8087–8095. doi: 10.1128/jvi.71.11.8087-8095.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra M.A., Jones S.J.M., Astell C.R.…Roper R.L. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- Mateos-Gomez P.A., Morales L., Zuñiga S., Enjuanes L., Sola I. Long distance RNA-RNA interactions in the coronavirus genome form high-order structures promoting discontinuous RNA synthesis during transcription. J. Virol. 2013;87:177–186. doi: 10.1128/JVI.01782-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales L., Oliveros J.C., Fernandez-Delgado R., tenOever B.R., Enjuanes L., Sola I. SARS-CoV-Encoded small RNAs contribute to infection-associated lung pathology. Cell Host Microbe. 2017;21:344–355. doi: 10.1016/j.chom.2017.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish S., Fleenor J., Xu S., Mello C., Fire A. Functional anatomy of a dsRNA trigger: differential requirement for the two trigger strands in RNA interference. Mol. Cell. 2000;6:1077–1087. doi: 10.1016/s1097-2765(00)00106-4. [DOI] [PubMed] [Google Scholar]
- Pasquier C., Agnel S., Robichon A. Transcriptome-wide-scale-predicted dsRNAs potentially involved in RNA homoeostasis are remarkably excluded from genes with no/very low expression in all developmental stages. RNA Biol. 2020;17:554–570. doi: 10.1080/15476286.2020.1717154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017;14(4):417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul D., Bartenschlager R. Architecture and biogenesis of plus-strand RNA virus replication factories. World J. Virol. 2013;2:32–48. doi: 10.5501/wjv.v2.i2.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez J.T., Varble A., Sachidanandam R., Zlatev I., Manoharan M., Garcı´a- Sastre A., tenOever B.R. Influenza A virus-generated small RNAs regulate the switch from transcription to replication. Proc. Natl. Acad. Sci. U.S.A. 2010;107:11525–11530. doi: 10.1073/pnas.1001984107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y., Xu Y., Zhang Y., Zhou H., Deng Y.-Q., Li X.-F., Miao M., Zhang Q., Zhong B., Hu Y., Zhang F.-C., Wu L., Qin C.-F., Zhou X. Human virus-derived small RNAs can confer antiviral immunity in mammals. Immunity. 2017;46:992–1004. doi: 10.1016/j.immuni.2017.05.006. [DOI] [PubMed] [Google Scholar]
- Qiu Y., Xu Y.-P., Wang M., Miao M., Zhou H., Xu J., Kong J., Zheng D., Li R.-T., Zhang R.-R., Guo Y., Li X.-F., Cui J., Qin C.-F., Zhou X. Flavivirus induces and antagonizes antiviral RNA interference in both mammals and mosquitoes. Sci. Adv. 2020;6 doi: 10.1126/sciadv.aax7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajsbaum R., Giraldo M.I., Bharaj P., Atkins C., Xia H., Rossi S.L., Lee B., Shi P.Y., Freiberg A.N. The host ubiquitin system in innate immunity and virus replication: proviral and antiviral functions of the host E3-ubiquitin ligase TRIM family. J. Immunol. 2018;200(1 Supplement):50–53. [Google Scholar]
- Rambaut A., Loman N., Pybus O., Barclay W., Barrett J., Carabelli A., Connor T., Peacock T., Robertson D.L., Volz E., on behalf of COVID-19 Genomics Consortium UK (CoG-UK) Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. Gen. Epidemiol. 2020 [Google Scholar]
- Riley K.J., Rabinowitz G.S., Steitz J.A. Comprehensive analysis of Rhesus lymphocryptovirus microRNA expression. J. Virol. 2010;84:5148–5157. doi: 10.1128/JVI.00110-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley K.J., Rabinowitz G.S., Yario T.A., Luna J.M., Darnell R.B., Steitz J.A. EBV and human microRNAs co-target oncogenic and apoptotic viral and human genes during latency. EMBO J. 2012;31:2207–2221. doi: 10.1038/emboj.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler A.J., Williams B.R. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha N.K., Iwasa J., Shen P.S., Bass B.L. Dicer uses distinct modules for recognizing dsRNA termini. Science. 2018;359:329–334. doi: 10.1126/science.aaq0921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siva N. 1000 Genomes project. Nat. Biotechnol. 2008;26:256. doi: 10.1038/nbt0308-256b. [DOI] [PubMed] [Google Scholar]
- Snead N.M.J.J., Rossi J.J. RNA interference trigger variants: getting the most out of RNA for RNA interference-based therapeutics. Nucleic Acid Therapeut. 2012;22:139–146. doi: 10.1089/nat.2012.0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snead N.M., Wu X., Li A., Cui Q., Sakurai K., Burnett J.C., Rossi J.J. Molecular basis for improved gene silencing by Dicer substrate interfering RNA compared with other siRNA variants. Nucleic Acids Res. 2013;41:6209–6221. doi: 10.1093/nar/gkt200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola I., Almazán F., Zúñiga S., Enjuanes L. Continuous and discontinuous RNA synthesis in coronaviruses. Annu. Rev. Virol. 2015;2:265–288. doi: 10.1146/annurev-virology-100114-055218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soneson C., Love M.I., Robinson M.D. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Research. 2015:4. doi: 10.12688/f1000research.7563.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Jean J.R., Jacomy H., Desforges M., Vabret A., Freymuth F., Talbot P.J. Human respiratory coronavirus OC43: genetic stability and neuroinvasion. J. Virol. 2004;78:8824–8834. doi: 10.1128/JVI.78.16.8824-8834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., Sun L., Liu H.H., Chen X., Seth R.B., Forman J., Chen Z.J. The specific and essential role of MAVS in antiviral innate immune responses. Immunity. 2006;24:633–642. doi: 10.1016/j.immuni.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Sun X., Rogoff H.A., Li C.J. Asymmetric RNA duplexes mediate RNA interference in mammalian cells. Nat. Biotechnol. 2008;26:1379–1382. doi: 10.1038/nbt.1512. [DOI] [PubMed] [Google Scholar]
- Sun J., Ye F., Wu A., Yang R., Pan M., Sheng J., Zhu W., Mao L., Wang M., Xia Z., Huang B., Tan W., Jiang T. Comparative transcriptome analysis reveals the intensive early stage responses of host cells to SARS-CoV-2 infection. Front. Microbiol. 2020;11 doi: 10.3389/fmicb.2020.593857. article 593857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahamtan A., Inchley C.S., Marzban M., Tavakoli-Yaraki M., Teymoori-Rad M., Nakstad B., Salimi V. The role of microRNAs in respiratory viral infection: friend or foe? Rev. Med. Virol. 2016;26:389–407. doi: 10.1002/rmv.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O., Akira S. Innate immunity to virus infection. Immunol. Rev. 2009;227:75–86. doi: 10.1111/j.1600-065X.2008.00737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J.…de Oliveira T. medRxiv; 2020. Emergence and Rapid Spread of a New Severe Acute Respiratory Syndrome-Related Coronavirus 2 (SARS-CoV-2) Lineage with Multiple Spike Mutations in South Africa. [Google Scholar]
- tenOever B.R. RNA viruses and the host microRNA machinery. Nat. Rev. Microbiol. 2013;11:169–180. doi: 10.1038/nrmicro2971. [DOI] [PubMed] [Google Scholar]
- Theotokis P.I., Usher L., Kortschak C.K., Schwalbe E., Moschos S.A. Profiling the mismatch tolerance of Argonaute 2 through deep sequencing of sliced polymorphic viral RNAs. Mol. Ther. Nucleic Acids. 2017;9:22–33. doi: 10.1016/j.omtn.2017.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel V., Herold J., Schelle B., Siddell S.G. Infectious RNA transcribed in vitro from a cDNA copy of the human coronavirus genome cloned in vaccinia virus. J. Gen. Virol. 2001;82:1273–1281. doi: 10.1099/0022-1317-82-6-1273. [DOI] [PubMed] [Google Scholar]
- Trobaugh D.W.C.L., Sun Chengqun, Haddow A.D., Wang E., Chapnik E., Mildner A., Weaver S.C., Ryman K.D., Klimstra W.B. RNA viruses can hijack vertebrate microRNAs to suppress innate immunity. Nature. 2014;506:245–248. doi: 10.1038/nature12869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski K.T., Guo Y.E., Lee N., Moss W.N., Vallery T.K., Xie M., Steitz J.A. Viral noncoding RNAs: more surprises. Genes Dev. 2015;29:567–584. doi: 10.1101/gad.259077.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzimas C., Michailidou G., Arsenakis M., Kieff E., Mosialos G., Hatzivassiliou E.G. Human ubiquitin specific protease 31 is a deubiquitinating enzyme implicated in activation of nuclear factor-kappaB. Cell. Signal. 2006;18:83–92. doi: 10.1016/j.cellsig.2005.03.017. [DOI] [PubMed] [Google Scholar]
- Umbach J.L., Cullen B.R. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev. 2009;23:1151–1164. doi: 10.1101/gad.1793309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Boheemen S., de Graaf M., Lauber C., Bestebroer T.M., Raj V.S., Zaki A.M., Osterhaus A.D., Haagmans B.L., Gorbalenya A.E., Snijder E.J., Fouchier R.A. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio. 2012;3:e00473. doi: 10.1128/mBio.00473-12. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voloch C.M., da Silva Francisco R., de Almeida L.G., Cardoso C.C., Brustolini O.J., Gerber A.L.…de Vasconcelos A.T.R. Genomic characterization of a novel SARS-CoV-2 lineage from Rio de Janeiro, Brazil. J. Virol. 2021 doi: 10.1128/JVI.00119-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng K.F., Hung C.T., Hsieh P.T., Li M.L., Chen G.W., Kung Y.A., Huang P.N., Kuo R.L., Chen L.L., Lin J.Y. A cytoplasmic RNA virus generates functional viral small RNAs and regulates viral IRES activity in mammalian cells. Nucleic Acids Res. 2014;42:12789–12805. doi: 10.1093/nar/gku952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojnicka M., Szczepanska A., Kurzynska-Kokorniak A. Unknown areas of activity of human ribonuclease dicer: a putative deoxyribonuclease activity. Molecules. 2020;25:1414. doi: 10.3390/molecules25061414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.Y., Brian D.A. Subgenomic messenger RNA amplification in coronaviruses. Proc. Natl. Acad. Sci. Unit. States Am. 2010;107:12257–12262. doi: 10.1073/pnas.1000378107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C. E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S., Hasegawa T., Suzuki M., Sugeno-N-Baba T., Takeda A., Mochizuki H., Nagai Y., Aoki M. Parkinson's disease-linked DNAJC13 mutation aggravates alpha-synuclein-induced neurotoxicity through perturbation of endosomal trafficking. Hum. Mol. Genet. 2018;27:823–836. doi: 10.1093/hmg/ddy003. [DOI] [PubMed] [Google Scholar]
- Yu Z., Chen T., Li X., Yang M., Tang S., Zhu X., Gu Y., Su X., Xia M., Li W., Zhang X., Wang Q., Cao X., Wang J. Lys29-linkage of ASK1 by Skp1-Cullin 1-Fbxo21 ubiquitin ligase complex is required for antiviral innate response. Elife. 2016;5 doi: 10.7554/eLife.14087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv O., Gabryelska M.M., Lun A.T.L., Gebert L.F.R., Sheu-Gruttadauria J., Meredith L.W., Liu Z.Y., Kwok C.K., Qin C.F., MacRae I.J., Goodfellow I., Marioni J.C., Kudla G., Miska E.A. COMRADES determines in vivo RNA structures and interactions. Nat. Methods. 2018;15:785–788. doi: 10.1038/s41592-018-0121-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv O., Price J., Shalamova L., Kamenova T., Goodfellow I., Weber F., Miska E.A. The short- and long-range RNA-RNA Interactome of SARS-CoV-2. Mol. Cell. 2020;80:1067–1077. doi: 10.1016/j.molcel.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in article/supp. material/referenced in article.