Abstract
Aim
Klebsiella pneumoniae (K. pneumoniae) is an encapsulated Gram-negative bacterium that can lead to 14–20% of nosocomial infections. The ability of biofilm formation in this bacterium decreases the host immune response and antibiotic efficacy. This may impose a huge impact on patients and healthcare settings. This study aimed to evaluate the antibiotic resistance pattern and biofilm formation in K. pneumoniae strains isolated from two major Hamadan hospitals, west of Iran.
Methods
A total of 83 K. pneumoniae strains were isolated from clinical samples of patients in different wards of Hamadan hospitals from September 2018 to March 2019. Determination of antimicrobial susceptibility was performed using the disk diffusion method. Biofilm formation was evaluated by the crystal violet method. Data were analyzed by the SPSS software and chi-square test.
Results
The results showed that clinical samples included 18 urinary tract samples (22%), 6 wound samples (7%), 6 blood samples (7%), 17 tracheal tube aspiration samples (20%), 32 throat cultures (38%), 2 sputum samples (2.5%), and 2 abscess drain cultures (2.5%). High-level resistance to cefotaxime was detected in 92%, and all of isolates were susceptible to colistin. Biofilm formation was seen in 62 (75%) isolates. Strong biofilm formation was observed in 17 (20%) strains. A significant correlation was seen between biofilm formation and antibiotic resistance (P value <0.05).
Conclusion
Our findings emphasize the need for proper diagnosis, control, and treatment of infections caused by K. pneumoniae especially in respiratory tract infections due to the strong biofilm formation and high antibiotic resistance in these strains.
1. Introduction
Klebsiella pneumoniae (K. pneumoniae) is a Gram-negative, encapsulated, nonmotile, rod-shaped bacterium and an important member of Enterobacteriaceae which can lead to various infections including gastrointestinal, skin, nasopharyngeal, osteomyelitis, biliary and urinary tract infections, and bacteremia. K. pneumoniae virulence factors including polysaccharide capsule which protects against bactericidal serum factors, type I and III pili that adhere to the surfaces, adhesins, and siderophore, play a key role in its pathogenicity [1, 2]. K. pneumoniae strain is more common in immunocompromised individuals such as diabetics and the elderly and children and widely colonized in hospitalized patients [3]. Urinary tract infection (UTI) is known to be the most prevalent type of nosocomial infection, and K. pneumonia is the second cause of urinary tract infections among other Gram-negative bacteria [4]. The increasing emergence of multidrug-resistant (MDR) bacterial pathogens is known as a major public health challenge worldwide [5]. The overuse of antibiotics has resulted in difficulties in the treatment of K. pneumoniae and limitations in our available options for effective management of this bacterial infection [4–6]. In Gram-negative bacteria, beta-lactam resistance has been detected through chromosomal or plasmid genes, but in clinical specimens, resistance is usually dependent on plasmid R genes [7, 8]. Extended-spectrum β-lactamase (ESBLs), part of group A beta-lactamases, can lead to the hydrolysis of broad-spectrum cephalosporin and lead to resistance to penicillin and cephalosporins. However, they can be inhibited by beta-lactamase inhibitors such as clavulanic acid [9, 10]. Previous studies have shown that K. pneumoniae strains that are resistant to a broad spectrum of antibiotics are rapidly expanding, especially when the bacteria are capable of forming biofilm [2]. These bacteria can form a thick layer of extracellular biofilm which helps them attach to living and abiotic surfaces [11]. Treatment of infections caused by biofilm-forming K. pneumoniae strains is more difficult than other strains [12]. The antibiotic resistance and bacterial tendency to biofilm formation may play a key role in the emergence of MDR-K. pneumoniae strains [13]. Due to the antiphagocytic feature of biofilm, it is more challenging for the host immunity to eliminate this kind of bacterial pathogens [14]. The use of antibiotics in patients with bacterial infections can lead to bacterial elimination and accelerate the treatment process. Consequently, the increasing incidence of drug resistance causes complications in patients and higher medical costs [15]. This study was designed to investigate the biofilm formation and antibiotic resistance patterns in clinical isolates of K. pneumoniae isolated from Hamadan hospitals, west of Iran.
2. Methods
This study was performed on 83 K. pneumoniae strains isolated from clinical samples of patients with various infections in different wards of Hamadan hospitals from September 2018 to March 2019. The isolates were identified as K. pneumoniae, using conventional microbiological tests [16]. After recognition, the K. pneumoniae strains were stored in trypticase soy broth (TSB) containing 18% glycerol at −70°C.
Antimicrobial susceptibility to 15 different antibiotics including amikacin (30 µg), ampicillin-sulbactam (10/10 µg), trimethoprim-sulfamethoxazole (1.25 + 23.75 µg), ceftriaxone (30 µg), cefotaxime (30 µg), ceftazidime (30 µg), ciprofloxacin (5 µg), levofloxacin (5 µg), imipenem (10 µg), meropenem (10 µg), colistin (10 µg), gentamicin (10 µg), piperacillin-tazobactam (100/10 µg), tobramycin (10 µg), and nitrofurantoin (300 µg) was detected by the disk diffusion method according CLSI criteria [17]. The antibiotic disks were supplied by (Rosco Co., Denmark). Escherichia coli ATCC 25922 was used as the quality control strain. MICs of colistin in K. pneumoniae isolates were determined by the broth microdilution method over a range of dilutions from 0.125 to 128 μg/ml using colistin sulfate (Sigma–Aldrich). According to European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines [18, 19], breakpoints were used for interpretation of colistin MIC results (susceptible, ≤2 µg/ml; resistant, >2 µg/ml).
The biofilm formation was performed by the microtiter plate method (MTP) as described previously. The ability of the biofilm formation in each test isolate was compared with the negative and positive controls by analyzing the absorbance of the crystal violet stain applied for each biofilm. The following values were allocated for definition of the biofilm formation: nonbiofilm producer: OD550 ≤ 1, weak biofilm producer: 1 < OD550 ≤ 2, medium biofilm producer: 2 < OD550 ≤ 3, and strong biofilm producer: OD595 > 3 [20, 21].
SPSS software (version 22) was used for statistical analysis. Chi-square analysis was used for comparisons between the capacity of biofilm production and antibiotic resistance.
3. Results
Out of 83 K. pneumoniae clinical isolates, 32 (38.5%) were collected from the throat culture, 18 (21.6%) from the urine, 17 (20.4%) from the trachea, 6 (7.25%) from the blood culture, 6 (7.25%) from the wound, 2 (2.5%) from the sputum, and 2 (2.5%) from the abscess drain samples. About 57 (68%) of K. pneumoniae strains were obtained from male and 26 (32%) from female patients. Among all clinical isolates, 9 (11%) were taken from outpatients and 74 (89%) from inpatients specimens.
According to antibiogram results of 83 clinical isolates of K. pneumoniae, the highest antibiotic resistance was related to cefotaxime (92%), and all isolates were susceptible to colistin. After cefotaxime, the most resistance was to piperacillin-tazobactam (91%) and ampicillin-sulbactam (87%). Resistance to levofloxacin and tobramycin was detected in 85% of isolates. The prevalence of resistance to ceftazidime, trimethoprim-sulfamethoxazole, amikacin, ciprofloxacin, meropenem, imipenem, gentamicin, and ceftriaxone are shown in Figure 1. Different antibiotic resistance patterns were detected in various clinical samples, for example, the highest resistance to amikacin, ceftazidime, and gentamicin was found in wound and blood samples and the highest antibiotic resistance to trimethoprim-sulfamethoxazole, tobramycin, cefotaxime, piperacillin-tazobactam, ampicillin-sulbactam, and meropenem was observed in urine samples. Table 1 provides the prevalence of antibiotic resistance in different clinical specimens. A significant correlation was seen between the type of specimen and antibiotic resistance (P value <0.05). All K. pneumoniae isolates were susceptible to colistin, and the colistin MICs of 63 (75%), 11 (%13), and 9 (12%) of K. pneumoniae isolates were 0.125 µg/ml, 0.5 µg/ml, and 1 µg/ml, respectively.
Figure 1.
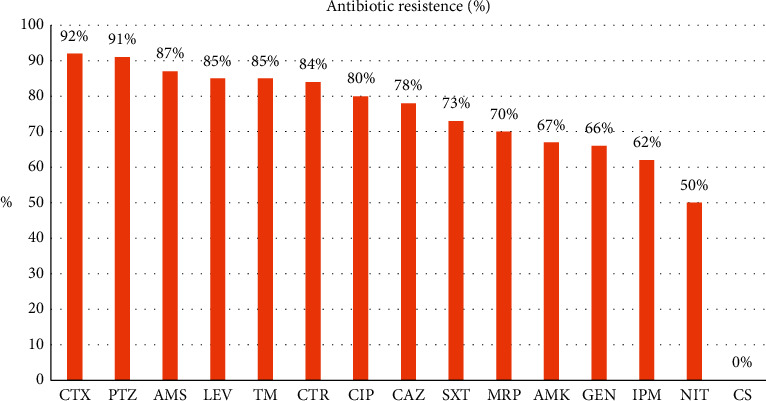
The prevalence (%) of resistance to antibiotics in K. pneumoniae isolates. CS, colistin; NIT, nitrofurantoin; IPM, imipenem; GEN, gentamicin; AMK, amikacin; MRP, meropenem; SXT, trimethoprim-sulfamethoxazole; CAZ, ceftazidime; CIP, ciprofloxacin; CTR, ceftriaxone; TM, tobramycin; LEV, levofloxacin; AMS, ampicillin-sulbactam; PTZ, piperacillin-tazobactam; CTX, cefotaxime.
Table 1.
Prevalence of antibiotic resistance in K. pneumoniae isolated from different clinical samples.
| Antibiotic | CS (%) | NIT (%) | IPM (%) | GEN (%) | AMK (%) | MRP (%) | SXT (%) | CAZ (%) | CIP (%) | CTR (%) | TM (%) | LEV (%) | AMS (%) | PTZ (%) | CTX (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample type | |||||||||||||||
| Urine | 0 | 50 | 40 | 45 | 42 | 100 | 100 | 75 | 62 | 87 | 100 | — | 100 | 100 | 100 |
| Wound | 0 | — | 83 | 100 | 83 | 83 | 66 | 83 | 100 | — | 83 | 100 | 80 | 80 | 66 |
| Blood | 0 | — | 50 | 100 | 100 | 50 | 60 | 100 | 80 | — | 100 | 75 | 100 | 100 | — |
| Throat | 0 | — | 65 | 100 | 70 | 64 | 62.1 | 70 | 83 | — | 82 | 81 | 93 | 93 | 100 |
| Sputum | 0 | — | 100 | — | 0 | 100 | — | — | 0 | 0 | — | — | 0 | — | — |
| Trachea | 0 | — | 100 | 75 | 80 | 100 | 100 | 100 | 90 | 100 | 100 | 100 | 92 | 75 | — |
| Abscess | 0 | — | 0 | — | — | 0 | 100 | 0 | 100 | — | 0 | 100 | 0 | 100 | — |
CS, colistin; NIT, nitrofurantoin; IPM, imipenem; GEN, gentamicin; AMK, amikacin; MRP, meropenem; SXT, trimethoprim-sulfamethoxazole; CAZ, ceftazidime; CIP, ciprofloxacin; CTR, ceftriaxone; TM, tobramycin; LEV, levofloxacin; AMS, ampicillin-sulbactam; PTZ, piperacillin-tazobactam; CTX, cefotaxime.
Our findings indicate that 62 (74.5%) K. pneumoniae isolates formed biofilm. The strains were classified into four categories as described above. Based on the biofilm analysis, 27 (32.5%) K. pneumoniae isolates formed biofilm weakly, 18 (21.6%) isolates created moderately, and 17 (20.4%) isolates were strong producers of biofilms. Among different clinical specimens, the lowest biofilm formation was seen in drainage abscess, and sputum culture and endotracheal aspiration samples had the highest biofilm formation. Additionally, blood culture samples formed biofilm weakly more than others and aspiration of the endotracheal tube showed the highest moderate biofilm formation. A significant correlation was seen between the type of clinical sample and the rate of biofilm formation (P value = 0.000), and the results are given in Table 2. Statistical analysis showed that there is a meaningful correlation between biofilm formation and resistance to antibiotics (P value <0.05). The results are given in Table 3.
Table 2.
Biofilm formation in K. pneumoniae isolated from different clinical samples.
| Sample type | Biofilm formation strength | |||
|---|---|---|---|---|
| No biofilm (%) | Weak (%) | Moderate (%) | Strong (%) | |
| Urine | 29.0 | 35.0 | 11.0 | 23.0 |
| Wound | 33.0 | 50.0 | 00.0 | 16.0 |
| Blood | 20.0 | 60.0 | 00.0 | 20.0 |
| Throat | 25.0 | 35.0 | 25.0 | 13.0 |
| Sputum | 100 | 00.0 | 00.0 | 00.0 |
| Trachea | 12.0 | 18.0 | 31.0 | 37.0 |
| Abscess | 100 | 00.0 | 00.0 | 00.0 |
Table 3.
Comparison of biofilm formation in antibiotic-resistant K. pneumoniae strains.
| Antibiotics | Strength of biofilm formation | ||||||
|---|---|---|---|---|---|---|---|
| Number | Negative | Poor | Moderate | Strong | P value | ||
| Ampicillin-sulbactam | Resistant | 54 | 10 | 17 | 15 | 12 | 0.038 |
| Susceptible | 8 | 5 | 2 | 0 | 1 | ||
|
| |||||||
| Cefotaxime | Resistant | 12 | 4 | 5 | 0 | 3 | <0.001 |
| Susceptible | 1 | 0 | 1 | 0 | 0 | ||
|
| |||||||
| Ceftazidime | Resistant | 45 | 10 | 15 | 11 | 9 | <0.001 |
| Susceptible | 11 | 5 | 4 | 1 | 1 | ||
|
| |||||||
| Ceftriaxone | Resistant | 11 | 4 | 2 | 2 | 3 | <0.001 |
| Susceptible | 1 | 1 | 0 | 0 | 0 | ||
|
| |||||||
| Piperacillin-tazobactam | Resistant | 45 | 11 | 20 | 7 | 7 | <0.001 |
| Susceptible | 3 | 1 | 2 | 0 | 0 | ||
|
| |||||||
| Tobramycin | Resistant | 41 | 8 | 16 | 8 | 9 | 0.035 |
| Susceptible | 7 | 5 | 1 | 1 | 0 | ||
|
| |||||||
| Ciprofloxacin | Resistant | 62 | 14 | 19 | 17 | 12 | <0.001 |
| Susceptible | 14 | 5 | 7 | 0 | 2 | ||
|
| |||||||
| Levofloxacin | Resistant | 35 | 9 | 11 | 10 | 5 | <0.001 |
| Susceptible | 6 | 3 | 3 | 0 | 0 | ||
|
| |||||||
| Trimethoprim-sulfamethoxazole | Resistant | 8 | 4 | 5 | 0 | 3 | <0.001 |
| Susceptible | 15 | 5 | 4 | 5 | 1 | ||
|
| |||||||
| Imipenem | Resistant | 39 | 9 | 15 | 10 | 5 | <0.001 |
| Susceptible | 22 | 10 | 7 | 1 | 4 | ||
|
| |||||||
| Meropenem | Resistant | 36 | 8 | 14 | 9 | 5 | <0.001 |
| Susceptible | 15 | 6 | 5 | 1 | 3 | ||
|
| |||||||
| Gentamicin | Resistant | 14 | 3 | 8 | 2 | 1 | <0.001 |
| Susceptible | 7 | 3 | 2 | 0 | 2 | ||
|
| |||||||
| Amikacin | Resistant | 44 | 5 | 16 | 10 | 13 | 0.024 |
| Susceptible | 19 | 10 | 5 | 1 | 3 | ||
|
| |||||||
| Nitrofurantoin | Resistant | 4 | 3 | 1 | 0 | 0 | <0.001 |
| Susceptible | 3 | 2 | 1 | 0 | 0 | ||
4. Discussion
In this current research, the ability of biofilm formation in K. pneumoniae isolates obtained from the clinical specimens and the correlation between the strength of biofilm formation and patterns of antibiotic resistance, sites of infection, or type of clinical samples were assayed. Our findings indicated that high percentages (74.5%) of K. pneumoniae isolates were able to form biofilms, and 20.4% of them formed biofilm strongly. It has already been determined that the formation of biofilms by bacteria is linked to the development of infections associated with implants and catheters which can even threaten the lives of patients with cystic fibrosis and chronic wound infections [22]. K. pneumoniae is now considered a biofilm-forming bacterium that can lead to nosocomial opportunistic infections and also affects the efficacy of antibiotic treatments [23, 24].
There are different findings of the correlation between biofilm formation and site of infection. Yang and colleagues conducted an investigation on biofilm formation by K. pneumoniae strains isolated from blood samples, wounds, swabs, urine, and sputum samples. Their study indicated that 62.5% of the isolates generated biofilms, which is less than our results [24]. Seifi et al. in Iran reported that 93.6% of K. pneumoniae isolates had the ability of biofilm formation and 33% of them could produce biofilm strongly which is more than the results of our study. They also reported that strong biofilms were more common in wound and sputum specimens, while in our study, the samples collected from the tracheal tube were able to form stronger biofilms, and sputum specimens formed weaker biofilm compared to them [11]. Diversities in the results of different studies can be related to the geographical area, type, and number of samples or characteristics of bacterial isolates, including antibiotic resistance patterns.
Another result of this study was that the amount of K. pneumoniae isolates in men was higher than in women (68%). Some studies, such as one by Nirwati et al. revealed that in 64% of cases, K. pneumoniae was isolated from male patients [25], and also, almost the same results were obtained in studies conducted by Osagei et al. from Nigeria and Akter et al. from Bangladesh [26, 27]. A possible reason for the high risk of developing K. pneumoniae infections in men could be the higher consumption of cigarettes and alcohol in men compared to women [25].
In the present study, isolates related to the respiratory system were more than isolates extracted from urine culture and blood samples, consistent with the studies of Yang et al. from China and Nirwaty et al. from Indonesia [24, 25]. However, according to research by Seifi et al. from Iran, the isolates from urine culture (61.7%) were higher than the wound, blood, and sputum samples [11].
Our findings reveal that over 90% of isolates were resistant to cefotaxime and piperacillin-tazobactam, and also, more than 80% of strains were resistant to ampicillin-sulbactam, levofloxacin, tobramycin, ceftriaxone, and ciprofloxacin. Furthermore, more than 70% of isolates were resistant to ceftazidime and trimethoprim-sulfamethoxazole. Colistin was the only effective antibiotic. This is in contrast to the findings of Borges et al. who have identified meropenem and piperacillin-tazobactam as effective antibiotics against K. pneumoniae [23]. In our study, the resistance to ciprofloxacin is higher than the resistance reported by Madahiah et al. and Cepas et al. [28, 29]. According to the results of a research from Iran, the highest antibiotic resistance in K. pneumoniae isolates belongs to ampicillin (91%), the lowest resistance is related to imipenem (5.5%), and more than 50% of the strains were resistant to ceftazidime and cefalotin [30]. However, our results indicate that 62% of isolates were resistant to imipenem, and almost 73% of specimens were resistant to ceftazidime.
Another finding of the study was that the strength of biofilm formation in antibiotic-resistant strains was higher than the sensitive strains, and a remarkable correlation was observed between antibiotic resistance and biofilm formation. However, among sensitive isolates, biofilm formation has been observed, or in resistant strains, nonbiofilm producers and weak and moderate biofilm former have been observed. Different results have been reported in this regard. Zheng et al. have demonstrated no correlation between biofilm formation and the production of broad-spectrum beta-lactamase (ESBL) enzymes in the K. pneumoniae strains [14]. According to Zheng et al. and Yang's and Zhang study, about 80% percent of the K. pneumoniae strains, which were extracted from urine and sputum samples and tested positive for biofilm, were also able to produce ESBL [14, 24]. It has also been claimed that there may be a correlation between the strength of biofilm formation in bacterial strains and different geographical areas. One of the most important limitations of this study was the almost insufficient number of K. pneumoniae isolates from patients, due to limited supply of materials for laboratory tests and limitations in financial support. More sampling and molecular epidemiological studies are recommended to obtain stronger results.
In conclusion, results of this study confirm the role of biofilm formation in resistance to clinical isolates of K. pneumoniae isolated from Hamadan hospitals, west of Iran. The strength of biofilm formation was different in clinical samples. So, characterization of nosocomial pathogens is very useful to control and treat infections caused by these pathogens.
Acknowledgments
The authors would like to thank all members of all staffs of Microbiology laboratory of Besat and Sina hospitals in Hamadan. This research was supported by Student Research Committee, Hamadan University of Medical Sciences, Hamadan, Iran (9711237178).
Abbreviations
- K. pneumoniae:
Klebsiella pneumoniae
- MDR:
Multidrug-resistant
- ESBL:
Extended-spectrum β-lactamase
- TSB:
Trypticase soy broth
- MTP:
Microtiter plate method
- CTX:
Cefotaxime
- PTZ:
Piperacillin-tazobactam
- AMS:
Ampicillin-sulbactam
- LEV:
Levofloxacin
- TM:
Tobramycin
- CTR:
Ceftriaxone
- CIP:
Ciprofloxacin
- CAZ:
Ceftazidime
- SXT:
Trimethoprim-sulfamethoxazole
- MRP:
Meropenem
- AMK:
Amikacin
- GEN:
Gentamicin
- IMP:
Imipenem
- NIT:
Nitrofurantoin
- CS:
Colistin.
Data Availability
The data used to support the findings of this study are included within the article.
Ethical Approval
This study was approved by the Ethics Committee of Hamadan University of Medical Sciences (IR.UMSHA.REC.1397.749).
Conflicts of Interest
The authors declare that there are no conflicts of interest.
References
- 1.Ryan K. J., Ray C. G. Medical Microbiology. New York, NY, USA: McGraw-Hill; 2004. [Google Scholar]
- 2.Vuotto C., Longo F., Balice M., Donelli G., Varaldo P. Antibiotic resistance related to biofilm formation in Klebsiella pneumoniae. Pathogens. 2014;3(3):743–758. doi: 10.3390/pathogens3030743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ripabelli G., Sammarco M. L., Salzo A., Scutellà M., Felice V., Tamburro M. New Delhi metallo‐β‐lactamase (NDM‐1)‐producing Klebsiella pneumoniae of sequence type ST11: first identification in a hospital of central Italy. Letters in Applied Microbiology. 2020;71(6):652–659. doi: 10.1111/lam.13384. [DOI] [PubMed] [Google Scholar]
- 4.Ding Y., Wang H., Pu S., Huang S., Niu S. Resistance trends of Klebsiella pneumoniae causing urinary tract infections in chongqing, 2011–2019. Infection and Drug Resistance. 2021;14:475–481. doi: 10.2147/idr.s295870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kidd T. J., Mills G., Sá‐Pessoa J., et al. A Klebsiella pneumoniae antibiotic resistance mechanism that subdues host defences and promotes virulence. EMBO Molecular Medicine. 2017;9(4):430–447. doi: 10.15252/emmm.201607336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Di Tella D., Tamburro M., Guerrizio G., Fanelli I., Sammarco M. L., Ripabelli G. Molecular epidemiological insights into colistin-resistant and carbapenemases-producing clinical Klebsiella pneumoniae isolates. Infection and Drug Resistance. 2019;12:3783–3795. doi: 10.2147/idr.s226416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moradigaravand D., Martin V., Peacock S. J., Parkhill J. Evolution and epidemiology of multidrug-resistant Klebsiella pneumoniae in the United Kingdom and Ireland. MBio. 2017;8(1):e01976–e02016. doi: 10.1128/mBio.01976-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mohamedalagamy M., Eldinashour M., Wiegand I. First description of CTX-M β-lactamase-producing clinical Escherichia coli isolates from Egypt. International Journal of Antimicrobial Agents. 2006;27(6):545–548. doi: 10.1016/j.ijantimicag.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 9.Mocktar C., Govinden U., Sturm A. W., Essack S. TEM-145 and TEM-146 ß-lactamases produced by Escherichia coli isolates from state hospitals in KwaZulu-Natal, South Africa. African Journal of Biotechnology. 2007;6(5) doi: 10.4314/ajb.v6i5.56857. [DOI] [Google Scholar]
- 10.Mesa R. J., Blanc V., Blanch A. R., et al. Extended-spectrum -lactamase-producing enterobacteriaceae in different environments (humans, food, animal farms and sewage) Journal of Antimicrobial Chemotherapy. 2006;58(1):211–215. doi: 10.1093/jac/dkl211. [DOI] [PubMed] [Google Scholar]
- 11.Seifi K., Kazemian H., Heidari H., et al. Evaluation of biofilm formation among Klebsiella pneumoniae isolates and molecular characterization by ERIC-PCR. Jundishapur Journal of Microbiology. 2016;9(1) doi: 10.5812/jjm.30682.e30682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shadkam S., Goli H. R., Mirzaei B., Gholami M., Ahanjan M. Correlation between antimicrobial resistance and biofilm formation capability among Klebsiella pneumoniae strains isolated from hospitalized patients in Iran. Annals of Clinical Microbiology and Antimicrobials. 2021;20(1):13–17. doi: 10.1186/s12941-021-00418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ribeiro S. M., Fuente-Núñez C. d. l., Baquir B., Faria-Junior C., Franco O. L., Hancock R. E. W. Antibiofilm peptides increase the susceptibility of carbapenemase-producing Klebsiella pneumoniae clinical isolates to β-lactam antibiotics. Antimicrobial Agents and Chemotherapy. 2015;59(7):3906–3912. doi: 10.1128/aac.00092-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng J.-X., Lin Z.-W., Chen C., et al. Biofilm formation in Klebsiella pneumoniae bacteremia strains was found to be associated with CC23 and the presence of wcaG. Frontiers in Cellular and Infection Microbiology. 2018;8(8):p. 21. doi: 10.3389/fcimb.2018.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lebeaux D., Ghigo J.-M., Beloin C. Biofilm-related infections: bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiology and Molecular Biology Reviews. 2014;78(3):510–543. doi: 10.1128/mmbr.00013-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ewing W. H. Edwards and Ewing’s Identification of Enterobacteriaceae. Amsterdam, Netherlands: Elsevier Science Publishing Co. Inc.; 1986. [Google Scholar]
- 17.Clinical Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. CLSI Supplement M100. 28th. Pittsburgh, PA, USA: Clinical Laboratory Standards Institute; 2018. [Google Scholar]
- 18.Haeili M., Javani A., Moradi J., Jafari Z., Feizabadi M. M., Babaei E. MgrB alterations mediate colistin resistance in Klebsiella pneumoniae isolates from Iran. Frontiers in Microbiology. 2017;8:p. 2470. doi: 10.3389/fmicb.2017.02470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wiegand I., Hilpert K., Hancock R. E. W. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nature Protocols. 2008;3(2):163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 20.O’Toole G. A. Microtiter dish biofilm formation assay. Journal of Visualized Experiments : JoVE. 2011;30(47) doi: 10.3791/2437.e2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zarei O., Arabestan M. R., Majlesi A., Mohammadi Y., Alikhani M. Y. Determination of virulence determinants of Escherichia coli strains isolated from patients with colorectal cancer compared to the healthy subjects. Gastroenterology and Hepatology from Bed to Bench. 2019;12(1):52–59. [PMC free article] [PubMed] [Google Scholar]
- 22.Bjarnsholt T. The role of bacterial biofilms in chronic infections. APMIS. 2013;121:1–58. doi: 10.1111/apm.12099. [DOI] [PubMed] [Google Scholar]
- 23.Borges A., Saavedra M., Simoes M. Insights on antimicrobial resistance, biofilms and the use of phytochemicals as new antimicrobial agents. Current Medicinal Chemistry. 2015;22(21):2590–2614. doi: 10.2174/0929867322666150530210522. [DOI] [PubMed] [Google Scholar]
- 24.Yang D., Zhang Z. Biofilm-forming Klebsiella pneumoniae strains have greater likelihood of producing extended-spectrum β-lactamases. Journal of Hospital Infection. 2008;68(4):369–371. doi: 10.1016/j.jhin.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 25.Nirwati H., Sinanjung K., Fahrunissa F., et al. Biofilm formation and antibiotic resistance of Klebsiella pneumoniae isolated from clinical samples in a tertiary care hospital, Klaten, Indonesia. BMC Proceedings. 2019;13(11):20–28. doi: 10.1186/s12919-019-0176-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osagie R. N. E. A., Iserhienrhien O., Okodua M., Onuabonah F., Daibo O. O. Antibiotic suscepbility profile of Klebsiella pneumoniae isolated from sputum samples amongst hospitalized adults in parts of Edo state, south-south. Niger Merit Research Journals. 2017;5(8):378–383. [Google Scholar]
- 27.Akter J., Chowd A. M. M. A., Al Forkan M. Study on prevalence and antibiotic resistance pattern of Klebsiella isolated from clinical samples in south east region of Bangladesh. American Journal of Drug Discovery and Development. 2013;4(1):73–79. doi: 10.3923/ajdd.2014.73.79. [DOI] [Google Scholar]
- 28.Madahiah B. M., Saba N. U., Abdul S., Dan Ali A. Q. Klebsiella pneumoniae urinary tract infections associated with long-term catheterization and spinal cord injuries. International Journal of Medical Sciences. 2002;2:227–229. doi: 10.3923/jms.2002.227.229. [DOI] [Google Scholar]
- 29.Cepas V., López Y., Muñoz E., et al. Relationship between biofilm formation and antimicrobial resistance in gram-negative bacteria. Microbial Drug Resistance. 2019;25(1):72–79. doi: 10.1089/mdr.2018.0027. [DOI] [PubMed] [Google Scholar]
- 30.Mehr V. P., Shokoohizadeh L., Mirzaee M., Savari M. Molecular typing of Klebsiella pneumoniae isolates by enterobacterial repetitive intergenic consensus (ERIC)–PCR. Infection Epidemiology and Microbiology. 2017;3(4):112–116. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are included within the article.


