Abstract
Human 4-hydroxyphenylpyruvate dioxygenase-like (HPDL) is a putative iron-containing non-heme oxygenase of unknown specificity and biological significance. We report 25 families containing 34 individuals with neurological disease associated with biallelic HPDL variants. Phenotypes ranged from juvenile-onset pure hereditary spastic paraplegia to infantile-onset spasticity and global developmental delays, sometimes complicated by episodes of neurological and respiratory decompensation. Variants included bona fide pathogenic truncating changes, although most were missense substitutions. Functionality of variants could not be determined directly as the enzymatic specificity of HPDL is unknown; however, when HPDL missense substitutions were introduced into 4-hydroxyphenylpyruvate dioxygenase (HPPD, an HPDL orthologue), they impaired the ability of HPPD to convert 4-hydroxyphenylpyruvate into homogentisate. Moreover, three additional sets of experiments provided evidence for a role of HPDL in the nervous system and further supported its link to neurological disease: (i) HPDL was expressed in the nervous system and expression increased during neural differentiation; (ii) knockdown of zebrafish hpdl led to abnormal motor behaviour, replicating aspects of the human disease; and (iii) HPDL localized to mitochondria, consistent with mitochondrial disease that is often associated with neurological manifestations. Our findings suggest that biallelic HPDL variants cause a syndrome varying from juvenile-onset pure hereditary spastic paraplegia to infantile-onset spastic tetraplegia associated with global developmental delays.
Keywords: hereditary spastic paraplegia, HSP, autosomal recessive, mitochondrial disorder, HPDL
Wiessner et al. establish HPDL as a new hereditary spastic paraplegia (HSP) disease gene. Biallelic variants in HPDL cause a syndrome varying from juvenile-onset pure HSP to infantile-onset spastic tetraplegia associated with global developmental delay.
Introduction
The hereditary spastic paraplegias (HSPs) are a group of inherited neurodegenerative conditions associated with progressive weakness, atrophy and spasticity as a result of upper motor neuron dysfunction. ‘Pure’ HSPs manifest with isolated lower limb spasticity, while ‘complicated’ HSPs are associated with additional neurological and non-neurological features (e.g. upper limb involvement, cerebellar ataxia, intellectual disability, seizures, retinopathy and liver disease).1 Remarkable progress has been made in identifying many of the genetic causes of these syndromes; however, the presence of a substantial number of HSP subjects without genetic diagnoses suggests the existence of further genetic heterogeneity.2 Currently, there are over 80 different types of HSP3 that can be inherited in an autosomal dominant, recessive or X-linked pattern and can involve a wide range of molecular and metabolic aetiologies4 that include metabolic dysfunction (e.g. FA2H/SPG35; ALDH18A1/SPG99), myelin abnormalities (e.g. PLP1/SPG2) and organellar dysfunction that can affect the endoplasmic reticulum (e.g. ATL1/SPG3; REEP1/SPG31), lysosomes (e.g. SPG11/SPG11; ZFYVE26/SPG15) and mitochondria (e.g. SPG7/SPG7; FARS2/SPG77). Spastic paraplegias due to mitochondrial dysfunction can present with a range of pure to complicated phenotypes, often involving the same gene.5-7
Here we report a series of subjects affected with autosomal recessive pure or complicated HSP associated with biallelic variants in the gene encoding a putative iron-containing oxygenase, 4-hydroxyphenylpyruvate dioxygenase-like (HPDL). Furthermore, our investigations indicate that HPDL is expressed within the nervous system, is localized to mitochondria and plays a role in motor function in zebrafish.
Materials and methods
Participants’ informed consent
Participants and their families consented to participation in the study and research protocols were approved by Institutional Review Boards at LMU Munich, University of Tübingen, University College London and Cedars-Sinai Medical Center.
Animals, cells and reagents
Animals (rats, mice and zebrafish) were kept and handled according to relevant legislation. Cell lines and human primary dermal fibroblasts were kept in Dulbecco’s modified Eagle medium with 10% foetal calf serum, 2 mM glutamine and antibiotics. Transient transfections were performed with jetPEI® (Polyplus) unless stated otherwise. Culture and genetic manipulation of bacterial cells followed standard protocols.8 All reagents were from Sigma-Aldrich if not indicated otherwise. Antibodies used in this study are listed in Supplementary Table 1.
Genome-wide linkage analysis
DNA samples were hybridized to Affymetrix GeneChip Human Mapping NspI 250 K (Family A) or Xba 142 10 K Arrays (Family B). Genotypes were called with GeneChip Genotyping Analysis Software and default thresholds. Parametric multipoint logarithm of the odds (LOD) scores were calculated with MERLIN (Family A)9 or ALLEGRO (Family B)10 assuming autosomal recessive inheritance with full penetrance, consanguinity and a disease gene frequency of 0.0001.
Identification of HPDL variants
Exome sequencing in Families A, B, C, CS1, CS2, CS3, CS4, D, E, F, FRL, FRN, T, ZA, ZB, ZD, ZE, ZH, ZI, ZJ and ZK was performed as described earlier.11–20 Genome sequencing in Families ZF and ZG was part of the 100k Genomes Project.21 In Family TUE, the genome sequencing protocol involved library preparation with a TruSeq DNA PCR-Free Kit (Illumina), 2 × 150 bp paired-end read sequencing on an Illumina NovaSeq 6000 instrument and data analysis with megSAP (https://github.com/imgag). Sanger sequencing was used to screen the HPDL gene in Family ZC.
Measuring Hpdl expression during mouse development
Tissues were harvested from C57BL/6 mouse embryos and postnatal stages. RNA was isolated with the RNeasy® Lipid Tissue Kit (Qiagen) and reverse transcribed with the PrimeScriptTM RT Kit (Takara). Quantitative PCR was performed with the SYBR® Green system (Applied Biosystems). Hpdl levels were normalized to Actb and results are expressed as means and standard deviations (SD) of three experiments.
Analysis of HPDL protein levels
Tissues were harvested from postnatal Day 56 (P56) C57BL/6 mice and total protein was extracted with a rotor-stator homogenizer and RIPA buffer. Proteins were separated by SDS-PAGE and transferred to polyvinylidene fluoride (PVDF) membranes. Specific bands were visualized by immunodetection with primary (rabbit anti-HPDL, mouse anti-GAPDH) and secondary [IRDye-conjugated anti-immunoglobulin G (IgG)] antibodies and infrared fluorescence imaging with an Odyssey Imager (LI-COR).
Localization of HPDL in rat brain neurons
Tissues were harvested from P56 Sprague Dawley rats, fixed in 4% paraformaldehyde in PBS for 4 h, incubated in sucrose (30% in PBS), frozen in O.C.T. (Thermo Fisher) and cryosectioned. Sections (14 µm) were permeabilized with methanol for 20 min at −20°C, blocked in 5% bovine serum albumin, 1% normal goat serum, 0.3% TritonTM X-100 in PBS for 1 h at room temperature and incubated with rabbit anti-HPDL and mouse anti-calbindin primary antibodies for 16 h at 4°C. Alexa Fluor-conjugated anti-IgG secondary antibodies were used for fluorescent detection with a Zeiss LSM 800 microscope.
HPDL levels during neural differentiation of Neuro2a cells
Cells were cultured in low-serum (0.5% foetal calf serum) medium with 6.25 µM retinoic acid for 48 h. Total protein was extracted with cell lysis buffer (1% SDS, 10 mM Tris-HCl, pH 7.4), run on SDS gels and blotted onto PVDF membranes. Protein bands were visualized by immunodetection with primary (rabbit anti-HPDL, rabbit anti-α-tubulin) and secondary (IRDye-conjugated anti-IgG) antibodies and infrared fluorescence imaging. Data were given as means and SD of four experiments. The Student’s two-tailed t-test was used for statistical analysis.
Fractionation of HeLa cells
Cytosolic, microsomal and mitochondrial fractions were obtained with the Qproteome Mitochondria Isolation Kit (Qiagen), run on SDS gels and transferred to PVDF membranes. Specific bands were visualized by immunodetection with primary (rabbit anti-HPDL, rabbit anti-α-tubulin, mouse anti-PDI, rabbit anti-Cox IV) and secondary (IRDye-conjugated anti-IgG) antibodies and infrared fluorescence imaging.
Subcellular localization of HPDL
Untreated or HPDL-myc-transfected HeLa cells were labelled with MitoTrackerTM Red (Invitrogen), fixed in 4% paraformaldehyde in PBS for 10 min, permeabilized with 0.1% TritonTM X-100 in PBS for 15 min and blocked with 5% horse serum in PBS for 1 h. Cells were stained with primary (rabbit anti-HPDL, mouse anti-myc) and secondary (Alexa Fluor-conjugated anti-IgG) antibodies. Images were captured with a Zeiss Axiovert 200 M microscope.
Fractionation of mitochondrial compartments from HeLa cells
Mitochondria were isolated with the Qproteome kit, incubated in MTiso buffer22 with 0.3 mg/ml digitonin for 20 min and spun at 10 000g for 10 min at 4°C to separate mitoplasts (inner membrane and matrix) from the outer membrane and intermembrane space. Fractions were run on SDS gels and blotted onto PVDF membranes. Protein bands were visualized by immunodetection with primary (rabbit anti-HPDL, mouse anti-Cyt C, rabbit anti-Cox IV) and secondary (IRDye-conjugated anti-IgG) antibodies and infrared fluorescence imaging.
Proteinase K digest of mitochondria isolated from HeLa cells
Mitochondria were isolated with the Qproteome Kit and treated with proteinase K (100 µg/ml) with or without 0.02% SDS for 30 min at 4°C. The digest was stopped with 1 mM PMSF and samples were run on SDS gels and blotted onto PVDF membranes. Protein bands were visualized by immunodetection with primary (rabbit anti-HPDL, mouse anti-Tom20, rabbit anti-Cox IV) and secondary (IRDye-conjugated anti-IgG) antibodies and infrared fluorescence imaging.
Bacterial expression of recombinant proteins
Escherichia coli [strain BL21(DE3)pLysS] were transformed with 4-hydroxyphenylpyruvate dioxygenase (HPPD) expression constructs (in pTRC vector) and grown in LB medium with 0.03 mg/ml chloramphenicol. Total protein was extracted by sonication of cells in lysis buffer (50 mM Tris-HCl, 1 mM EDTA, 1 mM DTT, pH 7.5), quantified with the bicinchoninic acid assay (Thermo Fisher) and run on SDS gels (41 μg/well). Protein bands were visualized by transfer to PVDF membranes and immunodetection with rabbit anti-HPPD primary and horseradish peroxidase-conjugated anti-IgG secondary antibodies and recording of enhanced chemiluminescence.
Colorimetric assay for HPPD enzyme activity
Recombinant E. coli expressing HPPD species were grown in LB medium with 0.6 mg/ml tyrosine and 0.03 mg/ml chloramphenicol. Enzyme activity was assessed by pigment formation and absorbance at 410 nm in the culture supernatant. Results are expressed as means and SD of three experiments. One-way ANOVA followed by Tukey’s post hoc correction for multiple comparisons was used for statistical analysis.
Behavioural studies in zebrafish embryos
Zebrafish (Danio rerio) embryos (strain Tübingen) were injected at the single-cell stage with 1.5 nl injection buffer (3 µM spermine, 0.7 mM spermidine, 0.1% phenol red in PBS) containing morpholinos (MO) targeting hpdl or a standard control MO (Genetools) at concentrations of 75 or 150 µM. Larvae were kept in E3 medium at 28.5°C for 5 days, then placed in 48-well plates (one embryo per well in 1 ml medium) and left to acclimatize for 20 min. Locomotor responses to standardized stimuli were studied with the DanioVision system (Noldus). During a 10 min interval, the system generated a vibrational stimulus every 60 s and the locomotor behaviour of larvae (response to stimuli, swimming distance and speed) was recorded at 50 frames/s with EthoVision XT tracking software. Data were analysed in MATLAB (MathWorks) and results are expressed as means and standard errors of the mean (SEM) from four experiments each involving 16 embryos per condition. One-way ANOVA was used for statistical analysis.
Data availability
PCR primer, siRNA and morpholino sequences as well as procedures for cloning of DNA constructs are available from the authors. Anonymized data will be shared upon reasonable request as long as data transfer is in agreement with relevant legislation and approved by Institutional Review Boards.
Results
Identification of HPDL as a new HSP gene
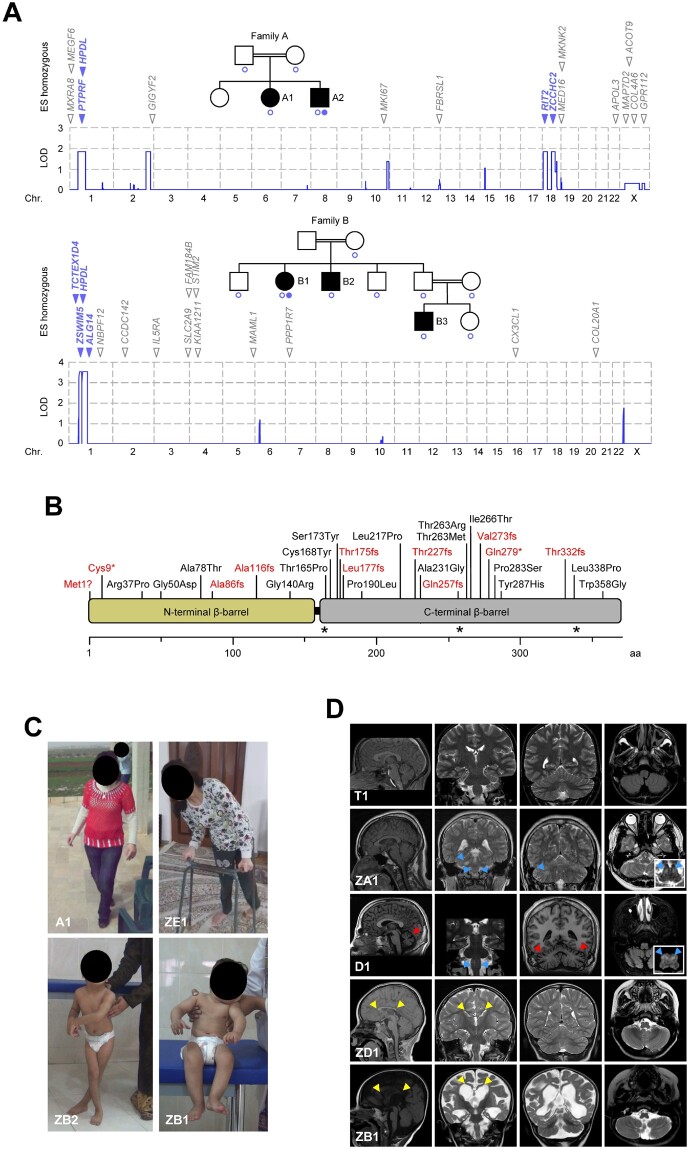
We performed genome-wide linkage analysis and exome sequencing on two unrelated consanguineous families with genetically undiagnosed juvenile-onset pure HSP. Linkage analysis using high-density single nucleotide polymorphism (SNP) arrays resulted in a single linkage peak on chromosome 1p34.1 in Family B (LOD score 3.56) that overlapped with one of four chromosomal regions that reached the maximum theoretical LOD score in Family A (LOD score 1.81). Exome sequencing on one affected individual from each family disclosed several rare non-synonymous homozygous variants [gnomAD minor allele frequencies (MAF) <0.0123] that were located in regions of interest as defined by linkage analysis; however, HPDL was the only gene residing in the linked region that carried different rare homozygous missense variants in both subjects (Fig. 1A and Supplementary Table 2). Subsequent data exchange with other research groups using informal enquiries and genetic collaboration platforms such as GeneMatcher24 and GENESIS12 resulted in the identification of 23 additional index cases with spastic para- or tetraparesis associated with rare non-synonymous biallelic HPDL variants (Table 1, Supplementary Fig. 1 and Supplementary Table 3). In 22 subjects, exome sequencing data (Families C, CS1, CS2, CS3, CS4, D, E, F, FRL, FRN, T, ZA, ZB, ZD, ZE, ZH, ZI, ZJ and ZK) or genome sequencing data (Families TUE, ZF and ZG) had been screened previously for variants in all disease genes annotated in OMIM; however, no variants likely or possibly related to their phenotypes had been observed. In one subject with pure HSP (Family ZC), HPDL was analysed by Sanger sequencing after multi-gene panel testing had not revealed convincing variants in >300 genes associated with HSP, motor neuron diseases, peripheral neuropathies and spinocerebellar ataxias. Whenever tested, HPDL variants segregated in agreement with an autosomal recessive pattern of transmission (Supplementary Fig. 1).
Figure 1.
Biallelic HPDL variants and associated phenotypes. (A) Genome-wide linkage analysis and exome sequencing in two consanguineous HSP families. SNP-based linkage analysis pinpointed four chromosomal regions where the maximum theoretical LOD score was obtained in Family A (LOD 1.81) and a single linkage interval reaching genome-wide significance in Family B (LOD 3.56). Blue circles indicate individuals included in linkage studies. Exome sequencing on one subject from each family identified several rare non-synonymous homozygous variants that were located in regions of interest as defined by linkage analysis (blue) or outside these regions (grey). HPDL was the only gene residing in the linked region on chromosome 1 that harboured rare homozygous non-synonymous variants in both families. Filled blue circles indicate individuals whose DNA was analysed by exome sequencing. (B) Schematic representation of the human HPDL protein and distribution of truncating (red) and missense (black) variants. By analogy to the crystallographic structures of HPPD, the structure of HPDL is predicted to contain two open β-barrels. The carboxy-terminal domain (grey) would possess the putative catalytic centre, while the amino-terminal domain (yellow) may not have a direct catalytic function but may play a role in dimer formation. Amino acid numbering is shown along the bottom. Asterisks mark the position of residues predicted to coordinate an iron ion to form the active site (similar to HPPD). (C) Variable severity of clinical presentation of individuals with HPDL variants. Subject A1 is able to walk without support but with scissoring gait (age 16 years). Subject ZE1 can only ambulate with a walker (age 15 years). Subject ZB2 needs support to stand with legs crossed at age 6 years. Subject ZB1 cannot sit independently at age 2.5 years. (D) Brain MRI findings. Subject T1 (age 17 years) had mild juvenile-onset pure HSP and normal neuroimaging. Subjects ZA1 (age 15 years) and D1 (age 21 years) had earlier-onset, intermediate disease associated with cerebellar atrophy (red arrowheads, Subject D1) and T2 hyperintensities (blue arrowheads) in the medulla oblongata (Subject ZA1 and D1) and middle cerebellar peduncle (Subject ZA1). Subjects ZD1 (age 5 years) and ZB1 (age 19 months) had severe disease associated with a hypoplastic corpus callosum (yellow arrowheads, Subjects ZD1 and ZB1), generalized reduction of white matter volume (Subjects ZD1 and ZB1) as well as ventriculomegaly, global cerebral atrophy and a simplified gyral pattern (Subject ZB1).
Table 1.
Genetic, clinical, neuroimaging and laboratory findings in patients with biallelic HPDL variants
| Individuals with mild disease (n = 10) | Individuals with intermediate disease (n = 9) | Individuals with severe disease (n = 15) | |
|---|---|---|---|
| HPDL variants | |||
| Loss-of-function alleles | 2/20 | 5/18 | 14/30 |
| Missense alleles | 18/20 | 13/18 | 16/30 |
| Initial presentation | |||
| Age at onset: range, median (n) | 8–17 y, 12.5 y (10) | 3–7 y, 6 y (7) | 0–12 m, 5 m (12) |
| Lower limb spasticity | 10/10 | 9/9 | 1/14 |
| Developmental delay | 0/10 | 0/9 | 6/14 |
| Encephalopathy | 0/10 | 0/9 | 12/14 |
| Clinical features | |||
| Motor delay | 0/10 | 2/9 | 15/15 |
| Cognitive delay | 2/10a | 1/9 | 15/15 |
| Spastic gait | 10/10 | 6/6c | n.a.e |
| Upper limb UMN involvement | 3/10b | 7/9d | 14/15 |
| Lower limb UMN involvement | 10/10 | 9/9 | 15/15 |
| Pseudobulbar signs | 0/7 | 2/9 | 6/12 |
| Bladder dysfunction | 0/6 | 3/8 | 2/3f |
| Ataxia | 1/8 | 6/9 | 2/12 |
| Encephalopathic episodes | 0/10 | 1/9 | 6/15 |
| Contractures | 2/10 | 6/8 | 11/15 |
| Seizures | 0/8 | 2/9 | 11/15 |
| Oculomotor abnormalities | 1/8 | 6/9 | 4/14 |
| Neuroimaging studies | |||
| Cerebellar atrophy | 0/7 | 5/8 | 0/14 |
| Callosal abnormalities | 0/7 | 4/8 | 11/14 |
| Olival T2-hyperintense lesions | 0/7 | 5/8 | 0/14 |
| Global cerebral atrophy | 0/7 | 0/8 | 7/14 |
| Abnormal gyration | 0/7 | 0/8 | 4/14 |
| Generalized white matter changes | 0/7 | 0/8 | 11/14 |
| Spinal cord abnormalities | 0/6 | 0/4 | 0/2 |
| Laboratory studies | |||
| High blood or CSF lactate | 0/3 | 0/4 | 0/4 |
| High blood tyrosine | 0/3 | 0/2 | 0/9 |
| Abnormal routine UOA profile | 0/1 | 0/3 | 0/8 |
Not all data were available for all patients because of incomplete examination or documentation or because features were not applicable (e.g. spastic gait in patients who were unable to ambulate). Sample sizes are indicated in each cell. m = month(s); n.a. = not applicable; UMN = upper motor neuron; UOA = urine organic acids; y = year(s).
Both patients had learning disability and were siblings from the same consanguineous family.
Patients had upper limb pyramidal signs only. There was no overt spasticity or weakness.
The remaining three patients had lost the ability to walk and were wheelchair-bound.
Four patients had pyramidal signs only. There was no overt spasticity or weakness.
Nine patients had never walked, three patients were too young (<18 m) at the time of last follow-up and three patients had insufficient clinical data.
This feature was not considered in seven patients who were too young (<4 years) and in one patient who had severe global developmental delays. No information was available for four patients.
Human HPDL is a 1.8-kb single-exon gene on chromosome 1p34.1 and is expressed as one transcript (RefSeq NM_032756.3). HPDL encodes the 371 amino acid protein HPDL (UniProt Q96IR7). The name is the result of its homology with the previously identified enzyme HPPD. We used crystallographic structures of HPPDs from several species (available through the Protein Data Bank25) to predict that the structure of HPDL may contain two open β-barrels. By further analogy to HPPD, the carboxy-terminal domain would possess the putative catalytic centre, while the amino-terminal domain does not seem to have a direct catalytic function but may play a role in dimer formation. In our study, we observed 28 distinct HPDL variants in our 25 families. Most variants were unique within single families, while four variants were shared between two or more independent kindreds [p.Gly50Asp (4×), p.Ala86fs (3×), p.Thr263Ile (2×), and p.Ile266Thr (2×)]. Variants were distributed throughout the gene with most appearing within the putative carboxy-terminal catalytic domain (Fig. 1B). Variants were either loss-of-function (start-loss, frameshift or stop-gain) or missense substitutions involving residues that are conserved across several species (Supplementary Fig. 2 and Supplementary Table 4). Further supporting the likely pathogenicity of these variants were the absence or extremely low MAF in public databases covering human genome variation (Supplementary Table 5) and the results from in silico algorithms for missense changes (Supplementary Table 6).
Phenotypic spectrum of subjects with biallelic HPDL variants
Specific phenotypic information was available for 34 affected individuals (Fig. 1C, Table 1 and Supplementary Table 3). Spasticity was a consistent feature of all subjects; some individuals presented with pure spastic paraplegia, while others had a more complicated presentation. Several subjects (Subjects A2, CS1, CS3, F1, T1, T2, T3, TUE1, ZF1 and ZG1; mild phenotype) had normal motor development but developed spastic paraparesis during late childhood or adolescence. Some of these subjects also had upper limb involvement (3/10) that usually manifested with pyramidal tract signs. Another group of subjects (Subjects A1, B1, B2, B3, C1, D1, ZA1, ZC1 and ZE1; intermediate phenotype) also achieved independent walking (though delayed in 2/9), but had disease onset in early or middle childhood. Several subjects with this intermediate phenotype presented with additional manifestations including upper limb involvement (7/9), ataxia (6/9) and oculomotor abnormalities (6/9). A third group of subjects (Subjects CS2, CS4, E1, FRL1, FRN1, ZB1, ZB2, ZD1, ZD2, ZH1, ZH2, ZH3, ZI1, ZJ1 and ZK1; severe phenotype) had a much more serious condition consisting of global developmental delays and infantile-onset spasticity, which affected all four limbs in all but one subject (Subject E1). In this group, the disease was associated with variable combinations of additional features including seizures (11/15) and encephalopathic episodes of acute neurological and respiratory decompensation (6/15). Of note, one subject with an intermediate phenotype (Subject C1) had a past medical history of a single episode of acute neurological deterioration with brain MRI indicating bilateral T2 hyperintensities of the inferior olive. This event had been diagnosed as acute disseminated encephalomyelitis (ADEM) and treated with corticosteroids leading to an incomplete clinical recovery.
Neuroimaging was performed in 29 subjects (Fig. 1D, Table 1, Supplementary Fig. 3 and Supplementary Table 3). Brain MRI was normal in subjects within the mild phenotype cohort (7/7). Corpus callosal abnormalities were distributed across both the intermediate (4/8) and severe (11/14) phenotype cohorts; otherwise, the observed radiological features largely segregated with clinical severity. Cerebellar atrophy (5/8) and bilateral inferior olivary hyperintensities on T2-weighted MRI images (5/8) were exclusively seen with intermediate presentations. Alternatively, generalized white matter changes (11/14), ventriculomegaly and global cerebral atrophy (7/14) as well as gyral abnormalities (4/14) were restricted to children with severe presentations. In one subject with the severe phenotype (Subject CS4), brain MRI was indicative of Leigh syndrome and magnetic resonance spectroscopy showed a high lactate peak in several locations. Spinal MRI studies were normal in 12 individuals for whom sufficient information could be retrieved from medical records (six subjects with mild, four with intermediate and two with severe disease). Overall, clinical and neuroimaging results suggested that individuals with HPDL-associated spastic paraplegia had a range of phenotypes consisting of severely-affected subjects with infantile-onset diffuse cortical disease to more mildly-affected subjects with later-onset upper motor neuron dysfunction.
Laboratory work-up for inborn errors of metabolism was either normal or yielded non-specific results that were not diagnostic (Table 1 and Supplementary Table 3). In particular, lactate concentrations in blood (11/11) and CSF (4/4) as well as tyrosine levels in blood (14/14) were always within normal ranges. Urine organic acids were also normal or non-specifically altered whenever measured (12/12). Tyrosine derivatives 4-hydroxyphenylpyruvate (4-HPP), 4-hydroxyphenyllactate (4-HPL) and 4-hydroxyphenylacetate (4-HPA) were usually not explicitly mentioned in laboratory reports; however, detailed analysis in two subjects revealed normal values for all three metabolites except for borderline high 4-HPA in one subject (Subject CS1).
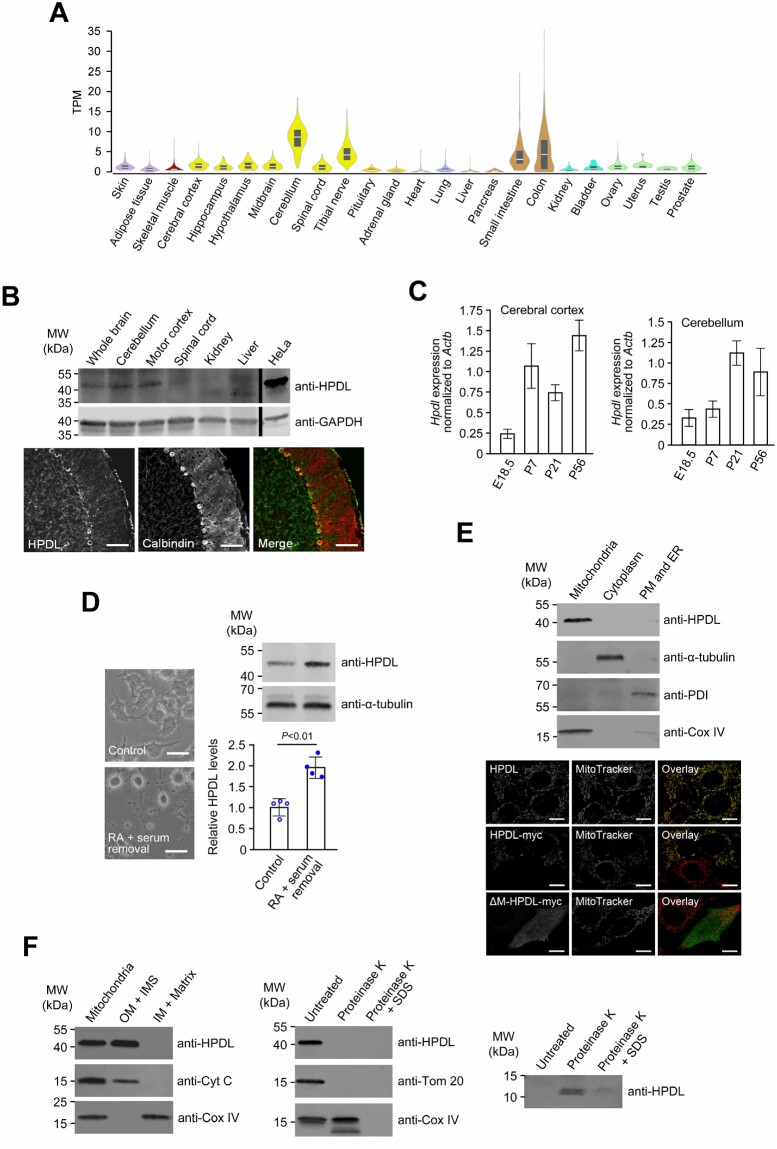
Detection of HPDL mRNA and the encoded protein in the nervous system
Human RNA-seq data26 indicated that HPDL mRNA levels were generally low, with the highest expression observed within the nervous system (Fig. 2A). Using a specific anti-HPDL antibody (Supplementary Fig. 4), we confirmed the presence of HPDL in adult mouse neural tissues including cerebellum and cerebral cortex (Fig. 2B). At the cellular level, neurons (Purkinje cells) in adult rat cerebellum had positive signals for HPDL (Fig. 2B). Analysis of Hpdl mRNA levels in murine neural tissues during late embryonic and early postnatal stages showed that expression was upregulated during nervous system maturation (Fig. 2C). In agreement with this observation, we noted a significant increase in HPDL levels during in vitro neuronal differentiation of murine neural cell line Neuro2a (Fig. 2D). Altogether, the spatiotemporal expression profile of HPDL was consistent with a continual requirement in the nervous system and its assumed aetiological role in a neurological condition when mutated.
Figure 2.
Expression and subcellular localization of HPDL. (A) Gene expression for HPDL across 24 human tissues. Expression values are shown as transcripts per million (TPM). Box plots represent median and 25th and 75th percentiles. Data were retrieved from the GTEx web resource [http://www.gtexportal.org/home/ (date accessed 20/06/2019)]. (B) Detection of the HPDL protein in young adult mice and rats (both P56). Top: HPDL levels in mouse neural and non-neural tissues were analysed by immunoblotting using an anti-HPDL antibody. Levels of the housekeeping protein GAPDH were determined to ensure equal loading of samples. Bottom: Immunofluorescence microscopy with an anti-HPDL antibody demonstrated presence of HPDL in rat cerebellar neurons (Purkinje cells). Calbindin was used as a marker to visualize Purkinje cells. Merge represents the overlay of calbindin and HPDL signals. Scale bar = 100 µm. (C) Hpdl expression during development of the mouse nervous system. Hpdl mRNA levels in embryonic and postnatal stages were determined by quantitative RT-PCR and normalized to Actb expression. Bars correspond to means of three biological and technical replicates and error bars represent SD. (D) HPDL expression in an in vitro model of neural differentiation. HPDL protein levels in undifferentiated (control) and retinoic acid-differentiated (RA + serum removal) Neuro2a murine neuroblastoma cells were analysed by immunoblotting. Bars represent the mean of four independent experiments and error bars indicate SD. The P-value was calculated by Student’s unpaired two-tailed t-test. Changes of cell morphology were recorded to document successful differentiation of cells. Scale bars = 50 µm. (E) Association of HPDL with mitochondria. Top: Fractionation of HeLa cells using the Qproteome Mitochondria Isolation Kit (Qiagen) showed co-isolation of HPDL with mitochondrial marker Cox IV. ER = endoplasmic reticulum; PM = plasma membrane. Bottom: Immunofluorescence microscopy of HeLa cells showed co-localization of endogenous HPDL with the mitochondrial stain MitoTracker Red. Myc-tagged HPDL showed an identical pattern of expression, while an artificial myc-tagged mutant lacking the mitochondrial targeting signal (Δ-M-HPDL-myc) displayed a diffuse cytoplasmic signal. Scale bars = 10 µm. (F) Submitochondrial localization of HPDL. Left: Upon subfractionation of HeLa mitochondria, HPDL was found within the fraction containing the outer membrane (OM) and intermembrane space (IMS) but was not detectable in the fraction consisting of inner membrane (IM) and the mitochondrial matrix. Middle: Following proteinase K digestion of intact HeLa mitochondria, signals for HPDL and OM protein Tom20 but not for IMS protein cytochrome C (Cyt C) were lost. Right: Using a high-resolution gel and short blotting time, a low molecular weight band was detected by the anti-HPDL antibody after proteinase K treatment, possibly corresponding to the region that is N-terminal of a predicted transmembrane domain and projects into the IMS. Uncropped versions of blots in B and D–F are shown in Supplementary Fig. 15.
Localization of HPDL to the outer mitochondrial membrane
Several in silico algorithms predicted that HPDL possessed an amino-terminal mitochondrial targeting signal (Supplementary Table 7). Separation of cellular compartments from HeLa cells, which had readily detectable levels of HPDL (Supplementary Fig. 5), showed endogenous HPDL resided in the mitochondrial fraction (Fig. 2E). Further confirmation was revealed with cytochemical co-localization of the HPDL signal with MitoTrackerTM, a mitochondrial marker (Fig. 2E), while no major overlap was seen for HPDL and PDI or golgin 97 (markers for the endoplasmic reticulum and Golgi apparatus, respectively, Supplementary Fig. 6). Similarly, forced expression of myc-tagged wild-type HPDL showed an identical pattern of localization; however, removal of the predicted mitochondrial targeting signal from myc-tagged HPDL resulted in a diffuse cytoplasmic signal (Fig. 2E). Subfractionation and proteinase K digestion of isolated intact mitochondria from HeLa cells indicated HPDL’s preferential association with the outer mitochondrial membrane (Fig. 2F), consistent with in silico predictions of the amino-terminal portion of the protein possessing a single membrane-spanning domain (Supplementary Table 8).
Functional effects of decreased HPDL were not overt, as siRNA-mediated knockdown of HPDL did not alter levels of mitochondrial respiratory chain complex proteins (Supplementary Fig. 7) or have a detectable effect on cellular oxygen consumption (Supplementary Fig. 8) in HeLa cells. We also did not find evidence for a role of HPDL as a critical regulator of mitochondrial dynamics (fission and fusion): mitochondrial morphology in cells with decreased HPDL levels was largely indistinguishable from controls. Similarly, increasing HPDL levels by transfection of HeLa cells with a plasmid expressing wild-type myc-tagged HPDL did not alter mitochondrial shape or numbers compared to control cells (Supplementary Fig. 9).
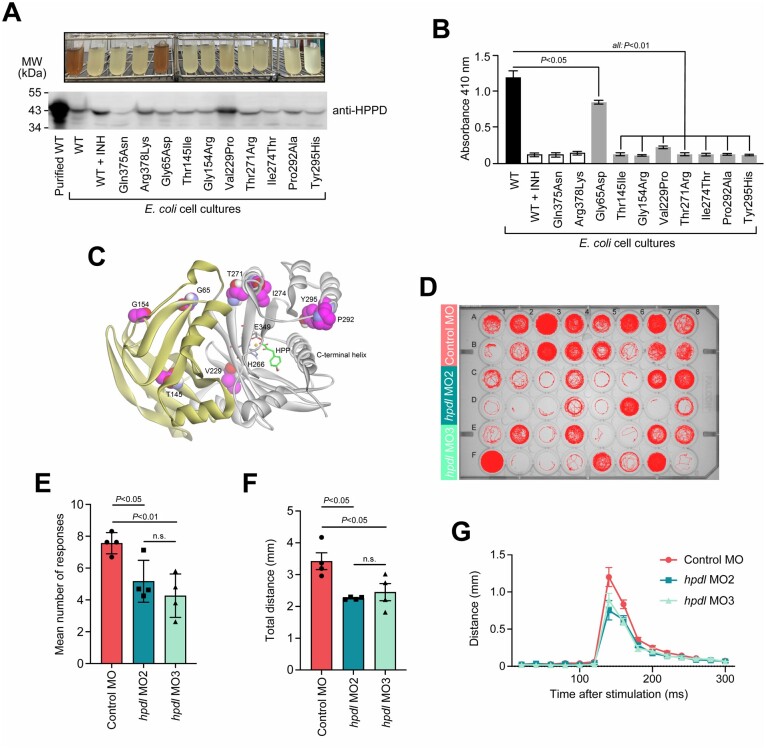
Interference of HPDL missense mutants with requirements for catalytic activity
Transfection studies with expression vectors for wild-type and mutant HPDL proteins did not show any major effects of the variants on protein levels or localizations (Supplementary Fig. 10). The functionality of variants could not be determined directly as the enzymatic specificity of HPDL is unknown; however, sequence and structural similarities between HPDL and its functionally well-characterized orthologue HPPD imply that these enzymes metabolize the same class of substrates (α-keto acids) and share a conserved reaction mechanism,27 even though they are probably not entirely functionally interchangeable (Supplementary Fig. 11). We thus adapted an assay that monitors HPPD’s ability to convert 4-HPP into homogentisate via the detection of a brownish pigment (formed by further metabolism of homogentisate28) and tested the effect HPDL missense variants had on pigment production when placed in homologous sites of the HPPD protein (Supplementary Fig. 12). Mutants corresponding to HPDL missense variants (Leu217Pro, Thr263Arg, Ile266Thr, Tyr287His) affecting the carboxy-terminal domain all had drastically reduced pigment formation (Fig. 3A and B). According to the known crystal structure of human HPPD (Protein Data Bank accession number 3ISQ), most of these residues were in proximity to the predicted catalytic centre (Fig. 3C), consistent with the observed decrease in HPPD enzymatic activity. One variant (Gly140Arg) located in the amino-terminal domain (which likely has no direct catalytic function) also affected the HPPD reaction, potentially through alteration of the overall protein structure or impaired dimerization. Another amino-terminal variant, Gly50Asp, had only a modest though still significant effect on HPPD activity (Fig. 3A and B).
Figure 3.
Functional analysis of HPDL. (A) Enzymatic activity of recombinant E. coli expressing HPDL missense variants on a HPPD backbone. The variants tested included artificial variants known to render HPPD functionless (Gln375Asn, Arg378Lys29) or affecting amino acid residues invariant in HPDL and HPPD amino-terminal (HPDL-Thr131 = HPPD-Thr145) and carboxy-terminal domains (HPDL-Pro284 = HPPD-Pro292) as well as six amino acid changes corresponding to disease-related HPDL variants (HPDL-Gly50 = HPPD-Gly65, HPDL-Gly140 = HPPD-Gly154, HPDL-Leu217 = HPDD-Val229, HPDL-Thr263 = HPPD-Thr271, HPDL-Ile266 = HPPD-Ile274, and HPDL-Tyr287 = HPPD-Tyr295). Catalytic activity of HPPD species in growth medium was reflected by formation of a brown pigment (top). Expression of HPPD species was confirmed by immunoblotting of total bacterial protein extracts (bottom, wild-type HPPD purified from bacteria served as a size control). An uncropped version of the blot is shown in Supplementary Fig. 15. (B) Pigment formation reflecting HPPD activity was quantified by measuring the absorbance at 410 nm. Bars represent the mean of three independent experiments and error bars indicate SD. The P-values were calculated by one-way ANOVA followed by Tukey’s post hoc correction for multiple comparisons. (C) Structural model of human HPPD (Protein Data Bank accession number 3ISQ) with bound 4-HPP. The protein folds into an amino-terminal (yellow) and a carboxy-terminal β-barrel (grey), which are represented as a ribbon backbone trace. Residues that bind to the substrate 4-HPP (green) and coordinate the iron ion within the catalytic centre are displayed in ball and stick model format (grey). Residues affected by substitutions that were tested in the enzymatic assay (A and B) are drawn as a CPK model (magenta). (D) Behavioural analysis of hpdl zebrafish morphants. MO2 and MO3 were morpholinos targeting zebrafish hpdl. The control morpholino was a random sequence not predicted to target any known gene. A single fish was placed in each well of the plate and movements were recorded automatically. The image shows representative traces (red) of the entire path the animals swam during the whole experiment (20-min acclimation phase followed by 10-min experimental phase, 1 vibratory stimulus/min). (E) Average number of zebrafish’s positive responses to stimulation. The maximum reachable number of responses was 10 responses to 10 stimuli. (F) Average distance zebrafish larvae swam in response to stimulation. Data were only included for measurements where fish responded to stimulation. (G) Representation of the dislocation of fish over time. The graph shows the Euclidian distance between the positions of fish in consecutive frames (50 frames/s). In all three experimental groups, maximum motor response was recorded at ∼140 ms (time point 0 ms corresponds to the time of stimulation). Data were only included for measurements where fishes responded to stimulation. Data-points in E–G correspond to means from four independent experiments each involving 16 embryos per condition. Error bars represent SEM. The P-values were calculated by one-way ANOVA. INH = inhibitor sulcotrione; WT = wild-type.
Abnormal locomotor behaviour in HPDL-deficient zebrafish
The zebrafish HPDL orthologue hpdl (NM_001109708) displays 40% sequence identity and 56% sequence similarity to the human protein (Supplementary Fig. 2). In a reverse genetic approach, we designed two separate morpholinos to silence hpdl expression in zebrafish larvae (Supplementary Fig. 13). Gross morphology of morphant zebrafish appeared normal with no detectable difference compared to control larvae (Supplementary Fig. 14). At 5 days post-fertilization, we compared the motor performance of hpdl-morphants and control fish using an automatized system that generates mechanical stimuli (vibration) and records the zebrafish’s subsequent movements in a reproducible way (Fig. 3D). We found that hpdl-morphants responded less frequently to stimulation (Fig. 3E) and moved a shorter distance when responding (Fig. 3F). Alternatively, for larvae that responded to stimuli, the time from stimulus to maximum motor activity was similar in all three experimental groups (Fig. 3G), which implies an intact and functional sensory system was present and allowed appropriate detection of stimuli. Altogether, these results suggested that hpdl-morphant zebrafish had a primary motor impairment in general agreement with the findings in humans with biallelic HPDL variants.
Discussion
We report the genetic and clinical features of HPDL-associated spastic paraplegia. We have identified 34 individuals with spasticity being their most consistent clinical feature as a result of biallelic HPDL variants. This disorder has a range of phenotypic severity from severe early-infantile encephalopathy to milder adolescent-onset pure spastic paraparesis. Further supporting the role of HPDL in our subjects’ neurological phenotypes was that HPDL was predominantly (though not exclusively) expressed in the nervous system and was upregulated in an in vitro model of neuronal differentiation. Moreover, silencing of the zebrafish orthologue, hpdl, resulted in a motor phenotype that replicated aspects of the human disease. Database searches also revealed comparably high HPDL mRNA levels in colon and intestine; however, individuals with biallelic HPDL variants had no clinical manifestations related to the digestive system and little or no data have been reported regarding the function of HPDL in these organs.
Identified disease-related HPDL variants included bona fide pathogenic truncating changes as well as missense variants. Protein levels and subcellular localizations of missense mutants were comparable to wild-type HPDL, suggesting that impaired enzyme function could be the relevant pathomechanism rather than premature degradation or mislocalization of the protein. This would be consistent with the finding that missense variants clustered within the predicted catalytic domain and often altered residues that were conserved or had similar chemical characteristics to the corresponding residues within the functionally well-characterized catalytic domain of HPDL’s orthologue, HPPD.27 As HPDL’s substrate specificity is unknown, effects of missense variants on enzyme function could not be examined directly; however, expression of HPPD possessing disease-associated substitutions resulted in significant impairment of enzyme activity in an established colorimetric assay in recombinant bacteria.
The human HPPD gene is related to two distinct diseases, autosomal dominant hawkinsinuria30 and autosomal recessive tyrosinaemia type 3.31 Tyrosinaemia type 3 is associated with variable and generally mild neurological symptoms32 along with the accumulation of tyrosine in bodily fluids and massive excretion of tyrosine derivatives 4-HPP, 4-HPL and 4-HPA in the urine.33 Interestingly, subjects with biallelic HPDL variants had normal tyrosine levels in bodily fluids (including CSF) and normal levels of urine organic acids except for one subject (borderline high 4-HPA). It may thus be that HPDL does not catalyse the same reaction as HPPD, but may have oxygenase activity for another molecule, most likely another α-keto acid. The observation that substitution of wild-type HPDL for HPPD abolished pigment formation in the bacterial bioassay seems consistent with this hypothesis; however, interpretation of this finding requires some caution: Eukaryotic HPDL might be unable to degrade 4-HPP in bacteria because of technical constraints (perhaps as a result of impaired oligomerization or lack of specific cofactors in prokaryotes) but may display this activity under more physiological conditions.
Endogenous and overexpressed HPDL was localized at the outer mitochondrial membrane and HPDL-associated HSP shared clinical features with other known mitochondrial syndromes in terms of having a broad phenotypic spectrum, neurological dysfunction, and intermittent decompensation of chronic disease.34 Moreover, neuroimaging revealed abnormalities found with several other mitochondrial disorders including bilateral symmetric hyperintensities of the inferior olive35 in five subjects as well as Leigh syndrome associated with elevated lactate on magnetic resonance spectroscopy in one subject. These observations prompted us to investigate a potential role for HPDL as a regulator of the properties and functions of mitochondria. Of note, we did not observe any altered bioenergetics or dynamics in HPDL-depleted cells, which suggested that either our assay techniques and non-neuronal cell models were not suitable for detecting such alterations or that HPDL may regulate other structural or functional properties of mitochondria.36–39 We noted that truncating HPDL variants tended to be more frequent in subjects with severe phenotypes (47% of alleles) compared to subjects with intermediate (28%) or mild disease (10%), which may suggest a correlation between residual protein activity and phenotypic severity similar to other syndromes related to abnormal nuclear-encoded mitochondrial proteins (e.g. SARS2, FARS2).7,40
In summary, our findings are consistent with the conclusion that biallelic HPDL variants are a previously unrecognized cause of neurological disease and provide a reasonable basis for initiating in-depth analysis of the underlying disease mechanisms.
Supplementary Material
Acknowledgements
We thank the families for participating in this study. We are grateful to J. van Gaalen and M. Schouten (Radboud UMC) for their help in the clinic. Several authors of this publication are members of the European Reference Network for Rare Neurological Diseases (ERN-RND, Project ID No 739510). Part of this research was conducted by the Queen Square Genomics group at University College London (UCL), supported by the National Institute for Health Research (NIHR) UCL Hospitals Biomedical Research Centre. This study uses data of the 100k Genomes Project, managed by Genomics England, a wholly owned company of the Department of Health and Social Care. The 100k Genomes Project is funded by the NIHR and NHS England. The Wellcome Trust, Cancer Research UK and the MRC funded research infrastructure. The 100k Genomes Project uses data provided by patients and collected by the NHS as part of their care and support. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Neurological Diseases and Stroke or the National Institutes of Health. The opinions expressed here are those of the authors and do not reflect those of the Navy, the Department of Defense, or the United States government. The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health and by NCI, NHGRI, NHLBI, NIDA, NIMH and NINDS.
Funding
This work was supported by Wellcome Trust and strategic award (Synaptopathies) funding of the SYNaPS Study Group (WT093205 MA, WT104033AIA to H.H.), the Ministry of Science and Technology, Taiwan (MOST 107-2320-B-016-014 to H.J.L.), the National Defense Medical Bureau, Taiwan (MAB-108-070 to H.J.L.), the Cedars-Sinai institutional funding program (to T.M.P.), the Cedars-Sinai Diana and Steve Marienhoff Fashion Industries Guild Endowed Fellowship in Pediatric Neuromuscular Diseases (to T.M.P.), the Fashion Industries Guild Endowed Fellowship for the Undiagnosed Diseases Program (to T.M.P.), the Fritz-Thyssen-Stiftung (10.15.1.021MN to J.S.) and the Federal Ministry of Education and Research, Germany, through the CMT-NET network (01GM1511 to J.S.), the TreatHSP network (01GM1905 to R.Sc. and L.S.) and the E-Rare-3 network PREPARE (01GM1607 to M.Sy., B.P.vdW., F.M.S., P.D.J., S.Z., and R.H.). Funding was also obtained from the Medical Research Council (MR/N025431/1 to R.H.), the Wellcome Trust (109915/Z/15/Z to R.H.), the Newton Fund (MR/N027302/1 to R.H.), Radboud UMC (to B.P.vdW.), ZonMW (to B.P.vdW.), the Hersenstichting (to B.P.vdW.), uniQure (to B.P.vdW.), the Gossweiler Foundation (to B.P.vdW.), the National Institute of Neurological Diseases and Stroke (R01NS072248 to S.Z. and R.Sc.), the Research Foundation Flanders (1805016 N to J.B.), the Japan Agency for Medical Research and Development (19ek0109279h0003 to S.T.), the Ministry of Health, Czech Republic (AZV NU20-04-00279, DRO 00064203 to A.U.M. and P.S.), the Henan Province Major Science and Technology Project (171100310200 to C.Z.), the Swedish Research Council (2018-02667 to C.Z.), the Swedish governmental grants to scientists working in health care (ALFGBG-717791 to C.Z.), the Research Committee for Ataxic Disease (to Y.T.), the Ministry of Health, Labor and Welfare, Japan (JP18K07495 to Y.T.), the Ministry of Education, Culture, Sports, Science, and Technology, Japan (to Y.T.), the German Research Foundation and the Ministry of Health, Italy, through the PROSPAX consortium under the frame of the European Joint Program on Rare Diseases-EJP RD COFUND-EJP N° 825575 (441409627 to M.Sy., R.Sc., B.P.vdW., R.H., F.M.S. and I.R.), the Italian Ministry of Health (RF-2016-02361610, RC5X1000 to F.M.S.), the EU Horizon 2020 project Solve-RD (779257 to M.Sy., R.Sc., B.P.vdW., E.J.K., R.H. and H.H.), and the Tom-Wahlig-Stiftung (to C.Bee., R.Sc. and I.R.).
Competing interests
C.Bee. and M.N. are employees of Centogene AG, Rostock, Germany, C.Ber. is an employee of Limbach Genetics, Mainz, Germany, D.G. is an employee of genetikum, Neu-Ulm, Germany and F.M. is an employee of GeneDx Inc., Gaithersburg, USA. Centogene AG, Limbach Genetics, genetikum and Genedx are private companies which generate revenue from clinical genetic testing. The remaining authors declare no conflict of interest related to the content of this manuscript.
Supplementary material
Supplementary material is available at Brain online.
Glossary
- 4-HPP
4-hydroxyphenylpyruvate
- HPPD
4-hydroxyphenylpyruvate dioxygenase
- HSP
hereditary spastic paraplegia
References
- 1. Harding AE. Classification of the hereditary ataxias and paraplegias. Lancet. 1983;321:1151–1155. [DOI] [PubMed] [Google Scholar]
- 2. Hedera P. Hereditary spastic paraplegia overview. In: Adam MP, Ardinger HH, Pagon RA, et al., editors. GeneReviews ®. Seattle (WA): University of Washington, Seattle; 2000.
- 3. Trummer B, Haubenberger D, Blackstone C.. Clinical trial designs and measures in hereditary spastic paraplegias. Front Neurol. 2018;9:1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shribman S, Reid E, Crosby AH, Houlden H, Warner TT.. Hereditary spastic paraplegia: From diagnosis to emerging therapeutic approaches. Lancet Neurol. 2019;18:1136–1146. [DOI] [PubMed] [Google Scholar]
- 5. Casari G, De Fusco M, Ciarmatori S, et al. Spastic paraplegia and OXPHOS impairment caused by mutations in paraplegin, a nuclear-encoded mitochondrial metalloprotease. Cell. 1998;93:973–983. [DOI] [PubMed] [Google Scholar]
- 6. Pierson TM, Adams D, Bonn F, et al. Whole-exome sequencing identifies homozygous AFG3L2 mutations in a spastic ataxia-neuropathy syndrome linked to mitochondrial m-AAA proteases. PLoS Genet. 2011;7:e1002325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sahai SK, Steiner RE, Au MG, et al. FARS2 mutations presenting with pure spastic paraplegia and lesions of the dentate nuclei. Ann Clin Transl Neurol. 2018;5:1128–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sambrook J, Maniatis T, Fritsch EF.. Molecular cloning: A laboratory manual. New York: Cold Spring Harbour Laboratory Press, Cold Spring Harbour; 1989. [Google Scholar]
- 9. Abecasis GR, Cherny SS, Cookson WO, Cardon LR.. Merlin-rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. [DOI] [PubMed] [Google Scholar]
- 10. Gudbjartsson DF, Jonasson K, Frigge ML, Kong A.. Allegro, a new computer program for multipoint linkage analysis. Nat Genet. 2000;25:12–13. [DOI] [PubMed] [Google Scholar]
- 11. Alfares A, Alfadhel M, Wani T, et al. A multicenter clinical exome study in unselected cohorts from a consanguineous population of Saudi Arabia demonstrated a high diagnostic yield. Mol Genet Metab. 2017;121:91–95. [DOI] [PubMed] [Google Scholar]
- 12. Gonzalez M, Falk MJ, Gai X, Postrel R, Schüle R, Zuchner S.. Innovative genomic collaboration using the GENESIS (GEM.app) platform. Hum Mutat. 2015;36:950–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hanein S, Martin E, Boukhris A, et al. Identification of the SPG15 gene, encoding spastizin, as a frequent cause of complicated autosomal-recessive spastic paraplegia, including Kjellin syndrome. Am J Hum Genet. 2008;82:992–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Koh K, Ishiura H, Tsuji S, Takiyama Y.. JASPAC: Japan Spastic Paraplegia Research Consortium. Brain Sci. 2018;8: 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Makrythanasis P, Maroofian R, Stray-Pedersen A, et al. Biallelic variants in KIF14 cause intellectual disability with microcephaly. Eur J Hum Genet. 2018;26:330–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Neveling K, Feenstra I, Gilissen C, et al. A post-hoc comparison of the utility of sanger sequencing and exome sequencing for the diagnosis of heterogeneous diseases. Hum Mutat. 2013;34:1721–1726. [DOI] [PubMed] [Google Scholar]
- 17. Salih MA, Seidahmed MZ, El Khashab HY, et al. Mutation in GM2A leads to a progressive chorea-dementia syndrome. Tremor Other Hyperkinet Mov (N Y). 2015;5:306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shashi V, Magiera MM, Klein D, et al. Loss of tubulin deglutamylase CCP1 causes infantile-onset neurodegeneration. EMBO J. 2018;37:e100540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tanaka AJ, Cho MT, Millan F, et al. Mutations in SPATA5 are associated with microcephaly intellectual disability, seizures, and hearing loss. Am J Hum Genet. 2015;97:457–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tran Mau-Them F, Moutton S, Racine C, et al. Second-tier trio exome sequencing after negative solo clinical exome sequencing: An efficient strategy to increase diagnostic yield and decipher molecular bases in undiagnosed developmental disorders. Hum Genet. 2020;139:1381–1390. [DOI] [PubMed] [Google Scholar]
- 21.Genomics England. The National Genomics Research and Healthcare Knowledgebase v5. 2019. doi:10.6084/m9.figshare.4530893.v5
- 22. Nishimura N, Yano M.. Separation of the inner and outer mitochondrial membrane in HeLa cells. Bio-protocol. 2014;4:e1299. [Google Scholar]
- 23. Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sobreira N, Schiettecatte F, Valle D, Hamosh A.. GeneMatcher: A matching tool for connecting investigators with an interest in the same gene. Hum Mutat. 2015;36:928–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Berman HM, Westbrook J, Feng Z, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moran GR. 4-Hydroxyphenylpyruvate dioxygenase. Arch Biochem Biophys. 2005;433:117–128. [DOI] [PubMed] [Google Scholar]
- 28. Denoya CD, Skinner DD, Morgenstern MR.. A Streptomyces avermitilis gene encoding a 4-hydroxyphenylpyruvic acid dioxygenase-like protein that directs the production of homogentisic acid and an ochronotic pigment in Escherichia coli. J Bacteriol. 1994;176:5312–5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lin JF, Sheih YL, Chang TC, et al. The interactions in the carboxyl terminus of human 4-hydroxyphenylpyruvate dioxygenase are critical to mediate the conformation of the final helix and the tail to shield the active site for catalysis. PLoS One. 2013;8:e69733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tomoeda K, Awata H, Matsuura T, et al. Mutations in the 4-hydroxyphenylpyruvic acid dioxygenase gene are responsible for tyrosinemia type III and hawkinsinuria. Mol Genet Metab. 2000;71:506–510. [DOI] [PubMed] [Google Scholar]
- 31. Endo F, Kitano A, Uehara I, et al. Four-hydroxyphenylpyruvic acid oxidase deficiency with normal fumarylacetoacetase: A new variant form of hereditary hypertyrosinemia. Pediatr Res. 1983;17:92–96. [DOI] [PubMed] [Google Scholar]
- 32. Ellaway CJ, Holme E, Standing S, et al. Outcome of tyrosinaemia type III. J Inherit Metab Dis. 2001;24:824–832. [DOI] [PubMed] [Google Scholar]
- 33. Ruetschi U, Cerone R, Perez-Cerda C, et al. Mutations in the 4-hydroxyphenylpyruvate dioxygenase gene (HPD) in patients with tyrosinemia type III. Hum Genet. 2000;106:654–662. [DOI] [PubMed] [Google Scholar]
- 34. Gorman GS, Chinnery PF, DiMauro S, et al. Mitochondrial diseases. Nat Rev Dis Primers. 2016;2:16080. [DOI] [PubMed] [Google Scholar]
- 35. Bindu PS, Taly AB, Sonam K, et al. Bilateral hypertrophic olivary nucleus degeneration on magnetic resonance imaging in children with Leigh and Leigh-like syndrome. Br J Radiol. 2014;87:20130478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Eisner V, Picard M, Hajnoczky G.. Mitochondrial dynamics in adaptive and maladaptive cellular stress responses. Nat Cell Biol. 2018;20:755–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Koenig MK. Presentation and diagnosis of mitochondrial disorders in children. Pediatr Neurol. 2008;38:305–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Spinelli JB, Haigis MC.. The multifaceted contributions of mitochondria to cellular metabolism. Nat Cell Biol. 2018;20:745–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Thorburn DR, Chow CW, Kirby DM.. Respiratory chain enzyme analysis in muscle and liver. Mitochondrion. 2004;4:363–375. [DOI] [PubMed] [Google Scholar]
- 40. Linnankivi T, Neupane N, Richter U, Isohanni P, Tyynismaa H.. Splicing defect in mitochondrial seryl-tRNA synthetase gene causes progressive spastic paresis instead of HUPRA syndrome. Hum Mutat. 2016;37:884–888. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
PCR primer, siRNA and morpholino sequences as well as procedures for cloning of DNA constructs are available from the authors. Anonymized data will be shared upon reasonable request as long as data transfer is in agreement with relevant legislation and approved by Institutional Review Boards.