Abstract
Cell migration is essential for normal development, neural patterning, pathogen eradication, and cancer metastasis. Pre-mRNA processing events such as alternative splicing and alternative polyadenylation result in greater transcript and protein diversity as well as function and activity. A critical role for alternative pre-mRNA processing in cell migration has emerged in axon outgrowth during neuronal development, immune cell migration, and cancer metastasis. These findings suggest that migratory signals result in expression changes of post translational modifications of splicing or polyadenylation factors, leading to splicing events that generate pro-migratory isoforms. We summarize this recent progress and suggest emerging technologies that may facilitate a deeper understanding of the role of alternative splicing and polyadenylation in cell migration.
Keywords: Splicing, migration, cleavage and polyadenylation, metastasis, axons
Alternative pre-mRNA processing and cell migration
Cell migration is important for proper development of embryos, functioning of immune system, and wound healing [1]. Coordination of cell migration involves relaying signals from outside the cell to inside the cell, and spatiotemporal information exchange between the front and rear of a cell [2]. In mammalian systems, migration-inducing stimuli including chemokines, cytokines and growth factors serve as chemoattractants [3] (Figure 1). These external factors activate signaling pathways such as the mitogen activated protein (MAP) kinase pathway, the phosphatidylinositol 3 kinase (PI3K) pathway, and mammalian Target of Rapamycin Complex 2 (mTORC2), which modulate the activity of GTP-binding proteins that organize cell migration. Three GTP-binding proteins, Ras homologue gene family (Rho), Ras-related C3 botulinum toxin substrate 1 (Rac) and Cell division control protein 42 (Cdc42), in turn, regulate the actin cytoskeleton to promote cytoskeletal-mediated motility. Formins, members of the Wiskott-Aldrich Syndrome Protein (WASP) family, the actin related protein (Arp2/3) complex, and myosin can be modulated by GTP-binding proteins to generate polymerized actin filaments that push the leading edge of the cell forward and contract the rear of the cell. These same mechanisms are used in multiple physiological contexts. During development, individual cells detach from epithelial sheets and migrate to new locations. In adult organisms, immune cells migrate toward microbes to promote their elimination. In a related process, extracellular protein attractants and repellants stimulate or repress the directed growth of specialized protruding extensions such as axon growth cones [4].
Figure 1:
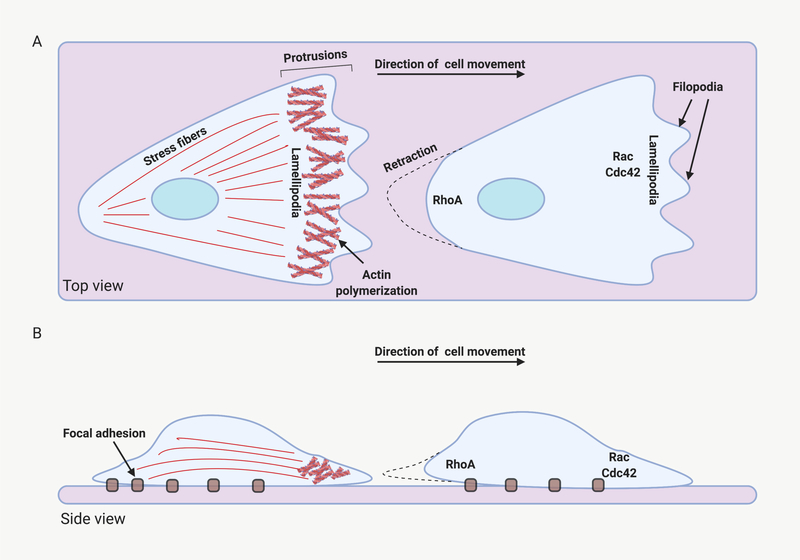
Molecular basis for cell migration. Some of the molecules that contribute to cell migration are depicted (A) Top view of a migrating cell. Actin filaments at the leading edge propel the plasma membrane forward in the form of lamellipodia and filopodia. The GTP-binding proteins Rac and Cdc42 organize these processes. Actin-myosin-based stress fibers promote contraction of the rear of the cell. The Rho GTP-binding protein is an important regulator of this process. (B) Side view of a moving cell. Focal adhesions along the bottom of the cell anchor stress fibers and contribute to the generation of force needed for forward movement. Created with BioRender
Recent studies have described model systems in which a signal for cell migration results in pre-mRNA processing mechanisms of alternative splicing - inclusion or exclusion of exons in the resulting mRNA product - and alternative polyadenylation - use of multiple polyadenylation sites for the same gene [5, 6] (Box 1). These mechanisms alter the expression of transcript isoforms involved in cell migration (Table 1). The transcript isoforms produced can result in protein products with distinct or even opposing functions. This diversity of proteins generated from a limited number of genes enables complex multicellular organisms to finely tune their developmental and homeostatic maintenance mechanisms temporally and in a tissue- or cell type-specific manner [7]. Nearly all human genes are estimated to be alternatively spliced in some context [6] and 15% of genetic diseases are predicted to result from aberrant splicing [8].
Text box 1: Mechanisms of alternative splicing and alternative polyadenylation.
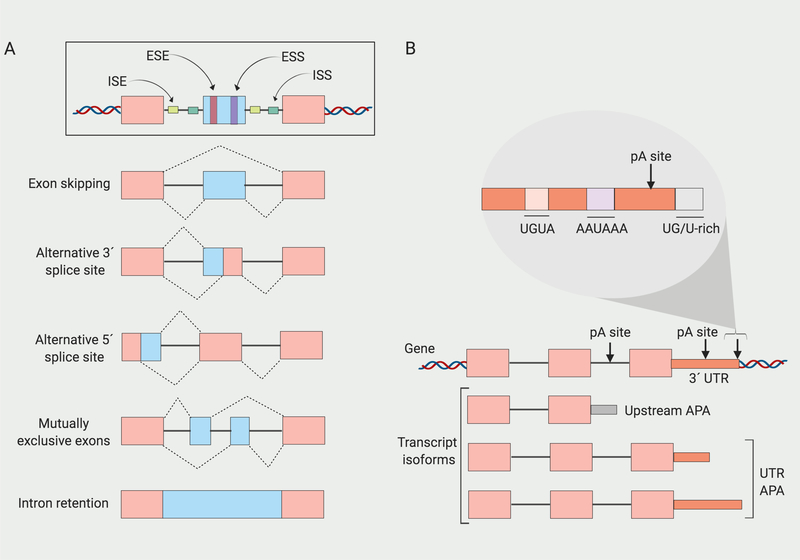
An average human gene is composed of eight exons, seven introns and 8.2 polyadenylation sites [53, 54]. During pre-mRNA splicing, removal of introns from immature pre-mRNA transcripts is performed by the spliceosome, a large ribonucleoprotein complex [6]. Spliceosomes contain five small nuclear RNAs and many associated proteins to form a complex that recognizes splice sites at the exon/intron boundaries in pre-mRNA. The spliceosome catalyzes the enzymatic steps involved in intron removal [55]. Formation of the spliceosome is facilitated or impeded by non-snRNP proteins (auxilliary splicing factors) that can bind at sequence elements (enhancers or silencers) in pre-mRNA exons or introns of pre-mRNA (Figure IA). Most well-known of these splicing factors are SR and hnRNP family of proteins that generally promote and repress exon inclusion, respectively. SR proteins have RNA binding domains (RBDs) containing RNA recognition motifs (RRM) that bind exonic splicing elements and also domains rich in arginine/serine dipeptides (RS domains) that participate in protein-protein interactions [56]. HnRNP family members contain four unique RBDs (RRM, quasi-RRM, KH K-homology, and RGG containing Arginine-glycine-glycine repeats) [57]. An excess of SR protein splicing factors tends to favor the use of proximal splicing sites [58]. One model is that higher levels of SR factors results in full occupancy of all of the 5’ splice sites by the spliceosome and under these conditions, the closest site is selected [58]. In contrast, hnRNPs tend to antagonize the SR proteins by blocking their access to 5’ splice sites [58]. Binding of hnRNPs to splicing silencers reduces the selection of nearby 3’ splice sites [58]. The differential activity of SR proteins, hnRNPs and other splicing factors can result in multiple different kinds of alternative splicing. Entire exons (exon skipping) or portions of exons (alternative 5’ or 3’ splice site) can be either included or excluded, introns can be retained (intron retention), and in some cases, one isoform will contain exon A, while a different isoform will contain exon B (mutually exclusive exons).
Maturation of pre-mRNA transcripts also requires cleavage of their 3’ end from the nascent pre-mRNA strand followed by synthesis of a poly(A) tail by poly(A)polymerase (cleavage and polyadenylation) [5]. (Figure IB). Sequences within the transcript including the hexamer A[A/U]UAAA and close variants serve as signals for the multi-protein cleavage and polyadenylation complexes that perform the cleavage reaction. Additional sequences that support the cleavage include U-rich elements and UGUA motifs upstream, and U-rich elements and GU-rich elements downstream [59]. The most 3’ polyadenylation site is usually the strongest, with the strongest hexamer, AAUAAA. Modulating the levels of these factors that participate in these complexes can affect the choice of polyadenylation site. The Cleavage Stimulating Factor-64 (CstF-64) core polyadenylation factor recognizes U-rich or G/U-rich sequences located 20–40 bps downstream of the cleavage site in mRNA transcripts (CstF)-64 favors use of proximal polyadenylation sites. Splicing U1 snRNP can also affect the use of proximal polyadenylation sites in 3’ UTRs and alternative terminal exons [60]. Changes in the levels and activity of cleavage and polyadenylation factors can result in transcripts that terminate early, either at polyadenylation sites within the 3’ UTR (UTR APA), or at polyadenylation sites within genes (Upstream APA). Upstream APA can change the coding sequence of a transcript and result in the synthesis of a distinct protein.
Figure I. Mechanisms of alternative splicing and alternative cleavage and polyadenylation.
(A) Exonic and intronic splicing enhancers (ESE and ISE) and splicing silencers (ESS and ISS) can bind to sequences in the 3’ and 5’ portions of exons and introns to regulate alternative splicing. Alternative splicing can result in exon skipping, alternative 5’ or 3’ splice site use, mutually exclusive exons, or intron retention. (B) Alternative cleavage and polyadenylation can result in transcripts of different lengths from the same gene by alternative selection of polyadenylation (pA) sites. Transcripts can terminate at different positions within the 3’ UTR (UTR-APA) or even within the gene (Upstream APA). Motifs within 3’ UTRs are recognize d by protein complexes that perform the cleavage and polyadenylation reactions. Created with BioRender.
Table 1.
Connections between cell migration and alternative isoform use.
| splicing APA factor | gene target | Isoform migration effect | Ref |
|---|---|---|---|
| Alternative Splicing | |||
| hnRNPA1/SR protein | Rac1 | Increased levels of Rac1b isoform result in increased migration | [13] |
| hnRNPM acetylation | CD44 | p300 binding promotes CD44v exon inclusion and reduces cell motility | [61] |
| hnRNPA2 | TP53INP2 | Knockdown of exon 2-containing TP53INP2 isoforms reduced migration speed and invasion | [62] |
| SRSF1 | Ron | Increased levels of ΔRon isoform lead to increased migration | [63] |
| RBM4 | DAB1 | Increased levels of DAB1 isoform lacking exons 7/8 lead to neuronal migration defect | [16] |
| RBM4 | TEAD4 | TEAD4-S leads to reduced cell migration | [64] |
| ESRP1 | LYN | Increased levels of Long isoform of LYN (LYNA) result in increased migration | [65] |
| ESRP1 | CD44 | ESRP1 suppresses cell motility mainly by repressing CD44 isoform switching from CD44v to CD44s | [35] |
| Nova1/2 | DCC | Increased levels of long isoform of DCC lead to increased axon outgrowth | [17] |
| PRPF4B and BUD31 | Splicing factors PRPF4B and BUD31 are required for cancer cell migration | [32] | |
| Rac1 | Increased levels of Rac1b isoform result in increased migration | [12] | |
| Cortactin | Cortactin variants were less effective at promoting migration than wild-type cortactin | [30] | |
| alpha-tropomyosin | TmBr1 expressing cells had shorter tracks and reduced speed. TmBr3 expressing cells had slower speed. | [31] | |
| RHAMM | Increased levels of RHAMMB isoform lacking exon 4 lead to increased metastasis | [15] | |
| Cyclin D1 | Cyclin D1a promotes migration but D1b fails to promote migration | [66] | |
| GLI1 | Increased levels of tGLI1 isoform leads to increased migration | [67] | |
| Cytohesin-1 | Diglycine isoform promote cell migration | [26] | |
| Alternative Polyadenylation | |||
| ELAV | DSCAM1 | DSCAM1-L promotes neuronal axon outgrowth | [50] |
| CstF64 | Reduced levels of CstF64 lead to decreased migration | [41] | |
| RECK | Reduced levels of short isoform of RECK lead to impaired migration | [42] | |
| BIRC3 | BIRC3-LU isoform is involved in regulating B cell migration | [46] | |
Splicing factors are proteins that bind to recognition sites at the beginning or end of an intron and affect the likelihood that and intron will be spliced out of the final transcript. Many cell signaling pathways, including the mitogen-activated protein (MAP) Kinase pathway, PI3K pathway and the wingless (Wnt) pathway [9], affect the synthesis or degradation rates of splicing factors. For example, both SR and heterogeneous nuclear ribonucleoproteins (hnRNP) proteins can bind to transcripts. SR proteins tend to promote binding of the spliceosome while hnRNP proteins usually inhibit spliceosome binding. SR protein binding promotes the inclusion of specific exons while binding of hnRNP proteins often results in exon exclusion [8]. In addition to regulation of splicing factor synthesis and degradation, splicing factors can be deleted or acquire mutations in tumors, which can also affect their activity [10]. Changes in the activity of splicing factors can result in changes in the expression of transcript isoforms encoding proteins that play a role in cell migration [11, 12]. In some cases, changes in exon or polyadenylation site use results in variations in isoform levels that are pro-migratory or anti-migratory.
We describe recent findings performed in vitro and in vivo as well as in cancer and neuronal cells to shed light on the central role that alternative splicing and polyadenylation plays in the regulation of cell motility and cellular protrusions across multiple contexts.
Alternative splicing in cell migration
Alterations in the levels of splicing factors involved in cell migration have been shown to regulated by external chemotactic signals, molecules that promote cell migration, including receptor tyrosine kinases (RTKs) in normal cells [13], hyaluronic acid receptor in certain types of cancer [14, 15], and receptors for attractant and repellant molecules in neurons [16, 17].
Receptor tyrosine kinase signaling-dependent splicing promotes migration
Chemotactic signaling by epidermal growth factor (EGF) modulates the activity of splicing factors through two parallel pathways that affect post-translational modification of splicing factors: (i) ubiquitination of hnRNP proteins, and (ii) phosphorylation of SR proteins [13] (Figure 2A). EGF signaling upregulates SPRY domain-containing SOCS box protein 1 (SPSB1), which is involved in the ubiquitination and degradation of the repressive splicing factor hnRNP A1. In parallel, activation of the PI3K/Akt branch of the EGF pathway results in nuclear translocation of SR protein kinases that phosphorylate SR proteins [18], which act as splicing enhancers [19]. Changes in the levels or localization of SR and hnRNP splicing modulators were shown to be important for the effect of EGF on cell migration as knockdown of SPSB1, expression of a ubiquitination-defective hnRNP mutant, or deletion of SR kinases resulted in slower cell migr ation in the presence of EGF [13].
Figure 2.
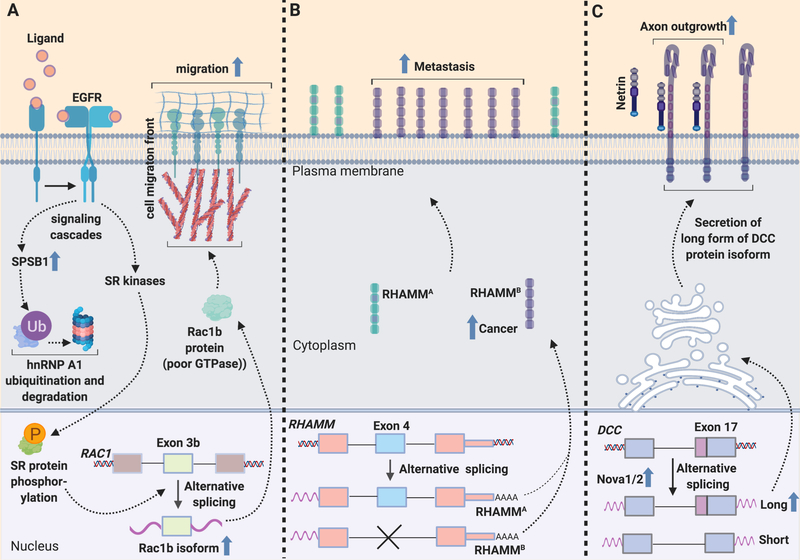
Effects of chemotactic cues on alternative splicing events. Many chemotactic factors and extracellular matrix molecules that promote cell migration in normal and cancer cells activate splicing factors. These splicing factors then modulate the incorporation of specific exons into processed transcripts. In many cases, the alternatively spliced transcript gains or loses functionality that serves to increase the cell’s motility. (A) In response to promigratory EGF signaling, SPSB1 increases, leading to ubiquitination and degradation of hnRNPA1. Also in response to EGF signaling, SR kinases phosphorylate SR proteins. Both of these pathways culminate in increased formation of a highly active alternative isoform of Rac1 containing exon 3b, Rac1b, that promotes migration. (B) Alternative splicing of exon 4 in pancreatic neuroendocrine tumors results in higher levels of a promigratory isoform of RHAMM, RHAMMB. RHAMMB, but not RHAMMA, is critical for in vivo metastatic capacity of pancreatic neuroendocrine tumors to the liver. (C) In neurons, the splicing factor Nova1/2 can promote alternative splicing of DCC to generate a long isoform that promotes neuron migration and the growth of axons from neuronal progenitors. Created with BioRender.
Further studies demonstrated that hnRNP A1 degradation and phosphorylation of SR proteins cooperate to promote the production of a splicing variant of Rac, Rac1b [13] (Figure 2A). Rac1 is a central organizer of the changes in the cytoskeleton that occur at the lamellipodia that define the leading edge of a moving cell. Rac1b is formed by the inclusion of an additional exon, exon3b, which adds 19 in-frame amino acids to Rac1 [11]. Rac1b, compared with the canonical protein, is more frequently in an active GTP-bound state and has different activities than the canonical isoform [20]. Knockdown of SPSB1, mutation of hnRNP A1 or knockdown of SR kinases SRPKs led to reduced levels of splice variant Rac1b [13]. Further, knockdown of SR kinases or Rac1b reduced cell migration upon EGF treatment, while higher Rac1b levels resulting from ubiquitination of hnRNP A1 and activation of SR kinases increased EGF-induced cell migration [13].
Additional studies have demonstrated a physiological role for Rac1b in promoting cell migration. Rac1b is overexpressed in a subset of aggressive colorectal cancers [11, 21], breast [22], ovarian tumors [23] compared with normal tissue. In colon cancer, activation of oncogenes RAS and MYC was associated with higher levels of the splicing factor polyprimidine tract binding protein 1 (PTBP1), which was shown to promote Rac1b expression [24]. Rac1b is also expressed in human inflammatory colon mucosa and mouse models of colon inflammation [25]. Rac1b overexpression in intestinal cells was associated with an increase in the number of Paneth cells at the bottom of intestinal crypts, in the proliferative index of the colonic epithelial cells, and in the migration of cells from the bottom to the top of crypts than controls [12]. When inflammation of the colon was experimentally induced, mice overexpressing Rac1b healed more rapidly, possibly as a result of increased proliferation and migration of epithelial intestinal cells [12]. These results support the importance of Rac1b as a mediator of cell proliferation, migration, and healing after injury.
Alternative splicing in response to RTK signaling is also associated with changes in subcellular localization of key proteins for cell migration. Hepatocyte growth factor interaction with its tyrosine kinase receptor results in increased migration. This pro-migratory effect was shown to depend on cytohesin, an ADP ribosylation factor 6 (Arf6) guanine nucleotide exchange factor [26]. Arf6 plays an important role in endocytosis of plasma membrane proteins and recycling of endocytic vesicles to the membrane that is required for the forward migration of the leading edge of a moving cell [27]. Cytohesin isoforms vary depending on the inclusion of a 3 nucleotide microexon that results in an additional glycine, generating a triglycine isoform, while exclusion of the exon results in a diglycine form. The diglycine form of cytohesin associates with phosphoinositol diphosphate PI(4,5)P2, leading to cytohesin localization at the plasma membrane [26]. Because phosphoinositol triphosphate (PI3P) is abundant at the leading edge of the cell, the effect is that cytohesin with the diglycine motif localizes at the leading edge. Diglycine cytohesin results in a local and polarized increase in Arf6 activity, receptor recycling and actin cytoskeletal rearrangement [27]. Consistent with this model, reintroduction of cytohesin with the diglycine sequence, but not the triglycine motif generated by inclusion of the microexon, in a cytohesin knockout cell line, rescued the reduced migration and invasion associated with cytohesin knockout [26].
Downstream of RTK signaling, molecules that mediate rearrangements in the actin cytoskeleton are critical for cell migration. Alternative splicing of actin-regulatory factors has been demonstrated to be important for cell motility. In invasive carcinomas, hepatocyte growth factor signaling results in the formation of invadopodia through the actin regulatory protein cortactin [28]. Cortactin binds to F-actin, regulates vesicle trafficking, and enhances cell migration, invasion and metastasis [29]. However, cortactin splice variants, one lacks the 6th cortactin repeat (exon 11) and the other lacks both the 5th and 6th repeats [30], were less effective at promoting cell migration than wild-type cortactin [30]. Finally, tropomyosins are important for the contractile forces that propel cells forward during cell migration. The tropomyosin family of proteins are actin-binding proteins that regulate the specialization of actin functions. Two splice variants of the alpha-tropomyosin gene TmBr1 and TmBr3 are not expressed in undifferentiated neuronal cells and are induced as neurons mature. Compared with the canonical isoform, either of the splice variants reduced the size of focal adhesions, physical structures that mediate the interaction between a cell and its extracellular matrix, and the structures that originate the tension-inducing stress fibers [31]. TmBr1 expression resulted in smaller, less developed focal adhesions while TmBr3 expression results in almost no focal adhesions and significantly less structured actin filaments. Expression of either TmBr1 or TmBr3 also reduced cell migration rates as cells expressing either isoform migrated more slowly into a denuded area of a plate, consistent with decreased tension from a smaller focal adhesion area. While the two alternative isoforms both reduced migration rate, they had opposing effects on migration persistence: cells expressing TmBr1 exhibited decreased persistence while TmBr3 cells showed increased persistence compared with controls. The studies taken together demonstrate that alternative splicing is emerging as a central player in the response to chemotactic signals and the cytoskeletal changes that result.
Alternative splicing promotes cancer cell invasion and metastasis
Alternative splicing has been demonstrated to be particularly important for cell migration in the context of metastasis. An unbiased RNAi-based screen of motile triple-negative breast cancer cells identified two splicing factors, PRPF4B and BUD31, among the genes essential for cancer cell migration [32]. Depletion of either of the splicing factors reduced the formation of focal adhesions and interactions with the extracellular matrix [32], emphasizing the importance of splicing for cancer cell metastasis.
One specific molecule in the extracellular matrix that controls migration and invasion is the glycosaminoglycan hyaluronic acid (HA). HA is associated with increased migration and invasion in aggressive cancers [33]. The HA receptor, receptor for hyaluronic acid -mediated motility (RHAMM), is upregulated in multiple types of cancer [14, 34] (Figure 2b). Splice variants of RHAMM consists of RHAMMA which includes all 18 exons and RHAMMB which lacks the 15 amino acid-long exon 4. Using RNA-Seq, RHAMM levels were found to be low in normal pancreatic islets, while splice variants were high in pancreatic neuroendocrine tumors with. RHAMMB being the highest expressed [15]. Moreover, RHAMMB levels, but not RHAMMA levels, were significantly higher in liver metastases than primary pancreatic tumors [15]. Tail vein introduction of mouse pancreatic neuroendocrine tumor cells overexpressing RHAMMB, but not RHAMMA, resulted in large liver metastases [15]. In pancreatic ductal carcinomas, patients with increased RHAMMB levels had worse survival compared with patients with high levels of RHAMMA. The introduction of RHAMMB-overexpressing human pancreatic cancer cells via intracardiac injection, increased metastasis throughout the body. Interestingly, alternative splicing of the other major hyaluronan receptor, CD44, is associated with altered migration and invasion of epithelial ovarian cancer cells [35]. Downregulation of the epithelial splicing regulatory protein 1 (ESRP1) splicing factor increased motility of epithelial ovarian cancer cells. This effect relied on its role in inducing a switch from the variant isoform generated by alternative splicing, CD44v to the standard isoform, CD44s. ESRP1 is expressed at higher levels in nuclei of ovarian cancer cells and malignant lesions compared to normal ovarian tissue. Poor clinical outcomes correlate with high ESRP1 expression in ovarian cancer cells, suggesting ESRP1 is a cancer biomarker. Together these findings indicate that splice variants of extracellular matrix molecule receptors such as HA receptors can promote a more invasive phenotype in a subset of cancers and reduce overall patient survival.
Alternative splicing affects neuronal migration and axon growth
Alternative splicing is important for many aspects of neuronal function including neurogenesis, synaptogenesis, synaptic plasticity and neuronal migration [36]. In the developing brain, there are two major types of cell migration: radial and tangential. Radial migration refers to neurons that migrate perpendicular to the surface of the brain from the ventricular zones to the cortex, while neurons generated in the telencephalon migrate tangentially (parallel to the surface of the cortex) to the cortical plate. Reelin, a large extracellular matrix glycoprotein secreted by cells in the prenatal neocortex, is essential for proper migration of neurons in the brain. In the absence of Reelin, neurons begin to migrate but fail to achieve the proper final location and orientation [37]. Disabled 1 (DAB1) is a cytoplasmic protein that acts an intracellular effector when cell surface receptors bind Reelin. Mutations in Dab1 result in similar phenotypes to loss of Reelin [37]. During brain development, Dab1 is expressed as isoforms with and without exons 7 and 8 [38]. Exons 7 and 8 encode tyrosine residues that are phosphorylated by Reelin, leading to degradation of Dab1 [39] (Figure 2c). Dab1 lacking exons 7/8 is expressed predominantly early in development and, as RNA binding protein and alternative splicing regulator RNA-binding motif 4 (RBM4) increases at embryonic day 14.5, the full-length protein becomes more prominent [16]. Mice lacking RBM4a, one of two adjacent copies of RBM in the mouse genome, have an increase in Dab1 transcripts lacking exons 7 or 8 and a decrease in full-length Dab1 transcripts that include these exons [16]. Re-introducing full-length Dab1, but not Dab1 lacking exons 7/8, rescued the neuronal migration defects observed in mice with RBM4a knockout [16]. The results demonstrate the importance of induction of the RBM4 splicing factor and its effects on DAB-1 alternative splicing for neuronal migration during development.
In addition to the movement of neuronal progenitor cells, the cytoskeletal machinery is also important for the projection of axons from neurons. Many of the molecules that serve as guidance cues for axons and their receptors undergo alternative splicing [17]. Dorsal neuronal progenitors undergo lateral migration to exit the ventricular zone [17]. These cells then differentiate into commissural interneurons and extend axons to and across the ventral midline. Knockdown of neural-specific RNA binding proteins that regulate alternative splicing, Neuro-oncological ventral antigen (Nova)1 or Nova2, resulted in defects in the migration of spinal cord interneurons [17]. Simultaneous knockdown of Nova1 and Nova2 reduced migration of spinal cord interneuron progenitors out of the subventricular zone, that resulted in fewer ventral ly projecting axons and axons that reached the midline, demonstrating a defect in commissural axon growth or guidance [17]. Reduced axon ventral projection was also observed with loss of function of Deleted in Colorectal Carcinoma (DCC) (Figure 2C) [17]. DCC can be expressed as a long or short isoform based on the presence or absence of 20 amino acids in the extracellular loop between fibronectin repeats 4 and 5. These two isoforms bind with similar affinities to netrin, but are reported to have different conformations after binding [40]. Nova was found to regulate DCC splicing in vitro, and DCClong levels were reduced while DCCshort levels increased in Nova knockout mouse embryos [17]. Re-introducing DCClong, but not DCCshort, into Nova knockout mice restored activity in axon outgrowth assays and rescued the neuronal migration and axon projection phenotypes in mouse embryos. The results define this alternative splicing event in DCC as the mechanism whereby Nova knockout reduces neuron migration and axon outgrowth. The data establish an instance in which migration in neurons and axonal outgrowth are both regulated by alternative splicing of a key protein that has a specific migration-inducing isoform.
Alternative polyadenylation affects cell migration
Proliferating primary human fibroblasts are more migratory than quiescent, serum-starved human fibroblasts and this effect is dependent on a shift in the use of polyadenylation sites of genes involved in splicing and cell migration [41]. Levels of cleavage and polyadenylation factor 3’ pre-RNA subunit 2, 64kDa (CstF-64) increase when fibroblasts proliferate or migrate into a denuded area [41]. Using polyadenylation site-enriched RNA-Sequencing, knockdown of CstF-64 was shown to affect the choice of polyadenylation sites and gene expression in multiple genes associated with cell migration [41]. In addition, the knockdown of CstF64 reduced the migration of both fibroblasts and triple-negative breast cancer cells into the denuded area, thus, establishing a molecular connection between pre-mRNA processing and cell migration.
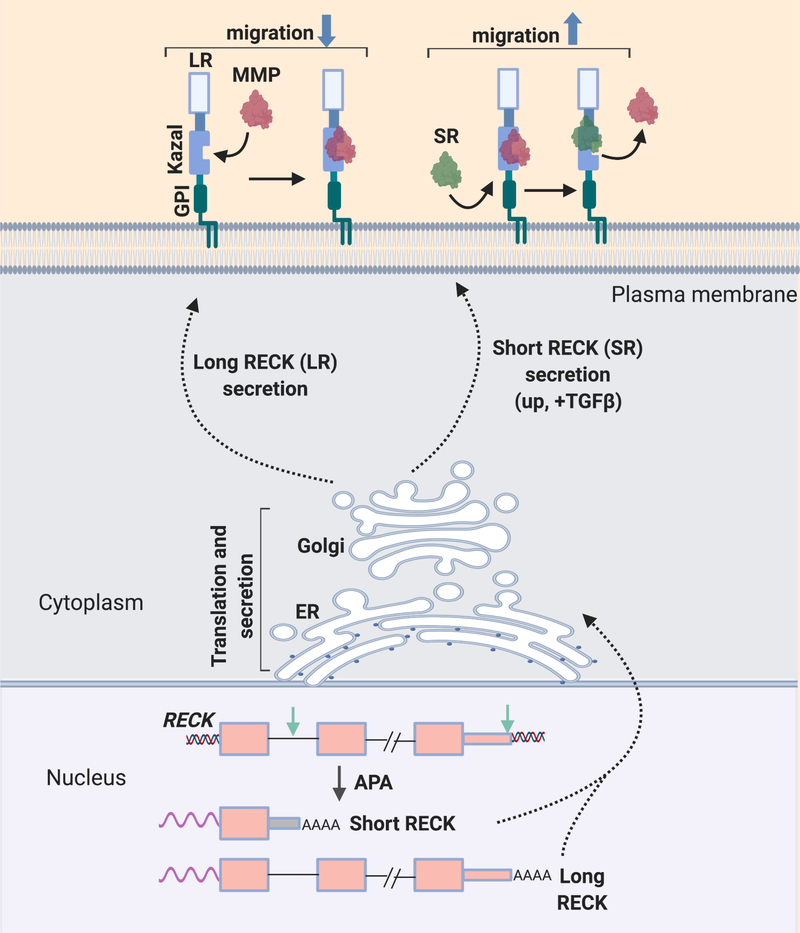
To investigate the mechanism by which alternative polyadenylation affects cell migration in fibroblasts, we investigated the tumor suppressor and matrix metalloproteinase inhibitor reversion-inducing-cysteine-rich protein with Kazal motifs (RECK) protein [42]. RECK was selected for further investigation as it was the genes that exhibited the greatest change in isoform expression between proliferating and quiescent fibroblasts [41]. The RECK gene undergoes a combined alternative splicing and alternative polyadenylation event to generate a short RECK isoform that lacks the Kazal motifs implicated in matrix metalloproteinase binding (Figure 3). The expression of the short RECK isoform increases in fibroblasts exposed to the pro-migratory stimulus, transforming growth factor-b (TGF-b) [42]. Moreover, silencing of the short RECK isoform impaired cell migration and invasion [42], while knockdown of the long RECK isoform promoted cell migration and invasion [42], in agreement with its role in inhibiting cell migration [43]. The short RECK isoform competes with matrix metalloproteinase-9 (MMP9) for binding to the long RECK isoform. Since the canonical long RECK protein inhibits the expression of MMPs, this competition by a shorter RECK variant would, in turn, increase the levels of pro-invasive MMPs [6]. The long and short RECK isoform also differentially affect tubulin post-translational modifications involved in cell migration [22]. Knockdown of the short RECK isoform decreased the levels of pro-migratory acetyl-tubulin, but increased the levels of anti-migratory detyrosinated-tubulin [44]. The opposite effects were observed when the long RECK isoform was silenced.
Figure 3.
Effect of chemotactic cues on an alternative polyadenylation event. In response to signals such as serum and TGF-b treatment, a short isoform of the GPI-anchored protein RECK is produced via Upstream APA. The short RECK isoform binds to the long RECK isoform at its Kazal motif region, and prevents the long isoform from binding to and inhibiting matrix metalloproteinases. The short RECK isoform promotes migration.
The importance of alternative polyadenylation for cell migration was also demonstrated in a recent study in immune cells [45]. A long isoform of baculoviral IAP repeat containing 3 (BIRC3) is upregulated in leukemia. The 3’ UTR of the long BIRC3 isoform was required for RNA binding proteins to bind to the transcript [46]. The long 3’ UTR resulted in recruitment of RNA binding proteins Staufen and Hur, which recruited IQGAP1 and RALA to the transcript. When BIRC3 was translated from the long, but not the short, isoform, the synthesized BIRC3 protein became part of a complex that included IQ Motif Containing GTPase Activating Protein 1 (IQGAP1) and Ras related protein A (RALA). This complex proved to be critical for the homing of B cells to bone marrow niches. Formation of these different complexes enabled B cells to migrate in response to chemokine receptor type 4 (CXCR4), as the short isoform of BIRC3 did not confer this capacity [45]. The findings suggest that manipulation of isoform length is a common mechanism that affects the migration of multiple cell types. Further experimentation will clarify which cell types and specific conditions invoke alternative polyadenylation to achieve a change in cell migration rate.
Concluding remarks
A stimulus for a cell to migrate often reflects a dynamic and rapidly changing situation—cells leaving an epithelial sheet, healing a wound or catching an invading bacterium. Under these conditions, the studies summarized here demonstrate that many different types of cells will take advantage of the enormous diversity of gene isoforms that are available based on the alternative inclusion or exclusion of specific exons, alternative start sites and alternative termination sites. The result is that cells have a much greater repertoire of molecules with different functional capacities. Changes in isoform use can affect the encoded protein’s association with other proteins or lipids, its effect on downstream signaling pathways or its subcellular localization. These changes in isoform expression can have a significant effect on the ability of cells to migrate and invade. Tumors, like migrating cells also take advantage of this rich source of diversity, as there is 30% more alternative splicing in a tumor than normal tissue and splicing factors such as Splicing Factor 3b Subunit 1 (SF3B1) and U2 small nuclear RNA auxiliary factor 1 (U2AF1) are oncogenic [47].
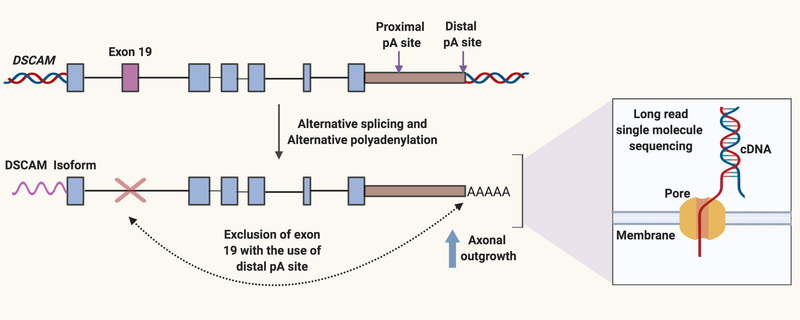
Many outstanding questions remain (See Outstanding Questions) and we anticipate that new technologies, improved bioinformatics tools and cell and molecular biology methodologies will allow researchers to gain greater insight into the role of alternative splicing and polyadenylation as an important mechanism to control cell migration. In particular, new technologies for single molecule long-read sequencing are likely to provide information about crosstalk between RNA processing events in different regions of a transcript as long reads can cover multiple exonic regions [48]. Emerging long read technologies offered by Pacific Biosciences or Oxford Nanopore have the advantage that the read lengths are longer. With Nanopore sequencing, direct RNA sequencing is possible, which can allow for a correlation between the length of the polyA tail and intron retention [49] (Figure 4). These approaches also have the advantage that they can represent an approach for the discovery of novel isoforms. Disadvantages of these technologies include relatively higher error rates compared with Illumina-based short reads and fewer reads. Recently, application of long-read technology revealed that for Down Syndrome Cell Adhesion Molecule 1 (DSCAM1), a transmembrane receptor involved in axon guidance, transcripts with long 3´ UTRs, but not short 3´ UTRs, preferentially excluded upstream exon 19 [50] (Figure 4). The presence of long 3´ UTRs was found to be required for exon 19 splicing. Knockdown or CRISPR deletion of the long, but not the short isoform of DSCAM1 impaired axon growth in Drosophila. These results demonstrate how alternative polyadenylation can regulate alternative splicing and thereby affect the guidance of cell protrusions.
Outstanding questions.
What are the mechanisms whereby extracellular, pro and anti-migratory signals affect the levels and activity of splicing factors?
How can the presence or absence of one or multiple exons or 3’ UTR sequences impact a cell’s migration?
How does alternative splicing affect migration during development and in disease, temporally and spatially?
How are different alternative splicing and alternative polyadenylation events coordinated and what is their functional role?
How did the close relationship between alternative splicing and polyadenylation and migration evolve?
Figure 4.
Long-read sequencing technologies provide information on crosstalk among processing events. Long-read sequencing technology involves sequencing of individual molecules rather than populations of molecules. With Oxford Nanopore Technology, the sequence of a single stranded DNA molecule is determined by measuring changes in current as the bases pass through a nanopore. The presence of a long 3’ UTR in the DSCAM transcript promotes the exclusion of exon 19, which results in a form of the protein that promotes axonal outgrowth. Created with BioRender.
Other emerging technologies also have the potential to improve our understanding of the relationship between alternative splicing and cell migration. A circular RNA for Nuclear Factor of Activated T cells 3 (NFATc3) was recently implicated in cancer cell properties including migration [51]. Methodologies to isolate, sequence and manipulate circular RNAs are likely to identify additional instances in which circular RNAs have effects on cell migration not shared by their linear counterparts. In addition, the application of CRISPR technology raises numerous new possibilities for understanding the functional role of alternative splicing. CRISPR knockout combined with fluorescent reporters for alternative splicing events [52] has the potential to identify new splicing regulators with important functional roles.
Highlights.
pre-mRNA processing can significantly impact cell migration at different stages of development, in response to pathogens, and in the context of cancer cell metastasis.
In response to external migratory signals, multiple factors that govern alternative splicing and cleavage and polyadenylation undergo changes in expression or gain post-translational modifications.
Changes in the levels, localization and activity of splicing and cleavage and polyadenylation factors can result in changes in the use of exons or polyadenylation sites in genes important for cell migration.
Alternative isoforms expressed more highly in pro-migratory conditions can promote cytoskeletal reorganization and other changes that direct the extension of protrusions and lamellipodia.
Recent technological advances in high-throughput sequencing have allowed the discovery of novel isoforms that affect cell migration.
Acknowledgments
The authors would like to thank Daniel Tan and all of the members of the Coller laboratory for helpful discussions. We acknowledge funding from NIH/NCI 1 R01 CA221296-01A1, NIH 1 R01 AR070245-01A1, Melanoma Research Alliance Team Science Award, Cancer Research Institute Clinical Laboratory Integration Program Award, University of California Cancer Research Coordinating Committee, David Geffen School of Medicine Metabolism Theme Award, the Clinical Translational Science Institute and Jonsson Comprehensive Cancer Center, the Prostate Cancer Spore at UCLA, and the Broad Stem Cell Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Reig G et al. (2014) Cell migration: from tissue culture to embryos. Development 141 (10), 1999–2013. [DOI] [PubMed] [Google Scholar]
- 2.Vicente-Manzanares M et al. (2005) Cell migration at a glance. J Cell Sci 118 (Pt 21), 4917–9. [DOI] [PubMed] [Google Scholar]
- 3.Devreotes P and Horwitz AR (2015) Signaling networks that regulate cell migration. Cold Spring Harb Perspect Biol 7 (8), a005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O'Donnell M et al. (2009) Axon growth and guidance: receptor regulation and signal transduction. Annu Rev Neurosci 32, 383–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tian B and Manley JL (2017) Alternative polyadenylation of mRNA precursors. Nat Rev Mol Cell Biol 18 (1), 18–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee Y and Rio DC (2015) Mechanisms and Regulation of Alternative Pre-mRNA Splicing. Annu Rev Biochem 84, 291–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baralle FE and Giudice J (2017) Alternative splicing as a regulator of development and tissue identity. Nat Rev Mol Cell Biol 18 (7), 437–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caceres JF and Kornblihtt AR (2002) Alternative splicing: multiple control mechanisms and involvement in human disease. Trends Genet 18 (4), 186–93. [DOI] [PubMed] [Google Scholar]
- 9.Goncalves V et al. (2017) Signaling Pathways Driving Aberrant Splicing in Cancer Cells. Genes (Basel) 9 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El Marabti E and Younis I (2018) The Cancer Spliceome: Reprograming of Alternative Splicing in Cancer. Front Mol Biosci 5, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jordan P et al. (1999) Cloning of a novel human Rac1b splice variant with increased expression in colorectal tumors. Oncogene 18 (48), 6835–9. [DOI] [PubMed] [Google Scholar]
- 12.Kotelevets L et al. (2018) The Rac1 splice form Rac1b favors mouse colonic mucosa regeneration and contributes to intestinal cancer progression. Oncogene 37 (46), 6054–6068. [DOI] [PubMed] [Google Scholar]
- 13.Wang F et al. (2017) SPSB1-mediated HnRNP A1 ubiquitylation regulates alternative splicing and cell migration in EGF signaling. Cell Res 27 (4), 540–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen YT et al. (2018) Immunohistochemical analysis of RHAMM expression in normal and neoplastic human tissues: a cell cycle protein with distinctive expression in mitotic cells and testicular germ cells. Oncotarget 9 (30), 20941–20952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi S et al. (2019) Function and clinical relevance of RHAMM isoforms in pancreatic tumor progression. Molecular Cancer 18 (1), 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.D. D et al. (2018) RBM4 Modulates Radial Migration via Alternative Splicing of Dab1 during Cortex Development. Mol Cell Biol 38 (12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leggere JC et al. (2016) NOVA regulates Dcc alternative splicing during neuronal migration and axon guidance in the spinal cord. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou Z et al. (2012) The Akt-SRPK-SR axis constitutes a major pathway in transducing EGF signaling to regulate alternative splicing in the nucleus. Mol Cell 47 (3), 422–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou Z and Fu XD (2013) Regulation of splicing by SR proteins and SR protein-specific kinases. Chromosoma 122 (3), 191–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matos P et al. (2003) Tumor-related alternatively spliced Rac1b is not regulated by Rho-GDP dissociation inhibitors and exhibits selective downstream signaling. J Biol Chem 278 (50), 50442–8. [DOI] [PubMed] [Google Scholar]
- 21.Alonso-Espinaco V et al. (2014) RAC1b overexpression correlates with poor prognosis in KRAS/BRAF WT metastatic colorectal cancer patients treated with first-line FOLFOX/XELOX chemotherapy. Eur J Cancer 50 (11), 1973–81. [DOI] [PubMed] [Google Scholar]
- 22.Schnelzer A et al. (2000) Rac1 in human breast cancer: overexpression, mutation analysis, and characterization of a new isoform, Rac1b. Oncogene 19 (26), 3013–20. [DOI] [PubMed] [Google Scholar]
- 23.Guo Y et al. (2015) R-Ketorolac Targets Cdc42 and Rac1 and Alters Ovarian Cancer Cell Behaviors Critical for Invasion and Metastasis. Mol Cancer Ther 14 (10), 2215–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hollander D et al. (2016) A network-based analysis of colon cancer splicing changes reveals a tumorigenesis-favoring regulatory pathway emanating from ELK1. Genome Res 26 (4), 541–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matos P et al. (2013) Ibuprofen inhibits colitis-induced overexpression of tumor-related Rac1b. Neoplasia 15 (1), 102–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ratcliffe CDH et al. (2019) HGF-induced migration depends on the PI(3,4,5)P3-binding microexon-spliced variant of the Arf6 exchange factor cytohesin-1. J Cell Biol 218 (1), 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D'Souza-Schorey C and Chavrier P (2006) ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol 7 (5), 347–58. [DOI] [PubMed] [Google Scholar]
- 28.Rajadurai CV et al. (2012) Met receptor tyrosine kinase signals through a cortactin-Gab1 scaffold complex, to mediate invadopodia. J Cell Sci 125 (Pt 12), 2940–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schnoor M et al. (2018) Cortactin: Cell Functions of A Multifaceted Actin-Binding Protein. Trends Cell Biol 28 (2), 79–98. [DOI] [PubMed] [Google Scholar]
- 30.van Rossum AG et al. (2003) Alternative splicing of the actin binding domain of human cortactin affects cell migration. J Biol Chem 278 (46), 45672–9. [DOI] [PubMed] [Google Scholar]
- 31.Bach CT et al. (2010) Tropomyosin isoform modulation of focal adhesion structure and cell migration. Cell Adh Migr 4 (2), 226–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koedoot E et al. (2019) Uncovering the signaling landscape controlling breast cancer cell migration identifies novel metastasis driver genes. Nat Commun 10 (1), 2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sironen RK et al. (2011) Hyaluronan in human malignancies. Exp Cell Res 317 (4), 383–91. [DOI] [PubMed] [Google Scholar]
- 34.Greiner J et al. (2002) Receptor for hyaluronan acid-mediated motility (RHAMM) is a new immunogenic leukemia-associated antigen in acute and chronic myeloid leukemia. Exp Hematol 30 (9), 1029–35. [DOI] [PubMed] [Google Scholar]
- 35.Chen L et al. (2017) Snail Driving Alternative Splicing of CD44 by ESRP1 Enhances Invasion and Migration in Epithelial Ovarian Cancer. Cell Physiol Biochem 43 (6), 2489–2504. [DOI] [PubMed] [Google Scholar]
- 36.Vuong CK et al. (2016) The neurogenetics of alternative splicing. Nat Rev Neurosci 17 (5), 265–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tissir F and Goffinet AM (2003) Reelin and brain development. Nat Rev Neurosci 4 (6), 496–505. [DOI] [PubMed] [Google Scholar]
- 38.Yano M et al. (2010) Nova2 regulates neuronal migration through an RNA switch in disabled-1 signaling. Neuron 66 (6), 848–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sekine K et al. (2014) How does Reelin control neuronal migration and layer formation in the developing mammalian neocortex? Neurosci Res 86, 50–8. [DOI] [PubMed] [Google Scholar]
- 40.Xu K et al. (2014) Neural migration. Structur es of netrin-1 bound to two receptors provide insight into its axon guidance mechanism. Science 344 (6189),1275–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mitra M et al. (2018) Alternative polyadenylation factors link cell cycle to migration. Genome Biol 19 (1), 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee HN et al. (2018) RECK isoforms have opposing effects on cell migration. Mol Biol Cell 29 (15), 1825–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oh J et al. (2001) The membrane-anchored MMP inhibitor RECK is a key regulator of extracellular matrix integrity and angiogenesis. Cell 107 (6), 789–800. [DOI] [PubMed] [Google Scholar]
- 44.Bance B et al. (2019) Microtubule acetylation but not detyrosination promotes focal adhesion dynamics and astrocyte migration. J Cell Sci 132 (7). [DOI] [PubMed] [Google Scholar]
- 45.Lee SH and Mayr C (2019) Gain of Additional BIRC3 Protein Functions through 3'-UTR-Mediated Protein Complex Formation. Mol Cell. [DOI] [PMC free article] [PubMed]
- 46.Lee SH and Mayr C (2019) Gain of Additional BIRC3 Protein Functions through 3'-UTR-Mediated Protein Complex Formation. Mol Cell 74 (4), 701–712 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kahles A et al. (2018) Comprehensive Analysis of Alternative Splicing Across Tumors from 8,705 Patients. Cancer Cell 34 (2), 211–224 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stark R et al. (2019) RNA sequencing: the teenage years. Nat Rev Genet. [DOI] [PubMed] [Google Scholar]
- 49.Workman RE et al. (2018) Nanopore native RNA sequencing of a human poly(A) transcriptome. bioRxiv. [DOI] [PMC free article] [PubMed]
- 50.Zhang Z et al. (2019) Elav-Mediated Exon Skipping and Alternative Polyadenylation of the Dscam1 Gene Are Required for Axon Outgrowth. Cell Rep 27 (13), 3808–3817 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karedath T et al. (2019) Regulation of circular RNA cirNFAT c3 in cancer cells alters proliferation, migration and oxidative phosphorylation. bioRxiv. [DOI] [PMC free article] [PubMed]
- 52.Gonatopoulos-Pournatzis T et al. (2018) Genome-wide CRISPR-Cas9 Interrogation of Splicing Networks Reveals a Mechanism for Recognition of Autism-Misregulated Neuronal Microexons. Mol Cell 72 (3), 510–524 e12. [DOI] [PubMed] [Google Scholar]
- 53.Wang R et al. (2018) A compendium of conserved cleavage and polyadenylation events in mammalian genes. Genome Res 28 (10), 1427–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pertea M et al. (2018) CHESS: a new human gene catalog curated from thousands of large-scale RNA sequencing experiments reveals extensive transcriptional noise. Genome Biol 19 (1), 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.House AE and Lynch KW (2008) Regulation of alternative splicing: more than just the ABCs. J Biol Chem 283 (3), 1217–21. [DOI] [PubMed] [Google Scholar]
- 56.Shepard PJ and Hertel KJ (2009) The SR protein family. Genome Biol 10 (10), 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Geuens T et al. (2016) The hnRNP family: insights into their role in health and disease. Hum Genet 135 (8), 851–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith CW and Valcarcel J (2000) Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem Sci 25 (8), 381–8. [DOI] [PubMed] [Google Scholar]
- 59.Tian B and Manley JL (2013) Alternative cleavage and polyadenylation: the long and short of it. Trends Biochem Sci 38 (6), 312–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Berg MG et al. (2012) U1 snRNP determines mRNA length and regulates isoform expression. Cell 150 (1), 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Siam A et al. (2019) Regulation of alternative splicing by p300-mediated acetylation of splicing factors. RNA. [DOI] [PMC free article] [PubMed]
- 62.Moran-Jones K et al. (2009) hnRNP A2 regulates alternative mRNA splicing of TP53INP2 to control invasive cell migration. Cancer Res 69 (24), 9219–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ghigna C et al. (2005) Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol Cell 20 (6), 881–90. [DOI] [PubMed] [Google Scholar]
- 64.Qi Y et al. (2016) A splicing isoform of TEAD4 attenuates the Hippo-YAP signalling to inhibit tumour proliferation. Nat Commun 7, ncomms11840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tornillo G et al. (2018) Dual Mechanisms of LYN Kinase Dysregulation Drive Aggressive Behavior in Breast Cancer Cells. Cell Rep 25 (13), 3674–3692 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li Z et al. (2008) Alternate cyclin D1 mRNA splicing modulates p27KIP1 binding and cell migration. J Biol Chem 283 (11), 7007–15. [DOI] [PubMed] [Google Scholar]
- 67.Lo HW et al. (2009) A novel splice variant of GLI1 that promotes glioblastoma cell migration and invasion. Cancer Res 69 (17), 6790–8. [DOI] [PMC free article] [PubMed] [Google Scholar]